Abstract
Macrophages, differentiation from monocytes infiltrated in the wound, have been suggested to be involved and to play an important role in the pathogenesis of wound healing. Nevertheless, no evidence has been established regarding M1 and M2 type macrophages in Keloid. To understand the status of M1 and M2 type macrophages in keloid, immunohistochemistry was performed on 30 cases of Keloid tissues and normal controls, with CD68, typical surface marker for M1 and CD163, well-accepted marker for M2 being immunostained. Meanwhile, the glucocorticoid receptor NR3C1 was also detected. As further confirmation, quantitative real-time PCR was utilized to verify the expression of CD68, CD163 and NR3C1 on mRNA level. It was consistently shown that infiltrated M2 macrophages pronouncedly outnumbered M1 macrophages in the dermis of keloids; and that NR3C1 expression was significantly up-regulated in keloids than that in normal controls. In addition, there was a marked correlation between CD163 and NR3C1 expression. Our results suggest that the number of infiltrated M2 macrophages in the dermis of keloids may be linked to the responsiveness to glucocorticoids in the pathogenesis of keloid.
Keywords: Keloid, M2 macrophage, NR3C1, CD163
Introduction
Keloids are fibroproliferative disorders of the skin whose pathophysiological mechanisms remains not well understood despite it were caused by abnormal healing of injured or irritated skin. Keloids, marked by excessive extracellular matrix accumulation, sometimes are painful and itchy and, along with their aesthetic burden, are not only aesthetically displeasing but can also be both painful and psychological distress for patients [1]. Although much has been reported regarding the pathophysiology of keloid, the pathology of keloid is not well understood and is still under investigation.
Generally, there’ve been two kinds of considerations as to the pathogenesis of keloid [1]. Some held that keloid was an abnormal cellular proliferation of fibroblasts as the key reason of keloid scar formation in that keloid fibroblasts proliferate faster than the fibroblasts of hypertrophic scars producing greater amounts of collagen and matrix metalloproteinases than that in hypertrophic scars. The others argued that the proinflammatory genes are up-regulated through an inflammatory response in the microenvironment where immune cells infiltrated was a primary pathological hallmark [1]. In the pathological process, immune cells infiltration was commonly present in keloid [2]. In the activation of fibroblasts, macrophages were a major source of TGF-1, while T and B lymphocytes can facilitate secretion of other fibrogenic cytokines [3]. Thus, these findings suggest that macrophages were involved in the pathogenesis of keloid. Considering that the macrophages were used to being generally classified into two main types [4], that is, M1 and M2 subtype macrophages; the status of infiltrated M1 and M2 macrophages in keloid remains unreported till now.
In the present investigation, to understand the status of infiltrated of M1 and M2 macrophages in keloid tissues, immunohistochemistry was carried out with 30 cases of keloid tissues and healthy normal controls. As confirmation, quantitative real-time PCR technique was employed to verify the expression on mRNA level. It was consistently shown that infiltrated M2 macrophages were significantly more common present than M1 macrophages were in keloid, indicating that M2 macrophage may play a key role in the pathogenesis of keloid relative to M1 macrophages. Meanwhile, we have also evaluated the NR3C1, the glucocorticoid receptor in the keloid tissues both on mRNA and protein level. The correlation between the expression of NR3C1 and status of M2 macrophage was analyzed. It exhibited that there was marked correlation between them. Taken as a whole, all the results we obtained suggest that the number of infiltrated M2 macrophages in the dermis section of keloid may be linked to the sensitivity for response to glucocorticoid.
Materials and methods
Keloid tissues
The current study gets approved by the Medical Ethics Committee of Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University; and the written informed consent was obtained from each participant involved. Thirty patients (10 men and 20 women; mean age 32 years; range from 20 to 51 years) with untreated keloids were recruited from the department of Burn and Plastic Surgery. They all met the currently accepted criteria for keloid, defined as the presence of typical skin lesions confirmed by two independent plastic surgeons. Duration of the disease ranged from 1.8 to 15 years. Lesions were located on the earlobe (4 cases), abdomen (3 cases), neck (4 cases), arm (5 cases), shoulder (2 cases) and chin (3 cases), and two or more sites (9 cases). Pain or itching caused by the lesions were complained in 20 patients and presented redness in 18 patients. All the patients didn’t receive any laser, radiation, cryotherapy or intralesional treatment in the previous 6 months at the time of diagnosis. In addition, 30 age- and gender-matched healthy subjects who presented no keloid, hypertrophic scars or current infections were enrolled from the department of Burn and Plastic Surgery as normal control. No subjects were received hormone, immunosuppressant, or antitumor regimens in the last year. A full medical history was taken from each patient with keloid. Keloid specimens were harvested from each patient at the time of surgical resection under local anaesthetic condition, whereas the normal skin specimens were obtained from patients undergoing surgical procedures for cosmetic reasons.
