Abstract
The gut microbiome is a key factor in determining inter-individual variability in response to diet. Thus, far, research in this area has focused on metabolic health outcomes such as obesity and type 2 diabetes. However, understanding the role of the gut microbiome in determining response to diet may also lead to improved personalization of sports nutrition for athletic performance. The gut microbiome has been shown to modify the effect of both diet and exercise, making it relevant to the athlete's pursuit of optimal performance. This area of research can benefit from recent developments in the general field of personalized nutrition and has the potential to expand our knowledge of the nexus between the gut microbiome, lifestyle, and individual physiology.
Keywords: gut microbiome, exercise, personalized nutrition, sports nutrition, performance, metabolism, athletes, optimization
Introduction
The gut microbiome has been implicated in the modulation of human health and metabolism (1, 2). This microbial “organ” has been linked to nutrition-related chronic diseases such as obesity and diabetes (3–6) and has also been shown to influence systemic functions including immunity (7, 8) and brain function (9, 10). The gut microbiome may influence health via mechanisms such as the production of metabolites (2, 11) [e.g., short-chain fatty acids (SCFAs)] that can influence a wide array of host systems and metabolic pathways (12, 13).
However, the gut microbiome is not a fixed trait, but instead responds to environmental stimuli and is a malleable part of the human supraorganism (14) (Figure 1). Much of microbiome research has focused on the effect of lifestyle factors, such as diet (15–17) and exercise (18, 19), on the gut microbiota. Variability in the composition and function of the gut microbiome (20, 21) has also fueled research on the relationship between features of the gut microbiota, such as diversity or the presence, absence, or amount of certain taxa, and host health. Precision nutrition studies are now investigating how to predict individual differences in glycemic response, triglycerides, cholesterol levels, and other indicators of health as a way to personalize nutrition recommendations and prevent diet-related chronic diseases such as obesity and type 2 diabetes. Our previous two-part review (22, 23) explored the effect of the gut microbiome on inter-individual variability in response to diet and how this may contribute to metabolic health.
Figure 1.
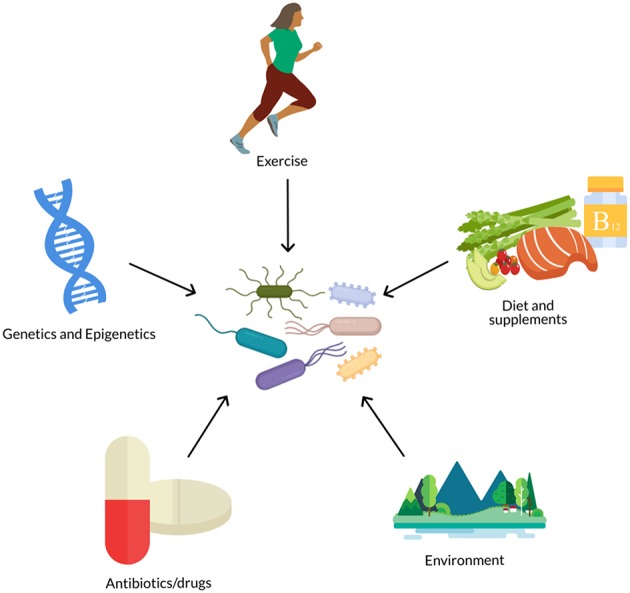
The gut microbiome is influenced by numerous biological and lifestyle factors such as diet, genetics, antibiotics, exercise, and environment (e.g., pollutants, urban vs. rural, etc.).
Alternatively, we may consider the potential effect of the gut microbiome on measures of athletic performance. Successful performance in training, such as a time trial or rep max load, and general metabolic health are two distinct aspects of metabolic response that are not necessarily directly coupled (24). While nutrition is an important part of general health and well-being, it is also an important tool in an athlete's arsenal to optimize performance (25).
Variability in the physiological response to training and nutrition has been attributed to factors such as age, sex, training history, initial training status, psychological factors, and the mode, duration, intensity, and frequency of training (26). Genetics has also become a large topic of research in the area of variability in response to exercise training and potentially ergogenic dietary components (27–38). It is possible that variability in the gut microbiome may also influence gains in performance in response to training and nutrition. Despite the growing interest in the gut microbiome and personalized nutrition, very few studies have combined these fields with that of athletic performance. This is surprising as athletes are extremely motivated to capitalize on any advantage, however small, that could increase their performance. This review focuses on several topics related to the question of whether the gut microbiome may be used to predict performance response to dietary and/or training interventions. This includes topics such as (1) the effect of exercise on the gut microbiome, (2) the effect of dietary components or patterns relevant to athletic nutrition on the gut microbiome, and (3) the effect of the gut microbiome on performance response to diet and exercise. Each of these related topics will be discussed, as will gaps in the research and future directions.
Methods
Numerous reviews have been published highlighting the effect of exercise on the gut microbiota (39–56). However, the primary focus of these reviews has been the implications for aspects of host health, such as the immune system and risk of chronic diseases. Only a few have discussed the implications for athletic performance (41, 48, 52). This review aims to provide a more in-depth discussion of the interactive effect between the gut microbiota and diet on athletic performance and highlight the need for further research in this area. A literature search in PubMed and Google Scholar, including combinations of key words “gut microbiota”, “exercise”, “performance,” “variability,” and “effect,” was used to identify relevant studies. References were also obtained from the above review articles.
The earliest study found was published in 2008 by Matsumoto et al. (57) but was followed by a host of studies aiming to identify the effects of exercise on the gut microbiota (19, 58–83). The majority of these studies have investigated the effect of exercise on the gut microbiota in rodents (57–75), though some have studied humans in intervention trials (18, 76–78, 84) and in cross-sectional or observational comparisons of athletes or active individuals and sedentary individuals (19, 79–83, 85, 86).
Results
The Effect of Exercise on the Gut Microbiome
Microbiota Features Affected by Exercise
Table 1 summarizes the studies listed above and their findings of the effect of exercise on the gut microbiota.
Table 1.
Summary of effect of exercise on the gut microbiome.
| References | Subjects | Type of exercise | Type of study | Diet | Microbiota method | Microbial diversity | Firmicutes (phylum) | Bacteroidetes (phylum) | Lactobacillaceae (family) | Bifidobacteriaceae (family) | Other microbiota factors |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rodent studies | |||||||||||
| Allen et al. (59) |
Mice (C57BL/6J, 6 wk, male) | Voluntary wheel running (VWR) vs. forced treadmill running (FTR) for 6 wk | Intervention | Commercial diet | Composition (16S) | ↓ in VWR (Chao1) ↔ (Shannon) |
↓ Turicibacter in VWR | ||||
| Lambert et al. (62) |
Mice (diabetic db/db C57BL/KsJ-leprdb/leprdb and normal db/+, 6 wk, male) | Treadmill | Intervention | Chow diet | Composition (qPCR) | ↑ Firmicutes, Clostridium | ↓ Bacteroides, Prevotella | ↑ Lactobacillus (not after adjustment for body weight and blood glucose) | ↑ Bifidobacterium in exercised normal vs. sedentary normal ↓ Bifidobacterium in exercised diabetic |
↓ Enterobacteriaceae in exercised diabetic vs. sedentary diabetic | |
| Lamoureux et al. (63) |
Mice (C57BL/6, 6–10 wks, 11 male and 31 female) | Voluntary exercise (VE) vs. moderate forced exercise (treadmill) (FE) for 8 wk | Intervention | Normal diet | Composition (16S) | ↔α-diversity (species richness) or β-diversity (weighted and unweighted UniFrac, Bray-Curtis) | Random forest predicted voluntary exercise with 97% accuracy using Bacteroides, Lactobacillus, Rikenellaceae, Lachnospiraceae; predicted forced exercise with 86% accuracy using Bacteroides, Clostridiales, and Lactobacillales | ||||
| Liu et al. (75) |
Mice (C57BL/6J, 4 wk, male; myocardial infarction (MI), sham, or no-surgery) | Treadmill for 4 wk | Intervention | None | Composition (16S) | ↑ α-diversity (Shannon, PD_whole_tree) | ↑ Butyricimonas, Prevotella, Akkermansia in exercise/non-surgery mice ↑ Parasutterella in control/non-surgery mice ↑ Erysipelotrichaceae, Sphingobacteriales, Akkermansia in exercise/sham mice ↑ Corynebacterium, Staphylococcus, Enterobacteriaceae in control/sham mice ↑ Phenylobacterium and Roseateles in exercise/MI mice |
||||
| Brandt et al. (73) |
Mice (C57BL/6N,8–10 wk, male, loxP insertions in Ppargc1a gene) | Voluntary wheel running (VWR) for 16 wk | Intervention | Standard rodent chow (CON) vs. High-fat diet (HFD) vs. HFD + resveratrol | Composition (16S) | ↓ α-diversity in HFD mice vs. CON ↑ β-diversity in HFD mice vs. CON |
↑ Bacteroidetes in HFD plus exercise vs. HFD | ↓ Actinobacteria in HFD plus exercise vs. HFD | ↓ Erysipelotrichaceae, Verrumicrobioa in HFD plus exercise vs. HFD ↑ Alistipes in HFD plus exercise vs. HFD |
||
| Campbell et al. (67) |
Mice (C57BL/6NT, 8 wk, male) | Voluntary wheel running for 12 wk | Intervention | Normal diet vs. High-fat diet | Composition (TRFLP, 16S) | ↑ Allobaculum, Clostridiales, Faecalibacterium prausnitzii | |||||
| Evans et al. (64) |
Mice (C57BL/6J, 6 wk, male) | Voluntary wheel running for 12 wk | Intervention | Low-fat vs. High-fat diet | Composition (16S, qPCR, TRFLP) | ↑ α-diversity (Shannon) with high-fat diet and exercise | ↓ Firmicutes (16S), Turicibacteraceae, Erysipelotrichaceae (qPCR) ↔Firmicutes (qPCR) ↑ Bacteroidetes/ Firmicutes (qPCR) ↓ Bacteroidetes/ Firmicutes (qPCR) |
↑ Bacteroidetes (16S), Bacteroidetes/ Firmicutes (qPCR) ↔Bacteroidetes (qPCR) ↓ Bacteroidetes/ Firmicutes (qPCR) |
↓ Lactobacillaceae (qPCR) | ↓ Actinobacteria (16S), Bifidobacteriaceae (qPCR) | ↑ butyrate-producing taxa |
| McCabe et al. (74) |
Mice (C57BL/6J, 6 wk, male) | Voluntary wheel running for 14 wk | Intervention | Low-fat vs. High-fat diet | Composition (16S) | ↓ Firmicutes/ Bacteroides in HF-exercise |
↓ Firmicutes/ Bacteroides in HF-exercise |
||||
| Kang et al. (61) |
Mice (C57BL/6J, 8 wk, male) | Motorized wheel running for 16 wk | Intervention | Normal diet vs. High-fat diet | Composition (16S) | ↑ Firmicutes, Lachnospiraceae ↓ Streptococcaceae |
↓ Bacteroidetes | ↓ Tenericutes | |||
| Denou et al. (70) |
Mice (C57BL/6J, 8 wk, male) | High-intensity interval training (HIIT) on treadmill for 6 wk | Intervention | Chow diet vs. High-fat diet | Composition (16S) and function (PICRUSt) | ↑ α-diversity (Shannon) | ↑ Bacteroidetes/ Firmicutes |
↑ Bacteroidetes/ Firmicutes, Bacteroidales |
↑ KEGG-annotated metabolism genes | ||
| Choi et al. (60) |
Mice (C56BL/6NT, 11–13 mo, male) | Voluntary wheel running for 5 wk | Intervention | Polychlorinated biphenyls (PCBs) | Composition (16S) | ↑ abundance | ↑ Firmicutes (mostly Lactobacillales) | ↑ Lactobacillales | ↓ Tenericutes (Erysipelotichaceae) ↑ Proteobacteria (prevented PCB-induced decrease) |
||
| Liu et al. (65) |
Rats (ovariectomized (OVX) high capacity (HCR) and low capacity (LCR) runners, 27 wk, females) | Voluntary wheel running for 11 wk | Intervention | Chow diet | Composition (16S) | ↔α-diversity (Chao1) | ↑ Firmicutes in HCR ↓ Firmicutes in LCR |
↓ Proteobacteria, Cyanobacteria in HCR ↑ Proteobacteria, Cyanobacteria in LCR |
|||
| Mika et al. (50) |
Rats (F344, day 24 vs. day 70, male) | Voluntary wheel running for 6 wk | Intervention | Standard diet | Composition (16S) | ↓ α-diversity (Shannon entropy, species richness) in young rats ↑ β-diversity (unweighted UniFrac) in young rats |
↓ Firmicutes in young rats ↑ Blautia, Anaerostipes in young rats ↑ Turicibacter in adult rats |
↑ Bacteroidetes in young rats | ↑ Euryarchaeota (Methanosphaera) in young rats ↓ Proteobacteria (Desulfovibrio) in young rats ↓Rikenellaceae in young rats ↑ Rikenellaceae in adult rats Bacteroides, Bifidobacterium, Ruminococcus, Rikenellaceae, Parabacteroides, Christensenellaceae, Methanosphaera predict time point in young rats |
||
| Matsumoto et al. (57) |
Rats (Wistar, 7 wk, male) | Voluntary wheel running for 5 wk | Intervention | Casein-sucrose diet | Composition (PCR-TGGE) | Differential clustering between exercise and controls ↑ butyrate-producing taxa |
|||||
| Queipo-Ortuno et al. (58) |
Rats (Sprague Dawley, 5 wk, male) | Voluntary wheel running for 6 d | Intervention | Activity based anorexia (ABA, 1 h food intake w/ exercise), ABA control (sedendary), Exercise (ad lib w/ exercise), Ad lib (ad lib sedentary) | Composition (PCR-DGGE, qPCR) | ↓ α-diversity (band richness) | ↓ Firmicutes in ABA vs. Exercise and Ad lib; in Exercise vs. Ad lib ↑ Clostridium in ABA ↑ B. Coccoides-E. rectale group in Exercise vs. Ad lib ↓ Clostridium, Enterococcus in Exercise vs. Ad lib |
