Graphical abstract
Keywords: Polynucleotide phosphorylase (PNPase); PNPase structure; PNPase recognition; RNA oxidation; 8-oxo-7,8-dihydroguanosine; Oxidative stress; MD simulations
Abstract
The oxidation of RNA has been implicated in the development of many diseases. Among the four ribonucleotides, guanosine is the most susceptible to oxidation, resulting in the formation of 8-oxo-7,8-dihydroguanosine (8-oxoG). Despite the limited knowledge about how cells regulate the detrimental effects of oxidized RNA, cellular factors involved in its regulation have begun to be identified. One of these factors is polynucleotide phosphorylase (PNPase), a multifunctional enzyme implicated in RNA turnover. In the present study, we have examined the interaction of PNPase with 8-oxoG in atomic detail to provide insights into the mechanism of 8-oxoG discrimination. We hypothesized that PNPase subunits cooperate to form a binding site using the dynamic SFF loop within the central channel of the PNPase homotrimer. We evolved this site using a novel approach that initially screened mutants from a library of beneficial mutations and assessed their interactions using multi-nanosecond Molecular Dynamics simulations. We found that evolving this single site resulted in a fold change increase in 8-oxoG affinity between 1.2 and 1.5 and/or selectivity between 1.5 and 1.9. In addition to the improvement in 8-oxoG binding, complementation of K12 Δpnp with plasmids expressing mutant PNPases caused increased cell tolerance to H2O2. This observation provides a clear link between molecular discrimination of RNA oxidation and cell survival. Moreover, this study provides a framework for the manipulation of modified-RNA protein readers, which has potential application in synthetic biology and epitranscriptomics.
1. Introduction
Chemical modifications occur in all classes of RNA [1]. These modifications contribute to RNA function by affecting their coding, folding and stability and can regulate their interactions with other cellular factors (i.e. RNAs, DNAs, and proteins) [2], [3]]. Some modifications are reversible and dynamically regulate gene expression in response to environmental conditions. For instance, N6-methyladenosine (m6A) modifications show increased levels in mRNAs near damaged DNA sites in U2OS cells exposed to UV radiation, promoting the recruitment of the DNA repair machinery to facilitate cell survival [4]. Moreover, certain chemical modifications can also result from interactions with reactive oxygen species (ROS) formed by cellular metabolism or introduced by exogenous factors (for example, UV radiation, hypoxia, nutrient deprivation). These reactive species can hydroxylate guanosine to produce 8-oxo-7,8-dihydroguanosine (8-oxoG) oxidative modifications in RNA. 8-oxoG modifications are implicated in deregulation of cellular processes by direct alteration of RNA function, stability, and processability [5], [6]]. Recent findings suggest that this modification also plays signaling and regulatory roles during the rapid reprogramming of cellular function in response to oxidative stress [7], [8]]. At the molecular level, 8-oxoG can pair with both cytidine and adenosine [9], causing alterations in mRNA decoding and a reduction of protein expression [10], [11]]. When present in miRNAs, 8-oxoG affects recognition of non-canonical targets in heart cells, signaling the cells to undergo apoptosis [12]. Remarkably, accumulation of certain 8-oxoG-modified mRNAs could contribute to pathogenesis, especially in neurological conditions such as Parkinson’s disease and Alzheimer’s disease [13].
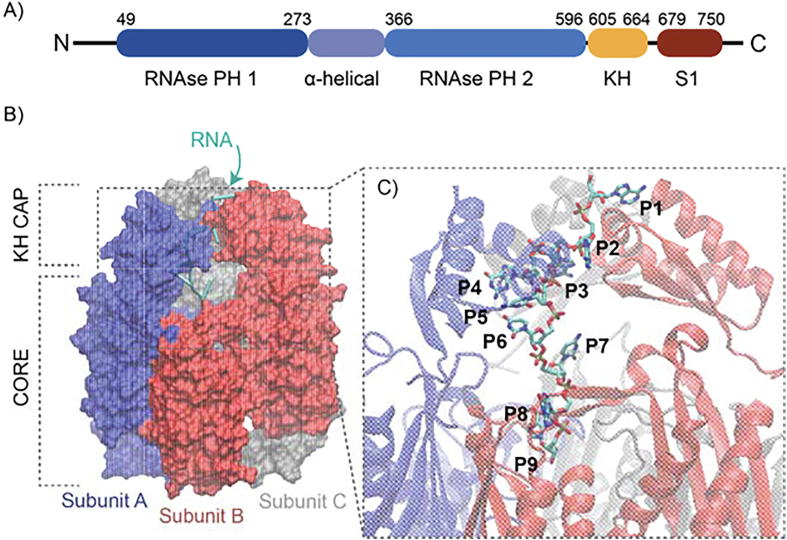
Oxidized RNA appears to turn over faster than its intact counterpart [14], suggesting that cells have evolved mechanisms for its recognition and processing. To this end, surveillance proteins that have evolved to play a role in RNA quality control may directly recognize oxidized RNAs [15]. One protein shown to specifically interact with 8-oxoG in RNA is polynucleotide phosphorylase (PNPase), a highly conserved 3′ to 5′ exoribonuclease in bacteria and eukaryotes that modulates steps in RNA metabolism and degradation of RNAs [16]. PNPase is a multi-domain protein composed of two homologous RNase PH-like domains and two RNA binding domains; a K homology (KH) domain and an S1 domain (Fig. 1A). The enzyme is composed of three identical PNPase subunits assembled into a torus-shape core composed of the RNase PH-like domains, from which the KH and S1 domains extend (Fig. 1B). Notably, PNPase plays an important role in supporting cellular tolerance to oxidative stress. The deletion of the pnp gene (encoding PNPase) in Escherichia coli – and the resulting lack of PNPase expression – has been reported to significantly decrease cell viability under H2O2 exposure [17]. A similar effect on cell viability was later described in a PNPase knockdown in HeLa cells exposed to H2O2 [18]. Human and bacterial PNPases specifically bind to oxidized RNA [19], [20]]; however, in vitro evidence suggests that PNPase does not directly degrade 8-oxoG-containing RNA substrates through its catalytic site [21]. How PNPase discriminates 8-oxoG-modified RNAs and how 8-oxoG modifications prevent degradation of RNA by PNPase require further investigation.
Fig. 1.
Domains and structure of E. coli PNPase bound to single-stranded RNA (ssRNA). A) Domain organization of E. coli PNPase. B) Structure of the modeled ssRNA-PNPase complex. The ssRNA-protein structure was truncated to the amino acids surrounding the RNA to reduce the computational time required to investigate the complex through MD simulations and free energy calculations. The three PNPase subunits are shown in blue, red, and grey surface representation. The ssRNA is shown in cartoon representation. C) Magnified structure of the ssRNA within the PNPase tunnel. The RNA strand is shown in licorice representation. PNPase subunits A, B, and C are shown in blue, red and grey cartoon representation, respectively. The RNA nucleotide positions P1 – P9 are labeled in black. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As part of the momentum surrounding the field of RNA modifications, new approaches that aid in deciphering the unique functions of these marks in regulation of gene expression and their connection to human disease are currently sought. Specifically, attention needs to be placed on expanding the limited number of known RNA modification effectors (readers, writers, and erasers) and on elucidating the roles of RNA modifications. Molecular dynamics (MD) simulations have become a powerful tool for characterizing structural interactions of macromolecular complexes at an atomic level, revealing functional mechanisms [22]. However, the application of MD simulations in studies of modified RNA structures is challenging, in part, due to the lack of physical models available for modified nucleotides [23]. Recent efforts led to the empirical parametrization of more than 100 RNA modifications in existing force fields such as AMBER [24] and CHARMM [25]. More researchers are starting to adopt MD simulations as a tool to gain insights into the biochemistry of RNA modifications [26], [27]] and into the recognition and specificity of protein readers [28], [29]].
In this work, we used the E. coli PNPase molecular model bound to 8-oxoG RNA reported in our previous study [28] to identify amino acids at the 8-oxoG binding site and evolved them to improve 8-oxoG affinity using a semi-rational approach. This approach implemented computational biophysical constraints and a multi-stage computational screening strategy to select the most promising mutants for 8-oxoG binding experiments. Through in vitro binding assays, we validated that the generated mutant PNPases increased affinity and/or selectivity for 8-oxoG-containing RNA. Additionally, we evaluated strains expressing mutant PNPases on cellular tolerance to oxidative stress by complementation assays in E. coli K12 Δpnp strains, demonstrating the contribution of several mutants towards increased cell viability under H2O2 exposure. Importantly, as part of this work, we established a protein evolution methodology based on MD simulations and free energy calculations that resulted in enhanced protein affinity to modified RNAs. Together, this study provides a framework for the programmable manipulation of protein readers that act on RNA modifications. The principles established here are of broad relevance for synthetic biology applications in epitranscriptomics, given the importance of being able to alter RNA modification systems for therapeutics [30], [31]].
2. Materials and methods
2.1. Modeling of E. coli PNPase in complex with an ssRNA
We used the molecular model of E. coli PNPase bound to a single-stranded RNA (ssRNA) reported in our previous study [28] as the structural basis for this new study. This model was generated through homology modeling of positions 517–549 of the unbound E. coli PNPase structure previously resolved by X-ray crystallography [32] (PDB ID : 3GCM) to the RNA-bound structure of Caulobacter crescentus PNPase resolved by X-ray crystallography [33] (PDB ID : 4AM3). To model the conformation of the bound structure, we docked a nine nucleotide long ssRNA with an identical sequence used for the crystal structure of C. crescentus PNPase in complex with RNA (PDB ID: 4AM3): 5′ – AAAGCUCGG – 3′, with guanosines introduced to positions P4, P8, and P9 (Fig. 1C). The guanosines were introduced to provide molecular and energetic references for the analysis of the 8-oxoG interactions. We then introduced energy minimization steps (comprising of steepest descent and adopted basis Newton-Raphson) to alleviate any steric clashes within the complex structure and subsequently simulated the complex in explicit solvent for 5 ns to produce the starting structure for the 50 ns explicit solvent MD simulations using CHARMM [34] (see Section 2.3). The simulation setup and parameters are the same as those described for the 50 ns MD simulations (see Section 2.3). The short simulation was sufficient to alleviate unfavorable interactions within the complex structure without deviating significantly from its initial structure. To model the E. coli PNPase structure bound to the 8-oxoG-containing ssRNA, we introduced 8-oxoG using the procedure detailed in our previous study [28]. Briefly, we parametrized 8-oxoG using CGenFF [35], and then replaced the guanosines with 8-oxoGs at positions P4, P8 or P9 in CHARMM, producing the following modified RNAs: P4: 5′ – AAA(8-oxoG)CUCGG – 3′, P8: 5′ – AAAGCUC(8-oxoG)G – 3′ and P9: 5′ – AAAGCUCG(8-oxoG) – 3′ [34]. Subsequently, the structure was energetically minimized to alleviate any steric clashes and used as the initial structure for mutagenesis simulations.
2.2. Semi-rational computational evolution of RNA-protein interactions
We combined biophysical (structural and energetic) and bioinformatics analyses to identify the key residues interacting with 8-oxoG in the PNPase binding site. In the biophysical analysis, we analyzed the structures obtained from triplicate 50 ns explicit solvent MD simulations of the RNA strand (with sequence 5′ – AAAGCUCGG –3′) in complex with the protein, to study the interactions between each nucleotide position and neighboring PNPase residues in the protein. The simulation setup, parameters and procedure are detailed in the Section 2.3 and the all-atom evaluation and rating stage of our previous study, in which the full system (i.e., residues 27–142, 336–453, 517–549) was investigated in explicit solvent all-atom representation. Upon completion of the 50 ns explicit solvent MD simulations, we performed a per-nucleotide interaction free energy analysis to identify the key interacting nucleotide positions (see Section 2.5). We subsequently introduced an 8-oxoG at the selected position and then performed triplicate 50 ns explicit solvent simulations. Upon completion of the 50 ns explicit solvent MD simulations, a per-residue interaction free energy analysis was performed (see Section 2.5).