Immunohistochemistry
Skin samples were fixed in 4% paraformaldehyde solution and then subjected to paraffin sectioning. Tissue sections were cut at 4 mm thickness, deparaffinized and subjected to antigen retrieval using 10 mM citrate buffer at 100°C for 20 minutes. Immunohistochemistry was performed on all cases using mouse monoclonal antibody to human CD68 (dilution at 1:200; catalogue number: ab955; Abcam, Cambridge, USA); mouse monoclonal antibody to human CD163 (dilution at 1:100; catalogue number: ab192666; Abcam, Cambridge, USA); mouse monoclonal antibody to human NR3C1 (dilution at 1:500; catalogue number: ab2768; Abcam, Cambridge, USA). Primary antibody replaced with normal mouse IgG antibody was used as negative control when performing immunohistochemistry. Horseradish peroxidase-conjugated rabbit anti-mouse IgG acted as the secondary antibody and was purchased from Abcam (dilution at 1:500; catalogue number: ab97046; Abcam, Cambridge, USA). Staining was performed using a DAB Stain kit (catalogue number: ZLI-9017; Zhongshanjinqiao, Beijing, China). Sections were counterstained with hematoxylin. The numbers of M1 macrophages (CD68 positive immunostaining) and M2 macrophages (CD163 positive immunostaining) were counted in one random high power field (200× magnification) five times.
Evaluation of IHC staining
Briefly, positive reaction was defined as those showing brown signals in the cell cytoplasm. Each separate tissue core was scored on the basis of the intensity and area of the positive staining. The intensity of positive staining was scored as follows: 0, negative; 1, weak staining; 2, moderate staining; and 3, strong staining. The rate of positive cells was scored on a 0 to 4 scale: 0, 0-5%; 1, 6-25%; 2, 26-50%; 3, 51-75%; and 4, > 75%. If the positive staining was homogeneous, a final score was achieved by multiplication of the two scores above, giving birth to a total range from 0 to 12. Whereas as the staining was heterogeneous, we scored it as follows: each component scored independently and summed for the results. Take, for example, a specimen containing 25% tumor cells with moderate intensity (1×2=2), 25% tumor cells with weak intensity (1×1=1), and 50% tumor cells without reactivity received a final score of 2+1+0=3. For statistical analysis, we divided all the samples of keloids into two groups according to positive intensity as follows: scores of 0 to 8 as low (infiltration for M1/M2 macropahges, or low expression for NR3C1) and scores of 9 to 12 as high (infiltration for M1/M2 macropahges, or high expression for NR3C1).
Quantitative real time-PCR (qRT-PCR)
Total RNA of both keloid and normal tissues were extracted using Trizol method and were reversely transcribed into 1 μg cDNA with RevertAid First Strand cDNA Synthesis Kit (Catalogue number: #K1622, Thermo Fisher Scientific, USA). Real-time PCR was performed using SYBR Green Premix PCR Master Mix (Roche, Mannheim, Germany) according to the manufacturer protocols accompanied. Relative mRNA expression of CD68, CD163 and NR3C1 was calculated using Ct method (2-ΔΔCt) after being normalized to β-actin which was used as internal controls. All reactions were performed independently in triplicate. Sequence of the all primers involved was listed below: homo sapiens CD68 (NM_001251.2) F-primer 5’-CTTCTCTCATTCCCCTATGGACA-3’; R-primer 5’-GAAGGACACATTGTACTCCACC-3’, the product size of which was 105 base pair (bp); CD163 (DQ058615.1) F-primer 5’-TTTGTCAACTTGAGTCCCTTCAC-3’; R-primer 5’-TCCCGCTACACTTGTTTTCAC-3’, the product size was 127 bp; NR3C1 (NM_001204262.1) F-primer 5’-ATAGCTCTGTTCCAGACTCAACT-3’; R-primer 5’-TCCTGAAACCTGGTATTGCCT-3’, the product size 111 bp; β-actin (M28424.1) F-primer 5’-CATGTACGTTGCTATCCAGGC-3’; R-primer 5’-CTCCTTAATGTCACGCACGAT-3’, the product size 200 bp. The annealing temperature for the four genes was 57°C.
