Highlights
-
•
The structure of a child brain is significantly different from an adult brain.
-
•
Standard software tools for processing brain MRI data might not be appropriate for analyzing pediatric neuroimaging data.
-
•
Age-specific and 4D brain MRI atlases have shown to improve the results for brain extraction, normalization, and segmentation.
-
•
Image quality enhancement and longitudinal registration are important processing steps for analysis of pediatric samples.
Keywords: Structural MRI, Children, Neural development, Neuroimaging methods, Brain atlas, Longitudinal analysis
Abstract
The structure of the brain is subject to very rapid developmental changes during early childhood. Pediatric studies based on Magnetic Resonance Imaging (MRI) over this age range have recently become more frequent, with the advantage of providing in vivo and non-invasive high-resolution images of the developing brain, toward understanding typical and atypical trajectories. However, it has also been demonstrated that application of currently standard MRI processing methods that have been developed with datasets from adults may not be appropriate for use with pediatric datasets. In this review, we examine the approaches currently used in MRI studies involving young children, including an overview of the rationale for new MRI processing methods that have been designed specifically for pediatric investigations. These methods are mainly related to the use of age-specific or 4D brain atlases, improved methods for quantifying and optimizing image quality, and provision for registration of developmental data obtained with longitudinal designs. The overall goal is to raise awareness of the existence of these methods and the possibilities for implementing them in developmental neuroimaging studies.
1. Introduction
Neuroimaging has gained importance and achieved widespread use in research on neurodevelopmental disorders during the past decade (Dennis and Thompson, 2013). Neuroimaging studies have traditionally focused on understanding the structure and the function of the (ab)normal adult brain. At present, research designed to study typical and atypical development also consists of investigating the brain before or at the onset of the disorder and combines this with neurobehavioral follow-up longitudinally. Although this approach is demanding, both in terms of feasibility and time investment, this type of study is needed to gain deeper insight in the underlying mechanisms of neurodevelopmental disorders. These studies can elucidate how the human brain changes throughout typical and atypical development and how these changes relate to cognitive, social and perceptual abilities. In the long-term, these designs can enhance the early identification and remediation of neurodevelopmental disorders, toward improving quality of life outcomes.
In contrast to electroencephalography (EEG) that has been applied in young children for decades (Brown and Jernigan, 2012, Holmes and Lombroso, 1993, Shaul, 2008, Vanhatalo and Kaila, 2006), investigations based on Magnetic Resonance Imaging (MRI) have only begun to increase in frequency. At present, MRI can be readily applied for use with young children thanks to the development of child-friendly protocols and technological advances that decrease the scanning time needed to achieve acceptable signal-to-noise ratios (Greene et al., 2016, Raschle et al., 2012, Vogel et al., 2016). The particular advantage of MRI over other techniques is associated with its ability to localize neurobiological deficits non-invasively and with high spatial precision, thereby providing high-resolution images of the brain in vivo. Because MRI does not apply ionizing radiation, repeated scans can typically be made on the same individual, enabling visualization of longitudinal changes in brain development and/or anomalies over time.
Given the importance of MRI research to investigations of children and throughout the timecourse of development, we aim to provide a review of MRI processing methods that have been developed for analyzing the young, developing brain. Previous review papers have focused on MRI acquisition in young children, and especially on child-friendly protocols (Greene et al., 2016), but to date have not been complemented by a similar synthesis on data processing in young children (here, defined as children under 6 years of age). Here, we summarize: 1. why child-adjusted MRI processing techniques are necessary (in Section 2); 2. which MRI processing approaches are currently applied in studies involving young children and which ones are particularly problematic (in Section 3); and 3. which MRI processing techniques may be best suited and adjusted to examine the pediatric brain (in Sections 4–6). Through this review, we aim to raise the awareness of the emergence of child-adjusted methods in the clinical and neuroscientific community and to encourage the use of such methods to achieve more accurate and reliable results in relevant studies.
2. The rationale behind child-adjusted MRI processing
2.1. Neuroanatomical specifics of children’s brains
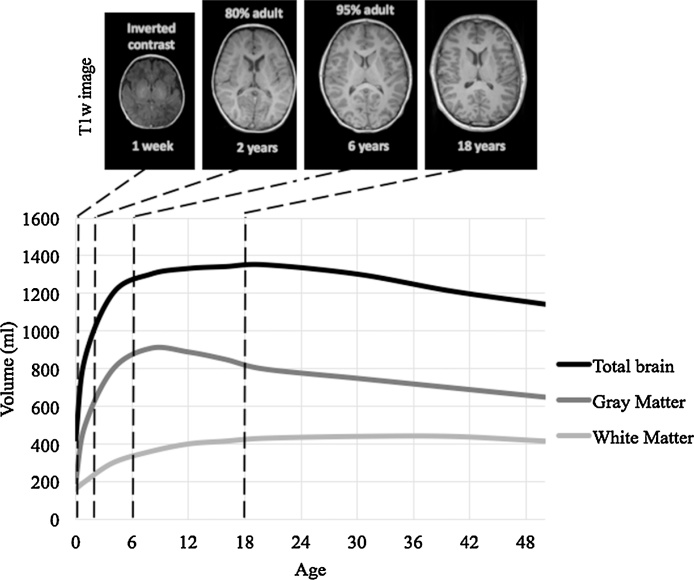
A child’s brain is not simply a scaled-down version of an adult brain, as it undergoes a series of non-linear changes throughout its development (Wilke et al., 2003b). As illustrated in Fig. 1, anatomical changes in the brain are cascaded over time and with trajectories of development that differ substantially over each type of brain tissue.
Fig. 1.
Schematic developmental trajectories for mean volumes of brain tissues in the normative population across age (adapted from Figs. 2–3 of (Courchesne et al., 2000) and Figs. 2–3 of (Knickmeyer et al., 2008)). Children’s brains are significantly different from the adult brain as the organization and properties of brain structures change with age.
The volume of the brain as a whole increases significantly between early childhood (1–3 years old) and adolescence (12–15 years old) when it reaches its maximum (Courchesne et al., 2000). The rate of increase is particularly substantial from birth to two years old, achieving 80% of its adult size at the age of 2 years and increasing to 95% at approximately 6 years of age (Knickmeyer et al., 2008, Lenroot and Giedd, 2006). Subsequently, the whole brain volume remains roughly stable from mid-adolescence until ages in the mid-thirties, when it starts to decrease considerably (Hedman et al., 2012). This volumetric stability between adolescence and mid-thirties suggests the absence of morphological changes, but instead changes in gray and in white matter tend to counterbalance each other, resulting in minimal changes in whole brain volumes during this period (Lebel and Beaulieu, 2011). Despite these consistencies, the brain size and developmental trajectories of brain structure are highly variable between persons, even for those with similar age and the same gender (Aubert-Broche et al., 2013, Dekaban and Sadowsky, 1978, Lenroot and Giedd, 2006).
Gray matter volume mainly increases from birth to later childhood (6–9 years old), with a developmental trajectory across the lifespan that follows an inverted U-shaped curve during the two first decades of the lifespan, with regionally specific rate of changes (Brain Development Cooperative Group, 2012a, Giedd et al., 2015), resulting in local maxima that are reached at different time points (Lenroot and Giedd, 2006). For example, maturation of the gray matter is completed first in primary sensorimotor and occipital visual areas and the latest in higher-order association cortex (Deoni et al., 2015, Gogtay et al., 2004, Westlye et al., 2010a). Previous studies have reported that cortical gray matter volumes peak near the onset of puberty (Giedd et al., 1999; Lenroot et al., 2007). However, more recent studies based on large longitudinal datasets suggested that such gray matter volume reaches a maximum during childhood and then decreases during adolescence and adulthood beginning in the mid-twenties (Aubert-Broche et al., 2013, Courchesne et al., 2000, Hedman et al., 2012, Mills et al., 2016, Mills and Tamnes, 2014). The mechanism by which cortical gray matter is reduced after childhood is linked to synaptic pruning (Petanjek et al., 2011, Webster et al., 2011, Whitford et al., 2007), although a direct causal relationship between synaptic anatomy and gray matter changes from MRI measurements has not been verified (Mills et al., 2016, Mills and Tamnes, 2014). Subcortical gray matter structures, such as in the striatum and thalamus, appear to attain maximum volume later than do cortical structures (Raznahan et al., 2014), and are substantially variable in their developmental trajectories. Such variability is associated, with hemisphere and gender effects, as well as puberty-related changes (Goddings et al., 2014, Mills and Tamnes, 2014).
In contrast to gray matter, volumetric measures of white matter increase at approximately 1–2% per year between childhood and adolescence, only reaching asymptote in mid-adulthood (Giedd et al., 2015, Mills and Tamnes, 2014). More recent longitudinal studies suggested that white matter volume increases until 10–15 years old, then decreases in the early twenties, and increases again before the plateau in the fourth decade (Hedman et al., 2012, Mills et al., 2016). Beyond this, a fall off starts from the sixth decade onwards (Courchesne et al., 2000, Westlye et al., 2010b). White matter maturation is mainly influenced by myelination, the mechanism for which consists of the deposition of fatty acid by Schwann cells around the axonal membrane. Myelination is more active during development but continues during adulthood at a slower rate. For example, the association cortices still myelinate during the second and third decades (Yakovlev and Lecours, 1967). Synaptic pruning acts to eliminate redundant neurons, so this process may also influence measures of white matter maturation (Yeatman et al., 2012), such as the decelerating increase in late childhood (6–9 years) when pruning seems to be particularly active. The developmental trajectory for white matter is relatively consistent across the major lobes in cortex (i.e. frontal, temporal and parietal lobes), but with some sub-regional variability (Thompson et al., 2000). Maturation of white matter starts in the proximal, sensory and projection pathways and initiates latest in more distal, motor and association pathways (Volpe, 2000). Changes in white matter organization is relatively complete by late adolescence for projection and commissural tracts, yet continues after adolescence for projection tracts (Lebel and Beaulieu, 2011, Westlye et al., 2010b).
As the mechanisms of myelination and pruning affect tissue properties (e.g. T1 and T2 relaxation time), measurements of gray matter and white matter volume are likely to be impacted by these mechanisms (Grydeland et al., 2013, Sowell et al., 2001, Westlye et al., 2010b). Consequently, the signal intensity values and thus, the contrast in MRI scans, change with age. With unmyelinated white matter during the first 6 months (Prastawa et al., 2005, Tau and Peterson, 2010), the brain appears with inverted contrast on MRI scans compared to the adult brain. By the age of two years, the contrast gradually becomes adult-like, with the majority of white matter structures being myelinated. Hence, MRI images from young children demonstrate smaller gray to white matter contrast than do images from adults (see Fig. 1). A lower contrast renders it more difficult to distinguish the boundaries between tissues, which is prerequisite to further MRI-analysis steps, particularly in segmentation (Murgasova et al., 2007, Prastawa et al., 2005).
To summarize, very drastic changes in brain development take place within the first six years, after which brain changes continue but at a slower rate. Due to the more prominent changes in the brain early in life, neuroimaging analyses on data collected from young children must be carried out with special considerations regarding the different image contrast and brain morphology compared to adults.
2.2. Quality of MRI data in children
Overcoming poor quality MRI data is another important issue for the analysis of neuroimaging data obtained from pediatric populations. The scanning environment provides a new and unusual experience for most children, who may feel uncomfortable for a number of reasons including, the confined space, fear of the unknown, difficulty for lying still during a long period and the ambient noise produced by the switching of gradients (Marshall et al., 1995). Hence, the risk of obtaining low quality images, particularly due to motion artifacts, is higher in young children than in the older population (Brown et al., 2010, Davidson et al., 2003, Wilke et al., 2003a). For example, Theys et al. reported that 11 percent of scans obtained from young children displayed extreme movement compared to 6 percent for adults, with much higher motion displacement in children (Theys et al., 2014).
Motion artifacts occur as blurring and/or ghosting artifacts of the image, hindering the identification of borders between brain structures and tissue types. Typically, MRI data with obvious motion artifacts are excluded from the analyses. With motion artifacts as the main reason for data exclusion in non-sedated young children, the success rate for data processing was reported to be 92% for participants between 5 and 19 years old (Castellanos et al., 2002), 58–82% for children between 5 and 11 years old (Bora et al., 2014, Sowell, 2004) and 60% for babies of 2–4 weeks (Knickmeyer et al., 2008). Moreover, a higher risk of failure is expected when studying children with developmental neurocognitive disorders, such as autism or attention deficit hyperactivity disorder (ADHD), which may affect their ability to comply to the demands of a particular scanning protocol (Castellanos et al., 2002).
Data acquisition is already difficult in cross-sectional studies involving young children, and is even more difficult for longitudinal designs which depend on data quality for the same participants across multiple time points. In longitudinal designs, the willingness to participate in several scanning sessions is not guaranteed and the number of participants tends to decrease with the number of times that data are collected (Castellanos et al., 2002, Choe et al., 2013, Knickmeyer et al., 2008). Participant dropout might be more critical for longitudinal studies on neurodevelopmental disorders for which children are recruited before diagnosis and atypical groups start with a relatively small number of participants, leading to statistical power issues (Clark et al., 2014).
Solutions toward minimizing head motion and participant dropout when scanning young children are used to ensure that age-appropriate experimental designs and child-friendly protocols are employed when possible (Greene et al., 2016, Raschle et al., 2012, Vogel et al., 2016). However, even with these solutions, motion artifacts remain a problem inherent to neuroimaging studies of pediatric populations. Even motion that is not readily visible by visual inspection may lead to systematic and regionally specific biases in structural measurements (Alexander-Bloch et al., 2016, Blumenthal et al., 2002, Reuter et al., 2015). Motion artifacts should therefore always be considered in analyses to avoid spurious interpretation, such as genuine group differences that are hidden, or conversely, observed group differences that are in fact not actually present (Yendiki et al., 2014).
3. Current practice in developmental studies using MRI
In this section, we review the processing methods that are currently the standard in developmental MRI-studies on typical and atypical brain development in young children. We define “young children” as children under 6 years of age because drastic structural changes in brain take place until this age (see Section 2.1, Fig. 1).
3.1. Selection of MRI-studies on (a)typical brain development in young children
For our review, we selected relevant MRI-studies of typical brain development (see Table 1) and of developmental neurocognitive disorders (see Table 2), namely autism, attention deficit/hyperactivity disorder (ADHD) and dyslexia. These studies were first identified through four review papers: in Dennis and Thompson (2013) regarding typical development, autism and ADHD, in Hoogman et al. (2017) for ADHD, in Ozernov-Palchik and Gaab (2016) and Vandermosten et al. (2016) for dyslexia. We complemented them with recent studies (between 2007 and 2017) found in the search engine, PubMed, with the combination of the following key words: (brain) and (development or maturation or growth) and (pediatric or children or infant or neonate or newborns) and (normal or typical or healthy)/(dyslexia or autism or attention deficit hyperactivity disorder) and (MRI or magnetic resonance and T1 or T2 or structural or morphometry or volume or surface area or cortical thickness).
Table 1.