In the bioinformatics analysis, we extracted 782 PNPase protein sequences from the UNIPROT database. Based on the analysis, we allowed the placement of the following sets of amino acids at the three residue positions investigated: G, N, S, or T at position 76, F or Y at position at 77, and F, H, I, K, L, M, N, Q, S, T, or V at position 78. Due to the initial screening nature of this approach, the entire ssRNA-protein complex (described in Section 2.1) was truncated to include only the binding site (i.e., residues within 10 Å of any atom of the RNA strand), and then short (5 ns) simulations were implemented in implicit solvent. The simulation setup, parameters and procedure were the same as those used in our previous study [28]. The mutations to the three investigated residues in the binding site and the 8-oxoG modification at the examined position of the RNA substrate were introduced to the truncated structure in CHARMM such that the original torsion angles and orientation of the residues were preserved during the modeling. Upon completion of the short implicit solvent MD simulations, we performed interaction energy calculations between the mutated site and the RNA strand containing 8-oxoG (as previously detailed in [28]), which served to screen out any combinations of mutants that did not predict more energetically favorable conditions with the RNA modification relative to the wild type PNPase. A relaxed criterion was preferred to reduce the number of false negatives (i.e., combinations of mutants which could presumably be worthy of further investigation). Any false positives selected in the initial screening due to the relaxed criterion were additionally evaluated using longer simulations and free energy calculations (described below) to screen them out and select only the most promising mutants for experimental testing.
As a final assessment, single 50 ns explicit solvent MD simulations were performed on the selected PNPase mutants from the initial screening in order to refine the mutant PNPase-RNA complex structures and the intermolecular interactions therein. These refined complexes were then used to assess the most energetically favored PNPase mutants for 8-oxoG binding. We then calculated the average association free energy of the PNPase mutants binding to the RNA strand containing 8-oxoG and selected PNPase mutants with improved average association free energies for 8-oxoG compared to the wild type E. coli PNPase motif at that location (SFF). An additional two 50 ns explicit solvent MD simulations were performed for the selected mutants binding to 8-oxoG such that each of the selected mutants were simulated in triplicate 50 ns explicit solvent MD simulations to ensure reproducibility of the single runs.
2.3. Molecular dynamics simulations
The 50 ns explicit solvent MD simulations described in the above sections were performed in CHARMM [34] using the CHARMM36 force field [36] as described in our previous study [28]. Additional topologies and parameters for 8-oxoG were generated using CGenFF [35]. The entire PNPase – RNA strand complex system, described in Section 2.1, was used as the initial structure for all 50 ns explicit solvent MD simulations. For the 50 ns explicit solvent MD simulations of PNPase in complex with an RNA strand containing 8-oxoG, the modification was introduced in CHARMM [34] such that the original torsion angles and orientation of the residues were preserved during the modeling. Likewise, for the 50 ns explicit solvent MD simulations of all the PNPase mutants, the amino acid substitutions were introduced in CHARMM [34] such that the original torsion angles and orientation of the residues were preserved during the modeling. Upon the introduction of the 8-oxoG or the amino acid substitutions, we introduced energy minimizations to alleviate any steric clashes that may have occurred during their substitution. Prior to the 50 ns explicit solvent MD simulation, the complex PNPase – RNA structure was solvated in a 120 Å3 water box. All protein and RNA backbone atoms were constrained using a harmonic force of 1.0 kcal/(mol·Å2) and all heavy side-chain atoms were constrained using 0.1 kcal/(mol·Å2) for 1 ns. After equilibration, all constraints were released and PNPase residues outside of 20 Å from any atom of the initial RNA fragment were subjected to 1.0 kcal/(mol·Å2) for backbone atoms and 0.1 kcal/(mol·Å2) for heavy side chain atoms. In this stage, each complex was simulated for 50 ns with simulation snapshots extracted every 20 ps. The 50 ns explicit solvent MD simulations were performed using the Leap-Frog Verlet algorithm under isobaric and isothermal conditions with the pressure set to 1.0 atm and the temperature held at 300 K using the Hoover thermostat. We applied fast table lookup routines [37] for nonbonded interactions and implemented the SHAKE algorithm [38] to constrain the bond lengths to hydrogen atoms.
2.4. Association free energy calculations
To identify the most energetically favorable PNPase mutants binding to 8-oxoG, we calculated the association free energy of each PNPase mutant bound to 8-oxoG over the entire 50 ns production run using the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) approximation [39], [40]]. The association free energies were calculated for each simulation snapshot extracted every 20 ps in each of the 50 ns explicit solvent MD simulations. Subsequently, we calculated the block average association free energy every 12.5 ns. Thus, the reported average and standard deviation association free energy values for the single runs are calculated over 4 measurements, where the first, second, third, and fourth measurement corresponds to the individual average association free energy values of the first, second, third, and fourth 12.5 ns segment of the 50 ns explicit solvent MD simulation production runs. For the triplicate 50 ns explicit solvent MD simulations of the promising PNPase mutants binding to 8-oxoG, the association free energy calculations were performed to ensure reproducibility, and the reported average and standard deviation association free energy values for the triplicate runs are calculated over three “measurements” corresponding to the individual average association free energy values of the first, second, and third 50 ns explicit solvent MD simulation. Additional information on the MM-GBSA association free energy calculations is provided in our previous study [28].
2.5. Interaction free energy analysis of residue-nucleotide pairs and independent groups (residues, nucleotides, nucleobases)
After conducting MD simulations of the RNA-protein complex, we applied the MM-GBSA approximation [39], [40]] to evaluate the interaction free energy of all possible interacting residue-nucleotide pairs using Equation [1], analogously to previous studies [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51]]. The pair-wise interaction free energy values between each PNPase residue and each RNA nucleotide were subsequently used to calculate per-residue interaction free energies (interaction free energy contribution of each PNPase residue to an RNA nucleotide) and per-nucleotide interaction free energies (interaction free energy contribution of each RNA nucleotide to the entire PNPase binding site).
| (1) |
The first, second and third components of the equation above represent the polar, van der Waals and non-polar solvation interactions free energies between P and R, respectively. The variable P corresponds to a given amino acid in the protein and R corresponds to the nucleotide at a given position in the ssRNA. The variable PR corresponds to the amino acid – nucleotide complex. The interaction-free energies of m = 1 to f (=2500) frames were summed and averaged.
The polar component of the total interaction free energy is comprised of electrostatic interaction () and generalized-Born () energy contributions between the residue P and nucleotide R. The polar component represents the interaction between the residue P and nucleotide R, and the interaction between residue P and the solvent polarization potential induced by the nucleotide R. The non-polar component (sum of the second and third term) consists of the van der Waals interactions between the residue P and the nucleotide R, in addition to the change in the non-polar solvation free energy due to binding (). The non-polar interaction free energy term represents the non-polar interactions with the surrounding solvent and cavity contributions.
The solvation terms were determined using the grid-based GBMV implicit solvent model [52]. These calculations were executed with the non-polar surface tension coefficient, γ, set to 0.03 kcal mol−1 Å−2. The generalized-Born energy contribution () and solvent accessible surface area () are affected by the location of P and R in the complex. To compute the term in Eq. (1), all atoms were included, and the charges of atoms outside the groups PR, P, and R were set to zero in each calculation of the terms , , and , respectively. The term expresses the difference in solvent accessible surface areas of the residue P and nucleotide R within the complex and in unbound states. For these calculations, we used infinite cutoff values.
Using the individual residue-nucleotide pairwise interaction free energy values, we calculated per-residue and per-nucleotide interaction free energies. We define the per-residue interaction free energy as the sum of all the energetic contributions of each residue interacting with a given nucleotide, and we define the per-nucleotide interaction free energy as the sum of all the per-residue interactions occurring with the given nucleotide.
In addition, we also performed per-nucleobase interaction free energy calculations, analogously to Ref. [53], in which we calculated the interaction free energy contributions of the PNPase residues to the nucleobase, rather than the entire nucleotide. For these calculations, in Eq. (1), the variable P corresponds to a given amino acid in the protein and R corresponds to the nucleobase at a given position in the ssRNA. The variable PR corresponds to the amino acid – nucleobase complex. To compute the term in Eq. (1), all atoms were included, and the charges of atoms outside the groups PR, P, and R (corresponding to the nucleobase and not the sugar or phosphate group of the given nucleotide) were set to zero in each calculation of the terms , , and , respectively. The phosphate and sugar group atoms were included in all energy calculations with zero charge, aiming at including the backbone screening effect between interacting atoms [53].
2.6. Reagents, bacterial strains and plasmids
The 8-oxoG RNA oligonucleotide 24-mer (with sequence: [NN(8-oxoG)N]6, where N is A, G, C or U) and the 24-mer unmodified RNA oligo (with sequence: [NNGN]6) were custom synthesized by GeneLink (Orlando, FL). The constitutive promoter plasmid (containing a lpp promoter and a synthetic RBS B31, with sequence CCCATCAAAAAAATATTCTCAACATAAAAAACTTTGTGTAATACTTGTAACGCTTCTAGAGTCACACAGGAAACCTACTAG) was kindly provided by Hal Alper’s group at UT Austin. ATP [γ-32P] (3000 Ci/mmol 10 mCi/ml, 100 µCi) for 5′-end labeling of RNA oligos was purchased from PerkinElmer (Waltham, MA). The E. coli K12 BW25113 Δpnp strain from the Keio collection [54] and the E. coli K12 MG1655 were kindly provided by Jeffrey Barrick group at UT Austin. Primers and plasmids used in this study are listed in Supplementary Tables S1 and S2.
2.7. FLP recombination of E. coli strain from Keio collection
To eliminate the kanamycin resistance cassette from the E. coli K12 BW25113 Δpnp strain from the Keio collection, we used FLP recombination based on the protocol adapted from [54], [55]]. Briefly, electrocompetent cells of the E. coli Δpnp strain were generated and then transformed with the plasmid pCP20 [56]. pCP20 has a temperature-sensitive origin of replication, confers ampicillin and chloramphenicol resistance and encodes the FLP recombinase. Cells were plated on a LB (Fischer Scientific, Hampton, NH) + ampicillin (50 μg/ml, VWT, Radnor, PA) plate overnight at 30 °C. Recombination was induced from a single colony that was inoculated in LB overnight at 43 °C. This step allows the induction of expression of the FLP recombinase and selects for loss of pCP20. Then, a 100fold dilution of the overnight culture was made using fresh LB and plated on a LB plate overnight at 30 °C. Ten individual colonies were patched onto LB + kanamycin (VWR, Radnor, PA), LB + ampicillin and LB plates, and grew overnight at 37 °C (for LB and LB + kanamycin plates) and at 30 °C for LB + ampicillin. Successful colony candidates in the LB plate that demonstrated sensitivity to both kanamycin and ampicillin were incubated in LB overnight at 37 °C.
