Abstract
The ubiquitin system, known as a common feature in eukaryotes, participates in multiple cellular processes, such as signal transduction, cell-cycle progression, receptor trafficking and endocytosis, and even the immune response. In lung cancer, evidence has revealed that aberrant events in ubiquitin-mediated processes can cause a variety of pathological outcomes including tumorigenesis and metastasis. Likewise, ubiquitination on the core components contributing to the activity of cell signaling controls bio-signal turnover and cell final destination. Given this, inhibitors targeting the ubiquitin system have been developed for lung cancer therapies and have shown great prospects for clinical application. However, the exact biological effects and physiological role of the drugs used in lung cancer therapies are still not clearly elucidated, which might seriously impede the progress of treatment. In this work, we summarize current research advances in cell signal regulation processes mediated through the ubiquitin system during the development of lung cancer, with the hope of improving the therapeutic effects by means of aiming at efficient targets.
Keywords: Ubiquitin, Ubiquitination, Deubiquitination, Cell signaling, Lung cancer
Background
Lung cancer is one of the most common malignant tumors and the leading cause of cancer-related mortality in the worldwide [1]. Non-small-cell lung carcinoma (NSCLC) represents 85% of all lung cancers and patients’ 5-year survival rate is only about 18% [2, 3]. The major challenges in the treatment of lung cancer are metastasis and drug resistance [4, 5]. At present, there is still no effective solution for them because of our poor understanding of the molecular mechanisms of lung cancer. Increasing evidence indicates that proteasome inhibition has become an attractive and potential anticancer therapy due to the UPS, like other cellular pathways, being critical for the proliferation and survival of cancer cells [6, 7]. For developing novel therapeutic approaches to treat lung cancer, it is important to deeply comprehend the different cell signaling and intricate mechanisms orchestrated via the ubiquitin pathway in association with lung cancer [4].
The UPS, which consists of a 26S proteasome and a small ubiquitin molecule, is a major protein degradation system which regulates a number of cellular functions, and is implicated in most of the cell signaling activities [8–10]. In recent years, dysregulation of various UPS components has been observed in cancer diseases including lung cancer [11–14]. Ubiquitination (and its reversal, deubiquitination) is one of the PTMs and plays important roles in regulation of a large number of cellular processes [15, 16], including cell cycle regulation [17, 18], apoptosis [19, 20], DNA damage [21–23] and immune functions [20, 24, 25]. Likewise, dysregulation of ubiquitination also results in aberrant activation or deactivation of signaling pathways. Thus, better understanding of the regulation mechanisms may ultimately lead to novel therapeutic modalities in lung cancer by targeting the ubiquitin pathway.
Here, we provide a comprehensive overview about ubiquitination and deubiquitination of the main components in cell signaling (i.e., PI3K-AKT-mTOR and RAS-RAF-MEK-ERK), which have been found to be regulated primarily in lung cancer. We also highlight the recent progress in our understanding of the molecular mechanisms by which cancer-associated proteins mediate cell signaling networks through the ubiquitin system.
Ubiquitin, ubiquitination and deubiquitination
Ubiquitin is found in all known eukaryotic organisms and it has a highly conserved 76 amino acid sequence that undergoes covalent attachment to lysine residues in target proteins via isopeptide linkage [15]. A single ubiquitin molecule contains 7 lysine residues (K6, K11, K27, K29, K33, K48, and K63) to which another ubiquitin can be ligated, resulting in different types of poly-ubiquitin chains involved in diverse cell processes [26, 27]. However, the poly-ubiquitin chain in which each additional ubiquitin molecule is linked to lysine 48 (K48) of the previous ubiquitin plays a main role in proteasome degradation [27], whereas K63-linked ubiquitin chains have been thought to participate in regulating various proteasome-independent cellular functions, including NF-κB signaling, DNA damage repair, ribosomal function, and intracellular trafficking [28, 29].
The process of ubiquitination requires the help of at least three different enzymes: ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin ligase E3 [30–32]. E3 ubiquitin ligases are critical in the UPS, as they mediate the specificity of substrate recognition and allow the transfer of activated ubiquitin from E2 enzymes to the target protein [33, 34]. Structurally, E3s can be divided into HECT-type E3s with a HECT domain which forms a thiolester bond with ubiquitin and then conjugates it to the substrate [35, 36], RING finger-containing E3s containing RING and U-box domains [37–39], RING-between-RING family members which have a RING1-in-between RING-RING2 motif [40]. Moreover, deubiquitination, known as a reverse process of ubiquitination, is also a complex enzymatic system responsible for removing ubiquitin from a substrate [41]. The isopeptide bond between ubiquitin and its substrate can be cleaved by the specific DUBs to produce monoubiquitin for recycling [16, 42–44]. Recently, more than 100 DUBs have been found that can be divided into five subfamilies: USP, OTU, MJD, UCH and JAMM/MPN metalloproteases [16, 44].
To date, increasing evidence has shown that ubiquitin E3 ligase and deubiquitination enzymes are directly involved in the regulation of tumor formation and metastasis of lung cancer, especially through the RAS-RAF-MEK-ERK and PI3K-AKT-mTOR signaling pathways, in which the ubiquitination of key signal nodes determines the biological and biochemical processes of tumor cells (Table 1). Below, we summarize the molecular mechanism mediated through the ubiquitin system in the development of lung cancer, and hope to supply more cues for the therapeutic strategy.
Table 1.
A list of ubiquitin E3 ligases and deubiquitination enzymes that have been found in regulation of associated genes in NSCLC
| Genes | E3 Ligases | DUBs | Main function in lung cancer through the ubiquitin system | Ref |
|---|---|---|---|---|
| EGFR | c-Cbl, Cbl-b | USP8, USP2a, AMSH | EGFR is often overexpressed and mutated in lung cancer, especially in non-small cell lung cancer. Mono-ubiquitinated or poly-ubiquitinated EGFR upon EGF stimulation do not affect tyrosine kinase activity or signaling capacity but play a critical role in lysosomal targeting. Ubiquitination of EGFR can facilitate its endocytosis and degradation. Disruption of the negative regulatory system is associated with lung carcinogenesis, while deubiquitinating enzymes can reverse this modification and hence oppose endosomal sorting and lysosomal degradation in non-small cell lung cancers. | [45–48] |
| Ras | NEDD4–1, Rabex5, βTrCP1 | – | RAS proteins are central mediators downstream of growth factor receptor signaling and therefore are critical for cell proliferation, survival, and differentiation. KRAS and NRAS mutations are more commonly found in lung cancers with adenocarcinoma histology. However, the ubiquitin system controls the subcellular localization and stability of Ras family protein in cancer cells, thereby contributing to the occurrences of tumor formation or metastasis. | [49–51] |
| Raf | RNF149, CHIP, TRAF2 | – | Function as protein kinase to phosphorylate MAP 2 K1 directly, and thereby contributes to the MAPK signal transduction. Somatic mutations in BRAF, especially at valine 600 (V600), commonly occurred in non-small cell lung cancer, and this mutated BRAF can escape from degradation via the ubiquitin proteasome system. However, K63-linkaged poly-ubiquitination of BRAF impairs the activity in BRAF-mediated ERK activation, which induced by RNF149 E3 ligase. TRAF2 is a novel E3 ligase of BRAF K48-linkaged poly-ubiquitination which affects its stability. | [52–54] |
| PTEN | NEDD4–1, WWP2, XIAP, CHIP | USP7, USP13 | It functions as a tumor suppressor that is mutated in a large number of cancers at high frequency and subcellular localization has been strongly implicated in the regulation of the PI3K/AKT pathway. Loss of PTEN expression is common in lung adenocarcinoma. Mono-ubiquitination and poly-ubiquitination of PTEN can be induced by NEDD4–1, WWP2, XIAP or CHIP, and deubiquitinated by USP7 or USP13 leading to its nuclear exclusion. Mono-ubiquitination of one of either Lys-13 or Lys-289 amino acid is sufficient to modulate PTEN compartmentalization. Phosphorylation of PTEN at Tyr-336 protects it from ubiquitin-mediated degradation probably by inhibiting its binding to NEDD4–1. | [55–58] |
| AKT | CHIP, BRCA1, NEDD4–1, Skp2, TRAF6, TRAF4 | CYLD | The AKT kinase, which regulates many cell processes, plays a critical role in the development of multiple cancer types and tumorigenesis. In lung cancer, it is abnormally activated via the post-translational modification, including phosphorylation, ubiquitination and sumoylation, thereby impairing its subcellular location and cell signaling transduction. The ubiquitin system induces the ubiquitination of AKT with different lysine-linkage types either for the proteasome degradation or for the function alteration to affect the cell signaling transduction and control the evolution of cancer cells. | [59–61] |
| mTOR | Fbxw7, TRAF6 | – | It functions as the main component of mTORC1 and mTORC2, and mTORC1 exerts a feedback control on upstream growth factor signaling including PI3K/AKT and MAPK signaling. Upon the stimuli of amino acid, the K63-likaged poly-ubiquitination of mTOR indirectly induced by TRAF6 promotes the mTORC1 activation to accelerate downstream signaling in non-small cell lung cancer. However, the K48-linkaged poly-ubiquitination of mTOR impacts both of mTORC1 and mTORC2 activities due to the proteasome degradation induced by Fbxw7. | [62–65] |
Ubiquitination in RAS-RAF-MEK-ERK pathway
The RAS-RAF-MEK-ERK pathway, mainly composed of Raf kinase, Ras-GTPase, MEK, and ERK, is the most characteristic pathway in cell biology involved in regulating cell proliferation, differentiation and apoptosis [66]. This signaling pathway is usually activated by a variety of growth factors, chemokines, polypeptide hormones, neurotransmitters, and phorbol esters through their cognate RTKs [67] and GPCRs [66], or by direct activation of PKC [68, 69]. Dysregulation of the ERK pathway, mainly caused by constitutive activation of Ras and Raf, has been well established in human malignancies [70] (Fig. 1). The activation of ERK1/2 promotes cell survival and chemotherapeutic resistance in lung cancer and greatly contributes to the development of NSCLC [71]. Likewise, ERK-dependent serine/threonine phosphorylation of specific substrates is essential for the ubiquitination and degradation process [72].