Statistical analysis
Statistical analysis was performed using the statistical package SPSS version 17.0 (SPSS, Inc, Chicago, IL, USA). Data were expressed as mean ± standard error of mean (SEM). Association was analyzed using the Chi-square test or Fisher’s exact test (when expected numbers were less than 5) when appropriate between high and low expression of biomarkers involved in keloid tissues based on clinicopathological variables, including age, gender. The correlation between CD163 and NR3C1 was analyzed using Spearman Correlation analysis method; Independent sample T test was used to analyze the difference between groups where the data were normal distribution. P values of less than 0.05 were taken to be statistically significant.
Results
Infiltrated M2 macrophages were more commonly present than M1 macrophages in keloid tissues compared to normal tissues
To understand the infiltrative status of both M1 and M2 macrophages in keloid tissues and normal controls, immunohistochemistry was carried out on keloid tissues and normal controls, with the cell surface makers CD68 and CD163 being immunostained. Parenthetically, considering that the quality of primary antibody was thought of being deadly important for the biomedical research, both specificity and correctness of the primary antibodies to CD68 and CD163 that were commercially available were pre-evaluated using antigen peptide pre-adsorption approach recommended previously [5,6]. Pre-test results exhibited that the specificity and correctness of the primary antibodies to CD68, CD163 and NR3C1 were sufficient to work (data not shown). Based on which, we’ve come to perform the immunohistochemical detection of expression of CD68, CD163 and NR3C1 on keloids and normal tissues, after routine histopathological review of the corresponding haematoxylin and eosin (HE) slides under the help of dermatologic pathologists (Figure 1A). The results showed that the positive immunostaining of both CD68 and CD163, if any, were mainly present in the dermis section, rather than in epidermis where they were hardly detectable (Figure 1B and 1C). Additionally, the immunostaining of CD68 and CD163 was mainly membranous and cytoplasmic. In terms of type of cells expressing CD68 and CD163, exactly, the positive immunostaining occasionally can be present on some fibroblasts as well in the dermis section, in addition to the histiocytes chiefly expressing CD68 and CD163. Few positive immunostaining of CD68 and CD163 can be detected and found on langerhans, keratinocytes or melanocytes scattered in the epidermis. By contrast, the immunostaining of NR3C1 was predominantly abundant in the epidermis section, with a little being present in dermis section (Figure 1D). What’s different from cells expressing CD68 and CD163, a few keratinocytes, langerhans and kelanocytes located in epidermis and fibroblasts found in dermis section can be observed to be positive immunostaining for NR3C1. The cellular sublocalization of NR3C1 was indisputably nuclear. In the case of expression of CD68, CD163, or NR3C1, they were readily observed to vary greatly in both number and shape from case to case of keloids we’ve enrolled, actually. Nonetheless, few CD163 can be detectable in the dermis section of normal tissues. In addition, it was easy to find that the infiltration of M2 macrophages was more commonly present in keloid than that was in normal tissues, whereas no remarkable difference of infiltrated M1 can be seen after overall statistical analysis, suggesting that M2 macrophages may be implicated in the pathogenesis of keloid compared to M1 macrophages. So did the NR3C1 expression, which was found to be markedly higher in keloid than in normal tissues. To avoid the potentially artificial bias toward detection of CD68, CD163 and NR3C1 caused by immunohistochemistry as such, quantitative real-time PCR (qRT-PCR) technique was employed to confirm the expression trend of CD68, CD163 and NR3C1 in keloid and normal tissues, on mRNA level. It was consistently shown that there was significant difference of CD163 and NR3C1 expression on mRNA level between keloid and normal tissues, but not for CD68 whose mRNA level, despite tends to be higher in keloid, was factually non-significant between keloid and normal tissues (Figure 1E).