Studies contributing to the review of typical brain development of young children (under 6 years old).
| Study | N | Ages | Measures | Study type |
|---|---|---|---|---|
| Typical Brain development (TD) | ||||
| Sowell, 2004 | 45 TD (22F) | 5–12yo | Cortical thickness | Longitudinal |
| Knickmeyer et al., 2008 | 98 TD (49F) | 0–2yo | Brain volumes | Longitudinal |
| Gilmore et al., 2012 | 72 TD | 0–2yo | Brain volumes | Longitudinal |
| Lebel and Beaulieu, 2011 | 103 TD (52F) | 5–30yo | Brain volumes | Longitudinal |
| Diffusion measures | ||||
| Brain Development Cooperative Group, 2012a, Brain Development Cooperative Group, 2012b | 325 TD | 4.5–18yo | Brain volumes | Cross-sectional |
| Longitudinal | ||||
| Brown et al., 2012 | 885 TD (423F) | 3–20yo | Brain volumes | Cross-sectional |
| Cortical thickness | ||||
| Surface area | ||||
| Diffusion measures | ||||
| Choe et al., 2013 | 27 TD | 3–12mo | Brain volumes | Longitudinal |
| Hu et al., 2013 | 306 TD | 4–18yo | Brain volumes | Cross-sectional |
| Nie et al., 2013 | 445 TD | 3–20yo | Cortical thickness | Cross-sectional |
| Curvature | ||||
| Connectivity | ||||
| Holland et al., 2014 | 87 TD (48F) | 0–3mo | Brain volumes | Cross-sectional |
| Krogsrud et al., 2014 | 244 (218F) | 4–22yo | Brain volumes | Cross-sectional |
| Li et al., 2014b | 73 TD | 0–2yo | Cortical gyrification | Longitudinal |
| Lyall et al., 2015 | 71 TD | 0–2yo | Cortical thickness | Longitudinal |
| Surface area | ||||
TD, typical development; F, female; yo, years old; mo, months
Table 2.
Studies of developmental neurocognitive disorders including young children (under 6 years old, that contributed to the review.
| Study | N | Ages | Measures | Study type |
|---|---|---|---|---|
| Autism Spectrum Disorder (ASD) | ||||
| Nordahl et al., 2011 | 114 ASD (22F) | 2–4yo | Brain volumes | Cross-sectional |
| 66 TD (24F) | ||||
| Shen et al., 2013 | 41 HR | 6–36mo | Brain volumes | Longitudinal |
| 23 LR | ||||
| Hazlett et al., 2012 | 98 HR | 6 mo | Brain volumes | Cross-sectional |
| 36 LR | ||||
| Hazlett et al., 2017 | 106 HR | 6–24mo | Brain volumes | Longitudinal |
| 42 LR | Cortical thickness | |||
| Surface area | ||||
| Attention deficit hyperactivity disorder (ADHD) | ||||
| Castellanos et al., 2002 | 151 ADHD (63F) | 4.5–19yo | Brain volumes | Longitudinal |
| 139 TD (56F) | ||||
| Shaw et al., 2007 | 223 ADHD | 4–25yo | Cortical thickness | Longitudinal |
| 223 TD | ||||
| Shaw et al., 2009 | 218 ADHD | 3–22yo | Cortical thickness | Longitudinal |
| 358 TD | ||||
| Bora et al., 2014 | 110 preterm | 4–9yo | Brain volumes | Cross-sectional |
| 113 full term | ||||
| Yang et al., 2015 | 25 ADHD | 5–12yo | Cortical thickness | Cross-sectional |
| 25 TD | ||||
| Dyslexia (DYS) | ||||
| Raschle et al., 2011 | 10 HR | 5–6yo | Brain volumes | Cross-sectional |
| 10 LR | ||||
| Black et al., 2012 | 27 HR (12F) | 5–6yo | Brain volumes | Cross-sectional |
| 24 LR (10F) | Cortical thickness | |||
| Surface area | ||||
| Hosseini et al., 2013 | 22 HR (9F) | 5–6yo | Brain volumes | Cross-sectional |
| 20 LR (11F) | Cortical thickness | |||
| Surface area | ||||
| Connectivity | ||||
| Clark et al., 2014 | 11 DYS | 5–11 yo | Cortical thickness | Longitudinal |
| 16 TD | ||||
| Im et al., 2016 | 15 HR | 4–13 yo | Sulcal graph | Cross-sectional |
| 16 LR | ||||
| 15 DYS | ||||
| 13 TD | ||||
| Vanderauwera et al., 2016 | 36 HR | 5–6 yo | Surface area | Cross-sectional |
| 35 LR | ||||
TD, typical development; F, female; ASD, autism spectrum disorder; ADHD, attention deficit hyperactivity disorder; DYS, dyslexia; HR, high risk; LR, low risk; yo, years old; mo, months
From this initial sample of studies, we selected the sample for review using the following criteria for inclusion: (1) the studies contained at least five children below 6 years old, (2) the studies includeed children who were not sedated during scanning and (3) the studies described processing methods of structural T1 or T2 MRI-analyses. These processing methods are of specific interest on their own and are also relevant- and often crucial- for co-registration and normalization of other modalities, such as diffusion MRI and functional MRI. On the other hand, studies were excluded when (1) the main focus was on genetic or environmental influences on brain structures, when (2) the paper comprised a review or meta-analysis or when (3) studies applied diffusion MRI (to assess tissue microstructure) or functional MRI (to assess activation patterns) as a primary focus.
3.2. Current practice of MRI data processing for pediatric samples
Following acquisition, there are a number of different options for statistical analysis of the structural properties extracted from MRI scans. First, published neurodevelopmental studies have typically drawn associations between morphometric changes in brain and age reported with graphical illustrations of developmental trajectory curves or through summary brain maps reflecting percent changes at different spatial locations (Brain Development Cooperative Group, 2012a, Choe et al., 2013, Gilmore et al., 2012, Holland et al., 2014, Knickmeyer et al., 2008, Krogsrud et al., 2014, Lebel and Beaulieu, 2011, Lyall et al., 2015). Secondly, studies of neurodevelopmental disorders have generally compared results between control and clinical groups to highlight disorder-specific differences, which are then correlated with measures of cognitive ability that assess the degree of behavioral impairment (Bora et al., 2014, Castellanos et al., 2002, Hazlett et al., 2017, Nordahl et al., 2011, Raschle et al., 2011, Shen et al., 2013, Yang et al., 2015). Third, analyses of regional or whole-brain networks have been applied to investigate structural connectivity patterns, inferred from correlations between brain regions based on MRI measures (Hosseini et al., 2013, Im et al., 2016, Nie et al., 2013).
Prior to statistical analysis (e.g. group comparisons, correlations), data processing is a pre-requisite step for obtaining relevant structural measures such as brain volumes, cortical thickness or surface area. Data processing for structural analysis generally consists of brain extraction, segmentation and normalization. As manual measurement of structural parameters is laborious, time-consuming and subject to rater-biases, automated processing methods were developed to streamline these processes. Software tools for automated processing are publicly available and many of them have been applied for analysis of data in pediatric samples, as shown in Table 3.
Table 3.
Standard software tools for MRI processing and their application for pediatric data.
| Software tool | MRI processing | Application in studies on young children |
|---|---|---|
| Autoseg (Wang et al., 2014) – https://www.nitrc.org/projects/autoseg/ | N4 bias field correction, noise reduction, rigid registration, skull-stripping, intensity rescaling, multi-atlas segmentation | Group comparisons based on brain volumes (Hazlett et al., 2017, Hazlett et al., 2012) |
| BrainVISA/Anatomist (Rivière et al., 2003) – http://brainvisa.info | Data visualization, manual tissue labeling | Segmentation of planum temporale (Vanderauwera et al., 2016) |
| Freesurfer (Dale et al., 1999, Fischl et al., 1999) – http://freesurfer.net/ | Motion correction, brain extraction, volumetric segmentation, surface extraction, cortical parcellation and reconstruction longitudinal processing and analysis | Morphometric analysis (Brown et al., 2012) Graph theory analysis based on structural correlation network (Hosseini et al., 2013, Im et al., 2016) |
| Correlation between structural measures and risk factors (Black et al., 2012) | ||
| Group comparisons based on cortical thickness (Clark et al., 2014, Yang et al., 2015) | ||
| Hippocampus segmentation (Krogsrud et al., 2014) | ||
| Longitudinal volumetric analysis (Lebel and Beaulieu, 2011) | ||
| FSL (Jenkinson et al., 2012, Smith et al., 2004) https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/ | ||
| - BET (Smith, 2002) | Brain extraction | Brain masking (Brain Development Cooperative Group, 2012b, Nordahl et al., 2011, Shaw et al., 2009, Shen et al., 2013) |
| - FAST (Zhang et al., 2001) | Brain segmentation | Structural correlation network based on cortical thickness (Nie et al., 2013) |
| - SIENA (Smith et al., 2002) | Cross-sectional and longitudinal analysis of brain changes | Morphometric analysis (Clark et al., 2014) |
| ITK-SNAP (Yushkevich et al., 2006) – http://www.itksnap.org/pmwiki/pmwiki.php | Manual and semi-automated brain segmentation | Segmentation of lateral ventricles and caudate (Hazlett et al., 2012, Knickmeyer et al., 2008) |
| The McConnell Brain Imaging Centre software toolboxhttp://www.bic.mni.mcgill.ca/software/ | ||
| - ANIMAL (Collins et al., 1995, Collins et al., 1994) | Anatomical labeling | Morphometric analysis (Brain Development Cooperative Group, 2012a, Castellanos et al., 2002, Hazlett et al., 2017) |
| - CIVET | Analysis of cortical surfaces | Group comparison based on cortical thickness (Shaw et al., 2009, Shaw et al., 2007) |
| - INSECT (Collins et al., 1999) | Multi-modal tissue classification | |
| - mni_autoreg (Collins et al., 1994) | MRI linear registration | |
| Statistical Parametric Mapping (SPM) (Ashburner and Friston, 1997) – http://www.fil.ion.ucl.ac.uk/spm/ | Brain segmentation, bias correction, spatial normalization | Voxel-based morphometry (Black et al., 2012, Raschle et al., 2011) |
Standard software tools for automated processing of MRI data such as mni_autoreg (Collins et al., 1994), SPM (Ashburner and Friston, 1997), Freesurfer (Dale et al., 1999, Fischl et al., 1999) and FSL (Jenkinson et al., 2012, Smith et al., 2004) use a brain template. These templates have typically been created using data from adults, which is not optimized for use with pediatric samples however. As reviewed in Section 2.1, the morphology of the child brain can be significantly different from that of adults with respect to a number of structural parameters. Therefore, using an adult template within the pediatric analysis risks introducing inaccuracies to the processing pipeline (Serag et al., 2016, Shi et al., 2012, Wilke et al., 2003b). In this section, we will discuss the most common approaches of processing MRI data used in pediatric studies and illustrate the downsides of applying adult-based approaches on young children data for the main processing steps used in neuroimaging: preprocessing, brain extraction, brain segmentation and normalization.
3.2.1. Quality control and preprocessing
Noise and artifacts degrade image quality, which can lead to biases and inaccuracies in processing. Nevertheless, low image quality can typically be identified with careful visual inspection and post-processing quality control, and handled using preprocessing for quality enhancement (Ducharme et al., 2016). In this manuscript, we refer “preprocessing” as any manipulations used to enhance image quality and whichoptimally prepares data for the processing pipeline, in contrast to “processing” that corresponds to any manipulations used to extract MRI measures.
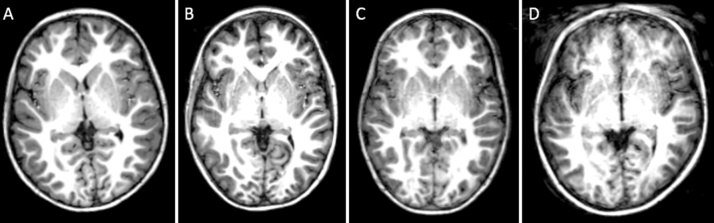
Following completion of data acquisition, visual inspection provides a first stage of image quality screening toward determining whether or not data should be included in a study, based on examination of the raw data. Out of the 28 reviewed studies, three studies performed visual inspection of the general data quality, such as the noise, contrast, intensity inhomogeneity, distortion artifacts and motion artifacts, (Brain Development Cooperative Group, 2012a, Hazlett et al., 2012). For motion artifacts, three of the reviewed studies excluded images based on experimenter ratings of severe motion (Krogsrud et al., 2014, Vanderauwera et al., 2016, Yang et al., 2015). Blumenthal et al. suggested to grade motion into four categories (Blumenthal et al., 2002): “none” which corresponds to little or no visible motion artifacts, “mild” to enough detectable motion shown as subtle ringing, “moderate” to significant ringing and “severe” to extreme motion that makes the scan unusable (see Fig. 2). Out of the reviewed studies, this grading scale was used by Lyall et al. (2015) who included images with mild and moderate motion artifacts in the analysis, and Shaw et al., 2009, Shaw et al., 2007 who included images with mild motion artifacts. However, mild and moderate motion can already affect processing reliability, which can lead, for example, to an underestimation of brain tissue volume (Alexander-Bloch et al., 2016, Blumenthal et al., 2002)
Fig. 2.
Examples of T1-weighted images with each motion type (and their frequency) in a dataset of 72 children at 5–6 years of age (Theys et al., 2014): (A) none (43%), (B) mild motion (24%), (C) moderate motion (18%) and (D) severe motion (15%).
Visual inspection can take place subsequent to the processing step as post-processing quality control. This enables assessment of the impact of systematic error introduced at the initial data processing stage. Out of 28 reviewed pediatric studies, only six studies reported to have performed post-processing quality control to strengthen the reliability of their results (Bora et al., 2014, Castellanos et al., 2002, Hosseini et al., 2013, Knickmeyer et al., 2008, Krogsrud et al., 2014, Li et al., 2014b). In two studies, results obtained from this processing step were compared with manual segmentations to assess their quality (Brown et al., 2012, Hazlett et al., 2012).
In contrast to qualitative assessment of data quality that is rather subjective, quantitative measures enable more objective assessment of noise, motion or susceptibility artifacts contained in the image. The main advantage of these quantitative measures is that the variance introduced by subjective ratings from different observers is reduced, which might lead to standardization of procedures across studies. Although quantitative measures could help to assess quality more precisely and to estimate the correction to be applied, these measures are not widely used and were only reported in one pediatric study (Brain Development Cooperative Group, 2012a).
To improve image quality before processing, most standard software tools include preprocessing in their pipeline, such as Freesurfer, SPM and Autoseg. Six of the reviewed studies applied preprocessing and examples of preprocessing used were noise reduction, bias field correction and distortion correction (Brain Development Cooperative Group, 2012a, Choe et al., 2013, Hazlett et al., 2017, Hazlett et al., 2012, Li et al., 2014a, Nordahl et al., 2011, Shen et al., 2013).
