Abstract
Studies have found that mutant, misfolded superoxide dismutase [Cu–Zn] (SOD1) can convert wild type SOD1 (wtSOD1) in a prion-like fashion, and that misfolded wtSOD1 can be propagated by release and uptake of protein aggregates. In developing a prion-like mechanism for this propagation of SOD1 misfolding we have previously shown how enervation of the SOD1 electrostatic loop (ESL), caused by the formation of transient non-obligate SOD1 oligomers, can lead to an experimentally observed gain of interaction (GOI) that results in the formation of SOD1 amyloid-like filaments. It has also been shown that freedom of ESL motion is essential to catalytic function. This work investigates the possibility that restricting ESL mobility might not only compromise superoxide catalytic activity but also serve to promote the peroxidase activity of SOD1, thus implicating the formation of SOD1 oligomers in both protein misfolding and in protein oxidation.
Keywords: Superoxide dismutase, Amyotrophic lateral sclerosis, SOD1 gain of function, SOD1 dismutase mechanism, sod1 peroxidase mechanism
Graphical abstract
Highlights
-
•
A free energy surface for the peroxidase mechanism of superoxide dismutase (SOD1) has been calculated.
-
•
A mechanism that implicates the restriction of mobility in the SOD1 electrostatic loop in protein oxidation is proposed.
-
•
The proxidant nature of bicarbonate, or dissolved carbon dioxide, is investigated.
1. Introduction
The misfolding and aggregation of Superoxide dismutase [Cu–Zn] (SOD1), a homo-dimeric protein that functions as an antioxidant by scavenging for superoxide, is linked to inherited, or familial, amyotrophic lateral sclerosis (fALS). Aberrant SOD1 folding has also been strongly implicated in disease causation for sporadic ALS, or sALS, which accounts for ~90% of ALS cases [1]. However there is a growing body of evidence to suggest that fibrillary aggregates may be just end-stage products, leading to a shift in focus away from deposition and towards aggregation and oligomerization [2]. In addition to being classified as a proteinopathy, studies have found that mutant and misfolded SOD1 can convert wtSOD1 in a prion-like fashion [3], and that misfolded wtSOD1 can be propagated by release and uptake of protein aggregates [4]. Today it is accepted that loss of function is not causative for ALS, though there is evidence that gain of function may play a modifying role in SOD1-related ALS. Not surprisingly, given the antioxidant function, there are also indications that oxidative stress may be relevant to disease pathogenesis, including the possibility that aberrant misfolding and aggregation of wtSOD1 is enhanced by exposure to such stress.
Oxidative stress and the accompanying mitochondrial damage have long been identified as important factors in non-SOD1 ALS [5], and an over-oxidized form of wtSOD1 has recently been characterized in a subset of sALS patients [6]. This iper-oxidized species was found to be toxic only upon exposure to additional pro-oxidant stressors, whereupon it adopts a pathogenic mechanism similar to that of mutant SOD1. SOD1 mutants have been identified with lower KM values for binding H2O2 [7], while the H43R mutant acquires a pro-oxidant potential that enhances oxidative stress [8]. For sALS it has been postulated that oxidation can trigger misfolding of wtSOD1 while wtSOD1 oxidized with hydrogen peroxide shares conformational epitopes to the C4F6 antibody with mutants implicated in fALS [9]. These results indicate the possibility that transient SOD1 oligomers could be implicated, not just in protein misfolding, but also in protein oxidation, thereby increasing the propensity for each of the components to aggregate. Such a scenario would provide a mechanism whereby loss of normal SOD1 function, or gain of aberrant function, could play a role in sALS.