Figure 1.
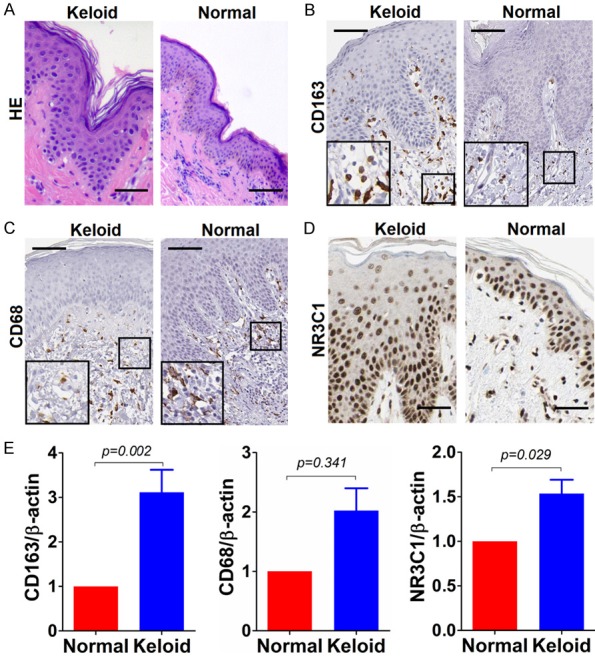
Immunostaining of CD163, CD68 and NR3C1 in keloid and normal tissues. A. Haematoxylin-eosin (HE) staining of keloid and normal control tissue; B. Immunostaining of CD163 in keloid and normal control tissue; C. Likewise, CD68 staining in keloid and normal control tissue; D. NR3C1 immunostaining in keloid and normal control tissue. Shown were representative figures selected among all cases subjected to immunostaining detection analysis. Scale bar stands for 100 μm. E. Detection of relative expression of CD163, CD68 and NR3C1 on mRNA level using qRT-PCR method in 30 cases of keloids and normal controls. Independent sample T test was used to analyze the difference between keloid and normal control group.
NR3C1 was remarkably up-regulated in keloid relative to normal tissues and was markedly correlated with infiltrated M2 macrophages
Having understood the status of infiltrated M1 and M2 macrophages and NR3C1 expression in keloid, next, we sought to analyze whether there was the possible correlation between the three makers. As described in the preceding, although NR3C1 can be detectable in both keloids and normal tissues, NR3C1 was displayed to be pronouncedly up-regulated in keloids compared with normal tissues. Noticeably, for the majority of normal cases enrolled as control, in which it’s hardly detectable for CD68 and CD163 expression that was commonly seen. No difference of CD68 expression, whatever on protein or mRNA level, was observed between keloids and normal tissues. However, CD163 was shown to be significantly up-regulated in keloids in comparison with normal controls (Table 1). Based on these observations we made, we postulated that there could be correlation between CD163 and NR3C1 expression. To test the hypothesis, we’ve used the Spearman correlation analysis (Table 2) and Pearson correlation analysis (Figure 2) to analyze the possible correlation we proposed based on our findings. It was expectedly found that there was significantly positive correlation between expression of CD163 and NR3C1, as analyzed by Spearman and Pearson correlation analysis, indicating that there might be interplay between M2 macrophages and NR3C1 expression in keloids.
Table 1.
Association of CD68, CD163 and NR3C1 expression was analyzed in keloid and normal controls
| Total | CD68 expression | χ2 | P | CD163 expression | χ2 | P | NR3C1 expression | χ2 | P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||
| Negative | Positive | Negative | Positive | Low | High | ||||||||
| Keloid | 30 | 14 | 16 | 1.071 | 0.438 | 8 | 22 | 11.279 | 0.002 | 10 | 20 | 8.076 | 0.009 |
| Normal | 30 | 18 | 12 | 21 | 9 | 21 | 9 | ||||||
Table 2.
Spearman correlation was carried out between CD163 and NR3C1 expression in keloid
| NR3C1 expression | Spearman value | P | |||
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| CD163 | Negative | 6 | 2 | 0.533 | 0.002 |
| Positive | 4 | 18 | |||
Figure 2.
