Abstract
Genetic lineage tracing is widely used to study organ development and tissue regeneration. Multicolor reporters are a powerful platform for simultaneously tracking discrete cell populations. Here, combining Dre-rox and Cre-loxP systems, we generated a new dual-recombinase reporter system, called Rosa26 traffic light reporter (R26-TLR), to monitor red, green, and yellow fluorescence. Using this new reporter system with the three distinct fluorescent reporters combined on one allele, we found that the readouts of the two recombinases Cre and Dre simultaneously reflect Cre+Dre−, Cre−Dre+, and Cre+Dre+ cell lineages. As proof of principle, we show specific labeling in three distinct progenitor/stem cell populations, including club cells, AT2 cells, and bronchoalveolar stem cells, in Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice. By using this new dual-recombinase reporter system, we simultaneously traced the cell fate of these three distinct cell populations during lung repair and regeneration, providing a more comprehensive picture of stem cell function in distal airway repair and regeneration. We propose that this new reporter system will advance developmental and regenerative research by facilitating a more sophisticated genetic approach to studying in vivo cell fate plasticity.
Keywords: lung, lung injury, cardiomyocyte, cardiovascular, stem cells, regeneration, development, bronchioalveolar stem cells (BASCs), Cre-loxP, Dre-rox, heart development, lineage tracing
Introduction
Mammalian organs are complex, dynamic tissues whose maintenance and repair depend on diverse stem/progenitor cell populations. During multiple types of organogenesis, such as heart, lung, and intestine, one stem/progenitor cell population can contribute to several cell types. For example, the epicardium and endocardium of the heart (1–4), basal cells of the lungs (5–7), and Lgr5+ stem cells of intestinal crypts (8) can give rise to multiple cell types during homeostasis, growth, and regeneration. Some differentiated cell types can also originate from multiple stem/progenitor cell sources. For instance, during heart development, coronary vascular endothelial cells, fibroblasts, pericytes, and adipocytes are derived from both epithelial and endocardial cells, albeit with different proportions of contribution (9–12). Additionally, cardiomyocytes in the developing heart are derived from both the primary and secondary heart field (13, 14). Multiple different epithelial cells in the distal airway are derived from club cells or alveolar type 2 cells and bronchioalveolar stem cells (BASCs)2 after lung injury (15, 16). Fate mapping of multiple cellular origins for their contribution of discrete cell lineages provides critical information for understanding tissue homeostasis renewal and regeneration after injury.
The ability to study multiple cell populations in one tissue is important to understand the complex biological process underlying development, tissue homeostasis, and regeneration. Genetic lineage tracing mediated by DNA site-specific recombination systems has been widely used for cell lineage and fate studies (17). Similar to the Cre-loxP system, multiple site-specific recombination systems, such as Flpe-Frt, Dre-rox, and Nigri-nox, have been developed and used for cell fate tracing studies (17–21). Two types of reporter systems have been developed and used for cell fates analysis. One type is conventional single-color reporter systems, such as Rosa26-tdTomato, Rosa26-LacZ, or Rosa26-GFP, which are used to track one cell lineage in one color (22–24), or multicolor systems, such as Brainbow or Confetti, used for clonal analysis of one cell lineage (25, 26). Another type is multicolor reporter systems that combine two or more recombination systems for genetic targeting of more than one cell population (usually two). Intersectional dual-reporter systems, such as RC::Fela, R26::FLAP, RC::RLTG, and R26NZG, can be used to label two distinct cell populations (27–30). In addition, sequential and exclusive double-reporter systems are also explored for labeling specific cell types more precisely (31–33). However, because of the exclusive nature of recombination design, these genetic tools only allow simultaneous labeling of two (sub)populations in tissue. Development of a multicolor reporter system capable of labeling three distinct cell populations by noninterfering recombination is useful for studying the behavior of more diverse cell types simultaneously in vivo.
In this study, we generated a new dual genetic system that incorporates both the Cre-loxP and Dre-rox recombination systems. Different from previous dual-recombinase reporters that reflect combination readout of Cre/Dre recombinases, the current reporters are parallel for two recombination readouts. Thus, this new dual system has the capacity to label three distinct cell populations simultaneously: Dre+Cre−, Dre−Cre+, and Dre+Cre+ cell populations. As proof of principle, we traced cardiomyocytes and endothelial cells simultaneously during heart development. Additionally, we used the system to label three stem cell populations (club cells, AT2 cells, and BASCs) in the lungs at homeostasis and to track their cell fate after injury. Our new mouse reporter R26-TLR extents the scope of cell type labeling with one reporter allele, which could be broadly used for diverse cell origin and cell fate studies in development, disease, and regeneration.
Results
Generation and characterization of R26-TLR
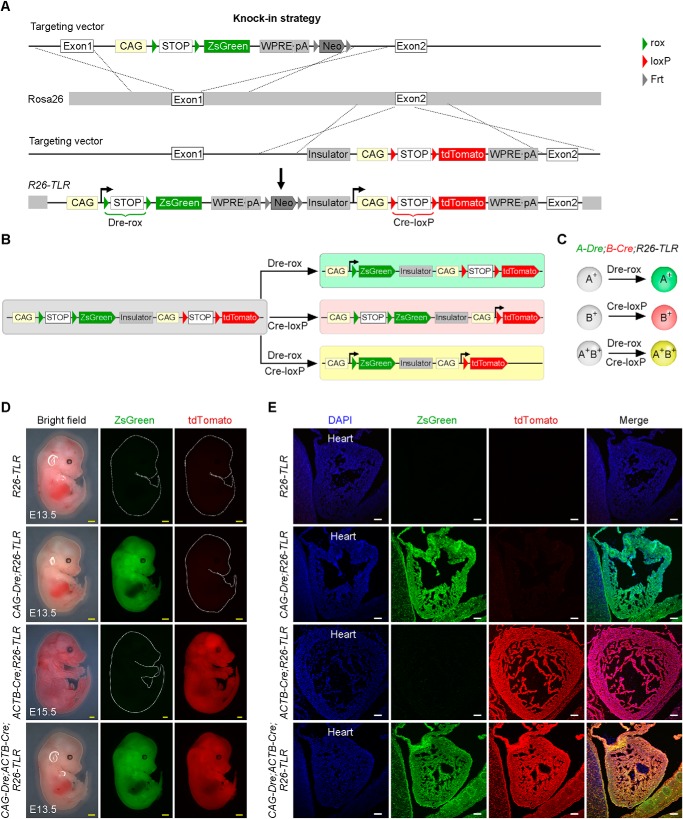
The Cre-loxP system is a widely used site-specific recombinase-based system. Like the Cre-loxP system, Dre recombinase specifically targets its recombination site rox (19). For genetic lineage studies, cell-specific promoter-derived Cre (or Dre) removes a loxP- or rox-flanked transcriptional stop cassette (stop) after the Rosa26 promoter, leading to permanent constitutively active expression of the following reporter in Cre- or Dre-expressing cells and their descendants (4). To genetically label multiple cell lineages, we generated a new lineage tracing reporter system that incorporates both the Cre-loxP and Dre-rox recombinations. This reporter was generated by knocking the CAG-rox-stop-rox-ZsGreen-WPRE-pA-Frt-Neo-Frt sequence into exon1 of the Rosa26 gene locus and knocking the insulator-CAG-loxP-stop-loxP-tdTomato-WPRE-pA sequence into exon2 of the Rosa26 gene locus through two homologous recombinations (Fig. 1A). Whether ZsGreen-WPRE-poly(A) after the Dre-rox recombination event can affect tdTomato gene expression by Cre-loxP recombination in the same allele is unknown. We therefore intentionally inserted the insulator between them to block the potential influence between them and ensure that the readouts of Dre-rox and Cre-loxP are specific and efficient. The insulator sequence is in Table S1. Dre and Cre recombinases driven by two different promoters recombine rox or loxP sites, resulting in ZsGreen or tdTomato reporter expression, respectively. Notably, when the two promoters are both active in a cell, the cell expresses ZsGreen and tdTomato at the same time, yielding yellow fluorescence (Fig. 1, B and C). We named this reporter line Rosa26 traffic light reporter (R26-TLR).
