Abstract
The liver is constantly exposed to dietary antigens, viruses, and bacterial products with inflammatory potential. For decades cellular uptake of virus has been studied in connection with infection, while the few studies designed to look into clearance mechanisms focused mainly on the role of macrophages. In recent years, attention has been directed towards the liver sinusoidal endothelial cells (LSECs), which play a central role in liver innate immunity by their ability to scavenge pathogen- and damage-associated molecular patterns. Every day our bodies are exposed to billions of gut-derived pathogens which must be efficiently removed from the circulation to prevent inflammatory and/or immune reactions in other vascular beds. Here, we have used GFP-labelled Enterobacteria phage T4 (GFP-T4-phage) as a model virus to study the viral scavenging function and metabolism in LSECs. The uptake of GFP-T4-phages was followed in real-time using deconvolution microscopy, and LSEC identity confirmed by visualization of fenestrae using structured illumination microscopy. By combining these imaging modalities with quantitative uptake and inhibition studies of radiolabelled GFP-T4-phages, we demonstrate that the bacteriophages are effectively degraded in the lysosomal compartment. Due to their high ability to take up and degrade circulating bacteriophages the LSECs may act as a primary anti-viral defence mechanism.
Subject terms: Biological techniques, Cell biology
Introduction
Bacterial viruses, or bacteriophages are among the most numerous biological entities within the human body, capable of stimulating humoral immune responses and inducing anti-phage antibodies1,2. Containing an impressive 2 × 1012 phage particles3 the gastrointestinal tract is the natural reservoir of phages in humans4,5. From here, they can spread throughout the body via different mechanisms, e.g. intestinal epithelial damage or “leaky gut”6, and transcytosis7. It is estimated that 31 × 109 phage particles are transcytosed across the epithelial cell layers of the gut into the human body every day7, and this number rises in patients with increased gut permeability. Aberrant transcytosis may contribute to enhanced immune responses, allergic reactions, and inflammatory diseases. In these conditions, the phages that leak into the blood stream must be efficiently removed to prevent inflammatory and/or immune reactions in other vascular beds. While cellular entrance of virus and virus-like particles in host cells have been much studied, few studies were designed to look into general clearance mechanisms for circulating virus particles, including those viruses that gain access to the general circulation from the gut in states such as inflammation/epithelial damage8. Previous studies have identified the liver as a central player in clearance of blood-borne pathogens9–11. Due to the traditional view that the clearance of material from the circulation is mainly a function of the liver macrophages (or Kupffer cells), most reports on liver-mediated clearance of blood-borne virus and phage particles have uncritically claimed that the hepatic uptake of blood-borne virus/phages is by phagocytosis in Kupffer cells9,12–14. However, recent studies have shown that the liver sinusoidal endothelial cells (LSECs), lining the liver sinusoids and outnumbering the Kupffer cells in the sinusoids, are geared to clathrin-mediated endocytosis of small particles of the same size as most viral/phage particles, and thus play a major role in the elimination of blood-borne viral particles10,15–18. In fact, it has recently been shown that Adenovirus 1 (AdV), BK and JC polyomavirus-like particles, and HIV-like particles are rapidly cleared from the mouse circulation by uptake mainly in the LSECs10,16,18. However, the intracellular processing of virus and viral-like particles in these cells is currently unknown.
Knowing that phages easily gain access to the blood circulation, and that intravenously administered phages and other viruses are quickly and effectively cleared by the liver9,10,16,17, we hypothesize that bacteriophages that reach the liver are cleared via uptake and degradation by the LSECs. To test this hypothesis, we used GFP-labelled lysozyme-inactivated T4 bacteriophages (GFP-T4-phages)19 as a model virus to study viral endocytosis and metabolism in the LSECs. The T4 phage is a structurally simple, double-stranded DNA virus, and a perfect tool for various applications including cellular imaging20,21. Here, we studied the uptake and degradation of T4-phages in primary cultures of freshly isolated rat LSECs, demonstrating the transport of the particles to the lysosomes in real-time using deconvolution microscopy. The cellular binding and degradation of the phages were assessed by quantitative uptake studies using radiolabeled GFP-T4-phages. This is the first report showing in vitro uptake and lysosomal degradation of bacteriophages by LSECs, providing a direct evidence for the role of LSECs in bacteriophage clearance, entailing their contribution to the anti-viral defence mechanism.