3.2.2. Brain extraction
Brain extraction, or skull stripping, is the processing step where non-neural tissue is removed from anatomical scans. Generally applied to extract whole brain volumes, this step has also become a standard procedure that improves the accuracy and efficiency in a number of processing steps, including brain segmentation and normalization. For this purpose, most standard software packages include brain extraction as one of the initial steps in the processing pipeline (see Table 3). The Brain Extraction Tool (BET) from FSL software is one of the most widely adopted methods used for brain extraction, providing an intensity-based approach that deforms a surface model to fit brain boundaries (Smith, 2002). Five of the reviewed studies applied the BET procedure on data of young children (Brain Development Cooperative Group, 2012b, Nie et al., 2013, Nordahl et al., 2011, Shaw et al., 2009, Shen et al., 2013)
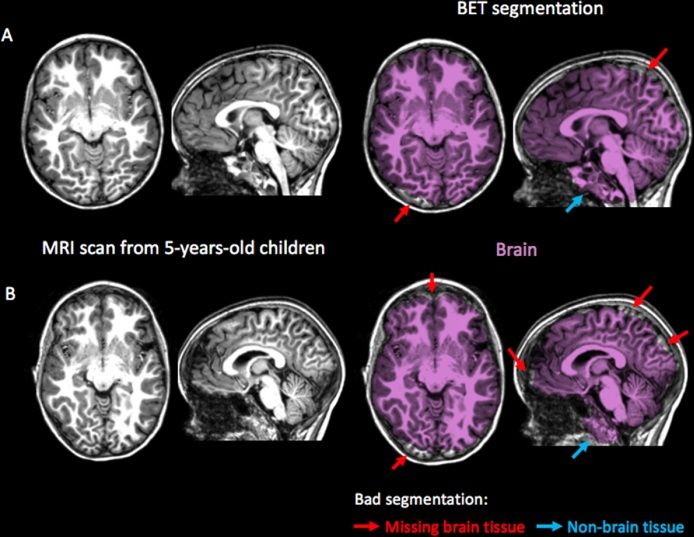
BET and similar approaches, such as 3DSkullStrip from AFNI toolkit (Cox, 1996) and Brain Surface Extractor from Brainsuite (Shattuck et al., 2001), are less accurate when applied in pediatric populations, as they tend to remove brain tissue or including non-brain tissue erroneously (Serag et al., 2016, Shi et al., 2012). This type of inaccuracy is illustrated in Fig. 3, where the BET procedure was applied on T1-weighted brain images from 5-year-old children. A potential mechanism of these errors may be linked to the narrower boundary between brain and non-brain tissues compared to adults (Fennema-Notestine et al., 2006, Lee et al., 2003, Ségonne et al., 2004).
Fig. 3.
Brain segmentation obtained with the Brain Extraction Tool (BET) from the FSL 5.0.9. software packages, applied on MRI scans from 5-year-old children (Theys et al., 2014). Systematic errors in brain extraction occurred with brain tissue removed erroneously (red arrows) and with non-brain tissue included (blue arrows) in both (A) good quality image and (B) image with Brain segmentation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The segmentation step of processing consists of the assignment of a tissue type to each voxel of the MRI scan. As a starting point, the intra-cranial space is generally segmented into one of the three main tissue types, comprising the gray matter, the white matter and the cerebrospinal fluid (CSF). For this purpose, computer learning-based approaches based on a priori anatomical information are usually used, including atlas-based expectation-maximization segmentation (e.g. in Autoseg, FAST and SPM), surface model-based segmentation (e.g. in Freesurfer) or artificial neural network approaches (e.g. in INSECT). Following the main tissue segmentation, the anatomical image is segmented into smaller sub-regions, as is commonly applied for cortical parcellation using label propagation methods (e.g. in Autoseg, Freesurfer and CIVET).
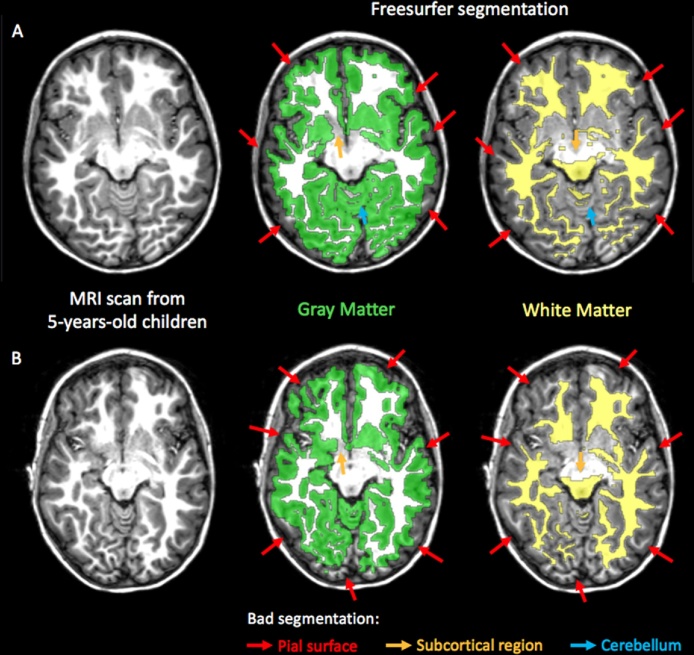
Standard software tools based on adult brain templates, such as Freesurfer and FSL, have been shown to provide inaccurate segmentation for pediatric brains (Schoemaker et al., 2016, Schumann et al., 2010). Representative examples of the failure to achieve accurate segmentation using Freesurfer without manual editing on scans from 5-year-old children are illustrated in Fig. 4. In four of the pediatric studies reviewed, manual or semi-automated methods were applied for the segmentation of specific brain tissues of interest, for example, planum temporale, lateral ventricles and caudate (Hazlett et al., 2012, Shen et al., 2013, Sowell, 2004, Vanderauwera et al., 2016). The authors argued that for these specific brain tissues, segmentation is particularly difficult due to high variability in their shape among the same population.
Fig. 4.
Gray matter and white matter segmentation obtained using Freesurfer software tools without manual editing and applied on MRI scans obtained from 5-year-old children (Theys et al., 2014). Although image quality was good (A), systematic errors were found in segmentation near the pial surface (red arrows), in subcortical regions (orange arrows) and in cerebellum (blue arrows). Segmentation in Freesurfer is highly sensitive to motion as it is already degraded in images with mild motion artifact (B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The main causes underlying failures of segmentation accuracy in children stem from the low contrast between gray and white matter (Schumann et al., 2010) and coupled with substantial differences in shape, especially in subcortical regions such as the hippocampus and amygdala (Schoemaker et al., 2016), between adults and children. For segmentation of medial temporal lobe structures, including the hippocampus and amygdala, Hu et al. reported that an appearance-model based method, based on the use of young adult data as training samples, was able to reliably segment pediatric MRI data (Hu et al., 2013, Hu et al., 2011). However, there is currently a lack of validation to confirm the reliability of these sub-regional segmentation methods (including Autoseg and CIVET) for use with pediatric MRI data.
3.2.3. Spatial normalization
Spatial normalization acts to deform brain images to achieve spatial correspondence between the same brain areas across a sample of participants. As human brains differ substantially in size and shape, even among the same age group, this processing step ensures that the same anatomical structures are compared, thereby facilitating statistical comparisons across groups of individuals. Spatial normalization depends on the registration of each individual brain image to a common reference with known spatial dimensions.
In three pediatric studies that performed spatial normalization (Black et al., 2012, Brain Development Cooperative Group, 2012a, Castellanos et al., 2002), data were aligned to a standardized stereotaxic atlas such as compiled by Talairach, SPM96 (Statistical Parametric Mapping 96), ICBM152 (International Consortium for Brain Mapping) or the MNI305 (Montreal Neurological Institute) template. However, when using these adult brain templates, the resulting registration of pediatric data was more variable and less robust than for independent samples of adults (Ghosh et al., 2010, Hoeksma et al., 2005, Machilsen et al., 2007, Muzik et al., 2000, Wilke et al., 2003b). Poor spatial normalization in pediatric samples can increase the probability of observing group differences resulting from registration errors, instead of true anatomical variation in the population of interest.
3.3. Current practice of MRI-processing in longitudinal studies
Longitudinal designs offer clear advantages for studies on development compared to cross-sectional designs, because changes are measured at multiple time points in the same individuals rather than across separate samples of individuals at different ages. When longitudinal data are collected for modeling brain development, the trajectory curves are more meaningful in terms of changes in the brain structure than when cross-sectional data would have been acquired, and inter- and intra-subject variability is better assessed (Mills and Tamnes, 2014).
Few longitudinal MRI-studies have been carried out in young pediatric populations, in contrast to those for older populations (Hedman et al., 2012, Mills and Tamnes, 2014). Although longitudinal designs are more difficult to implement than cross-sectional designs, the number of longitudinal studies of young children has recently increased, particularly in the context of studying structural changes between birth and 2 years of age (see Table 1, Table 2). Nevertheless, involving young children in longitudinal studies can add challenges in the implementation of processing and of analysis.
In 11 out of the 14 of the longitudinal studies reviewed, developmental growth patterns of brain structures were estimated using mixed-effects statistical modeling (Brain Development Cooperative Group, 2012a, Castellanos et al., 2002, Clark et al., 2014, Gilmore et al., 2012, Hazlett et al., 2017, Holland et al., 2014, Knickmeyer et al., 2008, Lebel and Beaulieu, 2011, Lyall et al., 2015, Shaw et al., 2007, Shaw et al., 2009, Shen et al., 2013). Mixed model analyses estimate the effects of a chosen variable, typically the age, on a dependent measure of interest while taking into account the dependence of the data within the subject (Singer and Willett, 2009). These statistical analyses allow the modelling of data collected at uneven intervals and with unequal numbers of data points (Mills and Tamnes, 2014).
However, already during the processing steps (i.e. prior to statistical analyses), longitudinal data can be exploited to improve segmentation and to extract growth patterns of targeted brain structures over time. Concerning segmentation, longitudinal data can help to extract structural properties with more efficiency, by jointly segmenting scans at different time points. It consists of propagating the tissue labels toward each time point. For example, the longitudinal pipeline of Freesurfer performs segmentation by building an unbiased template for each subject using information across all time points so that longitudinal consistency is respected. Assuming a fixed head over time, this pipeline may fail in pediatric data for which head size is changing substantially. Concerning growth patterns, longitudinal data processing enables us to measure structural changes between scans at different time points and to identify the location where the changes occur at the individual level. The changes between different time points are compared by using either, (1) methods based on the shift of the image intensity profile or (2) methods based on the deformation field between scan pairs. An example of methods based on the intensity profile is SIENA, the longitudinal pipeline of FSL (Smith et al., 2002). Displacement between two time points is estimated by aligning the peaks of the spatial derivatives of intensity profiles measured in both images. SIENA is mainly used to assess gray matter atrophy, but it cannot take into account more than two time points, in contrast to the longitudinal processing pipeline from Freesurfer (Reuter et al., 2012). An example of methods based on deformation field is the deformation-based morphometry in SPM (Ashburner and Friston, 2000). It consists of computing the non-linear transformations from a scan at one time point towards the baseline image. The brain growth is then quantified with the Jacobian (matrix of all first-order partial derivatives) of this deformation. The Jacobian determinants then represent the fractional volume expansion and contraction of each voxel needed to align the scans from different time points. Out of the longitudinal studies that we reviewed, only the study by Sowell et al. applied deformation-based morphometry to measure the cortical thickness changes in children between the ages of 5–12 years (Sowell, 2004). Using longitudinal registration, a straightforward application of deformation-based morphometry can lead to substantial biases in the estimation of the changes, such as the inconsistency of rigid registration, the interpolation asymmetry or the differential bias field (Lewis and Fox, 2004, Reuter and Fischl, 2011, Yushkevich et al., 2010).
Although processing pipelines for longitudinal data are proposed in standard software tools, such as in Freesurfer and SIENA, they are typically used for segmentation and longitudinal analysis at the group level but rarelyfor measuring within subject changes over time. For example, a study by Clark et al. used both FSL and Freesurfer for segmentation only, and analyzed longitudinal data from children with dyslexia between 5 and 11 years with SPSS (Clark et al., 2014). Only four longitudinal studies on young children have used processing methods that make full use of longitudinal information to extract MRI measures (Gilmore et al., 2012, Lebel and Beaulieu, 2011, Li et al., 2014b, Lyall et al., 2015).
3.4. Conclusions on current practice
As reviewed in Section 2.2, image quality affects the reliability of neuroimaging analyses at all subsequent processing stages. To fully control the potential impact of data quality, pediatric studies have reported to have performed either visual inspection (one tenth of the reviewed studies), preprocessing (one tenth of the reviewed studies) or post-processing quality control (one fifth of the reviewed studies). However, these three steps were not systematically performed in all studies, despite the fact that standard software tools have implemented pipelines for quality control and preprocessing (motion correction, distortion correction, bias correction). Therefore, we want to emphasize the quantification and optimization of data quality toward improving the data validity and the success rate of data processing (see Section 4).
In Section 2.1, we highlighted the principle that organization of brain structure is constantly changing with age and with the presence of variable trajectories of development across different brain regions during maturation. As a consequence, the brain of a young child cannot successfully be modeled using a version of adult brain that is simply scaled to size. Most of standard software tools (e.g. Freesurfer) that have been deployed in samples of young children have nevertheless based their analyses on an adult atlas, which necessitated post-processing correction of inaccuracies that needed to be implemented manually (Choe et al., 2013, Li et al., 2014a). In contrast, studies of infants (i.e. under 2 years of age) have generally adopted bespoke methods to circumvent the limitations of standard software tools for use with this population (Bora et al., 2014, Gilmore et al., 2012, Holland et al., 2014, Knickmeyer et al., 2008, Li et al., 2014b, Lyall et al., 2015, Nordahl et al., 2011, Shen et al., 2013). We suggest that it is crucial to adopt child-adjusted methods for children up to at least 6 years of age (corresponding to the age until which drastic structural changes take place). We explain in more detail the possible child-adjusted techniques in Section 5.
Longitudinal processing and analysis in young children requires attention on two main aspects. Firstly, longitudinal registration helps in measuring brain growth pattern by computing the transformations between different time points but also introduce substantial biases in the estimation of the changes, owing to inconsistency of rigid registration, the interpolation asymmetry or the differential bias field. Therefore, additional processing steps should be performed to avoid biases and are explained in more detailed in Section 6.1. Secondly, early brain development might require more complex modeling due to the drastic structural changes that occur during early life. Therefore, additional considerations and challenges might be introduced in the analysis of longitudinal data of young children and are discussed in Section 6.2.
4. Quantification and optimization of data quality
Data from young children are typically of lower contrast- and of reduced signal-to-noise ratio compared to adult data. It is therefore important to quantify and optimize data quality to improve the amount of usable data, as well as their validity and reproducibility. The following sections review neuroimaging preprocessing methods that, while not specific to pediatric protocols, deserve particular attention due to the increased presence of noise and artifact in pediatric data.
4.1. Quality quantification
As a first step, quality inspection of data is an essential procedure because processing may be substantially affected by low image quality. Visual inspection is the most common method used to check for data quality of structural MRI-scans, but quantitative measures might be more useful since they enable more precise grading of image quality.
Typical quantitative measures include evaluation of the signal-to-noise ratio (SNR)- the amount of signal-of-interest compared to the noise, and the contrast-to-noise ratio (CNR)- the differences in signal between different regions-of-interest relative to the noise. For anatomical brain MRI, Gedamu et al. proposed to define SNR as the mean signal intensity distribution (measured in white matter for T1-weighted images and in CSF for T2-weighted images) divided by the standard deviation of the noise intensity distribution (measured in the background) (Gedamu et al., 2008). In brain MRI, the contrast should be high enough to distinguish gray matter from white matter. Magnotta et al. proposed to define CNR as the difference in intensity distribution between gray matter and white matter divided by the standard deviation of the noise intensity distribution (Magnotta et al., 2006). The higher the SNR and CNR, the better is the image quality.
