Abstract
Tobacco use is the leading preventable cause of mortality in the world. The limited number of smoking cessation aids currently available are minimally effective, highlighting the need for novel therapeutic interventions. We describe a genome-wide approach to identify potential candidates for such interventions. Next-generation sequencing was performed using RNA isolated from the habenulo-interpeduncular circuit of male mice withdrawn from chronic nicotine treatment. This circuit plays a central role in the nicotine withdrawal response. Differentially expressed miRNAs and mRNAs were validated using RT-qPCR. Many of the differentially expressed mRNAs are predicted targets of reciprocally expressed miRNAs. We illustrate the utility of the dataset by demonstrating that knockdown in the interpeduncular nucleus of a differentially expressed mRNA, that encoding profilin 2, is sufficient to induce anxiety-related behavior. Importantly, profilin 2 knockdown in the ventral tegmental area did not affect anxiety behavior. Our data reveal wide-spread changes in gene expression within the habenulo-interpeduncular circuit during nicotine withdrawal. This dataset should prove to be a valuable resource leading to the identification of substrates for the design of innovative smoking cessation aids.
Subject terms: Molecular neuroscience, Addiction
Introduction
Tobacco use is a prevalent health problem worldwide, with more than 900 million daily smokers1 and an estimated 6 million deaths due to tobacco-related illnesses annually2. The few pharmacological cessation aids currently available have limited efficacy3–6. Without the intervention of new effective treatments, tobacco-related mortality is anticipated to rise to 8 million deaths per year2.
The addictive component of tobacco is nicotine, an agonist of nicotinic acetylcholine receptors (nAChRs), ligand-gated ion channels endogenously activated by acetylcholine7. nAChRs are enriched in the mesolimbic and habenulo-interpeduncular circuitries involved in nicotine reward and withdrawal, respectively8–10. Cessation of nicotine induces a withdrawal syndrome consisting of negative somatic, affective and cognitive symptoms. Affective symptoms include depressed mood, irritability, craving and anxiety11,12. While the rewarding properties of nicotine dominate initial drug-taking, it is largely avoidance of affective withdrawal symptoms that promotes relapse and habitual use13,14.
The habenulo-interpeduncular circuitry consists primarily of neurons projecting from the medial habenulae (MHb) to the interpeduncular nucleus (IPN) via the fasciculus retroflexus. In mice experiencing spontaneous nicotine withdrawal, there is increased glutamate release from habenular axon terminals, activating GABAergic neurons in the IPN15. Activation of the IPN results in the increased somatic signs and anxiety observed during nicotine withdrawal15,16. Optogenetic investigation of this circuit has revealed that silencing the MHb decreases activation of IPN neurons and decreases anxiety during nicotine withdrawal16.
While much is known about the neurocircuitry responsible for nicotine withdrawal-associated behaviors, the mechanisms underlying the induction of these behavioral alterations are less clear. Chronic drug exposure induces stable neuroadaptations underlying the compulsion for continued use and susceptibility to relapse. These long-lasting changes in the function of the addicted brain are thought to require alterations of gene expression through regulation of transcription factors, epigenetic mechanisms and non-coding RNAs17,18.
microRNAs (miRNAs) are non-coding RNA molecules ~22 nucleotides in length that repress the expression of mRNA targets by promoting mRNA decay or translational repression19. miRNAs recognize their targets through imperfect base-pairing of the seed region (nucleotides 2–7) with miRNA response elements (MREs) within the 3′-untranslated regions (UTRs) of mRNAs20,21. Tobacco smoke and nicotine have been shown to regulate the expression of miRNAs in various cell lines, organisms and tissues22–26.
Dysregulation of miRNAs in the brain have been shown to contribute to neurodevelopmental and neuropsychiatric disorders, including drug addiction27,28. However, little is known about the role of miRNAs in the mechanisms underlying nicotine dependence and withdrawal. Few studies have examined the regulation of miRNAs by nicotine in specific regions of the mammalian brain29–31. Previously, we showed that the expression of miRNAs predicted to target MREs within the 3′-UTRs of nAChR genes was altered after chronic nicotine treatment in whole brain or a specific brain region30. Nicotine has been reported to regulate miRNAs and mRNAs in the MHb, with microarrays detecting expression changes in mice self-administering nicotine31. However, the targets of these miRNAs and their functions in the context of nicotine dependence have not been elucidated.
Using integrated miRNA- and mRNA-Seq, we asked whether there are changes in miRNA and mRNA expression in the MHb and IPN during acute nicotine withdrawal. We hypothesize that using these sequencing datasets as valuable resources, it will be possible to identify a multitude of miRNAs/genes playing a role(s) in the mechanisms underlying the neuroadaptations involved in nicotine dependence.
Results
Differential expression of miRNAs and mRNAs in the habenulo-interpeduncular axis during acute withdrawal from nicotine
To assess the regulation of miRNAs and mRNAs by chronic nicotine treatment (Nic) and acute withdrawal (NAWD), we sequenced and performed differential expression analysis of RNAs isolated from the IPN and MHb (Fig. 1B–M; Figs. S2 and S3). As a reference brain region outside of the habenulo-interpeduncular circuit, we sequenced RNA from the nucleus accumbens (NAc), a prominent mesolimbic reward region (Fig. 1N–S; Figs. S2 and S3).
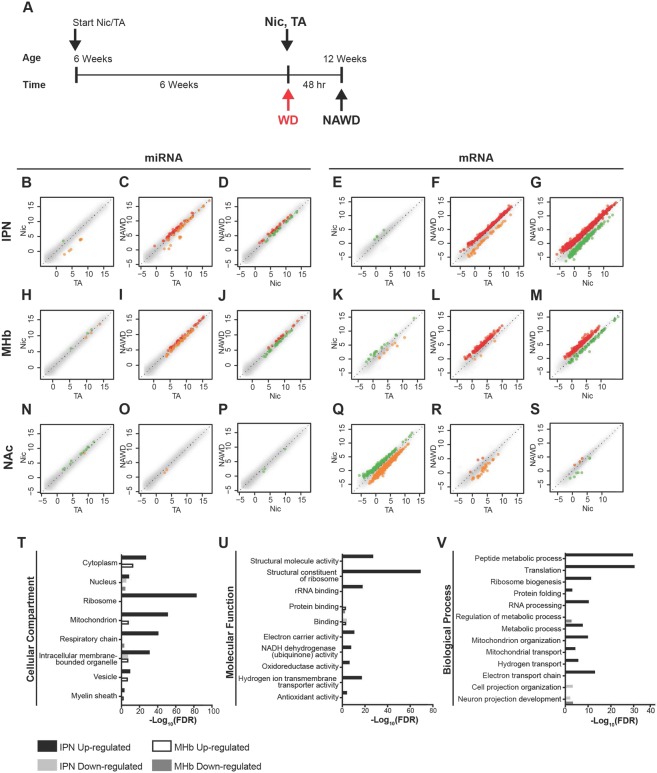
Figure 1.
Differential expression of miRNAs and mRNAs within the MHb-IPN withdrawal axis and NAc during acute nicotine withdrawal. (A) Schematic of nicotine treatment and spontaneous withdrawal of mice used for miRNA- and mRNA-Seq. Mice were treated chronically with nicotine (Nic) or tartaric acid (TA) for 6 weeks, then either sacrificed or spontaneously withdrawn (WD) for 48 hours before sacrifice (NAWD). (B–S) Scatter plots displaying differential expression of miRNAs or mRNAs in the IPN, MHb, and NAc measured by deep sequencing. miRNAs isolated from tissue punches of nicotine-treated (B,H,N) and NAWD-treated mice are compared to TA-treated controls (C,I,O) or nicotine alone samples (D,J,P). mRNAs of nicotine-treated (E,K,Q) and NAWD-treated mice are compared to TA-treated controls (F,L,R) or nicotine alone samples (G,M,S). Each dot represents an individual miRNA or mRNA plotted as log2(RPM) or log2(TPM), respectively. All miRNAs with an FDR < 0.01 and mRNAs with FDR < 0.01 and FC > 2 are highlighted in color. Orange dots are up-regulated in TA compared to Nic or NAWD. Green dots are up-regulated in Nic compared to TA controls. Red dots are up-regulated in NAWD compared to TA controls. Only miRNAs with RPM > 0 and mRNAs with TPM > 0 in at least half of the replicates in each treatment group are shown. n = 4–5, with each library synthesized using pooled RNA from 4 mice. (T–V) Genes differentially expressed during acute nicotine withdrawal are enriched in gene ontology terms related to energetics, translation, and cell projection organization in the IPN and MHb. The vertical axes define the selected GO terms describing the cellular compartment (T), molecular function (U), or biological process (V). The number of GO terms was reduced combining similar terms using a simRel cutoff of 0.4 as described in the Materials and Methods. The horizontal axis plots the significance of enrichment of the GO term, plotted as −log10(FDR).
In the habenulo-interpeduncular circuit, there were few miRNAs differentially expressed after chronic nicotine treatment. In the IPN, only 8 miRNAs were significantly altered after nicotine treatment alone (Fig. 1B). The 7 down-regulated miRNAs remained similarly repressed after a 48-hr withdrawal (see GEO upload). In the MHb, there were only 6 differentially expressed miRNAs in Nic mice compared to TA controls (Fig. 1H), and only 1 remained similarly regulated after nicotine was discontinued (see GEO upload).