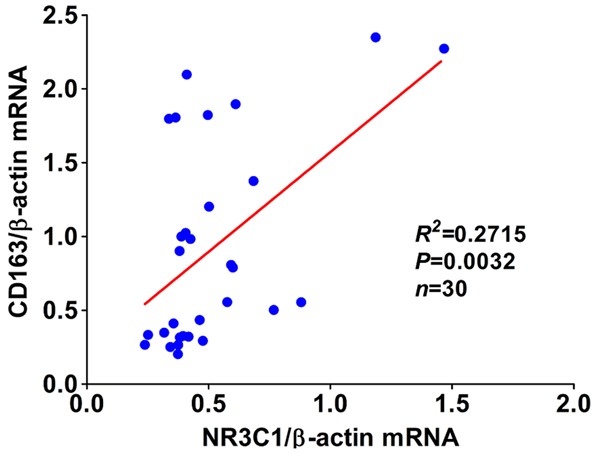
Pearson correlation analysis was performed between CD163 and NR3C1 mRNA expression in 30 cases of keloid and normal tissues.
Discussion
This is the first report presenting the status of infiltrated M1 and M2 macrophages as well as expression of NR3C1, the glucocorticoid receptor, in the keloids, to the best of our knowledge. Infiltrated M2 macrophages were more commonly seen than M1 macrophages were in keloids compared to normal tissues. Exactly, M2 macrophages were demonstrated to be remarkably infiltrated in keloid tissues relative to normal controls, whereas no marked difference of infiltrated M1 macrophages was observed between keloid and normal tissues. In addition, despite immunostaining of NR3C1 can be both present in keloid and normal tissues, NR3C1 expression was shown to be overall pronouncedly up-regulated in keloid in comparison with normal control. Subsequent statistical analysis exhibited that there was significantly positive correlation between CD163 and NR3C1 expression in keloid, suggesting that there may be interplay between infiltrated M2 macrophages and NR3C1 expression in the pathogenesis of keloid.
Despite numbers of studies have been reported regarding keloid, the pathogenesis of keloid remains not well understood. In the pathogenesis of keloid, infiltrated inflammatory cells can be commonly observed in keloid tissues [3]. Several major immune cell populations involved in the wound healing have been suggested to be responsible for in the physiopathological process of keloid, including mast cells [7], macrophages [7,8], lymphocytes [7], in addition to keratinocytes and fibroblasts [7], the two major cells that abound in keloids. Out of these cell types observed in keloid, the macrophages may have received the relatively less attention than other kind of cells in the study of keloid to date. This is not surprising in consideration of the dual role of macrophages that developed from monocytes [8], which is to say the phagocytosis of any remaining cell debris and remodeling of new tissue. The original study mentioning infiltrated macrophages found in keloid was performed by Boyce DE et al [9] who clearly reported there were high numbers of macrophages found in keloid. Nevertheless, they failed to specify the macrophages in light of the macrophages were used to being broadly classified into two different subtypes [4], namely M1 subtype and M2 subtype. On the basis of the study, Jiao H and colleagues [2] subsequently analyzed the infiltrated immune cells, including M1 macrophages (they detected only CD68) in keloid proposing that keloid might be mediated by autoimmune responses. Regrettably, they also failed to analyze and detect the infiltration of M2 type macrophages in keloid meanwhile. Thus, it were the two earlier studies referenced above that prompted us to initiate the current investigation. Our study was absolutely not simple reproduction of these previous studies in our own cases but verification and extention of them. It was shown that the infiltration of M2 type macrophages was remarkably more common present in keloid than that in normal controls, whereas no significant difference of infiltrated M1 macrophages can be found between keloid and normal controls, which distinctively differs from the observation made by Jiao H and co-workers [2] using immunohistochemistry that the number of macrophages with CD68 was found to be remarkably increased in keloid compared to normal skins. The underlying reason leading to the discrepancy between ours and Jiao H et al’s finding was unknown but it may be owing to the different primary antibodies to CD68 employed. In terms of infiltration of M1 type macrophages in keloid, another previous study immunophenotying the M1 and M2 macrophages in keloid by Bagabir R and associates [10] was partly consistent with our observation. In their study, both infiltrated M1 and M2 type macrophages were revealed to be significantly increased in keloid compared to normal controls, under the help of quantitative immunohistomorphometry. In contrast, only infiltrated M2 type macrophages were observed to