By quantifying motion, a threshold can be set to define the acceptable level of motion in the study (Theys et al., 2014). However, this quantification is not straightforward for anatomical MRI. Generally, only one image is acquired, and therefore motion across multiple images cannot be calculated. In anatomical MRI, motion can be quantified through the artifacts caused by it, for example through ringing/ghosting artifacts that generally appear in phase encode direction and that also cause non-uniform intensities in the brain and in the surrounding background. Gedamu et al. proposed using a ghosting ratio based on the standard deviation of noise in anterior-posterior regions of the head divided by the standard deviation of noise in lateral regions next to the head (Gedamu et al., 2008). A ghosting ratio around 1 corresponds to an image without ghosting artifacts, under 1 to ghosting artifacts in lateral directions, and above 1 to ghosting artifacts in anterior-posterior direction.
Quantitative measures for image quality (SNR, CNR and ghosting ratio) are often based on the image intensity that is generally not normalized to the same values. As a consequence, the minimum threshold to determine whether the image is of good or low quality is study-dependent and should be validated to the performance of processing methods.
4.2. Motion correction
Motion correction is a preprocessing step that reduces motion artifacts in the image. In diffusion and functional MRI, these techniques correct for motion artifacts by comparing volumes acquired in different directions and at different time points respectively (Dubois et al., 2014, Liu et al., 2015, Rohde et al., 2004, Smith et al., 2004). For anatomical MRI modalities, such as T1-weighted images, motion correction is less applicable because multiple volumes are rarely acquired for this purpose and the comparison of these high resolution images is computationally intensive (Kochunov et al., 2006).
Although total scanning time would be extended, an advantage of acquiring several T1-weighted images (e.g. test-retest scans) is the opportunity to select the best quality scan or, alternatively, to average scans following registration, in order to improve image quality. Included in standard software tools such as Freesurfer and FSL, averaging several T1-weighted images has become a common preprocessing step that increases the SNR, which can lead to improved segmentation. However, it might also reduce the contrast and fail to eliminate motion if one of the scans is affected by significant artifact (Han et al., 2006, Jovicich et al., 2006). Moreover, it might be problematic to mix averaged and single acquisitions when comparing across individuals or time points. In most cases when only one anatomical image is available, head motion remains an unsolved issue where few solutions are proposed once the scan is already acquired. Therefore, it is important to correct motion artifacts with methods that are applied during the scanning session, so called 'prospective motion correction' (Maclaren et al., 2013).
Prospective motion correction methods account for motion artifacts by updating the pulse sequence, depending on the head motion measured with sensors. Some examples include the orbital navigator echoes (Fu et al., 1995), BLADE/PROPELLER MRI (Pipe, 1999), radial imaging (McLeish et al., 2004) or PROMO (White et al., 2010). It has been demonstrated that prospective motion correction such as BLADE and PROMO improves accuracy of measurements in T1-weighted images acquired on pediatric populations, with an acceptable additional scanning time (around 10 s, depending on the motion degree) for clinical practice (Alibek et al., 2008, Brown et al., 2010, Kuperman et al., 2011). More recently, pediatric studies have started to use prospective motion correction that is integrated in the conventional multi-echo MPRAGE sequence used to acquire T1-weighted images (Silk et al., 2016, Wang et al., 2016). Although prospective motion correction methods are currently made available by most vendors, these methods are not yet well disseminated in current practice, with only one of the reviewed studies having applied prospective motion correction (Brown et al., 2012). Given the advantage that it enables without significantly extending scanning time, motion correction at the source will likely become an important and standardly applied tool for future studies.
4.3. Distortion correction
Distortion artifacts associated with scanner hardware or head placement can be corrected using the 3D image distortion map acquired with a calibration phantom to ensure accuracy in the measurement (Evans, 2006, Nordahl et al., 2011, Shen et al., 2013). When the 3D distortion map is not available, an alternative solution is to use gradient non-linearity distortion correction methods to provide distortion correction based on the knowledge of the spherical harmonic coefficients from the imaging gradients (Jovicich et al., 2006). This information can be obtained from the scanner’s vendor and enables estimation of the displacement and intensity correction to be applied. For images acquired with Echo Planar Imaging (EPI) sequences, spatial distortion displacement can be estimated with a fast nonlinear registration of scans acquired with opposite phase encoding polarities (Holland et al., 2010). Distortion correction methods are often included in standard software packages, such as the gradient unwarping in Freesurfer or the Fieldmap toolbox of SPM, and can be applied as long as the information on distortion is known (e.g. field maps, spherical harmonic coefficients, etc.).
4.4. Bias field correction
Bias field correction consists of removing intensity inhomogeneities resulting from non-uniformity in the field coils or due to magnetic susceptibility changes at the boundaries between brain tissue and air (see Fig. 5). At present, bias field correction is performed at the same time as the segmentation step, such as in the FAST segmentation method from FSL (Zhang et al., 2001), in recent versions of SPM (Ashburner and Friston, 2005) or in similar techniques (Jain et al., 2015, Van Leemput et al., 1999). The bias field should first be estimated from the scans before it can be removed. In these methods, the bias field is estimated both from scans and from a model of the brain tissues that is improved by segmentation. Bias field correction has been demonstrated to result in more accurate brain segmentation (Gousias et al., 2013, Van Leemput et al., 1999) and more robust measurement of the deformation of brain structures (Leow et al., 2006). Because bias field correction is integrated within the segmentation algorithm, the quality of the correction and segmentation depends mainly on the model used to represent brain tissues (i.e. the brain atlas). The choice of model is especially important for work with young children as reviewed in Section 5.
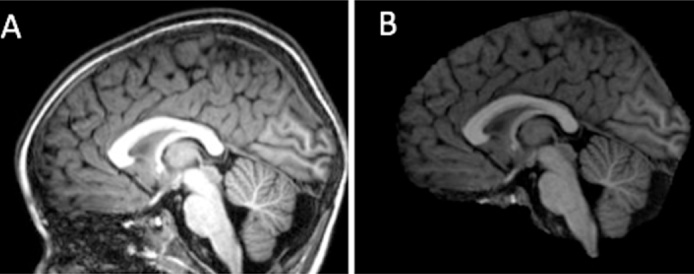
Fig. 5.
Effect of bias field correction on (A) MR scan with intensity inhomogeneity (higher intensity in the center than at the top of the head) compared with (B) the bias corrected image after skull-stripping.
5. Brain MRI processing adjusted to children with age-specific atlases
Neuroimaging processing methods need to be adapted for use with pediatric populations. Accurate image registration and segmentation of the pediatric brains is driven by the age range of the subjects used to construct the brain template. The use of an age-specific atlas has been shown to increase reliability in the processing of pediatric data, validating their utility (Li et al., 2015, Murgasova et al., 2007, Shi et al., 2011, Yoon et al., 2009). This section explains how data processing has been adapted for use with young children, including the provision of age-specific brain atlases and spatio-temporal atlases (more specific to infants). We demonstrate their use within standard software tools, comment on the improvements conferred by child-adjusted approaches compared to standard methods and review the challenges in creating/selecting age-specific atlases.
5.1. Age-specific brain atlases
Age-specific brain atlases are built from averaging (in intensity and shape) brain images of children in the targeted age-range (Fonov et al., 2009, Kuklisova-Murgasova et al., 2011, Richards et al., 2016, Sanchez et al., 2012). A brain atlas refers to a brain template (i.e. grayscale average image) combined with maps of the brain anatomy (e.g. anatomical parcellation maps or tissue probability maps).
Anatomical labeling of the atlas assigns a specific tissue type to each voxel of the atlas and is mainly applied for the delineation of non-overlapping regions-of-interests. Anatomical parcellation maps are usually obtained by manual delineation of a single-subject atlas. This might not represent all population variability but has the advantage of segmenting the brain into any number of structures. If the subject's anatomy is close to that represented by the atlas, label propagation is one of the easiest and fastest ways to segment the brain. However, registration errors are larger if differences between the subject and the atlas are important (Cabezas et al., 2011). To construct anatomical parcellation maps specific to young children, a strategy used was to propagate manual parcellation maps from adults to infant brain scans and to fuse them to create the atlas (Fillmore et al., 2015, Gousias et al., 2008, Oishi et al., 2008). In contrast, Gousias et al. manually segmented brain images from neonates into 50 regions-of-interest, prior to label propagation and fusion, to create the atlas (Gousias et al., 2013). The results for anatomical labeling obtained with both methods (maps from adults vs. maps from neonates) were similar, leading to the possible conclusion that methods for label propagation and fusion contribute more to the improvements to greater extent than do the input maps to be propagated.
Tissue probability maps represent the frequency of each voxel that belongs to a specific structure in a population and are generally built for segmentation of main brain tissues (i.e. gray matter, white matter and cerebrospinal fluid). If manual segmentations are available for each scan contributing to the atlas, tissue probability maps can be obtained with the same averaging procedure as used to build the brain template. However, there is a noted lack of manual segmentations available from young children and obtaining a sufficient number of manual segmentations is extremely laborious and time-consuming. Therefore, other strategies were implemented to build age-specific tissue probability maps including: (1) averaging of segmentations that are first obtained by manually segmenting one child and then propagating the segmentation to others (Murgasova et al., 2007), (2) using adult prior information with spatial constraints (Altaye et al., 2008) or (3) using unsupervised tissue classification based on intensity (Fonov et al., 2009, Kuklisova-Murgasova et al., 2011, Shi et al., 2011).
5.2. Four-dimensional spatio-temporal brain atlases
For analysis of infant brains, significant challenges arise owing to the rapid changes in T1 and T2 contrasts during early development. To overcome these issues, four-dimensional (4D) spatio-temporal brain atlases have been implemented and consist of a series of age-dependent averaged brain references (3D atlases) that summarize the age-specific details of the brain structures (Kuklisova-Murgasova et al., 2011, Makropoulos et al., 2016, Serag et al., 2012a). The difference in 4D solutions compared to 3D age-specific atlases is that the brain images are averaged not only over intensity and shape but also over time. There are two types of 4D atlases that have been constructed for pediatric datasets: longitudinal atlases and dynamic probabilistic atlases.
Longitudinal atlases are constructed by performing the segmentation at one time point and subsequently propagating this segmentation to the other time points with longitudinal deformation fields. For example, Shi et al. propagated first the adult parcellation maps to the 2-year-old child atlas before propagating to the neonatal atlas, instead of a direct propagation (Shi et al., 2011). This indirect propagation minimized registration errors by taking into account the longitudinal correspondence between time points and resulted in more accurate brain segmentation.
Dynamic probabilistic atlases are obtained by constructing the template from the average transformation, weighted by age with a Gaussian kernel (Kuklisova-Murgasova et al., 2011, Makropoulos et al., 2016, Serag et al., 2012a). The brain images acquired at similar age to the time point of interest contributed more in the average transformation, which is used to warp all images in the same space where they were averaged to form the age-dependent template anatomy.
5.3. Use of young pediatric brain atlases in practice
Publicly available brain atlases specifically for use with pediatric datasets are listed in Table 4, and include both 3D and 4D atlases. In the processing workflow, these atlases should be used as reference in place of the standard adult atlas.
Table 4.
Publicly available brain atlases.
| Research group | Age range | Subjects | Modality | Brain regions |
|---|---|---|---|---|
| Brain-development.orghttp://brain-development.org/ | ||||
| (Gousias et al., 2008) | 2 yo | 33 TD | T1w, T2w | 83 brain regions |
| (Kuklisova-Murgasova et al., 2011) | 29–44 gw | 142 TD | T2w | cortex, WM, subcortical GM, brainstem and cerebellum |
| (Serag et al., 2012a) | 28–44 gw | 204 Preterms | T1w | cortex, WM, subcortical GM, brainstem and cerebellum |
| (Gousias et al., 2013) | 24–45 gw | 5 TD | T1w | 50 brain regions |
| 15 Preterms | ||||
| (Makropoulos et al., 2016) | 27–44 gw | 40 TD | T2w | 82 brain regions |
| 380 Preterms | ||||
| Cincinnati children’s hospital medical centerhttps://irc.cchmc.org/index.php | ||||
| (Wilke et al., 2003b) | 5–18 yo | 200 TD | T1w | GM, WM, CSF |
| (Altaye et al., 2008) | 9–15 mo | 76 TD | T1w | GM. WM, CSF |
| IDEAgrouphttp://bric.unc.edu/ideagroup/free-softwares | ||||
| (Shi et al., 2011) | 0–2 yo | 95 TD | T1w, T2w | GM, WM, CSF and 90 brain regions |
| Johns Hopkins Universityhttp://lbam.med.jhmi.edu/ | ||||
| (Oishi et al., 2011) | 37–41 gw | 25 TD | T1w, T2w, dMRI | 122 brain regions |
| NeuroImaging & Surgical Technologies labhttp://nist.mni.mcgill.ca/ | ||||
| (Fonov et al., 2009) | 0–4.5 yo | 108 TD | T1w, T2w, and PDw | Only brain template |
| (Fonov et al., 2011) | 4.5–18.5 yo (including: 4.5–8.5yo) | 324 TD | T1w, T2 w and PDw | GM, WM, CSF |
| Neurodevelopmental MRI database by John E. Richardshttp://jerlab.psych.sc.edu/neurodevelopmentalmridatabase/ | ||||
| (Sanchez et al., 2012) | 2w–4 yo | 6–32 TD | T1w, T2w | Only brain template |
| (Fillmore et al., 2015) | 3–12 mo | 11–36 TD | T1w | 14 brain regions |
TD, typical developmentd, gestional weeks; mo, months; yo, years old; T1w, T1-weighted images; T2w, T2-weighted images; PDw: proton density weighted images; dMRI: diffusion MRI; dMRI, diffusion Magneitc Resonance Imaging; GM, gray matter; WM, white matter; CSF, cerebrospinal fluid.
Other than using existing brain atlases, there are also automatic tools for building population-specific atlases. The “Template-O-matix” toolbox creates intensity average template that is specific to a given study population with linear registration (Wilke et al., 2008). However, the template might contain blurred anatomical details, because linear registration alone cannot handle high variability in brain regions (Fonov et al., 2011). In contrast, ANTs Template Creation and Labeling pipeline uses diffeomorphic transformations for registration and enables a labeled brain template to be constructed in a common space to which the smallest (affine and non-rigid) transformations are required to warp the data from every subject (Avants et al., 2011). The IDEAgroup (UNC School of Medicine) has also distributed freely available software packages for atlas construction, including longitudinal atlases (http://www.med.unc.edu/bric/ideagroup/free-softwares). Hence, ANTS and IDEAgroup software packages, in particular, may find utility in building pediatric brain atlases in order to adjust data processing for children.
Following selection of the appropriate brain atlas, the issue turns to decisions about how to integrate the age-specific brain atlas within the current processing pipeline. In the standard software tools listed in Table 5, it is possible to integrate an age-specific brain atlas in the processing. For example, a specific atlas can be specified in SPM during the normalization step in voxel-based analyses. In addition, a specific type of atlas (probabilistic atlas or labeled atlas) may be required depending on the processing algorithm used in the software tools. For example, labeled atlases can be used if there is a registration pipeline available in the software packages (e.g. ANTs, NiftyReg, SPM, FSL). Conversely, probabilistic atlases are preferable if the segmentation pipeline is based on a expectation-maximization algorithm (e.g. NiftySeg, SPM). Unfortunately, most of standard software tools cannot accommodate 4D spatio-temporal atlases at present.
