Abstract
Rap2A is overexpressed in a multitude of human cancers and plays an important role in cytoskeleton rearrangement, arteriogenesis and cell migration. However, its role and function in hepatocellular carcinoma (HCC) has not yet been explored. Here, we aimed to investigate the expression of Rap2A in HCC and the relationship between Rap2A and clinicopathologic features of patients. Western blot and quantitative real-time PCR (qRT-PCR) showed that Rap2A was remarkably upregulated in HCC tissues compared to adjacent normal liver tissues. Immunohistochemistry (IHC) showed that Rap2A was mainly localized in the cytoplasm rather than nuclei in HCC tissues. Overexpression of Rap2A was significantly correlated with tumor size (P=0.019), metastasis (P=0.002), pathological differentiation (P=0.028) and vascular invasion (P=0.017) in HCC. Kaplan-Meier analysis demonstrated that HCC patients with high Rap2A expression had shorter overall survival time than those with low Rap2A expression (P=0.011). Furthermore, High level of Rap2A was a risk factor for HCC patients according to Cox’s proportional hazard regression (P=0.026). In summary, our results suggest that high expression of Rap2A is involved in HCC progression and might be a novel prognostic indicator for patients.
Keywords: Rap2A, hepatocellular carcinoma, prognosis, survival time
Introduction
Hepatocellular carcinoma (HCC) ranks sixth among malignant tumors in the world and is the third leading cause of cancer death [1-3]. Although there are numerous therapy methods available now, surgical resection for HCC is still the best choice for patients. Most HCC patients usually have advanced-stage cancer at the time of initial diagnosis and lose their chance for tumor resection. Even though HCC patients have undergone standardized treatment, they will still have a worse outcome with local recurrence and distant metastasis in a short time after surgical operation. Hence, it is extremely necessary to seek new molecular markers and invent efficient imaging technologies which can better diagnose patients with early-stage HCC and predict their survival time.
Rap2A is one member of the RAS oncogene family. The full-length cDNA of the Rap2A gene is located at 13q34 and contains an open reading frame of 549 bp, encoding 183 amino acids with a molecular mass of 20,434 [4]. Previous studies have shown that Rap2A plays a critical role in regulation of various cellular processes such as cytoskeletal reorganization, brush border formation and cell migration [5-7]. It has been reported that Rap2A is upregulated in several types of human cancers, including prostate cancer, follicular thyroid cancer, nasopharyngeal carcinoma [8-10]. However, no reports relate to Rap2A expression in HCC except one study which examined the high expression of Rap2A in human hepatoblastoma HepG2 cells [7].
Here, we speculated that Rap2A may take part in the progression and development of HCC. To verify our hypothesis, we adopted different approaches including western blot, qRT-RCR and immunohistochemistry to detect Rap2A in HCC and gathered patients’ pathological information for statistical analysis. In the present study, we showed that Rap2A is obviously upregulated in HCC and maybe regarded as a novel biomarker for HCC patients’ prognosis.
Materials and methods
Clinical samples collection and follow-up
Clinical samples were achieved from the Chronic Liver Disease Biological Sample Bank, Department of Hepatobiliary Surgery, Zhongshan Hospital Xiamen University. Detailed procedures are described in our previous study [11]. The clinical samples were obtained from patients with HCC who were treated at Zhongshan Hospital between 2011 and 2014. Specimen collection was performed after obtaining informed consent from each patient, and the study was approved by the Ethics Committee of the hospital. Paired tumor and nontumor (with more than 2 cm distance from the primary tumor’s edge) tissue samples from the same patient were frozen quickly in liquid nitrogen postoperatively and stored at -80°C for western blot and qRT-PCR. None of the patients had received chemotherapy or radiotherapy before surgical operation resection. The median age of the patients included was 55 years. In addition, complete follow-up information of 82 patients was obtained by reviewing the patients’ medical records.