↓ Bacteroidetes in ABA vs. Exercise and Ad lib ↑ Bacteroides, Prevotella in ABA vs. ABA control ↓ Bacteroides, Prevotella in Exercise vs. Ad lib |
↑ Lactobacillus in Exercise vs. Ad lib | ↓ Actinobacteria in ABA ↑ Actinobacteria in Exercise vs. Ad lib ↑ Bifidobacterium in Exercise |
↑ Proteobacteria in ABA vs. Exercise and Ad lib |
| Welly et al. (69) |
Rats (obesity prone OP-CD, 4 wk, male) | Voluntary wheel running | Intervention | High-fat diet (HFD; groups: sedentary, w/ exercise, weight matched to exercise) | Composition (qPCR) | ↔α-diversity (species richness) | ↑ Streptococcaceae in Exercise ↔Firmicutes/ Bacteroidetes ratio |
↓ S24–7 in Exercise ↓ Bacteroidetes in Exercise and weight-matched (trending, not significant) ↔Firmicutes/ Bacteroidetes ratio |
↓ Rikenellaceae in Exercise | ||
| Feng et al. (71) |
Rats (high capacity (HCR) and low capacity (LCR) runners, sugery or sham) | Treadmill for 6 wk | Intervention | None | Composition (16S) | ↑ α-diversity (Shannon) in LCR rats ↑ β-diversity in LCR and HCR rats |
↑ Firmicutes in HCR rats | ↓ Bacteroidetes in HCR rats | |||
| Petriz et al. (68) |
Rats (Zucker (obese), Zucker (spotaneously hypertensive), and Wistar (non-obese, control), 20 wk, male/female?) | Forced treadmill running for 4 wk | Intervention | Not reported | Composition (16S) | ↑ α-diversity (Shannon, rarefaction) | ↑ Firmicutes ↓ Streptococcus in non-obese Wistar ↑ Allobaculum in hypertensive |
↓ Bacteroidetes in non-obese Wistar | ↑ Lactobacillus in obese Zucker | ↓ Proteobacteria ↓ Sutterella, Aggregatibacter in hypertensive |
|
| Batacan et al. (72) |
Rats (Wistar, 12 wk, male) | Control (CTL), sedentary (SED), light-intensity trained (LIT), and high-intensity interval trained (HIIT) for 12 wk | Intervention | Standard chow (SC) versis high-fat high-fructose (HF) diet | Composition (16S) | ↔α-diversity between activity groups regardless of diet (Chao1, observed species, Shannon, Simpson, | ↓ Firmicutes in LIT-SC ↑ Clostridiaceae in HF ↑ Lachnospiraceae in HIIT-SC |
↑ Lactobacillus johnsonii increased in LIT-SC | ↑ Actinobacteria in LIT-SC ↓ Bifidobacterium in HF |
↑ Tenericutes, Prevotella excrementihominis, Erysipelotrichaceae in LIT-SC (Erysipelotrichaceae higher in HF-fed rats) ↓ Turicibacteraceae in SC ↑ polysaccharide degraders and SCFA producers with exercise in both SC and HF (effects weaker in HF) |
|
| dominance, richness, equitability, evenness)↑β-diversity (weighted and/or unweighted UniFrac) | |||||||||||
| Human intervention studies | |||||||||||
| Allen et al. (18) |
Humans (32 previously sedentary subjects, 18 lean 14 obese) | Endurance exercise for 6 wk progressed from moderate to vigorous; followed by 6 wk sedentary | Intervention | Habitual diet | Composition (16S) and function (qPCR of select functional genes) | ↔α-diversity (Chao1) ↔β-diversity after exercise/ washout (weighted and unweighted UniFrac) β-diversity different between lean/obese at baseline |
↑ butyrate-regulating group in lean and obese Butyrate group explained 61.2% of variance in microbiota response and 84% of VO2max response ΔButyrate-producers ~Δlean mass |
||||
| Munukka et al. (76) |
Humans (19 overweight, sedentary women) | Endurance exercise (bike erg) for 6 wk | Intervention | None | Composition and function (16S, metagenomics) | ↔α-diversity (not reported) ↑ β-diversity (Jaccard) |
↔Firmicutes | ↔Bacteroidetes | ↑ Bifidobacteriaceae (dependent on weight, body fat %, android fat %, total energy intake, sucrose, fiber) | ↓ Proteobacteria ↑ Verrumicrobiaceae, Akkermansia ↓ genes in fructose, mannose, alanine, aromatic amino acid metabolism |
|
| Taniguchi et al. (77) |
Humans (31 Japanese adult males, >60 years old) | Cycling for 5 wk, no washout between intervention and 5 wk control period | Intervention | Habitual diet | Composition and function (16S, metagenomics) | ↔α-diversity (Shannon, observed OTUs) |
↓ Clostridium difficile during exercise | ↑ Oscillospira during exercise (no longer significant after adjusting for dietary changes and treatment sequence) ↑ Metagenomic functions belonging to “Genetic Information Processing” and “Nucleotide Metabolism” during exercise |
|||
| Morita et al. (84) |
Humans (32 Japanese sedentary adult women, >65 years old) | Aerobic exercise (AE) or trunk muscle training (TM) for 12 wk | Intervention | Habitual diet | Composition (TRFLP) | ↓ Clostridium subcluster XIVa decreased in AE ↑ Clostridium IX in TM |
↑ Bacteroides (negatively correlated with pre-Bacteroides) | ||||
| Cronin et al. (78) |
Humans (74 healthy Irish adults) | Mixed aerobic and resistance exercise training program for 8 wk | Intervention | Whey Protein+ Exercise (EP) vs. Exercise (E) vs. Whey Protein (P) |
Composition and function (metagenomics) | ↔α-diversity from baseline but higher in EP vs. P group | Differential abundance of virus species between groups | ||||
| after intervention ↓ Archaea diversity in E group alteration in β-diversity of the gut virome in P and EP groups |
|||||||||||
| Human cross-sectional studies | |||||||||||
| Bressa et al. (82) |
Humans [40 premenopausal Caucasian women; 19 active (ACT), 21 sedentary (SED)] | General physical activity (measured for 1 wk) | Cross-sectional | Habitual diet | Composition (16S, qPCR) | ↔α-diversity (Chao1, Observed, Shannon) ↔β-diversity (unweighted or weighted UniFrac) Observed # species, Shannon, Simpson indices (+) ~ minimum time per sedentary bout (work days) |
↑ Firmicutes (trending) ↔ F/B ratio |
↓ Bacteroidetes (trending) ↔ F/B ratio |
↑ Bifidobacterium in ACT women | ↑ Haemophilus, Paraprevotella, Coprococcus, Ruminococaceae unclassified 1 in ACT women (16S) ↓ Desulfovibrionaceae unclassified, Turicibacter, Barnesiellaceae, Odoribacteriaceae, Ruminococcaceae unclassified 2, Ruminococcus in ACT women (16S) ↑ Faecalibacterium prausnitzii, Roseburia hominis, Akkermansia mucinipihila in ACT women (qPCR) |
|
| Karl et al. (86) |
Humans (73 Norwegian soldiers, 26 provided pre- and post- stool samples) | 4-day cross country ski-march (STRESS) | Cross-setional | Rations with or without protein- or carbohydrate-based supplements | Composition (16S) | ↑ α-diversity post-STRESS (Shannon) | ↑ Firmicutes | ↓ Bacteroidetes | Random forest using microbiota predicted pre- and post-STRESS samples with 100% accuracy | ||
| Shukla et al. (81) |
Humans (10 myalgic encephalomyelitis/ chronic fatigue syndrome (ME/CFS) patients, 10 healthy controls) | Cycling (max test) | Cross-sectional | Habitual diet | Composition (16S) in blood and stool | ↑ Firmicutes in ME/CFS patients after exercise ↓ Firmicutes in healthy controls after exercise ↑ Firmicutes/ Bacilli and Clostridium in blood of ME/CFS patients after exercise (not in healthy controls) |
↓ Bacteroidetes in ME/CFS patients after exercise ↑ Bacteroidetes in healthy controls after exercise |
||||
| Barton et al. (79) |
Humans (40 professional rugby players, 46 controls) | Rugby | Cross-sectional | None | Function (metagenomics) | ↑ α-diversity in athletes vs. high-BMI controls (Shannon, Simpson) or all controls (Phylogenetic diversity, Chao1, Observed species) | ↑ Akkermansia pathways in athletes vs. high-BMI controls ↑ pathways (amino acid and antibiotic biosynthesis, carbohydrate metabolism) in athletes |
||||
| Clarke et al. (19) |
Humans (40 professional rugby players, 46 controls) | Rugby | Cross-sectional | Habitual diet | Composition (16S) | ↑ α-diversity in athletes vs. high-BMI controls (Shannon, Simpson) or all controls (Phylogenetic diversity, Chao1, Observed species) | ↑ Firmicutes in athletes vs. high-BMI controls | ↓ Bacteroidetes in athletes vs. high-BMI controls | ↑ Akkermansia in low-BMI athletes vs. high-BMI controls | ||
| O'Donovan et al. (83) |
Humans (37 professional Irish athletes) | 16 different sports across varying sports classification groups (SCGs) | Cross-sectional | Habitual diet | Composition and function (metagenomics) | ↔α-diversity (Shannon and Simpson) between SCGs | ↑ Bacteroides caccae ~ SCGs with high static/high dynamic components | ↑ Bifidobacterium animalis ~ SCGs with low static/high dynamic components | ↑ Streptococcus suis, Clostridium bolteae, Lactobacillus phage LfeInf, Anaerostipes hadrus, flavin biosynthesis and fermentation pathways ~ SCGs with moderate dynamic component ↑ Lactobacillus acidophilus, Prevotella intermedia, Faecalibacterium prausnitzii ~ SCGs with low static/high dynamic components ↑ folate and amino acid biosynthesis pathways ~ SCGs with high static/high dynamic components ↑ Nucleotide biosynthesis ~ SCGs with high static/low dynamic components No species associated with SCGs with high static/low dynamic components or high dynamic/moderate static components |
||
| Petersen et al. (80) |
Humans (33 amateur and professional cyclists) | Cycling | Cross-sectional | None | Composition and function (16S, metagenomics, RNA-Seq) | ↑ α-diversity (Shannon, # of genera) in Cluster 3 (contains more professional cyclists vs. amateur) | ↑ Prevotella with increased exercise load (hrs per week) ↓ Bacteroides in athletes (though no non-athlete control for comparison) |
↑ Methanobrevibacter smithii gene expression in professional cyclists | |||
ABA, activity-based anorexia; ACT, active [designated group in Bressa et al. (82)]; AE, aerobic exercise training; BMI, body mass index; FE, forced exercise; FTR, forced treadmill running; HCR, high-capacity running; HFD, high-fat diet; HIIT, High intensity interval training; KEGG, Kyoto Encyclopedia of Genes and Genomes; LCR, low-capacity running; OTU, operational taxonomic unit; OVX, ovariectomized; PCB, polychlorinated biphenyls; PCR-DGGE, polymerase chain reaction denaturing gradient gel electrophoresis; PCR-TGGE, polymerase chain reaction temperature gradient gel electrophoresis; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; qPCR, quantitative polymerase chain reaction; SCG, sports classification group; SED, sedentary [designated group in Bressa et al. (82)]; TM, truck muscle training; TRFLP, Terminal restriction fragment length polymorphism; VE, voluntary exercise; VO2max, maximal (O2) oxygen uptake; VWR, voluntary wheel running.
Although there are similarities in microbial factors shown to be affected by exercise within the literature, directions of the effects are inconsistent, and some studies show contradictory results. For example, while some studies show a reduction in Firmicutes and/or an increase in Bacteroidetes as a result of exercise (58, 64, 66, 70, 73, 74, 81, 84), others show the opposite effect (19, 60–62, 68, 71, 81, 86), and others show no effect (69, 76, 82).
Findings on the effect of exercise on measures of diversity are also highly variable, some showing increases in α-diversity (19, 60, 64, 68, 70, 71, 75, 79, 86), some showing decreases (58, 59, 66), and others reporting no difference (18, 59, 63, 65, 69, 76–78, 82, 83). Brandt et al. (73) also found that exercise attenuated the decrease in α-diversity that occurred when mice were fed a high-fat diet.
Bacterial taxa commonly shown to respond to exercise training include Lactobacillus (typically increased) (58, 60, 62, 68), Bifidobacterium (typically increased) (58, 62, 76, 82), Proteobacteria (typically decreased) (58, 65, 66, 68, 76), Akkermansia (typically increased) (19, 75, 76, 79, 82), Streptococcus (variable effects) (61, 68, 69), Clostridium (variable effects) (58, 62, 63, 67, 77), Turicibacter (typically decreased) (59, 64), and Rikenellaceae (typically decreased) (63, 66, 69) as well as measures of α- and β-diversity (variable effects) (19, 58, 60, 64, 66, 68, 70, 76, 80).
However, in some studies, changes in taxa are dependent on other factors such as changes in weight, body fat, and blood glucose (64, 76). This suggests, that the associated metabolic effects of the exercise regime may be the proximal cause, while exercise is the ultimate cause. Related variables, SCFA production and butyrate-producing taxa, have been consistently shown to increase in response to exercise (18, 57, 79) and have also been positively correlated to changes in lean muscle mass, also suggesting that SCFAs may play an important role in mediating the effects of exercise and the gut microbiome on host response (18). Table 2 summarizes additional effects of exercise on microbial metabolites, host health, and dietary interactions.
Table 2.