The validation of the removal of the kanamycin resistance cassette was performed by PCR of genomic DNA extracted using Wizard Genomic DNA purification kit (Promega, Madison, WI). Sanger sequencing was used to validate that no frame shifts were introduced. Primers were designed flanking the pnp gene, 240 bp upstream (from E. coli strain K12, accession U00096.3, region 3,311,408 – 3,311,427) and 258 bp downstream of the target gene (region 3,308,756 – 3,308,775) (see Supplementary Table S1 for sequence information). Correct removal of the cassette was detected by the length of the amplicon in 1% agarose gel electrophoresis stained with ethidium bromide (Invitrogen, Carlsbad, CA). The WT E. coli K12 MG1655 strain was used as positive control for gene presence.
2.8. Cloning and site-directed mutagenesis
The pnp sequence (from E. coli strain K12, accession U00096.3, region: 3,309,033 – 3,311,168) was synthesized by GenScript (Piscataway, NJ) and then cloned into the pET28a vector between NdeI and BamHI restriction sites, resulting in the pET28a-pnp construct. To introduce mutations in the Ser76-Phe77-Phe78 site, we used the Q5 Site Directed Mutagenesis Kit (NEB, Ipswich, MA) and NEBase Changer for primer design (https://nebasechanger.neb.com/). The primers used for mutagenesis are listed in Supplementary Table S1. We transformed the ElectroMAX DH5α-E Cells (Invitrogen, Carlsbad, CA) with the mutagenized plasmid by electroporation using a GenePulser Xcell electroporation system (Biorad, Hercules, CA), followed by an hour incubation in an I26 rotatory shaker (New Brunswick Scientific, Edison, NJ) at 37 °C in SOC media.
The cells were then plated on BD Difco LB Broth (Fischer Scientific, Hampton, NH) and agar (Fischer Scientific, Hampton, NH) plates supplemented with 50 μg/ml kanamycin sulfate (VWR, Radnor, PA) for selection. Individual colonies were inoculated into liquid LB medium and grown overnight for plasmid isolation the following day. Plasmid preparations were then submitted to the Genomic Sequencing and Analysis Facility at the University of Texas at Austin and confirmed by Sanger sequencing using primers listed in Supplementary Table S1. Once confirmed, miniprep DNA was used to transform E. coli BL21(DE3) competent cells (NEB, Ipswich, MA) following the supplier protocol.
To generate the strain used for oxidative stress assays, a pnp gene amplicon from CML366 (see Supplementary Table S2) was cloned into a plasmid containing a lpp constitutive promoter with a synthetic RBS B31 by Gibson Assembly (Primers for PCR amplification in Supplementary Table S1). Mutagenesis of the SFF site was performed in the constitutive plasmid as described above to introduce the modeled variants. The selection of colonies was performed using LB and agar plates supplemented with 25 μg/ml of chloramphenicol (Sigma-Aldrich, St. Louis, MO). Plasmids harbored by transformants were isolated and sequence confirmed by Sanger sequencing using primers listed in Supplementary Table S1.
2.9. Protein expression and purification
Frozen BL21(DE3) cells containing the pET28a-pnp mutants were used to start cultures for protein expression. Cells were grown in LB media with 50 μg/ml kanamycin sulfate until an OD600 of 0.6 was reached. The OD600 was measured in duplicate using 200 µl of sample in a 96-well clear plate and analyzed in a plate reader (BioTek, Winooski, VT). Then, protein expression was induced by addition of IPTG (MilliporeSigma, Burlington, MA) to a final concentration of 1 mM for 3 hrs at 37 °C with constant shaking. Cells were centrifuged and then resuspended in lysis buffer in 50 mM NaH2PO4, 300 mM NaCl, 5 mM MgCl2, and 15 mM imidazole (Fischer Scientific, Hampton, NH) before lysing via sonication (Q125 Sonicator, QSonica, Newton, CA). The lysate was centrifuged at 3,320 g for 30 min at 4 °C. The supernatant (soluble fraction) was collected and stored for protein purification.
Mutant PNPase variants were purified by affinity purification of the 6x-his-tagged protein using Ni-NTA Agarose beads following the protocol of the supplier (Qiagen, Hilden, Germany). Briefly, 1 mL of pre-washed Ni-NTA beads was mixed with 10–50 mg of the soluble fraction of the lysate followed by incubation on a rotator at 4 °C for 1 hr. After incubation, three washes were performed with increasing imidazole concentrations (25 mM, 35 mM, 50 mM). Next, the His-tagged protein was eluted in a solution containing 250 mM imidazole. The protein was then concentrated 10-50X using Amicon Ultra-15 centrifugal filters (with a cutoff of 30 kDa, MilliporeSigma, Burlington, MA) at 4 °C for 10-minute intervals (re-homogenizing each time) and buffer exchanged to a buffer containing 20 mM Tris Buffer (pH 7.0) and 100 mM NaCl. The resulting protein samples were diluted in one volume of 80% glycerol and stored at −20 °C. The purity of the proteins was evaluated by SDS-PAGE, and detection of the proteins was confirmed by Western blotting using anti-6x-his-tag monoclonal antibody (C-terminus, clone 3D5, Thermo Fisher, Waltham, MA).
2.10. Preparation of 32P-end-labeled RNA
The 8-oxoG containing oligomer and the unmodified oligomer were radiolabeled using T4 polynucleotide kinase (NEB, Ipswich, MA) as described by the manufacturer. After labeling, RNA was cleaned up by ethanol precipitation. This was done by first adding 1 M Tris buffer (pH 8.0) and 1 M sodium acetate (pH 5.2) to the reaction mixture to bring the final concentrations to 50 mM and 0.3 M respectively. Two volumes of phenol/chloroform/isoamyl alcohol (25:24:1) (Fisher Scientific, Hampton, NH) were then added and the solution was vortexed for one minute followed by centrifugation at 15,000 g for 2 min to achieve phase separation. The aqueous (top) phase was collected, and 1 µl of GlycoBlue Coprecipitant (Thermo Fisher, Waltham, MA) and 2.5 volumes of chilled 100% absolute ethanol (OmniPur, 200 Proof, Millipore Sigma, Burlington, MA) were added. The solution was mixed and then incubated overnight at −20 °C. The following day, the solution was centrifuged at 4 °C at 15,000 g for 15 min. The supernatant was removed and then washed with 95% ethanol followed by centrifugation at 15,000 g for 5 min. The supernatant was discarded, and the pellet was dried in a vacufuge plus (Eppendorf, Hamburg, Germany) for 5 min before resuspension in Molecular Biology Grade Water (Quality Biological, Gaithersburg, MD).
2.11. Electrophoretic mobility shift assays and KD determination
RNA-protein interactions were evaluated by EMSAs following the protocol by Hellman and Fried [57] with a few modifications. Running conditions were performed as described in ref. [58]. The binding reactions were conducted in 12 µl containing 1X TMK buffer [50 mM Tris-HCl pH 7.5, 50 mM KCl, and 10 mM (CH3COO)2 Mg], 10% glycerol, and 500 nM heparin (Sigma Aldrich, St. Louis, MO, F.W. ~ 6000 g/mol). 1.2 nmol of radiolabeled RNA (3,000 cpm/ul when labeled) was mixed with varying amounts of PNPase. The reactions were incubated for 1 hr at 37 °C and resolved via native electrophoresis in 5% glycerol and 5% polyacrylamide (VWR, Radnor, PA) gels in 0.5x TBE (VWR, Radnor, PA) at 4 °C for 2 h at 180 V. The gel was dried using a model 583 gel dryer (Bio-Rad, Hercules, CA) and exposed to a storage phosphor screen (GE Healthcare, Chicago, IL) overnight. The phosporimage was acquired using a Typhoon 9500 (GE, Marlborough, MA) and the bands were quantified using CLIQS (TotalLab, Newcastle upon Tyne, England). KD values were derived using the modified Hill equation [59] and solved using the lsqcurvefit function in MATLAB (Version R2019A, MATHWORKS, Natick, MA).
2.12. Hydrogen peroxide survival assays
E. coli K12 Δpnp strains containing mutagenized variants of the PNPase SFF motif (NYT, NYM, GFT, NFH and SYH mutant PNPases, see Supplementary Table S2) and E. coli K12 MG1655 were grown overnight in 5 mL LB with 50 µg/ml chloramphenicol or LB respectively in a shaking incubator at 37 °C. Five replicates were inoculated for assessment. The following day, 500 µl of each culture was passaged into 5 mL LB + chloramphenicol or LB and grown for one hour at 37 °C. The ODs were normalized with LB to the lowest OD of all cultures used in the experiment (0.5 – 0.6). 150 µl of each culture was then mixed with 150ul of 40 mM H2O2 in 1X PBS (pH 7.4) in a sterile 96-well plate and incubated at room temperature for 20 min. 20ul of each cell mixture was then serially diluted in 180 µl of PBS down to the 10-7 dilution. 100 µl of the 10−3 and 10−4 dilutions were plated and incubated overnight at 37 °C for CFU counts. Spot plates were also made with 10 µl from each dilution.
2.13. Bioinformatics analysis
To analyze the biological relevance of mutations in the SFF binding site, we obtained 782 non-redundant PNPase sequences from the UNIPROT database that were queried with the search term “polynucleotide phosphorylase” and filtered to “reviewed sequences” (manually curated). A multiple sequence alignment was conducted with Clustal Omega version 1.2.4 [60] to generate a sequence consensus using WebLogo3 [61]. Taxonomy distribution of the sequences was visualized with the module matplotlib (version 3.0.2) in Python (version 3.7.2).
We analyzed the occurrence of the characterized PNPase motifs in ~26,000 PNPase sequences collected from querying the NCBI protein database for “polynucleotide phosphorylase”. We limited the output to full-length sequences or longer than 600 amino acids in the RefSeq database of non-redundant, well-annotated protein sequences. Before aligning the sequences, we split the fasta file into seven approximately equally sized files to provide input files below the limit of 4,000 sequences permitted by the multiple alignment program Clustal Omega. The E. coli PNPase sequence from UNIPROT was included in each analysis to be used as reference. The resulting alignments were saved as CLUSTAL files and were manually checked for the highly conserved “R/K-R-E” region immediately downstream of the SFF site. The block with the R/K-R-E region on each of the seven CLUSTAL files was combined into a single CLUSTAL file. Given the list of characterized mutant PNPases, we searched for the presence of the mutant amino acid motifs in the combined CLUSTAL file using an in-house script. To obtain the species name, taxonomic lineage, and full PNPase sequence from the CLUSTAL files (initially annotated with GenInfo (gi) identifiers), we queried gi’s using Biopython Entrez Package (version 1.73).
2.14. Area analysis of spot plates
Analysis of cell spots was conducted using ImageJ. The color threshold was manipulated to highlight areas of high saturation of cell spots. Then, the rectangular selection tool was used to isolate each spot, and a particle analysis was used to obtain the total area occupied by cells. This data was compiled for each of the three exposed trials and normalized using the PBS control, which was analyzed using identical methods.
2.15. Statistical analysis
In single 50 ns explicit solvent MD simulation runs, the reported average and standard deviation values were calculated over four measurements, each corresponding to one fourth of the 50 ns explicit solvent MD simulation run. In the triplicate 50 ns explicit solvent MD simulation runs, the reported average and standard deviation values are calculated over three measurements, corresponding to the individual average values of all the 50 ns explicit solvent MD simulation.
We conducted all described experimental measurements as either triplicates or duplicates. All data were presented as the mean ± one standard deviation. Statistical analysis between groups was determined by student’s t-test in JMP (SAS, Cary, NC) with a significance of 0.05.