The dismutase mechanism of SOD1 involves cyclic reduction and oxidation of the copper by successive molecules of superoxide [10]. This produces one molecule each of oxygen and hydrogen peroxide, providing a cellular defense by scavenging the more toxic superoxide radical anion. For the CuZnSOD family three histidine residues coordinate copper, two histidines and an aspartate coordinate the zinc, and an exceptional histidine (His63 in human SOD1 and His61 in bovine) coordinates both the copper and the zinc through Nε and Nδ respectively. In the resting state the Cu2+ ion forms a tetra-coordinated distorted tetrahedron (td) due to deprotonation of the bridging histidine. In the first catalytic stage reduction to Cu+ is accompanied by protonation of the imidazolate bridge, breaking the link between the Nε and the copper and resulting in a trigonal planar (tp) coordinated Cu+. Oxidation of Cu+ is accompanied by proton transfer to the substrate and the reformation of the Nε−Cu2+ link. A structural and energetic characterization of this cycle obtained using high level DFT calculations of a model system [11], is shown in Fig. 1. The reverse of the dismutase oxidative cycle, i.e. the redox reaction of Cu2+ with hydrogen peroxide to generate the superoxide anion and Cu+, serves as the template for the first catalytic stage of the SOD1 peroxidase mechanism.
Fig. 1.
The characterization of the dismutase cycle obtained using high level DFT calculations of a model system from reference 11.
In addition to superoxide dismutase activity SOD1 also exhibits peroxidase activity. In a similar fashion to the dismutase cycle the peroxidase mechanism involves cyclic reduction and reoxidation of the copper by successive molecules of hydrogen peroxide [12]. While the reductive cycle regenerates superoxide the oxidative step involves a Fenton-type reaction where the reduced Cu+ ion cleaves the O–O bond to generate a bound hydroxyl radical [10]. Studies have demonstrated that the radical does not diffuse readily but instead attacks the copper binding histidines [13,14], ultimately leading to copper loss and enzyme deactivation [15]. Oxidative damage to the zinc binding site is also observed at a rate more rapid than inactivation due to copper demetallation [16].
Crystallographic structures of bovine SOD1 have revealed a notable asymmetry characterized by a fully oxidized Cu2+ ion in one subunit with the other adopting a reduced configuration [17]. This asymmetry is associated with a lack of mobility of residues in the electrostatic loop (ESL) of the subunit with the fully oxidized Cu2+ ion, caused in turn by an abundance of crystal contacts in the ESL. In addition to exhibiting larger B-factors, signifying greater mobility, for the ESL residues the reduced subunit also exhibits a range of conformations for the copper site, representing geometries intermediate between those expected for Cu2+ and Cu+,suggesting that freedom of ESL motion is essential to catalytic function [18]. In developing a prion-like mechanism for the propagation of SOD1 misfolding [19] we have previously shown how restricting the mobility of the SOD1 ESL can lead to an experimentally observed gain of interaction (GOI) that results in the formation of SOD1 amyloid-like filaments [20]. This restriction of the ESL in turn is postulated to be caused through the formation of a transient, non-obligate oligomer between pathogenic and wild type SOD1. Here we investigate the possibility that restricting ESL mobility might not only compromise superoxide catalytic activity but also serve to promote the peroxidase activity of SOD1.
2. Materials and methods
The crystal structures for human and bovine wild type (wt) superoxide dismutase are available from the RCSB (www.rcsb.org) as PDB entries 2C9V [21] and 1CBJ [17]. The structure for SOD1 with the bicarbonate anion bound in the active site is available as 4B3E [22]. After adding hydrogens all proteins were subjected to a minimization by steepest descent and conjugate gradient using the CHARMm force field [23]. Protein alignments and superimposition were done using the MODELER protocol.