Results and Discussion
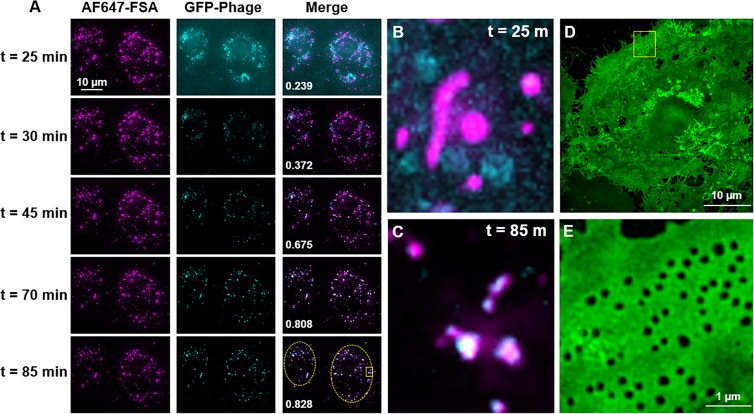
Viruses are quickly (minutes) and extensively (>90%) eliminated by the liver, with LSECs in particular being the primary site of uptake, leaving only a small fraction of circulating virus to infect the body9,10,16,17. However, little is known about what these nanoparticles undergo once they enter the scavenging LSECs. Here we investigated the uptake of bacteriophages by primary cultures of rat LSECs, focusing on the clearance ability of the LSECs rather than viral infection. T4 bacteriophages were used as a model virus, which we genetically engineered to express the green fluorescent protein (GFP) in the capsid19 in order to allow live cell imaging of interaction of the phages with the cells. The integrity of the phages was confirmed by negative staining and transmission electron microscopy showing that the head of the phage was attached to the contractile tail (Fig. 1). Importantly, the phages were not aggregated, but found as single particles, a critical prerequisite for their recognition by LSECs, and not by the Kupffer cells which engulf larger complexes (>200 nm)22,23. Freshly isolated rat LSECs in culture were pulsed for 15 min with a low concentration of Alexa Fluor-647-formaldehyde treated bovine serum albumin (AF647-FSA) (5 µg/ml), non-attached ligand washed off, and the cells further incubated for another 1.5 h to functionally mark the late endosomal and lysosomal compartments24. The cells were then challenged with GFP-T4-phages and imaged in real time using deconvolution microscopy (DV), with 5-min intervals for 60 min from the time the phages were detected intracellularly (Fig. 2 and Supplementary Video 1). Due to low GFP-fluorescence intensity per phage particle, the DV was unable to resolve individual phage particles. As a consequence, the phages could not be detected during the initial phase of uptake (not shown). It was only after 20–25 min, when phages had clustered in the endosomal compartment, that the accumulated fluorescence signal was sufficient to resolve and thus visualize the phages. Few GFP-T4-phages were found colocalized in the same vesicles as AF647-FSA at 25 min post-incubation (Fig. 2A,B), after which the phages displayed a gradual accumulation in the same compartments as the AF647-FSA (Fig. 2A–C). These compartments are late endosomes/lysosomes (Fig. 3). These findings are in line with previously reported endocytosis of FITC-labelled ligands in rat and pig LSECs, where shortly after internalization and during the first 20 min, the ligands were found mostly in early endosomes, and some (approximately 23% in rat) in late endosomes, and by 2 h all were transferred to late endosomes25–27.
Figure 1.

Transmission electron micrograph of T4-GFP. GFP-T4-phages diluted in PBS were placed on formvar-coated copper grids and negatively stained using uranyl acetate, prior to imaging using transmission electron microscopy. The phages were intact and found as single particles. Scale bar = 200 nm.
Figure 2.
Time-lapse endocytosis of GFP-T4-phages by LSECs. (A) Rat LSECs preincubated with AF647-FSA were challenged with GFP-T4-phages, and endocytosis imaged in real-time by deconvolution microscopy (DV), every 5 min for about 90 min. Images represent maximum intensity projections of an 8 µm 3D z-stack. Colocalization values for FSA and phages for each time point (shown in merged images) are Pearson’s correlation coefficients calculated using the Costes threshold in Volocity Quantitation software. Only cell-containing regions of the image were used for colocalization analysis (dashed lines in t = 85 m, merged image). The results are representative of experiments performed with cells isolated from 2 animals, 3 culture dishes, and 10 fields of view including an approximate total of 150 cells. (B,C) DV maximum intensity projections of the highlighted inset at min 25 and min 85 of acquisition, respectively. (D,E) SIM image and its corresponding magnified inset of the plasma membrane of a representative live LSEC labelled with CellMask Green at the end of the acquisition time. Images represent maximum intensity projections of a 2 µm 3D SIM z-stack.
Figure 3.
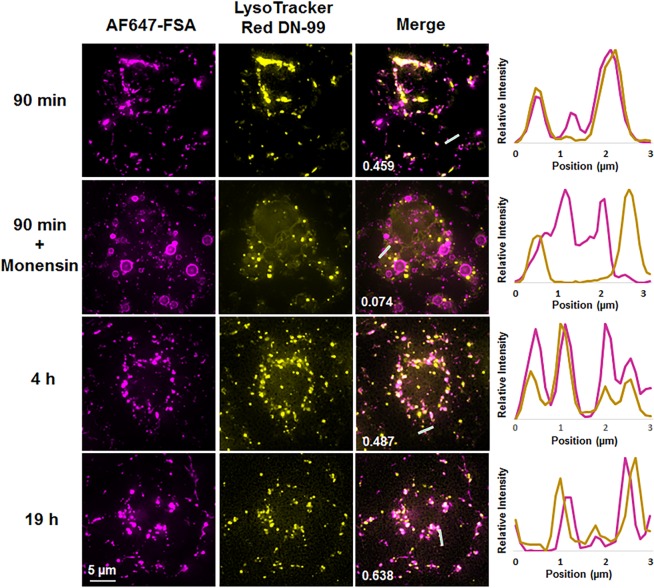
Time-lapse endocytosis of AF747-FSA in the presence or absence of monensin and LysoTracker. LSEC cultures were preincubated with LysoTracker (yellow) and challenged with AF647-FSA (magenta) in the absence or presence of monensin (10 µM). The uptake was monitored in real-time by DV overnight. Colocalization values for FSA and LysoTracker (shown in merged images) are Pearson’s correlation coefficients calculated using the Costes threshold in Volocity Quantitation software. Images represent maximum intensity projections of 4–9 µm 3D z-stacks. Right panel shows randomly selected line profiles within the cells, indicating the degree of colocalization of the two probes. The results are representative of experiments performed with cells isolated from 4 animals, and included 7 culture dishes, 49 fields of view, and approximately 350 cells; of which 2 animals, 2 culture dishes, 17 fields of view, and approximately 100 cells were in the presence of monensin).