3. Results
3.1. The S76-F77-F78 grooves from two PNPase subunits cooperate to form an 8-oxoG binding site
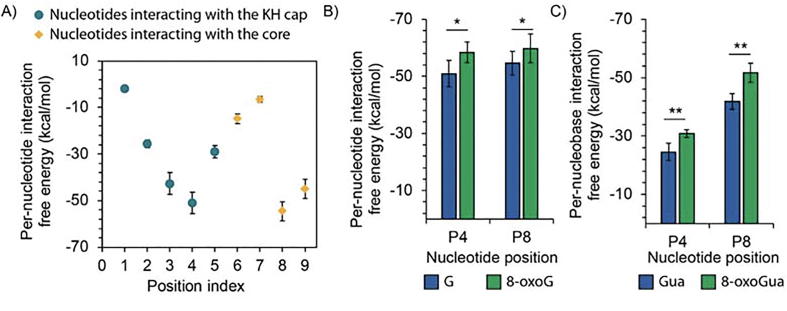
To determine the binding site for 8-oxoG binding, we investigated the interaction free energy in the section of the RNA path that is resolved in the model structure of the bound E. coli PNPase with unmodified RNA (Fig. 1B). We performed triplicate 50 ns explicit solvent MD simulations and subsequently calculated the per-nucleotide interaction free energies, defined as the sum of the average polar and non-polar energetic contributions of all the residues interacting with a single nucleotide position (see Section 2.5). This analysis indicates two separated regions in the ssRNA-protein complex with minimum values in the interaction free energy, one involving the KH domain and position P4 of the ssRNA and one involving the RNase PH-1 core domain and position P8 (Fig. 2A).
Fig. 2.
Per-nucleotide interaction of single-stranded RNA (ssRNA) within the tunnel of PNPase. A) Interaction free energy between each individual RNA nucleotide and PNPase residues. The position indices correspond to those shown in Fig. 1C. B) Interaction free energy between PNPase residues and either guanosine (G) or 8-oxo-7,8-dihydroguanosine (8-oxoG) (base + sugar + phosphate groups) individually introduced at the indicated position of the ssRNA. C) Interaction free energy of the isolated nucleobase, either guanine (Gua) or 8-oxo-7,8-dihydroguanine (8-oxoGua). The average and standard deviation interaction free energy values in panel A – C are calculated over triplicate 50 ns explicit solvent MD simulations of the RNA-protein complex. Error bars plotted as ± one standard deviation. Statistical analysis conducted using one-tailed homoscedastic t-test, * refers to p-value < 0.05, and ** refers to p-value < 0.001.
The first binding region is located in the KH domain, which is well known to participate in RNA binding [17]. Genomic deletion of the KH domain reduces the RNA affinity of PNPase by 28-fold, which is paralleled by loss of catalytic activity [62]. The second site is located in the dynamic FFRR loop of the RNase PH-1, previously implicated in the binding of unmodified RNA [32], [63]]. It is worth noting, that both sites are located in highly conserved regions [64], for example, the FFRR loop groups most of the conserved residues in the first core domain [64], [65]]. Therefore, we hypothesized that these sites are potential binding pockets for 8-oxoG.
We next introduced 8-oxoG in the ssRNA at either position P4 or P8 of the complex, and then conducted triplicate 50 ns explicit solvent simulations of the entire ssRNA-protein complex. As seen in Fig. 2B, the per-nucleotide energy calculations indicate that PNPase interacts more favorably with 8-oxoG than with guanosine at both position P4 and position P8. To provide further insights into how PNPase discriminates 8-oxoG RNA from normal RNA, we conducted energetic calculations to determine the driving forces at play on these binding sites.
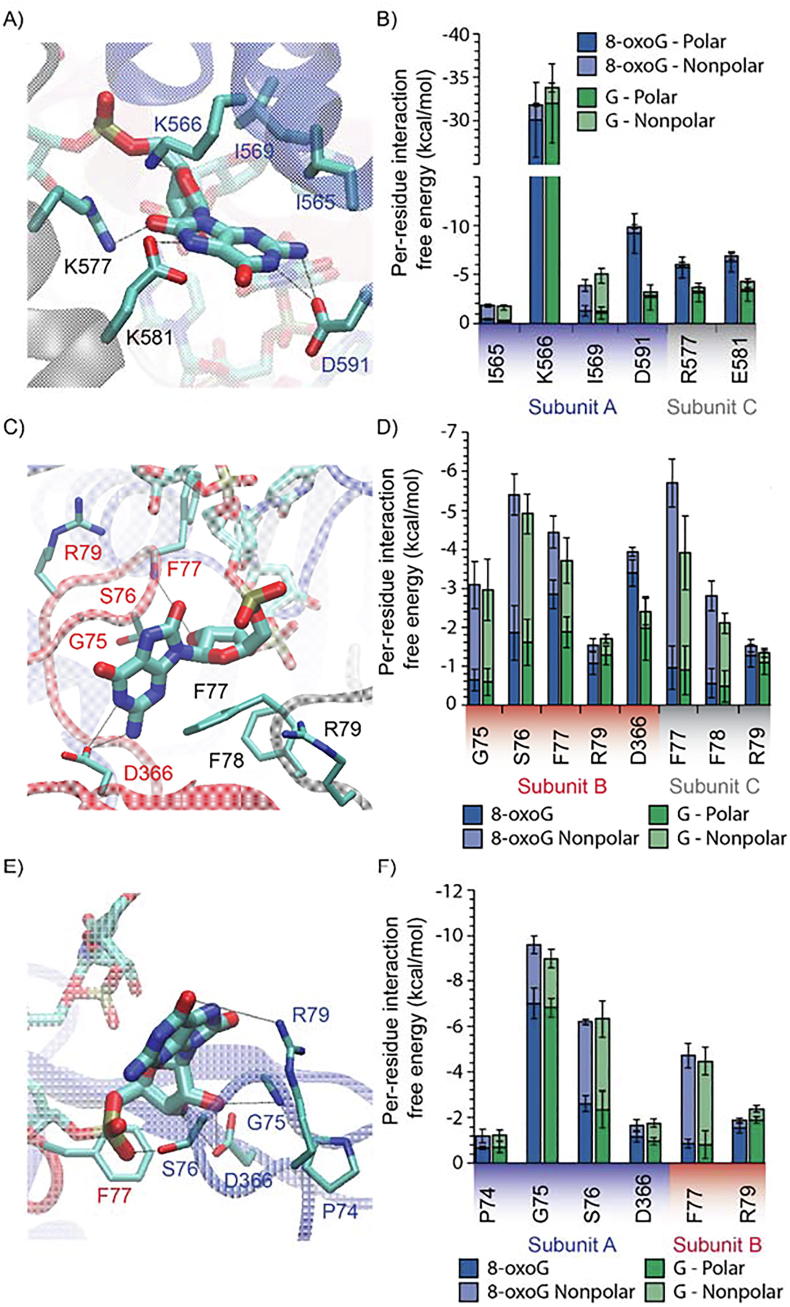
At position P4, the binding of guanosine and 8-oxoG to the PNPase binding site is largely dominated by the charged interaction between K566 of subunit A and the phosphate group of the RNA strand (Fig. 3A and 3B). For guanosine at position 4, the positively charged group of R577 forms a hydrogen bond with the 2′-OH of guanosine, whereas for 8-oxoG, the positively charged group of R577 forms hydrogen bonds with both the 2′-OH and the C-8 carbonyl group of 8-oxoG. Due to the additional carbonyl group in 8-oxoG in comparison with guanosine, the 8-oxoG forms a more stable hydrogen bond with R577. This interaction stabilizes the orientation of the 8-oxoG and the hydrogen bond between 8-oxoG and D591 as well as E581 (Fig. 3B). The interactions at this site are dominated by polar interactions of amino acids with electrically charged side chains, such as basic residues K566 and R577 and acidic residues E581 and D591. Owing to the negative electrostatic field associated with the RNA phosphate backbone, basic residues predominately favor electrostatic interactions with the ssRNA backbone [66], [67]]. However, our analysis indicates that the charged residues are also involved in interactions with 8-oxoG due to hydrogen bonding (Fig. 3A).
Fig. 3.
Molecular interactions of 8-oxo-7,8-dihydroguanosine (8-oxoG) in the active binding tunnel of PNPase. A) Key interactions between PNPase residues and 8-oxoG at position P4. B) Interaction free energy between each residue and guanosine (G) or 8-oxoG at position P4. Interaction is decomposed into polar (dark shade) and nonpolar (light shade) contributions. Only residues with an average interaction free energy less than −1.0 kcal/mol are shown. C) Key interactions between PNPase residues and 8-oxoG at position P8. D) Interaction free energy between each residue and G or 8-oxoG at position P8. E) Key interactions between PNPase residues and 8-oxoG at position P9. F) Interaction free energy between each residue and G or 8-oxoG at position P9. The interaction-free energies are obtained from three 50 ns MD simulations of the PNPase – RNA complex. Error bars plotted as ± one standard deviation.
At position P8, both guanosine and 8-oxoG stack between the G75-S76 peptide backbone and the F77 and F78 of subunit C (Fig. 3C and D). Specifically, they form a π — π interaction with the F77 benzyl group of subunit C and van der Walls interaction with the F78 of subunit C. On the opposite face of the RNA base, they form van der Waals interaction with the G75-S76 backbone. Compared to guanosine, 8-oxoG forms a more stable stacking contact with F77 of subunit C (Fig. 3D). In addition, 8-oxoG has a stronger hydrogen bond with its 2′-OH and the backbone amide group of F77 from subunit B, as well as a slightly stronger long-range electrostatic contact with its phosphate backbone and the guanidinium group of R79 in subunit C (Fig. 3D). Moreover, the hydrogen bond formed between D366 side chain with the C-2 carbonyl group of 8-oxoG is more stable in 8-oxoG as compared to guanosine (Fig. 3D). We observed that the added C-8 carbonyl of 8-oxoG forms a stable long-range electrostatic interaction with the positively charged guanidinium group of R79 in subunit B.
We observed that the interaction free energy at position P4 is attributed to backbone interactions (mostly through K566) whereas the interaction free energy at position P8 is attributed to a multitude of contacts with atoms located in the base or the backbone of the nucleotide. To determine the extent to which the two binding sites interact with the nucleobase (either guanine or 8-oxo-7,8-dihydroguanine (8-oxoGua)), we conducted per-nucleobase interaction free energy calculations in which we only considered the interactions between the protein residues and the base. In line with our initial observation, the binding of guanine and 8-oxoGua at position P8 is significantly more energetically favorable than at position P4 (Fig. 2). Given that interactions with the nucleobase are described to provide sequence specificity [68] and that the carbonyl (8–oxo) group occurs at the base and not the phosphate or sugar groups, position P8 may be more critical for 8-oxoG discrimination than position P4.
We observed that the binding of nucleotides at position P9 involves similar residues as at position P8 from the PNPase core, specifically, of the groove S76, F77 and F78. Contrarily, nucleotides at position P3 and P4 are contacted by divergent residues of the KH cap. Thus, we introduced the 8-oxoG in the ssRNA at position P9 to study whether it can contribute to 8-oxoG binding. Results from this analysis suggest that PNPase can also bind to 8-oxoG with higher affinity than guanosine at position P9 (Supplementary Fig. S1). Remarkably, the interaction involves almost identical residues at position P8 but from a different pair of neighboring PNPase subunits (Fig. 3F). As such, a combination of residues from subunit B and C yields a strong interaction with 8-oxoG at position P8, and a similar combination of residues from subunit A and B yields a strong interaction with 8-oxoG at position P9 (Fig. 3D and F). At position 9, the backbone of both 8-oxoG and guanosine are stacked over the benzyl group of F77 from subunit B, and either the 2′-OH or 3′-OH groups in the ribose formed a hydrogen bond with the negatively charged side-chain of D366 (Fig. 3E and F). As seen in position P8, the nucleobase interacts with the peptide bond of the stretch P74-G75-S76 via hydrogen bonding and van der Waals interactions. Compared to guanosine, the 8-oxoG exhibits a pronounced non-planarity of its 3D structure, which allows its phosphate group to contact the OH side group of S76 by hydrogen bonding. In addition, the twisting of the 8-oxoG base plane allows a more stable van der Waals interaction with the backbone amide group of S76 and the OH group of G75 (Fig. 3F).