be pronouncedly present in keloid in our own investigation. The different methodologies and primary antibodies employed might account for the inconsistency between our study and Bagabir R et al’s report [10]. Considering that potential bias of immunostaining could be brought about by immunohistochemistry itself, we’ve confirmed using qRT-PCR technique as complementary method, the expression trend of CD68 and CD163 on mRNA level. The results obtained from qRT-PCR were totally in congruent with that from immunohistochemistry, thus ruling out the possibility of underlying bias of our results. There, in fact, has been little reported concerning M1 and/or M2 macrophages in keloid other than the previous relevant studies mentioned above. In another keloid study, Uppal RS et al [11] analyzed the variation of CD68 expression before and after the keloid patients subjected to 5-fluorouracil therapy. No variation was found regarding the expression of CD68 in the keloid treated with 5-fluorouracil. From the single literature, it would be hard to draw any conclusion that disinvolvement of M1 or M2 type macrophages in the reaction to 5-fluorouracil therapy. More evidence obviously would be needed to determine in this regard. Furthermore, as for the underlying mechanism by which M1 or M2 type macrophages involved in the pathogenesis of keloid, it remains unclear that deserves to be further investigated.
NR3C1 (abbreviated for Nuclear Receptor Subfamily 3 Group C Member 1) encodes glucocorticoid receptor [12]. In view of the importance of expression of glucocorticoid receptor in the steroid therapy of keloid [13,14] and the evidence concerning expression of NR3C1 has been unestablished in keloid to date, we attempted to understand the status of NR3C1 in keloid. Actually, original report regarding NR3C1 in the setting of steroid therapy came from its genetic polymorphism study revealing that genetic variant in NR3C1 gene may be able to account for the variability in glucocorticoid responsiveness [15]. Then, it was extended to bronchial asthma study [16,17] where polymorphism of the NR3C1 gene was found to be markedly associated with an increased sensitivity to glucococorticoid and susceptibility to the development of bronchial asthma. Till now, there was only one study mentioning the expression of NR3C1 in the context of skin was from epidermal keratinocyte [18], which was fundamentally in agreement with our finding that the immunostaining of NR3C1 was nuclear mainly in the epidermis section. It was for the first time, to the best of our knowledge, observed that NR3C1 was markedly up-regulated in keloid relative to normal controls. The reason why NR3C1 was observed to be pronouncedly up-regulated in keloid compared to normal control was unknown. Several lines of evidence, however, suggested that corticoid receptor played an important role in the polarization and regulation of macrophage [19,20]. Based on these previous evidence, we therefore hypothesized that there could be correlation between expression of CD163 and NR3C1. To test the postulation, we tried to analyze the correlation between them using statistical analysis. It was expectedly found that, there was significantly positive correlation between expression of CD163 and NR3C1 in keloid tissues, suggesting that there may be association between NR3C1 expression and M2 macrophages. As to the underlying mechanism between CD163 and NR3C1, nevertheless, remains poorly understood. But our observation could be indirectly accounted for by one earlier study reporting that glucocorticoids increased CD163 expression in placental Hofbauer cells [21], which leads to the suggestion that the number of infiltrated M2 macrophages in the dermis section could be indicative of the sensitivity for response to glucocorticoid. To support this notion, Yeager MP and colleagues [22] subsequently found that glucocorticoids enhanced the in vivo migratory response of human monocytes, which further developed into macrophages. Further work, therefore would be needed to elucidate the correlation between the number of infiltrated M2 macrophages and NR3C1 expression in keloid.