At the end of the incubation period, the plasma membrane of the live cell culture was labelled with CellMask Green (CMG), and a subset of the same cells that were studied using DV were then imaged at super resolution using structured illumination microscopy (SIM). This allowed us to confirm that the GFP-T4-phage containing cells were indeed LSECs due to the expression of fenestrations (Fig. 2D,E), the signature morphological characteristic of LSECs28. The fenestrations are small transmembrane pores with approximate diameters of 50 to 150 nm which provide open channels between the sinusoidal blood and the subendothelial space, facilitating the transfer of substrates between the blood and the parenchymal hepatocytes.
FSA is a frequently used and highly specific endocytic ligand for the LSEC scavenger receptors stabilin-1 and stabilin-229,30, and we chose to use AF647-conjugated FSA here as a late endosome/lysosomal marker for live cell imaging. Previous studies using unconjugated and fluorescently conjugated FSA have shown that the ligand is avidly endocytosed by LSECs in vivo and in vitro, and transported to the lysosomes where it starts to be degraded within 10 min of cellular uptake31,32. When conjugated with fluorescent adducts (i.e. FITC or TRITC), or with gold particles, FSA follows the same route of uptake and degradation as unconjugated FSA, except that the non-degradable adducts remain trapped intralysosomally in LSECs for at least 24 h after uptake25–27. AF647 adduct acts in a similar way when bound to carrier ligands (i.e. albumin or dextran), as shown previously using other cell types33,34. Of note, in contrast to the present study, all previous studies assessing intracellular localization of endocytosed ligands in LSECs were performed by fixing the cells after various times of incubation with the ligands (pulse-chase studies). Currently, only two types of markers have been used in live cells to specifically label lysosomes for time lapse imaging. These are (i) LysoTracker probes, which are weak basic amines that accumulate in the acidic lumen of lysosomes35, and (ii) fluorescently labelled dextran and gold-labelled bovine serum albumin, both ligands that enter the cells via endocytosis and accumulate in the late endosomes/lysosomes36,37. In our setup, we found that LysoTracker® Red DND-99 (LT) used at the recommended concentration of 50–75 nM was susceptible to rapid photobleaching (data not shown). To bypass this limitation, we increased five-fold the LT concentration, allowing us to capture approximately 5–6 images of the lysosomal dye before photobleaching. However, high concentrations of LT and prolonged live cell imaging are known to affect the lysosomal pH that will sometimes radically disturb the intensity and spectral characteristics of a fluorophore35,38. In contrast, FSA, the formaldehyde-modified BSA, is a non-toxic and frequently used marker for LSECs, that is rapidly taken up by these cells at high specificity and efficiency39. Furthermore, FSA is easily tagged with fluorescent dyes. On this basis we incubated LSECs with a low concentration of AF647-FSA (5 µg/ml) for only 15 min, after which the non-bound ligand was washed off and the cells imaged. Given the very high endocytic efficiency of LSECs, visible uptake of this marker ligand is achieved at low ligand dose and short incubation period, avoiding non-specific uptake in all cells other than LSECs. In the present work this was a prerequisite for optimal imaging, avoiding overloading of the cells with fluorescence. AF647-FSA gradually accumulated in the lysosomes and remained colocalized with the LT up to 20 h of live imaging (Fig. 3). To further confirm that AF647-FSA can serve as a functional marker for different endocytic compartments, we used monensin as a tool to stop the internalization of the FSA at the early endosomes. Monensin is an ionophore that exchanges protons for Na+ and K+ thus perturbing gradients across cellular membranes, causing multiple effects on the intracellular transport of a variety of ligands, e.g. acidification of early endosomes, inhibition of receptor recycling back to the cell surface, inhibition of receptor-ligand dissociation and further transport of ligands to late endosomes/lysosomes40. In the presence of monensin, the AF647-FSA was arrested in fluorescent ring-like structures indicative of early endosomes25 (Fig. 3). No colocalization of the ligand with the lysosomes was further observed. This is consistent with previous reports of uptake of other ligands by LSECs in the presence of monensin24,41,42. With these experiments, we demonstrated that AF647-FSA used in live cell imaging could serve three purposes: i) as a specific functional marker for endocytically active LSECs, ii) as a marker for endosomal/lysosomal integrity, and iii) as a non-toxic and highly photostable marker for time-lapse imaging in live LSECs.
The final step of intracellular processing of ligands that have been taken up by LSECs is normally degradation in the endo-lysosomal compartment39. However, current fluorescence microscopy approaches lack the ability to probe the degradation of cargo within the cell. A previous study on peritoneal macrophages challenged with T2 phages, a close relative of T4 phages, attempted to detect degraded phages using transmission electron microscopy43. This “searching for the needle in the haystack” approach in the referred study did not succeed to show evidence of phage degradation. Other studies have used radioactive isotopes such as 51Cr, 131I, 35S or 125I to label T4 and M13 bacteriophages and polyoma JC and BK virus-like particles (VLPs)9,10,44. While these studies investigated the in vivo clearance mechanisms of these phages and viruses, showing that the liver is the major organ for uptake and degradation, they did not provide details on degradation at the cellular level. However, Simon-Santamaria et al.10 found degradation products in the blood circulation after intravenous injection of 125I-labeled JC polyoma VLPs, as well as uptake of non-labelled VLPs and intact virus in mouse LSECs in vitro, strongly suggesting that the LSECs were responsible for their degradation.