Overall our biophysical analysis suggests that the groove formed by S76, F77, and F78 (SFF) in the three PNPase subunits is involved in the binding and discrimination of 8-oxoG at either position P8 or P9 of the RNA substrate. In contrast to charged amino acids seen in position P4, hydrogen bonding and hydrophobic interactions predominate at the SFF groove, which typically contribute to sequence and structure specificity that is attributed to many RNA-binding proteins [68], [69], [70], [71]]. Specifically, aromatic residues are more often involved in base recognition [68], [72]]. Therefore, we further study the SFF binding site given the predominance of interactions that we hypothesize are likely implicated on discrimination of 8-oxoG. Despite the high conservation of the FFRR loop, previous mutation studies have only focused on the arginine residues, thus no similar analysis has been conducted yet on the SFF groove.
3.2. Computational evolution of the S76-F77-F78 binding site yields mutants with differential 8-oxoG binding
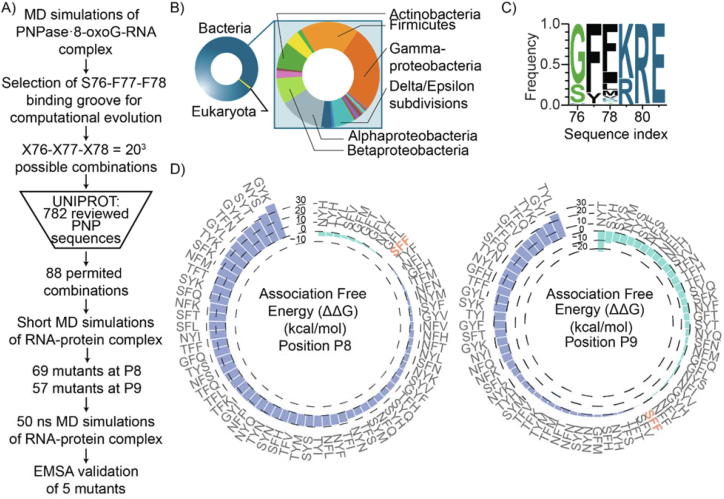
Once we identified the SFF groove as a specific site of 8-oxoG interaction, we attempted to vary its sequence to create mutations that exhibit higher 8-oxoG binding using a semi-rational computational approach (Fig. 4A). We hypothesized that mutations on this site could provide a range of 8-oxoG binding affinities. Given the high conservation of this region, we constrained mutations within the groove to only amino acids that naturally occur at each position, as determined from bioinformatics analysis of PNPase sequences in the UNIPROT database, analogous to a previous study [73]. Our rationale was that residues persisting through evolution were more likely to favor protein function and preserve tertiary and quaternary structure; computationally, this strategy minimized the number of necessary simulations from 203 combinations to less than 100. Specifically, to limit the number of combinations, we analyzed the residue frequency in 782 PNPase sequences in 777 species available in UNIPROT. The analyzed sequences are highly dominated by bacteria (98.8% of the sequences), of which 26% corresponds to Gram-negative Gammaproteobacteria (i.e. pathogens such as Salmonella, Yersina, Vibrio, and Pseudomonas species), 18% to Gram-positive Firmicutes (i.e., gut bacteria such as Clostridium, Streptococcus and Staphylococcus species, and Bacillus species) and 14% to Alphaproteobacteria (i.e., Zymomonas mobilis and members of Nitrobacter genus and Methylobacterium genus) (Fig. 4B). Within the sequences analyzed, glycine and serine residues predominate at the X76 amino acid position, while two aromatic residues (phenylalanine and tyrosine) were most frequently identified at position X77 (Fig. 3C). Despite the X78 position containing more diversity (11differentaminoacids) aliphatic or non-polar aromatic residues prevail at this position. This analysis yielded 88 beneficial mutations, which were subsequently studied to screen their energetic favorability for binding 8-oxoG by simulations.
Fig. 4.
Computational evolution of PNPase SFF groove identifies variants with differential 8-oxoG binding affinity. A) Schematic illustrating the design workflow of the PNPase mutants. B) Distribution of the most enriched motif by domains and within bacterial phylum from the 782 PNPase sequences in UNIPROT. C) Sequence logo of motif enriched among the 782 PNPase sequences. The motif downstream the SFF is highly conserved. D) Total MM-GBSA association free energy (ΔΔG) for the 69 PNPase mutants in complex with the 8-oxoG-RNA. The total MM GBSA association free energies are obtained from three 50 ns explicit solvent MD simulations of the mutant PNPase − 8-oxoG-RNA complex.
We initially performed the screening of each of the 88 mutations in PNPase with the RNA strand containing 8-oxoG at either position P8 or P9, through short implicit solvent MD simulations and interaction energy calculations. The two positions were investigated separately to reflect the stepwise interaction of PNPase with its RNA substrate and the cooperativity seen by the SFF groove from each PNPase subunit. For the initial screening, we modeled the residues within 10 Å of any atom of the 8-oxoG RNA fragment through short 5 ns simulations in implicit solvent to reduce computational load [28]. The mutations resulting in more favorable interaction energies from this step were further investigated. A lenient interaction energy cutoff was favored over a stricter cutoff to reduce the possibility of removing false negatives from the selected mutant PNPases. This analysis yielded 69 and 57 mutants at positions P8 and P9, respectively, that were subsequently assayed by 50 ns explicit solvent MD simulations. As shown in Fig. 3D, we observed variations in the association free energy for both positions P8 and P9. Out of these, nine combinations improved 8-oxoG association free energy (calculated by the MM-GBSA approximation) when mutation combinations were introduced at position P8 (ranging from −4.9 ± 7.18 kcal/mol (NYH) to −0.58 ± 15.79 kcal/mol (GFL) gain in average association free energy compared to SFF PNPase). Additionally, 30 mutation combinations improved 8-oxoG association free energy when introduced at position P9 (ranging from −25.45 ± 7.75 kcal/mol (NYT) to −4.86 ± 7.65 kcal/mol (NFF) gain in average association free energy compared to SFF PNPase). Because of the low overlap between positions P8 and P9, these results suggest that different combinations can modulate the specific contribution of each pair of PNPase subunits in 8-oxoG binding.
Based on the association free energy values (Fig. 4D), NYH and TYH have the 1st and 2nd lowest ΔΔG association free energy for 8-oxoG at position P8, respectively. NYT, and SYH show the 1st and 2nd lowest ΔΔG association free energy for 8-oxoG at position P9, respectively. GFT has moderately improved binding to 8-oxoG at both position P8 and P9; NYM and NFH have significantly improved binding to 8-oxoG at position P9 and similar affinity to 8-oxoG at position P8. Notably, NYT has significantly improved binding to 8-oxoG at both position P8 and P9. These mutations (NYT, NYM, GFT, NFH, and SYH) were evaluated in triplicate 50 ns explicit solvent MD simulations, confirming that each simulation converged towards reproducible values of the association free energies for each RNA-protein complex (Supplementary Fig. S2).
3.3. Computationally designed PNPase mutants improve 8-oxoG binding affinity and selectivity in vitro
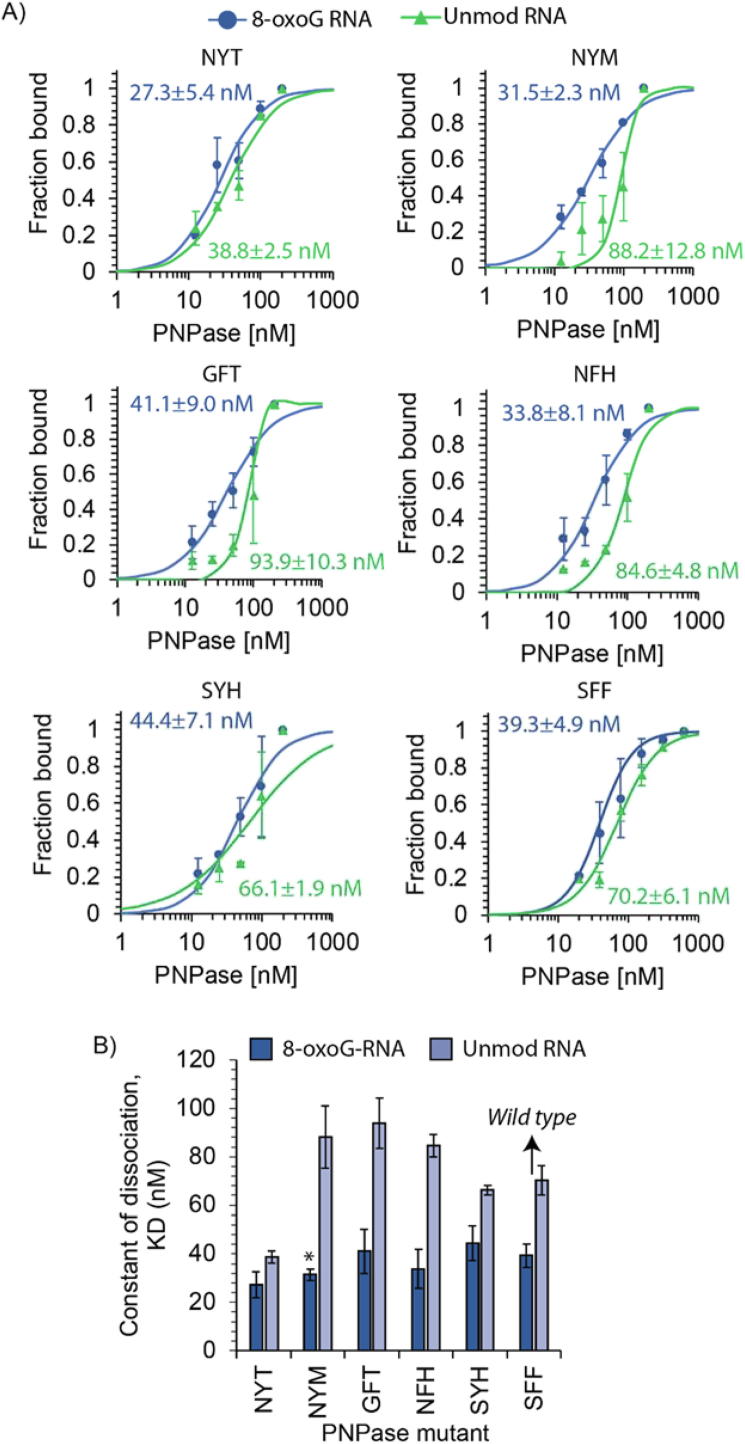
To corroborate our computational predictions, we constructed expression plasmids of the selected PNPase mutants (NYT, NYM, GFT, NFH and SYH) that showed the largest improvements in the interaction free energy compared to the wild type sequence (SFF) at either position P8 or P9 (Fig. 5). We purified these variants using His-tag affinity purification and then conducted in vitro binding assays. Specifically, we used increasing levels of each individually purified protein with an RNA oligo containing 8-oxoG (oligo sequence: 5′ – [NN(8-oxoG)N]6 – 3′, where N is A, C, G or U). In parallel, we conducted assays with a control unmodified oligo that lacked the 8-oxoG modification (sequence: 5′ – [NNGN]6 – 3′) to assess for varying binding selectivity towards 8-oxoG.