However, after acute nicotine withdrawal, the expression of a multitude of miRNAs was altered in the habenulo-interpeduncular circuit. In the IPN of NAWD mice, there were 42 up-regulated and 45 down-regulated miRNAs in NAWD mice compared to TA controls (Fig. 1C). Twenty-nine of these miRNAs were similarly differentially expressed when NAWD mice are compared to either TA or Nic mice (Fig. 1D), suggesting their regulation in the IPN is due entirely to withdrawal and not an effect of nicotine exposure alone.
In the MHb, there were 35 up-regulated and 50 down-regulated miRNAs in NAWD mice compared to TA controls (Fig. 1I). Forty miRNAs were similarly differentially expressed when NAWD mice are compared to either TA or Nic (Fig. 1J), suggesting their expression was altered only after withdrawal, and was not simply an effect of the nicotine treatment.
To determine the persistence of the alterations in miRNA expression following acute nicotine withdrawal, another group of mice underwent a long (4-week) withdrawal after chronic nicotine treatment (NLWD) prior to sequencing. Interestingly, there were no differentially expressed miRNAs in either the IPN or MHb of NLWD mice compared to age-matched TA-treated (TAL) controls (Fig. S2S–V).
In the NAc, expression of 12 miRNAs was altered in Nic mice (Fig. 1N). However, in NAWD mice only 2 miRNAs were differentially expressed (Fig. 1O). Twenty-six miRNAs were differentially expressed in NLWD mice compared to TAL controls (Fig. S2W,X), and none of these were similarly altered in acute withdrawal (see GEO upload).
With respect to the mRNAs, few were regulated by chronic nicotine treatment with a fold change >2 in the IPN and MHb. In the IPN, there were 4 differentially expressed mRNAs in Nic mice compared to controls (Fig. 1E) and all of these returned to control expression levels in NAWD mice (see GEO upload). In the MHb, there were 18 mRNAs up- and 8 mRNAs down-regulated by chronic nicotine treatment compared to TA controls (Fig. 1K) and only 2 remained differentially expressed after acute withdrawal (see GEO upload).
In contrast to chronic nicotine treatment alone, there were 292 up- and 43 down-regulated mRNAs in the IPN of NAWD mice compared to TA controls (Fig. 1F). Of these mRNAs, 303 were similarly altered when NAWD mice are compared to either TA or Nic mice (Fig. 1G), suggesting their regulation was an effect of acute nicotine withdrawal and not an effect of nicotine treatment alone.
In the MHb, there were 151 up- and 3 down-regulated mRNAs in NAWD mice compared to TA controls (Fig. 1L). When NAWD mice were compared to either TA or Nic, the expression of 122 mRNAs was similarly altered (Fig. 1M), suggesting their regulation was an effect of acute withdrawal from nicotine and not an effect of nicotine treatment alone.
In the NAc, in contrast to the habenulo-interpeduncular circuit, there were a total of 852 genes differentially expressed in Nic compared to TA mice (Fig. 1Q). After acute withdrawal, there were only 31 mRNAs differentially expressed in the NAc of NAWD mice compared to TA (Fig. 1R). Of these mRNAs, 25 were also altered after nicotine-treatment alone, suggesting this was largely an effect of the chronic nicotine treatment (Fig. 1S).
To determine the stability of altered mRNA expression induced by nicotine withdrawal, NLWD mice were compared to TAL controls. In the IPN, 39 genes were differentially expressed in NLWD mice compared to TAL (Fig. S3S,T). However, none of these genes were differentially expressed after acute withdrawal (see GEO upload). In the MHb, all mRNAs were expressed at control levels in NLWD mice (Fig. S3U,V).
Gene ontology analysis was performed on differentially expressed mRNAs for all treatment groups and brain regions (Fig. 1T–V, Table S1). For genes up- or down-regulated in the IPN and MHb of NAWD mice compared to TA controls, we determined enrichment of reduced terms describing the cellular compartment (CC, Fig. 1T), molecular function (MF, Fig. 1U), and biological process (BP, Fig. 1V). CC analysis determined there was significant enrichment of genes up-regulated in the IPN related to the mitochondrion. There was also enrichment of MF and BP terms related to mitochondrion, such as electron carrier activity, NADH dehydrogenase activity, hydrogen ion transport, and regulation of metabolic processes. There was also significant enrichment of genes up-regulated in the IPN related to ribosomal terms, including structural constituent of the ribosome, rRNA binding, ribosome biogenesis, and translation. In both the IPN and the MHb there was significant enrichment of down-regulated genes related to neuron projection development.
Differentially expressed miRNAs are predicted to target reciprocally expressed mRNAs in the IPN and MHb during acute nicotine withdrawal
TargetScan Mouse 7.1 was used to identify transcripts containing conserved MREs potentially targeted by miRNAs in all treatment groups and brain regions (Table S2).
In the IPN, 38 mRNAs that were differentially expressed in NAWD mice compared to controls are predicted targets of at least one of 35 inversely expressed miRNAs (Table 1). For example, expression of Profilin 2 (Pfn2), a regulator of cytoskeletal dynamics enriched in the brain32, was down-regulated by ~50% in the IPN of NAWD mice. miR-106b-5p is up-regulated by ~80% and is predicted to target two MREs within the 3′-UTR of Pfn2.
Table 1.
Predicted miRNA targets differentially expressed in the IPN during acute nicotine withdrawal.
| mRNA | mRNA FC | mRNA FDR | miRNA | miRNA FC | miRNA FDR | Seed Type | Position in 3′-UTR |
|---|---|---|---|---|---|---|---|
| Hbq1a | 3.002 | 2.57E-07 | miR-377-3p | 0.592 | 1.08E-03 | 7mer-m8 | 252–258 |
| Rpp21 | 2.852 | 2.35E-20 | miR-136-5p | 0.613 | 9.18E-06 | 8mer | 3537–3544 |
| Gchfr | 2.696 | 9.55E-06 | miR-129-1-3p | 0.570 | 4.18E-04 | 7mer-m8 | 298–304 |
| miR-129-2-3p | 0.590 | 1.62E-05 | 7mer-m8 | 298–304 | |||
| Gm20390 | 2.669 | 2.25E-23 | miR-429-3p | 0.329 | 3.53E-12 | 7mer-m8 | 3377–3383 |
| miR-429-3p | 0.329 | 3.53E-12 | 8mer | 4329–4336 | |||