The free energy of the oxidized Cu2+ state, relative to a Cu+ resting state, was calculated by coupling the redox potential versus normal hydrogen electrode (NHE) for CuZnSOD1(Cu2+/Cu+) of + of 0.4 eV [24] with the Eo for (O2-.,2H+/H2O2) of 0.94 eV [25] to give:
| Cu+(HNHis) + O2-./H+ → Cu2+ (−NHis) + H2O2 ΔGo = -12.4 kcal mol-1 | (1) |
Hydrogen peroxide was docked to the fully oxidized subunit of bovine SOD1 using the CDOCKER algorithm [26]. The average binding energy for the five docked poses was calculated using the CHARMm force field and including the Generalized Born with simple SWitching (GBSW) implicit solvent model [27]. The principle moments of inertia calculated for the complex, protein and ligand were used to calculate the translational and rotational entropies for each at 298 K, using the CHARMm force field. The loss of conformational free energy for the bound ligand was calculated using the principle of conformer focusing [28]. The resulting binding free energy was calculated to be +2.4 kcal mol-1.
The following evaluation was used to calculate the free energy of proton donation from hydrogen peroxide to the bridging imidazole:
| (2) |
| ΔGo = +1.4 kcal mol-1 |
The free energy for protonation of the superoxide radical anion was calculated by combining Eo for (O2-.,2H+/H2O2) with the Eo for (HO2.,H+/H2O2) of 1.06 eV [25] to give:
| O2-. + H+ → HO2. ΔGo = +2.8 kcal mol-1 | (3) |
For the oxidative phase hydrogen peroxide was successfully docked to a modified subunit A of bovine SOD1 using CDOCKER and the CHARMm force field. Selecting the highest ranked pose QM/MM energies were calculated at the DFT (B3LYP//DNP)/CHARMm level, using the Becke, three-parameter, Lee-Yang-Parr (B3LYP) exchange-correlation functional, the double numerical polarized (DNP) basis set level, the CHARMm force field, and including GBSW solvent model. The QM region was defined as the copper and the bound ligand. The principle moments of inertia calculated for the complex, protein and ligand were used to calculate the translational and rotational entropies for each at 298 K, using the CHARMm force field.
Docking the H2O2 ligand to this modified reduced subunit of the bovine SOD1 crystal structure yields an initial complex with the copper nearly equidistant to both hydroperoxyl oxygens and a binding free energy of +11.3 kcal mol-1, Fig. 2. Single point QM/MM energy calculations along these Cu–O coordinates shows this to be a metastable structure, with an energy minimum structure at Cu–O distances of 2.05 and 3.08 Å, and a binding free energy of +7 kcal mol-1.
Fig. 2.
A Free energy profile for the peroxidative cycle of SOD1; inset FMO and charge distributions for the side-on dock of H2O2 to SOD1, for the end-on dock of H2O2 to SOD, and for the dock of HCO4- to SOD1.
The following evaluation was used to calculate the free energy for the loss of a proton from hydrogen peroxide in solution:
| (4) |
| ΔGo = +6.4 kcal mol-1 |
Combining the CuZnSOD1(Cu2+/Cu+) potential with the Eo for (H2O2,2H+/2H2O) of 1.77 eV and Eo for (.OH,H+/H2O) [25] of 2.31 eV gives:
| Cu+(HNHis) + H2O2 → Cu2+(−NHis) + .OH + H2O ΔGo = -19.0 kcal mol-1 | (5) |
Peroxomonocarbonate was also successfully docked to the modified subunit A of bovine SOD1 using CDOCKER and the CHARMm force field. Selecting the highest ranked pose and calculating QM/MM energies and translational and rotational entropies as before gave a binding free energy of +1.6 kcal mol-1. The free energy for the Cu2+ state with a freely diffusible carbonate anion radical was calculated by coupling the redox potentials for CuZnSOD1(Cu2+/Cu+) with Eo for (HCO4-,2H+/HCO3-, H20) of 1.8 eV [29] and the Eo for (CO3-.,H+/HCO3-) of 1.78 eV [25] to give:
| Cu+(HNHis) + HCO4- → Cu2+(−NHis) + H2O + CO3-. ΔGo = -32.6 kcal mol-1 | (6) |
For the Molecular dynamic (MD) simulation of apo SOD1 the dimer was immersed in an orthorhombic cell of TIP3P explicit water molecules and neutralized with counterions, and the system was minimized by steepest descent and conjugate gradient. The complex was heated to 300 K over 100 ps, and equilibrated at 300 K for 300 ps A production run for 4ns was obtained for human wtSOD1 at 300 K, and the SHAKE algorithm was employed to keep bonds involving hydrogen atoms at their equilibrium length, allowing the use of a 2 fs time step.