Fig. 5.
Electrophoretic mobility shift assays (EMSAs) of E. coli PNPase and 8-oxoG RNA. A) Profiles illustrate the behavior of the fraction of RNA bound as a function of protein concentration (2-fold concentration increase from 12.5 to 200 nM). Constant of dissociation (KD) values were calculated for each mutant with 8-oxoG-modified oligonucleotides (sequence: [NN8-oxoGN]6) and unmodified oligonucleotides (sequence: [NNGN]6) conducted in duplicate. B) Comparison of the constant of dissociation for the mutant PNPases. Error bars plotted as ± one standard deviation. Statistical analysis conducted using one-tailed homoscedastic t-test, * refers to p-value < 0.05.
Of the five mutants screened by in vitro binding assays, four show increased 8-oxoG binding affinity or selectivity (Fig. 5A). Three mutants show higher 8-oxoG binding affinity compared to the wild type PNPase as measured by KD values (NYT, NYM, and NFH). Of these, only the NYM mutant PNPase causes significant increase in 8-oxoG binding affinity (one-tailed heteroscedastic t-test, p-value < 0.05) (Fig. 5B). Remarkably, we observed a reduction in the binding affinity of several of these mutants to unmodified RNA (e.g., not containing 8-oxoG). For example, the selectivity (determined as the ratio of KD 8-oxoG and KD unmodified) for the NYM mutant is 2.8 and for the NFH mutant is 2.5, while the wild type is 1.8. As seen in Fig. 5B, the remaining mutants display a gradient of binding affinities. Notably, the GFT mutant conserves similar binding affinity to the wild type motif but with increased selectivity (SGFT = 2.3 vs SSFF = 1.8). These data confirm that minor changes in the conserved SFF binding groove can influence PNPase substrate binding activity.
3.4. Biophysical insights of the mutant PNPases with enhanced 8-oxoG affinity and selectivity
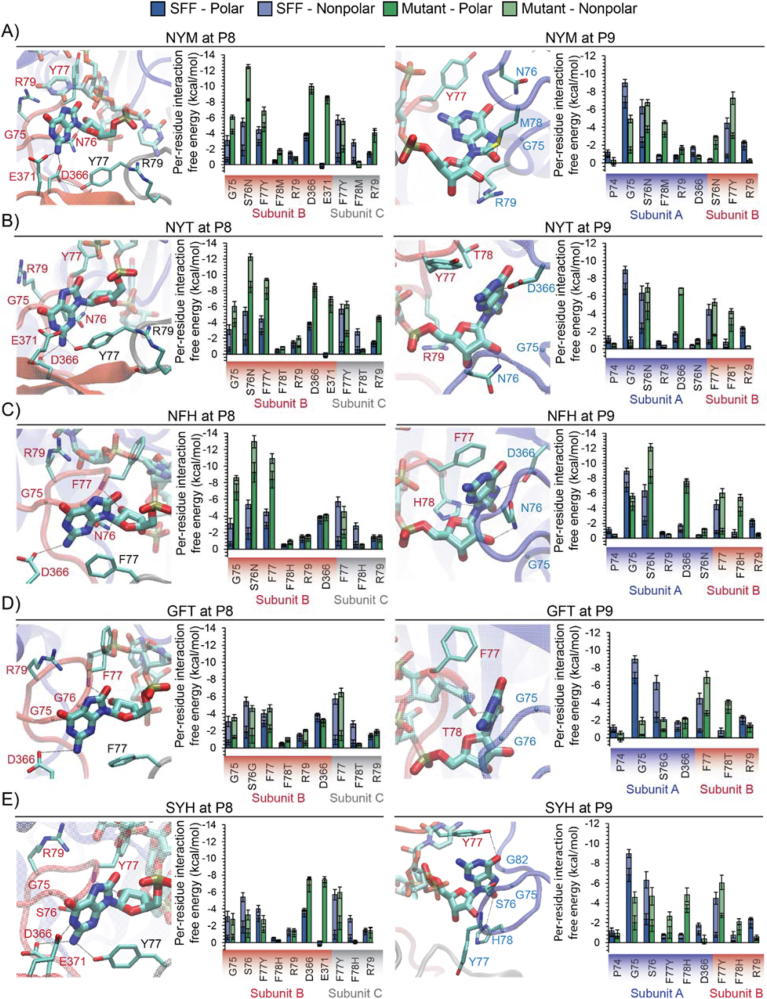
We computationally examined the NYM, NYT, NFH, GFT, and SYH mutant PNPases in complex with an RNA strand containing 8-oxoG at position P8 or P9 to determine their effect on affinity and/or specificity of 8-oxoG RNA. We observed a few key trends: (1) mutations involving S76N and/or Y77F substitutions provide higher 8-oxoG affinity in the NYM, NYT and NFH mutants, attributed to new hydrogen bond interactions with 8-oxoG. (2) a unique trend seen in the GFT mutant is that the F78T substitution directly contacts the 8-oxoG modification, which can be linked to the increased 8-oxoG selectivity in this mutant. And (3), for the SYH mutant, we observed a balance between diminished 8-oxoG affinity at position P8 and increased 8-oxoG affinity at position P9.
For the S76N mutation in the NYM, NYT and NFH mutants, the increased 8-oxoG affinity may be attributed to stabilized hydrogen bond interactions to 8-oxoG at either position P8 or P9 (Fig. 6A, B and C). At position P8, we observed that the longer side chain of N76 (compared to the wild type S76) induces a more stable hydrogen bond between the carboxamide group of N76 in subunit B and the 2′-OH of 8-oxoG. This interaction permits stronger hydrogen bonds between the adjacent residue (either Y or F) in subunit B and the 2′-OH and C-8 carbonyl groups of 8-oxoG. At position P9, the longer side chain of N76 allows the formation of a hydrogen bond between the carboxamide group of N76 in subunit B and either the phosphate group oxygens or the 3′-OH of 8-oxoG. With regard to the Y77F mutations (present in the NYM and NYT), the increased 8-oxoG affinity may be attributed to the formation of new hydrogen bonds that stabilize negatively charged residues interacting with 8-oxoG (Fig. 6A and B). In NYM, we observed that the OH group of Y77 in subunit C, allows the formation of an intramolecular hydrogen bond to D366 in subunit B, which stabilizes the hydrogen bond between the carboxyl group of D366 and the C-2 amide group of 8-oxoG. In NYT, we observed that OH group of Y77 in subunit C, allows the formation of an intramolecular hydrogen bond to E371, which stabilizes a new hydrogen bond between the carboxyl group of E371 in subunit B and the N-1 amide group of 8-oxoG. Overall, the formation of new interactions could potentially explain the enhanced 8-oxoG affinity attributed to the mutants NYM and NYT.
Fig. 6.
Molecular interactions of 8-oxoG with the mutant PNPases: NYM (panel A for position P8 and panel B for position P9); NYT (panel C for position P8 and panel D for position P9); NFH (panel E for position P8 and panel F for position P9); GFT (panel G for position P8 and panel H for position P9) and SYH (panel I for position P8 and panel J for position P9). The interaction-free energies are obtained from three 50 ns MD simulations of the RNA-protein complex. Error bars plotted as ± one standard deviation.
The increased 8-oxoG selectivity of the GFT mutant can primarily be attributed to interactions occurring at position P9 (Fig. 6D). At this position, the F78T mutation allows for the formation of a hydrogen bond between T78 in subunit B and the C-8 carbonyl group of 8-oxoG. Although the affinity of the GFT mutant to the unmodified RNA is less than that of the wild type SFF, the interactions at position P9 could contribute to the similar affinity of the GFT mutant to 8-oxoG compared to that of the wild type SFF (Fig. 5B), thereby enhancing the selectivity of the GFT mutant for 8-oxoG.
The similar affinity of the SYH mutant to 8-oxoG compared to the wild type PNPase SFF could potentially be attributed to the balance between diminished interaction energies occurring at position P8 and enhanced interaction energies occurring at position P9 (Fig. 6E). As seen in NYM and NYH mutants, the F77Y mutation allows for the formation of a new hydrogen bond between the OH group of Y77 in subunit C and the C-2 amide group of 8-oxoG. This hydrogen bond stabilizes the orientation of the 8-oxoG base promoting hydrogen bonds between D366 and E371 with 8-oxoG. However, given that SYH has a shorter residue side chain at position 76 than NYM and NYT, the nucleoside is drawn away from S76 in subunit B due to the Y77, D366 and E371 interactions. As the nucleoside is positioned away from residues 75–78 in subunit B, the interactions are overall diminished. On the contrary, for 8-oxoG in position P9, the F77Y and F78H mutations allow for the formation of new hydrogen bonds, which enhance the binding of SYH to 8-oxoG. The hydroxyl group of Y77 in subunit B allows for the formation of a hydrogen bond to the C-6 carbonyl group of 8-oxoG and the protonated nitrogen in the imidazole group of H78 in subunit A allows for the formation of a hydrogen bond to the C-8 carbonyl group of 8-oxoG or the 3′-OH of 8-oxoG.
3.5. Computationally designed PNPase variants complement cell survival under oxidative stress
PNPase-deficient E. coli cells have been shown to be more sensitive under oxidative stress conditions compared to wild type E. coli cells, implying that PNPase has an important role in oxidative stress resistance [17]. To gain insight into the effect that increased 8-oxoG RNA binding in PNPase could have in cellular tolerance to H2O2, we performed cell viability assays in PNPase-deficient E. coli cells complemented with one of the five characterized PNPase mutants (NYT, NYM, GFT, NFH, and SYH).
We verified the genomic deletion of pnp (Δpnp) in E. coli K12 BW25113 strain from the Keio collection by PCR (Supplementary Fig. S3). Complementation of the Δpnp strain with a constitutive lpp promoter expressing each mutant PNPase or the wild type sequence (denoted as SFF strain) does not provoke a significant impact on cell growth and viability (Supplementary Fig. S4A and S4B). Moreover, native western blotting analysis indicates that the different mutant PNPases have similar relative levels of expression (Supplementary Fig. S4C). And most importantly, given the similar migration of the mutants to wild type PNPase in native gels, this assay also suggests that the mutations do not produce distinct affections in tertiary and quaternary structure.
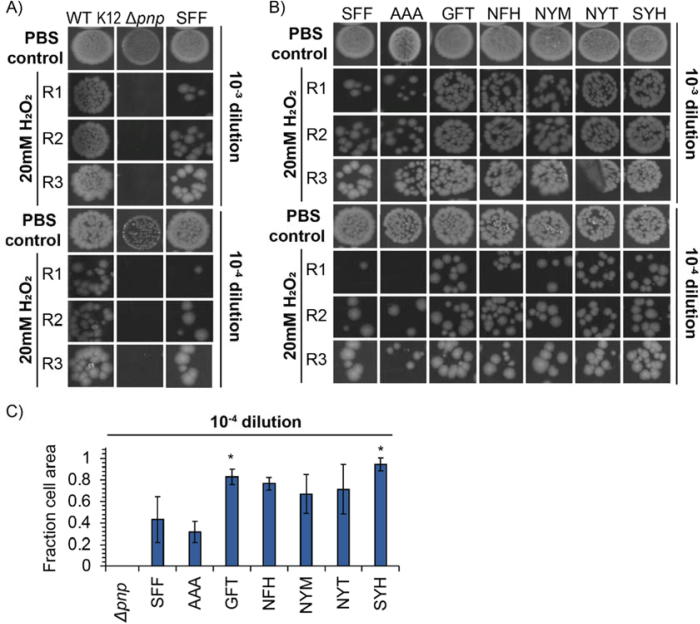
We exposed cells to 20 mM H2O2 for 20 min at room temperature, and 10−3 and 10−4 cell dilutions post-exposure were plated and incubated overnight at 37 °C for colony-forming counts and spot plate analysis. As shown in Fig. 7A, the Δpnp strain has decreased cell viability relative to E. coli K12 MG1655 WT cells. We then complemented the Δpnp strain with the wild type pnp gene using the constitutive lpp promoter plasmid (SFF strain). Our results suggest that this constitutive expression rescued survivability compared to the Δpnp strain (Fig. 7A).
