Abstract
Course-based undergraduate research experiences (CUREs) using inquiry-based methodology provide a range of positive benefits to undergraduates and instructors. Yet, the required time and cost in designing and running CUREs with detailed data acquisition steps can lead to barriers in CURE implementation. This report describes an alternative approach to CUREs that utilizes free, open access 3D image volumes as data-rich resources for neurobiology CUREs. These open access image volume CUREs (ivCUREs) effectively combine the data acquisition and analysis steps within the course, allowing more time for students to critically evaluate their hypotheses and results, compare data with peers, and reflect on their experiences. Undergraduates in this 10-week ivCURE analyzed >670 excitatory synapses across two brain areas for the presence and origins of spinules within presynaptic boutons, and fully reconstructed 13 of these synapses in 3D. These data highlight the prevalence of these enigmatic synaptic features within excitatory presynaptic boutons, and their potential importance to neuronal function. Moreover, these results underscore key benefits to ivCURE implementation, including the (1) low-cost of experimental design and implementation, (2) ability to utilize the same data-rich image volume across multiple ivCUREs, (3) potential to generate publishable analyses, and (4) flexibility to scale projects and class sizes up at little to no cost. Opportunities for undergraduates to engage in inquiry-based ivCUREs that examine a host of unexplored questions in neurobiology will continue to grow, in parallel with rapid advances in 3D microscopy techniques and the increased availability and diversity of open access image volumes and analytical tools.
Keywords: neuroanatomy, three-dimensional (3D) reconstruction, presynaptic bouton, spinule, spinule-bearing bouton (SBB), CURE, serial section electron microscopy (ssEM), image analysis, cortex, hippocampus, ImageJ
Course-based undergraduate research experiences (CUREs) provide quantifiable benefits in critical thinking, scientific analysis, and confidence to students across diverse STEM disciplines and collegiate settings (Lopatto, 2004; Seymour et al., 2004; Thiry et al., 2012; Corwin et al., 2015; Staub et al., 2016), and national scientific panels have recommended increased inquiry-based CURE implementation in order to transcend barriers to STEM excellence in the U.S. (American Association for the Advancement of Science [AAAS], 2011; President’s Council of Advisors on Science and Technology, 2012; National Academies of Sciences [NAS], 2015). Yet, the required time and cost in designing and running CUREs can lead to barriers in CURE implementation, especially at primarily undergraduate institutions where faculty may have minimal research infrastructure, laboratory staff, and/or have heavier teaching responsibilities (Kowalski et al., 2016; Shortlidge et al., 2016; Staub et al., 2016). Moreover, the time required for data acquisition during CUREs can limit the available time for CURE elements deemed crucial to realizing student benefits, such as discovery-based literature review, analytical iteration, and data interpretation (AAAS, 2011; Auchincloss et al., 2014; Staub et al., 2016).
Here I present an alternative approach to CURE implementation that utilizes free, open access image volumes as flexible and data rich resources for neurobiology CUREs. These downloadable image volumes span a range of scales (e.g., synapses - circuits), imaging modalities (e.g., fluorescence microscopy, electron microcopy), and targets (e.g., retinal amacrine cells, CA1 hippocampal synapses and circuits). By using these image volumes, students are able to: (1) Propose a hypothesis based on outstanding questions within the relevant literature, (2) Develop their own analytical protocol, (3) Iteratively analyze their image volume for multiple criteria, and (4) Interpret, present, and reflect on their data with reference to foundational literature. By effectively combining the data acquisition and analysis steps, CUREs using open access image volumes (ivCUREs) purposefully allow more course time for students to critically evaluate their underlying hypothesis and results. Thus, ivCUREs may encourage students to develop a deeper understanding of the rationale behind their analyses and their relevance to open questions within the literature (Auchincloss et al., 2014). Importantly, while many of these image volumes have been used in publications, instructors can guide undergraduates in ivCUREs toward a host of outstanding questions that remain unexamined within each data-rich set of images.
This report details the flexible course structure, methodology, and range of available resources for running ivCUREs, with a particular emphasis on courses utilizing serial section electron microscopy (ssEM) image volumes. In addition, I describe the learning objectives, assessments, outcomes, and student evaluations from an ongoing ssEM ivCURE with upper-level biology majors at a primarily undergraduate institution. Major benefits to utilizing open access image volumes in ivCUREs include increasing the availability of course time to develop critical thinking skills, the low-cost of experimental design and implementation, the potential to generate publishable analyses and 3D models, and the flexibility to scale projects and class sizes up at little to no cost. A potential challenge to ivCURE implementation can be the time necessary to train students on image visualization and analysis software, however this challenge can be mitigated by selecting simpler and more user-friendly image visualization/analysis software platforms such as those suggested in this report. All of the image analysis and 3D visualization software used in the ivCURE described in this report are open source platforms amenable to undergraduate training and use within the time constraints of most quarter or semester-long CUREs.
COURSE STRUCTURE & RESOURCES
Course Enrollment and Length
ivCUREs at UW Tacoma (a primarily undergraduate institution with a majority of first-generation students) are restricted to six (academic year) or seven (summer) junior and senior undergraduate Biomedical Sciences majors (includes all Biology, Chemistry, Physics, and Health Sciences interested students). Enrollment in these 10-week (i.e., one quarter), three credit ivCUREs is by instructor permission, allowing an instructor to screen undergraduate students for their level of interest and experience before admittance. However, most students who enroll in ivCUREs at UW Tacoma have little neurobiology background other than that gained in a few class sessions in an Introductory Biology and Human Physiology course. In addition, given the low-cost (essentially computer lab time and data storage) of running ivCUREs, these courses can be easily scaled up based on the specific constraints of the institutional unit, instructor, and ivCURE goals.
In addition to flexibility in course size, ivCUREs can be run as shorter 8 to 10-week courses, or semester-length 14 to 15-week courses based on the goals of each particular ivCURE. UW Tacoma ivCUREs meet once a week for 6 hours each session, with meetings taking place in a campus computer lab with presentation capabilities (Figure 1). Longer course times allow students to learn and practice image analysis skills and software routines while under instructor supervision. That is, a central goal for ivCURE course sessions is for students to acquire these skills, pinpoint their specific challenges in the analysis pipeline, and receive iterative instructor feedback. Thus, shorter (e.g., two – three hour) course periods that repeat two to three times a week would also be amenable to an ivCURE design. As the course progresses students are able to do more of their analyses on their own and course time can become more flexible. For our 3 credit, 6-hours/week ivCUREs, students are expected to do at least 3 additional hours/week of course work on their own outside of class time (e.g., literature review, image analysis).