Summary of effect of exercise on the microbial metabolites, host health, and dietary interactions.
| References | SCFAs | Other metabolites | Host health | GI tract physiology | Diet interactions |
|---|---|---|---|---|---|
| Rodent studies | |||||
| Allen et al. (59) | ↓ gastrointestinal inflammation in VWR, ↑ in FTR |
||||
| Lambert et al. (62) | ↑ glucose in exercised normal vs. sedentary normal ↔ glucose in exercised diabetic vs. sedentary diabetic |
||||
| Liu et al. (75) | ↑ Left ventricular ejection fraction (EF), fractional shortening (FS), cardiac output (CO), and stroke volume (SV) FS, EF, CO, SV, and left ventricular end systolic diameter (LVESD) correlated with gut microbiota taxa | ||||
| Brandt et al. (73) | ↑ body weight in HFD vs. CON, HFD plus resveratrol ↔ body weight in HFD vs. HFD plus exercise ↑ body fat %, subcutaneous, and visceral adipose tissue in HFD and HFD plus exercise vs. CON ↓ lean body mass in HFD and HFD plus exercise vs. CON ↑ serum amyloid A (SAA) in all HFD groups vs. CON |
||||
| Campbell et al. (67) | Body fat %: High-fat sedentary > High-fat exercise > low-fat sedentary > low-fat exercise | ↓ inflammatory infiltrate, COX-2, and high-fat diet-induced morphological changes in exercise | |||
| McCabe et al. (74) | ↑ Bone volume fraction with exercise [~ Firmicutes/Bacteroides (–), Clostridia (–), Lachnospiraceae (–), Actinobacteria (+)] ↓ Trabecular bond volume with high-fat diet ↑ Marrow adiposity with high-fat diet ↓ Body weight, fat pad mass, fasting glucose with exercise and low-fat diet |
||||
| Kang et al. (61) | ↔ high-fat diet-induced anxiety ↑ cognitive abilities |
||||
| Denou et al. (70) | High-fat diet with exercise vs. high-fat diet: ↔ body mass, fasting blood glucose ↑ insulin tolerance, RER, food intake, time to exhaustion with exercise |
||||
| Liu et al. (65) | ↓ non-esterified fatty acids (NEFAs) and triglycerides in LCR rats ↑ non-esterified fatty acids (NEFAs) and triglycerides in HCR rats |
↓ body weight, fat mass, feed efficiency in LCR rats ↑ body weight, fat mass, food intake, feed efficiency of HCR rats |
Christensenellaceae ~ food intake | ||
| Mika et al. (50) | ↓ weight in adult rats ↑ weight, lean mass in young rats |
||||
| Matsumoto et al. (57) | ↑ cecal n-butyrate | ↑ cecum size/weight | |||
| Queipo-Ortuno et al. (58) | ↑ body weight in Exercise and Ad lib ↓ body weight in ABA and ABA control ↑ ghrelin [~Lactobacillus (–), Bifidobacterium (–)] |
||||
| ↓ leptin [~Lactobacillus (+), Bifidobacterium (+)] | |||||
| Welly et al. (69) | ↓ total cholesterol, adiposity, inflammation in Exercise and weight-matched sedentary ↑ total energy expenditure in Exercise and weight-matched sedentary ↓ RQ, insulin resistance, LDL, liver mass in Exercise ↑ mitochondrial function in brown adipose in Exercise |
||||
| Feng et al. (71) | Exercise improved preoperative cognitive impairment in LCR rats | ||||
| Petriz et al. (68) | ↑ velocity ↓ lactate [~Clostridiaceae (–), Bacteroidaceae (–), Osillospira (+), Ruminococcus (–)] |
||||
| Batacan et al. (72) | Samples grouped by diet (not activity group) | ||||
| Human intervention studies | |||||
| Allen et al. (18) | ↑ SCFAs in lean ΔButyrate ~Δlean mass |
↑ lean body mass, bone mineral density, VO2max ↓ body fat % ΔButyrate and butyrate-producers ~Δlean mass |
|||
| Munukka et al. (76) | ↑ max power, VO2max, glucose ↓ lactate, HDL, LDL, large VLDL ↔ weight, blood pressure, weight circumfrence, BMI, fat mass/% |
||||
| Taniguchi et al. (77) | ↑ VO2peak [~C. difficile (–)], HDL [~Oscillospira (+)], total cholesterol [~C. difficile (–)] during exercise ↓ CAVI [~C. difficile (+)], intrahepatic fat %, HbA1c [~Oscillospira (–), ~C. difficile (+)] during exercise ↔ body fat % [~Oscillospira (–)]; visceral fat area, SBP, AST, ALT [~C. difficile (+)]; LDL [~C. difficile (–)] |
Oscillospira ~ changes in light-colored vegetable, seaweed, and rice consumption | |||
| Cronin et al. (78) | ↑ VO2max, lean mass in E and EP groups ↓ resting heart rate, % body fat, total fat mass, truck fat mass in E and EP groups ↔ pro-inflammatory markers |
||||
| Human cross-sectional studies | |||||
| Bressa et al. (82) | ↑ cysteine aminopeptidase in ACT women [~Bacteroides (–)] ↔α-fucosidase [~Bifidobacterium (+), Odoribacter (+)], alkaline phosphatase [~Desulfovibrio (–)] |
↔ BMI, weight, adiposity and muscle parameters Turicibacter (–) ~ BMI Barnesiellaceae (+) ~ % body fat Odoribacter (+), Haemophilus (–) ~ adiposity index, estimated visceral fat, % body fat Faecalibacterium (+) ~ muscle mass index, appendicular muscle mass index Coprococcus (+), Lachnospiraceae unclassified 1 (+) ~ appendicular muscle mass index | Turicibacter ~ dairy products, cereals Bifidobacterium ~ protein intake % Odoribacter ~ fiber, fat intake % Ruminococcaceae unclassified 1&2 ~ fat intake % | ||
| Karl et al. (86) | Random forest using stool metabolites predicted pre- and post-STRESS samples with 84% accuracy | ↑ intestinal permeability (IP) Pre-STRESS Actinobacteria |
|||
| 81% of stool metabolites decreased during STRESS 478 plasma metabolites significantly changed during STRESS, including metabolites partially or fully derived from microbial metabolism |
and changes in serum IL-6 and stool cysteine accounted for 84% of variability in change in IP | ||||
| Barton et al. (79) | ↑ SCFAs in athletes | In athletes: ↑ TMA, TMAO, L-carnitine, dimethylglycine, O-acetyl carnitine, proline betaine, creatine, acetoacetate, 3-hydroxy-isovaleric acid, acetone, N-methylnicotinate, N-methylnicotinamide, phenylacetylglutamine (PAG), 3-methylhistidine, lysine, and methylamine ↓ glycerate, allantoin, succinate, glycine, tyrosine |
Propionate ~ protein Butyrate ~ dietary fiber | ||
| Clarke et al. (19) | Diversity ~ protein intake and creatine kinase | ||||
| O'Donovan et al. (83) | 21 metabolites significantly different between SCGs (4 with significant pairwise differences: succinic acid, cis-aconitate, lactate, and creatinine) ↑ cis-aconitate, succinic acid in SCG with moderate static/high dynamic vs. low static/high dynamic ↑ fecal creatinine in SCG with low static/high dynamic vs. high static/low dynamic and moderate/high static and high dynamic ↓ lactate in SCG with low static/high dynamic vs. moderate/high static and high dynamic |
||||
ABA, activity-based anorexia; ACT, active [designated group in Bressa et al. (82)]; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; CAVI, cardio-ankle vascular index; CO, cardiac output; CON, control; E, Exercise [designated group in Cronin et al. (78)]; EF, left ventricular ejection fraction; EP, Whey protein + Exercise [designated group in Cronin et al. (78)]; FS, fractional shortening; FTR, forced treadmill running; HCR, high-capacity running; HDL, high-density lipoprotein; HFD, high-fat diet; LCR, low-capacity running; IP, intestinal permeability; LDL, low-density lipoprotein; LVESD, left ventricular end systolic diameter; NEFA, non-esterified fatty acid; PAG, phenylacetylglutamine; RER, respiratory exchange ratio; RQ, respiratory quotient; SAA, serum amyloid A; SBP, systolic blood pressure; SCFA, short-chain fatty acid; SV, stroke volume; TMA, trimethylamine; TMAO, trimethylamine-N-oxide; TRFLP, VLDL, very low-density lipoprotein; VO2max, maximal (O2) oxygen uptake; VO2peak, highest value of VO2 achieved during high-intensity exercise test.
Potential Causes of Discrepancies Between Studies
Potential reasons for the disparate results of these studies include study design factors as well as analytic methods (Figure 2). Study design factors include the choice of model (e.g., humans, mice, rats), the strain of mouse/rat (e.g., C57BL/6J, Zucker, Wistar, Sprague Dawley, etc.) (87), choice of diet (72), health or disease status (75, 81), age (66), gender, and the mode, duration, and intensity of training (59, 63, 72, 83, 84) as well as analytic methods such as DNA extraction and PCR primer biases (88–90), choice of microbiome sequencing methods (e.g., 16S rRNA gene sequencing, qPCR, metagenomics, etc.) (64), bioinformatic pipelines (90), and choice of diversity metrics (e.g., Shannon, Chao1, Simpson, etc.). For example, mouse models often use forced treadmill running or voluntary wheel running as modes of training. However, forced treadmill exercise often uses aversive motivation, such as shocks, which could induce negative stress responses (91–93) that may also affect intestinal permeability and the gut microbiome (59, 94, 95). In humans, exercise or sport is a broad term that can apply to a wide range of modes, durations, and intensities of activity. O'Donovan et al. (83) attempted to determine differential effects of different modes of exercise on the gut metagenome by doing a cross-sectional analysis of professional athletes from different sports with varying degrees of static and dynamic components. In this analysis, O'Donovan etal. found some differences in bacterial taxa and metabolites between sports classification groups (SCGs) that did not correlate with any other metadata (e.g., diet, sex, etc.) (83).
Figure 2.
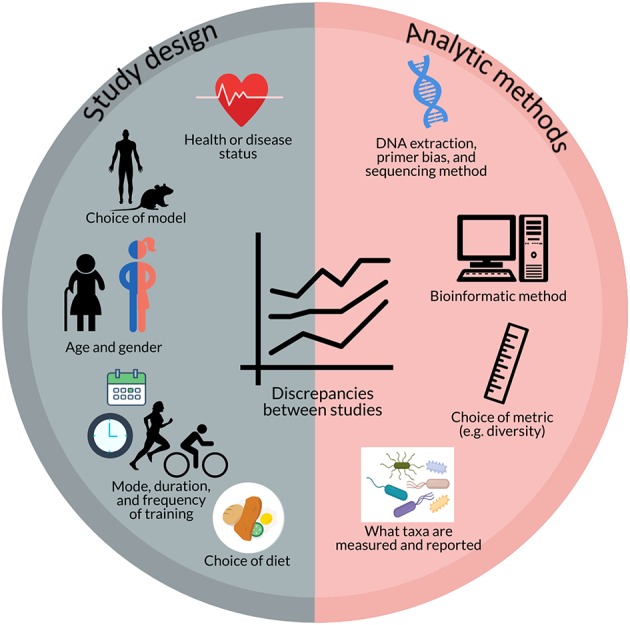
Potential factors contributing to discrepancies between studies investigating the effect of exercise on the gut microbiome include aspects of study design (e.g., health or disease status; choice of model; age and gender; mode, duration, and frequency of training; and choice of diet) and analytic methods (e.g., DNA extraction, primer bias, and sequencing method; bioinformatic method; choice of metrics; and what taxa are measured and reported).
In addition to differences in how results are obtained or measured, there is also a great deal of heterogeneity in what results are measured, or reported, that make it difficult to determine the full extent of variability in response between studies. In order to gain better insight into the potential effects and pathways by which exercise exerts its effect on the gut microbiome, it would be beneficial for studies to report effects on at least a certain standard set of microbiota variables that have already been shown to be relevant by multiple studies such as Firmicutes, Bacteroidetes, Lactobacillus, Bifidobacterium, Akkermansia, Clostridium, and Proteobacteria as well as diversity (though a standard metric has yet to be determined), butyrate-producing taxa (96), and SCFA production, even if the result is no change/difference. Munukka et al. (76) reported a lack of consistent effects due to inter-individual variability in response of the gut microbiota to exercise. This too is an important finding that should be reported and explored to determine factors that contribute to this variability in response and whether these differences in microbial response translate to differences in physiological response. These reporting standards would allow for better comparison between studies and potentially enable researchers to determine how different methods impact the results and elucidate factors that may contribute to variability in response.
Effect of Dietary Components Relevant to Exercise Nutrition on the Gut Microbiome
Confounding Effects of Diet
Diet is also a major factor that influences and shapes the gut microbiome (15–17). Kang et al. report that diet and exercise both cause shifts in the gut microbiome but that these changes are orthogonal (61). However, some of the studies above reported that dietary factors influenced the gut microbiota independently of, or in combination with, exercise. Dietary factors found in the studies presented here to associate with gut microbiome differences or changes include dairy products (82), light-colored vegetables (77), seaweed (77), rice (77), cereals (82), sucrose (76), fiber (76, 79, 82), protein intake (19, 79, 82), fat intake (82), and total food intake (65, 76) (Figure 3). Some differences or changes in the gut microbiota that seem to be associated with exercise may therefore be due to differences or changes in dietary intake, especially plants and carbohydrates, rather than exercise itself. There is therefore a need for studies investigating the link between the gut microbiome and exercise that control and standardize the dietary intake of participants.
Figure 3.
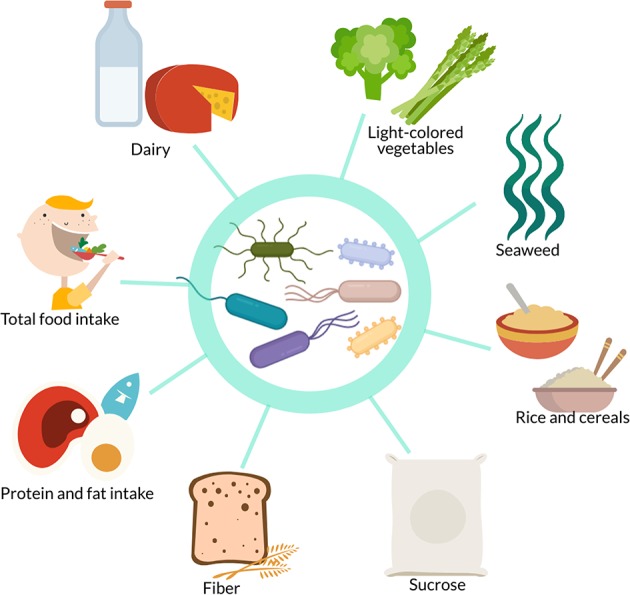
In studies investigating the effect of exercise on the gut microbiome, confounding dietary factors include dairy, light-colored vegetables, seaweed, rice, cereals, sucrose, fiber, protein intake, fat intake, and total food intake.
Effects of Supplements and Dietary Patterns on the Gut Microbiome
Although studies have shown some dietary interactions with the gut microbiome in athletes, it is unclear the extent to which the gut microbiome might be affected by supplements or dietary patterns commonly used by athletes, and the potential effects of this on the host. A review by Kårlund et al. (97) comprehensively discusses the topic of protein supplementation in athletes and the potential unknown effects on the gut microbiome. Excess protein may be fermented in the large intestine by various species from the genera Clostridium, Bacteroides, and others from the Proteobacteria phylum (98, 99), resulting in end products such as ammonia, amines, phenols, and sulfides as well as some SCFAs that may have systemic and metabolic effects on the host (100, 101). Different protein types have been shown to have differential effects on the gut microbiome (102–104) and plant-based vs. animal-based diets have also been shown to induce differences in the gut microbiome composition in humans (16). Additionally, different protein types have been assessed in the context of the anabolic response to exercise (105). However, there are no studies evaluating the impact of different types of protein supplements or whole-food protein sources on the gut microbiome and amino acid fermentation in athletes (97). As protein and protein supplements are widely advertised and recommended to athletes, this is an important gap in the research that should be addressed. Future research should also be sure to compare the effects of different protein sources as both isolated supplements as well as in their whole-food form as the matrix of the whole food has been shown to play an important role in the anabolic response to exercise and may alter effects based on factors such as the type and amount of fat (106, 107). Additionally, it would be interesting to know whether supplementation of plant-based proteins with amino acids such as leucine, lysine, and methionine, which is a strategy that has been shown by a couple studies to augment the anabolic effect of plant proteins (105), alters the effect of these proteins on the gut microbiome.