Western blot
Tissue lysates or cell lysates were subjected to SDS-PAGE, and then proteins were transferred onto polyvinylidene fluoride membranes (Millipore, Germany). The membranes were blocked in Tris-buffered saline (TBS; pH 7.4) containing 5% non-fat milk and 0.1% Tween-20, incubated with the primary antibody against Rap2 (1:1000, 610215, BD Transduction Laboratories) and β-Actin (1:1000, AT0001, CMCTAG) overnight at 4°C, washed three times in TBS containing 0.1% Tween-20, and followed by the secondary antibody labeled with horseradish peroxidase (Jackson, the USA) for 1 h at room temperature. Antibody binding was visualized using enhanced chemiluminescence reagents (Advansta, the USA). Quantification of band densitometry was performed using ImageJ with normalization to the loading control β-Actin. Relative expression of Rap2A is calculated as Rap2A/β-Actin (Figure S1 and Table S1).
Quantitative real-time PCR
Total RNA was extracted from tissues or cell samples using the Trizol reagent (Ambion/Life Technologies, the USA) according to the manufacturer’s instructions. Quantitative real-time PCR for Rap2A mRNA detection was performed as follows, with primers designed and synthesized by BGI-Tech (BGI, China): reverse transcription of 1 μg RNA was performed with the prime ScriptTM RT reagent kit with gDNA eraser (TaKaRa Biotechnology Inc, Japan) and qRT-PCR was performed using FastStart Universal SYBR Green Master ROX (Roche, Switzerland) in a total volume of 20 μl with LightCycler® 96 Real-Time PCR System (Roche, Switzerland). The sequences of the primer pairs were as follows: Rap2A forward, 5’-ACAATGGTGGACGAACTCTTT-3’, reverse, 5’-CAGAACAGCATGGGTCATCT T-3’; β-Actin forward, 5’-ATAGCACAGCCTGGATAGCAACGTAC-3’, reverse, 5’-CACCTTCTACAATGAGCTGCGTGTG-3’. A dissociation procedure was performed to generate a melting curve for confirmation of amplification specificity. β-Actin mRNA was quantified in parallel as the reference control. Relative gene expression was calculated using LightCycler® 96 Software. All experiments were replicated three times independently.
Haematoxylin and eosin (H&E) and immunohistochemistry staining
Tissues were fixed with 10% neutral formalin, embedded in paraffin, and 3-μm-thick sections were prepared by the pathology technologist. For H&E staining, sections were deparaffinized and hydrated with a gradient of alcohols. After soaking in phosphate-buffered saline (PBS), sections were stained with H&E. For IHC, sections were stained using the streptavidin-peroxidase method. In short, sections were deparaffinized, hydrated and soaked in 3% H2O2 for 10 min at room temperature, and then incubated with Rap2(12) monoclonal antibody (1:500, sc-136138; SANTA CRUZ) at 4°C overnight. Biotinylated secondary antibody and diaminobenzidine (DAB) were purchased from Maixin Biotechnology (Fuzhou, China). Evaluation of Rap2A staining in HCC tissue sections was performed with semi-quantitative IHC assessment method [12].
Cell culture
The normal human hepatocytes cell line LO2, human hepatoblastoma cell line HepG2 and HCC cell lines including Huh7, MHCC97H, SK-Hep1 and SMCC-7721 were purchased from the cell bank of Shanghai Institute of Cell Biology. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (HyClone, the USA) supplemented with 10% fetal bovine serum (Gibco, the USA) and 100 IU/ml penicillin and 100 μg/ml streptomycin (Millipore, Germany) at 37°C, 5% CO2.
Statistical analysis
Data was analyzed using SPSS version 22.0 for Windows (IBM Corporation, New York, USA). The two-related samples Wilcoxon nonparametric test was performed to evaluate the difference of Rap2A expression between HCC tissues and adjacent normal liver tissues. The chi-squared test and Fisher’s exact tests were used to examine possible correlations between Rap2A expression and clinicopathological features. Survival analysis was performed using the Kaplan-Meier test, and differences between curves were compared by the log-rank test. Multivariate analysis was performed to analyze the risk factors on postoperative survival. All P values were two-sided, and a P value of <0.05 was considered statistically significant.