Figure 1.
ivCURE classroom at the UW Tacoma. This particular computer lab contains 40 student desktop computers and a presentation-capable computer for the instructor. ivCUREs can be scaled up or down based on computer lab constraints and the requirements of the individual instructor and academic unit.
Classroom and Equipment Requirements
ivCUREs at UW Tacoma are run in computer labs containing a desktop computer for each student and an instructor computer with presentation capabilities (Figure 1). The scope of ivCURE topics will dictate the specific requirements for computer processing speed, graphics cards, and random-access memory (RAM). However, most courses will not require specialized computers or large alterations to existing infrastructure. The desktop computers we use in our ivCUREs run on Intel Core i7-6700 3.40 GHz CPUs with 16 GB of internal RAM, 1 TB hard drives, and standard graphics cards. For reference, students often analyze 0.5 – 3 GB image volumes, and the 3D reconstruction software we run has a minimum requirement of 128 MB of RAM. It is recommended that these computers run Microsoft Windows 10 or later operating systems, as some open source reconstruction software platforms (e.g., Reconstruct) are only available for Windows-based systems. In addition, students in our ivCUREs save and access their data on a UW Tacoma administered cloud server (i.e., Microsoft OneDrive), allowing them to continue their data analyses from home. Other free options for real-time data sharing exist (e.g., Google Drive, Dropbox), but there can be limits on available free space in these alternative. It may also be possible to store and share data in the course’s course management system site (e.g., Canvas, Blackboard), but this option may also be space and speed restricted. It is recommended that students and instructors have real-time access to course analyses as it allows students to stop and start their work to accommodate their schedules and permits students and instructors to iteratively correct and amend analyses.
ssEM Image Primers, Repositories, and Project Ideas
Importantly, instructors need not be experts in electron microscopy to run an ssEM ivCURE as multiple ssEM image primers can guide first-time instructors and their students in ssEM analysis. For example, Microscopy Australia has a wonderful primer on the fundamentals of transmission and scanning electron microscopy (https://myscope.training). In addition, Dr. Kristen Harris has developed an accessible tutorial on neuronal morphology within ssEM images and 3D neurite reconstructions on her lab’s SynapseWeb site (http://synapseweb.clm.utexas.edu/tutorials) and this site also hosts Dr. Josef Spacek’s Atlas of Ultrastructural Neurocytology, a comprehensive primer for learning how to recognize neurites, glia, synapses, and subcellular organelles within electron micrographs (http://synapseweb.clm.utexas.edu/atlas).
In designing ivCUREs instructors have an array of image repositories to choose from, each hosting a diverse collection of image stacks from a range of model species, imaging modalities, and scales. This report focuses on using ssEM image volumes to investigate the subcellular ultrastructure of the brain. ssEM images contain a host of nanoscale organelles and ultrastructural components that can encourage undergraduate hypotheses and analytical strategies. Moreover, the sizes and morphological characteristics of many of these structures correlate with measures of activity, neuronal development, and neuronal subtype. A listing of some potential ssEM ivCURE topics and quantitative criteria are listed in Table 1 and an abridged listing of open access image volume repository sites can be found in Table 2. Importantly, the selection of course image volumes and topics can be driven by instructor interest or area(s) of research, fueling quality data analyses toward publishable manuscripts.
Table 1.
Sample of potential ssEM ivCURE topics and the quantifiable subcellular structures that can be analyzed and 3D reconstructed within each topic area. Correlative measures of synaptic strength and plasticity can be investigated within unexplored brain areas or neuronal subtypes, while measures of synapse specific morphology and many other 3D subcellular relationships remain largely uncharacterized and ripe for ‘authentic’ ivCURE research projects.
| Course Focus | Quantitative Criteria |
|---|---|
| Correlates of Synaptic Plasticity, Strength, and Stability |
|
| Synapse Specific Morphology |
|
| Underexplored 3D Subcellular Relationships |
|
Table 2.
Abridged Listing of Cellular and Subcellular Neuronal Image Volume Repositories.
| Image Repository | Model Species | Available Projects* | Imaging Modalities | Website |
|---|---|---|---|---|
| Open Connectome Project | Human, Rat, Mouse, Zebrafish, Fly, Worm | 24 | CM; COLM; AT; ssEM; XMT; ATUM SEM; SBEM | https://neurodata.io/ocp |
| UCSD Cell Image Library | Human, Cat, Chicken, Mouse, Rat, Fly | 40 | CM; CryoTM; CLEM; MRI; ssEM; SBSEM | http://ccdb.ucsd.edu/pages/datasets |
| Allen Brain Connectivity Atlas | Mouse | 3 | CM Tract tracing; InSitu | http://connectivity.brain-map.org/projection |
| CA1 Hippocampus Dataset | Rat | 1 | ssEM | https://www.epfl.ch/labs/cvlab/data/data-em/ |
| FlyEM | Fly | 7 | ssEM | https://www.janelia.org/project-team/flyem/tools-and-data-release |
| WormImage | Worm | Numerous | LM; ssEM; SEM | https://www.wormimage.org/ |
| Neuroscience Information Network (NIH-funded resource catalog) | Various | Numerous | Various | https://neuinfo.org/ |
LM = Light Microscopy; CM = Confocal Microscopy; COLM = CLARITY-Optimized Light-sheet Microscopy; AT = Array Tomography; ssEM = Serial Section Electron Microscopy; XMT = X-ray Micro-Tomography; ATUM SEM = Automated Tape-collecting Ultramicrotome Scanning Electron Microscopy; SBSEM = Serial Block-face Scanning Electron Microscopy; CryoTM = Cryogenic Tomography Electron Microscopy; InSitu = In situ Hybridization;
= most projects have multiple open access image volumes.
