Abstract
Forward genetics is a powerful approach to understand the molecular basis of animal behaviors. Fruit flies were the first animal to which this genetic approach was applied systematically and have provided major discoveries on behaviors including sexual, learning, circadian, and sleep-like behaviors. The development of different classes of model organism such as nematodes, zebrafish, and mice has enabled genetic research to be conducted using more-suitable organisms. The unprecedented success of forward genetic approaches was the identification of the transcription–translation negative feedback loop composed of clock genes as a fundamental and conserved mechanism of circadian rhythm. This approach has now expanded to sleep/wakefulness in mice. A conventional strategy such as dominant and recessive screenings can be modified with advances in DNA sequencing and genome editing technologies.
Keywords: forward genetics, animal behavior, sleep, circadian rhythm, mutagenesis, model animal
1. Introduction
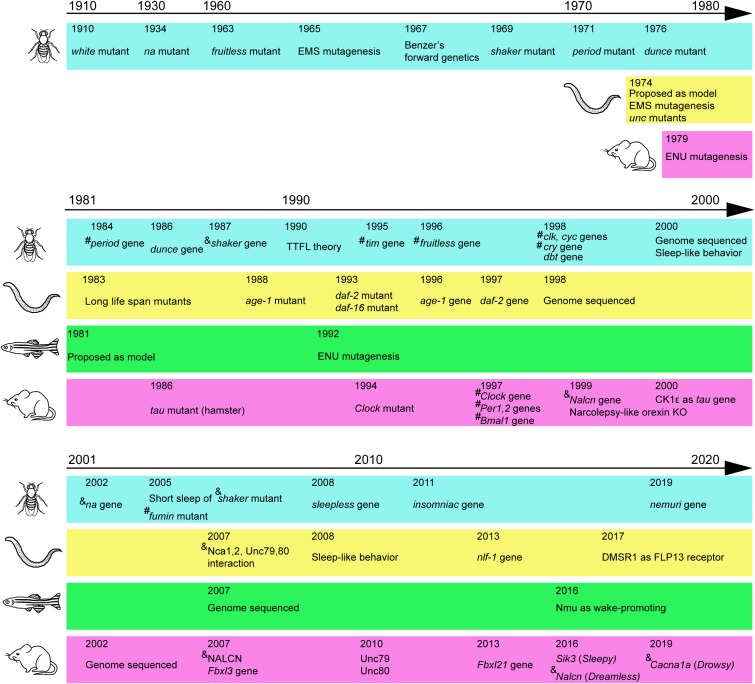
Forward genetics is applied to understand the genetic basis of the function, structure, and development of living organisms through a phenotype-based approach. Forward genetic research does not require a specific working hypothesis; instead, a heritable phenotype leads to the identification of the underlying genetic change. To determine the genetic basis of animal behavior, this approach requires appropriate animal models. In fact, advances in biology and medical research over the past hundred years have been largely dependent on model organisms. The fruit fly Drosophila melanogaster, nematode Caenorhabditis elegans, zebrafish Danio rerio, and house mouse Mus musculus are four major model animals in which genetic approaches have been applied (Fig. 1). Fruit flies are invertebrates with an elaborate brain and body structure and behavior. Nematodes are invertebrates with a simpler brain and body structure composed of approximately 950 cells. Zebrafish are vertebrates with small transparent bodies. Mice are mammals and constitute a closer model to humans. Each animal model has advantages and disadvantages over other models. These animal models have led to the elucidation of the genetic basis of complex behaviors such as circadian behavior, sleep, reproductive behavior, and learning. This review mainly focuses on circadian and sleep behaviors that are behaviorally well conserved among different model organisms, showing that a finding in one animal model can be confirmed in another, enabling us to obtain a more general and deeper understanding of conserved animal behaviors.
Figure 1.
(Color online) Major discoveries mainly due to forward genetics. Major mutants and cloned genes related to behaviors are indicated for fruit fly, nematode, zebrafish, and mouse in chronological order. Several technical advances are also indicated. # indicates transcription factors. & indicates channels or transporters. Before 1990, however, the function of the gene was usually unknown at the time of gene identification. TTFL: transcription–translation feedback loop.
2. Phenotype-based screening
The basic scheme for forward genetic research is to identify a gene mutation that alters a target phenotype through phenotypic screening of randomly mutagenized organisms. Chemical mutagens such as ethyl nitrosourea (ENU) or ethyl methanesulfonate (EMS) are used to induce random point mutations. There are several screening strategies.1–3)
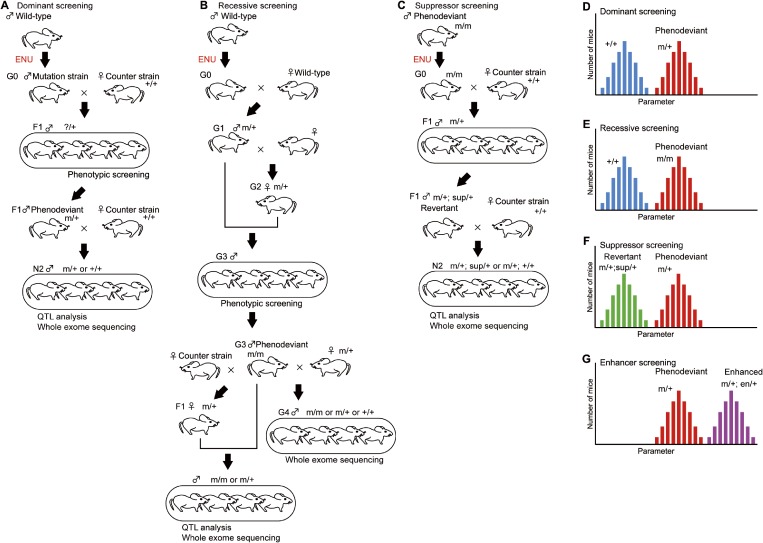
The dominant screening strategy tries to identify a heterozygous mutation that leads to phenotypic changes (Fig. 2A, D).4) In the case of mice, ENU is intraperitoneally administered to male mice (called generation 0 [G0]), which produces numerous point mutations in mouse spermatogonia and, subsequently, sperm. The next generation is produced by natural mating or in vitro fertilization with wild-type females, and the offspring are evaluated for the target phenotype to establish animal pedigrees showing heritable abnormalities. When the pedigree is maintained in a parental inbred strain such as C57BL/6J (B6J), each generation after G0 is called G1, G2, etc.
Figure 2.
(Color online) Forward genetic screening schemes. A. Dominant screening identifies a heterozygous mutation (m/+) that causes phenotypic changes. B. Recessive screening identifies a homozygous mutation (m/m) that causes phenotypic changes. Compared with dominant screening, recessive screening takes a larger number of generations to identify a mutation. C. Suppressor screening begins with the injection of ENU to the phenodeviants (m/m). The effect of the suppressor mutation on the phenotype was assessed using heterozygous mutant mice (m/+). D. Histogram of a pedigree with a strong dominant phenotype. The two peaks are mice heterozygous for the mutant gene (m/+; red bars) and mice without the mutant gene (+/+; blue bars). E. Histogram of a pedigree with a strong recessive phenotype. The two peaks are mice homozygous for the mutant gene (m/m; red bars) and mice without the mutant gene (+/+; blue bars). Heterozygous mice (m/+) are omitted from this histogram. F. Histogram of heterozygous mutant mice (m/+) with a suppressor mutation (sup/+; green bars) or without the mutation (red bars). G. Histogram of heterozygous mutant mice (m/+) with an enhancer mutation (en/+; purple bars) or without the mutation (red bars).
To map the chromosomal regions associated with the abnormal phenotypes, the mutated inbred strain needs to be backcrossed to a different strain. Currently, we have used C57BL/6J (B6J) and C57BL/6N (B6N) as a mutagenized strain and counter strain, respectively.4) The first generation crossed with a different strain are the first filial (F1) mice. We usually used the second backcross generation (N2) for quantitative trait locus (QTL) analysis.4–6) The dominant screening scheme shown in Fig. 2A enables us to identify gene mutations with the least number of generations. Male F1 mice were evaluated to identify mice showing a deviated phenotype, phenodeviants. F1 phenodeviants were then mated with wild-type female mice to obtain the next generation, N2 mice, which were used to examine whether the abnormal phenotype was heritable or not. N2 mice showing heritable abnormalities were subjected to QTL analysis and whole exome sequencing to identify the gene mutations responsible for the altered phenotype. Dominant screening identifies gain-of-function mutations including hypermorphic, antimorphic, and neomorphic mutations, and loss-of-function mutations showing haploinsufficiency.