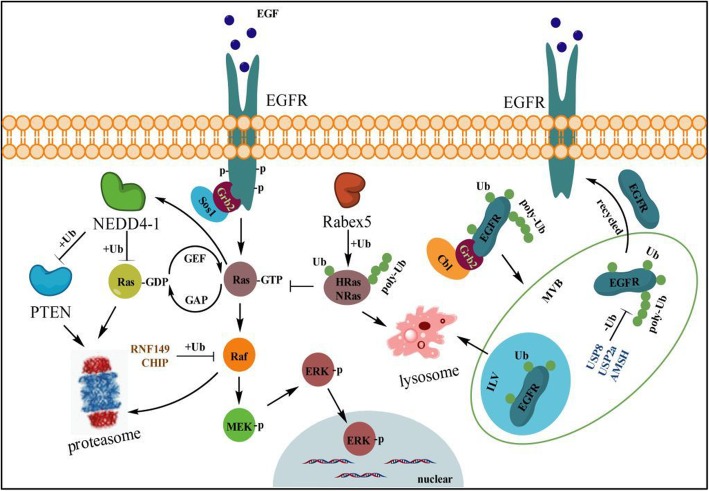
Fig. 1.
Ubiquitination on RAS-RAF-MEK signal. Upon EGF or other growth factors stimulation, activated Ras-GTP enhances the transcripts of NEDD4–1 which promotes the ubiquitination on all three forms of Ras-GTP and PTEN for the proteasome degradation to accelerate the downstream signaling activation. Rabex, to some extent as a ‘break’, can ligate mono- or poly-ubiquitin (K63-link) to HRas or NRas, but not KRas, which promotes their endosome localization and lysosome degradation, thereby limiting the transformation of Ras-GTP from Ras-GDP to suppress the phosphorylation activation of ERK. Likewise, BRAF and CRAF can be ubiquitinated by RNF149 and CHIP, respectively, and be degraded by the proteasome to decelerate MEK/ERK activation, which attenuates the increase in cell growth. Following EGF stimulation, RING domain E3 ubiquitin ligases c-Cbl or Cbl-b, with the assistance of Grb2, induces mono- or poly-ubiquitination (K63-link) of EGFR and mediates the endosomal sorting and trafficking events, in which process the mono-ubiquitinated EGFR is trapped within ILVs of multivesicular bodies (MVBs), whereas poly-ubiquitinated forms remaining in the MVBs are deubiquitinated by DUB enzymes USP8, USP2a or AMSH to escape the ILVs trapping and lysosome degradation. Instead, the non-ubiquitinated EGFR forms are recycled to the cell membrane for the downstream signal activation
Ubiquitination controls Ras protein level and subcellular localization
Ras is a small GTPase that is activated by various cell-surface molecules, and membrane localization is essential for its activation [73]. All three Ras isoforms, H-Ras, K-Ras (two splice variants, K-Ras4A and K-Ras4B), and N-Ras reside in the plasma membrane and switch on/off for the downstream signal transduction [74], whereas the ubiquitination of Ras has been shown to control Ras protein turnover as well as its subcellular localization [50]. Rabex-5 (also known as RabGEF1) functions as an E3 ligase for mediating Ras (H-Ras and N-Ras, but not K-Ras) ubiquitination to promote Ras endosomal localization, and further leads to the suppression of ERK activation [75]. Meanwhile NEDD4–1 can regulate Ras-GDP level of all three forms and subsequently drives PTEN degradation, leading to tumor processes [76] (Fig. 1). Smurf2 and UbcH5 as a critical E3 and E2, respectively, are important in maintaining K-Ras protein stability, and targeting such a complex was supposed to be a unique strategy to degrade mutant K-RasG12/V or C to kill cancer cells [51]. However, knock-down Smurf2 can accumulate the F-box protein βTrCP1 which mediates poly-ubiquitination and proteasome-dependent degradation of Ras [51, 77]. H- and N-Ras, but not K-Ras, are subjected to mono- and K63-linked di-ubiquitination, and stabilize their associations with the endosome, resulting in a change in the signaling output [49], while K-Ras shows only minor or transient association with the endosome [78]. However, K147 in K-Ras or H-Ras was identified as one of the major ubiquitination sites; the modification of it increases the fraction of GTP-bound Ras and more efficiently activates Raf and PI3K to enhance its tumorigenic activity [50]. Intriguingly, monoubiquitination on K147 in K-Ras does not affect protein localization, but rather impedes GAP-mediated GTP hydrolysis and promotes the association with downstream effectors [79, 80]. In lung cancer disease, the sustained activation of ERK is a common event and frequently contributes the tumor growth and even the metastatic processes; therefore it might be an efficient way to abolish the enhanced Ras protein level or Ras-GTP activity by inhibiting its E3 ubiquitin ligase (i.e. NEDD4–1) to suppress the tumor progress.
The direct deubiquitination process of Ras has not been described in any cancer or other diseases. As reported, carboxyl-terminal CAAX motifs in Ras are essential for its activity and proper membrane localization, and can be cleaved by RCE1 [81], whereas RCE1 can be down-regulated by the deubiquitinating enzyme USP17, a process that occurs in the ER, through removing the functional K63 polyubiquitin chains of RCE1 [81, 82]. As a consequence, it blocks Ras membrane localization and activation, thereby inhibiting phosphorylation of the downstream kinases MEK and ERK [81]. Intriguingly, USP17 impedes EGF-induced H-Ras and N-Ras but not K-Ras membrane trafficking, no matter whether wild-type Ras or oncogenic mutants [81–83]. Suppression of USP17 inhibits the abilities of tumorigenesis and invasion of NSCLC cells in vitro and in vivo [84]. In clinical practice, USP17 was always observed over-expressed in both squamous and adenocarcinoma NSCLC tissues. Patients with USP17-positive tumors had significantly reduced recurrence-free survival and USP17 mRNA level positively correlated with NSCLC distant metastasis [85]. USP17 depletion can not only block the proliferation of NSCLC cells with EGFR wild-type, but also those bearing active mutations of EGFR or TKI resistant mutations [86]. This evidence suggests that USP17 may ultimately enhance the Ras activity to promote the tumor processes in NSCLC and could be a great potential therapy target for drug development for the treatment of NSCLC.
Different Raf proteins play a distinct role in the cell signaling pathway
As the receptor tyrosine kinase effecter in the ERK pathway, Raf consisting of ARAF, BRAF, and CRAF shows a serine/threonine kinase activity, relevant to tumorigenesis, including cell proliferation, survival, invasion, and angiogenesis [54]. The three Raf proteins have a similar structure and all are considered to be oncogenic, but they execute distinct properties for MEK phosphorylation and activation [74]. Intriguingly, complex formation by these different isoforms play a critical role in their activation, particularly in response to RAF inhibitors, and BRAF/CRAF complexes seem to be stabilized by ARAF in cells, thereby regulating cell signaling to ensure signaling efficiency [87].
BRAF is modified by K63-linked polyubiquitination at K578 through gain of a constitutively active mutation (V600E, which renders the constitutive activation of BRAF and is responsible for more than 90% of somatic mutations in human tumors) under EGF stimulation [52]. Substitution of BRAF lysine 578 with arginine (K578R) weakened K63 polyubiquitination and inhibited BRAF-mediated ERK activation [88]. However, the specific E3 ligase(s) and deubiquitinating enzyme(s) controlling the positive and negative regulation of BRAF K63-linked polyubiquitination still need to be further identified [52]. RNF149, as a RING domain-containing E3 ubiquitin ligase, is involved in control of gene transcription, translation, cell adhesion, cytoskeletal organization or epithelial development. It is an authentic E3 ligase of wild-type BRAF, but not of mutant BRAF (V600E), and induces BRAF degradation through the ubiquitin proteasome system and thereby reduction of MEK/ERK activity [53] (Fig. 1). TRAF2 is a novel E3 ligase of BRAF K48-linked ubiquitination. TRAF1 binding with TRAF2 could decrease BRAF K48-linked ubiquitination but not affect K63-linked ubiquitination. TRAF1 seems to serve as a ‘break’ for TRAF2 driving BRAF degradation, which activates MEK and ERK mediation of lung cancer cell growth, apoptosis or lung tumorigenesis [89] (Fig. 1). Therefore, blocking TRAF1 using appropriate chemical drugs to release TRAF2 might also be an ideal way to inhibit the sustained MEK/ERK activation in NSCLC.