| miR-429-3p | 0.329 | 3.53E-12 | 8mer | 4714–4721 | |||
| miR-200b-3p | 0.375 | 6.58E-08 | 7mer-m8 | 3377–3383 | |||
| miR-200b-3p | 0.375 | 6.58E-08 | 8mer | 4329–4336 | |||
| miR-200b-3p | 0.375 | 6.58E-08 | 8mer | 4714–4721 | |||
| miR-344d-3p | 0.741 | 4.47E-03 | 7mer-A1 | 3316–3122 | |||
| Gm9726 | 2.627 | 2.76E-04 | miR-29b-3p | 0.726 | 4.41E-03 | 8mer | 326–333 |
| Hist1h4h | 2.505 | 1.18E-15 | miR-9-5p | 0.753 | 5.60E-03 | 7mer-m8 | 26–32 |
| Serf2 | 2.421 | 7.81E-20 | miR-200a-3p | 0.381 | 1.24E-13 | 7mer-m8 | 213–219 |
| miR-141-3p | 0.458 | 3.07E-04 | 7mer-m8 | 213–219 | |||
| let-7f-5p | 0.735 | 8.04E-04 | 8mer | 2110–2117 | |||
| let-7b-5p | 0.741 | 6.40E-03 | 8mer | 2110–2117 | |||
| let-7c-5p | 0.766 | 9.52E-03 | 8mer | 2110–2117 | |||
| Mrps16 | 2.418 | 2.40E-15 | miR-136-5p | 0.613 | 9.18E-06 | 7mer-m8 | 86–92 |
| Zfp46 | 2.409 | 3.91E-12 | miR-129-1-3p | 0.570 | 4.18E-04 | 7mer-m8 | 464–470 |
| miR-129-2-3p | 0.590 | 1.62E-05 | 7mer-m8 | 464–470 | |||
| Ndufb2 | 2.368 | 2.31E-22 | miR-429-3p | 0.329 | 3.53E-12 | 7mer-m8 | 3896–3902 |
| miR-200b-3p | 0.375 | 6.58E-08 | 7mer-m8 | 3896–3902 | |||
| Esrrb | 2.332 | 7.67E-08 | let-7f-5p | 0.735 | 8.04E-04 | 8mer | 617–624 |
| let-7b-5p | 0.741 | 6.40E-03 | 8mer | 617–624 | |||
| miR-485-5p | 0.753 | 4.04E-03 | 7mer-m8 | 55–61 | |||
| miR-9-5p | 0.753 | 5.60E-03 | 8mer | 2337–2344 | |||
| let-7c-5p | 0.766 | 9.52E-03 | 8mer | 617–624 | |||
| Polr2f | 2.323 | 2.02E-26 | miR-488-3p | 0.752 | 8.56E-03 | 7mer-m8 | 485–491 |
| Nme1 | 2.305 | 3.32E-21 | miR-200a-3p | 0.381 | 1.24E-13 | 8mer | 123–130 |
| miR-141-3p | 0.458 | 3.07E-04 | 8mer | 123–130 | |||
| Gm9844 | 2.296 | 1.27E-04 | miR-129-5p | 0.721 | 1.46E-03 | 8mer | 26–33 |
| Diras1 | 2.234 | 1.93E-12 | miR-7b-5p | 0.715 | 1.25E-03 | 7mer-m8 | 2200–2206 |
| Myl6 | 2.234 | 8.56E-15 | miR-340-5p | 0.741 | 6.55E-03 | 7mer-A1 | 851–857 |
| Psma2 | 2.174 | 3.46E-21 | miR-132-3p | 0.726 | 6.55E-03 | 7mer-A1 | 111–117 |
| Mrpl14 | 2.172 | 2.53E-14 | miR-340-5p | 0.741 | 6.55E-03 | 7mer-A1 | 2823–2829 |
| Ptrhd1 | 2.170 | 1.90E-13 | miR-181d-5p | 0.676 | 2.00E-04 | 7mer-m8 | 2022–2028 |
| miR-181b-5p | 0.680 | 5.42E-05 | 7mer-m8 | 2022–2028 | |||
| miR-488-3p | 0.752 | 8.56E-03 | 8mer | 76–83 | |||
| Dynlt1f | 2.169 | 1.11E-08 | miR-29b-3p | 0.726 | 4.41E-03 | 7mer-A1 | 215–221 |
| Sod1 | 2.162 | 7.65E-13 | miR-377-3p | 0.592 | 1.08E-03 | 7mer-m8 | 78–84 |
| Cst6 | 2.158 | 6.60E-14 | miR-377-3p | 0.592 | 1.08E-03 | 7mer-m8 | 2027–2033 |
| miR-181d-5p | 0.676 | 2.00E-04 | 7mer-m8 | 2023–2029 | |||
| miR-181b-5p | 0.680 | 5.42E-05 | 7mer-m8 | 2023–2029 | |||
| Rpl31 | 2.153 | 6.45E-09 | miR-653-5p | 0.616 | 4.04E-03 | 7mer-A1 | 639–645 |
| miR-129-5p | 0.721 | 1.46E-03 | 8mer | 671–678 | |||
| Tmsb4x | 2.126 | 2.94E-14 | miR-183-5p | 0.319 | 3.96E-21 | 7mer-m8 | 327–333 |
| Ndufb9 | 2.104 | 7.81E-20 | miR-200a-3p | 0.381 | 1.24E-13 | 8mer | 2453–2460 |
| miR-141-3p | 0.458 | 3.07E-04 | 8mer | 2453–2460 | |||
| miR-365-3p | 0.654 | 9.94E-03 | 7mer-m8 | 2718–2724 | |||
| miR-532-5p | 0.740 | 2.06E-03 | 7mer-m8 | 3048–3054 | |||
| miR-340-5p | 0.741 | 6.55E-03 | 7mer-A1 | 1850–1856 | |||
| miR-379-5p | 0.752 | 6.55E-03 | 7mer-m8 | 3186–3192 | |||
| Rps12 | 2.097 | 2.19E-06 | miR-96-5p | 0.235 | 1.36E-17 | 7mer-m8 | 2285–2291 |
| Acot13 | 2.097 | 2.94E-07 | miR-429-3p | 0.329 | 3.53E-12 | 7mer-m8 | 4417–423 |
| miR-200b-3p | 0.375 | 6.58E-08 | 7mer-m8 | 4417–423 | |||
| miR-653-5p | 0.616 | 4.04E-03 | 7mer-A1 | 5139–5145 | |||
| miR-181d-5p | 0.676 | 2.00E-04 | 7mer-m8 | 4525–4531 | |||
| miR-181b-5p | 0.680 | 5.42E-05 | 7mer-m8 | 4525–4532 | |||
| Aamdc | 2.094 | 9.88E-15 | miR-485-5p | 0.753 | 4.04E-03 | 7mer-m8 | 64–70 |
| Cisd3 | 2.093 | 9.98E-20 | miR-9-5p | 0.753 | 5.60E-03 | 7mer-A1 | 358–364 |
| Tmem151b | 2.035 | 2.86E-07 | miR-129-1-3p | 0.570 | 4.18E-04 | 7mer-m8 | 2722–2728 |
| miR-129-2-3p | 0.590 | 1.62E-05 | 7mer-m8 | 2722–2728 | |||
| miR-181d-5p | 0.676 | 2.00E-04 | 7mer-m8 | 2226–2232 | |||
| miR-181b-5p | 0.680 | 5.42E-05 | 7mer-m8 | 2226–2233 | |||
| miR-132-3p | 0.726 | 6.55E-03 | 8mer | 339–346 | |||
| miR-29b-3p | 0.726 | 4.41E-03 | 7mer-m8 | 2452–2458 | |||
| miR-485-5p | 0.753 | 4.04E-03 | 8mer | 2089–2096 | |||
| Erh | 2.031 | 1.69E-09 | miR-181d-5p | 0.676 | 2.00E-04 | 7mer-m8 | 118–124 |
| miR-181b-5p | 0.680 | 5.42E-05 | 7mer-m8 | 118–124 | |||
| Tomm5 | 2.011 | 4.37E-19 | miR-151-3p | 0.770 | 6.36E-03 | 7mer-m8 | 383–389 |
| Gosr2 | 0.374 | 1.22E-11 | miR-27a-3p | 1.421 | 8.66E-04 | 8mer | 94–101 |
| Nkx2-1 | 0.459 | 1.19E-03 | miR-503-5p | 1.581 | 1.48E-03 | 7mer-m8 | 74–80 |
| Prkag1 | 0.488 | 8.60E-03 | miR-34a-5p | 1.941 | 7.79E-08 | 7mer-m8 | 42–48 |
| miR-34b-5p | 1.893 | 1.43E-06 | 7mer-m8 | 42–48 | |||
| Lurap1l | 0.489 | 1.18E-06 | miR-497a-5p | 2.053 | 3.09E-13 | 8mer | 60–67 |
| miR-503-5p | 1.581 | 1.48E-03 | 7mer-A1 | 61–67 | |||
| Pfn2 | 0.496 | 8.36E-09 | miR-106b-5p | 1.789 | 9.32E-11 | 7mer-m8 | 452–458 |
| miR-106b-5p | 1.789 | 9.32E-11 | 7mer-m8 | 1197–1203 | |||
| Zbtb9 | 0.496 | 3.34E-05 | miR-497a-5p | 2.053 | 3.09E-13 | 7mer-m8 | 42–48 |
| miR-497a-5p | 2.053 | 3.09E-13 | 7mer-m8 | 45–51 | |||
| miR-106b-5p | 1.789 | 9.32E-11 | 8mer | 822–829 |
TargetScan Mouse 7.1 predicted conserved MREs within the 3′-UTRs of a subset of mRNAs differentially expressed in the IPN of NAWD mice compared to TA controls. miRNAs conserved in vertebrates and mammals that were inversely expressed (FDR < 0.01) and predicted to target these MREs are listed. Only mRNAs with a FC > 2 and an FDR < 0.01 were included in target prediction analysis. Seed type of the predicted MRE and its position within the 3′-UTR are listed.
In the MHb, 50 mRNAs that were differentially expressed in NAWD mice compared to TA controls are predicted targets of at least one of 32 inversely expressed miRNAs (Table 2). The up-regulation of at least four mRNAs (Arf5, Atg7, Bcl2l1, and Eif5a) that are predicted to be targeted by down-regulated miRNAs (Table 2), was verified by RT-qPCR (Fig. S4D–G).
Table 2.
Predicted miRNA targets differentially expressed in the MHb during acute nicotine withdrawal.
