Abstract
Objective
Rho GTPase-activating protein 4 (ARHGAP4) is a GTPase‐activating protein for the small GTPases of the Rho family that is involved in tumorigenesis. We recently reported that ARHGAP4 can mediate Warburg effect and malignant phenotype of pancreatic cancer. However, the regulation of ARHGAP4 remains unclear.
Methods
ARHGAP4 and miR-939-5p expressions in pancreatic cancer tissues and cell lines were measured by real-time PCR or Western blotting. Pancreatic cancer cells were transfected with miR-939-5p inhibitor, miR-939-5p mimic and/or lentivirus expressing ARHGAP4, and the cell viability, invasion and migration were measured by CCK-8 and Transwell assay, respectively. The suppression of ARHGAP4 expression by miR-939-5p was revealed by luciferase reporter assay, real-time PCR or Western blotting.
Results
ARHGAP4 expression was decreased, while miR-939-5p was increased in pancreatic cancer tissues compared with adjacent-normal pancreatic tissues. Higher miR-939-5p expression was correlated with advanced pathological stages and poor prognosis of pancreatic cancer patients. miR-939-5p directly targeted ARHGAP4. Either miR-939-5p down-regulation or ARHGAP4 overexpression inhibited viability, invasion and migration of pancreatic cancer cells. However, ARHGAP4 overexpression markedly inhibited the increased viability, migration, and invasion induced by miR-939-5p up-regulation in pancreatic cancer cells.
Conclusion
These observations suggested that miR-939-5p regulates the malignant phenotype of pancreatic cancer cells by targeting ARHGAP4, establishing miR-939-5p as a novel regulator of ARHGAP4 with a critical role in tumorigenesis in pancreatic cancer.
Keywords: miR-939-5p, ARHGAP4, pancreatic cancer, carcinogenesis
Introduction
Pancreatic cancer is one of the digestive system tumors in the world, with high invasiveness and malignancy.1 Although the comprehensive treatment of cancer has made great progress over the decade, the incidence and mortality of pancreatic cancer are increasing worldwide, seriously endangering human life and health. Currently, the treatment is not effective, and the 5-year survival rate is still very low, only about 5%.2 It is predicted that pancreatic cancer will surpass colorectal, prostate, and breast cancers to become the second leading cause of cancer-related death by 2030.3 As the early clinical symptoms of pancreatic cancer are not typical, most of the patients were diagnosed with advanced stage or distant metastasis, so the optimal time for surgical treatment is missed.4 Meanwhile, only 10% to 20% of the patients have the opportunity to receive radical resection,4 and 80% of patients will have recurrence and metastasis within 1–2 years after surgery.5 Consequently, it is sensible to the importance of studying the pathogenesis of pancreatic cancer and finding new markers closely implicated in the malignant progression and prognosis of pancreatic cancer, so as to improve the early diagnosis and improve the prognosis.
Recently, Rho GTPase activating proteins (RhoGAPs) have been identified as tumor suppressors in some human cancers, including Rho GTPase‐activating protein 17 (ARHGAP17),6 ARHGAP6,7 ARHGAP24,8 and ARHGAP30.9 Besides, evidences have been reported that ARHGAP4 inhibits cell motility and axon outgrowth,10 associates with nephrogenic diabetes insipidus and intellectual disability,11 correlates with pathological stages, vascular invasion and prognosis of pancreatic cancer patients, and regulates glycolysis, invasion and migration in pancreatic cancer.12,13 Nevertheless, the regulation of ARHGAP4 in pancreatic cancer is still undiscovered microRNAs (miRNA) are small endogenous non-coding RNA molecules with a length of about 18–25 nucleotides that repress protein translation through binding to the 3ʹ-untranlated region (UTR) of their target mRNA and are significantly involved in various cancers, including pancreatic cancer.14–16 High expression of miR-196a, miR-27a, miR-221, miR-143, miR-135b, miR-199b-5p and miR-21, but low expression of miR-744, miR-455-3p and miR-655, in pancreatic cancer samples were found compared with normal samples and associated with poor prognosis.17 In addition, the expression profile of miRNAs varies even in the different locations,18 which further confirms that miRNAs have high application value in the diagnosis, prognosis evaluation and treatment of pancreatic cancer. Recently, a widely reported miRNA, miR-939, has been pointed out to be pivotal in the development and procession of cancers, such as hepatocellular carcinoma,19 tongue squamous cell carcinoma,20 gastric,21 lung,22 colorectal,23 and ovarian cancer.24 However, the clinical significances and roles of miR-939 in pancreatic cancer are still unknown.
In the present study, our data revealed that the expression of miR-939-5p had a negative pattern as ARHGAP4 in pancreatic cancer tissues and cell lines and was relevant to pathological stages and survival time of patients. Down-regulation of miR-939-5p in pancreatic cancer inhibited cell viability, invasion and migration, while its up-regulation demonstrated an inverse effects which was reversed by overexpression of ARHGAP4, suggesting that miR-939-5p contributes to the invasion and migration in pancreatic cancer by targeting ARHGAP4.
Materials and Methods
Clinical Samples
Pancreatic cancer tissues were collected from 70 pancreatic cancer patients who went through tumor resection at Fudan University Shanghai Cancer Center between October 12, 2014 and April 8, 2017. Fresh surgical specimens of primary pancreatic cancer and adjacent-normal pancreatic tissues were collected, stored in liquid nitrogen, and later used for Real-time PCR analysis. This study was approved by the ethic commitment of Fudan University Shanghai Cancer Center. Written informed consent was obtained from all patients, and that this was conducted in accordance with the Declaration of Helsinki.