Fig. 7.
PNPase mutants complement E. coli survival to H2O2 exposure. A) Spot plating of 10−3 and 10−4 cell culture dilutions after exposure to 20 mM H2O2 for 20 min. K12 Δpnp strain (from the Keio collection), E. coli K12 MG1655 WT, and the PNPase wild type rescue plasmid (SFF) transfected in the low expression plasmid are shown. 1X PBS in place of H2O2 solution was used to demonstrate cell growth under unstressed conditions. B) Spot plating of the 10−3 and 10−4 cell culture dilutions after exposure to 20 mM H2O2 for 20 min. K12 Δpnp cells were transfected with the low expression plasmid encoding each mutant PNPase. The alanine mutation (AAA) was used as a negative control. C) Cell area from spot plates of the 10−3 dilution was calculated using image J after normalization with the PBS control cell area. Statistical analysis conducted using a one-tailed homoscedastic t-test, * refers to a p-value < 0.05. Error bars plotted as ± one standard deviation.
We tested the viability of the five mutant PNPases using the same H2O2 exposure conditions described above (Fig. 7B). A PNPase with mutations to alanine (AAA motif) was included as a negative control. We measured the area covered by small colonies on each spot in the 10−3 and 10−4 dilution using image J and normalized to the same area on the PBS control to approximate the number of colonies in the spot plates (Fig. 7C and Supplementary Fig. S4C). As seen in Fig. 7C, all five mutants showed higher survivability to H2O2 exposure compared to the complemented wild type PNPase (SFF strain), increasing cell tolerance between 1.5 and 2.2 times. Although, only the GFT and SYH PNPase mutants were statistically significant using one-tailed heteroscedastic t-test (p-value < 0.05), these data demonstrate that PNPase variants with enhanced 8-oxoG affinity and selectivity differentially affect cellular tolerance to oxidative stress.
4. Discussions and conclusions
In this work, we studied the interaction of PNPase with 8-oxoG-containing RNA to better understand the mechanism of 8-oxoG discrimination. Analysis of the MD simulations and free energy calculations of the 8-oxoG RNA-protein complex identified two binding regions in the resolved RNA path within the PNPase channel. We evolved one of the identified binding sites for higher 8-oxoG affinity using a novel approach that initially screens mutants from a library of mutations and thoroughly assesses their interactions using multi-ns MD simulations and free energy calculations. As such, we found that evolving a single binding site in PNPase’s RNA interface increased 8-oxoG affinity between 1.2 and 1.5 times and/or selectivity between 1.5 and 1.9 times. Despite the slight improvement in 8-oxoG binding, these mutants caused increased cell tolerance to H2O2. This observation provides a clear link between molecular discrimination of RNA oxidation and cell survival.
We observed that the 8-oxoG binding interface is scattered on the central channel, mainly involving two regions (within the resolved section of the RNA path), which are spatially separated by three nucleotides of the RNA substrate (Fig. 2). We focus our study on the second binding site, located in the first core domain of PNPase, because our analysis indicates that it may have a more substantial role in the binding and discrimination of 8-oxoG. This site engages RNA with the highly conserved FFRR loop, which forms the aperture to the central channel of the core, to make aromatic stacking interactions with the bases of engaged RNA [33]. We found that two loops from neighboring subunits contact opposite faces of the RNA base providing a large interacting interface for 8-oxoG recognition. Previous studies indicate that adjacent positions to the SFF groove, specifically the residues R79 and R80, have a conformational role that regulates both RNA binding and catalytic degradation despite not directly interacting with the ssRNA [64]. Notably, our energetic calculations determined that R79 contributes to 8-oxoG binding at both position P8 and P9. Moreover, the FFRR loop is very dynamic. Structural studies have shown that the two phenylalanine residues from two neighboring subunits stack, making the channel more constrained in the apo structure than the structure containing RNA [32]. However, when PNPase binds the RNA substrate, the channel opens and structural changes propagate to the active site resulting in the proper orientation of the catalytic residues [63]. Specifically, R79 and R80 (upon contact of the phenylalanine residues with the RNA substrate) form a hydrogen bond with Y404 and the latter residue contacts T462, located in the active site [32]. Because FFRR loop is able to influence the catalytic site, it is possible that upon 8-oxoG binding, the loop’s new conformation influences changes in the active site that blocks the enzymatic degradation of RNA, explaining the inability of PNPase to process 8-oxoG-containing RNA [21].
Computational biophysical approaches have become important tools for understanding modified RNA-protein interactions by providing insight into structural features that help explain mechanistic increases in binding [26], [27], [28], [74], [75], [76], [77]]. To overcome the difficulties of designing a complex binding site, we used a combination of structural and bioinformatics strategies to establish a framework that facilitates the rapid sampling of binding affinities from protein mutants towards modified RNA substrates. To start, we only permitted naturally occurring amino acids in the selected mutable positions. Next, we implemented a two-step MD simulation screening, which involved an initial run of short implicit solvent MD simulations of the truncated RNA-protein complex to rapidly sample the mutations that improve 8-oxoG interaction compared to the wild type SFF sequence. Then, a second step of MD simulations consisting of all-atom 50 ns explicit solvent MD simulations was conducted to more accurately predict the interactions that passed the first step followed by free energy calculations. The use of biological constraints allowed for the rapid identification of biologically relevant mutations with favorable interactions to a modified RNA [73]. Finally, triplicate 50 ns explicit solvent MD simulations for the mutants acquiring the lowest association free energy were conducted to ensure reproducibility. We previously applied a similar multi-step approach to screen the interactions of PNPase with a library of 46 naturally occurring RNA modifications providing a reasonably high agreement between computational and experimental results [28]. Our study successfully combines the use of computational biophysics approaches assisted by biological constraints to understand RNA-protein interactions, as well as the use of multi-stage component strategy comprising an initial screening stage followed by more accurate simulations.
The approach shown here may be used as a tool in predicting PNPase mutants in the SFF groove, which would bind specifically to 8-oxoG-containing-RNA. One limitation of this study is the poor structural resolution of the electron density maps of the bound PNPase in proximity to the catalytic site [33]. Because of the high mobility of the S1 in the bound structure [32], this study was also unable to capture the contribution of S1 domain in 8-oxoG binding, which likely assists in recruitment of the RNA substrate. As such, some of the discrepancies observed between the predicted interaction free energies and the experimental KD values could be attributed to the lack of fully resolved crystal structure of the RNA-protein complex. The validation using in vitro binding assays revealed that the computational approach provided reasonable prediction ability of many mutant sequences.
Further studies are needed to shed light on the most distinct biochemical principles of modification-dependent binding of RNA by natural protein readers. We found that the introduction of asparagine (N) at X76 and tyrosine (Y) at X77 could potentially enhance the binding of mutant PNPases to 8-oxoG through primarily increased interactions to the nucleobase at position P8. We observed that asparagine at X76 primarily stabilizes hydrogen bonds to the sugar OH group of 8-oxoG at position P8 while tyrosine at X77 indirectly enhances the binding of 8-oxoG by stabilizing D366 and E371 through intramolecular hydrogen bonding. Interestingly, we also found that the introduction of a polar amino acid at X78 (such as threonine) with a sufficiently large sidechain such that it is in proximity to the C-8 carbonyl group of 8-oxoG could enhance 8-oxoG selectivity. We also observed that interactions to 8-oxoG at both positions P8 and P9 are important for the improved 8-oxoG binding. Intriguingly, we observed mutant PNPases with reduced affinity for 8-oxoG at one RNA position (either P8 or P9) while having significantly improved affinity for 8-oxoG at the other position, for instance in the SYH mutant relative to SFF. It is worth noting, that for this example, we observed no overall improvement in 8-oxoG binding experimentally. This analysis raised the question of whether certain amino acids can favor interactions with modified nucleotides, which has been fairly unexplored in the epitranscriptomics field [71]. For instance, one of the few studies exploring the basic principles of protein recognition of RNA modifications examined a model peptide chain using MD simulations to describe the mechanism of binding with the anticodon stem-loop of the human tRNALys3, which is the primer for HIV replication [74]. Importantly, while examining the loop of this tRNA, which contains two highly chemically modified bases (one 5-methylmethoxymethul-2-thuiuridine (mcm5s2U) and one 2-methylthio-N6-threonylcarbamoyladenine (ms2t6A)), their results highlighted modification-dependent binding of the peptide ligand; in this case, they demonstrated preferential interactions between the hydrophobic phenylalanine and the anticodon loop, while also showing preferential interactions between basic arginines and the RNA phosphate backbone [72], [74]]. More importantly, these principles were later applied to design peptides that mimic the native binding, resulting in a drug candidate for HIV therapeutics [72].
To better understand the physiological functions of the epitranscriptome, advances in the toolbox that facilitate the manipulation of the enzymes that recognize and/or edit RNA modifications are required [78]. Engineering efforts using these enzymes could provide enhanced control over gene expression of specific transcripts and/or modified sites beyond what the current approaches investigating global perturbation of these factors allow, providing more sensitive and reproducible approaches. For example, CRISPR-Cas9 technology has provided ways to deliver a range of RNA enzymes to specific transcripts to study mechanisms of epitranscriptomic regulation [30], [31]]. These tools are important because it may allow controlled modulation of the modifications at individual transcripts or sites to elucidate their functions, as well as for potential therapeutic and diagnostic applications derived from RNA modification studies [79].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank all members of the Contreras and Tamamis laboratories for discussions, suggestions, and support. We also acknowledge that the MD simulations and energy calculations were performed on the Ada and Curie supercomputing clusters at the Texas A&M High Performance Research Computing. This work is funded by the Health Effects Institute Grant 4949-RFA14-2/15-3 (L.M.C.) and Welch Foundation Grant F-1756 (L.M.C.) and the Artie McFerrin Department of Chemical Engineering at Texas A&M (P.T.). J.C.G-R. was supported by the Administrative Department of Science, Technology, and Innovation (COLCIENCIAS) of Colombia and Fulbright (Grant 479). A.A.O. was supported by the Texas A&M University Graduate Diversity Fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.12.003.
Contributor Information
Lydia M. Contreras, Email: lcontrer@che.utexas.edu.