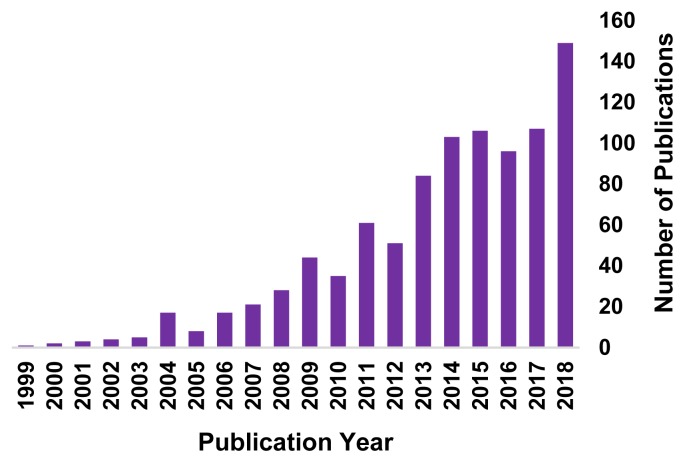
Currently, the Open Connectome Project and UCSD Cell Image Library repositories (see Table 2) contain some of the largest collections of ssEM neuronal image volumes. Most of these are image volumes from rodent cortex (i.e., primary visual and somatosensory cortex), rodent CA1 hippocampus, and C. elegans head. All of the example projects listed in Table 1 will work with one or more of the image volumes currently available in these repositories. However, given the rising popularity of 3D ssEM techniques (Figure 2), the number and diversity of open access ssEM image volumes available in image repositories seems poised to increase.
Figure 2.
Number of publications per year (1999–2018) using either focused ion beam scanning electron microscopy (FIBSEM) or serial block face scanning electron microscopy (SBFSEM), techniques that generate ssEM image volumes. Data from pubmed.gov.
Visualizing and Downloading Open Connectome Project 3D Image Stacks
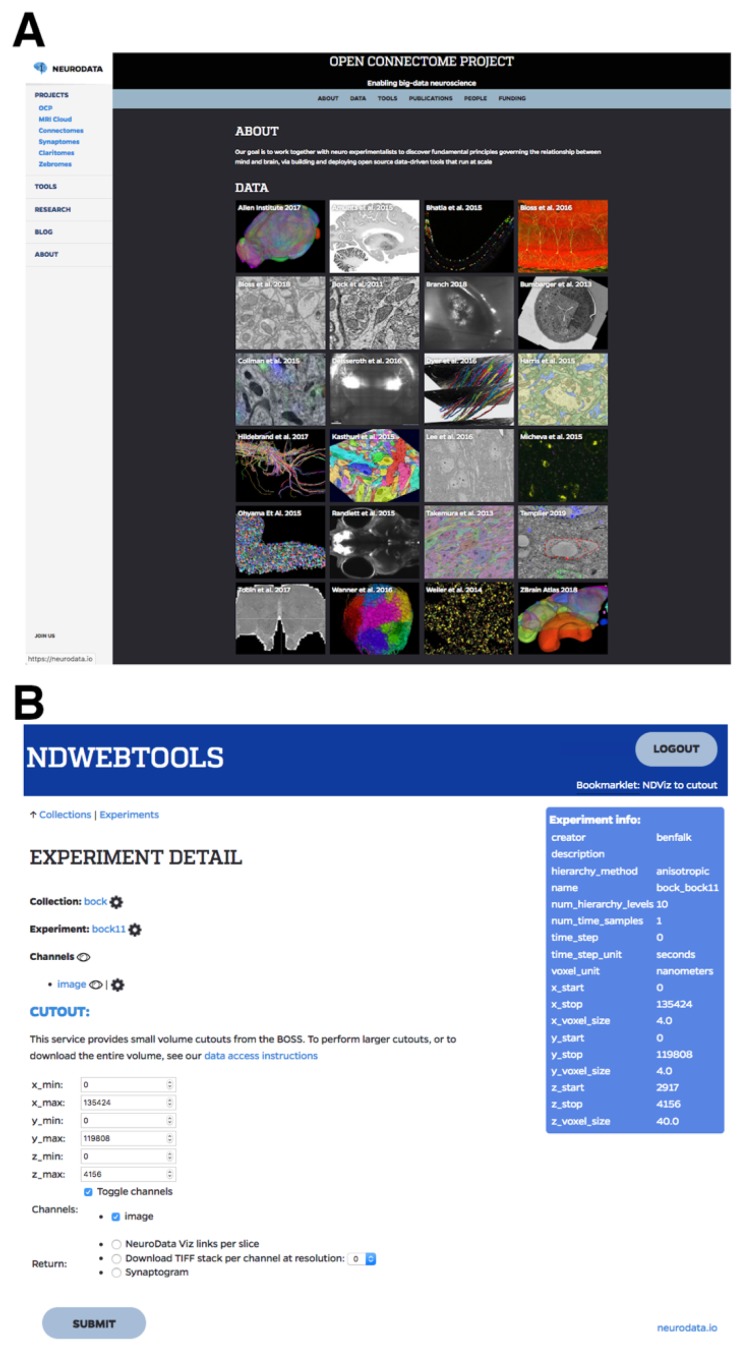
Since a majority of the available high-resolution 3D ssEM image volumes are housed on the Open Connectome Project site, a brief introduction to using this repository follows. First, instructors should sign up for a free account (https://api.boss.neurodata.io) in order to visualize and download the available image volumes. Next, image volumes can be visualized or downloaded by following instructions on the neurodata help page (https://neurodata.io/help/download/). Instructors can use the neuroglancer tool (https://neurodata.io/help/visualization/) to visualize a stack and determine the x/y/z coordinates that contain the data of interest. Once logged in to the site, a simple way to visualize any stack directly is to click on the project of interest on the open connectome home page (https://neurodata.io/ocp/), and then click on the ‘eye’ icon (Figure 3). Once ported to the neuroglancer tool, instructors often have to click the “Request Authorization” button in the lower-left corner of the window. When using neuroglancer, pressing the ‘H’ key will bring up a list of helpful keyboard shortcuts and pressing the ‘esc’ key will make this list disappear. Once x/y/z coordinates have been determined, instructors can download small (< 1 GB) stacks using the ndwebtools tool (Figure 3) by clicking on the project of interest on the open connectome home page, and then clicking on the name of the stack (next to the eye icon). If desired, larger image volumes can be downloaded by either, (1) iteratively downloading small image volumes and concatenating them in the Fiji (Schindelin et al., 2012) (i.e., Image>Stacks>Tools>Concatenate), or (2) using the ndex Python command-line tool to download entire image volumes (https://github.com/neurodata/ndex/).