Carbohydrate is a primary fuel source for exercise and is therefore also a primary focus of athletes' dietary intake (108, 109). In addition to whole food forms of carbohydrates, such as bread, pasta, fruit, and potatoes, there is also a wide array of carbohydrate supplements that may be used before, during, or after exercise to enhance performance and recovery (110). The effects of whole food carbohydrates on the gut microbiome differs widely as a function of fiber content and type (111–113), though generally fiber tends to increase SCFA producing bacteria such as Bacteroidetes and Actinobacteria and decrease Firmicutes (114). However, the effects of frequent use of carbohydrate supplements, which are typically high in sugar and low in microbiota-accessible carbohydrates, on the gut microbiome is unknown. It is may therefore be of interest to develop carbohydrate supplements that also target the gut microbiome.
Though carbohydrates and protein are made the primary focus of athlete nutrition, fat is also an important fuel source during prolonged exercise and the popularity of high-fat diets such as the ketogenic diet has prompted athletes and scientists to investigate its potential for sports performance (115). However, evidence suggests that a high-fat diet does not improve exercise performance more than or as much as a high-carbohydrate diet (116). Additionally, the lack of microbiota-accessible carbohydrates on the ketogenic diet makes it questionable whether or how it would benefit the gut microbiome, though there has been little research in this area and none of it has been in athletes (117, 118).
Caffeine is also a widely used ergogenic aid among athletes. Coffee, one of the primary dietary sources of caffeine, has been linked to increases in Bifidobacterium and protection against high-fat diet-induced decreases in Lactobacillus, though these effects may be due to other bioactive compounds present in coffee such as chlorogenic acid (119). The effect of these aspects of dietary intake on the gut microbiome in athletes is only one half of the story. The other is the effect of the gut microbiome on the overall response, in terms of performance and training adaptation, of the athlete.
Personalized Sports Nutrition and the Potential Effect of the Gut Microbiome on Response to Diet and Exercise
The Gut Microbiome in Personalized Sports Nutrition
As discussed in Hughes et al. (22, 23), the gut microbiome is a potential predictor of response to diet. However, that review focused on predictors of response relevant to general health and prevention of chronic disease. Here, the evidence that the gut microbiome may be a predictor of athletic performance is reviewed. Personalized sports nutrition has incorporated the type of sport or activity, the training status of the individual, the athlete's goals, the time of the competitive season, and the athlete's food preferences (120, 121) as well as biological traits such as genetic polymorphisms, RNA expression, and epigenetic modifications (28–30, 34–38) in the attempt to optimize athletic performance and response to training programs.
The gut microbiome should be incorporated into this system as it modulates metabolism of diet and dietary supplements, and therefore has the potential to contribute to variability in response. Inter-individual variability among athletes in response to dietary supplements, such as caffeine and antioxidants, has been attributed to genetic polymorphisms and baseline antioxidant concentrations (35, 122). However, the gut microbiome has been identified as an important factor in the bioavailability and metabolism of antioxidants (123–125) and may be involved in caffeine metabolism via mechanisms such as modulation of the expression of the N-acetyltransferase 2 (NAT2) gene (126). Variability in the gut microbiome has been linked to variability in serum carotenoid concentrations (127), which suggests that the gut microbiome does indeed play a role in modulating antioxidant metabolism. In short, the gut microbiome may affect the metabolism of dietary components, supplements, and dietary patterns marketed to and used by athletes, but this is an area of research that has not yet been adequately explored.
The Effect of the Gut Microbiome on Performance
Though the gut microbiome has been shown to modulate metabolism of relevant dietary components, as discussed above, the implications of this for performance are still unclear.
Mostly cross-sectional, studies have examined the correlation between measures of fitness, such as VO2max and VO2peak, and the gut microbiota (18, 77, 85, 128–130) (Table 3). Butyrate-producing bacteria have been shown in both Allen et al. (18) and Estaki et al. (129) to correlate positively with VO2max and VO2peak, respectively. Bacteroides and the Firmicutes/Bacteroidetes ratio have also been shown to correlate with VO2max (85, 128, 130), although studies have shown contrasting results. Durk et al. (128) found the Firmicutes/Bacteroidetes ratio to correlate positively with VO2max. Conversely, Yu et al. (85) found a lower F/B ratio in elderly adults with higher exercise capacity and Yang et al. (130) found that the high VO2max group had lower Eubacterium rectale-Clostridium coccoides (Erec), which are members of the Firmicutes phylum, and higher Bacteroides. Yu et al. (85) also identified several other taxa that were correlated with VO2peak in their elderly population such as Lactobacillales, Blautia, Ruminococcus, E. coli, and Alcaligenaceae. Taniguchi et al. (77) found an inverse correlation between Clostridium difficile and changes in VO2peak in elderly Japanese men during a cycling intervention.
Table 3.
Summary of studies investigating the correlation between gut microbiota composition and measures of fitness.
| References | Subjects | Type of exercise | Type of study | Microbiota method | Performance metric(s) | Results |
|---|---|---|---|---|---|---|
| Allen et al. (18) | Humans (32 previously sedentary subjects, 18 lean 14 obese) | Endurance for 6 wk progressed from moderate to vigorous; followed by 6 wk sedentary | Intervention | Composition (16S) and function (qPCR of select functional genes) | VO2max | Butyrate-regulating bacteria group explained 61.2% of variance in microbiota response and 84% of VO2max response |
| Durk et al. (128) | Humans (healthy young adults) | Treadmill test | Cross-sectional | Composition (qPCR) | VO2max | Firmicutes/Bacteroidetes (F/B) ratio positively correlated to VO2max VO2max explained ~22% of variance in F/B ratio |
| Estaki et al. (129) | Humans (varying cardiorespiratory fitness levels) | Cycle ergometer | Cross-sectional | Composition (16S) and function (PICRUSt) | VO2peak | VO2peak accounted for ~20% of variation in α-diversity and positively correlated with abundance of butryate-producing taxa VO2peak + sex, fiber, and sugar intake explained 15.5% of variation in functional categories Protein associated with Bacteroides and explained 12.7% of taxonomic composition |
| Taniguchi et al. (77) | Humans (31 Japanese adult males, >60 years old) | Cycling for 5 wk, no washout between intervention and 5 wk control period | Intervention | Composition and function (16S, metagenomics) | VO2peak | Abundance of Clostridium difficile was negatively correlated with the increase in VO2peak achieved during the exercise intervention |
| Morita et al. (84) | Humans (32 Japanese sedentary adult women, >65 years old) | Aerobic exercise (AE) or trunk muscle training (TM) | Intervention | Composition (TRFLP) | Trunk muscle strength (Kraus-Weber test), 6-min walk test (6MWT) | Abundance of Bacteroides positively correlated with increases in distance during 6MWT |
| Yang et al. (130) | Humans (premenopausal mostly overweight/obese Finnish women with low fitness levels) | Cycle ergometer | Cross-sectional | Composition (flow cytometry, 16S rRNA gene hybridization, DNA-staining) | VO2max | High VO2max group had higher Bacteroides and lower Eubacterium rectale-Clostridium coccoides (EreC) Association between VO2max and EreC disappeared after correction for fat % (association mediated by body fatness) |
| Yu et al. (85) | Humans (56 hypertensive Chinese adults, 65–80 years old) | Cardiopulmonary treadmill exercise test | Intervention | Composition (16S) | VO2peak | 3 groups based on VO2peak: Weber A (normal exercise capacity), Weber B (mildly impaired exercise capacity), Weber C (moderately impaired exercise capacity) Lower F/B ratio in Weber A group (not statistically significant) No difference in α-diversity Betaproteobacteria, Ruminococcaceae, Faecalibacterium increased in Weber A group Blautia and Eubacterium increased in Weber B Escherichia increased in Weber C Eubacterium and Blautia positively correlated with CRP; Alcaligenaceae negatively correlated with CRP Lactobacillales, Blautia, Ruminococcus, and E. coli negatively correlated with VO2peak; Alcaligenaceae positively correlated with VO2peak |
6MWT, 6-minute walk test; DNA, deoxyribonucleic acid; EreC, Eubacterium rectale-Clostridium coccoides; F/B ratio, Firmicutes-to-Bacteroidetes ratio; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; qPCR, quantitative polymerase chain reaction; rRNA, ribosomal ribonucleic acid; VO2max, maximal (O2) oxygen uptake; VO2peak, highest value of VO2 achieved during high-intensity exercise test.
Few studies have directly investigated the effect of the gut microbiome on athletic performance (Table 4). Hsu et al. (131) and Huang et al. (132) both used germ free (GF) mice (C57BL/6JNarl) and compared these to mice colonized with bacteria to determine potential effects of the presence of the microbiome as well as specific bacteria on physical performance. Specific pathogen free (SPF) mice were found to have the highest exercise capacity and germ-free mice the lowest (131, 132). Mice colonized with individual bacterial taxa showed improvements in exercise capacity compared to their GF counterparts (131), though not all bacteria showed the same degree of impact (132). Hsu et al. (131), compared germ free (GF) mice, gnotobiotic mice colonized with Bacteroides fragilis (BF), and specific pathogen free (SPF) mice in a test of endurance swimming. In a similar study, Huang et al. (132) compared germ-free mice to gnotobiotic mice monocolonized with either Eubacterium rectale, Clostridium coccoides, or Lactobacillus plantarum TWK10 on performance in a swim-to-exhaustion test. In Hsu et al. (131), swim-to-exhaustion time was significantly different among all groups, with SPF mice having the greatest endurance, followed by BF mice, with GF mice having the least endurance capacity. In Huang et al. (132), gnotobiotic mice colonized with E. rectale showed significantly higher performance, both with and without aerobic training, than the GF mice as well as the mice colonized with C. coccoides and L. plantarum.
Table 4.
Summary of studies investigating the effect of the gut microbiota or probiotic supplementation on exercise performance.
| References | Subjects | Type of exercise | Microbiota analysis/supplementation method | Performance metric(s) | Performance results | Host health results |
|---|---|---|---|---|---|---|
| Hsu et al. (131) | Mice (C57BL/6JNarl, specific pathogen free (SPF), germ free (GF), gnotobiotic Bacteroides fragilis (BF); 12 wk, male) | Endurance swimming | N/A | Swim-to-exhaustion time | Swim-to-exhaustion time SPF > BF > GF Antioxidant systems glutathione peroxidase (GPx) and catalase (CAT) SPF > GF and BF; superoxide dismutase (SOD) activity SPF and GF > BF | Levels of antioxidant exyme activity GPx, SOD, and CAT: SPF > BF > GF % weight of liver, muscle, brown adipose tissue, and epididymal fat pad SPF > BF & GF |
| Huang et al. (132) | Mice (C57BL/6JNarl; germ free (GF), maintained germ free or colonized with Eubacterium rectale, Clostridium coccoides, or Lactobacillus plantarum TWK10; 6 wk, male) | Endurance swimming | N/A | Swim-to-exhaustion time | Swim-to-exhaustion time in specific pathogen free (SPF) > GF mice both before and after training (time increased in both SPF and GF mice after training) After training, swim-to-exhaustion time in E. rectale-colonized mice > GF, C. coccoides, and L. plantarum | E. rectale and C. coccoides mice showed higher lactate level vs. GF and L. plantarum Ammonia level increased more in GF group Creatine kinase (CK) lower in E. rectale vs. C. coccoides (no difference between GF and L. plantarum) Glucose levels higher in E. rectale and C. coccoides vs. GF and L. plantarum (GLUT4 higher in E. rectale vs. GF and L. plantarum) Hepatic glycogen higher in GF vs. SPF, E. rectale, L. plantarum, and C. coccoides Basal metabolic rate (BMR) higher in L. plantarum and C. coccoides mice vs. GF and E. rectale Wheel running distance higher in gnotobiotic mice (E. rectale > L. plantarum and C. coccoides > GF) Growth curve higher in GF and E. rectale vs. L. plantarum and C. coccoides |
| Chen et al. (133) | Mice (ICR, specific pathogen free (SPF), 6 wk, male) | Grip strength and endurance swimming | N/A Probiotic supplementation (Lactobacillus plantarum TWK10—dose-response in mice) | Grip strength and swim-to-exhaustion time | Probiotic supplementation increased grip strength and endurance swimming time after exercise | Probiotic supplementation decreased body weight, serum lactate, ammonia, urea nitrogen, albumin, CK, creatinine, triacylglycerol (TAG), and glucose and increased relative muscle weight, number of type I muscle fibers in gastrocnemius muscle |
| Huang et al. (134) | Humans (16 male runners) | Running (treadmill test) | N/A Probiotic supplementation (1 ×1011 CFU Lactobacillus plantarum TWK10) | Run time-to-fatigue | Probiotic supplementation increased run time-to-fatigue but not VO2max | Blood glucose levels higher in TWK10 group vs. placebo after exercise No significant differences in lactate, ammonia, free fatty acids (FFAs), or CK |
| Huang et al. (132) | Humans (54 healthy adults with no prior training; 27 men, 27 women) | Running (treadmill test) | N/A Probiotic supplementation (Lactobacillus plantarum TWK10 - placebo, low dose 3 ×10∧10 CFU, high dose 9 ×10∧10 CFU) |
Run time-to-fatigue | Probiotic supplementation increased time to exhaustion in both TWK10 groups but was significantly higher in the high-dose compared to low-dose group | Lactate accumulation and ammonia production improved in the TWK10 groups during exercise and recovery phase. Blood glucose higher in high-dose group during exercise. Decrease in body fat and increase in muscle mass in high-dose group. |
| Jäger et al. (135) | Humans (29 recreationally-trained men) | Resistance training | N/A Probiotic supplementation (1 ×1010 CFU Bacillus coagulans GBI-30, | 1 rep max (RM) one-legged leg press Vertical jump power Wingate power | Probiotic supplementation did not improve 1 RM or vertical jump power though a decrease in Wingate power was attenuated in the probiotic group | Probiotic supplementation increased perceived recovery and decreased perceived muscle soreness and measured muscle damage as indicated by CK |
| Lamprecht et al. (136) | Humans (23 trained men) | Cycling (cycle ergometer test) | N/A Probiotic supplementation (1 ×1010 CFU Bifidobacterium bifidum W23, Bif. lactis W51, Enterococcus faecium W54, L. acidophilus W22, L. brevis W63, L. lactis W58) | Incremental cycle ergometer exercise test | Probiotic supplementation did not improve VO2max or VO2max relative to body weight | Probiotic supplementation decreased zonulin and tendentially decreased carbonyl proteins and TNF-a but had no significant effects on a1-antitrypsin, malondialdehyde, total oxidation status of lipids, or IL-6 |
| Martarelli et al. (137) | Humans (24 male cyclists) | Cycling | Plate and Randomly Amplified Polymorphic DNA (RAPD) Probiotic supplementation (1 ×10∧9 CFU/g 1:1 Lactobacillus rhamnosus IMC 501 and Lactobacillus paracasei IMC 502) vs. control |
Intense exercise training | No performance results reported. Probiotic supplementation increased counts of Lactobacillus in stool (in different proportions for each subject) |
Reactive oxygen metabolite (ROM) concentrations significantly increased after exercise in control group but not in probiotic group (though ROM levels not significantly different between the two groups) Biological antioxidant potential (BAP) increased after probiotic supplementation and were higher in probiotic group vs. control group |
| Salarkia et al. (138) | Humans (46 adolescent females) | Swimming | N/A Probiotic supplementation (4 ×1010 CFU/ml Lactobacillus acidophilus spp, L. delbrueckii bulgaricus, Bif. bifidum, Streptococcus salivarus thermnophilus) vs. ordinary yogurt | 400m swim time Harvard step test | Probiotic supplementation increased VO2max but did not improve 400m swim time | Probiotic supplementation reduced frequency and duration of respiratory infections and some symptoms (dyspnea and ear pain) |
| Shing et al. (139) | Humans (10 male runners) | Running (treadmill test) | N/A Probiotic supplementation (4.5 ×1010 Lactobacillus acidophilus, L. rhamnosus, L. casei, L. plantarum, L. fermentum, Bifidobacterium lactis, Bif. breve, Bif. Bifidum, Streptococcus thermophilus) | Run time-to-fatigue | Probiotic supplementation increased run time-to-fatigue | Probiotic supplementation reduced serum lipopolysaccharide (LPS), slightly reduced lactulose:rhamnose (gastrointestinal permeability), and gastrointestinal discomfort |
| Townsend et al. (140) | Humans (25 male baseball athletes) | Off-season training | N/A Probiotic supplementation (1 ×109 CFU/day Bacillus subtilis DE111) vs. placebo |
1 rep max (RM) squat and deadlift, 10 yd sprint, standing long jump | No significant differences in performance between probiotic and placebo groups | TNF-α concentrations significantly lower in probiotic group (not in any other biochemical markers) No significant differences in body composition, testosterone, cortisol, IL-10, zonulin, salivary immunoglobulin A (SIgA), and SIgM |
| Scheiman et al. (141) | Humans (15 athletes pre- and post- marathon, 87 ultramarathoners and olympic trial rowers pre- and post-exercise (validation cohort) vs. 10 sedentary controls) | Running | Composition (16S) | Marathon run | No performance results reported. Veillonella increased post-exercise in athletes | |
| Mice (CL57BL/6, 12 wk, male/female?) | N/A Probiotic supplementation (Lactobacillus bulgaricus (control) or Veillonella atypica 5 ×109 CFU/ml) | Run-to-exhaustion time | Veillonella atypica increased run-to-exhaustion time (via lactate → propionate) | Decreased inflammatory cytokines in Veillonella-treated mice | ||
| Soares et al. (142) | Rats (Wistar, 11 wk, male) | Running (treadmill test) | N/A Probiotic supplementation (Saccharomyces boulardii 1 ×108 CFU/ml) | VO2max, run time-to-fatigue | Saccharomyces boulardii increased VO2max, run time-to-fatigue, max speed attained, and total work | Yeast supplementation had no effect on body mass gain or food intake |
BF, gnotobiotic colonized with Bacteroides fragilis; BMR, basal metabolic rate; CAT, catalase; CFU, colony forming unit; CK, creatine kinase; FFA, free fatty acid; GF, germ-free; GPx, glutathione peroxidase; ICR, Institute of Cancer Research; IL-6, interleukin-6; LPS, lipopolysaccharide; RM, rep max; SOD, superoxide dismutase; SPF, specific pathogen free; TAG, triacylglycerol; TNF-α, tumor necrosis factor alpha; VO2max, maximal (O2) oxygen uptake.