Results
Rap2A is overexpressed in human HCC tissues and cell lines
Western blot for Rap2A in 42 paired HCC tissues and the quantification results are shown in supplementary Figure S1 and Table S1. Rap2A protein was upregulated in HCC tissues (27 of 42=64.3%, P<0.01, Figure 1A). The mRNA expression of Rap2A was detected by qRT-PCR after extracting total RNA from 50 paired HCC tissues and neighboring non-cancerous tissues. As shown in Figure 1B, the expression level of Rap2A mRNA was higher in HCC than that in normal liver tissues (34 of 50=68.0%, P<0.001). We also found that Rap2A was overexpressed at protein and mRNA levels in Hep G2, MHCC97H and Sk-hep1 cells compared to weak expression in normal hepatocytes LO2 cells (Figure 1C, 1D).
Figure 1.
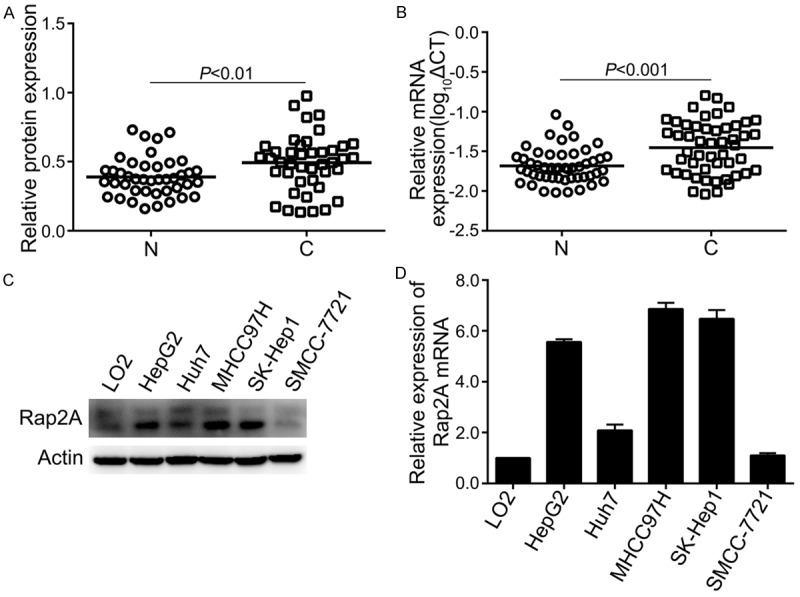
Upregulation of Rap2A protein and mRNA in HCC tissues and cell lines. A. Quantitative western blot analysis of Rap2A protein from 42 paired HCCs and their corresponding non-cancerous tissues. B. Analysis of Rap2A mRNA in 50 paired HCCs with their adjacent non-cancerous tissues by qRT-PCR method. Log10ΔCT of Rap2A mRNA were normalized to β-Actin mRNA. N, Non-cancer; C, Cancer. C, D. Rap2A protein and mRNA expression level in one normal hepatocytes (L02), one hepatoblastoma cell line (HepG2) and 4 HCC cell lines (Huh7, MHCC97H, SK-Hep1, SMCC-7721).
Immunohistochemical staining of Rap2A protein in HCC
IHC was adopted to identify the localization and expression pattern of Rap2A in 82 postoperative HCC samples. Rap2A was mainly detected in the cytoplasm and almost absent in the nuclei for HCC tissues (Figure 2). In addition, much stronger Rap2A staining was observed in HCC tissues (54 of 82=65.9%, P<0.001), while relatively weak Rap2A staining was examined in the majority of adjacent liver tissues.
Figure 2.