Here we firstly presented the status of infiltrated M2 type macrophages in keloid, revealing that there was significant correlation between NR3C1 expression and M2 macrophages in keloid; though, there have still been several limitations that have to be noted. Firstly, our observations shown here were made on the rather limited number of cases that could be a source of potential bias; thus, our study needs to be warranted in the larger sample size; secondly, despite we’ve pre-tested the specificity of primary antibodies to CD68 and CD163 from the outset, basically ruling out the potential problems brought about by the antibodies we used; Interestingly, we did observe that even a few positive immunostaining of CD68 and CD163 can be also found on some of fibroblasts and large lymphocytes as well in the dermis section of keloid. Consequently, we didn’t mean that the expression of CD68 and CD163 presented in our study can entirely and accurately reflect the real status of infiltrated M1 and M2 macrophages in keloids. But, our histological findings at least were able to suggest the extent to which M1 and M2 macrophages infiltrated and involved. Given that both M1 and M2 type macrophages had other specific cell surface makers as well (for instance of iNOS for M1 macrophages and CD206 for M2 macrophages), other than the CD68 and CD163 we selected in the study; Our conclusion may therefore need to be warranted using other kinds of cell surface markers. At last, we just focused on M1 and M2 type macrophages in keloid not in hypertrophic scar, which was readily confused in clinic but in actuality different from keloid [23,24]. Thus, the status of M1 and M2 type macrophages in hypertrophic scar remains to be studied.
In summary, our study for the first time exhibited that infiltrated M2 macrophages not M1 macrophages were predominantly present in the dermis section of keloid relative to normal control; and that there was significant correlation between NR3C1 expression and M2 macrophages in keloid, suggesting that the number of infiltrated M2 macrophages in the dermis section may be linked to the sensitivity for response to glucocorticoid.
Acknowledgements
The present work was supported by the donations from the Department of Burn and Plastic Surgery, Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University.
Disclosure of conflict of interest
None.
References
- 1.Mari W, Alsabri SG, Tabal N, Younes S, Sherif A, Simman R. Novel insights on understanding of keloid scar: article review. J Am Coll Clin Wound Spec. 2015;7:1–7. doi: 10.1016/j.jccw.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiao H, Fan J, Cai J, Pan B, Yan L, Dong P, Zong X, Gan C, Xiao R. Analysis of characteristics similar to autoimmune disease in keloid patients. Aesthetic Plast Surg. 2015;39:818–25. doi: 10.1007/s00266-015-0542-4. [DOI] [PubMed] [Google Scholar]
- 3.Shih B, Garside E, McGrouther DA, Bayat A. Molecular dissection of abnormal wound healing processes resulting in keloid disease. Wound Repair Regen. 2010;18:139–53. doi: 10.1111/j.1524-475X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 5.Burry RW. Controls for immunocytochemistry: an update. J Histochem Cytochem. 2011;59:6–12. doi: 10.1369/jhc.2010.956920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hewitt SM, Baskin DG, Frevert CW, Stahl WL, Rosa-Molinar E. Controls for immunohistochemistry: the Histochemical Society’s standards of practice for validation of immunohistochemical assays. J Histochem Cytochem. 2014;62:693–7. doi: 10.1369/0022155414545224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaker SA, Ayuob NN, Hajrah NH. Cell talk: a phenomenon observed in the keloid scar by immunohistochemical study. Appl Immunohistochem Mol Morphol. 2011;19:153–9. doi: 10.1097/PAI.0b013e3181efa2ef. [DOI] [PubMed] [Google Scholar]
- 8.Mahdavian Delavary B, van der Veer WM, van Egmond M, Niessen FB, Beelen RH. Macrophages in skin injury and repair. Immunobiology. 2011;216:753–62. doi: 10.1016/j.imbio.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Boyce DE, Ciampolini J, Ruge F, Murison MS, Harding KG. Inflammatory-cell subpopulations in keloid scars. Br J Plast Surg. 2001;54:511–6. doi: 10.1054/bjps.2001.3638. [DOI] [PubMed] [Google Scholar]