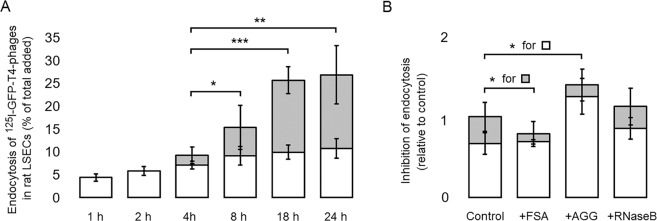
To investigate the intracellular processing of the bacteriophages in LSECs in the present study, we challenged the cells in vitro with trace amounts of 125I-GFP-T4-phages. This enabled us to measure the rate of uptake and degradation over a time interval of 24 h. The functional integrity of LSECs was confirmed in these experiments by the ability of the cells to endocytose and degrade 125I-FSA (not shown). We found that the LSECs rapidly internalized the 125I-labeled GFP-T4 phages, supporting our imaging data presented in Fig. 2, and that increasing amounts of degradation products were released to the spent culture medium shortly after the phages had reached the late endosomes/lysosomes (Fig. 4A). Bacteriophages are relatively large viral particles, with a complex structure composed of hundreds of proteins, and highly stable under a variety of harsh environmental conditions. Their catabolism undoubtedly takes longer time than single proteins e.g. modified albumin and collagen alpha chains, known to be efficiently eliminated by LSECs24,32 (Fig. S4), and is more dependent on the high concentration and specific activity of enzymes in the lysosomes, the terminal degradative compartment24,45. Nevertheless, during 24 h post incubation of LSECs with T4-phages, 27% and 16% of total added phages were internalized and degraded by the cells, respectively, demonstrating a substantial capacity of LSECs to take up and degrade the phages.
Figure 4.
Endocytosis of T4-phages in LSECs in vitro. (A) Time course endocytosis of 125I-GFP-T4-phages by LSECs. LSEC cultures were incubated with radiolabelled phages for various time periods. For each time point, 3 separate wells containing cells, and 1 cell-free well were used. After each time period, the supernatant from the cells and cell-free well was collected along with one 0.5 ml washing volume of PBS. Trichloroacetic acid (TCA) precipitation was then used to differentiate between free iodine = degraded phages (TCA soluble) (grey columns), and unbound, intact phages (TCA precipitable). Cell bound and internalized phages were quantified in the cell lysates, after solubilizing the cells in 1% SDS (white columns). The results were normalized by subtracting the amount of radioactivity corresponding to the non-specific binding and free 125I in cell-free wells. Each experiment was performed in triplicates, on cells isolated from four animals (Total N = 12 cell cultures for each time point). Bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 represent the statistical differences between the total endocytosis at 18 h and 24 h as compared to 4 h. (B) The specificity of uptake was studied by incubating the cells with 125I-GFP-T4-phages in the absence (Control) or presence of blocking concentrations (0.1 mg/ml) of FSA, ribonuclease B (RNaseB) and aggregated gamma globulin (AGG), inhibitors for stabilin1/2, mannose receptor and FcγRIIb2, respectively. Cell association and degradation were assessed as above. The results are presented as relative uptake compared to the control which was set to 1. Each experiment was performed in triplicates, on cells isolated from 3 animals (Total N = 9 cell cultures for each time point). Bars represent mean ± SD.
Using a variety of receptors (mannose receptor, scavenger receptors, and the endocytic Fc-gamma receptor IIb2), the LSECs can directly recognize and internalize pathogen associated molecular patterns, cellular debris and immune complexes, contributing thus to the liver’s immune tolerance46. To gain further insights into the uptake mechanism of the T4-phages, we investigated whether the stabilin1/2, mannose receptor, or the FcγRIIb2 are involved in the uptake of T4-phages by incubating the cells with radiolabeled phages in the absence or presence of blocking concentrations of native, unlabeled FSA, RNaseB, and AGG, inhibitors for stabilin1/2, mannose receptor, and FcγRIIb2, respectively31,47,48. The results presented in Fig. 4B show a borderline significant (p = 0.0497) decrease of phage degradation in the presence of FSA as compared to control. Decreased degradation was also noted in the presence of AGG, although not significant (p = 0.17). Since these ligands had no inhibitory effects on the binding (rather the opposite in the presence of AGG), the results suggest that stabilin1/2 and FcγRIIb2 are probably not involved in the internalization of T4 phages in LSECs. The observed decrease in phage degradation is most probably due to FSA and AGG overloading the later stages of the endocytic pathway. No effect on the uptake and degradation was observed in the presence of RNase B either, suggesting that the mannose receptor is also not involved in the uptake T4 phages. Further studies are needed to investigate the mechanisms by which these cells recognize and internalize bacteriophages.
In conclusion, we have demonstrated that freshly isolated and functionally intact LSECs efficiently take up and degrade T4 bacteriophages at high capacity, indicating that these cells act as a primary anti-viral defence system, adding to the role that these cells have in innate immunity. In vivo studies have reported that injected bacteriophages used in phage therapy are quickly removed from the circulation, thus resulting in lower efficiency of the therapy49–51. Administration of huge doses of phages is frequently required to saturate unwanted liver uptake, showing the great virus elimination capacity of the liver9,17. Our study showing that the LSECs take up and degrade phages may explain some of the challenges with using bacteriophages both in phage therapy and as gene delivery vehicles.
Materials and Methods
Animals and ethics statement
Sprague Dawley, Crl:CD(SD), male rats (Charles River, Sulzfeld, Germany) were group housed (2–3 rats per cage) in conventional Eurostandard type IV cages with aspen bedding (Tapvei, Estonia) and with nesting material (Sizzelnest®, Datesand, UK), rat tunnels (Scanbur, Norway) and aspen chew blocks (Scanbur, Norway) as environmental enrichment. The rats were housed under controlled environmental conditions (21°C ± 1°, relative humidity 55% ± 5% and 12 h light/12 h dark cycle). They were fed a standard chow ad libitum (RM1-E, Special Diet Service, UK) and tap water ad libitum. All methods were carried out in accordance with relevant guidelines and regulation protocols, and approved by the Norwegian Food Safety Authority (approval ID: 8455). All experimental protocols and animal handling were approved by and carried out according to local authorities (Department of Comparative Medicine). The rats (body weight 150–300 g) were anesthetized with a mixture (ZRF-mix) of zolazepam /tiletamine hydrochloride 12.9/12.9 mg/mL (Zoletil forte vet, Virbac, Norway), xylazine 1.8 mg/mL (Rompun, Bayer Nordic, Norway) and fentanyl 10.3 µg/mL (Actavis, Norway).