Recessive screening identifies a homozygous mutation that causes a phenotypic change (Fig. 2B, E). Compared with dominant screening, recessive screening requires a larger number of generations to identify a mutation. However, advances in DNA sequencing technology have enabled whole exome sequencing as early as G1 mice, which changes the screening scheme completely.7) Because loss-of-function or null alleles are commonly recessive to the wild-type allele, recessive screening usually leads to the identification of loss-of-function mutants. Whereas both dominant and recessive screenings induce random mutations in wild-type mice, suppressor and enhancer screenings induce mutations in phenodeviant animals. Suppressor screening identifies a gene mutation that suppresses the effect of original mutation, which turns a phenodeviant into a revertant (Fig. 2C, F). Enhancer screening identifies a gene mutation that enhances the phenotype of phenodeviants (Fig. 2G). In fact, these basic screening strategies can be modified with a variety of technical advances such as sequencing and genome editing technologies.
3. The fruit fly Drosophila melanogaster
3.1. Drosophila as a model organism for genetic research.
In 1900, Mendel’s laws of inheritance were rediscovered independently by Hugo De Vries, Carl Correns, and Erich von Tscherma. De Vries was a botanist who had been interested in the mechanisms of evolution and the genesis of new species. He originated the novel term “mutation” when he identified a new form of evening primrose, Oenothera lamarckiana, and he hypothesized that a new form suddenly arises at random and then is stable for generations, leading him to develop his “mutation theory”.8)
Thomas Hunt Morgan, who originally trained as an embryologist, was determined to prove the mutation theory with experimental evidence using Drosophila melanogaster, which was a newly recognized animal model for genetic research according to Charles William Woodworth’s proposal. Drosophila can be readily reared in a laboratory without special equipment or a large space, presents a short generation time of 10 days, lays many eggs and has only four pairs of chromosomes, including three pairs of autosomal chromosomes and one pair of sex chromosomes. The presence of large chromosomes, polytene chromosomes, in the salivary glands in the mature Drosophila larva made it possible to visually recognize physical chromosomal changes using a microscope, which allowed researchers to link morphological and functional changes in individual flies with chromosomal changes.
After many rounds of trials, Morgan found that a white-eyed mutant white suddenly appeared among red-eyed flies and that, once the white-eye trait was identified, it was transmitted to the next generation according to Mendelian inheritance and was linked to the female chromosome, which suggested that a certain region of the chromosome was associated with the genetic determinant for eye color (Fig. 1).9,10) Around that time, Wilhelm Johannsen introduced the term ‘gene’ to indicate the physical carrier of a genetic trait. Morgan’s student Alfred Sturtevant constructed the first genetic map of a chromosome,11) proving the principle of the concept of “genetic mapping” for diverse phenotypes. Sturtevant reported that yellow and white mutants showed some abnormalities in mating behaviors,12) which implied that mating behavior is affected by certain genes. Many years later, it turned out that the white gene encodes an ATP-binding cassette (ABC) transporter family protein required for the transport of guanine and tryptophan.13)
Donald Poulson, one of Morgan’s students, launched a study focusing on embryogenesis and developmental biology using mutant fruit flies such as Notch mutants,14) which eventually led to the discovery of morphological pattern mutants through the systematic screening of embryonic lethal mutants by Christiane Nüsslein-Volhard and Eric Wieschaus, a student of Poulson.15)
3.2. Courtship behavior in Drosophila.
In contrast to the consensus that the morphology and structure of living organisms are largely determined by genes, it was previously generally thought that animal behaviors are too complex to be immediately determined by genetic factors. For example, male courtship behavior in Drosophila is a ritual-like behavior, in which a male follows a female, taps the female, sings a song by vibrating one wing, licks the female’s genitalia and curls the abdomen to attempt copulation,16) which is an obviously complex behavior. However, the courtship behavior was one of the first behaviors in which researchers found that a single gene specifies a behavior.
A graduate student, Kulbir S. Gill, mentored by Poulson, conducted his thesis research on the development of the sexual organs using fruit flies with random mutations induced by X-rays.17) Because Gill focused on mutant fly pedigrees showing sex-specific sterility, his research revealed morphological as well as functional changes, including sexual behaviors, causing sterility. He established a male-specific infertile mutant that was later referred to as the fruitless mutant.18,19) Male homozygous fruitless mutants have normal genitalia but do not exhibit courtship behavior toward female flies. Homozygotes fruitless females are healthy and fertile. In addition to fruitless mutants, Florian von Schilcher performed screening of EMS-induced mutant flies to identify mutants showing an abnormal courtship song20) following the forward genetic study strategy that was developed by Seymour Benzer (see later).21)
In 1996, more than 30 years after the first report of the fruitless mutant, the fruitless gene was isolated. The fruitless gene produces sex-specific transcripts resulting in the production of a transcription factor with sex-specific differences in the amino acid sequence.22,23) Male-specific splicing is crucial for male courtship behavior and sexual preference toward females. Female flies with the male-specific transcript showed male courtship behavior,24) suggesting that the male-specific transcript is crucial for the expression of male courtship. The Fruitless protein forms a complex with histone deacetylase 1, heterochromatin protein 1a, and Bonus.16) The male-specific form of the Fruitless protein transrepresses the robo1 gene, which encodes the receptor for a repulsive axon guidance signal. Female-specific Robo1 expression is crucial for the formation of the sexually dimorphic neural circuit that is required for normal courtship behavior.25) Thus, the genetic approach for examining courtship behavior elucidated the developmental mechanism of the sexually dimorphic neural circuit responsible for sex-specific behaviors. Because the fruitless gene does not have a mammalian homologue, this finding cannot be applied to mammals, but it suggests that a similar genetic mechanism exists for the formation of the sexually dimorphic structure that underlies innate sex-specific behaviors in mammals.
3.3. Circadian behavior in Drosophila.
When EMS was recognized as a highly potent mutagen for Drosophila,26) Benzer started his visionary research on behavior using inbred flies that were randomly mutagenized with this chemical mutagen to identify genes regulating a target phenotype through systematic screening.21) Benzer established the basic strategy of forward genetics that has led to novel discoveries in neuroscience, developmental biology, immunology, metabolism and other research areas. We still use the Benzerian scheme for forward genetics today.
Circadian behavior is defined as behavior internally entrained to the circadian rhythm that is robust without an external signal, zeitgeber, such as the light–dark change. Ronald Konopka who studied circadian rhythm in plants, joined the Benzer lab as a graduate student and worked on a project aimed at identifying fly mutants showing abnormal circadian behavior. Konopka first examined eclosion rhythm in constant darkness as a readout of circadian behavior. However, eclosion is a single event within the entire life cycle, so Konopka started to examine the locomotor activity of each fly as an index of circadian rhythm. He successfully established three fruit fly pedigrees showing behavioral abnormalities related to circadian rhythm. Mapping and complementation tests suggested that the three mutants showed alterations in the same gene,27) which was later named period (per). Jeffrey Hall, who was a postdoc in the Benzer lab, continued to try to identify the per gene in a collaboration with Michael Rosbash. They and Michael Young independently, finally succeeded in identifying the per gene,28–31) followed by a report on per transcripts.32)
Mechanistic insight into the circadian rhythm generation was obtained from the critical finding of a circadian change in the abundance of PER protein, showing a peak during the night, which was not recognized in the mutant pedigrees.33) The abundance of per mRNA also showed circadian rhythmicity, with a peak several hours before the peak in PER protein,34) which led Rosbash et al. to propose a negative feedback loop of clock genes at the transcriptional and translational levels.34) PER protein has an approximately 270-amino-acid sequence called the PER-ARNT-SIM (PAS) domain.35) In line with their hypothesis, Rosbash and colleagues showed that the PER protein belongs to the basic-helix-loop-helix PAS family of transcription regulators.36) Genetic screening identified a second core clock gene, timeless (tim), which also encodes a basic-helix-loop-helix PAS protein and exhibits circadian changes in abundance.37,38) TIM protein forms a complex with PER protein,39,40) which further supports the transcription–translation feedback loop as the basic mechanism for counting time. Moreover, continued forward genetic screening of randomly mutagenized fruit flies identified Clock (Clk),41) cryptochrome (cry),42) and cycle (cyc).43) Clk is the Drosophila homologue of mammalian Clock.44) Both CLK and CYC are basic helix-loop-helix PAS transcription factors and form a dimer to transactivate per and tim. Subsequently, the PER/TIM complex blocks the transcriptional activity of CLK and CYC.45,46) Thus, the four core clock proteins form a closed feedback loop with a certain time lag.