- 10.Bagabir R, Byers RJ, Chaudhry IH, Muller W, Paus R, Bayat A. Site-specific immunophenotyping of keloid disease demonstrates immune upregulation and the presence of lymphoid aggregates. Br J Dermatol. 2012;167:1053–66. doi: 10.1111/j.1365-2133.2012.11190.x. [DOI] [PubMed] [Google Scholar]
- 11.Uppal RS, Khan U, Kakar S, Talas G, Chapman P, McGrouther AD. The effects of a single dose of 5-fluorouracil on keloid scars: a clinical trial of timed wound irrigation after extralesional excision. Plast Reconstr Surg. 2001;108:1218–24. doi: 10.1097/00006534-200110000-00018. [DOI] [PubMed] [Google Scholar]
- 12.Pascussi JM, Gerbal-Chaloin S, Drocourt L, Maurel P, Vilarem MJ. The expression of CYP2B6, CYP2C9 and CYP3A4 genes: a tangle of networks of nuclear and steroid receptors. Biochim Biophys Acta. 2003;1619:243–53. doi: 10.1016/s0304-4165(02)00483-x. [DOI] [PubMed] [Google Scholar]
- 13.Rutkowski D, Syed F, Matthews LC, Ray DW, McGrouther DA, Watson RE, Bayat A. An abnormality in glucocorticoid receptor expression differentiates steroid responders from nonresponders in keloid disease. Br J Dermatol. 2015;173:690–700. doi: 10.1111/bjd.13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed F, Singh S, Bayat A. Superior effect of combination vs. single steroid therapy in keloid disease: a comparative in vitro analysis of glucocorticoids. Wound Repair Regen. 2013;21:88–102. doi: 10.1111/j.1524-475X.2012.00862.x. [DOI] [PubMed] [Google Scholar]
- 15.Kumsta R, Entringer S, Koper JW, van Rossum EF, Hellhammer DH, Wust S. Glucocorticoid receptor gene polymorphisms and glucocorticoid sensitivity of subdermal blood vessels and leukocytes. Biol Psychol. 2008;79:179–84. doi: 10.1016/j.biopsycho.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Pietras T, Panek M, Tworek D, Oszajca K, Wujcik R, Gorski P, Kuna P, Szemraj J. The Bcl I single nucleotide polymorphism of the human glucocorticoid receptor gene h-GR/NR3C1 promoter in patients with bronchial asthma: pilot study. Mol Biol Rep. 2011;38:3953–8. doi: 10.1007/s11033-010-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panek M, Pietras T, Antczak A, Fabijan A, Przemecka M, Gorski P, Kuna P, Szemraj J. The N363S and I559N single nucleotide polymorphisms of the h-GR/NR3C1 gene in patients with bronchial asthma. Int J Mol Med. 2012;30:142–50. doi: 10.3892/ijmm.2012.956. [DOI] [PubMed] [Google Scholar]
- 18.Wierzbicka JM, Zmijewski MA, Antoniewicz J, Sobjanek M, Slominski AT. Differentiation of keratinocytes modulates skin HPA analog. J Cell Physiol. 2017;232:154–66. doi: 10.1002/jcp.25400. [DOI] [PubMed] [Google Scholar]
- 19.Usher MG, Duan SZ, Ivaschenko CY, Frieler RA, Berger S, Schutz G, Lumeng CN, Mortensen RM. Myeloid mineralocorticoid receptor controls macrophage polarization and cardiovascular hypertrophy and remodeling in mice. J Clin Invest. 2010;120:3350–64. doi: 10.1172/JCI41080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coppo M, Chinenov Y, Sacta MA, Rogatsky I. The transcriptional coregulator GRIP1 controls macrophage polarization and metabolic homeostasis. Nat Commun. 2016;7:12254. doi: 10.1038/ncomms12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Niven-Fairchild T, Tadesse S, Norwitz ER, Buhimschi CS, Buhimschi IA, Guller S. Glucocorticoids enhance CD163 expression in placental Hofbauer cells. Endocrinology. 2013;154:471–82. doi: 10.1210/en.2012-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeager MP, Pioli PA, Collins J, Barr F, Metzler S, Sites BD, Guyre PM. Glucocorticoids enhance the in vivo migratory response of human monocytes. Brain Behav Immun. 2016;54:86–94. doi: 10.1016/j.bbi.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kose O, Waseem A. Keloids and hypertrophic scars: are they two different sides of the same coin? Dermatol Surg. 2008;34:336–46. doi: 10.1111/j.1524-4725.2007.34067.x. [DOI] [PubMed] [Google Scholar]
- 24.Burd A, Huang L. Hypertrophic response and keloid diathesis: two very different forms of scar. Plast Reconstr Surg. 2005;116:150e–7e. doi: 10.1097/01.prs.0000191977.51206.43. [DOI] [PubMed] [Google Scholar]