Cell Culture
Pancreatic cancer cell lines BxPC-3, Capan-1, PANC-1, MIA PaCa-2 and SW1990 were purchased from Shanghai Institute of Pharmaceutical Industry (Shanghai, China). SW1990, MIA PaCa-2 and PANC-1 were cultivated in DMEM (Gibco, USA), while BxPC-3 and Capan-1 cells were cultivated in RPMI-1640, complementing with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 U/mL penicillin under humidified conditions (37°C, 5% CO2).
Cell Transfection
The miR-3150a-3p, miR-939-5p, or miR-762 inhibitor, miR-939-5p mimic and negative control (NC) were purchased from RiboBio (Guangzhou, China). Transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. ARHGAP4 overexpression constructed by cloning full-length human ARHGAP4 into the lentiviral expression vector pLVX-Puro (Addgen, USA) or blank lentivirus pLVX-Puro (Vector) was transfected into 293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and then transduced into pancreatic cancer cells.
Luciferase Reporter Assay
The 3ʹ-UTR of ARHGAP4 and the mutated target site of ARHGAP4 3ʹ-UTR were cloned into a pGL3-Promoter vector (Promega, Madison, WI, USA) to constitute ARHGAP4-wild-type (ARHGAP4-WT) and ARHGAP4-mutated-type (ARHGAP4-Mut) reporter plasmid. Pancreatic cancer cells were co-transfected with the reporter plasmid and miR-939-5p mimic, inhibitor, or negative control (NC) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The Dual-Luciferase Reporter Assay System (Promega) was used to assay for luciferase activity at 48 h after transfection. Relative luciferase activity was calculated as the ratio of Firefly/Renilla luciferase activity.
Cell Viability Assay
Cell viability was examined using a Cell Counting Kit (CCK)-8 (Dojindo Lab, Kumamoto, Japan). Cells were transfected with miR-939-5p inhibitor, mimic, or negative control (NC) and/or transduced with lentivirus expressing ARHGAP4 or blank lentivirus (Vector). After treatment, 10 μL/well Cell Counting Kit-8 (CCK-8) solution was added, and cells were further incubated for 1 h at 37°C. Absorbance values at 450 nm were measured using a microplate reader (Tecan Safire, Crailsheim, Germany).
Transwell Assay
The cell migration and invasion were measured by Transwell assay as previously described.12
Real-Time PCR
Total RNA was extracted using RNeasy Plus Universal Kits (QIAGEN, USA). cDNA was synthesized from 1 μg of total RNA via thermal cycler system (Bio-Rad, USA). The Real-time PCR was carried out with a LightCycler (Roche Diagnostics GmbH, Manheim, Germany) using SYBR Green Supermix (Bio-Rad). ARHGAP4 expression was normalized to GAPDH, and miRNAs were normalized to U6. The primer sequences are present as follows: ARHGAP4-forward 5ʹ-CTACAACCTGGCCGTGTGCTTC-3ʹ; ARHGAP4-reverse 5ʹ-CTTCCAGCTCCGGCTCATTGTC-3ʹ; GAPDH-forward 5ʹ-AATCCCATCACCATCTTC-3ʹ; GAPDH-reverse 5ʹ-AGGCTGTTGTCATACTTC-3ʹ; miR-3150a-3p-forward 5ʹ-CGCTGGGGAGATCCTCGA-3ʹ; miR-3150a-3p-reverse 5ʹ-AGTGCAGGGTCCGAGGTATT-3ʹ; miR-939-5p-forward 5ʹ-TGGGGAGCTGAGGCTCTG-3ʹ; miR-939-5p-reverse 5ʹ-AGTGCAGGGTCCGAGGTATT-3ʹ; miR-762-forward 5ʹ-GGGGCTGGGGCCGGGG-3ʹ; miR-762-reverse 5ʹ-AGTGCAGGGTCCGAGGTATT-3ʹ; U6-forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; U6-reverse 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. Analyses of relative gene expression were performed using the 2−ΔΔCT method.
Western Blot
Total protein was extracted and separated by electrophoresis on 10% sodium dodecyl sulfonate (SDS) polyacrylamide gels and transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore Corp., USA). Primary antibodies were added to the blocking solution, diluted to the desired concentration according to the instructions (ARHGAP4 1: 500; GAPDH 1: 2000), and incubated overnight at 4°C. Finally, the protein bands were visualized using enhanced chemiluminescence system (Bio-Rad).
Statistical Analysis
Data were expressed as mean ± standard deviations (SD). Statistical comparisons were conducted using the GraphPad Prism (version 7.0; GraphPad Software, USA) through paired two-sided Student’s t-test for two-group or one-way analysis of variance (ANOVA) followed by post-hoc Tukey’s test for multiple comparisons. Statistical significances were defined as P<0.05.