Figure 3.
Open Connectome Project (OCP) Website Interfaces. A. OCP site for the selection of an open access 3D image volume project (https://neurodata.io/ocp). B. OCP tool (ndwebtools) interface for downloading small sections (< 1 GB) of a larger image volume.
ivCURE CASE STUDY: SYNAPTIC SPINULES
Course Topic and Schedule
This specific ivCURE centered on characterizing the prevalence and origins of synaptic spinules within cortical and hippocampal presynaptic boutons. Spinules are finger-like projections that originate from neurites and glia and can become enveloped by a synaptic bouton (Tarrant and Routtenberg, 1977; Spacek and Harris, 2004), greatly increasing the membrane interface and potential for communication and stable anchoring between these structures. While the percentages of neurites that project spinules have been characterized in CA1 hippocampus (Spacek and Harris, 2004), the relative prevalence and origins of spinules embedded within cortical and hippocampal presynaptic boutons remained unknown.
The rationale for choosing spinules as a course topic for this ivCURE were that, (1) the instructor has an ongoing research program investigating spinules and the ability to analyze and reconstruct these fine structures in 3D was highly desired, (2) since there are so few investigations (and no published 3D accounts) into these potentially ubiquitous synaptic structures within excitatory boutons, undergraduates had the opportunity to form their own hypotheses on the nature of spinules and ‘test’ them through their 3D analyses, and (3) based on the previous two factors, motivated undergraduates had the potential to make substantial contributions and earn authorship on published manuscripts.
In this ivCURE, students were first able to choose a brain area of interest (i.e., primary visual cortex or CA1 hippocampus) to analyze and then performed a guided literature search to determine what was known about SBBs in that brain area. Next, students worked in pairs to define the open questions in the literature on which to formulate their hypotheses. Students were then trained on how to analyze ssEM images for the presence of these structures (see “Undergraduate Analysis and Reconstruction Training” below) and how to measure and reconstruct these elements using image analysis and 3D reconstruction software. Next, students were given their own image stacks to analyze and tasked with interpreting their findings in light of the relevant literature. Student projects included determining: (1) the percentages of presynaptic boutons from primary visual cortex and/or CA1 hippocampus that contain spinules, (2) the percentages of neurites/glia that send spinules into these presynaptic boutons, and (3) the 3D relationship of spinule-bearing boutons (SBBs) to their spinules and postsynaptic partners. Examples of future projects that emerged from these analyses are the percentage of spinules within inhibitory boutons, the potential preference for individual axons to contain more SBBs than other axons, and the range of morphologies (e.g., % volume, depth of penetration, tortuosity) of spinules within different sized boutons.
This ivCURE was run as a 3 credit 10-week course that met once per week for 6 hours per meeting. The course schedule was as follows: Weeks 1 – 2: Introduction to neuroanatomy, ssEM analysis, and general course topic; Weeks 3 – 4: ssEM analysis and interrater reliability assessment, 3D reconstruction software training, and course hypothesis formulation; Weeks 5 – 6: Troubleshooting individual analyses; Weeks 7 – 8: Self-directed analysis and guided data interpretation; Weeks 9 – 10: Data interpretation and Final Project Preparation. In addition, ivCUREs were linked with a Biomedical Sciences major bookend course in which students were required to prepare scientific abstracts and posters of their ivCURE analyses, culminating in a campus-wide research symposium where UW Tacoma Sciences and Mathematics majors presented their posters to the community.
Learning Objectives and Formative Assessments
This course’s three main learning objectives were for students to: (1) Demonstrate a detailed understanding of neuronal anatomy, with an emphasis on the ultrastructure of excitatory and inhibitory synapses and synaptic spinules, (2) Acquire practical skills in quantitative image analysis and 3-D reconstructions by learning how to use ubiquitous image analysis platforms (i.e. Fiji and Reconstruct) to analyze neuronal anatomy, and (3) Logically interpret ssEM image analyses with reference to the published literature. These learning objectives were aligned with the course’s three main formative assessments.
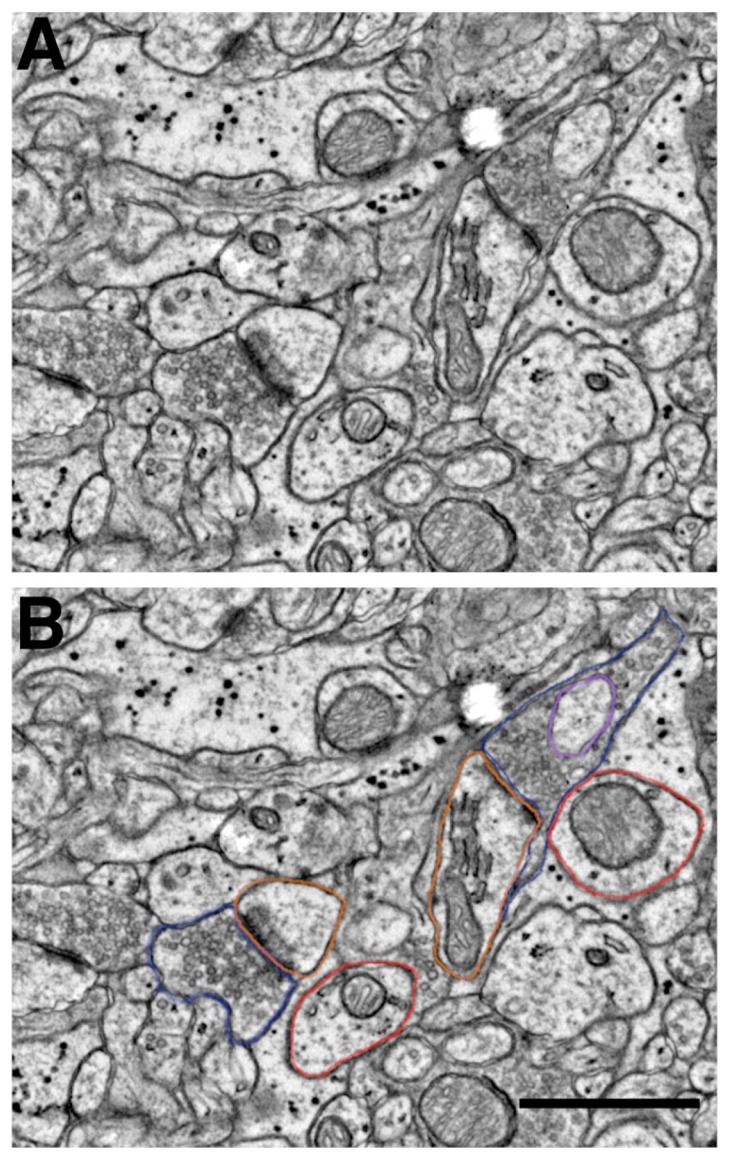
Specifically, students received 15% of their grade for in-class participation and peer discussion of image analyses. In the first three to four weeks of the course students had to learn to interpret and analyze ssEM images for the presence of synaptic structures. During this training period, students were given a series of training images (i.e. single sections from an ssEM image stack) to analyze in class and were asked to discuss the similarities and differences they encountered in their analyses (Figure 4). By comparing their results to the annotated image ‘answer key’ provided by the instructor, students began to fine tune their analytical skills and increase their image analysis interrater reliability. Therefore, in-class participation in this ivCURE was weighted relatively heavily in terms of the total course grade. Missing a classroom analysis peer discussion without a valid excuse resulted in a 3% loss of this participation grade per instance.