Table 5.
A non-exhaustive list of commonly used software tools for MRI processing using atlas based methods.
| Software | Atlas building pipeline | Atlas-based processing | Type of atlas accepted in the processing |
|---|---|---|---|
| ANTs (Avants et al., 2011) – http://stnava.github.io/ANTs/ | Template Creation and Labeling pipeline | Brain extraction: ✓ | Labeled and probabilistic atlas |
| Normalization: ✓ | |||
| Brain segmentation: ✓ | |||
| NiftyReg/NiftySeg (Cardoso et al., 2013a, Ourselin et al., 2000) – http://cmictig.cs.ucl.ac.uk/wiki/index.php/Main_Page | Niftyreg Groupwise | Normalization: ✓ | Labeled, probabilistic and 4D atlas |
| Brain segmentation: ✓ | |||
| Freesurfer (Dale et al., 1999, Fischl et al., 1999) – http://freesurfer.net/ | FsAverage (only intensity average) | Cortical and subcortical segmentation: − | Labeled and probabilistic atlas |
| FSL (Jenkinson et al., 2012, Smith et al., 2004) | / | Brain extraction: – | Brain template |
| Normalization: ✓ | |||
| Brain segmentation: − | |||
| Statistical Parametric Mapping (SPM) (Ashburner and Friston, 1997) – http://www.fil.ion.ucl.ac.uk/spm/ | / | Normalization: ✓ | Labeled and probabilistic atlas |
| Brain segmentation: ✓ | Probabilistic atlas | ||
Brain atlas non-specific to the software can be integrated (✓) or not (−).
5.4. Multi-atlas based methods for processing
Instead of selecting only one model-based, average representation, multi-atlas based methods automatically select the best-matching atlases for the subject (Iglesias and Sabuncu, 2015). Following atlas selection, a tissue type is assigned to each voxel by the fusion of segmentations from the selected atlases. Multi-atlas based methods enable anatomical variation to be better captured and circumvents inter-rater segmentation variability and registration errors obtained from single atlas. In this way, higher segmentation accuracy is generally obtained, compared to the accuracy obtained when using a single atlas. Examples of multi-atlas based methods applied on young children are LABEL (Shi et al., 2012) included in iBEAT software (Dai et al., 2013), and ALFA (Serag et al., 2016), with both performing brain extraction for neonatal brains. Serag et al. demonstrated a higher overlap with manual segmentation of 14–19% for LABEL and ALFA compared to results obtained with BET for brain extraction in T2-weighted images (Serag et al., 2016).
Although learning-based methods are promising and tend to be best options in terms of performance, those methods are more difficult to implement and the processing is more computationally demanding. To segment one subject, each atlas needs to be registered towards the subject image, and is particularly intensive when non-rigid registration is involved. In the same way as building a population-based atlas, significant resource should be spent in the creation and optimization of the multi-atlas library, and high memory cost is required if the library contains a large number of atlases. Moreover, the settings of these methods are more complex, such as, choosing the similarity metric, number of selected atlases and fusion algorithm. Further validations are needed to determine the best setting to adjust brain segmentation methods to young children brain.
5.5. Impact of using age-specific atlases in pediatric studies
A first important impact of age-specific atlases is more efficient processing of data from young children, because the atlases are more representative of this population. To illustrate this, Fonov et al. built unbiased pediatric brain templates for relevant age groups in the development and fewer deformations were required when the data of subjects (5–7 years old) were normalized to the age-matching template, indicating smaller bias in the estimation of tissue growth or shrinkage (Fonov et al., 2011, Fonov et al., 2009).
Secondly, when age-specific atlases were used, more accurate segmentation of brain structures has been demonstrated, in terms of improved overlap with the manual segmentation (considered the gold standard). Murgasova et al. reported a 2 percent higher overlap in gray and white matter segmentation, and 22 percent higher for thalamus segmentation, by using an age-specific atlas (validated on four 2-years old subjects), compared to results using an adult atlas (Murgasova et al., 2007). Validated in infants between 3 and 12 months from the NIHPD dataset, Fillmore et al. reported improvements after using age-appropriate atlases (mean overlap of 85.1%) compared with using a 2-years old atlas (mean overlap of 81,2%) in the segmentation of cerebellum, brainstem, thalamus, frontal lobe, occipital lobe and temporal lobe (Fillmore et al., 2015).
Even better results appear to be obtained with 4D spatio-temporal atlases. Longitudinal 4D infant atlases improved accuracy for spatial normalization and segmentation across individual infants. Using a longitudinal 4D atlas, Shi et al. showed 2–5% higher overlap with manual segmentation for gray matter, 1–4% for white matter and 2–13% for CSF, in comparison with results obtained with standard adult- and infant- atlases without longitudinal consistency (Shi et al., 2011). Li et al. measured higher overlap ratios for sulcal and gyral regions for the 4D infant surface atlas (mean overlap of 69.7–73.2%) than both independent infant surface atlases (mean overlap of 69–72,1%) and adult atlases from Freesurfer (mean overlap of 67.4–71,2%) (Li et al., 2015).
4D dynamic probabilistic atlases improve atlas-based segmentation in the studies of newborns by matching the age-corresponding template and tissue probability maps (Serag et al., 2012b). Using 4D anatomical neonatal priors (Kuklisova-Murgasova et al., 2011) and an iterative relaxation strategy, AdAPT provides an expectation-maximization segmentation algorithm adapted to the preterm brain. Validated on pathological and normal neonates, the AdAPT method showed higher overlap with manual segmentation for gray matter (mean higher overlap of 15%), cerebellum (14%), white matter (23%) and ventricular volumes (55%) than the widely-used expectation-maximization segmentation (Cardoso et al., 2013b).
Improved results from using age-specific atlases might affect the analysis, notably on the developmental trajectory curves obtained from MRI measures. Developmental trajectory curves observed in prior studies might be biased, as failed segmentation generally leads to inaccurate estimation of brain properties (volume, cortical thickness, surface area). By using child-adjusted methods, the shape of developmental trajectory curves, as well as the percent change per year, might differ. Future studies should further investigate the impact of age-specific atlas in the analysis of typical and atypical brain development, especially for older and broader pediatric age ranges (between 2 and 6 years old).
5.6. Considerations and challenges in creating and using age-specific atlases
There are several considerations and challenges in creating and selecting an age-specific atlas. The specificity of the atlas should be considered owing to the age, the type of population, and the regions-of-interest in brain. As a general rule, we recommend using the atlas that is the most similar to the study population (e.g. similar age, same modality, same disease).
Although the utility of age-specific atlases has been demonstrated, it is not clear how specific the atlas should be with respect to age. Defining the age range represented by the atlas is therefore an important consideration because changes with age are typically non-linear. The age-range should be specific enough to follow age-related changes and thus, should correspond to a period in which the expected changes occur. In infant populations, the atlas age range was set in weeks for neonates (Kuklisova-Murgasova et al., 2011, Oishi et al., 2011, Serag et al., 2012b), in months for children under 1 year old (Altaye et al., 2008, Li et al., 2015), and in years between 1 and 2 years old (Shi et al., 2011). Even though a restricted age range clearly showed an association with more accurate results in infant populations, the benefit of restricted age range has not been well studied in older pediatric populations. For cross-sectional studies covering a wide age range and longitudinal studies with a long period of follow-up, it is currently unknown whether it is best to use one atlas covering the whole period or several age-specific atlases. Using several age-specific atlases might introduce bias in the comparison across subjects that have been analyzed with different atlases.
Other than the consideration of age, it is also uncertain how specific the atlas should be to the population or the studied group in order to account for differences between pathological and control cases. In the same way as applied to age-specific atlases, disease-specific atlases seem to improve data processing in populations with pathologies for which the anatomical structures significantly differ from the normative population. Thompson et al. demonstrated that for subjects with Alzheimer’s disease that a disease-specific population-based atlas was necessary for accurate brain segmentation. As the disease can be very heterogeneous in presentation (e.g. different forms or grades), they recommended building the atlas based on constituent homogeneous groups to ensure more accurate data processing (Thompson et al., 2004). To the best of our knowledge, group-specific atlases have not been extensively validated on pediatric populations.
The choice on the type of atlas (probabilistic or labeled) is mainly determined by the regions-of-interest to be segmented. Murgasova et al. reported that expectation-maximization methods using prior probability maps performed better for small complex structures in the cortex, with label propagation methods achieving better results for the central brain structures (Murgasova et al., 2007). Therefore, an atlas combined with both probabilistic and parcellation maps, such as proposed by Shi et al. (2011), might find more utility in building segmentation accurately for all brain structures. If other modalities (for example, T2-weighted images, diffusion and functional MRI) are available, normalization and segmentation can also be guided by such multi-modality information (Glasser et al., 2016). Oishi et al. reported that multi-contrast neonatal brain atlases enable a segmentation of neonatal brains into 122 regions with the same accuracy as manual segmentation (Oishi et al., 2011).
Because of all the possibilities to build age-, disease- and group-specific population-based atlas, concordance between these different atlases might therefore decrease. As standard adult atlases already have demonstrated substantial variability (Bohland et al., 2009), this might even be increased in age/disease/group-specific atlases, potentially leading to reduced reproducibility across sites or studies. As a result, interpretations and conclusions might be more difficult to generalize across studies.
6. Longitudinal data processing in young children
Whereas the advantages of longitudinal designs are well-known in statistical analyses (e.g. improved assessment of inter- and intra-subject variability), longitudinal data processing is less highlighted as good practice for measuring brain growth, particularly with appropriate longitudinal registration. Moreover, there are additional considerations for modeling the brain from longitudinal data when young children are involved in the study.
6.1. Longitudinal registration for measuring brain growth
In order to compare scans of the same subject at different time points, the scans need to be registered in the same space. An initial step toward removing differences due to the head position and orientation in the scanner is to apply a rigid transformation or an affine transformation that additionally compensates for geometric distortions. Bias may be introduced by the inverse inconsistency, which means that the inverse transform from the baseline scan (generally the first-time point) toward a follow-up scan does not yield the same result as applying the direct transform from the follow-up scan to the baseline scan. To avoid this type of bias, it is important to use a robust symmetric rigid registration (Reuter and Fischl, 2011).
When scans at multiple time points are available, the follow-up scans are generally registered with the baseline scan considered as the reference. However, this might introduce bias resulting from the interpolation asymmetry because the first-time point is treated differently than subsequent time points. To avoid this asymmetry effect, all time points should be treated identically, either by using an intermediate within-subject template space (Yushkevich et al., 2010) or by performing registrations in both directions (Smeets et al., 2016). The registration inconsistency and the interpolation asymmetry are taken into account in the longitudinal processing pipeline of Freesurfer (Reuter et al., 2012) and in icobrain (Smeets et al., 2016).
Once all time points are rigidly aligned, they are then non-rigidly warped toward each other. Non-rigid registration can be affected by differences in the bias field across time points, which results in unrealistic deformations and consequently, in inaccurate quantification of growth/shrinkage. To correct for this bias, a solution is to perform an intensity normalization that ensures that the corresponding tissues in both scans have the same intensity. After it is estimated, the differential bias field between scan pairs is corrected with the appropriate filter (Lewis and Fox, 2004).
Although these registration steps enable an unbiased evaluation of age-related changes in individuals, they assume that the head size does not change across time. This assumption might be true in adult and elderly populations, but not for pediatric populations. It has not yet been well studied how an unbiased template can be developed to assess changes over time when head size is also changing.
6.2. Considerations for brain modeling based on longitudinal data of young children
As discussed in Mills and Tamnes’s review paper, there are some important considerations for brain modeling based on longitudinal data, including: (1) the growth model should be selected with consideration of the physiological plausibility within a certain age range; (2) any relevant group comparisons can be difficult because the developmental trajectories of brain structures are mainly non-linear and differences between groups might not be constant over time; (3) the impact of correcting brain measures for brain size in longitudinal data is still an open question, as little has been studied on the impact of such correction (Mills and Tamnes, 2014).
Applying correction for brain size may depend more on the research questions. If the questions focus on size-related changes, raw measures are generally used. Alternatively, if they are about differences that are largely invariant with brain size, corrected measures are used in preference. Mills et al. observed that correcting for the overall brain size could change the shape of the developmental curve of regional brain volumes but they could not conclude if this correction is suggested or not. They have also shown that the interpretation and consequently, the understanding of brain development (e.g. in the case of sexual dimorphism), might be driven by methodological differences (Mills et al., 2016). Therefore, it is important to be aware of this caveat and to understand how the overall brain size is related to the brain regions-of-interest to decide if corrected measures should be used or not.
Involving young children in longitudinal studies might raise further considerations, the first of which concerns the follow up frequency and whether it should be in weeks, months or years, given that the changes occur more rapidly in early development. Before starting longitudinal data collection, it might be useful to establish a priori when the different time points are acquired. This is mainly determined by the time intervals in which structural changes are expected to be assessed (e.g. in an infant population, significant structural changes might occur after weeks or months, and in older pediatric populations after years). This also depends on the feasibility of the data collection at these different sampling intervals. Large numbers of repeated scans help to improve estimates of average developmental trajectories and in capturing more subtle changes associated with development. However, acquiring a high number of samples also over-represents some individuals in the population and accommodation of numerous follow up scans at different time points is not always feasible in practice, with time and resources constraints.
Another consideration concerns the choice of transformations used for measuring brain growth. Considering the drastic changes that occur at early ages, it is possible that current methods for image registration are not effective in registering with the morphological changes that occur during this period of development. Diffeomorphic transformations are generally used in deformation-based morphometry for evaluating the deformation field, but it is still unknown if this type of transformation is sufficient for estimating the brain changes that occur in young children.
7. Limitations and future perspectives for young pediatric MRI
With the increasing number of studies focusing on young children, it became necessary to adapt the analysis methods because most of the existing standard software tools were not appropriate for the analysis of the pediatric brain. Child-adjusted methods offer elegant solutions that provide more accurate and less biased quantitative measures for MRI analysis. This field is still in its infancy, however, leaving room for additional advancement. This section lists the current limitations and future perspective for improvement of structural MRI analyses of the young pediatric brain.
7.1. Validations of novel child-adjusted methods
Novel methods are being continuously developed to overcome the limitations of current methods. For example, deep learning has recently become one of the trending methods in the field toward achieving brain segmentation. It consists of deep and recurrent networks using multiple processing layers to extract features, with backpropagation of errors to optimize the process to be learned (LeCun et al., 2015). Deep learning has been shown to outperform other segmentation methods applied for the analysis of brain aging (Chen et al., 2016, Mendrik et al., 2015) and this has been also demonstrated for brain segmentation in the infant population (Zhang et al., 2015). However, novel child-adjusted methods have yet to be extensively validated on pediatric data before being applied to clinical research questions.