3. Results and discussion
Docking hydrogen peroxide to fully reduced wt SOD1first yields a metastable state where the H2O2 is loosely bound at a Cu–O distance of 2.35 Å. Proton transfer from the peroxide gives a stable intermediate where coordination to the bridging histidine is lost due to protonation of the histidine Nε. Complete reversal of the dismutase half-reaction produces the fully reduced, trigonal planar Cu+ state. The relative free energies in Fig. 2 are in good agreement with those calculated for the model system with one exception [11]. Whereas the DFT calculations for the dismutase reaction of the model system predicted that the copper-bound hydroperoxyl anion is a global minimum on the energy surface, our calculations based on experimental reduction potentials indicate that its formation from the Cu2+ resting state is endergonic. The fact that the model system underestimated the polarity of the SOD1 active site was cited as the reason for why the model system calculation gave a value at odds with the observed half-cell potentials.
To characterize the second stage of the peroxidase cycle a H2O2 ligand was docked to the fully reduced subunit of the bovine SOD1 crystal structure, however the results revealed no functional Cu+ –H2O2 complexes with all Cu–O distances in excess of 3 Å. In an investigation of patterns of flexibility of wild type and SOD1 mutants using principal coordinate analysis (PCA) of MD trajectories [30] it was demonstrated that a correlated movement of the electrostatic loops of the dimer behaves much like breathing motion, with the ESL opening and closing in a coupled fashion. PCA analysis of our 4 ns MD trajectory of wtSOD1 (pdb id 2C9V) (the eigenmode is available as movies in the Supporting Information) demonstrates how this breathing motion correlates with mobility within the copper binding site. Positioning the copper equidistant from the His46, His48 and His120 residues (His44, His46 and His118 in bovine SOD1) it can be seen that movement is dominated by an asymmetric stretch of the putative Cu- His46 and Cu- His120 bonds, with the Cu- His46 distance elongating as the Cu- His120 distance shortens, and vice versa. This is the pattern observed for the series of bovine SOD1 crystal structures representing conformations intermediate between those expected for Cu2+ and Cu+, where the Cu- His44 distance extends from 2.00, through 2.12 to 2.14 and 2.19 Å as Cu- His118 contracts from 2.19, through 2.18 to 2.03 and 2.02 Å. To model the effect of loss of ESL mobility metal ligand distances were set to the median values from our PCA analysis of the eigenmode and harmonic restraints were applied. Minimization using the CHARMm force field with implicit solvent yields the conformation shown in Fig. 3B. Alignment with the crystal structure for the fully reduced subunit shows a marked withdrawal of the His44 and His118 residues from a trigonal planar orientation, thin bond in Fig. 3B, resulting in a geometry best described as distorted td, thick bond in Fig. 3B.
Fig. 3.
A. Fenton chemistry of the pyridyl pendant dibenzylamine-Cu(I)–OOH species from reference 48; B Alignment and superimposition of our Cu(I) modified geometry (thick bond) with the crystal structure for the fully reduced bovine SOD1 subunit (thin bond); C alignment of our modified SOD1 conformation, the DFT-generated Cu(I) structure from reference 37; D alignment with the structure of the reduced SOD1 subunit with a bound bicarbonate anion from reference 22.