Chemicals and materials
Collagenase was from Worthington Biochemical Corporation (Lakewood, NJ, USA). Iodogen was from Pierce Chemicals (Rockford, IL, USA), and carrier-free Na125I from PerkinElmer Norge AS (Oslo, Norway). Bovine serum albumin (BSA) was from MP Biomedicals (Solon, OH, USA). Fibronectin was purified from human plasma by affinity chromatography on Gelatin Sepharose 4B as described by the manufacturer (GE Healthcare, Uppsala, Sweden). Percoll and PD-10 columns (Sephadex G-25) were from GE Healthcare. Serum-free Roswell Park Memorial Institute (RPMI-1640) cell culture medium (supplemented with 20 mM sodium bicarbonate, 0.006% penicillin, and 0.01% streptomycin) and monensin were from Sigma-Aldrich (Oslo, Norway). Culture dishes of 35 mm diameter and #1.5 glass coverslip-bottom were from MatTek Corp. (Ashland, MA, USA). LysoTracker Red DND-99, CellMask Green, and AlexaFluor-647 carboxylic acid were from ThermoFisher Scientific (Oslo, Norway). Trichloroacetic acid (TCA) was from Merck (Darmstadt, Germany). Formaldehyde-treated bovine serum albumin (FSA) was prepared by treating BSA with 10% formaldehyde in 0.2 M carbonate buffer, pH 10, for 3 days as described52.
Preparation and GFP labelling of T4 phages
GFP-Enterobacteria phages T4 (GFP-T4-phages) were prepared as previously described19, dialyzed against phosphate buffer saline (PBS) and stored aseptically at 4 °C at a titre of 108 PFU/ml.
Isolation of liver sinusoidal endothelial cells (LSECs), and assessment of endocytosis and specificity of uptake
LSECs were isolated and purified from anesthetized rats as described53, and seeded at a density of 0.5 × 106 cells/cm2 on fibronectin coated coverslip-bottom dishes (MatTek In Vitro Life Science Laboratories, Bratislava, Slovak Republic), or 24 well tissue culture plates (Sarstedt, Nümbrecht, Germany) in serum-free RPMI-1640. All experiments started 2–3 h after isolation and seeding. For quantitative studies of uptake and degradation, confluent cultures of freshly isolated LSECs established in 24-well culture dishes coated with fibronectin were incubated in 0.2 mL RPMI containing 3–4 × 104 cpm 125I-GFP-T4-phages. Time-course endocytosis was performed by incubating the cells for 1, 2, 4, 8, 18, and 24 h. For each time point, 3 separate wells containing cells, and 1 cell-free well were used. At the end of each time point, the percent of degraded phages was measured by collecting the spent medium together with one wash volume of 0.5 mL PBS. TCA (0.75 mL, 20%) was added to precipitate intact phages. The amount of TCA-soluble radioactivity measured in the supernatant after centrifugation represented degraded phages. To determine the amount of cell bound and internalized phages, the cells were lysed in 2 × 0.5 mL of 0.1% sodium dodecyl sulfate (SDS) in PBS. The radioactivity was measured in all three fractions using a Cobra II, Auto-Gamma detector (Packard Instruments, Laborel, Oslo, Norway). The results were normalized by subtracting the amount of radioactivity corresponding to the non-specific binding and free 125I in cell-free wells. The specificity of uptake was studied by incubating the cells with 125I-GFP-T4-phages in the absence or presence of blocking concentrations (0.1 mg/ml) of FSA, ribonuclease B (RNaseB) or aggregated gamma globulin (AGG), inhibitors for stabilin1/2, mannose receptor and FcγRIIb2, respectively.
Fluorescence labelling and radioiodination
GFP-T4-phages in PBS were labelled with carrier-free Na125I, using Iodogen as described by the manufacturer (Pierce Chemicals), and separated from unbound 125I on a PD-10 column. The resulting specific radioactivity was approximately 40 × 106 cpm per 106 PFU phages. FSA was conjugated with Alexa Fluor-647 carboxylic acid succinimidyl ester according to the manufacturer’s instructions (ThermoFisher Scientific). Free AF647 was separated from FSA using a Vivaspin 6, 10.000 kDa cutoff (ThermoFisher Scientific). The final concentration of the protein was measured with a NANODROP 2000 spectrophotometer (ThermoFisher Scientific).
Imaging methods and analyses
Transmission electron microscopy (TEM)
Glow discharged formvar-coated copper grids were placed on top of 5 µl droplets of GFP-T4-phages (stock solution of 108 PFU/ml in PBS was diluted 1/10 in 0.9% sodium chloride), for 5 min in a moist chamber. The grids were gently washed on droplets of double distilled water and incubated for 20 seconds on a droplet of freshly made 1% uranyl acetate in double distilled water. The uranyl acetate solution was partly removed by filter paper, and the grids air dried, with the remaining uranyl acetate solution faced up, before electron microscopy. The images were recorded in a JEOL JEM 1010 transmission electron microscope (TEM) (JEOL ltd, Tokyo, Japan), operating at 80 kV, and equipped with Morada digital CCD camera (Olympus, Tokyo, Japan). The analysis of the negatively stained GFP-T4-phages showed that the phages were intact and found as single particles (Fig. 1).