Figure 4.
ssEM Student Training Image with ROIs. A. Representative area within a ssEM training image, showing two prominent excitatory synapses. B. Identical training image area with ROIs shown around excitatory presynaptic boutons (blue), postsynaptic spines (orange), spinule (purple) within SBB, and putative dendrites (red). Note the large ‘flattened pancake-shaped’ spine apparatus in the larger spine toward the top of the image. Scale bar = 0.25 μm (for A & B); Raw data from Bock et al. (2011).
In addition, students were assessed on their ability to complete their assigned training image analyses or assigned number of synapses per week and upload an excel sheet containing these results to our course site. Completing and uploading these analyses were worth 25% of a student’s total grade, and failure to complete or upload an analysis resulted in a 5% loss in a student’s grade per event. The culmination of the course assessment was the completion of the final analysis project. For this specific ivCURE, the final project included analyzing ≥ 90 excitatory synapses in a student’s chosen image volume, and fully reconstructing two of these synapses in 3D. This final project was worth 60% of a student’s grade. However, students were able to continually correct and amend their analyses and reconstructions over the course of ~6 – 7 weeks with instructor supervision, such that in practice the final project became more of a formative assessment rather than a summative assessment. Students could lose 5% per day of their total grade for turning in their final projects after the final due date and could be assessed a 5 – 20% reduction in their grade based on a failure to analyze a commensurate portion of their analysis or failure to complete their 3D reconstructions.
Undergraduate Analysis and Reconstruction Training
Upper-level Biomedical Science majors in this ivCURE, most without any direct neurobiology experience, were first trained to recognize neurite and synapse cross sections in digital ssEM images. On the first day of class, the instructor gave a brief overview of basic neuroanatomy and students participated in informal quizzes that gauged their previous knowledge of neuronal and synaptic components. For example, one question asked students to “Draw a synapse and label the major components”, and another question asked students to “Describe the flow of information from one neuron to another in your own words”. These quizzes were think-pair-share activities, wherein students completed these quizzes individually, compared their answers with their neighbor, and then came up in pairs to draw/write their answers on the board. These quizzes unmasked latent misunderstandings that were corrected in interactive, informal settings.
After reviewing neuroanatomy, students were introduced to a 2D ssEM training image and the image analysis software platform Fiji (Schindelin et al., 2012). First, students were shown various cross sections of neurites within a digital electron micrograph that were surrounded by a region of interest (ROI) in Fiji (Figure 4). Students were asked to guess the identity of individual neuroanatomical components (e.g., myelinated axon, bouton, dendrite, dendritic spine, soma) within the ROIs. Learning to identify neurites from their cross-sections and being able to grasp how these 3D neurites would project from their 2D cross-sections was one of the central goals of the first two course meetings. Toward this end, all students were given the same ssEM training images to analyze during the first two course sessions.
Briefly, students were shown how to use the ‘peanut’, aka freehand tool in Fiji to draw ROIs around the presynaptic bouton and postsynaptic objects forming excitatory synapses. Students were given criteria for positively identifying excitatory Gray’s Type I synapses (Gray, 1959) within the neuropil, namely that, (1) the pre- and postsynaptic compartments had parallel membranes, (2) there was a prominent asymmetric postsynaptic density, and (3) there were at least three synaptic vesicles opposite the postsynaptic density within 120 nm of the presynaptic membrane. In addition, students were shown how to use the freehand line tool in Fiji to measure the postsynaptic density length, and the straight line tool to draw lines across the postsynaptic density and adjacent membranes to compare their line profile plots (i.e., control/command + K in Fiji). Students used these profile plots to determine the width and electron opaque ‘darkness’ of putative asymmetric densities and compare them to either nearby negative control membranes, or to putative inhibitory symmetric postsynaptic densities. Asymmetric postsynaptic densities that measured ≥ 2-fold wider than putative symmetric densities and ≥ 1.5-fold darker at their centroid than negative control membranes were positively identified as asymmetric postsynaptic densities. Thus, students were able to double check each putative excitatory synapse against quantitative criteria, and mark down their questions regarding these assertions for discussion during class.
Students were also taught how to recognize the double membrane-bound cross-sections of spinules within presynaptic boutons and then how to ‘follow’ these spinules back to their parent neurites in 3D ssEM image volumes. Within 2D cross-sections and 3D image stacks, dendrites were identified based on their orderly arrangement of microtubules and the presence of mitochondria. Axons were distinguished based on their smaller size (versus dendrites), presence of mitochondria, potential presence of synaptic vesicles, and potential presence of electron dense myelin. Spine 2D cross-sections were distinguished from dendritic shaft cross-sections based on their diffuse ‘fuzzy’-looking actin, lack of mitochondria (with some exceptions), and potential presence of a spine apparatus (Figure 4).