Figure 1.
Generation and characterization of the R26-TLR mouse line. A, schematic showing the knock-in strategy of R26-TLR by homologous recombination. B and C, schematics showing the results after Dre-rox or Cre-loxP recombination. D, whole-mount bright-field and epifluorescence images of E13.5 embryos of R26-TLR, CAG-Dre;R26-TLR, CAG-Dre;ACTB-Cre;R26-TLR, and E15.5 embryos of ACTB-Cre;R26-TLR. E, immunostaining for ZsGreen and tdTomato on the indicated embryos sections in D. Yellow scale bars = 1 mm; white scale bars = 100 μm. Each image is representative of five individual samples.
To characterize R26-TLR, we crossed the R26-TLR mouse line with CAG-Dre and ACTB-Cre lines. CAG and ACTB are gene promoters broadly active in almost all tissues. We acquired four littermate genotypes: R26-TLR, CAG-Dre;R26-TLR, ACTB-Cre;R26-TLR, and CAG-Dre;ACTB-Cre;R26-TLR. As expected by design (Fig. 1A), whole-mount epifluorescence analysis showed that neither ZsGreen nor tdTomato was detected in embryonic day 13.5 (E13.5) R26-TLR embryos, indicating no leakiness of R26-TLR without any recombinase. ZsGreen+ but not tdTomato+ signals were detected in E13.5 CAG-Dre;R26-TLR embryos, whereas tdTomato+ but not ZsGreen+ signals were detected in E15.5 ACTB-Cre;R26-TLR embryos. In CAG-Dre;ACTB-Cre;R26-TLR triple-positive embryos, both ZsGreen+ and tdTomato+ signals were detected (Fig. 1D). Immunostaining of ZsGreen and tdTomato on four genotyped embryonic sections showed that no ZsGreen or tdTomato was detected in cells in R26-TLR embryonic sections. All cells were ZsGreen+tdTomato− in the CAG-Dre;R26-TLR line, whereas all cells were ZsGreen−tdTomato+ in the ACTB-Cre;R26-TLR line, indicating that Dre or Cre recombinase specifically recombined its own target site (Fig. 1E). In the CAG-Dre;ACTB-Cre;R26-TLR triple-positive line, all cells expressed both ZsGreen and tdTomato, yielding yellow fluorescence in those Dre+Cre+ cells. Taken together, these data demonstrated that the new reporter R26-TLR could be used for tracking different cell populations simultaneously in vivo.
Tracing distinct cell populations simultaneously in the heart
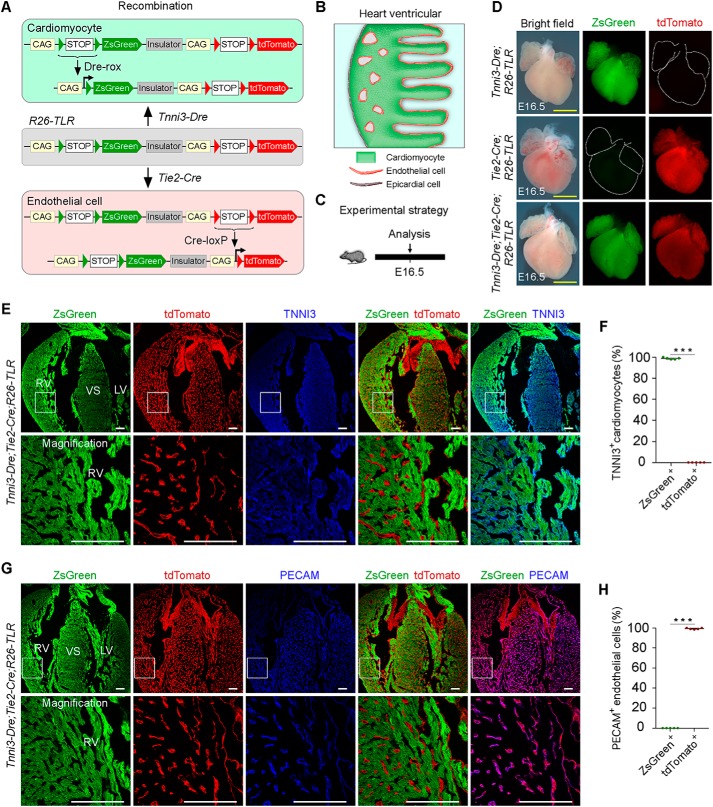
To further prove that R26-TLR can be used to trace different/diverse cell populations simultaneously, we crossed the R26-TLR mouse line with the Tnni3-Dre and Tie2-Cre mouse lines, which specifically target cardiomyocytes and endothelial cell lineages, respectively (32, 35). As shown in the design, Dre expression driven by the Tnni3 promoter resulted in ZsGreen gene expression, and Tie2-derived Cre led to tdTomato gene expression (Fig. 2, A and B). For analysis, we collected hearts from E16.5 embryos of three genotypes: Tnni3-Dre;R26-TLR, Tie2-Cre;R26-TLR, and Tnni3-Dre;Tie2-Cre;R26-TLR (Fig. 2C). Whole-mount epifluorescence imaging showed ZsGreen+ but not tdTomato+ signals detected in Tnni3-Dre;R26-TLR hearts. In contrast, tdTomato+ but not ZsGreen+ signals in vascular patterns were detected in Tie2-Cre;R26-TLR hearts. As expected, both ZsGreen+ and tdTomato+ signals were detected in Tnni3-Dre;Tie2-Cre;R26-TLR triple-positive hearts (Fig. 2D). To further verify the labeling specificity, we performed immunostaining on heart sections with the cardiomyocyte marker Tnni3 and the endothelial cell marker Pecam. Immunostaining for ZsGreen, tdTomato, and Tnni3 on Tnni3-Dre;Tie2-Cre;R26-TLR triple-positive heart sections showed that over 99% of all cardiomyocytes were ZsGreen+ tdTomato− (Fig. 2, E and F). We also performed immunostaining for ZsGreen, tdTomato, and Pecam on triple-positive heart sections and detected that more than 99% of endothelial cells were tdTomato+ZsGreen− (Fig. 2, G and H). The distributions of ZsGreen+ cardiomyocytes and tdTomato+ endothelial cells were interleaved and connected to each other. Furthermore, we could hardly detect any yellow signals, indicating that the promoters of Tnni3 and Tie2 were less likely to be active in the same cell types (Fig. 2, E and G). Additionally, immunostaining data from the Tnni3-Dre;R26-TLR and Tie2-Cre;R26-TLR double-positive line confirmed no cross-talk between these two Dre-rox and Cre-loxP recombinations (Fig. S1, A and B). Together, these data demonstrate that R26-TLR can be used for simultaneous labeling of distinct cell populations.