Deconvolution and structured illumination microscopy
After washing to remove non-adherent cells, cultures on coverslips were pre-incubated for 30 min with 0.25 μM LysoTracker in RPMI, then with 5 μg/mL AF647-FSA for 15 min, in the presence or absence of 10 μM monensin. The non-bound FSA was removed by washes with PBS, and the cells allowed to endocytose the FSA for another hour in culture medium containing LysoTracker with or without monensin. For live cell imaging, the LSECs were incubated with 0.3 mL RPMI containing 5 × 106 PFU GFP-T4-phages, and the uptake followed by deconvolution (DV) microscopy with 5 min intervals between acquisitions, over a period of 90 min. The experiments were terminated by live staining of the plasma membrane with CellMask Green (1:1000 in RPMI) and imaging using structured illumination microscopy (SIM). Live cell imaging was performed using a DeltaVision OMX V4 Blaze imaging system (GE Healthcare) equipped with a 60 × 1.42 NA oil-immersion objective (Olympus), 3 sCMOS cameras, a solid-state illumination source for widefield deconvolution imaging, and a 488 nm laser for structured illumination imaging. Additional widefield deconvolution imaging was performed on a DeltaVision Elite microscope (GE Healthcare) equipped with an identical 60 × 1.42 NA oil-immersion objective (Olympus), single sCMOS camera, and a solid-state illumination source. Widefield deconvolution images were acquired in 4–8 μm z-stacks with one image taken per 0.20–0.25 μm z-step. SIM image stacks of 2 μm were acquired with 15 raw images (five phases, three angles) acquired at each plane with a z-step size of 0.125 μm. During live imaging, the cells were kept under an incubator/control chamber supplied by GE Healthcare, calibrated to approximately 5% humidified CO2, and a temperature set at approximately 36 °C. This paper incorporates the results of imaging experiments from approximately 20 different cell isolations. Over the course of the study, experimental procedures were refined and/or had to be repeated/optimized due to high fluorescence background, improvement of staining, quality of the primary cell cultures, etc. Typically, 5–10 fields of view were imaged for each experiment with 2 (for SIM) or 10 (for DV) cells per field of view. 1–6 imaging experiments were conducted for each cell isolation, including controls and photobleaching assessments (Supplementary Fig. S3).
The experimental conditions are listed in Supplementary Table. Raw datasets were computationally reconstructed and colour alignment was performed using SoftWoRx software (GE Healthcare) for both deconvolution and structured illumination images. Colocalization was analysed in Volocity 6.3 (PerkinElmer) using the Costes threshold to compute the Pearson’s correlation coefficient, for hand-selected image regions which only contained a cell (no cell-free background regions). For clarity of display, figure images were linearly adjusted for brightness and contrast using Fiji (https://fiji.sc, version 2.0.2.)54.
Statistical analysis
For all endocytosis experiments, the statistical analyses were performed using the Excel 2016 software (Microsoft, USA). Student’s t test (two tailed, unpaired) was used for statistical analysis. All experiments were performed in triplicate, on cells isolated from 3 animals (Total N = 9 cell cultures for each time point and/or inhibitor) and the results were presented as mean ± standard deviation. The significance was set at: non significant p > 0.05, significant *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary information
Acknowledgements
This work was financially supported by European Research Council Starting Grant (B.S.A. #336716), UiT Strategic Funding (B.S.A.), the Tromsø Research Foundation, and the Research Council of Norway (#262538). The publication charges for this article have been funded by a grant from the publication fund of UiT-The Arctic University of Norway”.
Author contributions
C.I.Ø. designed the study and wrote the manuscript. C.I.Ø., D.L.W. and G.D. carried out the experiments. D.L.W. designed and carried out the imaging experiments and data analysis. T.Y. constructed and provided the GFP-T4-phages. K.S. performed the TEM experiments. K.S., P.M. B.S.A. and B.S. developed the theoretical framework and provided valuable scientific input to the overall study. All authors provided feedback on the manuscript and approved the final version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cristina I. Øie, Deanna L. Wolfson, Balpreet S. Ahluwalia and Bård Smedsrød.
Supplementary information
is available for this paper at 10.1038/s41598-020-57652-0.