CRAF (also termed Raf-1), normally residing in the cytoplasm as an inactive kinase, is activated by GTP-Ras and recruited to the cell membrane [90, 91]. This activation process is tightly regulated by various factors including kinases (e.g. ERK, Src, AKT, PKC), phosphatases (e.g. PP2A, PP1, PP5) and proteins that directly bind to CRAF (e.g. 14–3-3, RKIP, Hsp90, KSR) [90, 92]. Nevertheless, autophosphorylation of serine 621 (S621) is essential to ensure the correct folding and stability of the CRAF protein, which prevents it from being degraded by CHIP (carboxy terminus of Hsc70 interacting protein) [91]. Although CHIP is an identified E3 ubiquitin ligase of CRAF, it is not unique to induce the degradation of S621 non-phosphorylated CRAF [91].
Ubiquitination of EGFR is involved in endosomal sorting and lysosome degradation
EGFR (also termed as HER1), one of RTKs of the ErbB family, is a transmembrane glycoprotein with cytoplasmic kinase activity that regulates signaling pathways to control cellular proliferation [93]. Mutation of EGFR has been strongly implicated in the pathogenesis of many human malignancies, especially in NSCLC [45, 94, 95]. The basic signal mechanism is that growth factors (e.g. EGF, FGF, PDGF and TGF-α) trigger membrane EGFR homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues, leading to receptor hyperactivity [93, 96]. Further, the phosphorylated EGFR recruits adapter proteins such as GRB2, which in turn activates complex downstream signaling cascades [97, 98], including the RAS-RAF-MEK-ERK, PI3K-AKT-mTOR, and probably the NF-κB signaling cascade [96, 98].
As reported, most cell surface receptors, including RTKs such as EGFR and β2AR, can be internalized and undergo rapid clathrin-dependent or -independent endocytosis which is required for maintenance of regulated receptor trafficking and kinase signaling [99, 100]. More compelling evidence has proved that ubiquitination is a major posttranslational modification of EGFR that controls the endosomal sorting and trafficking of diverse signaling receptors after endocytosis [46, 99]. In that process, endosomal protein ESCRT complexes I, II, and III, each containing a ubiquitin binding domain, and Hrs, which might deliver ubiquitinated cargo to the outer membrane of the late endosome, are thought to participate in trapping EGFR within ILVs of multivesicular bodies (MVBs) that are destined for subsequent degradation in the lysosome, thereby preventing their recycling to the plasma membrane [47, 48] (Fig. 1).
RING domain E3 ubiquitin ligases c-Cbl and Cbl-b, with the assistance of Grb2, induce ubiquitination of EGFR following EGF stimulation, which has been implicated in regulation of both its localization and stability [48, 101]. Multiple monoubiquitination of activated EGFR is thought to be involved in endocytic trafficking, and even a single ubiquitin is sufficient for both receptor internalization and degradation [47]. However, quantitative mass spectrometry demonstrated that multiple lysines in the kinase domain of EGFR could be primarily conjugated to K63-linked polyubiquitin chains for the endosomal sorting process and lysosome degradation [48]. Nevertheless, the further experiments implied that EGFR ubiquitination is not necessary for clathrin-mediated internalization, which might be controlled by multiple kinase- and ubiquitination-dependent and -independent mechanisms [102]. Conversely, endosomal DUBs such as USP2a, USP8/UBPy and AMSH reverse this modification and hence oppose endosomal sorting and lysosomal degradation [103–105]. Controversially, UBPy and AMSH may either accelerate or inhibit degradation of EGFR upon EGF stimulation, which due to the distinct catalytic or regulatory domain in their structures differentially control the EGFR turnover [45, 106, 107]. Moreover, USP2a, which localizes to early endosomes, is over-expressed in NSCLC and shows an oncogenic property through increasing the plasma membrane-localized EGFR, as well as decreasing the internalized and ubiquitinated EGFR [104] (Fig. 1).
In addition, the functional defects of EGFR with specific mutations (L858R, del746–750 or L858R + T790 M) in NSCLC cells can be impaired, probably due to a propensity of the mutants to heterodimerize with HER2, thereby evading c-Cbl-mediated ubiquitination and subsequent sorting to degradation in lysosomes [108]. Intriguingly, the E3 ligase CHIP could selectively interact with and degrade the EGFR mutants such as G719S, L747_E749del A750P or L858R and inhibited tumor cell proliferation and xenograft growth of EGFR mutant but not EGFR WT cell lines, which might provide novel therapeutic strategies for overcoming the EGFR-TKI resistance in lung adenocarcinoma [109]. Taken together, this evidence suggested that it could be an efficient manner to promote WT- or Mut- EGFR internalization and ubiquitination to impede its membrane localization via targeting USP2a, which could attenuate the activation of ERK signaling and thereby suppress the cancer processes in NSCLC.
Ubiquitination in PI3K-AKT-mTOR pathway
The PI3K-AKT-mTOR signal is an intracellular signaling pathway and has a critical role in the regulation of the ubiquitin-proteasomal system and autophagy in mammalian cells [110, 111]. PI3K-AKT-mTOR pathway activation can be mediated by specific aberrations in PIK3CA, PIK3R1, AKT, LKB1, TSC1/2, EGFR or PTEN [112]. Many known factors can also enhance the PI3K-AKT-mTOR pathway including EGF [113], shh [114, 115], insulin [116], and IGF-1 [114]. Under the condition of stimuli, PI3K phosphorylates PIP2 to create PIP3 and recruits AKT to the plasma membrane to active PDK1 and mTOR complex which phosphorylates 4E-BP1 and p70 ribosomal S6 kinase that trigger ribosome biogenesis and translation in cell growth and division [61, 117] (Figs. 2&3).
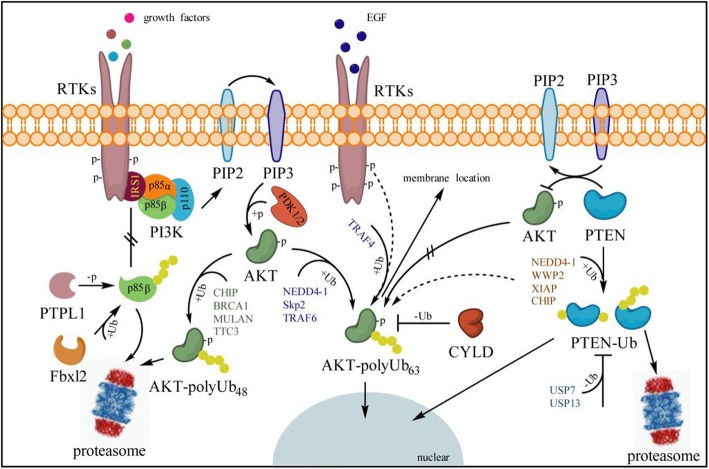
Fig. 2.