Figure 2.
Simultaneous labeling and tracing of cardiomyocytes and endothelial cells in Tnni3-Dre;Tie2-Cre;R26-TLR mice. A and B, schematics showing the results after Dre-rox and Cre-loxP recombination in Tnni3-Dre;Tie2-Cre;R26-TLR mice. C, schematic showing the time point for tissue analysis. D, whole-mount bright-field and epifluorescence images of E16.5 embryos of Tnni3-Dre;R26-TLR, Tie2-Cre;R26-TLR, and Tnni3-Dre;Tie2-Cre;R26-TLR. E, immunostaining for ZsGreen, tdTomato, and TNNI3 on E16.5 Tnni3-Dre;Tie2-Cre;R26-TLR heart sections shows that TNNI3+ cardiomyocytes are ZsGreen+ tdTomato−. F, quantification of the percentage of TNNI3+ cardiomyocytes labeled by Tnni3-Dre;Tie2-Cre;R26-TLR. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. G, immunostaining for ZsGreen, tdTomato, and PECAM on E16.5 Tnni3-Dre;Tie2-Cre;R26-TLR heat sections shows that PECAM+ endothelial cells are tdTomato+ ZsGreen−. Boxed regions are magnified in the bottom panels. H, quantification of the percentage of PECAM+ endothelial cells labeled by Tnni3-Dre;Tie2-Cre;R26-TLR. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. LV, left ventricle; RV, right ventricle; VS, ventricular septum. Yellow scale bars = 1 mm; white scale bars = 100 μm. Each image is representative of five individual samples.
Genetic tracing of diverse epithelial cell types simultaneously in the lungs
The lungs are a multifunctional organ consisting of pulmonary vasculature and a respiratory epithelial system that are essential for mammalian survival (36). Multiple stem/progenitor cells participate in the maintenance of lung function during homeostasis and repair (37). In the respiratory system, diverse resident epithelial stem cell populations have been identified from the proximal to the distal airway, which consists of four regions: the trachea, bronchi, bronchioles, and alveoli. The tracheal and bronchial epithelia predominantly consist of basal cells, club cells, ciliated cells, and goblet cells, whereas bronchioles mainly contain ciliated cells, club cells, and neuroendocrine cells (38). The alveolar epithelium includes squamous alveolar type I (AT1) and cuboidal alveolar type II (AT2) cells. These types of epithelial cells are vital for maintenance of the respiratory tract, and they are maintained by self-renewal and differentiation from stem cells (37, 39), e.g. club cells give rise to ciliated cells, and AT2 cells generate AT1 cells. Notably, a multipotent stem cell population has been reported to reside at bronchioalveolar duct junctions (BADJs); these BASCs coexpress the club cell maker CC10 and the AT2 cell maker SPC (40). In vivo genetic fate mapping studies have recently demonstrated that CC10+SPC+ BASCs can proliferate and differentiate into bronchiole epithelial club cells and ciliated cells and also alveolar epithelial AT1 and AT2 cells in distinct lung injury models (15, 16).
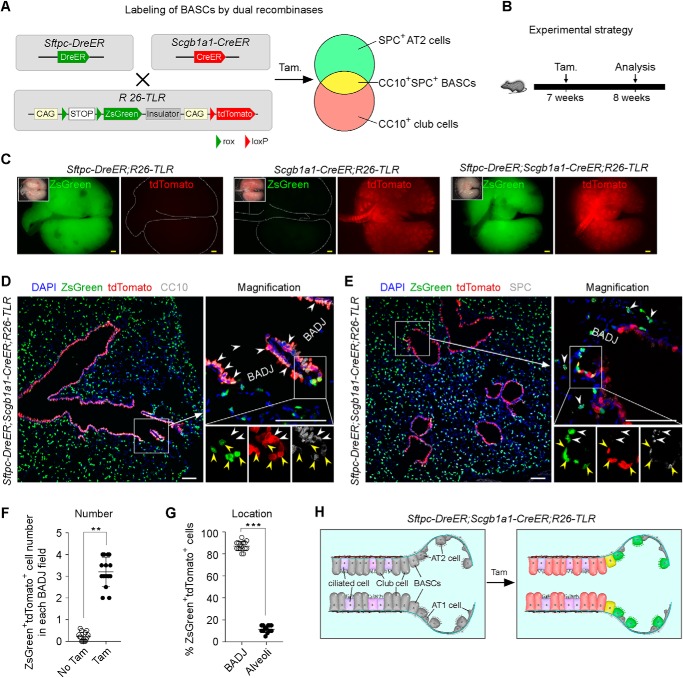
To understand the contribution of club cells, AT2 cells, and BASCs to lung repair in the same context, we employed R26-TLR to simultaneously label CC10+ club cells, SPC+ AT2 cells, and CC10+SPC+ BASCs in one mouse. We crossed R26-TLR with Sftpc-DreER and Scgb1a1-CreER lines to generate the Sftpc-DreER;Scgb1a1-CreER;R26-TLR triple-positive line. We expected tamoxifen treatment to induce ZsGreen gene expression in SPC+ AT2 cells by Dre-rox recombination, tdTomato gene expression in CC10+ club cells by Cre-loxP recombination, and ZsGreen and tdTomato gene expression in CC10+SPC+ BASCs by Dre-rox and Cre-loxP recombination (Fig. 3A). We administered tamoxifen to 7-week-old of Sftpc-DreER;Scgb1a1-CreER;R26-TLR triple-positive mice and collected the lungs after 1 week (Fig. 3B). Whole-mount epifluorescence data showed that ZsGreen+ and tdTomato+ signals were detected in triple-positive lungs. Furthermore, immunostaining for ZsGreen, tdTomato, and CC10 or SPC on triple-positive lung sections showed that tdTomato+ cells at bronchioles were CC10+ club cells, ZsGreen+ cells at alveolar region were SPC+ AT2 cells, and ZsGreen+tdTomato+ cells at BADJs were CC10+SPC+ BASCs (Fig. 3, D, E, and H). After 1 week of tamoxifen treatment (dose/weight, 0.2 mg/g), 2.94 ± 0.23 ZsGreen+tdTomato+ cells were detected in each BADJ field of tissue sections (Fig. 3F). Furthermore, 87.00% ± 1.71% of ZsGreen+tdTomato+ cells were located in BADJ regions, and 12.33% ± 1.26% of ZsGreen+tdTomato+ cells were located in alveolar regions (Fig. 3G), indicating that the majority of double-positive cells labeled by our system were specific. These data demonstrate that R26-TLR has the capability of labeling three distinct cell populations simultaneously in vivo.