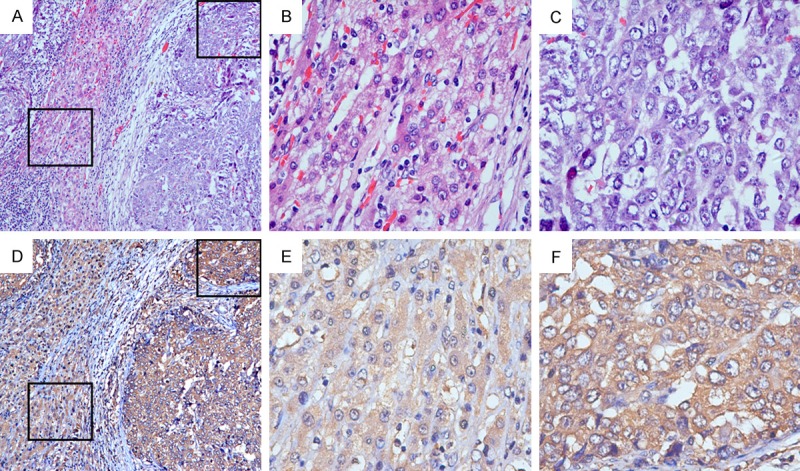
Representative immunostaining images of Rap2A in HCC. Stronger Rap2A expression was shown in HCC tissue (F) compared to relatively weak Rap2A expression in adjacent normal liver tissue (E). Rap2A was localized mainly in the cytoplasm and expressed little in the nuclei in HCC tissues (F). (A-C for H&E staining; D-F for IHC staining; Magnification: A, D 100×; B, C, E, F 400×).
Rap2A is correlated with HCC progression
After we detected Rap2A in 82 paired HCCs by IHC, we divided patients into a low Rap2A expression group (n=28) and a high Rap2A expression group (n=54). We then analyzed the clinical pathological correlation of Rap2A overexpression in HCC. High expression of Rap2A had strong correlation with tumor size (P=0.019), HCC metastasis (P=0.002), poor differentiation degree (P=0.028) and vascular invasion (P=0.017). On the contrary, there was no significant difference between high expression of Rap2A expression and patients’ age, gender, liver cirrhosis status, serum alphafetoprotein (AFP) and hepatitis B virus (HBV) level (Table 1). Furthermore, we found that HCC patients with high expression of Rap2A had shorter survival time than patients with low expression of Rap2A (P=0.011) (Figure 3). We applied multivariate Cox proportional hazards regression analysis to explore the association between high Rap2A expression and HCC patients’ prognosis. Our findings indicated that high expression of Rap2A was a risk factor related to the prognosis of patients with HCC (hazard ratio =3.090, with 95% confidence interval =1.142-8.362, P=0.026, Table 2). Meanwhile, patients’ prognosis was associated with tumor size, metastasis status, tumor differentiation and vascular invasion (P<0.05, Table 2). Age, gender, liver cirrhosis status, serum AFP and HBV level showed no significant correlation with the prognosis of HCC.
Table 1.
Correlation between Rap2A expression and clinicopathological features of HCC patients
| Variables | Rap2A expression level | χ2 | P | ||
|---|---|---|---|---|---|
|
| |||||
| Low (n=28) | High (n=54) | ||||
| Age (years) | <55 | 12 | 25 | 0.088 | 0.767 |
| ≥55 | 16 | 29 | |||
| Gender | Female | 6 | 9 | 0.280 | 0.597 |
| Male | 22 | 45 | |||
| Tumor size | <5 cm | 18 | 20 | 5.506 | 0.019* |
| ≥5 cm | 10 | 34 | |||
| Metastasis | No | 20 | 19 | 9.712 | 0.002* |
| Yes | 8 | 35 | |||
| Differentiation | Poor | 4 | 22 | 7.129 | 0.028* |
| Moderate | 21 | 30 | |||
| High | 3 | 2 | |||
| Liver cirrhosis status | No | 5 | 13 | 0.416 | 0.519 |
| Yes | 23 | 41 | |||
| Serum HBV level (cps/ml) | <1000 | 15 | 28 | 0.022 | 0.882 |
| ≥1000 | 13 | 26 | |||
| Serum AFP level (lg/l) | <400 | 19 | 37 | 0.004 | 0.951 |
| ≥400 | 9 | 17 | |||
| Vascular invasion | No | 17 | 18 | 5.651 | 0.017* |
| Yes | 11 | 36 | |||
HBV, hepatitis B virus; AFP, Alpha-fetoprotein.
representative statistically significant (P<0.05).
Figure 3.