Genetic studies also identified the kinase double-time (DBT), which is homologous to mammalian casein kinase Iε, to be important for the degradation of PER protein via phosphorylation.47,48) Slimb, an F-box/WD40 protein family of the ubiquitin ligase SCF complex is necessary for the degradation of the phosphorylated PER protein.49,50) Screening of 2300 Drosophila lines with UAS-GAL4-based mis/overexpression found that the overexpression of shaggy (sgg) shortened the circadian period.51) sgg encodes a Drosophila orthologue of glycogen synthase kinase-3, which alters the degradation of TIM through the phosphorylation of TIM protein.51) These discoveries based on genetic approaches and subsequent biochemical and molecular studies led to the current understanding of the molecular basis of circadian behavior in Drosophila.52)
It is important to note that the feedback loop of core clock genes requires many output pathways to entrain animal behavior to the circadian cycle. The narrow abdomen (na) mutant was originally identified on the basis of its abdominal morphology.53) The screening of EMS-induced mutagenized flies for resistance to halothane identified har38, har56, har63, and har85.54,55) har38 and har85 are allelic to na and exhibit circadian abnormalities.56) The locomotion of Drosophila shows two peaks per day, in the morning and evening, but na mutant flies show decreased locomotion during the morning peak. The na gene encodes an orthologue of NALCN that forms a voltage-independent, nonselective cation channel.57,58) UNC79 and UNC80, orthologues of C. elegans unc-79 and unc-80, are required for NA to properly function,59) similar to the situation in mammals (see below60)). In the morning, a sodium conductance via NA depolarizes DN1p neurons, which is required for the morning peak.61) NCA localizing factor-1 (NLF-1) is essential for the proper translocation of NA/NCA in Drosophila61) and C. elegans.62)
In addition to na, several other genes have been identified as downstream of the core clock, including the neuropeptide pigment-dispersing factor (PDF),63) a member of the BK family of voltage-dependent Ca2+-activated potassium channels including slowpoke (slo),64) and ebony.65) Behavioral screening of P-element insertion mutant lines identified the gene encoding the PDF receptor (PDFR).66) Independently, behavioral phenotyping of Drosophila ion channel mutants and expression screening of orphan G protein-coupled receptors for the PDF receptor led to the identification of the same gene.67,68) The PDF-PDFR signaling is crucial for synchronizing clock cells and orchestrating behaviors through the stabilization of PER.52) Knockdown of Nlf-1 in PDF neurons reduced free-running rhythm in constant darkness,61) suggesting that NLF-1-dependent NA localization is required for the proper function of PDF neurons.
3.4. Learning behavior in Drosophila.
Learning behavior is essential for animals to forage and survive in changing environments. Benzer’s student, Willian Quinn, established a screening system for odor-conditioned behavior,69) and obtained the fly pedigree dunce, which exhibited deficits in learning behavior.70) This approach further established the turnip and rutabaga mutants, which showed abnormal association learning similar to dunce mutant flies.71) cAMP phosphodiesterase is abnormal in dunce mutant flies.72) Subsequently, the dunce gene was found to encode calcium/calmodulin-dependent cAMP phosphodiesterase.73) Similarly, rutabaga mutant flies exhibited reduced adenylate cyclase activity,74) and the rutabaga gene product is a calcium-dependent adenylate cyclase.75) Thus, forward genetic studies using flies support the hypothesis that cAMP and calcium play an important role in learning and memory, and more specifically, at synapses. These findings are consistent with a series of pioneering works using sea slugs, Aplysia californica.76) Pharmacological and biochemical studies suggested the existence of receptors in the cell membrane that led to an increase in cAMP in response to the extracellular ligand.77) Subsequently, the cloning of G-protein-coupled receptors for neurotransmitters led to the elucidation of the molecular basis of learning and memory via cAMP signaling.78)
3.5. Sleep-like behavior in Drosophila.
In 2000, the behavioral definition of sleep was established and applied to Drosophila.79,80) Sleep-like behavior is quiescence that meets the following behavioral criteria: 1) an increased threshold to be aroused, i.e., decreased responsiveness to sensory stimuli; 2) rapid arousal in response to strong stimuli, different from conscious disturbance or anesthesia; 3) timing synchronized with the circadian rhythm; and 4) homeostatic regulation. Sleep-like behavior of Drosophila is commonly defined as an inactivity state longer than 5 min according to monitoring the breaking of an infrared beam.81,82) This simple monitoring system led sleep-like behavior to become a target of forward genetic research. Cirelli et al. succeeded in establishing a short sleeper pedigree and identifying the gene mutation responsible in a potassium channel Shaker.83) Previously, Shaker mutants were characterized by abnormal behaviors and the responsible gene was cloned in 1987.84–86) Accordingly, flies carrying a mutation in the Hyperkinetic gene, which encodes the β subunit of the Shaker channel, showed short sleep time.87) The investigation of another short sleep pedigree led to the identification of sleepless,88) which is a modulator of the Shaker channel.89) Sleepless also modulates the expression of nicotinic acetylcholine receptors.90) Another short sleep fly pedigree, redeye, was attributed to a loss-of-function mutant of nicotinic acetylcholine receptors.91) Although the Shaker channel is broadly expressed in the brain, knockdown of Shaker in sleep-promoting dorsal fan-shaped body neurons resulted in short sleep time accompanied by a longer interspike interval of the neurons,92) indicating that the dorsal fan-shaped body is the brain region responsible for the short sleep in Shaker mutants. The dorsal fan-shaped body is also responsible for short sleep in the fumin mutant,93) which exhibits a mutation in dopamine transporter.94)
Sleep screening of fruit flies with neuron-specific knockdown of genes using more than 4000 UAS-RNAi lines showed that knockdown of Rca1 (regulator of cyclin A1), a homologue of a mammalian early mitotic inhibitor 1, decreased sleep time.95) Consistent with the role of Rca1 in the proper expression and function of cyclin A, knockdown of cyclin A resulted in a decreased sleep time. Another forward genetic study independently found that Taranis (TARA), a Drosophila homologue of the Trip-Br (SERTAD) family of transcriptional coregulators, regulates sleep-like behavior.96) TARA regulates cyclin A protein levels and physically interacts with cyclin A to promote sleep through cyclin-dependent kinase 1 activity. Thus, two independent studies indicated the possibility that cell cycle regulation may be involved in sleep regulation.
Sleep screening of EMS-induced randomly mutagenized flies established the insomniac (inc) mutant line, which showed a short sleep time.97) The inc gene encodes a BTB/POZ superfamily protein that functions as an adaptor for the Cullin-3 ubiquitin ligase complex. Independently, reverse genetic screening of 1297 alleles covering 1015 genes led to the finding that loss of inc and Cullin-3 resulted in a short sleep.98)
Almost all Drosophila mutants for sleep-like behavior based on loss-of-function mutants show prolonged wakefulness, rather than an increase in sleep-like behavior. Toda et al. adopted an opposite approach. Gain-of-function screening using more than 12,000 fly lines that express different transcripts using a pan-neuronal driver identified nemuri.99) NEMURI is a secreted peptide and is involved in sleep homeostasis and sickness during infection.
These discoveries proved that sleep-like behavior is regulated by a subset of genes similar to circadian behavior. However, in contrast to circadian behavior, the discovery of sleep-like behavior-regulating genes did not lead to the elucidation of a basic and conserved mechanism similar to the transcription–translation feedback loop scheme. Thus, the basic regulatory mechanism of sleep remains unclear.