Figure 3.
Simultaneous labeling of CC10+ club cells, SPC+ AT2 cells, and CC10+SPC+ BASCs in Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice. A, schematic showing the strategy for simultaneous labeling of club cells, AT2 cells, and BASCs using Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice. B, schematic showing the experimental strategy. C, whole-mount bright-field and epifluorescence images of lungs collected from Sftpc-DreER;R26-TLR mice, Scgb1a1-CreER;R26-TLR mice, and Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice. D, immunostaining for ZsGreen, tdTomato, and the club cell maker CC10 on Sftpc-DreER;Scgb1a1-CreER;R26-TLR lung section shows that tdTomato+ cells specifically label CC10+ club cells (white arrowheads). Yellow arrowheads, BASCs. E, immunostaining for ZsGreen, tdTomato, and SPC on Sftpc-DreER;Scgb1a1-CreER;R26-TLR lung sections shows that SPC+ AT2 cells are ZsGreen+ (white arrowheads). Yellow arrowheads, tdTomato+ZsGreen+ BASCs. F, quantification of ZsGreen+tdTomato+ cells in each BADJ field. Data are mean ± S.D. *, p < 0.01 by two-tailed t test. G, quantification of the percentage of ZsGreen+tdTomato+ cells located in BADJs or alveolar regions. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. H, cartoon showing the labeling result after recombination by tamoxifen (Tam). Yellow scale bars = 1 mm, white scale bars = 100 μm. Each image is representative of five individual samples.
Tracing diverse epithelial cell types simultaneously in lung repair and regeneration
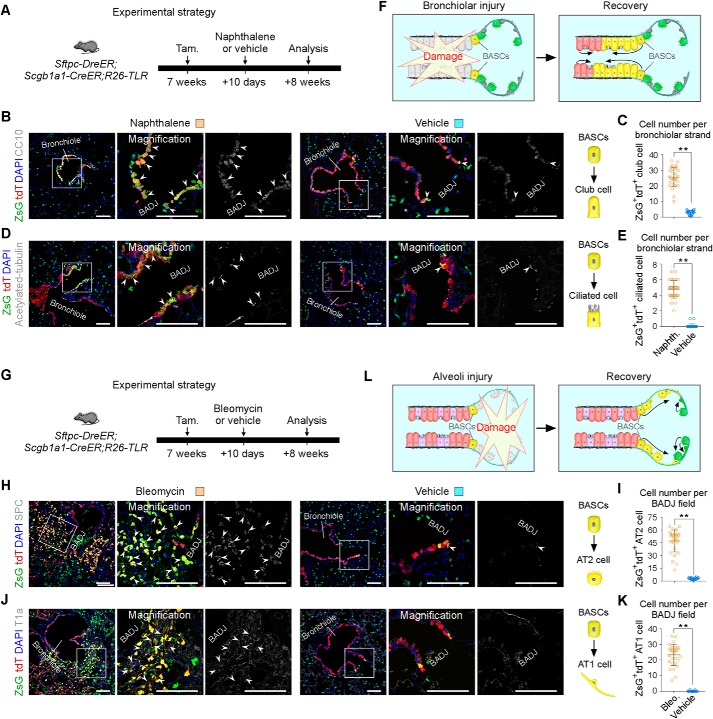
To monitor diverse epithelial cell behaviors labeled by R26-TLR after lung injury, we used two lung injury models: the naphthalene-induced bronchiolar injury model and the bleomycin-induced alveolar injury model. To study the fates of club cells, AT2 cells, and BASCs simultaneously in the bronchiolar injury model, Sftpc-DreER;Scgb1a1-CreER;R26-TLR triple-positive mice were treated with tamoxifen at 7 weeks and then with naphthalene or vehicle (corn oil) 10 days after tamoxifen induction (Fig. 4A). 8 weeks after injury, lung tissues were collected, and sectional immunostaining of ZsGreen, tdTomato, and the club cell maker CC10 or the ciliated cell maker Acetylated tubulin showed that both ZsGreen+tdTomato+ BASCs and ZsGreen−tdTomato+ club cells expanded and contributed to bronchiole epithelial cells, including club cells and ciliated cells, for repair of the distal airway (Fig. 4, B, D, and F). The alveolar region was largely undamaged, and we did not detect a significant contribution of ZsGreen+tdTomato− AT2 cells to ZsGreen−tdTomato+ club cells by naphthalene-induced injury (Fig. 4, B and D). Interestingly, ZsGreen+tdTomato+ BASCs were mainly responsible for repair of more terminal bronchioles. Quantification data showed a significant increase in bronchiolar ZsGreen+tdTomato+CC10+ cells (28.92 ± 2.07 naphthalene versus 3.72 ± 0.39 vehicle) and ZsGreen+tdTomato+ acetylated tubulin+ cells (4.80 ± 0.66 naphthalene versus 0.11 ± 0.07 vehicle) after bronchiolar injury (Fig. 4, C and E).
Figure 4.
Diverse types of epithelial stem cells respond distinctly after different lung injuries. A, schematic showing the strategy for bronchiolar injury. Tam, tamoxifen. B, immunostaining for CC10, ZsGreen, and tdTomato on lung tissue sections 8 weeks after injury. Boxed regions are magnified in the right panels. White arrowheads indicate CC10+ club cells derived from ZsGreen+tdTomato+ BASCs. C, quantification of the ZsGreen+tdTomato+CC10+ club cells in each bronchiolar strand in naphthalene- or vehicle-treated lungs. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. D, immunostaining for acetylated tubulin, ZsGreen, and tdTomato on lung tissue sections 8 weeks after injury. Arrowheads, acetylated tubulin+ ciliated cells derived from ZsGreen+tdTomato+ BASCs. E, quantification of the ZsGreen+tdTomato+acetylated tubulin+ ciliated cell in each bronchiolar strand. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. F, cartoon showing that club cells and BASCs mainly regenerate the terminal bronchiole after bronchiolar injury. G, schematic showing the strategy for alveolar injury. H, immunostaining for SPC, ZsGreen, and tdTomato on lung tissue sections 8 weeks after injury. Arrowheads, SPC+ AT2 cells derived from ZsGreen+tdTomato+ BASCs. I, quantification of ZsGreen+tdTomato+SPC+ AT2 cells in each BADJ field in bleomycin- or vehicle-treated lungs. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. J, immunostaining for T1a, ZsGreen, and tdTomato on lung tissue sections 8 weeks after injury. Arrowheads, T1a+ AT1 cells derived from ZsGreen+tdTomato+ BASCs. K, quantification of ZsGreen+tdTomato+T1a+ AT1 cells in each BADJ field. Data are mean ± S.D. *, p < 0.001 by two-tailed t test. L, cartoon showing that BASCs regenerate alveolar cells surrounding BADJs and that AT2 cells are responsible for alveolar regions away from BADJs after alveolar injury. ZsG, ZsGreen; tdT, tdTomato. Scale bars = 100 μm. Each image is representative of five individual samples.