Ubiquitination on PI3K-AKT signal. Under the conditions of growth factor stimuli, such as insulin, activated RTKs recruit IRS1 (insulin receptor substrate 1) for binding and activation of p85-p110 heterodimers (PI3K). With the assistance of phosphatase PTPL1, free p85β is ubiquitinated by Fbxl2 and degraded through the proteasome system to block its competition with p85-p110 heterodimers binding to IRS1, thereby promoting activation of the PI3K-AKT signal. The p110 subunit of PI3K catalyzes the conversion of PIP2 to PIP3, which recruits AKT to the plasma membrane for activation through PDK1 and PDK2. In that process, activation of AKT is inhibited by PTEN through conversion of PIP3 to PIP2 to restraint the downstream events, i.e., K48-linked polyubiquitination on phosphorylated AKT is triggered by E3 ligases CHIP, BRCA1, MULAN or TTC3 for proteasome degradation, but K63-linked AKT is induced by NEDD4–1, Skp2 and TRAF6 to mediate its nuclear localization for further activation of the PI3K-AKT signal. However, TRAF4 is the main effector for AKT K63-linked ubiquitination and promotes EGF-induced AKT membrane recruitment in human lung cancer cells to induce tumorigenic properties. Deubiquitinating enzyme CYLD plays a tumor suppressor role in inhibiting AKT activity by removing AKT K63-linked ubiquitin chains and serves as a negative regulator for AKT-mediated tumorigenesis. Ubiquitinated PTEN with poly-ub chains is usually degraded by the proteasome system, while partial mono-ubiquitinated forms translocate into the nucleus to escape proteasome degradation, mediated by NEDD4–1, WWP2, XIAP and CHIP E3 ligase. However, this progress can be reversed by deubiquitinating enzymes USP7 and USP13 to deactivate the PI3K-AKT signal
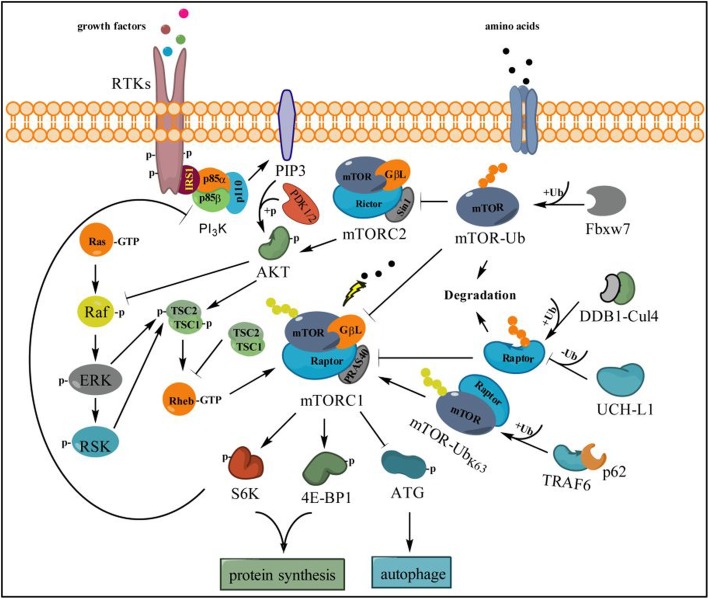
Fig. 3.
Ubiquitination on mTOR signal affects the cross-talk between RAS-RAF-MEK and PI3K-AKT-mTOR pathways. TSC2 and mTORC1 as the downstream sensor receive many inputs from both RAS-RAF-ERK and PI3K-AKT signaling to activate S6K and 4E-BP1 for mRNA translation and protein synthesis, as well for inhibition of autophagy. Meanwhile, activated mTORC1 can enhance the RAF-ERK signaling through feedback inhibition of PI3K but not of AKT or mTORC2. In this pathway, Fbxw7 is responsible for recognizing mTOR and executing the ubiquitination (K48-Ub chains) for further proteasome degradation to reduce the activity of mTORC1 and mTORC2. Upon the stimulation of amino acid, K63-linked ubiquitin chains on mTOR are essential for activation of mTORC1, which is triggered by TRAF6 with the assistance of p62 and adaptor protein Raptor. However, Raptor, an essential component of mTORC1, can be poly-ubiquitinated and deubiquitinated by the DDB1-Cul4 complex and UCH-L1, respectively, thereby impairing the activity of mTORC1 and downstream signaling
Ubiquitination on the p85 subunit contributes to the PI3K signaling cascade
In lung cancer, the PI3K pathway is frequently dysregulated due to genetic alterations [118]. PI3K is a family of lipid enzymes that specifically phosphorylate the 3′-hydroxyl group of phosphatidylinositols and phosphoinositides on membranes [119]. Classical PI3K is composed of a p85 (p85α, p85β and p55γ) regulatory subunit and a p110 catalytic subunit [120, 121]. A certain amount of p85, which lacks intrinsic kinase activity, is necessary for PI3K to bind phospho-tyrosine docking sites at the cell membrane [122], while excessive free p85 could compete with p85-p110 heterodimers binding to IRS1, a process which inhibits the PI3K activity and its downstream signaling. Fbxl2, a member of the F-box protein family which usually forms the ubiquitin E3 complex with Skp1, Rbx1 and Cul1, specifically binds p85α and p85β, but not p110 [122]. However, Fbxl2 can only induce the degradation of tyrosine-dephosphorylated p85β triggered by the phosphatase PTPL1, which inhibits the excessive free p85 contacting IRS1, leading to an increase in the binding of p85-p110 heterodimers to IRS1 and enhancement of the PI3K signaling cascade [122] (Fig. 2). Therefore, suppressing Fbxl2 activity to maintain free p85 forms in cells will help to inhibit the aberrant activation of PI3K/AKT signaling to some extent in NSCLC.
Different ubiquitin linkages of AKT mediate a diverse cell signaling pathway
AKT, also known as PKB, is a serine/threonine protein kinase involved in various signaling transduction pathways [123]. AKT, as one of the major downstream effectors of PI3K, plays a vital role in the promotion of cell proliferation and preventing the cell from entering the apoptotic pathway by interacting with caspase-9 and Bcl2 [124]. The inactivation of PTEN and RAS also can stimulate AKT activation, and excessive activation of AKT was suggested to be a poor prognostic factor for early stage NSCLC patients [61]. However, ubiquitination is completely essential for AKT signaling activation. For instance, K63-linked ubiquitination mediated by NEDD4–1 [125], Skp2 [59] and TRAF6 [59, 126], which itself could be negatively mediated by c-Cbl [127], induces AKT activation via promoting plasma membrane translocation and nuclear translocation. Converse evidence suggests that TRAF4, but not Skp2, is required for AKT K63 ubiquitination and promotes EGF-induced AKT membrane recruitment in human lung cancer cells to induce tumorigenic properties [60], but TRAF6 is unnecessary for EGF-induced AKT activation [59, 60]. In contrast, K48-linked ubiquitination mediated by CHIP, BRCA1, MULAN and TTC3 triggers the proteasomal degradation of phosphorylated AKT to terminate its activation [29, 111, 128–130]. Nevertheless, the ubiquitination of AKT can be reversed by CYLD, which is a deubiquitinating enzyme and plays a tumor suppressor role in inhibiting AKT activity by removing AKT K63-linked ubiquitin chains and serves as a negative regulator for AKT-mediated tumorigenesis or lung fibrosis [131] (Fig. 2). It is worth mentioning that deubiquitinase CYLD and E3 ubiquitin ligase Itch are able to form a complex by interaction through “WW-PPXY” motifs, and sequentially cleave K63-linked ubiquitin chains for catalyzing K48-linked ubiquitination on Tak1 to terminate inflammatory signaling via TNFs [132]. Deficiency in either Itch or CYLD will lead to the chronic production of cytokines specifically generated by tumor-associated macrophages, which further contributes to the aggressive growth of lung carcinoma [132]. Additionally, CYLD overexpression can block TRAIL-induced NF-κB activation directly, and consequently increase TRAIL-induced apoptosis in lung cancer cells [133]. This evidence revealed that TRAF4, SKP2, NEDD4–1 or TRAF6, but not CYLD, could be an ideal target for drug development and NSCLC therapy.
PTEN has always been targeted by the ubiquitin system
PTEN, as a famous tumor suppressor, directly dephosphorylates phosphoinositides to antagonize the PI3K-AKT/PKB signaling pathway and thereby modulates protein synthesis, cell cycle and cell survival [134, 135]. PTEN attenuates PI3K signaling by directly binding pleckstrin homology domains of specific signaling proteins to dephosphorylate PIP3 to PIP2 [136, 137]. Loss of PTEN increases the phosphorylation of AKT and deregulates PI3K signaling, which in turn enhances cell survival [138–141]. However, PTEN is not frequently targeted at the genetic level in the development of lung cancer, and mutations of the PTEN gene in patients harboring NSCLC have been reported in 8 to 17% [135]. On the other hand, PTMs of PTEN that regulate its enzymatic activity, interaction with other proteins and subcellular localization have been strongly implicated in the regulation of the PI3K/AKT pathway [57, 135, 142, 143]. The PTEN protein level is controlled to a large extent by E3 ligase-mediated UPS degradation [55]; therefore, stabilizing PTEN protein level is a promising therapeutic strategy for most cancer diseases including NSCLC. NEDD4–1 [58, 144], WWP2 [145], XIAP [146] and CHIP [55] have been reported to be responsible for PTEN turnover (Fig. 2). Among them, NEDD4–1 was first identified as an E3 ubiquitin ligase that regulates stability of PTEN [144] (Fig. 1 and Fig. 2), and plays a critical role during the development of NSCLC [135]. NEDD4–1 is over-expressed in 80% of NSCLC tumors and correlates with the deficiency of PTEN protein [56, 135, 147]. In the mechanism, NEDD4–1 physically interacts with PTEN and leads to both mono- and poly-ubiquitination of PTEN at K289 and K13 sites [57]. However, mono-ubiquitination of PTEN appears to be a limited step for proteasome degradation while it is crucial for its nuclear import [57] (Fig. 2).