| mRNA | mRNA FC | mRNA FDR | miRNA | miRNA FC | miRNA FDR | Seed Type | Position in 3′-UTR |
|---|---|---|---|---|---|---|---|
| Arf5 | 4.543 | 1.01E-24 | miR-29b-3p | 0.650 | 4.95E-06 | 8mer | 392–399 |
| Pcolce2 | 4.447 | 9.69E-19 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 329–335 |
| miR-146b-5p | 0.721 | 1.68E-04 | 7mer-A1 | 3039–3045 | |||
| Tom1l1 | 4.011 | 2.74E-21 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 397–403 |
| miR-340-5p | 0.618 | 2.19E-10 | 8mer | 2622–2629 | |||
| miR-181b-5p | 0.763 | 2.71E-03 | 8mer | 371–378 | |||
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-A1 | 2622–2628 | |||
| miR-181c-5p | 0.770 | 3.26E-03 | 8mer | 371–378 | |||
| Rasd1 | 3.629 | 3.25E-11 | miR-377-3p | 0.558 | 4.71E-04 | 7mer-m8 | 572–578 |
| miR-30c-5p | 0.710 | 2.01E-04 | 8mer | 465-–472 | |||
| miR-384-5p | 0.735 | 3.14E-04 | 8mer | 465–472 | |||
| Pcna | 3.195 | 6.07E-16 | miR-137-3p | 0.652 | 9.24E-08 | 8mer | 420–427 |
| Spns1 | 3.031 | 1.55E-11 | miR-29b-3p | 0.650 | 4.95E-06 | 8mer | 266–273 |
| Ppp2cb | 3.022 | 5.91E-20 | miR-183-5p | 0.551 | 2.97E-06 | 8mer | 152–159 |
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-A1 | 142–148 | |||
| Steap3 | 2.857 | 2.10E-06 | miR-377-3p | 0.558 | 4.71E-04 | 7mer-A1 | 704–710 |
| Dnajc9 | 2.787 | 7.78E-14 | miR-132-3p | 0.710 | 1.32E-04 | 7mer-m8 | 1040–1046 |
| Rpa1 | 2.704 | 2.98E-15 | miR-30c-5p | 0.710 | 2.01E-04 | 7mer-m8 | 1195–1201 |
| miR-384-5p | 0.735 | 3.14E-04 | 7mer-m8 | 1195–1201 | |||
| miR-361-5p | 0.762 | 2.72E-03 | 7mer-m8 | 591–597 | |||
| miR-361-5p | 0.762 | 2.72E-03 | 7mer-A1 | 627–633 | |||
| Net1 | 2.675 | 4.10E-07 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 31–37 |
| miR-135a-5p | 0.631 | 7.96E-09 | 8mer | 831–838 | |||
| miR-135b-5p | 0.645 | 4.36E-05 | 8mer | 831–838 | |||
| Gpx4 | 2.617 | 1.68E-17 | miR-374b-5p | 0.549 | 2.62E-05 | 7mer-A1 | 833–839 |
| Ogfod2 | 2.613 | 3.97E-11 | miR-340-5p | 0.618 | 2.19E-10 | 493-500 | 8mer |
| Sin3a | 2.598 | 2.68E-09 | miR-211-5p | 0.492 | 7.97E-12 | 7mer-m8 | 870–876 |
| miR-183-5p | 0.551 | 2.97E-06 | 7mer-m8 | 644–650 | |||
| miR-493-5p | 0.699 | 3.98E-03 | 7mer-m8 | 724–730 | |||
| miR-493-5p | 0.699 | 3.98E-03 | 8mer | 840–847 | |||
| Eif5a | 2.561 | 5.52E-19 | miR-495-3p | 0.472 | 8.00E-16 | 7mer-A1 | 589–595 |
| Gosr2 | 2.511 | 5.84E-16 | miR-493-5p | 0.699 | 3.98E-03 | 8mer | 1649–1656 |
| Chst15 | 2.455 | 2.63E-07 | miR-342-3p | 0.704 | 8.42E-05 | 8mer | 1364–1371 |
| Tmem159 | 2.443 | 1.65E-04 | miR-135a-5p | 0.631 | 7.96E-09 | 7mer-m8 | 607–613 |
| miR-135b-5p | 0.645 | 4.36E-05 | 7mer-m8 | 607–613 | |||
| Rab40b | 2.414 | 3.16E-05 | miR-211-5p | 0.492 | 7.97E-12 | 8mer | 336–343 |
| Rabl6 | 2.413 | 2.15E-08 | miR-30c-5p | 0.710 | 2.01E-04 | 8mer | 371–378 |
| miR-384-5p | 0.735 | 3.14E-04 | 8mer | 371–378 | |||
| Id3 | 2.400 | 5.82E-11 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 489–495 |
| Htra2 | 2.381 | 6.59E-05 | miR-410-3p | 0.667 | 4.11E-07 | 7mer-A1 | 160–166 |
| Kctd3 | 2.355 | 2.23E-08 | miR-29b-3p | 0.650 | 4.95E-06 | 7mer-A1 | 432–438 |
| miR-30c-5p | 0.710 | 2.01E-04 | 7mer-m8 | 411–417 | |||
| miR-384-5p | 0.735 | 3.14E-04 | 7mer-m8 | 411–417 | |||
| Atg16l1 | 2.346 | 3.04E-12 | let-7k | 0.615 | 1.81E-04 | 7mer-A1 | 235–241 |
| miR-410-3p | 0.667 | 4.11E-07 | 8mer | 1100–1107 | |||
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-m8 | 872–878 | |||
| Srl | 2.344 | 8.98E-04 | miR-211-5p | 0.492 | 7.97E-12 | 8mer | 2918–2925 |
| miR-136-5p | 0.582 | 2.87E-08 | 8mer | 818–825 | |||
| miR-136-5p | 0.582 | 2.87E-08 | 8mer | 4468–4475 | |||
| Marcksl1 | 2.271 | 1.11E-08 | miR-23b-3p | 0.740 | 1.75E-03 | 8mer | 576–583 |
| miR-23a-3p | 0.755 | 3.09E-03 | 8mer | 576–583 | |||
| Ccdc32 | 2.241 | 2.68E-07 | miR-377-3p | 0.558 | 4.71E-04 | 7mer-m8 | 165–171 |
| Prpf19 | 2.227 | 2.45E-17 | miR-377-3p | 0.558 | 4.71E-04 | 8mer | 2354–2361 |
| miR-421-3p | 0.701 | 6.33E-03 | 8mer | 2841–2848 | |||
| miR-203-3p | 0.732 | 7.63E-04 | 7mer-m8 | 120–126 | |||
| Tbc1d8b | 2.222 | 2.86E-03 | miR-340-5p | 0.618 | 2.19E-10 | 8mer | 2336–2343 |
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-m8 | 1016–1022 | |||
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-A1 | 2336–2341 | |||
| Kdelr1 | 2.222 | 7.47E-08 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 313–319 |
| miR-335-5p | 0.637 | 4.58E-06 | 7mer-A1 | 715–721 | |||
| Gm2a | 2.184 | 1.22E-07 | miR-6395 | 0.682 | 5.46E-03 | 8mer | 591–598 |
| Zfp622 | 2.170 | 1.71E-11 | miR-181b-5p | 0.763 | 2.71E-03 | 8mer | 410–417 |
| miR-181c-5p | 0.770 | 3.26E-03 | 8mer | 410–417 | |||
| Gap43 | 2.163 | 3.62E-03 | miR-23b-3p | 0.740 | 1.75E-03 | 7mer-m8 | 433–439 |
| miR-23a-3p | 0.755 | 3.09E-03 | 7mer-m8 | 433–439 | |||
| Grasp | 2.157 | 9.32E-04 | miR-132-3p | 0.710 | 1.32E-04 | 7mer-m8 | 668–674 |
| Gpr182 | 2.134 | 4.61E-03 | miR-30c-5p | 0.710 | 2.01E-04 | 7mer-A1 | 2658–2664 |
| miR-384-5p | 0.735 | 3.14E-04 | 7mer-A1 | 2658–2664 | |||
| Metap1 | 2.125 | 1.20E-06 | miR-211-5p | 0.492 | 7.97E-12 | 7mer-m8 | 1354–1360 |
| miR-382-3p | 0.528 | 3.46E-11 | 7mer-m8 | 1373–1379 | |||
| miR-181b-5p | 0.763 | 2.71E-03 | 7mer-m8 | 1334–1340 | |||
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-m8 | 1221–1227 | |||
| miR-181c-5p | 0.770 | 3.26E-03 | 7mer-m8 | 1334–1340 | |||
| Chid1 | 2.123 | 5.05E-07 | miR-142a-5p | 0.765 | 8.25E-03 | 7mer-m8 | 2673–2679 |
| Tmem184a | 2.121 | 3.51E-03 | miR-137-3p | 0.652 | 9.24E-08 | 8mer | 2889–2896 |
| miR-410-3p | 0.667 | 4.11E-07 | 7mer-A1 | 3706–3712 | |||
| miR-493-5p | 0.699 | 3.98E-03 | 7mer-m8 | 2883–2889 | |||
| miR-487b-3p | 0.702 | 4.08E-04 | 8mer | 3684–3691 | |||
| Htr2c | 2.106 | 2.43E-10 | miR-382-3p | 0.528 | 3.46E-11 | 8mer | 1230–1237 |
| miR-137-3p | 0.652 | 9.24E-08 | 8mer | 2964–2971 | |||
| miR-23b-3p | 0.740 | 1.75E-03 | 8mer | 1871–1878 | |||
| miR-23a-3p | 0.755 | 3.09E-03 | 8mer | 1871–1878 | |||
| Alpl | 2.103 | 3.66E-06 | miR-211-5p | 0.492 | 7.97E-12 | 8mer | 655–662 |
| Ddah2 | 2.093 | 1.58E-08 | miR-29b-3p | 0.650 | 4.95E-06 | 7mer-m8 | 69–75 |
| Zkscan4 | 2.087 | 1.94E-05 | miR-29b-3p | 0.650 | 4.95E-06 | 7mer-A1 | 390–396 |
| Atg7 | 2.087 | 1.98E-05 | miR-211-5p | 0.492 | 7.97E-12 | 8mer | 1454–1461 |
| miR-382-3p | 0.528 | 3.46E-11 | 7mer-m8 | 1438–1444 | |||
| miR-142a-5p | 0.765 | 8.25E-03 | 7mer-m8 | 879–885 | |||
| Galnt18 | 2.083 | 1.04E-04 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 245–251 |
| Thg1l | 2.060 | 1.05E-03 | miR-340-5p | 0.618 | 2.19E-10 | 7mer-A1 | 280–286 |
| Slc25a18 | 2.045 | 5.08E-05 | let-7k | 0.615 | 1.81E-04 | 8mer | 106–113 |
| Api5 | 2.023 | 3.22E-09 | miR-203-3p | 0.732 | 7.63E-04 | 8mer | 1699–1706 |
| Bcl2l1 | 2.020 | 8.98E-07 | miR-495-3p | 0.472 | 8.00E-16 | 7mer-A1 | 1219–1225 |
| miR-377-3p | 0.558 | 4.71E-04 | 7mer-m8 | 1242–1248 | |||
| miR-377-3p | 0.558 | 4.71E-04 | 8mer | 1424–1431 | |||
| let-7k | 0.615 | 1.81E-04 | 8mer | 947–954 | |||
| miR-342 | 0.704 | 8.42E-05 | 7mer-m8 | 1243–1249 | |||
| Cat | 2.014 | 3.45E-06 | miR-23b-3p | 0.740 | 1.75E-03 | 8mer | 793–800 |
| miR-23a-3p | 0.755 | 3.09E-03 | 8mer | 793–800 | |||
| Ccdc80 | 2.011 | 6.43E-03 | miR-29b-3p | 0.650 | 4.95E-06 | 8mer | 213–220 |
| miR-433-3p | 0.673 | 1.45E-05 | 8mer | 1243–1250 |
TargetScan Mouse 7.1 predicted conserved MREs within the 3′-UTRs of a subset of mRNAs differentially expressed in the MHb of NAWD mice compared to TA controls. miRNAs conserved in vertebrates and mammals that were inversely expressed (FDR < 0.01) and predicted to target these MREs are listed. Only mRNAs with a FC > 2 and an FDR < 0.01 were included in target prediction analysis. Seed type of the predicted MRE and its position within the 3′-UTR are listed.