References
- 1.Slopek S, Weber-Dabrowska B, Dabrowski M, Kucharewicz-Krukowska A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981–1986. Arch. Immunol. Ther. Exp. (Warsz). 1987;35:569–83. [PubMed] [Google Scholar]
- 2.Majewska J, et al. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses. 2015;7:4783–4799. doi: 10.3390/v7082845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016 doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Minot S, et al. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA. 2013;110:12450–5. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merril CR. Bacteriophage interactions with higher organisms. Transactions of the New York Academy of Sciences. 1974;36:265–72. doi: 10.1111/j.2164-0947.1974.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 6.Handley Scott A., Thackray Larissa B., Zhao Guoyan, Presti Rachel, Miller Andrew D., Droit Lindsay, Abbink Peter, Maxfield Lori F., Kambal Amal, Duan Erning, Stanley Kelly, Kramer Joshua, Macri Sheila C., Permar Sallie R., Schmitz Joern E., Mansfield Keith, Brenchley Jason M., Veazey Ronald S., Stappenbeck Thaddeus S., Wang David, Barouch Dan H., Virgin Herbert W. Pathogenic Simian Immunodeficiency Virus Infection Is Associated with Expansion of the Enteric Virome. Cell. 2012;151(2):253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen, S. et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio, 10.1128/mBio.01874-17 (2017). [DOI] [PMC free article] [PubMed]
- 8.Mu, Q., Kirby, J., Reilly, C. M. & Luo, X. M. Leaky gut as a danger signal for autoimmune diseases. Frontiers in Immunology, 10.3389/fimmu.2017.00598 (2017). [DOI] [PMC free article] [PubMed]
- 9.Inchley CJ. The actvity of mouse Kupffer cells following intravenous injection of T4 bacteriophage. Clin Exp Immunol. 1969;5:173–187. [PMC free article] [PubMed] [Google Scholar]
- 10.Simon-Santamaria J, et al. Efficient uptake of blood-borne BK and JC polyomavirus-like particles in endothelial cells of liver sinusoids and renal vasa recta. PLoS One. 2014;9:e111762. doi: 10.1371/journal.pone.0111762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynaud A., Cloastre L., Bernard J., Laveran H., Ackermann H.-W., Licois D., Joly B. Characteristics and diffusion in the rabbit of a phage for Escherichia coli 0103. Attempts to use this phage for therapy. Veterinary Microbiology. 1992;30(2-3):203–212. doi: 10.1016/0378-1135(92)90114-9. [DOI] [PubMed] [Google Scholar]
- 12.Persidsky, Y. et al. Permissiveness of Kupffer cells for simian immunodeficiency virus (SIV) and morphological changes in the liver of rhesus monkeys at different periods of SIV infection. Hepatology, 10.1016/0270-9139(95)90040-3 (1995). [PubMed]
- 13.He J. Q., Katschke K. J., Gribling P., Suto E., Lee W. P., Diehl L., Eastham-Anderson J., Ponakala A., Komuves L., Egen J. G., van Lookeren Campagne M. CRIg mediates early Kupffer cell responses to adenovirus. Journal of Leukocyte Biology. 2012;93(2):301–306. doi: 10.1189/jlb.0612311. [DOI] [PubMed] [Google Scholar]
- 14.Bilzer Manfred, Roggel Frigga, Gerbes Alexander L. Role of Kupffer cells in host defense and liver disease. Liver International. 2006;26(10):1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 15.Breiner KM, Schaller H, Knolle PA. Endothelial cell-mediated uptake of a hepatitis B virus: a new concept of liver targeting of hepatotropic microorganisms. Hepatology. 2001;34:803–808. doi: 10.1053/jhep.2001.27810. [DOI] [PubMed] [Google Scholar]
- 16.Ganesan LP, et al. Rapid and efficient clearance of blood-borne virus by liver sinusoidal endothelium. PLoS Pathog. 2011;7:e1002281. doi: 10.1371/journal.ppat.1002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alemany R, Suzuki K, Curiel DT. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 2000;81:2605–2609. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]
- 18.Mates JM, et al. Mouse Liver Sinusoidal Endothelium Eliminates HIV-Like Particles from Blood at a Rate of 100 Million per Minute by a Second-Order Kinetic Process. Front Immunol. 2017;8:35. doi: 10.3389/fimmu.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanji Y, et al. Escherichia coli detection by GFP-labeled lysozyme-inactivated T4 bacteriophage. J Biotechnol. 2004;114:11–20. doi: 10.1016/j.jbiotec.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Robertson Kelly L., Soto Carissa M., Archer Marie J., Odoemene Onyekachi, Liu Jinny L. Engineered T4 Viral Nanoparticles for Cellular Imaging and Flow Cytometry. Bioconjugate Chemistry. 2011;22(4):595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- 21.Liu Jinny L., Robertson Kelly L. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2013. Engineered Bacteriophage T4 Nanoparticles for Cellular Imaging; pp. 187–199. [DOI] [PubMed] [Google Scholar]
- 22.Elvevold K, Smedsrod B, Martinez I. The liver sinusoidal endothelial cell: a cell type of controversial and confusing identity. Am J Physiol Gastrointest Liver Physiol. 2008;294:G391–G400. doi: 10.1152/ajpgi.00167.2007. [DOI] [PubMed] [Google Scholar]
- 23.Seternes T, Sorensen K, Smedsrod B. Scavenger endothelial cells of vertebrates: a nonperipheral leukocyte system for high-capacity elimination of waste macromolecules. Proc Natl Acad Sci USA. 2002;99:7594–7597. doi: 10.1073/pnas.102173299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellevik T, Bondevik A, Smedsrod B. Intracellular fate of endocytosed collagen in rat liver endothelial cells. Exp Cell Res. 1996;223:39–49. doi: 10.1006/excr.1996.0056. [DOI] [PubMed] [Google Scholar]
- 25.Hellevik T, et al. Transport of residual endocytosed products into terminal lysosomes occurs slowly in rat liver endothelial cells. Hepatology. 1998;28:1378–1389. doi: 10.1002/hep.510280529. [DOI] [PubMed] [Google Scholar]
- 26.Elvevold KH, Nedredal GI, Revhaug A, Smedsrod B. Scavenger properties of cultivated pig liver endothelial cells. Comp Hepatol. 2004;3:4. doi: 10.1186/1476-5926-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oie CI, et al. Liver sinusoidal endothelial cells are the principal site for elimination of unfractionated heparin from the circulation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G520-8. doi: 10.1152/ajpgi.00489.2007. [DOI] [PubMed] [Google Scholar]