Next we performed alveolar injury to study the fates of these three types of epithelial cells (Fig. 4G). After bleomycin-induced alveolar injury, ZsGreen+tdTomato+ BASCs mainly gave rise to SPC+ AT2 cells and T1a+ AT1 cells surrounding BADJ regions (Fig. 4, H and J). Quantification data showed a significant increase in alveolar ZsGreen+tdTomato+SPC+ cells (49.60 ± 5.87 bleomycin versus 3.96 ± 0.51 vehicle) and ZsGreen+tdTomato+T1a+ cells (26.40 ± 1.97 bleomycin versus 0.09 ± 0.03 vehicle) after alveolar injury (Fig. 4, I and K). Conversely, AT2 cells and AT1 cells far from BADJ regions were derived from ZsGreen+tdTomato− AT2 cells (Fig. 4, H, J, and L). The fates of BASCs after distinct injury models were consistent with previous reports (15, 16). Taken together, using lung epithelial cells as an example, the above results demonstrate that R26-TLR can be used to study diverse cell origins and fates during development, disease, and regeneration.
Discussion
In this study, we developed a new genetic tool, R26-TLR, for readout of Cre-loxP and Dre-rox recombination. How different recombination sites are arranged with reporter genes is critical for its use in tracing multiple cell lineages in vivo. The arrangement between recombination sites and reporter genes of R26-TLR is different from all previously reported intersectional dual-reporter systems, such as RC::Fela, R26::FLAP, RC::RLTG, and R26NZG (27–30). The model of previous intersectional dual systems is CAG-site1-stop-site1-site2-reporter1-stop-site2-reporter2. Recombination of recombinase1-site1 results in reporter1 expression. Expression of reporter2 needs recombination of recombinase1-site1 and recombinase2-site2. Because of the exclusive nature of two reporters in recombinase-targeted cells, reporter2 can only reflect a subpopulation of reporter1-labeled populations (33). The model of our newly generated R26-TLR is CAG-site1-stop-site1-reporter1-CAG-site2-stop-site2-reporter2, which permits parallel expression of reporters by two different recombination events. We previously reported a dual system, R26-NLR, which was generated based on Nigri-nox and Cre-loxP systems (20). Driven by different cell-specific promoters, e.g. promoter A and promoter B, R26-NLR can be applied to label two distinct A+B− and A−B+ cell populations simultaneously in the same mouse. However, R26-NLR cannot clearly distinguish the double A+B+ cell population from the A+B− cell population or A−B+ cell population. In this study, because of the different design strategy, R26-TLR integrates the advantages of previously reported intersectional reporters and our previous R26-NLR tool. It can be used to label two distinct cell populations and also to label their intersectional subpopulations. The new dual R26-TLR system could be used to label all three cell populations: A+B−, A−B+, and A+B+ (double-positive population). It is a better system than our previous R26-NLR system and other previously reported intersectional reporters.
In this study, we used three examples to prove that R26-TLR works in vivo. In the first example, we used three mouse lines, CAG-Dre;R26-TLR, ACTB-Cre;R26-TLR, and CAG-Dre;ACTB-Cre;R26-TLR, to demonstrate that all cells were ZsGreen+tdTomato− after Dre-rox recombination, ZsGreen−tdTomato+ after Cre-loxP recombination, or ZsGreen+ tdTomato+ after both Dre-rox and Cre-loxP recombination, respectively. These data demonstrate that there is no cross-reaction between the Dre-rox and Cre-loxP systems. Further, we showed that the Tnni3-Dre;Tie2-Cre;R26-TLR triple-positive line can specifically and efficiently label cardiomyocytes and endothelial cells simultaneously in the heart. In the third example, we used the Sftpc-DreER;Scgb1a1-CreER;R26-TLR triple-positive line to demonstrate that R26-TLR can be used to genetically trace three cell populations, CC10−SPC+ AT2 cells, CC10+SPC− club cells, and the SPC+CC10+ BASCs population, which could not be achieved with previous tools. Their fates could also be clearly tracked after lung injury. In the same mouse, we observed the behavior of three types of cells in the same context after lung injury. BASCs could regenerate distal bronchioles and alveoli near BADJ regions, whereas club cells and AT2 cells could repair other distal airway regions after injury. We use this example to convincingly show that application of R26-TLR yields more comprehensive information than the previously reported intersectional reporters and our previous R26-NLR.
During organogenesis, multiple stem/progenitor cells were heterogeneous and could be identified by expressing more than one gene markers, such as epithelia and mesenchyme. The distribution and cell fate of these diverse cell populations could be well studied using this new tool. In several organs, in the border between two distinct types of epithelia, there existed transitional epithelium that coexpressed bilateral epithelial markers, including the squamous columnar junction of the gastroesophageal junction, cervix junction, and rectum–anus junction (41–43); BASCs at BADJs of lung epithelium (15, 16, 40); and limbal stem cells at the narrow zone between the conjunctiva and cornea of the eyes (44, 45). Moreover, many double-positive epithelial stem/progenitor cells appeared in addition to transitional epithelium during development. For example, during alveologenesis of the mouse, the epithelial progenitors of the distal tip are Sox9+Id2+ and responsible for giving rise to bronchiole and alveolar epithelial cells (37, 46, 47). Axin2+SPC+ AT2 cells were identified as alveolar epithelial progenitors in adult lung (48, 49). Basal cells, which can be identified by expression of P63 and Krt5 (double-positive), are multipotent progenitors of lung epithelia (50). On the other hand, mesenchymal cells were also heterogeneous. For instance, embryonic mesenchymal cells can be identified by expressing PDGFRa and PDGFRb, but they were considered to be fibroblasts and pericytes markers in adult tissues, respectively (9, 51, 52). Whether the expression of PDGFRa and PDGFRb is exclusive or whether the lineage of the PDGFRa+PDGFRb+ double-positive mesenchymal population represents a unique cell population that differs from other two single-positive populations remains unknown; their distinct roles in homeostasis and regeneration needed to be investigated further. Because of a lack of related mouse lines, we are still unable to answer these questions by using R26-TLR, but are they are worthy of discussion in the future. We used this new system to simultaneously track three distinct lung epithelial cell populations and examined their discrete contributions to different cell lineages during lung injury and repair. Given the capability of genetic tracing of diverse cell populations without the need of antibody staining, R26-TLR can be used broadly for diverse cell origin and cell fate studies and can also be combined with live imaging for more clear in vivo studies in the future. Moreover, using the strategy of an insulator to permit parallel expression of two recombination results, we can expand them into three or more and, in theory, provide an approach to genetically study multiple cell lineages simultaneously in vivo.