In the NAc, neither of the miRNAs differentially expressed in NAWD mice compared to TA-treated controls are conserved, and therefore were not considered in target analysis. However, after nicotine treatment alone, there were 204 differentially expressed mRNAs predicted to be targeted by at least one of 8 inversely expressed miRNAs (Table S2).
Down-regulation of Pfn2 in the IPN is sufficient to induce anxiety
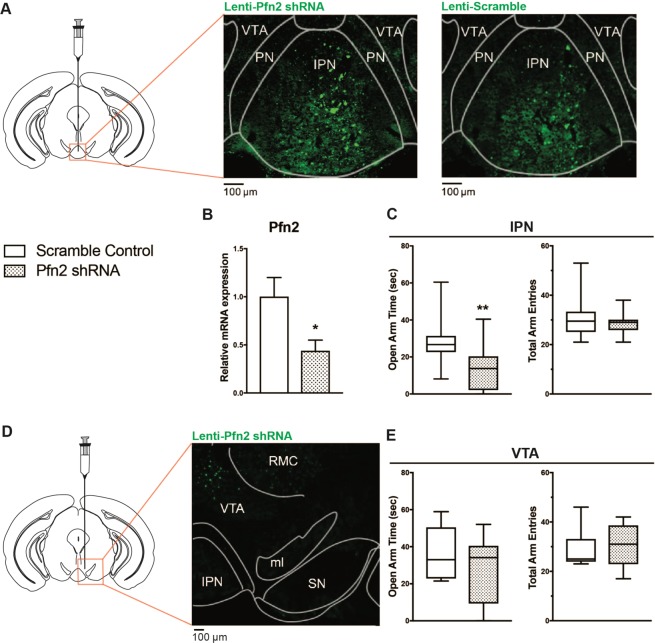
Withdrawal from nicotine induces anxiety in mice through activation of the IPN16. Using our treatment paradigm, NAWD mice display increased anxiety compared to nicotine- and TA-treated controls, as measured by the marble burying test and EPM (Fig. S4A–C). Because Pfn2-/- mice display increased synaptic vesicle exocytosis and hyperactivation of neurons33, we hypothesized that repression of Pfn2 in the IPN is sufficient to induce anxiety, mimicking the behaviors observed in NAWD mice. To knockdown Pfn2 expression, three different lentiviral shRNAs were tested in SN17 cells, revealing that lenti-pGIPZ-Pfn2-shRNA494369-tGFP produced robust knockdown (~81%) compared to virus expressing a scramble control shRNA (Fig. S5A). Injection of the IPN with lenti-pGIPZ-Pfn2-shRNA494369-tGFP (Fig. 2A) resulted in ~54% knockdown of Pfn2 expression in areas expressing tGFP compared to animals injected with control virus (Fig. 2B). Virus expression was limited to the IPN and adjacent brain areas, with no spread to distant midbrain regions, as detected by fluorescence microscopy (Fig. S5B). Quite strikingly, when Pfn2 was knocked down in the IPN, mice spent significantly less time in the open arms of the EPM compared to controls (Fig. 2C). Importantly, there was no significant difference in the number of total arm entries, indicating the results are not confounded by non-specific effects on locomotor activity (Fig. 2C). However, when Pfn2 was knocked down in the neighboring VTA (Fig. 2D), there was no significant effect on the amount of time spent in the open arms of the EPM or total arm entries (Fig. 2E). Because the IPN is also a prominent brain region in the circuitry underlying novelty preference34 and novelty seeking behaviors are altered in Pfn2 knockout mice33, we also tested the effect of Pfn2 knockdown on object novelty preference. Expression of lenti-pGIPZ-Pfn2-shRNA494369-tGFP in the IPN did not significantly affect the time spent exploring familiar and novel objects or the novel object preference ratio compared controls (Fig. S6).
Figure 2.
Knockdown of Pfn2 in the IPN is sufficient to increase anxiety. (A) Representative midbrain coronal section schematic and fluorescence microscopy images of WT mouse injected with either lenti-pGIPZ-Pfn2-shRNA-tGFP (left image) or lenti-pGIPZ-Scramble-tGFP (right image) in the IPN. Nuclei were labeled with DAPI (blue). (B) Knockdown of Pfn2 expression was measured by RT-qPCR using RNA from laser-captured IPN tissue infected with lenti-pGIPZ-Pfn2-shRNA-tGFP compared to lenti-pGIPZ-scramble-tGFP control. mRNA expression was normalized to β-2-microglobulin. Statistical analysis using an unpaired, two-tailed t-test compared lenti-Pfn2 shRNA mice to lenti-scramble controls (t11 = 2.272, P < 0.05). (*)P < 0.05; n = 6–7. (C) Average time spent in the open arms and total number of arm entries in the elevated plus maze of mice injected with either lenti-Pfn2 shRNA (stippled bars) or lenti-scramble shRNA control (open bars) in the IPN. Statistical analysis used an unpaired two-tailed t-test to compare time spent in open arms (t33 = 3.583, P = 0.0011) and total number of arm entries (t33 = 0.7355, P = 0.4672) of lenti-Pfn2 shRNA mice to lenti-scramble controls. (**)P < 0.01 n = 16–19. Box plot edges are 25th and 75th percentile, central line is the median and whiskers are max and min. (D) Representative midbrain coronal section schematic and fluorescence microscopy image of WT mouse injected with lenti-pGIPZ-Pfn2-shRNA-tGFP in the VTA. Nuclei were labeled with DAPI (blue). (E) Average time spent in the open arms and average total number of arm entries in the elevated plus maze of mice injected with either lenti-Pfn2 shRNA (stippled bars) or lenti-scramble shRNA control (open bars) in the VTA. Unpaired, two-tailed t-test was used for statistical analysis of time spent in open arms (t16 = 1.09, P = 0.2918) and total arm entries (t16 = 0.4153, P = 0.6835). n = 9. Box plot edges are 25th and 75th percentile, central line is the median and whiskers are max and min.
Discussion
Using RNA sequencing and differential expression analysis, we identified wide-spread changes in both miRNA and mRNA expression in the MHb-IPN withdrawal circuit during acute nicotine withdrawal. In contrast, there were a limited number of miRNAs/mRNAs altered by acute nicotine withdrawal in the NAc, a prominent reward center, suggesting that wide-spread changes in transcription during acute withdrawal occur in a brain region-specific manner, concentrated in the habenulo-interpeduncular circuitry.
In the MHb and IPN, miRNAs and mRNAs that were altered during acute nicotine withdrawal largely returned to control expression levels after a prolonged withdrawal. This is most striking in the MHb where there were no differentially expressed miRNAs or mRNAs 4 weeks after withdrawal. In the IPN, none of the mRNAs differentially expressed after a long withdrawal were similarly altered at the 48-hour withdrawal time point. This suggests that for a subset of genes in the IPN, changes in mRNA expression induced by nicotine withdrawal occur over a longer timescale and persist for at least 4 weeks. This subpopulation of genes stably dysregulated by nicotine treatment and withdrawal may contribute to the persistent susceptibility to relapse that plagues nicotine addicts.
Gene ontology analysis identified significant enrichment of up-regulated genes in the IPN related to ribosome structure and translation, further supporting the body of evidence suggesting that stable neuroadaptations associated with drug addiction require de novo protein synthesis17. Thus, it is possible that up-regulation of ribosomes and other regulators of translation fulfills the increased demand for protein products in the IPN during nicotine withdrawal. In addition, in the MHb and IPN, there is significant enrichment of up-regulated genes related to the mitochondria. The functions of activated neurons, including maintenance of membrane resting potential, intracellular calcium buffering, and synaptic vesicle recycling, are highly energy-demanding35,36. To meet the increased demand, neurons increase oxidative metabolism, but cannot up-regulate glycolysis37. The enrichment of up-regulated genes related to mitochondria and oxidative energy production reflect the highly active synapses of the IPN during nicotine withdrawal.
An interesting outcome of our work is the realization that some of the miRNAs we demonstrated to be differentially expressed as a result of nicotine withdrawal have been shown by others to regulate expression of mRNAs encoding cholinergic transcripts. Such miRNAs are referred to as CholinomiRs38 and include, for example, miR-132 that targets acetylcholinesterase39,40, as well as miR-211 that targets the gene encoding the alpha7 subunit of nAChRs41. Given that CholinomiRs function in a combinatorial manner such that their impact is a consequence of interactions between competing miRNAs and targets38, our dataset should prove useful in further elucidating the mechanisms underlying changes in cholinergic signaling that occur due to nicotine withdrawal.