4. The nematode, Caenorhabditis elegans
4.1. C. elegans as a model organism.
Sydney Brenner wrote that “Behavior is the result of a complex and ill-understood set of computations performed by nervous systems and it seems essential to decompose the problem into two: one concerned with the question of the genetic specification of nervous systems and the other with the way nervous systems work to produce behavior” in his seminal paper announcing C. elegans as a new animal model for genetics and neuroscience (Fig. 1).100) He considered fruit flies to be too large, too complicated, and to have too many neurons to understand. C. elegans are free-living nematodes with a 1-mm body length. Most C. elegans are self-reproducing hermaphrodites, in which each animal produces both sperm and eggs, and they can be easily maintained. An adult hermaphrodite is composed of 959 somatic cells, including 302 neurons. EMS-treatment enables the induction of random mutations in C. elegans and establishes C. elegans mutant pedigrees that exhibit characteristic abnormalities in morphology or movement. Brenner named any mutants showing abnormal behavior unc mutants, which led to the definition of more than 60 unc mutant genes.100) Brenner’s graduate student Jonathan Hodgkin continued to work on C. elegans from the early 1970s onward101) and registered many C. elegans mutants including unc-78, unc-79, and unc-80, which turned out to be crucial for the function of NALCN/NA.59,60)
4.2. Genetic studies using C. elegans.
C. elegans lives for 3 weeks, which is the shortest lifespan among major model animals. Long lifespan mutants were isolated,102) in which the gene responsible age-1 was mapped.103) This finding provided striking evidence that a single gene can lengthen lifespan. Subsequently, daf-2 and daf-16 mutants were reported to live twice as long.104) age-1 encodes a catalytic subunit of PI3 kinase.105) The daf-2 gene encodes a receptor for insulin-like growth factor.106) Subsequent studies identified other genes for long lifespan, which led to the finding that insulin-like growth factor and mTOR signaling play a crucial role in lifespan and aging.107,108)
The C. elegans unc-79 and unc-80 mutants show increased sensitivity to the volatile anesthetic halothane.109) Because Drosophila na mutants show an abnormal response to halothane,56) the na orthologues nca-1 and nca-2 can be associated with unc-79 in C. elegans.110) unc-80 was cloned, and the gene product was shown to be required for the proper localization of sodium leak channels, NCA-1 and NCA-2.111) Then, it was shown that both UNC-79 and UNC-80 were necessary for the proper localization and function of NCA-1/2.112,113) Genetic suppressor screening of gain-of-function nca mutants identified nlf-1 as a suppressor of nca.62) NLF-1 is localized at the endoplasmic reticulum and increases the translocation of NA to the cell membrane. These findings according to genetic studies of C. elegans unc-79, unc-80, and nlf-1 contributed to elucidate the role of their orthologs in circadian and sleep behaviors in fruit flies and mice.
4.3. Sleep-like behavior in C. elegans.
C. elegans lives in a habitat with circadian changes in light and temperature; therefore, this species may show circadian changes in locomotion,114,115) However, the rhythm is highly diverse and is not robust compared with those in other model organisms.116,117)
C. elegans exhibits several types of quiescent states that meet the behavior criteria for sleep. C. elegans larva has four periods of lethargus that lasts for approximately 2 hours, during which the worms show only a few movements and are less responsive to external stimuli.118) This quiescence is regulated in a homeostatic manner.118–120) Loss-of-function mutants of Lin-42, homologous to the clock gene period, show long and arrhythmic molts and lethargus stages.121) Sickness-induced sleep is distinct from lethargus,122,123) which is induced by the FMRFamide-like neuropeptide FLP-13, secreted from ALA neurons in response to stress or sickness.124) Suppressor screening of EMS-induced mutagenized worms with flp-13 overexpression identified a G-protein-coupled receptor, DMSR-1 as a receptor for FLP-13.125)
5. The zebrafish, D. rerio
5.1. Forward genetics research using zebrafish.
The zebrafish D. rerio is a tropical freshwater fish that originally inhabited small rivers, streams, and paddy fields in South Asia. In 1981, George Streisinger and his colleagues established zebrafish as a vertebrate model animal by producing homozygous diploid zebrafish (Fig. 1).126) Zebrafish have a short body length of 3 cm and a generation time of 3–4 months, lay several hundred eggs weekly and are easy to handle and care for. Their bodies are transparent from the early embryonic stage to adulthood, which is very useful for studying ontogeny. These characteristics have made zebrafish a popular genetic model.127) Initially, γ-rays were used to induce random mutations in zebrafish.128) However, irradiation often causes large deletions, translocations, or other gross chromosomal changes, which make it difficult to accurately identify the genes responsible for the abnormal phenotypes. Subsequently, ENU was shown to be a highly potent mutagen for inducing point mutations in zebrafish,129–131) which promoted forward genetic research using mutagenized zebrafish in the fields of developmental biology132–136) and tumorigenesis.137–139) ENU-based mutagenesis is a very simple and powerful tool, but it is not easy to pinpoint the exact mutation involved, and insertion mutagenesis using retroviral vectors140) or transposons141,142) has also been used.143)
The first behavioral screening in zebrafish was conducted using locomotion as a phenotype.144) For more complex behaviors such as addictive behavior, Darland and Dowling modified a conditioned place preference test, which was shown to be suitable for zebrafish and was used to establish several zebrafish pedigrees showing a blunt response to cocaine through behavioral screening of ENU-mutagenized zebrafish.145) Subsequent screening of zebrafish for the response to nicotine using insertion mutagenesis identified mutations in the chaperone containing protein 8 and Gaba-B receptor 1.2 genes.146) Although the number of behavioral studies in this model species has been rather small so far, advances in the visualization and assessment of zebrafish behavior147,148) will increase the number of forward genetic studies on zebrafish behavior.
5.2. Circadian and sleep-like behaviors in zebrafish.
Larval zebrafish show a robust diurnal activity pattern with small interindividual variability. Zebrafish show circadian behavior, similar to that of mammals, but the zebrafish body can directly sense and entrain light with no need for eyes or the suprachiasmatic nucleus (SCN).149) Zebrafish exhibited a core clock loop similar to mammals.150) Dominant screening of mutagenized zebrafish larvae led to the isolation of several pedigrees showing an altered circadian period.151) One of the pedigrees had a mutation in the Clock gene resulting in the substitution of a conserved isoleucine (I254N).152)
Larval and adult zebrafish exhibit a resting state that fulfills the behavioral criteria for sleep.153) Zebrafish larvae overexpressing one of 1268 human peptides were used for the large-scale screening of sleep abnormalities, which identified a neuropeptide, neuromedin U (Nmu), as a wakefulness-promoting factor.154) Nmu promotes wakefulness via Nmu receptor type 2 and corticotropin-releasing hormone receptor 1.
6. The mouse, M. musculus
6.1. Inbred mice.
Mice are the closest model to humans among commonly used animal models for genetic studies. In 1909, C. C. Little started mating mice to construct inbred mouse strains. Little was interested in the mouse’s response to transplanted tumors, which led him to conclude that genetically homogenous mice are necessary for genetic research using mice.155) He successfully established the first well-characterized inbred mouse strain DBA (Dilute, Brown and non-Agouti). In 1921, Little mated female 57 with male 52 that were obtained from Abbie Lathrop, eventually leading to the establishment of C57BL/6, which is the most popular mouse strain used in behavioral neuroscience and metabolism research worldwide.156) Little founded the Jackson Laboratory in 1929 and then started distributing inbred mice. Now, the genome sequences of major inbred mouse strains are available,157) which makes it easy to conduct QTL analysis.