Results
ARHGAP4 Expression in Pancreatic Cancer
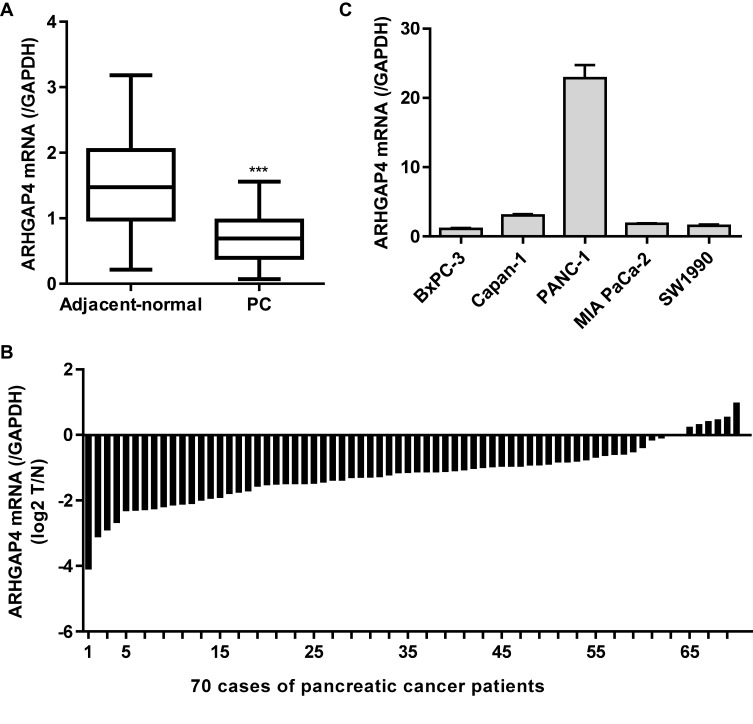
Our previous studies have shown decreased ARHGAP4 protein expression in pancreatic cancer tissues compared with the non-tumor tissues by immunohistochemistry.12,13 Here, the ARHGAP4 mRNA expression was also measured by Real-time PCR analysis in pancreatic cancer tissues. ARHGAP4 mRNA levels in pancreatic cancer tissues were markedly decreased compared with the adjacent-normal tissues (Figure 1A). The ARHGAP4 mRNA levels were also shown in 70 cases of pancreatic cancer patients, in which decreased ARHGAP4 mRNA expression levels (log2T/N) were found in 91.4% (64/70) of the pancreatic cancer patients (Figure 1B). Moreover, the level of ARHGAP4 mRNA in pancreatic cancer cell lines was also measured. It demonstrated that PANC-1 cells demonstrated highest ARHGAP4 mRNA level, whereas SW1990 and BxPC-3 cells demonstrated lower ARHGAP4 mRNA levels, compared with the other pancreatic cancer cell lines (Figure 1C).
Figure 1.
ARHGAP4 expression in pancreatic cancer tissues. (A) ARHGAP4 mRNA levels were decreased in pancreatic cancer tissues (n=70) compared with the adjacent-normal tissues (n=70). (B) ARHGAP4 mRNA levels in 70 cases of pancreatic cancer patients (log2 T/N). (C) ARHGAP4 mRNA expression in five pancreatic cancer cell lines, including BxPC-3, Capan-1, PANC-1, MIA PaCa-2 and SW1990. ***P<0.001 compared with adjacent-normal.
Abbreviations: PC, pancreatic cancer; N, adjacent-normal pancreatic tissues; T, pancreatic cancer tissues.
miR-939-5p Directly Targeted ARHGAP4 in Pancreatic Cancer Cells
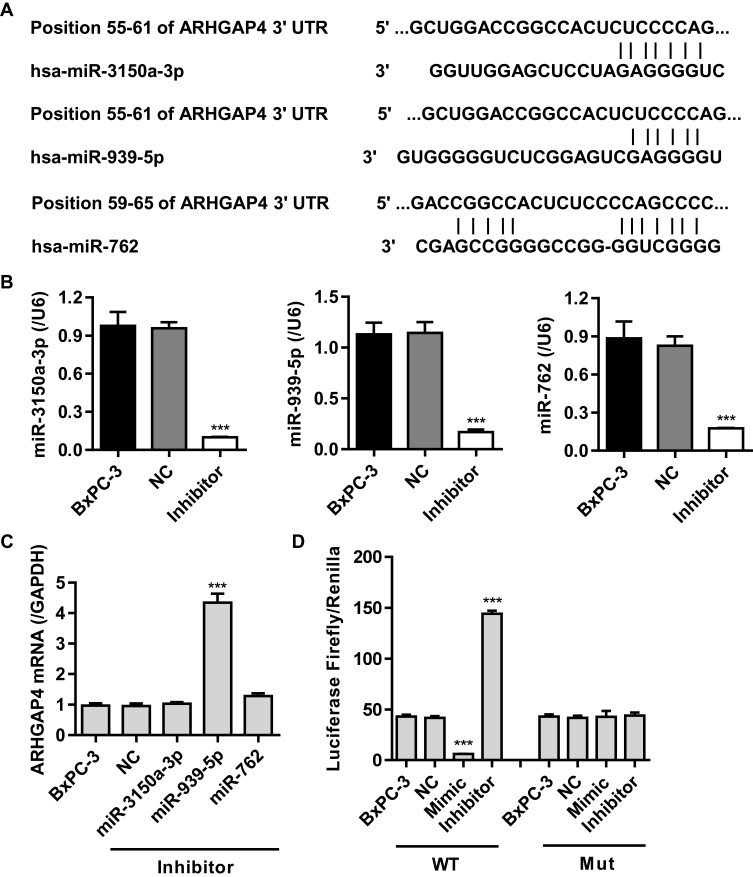
Studies have shown that miRNA can be used as a new biomarker for a variety of diseases through regulating gene expression by mediating gene silencing at transcriptional and post-transcriptional levels. According to TargetScan prediction, we found that miR-3150a-3p, miR-939-5p and miR-762 target ARHGAP4 (Figure 2A). To confirm the correlation of ARHGAP4 with miR-3150a-3p, miR-939-5p and miR-762 in pancreatic cancer, the corresponding inhibitor was transfected into the BxPC-3 cells which showed lowest ARHGAP4 mRNA levels (Figure 1B). And the expression of miR-3150a-3p, miR-939-5p, miR-762 and ARHGAP4 was measured (Figure 2B and C). Compared to miR-3150a-3p and miR-762 inhibitors, BxPC-3 cells transfected with miR-939-5p inhibitor significantly increased the expression of ARHGAP4 mRNA compared with negative control (NC). The similarly results were also found in SW1990 and PANC-1 cells which demonstrated lower and highest ARHGAP4 mRNA expression, respectively (Supplementary Figure S1A-C). We then constructed the wild-type and mutated 3ʹ-UTR of ARHGAP4, BxPC-3 cells were co-transfected with miR-939-5p mimic or inhibitor, and wild-type or mutated ARHGAP4. We found that up-regulation of miR-939-5p significantly decreased while down-regulation of miR-939-5p significantly increased the luciferase activity of wild-type ARHGAP4, but did not affect the luciferase activity of mutated ARHGAP4 (Figure 2D). The similarly results were also found in SW1990 and PANC-1 cells (Supplementary Figure S1D). These data indicated that miR-939-5p directly bound to the 3ʹ-UTR sequence of ARHGAP4 and regulated ARHGAP4 expression in pancreatic cancer cells.