As an example of the utility of our RNA-Seq data, we focused upon one example of a gene differentially regulated during acute nicotine withdrawal in the IPN. Pfn2 is a member of the profilin family of small, actin monomer-binding proteins42,43. While Pfn1 is ubiquitously expressed throughout the body, Pfn2 expression is highest in the brain32 and represents approximately 75% of the profilin in rodent brain32,44. In addition to its ability to bind actin monomers, Pfn2 has a poly-l-proline binding domain allowing it to interact with a plethora of ligands whose functions contribute to synaptic morphology and function42. Pfn2 is expressed pre- and post-synaptically33. Pre-synaptically, Pfn2 can influence synaptic vesicle release through interaction with regulators of membrane trafficking and endocytosis45,46. Interestingly, Pfn2 −/− mice display higher vesicle exocytosis and hyperactivation of the striatum33. Post-synaptically, Pfn2 accumulates in dendritic spines of activated neurons47,48, and plays a role in determining their complexity through the regulation of cytoskeletal dynamics49,50. Pfn2 regulates actin polymerization at the synapse by direct interaction with the WAVE-complex33, which in turn activates the Arp2/3 complex to promote actin nucleation. Interestingly, Arpc3 is a confirmed target of miR-29a/b, which is up-regulated in the mouse brain after acute nicotine injection, regulating dendritic spine morphology29. Furthermore, Pfn2 interacts directly with ROCK, a RhoA-GTPase effector, regulating neuritogenesis in response to physiological stimuli50. Interestingly, inhibition of ROCK in the mouse brain induces anxiety51. Our GO analysis revealed an enrichment of down-regulated genes related to cellular projection organization and neuron projection development in the IPN during nicotine withdrawal, possibly reflecting changes in neuron morphology regulated by the repression of Pfn2 expression.
In addition to various roles in mechanisms related to cytoskeletal dynamics underlying synaptic morphology, Pfn2 can also influence the composition of neurotransmitter receptors at post-synaptic surfaces42. For example, Pfn2 can decrease surface localization of excitatory kainate glutamate receptors through regulation of endocytosis and membrane trafficking52. Pfn2 also interacts with membrane scaffold proteins, such as gephyrin53, anchoring receptors to the cytoskeleton.
The synaptic roles of Pfn2 and its link to behaviors such as anxiety led us to hypothesize that down-regulation of Pfn2 during acute nicotine withdrawal contributes to the neural activation in the IPN during withdrawal, increasing anxiety-associated behaviors. Supporting this hypothesis, we show that knockdown of Pfn2 expression in the IPN of nicotine-naïve mice was sufficient to increase anxiety as measured by EPM, mimicking behaviors observed in WT mice during acute nicotine withdrawal. Effects on behavior are specific for anxiety, with no alterations in locomotor activity or object novelty preference. The effect of Pfn2 knockdown is also specific to the IPN, as knockdown within the neighboring VTA does not increase anxiety. Our results suggest a role for the down-regulation of Pfn2 in neuroadaptations of synapses induced by acute nicotine withdrawal in the IPN. Importantly, with respect to physiological relevance, Pfn2 is highly expressed in the IPN (expression data of all miRNAs and mRNAs are included in the GEO database associated with this article). Pfn2 ranks 180th in abundance (mean = 801.258 transcripts per million) amongst all genes detected in the IPN of at least one control replicate putting it in the top 1% of genes detected in at least one replicate (22,181 genes total). It seems likely that a significant reduction in expression of such a highly expressed gene whose product interacts with the cytoskeleton and proteins that regulate synaptic exo- and endocytosis will substantially impact neuronal function. Future studies should focus on determining the effect of withdrawal-induced repression of Pfn2 on neural activity and synaptic morphology in the IPN.
Our study uncovered wide-spread changes in gene expression of the habenulo-interpeduncular circuit during acute nicotine withdrawal. The multitude of genes and their predicted regulation by miRNAs represent potential novel mechanisms underlying the neuroadaptations of nicotine dependence. Hence, this dataset is a valuable resource, the use of which may lead to the identification of novel substrates for the design of innovative smoking cessation aids and the treatment of withdrawal symptoms, including anxiety. However, it is important to point out that, in addition to miRNA regulation of target genes, it is possible that the targets themselves regulate miRNA levels by regulating miRNA degradation19. This is particularly relevant to the nervous system where it has been shown that target RNA-directed miRNA degradation (TDMD) activity is substantially higher in primary neurons versus non-neuronal cells and established cell lines54. Thus, our dataset may also prove useful for deciphering TDMD pathways during withdrawal. However, there are limitations to our dataset that must be considered when analyzing miRNA/mRNA relationships following nicotine withdrawal. For example, some of the miRNAs identified using the mouse brain samples are not conserved in humans, raising obvious questions regarding their potential roles in the human condition. Further, while there are a plethora of miRNA/mRNA pairs conserved in all mammals, there are specific miRNA/mRNA pairs that are primate-specific38, which would obviously not be detected in our study utilizing mouse brain samples.
It is also of interest to utilize a similar sequencing strategy using other abused drugs (e.g., opioids) to determine whether a similar pattern of miRNA/mRNA regulation occurs in the habenulo-interpeduncular circuit, as it is likely that this circuit is involved in the withdrawal response to other drugs in addition to nicotine. Such a result would inform the rational development of therapeutic interventions.
Materials and Methods
Animals and drug treatment
All experiments were conducted in accordance with the guidelines for care and use of laboratory animals provided by the National Research Council as well as with an approved animal protocol from the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School (UMMS). Wild-type (WT) male C57BL/6 J (Jackson) were group-housed and were kept on a 12-h light/dark cycle (lights ON 7 A.M.) with food and water provided ad libitum. Mice aged 6 weeks were treated with nicotine tartrate (200 μg/ml nicotine base) or equivalent tartaric acid in their drinking water with saccharine (3 g/L) to increase palatability. After 6 weeks, mice in spontaneous withdrawal groups had nicotine bottles replaced with tartaric acid solution. After a 48-hour (Fig. 1A) or 4-week withdrawal period, mice were tested in behavioral assays or sacrificed for tissue collection.
Tissue collection and RNA isolation
Fresh frozen brains were coronally sliced (~1 mm) and regions were dissected using a circular tissue punch (0.75–1 mm diameter). Bregma ranges for each region were as follows: NAc: 1.8–0.8 mm; VTA: (−3.1) – (−3.9) mm; IPN: (−3.2) – (−3.9) mm. Punches were expelled in lysis buffer (miRVana miRNA isolation kit, Ambion). Punches from 4 or 2 brains were combined in each sample for sequencing and RT-qPCR experiments, respectively. Unless otherwise stated, total RNA was isolated using the miRVana kit according to manufacturer’s instructions. RNA concentration and A260/280 were assessed using a NanoDrop2000 (ThermoFisher).
Small RNA library synthesis
Total RNA (approximately 2–9 μg, A260/280 >1.8) was separated on a 15% polyacrylamide, 7 M urea, 1X TBE gel. Small RNA (18–30 nucleotides) was excised and eluted from the gel. Libraries were synthesized using a modified protocol for Illumina TruSeq Small RNA Cloning (April 2014, www.zamorlab.umassmed.edu/protocols/). Briefly, small RNAs were ligated to 3′- and 5′-adapters and cDNA was reverse transcribed by AMV Reverse Transcriptase (NEB). The cDNA libraries were amplified by Accuprime Pfx DNA Polymerase (Invitrogen) for 18 PCR cycles using a common primer and a primer containing a 6-nucleotide barcoding index. The libraries were gel-purified on a 2.5% Low-Range Ultra Agarose (BioRad) 1X TBE gel and eluted using a QIAquick Gel Extraction kit (Qiagen). Library sizes were assessed by Fragment Analyzer (Advanced Analytical, AATI). Library concentrations were determined using KAPA Library Quantification Kits (KAPA Biosystems).