USP7 (also known as HAUSP) and USP13, as specific DUBs of PTEN, reversely regulate the stability of PTEN. USP7 was first found to be able to stabilize p53 through its intrinsic deubiquitinating enzyme activity in the lung cancer cell line H1299 [148]. In acute promyelocytic leukemia, the removal of either K289 or K13 mono-ubiquitin from PTEN by USP7 restrains PTEN nuclear localization without affecting its protein level [149]. Despite that, PML opposes the activity of USP7 towards PTEN through a mechanism involving the adaptor protein DAXX (death domain-associated protein) [149]. USP13 functions as a tumor suppressor mainly through reversing PTEN poly-ubiquitination and stabilizing PTEN protein levels via its deubiquitination action [150]. However, the functions of USP13 in lung cancer have not yet been elucidated. More interestingly, ataxin-3, as one member of the Josephin family DUBs, can enhance the transcription level of PTEN probably through stabilizing its specific transcriptional activators to down-regulate AKT phosphorylation and PI3K signaling in NSCLC [143].
Cross-talk between PI3K-AKT-mTOR and RAS-RAF-MEK signal
The RAS-MEK-ERK and PI3K-AKT-mTOR pathways can negatively or positively regulate each other’s activities, in which the mammalian target of rapamycin (mTOR) is a core component sensor. mTOR, a serine/threonine protein kinase and the catalytic subunit of complexes including mTORC1 and mTORC2, has been identified as the downstream target of the PI3K/AKT pathway that regulates processes including mRNA translation, proliferation, and survival [151, 152]. The activity of mTORC1 can be regulated through the function of tuberous sclerosis complex (TSC1 and TSC2) activated by membrane localized AKT and enhances the GTPase activity of the mTOR activator Rheb [62, 153], thereby promoting cell growth via up-regulation of protein synthesis through activation of 4E-BP1 and p70S6 kinase [63, 154] (Fig. 3). However, inhibition of mTORC1 can lead to RAS-MEK-ERK activation through PI3K-dependent feedback but not mTORC2, AKT or targets of downstream of AKT in human cancer [62], which reveals an alternative signal whereby phosphorylation at Ser259 of Raf by AKT deactivated and inhibited the signal cascade of RAS-MEK-ERK [155] (Fig. 3). Evidence has shown that inhibition of the mTOR pathway represents a promising therapeutic approach for lung cancer [64, 156, 157].
Like AKT, ubiquitination also plays a key role in regulation of the mTOR pathway. E3 ubiquitin ligase TRAF6 is necessary for mTORC1 translocation to the lysosomes, and the TRAF6-catalyzed K63 ubiquitination of mTOR regulates mTORC1 activation through p62 upon amino acid stimulation [63]. Nonetheless, mTOR can be degraded by the ubiquitin proteasome pathway and Fbxw7 is an mTOR regulator that altered its expression in a manner opposite to mTOR, which affects the activity of both mTORC1 and mTORC2 [65, 158]. Moreover, Rictor as an essential component of mTORC2 could also be directly mediated by Fbxw7 for ubiquitination and proteasome degradation, which thereby impairs the AKT activation and downstream signaling [159]. Under mitochondrial stress, maintenance of mTORC1 activity requires the ubiquitination of mTOR at K2066 and K2306 catalyzed by Parkin, which in turn promotes cell survival and growth [160]. In addition, UCH-L1 as a ubiquitin hydrolase of Raptor has a critical role in regulation of the dichotomy between mTORC1 and mTORC2 signaling, and it impairs mTORC1 activity toward S6 kinase and 4E-BP1 while increasing mTORC2 activity toward AKT [161] (Fig. 3).
Therapies and prospects
Cell signaling responses play an important role in regulating cell characteristics. However, the key regulation nodes function as a signaling ‘switch’ to mediate cell processes. Meanwhile, the ubiquitination system intricately regulates activation and inactivation of these signaling pathways. The examples described here illustrate that the PI3K-AKT-mTOR and RAS-RAF-MEK pathways are subjected to the ubiquitin regulation in lung cancer, and the ubiquitination on those signaling nodes directly orchestrates the cell signal transduction positively or negatively. Although some of the E3s or DUBs mentioned above have shown some potential as drug targets for the treatment of NSCLC, more experimental evidence and clinical trials are needed to identify the effects.
Given this, inhibition of the ubiquitin system, including proteasome, E1, E2, E3 and DUB, has been developed and proven a very effective treatment in multiple malignancies in addition to NSCLC. For example, bortezomib (PS-341), the first proteasome inhibitor approved by the FDA for the treatment of multiple myeloma, has been tested in numerous NSCLC models in vitro and in vivo, and shows an active effect against NSCLC cells. In preclinical studies, bortezomib inhibits the proteasome activity and further affects the function of numerous proteins involved in processes such as cell-cycle control, apoptosis, angiogenesis, and chemoresistance [162–165]. Currently, phases I and II studies show promising results in combination therapy for NSCLC through combining bortezomib with available chemotherapeutic agents or targeted therapy [166–168], such as carboplatin/bevacizumab [166], paclitaxel/carboplaitn [167] and gemcitabine/carboplatin [168]. However, bortezomib in combination with erlotinib, which targeted EGFR mutations in NSCLC, did not show any survival benefit in patients with relapsed/refractory advanced NSCLC [169]. To date, bortezomib is not warranted for treating NSCLC patients in clinical practice; therefore further studies are needed to investigate the feasibility and validity in NSCLC patients.
TAK-243 (formerly known as MLN7243), as the primary mammalian E1 enzyme, is the first-in-class inhibitor of the UAE. TAK-243 treatment led to the depletion of cellular ubiquitin conjugates, resulting in disruption of signaling events in primary human xenograft [12]. Due to its specificity and potency, TAK-243 provides a new opportunity for UAE inhibition in cancer treatment. In addition, CC0651 is a small molecule inhibitor that selectively inhibits the E2 ubiquitin conjugating enzyme hCdc34 [170], but there is still a lack of preclinical or clinical information on CC0651 in lung cancer.
Likewise, DUB inhibitors targeting UPS have also become very attractive anticancer drugs and many of them have been investigated in preclinical studies. Pimozide and GW7647 are two potent and highly selective reversible inhibitors of the enzymatic activity of the USP1/UAF1 complex due to its involvement in translation synthesis and DNA damage response in NSCLC [22, 171, 172]. b-AP15 (also known as VLX1500) inhibited the activity of the deubiquitinases, ubiquitin C-terminal hydrolase 5 (UCHL5) and USP14, inducing tumor cells apoptosis and inhibiting tumor progression [173]. However, there still lacks information about drug efficacy and side effects to support the use of these inhibitors in clinical practice. Therefore, it is still too early to predict the therapeutic potential of DUBs in NSCLC and further ground-breaking developments might be obtained in the arenas of DUB biology and drug discovery in the future.
Despite these inhibitors showing promising prospects for clinical application, there are still a number of potential risks and problems to be solved. The ubiquitin system as an enormous biological regulator for thousands of genes plays a critical role in control of cellular signaling networks which affect a variety of phenotypes and biological process of tumor cells. Thus, in the development of new anti-cancer drugs in association with the ubiquitin system, the regulatory mechanisms of tumor related agents and the impacts on cell signaling still need to be described in-depth to effectively utilize the chemical inhibitors for therapy in cancer patients.
Acknowledgements
We acknowledge Dr. Jianxiu Yu at Shanghai Jiao Tong University School of Medicine for the first review of this manuscript and appreciate his kind suggestions on the whole work. We apologize to those authors whose primary work and outstanding contributions to this field that we did not reference directly due to space constraints.
Abbreviations
- 4E-BP1
4E binding protein 1
- APC/C
Anaphase-promoting complex/cyclosome
- CHIP
Carboxy terminus of Hsc70 interacting protein
- DAXX
Death domain-associated protein
- DUBs
Deubiquitinating enzymes
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- GPCRs
G protein-coupled receptors
- HECT
Homologous to E6-AP COOH terminus
- IGF-1
Insulin-like growth factor-1
- ILVs
Intraluminal vesicles
- MJD
Machado-Joseph disease proteases
- mTOR
Mammalian target of rapamycin
- MVBs
Multivesicular bodies
- NSCLC
Non-small-cell lung cancer
- OTU
Ovarian tumor-like proteases
- PI3K
Phosphoinositide-3-kinase
- PKB
Protein kinase B
- PKC
Protein kinase C
- PTMs
Post-translational modifications
- RBR
RING-between-RING
- RCE1
Ras-converting enzyme 1
- RING
Really Interesting New Gene
- RTKs
Receptor tyrosine kinases
- shh
Sonic hedgehog homolog
- Smurf2
Smad ubiquitination regulatory factor 2
- TNF
Tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- TSC
Tuberous sclerosis complex
- UAE
Ubiquitin activating enzyme
- UAF1
USP1-associated factor 1
- UCH
Ubiquitin carboxyl-terminal hydrolases
- UCHL5
Ubiquitin C-terminal hydrolase 5
- UPS
Ubiquitin-proteasome system
Authors’ contributions
MX and HY provided direction and guidance throughout the preparation of this manuscript. QF, QW, RC and HY conducted the literature review and drafted the manuscript. MX and HY reviewed the manuscript and made significant revisions of the drafts. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China to M.X. (81672708), grants from the National Natural Science Foundation of China to H.Y. (81802563), Fundamental research program funding of Ninth People’s Hospital affiliated to Shanghai Jiao Tong university School of Medicine to H.Y. (JYZZ008G) and Q.W. (JYZZ045).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiang Fan and Qian Wang contributed equally to this work.