Figure 2.
miR-939-5p targeted ARHGAP4 directly at the 3ʹ-UTR of ARHGAP4 in pancreatic cancer cells. (A) The binding sites of miR-3150a-3p, miR-939-5p and miR-762 on the 3ʹ-UTR of ARHGAP4 were predicted by Targetscan. (B) miR-3150a-3p, miR-939-5p and miR-762 levels in BxPC-3 cells transfected with their corresponding inhibitor or negative control (NC). (C) ARHGAP4 mRNA levels in BxPC-3 cells transfected with miR-3150a-3p, miR-939-5p, or miR-762 inhibitor or NC. (D) The luciferase activity was measured by the Dual-Luciferase Reporter Assay. ***P<0.001 compared with NC.
miR-939-5p Expression in Pancreatic Cancer and Its Correlation with Prognosis
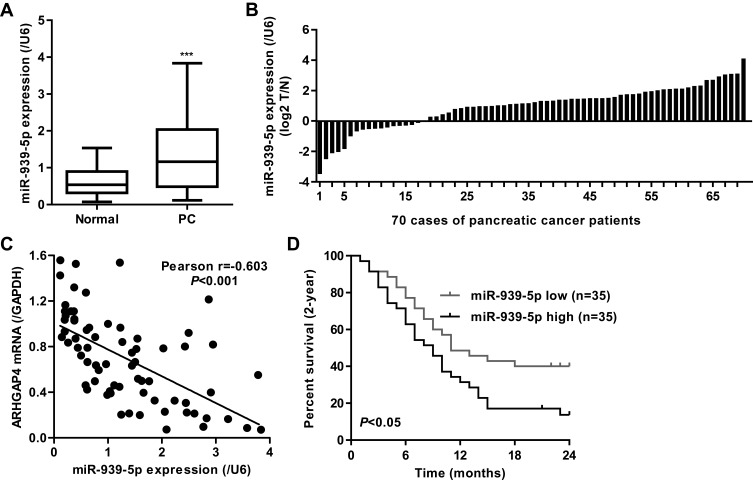
Our previous studies have shown that ARHGAP4 could regulate the invasion and migration in pancreatic cancer cells.12 Therefore, we suggested that miR-939-5p may also contribute to pancreatic cancer progression. We first investigated miR-939-5p expressions by Real-time PCR analysis in pancreatic cancer. miR-939-5p levels in pancreatic cancer tissues were markedly up-regulated compared with the adjacent-normal tissues (Figure 3A). The miR-939-5p levels were also shown in 70 cases of pancreatic cancer patients, in which increased miR-939-5p levels (log2T/N) were found in 75.7% (53/70) of the pancreatic cancer patients (Figure 3B). Pearson correlation analysis demonstrated that the expression of miR-939-5p in pancreatic cancer tissues was negatively correlated with the expression of ARHGAP4 (Figure 3C). To further analyze the correlation of miR-939-5p expression with clinical features of pancreatic cancer patients, 70 pancreatic cancer patients were divided into two groups (high vs low) according to the cutoff values (median value=0.151) measured by Real-time PCR. Chi-square test indicated that the miR-939-5p expression was correlated with the pathological stages, but not with tumor size and site, gender and age (Table 1). Kaplan-Meier analysis and log rank test showed that the overall survival time of patients with lower miR-939-5p expression were notably higher than that of patients with higher miR-939-5p expression, suggesting that increased miR-939-5p was significantly correlated with poor prognosis of pancreatic cancer patients (Figure 3D).
Figure 3.
miR-939-5p expression in pancreatic cancer tissues. (A) miR-939-5p level was increased in pancreatic cancer tissues (n=70) compared with the adjacent-normal tissues (n=70). (B) miR-939-5p levels in 70 cases of pancreatic cancer patients (log2 T/N). (C) Pearson correlation scatter plots in pancreatic tissues (n=70). (D) Kaplan-Meier curve of pancreatic patients with high or low miR-939-5p levels analyzed by Log-rank (Mantel-Cox) test. ***P<0.001 compared with adjacent-normal.
Abbreviations: PC, pancreatic cancer; N, adjacent-normal pancreatic tissues; T, pancreatic cancer tissues.
Table 1.