Adapter sequences
3′-Adapter: 5′-rApp/nnntggaattctcgggtgccaagg/ddC/-3′
5′-Adapter: 5′-guucagaguucuacaguccgacgauc-3′
Primers
RT primer: 5′-ccttggcacccgagaattcca-3′
PCR forward primer: 5′-aatgatacggcgaccaccgagatctacacgttcagagttctacagtccga-3′
PCR Index Primers (Barcode):
Index Primer 1: 5′-caagcagaagacggcatacgagatcgtgatgtgactggagttccttggcacccgagaattcca-3′
Index Primer 2: 5′-caagcagaagacggcatacgagatacatcggtgactggagttccttggcacccgagaattcca-3′
Index Primer 3: 5′-caagcagaagacggcatacgagatgcctaagtgactggagttccttggcacccgagaattcca-3′
Index Primer 4: 5′-caagcagaagacggcatacgagattggtcagtgactggagttccttggcacccgagaattcca-3′
Index Primer 5: 5′-caagcagaagacggcatacgagatcactgtgtgactggagttccttggcacccgagaattcca-3′
Index Primer 6: 5′-caagcagaagacggcatacgagatattggcgtgactggagttccttggcacccgagaattcca-3′
Index Primer 7: 5′-caagcagaagacggcatacgagatgatctggtgactggagttccttggcacccgagaattcca-3′
Index Primer 8: 5′-caagcagaagacggcatacgagattcaagtgtgactggagttccttggcacccgagaattcca-3′
Index Primer 9: 5′-caagcagaagacggcatacgagatctgatcgtgactggagttccttggcacccgagaattcca-3′
Index Primer 10: 5′-caagcagaagacggcatacgagataagctagtgactggagttccttggcacccgagaattcca-3′
Index Primer 11: 5′-caagcagaagacggcatacgagatgtagccgtgactggagttccttggcacccgagaattcca-3′
Index Primer 12: 5′-caagcagaagacggcatacgagattacaaggtgactggagttccttggcacccgagaattcca-3′
Index Primer 14: 5′-caagcagaagacggcatacgagatggaactgtgactggagttccttggcacccgagaattcca-3′
Index Primer 15: 5′-caagcagaagacggcatacgagattgacatgtgactggagttccttggcacccgagaattcca-3′
Index Primer 16: 5′-caagcagaagacggcatacgagatggacgggtgactggagttccttggcacccgagaattcca-3′
Small RNA sequencing and analysis
Small RNA libraries were pooled and subjected to single-end (75 cycles) sequencing using a NextSeq. 500 (Illumina) in the RNA Therapeutics Institute at UMMS. Following sequencing, the 3′-adapter sequence was removed using Cutadapt (version 1.11) with ‘–overlap’ option 7 bp. Reads between 15–27 nucleotides long (Fig. S1) were mapped to the UCSC mouse reference genome (mm10) using Bowtie (version 1.1.2) allowing for no mismatches. miRNA reads ±5 nucleotides from the mature, annotated 5′-start position were counted. Reads mapping to multiple locations on a single mature miRNA were recorded as a single count and reads mapped to multiple mature miRNAs were apportioned by the number of the aligned mature miRNAs. Prior to DESeq. 2, read counts were normalized using RUV-Seq55, removing 3–4 factors of variance. In each condition, miRNAs with 0 reads per million (RPM) in more than half the samples were omitted from the analysis. Samples with greater than 1 million mappable reads were used in differential expression (DE) analysis by DESeq. 2 (FDR < 0.01)56.
mRNA library synthesis
RNA >30 nucleotides long was eluted from the same denaturing gel as the small RNA. Libraries were synthesized using the TruSeq Stranded mRNA Library Prep kit (Illumina) according to manufacturer’s instructions. Libraries were gel-purified, assessed by Fragment Analyzer and their concentrations were quantified as described for the small RNA libraries.
mRNA sequencing and analysis
mRNA libraries were subjected to paired-end (76 cycles x 2) sequencing. Reads were mapped to the UCSC mouse reference genome (mm10) using STAR (version 2.5.3a). Following sequencing, mapped reads were quantified with RSEM (version 1.3.0) and RNA-seq libraries with >30 million mappable reads were used in DE analysis by DESeq. 2 (version 1.14.1; FDR < 0.01 and fold change >2). Prior to the DE analysis, read counts were normalized using RUV-Seq (version 1.8.0)55 to remove unwanted variables including batch effects. Genome Ontology (GO) term enrichment for biological processes, molecular function, and cellular component was performed on DE mRNAs using GOseq (version 1.28.0)57. REVIGO was used to reduce GO terms by combining closely related terms58. To provide a well-balanced representation of reduced GO terms, a simRel cutoff of 0.4 was used59. Raw and processed RNA-Seq data are available at the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI) of the National Institutes of Health, accession # GSE117069. Reviewers, please see ‘Data Availability’ section for access token.
Quantitative reverse transcription-PCR (RT-qPCR)
RT-qPCR for miRNAs and mRNAs was performed as previously described30 using TaqMan assays (ThermoFisher) for miR-106b-5p (Assay ID 000442), Pfn2 (Assay ID Mm01289572_m1), and Pfn1 (Assay ID Mm00726691_s1). Relative miRNA/gene expression was determined using the 2−ΔΔCt method60. miRNA and mRNA expression were normalized to snoRNA202 (Assay ID 001232) and β-glucuronidase (Assay ID Mm01197698_m1), respectively, unless otherwise noted.
Target prediction analysis
TargetScan Mouse 7.1 (http://www.targetscan.org/mmu_71/) was used to identify DE miRNAs that are predicted to target conserved MREs within the 3′-UTR of inversely expressed mRNAs. Analysis was limited to miRNAs conserved in vertebrates and mammals.
In Vitro Knockdown of Pfn2 with Lenti-pGIPZ-Pfn2-shRNA-tGFP
The mouse septal cholinergic cell line SN1761 was used to test distinct shRNAs for their abilities to knock down expression of Pfn2. SN17 cells were grown in Dulbecco’s Modified Eagle’s Medium (1X DMEM, Corning) supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. The cells were infected with three lots of commercially available lentiviral particles expressing one of three distinct shRNAs directed against Pfn2 or a scramble shRNA control (Lenti-pGIPZ-shRNA, Dharmacon). The shRNA source clones targeted the following sequences: V3LMM_494366 5′-TTTCCTACAATCATATCTA-3′, V3LMM_494367 5′-TATCCACGTAGCTCTGCCA-3′, V3LMM_494369 5′-CTGATCACAGAACACTTCT-3′. SN17 cells (passage 5) were seeded at a density of 2.5 × 104 cells per well in a 24-well plate and incubated overnight. Cells were transduced with lentivirus (multiplicity of infection 0.4) in technical duplicates using 8 μg/ml polybrene in a total of 250 μl in each well. After 5 hours, an additional 1 ml of complete growth medium was added to each well. Approximately 48 hours after transduction, medium was aspirated and replaced with 0.5 ml of complete growth medium containing 2.5 μg/ml puromycin to select for cells successfully transduced with lentivirus. Puromycin selection was performed for a total of 5 days, changing medium every other day. On day 5, cells were harvested and total RNA was isolated using the RNAqueous Total RNA Isolation Kit (ThermoFisher AM1912), eluting in 60 μl. RNA (125 ng) was reverse transcribed using M-MLV Reverse Transcriptase (ThermoFisher Cat No. 28025013) according to manufacturer’s instructions. Relative expression of Pfn2 was measured by RT-qPCR as described above.
shRNA-mediated inhibition of Pfn2
To inhibit Pfn2 expression in vivo, we employed lentiviral delivery of shRNA targeting Pfn2 (sense sequence 5′-AGAAGTGTTCTGTGATCAG-3′) to the IPN or VTA. Lenti-pGIPZ-Pfn2-shRNA-tGFP (2.37 × 108 TU/ml, V3LMM_494369, Dharmacon) or a control virus expressing a non-silencing shRNA (lenti-pGIPZ-scramble-tGFP, 9.06 × 108 TU/ml, RHS4348, Dharmacon) were injected into IPN or VTA and expressed for 4 weeks prior to all experiments. Stereotaxic injections were performed under aseptic conditions using mice aged 6 weeks as described by Molas et al.34. Virus was injected in the IPN or VTA (unilateral) at coordinates measured from Bregma (in mm, anterioposterior, mediolateral, dorsoventral at 0°): IPN (−3.47, ±0, −4.77), VTA (−3.45, ±0.5, −4.2) using a 26 s gauge 10-μl syringe (701RN, Hamilton) to deliver 300 nl of virus at a controlled rate of 60 nl/min.
Fresh frozen brains were cryosectioned (12-μm), fixed in ice-cold acetone, and dehydrated in ethanol (70%, 90%, 100%) and xylenes. Areas expressing tGFP were isolated by laser capture microdissection (LCM; Arcturus). RNA was isolated using RNAqueous-Micro Total RNA Isolation Kit (Ambion) for RT-qPCR.
Behavioral assays
All behavioral testing was performed during the light phase (8:00 A.M. to 5:00 P.M.) in dim white light after habituation to the testing room (≥30 min). On the first test day, mice underwent the marble burying test followed by the elevated plus maze (EPM) as described by Zhao-Shea et al.16. Approximately 48 hours later, mice underwent object novelty preference testing. Testing was performed in two cohorts of virus-injected mice, with balanced treatment groups by experimenters blind to virus injection. Lenti-virus expression was confirmed by fluorescence microscopy of tGFP. Mice with no virus expression in the brain region of interest were excluded from analysis.
Marble burying
For 2 days prior to testing, mice were habituated to individual standard mouse cages, filled 5–6 cm with bedding material, for approximately 1 hour per day. On the test day, 15 sterilized 1.5-cm diameter glass marbles were placed in the cages, equally spaced in a 3 × 5 grid arrangement. Mice were placed in the test cage for 30 min. then returned to their home cage. The number of marbles buried at least 75% with bedding was counted.
Object novelty preference test
A T-shaped maze (three arms, each 9 × 30 × 20 cm, connected through a 9 × 9 cm central zone) made of white Plexiglas was used to examine interest in familiar and novel inanimate objects under dim red light conditions. First, the mouse was habituated to the apparatus for 5 min and removed. Two identical plastic objects were placed in the ends of the T-maze, one in each end. The mouse was immediately placed back into the maze for 5 min and allowed to investigate the two identical novel objects (N1). The mouse was removed and one object was replaced by a novel object in the same location as the previous object (counter-balanced). The mouse was immediately placed in the T-maze and allowed to investigate the familiar (F1) and novel (N2) objects. All sessions were video recorded from above (HDR-CX4440 camera, Sony) and mouse behavior was tracked automatically using EthoVision XT 11.5 (Noldus Apparatus). A blind experimenter manually scored the times the mouse explored the familiar and novel objects. Exploration was defined as sniffing with the nose directed at the object from a distance of less than 2 cm. Simply sitting or resting next to the object was not counted as exploration. The novel object preference ratio was calculated as: (total novel object investigation – total investigation)/(total investigation) over the third 5-min session. The apparatus and objects were cleaned with Micro-90 solution (International Products Corporation) to eliminate olfactory traces after each session.