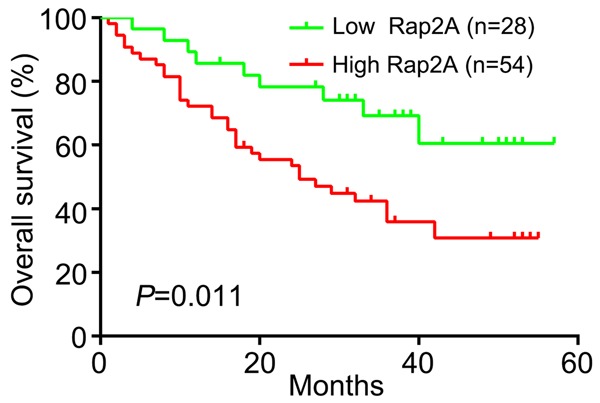
Kaplan-Meier analysis of overall survival between low and high Rap2A expression levels.
Table 2.
Cox regression model for prediction of 82 patients with HCC
| Variable | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Tumor size | 2.342 | 1.197-4.581 | 0.013* |
| Metastasis | 2.166 | 1.019-4.605 | 0.045* |
| Differentiation | 0.470 | 0.257-0.859 | 0.014* |
| Vascular invasion | 2.994 | 1.375-6.517 | 0.006* |
| High Rap2A expression | 3.090 | 1.142-8.362 | 0.026* |
CI, Confidence interval; SE, standard error.
representative statistically significant (P<0.05).
Discussion
HCC is the most common type of liver cancer and the third leading cause of cancer death worldwide [13]. Exploring the molecular mechanism of tumorigenesis and recurrence is extremely important for seeking better therapeutic strategies for patients with HCC. The RAS oncogene family members can regulate a range of biological functions such as proliferation, migration and signal transduction in human cells [14,15]. Rap2A belongs to the RAS oncogene family but its role and effect in hepatocellular carcinoma remain unknown.
We firstly examined the expression pattern of Rap2A in HCC tissues via western blot, qRT-PCR and immunohistochemistry. Rap2A was overexpressed in HCC tissues compared with neighboring nontumorous liver tissues. Rap2A was also upregulated in Hep G2, MHCC97H and Sk-hep1 cells at protein and mRNA levels. IHC showed that Pap2A was chiefly localized in the cytoplasm and significantly overexpressed in cancerous tissues. Then we analyzed the relationship between Rap2A expression and clinicopathological factors in HCC patients. Our results showed that high expression of Rap2A was of great clinical significance and correlated with tumor size, metastasis, differentiation and vascular invasion. The survival rate of high Rap2A expression group was much lower than that of low Rap2A expression group. Cox’s proportional hazard regression also indicated that high Rap2A expression, tumor size, metastasis status, pathological differentiation and vascular invasion could predict for HCC patients’ death.
Previous studies have proved that Rap2A can interact with and regulate several downstream effectors including MINK1, TNIK and MAP4K4 [5,16]. Besides, High expression of nuclear p-TNIK may be useful for predicting the postoperative survival of HCC patients [17]. MAP4K4 also promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma via activation of JNK and NF-κB signaling [18]. Pei et al demonstrated that Rap2A enhances the migration and invasion of osteosarcoma cells and increases activities of matrix metalloproteinase MMP2 and MMP9 [7]. Moreover, our statistical analysis on clinical data suggested that Rap2A was in connection with HCC patients’ tumor metastasis and vascular invasion. Given the results of aforementioned studies, we speculated that Rap2A might indeed participate in the development and progression of hepatocellular carcinoma.
According to our present study, Rap2A promotes HCC metastasis and brings a poor outcome for patients. On the other hand, Wu et al reported that Rap2A inhibits glioma cell migration and invasion by down-regulating p-AKT and probably serves as a tumor suppressor in the pathogenesis of glioma [19]. This study maybe controversial for the gene functions of Rap2A in carcinogenesis. The different effect of Rap2A on cancer might be due to cell-specific activity and diverse signal transduction. Thus, further studies about Rap2A in HCC are needed. For instance, we can examine the effect of Rap2A on HCC cell migration and invasion in nude mice using a heterotopic xenograft model.
In conclusion, our study demonstrated that high expression of Rap2A was correlated with poor prognosis of patients with HCC and Rap2A may be a predictive biomarker for HCC.