In summary, our newly generated R26-TLR eases the number of mice crossing of different Cre/Dre reporter lines and permits simultaneous tracing of three distinct cell lineages in vivo. Notably, this mouse line extends the capacity of previously reported dual-recombinase-mediated genetic cell labeling and can be used widely to study the origin and fate of diverse cell populations simultaneously in development, disease, and regeneration.
Experimental procedures
Mice
All animal procedures were reviewed and approved by the ethics committee of the Institutional Animal Care and Use Committee of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All experiments were performed strictly within the committee's guidelines. The ACTB-Cre, CAG-Dre, Tnni3-Dre, Tie2-Cre, Sftpc-DreER, and Scgb1a1-CreER mice lines were described previously (15, 19, 32, 34, 35). The R26-TLR mouse line was created by knocking the CAG-rox-stop-rox-ZsGreen-WPRE-pA-Frt-Neo-Frt sequence into exon1 of the Rosa26 locus and knocking the insulator-CAG-loxP-stop-loxP-tdTomato-WPRE-pA sequence into exon2 of the Rosa26 gene locus using traditional targeted mutation through homologous recombination in embryonic stem cells. The CAG promoter, consisting of the cytomegalovirus enhancer fused to the chicken β-actin promoter, is a strong synthetic promoter frequently used to drive high levels of gene expression in mammalian cells, such as mouse cells. Stop is a transcriptional stop cassette. rox and loxP are recombinases recognition sites. WPRE is a woodchuck hepatitis virus posttranscriptional regulatory element for enhancing the stability of RNA transcription. Neo is a screening gene in mouse generation. An insulator is used to block the potentially mutual influence between the two recombination events. ZsGreen and tdTomato are two kinds of fluorescent protein genes. The obtained chimeric mouse lines were crossed to C57BL/6 lines for germline transmission. Generation of R26-TLR was supported by Shanghai Model Organisms Center, Inc. All mice used in this study were kept at C57BL6 backgrounds. The dosage of tamoxifen (Sigma, T5648) treatment was 0.2 mg/g by oral gavage.
Genomic PCR
Genomic DNA was extracted from mouse tails. Briefly, the tail tissues were lysed in the lysis buffer for 8 h at 55 °C, and then the mixture was centrifuged at 15,000 rpm for 5 min to obtain a supernatant solution of genomic DNA. Next we used isopropanol to precipitate the DNA and then washed the DNA in 70% ethanol by centrifugation at 15,000 rpm for 3 min. Finally, the DNA was dissolved in double-distilled H2O.
Tissue collection and immunofluorescent staining
The immunostaining protocol was performed as described previously (20). Briefly, the tissues were collected after euthanasia of the mice and then fixed in 4% paraformaldehyde (Sigma) for 30∼60 min at 4 °C according to tissue size. The tissues were washed three times in PBS, and then the tissues were dehydrated at 30% sucrose (dissolved in PBS) at 4 °C. After the tissues sank to the bottom of the solution, they were embedded in optimum cutting temperature (Sakura) and stored at −80 °C. 10-μm frozen sections were collected on slides. For immunofluorescent staining, the tissue sections were first dried at room temperature and then placed in PBS to remove optimum cutting temperature. Next, the tissue sections were blocked in 5% PBSST (5% donkey serum in PBS and 0.1% Triton X-100 in PBS) for 30 min at room temperature. Then the blocking buffer was discarded, and tissue sections were incubated with primary antibodies at 4 °C overnight in the dark. The following primary antibodies and dilutions were used: PECAM (BD Biosciences, 553370, 1:500), ZsGreen (Clontech, 632474, 1:1000), tdTomato (Rockland, 600-401-379, 1:1000), CC10 (Santa Cruz Biotechnology, SC-9772, 1:200), acetylated tubulin (Sigma, T7451, 1:500), SPC (Millipore, AB3786, 1:200), TNNI3 (Abcam, ab56357; 1:200), and T1a (DSHB, 8.1.1, 1:200). The next day, the slides were washed in PBS to remove primary antibodies and then incubated with secondary antibodies for 40 min at room temperature in the dark. Next, the slides were washed in PBS several times. Then the slides were incubated with 4′,6-diamidino-2-phenylindole (Vector Laboratories) and mounted with mounting medium. The secondary antibodies were Alexa donkey anti-rabbit 555 (Invitrogen, A31572, 1:1000), donkey anti-rat 647 (Abcam, ab150155, 1:1000) Alexa donkey anti-mouse 647 (Invitrogen, A31571, 1:1000), biotin-sp-a-rabbit IgG (JIR, 711-065-152, 1:200), Biotin-sp-a-goat IgG (JIR, 705-065-147, 1:200), donkey anti-goat 647 (Invitrogen, A21447, 1:1000), biotin-sp-goat anti-Syrian hamster IgG(H+L) (JIR, 107-065-142, 1:200), Dylight 647-streptavidin (JIR, 016-490-084, 1:200), and Immpress goat-anti rat (Vector Laboratories, MP-7444, 1:3). An Olympus confocal microscope (FV1200) was used to acquire immunostaining images. Images were analyzed using ImageJ (National Institutes of Health) software.
Bronchiolar injury
Bronchiolar injury was achieved as described previously (15). Naphthalene (Sigma, 84679) was dissolved in sterile corn oil (25 mg ml−1). For bronchiolar injury, Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice were treated with tamoxifen at 7 weeks and then with 250 mg kg−1 naphthalene or vehicle (corn oil) by naphthalene intraperitoneal injection after 10 days. The lung tissues were collected after 8 weeks of recovery. For analysis, fluorescence+ CC10+ club cell number or fluorescence+ acetylated-tubulin+ ciliated cell number was quantified at BADJ field.
Alveolar injury
Alveolar injury was induced by bleomycin as described previously (15). Bleomycin (Sigma, B8416) was freshly dissolved in PBS (10 units ml−1) and stored at −80 °C. 10 units ml−1 bleomycin was diluted to 1 unit ml−1 with PBS before use. For alveolar injuries, Sftpc-DreER;Scgb1a1-CreER;R26-TLR mice were treated with tamoxifen at 7 weeks and then with 2 units kg−1 bleomycin or vehicle (PBS) by intratracheal instillation after 10 days. The lungs were collected after 8 weeks. For analysis, fluorescence+SPC+ AT2 cells or fluorescence+T1a+ AT1 cells were quantified at BADJ fields.