6.2. Forward genetics research using mice.
The circadian changes in locomotion are valid for the assessment of circadian behavior in rodents. A running wheel is a simple and robust way to assess circadian changes.158) When Martin Ralph, a graduate student in Michael Menaker lab, was examining the circadian behavior of outbred Syrian hamsters in a shipment from Charles River Laboratories, he found that one hamster showed a short free-running period in constant darkness. This phenotype turned out to be heritable and semidominant in 1988.159) This serendipitous discovery of the first mammalian circadian rhythm mutant, tau, proved that the circadian rhythm can be strongly influenced by a set of genes in mammals. The tau gene was identified as casein kinase Iε in 2000.160)
Joseph Takahashi, who was a graduate student in Michael Menaker lab, determined to take a Benzerian approach using mice even though mice are less suitable for forward genetic studies than the other model organisms discussed above because mice have a generation time of approximately 4–5 months, which is much longer than that of fruit flies, and mouse research required much more space and equipment. ENU is a potent mutagen in mice (Fig. 1),161,162) and the screening of hyperphenylalaninemia in ENU-induced mutagenized mice identified a gene mutation in the phenylalanine hydroxylase gene.163) Takahashi and colleagues injected ENU into C57BL mice with a tight period length164) and successfully identified a mutant mouse among 304 G1 mice showing a long circadian period in a very early phase of screening, 25th mice,165) which then became the founder of the Clock mutant pedigree. The Clock gene was the first core clock gene in mammal and encodes a basic-helix-loop-helix PAS transcription factor.44)
After the success of forward genetics for circadian behavior, ENU-mutagenized mice were used for the investigation of other behaviors, including memory using the fear-conditioning paradigm,166) open field activity, and psychostimulant responses.167) Because the scheme of dominant screening is rather simple, the scale of screening can be expanded. Large-scale mutagenesis in mice using ENU has been applied to generate mutant mouse resource, similar to work in other genetic model organisms such as C. elegans and Drosophila, for a variety of phenotypes, including behavior assessed by locomotion and prepulse inhibition.168–171) Large-scale recessive screening has also been performed for multiple phenotypes, including sleep.172)
6.3. Circadian rhythm research using mice.
In 1997, soon after the discovery of the Clock gene,44) Per1, a mammalian homologue of the Drosophila per gene was cloned,173,174) followed by the identification of Per2175,176) and Per3.175,177,178) PER proteins belong to the basic-helix-loop-helix PAS family of transcription regulators. Importantly, the abundance of Per1, Per2, and Per3 in the SCN showed circadian oscillation,173–176) which suggests the transcription–translation feedback of Per may work as the core mechanism of mammalian circadian rhythm, similar to fruit flies.
In 1997, brain and muscle Arnt-like 1 (Bmal1) (also known as aryl hydrocarbon receptor nuclear translocator like [Arntl]) was originally identified as a new member of basic helix-loop-helix PAS proteins through a database search on human EST.179,180) The next year, it was reported that BMAL1 forms a heterodimer with CLOCK and that the CLOCK/BMAL1 heterodimer binds to the E-box sites in the 5′ flanking region of the Per1 gene to transactivate Per1 in mice.181,182) Bmal1 is a mammalian orthologue of Drosophila cyc;45) thus, the CLOCK/BMAL complex in mammals corresponds to the CLK/CYC heterodimer in Drosophila. These heterodimers containing CLOCK or its orthologue commonly transactivates Per genes.
Mammalian Cry1 and Cry2 were cloned as genes that are homologous to the plant blue light receptor and Drosophila cry.183–185) CRY1 and CRY2 form a complex with PER to inhibit the CLOCK/BMAL1 complex186) and are also involved in the light entrainment of the circadian rhythm.187,188) Thus, the CLOCK/BMAL1 heterodimer transactivates Per1, which in turn inhibits the action of the CLOCK/BMAL1 complex through the PER/CRY heterodimer. Additionally, two basic-helix-loop-helix PAS transcription factors, DEC1 and DEC2, suppress CLOCK/BMAL1-induced expression of Per1.189) These transcription–translation feedback loops constitute a core clock mechanism in mammals.186,190) Thus, the mammalian circadian clock was elucidated in a very short period of time mainly through the advanced homology-based cloning method and biochemical assays to identify a binding partner, following the basic scheme according to Drosophila circadian research.
Takahashi and colleagues continued forward genetic research to identify mutations in casein kinase Iε gene of the hamster tau mutant pedigree,160) F-box and leucine-rich repeat proteins 3 (Fbxl3) gene of the Overtime mutant pedigree,191) and Fbxl21 gene of the Past-time mutant pedigree.192) Nolan and colleagues also found a mutation in the Fbxl3 gene of the After-hours mutant pedigree.193) Of note, biochemical analyses have also identified casein kinase Iε,194) FBXL3,195) and FBXL21196) as molecules required for the proper oscillation of clock genes. Casein kinase Iε was reported to phosphorylate PER proteins and regulate the abundance.197,198) An F-box protein, FBXL3, is a component of the SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligase complex and is involved in the degradation of CRY proteins.191,195) FBXL21, a paralogue of FBXL3, also regulates ubiquitin-dependent degradation of CRY proteins.195,196) These findings were consistent with earlier reports on the casein kinase Iε orthologue gene mutation in double-time (dbt) mutant,47,48) and an F-box protein of the SCF ubiquitin ligase complex, Slimb, in Drosophila.49,50)
Takahashi and colleagues conducted suppressor screening using mice to identify upstream transcription factor 1 (Usf1) as a suppressor of the gain-of-function Clock mutant mice.199) In contrast to strong circadian abnormality in the Clock mutant mice, Clock-deficient mice showed robust circadian behavior with mild periodicity changes.200) Apart from a classical genetic approach, a systems biological approach demonstrated that nuclear receptors, REV-ERBα and REV-ERBβ (encoded by NR1D1 and NR1D2, respectively) and retinoic acid-related orphan receptors (RORs) and their response element constitute another circadian loop.201,202) Biochemical analysis of the Bmal1 promoter also found that REV-ERBα and REV-ERBβ led to the repression of Bmal1 and Clock via the ROR response element.203)
Molecular understanding of the circadian rhythm has continued to advance.204) What is special about circadian behavior is that the molecular players involved and the basic scheme of the transcription–translation feedback loop are conserved among invertebrates and vertebrates. Thus, a finding in one model organism can be quickly examined and confirmed in another model animal.
6.4. Reverse genetic studies on sleep in mice.
The development of a method for modifying a specific gene, gene-targeting, enabled examination of the phenotypes of mice deficient in a certain gene.205) An approach that seeks to gain a better understanding of a target phenotype through gene-modified mice is called reverse genetics. The first behavior that was examined in gene-deficient mice was memory.206,207) The first application of the Cre-loxP system in the brain was to examine the role of the hippocampus in memory formation.208,209) Since then, memory and learning have been among the most commonly studied topics with further elaboration using genetic, electrophysiological, and imaging techniques.210)
The International Mouse Phenotyping Consortium (IMPC), which is a worldwide multicenter project, has produced and phenotyped more than 20,000 knockout mouse strains to generate a comprehensive catalog of mouse gene functions for every protein-coding gene.211,212) Multicenter large-scale phenotyping of knockout mice, including behavioral analysis213) have continued to produce results in the field of developmental,214) disease,215) and metabolism research.216)
In general, a gene to be modified is selected based on the expected function of the molecule. For example, most reverse genetic studies on sleep have been conducted in mice deficient in genes that are known to regulate sleep/wakefulness according to pharmacological and physiological studies such as the receptors for noradrenaline, histamine, 5-HT, dopamine, and acetylcholine.217) In some cases, gene-modified mice failed to show the predicted sleep changes. For example, a normal wake time was observed in mice deficient in histidine decarboxylase, a rate-limiting enzyme for a wake-promoting neurotransmitter, histamine,218) perhaps because of redundant regulation of sleep/wakefulness.
An exceptional success was provided by the serendipitous discovery by Yanagisawa and colleagues of narcolepsy-like sleep abnormalities in mice deficient in orexin,219) which was identified as a neuropeptide that promotes food intake.220) This surprising finding, together with studies in narcoleptic dogs,221) led to the identification of the orexin system as the core of narcolepsy pathogenesis.222) Of note, mice deficient in core clock genes frequently show sleep abnormalities in normal light–dark conditions in which mutant mice generally show normal circadian behavior. Clock mutant mice showed a shorter total non-rapid eye movement (NREM) sleep.223) Bmal1-deficient mice exhibited an increase in total NREM sleep.224) These findings may be important, but the underlying mechanisms remain unknown.