The potential mechanisms of these effects differed between the two studies. Differences in endurance capacity in Hsu et al. were accompanied by differences in antioxidant enzyme systems, with SPF mice showing greater serum and hepatic antioxidant enzyme activity, and physiological metrics, such as weight of muscle and brown adipose tissue (131). The gut microbiome modulates adipose tissue thermogenic pathways, including browning of white adipose and activity of brown adipose, via potential mechanisms such as bile acids and the endocannabinoid system (143). The gut microbiome may also modulate skeletal muscle anabolism and function via SCFA production and alteration of the availability of intramuscular fuels (55). Mice colonized with E. rectale and C. coccoides in Huang et al. showed higher lactate levels and higher glucose levels while mice colonized with E. rectale showed a lower creatine kinase (CK), a marker of muscular stress, and higher wheel running distance compared to both GF mice and the other gnotobiotic mice (132).
It is worth noting, however, in Huang et al. (132) that L. plantarum and C. coccoides did not colonize stably in the mice, fecal analysis showed no significant increases of these microbes, while E. rectale did colonize and increase over time. Therefore, it is unclear whether the ergogenic effect was due to the presence of E. rectale specifically, or simply due to the successful colonization by a microbe. These studies suggest that the gut microbiome may influence performance. They also indicate that a more diverse microbiome may be more beneficial as SPF mice performed better than monocolonized BF mice (131). While Huang et al. (132) suggests that individual taxa such as E. rectale may be partially responsible for performance effects, further research is needed to determine exactly what aspects or taxa contribute to this ergogenic effect. These studies also did not investigate responses to a dietary or training regimen, leaving room for further research on the potential of the gut microbiome to mediate or modify exercise performance response to diet.
The Effect of Probiotic Supplementation on Athlete Health and Performance
While there are a number of studies on probiotic supplementation in animals and human athletes, most focus on effects such as frequency of respiratory and gastrointestinal illness or biomarkers of inflammation and immune function (137, 144–146). Supplementation of probiotic bacteria to boost the abundance or activity of potentially beneficial taxa may also serve as a potential method of modifying performance response to training. Our review of the literature found eleven studies investigating the ergogenic effect of probiotic supplementation (133–142, 147) (Table 4). Common probiotic bacteria used were strains of Lactobacillus or Bifidobacterium (133, 134, 136–139, 147). Additional strains tested included those belonging to species Bacillus subtillis (140) or Bacillus coagulans (135), Veillonella atypica (141), or even yeast Saccharomyces boulardii (142).
The majority of studies investigated the effect of probiotic supplementation on aerobic exercise performance measures such as run time-to-fatigue, VO2max, max speed attained, 10-yard sprint, or 400-meter swim time (132–134, 136, 138, 139, 141, 142). However, some studies also investigated strength and anaerobic outcomes such as grip strength, vertical jump power, standing long jump, Wingate power, or 1 rep max (RM) lifts (133, 135, 140).
Effects on performance variables were highly mixed between studies, though a number of studies found beneficial effects on performance parameters such as time-to-fatigue (132–134, 139, 141, 142). However, some studies found no effects of probiotic supplementation on performance metrics (136, 140) while other studies found mixed effects, with probiotic supplementation improving some performance measures, but not others (134, 135, 138). For example, Huang et al. (134) found that probiotic supplementation with Lactobacillus plantarum TWK10 increased run time-to-fatigue but not VO2max. Thus, studies reporting effects of probiotic supplementation on only one performance outcome may not be providing a complete picture of the ergogenicity of probiotic bacteria. Additionally, all but one study (137) of probiotic supplementation of humans lacked confirmation of probiotic colonization and this study acknowledged that individuals showed different levels of colonization by the probiotic bacteria. It is important that future studies investigating probiotic supplementation also collect fecal samples from participants before and after the intervention to determine whether differences in probiotic colonization may contribute to inter-individual variability in the ergogenic effect of probiotic supplementation.
In addition to performance variables, many of these studies investigated effects on body composition and inflammation. Again, results were mixed, with some studies reporting significant effects of supplementation on outcomes such as fat mass and muscle mass (132, 133) or inflammatory markers (133, 137, 139, 141, 147), though results were often mixed with some biochemical markers showing no significant effect of probiotic treatment and some studies showing no significant effects at all on these outcomes (134, 142). However, as none of these variables were analyzed, further research is necessary to determine the mechanism of the effects as well as whether the same effects are seen in humans.
The Effect of Antibiotic Treatment on Exercise Performance
Conversely to the use of probiotics to determine the potential effect of the gut microbiome on athletic performance, the use of antibiotics in mouse models has recently been explored to determine the potential effects of a lack of gut microbes and their metabolites on exercise capacity and muscle function (148, 149). Table 5 displays the findings of these recent studies. In both studies, antibiotic treatment decreased the exercise capacity of the mice, tested using forced treadmill running. Additionally, this phenotype could be rescued by either natural reseeding (148) or acetate infusion (149). Nay et al. also found reduced gene expression of SCFA receptor G-protein coupled receptor 41 (GPR41) and sodium/glucose cotransporter 1 (SGLT1) as well as reduced muscle glycogen in antibiotic-treated mice, suggesting that the reduced exercise capacity in these mice may have been mediated by muscle glycogen availability (148). Okamoto et al. concluded that the reduced exercise capacity of antibiotic-treated mice was due to the lack of acetate available for use as a substrate during exercise as acetyl-CoA (149). With regards to changes in the gut microbial community, Okamoto et al. reported that relative abundance of Firmicutes was increased in antibiotic-treated mice while Bacteroidetes, α-diversity, and fecal bacterial DNA concentration was reduced (149). Nay et al. found that fecal bacterial DNA was reduced in antibiotic-treated mice but only reported differences in composition between control mice and mice treated with antibiotics but naturally reseeded, which showed no significant differences in α- and β-diversity, Bacteroides, and Firmicutes (148).
Table 5.
Summary of studies investigating the effect of antibiotics on exercise performance.
| References | Subjects | Type of exercise | Type of study | Diet and treatment groups | Microbiota method | Performance effects | Health effects | Microbiota effects |
|---|---|---|---|---|---|---|---|---|
| Nay et al. (148) | Mice (C57BL/6J mice, 14 wk, male) | Forced treadmill running | Intervention | control (CTL) vs. antibiotics (ATB) vs. antibiotics followed by natural reseeding (NAT) | Composition and function (RT-qPCR, 16S, metagenomics) | ↔ maximal aerobic velocity (MAV), extensor digitum longus (EDL) maximal strength in all groups ↓ time to exhaustion in ATB and NAT (restored in NAT after reseeding) ↓ EDL muscle fatigue index in ATB vs. CTL and NAT |
↑ cecum weight in ATB vs. CTL ↓ cecum weight in NAT vs. ATB ↔ muscle mass (gastrocnemius, quadriceps, EDL soleus) in ATB vs. CTL (body weight normalized without cecum weight) ↔ myofiber phenotype, mitochondrial metabolism, inflammatory signaling, Lat1 expression, GPR40, GPR120, blood glucose ↑ Fiaf expression in ATB vs. CTL and NAT ↓ GPR41 and Sglt1 expression, muscle glycogen in ATB vs. CTL and NAT |
↓ bacterial DNA in ATB and NAT (completely restored in NAT after reseeding) ↔α- and β-diversity, Bacteroides, Firmicutes between CTL and NAT |
| Okamoto et al. (149) | Mice (C57BL/6J mice, 10 wk, male) | Forced treadmill running | Intervention | antibiotic treatment (Abx) or antibiotic-free (Abx-free) group Acetate vs. saline infusion in Abx Butyrate infusion in Abx | Composition (16S) | ↓ treadmill running time in Abx ↑ treadmill running time in Abx+acetate ↔ treadmill running time in Abx+butyrate and Abx+saline |
↑ dietary intake, ceca size in Abx ↔ body mass gain, blood glucose ↓ muscle, white adipose, SCFA (fecal and plasma) in Abx ↔ body mass, muscle mass in Abx+acetate |
↑ Firmicutes in Abx ↓ Bacteroidetes, diversity (Shannon), fecal bacterial DNA concentration in Abx |
| Low microbiome-accessible carbohydrate (LMC) vs. high MC (HMC) diet FMT+inulin in LMC | ↓ treadmill running time in LMC group ↑ treadmill running time in LMC+FMT+inulin vs. LMC |
↓ muscle, fecal SCFA, plasma acetate and proprionate in LMC ↔ body mass gain, dietary intake ↑ white adipose in LMC ↔ body mass, tibialis anterior mass in LMC+FMT+inulin vs. LMC ↑ fecal SCFA in LMC+FMT+inulin vs. LMC |
↑ Firmicutes, F/B ratio, Lactococcus, Allobaculum in LMC ↓ Bacteroidetes, Prevotella, S24-7, diversity (Shannon) in LMC |
Abx, antibiotic; Abx-free, non-antibiotic treated; ATB, antibiotic; CTL, control; EDL, extensor digitum longus; F/B, Firmicutes/Bacteroidetes; FMT, fecal microbiota transplant; GPR, G-protein coupled receptor; HMC, high microbiome-accessible carbohydrate; LMC, low microbiota-accessible carbohydrate; MAV, maximal aerobic velocity; NAT, antibiotic-treated and naturally reseeded; RT-qPCR, real time quantitative polymerase chain reaction; SCFA, short-chain fatty acid.
Okamoto et al. additionally tested the effect of a low microbiota-available carbohydrate diet (LMC) vs. a high microbiota-available carbohydrate (HMC) diet to determine with substrate availability for the gut microbiome altered exercise capacity. In these treatment groups, treadmill running time was decreased in the LMC mice, concomitant with a decrease in muscle mass, fecal SCFA, and plasma acetate and proprionate as well as an increase in Firmicutes and decrease in Bacteroidetes and other SCFA producing bacterial taxa (149). This reduced exercise capacity phenotype was rescued when mice were given a fecal microbiota transplant (FMT) from HMC mice and a dose of inulin prior to exercise. The increased exercise capacity in LMC+FMT+inulin mice was not accompanied by changes in body mass or muscle mass but there was an increase in fecal SCFAs, again suggesting that SCFA concentration may act as a direct substrate or mediate substrate availability in such a way as to influence exercise capacity.
Summary of Findings and Putative Mechanisms of the Effect of the Gut Microbiome on Athletic Performance
These studies suggest that there may be an effect of the gut microbiome on exercise performance via mechanisms such as SCFA availability, muscle glycogen content, antioxidant enzyme activity, gastrointestinal permeability, and lactate metabolism (Figure 4). Additional speculative mechanisms may involve alterations in substrate utilization (142) as well as glycogen metabolism and storage (150), changes in neural function (142), as well as immune modulation (142) or cross-talk between the gut microbiome and mitochondria in energy production and inflammation (45). The gut microbiota has been hypothesized to impact skeletal muscle physiology and function via metabolites such as SCFAs, folate, tryptophan, glycine betaine, vitamins B2 and B12, and urolithins that may act through various pathways such as stimulation of insulin-like growth factor-1 (IGF-1), prevention of oxidative stress or inflammation, and promotion of mitochondrial biogenesis (151). However, these data also indicate that improvements in these functions do not always translate to improvements in performance. Further research is needed to investigate the effects of different probiotic strains, the interaction with dietary composition (e.g., differences of effect in athletes who have different overall dietary patterns), the use of dietary supplements, and in different modes of exercise, such as strength training.