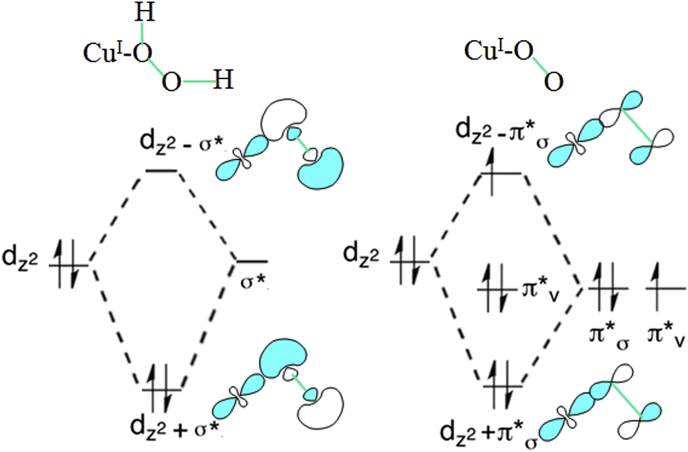
Docking the H2O2 ligand to this modified reduced subunit of the bovine SOD1 crystal structure yields an initial complex with the copper nearly equidistant to both hydroperoxyl oxygens and Cu–O distances of 2.29 and 2.37 Å. Frontier molecular orbital (FMO) analysis shows a side-on approach of the ligand to the Cu+ with the highest occupied molecular dxz orbital (HOMO) interacting with the HO–OH σ* antibonding lowest unoccupied molecular orbital (LUMO), Fig. 2 inset. Single point QM/MM energy calculations along these Cu–O coordinates shows this to be a metastable structure, with an energy minimum structure at Cu–O distances of 2.05 and 3.08 Å. FMO analysis of this structure indicates an end on interaction of the dz2 HOMO with the HO–OH σ* LUMO, and the orbital diagram for end-on bonding of H2O2 to Cu(I) is shown in Fig. 4. Generation of the experimentally observed bound radical species [10] requires heterolytic O–O bond cleavage of this complex to produce a free hydroxide that can abstract the Nε−Η proton of the bridging histidine to facilitate coordination with the fully oxidized copper. Polarization of the O–O bond, inset in Fig. 2 inset, indicates that deprotonation of the more acidic hydrogen on the proximate oxygen must precede bond scission, with bond cleavage then yielding an enzyme-bound oxidant that can be characterized as either (Cu-.O)+1 or (Cu-.OH) 2+. Based on rapid enzyme inactivation at pH > 9 it has been proposed that the reactive species is HO2- [31], with direct coordination of the hydroperoxyl anion also possible because of the electrostatic guidance for anions provided by a substrate channel that includes residues Lys134 and Arg141 (bovine SOD1 numbering) [32]. Further implicating the anion are the observations that enzyme inactivation by H2O2 occurred only in the presence of superoxide [33], and that oxidative degradation was much greater when cells were exposed to O2.-/H2O2 rather than just peroxide alone [34]. A comparison of the orbital diagrams for end-on bonding of hydrogen peroxide and superoxide to Cu+ in Fig. 4 highlights how for such a mixture binding to the peroxo ligand is favored.
Fig. 4.
Molecular orbital diagrams for end-on bonding of hydrogen peroxide and superoxide to Cu(I).
The mechanism whereby bicarbonate, or dissolved CO2, functions as a prooxidant, leading to extensive damage of both substrate and enzyme, remains in question. However the intermediacy of a peroxymonocarbonate species has been demonstrated by both NMR and EPR [35]. Docking HCO4- to our modified reduced SOD1 subunit yields a stable complex, Fig. 2 inset, where the copper center is bound to the oxygen proximate to the bridging histidine. Given the reaction scheme in Fig. 2 this complex can also be formed by nucleophilic attack of the deprotonated peroxide ligand on CO2, providing two pathways for mediation by peroxymonocarbonate. Cu+-catalyzed homolytic cleavage of this ligand yields a long-lived carbonate radical anion, allowing for diffusion away from the active site to facilitate oxidation of distant cellular targets. The reaction of Cu+-bound peroxide to form peroxymonocarbonate with subsequent cleavage to produce a diffusible carbonate radical anion has been previously proposed as a mechanism to explain bicarbonate-mediated peroxidation by SOD1 [36]. A crystal structure of a D125H SOD1 mutant from this study bound to HSO4- identifies the oxyanion as participating in a hydrogen bonding interaction with the Arg143 side chain (human numbering). The HCO4- docked complex from this work displays a similar interaction, Fig. 2 inset.