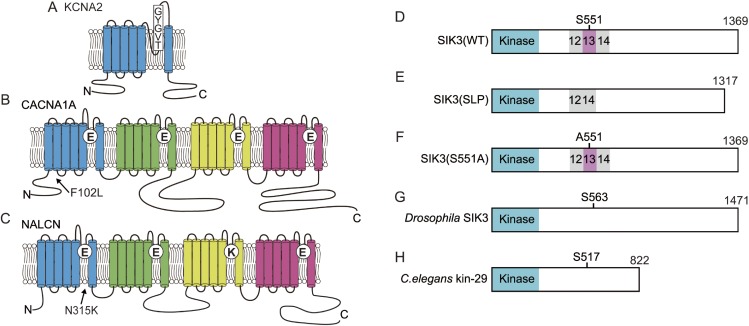
Many Drosophila short sleep mutants harbor mutations in Shaker or related molecules; therefore, potassium channels may play an important role in sleep regulation in mammals. The voltage-gated potassium channel has a structure in which a domain with six transmembrane segments is repeated four times. Mice exhibit approximately 40 voltage-gated potassium channels, which are divided into 12 subfamilies. Kv1.1 (KCNA1) to Kv1.8 (KCNA10) are mammalian Shaker-related channels. Among these channels, mice deficient in voltage-dependent potassium channel α-subunit Kv1.2 (KCNA2) (Fig. 3A) exhibited an increased awakening time, decreased NREM sleep time, and no change in REM sleep time.225) Thus, both Drosophila and mammals exhibit an increased arousal time due to Shaker-related channel deletion. Unfortunately, deficiency in many Shaker-related channel genes in mice causes embryonic lethality, developmental abnormalities, and epilepsy, so these mutants are not suitable for examining sleep/wakefulness. Recently, a systems biology approach together with the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 method revealed sleep changes in mice deficient in Ca2+-dependent potassium channels, voltage-dependent calcium channels and Ca2+/calmodulin-dependent kinase.226) The Cacna1a gene mutation was identified in the Drowsy mutant pedigree that shows long NREM sleep time (Fig. 3B).4)
Figure 3.
(Color online) SIK3 and ion channels related to sleep changes. A. KCNA2 has 6 transmembrane segments and the selective filter formed by a TVGYG sequence. B. CACNA1A is composed of four repeats of 6 transmembrane segments and has a selective filter motif of EEEE. The substitution of F102 to L decreased the time spent in wakefulness in Drowsy mutants. C. NALCN has four repeats of 6 transmembrane domains and has the selective filter motif of EEKE. The substitution of N315 to K caused abnormal REM sleep in Dreamless mutants. D. Wild-type murine SIK3 has a kinase domain and protein kinase A (PKA)-phosphorylation site, S551. E. Slp mutant SIK3 lacks the exon13-encoded region. F. Serine 551 of SIK3 is substituted with alanine. G. Drosophila SIK3 orthologue has a PKA-phosphorylation site, S563, equivalent to murine S551. H. C. elegans SIK3 orthologue, kin-29 has the PKA-phosphorylation site, S517.
6.5. Forward genetic studies on sleep in mice.
Considering that the size and structure of fruit fly brains are greatly different from those of mammals and REM sleep is not recognized in Drosophila, the mouse is an ideal genetic model for studying mammalian sleep. However, high-throughput screening based on electroencephalogram/electromyography recording is very challenging and labor-intensive. Moreover, mammalian sleep is thought to be controlled with high redundancy, which makes it difficult to establish families with heritable sleep abnormalities. Therefore, nobody has yet applied the Benzerian approach to mammalian sleep.
We conducted dominant screening for sleep as in Fig. 2A using B6J and B6N as mutagenized and counter strains, respectively. We successfully established the Sleepy (Slp) mutant pedigree showing a long NREM sleep time and then performed linkage analysis to obtain a large single peak logarithm of the odds score on chromosome 9. Whole-exome sequencing identified the responsible gene mutation in the Sik3 gene. The SIK3 protein is a kinase with a kinase domain at the amino terminus side (Fig. 3D) and belongs to the AMP kinase family.227) The mutation is located at the splice donor site of intron 13, which causes skipping of exon 13 and in-frame deletion (Fig. 3E). The exon 13-encoded region has a well conserved protein kinase A (PKA)-phosphorylated serine residue, serine 551 (S551). When S551 is substituted with alanine (S551A) (Fig. 3F) or aspartic acid (S551D), mice showed a decreased NREM sleep time similar to Sleepy mutant mice.228) SIK3 protein variants exhibited decreased binding to 14-3-3 proteins compared with the wild-type SIK3 protein.228) Although it is unclear whether binding to 14-3-3 protein itself is important for sleep control, changes in the biochemical properties of the SIK3 protein may lead to a reduced waking time.
The Drosophila Sik3 orthologue has a S551-equivalent residue (Fig. 3G). Overexpression of the alanine-substituted SIK3 orthologue increased sleep-like behavior in Drosophila.229) A loss-of-function mutant of the Drosophila Sik3 orthologue and the C. elegans Sik3 orthologue kin-29 (Fig. 3H) showed a decrease in the amount of time spent in sleep-like behavior or lethargus. These results indicated that PKA-SIK3 signaling plays a crucial and conserved role in sleep regulation.
Of note, Sleepy mutant mice were reported to exhibit circadian rhythm behavior similar to that of wild-type mice, whereas Sik3 gene-deficient mice exhibited prolonged periodicity in conditions of constant darkness.230) Gain-of-function mutations and loss-of-function mutations do not necessarily result in opposite phenotypes.
Phosphoproteomics analysis using Sleepy mutant mice and sleep-deprived wild-type mice identified increased phosphorylation of 80 phosphoproteins, which is thought to be a molecular signature of sleep need. Sixty nine of 80 phosphoproteins were associated with the function or structure of synapses.231) In addition to SIK3, ERK232) and Ca2+/calmodulin-dependent kinase226) were also reported to regulate sleep.
A forward genetic study established the Dreamless mutant pedigree that is characterized by short REM sleep episode duration and short REM sleep time. The causal mutation was found to be a point mutation in the Nalcn gene.229) The NALCN protein is a voltage-independent nonselective cation channel that is composed of four repeats of six transmembrane segments (Fig. 3C).233–235) The mutation results in asparagine-to-lysine substitution in segment 6 of domain I. The mutant NALCN protein showed increased ionic conductance compared with wild-type NALCN protein in HEK293 cells. Dreamless mutant mouse brain slices showed a higher membrane potential and higher firing rate of the midbrain deep nucleus neurons, which are REM-off neurons,236) than wild-type mice. However, such changes in the excitability of neurons are not necessarily specific to REM-off neurons, so it remains unknown how the mutant NALCN causes the characteristic REM sleep abnormalities. NALCN is abundantly expressed in the SCN, a master clock. Similar to the blunt peak in the Drosophila circadian pattern observed in the na mutant,61) Dreamless mutant mice showed a lower amplitude of rhythmic behavior with a normal circadian period length,229) suggesting that NALCN is not required for the determination of circadian rhythm but for generating output from the SCN, similar to na in Drosophila. Since studies using C. elegans and Drosophila have elucidated UNC-79, UNC-80 and NLF-1 as crucial components of proper NALCN function, these molecules are also thought to regulate sleep/wakefulness. Of note, a screen of ENU-mutagenized mice led to the identification of a heterozygous Unc79 null mutation responsible for abnormal ethanol response.237)
Although forward and reverse genetic studies identified many genes that affect sleep, most sleep-regulating genes are not functionally connected as seen in genes that are involved in circadian behavior. Connecting these sleep-regulating genes is the major challenge in the future.
7. New approaches using genome editing
A recent breakthrough in genome editing, CRISPR/Cas9 technology,238) will bring a new era in both forward and reverse genetics. The CRISPR/Cas9 method using multiplexed pools of single guide RNAs (sgRNAs) has been used for mutagenesis screening for organ development including electrical synapses in zebrafish.239–241) This approach facilitated reverse genetic screening of hundreds of genes of interest for a target phenotype that accelerates the screening of the selected genes and can examine the additive effect compared with a sgRNA. In addition, the use of tissue-specific Cas9 expression enables more spatially or cell-type targeted screening than conventional embryonic stem cell-based gene-targeting. Fruit flies that express Cas9 restricted in mushroom body γ neurons, together with the ubiquitous expression of sgRNAs, were used for the screening of genes that are involved in the remodeling of mushroom body γ neurons.242) This approach provided higher efficiency, higher phenotypic penetrance than RNAi-based gene knockdown. Thus, Cas9 with sgRNA is suitable for the multiple and systematic generation of loss-of-function mutations.