Publicly available pediatric MRI-databases have been assembled by several research groups, including that from the National Institutes of Health (NIH) (Evans, 2006), from Nathan Kline Institute (https://www.nki.rfmh.org/), from Imperial College London (http://brain-development.org/), from the Pediatric Imaging, Neurocognition, and Genetics (PING) Study database (http://pingstudy.ucsd.edu/), and the Human Connectome Project Lifespan pilot imaging data (http://lifespan.humanconnectome.org/). In order to validate novel analysis methods, these databases can be used as common data source for benchmarking the performance of different methods or as a training dataset for learning-based algorithms. However, most MRI measures (e.g. tissue segmentation, volumes, etc.) are currently obtained via manual delineations that are subject to inter- and intra-rater variability. As a consequence, quality of results and method performance are more difficult to assess.
Validation enables the assessment of the performance of methods based on several criteria. Generally, the accuracy or the measurement error (i.e. the closeness or difference between the computed values and the “true” values) is the main criterion to assess performance. In studies of brain development, the measurement error should be smaller than the expected changes over time or of group differences, in order to consider the method reliable. Another important criterion is the reproducibility (i.e. agreement between measurements) that is assessed from test-retest scans for which all measures should be the same. Therefore, reproducibility is not prone to the variability of the manual delineations in contrast to the accuracy. In studies of brain development, high reproducibility is important to measure the intra-subject changes over time. This means that intrinsic variations due to methods when measured at one time point should be smaller than expected changes between different time points. Finally, processing time and associated hardware and software costs are also important features that should be considered for the analysis of large quantities of data, for which fully automated methods are most applicable.
Future steps to improve the validity of new, child-adjusted methods for MRI analyses might be to assess the correlation between different anatomical measures and their biological determinants (e.g. symptoms, cognitive functions, genetic and/or environmental factors). Measures that are highly correlated with biological determinants are particularly relevant for clinical applications, which can lead to improved brain modeling and understanding of the factors that influence brain development.
7.2. Integration in the analysis workflow
Once child-adjusted methods have been developed and validated, challenges remain for their integration into existing processing workflows. At present, there are a wide variety of software package tools available for MRI processing. However, few software tools are specific for young children, such as iBEAT (Dai et al., 2013), and few offer flexibility to adapt processing to different populations (e.g. ethnicity, pathology, gender) and to include different processing tasks and file formats, which renders the compatibility between different packages more complicated. Tools such as Nipype or the Connectome Mapper (Daducci et al., 2012) were developed to merge the different software packages into one pipeline. Yet, to our knowledge, there are no similar tools that provide such an interface with integrated child-adjusted features (such as enabling the use or the creation of age-specific and 4D brain atlases) in the processing pipeline (from preprocessing to the statistical analysis).
7.3. Addressing conflation of developmental changes
With the development of novel methods to streamline the processing and analysis of pediatric data, results obtained from these methods might become more study-specific, possibly leading to reduced reproducibility across studies and thus, to inconsistencies in data interpretation (e.g. different timing in maturation of brain regions-of-interest).
Understanding the potential reasons for variance in findings across studies aids the assessment of the reliability and validity of the methods used, and ultimately in choosing the most plausible brain model. Main causes of inconsistent findings could arise from either data-dependent (sample heterogeneity and cohort effects) or method-dependent (curve fitting method, MRI measures definition and computation) sources.
One method for addressing conflicting findings is to assess the consistency of results across datasets by applying and replicating the same method on different datasets from the same population (e.g. similar age). By applying the same method and adjusting for overall brain size, Mills et al. demonstrated convergence of findings for the developmental trajectories of intracranial and whole brain volumes between four datasets (Mills et al., 2016).
Although datasets are necessarily heterogeneous across studies, variations (for e.g, due to size, age, or gender) can be corrected with normalized measures and with appropriate curve fitting methods. Thus, residual differences in findings may result mainly from the methods used to estimate brain anatomy and their developmental trajectories. By measuring this divergence resulting from imaging methods, Walhovd et al. encouraged multi-modal neuroimaging efforts to measure the same phenomenon, as well as data sharing to have independent groups working on the same data sets (Walhovd et al., 2016). Applying different methods on the same data enables quantification oftheir impact on the results and comparing to existing data enables the determinination of which methods are more likely to provide reliable results.
8. Conclusion
Given that the brain undergoes extremely rapid changes in early life, with non-linear and region-specific growth patterns, it is important to implement revised tools for studying brain development over this period. Substantial effort is currently being dedicated toward developing child-adjusted methods for MRI data processing, such as provision of age-specific brain atlases, 4D spatio-temporal atlases, as well as methods for improving image quality and processing of longitudinal data. These methods are still in their infancy however, and need to be further tested and validated to achieve more accurate and unbiased results.
Conflict of Interest
None.
References
- Alexander-Bloch A., Clasen L., Stockman M., Ronan L., Lalonde F., Giedd J., Raznahan A. Subtle in-scanner motion biases automated measurement of brain anatomy from in vivo MRI. Hum. Brain Mapp. 2016;37:2385–2397. doi: 10.1002/hbm.23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibek S., Adamietz B., Cavallaro A., Stemmer A., Anders K., Kramer M., Bautz W., Staatz G. Contrast-enhanced T1-weighted fluid-attenuated inversion-recovery BLADE magnetic resonance imaging of the brain. an alternative to spin-echo technique for detection of brain lesions in the unsedated pediatric patient? Acad. Radiol. 2008;15:986–995. doi: 10.1016/j.acra.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Altaye M., Holland S.K., Wilke M., Gaser C. Infant brain probability templates for MRI segmentation and normalization. Neuroimage. 2008;43:721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Multimodal image coregistration and partitioning – a unified framework. Neuroimage. 1997;6:209–217. doi: 10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Aubert-Broche B., Fonov V.S., García-Lorenzo D., Mouiha A., Guizard N., Coupé P., Eskildsen S.F., Collins D.L. A new method for structural volume analysis of longitudinal brain MRI data and its application in studying the growth trajectories of anatomical brain structures in childhood. Neuroimage. 2013;82:393–402. doi: 10.1016/j.neuroimage.2013.05.065. [DOI] [PubMed] [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.M., Tanaka H., Stanley L., Nagamine M., Zakerani N., Thurston A., Kesler S., Hulme C., Lyytinen H., Glover G.H., Serrone C., Raman M.M., Reiss A.L., Hoeft F. Maternal history of reading difficulty is associated with reduced language-related gray matter in beginning readers. Neuroimage. 2012;59:3021–3032. doi: 10.1016/j.neuroimage.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal J.D., Zijdenbos A., Molloy E., Giedd J.N. Motion artifact in magnetic resonance imaging: implications for automated analysis. Neuroimage. 2002;16:89–92. doi: 10.1006/nimg.2002.1076. [DOI] [PubMed] [Google Scholar]
- Bohland J.W., Bokil H., Allen C.B., Mitra P.P., Salamon G. The brain atlas concordance problem: quantitative comparison of anatomical parcellations. PLoS One. 2009;4:e7200. doi: 10.1371/journal.pone.0007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora S., Pritchard V.E., Chen Z., Inder T.E., Woodward L.J. Neonatal cerebral morphometry and later risk of persistent inattention/hyperactivity in children born very preterm. J. Child Psychol. Psychiatry Allied Discip. 2014;55:828–838. doi: 10.1111/jcpp.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain Development Cooperative Group, B.D.C Total and regional brain volumes in a population-based normative sample from 4 to 18 years: the NIH MRI study of normal brain development. Cereb. Cortex. 2012;22:1–12. doi: 10.1093/cercor/bhr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain Development Cooperative Group Total and regional brain volumes in a population-based normative sample from 4 to 18 years: the NIH MRI Study of Normal Brain Development. Cereb. Cortex. 2012;22:1–12. doi: 10.1093/cercor/bhr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T.T., Jernigan T.L. Brain development during the preschool years. Neuropsychol. Rev. 2012;22(2):313–333. doi: 10.1007/s11065-012-9214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T.T., Kuperman J.M., Erhart M., White N.S., Roddey J.C., Shankaranarayanan A., Han E.T., Rettmann D., Dale A.M. Prospective motion correction of high-resolution magnetic resonance imaging data in children. Neuroimage. 2010;53:139–145. doi: 10.1016/j.neuroimage.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T.T., Kuperman J.M., Chung Y., Erhart M., McCabe C., Hagler D.J., Venkatraman V.K., Akshoomoff N., Amaral D.G., Bloss C.S., Casey B.J., Chang L., Ernst T.M., Frazier J.A., Gruen J.R., Kaufmann W.E., Kenet T., Kennedy D.N., Murray S.S., Sowell E.R., Jernigan T.L., Dale A.M. Neuroanatomical assessment of biological maturity. Curr. Biol. 2012;22:1693–1698. doi: 10.1016/j.cub.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas M., Oliver A., Lladó X., Freixenet J., Bach Cuadra M. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Programs Biomed. 2011;104:e158–e177. doi: 10.1016/j.cmpb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Cardoso M.J., Leung K., Modat M., Keihaninejad S., Cash D., Barnes J., Fox N.C., Ourselin S. STEPS: similarity and truth estimation for propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 2013;17:671–684. doi: 10.1016/j.media.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Cardoso M.J., Melbourne A., Kendall G.S., Modat M., Robertson N.J., Marlow N., Ourselin S. AdaPT: an adaptive preterm segmentation algorithm for neonatal brain MRI. Neuroimage. 2013;65:97–108. doi: 10.1016/j.neuroimage.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Castellanos F.X., Lee P.P., Sharp W., Jeffries N.O., Greenstrein D.K., Clasen L.S., Blumenthal J.D., James R.S., Ebens C.L., Walter J.M., Zijdenbos A., Evans A.C., Giedd J.N., Rapoport J.L. Developmental trajectories of brain volume abnormalities in children and adolescents with attention-deficit/hyperactivity disorder. JAMA. 2002;288:1740–1748. doi: 10.1001/jama.288.14.1740. [DOI] [PubMed] [Google Scholar]
- Chen H., Dou Q., Yu L., Heng P.-A. VoxResNet: deep voxelwise residual networks for volumetric brain segmentation. arXiv. 2016;05895:1–9. doi: 10.1016/j.neuroimage.2017.04.041. [DOI] [PubMed] [Google Scholar]
- Choe M.S., Ortiz-Mantilla S., Makris N., Gregas M., Bacic J., Haehn D., Kennedy D., Pienaar R., Caviness V.S., Benasich A.A., Ellen Grant P. Regional infant brain development: an MRI-based morphometric analysis in 3–13 month olds. Cereb. Cortex. 2013;23:2100–2117. doi: 10.1093/cercor/bhs197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Helland T., Specht K., Narr K.L., Manis F.R., Toga A.W., Hugdahl K. Neuroanatomical precursors of dyslexia identified from pre-reading through to age 11. Brain. 2014;137:3136–3141. doi: 10.1093/brain/awu229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D.L., Neelin P., Peters T.M., Evans A.C. Automatic 3D intersuject registration fo MR volumetric data in standardized talairach space. J. Comput. Assist. Tomogr. 1994;18(2):192–205. [PubMed] [Google Scholar]
- Collins D.L., Holmes C.J., Peters T.M., Evans A.C. Automatic 3-D model-based neuroanatomical segmentation. Hum. Brain Mapp. 1995;3:190–208. [Google Scholar]
- Collins D.L., Zijdenbos A.P., Baaré W.F.C., Evans A.C. Springer; Berlin, Heidelberg: 1999. ANIMAL + INSECT: improved Cortical Structure Segmentation; pp. 210–223. [Google Scholar]
- Courchesne E., Chisum H.J., Townsend J., Cowles A., Covington J., Egaas B., Harwood M., Hinds S., Press G.A. Normal brain development and aging: quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology. 2000;216:672–682. doi: 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Daducci A., Gerhard S., Griffa A., Lemkaddem A., Cammoun L., Gigandet X., Meuli R., Hagmann P., Thiran J.P. The connectome mapper: an open-source processing pipeline to map connectomes with MRI. PLoS One. 2012;7:e48121. doi: 10.1371/journal.pone.0048121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Shi F., Wang L., Wu G., Shen D. IBEAT: a toolbox for infant brain magnetic resonance image processing. Neuroinformatics. 2013;11:211–225. doi: 10.1007/s12021-012-9164-z. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davidson M.C., Thomas K.M., Casey B.J. Imaging the developing brain with fMRI. Ment. Retard. Dev. Disabil. Res. Rev. 2003;9:161–167. doi: 10.1002/mrdd.10076. [DOI] [PubMed] [Google Scholar]
- Dekaban A.S., Sadowsky D. Changes in brain weights during the span of human life: relation of brain weights to body heights and body weights. Ann. Neurol. 1978;4:345–356. doi: 10.1002/ana.410040410. [DOI] [PubMed] [Google Scholar]
- Dennis E.L., Thompson P.M. Typical and atypical brain development: a review of neuroimaging studies. Dialogues Clin. Neurosci. 2013;15:359–384. doi: 10.31887/DCNS.2013.15.3/edennis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni S.C.L., Dean D.C., Remer J., Dirks H., O’Muircheartaigh J., O’Muircheartaigh J. Cortical maturation and myelination in healthy toddlers and young children. Neuroimage. 2015;115:147–161. doi: 10.1016/j.neuroimage.2015.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J., Kulikova S., Hertz-Pannier L., Mangin J.F., Dehaene-Lambertz G., Poupon C. Correction strategy for diffusion-weighted images corrupted with motion: application to the DTI evaluation of infants’ white matter. Magn. Reson. Imaging. 2014;32:981–992. doi: 10.1016/j.mri.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Ducharme S., Albaugh M.D., Nguyen T.-V., Hudziak J.J., Mateos-Pérez J.M., Labbe A., Evans A.C., Karama S. Trajectories of cortical thickness maturation in normal brain development — the importance of quality control procedures. Neuroimage. 2016;125:267–279. doi: 10.1016/j.neuroimage.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans A.C. The NIH MRI study of normal brain development. Neuroimage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Fennema-Notestine C., Ozyurt I.B., Clark C.P., Morris S., Bischoff-Grethe A., Bondi M.W., Jernigan T.L., Fischl B., Segonne F., Shattuck D.W., Leahy R.M., Rex D.E., Toga A.W., Zou K.H., Brown G.G. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: effects of diagnosis, bias correction, and slice location. Hum. Brain Mapp. 2006;27:99–113. doi: 10.1002/hbm.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore P.T., Richards J.E., Phillips-Meek M.C., Cryer A., Stevens M. Stereotaxic magnetic resonance imaging brain atlases for infants from 3 to 12 months. Dev. Neurosci. 2015;37:515–532. doi: 10.1159/000438749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Sereno M.I., Dale A.M. Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fonov V., Evans A.C., McKinstry R.C., Almli C.R., Collins D.L. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage. 2009;47:S102. [Google Scholar]