Statistical analysis
The data were acquired from five independent experiments and are presented as mean values ± S.D. Two-sided unpaired Student's t test was used to compare the difference between two groups. p < 0.001 was considered statistically significant.
Author contributions
K. L., Q. L., X. L., X. H., Y. L., L. Z., J. T., Z. L., H. W., H. Ji, and B. Z. resources; K. L. and M. T. data curation; K. L. and B. Z. software; K. L., M. T., and B. Z. formal analysis; K. L., H. Jin, L. H., H. Z., W. P., and B. Z. methodology; K. L. and B. Z. writing-original draft; K. L., M. T., and B. Z. writing-review and editing; L. H. and B. Z. supervision; B. Z. funding acquisition; B. Z. validation; B. Z. investigation; B. Z. visualization.
Supplementary Material
Acknowledgments
We thank Shanghai Model Organisms Center, Inc. for mouse generation and Baojin Wu, Guoyuan Chen, Zhonghui Weng, and Aimin Huang for animal husbandry. We also thank Wei Bian for technical assistance and members of National Center for Protein Science Shanghai for assistance with microscopy.
This work was supported by National Key Research and Development Program of China Grants 2018YFA0108100, 2019YFA0110400, 2018YFA0107900, 2016YFC1300600, and 2017YFC1001303; Strategic Priority Research Program of the Chinese Academy of Sciences Grants XDA16010507 and XDB19000000; and National Science Foundation of China Grants 31730112, 91639302, 31625019, 91849202, 81761138040, 81872241, 31701292, and 31922032. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1 and Table S1.
- BASC
- bronchioalveolar stem cell
- E13.5
- embryonic day 13.5
- BADJ
- bronchioalveolar duct junction
- WPRE
- woodchuck hepatitis virus posttranscriptional regulatory element
- CAG
- hybrid construct consisting of the cytomegalovirus enhancer fused to the chicken beta-actin promoter
- ACTB
- beta-actin
- PECAM
- platelet endothelial cell adhesion molecule
- PDGFRa
- platelet-derived growth factor receptor alpha
- PDGFRb
- platelet-derived growth factor receptor beta
- SPC
- surfactant protein C.
References
- 1. Chen Q., Zhang H., Liu Y., Adams S., Eilken H., Stehling M., Corada M., Dejana E., Zhou B., and Adams R. H. (2016) Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun. 7, 12422 10.1038/ncomms12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R., and Pu W. T. (2008) Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai C. L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X., Stallcup W. B., Denton C. P., McCulloch A., Chen J., and Evans S. M. (2008) A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104–108 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tian X., Pu W. T., and Zhou B. (2015) Cellular origin and developmental program of coronary angiogenesis. Circ. Res. 116, 515–530 10.1161/CIRCRESAHA.116.305097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynch T. J., Anderson P. J., Rotti P. G., Tyler S. R., Crooke A. K., Choi S. H., Montoro D. T., Silverman C. L., Shahin W., Zhao R., Jensen-Cody C. W., Adamcakova-Dodd A., Evans T. I. A., Xie W., Zhang Y., et al. (2018) Submucosal gland myoepithelial cells are reserve stem cells that can regenerate mouse tracheal epithelium. Cell Stem Cell 22, 653–667.e5 10.1016/j.stem.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tata A., Kobayashi Y., Chow R. D., Tran J., Desai A., Massri A. J., McCord T. J., Gunn M. D., and Tata P. R. (2018) Myoepithelial cells of submucosal glands can function as reserve stem cells to regenerate airways after injury. Cell Stem Cell 22, 668–683.e6 10.1016/j.stem.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y., Riccio P., Schotsaert M., Mori M., Lu J., Lee D. K., García-Sastre A., Xu J., and Cardoso W. V. (2018) Spatial-temporal lineage restrictions of embryonic p63+ progenitors establish distinct stem cell pools in adult airways. Dev. Cell 44, 752–761.e4 10.1016/j.devcel.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., and Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 9. Zhang H., Huang X., Liu K., Tang J., He L., Pu W., Liu Q., Li Y., Tian X., Wang Y., Zhang L., Yu Y., Wang H., Hu R., Wang F., et al. (2017) Fibroblasts in an endocardial fibroelastosis disease model mainly originate from mesenchymal derivatives of epicardium. Cell Res. 27, 1157–1177 10.1038/cr.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Pu W., Liu Q., He L., Huang X., Tian X., Zhang L., Nie Y., Hu S., Lui K. O., and Zhou B. (2016) Endocardium contributes to cardiac fat. Circ. Res. 118, 254–265 10.1161/CIRCRESAHA.115.307202 [DOI] [PubMed] [Google Scholar]
- 11. Liu Q. Z., Huang X., Oh J. H., Lin R. Z., Duan S., Yu Y., Yang R., Qiu J., Melero-Martin J. M., Pu W. T., and Zhou B. (2014) Epicardium-to-fat transition in injured heart. Cell Res. 24, 1367–1369 10.1038/cr.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Yan Y., Yang X., Zhong T. P., Pu W. T., and Zhou B. (2014) Vessel formation: de novo formation of a distinct coronary vascular population in neonatal heart. Science 345, 90–94 10.1126/science.1251487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu S. M., Chien K. R., and Mummery C. (2008) Origins and fates of cardiovascular progenitor cells. Cell 132, 537–543 10.1016/j.cell.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buckingham M., Meilhac S., and Zaffran S. (2005) Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826–835 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- 15. Liu Q., Liu K., Cui G., Huang X., Yao S., Guo W., Qin Z., Li Y., Yang R., Pu W., Zhang L., He L., Zhao H., Yu W., Tang M., et al. (2019) Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 51, 728–738 10.1038/s41588-019-0346-6 [DOI] [PubMed] [Google Scholar]
- 16. Salwig I., Spitznagel B., Vazquez-Armendariz A. I., Khalooghi K., Guenther S., Herold S., Szibor M., and Braun T. (2019) Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J. 38, e102099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kretzschmar K., and Watt F. M. (2012) Lineage tracing. Cell 148, 33–45 10.1016/j.cell.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 18. Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., and Dymecki S. M. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- 19. Anastassiadis K., Fu J., Patsch C., Hu S., Weidlich S., Duerschke K., Buchholz F., Edenhofer F., and Stewart A. F. (2009) Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2, 508–515 10.1242/dmm.003087 [DOI] [PubMed] [Google Scholar]
- 20. Liu K., Yu W., Tang M., Tang J., Liu X., Liu Q., Li Y., He L., Zhang L., Evans S. M., Tian X., Lui K. O., and Zhou B. (2018) A dual genetic tracing system identifies diverse and dynamic origins of cardiac valve mesenchyme. Development 145, dev167775 10.1242/dev.167775 [DOI] [PubMed] [Google Scholar]
- 21. Buckingham M. E., and Meilhac S. M. (2011) Tracing cells for tracking cell lineage and clonal behavior. Dev. Cell 21, 394–409 10.1016/j.devcel.2011.07.019 [DOI] [PubMed] [Google Scholar]