The use of homology-directed repair makes it possible to apply the CRISPR/Cas9 system for the generation of gain-of-function mutations in a thorough way. Saturated genome editing has been performed for functionally important exons of BRCA1 in cultured cells,243) which implies that saturated mutagenesis for any types of functional changes can in principle be conducted for animal behavior. Along this line, vSLENDER, which is an adeno-associated virus-mediated, homology-directed repair-based genome editing conducts neuron-specific genome editing.244) Advance in adeno-associated virus-mediated genome editing method will enable cell type-specific forward and reverse genetic research. Recently developed CRISPR-mediated base editing has further expanded the CRISPR-related tool box that can be applied for forward and reverse genetic research,245–247) where cytidine deaminases are fused to Cas9 nickase to substitute a single base without generating indels. Mutagenesis screening has identified a critical amino acid in DND1 protein for germ cell development.248)
Considering the randomness of mutations is a core strength of forward genetics, the combination of ENU-based random and multiple mutagenesis and genome editing-based more directed mutagenesis may lead to the identification of mutations that exert additive and interactive effects on a target behavior. Eventually, these efforts to elucidate a molecular basis for animal behavior will lead to a deeper understanding of psychiatric and neurological disorders and develop better therapeutic interventions for them.
Acknowledgements
The author thanks all members of Yanagisawa/Funato laboratory, WPI-IIIS, Univ. Tsukuba and Department of Anatomy, Toho Univ., and especially my long-time mentor, Prof. Masashi Yanagisawa, for introducing me to sleep research and forward genetic methods, and my Ph.D. mentor, Prof. Yasuo Ihara. Ms. Miyo Kakizaki drew the animals in Fig. 1. I also thank Prof. Nobutaka Hirokawa, M.J.A., who invited me to write this review.
Abbreviations
- CRISPR
clustered regularly interspaced short palindromic repeats
- EMS
ethyl methanesulfonate
- ENU
ethyl nitrosourea
- IMPC
International Mouse Phenotyping Consortium
- Nmu
neuromedin U
- NREM
non-rapid eye movement
- PAS
PER-ARNT-SIM
pigment-dispersing factor
- PKA
protein kinase A
- QTL
quantitative trait locus
- REM
rapid eye movement
- SCN
suprachiasmatic nucleus
- sgRNA
single guide RNA
Profile
Hiromasa Funato was born in Kobe in 1969 and grew up in Ehime, Chiba, Osaka, and Tokyo. He graduated from Tokyo Medical and Dental University School of Medicine in 1994. He received his Ph.D. degree in 1998 for a study on the neuropathology of human brain aging. He continued his research on the development and aging of the mammalian brain as a JSPS postdoctoral fellow. After clinical training as a psychiatrist at the University of Tokyo, Tokyo Metropolitan Bokutoh Hospital, and Yamaguchi University, he worked as a research fellow in the group led by Masashi Yanagisawa at the University of Texas Southwestern Medical Center. He became an Associate Professor at Toho University in 2011 and subsequently a Professor (WPI-IIIS) at Tsukuba University in 2013. He became a Professor at Toho University in 2018. His major accomplishments have been to identify several novel sleep-regulating genes through forward genetic research. His research interests focus on elucidating how the brain regulates an individual’s behavior and life and how intracellular events regulate animal behavior in a reciprocal manner. He has received the Bälz prize and is Councilor of the Japanese Association of Anatomists and the Japanese Society of Chronobiology.
References
- 1).Takahashi J.S., Shimomura K., Kumar V. (2008) Searching for genes underlying behavior: Lessons from circadian rhythms. Science 322, 909–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Beutler B., Du X., Xia Y. (2007) Precis on forward genetics in mice. Nat. Immunol. 8, 659–664. [DOI] [PubMed] [Google Scholar]
- 3).Acevedo-Arozena A., Wells S., Potter P., Kelly M., Cox R.D., Brown S.D.M. (2008) ENU mutagenesis, a way forward to understand gene function. Annu. Rev. Genomics Hum. Genet. 9, 49–69. [DOI] [PubMed] [Google Scholar]
- 4).Miyoshi C., Kim S.J., Ezaki T., Ikkyu A., Hotta-Hirashima N., Satomi K., et al. (2019) Methodology and theoretical basis of forward genetic screening for sleep/wakefulness in mice. Proc. Natl. Acad. Sci. U.S.A. 9, 16062–16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Broman K.W. (2001) Review of statistical methods for QTL mapping in experimental crosses. Lab Anim. (NY) 30, 44–52. [PubMed] [Google Scholar]
- 6).Sen S., Satagopan J.M., Broman K.W., Churchill G.A. (2007) R/qtlDesign: Inbred line cross experimental design. Mamm. Genome 18, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Wang T., Zhan X., Bu C.-H., Lyon S., Pratt D., Hildebrand S., et al. (2015) Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. U.S.A. 112, E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).De Vries, H. (1922) Age and area and the mutation theory. In A Study in Geographical Distribution and Origin of Species (ed. Willis, J.C.). Cambridge University Press, Cambridge, pp. 222–227. [Google Scholar]
- 9).Morgan T.H. (1910) Sex limited inheritance in Drosophila. Science 32, 120–122. [DOI] [PubMed] [Google Scholar]
- 10).Morgan T.H. (1911) The origin of five mutations in eye color in Drosophila and their modes of inheritance. Science 33, 534–537. [DOI] [PubMed] [Google Scholar]
- 11).Sturtevant A.H. (1913) A third group of linked genes in Drosophila ampelophila. Science 37, 990–992. [DOI] [PubMed] [Google Scholar]
- 12).Sturtevant A.H. (1915) Experiments on sex recognition and the problem of sex selection in Drosophila. J. Anim. Behav. 5, 351–366. [Google Scholar]
- 13).Mackenzie S.M., Brooker M.R., Gill T.R., Cox G.B., Howells A.J., Ewart G.D. (1999) Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim. Biophys. Acta 1419, 173–185. [DOI] [PubMed] [Google Scholar]
- 14).Poulson D.F. (1937) Chromosomal deficiencies and the embryonic development of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 23, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Nüsslein-Volhard C., Wieschaus E. (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- 16).Yamamoto D., Koganezawa M. (2013) Genes and circuits of courtship behaviour in Drosophila males. Nat. Rev. Neurosci. 14, 681–692. [DOI] [PubMed] [Google Scholar]
- 17).Gill K.S. (1963) Developmental genetic studies on oogenesis in Drosophila melanogaster. J. Exp. Zool. 152, 251–277. [Google Scholar]
- 18).Gill K.S. (1963) A mutation causing abnormal courtship and mating behavior in males of Drosophila melanogaster. Am. Zool. 3, 507. [Google Scholar]
- 19).Hall J.C. (2002) Courtship lite: A personal history of reproductive behavioral neurogenetics in Drosophila. J. Neurogenet. 16, 135–163. [DOI] [PubMed] [Google Scholar]
- 20).von Schilcher F. (1977) A mutation which changes courtship song in Drosophila melanogaster. Behav. Genet. 7, 251–259. [DOI] [PubMed] [Google Scholar]
- 21).Benzer S. (1967) Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. U.S.A. 58, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Ito H., Fujitani K., Usui K., Shimizu-Nishikawa K., Tanaka S., Yamamoto D. (1996) Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. U.S.A. 93, 9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Ryner L.C., Goodwin S.F., Castrillon D.H., Anand A., Villella A., Baker B.S., et al. (1996) Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 24).Demir E., Dickson B.J. (2005) fruitless splicing specifies male courtship behavior in Drosophila. Cell 121, 785–794. [DOI] [PubMed] [Google Scholar]
- 25).Ito H., Sato K., Kondo S., Ueda R., Yamamoto D. (2016) Fruitless represses robo1 transcription to shape male-specific neural morphology and behavior in Drosophila. Curr. Biol. 26, 1532–1542. [DOI] [PubMed] [Google Scholar]
- 26).Alderson T. (1965) Chemically induced delayed germinal mutation in Drosophila. Nature 207, 164–167. [DOI] [PubMed] [Google Scholar]
- 27).Konopka R.J., Benzer S. (1971) Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Bargiello T.A., Young M.W. (1984) Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 81, 2142–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Bargiello T.A., Jackson F.R., Young M.W. (1984) Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312, 752–754. [DOI] [PubMed] [Google Scholar]