Correlation of the Expression of miR-939-5p with Clinicopathological Parameters in Patients with PC
| Clinicopathological Parameters | miR-939-5p Expression | P-value | |
|---|---|---|---|
| Low High | |||
| (n=35, 50%) | (n=35, 50%) | ||
| Gender | |||
| Male | 19 (27.1%) | 21 (30.0%) | 0.6291 |
| Female | 16 (22.9%) | 14 (20.0%) | |
| Age (years) | |||
| <60 | 16 (22.9%) | 19 (27.1%) | 0.4733 |
| ≥60 | 19 (27.1%) | 16 (22.9%) | |
| Tumor size (cm) | |||
| ≤4 | 20 (28.6%) | 16 (22.9%) | 0.3388 |
| >4 | 15 (21.4%) | 19 (27.1%) | |
| Pathological stages | 0.0072 | ||
| I | 6 (8.6%) | 4 (5.7%) | |
| II | 20 (28.6%) | 8 (11.4%) | |
| III | 6 (8.6%) | 18 (25.7%) | |
| IV | 3 (4.3%) | 5 (7.1%) | |
| Tumor site | 0.5268 | ||
| Pancreatic head | 14 (20.0%) | 10 (14.3%) | |
| Pancreatic body | 11 (15.7%) | 15 (21.4%) | |
| Pancreatic tail | 10 (14.3%) | 10 (14.3%) | |
Note: Differences between groups were done by the Chi-square test.
miR-939-5p Down-Regulation Inhibited Invasion and Migration in Pancreatic Cancer Cells
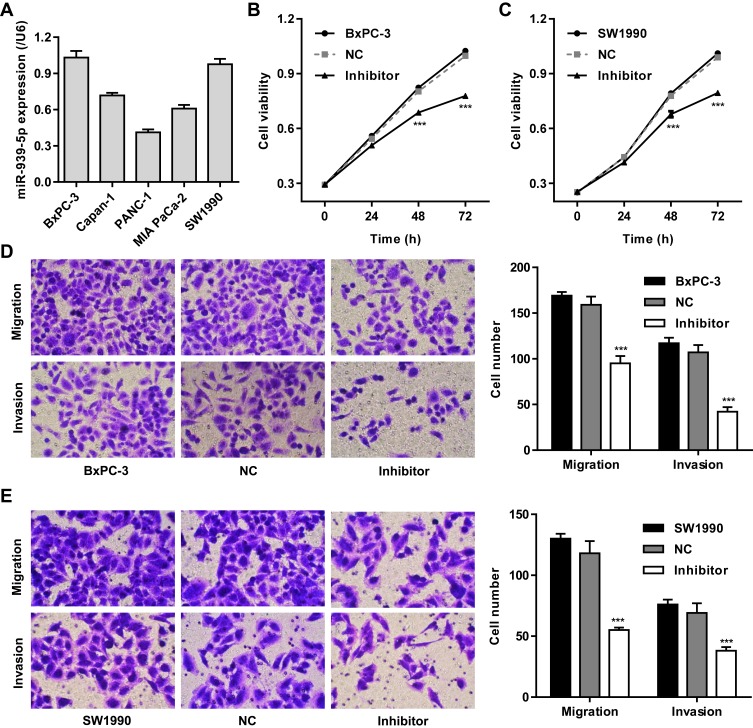
We further assumed the role of miR-939-5p in regulating cell invasion and migration of pancreatic cancer. Firstly, the expression of miR-939-5p in pancreatic cancer cell lines was measured. As shown in Figure 4A, PANC-1 cells showed lowest miR-939-5p level, whereas SW1990 and BxPC-3 cells showed higher miR-939-5p levels, compared with the other pancreatic cancer cell lines. SW1990 and BxPC-3 cells were therefore transfected with miR-939-5p inhibitor, and the cell viability, invasion and migration were measured. We found that miR-939-5p inhibitor markedly inhibited the viability of BxPC-3 and SW1990 cells by 14.4% and 12.9% at 48 h and by 21.9% and 19.7% at 72 h, respectively, compared with negative control (NC) (Figure 4B and C). Moreover, miR-939-5p inhibitor significantly inhibited BxPC-3 cell migration and invasion by 40.2% and 60.7% and that of SW1990 cells by 53.4% and 44.9%, respectively, compared with NC (Figure 4D and E).
Figure 4.
miR-939-5p down-regulation inhibited pancreatic cancer cell invasion and migration. (A) ARHGAP4 mRNA expression in five pancreatic cancer cell lines, including BxPC-3, Capan-1, PANC-1, MIA PaCa-2 and SW1990. After BxPC-3 (B, D) and SW1990 (C, E) cells were transfected with miR-939-5p inhibitor, the cell viability, invasion and migration were decreased than those transfected with negative control (NC). ***P<0.001 compared with NC.