- Fonov V., Evans A.C., Botteron K., Almli C.R., McKinstry R.C., Collins D.L. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54:313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.W., Wang Y., Grimm R.C., Rossman P.J., Felmlee J.P., Riederer S.J., Ehman R.L. Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magn. Reson. Med. 1995;34:746–753. doi: 10.1002/mrm.1910340514. [DOI] [PubMed] [Google Scholar]
- Gedamu E.L., Collins D.L., Arnold D.L. Automated quality control of brain MR images. J. Magn. Reson. Imaging. 2008;28:308–319. doi: 10.1002/jmri.21434. [DOI] [PubMed] [Google Scholar]
- Ghosh S.S., Kakunoori S., Augustinack J., Nieto-Castanon A., Kovelman I., Gaab N., Christodoulou J.A., Triantafyllou C., Gabrieli J.D.E., Fischl B. Evaluating the validity of volume-based and surface-based brain image registration for developmental cognitive neuroscience studies in children 4 to 11years of age. Neuroimage. 2010;53:85–93. doi: 10.1016/j.neuroimage.2010.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd J.N., Raznahan A., Alexander-Bloch A., Schmitt E., Gogtay N., Rapoport J.L. Child psychiatry branch of the national institute of mental health longitudinal structural magnetic resonance imaging study of human brain development. Neuropsychopharmacology. 2015;40:43–49. doi: 10.1038/npp.2014.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore J.H., Shi F., Woolson S.L., Knickmeyer R.C., Short S.J., Lin W., Zhu H., Hamer R.M., Styner M., Shen D. Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cereb. Cortex. 2012;22:2478–2485. doi: 10.1093/cercor/bhr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Coalson T.S., Robinson E.C., Hacker C.D., Harwell J., Yacoub E., Ugurbil K., Andersson J., Beckmann C.F., Jenkinson M., Smith S.M., Van Essen D.C. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddings A.L., Mills K.L., Clasen L.S., Giedd J.N., Viner R.M., Blakemore S.J. The influence of puberty on subcortical brain development. Neuroimage. 2014;88:242–251. doi: 10.1016/j.neuroimage.2013.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N., Giedd J.N., Lusk L., Hayashi K.M., Greenstein D., Vaituzis A.C., Nugent T.F., Herman D.H., Clasen L.S., Toga A.W., Rapoport J.L., Thompson P.M. Dynamic mapping of human cortical development during childhood through early adulthood. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousias I.S., Rueckert D., Heckemann R.A., Dyet L.E., Boardman J.P., Edwards A.D., Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40:672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Gousias I.S., Hammers A., Counsell S.J., Srinivasan L., Rutherford M.A., Heckemann R.A., Hajnal J.V., Rueckert D., Edwards A.D. Magnetic resonance imaging of the newborn brain: automatic segmentation of brain images into 50 anatomical regions. PLoS One. 2013:8. doi: 10.1371/journal.pone.0059990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene D.J., Black K.J., Schlaggar B.L. Considerations for MRI study design and implementation in pediatric and clinical populations. Dev. Cogn. Neurosci. 2016;18:101–112. doi: 10.1016/j.dcn.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grydeland H., Walhovd K.B., Tamnes C.K., Westlye L.T., Fjell A.M. Intracortical myelin links with performance variability across the human lifespan: results from T1- and T2-weighted MRI myelin mapping and diffusion tensor imaging. J. Neurosci. 2013;33:18618–18630. doi: 10.1523/JNEUROSCI.2811-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Jovicich J., Salat D., van der Kouwe A., Quinn B., Czanner S., Busa E., Pacheco J., Albert M., Killiany R., Maguire P., Rosas D., Makris N., Dale A., Dickerson B., Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32:180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Hazlett H.C., Gu H., McKinstry R.C., Shaw D.W.W., Botteron K.N., Dager S.R., Styner M., Vachet C., Gerig G., Paterson S.J., Schultz R.T., Estes A.M., Evans A.C., Piven J. Brain volume findings in 6-month-old infants at high familial risk for autism. Am. J. Psychiatry. 2012;169:601–608. doi: 10.1176/appi.ajp.2012.11091425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett H.C., Gu H., Munsell B.C., Kim S.H., Styner M., Wolff J.J., Elison J.T., Swanson M.R., Zhu H., Botteron K.N., Collins D.L., Constantino J.N., Dager S.R., Estes A.M., Evans A.C., Fonov V.S., Gerig G., Kostopoulos P., Mckinstry R.C., Pandey J., Paterson S., Pruett Jr J.R., Schultz R.T., Shaw D.W., Zwaigenbaum L., Piven J. Early brain development in infants at high risk for autism spectrum disorder. Nat. Publ. Gr. 2017;542 doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman A.M., van Haren N.E.M., Schnack H.G., Kahn R.S., Hulshoff Pol H.E. Human brain changes across the life span: a review of 56 longitudinal magnetic resonance imaging studies. Hum. Brain Mapp. 2012;33:1987–2002. doi: 10.1002/hbm.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeksma M.R., Kenemans J.L., Kemner C., van Engeland H. Variability in spatial normalization of pediatric and adult brain images. Clin. Neurophysiol. 2005;116:1188–1194. doi: 10.1016/j.clinph.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Holland D., Kuperman J.M., Dale A.M. Efficient correction of inhomogeneous static magnetic field-induced distortion in Echo Planar Imaging. Neuroimage. 2010;50:175–183. doi: 10.1016/j.neuroimage.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D., Chang L., Ernst T.M., Curran M., Buchthal S.D., Alicata D., Skranes J., Johansen H., Hernandez A., Yamakawa R., Kuperman J.M., Dale A.M. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol. 2014;71:1266. doi: 10.1001/jamaneurol.2014.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G.L., Lombroso C.T. Prognostic value of background patterns in the neonatal EEG. J. Clin. Neurophysiol. 1993;10:323–352. doi: 10.1097/00004691-199307000-00008. [DOI] [PubMed] [Google Scholar]
- Hoogman M., Bralten J., Hibar D.P., Mennes M., Zwiers M.P., Schweren L.S.J., van Hulzen K.J.E., Medland S.E., Shumskaya E., Jahanshad N., Zeeuw, de P., Szekely E., Sudre G., Wolfers T., Onnink A.M.H., Dammers J.T., Mostert J.C., Vives-Gilabert Y., Kohls G., Oberwelland E., Seitz J., Schulte-Rüther M., Ambrosino S., Doyle A.E., Høvik M.F., Dramsdahl M., Tamm L., van Erp T.G.M., Dale A., Schork A., Conzelmann A., Zierhut K., Baur R., McCarthy H., Yoncheva Y.N., Cubillo A., Chantiluke K., Mehta M.A., Paloyelis Y., Hohmann S., Baumeister S., Bramati I., Mattos P., Tovar-Moll F., Douglas P., Banaschewski T., Brandeis D., Kuntsi J., Asherson P., Rubia K., Kelly C., Martino A., Di, Milham M.P., Castellanos F.X., Frodl T., Zentis M., Lesch K.-P., Reif A., Pauli P., Jernigan T.L., Haavik J., Plessen K.J., Lundervold A.J., Hugdahl K., Seidman L.J., Biederman J., Rommelse N., Heslenfeld D.J., Hartman C.A., Hoekstra P.J., Oosterlaan J., Polier G., von, Konrad K., Vilarroya O., Ramos-Quiroga J.A., Soliva J.C., Durston S., Buitelaar J.K., Faraone S., V, Shaw P., Thompson P.M., Franke B. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry. 2017;4:310–319. doi: 10.1016/S2215-0366(17)30049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S.M.H., Black J.M., Soriano T., Bugescu N., Martinez R., Raman M.M., Kesler S.R., Hoeft F. Topological properties of large-scale structural brain networks in children with familial risk for reading difficulties. Neuroimage. 2013;71:260–274. doi: 10.1016/j.neuroimage.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Coupé P., Pruessner J., Collins L. 2011. Validation of Appearance-model Based Segmentation with Patch-based Refinement on Medial Temporal Lobe Structures; pp. 28–37. [Google Scholar]
- Hu S., Pruessner J.C., Coupe P., Collins D.L. Volumetric analysis of medial temporal lobe structures in brain development from childhood to adolescence. Neuroimage. 2013;74:276–287. doi: 10.1016/j.neuroimage.2013.02.032. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Sabuncu M.R. Multi-atlas segmentation of biomedical images: a survey. Med. Image Anal. 2015;24:205–219. doi: 10.1016/j.media.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im K., Raschle N.M., Smith S.A., Ellen Grant P., Gaab N. A typical sulcal pattern in children with developmental dyslexia and at-risk kindergarteners. Cereb. Cortex. 2016;26:1138–1148. doi: 10.1093/cercor/bhu305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Sima D.M., Ribbens A., Cambron M., Maertens A., Van Hecke W., De Mey J., Barkhof F., Steenwijk M.D., Daams M., Maes F., Van Huffel S., Vrenken H., Smeets D. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015;8:367–375. doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E.J., Woolrich M.W., Smith S.M. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R., Kennedy D., Schmitt F., Brown G., MacFall J., Fischl B., Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Knickmeyer R.C., Gouttard S., Kang C., Evans D., Wilber K., Smith J.K., Hamer R.M., Lin W., Gerig G., Gilmore J.H. A structural MRI study of human brain development from birth to 2 years. J. Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P., Lancaster J.L., Glahn D.C., Purdy D., Laird A.R., Gao F., Fox P. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum. Brain Mapp. 2006;27:957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsrud S.K., Tamnes C.K., Fjell A.M., Amlien I., Grydeland H., Sulutvedt U., Due-Tønnessen P., Bjørnerud A., Sølsnes A.E., Håberg A.K., Skrane J., Walhovd K.B. Development of hippocampal subfield volumes from 4 to 22 years. Hum. Brain Mapp. 2014;35:5646–5657. doi: 10.1002/hbm.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklisova-Murgasova M., Aljabar P., Srinivasan L., Counsell S.J., Doria V., Serag A., Gousias I.S., Boardman J.P., Rutherford M.A., Edwards A.D., Hajnal J.V., Rueckert D. A dynamic 4D probabilistic atlas of the developing brain. Neuroimage. 2011;54:2750–2763. doi: 10.1016/j.neuroimage.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Kuperman J.M., Brown T.T., Ahmadi M.E., Erhart M.J., White N.S., Roddey J.C., Shankaranarayanan A., Han E.T., Rettmann D., Dale A.M. Prospective motion correction improves diagnostic utility of pediatric MRI scans. Pediatr. Radiol. 2011;41:1578–1582. doi: 10.1007/s00247-011-2205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Lebel C., Beaulieu C. Longitudinal development of human brain wiring continues from childhood into adulthood. J. Neurosci. 2011;31:10937–10947. doi: 10.1523/JNEUROSCI.5302-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-M., Yoon U., Nam S.H., Kim J.-H., Kim I.-Y., Kim S.I. Evaluation of automated and semi-automated skull-stripping algorithms using similarity index and segmentation error. Comput. Biol. Med. 2003;33:495–507. doi: 10.1016/s0010-4825(03)00022-2. [DOI] [PubMed] [Google Scholar]
- Lenroot R.K., Giedd J.N. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci. Biobehav. Rev. 2006;30:718–729. doi: 10.1016/j.neubiorev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Leow A.D., Klunder A.D., Jack C.R., Toga A.W., Dale A.M., Bernstein M.A., Britson P.J., Gunter J.L., Ward C.P., Whitwell J.L., Borowski B.J., Fleisher A.S., Fox N.C., Harvey D., Kornak J., Schuff N., Studholme C., Alexander G.E., Weiner M.W., Thompson P.M. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B., Fox N.C. Correction of differential intensity inhomogeneity in longitudinal MR images. Neuroimage. 2004;23:75–83. doi: 10.1016/j.neuroimage.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Li G., Wang L., Shi F., Lin W., Shen D. Simultaneous and consistent labeling of longitudinal dynamic developing cortical surfaces in infants. Med. Image Anal. 2014;18:1274–1289. doi: 10.1016/j.media.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang L., Shi F., Lyall A.E., Lin W., Gilmore J.H., Shen D. Mapping longitudinal development of local cortical gyrification in infants from birth to 2 years of age. J. Neurosci. 2014;34:4228–4238. doi: 10.1523/JNEUROSCI.3976-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang L., Shi F., Gilmore J.H., Lin W., Shen D. Construction of 4D high-definition cortical surface atlases of infants: methods and applications. Med. Image Anal. 2015;25:22–36. doi: 10.1016/j.media.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhu T., Zhong J. Comparison of quality control software tools for diffusion tensor imaging. Magn. Reson. Imaging. 2015;33:276–285. doi: 10.1016/j.mri.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Lyall A.E., Shi F., Geng X., Woolson S., Li G., Wang L., Hamer R.M., Shen D., Gilmore J.H. Dynamic development of regional cortical thickness and surface area in early childhood. Cereb. Cortex. 2015;25:2204–2212. doi: 10.1093/cercor/bhu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machilsen B., d’Agostino E., Maes F., Vandermeulen D., Hahn H.K., Lagae L., Stiers P. Linear normalization of MR brain images in pediatric patients with periventricular leukomalacia. Neuroimage. 2007;35:686–697. doi: 10.1016/j.neuroimage.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Maclaren J., Herbst M., Speck O., Zaitsev M. Prospective motion correction in brain imaging: a review. Magn. Reson. Med. 2013;69:621–636. doi: 10.1002/mrm.24314. [DOI] [PubMed] [Google Scholar]
- Magnotta V.A., Friedman L., First Birn F. Measurement of signal-to-noise and contrast-to-noise in the fBIRN multicenter imaging study. J. Digit. Imaging. 2006;19:140–147. doi: 10.1007/s10278-006-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makropoulos A., Aljabar P., Wright R., Hüning B., Merchant N., Arichi T., Tusor N., Hajnal J.V., Edwards A.D., Counsell S.J., Rueckert D. Regional growth and atlasing of the developing human brain. Neuroimage. 2016;125:456–478. doi: 10.1016/j.neuroimage.2015.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S.P., Smith M.S., Weinberger E. Perceived anxiety of pediatric patients to magnetic resonance. Clin. Pediatr. (Phila) 1995;34:59–60. doi: 10.1177/000992289503400114. [DOI] [PubMed] [Google Scholar]
- McLeish K., Kozerke S., Crum W.R., Hill D.L.G. Free-breathing radial acquisitions of the heart. Magn. Reson. Med. 2004;52:1127–1135. doi: 10.1002/mrm.20252. [DOI] [PubMed] [Google Scholar]
- Mendrik A.M., Vincken K.L., Kuijf H.J., Breeuwer M., Bouvy W.H., de Bresser J., Alansary A., de Bruijne M., Carass A., El-Baz A., Jog A., Katyal R., Khan A.R., van der Lijn F., Mahmood Q., Mukherjee R., van Opbroek A., Paneri S., Pereira S., Persson M., Rajchl M., Sarikaya D., Smedby Ö., Silva C.A., Vrooman H.A., Vyas S., Wang C., Zhao L., Biessels G.J., Viergever M.A., Zhao L., Biessels G.J., Viergever M.A. MRBrainS challenge: online evaluation framework for brain image segmentation in 3T MRI scans. Comput. Intell. Neurosci. 2015;2015:1–16. doi: 10.1155/2015/813696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.L., Tamnes C.K. Methods and considerations for longitudinal structural brain imaging analysis across development. Dev. Cogn. Neurosci. 2014;9:172–190. doi: 10.1016/j.dcn.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.L., Goddings A.-L., Herting M.M., Meuwese R., Blakemore S.-J., Crone E.A., Dahl R.E., Güroğlu B., Raznahan A., Sowell E.R., Tamnes C.K. Structural brain development between childhood and adulthood: convergence across four longitudinal samples. Neuroimage. 2016;141:273–281. doi: 10.1016/j.neuroimage.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgasova M., Dyet L., Edwards D., Rutherford M., Hajnal J., Rueckert D. Segmentation of brain MRI in young children. Acad. Radiol. 2007;14:1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Muzik O., Chugani D.C., Juhász C., Shen C., Chugani H.T. Statistical parametric mapping: assessment of application in children. Neuroimage. 2000;12:538–549. doi: 10.1006/nimg.2000.0651. [DOI] [PubMed] [Google Scholar]