Acknowledgements
This work was supported by grants from the National Key Basic Research Program of China (2013CB933900), National Natural Science Foundation of China (81472231) and Projects of Xiamen Science and Technology Program (3502Z20130030 and 3502Z20144019).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Yao H, Liu X, Chen S, Xia W, Chen X. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:14830–14835. [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, Di J, Qu D, Gao Z, Zhang Y, Zheng J. Role of Rap2 and its downstream effectors in tumorigenesis. Anticancer Agents Med Chem. 2015;15:1269–1276. doi: 10.2174/1871520615666150518092840. [DOI] [PubMed] [Google Scholar]
- 5.Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem. 2004;279:49488–49496. doi: 10.1074/jbc.M406370200. [DOI] [PubMed] [Google Scholar]
- 6.Gloerich M, ten Klooster JP, Vliem MJ, Koorman T, Zwartkruis FJ, Clevers H, Bos JL. Rap2A links intestinal cell polarity to brush border formation. Nat Cell Biol. 2012;14:793–801. doi: 10.1038/ncb2537. [DOI] [PubMed] [Google Scholar]
- 7.Wu JX, Zhang DG, Zheng JN, Pei DS. Rap2a is a novel target gene of p53 and regulates cancer cell migration and invasion. Cell Signal. 2015;27:1198–1207. doi: 10.1016/j.cellsig.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Bigler D, Gioeli D, Conaway MR, Weber MJ, Theodorescu D. Rap2 regulates androgen sensitivity in human prostate cancer cells. Prostate. 2007;67:1590–1599. doi: 10.1002/pros.20644. [DOI] [PubMed] [Google Scholar]
- 9.Prabakaran I, Grau JR, Lewis R, Fraker DL, Guvakova MA. Rap2A is upregulated in invasive cells dissected from follicular thyroid cancer. J Thyroid Res. 2011;2011:979840. doi: 10.4061/2011/979840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YE, He HL, Chen TJ, Lee SW, Chang IW, Hsing CH, Li CF. The prognostic impact of RAP2A expression in patients with early and locoregionally advanced nasopharyngeal carcinoma in an endemic area. Am J Transl Res. 2015;7:912–921. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Li J, He F, Wang XM. Abnormal nuclear expression of Pygopus-2 in human primary hepatocellular carcinoma correlates with a poor prognosis. Histopathology. 2015;67:176–184. doi: 10.1111/his.12637. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S, Saiyin H, Wumaier R, Yang X, Wang C, Yu L. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016;372:101–109. doi: 10.1016/j.canlet.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 13.Bruix J, Reig M, Sherman M. Evidencebased diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Hattori M, Minato N. Rap1 GTPase: functions, regulation, and malignancy. J Biochem. 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- 15.Minato N. Rap G protein signal in normal and disordered lymphohematopoiesis. Exp Cell Res. 2013;319:2323–2328. doi: 10.1016/j.yexcr.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Nonaka H, Takei K, Umikawa M, Oshiro M, Kuninaka K, Bayarjargal M, Asato T, Yamashiro Y, Uechi Y, Endo S, Suzuki T, Kariya K. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem Biophys Res Commun. 2008;377:573–578. doi: 10.1016/j.bbrc.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 17.Jin J, Jung HY, Wang Y, Xie J, Yeom YI, Jang JJ, Lee KB. Nuclear expression of phosphorylated TRAF2- and NCK-interacting kinase in hepatocellular carcinoma is associated with poor prognosis. Pathol Res Pract. 2014;210:621–627. doi: 10.1016/j.prp.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Feng XJ, Pan Q, Wang SM, Pan YC, Wang Q, Zhang HH, Zhu MH, Zhang SH. MAP4K4 promotes epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. Tumour Biol. 2016;37:11457–11467. doi: 10.1007/s13277-016-5022-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Zhan W, Xie S, Hu J, Shi Q, Zhou X, Wu Y, Wang S, Fei Z, Yu R. Over-expression of Rap2a inhibits glioma migration and invasion by down-regulating p-AKT. Cell Biol Int. 2014;38:326–334. doi: 10.1002/cbin.10213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