After two weeks in the course, students were able to use Fiji to reliably assign ROIs to the components of excitatory synapses. During these course sessions, students’ ROIs were overlaid on each other within each training image in order to visualize ROI correspondence and divergence. Students were then encouraged to use the course’s quantitative rules in order to explain their rationale in identifying each neurite cross-section. These feedback sessions helped students reach ~90% interrater reliability for excitatory synapse, postsynaptic identity, and spinule origin identification after ~ 4 weeks. Students were able to reference previously annotated ssEM images for each of their four training images in order to check their analyses. It can also be helpful to offer a reference text (e.g., Peters et al., 1991) and/or a neuronal ultrastructure primer (e.g., http://synapseweb.clm.utexas.edu/atlas) for students to refer to while analyzing their assigned image volumes.
During the third week of class, students in this ivCURE began training on Reconstruct visualization and 3D reconstruction software (Fiala, 2005). Reconstruct is a user-friendly open source software platform (synapseweb.clm.utexas.edu/software-0/) where students can import a portion of a ssEM image stack containing objects of interest (e.g., excitatory SBBs), align these images (if necessary), draw ROIs around objects of interest, and reconstruct these objects in 3D. Notably, since students were already trained in how to identify neurites and synapses within the neuropil and create ROIs, Reconstruct training progressed relatively fast in this course. In addition, as most of the currently available open access image stacks are already well aligned, students only had to learn how to: (1) crop out an area of interest from their image stack in Fiji by using the rectangle tool, (2) import and scale this image stack in Reconstruct, (3) draw ROIs around their objects of interest, and (4) have the software automatically reconstruct their drawn ROIs into 3D models. The process of drawing ROIs in Reconstruct is very similar to that in Fiji, and the Reconstruct site has plentiful resources to teach beginners the basics and troubleshoot problems using the accessible manual (http://synapseweb.clm.utexas.edu/software-0/). In this ivCURE, students were able to reconstruct portions of their SBB-containing excitatory synapses in 3D within the first day of exposure to the program.
Hypothesis Formulation & Data Quantification
In the fourth week of this ivCURE, students were asked to read portions of peer-reviewed publications on spinules in hippocampus and cortex and come to class with at least five questions about spinules (e.g., origins, functions). Following postsynaptic spines vs. non-synaptic adjacent dendrites) that send spinules into these presynaptic boutons, and (3) the 3D relationship between an invaginating spinule and its SBB.
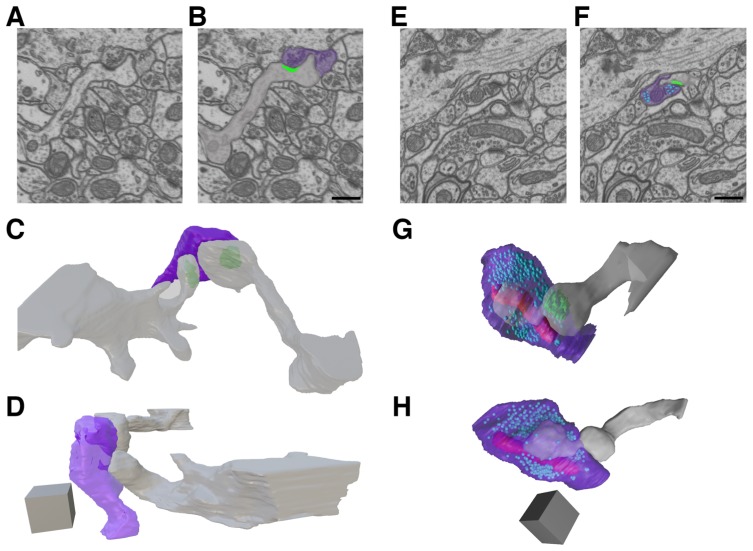
Students across the course were evenly split (three pairs per brain area) in their preference to analyze portions of ssEM image volumes from either primary visual cortex or CA1 hippocampus. Moreover following their discussion, students chose to analyze some key open questions in the spinule literature, namely: (1) the percentages of excitatory presynaptic boutons within these brain areas that contain spinules, (2) the percentages of specific neurites (e.g. Within this ten week course, students were able to analyze 678 synapses in 3D from portions of two previously an in-class discussion of student questions, students were paired up based on the similarity of their interests and asked to formulate hypotheses on one or more gaps in the spinule literature. Paring students up and assigning them smaller portions of a larger image volume permits a test of interrater reliability across measured variables and allows students to compare and troubleshoot their specific set of analyses with their peers. published primary visual cortex volumes (Bock et al., 2011; Lee et al., 2016) downloaded from the Open Connectome Project (https://neurodata.io/ocp/), and a portion of one previously published volume of rat CA1 hippocampus downloaded from the École Polytechnique Fédérale de Lausanne (EPFL; https://www.epfl.ch/labs/cvlab/data/data-em/). In addition, during this 10-week course students were able to fully reconstruct a total of 13 SBBs in 3D, including their postsynaptic partners and the parent neurite(s) that projected spinules into these SBBs (Figure 5).
Figure 5.
ivCURE Student 3D Reconstructions of SBBs in CA1 Hippocampus. A – B. A single ssEM section (A) and its pseudo-colored copy (B) showing a postsynaptic spine (gray) invaginating a spinule into its presynaptic bouton partner (purple). Note the postsynaptic density (green) at the edge of the spinule. C and D. Full reconstruction of the synapse in A – B from 311 serial ssEM sections, showing this SBB (purple) engulfing spinules from two postsynaptic spines (gray). E – F. A single ssEM section (E) and its pseudo-colored copy (F) showing a postsynaptic spine (gray) projecting a spinule into its presynaptic bouton (purple). Note the presynaptic vesicles (light blue), and the postsynaptic density (green) at the edge of the spinule. G – H. Full reconstruction of the synapse in E and F from 264 serial ssEM sections, showing this SBB (purple) with a large anchor-like spinule from its postsynaptic spine (gray). Note the mitochondria (red) and presynaptic vesicles (light blue) within this SBB. Scale bars for A, B, E, and F = 0.5 μm; Scale cubes for C, D, G, and H = 0.5 μm/side (0.125 μm3). Raw data from EPFL (https://www.epfl.ch/labs/cvlab/data/data-em/).