Contributor Information
Haihua Yuan, Email: ayuan790415@shsmu.edu.cn.
Ming Xu, Email: mingxu.msu@gmail.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ, Dobelbower M, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J National Compr Cancer Netw. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050. [DOI] [PubMed] [Google Scholar]
- 4.Perlikos F, Harrington KJ, Syrigos KN. Key molecular mechanisms in lung cancer invasion and metastasis: a comprehensive review. Crit Rev Oncol Hematol. 2013;87(1):1–11. doi: 10.1016/j.critrevonc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17(11):637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 6.Weathington NM, Mallampalli RK. Emerging therapies targeting the ubiquitin proteasome system in cancer. J Clin Invest. 2014;124(1):6–12. doi: 10.1172/JCI71602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Geng Y, Luo J, Shen W, Zhu W, Meng C, Li M, Zhou X, Zhang S, Cao J. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci Rep. 2015;5:9476. doi: 10.1038/srep09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson KD. The discovery of ubiquitin-dependent proteolysis. Proc Natl Acad Sci U S A. 2005;102(43):15280–15282. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6(8):599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 11.Hong SY, Kao YR, Lee TC, Wu CW. Upregulation of E3 ubiquitin ligase CBLC enhances EGFR dysregulation and signaling in lung adenocarcinoma. Cancer Res. 2018. [DOI] [PubMed]
- 12.Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, Huck J, Shi J, Gavin J, Brownell J, et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat Med. 2018;24(2):186–193. doi: 10.1038/nm.4474. [DOI] [PubMed] [Google Scholar]
- 13.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18(2):69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weathington NM, Mallampalli RK. New insights on the function of SCF ubiquitin E3 ligases in the lung. Cell Signal. 2013;25(9):1792–1798. doi: 10.1016/j.cellsig.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 16.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell. 2017;171(4):918–933. doi: 10.1016/j.cell.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heger K, Wickliffe KE, Ndoja A, Zhang J, Murthy A, Dugger DL, Maltzman A, de Sousa EMF, Hung J, Zeng Y, et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 2018;559(7712):120–124. doi: 10.1038/s41586-018-0256-2. [DOI] [PubMed] [Google Scholar]
- 20.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, Nakaya M, Zhou X, Cheng X, Yang P, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15(6):562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeman MK, Lin JR, Freire R, Cimprich KA. DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis. J Cell Biol. 2014;206(2):183–197. doi: 10.1083/jcb.201311063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T, Wu J, Fu M, Wu ZH. USP10 inhibits genotoxic NF-kappaB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32(24):3206–3219. doi: 10.1038/emboj.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Han C, Xie B, Wu Y, Liu S, Chen K, Xia M, Zhang Y, Song L, Li Z, et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat Immunol. 2014;15(7):612–622. doi: 10.1038/ni.2898. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3(4):255–261. [PubMed] [Google Scholar]
- 26.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65(15):2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, Wei W, Sun Y. Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases. Cell Res. 2013;23(5):599–619. doi: 10.1038/cr.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang WL, Wu CY, Wu J, Lin HK. Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 2010;9(3):487–497. doi: 10.4161/cc.9.3.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 31.Voutsadakis IA. Ubiquitin- and ubiquitin-like proteins-conjugating enzymes (E2s) in breast cancer. Mol Biol Rep. 2013;40(2):2019–2034. doi: 10.1007/s11033-012-2261-0. [DOI] [PubMed] [Google Scholar]
- 32.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10(1):29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu M, Zhu C, Zhao X, Chen C, Zhang H, Yuan H, Deng R, Dou J, Wang Y, Huang J, et al. Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget. 2015;6(2):979–994. doi: 10.18632/oncotarget.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You J, Pickart CM. A HECT domain E3 enzyme assembles novel polyubiquitin chains. J Biol Chem. 2001;276(23):19871–19878. doi: 10.1074/jbc.M100034200. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Kao WH, Howley PM. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272(21):13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 37.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47(6):933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. 2012;770:27–37. doi: 10.1007/978-1-4614-5398-7_3. [DOI] [PubMed] [Google Scholar]
- 39.Lee SJ, Choi JY, Sung YM, Park H, Rhim H, Kang S. E3 ligase activity of RING finger proteins that interact with Hip-2, a human ubiquitin-conjugating enzyme. FEBS Lett. 2001;503(1):61–64. doi: 10.1016/S0014-5793(01)02689-8. [DOI] [PubMed] [Google Scholar]
- 40.Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 41.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11(3):141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 43.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1-3):189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. J Biochem. 2003;134(1):9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 45.Alwan HA, van Leeuwen JE. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J Biol Chem. 2007;282(3):1658–1669. doi: 10.1074/jbc.M604711200. [DOI] [PubMed] [Google Scholar]
- 46.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490(3):142–152. doi: 10.1016/S0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 47.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 48.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21(6):737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21(5):679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 50.Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, Cantley LC. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal. 2011;4(163):ra13. doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K, Rumschlag M, Shankar S, Whitehead C, Schipper M, et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia. 2014;16(2):115–128. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cantwell-Dorris ER, O'Leary JJ, Sheils OM. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther. 2011;10(3):385–394. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- 53.Hong SW, Jin DH, Shin JS, Moon JH, Na YS, Jung KA, Kim SM, Kim JC, Kim KP, Hong YS, et al. Ring finger protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) J Biol Chem. 2012;287(28):24017–24025. doi: 10.1074/jbc.M111.319822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knight T, Irving JA. Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and Its Therapeutic Targeting. Front Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U, Ghosh MK. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J Biol Chem. 2012;287(19):15996–16006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23(11):1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 57.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Shi Y, Wang J, Huang G, Jiang X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem J. 2008;414(2):221–229. doi: 10.1042/BJ20080674. [DOI] [PubMed] [Google Scholar]
- 59.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149(5):1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y, Yang G, Sheng Y, Xiao L, Dong X, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73(23):6938–6950. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarris EG, Saif MW, Syrigos KN. The Biological Role of PI3K Pathway in Lung Cancer. Pharmaceuticals (Basel) 2012;5(11):1236–1264. doi: 10.3390/ph5111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51(3):283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Sun SY, Owonikoko TK, Sica GL, Curran WJ, Khuri FR, Deng X. Rapamycin induces Bad phosphorylation in association with its resistance to human lung cancer cells. Mol Cancer Ther. 2012;11(1):45–56. doi: 10.1158/1535-7163.MCT-11-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okazaki H, Matsunaga N, Fujioka T, Okazaki F, Akagawa Y, Tsurudome Y, Ono M, Kuwano M, Koyanagi S, Ohdo S. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74(2):543–551. doi: 10.1158/0008-5472.CAN-12-3241. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12(1):9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 67.Tamborini E, Virdis E, Negri T, Orsenigo M, Brich S, Conca E, Gronchi A, Stacchiotti S, Manenti G, Casali PG, et al. Analysis of receptor tyrosine kinases (RTKs) and downstream pathways in chordomas. Neuro Oncol. 2010;12(8):776–789. doi: 10.1093/neuonc/noq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leung CH, Lam W, Zhuang WJ, Wong NS, Yang MS, Fong WF. PKCdelta-dependent deubiquitination and stabilization of Gadd45 in A431 cells overexposed to EGF. Biochem Biophys Res Commun. 2001;285(2):283–288. doi: 10.1006/bbrc.2001.5164. [DOI] [PubMed] [Google Scholar]
- 69.Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X, Chen T. E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKC{zeta} J Exp Med. 2011;208(10):2099–2112. doi: 10.1084/jem.20102667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, Franklin RA, McCubrey JA. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17(7):1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- 71.Vicent S, Lopez-Picazo JM, Toledo G, Lozano MD, Torre W, Garcia-Corchon C, Quero C, Soria JC, Martin-Algarra S, Manzano RG, et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 2004;90(5):1047–1052. doi: 10.1038/sj.bjc.6601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J, Wei J, Mialki R, Zou C, Mallampalli RK, Zhao Y. Extracellular signal-regulated kinase (ERK) regulates cortactin ubiquitination and degradation in lung epithelial cells. J Biol Chem. 2012;287(23):19105–19114. doi: 10.1074/jbc.M112.339507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jura N, Bar-Sagi D. Mapping cellular routes of Ras: a ubiquitin trail. Cell Cycle. 2006;5(23):2744–2747. doi: 10.4161/cc.5.23.3532. [DOI] [PubMed] [Google Scholar]