Phanourios Tamamis, Email: tamamis@tamu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Boccaletto P. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. 2017 update. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mihailovic M.K., Chen A., Gonzalez-Rivera J.C., Contreras L.M. Defective Ribonucleoproteins, Mistakes in RNA Processing, and Diseases. Biochemistry. 2017;56(10):1367–1382. doi: 10.1021/acs.biochem.6b01134. [DOI] [PubMed] [Google Scholar]
- 4.Xiang Y. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543(7646):573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willi J. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res. 2018;46(4):1945–1957. doi: 10.1093/nar/gkx1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabretta A., Küpfer P.A., Leumann C.J. The effect of RNA base lesions on mRNA translation. Nucleic Acids Res. 2015;43(9):4713–4720. doi: 10.1093/nar/gkv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci USA. 2017;114(10):2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redstone S.C.J., Fleming A.M., Burrows C.J. Oxidative Modification of the Potential G-Quadruplex Sequence in the PCNA Gene Promoter Can Turn on Transcription. Chem Res Toxicol. 2019;32(3):437–446. doi: 10.1021/acs.chemrestox.8b00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu G.W., Ober M., Carell T., Beese L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431(7005):217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 10.Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell reports. 2014;9(4):1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka M., Chock P.B., Stadtman E.R. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci USA. 2007;104(1):66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J.X. Oxidative Modification of miR-184 Enables It to Target Bcl-xL and Bcl-w. Mol Cell. 2015;59(1):50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Nunomura A., Lee H.G., Zhu X., Perry G. Consequences of RNA oxidation on protein synthesis rate and fidelity: implications for the pathophysiology of neuropsychiatric disorders. Biochem Soc Trans. 2017;45(5):1053–1066. doi: 10.1042/BST20160433. [DOI] [PubMed] [Google Scholar]
- 14.Hofer T. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386(4):333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 15.Simms C.L., Zaher H.S. Quality control of chemically damaged RNA. Cellular Mol Life Sci: CMLS. 2016;73(19):3639–3653. doi: 10.1007/s00018-016-2261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cameron T.A., Matz L.M., De Lay N.R. Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 2018;14(10) doi: 10.1371/journal.pgen.1007654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48(9):2012–2020. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J., Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Biophys Res Commun. 2008;372(2):288–292. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayakawa H., Sekiguchi M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45(21):6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- 20.Hayakawa H. Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochem Biophys Res Commun. 2010;403(2):220–224. doi: 10.1016/j.bbrc.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Hayakawa H., Kuwano M., Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40(33):9977–9982. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- 22.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9(9):646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 23.Vangaveti S., Ranganathan S.V., Chen A.A. Advances in RNA molecular dynamics: a simulator's guide to RNA force fields. Wiley Interdiscip Rev RNA. 2017;8(2) doi: 10.1002/wrna.1396. [DOI] [PubMed] [Google Scholar]
- 24.Aduri R. AMBER Force Field Parameters for the Naturally Occurring Modified Nucleosides in RNA. J Chem Theory Comput. 2007;3(4):1464–1475. doi: 10.1021/ct600329w. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y., Vanommeslaeghe K., Aleksandrov A., MacKerell A.D., Jr., Nilsson L. Additive CHARMM force field for naturally occurring modified ribonucleotides. J Comput Chem. 2016;37(10):896–912. doi: 10.1002/jcc.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H. m(1)A and m(1)G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat Struct Mol Biol. 2016;23(9):803–810. doi: 10.1038/nsmb.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartono Y.D., Ito M., Villa A., Nilsson L. Computational Study of Uracil Tautomeric Forms in the Ribosome: The Case of Uracil and 5-Oxyacetic Acid Uracil in the First Anticodon Position of tRNA. J Phys Chem B. 2018;122(3):1152–1160. doi: 10.1021/acs.jpcb.7b10878. [DOI] [PubMed] [Google Scholar]
- 28.Orr A.A. A high-throughput and rapid computational method for screening of RNA post-transcriptional modifications that can be recognized by target proteins. Methods. 2018 doi: 10.1016/j.ymeth.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 29.deLorimier E. Pseudouridine Modification Inhibits Muscleblind-like 1 (MBNL1) Binding to CCUG Repeats and Minimally Structured RNA through Reduced RNA Flexibility. J Biol Chem. 2017;292(10):4350–4357. doi: 10.1074/jbc.M116.770768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauch S., He C., Dickinson B.C. Targeted m(6)A Reader Proteins To Study Epitranscriptomic Regulation of Single RNAs. J Am Chem Soc. 2018;140(38):11974–11981. doi: 10.1021/jacs.8b05012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauch S. Programmable RNA-Guided RNA Effector Proteins Built from Human Parts. Cell. 2019;178(1):122–134. doi: 10.1016/j.cell.2019.05.049. e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurmohamed S., Vaidialingam B., Callaghan A.J., Luisi B.F. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009;389(1):17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick S.W., Gubbey T., Hug I., Jenal U., Luisi B.F. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open biology. 2012;2(4) doi: 10.1098/rsob.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks B.R. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J., MacKerell A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J Comput Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hynninen A.P., Crowley M.F. New faster CHARMM molecular dynamics engine. J Comput Chem. 2014;35(5):406–413. doi: 10.1002/jcc.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 39.Gohlke H., Case D.A. Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J Comput Chem. 2004;25(2):238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 40.Hayes J.M., Archontis G. Biological Macromolecules:171–190; 2012. MM-GB (PB) SA calculations of protein-ligand binding free energies. Molecular Dynamics-Studies of Synthetic and. [Google Scholar]
- 41.Kieslich A.C. Exploring protein-protein and protein-ligand interactions in the immune system using molecular dynamics and continuum electrostatics. Current Physical Chemistry. 2012;2(4):324–343. [Google Scholar]
- 42.Tamamis P., Floudas C.A. Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop. Biophys J. 2013;105(6):1502–1514. doi: 10.1016/j.bpj.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamamis P., Floudas C.A. Molecular Recognition of CCR5 by an HIV-1 gp120 V3 Loop. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamamis P., Floudas C.A. Elucidating a Key Component of Cancer Metastasis: CXCL12 (SDF-1α) Binding to CXCR4. J Chem Inf Model. 2014;54(4):1174–1188. doi: 10.1021/ci500069y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamamis P., Floudas C.A. Elucidating a Key Anti-HIV-1 and Cancer-Associated Axis: The Structure of CCL5 (Rantes) in Complex with CCR5. Sci Rep. 2014;4:5447. doi: 10.1038/srep05447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamamis P. Insights into the mechanism of C5aR inhibition by PMX53 via implicit solvent molecular dynamics simulations and docking. BMC Biophysics. 2014;7(1):5. doi: 10.1186/2046-1682-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamamis P. Molecular Dynamics in Drug Design: New Generations of Compstatin Analogs. Chem Biol Drug Des. 2012;79(5):703–718. doi: 10.1111/j.1747-0285.2012.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamamis P., Morikis D., Floudas C.A., Archontis G. Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins: Struct, Funct, Bioinf. 2010;78(12):2655–2667. doi: 10.1002/prot.22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamamis P. Design of a modified mouse protein with ligand binding properties of its human analog by molecular dynamics simulations: The case of C3 inhibition by compstatin. Proteins Struct Funct Bioinf. 2011;79(11):3166–3179. doi: 10.1002/prot.23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orr A.A. Elucidating the multi-targeted anti-amyloid activity and enhanced islet amyloid polypeptide binding of beta-wrapins. Comput Chem Eng. 2018;116:322–332. doi: 10.1016/j.compchemeng.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orr A.A., Wördehoff M.M., Hoyer W., Tamamis P. Uncovering the Binding and Specificity of beta-Wrapins for Amyloid-beta and alpha-Synuclein. J Phys Chem B. 2016;120(50):12781–12794. doi: 10.1021/acs.jpcb.6b08485. [DOI] [PubMed] [Google Scholar]
- 52.Chocholoušová J., Feig M. Balancing an accurate representation of the molecular surface in generalized born formalisms with integrator stability in molecular dynamics simulations. J Comput Chem. 2006;27(6):719–729. doi: 10.1002/jcc.20387. [DOI] [PubMed] [Google Scholar]
- 53.López de Victoria A., Tamamis P., Kieslich C.A., Morikis D. Insights into the structure, correlated motions, and electrostatic properties of two HIV-1 gp120 V3 loops. PLoS ONE. 2012;7(11):e49925. doi: 10.1371/journal.pone.0049925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baba T. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2(2006):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 57.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández-Ramírez F., Bermúdez-Cruz R.M., Montañez C. Nucleic acid and protein factors involved in Escherichia coli polynucleotide phosphorylase function on RNA. Biochimie. 2010;92(5):445–454. doi: 10.1016/j.biochi.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 59.Ryder S.P., Recht M.I., Williamson J.R. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 61.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stickney L.M., Hankins J.S., Miao X., Mackie G.A. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J Bacteriol. 2005;187(21):7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carzaniga T. A conserved loop in polynucleotide phosphorylase (PNPase) essential for both RNA and ADP/phosphate binding. Biochimie. 2014;97:49–59. doi: 10.1016/j.biochi.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 64.Jarrige A., Bréchemier-Baey D., Mathy N., Duché O., Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol. 2002;321(3):397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- 65.Bermúdez-Cruz R.M., Fernández-Ramírez F., Kameyama-Kawabe L., Montañez C. Conserved domains in polynucleotide phosphorylase among eubacteria. Biochimie. 2005;87(8):737–745. doi: 10.1016/j.biochi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Jones S., Daley D.T., Luscombe N.M., Berman H.M., Thornton J.M. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29(4):943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moras D., Poterszman A. Getting into the major groove. Protein-RNA interactions. Curr Biol. 1996;6(5):530–532. doi: 10.1016/s0960-9822(02)00534-1. [DOI] [PubMed] [Google Scholar]
- 68.Ellis J.J., Broom M., Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins. 2007;66(4):903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 69.Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42(22):13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu C. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10(11):927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 71.Frohlich K.M. Post-Transcriptional Modifications of RNA: Impact on RNA Function and Human Health. Rna Technol. 2016:91–130. [Google Scholar]
- 72.Xiao X., Hall C.K., Agris P.F. The design of a peptide sequence to inhibit HIV replication: a search algorithm combining Monte Carlo and self-consistent mean field techniques. J Biomol Struct Dyn. 2014;32(10):1523–1536. doi: 10.1080/07391102.2013.825757. [DOI] [PubMed] [Google Scholar]
- 73.Smadbeck J., Peterson M.B., Khoury G.A., Taylor M.S., Floudas C.A. Protein WISDOM: a workbench for in silico de novo design of biomolecules. J Visualized Experiments: JoVE. 2013;77 doi: 10.3791/50476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao X., Agris P.F., Hall C.K. Molecular recognition mechanism of peptide chain bound to the tRNA(Lys3) anticodon loop in silico. J Biomol Struct Dyn. 2015;33(1):14–27. doi: 10.1080/07391102.2013.869660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spears J.L., Xiao X., Hall C.K., Agris P.F. Amino acid signature enables proteins to recognize modified tRNA. Biochemistry. 2014;53(7):1125–1133. doi: 10.1021/bi401174h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Väre V.Y., Eruysal E.R., Narendran A., Sarachan K.L., Agris P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017;7(1) doi: 10.3390/biom7010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao X., Agris P.F., Hall C.K. Introducing folding stability into the score function for computational design of RNA-binding peptides boosts the probability of success. Proteins. 2016;84(5):700–711. doi: 10.1002/prot.25021. [DOI] [PubMed] [Google Scholar]
- 78.Schaefer M., Kapoor U., Jantsch M.F. Understanding RNA modifications: the promises and technological bottlenecks of the 'epitranscriptome'. Open biology. 2017;7(5) doi: 10.1098/rsob.170077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rauch S., Dickinson B.C. Programmable RNA Binding Proteins for Imaging and Therapeutics. Biochemistry. 2018;57(4):363–364. doi: 10.1021/acs.biochem.7b01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.