Each pair of students analyzed half of a ~300–600 image stack. Cortical image stacks had 4 × 4 × 40 nm image resolution, whereas the CA1 hippocampus image stack had a 5 nm isotropic pixel resolution. In their analyses, each excitatory presynaptic SBB that students encountered was given a region-of-interest (ROI) name that described its identity. For example, the first SBB encountered that had a synapse onto a spine (“s”) and contained a spinule emanating from its postsynaptic spine (“PSs”) would be given the name: “SBB1s_spinPSs”. Students then compiled their results in an Excel spreadsheet template provided by their instructor. During each course session, the instructor met with students one-on-one in order to answer individualized questions as well as to randomly sample their 3D analyses for validity and reliability. Each student then revised their analyses for the next course session, and this random sampling was repeated until their analyses were finalized.
Results were quantified by calculating the number of excitatory SBBs divided by the total number of excitatory boutons within an image stack, and the number of spinules that emerged from each identified source divided by the total number of SBBs encountered. In addition, 3D reconstructions of excitatory SBBs, spinules, and postsynaptic partners were performed using the open source software Reconstruct (Figure 5). Following student generation of their reconstructions in Reconstruct, the instructor has the option of exporting these 3D object files to the open access software program Blender (www.blender.org) to remesh their surface profiles and augment their lighting if desired.
Students’ analyses revealed that between 26–66% of excitatory boutons within rodent cortex and CA1 hippocampus contained embedded spinules. These data represent one of the first reports on the prevalence of these enigmatic synaptic structures within the excitatory bouton population in the brain. Indeed, these data suggest that spinules are relatively common features of excitatory synapses across the mammalian brain. Furthermore, students found that a majority of spinules within cortical and hippocampal SBBs originate from postsynaptic spines and nearby axons/boutons, identifying these spinule-types as prime targets for future investigations. In addition, students’ 3D reconstructions revealed a range of spinule morphologies embedded within SBBs, from smaller clathrin coated invaginations (Figure 5A – D; Spacek and Harris, 2004), to large anchor-like structures emanating primarily from postsynaptic spines (Figure 5E – H).
Student Evaluations
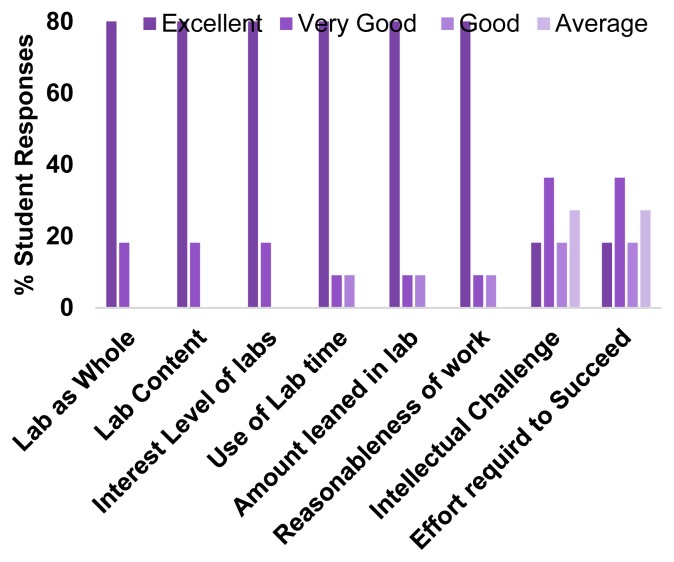
Salient quantitative responses from anonymous end of term student evaluations (11 responses out of 12 total students) from two ssEM ivCUREs were averaged in order to assess student experiences with ivCURE usefulness, intellectual engagement, and temporal efficiency (Figure 6). These evaluations indicated that students enjoyed these research experiences, found the content matter engaging, and acquired new skills in lab. In addition, a majority of students reported that the course’s intellectual challenge and effort required to succeed were “excellent” to “very good”. Overall, students rated ivCUREs a 4.9 out of 5 on an overall summative rating scale (≥ 90th percentile for course ratings at UW Tacoma), with an average challenge and engagement index of 5 (out of 7). The challenge and engagement index is a summative measure of the perceived course challenge and level of student engagement. Thus, ivCUREs involve undergraduates in research that they feel is engaging and intellectually stimulating. Furthermore, a majority of students report an “excellent” use of lab time, despite a relatively long once/week six-hour class session.
Figure 6.
Averaged Student Evaluations from two ivCUREs. End of term student evaluations on representative measures of overall course effectiveness, lab efficiency, and intellectual challenge.
Moreover, in response to the qualitative evaluation prompt “Was this class intellectually stimulating? Did it stretch your thinking?”, students’ responses backed up their quantitative assessments of the course. For example, some representative student responses to this prompt were:
“We were presented with a difficult material of analyzing synapses …and it required a lot of thinking at times because there were no set answers,”
“I learned a lot of new things like how to use a new program ‘fiji’ and how to identify and label neurons,”
“Yes – especially the paper we reviewed required us to interpret difficult figures.”
Thus, while ivCURE students are tasked with acquiring new skills in ssEM neurite identification and image analysis and challenged to place their research within the context of the existing literature, students find these aspects of the course both valuable and intellectually stimulating.
DISCUSSION
Given the gains in graduation rates (Rodenbusch et al., 2016), knowledge acquisition (NAS, 2015), and critical thinking skills (Brownell and Kloser, 2015) for undergraduates who participate in CUREs with ‘authentic’ research experiences, it is important to develop diverse sets of inquiry-based experimental protocols and resources that serve students across an array of collegiate environments. This report describes ivCUREs, inquiry-based research courses where students investigate their own hypotheses relating to morphological variables within open access image volumes.
Notably, this ivCURE was run using inquiry-based/research-based framework (Weaver et al., 2008), wherein students were guided to determine the best analytical strategies for their particular data sets, and where the results of the experiment were unknown. As such, undergraduates in these CUREs had to interpret their own data in light of the relevant literature, compare and contrast their data with their ivCURE peers, and reflect on their experience in light of their conceptions of the field and their future career paths – recommended components for realizing students gains across a range of inquiry-based CUREs (Weaver et al., 2008; Auchincloss et al., 2014; Corwin et al., 2015; NAS, 2015).
ivCUREs using ssEM open access image volumes have many advantages, including their evinced level of student engagement, low-cost of operation, and potential for generating publication-quality analyses. Students in these courses are excited to be the first to analyze and reconstruct particular synaptic elements within a circuit, especially since they can visualize the ‘fruits of their labors’ in the form of 3D models. Indeed, multiple ivCURE students have been motivated to print their 3D synapse reconstructions on 3D printers, and ~30% have transitioned to working as research assistants in neurobiology laboratories. However, some students with less exposure to software-based analysis can struggle in the initial weeks of an ivCURE to become fluent using the required software platforms. For this reason, it is particularly important to stress to students that asking questions early and often of the instructor is crucial for success, and that all questions are important and valid when learning these new skills. Students should be continually encouraged to ask questions of the instructor during class, and class discussion boards and FAQ lists on the course learning management software can help crowd source answers to common software issues.