miR-939-5p Up-Regulation Promoted Invasion and Migration in Pancreatic Cancer Cells Through Targeting ARHGAP4
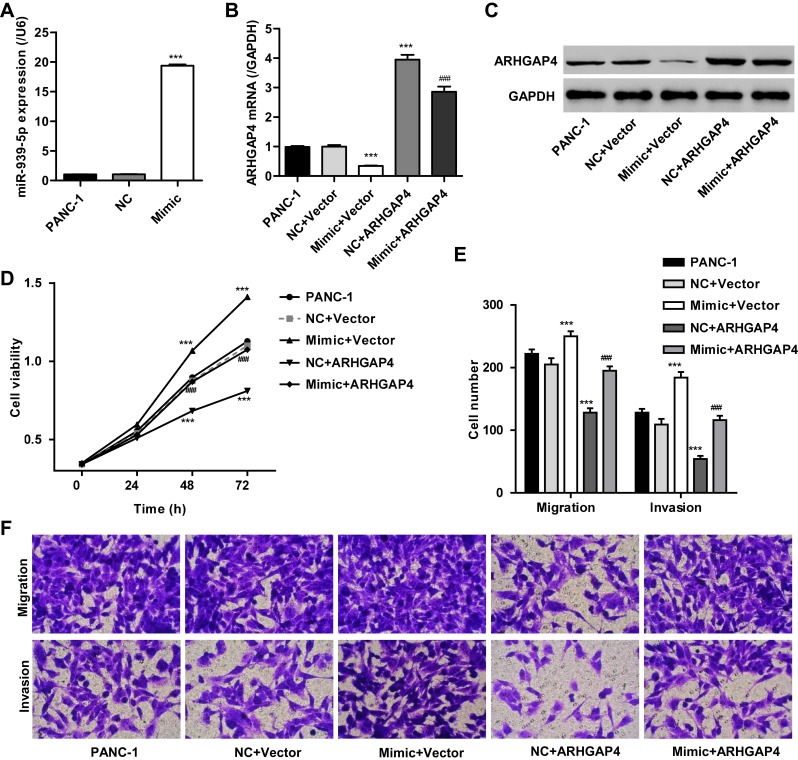
To investigate the mechanism of miR-939-5p affecting the invasion and migration in pancreatic cancer cells, PANC-1 cells which showed lowest miR-939-5p levels (Figure 4A) were transfected with miR-939-5p mimic and/or transduced with lentivirus expressing ARHGAP4. And the expression levels of miR-939-5p and ARHGAP4 were shown in Figure 5A–C. Up-regulation of miR-939-5p significantly increased the miR-939-5p levels and decreased ARHGAP4 levels in PANC-1 cells. Moreover, miR-939-5p mimic markedly increased while ARHGAP4 overexpression markedly suppressed the PANC-1 cell viability, invasion and migration (Figure 5D–F). Importantly, ARHGAP4 overexpression also inhibited the malignant phenotype of PANC-1 cells induced by miR-939-5p mimic, suggesting that miR-939-5p up-regulation promoted invasion and migration of pancreatic cancer cells through targeting ARHGAP4 (Figure 5D–F).
Figure 5.
miR-939-5p up-regulation promoted pancreatic cancer cell invasion and migration through targeting ARHGAP4. After PANC-1 cells were transfected with miR-939-5p mimic and/or transduced with lentivirus expressing ARHGAP4, miR-939-5p levels (A), ARHGAP4 mRNA levels (B, C) and cell viability (D), migration, and invasion (E, F) were measured. ***P<0.001 compared with NC+blank lentivirus vector (Vector). ###P<0.001 compared with mimic+Vector.
Discussion
In this study, we identified ARHGAP4 as a down-regulated gene while miR-939-5p as an up-regulated miRNA in pancreatic cancer based on Real-time PCR analysis. Meanwhile, we demonstrated a critical role of miR-939-5p in the progression of pancreatic cancer and proposed an ARHGAP4-mediated mechanism.
ARHGAP4 is a member of the RhoGAPs family that involves in the cancer progression by stimulating GTP hydrolysis to inactivate small GTPases. p190 RhoGAP reduced cell invasion and metastasis of pancreatic cancer.25 In addition to p190 RhoGAP, only ARHGAP4 has been found to correlate with pathological stages, vascular invasion and prognosis of pancreatic cancer patients and regulate Warburg effect as well as cell invasion and migration.12,13 Our previous study demonstrated decreased ARHGAP4 protein levels in pancreatic cancer tissues based on the immunohistochemical staining on tissue microarrays. Here, the decreased ARHGAP4 mRNA levels in pancreatic cancer tissues were also observed compared with the adjacent-normal tissues. However, ARHGAP4 was up-regulated in colorectal cancer tissues and cell lines, and its overexpression was correlated with T stage, N stage, clinical stage, metastasis and poor prognosis,26 which were inconsistent with our present and previous studies.12 These data suggest that the expression and clinical correlation of ARHGAP4 vary in different cancers.
Recently, increasing evidences have demonstrated an important role of miRNAs in the development and progression of pancreatic cancer. For example, miR-196a-5p promoted the invasion, migration and EMT markers in MIA PaCa-2 cells.27 Knockdown of miR-21 inhibited the malignant phenotype of PANC-1 cells, at least partially by regulating the VHL/HIF-1α/VEGF pathway and the expression of MMP-2 and MMP-9, and tumor growth in vivo.28 miR-129-5p suppressed the progression of pancreatic cancer by targeting PBX3, leading to the reduction of proliferation, migration and invasion, and induction of apoptosis of AsPC-1 and BxPC-3 cells.29 Knockdown of miR-155 increased apoptosis, inhibited proliferation and JAK2/STAT3 pathway in SW1990 cells by targeting SOCS3.30 Moreover, many ARHGAP family genes have also been found to play important role in the involvement of miRNAs in tumorigenesis. miR-200b suppressed the metastasis in triple negative breast cancer by targeting ARHGAP18.31 miR-3174 inhibited mitochondria-dependent apoptosis and autophagic cell death in gastric cancer by targeting ARHGAP10.32 miR-744 promoted the metastasis and progression of nasopharyngeal carcinoma by targeting ARHGAP5.33 These data suggest that the regulation of ARHGAP4 by miRNAs may also exist in pancreatic cancer. Indeed, miR-939-5p directly targeted ARHGAP4 in pancreatic cancer cells and was negatively correlated with ARHGAP4 in pancreatic cancer tissues and cell lines. miR-939 expression in ovarian and triple-negative breast cancers was increased and associated with worse disease-free survival of patients with triple-negative breast cancer,34,35 which were in line with our findings. In addition to survival, pathological stages were also correlated with miR-939-5p expression in pancreatic cancer patients. Similarly, miR-939-3p expression was up-regulated in lung cancer and associated with TNM stage, metastasis, and poor prognosis.22 However, miR-939-5p expression in gastric and bladder cancer was decreased.21,36 These data suggest that the expression and clinical correlation of miR-939 vary in different cancers.