- 28.Braet F, Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 2002;1:1. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nedredal GI, et al. Liver sinusoidal endothelial cells represents an important blood clearance system in pigs. Comp Hepatol. 2003;2:1. doi: 10.1186/1476-5926-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, et al. Role of liver sinusoidal endothelial cells and stabilins in elimination of oxidized low-density lipoproteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G71–81. doi: 10.1152/ajpgi.00215.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blomhoff R, Eskild W, Berg T. Endocytosis of formaldehyde-treated serum albumin via scavenger pathway in liver endothelial cells. Biochem J. 1984;218:81–86. doi: 10.1042/bj2180081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eskild W, Berg T. Endocytosis of formaldehyde-denatured serum albumin by nonparenchymal liver cells in vitro. Biochim Biophys Acta. 1984;803:63–70. doi: 10.1016/0167-4889(84)90055-7. [DOI] [PubMed] [Google Scholar]
- 33.Humphries WH, Payne CK. Imaging lysosomal enzyme activity in live cells using self-quenched substrates. Anal. Biochem. 2012;424:178–183. doi: 10.1016/j.ab.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson Danielle E., Ostrowski Philip, Jaumouillé Valentin, Grinstein Sergio. The position of lysosomes within the cell determines their luminal pH. The Journal of Cell Biology. 2016;212(6):677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierzyńska-Mach A, Janowski PA, Dobrucki JW. Evaluation of acridine orange, LysoTracker Red, and quinacrine as fluorescent probes for long-term tracking of acidic vesicles. Cytometry. A. 2014;85:729–37. doi: 10.1002/cyto.a.22495. [DOI] [PubMed] [Google Scholar]
- 36.Richardson SCW, et al. The use of fluorescence microscopy to define polymer localisation to the late endocytic compartments in cells that are targets for drug delivery. J. Control. Release. 2008;127:1–11. doi: 10.1016/j.jconrel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baravalle G, et al. Transferrin recycling and dextran transport to lysosomes is differentially affected by bafilomycin, nocodazole, and low temperature. Cell Tissue Res. 2005;320:99–113. doi: 10.1007/s00441-004-1060-x. [DOI] [PubMed] [Google Scholar]
- 38.Smith DG, McMahon BK, Pal R, Parker D. Live cell imaging of lysosomal pH changes with pH responsive ratiometric lanthanide probes. Chem. Commun. 2012;48:8520. doi: 10.1039/c2cc34267g. [DOI] [PubMed] [Google Scholar]
- 39.Sorensen KK, Simon-Santamaria J, McCuskey RS, Smedsrod B. Liver Sinusoidal Endothelial Cells. Compr. Physiol. 2015;5:1751–1774. doi: 10.1002/cphy.c140078. [DOI] [PubMed] [Google Scholar]
- 40.Misinzo G., Delputte P. L., Nauwynck H. J. Inhibition of Endosome-Lysosome System Acidification Enhances Porcine Circovirus 2 Infection of Porcine Epithelial Cells. Journal of Virology. 2007;82(3):1128–1135. doi: 10.1128/JVI.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen B, et al. Stabilin-1 and stabilin-2 are both directed into the early endocytic pathway in hepatic sinusoidal endothelium via interactions with clathrin/AP-2, independent of ligand binding. Exp Cell Res. 2005;303:160–173. doi: 10.1016/j.yexcr.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 42.McCourt PA, Smedsrod BH, Melkko J, Johansson S. Characterization of a hyaluronan receptor on rat sinusoidal liver endothelial cells and its functional relationship to scavenger receptors. Hepatology. 1999;30:1276–1286. doi: 10.1002/hep.510300521. [DOI] [PubMed] [Google Scholar]
- 43.Aronow R, Danon D, Shahar A, Aronson M. Electron microscopy of in vitro endocytosis of T2 phage by cells from rabbit peritoneal exudate. J. Exp. Med. 1964;120:943–954. doi: 10.1084/jem.120.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molenaar TJ, et al. Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology. 2002;293:182–191. doi: 10.1006/viro.2001.1254. [DOI] [PubMed] [Google Scholar]
- 45.Elvevold K, et al. Liver Sinusoidal Endothelial Cells Depend on Mannose Receptor-Mediated Recruitment of Lysosomal Enzymes for Normal Degradation Capacity. Hepatology. 2008;48:2007–2015. doi: 10.1002/hep.22527. [DOI] [PubMed] [Google Scholar]
- 46.Sorensen KK, et al. The scavenger endothelial cell: a new player in homeostasis and immunity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012;303:R1217–30. doi: 10.1152/ajpregu.00686.2011. [DOI] [PubMed] [Google Scholar]
- 47.Mousavi SA, et al. Receptor-mediated endocytosis of immune complexes in rat liver sinusoidal endothelial cells is mediated by FcgammaRIIb2. Hepatology. 2007;46:871–884. doi: 10.1002/hep.21748. [DOI] [PubMed] [Google Scholar]
- 48.Stahl PD, Rodman JS, Miller MJ, Schlesinger PH. Evidence for receptor-mediated binding of glycoproteins, glycoconjugates, and lysosomal glycosidases by alveolar macrophages. Proc Natl Acad Sci USA. 1978;75:1399–1403. doi: 10.1073/pnas.75.3.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worgall Stefan, Wolff Gerhard, Falck-Pedersen Erik, Crystal Ronald G. Innate Immune Mechanisms Dominate Elimination of Adenoviral Vectors Following In Vivo Administration. Human Gene Therapy. 1997;8(1):37–44. doi: 10.1089/hum.1997.8.1-37. [DOI] [PubMed] [Google Scholar]
- 50.Merril CR, et al. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA. 1996;93:3188–92. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlton RM. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. (Warsz). 1999;47:267–74. [PubMed] [Google Scholar]
- 52.Mego John L., McQueen J.Donald. Further studies on the degradation of injected [131I]albumin by secondary lysosomes of mouse liver. Biochimica et Biophysica Acta (BBA) - General Subjects. 1965;111(1):166–173. doi: 10.1016/0304-4165(65)90483-6. [DOI] [PubMed] [Google Scholar]
- 53.Smedsrod B, Pertoft H. Preparation of pure hepatocytes and reticuloendothelial cells in high yield from a single rat liver by means of Percoll centrifugation and selective adherence. J. Leukoc. Biol. 1985;38:213–230. doi: 10.1002/jlb.38.2.213. [DOI] [PubMed] [Google Scholar]
- 54.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.