Importantly, undergraduate analyses from ivCUREs can shape the questions and analytical frameworks for future ivCUREs at each institution. Indeed, consecutive ivCUREs can be run on a single topic (e.g. spinule prevalence and morphology) across multiple course sessions due to the rich set of 3D variables that can be quantified within each image volume. Since a published manuscript can only focus on some finite set of variables within an image stack, opportunities will persist for undergraduates to engage in inquiry-based ivCUREs that examine a range of unexplored questions within open access image volumes. Moreover, since the number and diversity of these image volumes within open access repositories and respective analytical tools are increasing (Figure 2; Berger et al., 2018; Scheffer, 2018), the potential to examine research-based neurobiological questions within ivCUREs in the coming years appears robust. It is hoped that in a positive feedback fashion, the data derived from ivCUREs can fuel research programs from like-minded investigators whose work will then generate a range of open access image volumes that benefit the ivCURE community.
Footnotes
This work was supported by The University of Washington Tacoma Alternate Royalty Research Fund, and Scholarship and Teaching Fund. The author thanks Ms. My Nguyen, and the students in TBIOMD 495 for their technical assistance, enthusiasm, and feedback on this ivCURE.
REFERENCES
- American Association for the Advancement of Science. Vision and Change in Undergraduate Biology Education. In: Brewer CA, Smith D, editors. Vision and Change: A Call to Action. Washington, D.C: AAAS; 2011. pp. 1–79. [Google Scholar]
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ. 2014;13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DR, Seung HS, Lichtman JW. VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Front Neural Circuits. 2018;12:88. doi: 10.3389/fncir.2018.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell SE, Kloser MJ. Toward a conceptual framework for measuring the effectiveness of course-based undergraduate research experiences in undergraduate biology. Studies in Higher Education. 2015;40:525–544. [Google Scholar]
- Corwin LA, Graham MJ, Dolan EL. Modeling Course-Based Undergraduate Research Experiences: An Agenda for Future Research and Evaluation. CBE Life Sci Educ. 2015;14:1–13. doi: 10.1187/cbe.14-10-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC. Reconstruct: a free editor for serial section microscopy. J Miscrosc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Gray EG. Axo-somatic and axo-dendrritic synapses of the cerebral cortex: An electron microscope study. J Anat. 1959;93:420–433. [PMC free article] [PubMed] [Google Scholar]
- Kowalski JR, Hoops GC, Johnson RJ. Implementation of a Collaborative Series of Classroom-Based Undergraduate Research Experiences Spanning Chemical Biology, Biochemistry, and Neurobiology. CBE Life Sci Educ. 2016;15:1–17. doi: 10.1187/cbe.16-02-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Bonin V, Reed M, Graham BJ, Hood G, Glattfelder K, Reid RC. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatto D. Survey of Undergraduate Research Experiences (SURE): First Findings. Cell Biology Education. 2004;3:270–277. doi: 10.1187/cbe.04-07-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. Integrating Discovery-Based Research into the Undergraduate Curriculum: Report of a Convocation. Washington, D.C: The National Academies Press; 2015. Available at . [DOI] [Google Scholar]
- President’s Council of Advisors on Science and Technology. Executive Report to the President. Washington, D.C: Executive Office of the President; 2012. Engage to Excel: Producing One Million Additional College Graduates with Degrees in Science, Technology, Engineering, and Mathematics. [Google Scholar]
- Peters A, Palay SL, Webster HD. Fine Structure of the Nervous System: Neurons and Their Supporting Cells. 3rd edition. New York, NY: Oxford University Press; 1991. [Google Scholar]
- Rodenbusch SE, Hernandez PR, Simmons SL, Dolan EL. Early Engagement in Course-Based Research Increases Graduation Rates and Completion of Science, Engineering, and Mathematics Degrees. CBE Life Sci Educ. 2016;15:1–10. doi: 10.1187/cbe.16-03-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer LK. Analysis Tools for Large Connectomes. Front Neural Circuits. 2018;12:85. doi: 10.3389/fncir.2018.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour E, Hunter A, Laursen SL, DeAntoni T. Establishing the Benefits of Research Experiences for Undergradutes in the Sciences: First Findings from a Three-Year Study. Science Education. 2004;88:493–534. [Google Scholar]
- Shortlidge EE, Bangera G, Brownell SE. Faculty Perspectives on Developing and Teaching Course-Based Undergraduate Research Experiences. BioScience. 2016;66:54–62. [Google Scholar]
- Spacek J, Harris KM. Trans-endocytosis via spinules in adult rat hippocampus. J Neurosci. 2004;24:4233–4241. doi: 10.1523/JNEUROSCI.0287-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub NL, Beck CW, Griffin GD, Hennington BS, White SL. Course-based Science Research Promotes Learning in Diverse Students at Diverse Institutions. Council on Undergraduate Research. 2016;38:36–46. [Google Scholar]
- Tarrant SB, Routtenberg A. The synaptic spinule in the dendritic spine: electron microscopic study of the hippocampal dentate gyrus. Tissue Cell. 1977;9:461–473. doi: 10.1016/0040-8166(77)90006-4. [DOI] [PubMed] [Google Scholar]
- Thiry H, Weston TJ, Laursen SL, Hunter A. The Benefrist of Multi-Year Research Experiences: Differences in Novice and Experienced Students’ Reported Gains from Undergraduate Research. CBE Life Sci Educ. 2012;11:260–272. doi: 10.1187/cbe.11-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver GC, Russell CB, Wink DJ. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol. 2008;4:577–580. doi: 10.1038/nchembio1008-577. [DOI] [PubMed] [Google Scholar]