miR-939-5p promoted cell proliferation and repressed cell apoptosis of tongue squamous cell carcinoma,20 and miR-939 also regulated epithelial-mesenchymal transition in epithelial ovarian cancer, implicating in tumorigenesis, tumor progression, metastasis and relapse.24 miR-939-3p promoted cell invasion, migration and proliferation of lung cancer.22 However, miR-939-5p suppressed invasion and migration of colorectal cancer cells through directly targeting LIMK2,23 and miR-939 overexpression inhibited gastric cancer cell metastasis, proliferation and chemosensitivity through directly targeting SLC34A2.21 Considering the expression of miR-939-5p is increased in pancreatic cancer tissues, we investigated whether miR-939-5p acts as an oncogene role in cell proliferation, migration, and invasion of pancreatic cancer. The results demonstrated that miR-939-5p down-regulation inhibited cell viability, migration and invasion in pancreatic cancer, while its up-regulation showed an inverse effect by targeting ARHGAP4. Previous studies showed that ARHGAP4 inhibited invasion and migration in pancreatic cancer cells through inhibiting β-catenin activation12 and regulated Warburg effect through HIF-1α and mTOR signaling pathways.13 β-catenin as well as its target C-myc expression was increased in ovarian cancer cells with miR-939 up-regulation, but decreased with miR-939 down-regulation.34 C-myc has also been reported to regulate aerobic glycolysis and promote tumor progression in pancreatic cancer.37 These results suggest that miR-939-5p may regulate Warburg effect and tumor progression in pancreatic cancer via the ARHGAP4/β-catenin/C-myc axis. HIF-1α and mTOR not only involved in the aerobic glycolysis but also regulated tumor growth and metastasis.38 However, further investigations on the functional role and mechanism by which miR-939-5p/ARHGAP4 regulates aerobic glycolysis and tumorigenesis are urgently needed, and whether miR-939-5p/ARHGAP4 regulates tumorigenicity through control of glucose metabolism also need further investigation.
Conclusion
In conclusion, our study illustrated that miR-939-5p was increased in pancreatic cancer tissues and associated with pathological stages and prognosis. miR-939-5p overexpression promoted pancreatic cancer cell invasion and migration by down-regulating ARHGAP4. miR-939-5p/ARHGAP4 axis might thus serve as a novel biomarker to predict the progression and prognosis of pancreatic cancer, as well as a potent therapeutic target for pancreatic cancer treatment.
Acknowledgment
This work was funded by the National Natural Science Foundation of China (81573757/H2902).
Disclosure
The authors declare that they have no competing interests.
References
- 1.Wu J, Liu J, Wei X, et al. A feature-based analysis identifies COL1A2 as a regulator in pancreatic cancer. J Enzyme Inhib Med Chem. 2019;34:420–428. doi: 10.1080/14756366.2018.1484734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J, Zhang Y, Yang J, et al. NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335-5p/c-met axis. Am J Cancer Res. 2016;6:2361–2374. [PMC free article] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 4.Matsuno S, Egawa S, Fukuyama S, et al. Pancreatic cancer registry in Japan: 20 years of experience. Pancreas. 2004;28(3):219–230. doi: 10.1097/00006676-200404000-00002 [DOI] [PubMed] [Google Scholar]
- 5.Schnelldorfer T, Ware AL, Sarr MG, et al. Long-term survival after pancreatoduodenectomy for pancreatic adenocarcinoma: is cure possible? Ann Surg. 2008;247:456–462. doi: 10.1097/SLA.0b013e3181613142 [DOI] [PubMed] [Google Scholar]
- 6.Guo Q, Xiong Y, Song Y, Hua K, Gao S. ARHGAP17 suppresses tumor progression and up-regulates P21 and P27 expression via inhibiting PI3K/AKT signaling pathway in cervical cancer. Gene. 2019;692:9–16. doi: 10.1016/j.gene.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 7.Wu Y, Xu M, He R, Xu K, Ma Y. ARHGAP6 regulates the proliferation, migration and invasion of lung cancer cells. Oncol Rep. 2019;41(4):2281–2888. doi: 10.3892/or.2019.7031 [DOI] [PubMed] [Google Scholar]
- 8.Dai X, Geng F, Dai J, Li M, Liu M. Rho GTPase activating protein 24 (ARHGAP24) regulates the anti-cancer activity of sorafenib against breast cancer MDA-MB-231 cells via the signal transducer and activator of transcription 3 (STAT3) signaling pathway. Med Sci Monit. 2018;24:8669–8677. doi: 10.12659/MSM.911394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao X, Tong J. ARHGAP30 suppressed lung cancer cell proliferation, migration, and invasion through inhibition of the Wnt/beta-catenin signaling pathway. Onco Targets Ther. 2018;11:7447–7457. doi: 10.2147/OTT.S175255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogt DL, Gray CD, Young WS 3rd, Orellana SA, Malouf AT. ARHGAP4 is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol Cell Neurosci. 2007;36:332–342. doi: 10.1016/j.mcn.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L, Poke G, Gecz J, Gibson K. A novel contiguous gene deletion of AVPR2 and ARHGAP4 genes in male dizygotic twins with nephrogenic diabetes insipidus and intellectual disability. Am J Med Genet A. 2012;158a:2511–2518. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Xu L, Ning Z, et al. ARHGAP4 regulates the cell migration and invasion of pancreatic cancer by the HDAC2/beta-catenin signaling pathway. Carcinogenesis. 2019;40:1405–1414. doi: 10.1093/carcin/bgz067 [DOI] [PubMed] [Google Scholar]