Supplementary information
Acknowledgements
We sincerely thank Drs. Phil Zamore and Jennifer Broderick for their help with library generation and Dr. Alper Kucukural for help with data analysis and uploading. This work was supported by award numbers F30DA036312 (APC), U24HG009446 (ZW), R21DA033543 (PDG), and R21DA031952 (PDG and ART). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute for Drug Abuse or the National Institutes of Health.
Author contributions
A.P.C., J.T., R.Z.-S., S.M., A.R.T., Z.W. and P.D.G. designed the experiments. A.P.C., J.T., R.Z.-S., C.B.S. and S.M. performed the experiments. A.P.C., J.T., R.Z.-S., C.B.S., S.M., A.R.T., Z.W. and P.D.G. analyzed the data. A.P.C. and P.D.G. wrote the manuscript. All authors reviewed the manuscript.
Data availability
Raw and processed RNA-Seq data are available at the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI) of the National Institutes of Health, accession # GSE117069.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57907-w.
References
- 1.Peacock Amy, Leung Janni, Larney Sarah, Colledge Samantha, Hickman Matthew, Rehm Jürgen, Giovino Gary A., West Robert, Hall Wayne, Griffiths Paul, Ali Robert, Gowing Linda, Marsden John, Ferrari Alize J., Grebely Jason, Farrell Michael, Degenhardt Louisa. Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction. 2018;113(10):1905–1926. doi: 10.1111/add.14234. [DOI] [PubMed] [Google Scholar]
- 2.Organization, W. H. WHO Report on the Global Tobacco Epidemic, 2011. (2011).
- 3.Kotz D, Brown J, West R. ‘Real-world’ effectiveness of smoking cessation treatments: a population study. Addiction. 2014;109:491–499. doi: 10.1111/add.12429. [DOI] [PubMed] [Google Scholar]
- 4.Jorenby DE, et al. Efficacy of varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, vs placebo or sustained-release bupropion for smoking cessation: a randomized controlled trial. JAMA. 2006;296:56–63. doi: 10.1001/jama.296.1.56. [DOI] [PubMed] [Google Scholar]
- 5.Koegelenberg CF, et al. Efficacy of varenicline combined with nicotine replacement therapy vs varenicline alone for smoking cessation: a randomized clinical trial. JAMA. 2014;312:155–161. doi: 10.1001/jama.2014.7195. [DOI] [PubMed] [Google Scholar]
- 6.Cinciripini PM, et al. Effects of varenicline and bupropion sustained-release use plus intensive smoking cessation counseling on prolonged abstinence from smoking and on depression, negative affect, and other symptoms of nicotine withdrawal. JAMA Psychiatry. 2013;70:522–533. doi: 10.1001/jamapsychiatry.2013.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picciotto MR, Mineur YS. Molecules and circuits involved in nicotine addiction: The many faces of smoking. Neuropharmacology. 2014;76(Pt B):545–553. doi: 10.1016/j.neuropharm.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dani JA, De Biasi M. Mesolimbic dopamine and habenulo-interpeduncular pathways in nicotine withdrawal. Cold Spring Harb Perspect Med. 2013;3:1–13. doi: 10.1101/cshperspect.a012138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molas S, DeGroot SR, Zhao-Shea R, Tapper AR. Anxiety and Nicotine Dependence: Emerging Role of the Habenulo-Interpeduncular Axis. Trends Pharmacol Sci. 2017;38:169–180. doi: 10.1016/j.tips.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes JR. Effects of abstinence from tobacco: valid symptoms and time course. Nicotine Tob Res. 2007;9:315–327. doi: 10.1080/14622200701188919. [DOI] [PubMed] [Google Scholar]
- 12.Jackson KJ, Muldoon PP, De Biasi M, Damaj MI. New mechanisms and perspectives in nicotine withdrawal. Neuropharmacology. 2015;96:223–234. doi: 10.1016/j.neuropharm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koob GF, Le Moal M. Review. Neurobiological mechanisms for opponent motivational processes in addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363:3113–3123. doi: 10.1098/rstb.2008.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen SS, Bade T, Hatsukami D, Center B. Craving, withdrawal, and smoking urges on days immediately prior to smoking relapse. Nicotine Tob Res. 2008;10:35–45. doi: 10.1080/14622200701705076. [DOI] [PubMed] [Google Scholar]
- 15.Zhao-Shea R, Liu L, Pang X, Gardner PD, Tapper AR. Activation of GABAergic neurons in the interpeduncular nucleus triggers physical nicotine withdrawal symptoms. Curr Biol. 2013;23:2327–2335. doi: 10.1016/j.cub.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao-Shea R, et al. Increased CRF signalling in a ventral tegmental area-interpeduncular nucleus-medial habenula circuit induces anxiety during nicotine withdrawal. Nat Commun. 2015;6:6770. doi: 10.1038/ncomms7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen HB, Brown RM, Lawrence AJ. Neuroplasticity in addiction: cellular and transcriptional perspectives. Front Mol Neurosci. 2012;5:99. doi: 10.3389/fnmol.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan H, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Pan T, Zhong X, Cheng C. Nicotine upregulates microRNA-21 and promotes TGF-beta-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014;35:7063–7072. doi: 10.1007/s13277-014-1968-z. [DOI] [PubMed] [Google Scholar]
- 24.Izzotti A, et al. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat Res. 2011;717:9–16. doi: 10.1016/j.mrfmmm.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Izzotti A, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, et al. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol. 2013;272:154–160. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollander JA, et al. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippi G, et al. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan EM, et al. miRNAome analysis of the mammalian neuronal nicotinic acetylcholine receptor gene family. RNA. 2014;20:1890–1899. doi: 10.1261/rna.034066.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Woo J, Kim YS, Im HI. Integrated miRNA-mRNA analysis in the habenula nuclei of mice intravenously self-administering nicotine. Sci Rep. 2015;5:12909. doi: 10.1038/srep12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilo Boyl P, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molas S, et al. A circuit-based mechanism underlying familiarity signaling and the preference for novelty. Nat Neurosci. 2017;20:1260–1268. doi: 10.1038/nn.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devine MJ, Kittler JT. Mitochondria at the neuronal presynapse in health and disease. Nat Rev Neurosci. 2018;19:63–80. doi: 10.1038/nrn.2017.170. [DOI] [PubMed] [Google Scholar]
- 36.Harris JJ, Jolivet R, Attwell D. Synaptic energy use and supply. Neuron. 2012;75:762–777. doi: 10.1016/j.neuron.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 38.Soreq H. Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci. 2015;38:448–458. doi: 10.1016/j.tins.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Shaked I, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Mishra N, et al. Antisense miR-132 blockade via the AChE-R splice variant mitigates cortical inflammation. Sci Rep. 2017;7:42755. doi: 10.1038/srep42755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekenstein U, et al. Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc Natl Acad Sci USA. 2017;114:E4996–E5005. doi: 10.1073/pnas.1701201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Honore B, Madsen P, Andersen AH, Leffers H. Cloning and expression of a novel human profilin variant, profilin II. FEBS Lett. 1993;330:151–155. doi: 10.1016/0014-5793(93)80262-S. [DOI] [PubMed] [Google Scholar]
- 44.Tariq N, Basharat Z, Butt S, Baig DN. Distribution analysis of profilin isoforms at transcript resolution with mRNA-seq and secondary structure in various organs of Rattus norvegicus. Gene. 2016;589:49–55. doi: 10.1016/j.gene.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 45.Gareus R, Di Nardo A, Rybin V, Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J Biol Chem. 2006;281:2803–2811. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- 46.Witke W, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 48.Lamprecht R, Farb CR, Rodrigues SM, LeDoux JE. Fear conditioning drives profilin into amygdala dendritic spines. Nat Neurosci. 2006;9:481–483. doi: 10.1038/nn1672. [DOI] [PubMed] [Google Scholar]
- 49.Michaelsen K, et al. Fine-tuning of neuronal architecture requires two profilin isoforms. Proc Natl Acad Sci USA. 2010;107:15780–15785. doi: 10.1073/pnas.1004406107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Da Silva JS, et al. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitoh A, et al. ROCK inhibition produces anxiety-related behaviors in mice. Psychopharmacology (Berl) 2006;188:1–11. doi: 10.1007/s00213-006-0466-4. [DOI] [PubMed] [Google Scholar]
- 52.Mondin M, Carta M, Normand E, Mulle C, Coussen F. Profilin II regulates the exocytosis of kainate glutamate receptors. J Biol Chem. 2010;285:40060–40071. doi: 10.1074/jbc.M110.140442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mammoto A, et al. Interactions of drebrin and gephyrin with profilin. Biochem Biophys Res Commun. 1998;243:86–89. doi: 10.1006/bbrc.1997.8068. [DOI] [PubMed] [Google Scholar]
- 54.de la Mata M, et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16:500–511. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlicker A, Domingues FS, Rahnenfuhrer J, Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Hammond DN, Lee HJ, Tonsgard JH, Wainer BH. Development and characterization of clonal cell lines derived from septal cholinergic neurons. Brain Res. 1990;512:190–200. doi: 10.1016/0006-8993(90)90626-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-Seq data are available at the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI) of the National Institutes of Health, accession # GSE117069.