- 74.Miller CR, Oliver KE, Farley JH. MEK1/2 inhibitors in the treatment of gynecologic malignancies. Gynecol Oncol. 2014;133(1):128–137. doi: 10.1016/j.ygyno.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 75.Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr Biol. 2010;20(15):1372–1377. doi: 10.1016/j.cub.2010.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng T, Wang Q, Fu J, Lin Q, Bi J, Ding W, Qiao Y, Zhang S, Zhao W, Lin H, et al. Impeded Nedd4-1-mediated Ras degradation underlies Ras-driven tumorigenesis. Cell Rep. 2014;7(3):871–882. doi: 10.1016/j.celrep.2014.03.045. [DOI] [PubMed] [Google Scholar]
- 77.Kim SE, Yoon JY, Jeong WJ, Jeon SH, Park Y, Yoon JB, Park YN, Kim H, Choi KY. H-Ras is degraded by Wnt/beta-catenin signaling via beta-TrCP-mediated polyubiquitylation. J Cell Sci. 2009;122(Pt 6):842–848. doi: 10.1242/jcs.040493. [DOI] [PubMed] [Google Scholar]
- 78.Arozarena I, Calvo F, Crespo P. Ras, an actor on many stages: posttranslational modifications, localization, and site-specified events. Genes Cancer. 2011;2(3):182–194. doi: 10.1177/1947601911409213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker R, Lewis SM, Sasaki AT, Wilkerson EM, Locasale JW, Cantley LC, Kuhlman B, Dohlman HG, Campbell SL. Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat Struct Mol Biol. 2013;20(1):46–52. doi: 10.1038/nsmb.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baker R, Wilkerson EM, Sumita K, Isom DG, Sasaki AT, Dohlman HG, Campbell SL. Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination. J Biol Chem. 2013;288(52):36856–36862. doi: 10.1074/jbc.C113.525691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burrows JF, Kelvin AA, McFarlane C, Burden RE, McGrattan MJ, De la Vega M, Govender U, Quinn DJ, Dib K, Gadina M, et al. USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J Biol Chem. 2009;284(14):9587–9595. doi: 10.1074/jbc.M807216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaworski J, Govender U, McFarlane C, de la Vega M, Greene MK, Rawlings ND, Johnston JA, Scott CJ, Burrows JF. A novel RCE1 isoform is required for H-Ras plasma membrane localization and is regulated by USP17. Biocheml J. 2014;457(2):289–300. doi: 10.1042/BJ20131213. [DOI] [PubMed] [Google Scholar]
- 83.de la Vega M, Burrows JF, McFarlane C, Govender U, Scott CJ, Johnston JA. The deubiquitinating enzyme USP17 blocks N-Ras membrane trafficking and activation but leaves K-Ras unaffected. J Biol Chem. 2010;285(16):12028–12036. doi: 10.1074/jbc.M109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang S, Yuan J, Zheng R. Suppression of Ubiquitin-Specific Peptidase 17 (USP17) Inhibits Tumorigenesis and Invasion in Non-Small Cell Lung Cancer Cells. Oncol Res. 2016;24(4):263–269. doi: 10.3727/096504016X14666990347392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McFarlane C, McFarlane S, Paul I, Arthur K, Scheaff M, Kerr K, Stevenson M, Fennell DA, Johnston JA. The deubiquitinating enzyme USP17 is associated with non-small cell lung cancer (NSCLC) recurrence and metastasis. Oncotarget. 2013;4(10):1836–1843. doi: 10.18632/oncotarget.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCann AP, Smyth P, Cogo F, McDaid WJ, Jiang L, Lin J, Evergren E, Burden RE, Van Schaeybroeck S, Scott CJ, et al. USP17 is required for trafficking and oncogenic signaling of mutant EGFR in NSCLC cells. Cell Commun Signal. 2018;16(1):77. doi: 10.1186/s12964-018-0291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rebocho AP, Marais R. ARAF acts as a scaffold to stabilize BRAF:CRAF heterodimers. Oncogene. 2013;32(26):3207–3212. doi: 10.1038/onc.2012.330. [DOI] [PubMed] [Google Scholar]
- 88.An L, Jia W, Yu Y, Zou N, Liang L, Zhao Y, Fan Y, Cheng J, Shi Z, Xu G, et al. Lys63-linked polyubiquitination of BRAF at lysine 578 is required for BRAF-mediated signaling. Sci Rep. 2013;3:2344. doi: 10.1038/srep02344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Q, Gao G, Zhang T, Yao K, Chen H, Park MH, Yamamoto H, Wang K, Ma W, Malakhova M, et al. TRAF1 Is Critical for Regulating the BRAF/MEK/ERK Pathway in Non-Small Cell Lung Carcinogenesis. Cancer Res. 2018;78(14):3982–3994. doi: 10.1158/0008-5472.CAN-18-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351(Pt 2):289–305. doi: 10.1042/bj3510289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noble C, Mercer K, Hussain J, Carragher L, Giblett S, Hayward R, Patterson C, Marais R, Pritchard CA. CRAF autophosphorylation of serine 621 is required to prevent its proteasome-mediated degradation. Mol Cell. 2008;31(6):862–872. doi: 10.1016/j.molcel.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaumot M, Hancock JF. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene. 2001;20(30):3949–3958. doi: 10.1038/sj.onc.1204526. [DOI] [PubMed] [Google Scholar]
- 93.Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278(37):35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 94.Dowell JE, Minna JD. EGFR mutations and molecularly targeted therapy: a new era in the treatment of lung cancer. Nat Clin Pract Oncol. 2006;3(4):170–171. doi: 10.1038/ncponc0476. [DOI] [PubMed] [Google Scholar]
- 95.Hung JJ, Jeng WJ, Hsu WH, Liu JS, Wu YC. EGFR mutations in non-small-cell lung cancer. Lancet Oncol. 2010;11(5):412–413. doi: 10.1016/S1470-2045(10)70065-6. [DOI] [PubMed] [Google Scholar]
- 96.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 97.da Cunha SG, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 98.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10(9):609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eden ER, Huang F, Sorkin A, Futter CE. The role of EGF receptor ubiquitination in regulating its intracellular traffic. Traffic. 2012;13(2):329–337. doi: 10.1111/j.1600-0854.2011.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12(23):3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci U S A. 2007;104(43):16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166(4):487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, Pyne S, Agostini M, Freeman MR, Loda M. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. 2013;32(13):1660–1669. doi: 10.1038/onc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Niendorf S, Oksche A, Kisser A, Lohler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch KP. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27(13):5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16(11):5163–5174. doi: 10.1091/mbc.e05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281(8):5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 108.Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben-Basat Y, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26(49):6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 109.Chung C, Yoo G, Kim T, Lee D, Lee CS, Cha HR, Park YH, Moon JY, Jung SS, Kim JO, et al. The E3 ubiquitin ligase CHIP selectively regulates mutant epidermal growth factor receptor by ubiquitination and degradation. Biochem Biophys Res Commun. 2016;479(2):152–158. doi: 10.1016/j.bbrc.2016.07.111. [DOI] [PubMed] [Google Scholar]
- 110.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120(2):237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 111.Suizu F, Hiramuki Y, Okumura F, Matsuda M, Okumura AJ, Hirata N, Narita M, Kohno T, Yokota J, Bohgaki M, et al. The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt. Dev Cell. 2009;17(6):800–810. doi: 10.1016/j.devcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 112.Janku F. Bringing target-matched PI3King from the bench to the clinic. Cell Cycle. 2013;12(12):1817–1818. doi: 10.4161/cc.25118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ojeda L, Gao J, Hooten KG, Wang E, Thonhoff JR, Dunn TJ, Gao T, Wu P. Critical role of PI3K/Akt/GSK3beta in motoneuron specification from human neural stem cells in response to FGF2 and EGF. PLoS One. 2011;6(8):e23414. doi: 10.1371/journal.pone.0023414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67(10):1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 115.Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6(1):21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 116.Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol. 2011;93(2):182–203. doi: 10.1016/j.pneurobio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 117.Markowska A, Pawalowska M, Lubin J, Markowska J. Signalling pathways in endometrial cancer. Contemp Oncol (Pozn) 2014;18(3):143–148. doi: 10.5114/wo.2014.43154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akinleye A, Avvaru P, Furqan M, Song Y, Liu D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J Hematol Oncol. 2013;6(1):88. doi: 10.1186/1756-8722-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fang D, Wang HY, Fang N, Altman Y, Elly C, Liu YC. Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J Biol Chem. 2001;276(7):4872–4878. doi: 10.1074/jbc.M008901200. [DOI] [PubMed] [Google Scholar]
- 121.Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274(13):8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 122.Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, Washburn MP, Pagano M. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15(5):472–480. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mayadagli A, Karabulut Gul S, Bilici A, Fatih Oruc A, Kocak M, Ozkan A, Aksu A, Ozdemir Barisik N, Gumus M. Prognostic significance of protein kinase B/Akt pathway in patients with non-small cell lung cancer. J BUON. 2014;19(1):157–163. [PubMed] [Google Scholar]