To understand the conditions under which SOD1 would catalyze Fenton chemistry it is instructive to study the pyridyl pendant dibenzylamine system, Fig. 3A [37]. Treatment of this Cu+2 complex with H2O2 yields a metastable Cu+2 complex results in cleavage of the Cu–O bond to yield the hydroperoxyl radical and a Cu+ center that binds a second equivalent of H2O2 to give a Cu+−OOH complex. Subsequent disproportionation of the ligand via Fenton chemistry yields a hydroxy radical. Alignment of our modified SOD1 complex with this Cu+ complex, Fig. 3C, highlights how in both cases the Cu+ arrangement is distorted from planarity, supporting the prediction that distortion of the normal reduced SOD1 conformation is required before the copper can catalyze Fenton chemistry. The alignment in Fig. 3D is with a reduced form of SOD1 where the copper was again observed to change from trigonal planar to td, this time as a consequence of binding a bicarbonate anion ligand [22]. Coordinating directly to the Cu+ center this ligand displaces a conserved crystallographic water, w0 in Fig. 3D, previously proposed as occupying the binding site for superoxide.
Given the coupling of the ESL “breathing motion” to the assymetric movement of the histidine ligands relative to the metal, these structural characteristics provide a potential causative link between ESL mobility and catalytic function at the Cu center. For wtSOD1 the trigonal planar geometry is normally associated with the reduced Cu+ form of the protein where the Nε on the bridging histidine is protonated. Meanwhile the oxidized form of SOD1 typically displays a tetra-coordinated Cu2+ in a td-like arrangement. Loss of ESL mobility increases the likelihood of stranding the Cu2+ center in a planar arrangement of His44, His46 and His118. Model chemistry confirms that this planar Cu2+ arrangement catalyzes the binding of H2O2, with subsequent generation of a Cu+ center in a distorted td conformation. Copper-catalyzed disproportionation of a second hydroperoxyl to yield a bound hydroxyl radical, or a diffusible carbonate anion radical through intermediation by peroxyomonocarbonate, facilitates SOD1 oxidation.
CRediT authorship contribution statement
Eamonn F. Healy: Conceptualization, Methodology, Software, Supervision, Visualization, Data curation, Writing - original draft. Analise Roth-Rodriguez: Investigation, Software. Santiago Toledo: Methodology, Formal analysis, Validation, Writing - review & editing, Funding acquisition.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors wish to acknowledge the support of the National Institute of General Medical Sciences (1K12GM102745), the National Science Foundation (#1832282), as well as Welch Foundation (Grant# BH-0018) for its continuing support of the Chemistry Department at St. Edward's University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100728.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bosco D.A., Morfini G., Karabacak N.M., Song Y., Gros-Louis F., Pasinelli P., Brown P.R.H., Jr. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redler R.L., Fee L., Fay J.M., Caplow M., M, Dokholyan N.V. Non-native soluble oligomers of Cu/Zn superoxide dismutase (SOD1) contain a conformational epitope linked to cytotoxicity in amyotrophic lateral sclerosis (ALS) Biochemistry. 2014;53:2423–2432. doi: 10.1021/bi500158w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grad L.I., Guest W.C., Yanai A., Pokrishevsky E., O'Neill M.A., Gibbs E. Intermolecular transmission of superoxide dismutase 1 misfolding in living cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16398–16403. doi: 10.1073/pnas.1102645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grad L.I., Yerbury J.J., Turner B.J., Guest W.C., Pokrishevsky E., O'Neill M.A. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and-independent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrì M.T., Valle C., Bozzo F., Cozzolino M. Oxidative stress and mitochondrial damage: importance in non-SOD1 ALS. Front. Cell. Neurosci. 2015;9:41. doi: 10.3389/fncel.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guareschi S., Cova E., Cereda C., Ceroni M., Donetti E., Bosco D.A. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5074–5079. doi: 10.1073/pnas.1115402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X.F., Crow J.P., Cashman NR N.R., Kondejewski L.R. J. Biol. Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura F., Fujimaki N., Okita W., Hiramatsu H., Takeuchi H. Structural instability and Cu-dependent pro-oxidant activity acquired by the apo form of mutant SOD1 associated with amyotrophic lateral sclerosis. Biochemistry. 2011;50:4242–4250. doi: 10.1021/bi200338h. [DOI] [PubMed] [Google Scholar]
- 9.Bosco D.A., Morfini G., Karabacak N.M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;11:1396. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abreu I.A., Cabelli D.E. Superoxide dismutases—a review of the metal- associated mechanistic variations. Biochim. Biophys. Acta. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Pelmenschikov V., Siegbahn P.E. Copper− Zinc superoxide dismutase: theoretical insights into the catalytic mechanism. Inorg. Chem. 2005;44:3311–3320. doi: 10.1021/ic050018g. [DOI] [PubMed] [Google Scholar]
- 12.Yim M.B., Chock P.B., Stadtman E.R. Copper, zinc superoxide dismutase catalyzes hydroxyl radical production from hydrogen peroxide. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5006–5010. doi: 10.1073/pnas.87.13.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgson E.K., Fridovich I. Interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide. Inactivation of the enzyme. Biochemistry. 1975;14:5294–5299. doi: 10.1021/bi00695a010. [DOI] [PubMed] [Google Scholar]
- 14.Kurahashi T., Miyazaki A., Suwan S., Isobe M. Extensive investigations on oxidized amino acid residues in H2O2-treated Cu, Zn-SOD protein with LC-ESI-Q-TOF-MS, MS/MS for the determination of the copper-binding site. J. Am. Chem. Soc. 2001;123:9268–9278. doi: 10.1021/ja015953r. [DOI] [PubMed] [Google Scholar]
- 15.Jewett S.L., Rocklin A.M., Ghanevati M., Abel J.M., Marach J.A. A new look at a time-worn system: oxidation of CuZn-SOD by H2O2. Free Radical Biol. Med. 1999;26:905–918. doi: 10.1016/s0891-5849(98)00274-3. [DOI] [PubMed] [Google Scholar]
- 16.Sampson J.B., Beckman J.S. Hydrogen peroxide damages the zinc-binding site of zinc-deficient Cu, Zn superoxide dismutase. Arch. Biochem. Biophys. 2001;392:8–13. doi: 10.1006/abbi.2001.2418. [DOI] [PubMed] [Google Scholar]
- 17.Hough M.A., Hasnain S.S. Crystallographic structures of bovine copper-zinc superoxide dismutase reveal asymmetry in two subunits: functionally important three and five coordinate copper sites captured in the same crystal1. J. Mol. Biol. 1999;287:579–592. doi: 10.1006/jmbi.1999.2610. [DOI] [PubMed] [Google Scholar]
- 18.Hough M.A., Strange R.W., Hasnain S.S. Conformational variability of the Cu site in one subunit of bovine CuZn superoxide dismutase: the importance of mobility in the Glu119-Leu142 loop region for catalytic function1. J. Mol. Biol. 2000;304:231–241. doi: 10.1006/jmbi.2000.4186. [DOI] [PubMed] [Google Scholar]