- Nie J., Li G., Shen D. Development of cortical anatomical properties from early childhood to early adulthood. Neuroimage. 2013;76:216–224. doi: 10.1016/j.neuroimage.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl C.W., Lange N., Li D.D., Barnett L.A., Lee A., Buonocore M.H., Simon T.J., Rogers S., Ozonoff S., Amaral D.G. Brain enlargement is associated with regression in preschool-age boys with autism spectrum disorders. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20195–21200. doi: 10.1073/pnas.1107560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Zilles K., Amunts K., Faria A., Jiang H., Li X., Akhter K., Hua K., Woods R., Toga A.W., Pike G.B., Rosa-Neto P., Evans A., Zhang J., Huang H., Miller M.I., Van Zijl P.C.M., Mazziotta J., Mori S. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. Neuroimage. 2008;43:447–457. doi: 10.1016/j.neuroimage.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Mori S., Donohue P.K., Ernst T., Anderson L., Buchthal S., Faria A., Jiang H., Li X., Miller M.I., van Zijl P.C.M., Chang L. Multi-contrast human neonatal brain atlas: application to normal neonate development analysis. Neuroimage. 2011;56:8–20. doi: 10.1016/j.neuroimage.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourselin S., Roche A., Prima S., Ayache N. Springer; Berlin Heidelberg: 2000. Block Matching: A General Framework to Improve Robustness of Rigid Registration of Medical Images; pp. 557–566. [Google Scholar]
- Ozernov-Palchik O., Gaab N. Tackling the dyslexia paradox: reading brain and behavior for early markers of developmental dyslexiax. Wiley Interdiscip. Rev. Cogn. Sci. 2016;7:156–176. doi: 10.1002/wcs.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z., Judaš M., Šimic G., Rasin M.R., Uylings H.B.M., Rakic P., Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipe J.G. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn. Reson. Med. 1999;42:963–969. doi: 10.1002/(sici)1522-2594(199911)42:5<963::aid-mrm17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Prastawa M., Gilmore J.H., Lin W., Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Raschle N.M., Chang M., Gaab N. Structural brain alterations associated with dyslexia predate reading onset. Neuroimage. 2011;57:742–749. doi: 10.1016/j.neuroimage.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle N.M., Zuk J., Ortiz-Mantilla S., Sliva D.D., Franceschi A., Grant P.E., Benasich A.A., Gaab N. Pediatric neuroimaging in early childhood and infancy: challenges and practical guidelines. Ann. N. Y. Acad. Sci. 2012;1252:43–50. doi: 10.1111/j.1749-6632.2012.06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A., Shaw P.W., Lerch J.P., Clasen L.S., Greenstein D., Berman R., Pipitone J., Chakravarty M.M., Giedd J.N. Longitudinal four-dimensional mapping of subcortical anatomy in human development. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1592–1597. doi: 10.1073/pnas.1316911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Fischl B. Avoiding asymmetry-induced bias in longitudinal image processing. Neuroimage. 2011;57(1):19–21. doi: 10.1016/j.neuroimage.2011.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmansky N.J., Rosas H.D., Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Tisdall M.D., Qureshi A., Buckner R.L., van der Kouwe A.J.W., Fischl B. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. Neuroimage. 2015;107:107–115. doi: 10.1016/j.neuroimage.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J.E., Sanchez C., Phillips-Meek M., Xie W. A database of age-appropriate average MRI templates. Neuroimage. 2016;124:1254–1259. doi: 10.1016/j.neuroimage.2015.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière D., Régis J., Cointepas Y., Ochiai T., Cachia A. A freely available Anatomist/BrainVISA package for structural morphometry of the cortical sulci. Neuroimage. 2003;19:934. [Google Scholar]
- Rohde G.K., Barnett A.S., Basser P.J., Marenco S., Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn. Reson. Med. 2004;51:103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- Ségonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Sanchez C.E., Richards J.E., Almli C.R. Neurodevelopmental MRI brain templates for children from 2 weeks to 4 years of age. Dev. Psychobiol. 2012;54:77–91. doi: 10.1002/dev.20579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker D., Buss C., Head K., Sandman C.A., Davis E.P., Chakravarty M.M., Gauthier S., Pruessner J.C. Hippocampus and amygdala volumes from magnetic resonance images in children: assessing accuracy of FreeSurfer and FSL against manual segmentation. Neuroimage. 2016;129:1–14. doi: 10.1016/j.neuroimage.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann C.M., Bloss C.S., Barnes C.C., Wideman G.M., Carper R.A., Akshoomoff N., Pierce K., Hagler D., Schork N., Lord C., Courchesne E. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J. Neurosci. 2010;30:4419–4427. doi: 10.1523/JNEUROSCI.5714-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serag A., Aljabar P., Ball G., Counsell S.J., Boardman J.P., Rutherford M.A., Edwards A.D., Hajnal J.V., Rueckert D. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage. 2012;59:2255–2265. doi: 10.1016/j.neuroimage.2011.09.062. [DOI] [PubMed] [Google Scholar]
- Serag A., Edwards A.D., Hajnal J.V., Counsell S.J., Boardman J.P., Rueckert D. A multi-channel 4D probabilistic atlas of the developing brain: application to fetuses and neonates. Ann. Br. 2012:1–14. [Google Scholar]
- Serag A., Blesa M., Moore E.J., Pataky R., Sparrow S.A., Wilkinson A.G., Macnaught G., Semple S.I., Boardman J.P. Accurate Learning with Few Atlases (ALFA): an algorithm for MRI neonatal brain extraction and comparison with 11 publicly available methods. Sci. Rep. 2016;6:23470. doi: 10.1038/srep23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck D.W., Sandor-Leahy S.R., Schaper K.A., Rottenberg D.A., Leahy R.M. Magnetic resonance image tissue classification using a partial volume model. Neuroimage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shaul S. Brain Research in Language. Springer; US, Boston, MA: 2008. Event-Related Potentials (ERPS) in the study of dyslexia; pp. 51–92. [Google Scholar]
- Shaw P., Eckstrand K., Sharp W., Blumenthal J., Lerch J.P., Greenstein D., Clasen L., Evans A., Giedd J., Rapoport J.L. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19649–19654. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Lalonde F., Lepage C., Rabin C., Eckstrand K., Sharp W., Greenstein D., Evans A., Giedd J.N., Rapoport J. Development of cortical asymmetry in typically developing children and its disruption in attention-deficit/hyperactivity disorder. Arch. Gen. Psychiatry. 2009;66:888–896. doi: 10.1001/archgenpsychiatry.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.D., Nordahl C.W., Young G.S., Wootton-Gorges S.L., Lee A., Liston S.E., Harrington K.R., Ozonoff S., Amaral D.G. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain. 2013;136:2825–2835. doi: 10.1093/brain/awt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Yap P.T., Wu G., Jia H., Gilmore J.H., Lin W., Shen D. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6:e18746. doi: 10.1371/journal.pone.0018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Wang L., Dai Y., Gilmore J.H., Lin W., Shen D. LABEL: Pediatric brain extraction using learning-based meta-algorithm. Neuroimage. 2012;62:1975–1986. doi: 10.1016/j.neuroimage.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk T.J., Genc S., Anderson V., Efron D., Hazell P., Nicholson J.M., Kean M., Malpas C.B., Sciberras E. Developmental brain trajectories in children with ADHD and controls: a longitudinal neuroimaging study. BMC Psychiatry. 2016;16:59. doi: 10.1186/s12888-016-0770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J.D., Willett J.B. Applied longitudinal data analysis: modeling change and event occurrence. Appl. Longitudinal Data Anal.: Model. Change Event Occurrence. 2009;15(1):1–644. [Google Scholar]
- Smeets D., Ribbens A., Sima D.M., Cambron M., Horakova D., Jain S., Maertens A., Van Vlierberghe E., Terzopoulos V., Van Binst A.-M., Vaneckova M., Krasensky J., Uher T., Seidl Z., De Keyser J., Nagels G., De Mey J., Havrdova E., Van Hecke W. Reliable measurements of brain atrophy in individual patients with multiple sclerosis. Brain Behav. 2016;6:e00518. doi: 10.1002/brb3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Zhang Y., Jenkinson M., Chen J., Matthews P.M., Federico A., De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E.J., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell E.R., Thompson P.M., Tessner K.D., Toga A.W. Mapping continued brain growth and gray matter density reduction in dorsal frontal cortex: inverse relationships during postadolescent brain maturation. J. Neurosci. 2001;21:8819–8829. doi: 10.1523/JNEUROSCI.21-22-08819.2001. (21/22/8819 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell E.R. Longitudinal mapping of cortical thickness and brain growth in normal children. J. Neurosci. 2004;24:8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau G.Z., Peterson B.S. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theys C., Wouters J., Ghesquiere P. Diffusion tensor imaging and resting-state functional MRI-scanning in 5- and 6-year-old children: training protocol and motion assessment. PLoS One. 2014;9:1–7. doi: 10.1371/journal.pone.0094019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M., Giedd J.N., Woods R.P., MacDonald D., Evans A.C., Toga A.W. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Thompson P.M., Hayashi K.M., Sowell E.R., Gogtay N., Giedd J.N., Rapoport J.L., De Zubicaray G.I., Janke A.L., Rose S.E., Semple J., Doddrell D.M., Wang Y., Van Erp T.G.M., Cannon T.D., Toga A.W. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage. 2004:S2–18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Van Leemput K., Maes F., Vandermeulen D., Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imaging. 1999;18:885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- Vanderauwera J., Altarelli I., Vandermosten M., Vos A., De Wouters J., Ghesquière P. Atypical structural asymmetry of the planum temporale is related to family history of dyslexia. Cereb. Cortex. 2016;57:1–10. doi: 10.1093/cercor/bhw348. [DOI] [PubMed] [Google Scholar]
- Vandermosten M., Hoeft F., Norton E.S. How MRI brain imaging studies of pre-reading children inform theories of the etiology of developmental dyslexia and educational practice. Curr. Opin. Behav. Sci. 2016;10:155–161. doi: 10.1016/j.cobeha.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhatalo S., Kaila K. Development of neonatal EEG activity: from phenomenology to physiology. Semin. Fetal Neonatal Med. 2006;11:471–478. doi: 10.1016/j.siny.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Vogel S.E., Matejko A.A., Ansari D. Imaging the developing human brain using functional and structural magnetic resonance imaging: methodological and practical guidelines. In: Prior J., Van Herwegen J., editors. Practical Research with Children. Psychology Press; 2016. pp. 46–69. [Google Scholar]
- Volpe J.J. Overview: normal and abnormal human brain development. Ment. Retard. Dev. Disabil. Res. Rev. 2000;6:1–5. doi: 10.1002/(SICI)1098-2779(2000)6:1<1::AID-MRDD1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Walhovd K.B., Fjell A.M., Giedd J., Dale A.M., Brown T.T. Through thick and thin: a need to reconcile contradictory results on trajectories in human cortical development. Cereb. Cortex. 2016;1989:bhv301. doi: 10.1093/cercor/bhv301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Vachet C., Rumple A., Gouttard S., Ouziel C., Perrot E., Du G., Huang X., Gerig G., Styner M. Multi-atlas segmentation of subcortical brain structures via the AutoSeg software pipeline. Front. Neuroinform. 2014;8:7. doi: 10.3389/fninf.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mauer M., Raney V., Peysakhovich T., Becker B., Sliva B.L.C., Gaab D.D. Development of tract-specific white matter pathways during early reading development in at-risk children and typical controls. Cereb. Cortex. 2016:1–17. doi: 10.1093/cercor/bhw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M.J., Elashoff M., Weickert C.S. Molecular evidence that cortical synaptic growth predominates during the first decade of life in humans. Int. J. Dev. Neurosci. 2011;29:225–236. doi: 10.1016/j.ijdevneu.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Westlye L.T., Walhovd K.B., Dale A.M., Bjørnerud A., Due-Tønnessen P., Engvig A., Grydeland H., Tamnes C.K., Østby Y., Fjell A.M. Differentiating maturational and aging-related changes of the cerebral cortex by use of thickness and signal intensity. Neuroimage. 2010;52:172–185. doi: 10.1016/j.neuroimage.2010.03.056. [DOI] [PubMed] [Google Scholar]
- Westlye L.T., Walhovd K.B., Dale A.M., Bjørnerud A., Due-Tønnessen P., Engvig A., Grydeland H., Tamnes C.K., Østby Y., Fjell A.M. Life-span changes of the human brain white matter: diffusion tensor imaging (DTI) and volumetry. Cereb. Cortex. 2010;20:2055–2068. doi: 10.1093/cercor/bhp280. [DOI] [PubMed] [Google Scholar]
- White N., Roddey C., Shankaranarayanan A., Han E., Rettmann D., Santos J., Kuperman J., Dale A. PROMO: real-time prospective motion correction in MRI using image-based tracking. Magn. Reson. Med. 2010;63:91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford T.J., Rennie C.J., Grieve S.M., Clark C.R., Gordon E., Williams L.M. Brain maturation in adolescence: concurrent changes in neuroanatomy and neurophysiology. Hum. Brain Mapp. 2007;28:228–237. doi: 10.1002/hbm.20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M., Holland S.K., Myseros J.S., Schmithorst V.J., Ball W.S. Functional magnetic resonance imaging in pediatrics. Neuropediatrics. 2003;34:225–233. doi: 10.1055/s-2003-43260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M., Schmithorst V.J., Holland S.K. Normative pediatric brain data for spatial normalization and segmentation differs from standard adult data. Magn. Reson. Med. 2003;50:749–757. doi: 10.1002/mrm.10606. [DOI] [PubMed] [Google Scholar]
- Wilke M., Holland S.K., Altaye M., Gaser C. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage. 2008;41:903–913. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Yakovlev P.I., Lecours A.-R. Regional Development of Brain in Early Life. 1967. The myelogenetic cycles of regional maturation of the brain; pp. 3–70. [Google Scholar]
- Yang X.-R., Carrey N., Bernier D., MacMaster F.P. Cortical thickness in young treatment-naive children with ADHD. J. Atten. Disord. 2015;19:925–930. doi: 10.1177/1087054712455501. [DOI] [PubMed] [Google Scholar]
- Yeatman J.D., Dougherty R.F., Ben-Shachar M., Wandell B.A. Development of white matter and reading skills. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3045–53. doi: 10.1073/pnas.1206792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yendiki A., Koldewyn K., Kakunoori S., Kanwisher N., Fischl B. Spurious group differences due to head motion in a diffusion MRI study. Neuroimage. 2014;88:79–90. doi: 10.1016/j.neuroimage.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon U., Fonov V.S., Perusse D., Evans A.C. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45:769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Avants B.B., Das S.R., Pluta J., Altinay M., Craige C. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 T MRI data. Neuroimage. 2010;50:434–445. doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhang W., Li R., Deng H., Wang L., Lin W., Ji S., Shen D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]