- 13.Shen Y, Chen G, Zhuang L, Xu L, Lin J, Liu L. ARHGAP4 mediates the Warburg effect in pancreatic cancer through the mTOR and HIF-1alpha signaling pathways. Onco Targets Ther. 2019;12:5003–5012. doi: 10.2147/OTT.S207560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, He C, Wang X, Wang H, Nan G, Fang L. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol. 2019;47:3163–3171. doi: 10.1080/21691401.2019.1642903 [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Kong D, Sun D, Li J. Long non-coding RNA CCAT2 acts as an oncogene in osteosarcoma through regulation of miR-200b/VEGF. Artif Cells Nanomed Biotechnol. 2019;47:2994–3003. doi: 10.1080/21691401.2019.1640229 [DOI] [PubMed] [Google Scholar]
- 16.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59 [DOI] [PubMed] [Google Scholar]
- 17.Zhou J, Hui X, Mao Y, Fan L. Identification of novel genes associated with a poor prognosis in pancreatic ductal adenocarcinoma via a bioinformatics analysis. Biosci Rep. 2019;39(8). doi: 10.1042/BSR20190625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Q, Zhu EC, Qu YL, et al. Serum level of co-expressed hub miRNAs as diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma. J Cancer. 2018;9:3991–3999. doi: 10.7150/jca.27697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fornari F, Ferracin M, Trere D, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10:e0141448. doi: 10.1371/journal.pone.0141448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Guo Y, Yan W. lncRNA RP5-916L7.2 correlates with advanced tumor stage, and promotes cells proliferation while inhibits cells apoptosis through targeting miR-328 and miR-939 in tongue squamous cell carcinoma. Clin Biochem. 2019;67:24–32. doi: 10.1016/j.clinbiochem.2019.02.013 [DOI] [PubMed] [Google Scholar]
- 21.Zhang JX, Xu Y, Gao Y, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16:18. doi: 10.1186/s12943-017-0586-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Han X, Du C, Chen Y, et al. Overexpression of miR-939-3p predicts poor prognosis and promotes progression in lung cancer. Cancer Biomark. 2019;25:325–332. doi: 10.3233/CBM-190271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manag Res. 2019;11:1779–1789. doi: 10.2147/CMAR.S192452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang M, Jiang L, Lin Y, et al. Platelet microparticle-mediated transfer of miR-939 to epithelial ovarian cancer cells promotes epithelial to mesenchymal transition. Oncotarget. 2017;8:97464–97475. doi: 10.18632/oncotarget.v8i57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusama T, Mukai M, Endo H, et al. Inactivation of Rho GTPases by p190 RhoGAP reduces human pancreatic cancer cell invasion and metastasis. Cancer Sci. 2006;97:848–853. doi: 10.1111/j.1349-7006.2006.00242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xuehu X, Yuandong X, Zhu Z, Shuling L, Xiaobing W, Liu X. Expression and clinical significance of ARHGAP4 in colorectal cancer. J Pract Med. 2017;33:705–708. [Google Scholar]
- 27.Belvedere R, Saggese P, Pessolano E, Memoli D, Bizzarro V, Rizzo F. miR-196a is able to restore the aggressive phenotype of annexin A1 knock-out in pancreatic cancer cells by CRISPR/Cas9 genome editing. Int J Mol Sci. 2018;19:pii: E1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Jiang Z, Li Y, Wang K, Chen X, Liu G. Downregulation of miR-21 inhibits the malignant phenotype of pancreatic cancer cells by targeting VHL. Onco Targets Ther. 2019;12:7215–7226. doi: 10.2147/OTT.S211535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu Z, Wang X, Shi Y, Da M. miR-129-5p suppresses proliferation, migration, and induces apoptosis in pancreatic cancer cells by targeting PBX3. Acta Biochim Biophys Sin. 2019;51:997–1007. doi: 10.1093/abbs/gmz096 [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Guo J, Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. 2019;23:5168–5175. doi: 10.26355/eurrev_201906_18181 [DOI] [PubMed] [Google Scholar]
- 31.Yang C, Humphries B, Li Y, et al. Abstract 1468: miR-200b targets ARHGAP18 and suppresses triple negative breast cancer metastasis. Cancer Res. 2017;77:1468. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Wang L, Li Z, et al. miR-3174 contributes to apoptosis and autophagic cell death defects in gastric cancer cells by targeting ARHGAP10. Mol Ther Nucleic Acids. 2017;9:294–311. doi: 10.1016/j.omtn.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Fang Y, Zhu X, Wang J, et al. MiR-744 functions as a proto-oncogene in nasopharyngeal carcinoma progression and metastasis via transcriptional control of ARHGAP5. Oncotarget. 2015;6:13164–13175. doi: 10.18632/oncotarget.v6i15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ying X, Li-ya Q, Feng Z, Yin W, Ji-hong L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed Pharmacother. 2015;71:64–69. doi: 10.1016/j.biopha.2015.02.020 [DOI] [PubMed] [Google Scholar]
- 35.Di Modica M, Regondi V, Sandri M, et al. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017;384:94–100. doi: 10.1016/j.canlet.2016.09.013 [DOI] [PubMed] [Google Scholar]
- 36.Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110:408–419. doi: 10.1111/cas.13856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He TL, Zhang YJ, Jiang H, Li XH, Zhu H, Zheng KL. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med Oncol. 2015;32:187. doi: 10.1007/s12032-015-0633-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Q, Chen X, Ma J, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108 [DOI] [PMC free article] [PubMed] [Google Scholar]