- 19.Healy E.F. A prion-like mechanism for the propagated misfolding of SOD1. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elam J.S., Taylor A.B., Strange R.W., Antonyuk S., Doucette P.A., Rodriguez J. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 21.Strange R.W., Antonyuk S.V., Hough M.A., Doucette P.A., Valentine J.S., Hasnain S.S. Variable metallation of human superoxide dismutase: atomic resolution crystal structures of Cu–Zn, Zn–Zn and as-isolated wild-type enzymes. J. Mol. Biol. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 22.Strange R.W., Hough M.A., Antonyuk S.V., Hasnain S.S. structural evidence for a copper-bound carbonate intermediate in the peroxidase and dismutase activities of superoxide dismutase. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks B.R., Bruccoleri B.E., Olafson B.D., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 24.Clair C.S. St, Gray H.B., Valentine J. Spectroelectrochemistry of copper-zinc superoxide dismutase. Inorg. Chem. 1992;31:925–927. [Google Scholar]
- 25.Buettner G.R. The pecking order of free radicals and antioxidants: lipid peroxidation, α-tocopherol, and ascorbate. Arch. Biochem. Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- 26.Wu G., Robertson D.H., Brooks C.L., III, Vieth M. Detailed analysis of grid-based molecular docking: a case study of CDOCKER - a CHARMm-based MD docking algorithm. J. Comput. Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 27.Im W., Lee M.S., Brooks C.L. Generalized born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 28.Tirado-Rives J., Jorgensen W.L. Contribution of conformer focusing to the uncertainty in predicting free energies for protein− ligand binding. J. Med. Chem. 2006;49:5880–5884. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 29.Richardson D.E., Yao H., Frank K.M., Equilibria D. A. Bennett. kinetics, and mechanism in the bicarbonate activation of hydrogen peroxide: oxidation of sulfides by peroxymonocarbonate. J. Am. Chem. Soc. 2000;122:1729–1739. [Google Scholar]
- 30.Khare S.D., Dokholyan N.V. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blech D.M., Borders C.L., Jr. Hydroperoxide anion, HO2−, is an affinity reagent for the inactivation of yeast Cu, Zn superoxide dismutase: modification of one histidine per subunit. Arch. Biochem. Biophys. 1983;224:579–586. doi: 10.1016/0003-9861(83)90245-x. [DOI] [PubMed] [Google Scholar]
- 32.Fisher C.L., Cabelli D.E., Hallewell R.A., Beroza P., Lo T.P., Getzoff E.D., Tainer J.A. Computational, pulse‐radiolytic, and structural investigations of lysine‐136 and its role in the electrostatic triad of human C u, Z n superoxide dismutase. Proteins: Struct. Funct. Bioinf. 1997;29:103–112. doi: 10.1002/(sici)1097-0134(199709)29:1<103::aid-prot8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 33.Sinet P.M., Garber P., Sinet P.M., Garber P. Inactivation of the human CuZn superoxide dismutase during exposure to O2− and H2O2. Arch. Biochem. Biophys. 1981;212:411–416. doi: 10.1016/0003-9861(81)90382-9. [DOI] [PubMed] [Google Scholar]
- 34.Salo D.C., Pacifici R.E., Lin S.W., Giulivi C., Davies K.J. Superoxide dismutase undergoes proteolysis and fragmentation following oxidative modification and inactivation. J. Biol. Chem. 1990;265(119):19–27. 1990. [PubMed] [Google Scholar]
- 35.Bonini M.G., Gabel S.A., Ranguelova K., Stadler K., DeRose E.F., London R.E., Mason R.P. Direct magnetic resonance evidence for peroxymonocarbonate involvement in the Cu, Zn-superoxide dismutase peroxidase catalytic cycle. J. Biol. Chem. 2009;284:14618–14627. doi: 10.1074/jbc.M804644200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medinas D.B., Cerchiaro G., Trindade D.F., Augusto O. The carbonate radical and related oxidants derived from bicarbonate buffer. IUBMB Life. 2007;59:255–262. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]
- 37.Kim S., Ginsbach J.W., Lee J.Y., Peterson R.L., Liu J.J., Siegler M.A., Sarjeant A.A., Solomon E.I., Karlin K.D. Amine oxidative N-dealkylation via cupric hydroperoxide Cu-OOH homolytic cleavage followed by site-specific Fenton chemistry. J. Am. Chem. Soc. 2015;137:2867–2874. doi: 10.1021/ja508371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.