Figure 4.
The gut microbiome may influence performance via mechanisms such as antioxidant enzyme activity, immune modulation, gastrointestinal permeability, substrate utilization and storage, mitochondria cross-talk, and/or the gut-brain axis.
Conclusions and Future Directions
The gut microbiome represents an open field of study in the realm of personalized sports nutrition. High interindividual variability in response to training and physical activity is regularly reported (152) and the gut microbiome may contribute to this variability by impacting individual metabolism of food components and/or adaptation to the homeostatic stress, or training load, of the exercise stimulus (153). More research is needed to determine whether the gut microbiome could be an important predictor of athletic performance in response to dietary and exercise interventions. Researchers should refer to guides such as Ross et al. (36), Hecksteden et al. (152), Mann et al. (153), Swinton et al. (154), and Hopkins et al. (155) for statistical frameworks to interpret inter-individual variability in response and identify factors that contribute to this variability.
Specific questions that could be addressed are the role of specific bacterial taxa or groups of taxa involved in gains in athletic performance in response to certain dietary factors (e.g., protein sources such as whey, casein, soy, etc.; macronutrient distribution; or supplements such as caffeine, beta-alanine, antioxidants) or exercise stimuli. This could be investigated by using a combined dietary-exercise intervention, measuring both baseline and final microbiome and performance variables, and using predictive machine learning algorithsms such as random forests (156) to determine whether baseline abundance or changes in certain bacterial taxa can predict an individual's physical performance response. Another question is whether different taxa are involved in different responses (e.g., VO2max, time-to-fatigue, rep max loads, etc.) and the mechanisms of these effects (e.g., SCFA production, antioxidant enzyme activity, muscle protein synthesis, glycogen formation, energy harvest and fuel utilization, inflammation, etc.). While the first question could be addressed by a study measuring multiple exercise performance outcomes within the same population, determining the mechanisms of these effects would require in vitro or animal models and measurement of potential mediating metabolites, such as SCFAs, and physiological variables, such as muscle mass or muscle glycogen content. Furthermore, larger and longer studies are needed to address whether effects or responses differ between demographics (e.g., gender, age, ethnicity, etc.) and whether modulation of the gut microbiome via probiotics and/or prebiotics or modulation of the dietary or exercise stimulus (e.g., amount or type of supplement; mode, duration, intensity of exercise) may serve to increase positive response to the stimulus, decreasing the number of “non-responders.” For example, a study using the same participants and measuring microbiome and performance responses to stepwise increases in the duration and/or intensity of exercise may serve to elucidate what type of exercise may be optimal for certain individuals and their microbiomes. Additionally, studies that have measured changes in performance in response to probiotic supplementation have not looked at individual's gut microbiota composition directly. This is a limitation of these studies as different strains of probiotic bacteria show differing rates of survival through the gastrointestinal tract (157) and the composition of an individual's gut microbiota may also influence the persistence and function of probiotic bacteria in the gut (158–160). Therefore, not all probiotic strains may survive in sufficient quantities to make it down to the gut microbiome and, even if the probiotic bacteria reach the gut microbiome, it may not last as long or have the same effect in each individual.
Additional challenges and limitations in the area of research are numerous and must be taken into consideration when making claims about the exercise-microbiome connection. While the effects of diet and exercise have been shown to be orthogonal to one another (61), diet can still be a confounding factor within and between studies. Thus, conclusive effects of exercise on the gut microbiome must standardize the diet of participants, which has not yet been done. In addition to diet, variables such as genetics (32, 161), epigenetics (162), sleep behavior (163, 164), gender (165, 166), age (66, 167), and a host of other factors contribute to variability in the gut microbiome as well as performance response. This variability makes it extremely difficult to draw concrete conclusions about the effects of the gut microbiome and should always be considered when designing or interpreting studies on the interaction of the gut microbiome and host.
A related body of research has developed investigating the “gut-muscle axis” as it relates to age-related changes in muscle mass (i.e., sarcopenia) and physical frailty (151, 168–174) as well as its potential role in the “muscle-gut-brain” axis and neurodegenerative diseases in aging (175, 176). This field of research has the potential to inform the research in the field of the gut microbiome and exercise performance. Though this research focuses on preservation of muscle mass rather than physical or athletic performance, it is extremely relevant to identifying the pathways that connect these systems and how they can be modulated. Taxa, such as Faecalibacterium prausnitzii (151), or supplementation with prebiotics (177), butyrate (178), or other microbial metabolites such as urolithin A (179, 180) have shown beneficial associations or effects on muscle function and protection against aging-related atrophy. It has also been postulated that the aging gut microbiome may play a role in the phenomenon of anabolic resistance, not by altering protein metabolism per se, but by mechanisms such as gut barrier function, inflammation, and mitochondrial dysfunction (168, 170). Thus, by looking at how age-related changes in the gut microbiome may contribute to sarcopenia and decreases in muscle function, we may better understand how to modify or supplement this community to both maintain health as well as potentially increase performance.
In conclusion, there are several different fields of research that have touched on the question of the role of the gut microbiome in exercise and athletic performance. However, there are many gaps and limitations in the research thus far that must still be addressed. While there have not yet been any conclusive findings, further research and collaboration among disciplines may help shed light on the connection between exercise and the gut microbiome and the potential implications on athletic performance.
Author Contributions
RH performed the literature review, conceived, and composed the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thank you to James P. Hughes and Nancy L. Keim who assisted in proofing this manuscript. Figures were generated using Piktochart.
Footnotes
Funding. Partial funding for this manuscript was provided by the UC Davis Open Access Fund.
References
- 1.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. (2012) 489:242–9. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 4.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. (2011) 62:361–80. 10.1146/annurev-med-012510-175505 [DOI] [PubMed] [Google Scholar]
- 5.de Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE. (2017) 12:e0188475. 10.1371/journal.pone.0188475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 7.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. (2011) 474:327–36. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. (2012) 336:1268–73. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinan TG, Cryan JF. The microbiome-gut-brain axis in health and disease. Gastroenterol Clin North Am. (2017) 46:77–89. 10.1016/j.gtc.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 10.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. (2012) 13:701–12. 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- 11.Lamichhane S, Sen P, Dickens AM, Oresic M, Bertram HC. Gut metabolome meets microbiome: a methodological perspective to understand the relationship between host and microbe. Methods. (2018) 149:3–12. 10.1016/j.ymeth.2018.04.029 [DOI] [PubMed] [Google Scholar]
- 12.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein–coupled receptor FFAR2. Diabetes. (2012) 61:364–71. 10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. (2015) 11:577–91. 10.1038/nrendo.2015.128 [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. (2010) 107:14691–6. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. (2014) 505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. (2011) 334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen JM, Mailing LJ, Niemiro GM, Moore R, Cook MD, White BA, et al. Exercise alters gut microbiota composition and function in lean and obese humans. Med Sci Sports Exerc. (2018) 50:747–57. 10.1249/MSS.0000000000001495 [DOI] [PubMed] [Google Scholar]
- 19.Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. (2014) 63:1913–20. 10.1136/gutjnl-2013-306541 [DOI] [PubMed] [Google Scholar]
- 20.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koppel N, Balskus EP. Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem Biol. (2016) 23:18–30. 10.1016/j.chembiol.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 22.Hughes RL, Kable ME, Marco M, Keim NL. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models. Part II: results. Adv Nutr. (2019) 10:979–98. 10.1093/advances/nmz049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes RL, Marco ML, Hughes JP, Keim NL, Kable ME. The role of the gut microbiome in predicting response to diet and the development of precision nutrition models—Part I: overview of current methods. Adv Nutr. (2019) 10:953–78. 10.1093/advances/nmz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmons JA. Variability in training-induced skeletal muscle adaptation. J Appl Physiol. (2011) 110:846–53. 10.1152/japplphysiol.00934.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas D, Erdman K, Burke L. American College of Sports Medicine Joint Position Statement. Nutrition and athletic performance. Med Sci Sport Exerc. (2016) 48:543–68. 10.1249/MSS.0000000000000852 [DOI] [PubMed] [Google Scholar]
- 26.Borresen J, Lambert MI. The quantification of training load, the training response and the effect on performance. Sports Med. (2009) 39:779–95. 10.2165/11317780-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 27.Pickering C, Kiely J. Exercise response efficiency: a novel way to enhance population health? Lifestyle Genom. (2019) 11:129–35. 10.1159/000501206. [DOI] [PubMed] [Google Scholar]
- 28.Jones N, Kiely J, Suraci B, Collins D, De Lorenzo D, Pickering C, et al. A genetic-based algorithm for personalized resistance training. Biol Sport. (2016) 33:117. 10.5604/20831862.1198210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickering C, Kiely J, Suraci B, Collins D. The magnitude of Yo-Yo test improvements following an aerobic training intervention are associated with total genotype score. PloS ONE. (2018) 13:e0207597. 10.1371/journal.pone.0207597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guest NS, Horne J, Vanderhout SM, El-Sohemy A. Sport nutrigenomics: personalized nutrition for athletic performance. Front Nutr. (2019) 6:8. 10.3389/fnut.2019.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Pérusse L, et al. Familial aggregation of Vo2 max response to exercise training: results from the HERITAGE Family Study. J Appl Physiol. (1999) 87:1003–8. 10.1152/jappl.1999.87.3.1003 [DOI] [PubMed] [Google Scholar]
- 32.Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and athletic performance: an update. Med Sport Sci. (2016) 61:41–54. 10.1159/000445240 [DOI] [PubMed] [Google Scholar]
- 33.Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, et al. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sport Exer. (2009) 41:34–72. 10.1249/MSS.0b013e3181844179 [DOI] [PubMed] [Google Scholar]
- 34.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. (2010) 108:1487–96. 10.1152/japplphysiol.01295.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickering C, Kiely J. Are the current guidelines on caffeine use in sport optimal for everyone? Inter-individual variation in caffeine ergogenicity, and a move towards personalised sports nutrition. Sports Med. (2018) 48:7–16. 10.1007/s40279-017-0776-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross R, Goodpaster BH, Koch LG, Sarzynski MA, Kohrt WM, Johannsen NM, et al. Precision exercise medicine: understanding exercise response variability. Br J Sports Med. (2019) 53:1141–53. 10.1136/bjsports-2018-100328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Cuervo M, Goni L, Martinez JA. Genetic and nongenetic factors explaining metabolically healthy and unhealthy phenotypes in participants with excessive adiposity: relevance for personalized nutrition. Ther Adv Endocrinol. (2019) 10:2042018819877303. 10.1177/2042018819877303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danaher J. Metabolic Mechanisms of the Fat Mass and Obesity-Associated (FTO) Gene. Melbourne, VIC: Victoria University; (2016). [Google Scholar]
- 39.Cook MD, Allen JM, Pence BD, Wallig MA, Gaskins HR, White BA, et al. Exercise and gut immune function: evidence of alterations in colon immune cell homeostasis and microbiome characteristics with exercise training. Immunol Cell Biol. (2016) 94:158–63. 10.1038/icb.2015.108 [DOI] [PubMed] [Google Scholar]
- 40.Cronin O, Molloy MG, Shanahan F. Exercise, fitness, and the gut. Curr Opin Gastroenterol. (2016) 32:67–73. 10.1097/MOG.0000000000000240 [DOI] [PubMed] [Google Scholar]
- 41.Cronin O, O'Sullivan O, Barton W, Cotter PD, Molloy MG, Shanahan F. Gut microbiota: implications for sports and exercise medicine. Br J Sports Med. (2017) 51:700–1. 10.1136/bjsports-2016-097225 [DOI] [PubMed] [Google Scholar]
- 42.Bermon S, Petriz B, Kajeniene A, Prestes J, Castell L, Franco OL. The microbiota: an exercise immunology perspective. Exerc Immunol Rev. (2015) 21:70–9. Available online at: https://www.researchgate.net/profile/Bernardo_Petriz/publication/272480734_The_Microbiota_an_Exercise_Immunology_Perspective/links/54ee51a00cf2e55866f2871a.pdf [PubMed] [Google Scholar]
- 43.Cerdá B, Pérez M, Pérez-Santiago JD, Tornero-Aguilera JF, González-Soltero R, Larrosa M. Gut microbiota modification: another piece in the puzzle of the benefits of physical exercise in health? Front Physiol. (2016) 7:51. 10.3389/fphys.2016.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Guo Y, Gui Y, Xu D. Physical exercise, gut, gut microbiota, and atherosclerotic cardiovascular diseases. Lipids Health Dis. (2018) 17:17. 10.1186/s12944-017-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark A, Mach N. The crosstalk between the gut microbiota and mitochondria during exercise. Front Physiol. (2017) 8:319. 10.3389/fphys.2017.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Codella R, Luzi L, Terruzzi I. Exercise has the guts: how physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig Liver Dis. (2018) 50:331–41. 10.1016/j.dld.2017.11.016 [DOI] [PubMed] [Google Scholar]