- 124.Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW. Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene. 2004;23(16):2934–2949. doi: 10.1038/sj.onc.1207515. [DOI] [PubMed] [Google Scholar]
- 125.Fan CD, Lum MA, Xu C, Black JD, Wang X. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J Biol Chem. 2013;288(3):1674–1684. doi: 10.1074/jbc.M112.416339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chan CH, Jo U, Kohrman A, Rezaeian AH, Chou PC, Logothetis C, Lin HK. Posttranslational regulation of Akt in human cancer. Cell Biosci. 2014;4(1):59. doi: 10.1186/2045-3701-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jang HD, Hwang HZ, Kim HS, Lee SY. C-Cbl negatively regulates TRAF6-mediated NF-kappaB activation by promoting K48-linked polyubiquitination of TRAF6. Cell Mol Biol Lett. 2019;24:29. doi: 10.1186/s11658-019-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Solit DB, Basso AD, Olshen AB, Scher HI, Rosen N. Inhibition of heat shock protein 90 function down-regulates Akt kinase and sensitizes tumors to Taxol. Cancer Res. 2003;63(9):2139–2144. [PubMed] [Google Scholar]
- 129.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN, Yang Q. Negative Regulation of AKT Activation by BRCA1. Cancer Res. 2008;68(24):10040–10044. doi: 10.1158/0008-5472.CAN-08-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bae S, Kim SY, Jung JH, Yoon Y, Cha HJ, Lee H, Kim K, Kim J, An IS, Um HD, et al. Akt is negatively regulated by the MULAN E3 ligase. Cell Res. 2012;22(5):873–885. doi: 10.1038/cr.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Massoumi R. CYLD: a deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7(2):285–297. doi: 10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]
- 132.Ahmed N, Zeng M, Sinha I, Polin L, Wei WZ, Rathinam C, Flavell R, Massoumi R, Venuprasad K. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 2011;12(12):1176–1183. doi: 10.1038/ni.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Deng LL, Shao YX, Lv HF, Deng HB, Lv FZ. Over-expressing CYLD augments antitumor activity of TRAIL by inhibiting the NF-kappaB survival signaling in lung cancer cells. Neoplasma. 2012;59(1):18–29. doi: 10.4149/neo_2012_003. [DOI] [PubMed] [Google Scholar]
- 134.Pfeifer M, Grau M, Lenze D, Wenzel SS, Wolf A, Wollert-Wulf B, Dietze K, Nogai H, Storek B, Madle H, et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(30):12420–12425. doi: 10.1073/pnas.1305656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Amodio N, Scrima M, Palaia L, Salman AN, Quintiero A, Franco R, Botti G, Pirozzi P, Rocco G, De Rosa N, et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am J Pathol. 2010;177(5):2622–2634. doi: 10.2353/ajpath.2010.091075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95(23):13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Makker A, Goel MM, Mahdi AA. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: an update. J Mol Endocrinol. 2014;53(3):R103–R118. doi: 10.1530/JME-14-0220. [DOI] [PubMed] [Google Scholar]
- 139.Carnero A, Paramio JM. The PTEN/PI3K/AKT Pathway in vivo. Cancer Mouse Models Front Oncol. 2014;4:252. doi: 10.3389/fonc.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/S0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 141.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1999;96(11):6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2) Curr Biol. 2001;11(4):263–267. doi: 10.1016/S0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- 143.Sacco JJ, Yau TY, Darling S, Patel V, Liu H, Urbe S, Clague MJ, Coulson JM. The deubiquitylase Ataxin-3 restricts PTEN transcription in lung cancer cells. Oncogene. 2014;33(33):4265–4272. doi: 10.1038/onc.2013.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128(1):129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, Chen J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13(6):728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Van Themsche C, Leblanc V, Parent S, Asselin E. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J Biol Chem. 2009;284(31):20462–20466. doi: 10.1074/jbc.C109.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, Stambolic V, Rotin D. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105(25):8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 149.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung MC, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15(12):1486–1494. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Davis NM, Sokolosky M, Stadelman K, Abrams SL, Libra M, Candido S, Nicoletti F, Polesel J, Maestro R, D'Assoro A, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget. 2014;5(13):4603–4650. doi: 10.18632/oncotarget.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ellis L, Ku SY, Ramakrishnan S, Lasorsa E, Azabdaftari G, Godoy A, Pili R. Combinatorial antitumor effect of HDAC and the PI3K-Akt-mTOR pathway inhibition in a Pten defecient model of prostate cancer. Oncotarget. 2013;4(12):2225–2236. doi: 10.18632/oncotarget.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Showkat M, Beigh MA, Andrabi KI. mTOR Signaling in Protein Translation Regulation: Implications in Cancer Genesis and Therapeutic Interventions. Mol Biol Int. 2014;2014:686984. doi: 10.1155/2014/686984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286(5445):1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 156.Qi H, Fan L. PI3K/Akt/mTOR signaling pathway and non-small cell lung cancer. Chin J Lung Cancer. 2010;13(12):1149–1154. doi: 10.3779/j.issn.1009-3419.2010.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ivanov SS, Roy CR. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol. 2013;14(12):1219–1228. doi: 10.1038/ni.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koo J, Wu X, Mao Z, Khuri FR, Sun SY. Rictor Undergoes Glycogen Synthase Kinase 3 (GSK3)-dependent, FBXW7-mediated Ubiquitination and Proteasomal Degradation. J Biol Chem. 2015;290(22):14120–14129. doi: 10.1074/jbc.M114.633057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Park D, Lee MN, Jeong H, Koh A, Yang YR, Suh PG, Ryu SH. Parkin ubiquitinates mTOR to regulate mTORC1 activity under mitochondrial stress. Cell Signal. 2014;26(10):2122–2130. doi: 10.1016/j.cellsig.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 161.Hussain S, Feldman AL, Das C, Ziesmer SC, Ansell SM, Galardy PJ. Ubiquitin hydrolase UCH-L1 destabilizes mTOR complex 1 by antagonizing DDB1-CUL4-mediated ubiquitination of raptor. Mol Cell Biol. 2013;33(6):1188–1197. doi: 10.1128/MCB.01389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bunn PA., Jr The potential role of proteasome inhibitors in the treatment of lung cancer. Clin Cancer Res. 2004;10(12 Pt 2):4263s–4265s. doi: 10.1158/1078-0432.CCR-040011. [DOI] [PubMed] [Google Scholar]
- 163.Scagliotti G. Proteasome inhibitors in lung cancer. Crit Rev Oncol Hematol. 2006;58(3):177–189. doi: 10.1016/j.critrevonc.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 164.Yuan BZ, Chapman J, Reynolds SH. Proteasome inhibitors induce apoptosis in human lung cancer cells through a positive feedback mechanism and the subsequent Mcl-1 protein cleavage. Oncogene. 2009;28(43):3775–3786. doi: 10.1038/onc.2009.240. [DOI] [PubMed] [Google Scholar]
- 165.Denlinger CE, Rundall BK, Keller MD, Jones DR. Proteasome inhibition sensitizes non-small-cell lung cancer to gemcitabine-induced apoptosis. Ann Thorac Surg. 2004;78(4):1207–1214. doi: 10.1016/j.athoracsur.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 166.Piperdi B, Walsh WV, Bradley K, Zhou Z, Bathini V, Hanrahan-Boshes M, Hutchinson L, Perez-Soler R. Phase-I/II study of bortezomib in combination with carboplatin and bevacizumab as first-line therapy in patients with advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7(6):1032–1040. doi: 10.1097/JTO.0b013e31824de2fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhao Y, Foster NR, Meyers JP, Thomas SP, Northfelt DW, Rowland KM, Jr, Mattar BI, Johnson DB, Molina JR, Mandrekar SJ, et al. A phase I/II study of bortezomib in combination with paclitaxel, carboplatin, and concurrent thoracic radiation therapy for non-small-cell lung cancer: North Central Cancer Treatment Group (NCCTG)-N0321. J Thorac Oncol. 2015;10(1):172–180. doi: 10.1097/JTO.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Davies AM, Chansky K, Lara PN, Jr, Gumerlock PH, Crowley J, Albain KS, Vogel SJ, Gandara DR, Southwest Oncology G. Bortezomib plus gemcitabine/carboplatin as first-line treatment of advanced non-small cell lung cancer: a phase II Southwest Oncology Group Study (S0339) J Thorac Oncol. 2009;4(1):87–92. doi: 10.1097/JTO.0b013e3181915052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lynch TJ, Fenton D, Hirsh V, Bodkin D, Middleman EL, Chiappori A, Halmos B, Favis R, Liu H, Trepicchio WL, et al. A randomized phase 2 study of erlotinib alone and in combination with bortezomib in previously treated advanced non-small cell lung cancer. J Thorac Oncol. 2009;4(8):1002–1009. doi: 10.1097/JTO.0b013e3181aba89f. [DOI] [PubMed] [Google Scholar]
- 170.Huang H, Ceccarelli DF, Orlicky S, St-Cyr DJ, Ziemba A, Garg P, Plamondon S, Auer M, Sidhu S, Marinier A, et al. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat Chem Biol. 2014;10(2):156–163. doi: 10.1038/nchembio.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011;18(11):1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Garcia-Santisteban I, Peters GJ, Giovannetti E, Rodriguez JA. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol Cancer. 2013;12:91. doi: 10.1186/1476-4598-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17(12):1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.