- 22. Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., and Zeng H. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- 24. Srinivas S., Watanabe T., Lin C.-S., William C. M., Tanabe Y., Jessell T. M., and Costantini F. (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., Bennis R. A., Sanes J. R., and Lichtman J. W. (2007) Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- 26. Snippert H. J., van der Flier L. G., Sato T., van Es J. H., van den Born M., Kroon-Veenboer C., Barker N., Klein A. M., van Rheenen J., Simons B. D., and Clevers H. (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 27. Jensen P., Farago A. F., Awatramani R. B., Scott M. M., Deneris E. S., and Dymecki S. M. (2008) Redefining the serotonergic system by genetic lineage. Nat. Neurosci. 11, 417–419 10.1038/nn2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Awatramani R., Soriano P., Rodriguez C., Mai J. J., and Dymecki S. M. (2003) Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat. Genet. 35, 70–75 10.1038/ng1228 [DOI] [PubMed] [Google Scholar]
- 29. Plummer N. W., Evsyukova I. Y., Robertson S. D., de Marchena J., Tucker C. J., and Jensen P. (2015) Expanding the power of recombinase-based labeling to uncover cellular diversity. Development 142, 4385–4393 10.1242/dev.129981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamamoto M., Shook N. A., Kanisicak O., Yamamoto S., Wosczyna M. N., Camp J. R., and Goldhamer D. J. (2009) A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis 47, 107–114 10.1002/dvg.20474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schönhuber N., Seidler B., Schuck K., Veltkamp C., Schachtler C., Zukowska M., Eser S., Feyerabend T. B., Paul M. C., Eser P., Klein S., Lowy A. M., Banerjee R., Yang F., Lee C. L., et al. (2014) A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat. Med. 20, 1340–1347 10.1038/nm.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He L., Li Y., Li Y., Pu W., Huang X., Tian X., Wang Y., Zhang H., Liu Q., Zhang L., Zhao H., Tang J., Ji H., Cai D., Han Z., et al. (2017) Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 23, 1488–1498 10.1038/nm.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao H., and Zhou B. (2019) Dual genetic approaches for deciphering cell fate plasticity in vivo: more than double. Curr. Opin. Cell Biol. 61, 101–109 10.1016/j.ceb.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 34. Rawlins E. L., Okubo T., Xue Y., Brass D. M., Auten R. L., Hasegawa H., Wang F., and Hogan B. L. (2009) The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534 10.1016/j.stem.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kisanuki Y. Y., Hammer R. E., Miyazaki J., Williams S. C., Richardson J. A., and Yanagisawa M. (2001) Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 10.1006/dbio.2000.0106 [DOI] [PubMed] [Google Scholar]
- 36. Glenny R. W. (2011) Emergence of matched airway and vascular trees from fractal rules. J. Appl. Physiol. 110, 1119–1129 10.1152/japplphysiol.01293.2010 [DOI] [PubMed] [Google Scholar]
- 37. Hogan B. L., Barkauskas C. E., Chapman H. A., Epstein J. A., Jain R., Hsia C. C., Niklason L., Calle E., Le A., Randell S. H., Rock J., Snitow M., Krummel M., Stripp B. R., Vu T., et al. (2014) Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 10.1016/j.stem.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morrisey E. E., and Hogan B. L. (2010) Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8–23 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zepp J. A., and Morrisey E. E. (2019) Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol. 20, 551–566 10.1038/s41580-019-0141-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim C. F., Jackson E. L., Woolfenden A. E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R. T., and Jacks T. (2005) Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 10.1016/j.cell.2005.03.032 [DOI] [PubMed] [Google Scholar]
- 41. Jiang M., Li H., Zhang Y., Yang Y., Lu R., Liu K., Lin S., Lan X., Wang H., Wu H., Zhu J., Zhou Z., Xu J., Lee D. K., Zhang L., et al. (2017) Transitional basal cells at the squamous-columnar junction generate Barrett's oesophagus. Nature 550, 529–533 10.1038/nature24269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mirkovic J., Howitt B. E., Roncarati P., Demoulin S., Suarez-Carmona M., Hubert P., McKeon F. D., Xian W., Li A., Delvenne P., Crum C. P., and Herfs M. (2015) Carcinogenic HPV infection in the cervical squamo-columnar junction. J. Pathol. 236, 265–271 10.1002/path.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang E. J., Quick M. C., Hanamornroongruang S., Lai K., Doyle L. A., McKeon F. D., Xian W., Crum C. P., and Herfs M. (2015) Microanatomy of the cervical and anorectal squamocolumnar junctions: a proposed model for anatomical differences in HPV-related cancer risk. Mod. Pathol. 28, 994–1000 10.1038/modpathol.2015.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ksander B. R., Kolovou P. E., Wilson B. J., Saab K. R., Guo Q., Ma J., McGuire S. P., Gregory M. S., Vincent W. J., Perez V. L., Cruz-Guilloty F., Kao W. W., Call M. K., Tucker B. A., Zhan Q., et al. (2014) ABCB5 is a limbal stem cell gene required for corneal development and repair. Nature 511, 353–357 10.1038/nature13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gonzalez G., Sasamoto Y., Ksander B. R., Frank M. H., and Frank N. Y. (2018) Limbal stem cells: identity, developmental origin, and therapeutic potential. Wiley Interdiscip. Rev. Dev. Biol. 10.1002/wdev.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rawlins E. L., Clark C. P., Xue Y., and Hogan B. L. (2009) The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 10.1242/dev.037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rockich B. E., Hrycaj S. M., Shih H. P., Nagy M. S., Ferguson M. A., Kopp J. L., Sander M., Wellik D. M., and Spence J. R. (2013) Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E4456–E4464 10.1073/pnas.1311847110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nabhan A. N., Brownfield D. G., Harbury P. B., Krasnow M. A., and Desai T. J. (2018) Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zacharias W. J., Frank D. B., Zepp J. A., Morley M. P., Alkhaleel F. A., Kong J., Zhou S., Cantu E., and Morrisey E. E. (2018) Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zuo W., Zhang T., Wu D. Z., Guan S. P., Liew A. A., Yamamoto Y., Wang X., Lim S. J., Vincent M., Lessard M., Crum C. P., Xian W., and McKeon F. (2015) p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature 517, 616–620 10.1038/nature13903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boström H., Willetts K., Pekny M., Levéen P., Lindahl P., Hedstrand H., Pekna M., Hellström M., Gebre-Medhin S., Schalling M., Nilsson M., Kurland S., Törnell J., Heath J. K., and Betsholtz C. (1996) PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85, 863–873 10.1016/S0092-8674(00)81270-2 [DOI] [PubMed] [Google Scholar]
- 52. Armulik A., Genové G., and Betsholtz C. (2011) Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 10.1016/j.devcel.2011.07.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.