- 47.Hamasaki H. Exercise and gut microbiota: clinical implications for the feasibility of Tai Chi. J Integr Med. (2017) 15:270–81. 10.1016/S2095-4964(17)60342-X [DOI] [PubMed] [Google Scholar]
- 48.Mach N, Fuster-Botella D. Endurance exercise and gut microbiota: a review. J Sport Health Sci. (2017) 6:179–97. 10.1016/j.jshs.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mailing LJ, Allen JM, Buford TW, Fields CJ, Woods JA. Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci Rev. (2019) 47:75–85. 10.1249/JES.0000000000000183 [DOI] [PubMed] [Google Scholar]
- 50.Mika A, Fleshner M. Early-life exercise may promote lasting brain and metabolic health through gut bacterial metabolites. Immunol Cell Biol. (2016) 94:151. 10.1038/icb.2015.113 [DOI] [PubMed] [Google Scholar]
- 51.O'Sullivan O, Cronin O, Clarke SF, Murphy EF, Molloy MG, Shanahan F, et al. Exercise and the microbiota. Gut Microbes. (2015) 6:131–6. 10.1080/19490976.2015.1011875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rankin A, O'Donovan C, Madigan SM, O'Sullivan O, Cotter PD. 'Microbes in sport' -the potential role of the gut microbiota in athlete health and performance. Br J Sports Med. (2017) 51:698–9. 10.1136/bjsports-2016-097227 [DOI] [PubMed] [Google Scholar]
- 53.Monda V, Villano I, Messina A, Valenzano A, Esposito T, Moscatelli F, et al. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev. (2017) 2017:3831972. 10.1155/2017/3831972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sohail MU, Yassine HM, Sohail A, Al Thani AA. Impact of physical exercise on gut microbiome, inflammation, and the pathobiology of metabolic disorders. Rev Diabet Stud. (2019) 15:35–48. 10.1900/RDS.2019.15.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawley JA. Microbiota and muscle highway—two way traffic. Nat Rev Endocrinol. (2019). [Epub ahead of print]. 10.1038/s41574-019-0291-6 [DOI] [PubMed] [Google Scholar]
- 56.Dalton A, Mermier C, Zuhl M. Exercise influence on the microbiome–gut–brain axis. Gut Microbes. (2019) 10:555–68. 10.1080/19490976.2018.1562268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto M, Inoue R, Tsukahara T, Ushida K, Chiji H, Matsubara N, et al. Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci Biotechnol Biochem. (2008) 72:572–6. 10.1271/bbb.70474 [DOI] [PubMed] [Google Scholar]
- 58.Queipo-Ortuno MI, Seoane LM, Murri M, Pardo M, Gomez-Zumaquero JM, Cardona F, et al. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS ONE. (2013) 8:e65465. 10.1371/journal.pone.0065465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen JM, Berg Miller ME, Pence BD, Whitlock K, Nehra V, Gaskins HR, et al. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J Appl Physiol. (2015) 118:1059–66. 10.1152/japplphysiol.01077.2014 [DOI] [PubMed] [Google Scholar]
- 60.Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ Health Perspect. (2013) 121:725–30. 10.1289/ehp.1306534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang SS, Jeraldo PR, Kurti A, Miller ME, Cook MD, Whitlock K, et al. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol Neurodegen. (2014) 9:36. 10.1186/1750-1326-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambert JE, Myslicki JP, Bomhof MR, Belke DD, Shearer J, Reimer RA. Exercise training modifies gut microbiota in normal and diabetic mice. Appl Physiol Nutr Metab. (2015) 40:749–52. 10.1139/apnm-2014-0452 [DOI] [PubMed] [Google Scholar]
- 63.Lamoureux EV, Grandy SA, Langille MGI. Moderate exercise has limited but distinguishable effects on the mouse microbiome. mSystems. (2017) 2:e00006-17. 10.1128/mSystems.00006-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS ONE. (2014) 9:e92193. 10.1371/journal.pone.0092193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu TW, Park YM, Holscher HD, Padilla J, Scroggins RJ, Welly R, et al. Physical activity differentially affects the cecal microbiota of ovariectomized female rats selectively bred for high and low aerobic capacity. PLoS ONE. (2015) 10:e0136150. 10.1371/journal.pone.0136150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mika A, Van Treuren W, Gonzalez A, Herrera JJ, Knight R, Fleshner M. Exercise is more effective at altering gut microbial composition and producing stable changes in lean mass in juvenile versus adult male F344 rats. PLoS ONE. (2015) 10:e0125889. 10.1371/journal.pone.0125889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell SC, Wisniewski PJ, Noji M, McGuinness LR, Häggblom MM, Lightfoot SA, et al. The effect of diet and exercise on intestinal integrity and microbial diversity in mice. PloS ONE. (2016) 11:e0150502. 10.1371/journal.pone.0150502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petriz BA, Castro AP, Almeida JA, Gomes CP, Fernandes GR, Kruger RH, et al. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genomics. (2014) 15:511. 10.1186/1471-2164-15-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welly RJ, Liu TW, Zidon TM, Rowles JL, III, Park YM, Smith TN, et al. Comparison of diet versus exercise on metabolic function and gut microbiota in obese rats. Med Sci Sports Exerc. (2016) 48:1688–98. 10.1249/MSS.0000000000000964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denou E, Marcinko K, Surette MG, Steinberg GR, Schertzer JD. High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. Am J Physiol-Endoc Metab. (2016) 310:E982–93. 10.1152/ajpendo.00537.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng X, Uchida Y, Koch L, Britton S, Hu J, Lutrin D, et al. Exercise prevents enhanced postoperative neuroinflammation and cognitive decline and rectifies the gut microbiome in a rat model of metabolic syndrome. Front Immunol. (2017) 8:1768. 10.3389/fimmu.2017.01768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batacan R, Fenning A, Dalbo V, Scanlan A, Duncan M, Moore R, et al. A gut reaction: the combined influence of exercise and diet on gastrointestinal microbiota in rats. J Appl Microbiol. (2017) 122:1627–38. 10.1111/jam.13442 [DOI] [PubMed] [Google Scholar]
- 73.Brandt N, Kotowska D, Kristensen CM, Olesen J, Lützhøft DO, Halling JF, et al. The impact of exercise training and resveratrol supplementation on gut microbiota composition in high-fat diet fed mice. Physiol Rep. (2018) 6:e13881. 10.14814/phy2.13881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCabe LR, Irwin R, Tekalur A, Evans C, Schepper JD, Parameswaran N, et al. Exercise prevents high fat diet-induced bone loss, marrow adiposity and dysbiosis in male mice. Bone. (2019) 118:20–31. 10.1016/j.bone.2018.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z, Liu HY, Zhou H, Zhan Q, Lai W, Zeng Q, et al. Moderate-intensity exercise affects gut microbiome composition and influences cardiac function in myocardial infarction mice. Front Microbiol. (2017) 8:1687. 10.3389/fmicb.2017.01687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Munukka E, Ahtiainen JP, Puigbo P, Jalkanen S, Pahkala K, Keskitalo A, et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front Microbiol. (2018) 9:2323 10.3389/fmicb.2018.02323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taniguchi H, Tanisawa K, Sun X, Kubo T, Hoshino Y, Hosokawa M, et al. Effects of short-term endurance exercise on gut microbiota in elderly men. Physiol Rep. (2018) 6:e13935. 10.14814/phy2.13935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cronin O, Barton W, Skuse P, Penney NC, Garcia-Perez I, Murphy EF, et al. A prospective metagenomic and metabolomic analysis of the impact of exercise and/or whey protein supplementation on the gut microbiome of sedentary adults. MSystems. (2018) 3:e00044–18. 10.1128/mSystems.00044-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, et al. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. (2018) 67:625–33. 10.1136/gutjnl-2016-313627 [DOI] [PubMed] [Google Scholar]
- 80.Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, et al. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome. (2017) 5:98. 10.1186/s40168-017-0320-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shukla SK, Cook D, Meyer J, Vernon SD, Le T, Clevidence D, et al. Changes in gut and plasma microbiome following exercise challenge in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). PLoS ONE. (2015) 10:e0145453. 10.1371/journal.pone.0145453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bressa C, Bailén-Andrino M, Pérez-Santiago J, González-Soltero R, Pérez M, Montalvo-Lominchar MG, et al. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE. (2017) 12:e0171352. 10.1371/journal.pone.0171352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Donovan CM, Madigan SM, Garcia-Perez I, Rankin A, O'Sullivan O, Cotter PD. Distinct microbiome composition and metabolome exists across subgroups of elite Irish athletes. J Sci Med Sport. (2019) 23:63–8. 10.1016/j.jsams.2019.08.290 [DOI] [PubMed] [Google Scholar]
- 84.Morita E, Yokoyama H, Imai D, Takeda R, Ota A, Kawai E, et al. Aerobic exercise training with brisk walking increases intestinal bacteroides in healthy elderly women. Nutrients. (2019) 11:868. 10.3390/nu11040868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu Y, Mao G, Wang J, Zhu L, Lv X, Tong Q, et al. Gut dysbiosis is associated with the reduced exercise capacity of elderly patients with hypertension. Hypertens Res. (2018) 41:1036. 10.1038/s41440-018-0110-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol-Gastr Liver Physiol. (2017) 312:G559–71. 10.1152/ajpgi.00066.2017 [DOI] [PubMed] [Google Scholar]
- 87.Lerman I, Harrison BC, Freeman K, Hewett TE, Allen DL, Robbins J, et al. Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. J Appl Physiol. (2002) 92:2245–55. 10.1152/japplphysiol.01045.2001 [DOI] [PubMed] [Google Scholar]
- 88.Zielinska S, Radkowski P, Blendowska A, Ludwig-Gałezowska A, Łoś JM, Łoś M. The choice of the DNA extraction method may influence the outcome of the soil microbial community structure analysis. Microbiol Open. (2017) 6:e00453. 10.1002/mbo3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. (2011) 12:R18. 10.1186/gb-2011-12-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol. (2017) 35:1077–86. 10.1038/nbt.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moraska A, Deak T, Spencer RL, Roth D, Fleshner M. Treadmill running produces both positive and negative physiological adaptations in Sprague-Dawley rats. Am J Physiol Regul Integr Comp Physiol. (2000) 279:R1321–9. 10.1152/ajpregu.2000.279.4.R1321 [DOI] [PubMed] [Google Scholar]
- 92.Noble EG, Moraska A, Mazzeo RS, Roth DA, Olsson MC, Moore RL, et al. Differential expression of stress proteins in rat myocardium after free wheel or treadmill run training. J Appl Physiol. (1999) 86:1696–701. 10.1152/jappl.1999.86.5.1696 [DOI] [PubMed] [Google Scholar]
- 93.Leasure J, Jones M. Forced and voluntary exercise differentially affect brain and behavior. Neuroscience. (2008) 156:456–65. 10.1016/j.neuroscience.2008.07.041 [DOI] [PubMed] [Google Scholar]
- 94.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front Cell Neurosci. (2015) 9:392. 10.3389/fncel.2015.00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clark A, Mach N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: a systematic review for athletes. J Int Soc Sport Nutr. (2016) 13:43. 10.1186/s12970-016-0155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. (2009) 294:1–8. 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- 97.Kårlund A, Gómez-Gallego C, Turpeinen AM, Palo-oja OM, El-Nezami H, Kolehmainen M. Protein supplements and their relation with nutrition, microbiota composition and health: is more protein always better for sportspeople? Nutrients. (2019) 11:E829. 10.3390/nu11040829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dallas DC, Sanctuary MR, Qu Y, Khajavi SH, Van Zandt AE, Dyandra M, et al. Personalizing protein nourishment. Crit Rev Food Sci. (2017) 57:3313–31. 10.1080/10408398.2015.1117412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diether NE, Willing BP. Microbial fermentation of dietary protein: an important factor in diet–microbe–host interaction. Microorganisms. (2019) 7:19 10.3390/microorganisms7010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Windey K, De Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol Nutr Food Res. (2012) 56:184–96. 10.1002/mnfr.201100542 [DOI] [PubMed] [Google Scholar]
- 101.Portune KJ, Beaumont M, Davila AM, Tomé D, Blachier F, Sanz Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: the two sides of the coin. Trends Food Sci Tech. (2016) 57:213–32. 10.1016/j.tifs.2016.08.011 [DOI] [Google Scholar]
- 102.Zhu Y, Lin X, Zhao F, Shi X, Li H, Li Y, et al. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci Rep. (2015) 5:15220 10.1038/srep15220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Butteiger DN, Hibberd AA, McGraw NJ, Napawan N, Hall-Porter JM, Krul ES. Soy protein compared with milk protein in a western diet increases gut microbial diversity and reduces serum lipids in golden syrian hamsters. J Nutr. (2015) 146:697–705. 10.3945/jn.115.224196 [DOI] [PubMed] [Google Scholar]
- 104.An C, Kuda T, Yazaki T, Takahashi H, Kimura B. Caecal fermentation, putrefaction and microbiotas in rats fed milk casein, soy protein or fish meal. Appl Microbiol Biol. (2014) 98:2779–87. 10.1007/s00253-013-5271-5 [DOI] [PubMed] [Google Scholar]
- 105.van Vliet S, Burd NA, van Loon LJ. The skeletal muscle anabolic response to plant-versus animal-based protein consumption. J Nutr. (2015) 145:1981–91. 10.3945/jn.114.204305 [DOI] [PubMed] [Google Scholar]
- 106.Burd NA, Beals JW, Martinez IG, Salvador AF, Skinner SK. Food-first approach to enhance the regulation of post-exercise skeletal muscle protein synthesis and remodeling. Sports Med. (2019) 49:59–68. 10.1007/s40279-018-1009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Vliet S, Beals JW, Martinez IG, Skinner SK, Burd NA. Achieving optimal post-exercise muscle protein remodeling in physically active adults through whole food consumption. Nutrients. (2018) 10:224 10.3390/nu10020224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burke LM, Cox GR, Cummings NK, Desbrow B. Guidelines for daily carbohydrate intake. Sports Med. (2001) 31:267–99. 10.2165/00007256-200131040-00003 [DOI] [PubMed] [Google Scholar]
- 109.Burke LM, Hawley JA, Wong SH, Jeukendrup AE. Carbohydrates for training and competition. J Sports Sci. (2011) 29(Suppl. 1):S17–27. 10.1080/02640414.2011.585473 [DOI] [PubMed] [Google Scholar]
- 110.Stellingwerff T, Cox GR. Systematic review: carbohydrate supplementation on exercise performance or capacity of varying durations. Appl Physiol Nutr Metab. (2014) 39:998–1011. 10.1139/apnm-2014-0027 [DOI] [PubMed] [Google Scholar]
- 111.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. (2014) 20:779–86. 10.1016/j.cmet.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chassard C, Lacroix C. Carbohydrates and the human gut microbiota. Curr Opin Clin Nutr Metab Care. (2013) 16:453–60. 10.1097/MCO.0b013e3283619e63 [DOI] [PubMed] [Google Scholar]
- 113.Hamaker BR, Tuncil YE. A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol. (2014) 426:3838–50. 10.1016/j.jmb.2014.07.028 [DOI] [PubMed] [Google Scholar]
- 114.Sharma P, Bhandari C, Kumar S, Sharma B, Bhadwal P, Agnihotri N. Dietary fibers: a way to a healthy microbiome. Diet Microbiome Health. (2018) 2018:299–345. 10.1016/B978-0-12-811440-7.00011-9 [DOI] [Google Scholar]
- 115.Paoli A, Bianco A, Grimaldi KA. The ketogenic diet and sport: a possible marriage? Exerc Sport Sci Rev. (2015) 43:153–62. 10.1249/JES.0000000000000050 [DOI] [PubMed] [Google Scholar]
- 116.Fritzen AM, Lundsgaard AM, Kiens B. Dietary fuels in athletic performance. Annu Rev Nutr. (2019) 39:45–73. 10.1146/annurev-nutr-082018-124337 [DOI] [PubMed] [Google Scholar]
- 117.Paoli A, Mancin L, Bianco A, Thomas E, Mota JF, Piccini F. Ketogenic diet and microbiota: friends or enemies? Genes. (2019) 10:534 10.3390/genes10070534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ellerbroek A. The effect of ketogenic diets on the gut microbiota. J Exerc Nutr. (2018) 1:534 Available online at: https://www.journalofexerciseandnutrition.com/ManuscriptUploadsPDF/65.pdf [Google Scholar]
- 119.Nishitsuji K, Watanabe S, Xiao J, Nagatomo R, Ogawa H, Tsunematsu T, et al. Effect of coffee or coffee components on gut microbiome and short-chain fatty acids in a mouse model of metabolic syndrome. Sci Rep. (2018) 8:16173. 10.1038/s41598-018-34571-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Casazza GA, Tovar AP, Richardson CE, Cortez AN, Davis BA. Energy availability, macronutrient intake, and nutritional supplementation for improving exercise performance in endurance athletes. Curr Sport Med Rep. (2018) 17:215–23. 10.1249/JSR.0000000000000494 [DOI] [PubMed] [Google Scholar]
- 121.Jeukendrup A. A step towards personalized sports nutrition: carbohydrate intake during exercise. Sports Med. (2014) 44(Suppl. 1): S25–33. 10.1007/s40279-014-0148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Margaritelis NV, Paschalis V, Theodorou AA, Kyparos A, Nikolaidis MG. Antioxidants in personalized nutrition and exercise. Adv Nutr. (2018) 9:813–23. 10.1093/advances/nmy052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Goñi I, Serrano J, Saura-Calixto F. Bioaccessibility of β-carotene, lutein, and lycopene from fruits and vegetables. J Agr Food Chem. (2006) 54:5382–7. 10.1021/jf0609835 [DOI] [PubMed] [Google Scholar]
- 124.Palafox-Carlos H, Ayala-Zavala JF, González-Aguilar GA. The role of dietary fiber in the bioaccessibility and bioavailability of fruit and vegetable antioxidants. J Food Sci. (2011) 76:R6–15. 10.1111/j.1750-3841.2010.01957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Selma MV, Espin JC, Tomas-Barberan FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. (2009) 57:6485–501. 10.1021/jf902107d [DOI] [PubMed] [Google Scholar]
- 126.Meinl W, Sczesny S, Brigelius-Flohe R, Blaut M, Glatt H. Impact of gut microbiota on intestinal and hepatic levels of phase 2 xenobiotic-metabolizing enzymes in the rat. Drug Metab Dispos. (2009) 37:1179–86. 10.1124/dmd.108.025916 [DOI] [PubMed] [Google Scholar]
- 127.Djuric Z, Bassis CM, Plegue MA, Ren J, Chan R, Sidahmed E, et al. Colonic mucosal bacteria are associated with inter-individual variability in serum carotenoid concentrations. J Acad Nutr Diet. (2018) 118:606–16. e3. 10.1016/j.jand.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Durk RP, Castillo E, Marquez-Magana L, Grosicki GJ, Bolter ND, Lee CM, et al. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int J Sport Nutr Exerc Metab. (2018) 29:249–53. 10.1123/ijsnem.2018-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. (2016) 4:42. 10.1186/s40168-016-0189-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang Y, Shi Y, Wiklund P, Tan X, Wu N, Zhang X, et al. The association between cardiorespiratory fitness and gut microbiota composition in premenopausal women. Nutrients. (2017) 9:792. 10.3390/nu9080792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hsu YJ, Chiu CC, Li YP, Huang WC, Huang YT, Huang CC, et al. Effect of intestinal microbiota on exercise performance in mice. J Strength Cond Res. (2015) 29:552–8. 10.1519/JSC.0000000000000644 [DOI] [PubMed] [Google Scholar]
- 132.Huang WC, Chen YH, Chuang HL, Chiu CC, Huang CC. Investigation of the effects of microbiota on exercise physiological adaption, performance, and energy utilization using a gnotobiotic animal model. Front Microbiol. (2019) 10:1906. 10.3389/fmicb.2019.01906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen YM, Wei L, Chiu YS, Hsu YJ, Tsai TY, Wang MF, et al. Lactobacillus plantarum TWK10 supplementation improves exercise performance and increases muscle mass in mice. Nutrients (2016) 8:205 10.3390/nu8040205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang WC, Hsu YJ, Li H, Kan NW, Chen YM, Lin JS, et al. Effect of Lactobacillus plantarum TWK10 on improving endurance performance in humans. Chin Physiol J. (2018) 61:163–70. 10.4077/CJP.2018.BAH587 [DOI] [PubMed] [Google Scholar]
- 135.Jäger R, Shields KA, Lowery RP, De Souza EO, Partl JM, Hollmer C, et al. Probiotic Bacillus coagulans GBI-30, 6086 reduces exercise-induced muscle damage and increases recovery. Peer J. (2016) 4:e2276. 10.7717/peerj.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lamprecht M, Bogner S, Schippinger G, Steinbauer K, Fankhauser F, Hallstroem S, et al. Probiotic supplementation affects markers of intestinal barrier, oxidation, and inflammation in trained men; a randomized, double-blinded, placebo-controlled trial. J Int Soc Sport Nutr. (2012) 9:45. 10.1186/1550-2783-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martarelli D, Verdenelli MC, Scuri S, Cocchioni M, Silvi S, Cecchini C, et al. Effect of a probiotic intake on oxidant and antioxidant parameters in plasma of athletes during intense exercise training. Curr Microbiol. (2011) 62:1689–96. 10.1007/s00284-011-9915-3 [DOI] [PubMed] [Google Scholar]
- 138.Salarkia N, Ghadamli L, Zaeri F, Rad LS. Effects of probiotic yogurt on performance, respiratory and digestive systems of young adult female endurance swimmers: a randomized controlled trial. Med J Islam Repub Iran. (2013) 27:141–6. [PMC free article] [PubMed] [Google Scholar]
- 139.Shing CM, Peake JM, Lim CL, Briskey D, Walsh NP, Fortes MB, et al. Effects of probiotics supplementation on gastrointestinal permeability, inflammation and exercise performance in the heat. Eur J Appl Physiol. (2014) 114:93–103. 10.1007/s00421-013-2748-y [DOI] [PubMed] [Google Scholar]
- 140.Townsend J, Bender D, Vantrease W, Sapp P, Toy A, Woods C, et al. Effects of Probiotic (Bacillus subtilis DE111) supplementation on immune function, hormonal status, and physical performance in division I baseball players. Sports. (2018) 6:70. 10.3390/sports6030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. (2019) 25:1104–9. 10.1038/s41591-019-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soares ADN, Wanner SP, Morais ESS, Hudson ASR, Martins FS, Cardoso VN. Supplementation with Saccharomyces boulardii increases the maximal oxygen consumption and maximal aerobic speed attained by rats subjected to an incremental-speed exercise. Nutrients. (2019) 11:2352. 10.3390/nu11102352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moreno-Navarrete JM, Fernandez-Real JM. The gut microbiota modulates both browning of white adipose tissue and the activity of brown adipose tissue. Rev Endocr Metab Disord. (2019). [Epub ahead of print]. 10.1007/s11154-019-09523-x [DOI] [PubMed] [Google Scholar]
- 144.Nichols AW. Probiotics and athletic performance: a systematic review. Curr Sport Med Rep. (2007) 6:269–73. 10.1007/s11932-007-0044-5 [DOI] [PubMed] [Google Scholar]
- 145.Pyne DB, West NP, Cox AJ, Cripps AW. Probiotics supplementation for athletes - clinical and physiological effects. Eur J Sport Sci. (2015) 15:63–72. 10.1080/17461391.2014.971879 [DOI] [PubMed] [Google Scholar]
- 146.Wosinska L, Cotter PD, O'Sullivan O, Guinane C. The potential impact of probiotics on the gut microbiome of athletes. Nutrients. (2019) 11:2270. 10.3390/nu11102270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang WC, Lee MC, Lee CC, Ng KS, Hsu YJ, Tsai TY, et al. Effect of Lactobacillus plantarum TWK10 on exercise physiological adaptation, performance, and body composition in healthy humans. Nutrients. (2019) 11:2836. 10.3390/nu11112836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nay K, Jollet M, Goustard B, Baati N, Vernus B, Pontones M, et al. Gut bacteria are critical for optimal muscle function: a potential link with glucose homeostasis. Am J Physiol Endocrinol Metab. (2019) 317:E158–71. 10.1152/ajpendo.00521.2018 [DOI] [PubMed] [Google Scholar]
- 149.Okamoto T, Morino K, Ugi S, Nakagawa F, Lemecha M, Ida S, et al. Microbiome potentiates endurance exercise through intestinal acetate production. Am J Physiol Endocrinol Metab. (2019) 316:E956–66. 10.1152/ajpendo.00510.2018 [DOI] [PubMed] [Google Scholar]
- 150.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. (2015) 22:971–82. 10.1016/j.cmet.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 151.Ticinesi A, Lauretani F, Milani C, Nouvenne A, Tana C, Del Rio D, et al. Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: is there a gut–muscle axis? Nutrients. (2017) 9:1303. 10.3390/nu9121303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hecksteden A, Kraushaar J, Scharhag-Rosenberger F, Theisen D, Senn S, Meyer T. Individual response to exercise training - a statistical perspective. J Appl Physiol. (2015) 118:1450–9. 10.1152/japplphysiol.00714.2014 [DOI] [PubMed] [Google Scholar]
- 153.Mann TN, Lamberts RP, Lambert MI. High responders and low responders: factors associated with individual variation in response to standardized training. Sports Med. (2014) 44:1113–24. 10.1007/s40279-014-0197-3 [DOI] [PubMed] [Google Scholar]
- 154.Swinton PA, Hemingway BS, Saunders B, Gualano B, Dolan E. A statistical framework to interpret individual response to intervention: paving the way for personalized nutrition and exercise prescription. Front Nutr. (2018) 5:41. 10.3389/fnut.2018.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hopkins WG. Individual responses made easy. J Appl Physiol. (2015) 118:1444–6. 10.1152/japplphysiol.00098.2015 [DOI] [PubMed] [Google Scholar]
- 156.Breiman L, Random forests. Mach Learn. (2001) 45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 157.Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. Am J Clin Nutr. (2001) 73:399s–405s. 10.1093/ajcn/73.2.399s [DOI] [PubMed] [Google Scholar]
- 158.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. (2018) 174:1388–405 e21. 10.1016/j.cell.2018.08.041 [DOI] [PubMed] [Google Scholar]
- 159.Zhang C, Derrien M, Levenez F, Brazeilles R, Ballal SA, Kim J, et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. (2016) 10:2235. 10.1038/ismej.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Senan S, Prajapati JB, Joshi CG, Sreeja V, Gohel MK, Trivedi S, et al. Geriatric respondents and non-respondents to probiotic intervention can be differentiated by inherent gut microbiome composition. Front Microbiol. (2015) 6:25. 10.3389/fmicb.2015.00944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. (2014) 159:789–99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics. (2016) 11:205–15. 10.1080/15592294.2016.1155011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Benedict C, Vogel H, Jonas W, Woting A, Blaut M, Schürmann A, et al. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol Metab. (2016) 5:1175–86. 10.1016/j.molmet.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fullagar HH, Skorski S, Duffield R, Hammes D, Coutts AJ, Meyer T. Sleep and athletic performance: the effects of sleep loss on exercise performance, and physiological and cognitive responses to exercise. Sports Med. (2015) 45:161–86. 10.1007/s40279-014-0260-0 [DOI] [PubMed] [Google Scholar]
- 165.Tarnopolsky L, MacDougall J, Atkinson S, Tarnopolsky M, Sutton J. Gender differences in substrate for endurance exercise. J Appl Physiol. (1990) 68:302–8. 10.1152/jappl.1990.68.1.302 [DOI] [PubMed] [Google Scholar]
- 166.Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PloS ONE. (2016) 11:e0154090. 10.1371/journal.pone.0154090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.O'Toole PW, Claesson MJ. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int Dairy J. (2010) 20:281–91. 10.1016/j.idairyj.2009.11.010 [DOI] [Google Scholar]
- 168.Ni Lochlainn M, Bowyer R, Steves C. Dietary protein and muscle in aging people: the potential role of the gut microbiome. Nutrients. (2018) 10:929. 10.3390/nu10070929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shin HE, Kwak SE, Lee JH, Zhang D, Bae JH, Song W. Exercise, the gut microbiome, and frailty. Ann Geriatr Med Res. (2019) 23:105–14. 10.4235/agmr.19.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Grosicki GJ, Fielding RA, Lustgarten MS. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: biological basis for a gut-muscle axis. Calcified Tissue Int. (2018) 102:433–42. 10.1007/s00223-017-0345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Claesson MJ, Jeffery IB, Conde S, Power SE, O'connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. (2012) 488:178. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 172.Ticinesi A, Tana C, Nouvenne A. The intestinal microbiome and its relevance for functionality in older persons. Curr Opin Clin Nutr Metab Care. (2019) 22:4–12. 10.1097/MCO.0000000000000521 [DOI] [PubMed] [Google Scholar]
- 173.Siddharth J, Chakrabarti A, Pannerec A, Karaz S, Morin-Rivron D, Masoodi M, et al. Aging and sarcopenia associate with specific interactions between gut microbes, serum biomarkers and host physiology in rats. Aging. (2017) 9:1698. 10.18632/aging.101262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ticinesi A, Lauretani F, Tana C, Nouvenne A, Ridolo E, Meschi T. Exercise and immune system as modulators of intestinal microbiome: implications for the gut-muscle axis hypothesis. Exerc Immunol Rev. (2019) 25:84–95. Available online at: http://eir-isei.de/2019/eir-2019-084-article.pdf [PubMed] [Google Scholar]
- 175.Gubert C, Kong G, Renoir T, Hannan AJ. Exercise, diet and stress as modulators of gut microbiota: implications for neurodegenerative diseases. Neurobiol Dis. (2019) 134:104621. 10.1016/j.nbd.2019.104621 [DOI] [PubMed] [Google Scholar]
- 176.Schlegel P, Novotny M, Klimova B, Valis M. “Muscle-gut-brain axis”: can physical activity help patients with Alzheimer's disease due to microbiome modulation? J Alzheimers Dis. (2019) 71:861–78. 10.3233/JAD-190460 [DOI] [PubMed] [Google Scholar]
- 177.Buigues C, Fernández-Garrido J, Pruimboom L, Hoogland A, Navarro-Martínez R, Martínez-Martínez M, et al. Effect of a prebiotic formulation on frailty syndrome: a randomized, double-blind clinical trial. Int J Mol Sci. (2016) 17:932. 10.3390/ijms17060932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Walsh ME, Bhattacharya A, Sataranatarajan K, Qaisar R, Sloane L, Rahman MM, et al. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. (2015) 14:957–70. 10.1111/acel.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-dit-Félix AA, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. (2016) 22:879. 10.1038/nm.4132 [DOI] [PubMed] [Google Scholar]
- 180.Andreux PA, Blanco-Bose W, Ryu D, Burdet F, Ibberson M, Aebischer P, et al. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat Metab. (2019) 1:595–603. 10.1038/s42255-019-0073-4 [DOI] [PubMed] [Google Scholar]