- 30).Reddy P., Zehring W.A., Wheeler D.A., Pirrotta V., Hadfield C., Hall J.C., et al. (1984) Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710. [DOI] [PubMed] [Google Scholar]
- 31).Zehring W.A., Wheeler D.A., Reddy P., Konopka R.J., Kyriacou C.P., Rosbash M., et al. (1984) P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39, 369–376. [DOI] [PubMed] [Google Scholar]
- 32).Citri Y., Colot H.V., Jacquier A.C., Yu Q., Hall J.C., Baltimore D., et al. (1987) A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326, 42–47. [DOI] [PubMed] [Google Scholar]
- 33).Siwicki K.K., Eastman C., Petersen G., Rosbash M., Hall J.C. (1988) Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150. [DOI] [PubMed] [Google Scholar]
- 34).Hardin P.E., Hall J.C., Rosbash M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540. [DOI] [PubMed] [Google Scholar]
- 35).Nambu J.R., Franks R.G., Hu S., Crews S.T. (1990) The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63, 63–75. [DOI] [PubMed] [Google Scholar]
- 36).Huang Z.J., Edery I., Rosbash M. (1993) PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364, 259–262. [DOI] [PubMed] [Google Scholar]
- 37).Myers M.P., Wager-Smith K., Wesley C.S., Young M.W., Sehgal A. (1995) Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270, 805–808. [DOI] [PubMed] [Google Scholar]
- 38).Sehgal A., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M.P., Young M.W. (1995) Rhythmic expression of timeless: A basis for promoting circadian cycles in period gene autoregulation. Science 270, 808–810. [DOI] [PubMed] [Google Scholar]
- 39).Gekakis N., Saez L., Delahaye-Brown A.M., Myers M.P., Sehgal A., Young M.W., et al. (1995) Isolation of timeless by PER protein interaction: Defective interaction between timeless protein and long-period mutant PERL. Science 270, 811–815. [DOI] [PubMed] [Google Scholar]
- 40).Zeng H., Qian Z., Myers M.P., Rosbash M. (1996) A light-entrainment mechanism for the Drosophila circadian clock. Nature 380, 129–135. [DOI] [PubMed] [Google Scholar]
- 41).Allada R., White N.E., So W.V., Hall J.C., Rosbash M. (1998) A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. [DOI] [PubMed] [Google Scholar]
- 42).Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. [DOI] [PubMed] [Google Scholar]
- 43).Rutila J.E., Suri V., Le M., So W.V., Rosbash M., Hall J.C. (1998) CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. [DOI] [PubMed] [Google Scholar]
- 44).King D.P., Zhao Y., Sangoram A.M., Wilsbacher L.D., Tanaka M., Antoch M.P., et al. (1997) Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D., et al. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- 46).Glossop N.R., Lyons L.C., Hardin P.E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768. [DOI] [PubMed] [Google Scholar]
- 47).Kloss B., Price J.L., Saez L., Blau J., Rothenfluh A., Wesley C.S., et al. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94, 97–107. [DOI] [PubMed] [Google Scholar]
- 48).Price J.L., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M.W. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. [DOI] [PubMed] [Google Scholar]
- 49).Grima B., Lamouroux A., Chélot E., Papin C., Limbourg-Bouchon B., Rouyer F. (2002) The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178–182. [DOI] [PubMed] [Google Scholar]
- 50).Ko H.W., Jiang J., Edery I. (2002) Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420, 673–678. [DOI] [PubMed] [Google Scholar]
- 51).Martinek S., Inonog S., Manoukian A.S., Young M.W. (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779. [DOI] [PubMed] [Google Scholar]
- 52).Dubowy C., Sehgal A. (2017) Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205, 1373–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Miller H.M. (1934) Sex-linked mutant characters induced by X-ray dosage of 5000 r-u. Dros. Inf. Serv. 2, 9. [Google Scholar]
- 54).Krishnan K.S., Nash H.A. (1990) A genetic study of the anesthetic response: Mutants of Drosophila melanogaster altered in sensitivity to halothane. Proc. Natl. Acad. Sci. U.S.A. 87, 8632–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Campbell D.B., Nash H.A. (1994) Use of Drosophila mutants to distinguish among volatile general anesthetics. Proc. Natl. Acad. Sci. U.S.A. 91, 2135–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Nash H.A., Scott R.L., Lear B.C., Allada R. (2002) An unusual cation channel mediates photic control of locomotion in Drosophila. Curr. Biol. 12, 2152–2158. [DOI] [PubMed] [Google Scholar]
- 57).Lee J., Cribbs L., Perez-Reyes E. (1999) Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 445, 231–236. [DOI] [PubMed] [Google Scholar]
- 58).Lu B., Su Y., Das S., Liu J., Xia J., Ren D. (2007) The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129, 371–383. [DOI] [PubMed] [Google Scholar]
- 59).Lear B.C., Darrah E.J., Aldrich B.T., Gebre S., Scott R.L., Nash H.A., et al. (2013) UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS One 8, e78147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Lu B., Zhang Q., Wang H., Wang Y., Nakayama M., Ren D. (2010) Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron 68, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Flourakis M., Kula-Eversole E., Hutchison A.L., Han T.H., Aranda K., Moose D.L., et al. (2015) A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Xie L., Gao S., Alcaire S.M., Aoyagi K., Wang Y., Griffin J.K., et al. (2013) NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron 77, 1069–1082. [DOI] [PubMed] [Google Scholar]
- 63).Renn S.C., Park J.H., Rosbash M., Hall J.C., Taghert P.H. (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. [DOI] [PubMed] [Google Scholar]
- 64).Ceriani M.F., Hogenesch J.B., Yanovsky M., Panda S., Straume M., Kay S.A. (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Suh J., Jackson F.R. (2007) Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Hyun S., Lee Y., Hong S.-T., Bang S., Paik D., Kang J., et al. (2005) Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron 48, 267–278. [DOI] [PubMed] [Google Scholar]
- 67).Lear B.C., Merrill C.E., Lin J.-M., Schroeder A., Zhang L., Allada R. (2005) A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48, 221–227. [DOI] [PubMed] [Google Scholar]
- 68).Mertens I., Vandingenen A., Johnson E.C., Shafer O.T., Li W., Trigg J.S., et al. (2005) PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48, 213–219. [DOI] [PubMed] [Google Scholar]
- 69).Quinn W.G., Harris W.A., Benzer S. (1974) Conditioned behavior in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 71, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Dudai Y., Jan Y.N., Byers D., Quinn W.G., Benzer S. (1976) dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. U.S.A. 73, 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Duerr J.S., Quinn W.G. (1982) Three Drosophila mutations that block associative learning also affect habituation and sensitization. Proc. Natl. Acad. Sci. U.S.A. 79, 3646–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Byers D., Davis R.L., Kiger J.A. (1981) Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature 289, 79–81. [DOI] [PubMed] [Google Scholar]
- 73).Chen C.N., Denome S., Davis R.L. (1986) Molecular analysis of cDNA clones and the corresponding genomic coding sequences of the Drosophila dunce+ gene, the structural gene for cAMP phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 83, 9313–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Livingstone M.S., Sziber P.P., Quinn W.G. (1984) Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell 37, 205–215. [DOI] [PubMed] [Google Scholar]
- 75).Levin L.R., Han P.L., Hwang P.M., Feinstein P.G., Davis R.L., Reed R.R. (1992) The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell 68, 479–489. [DOI] [PubMed] [Google Scholar]
- 76).Kandel E.R. (1981) Calcium and the control of synaptic strength by learning. Nature 293, 697–700. [DOI] [PubMed] [Google Scholar]
- 77).van Calker D., Müller M., Hamprecht B. (1979) Adenosine regulates via two different types of receptors, the accumulation of cyclic AMP in cultured brain cells. J. Neurochem. 33, 999–1005. [DOI] [PubMed] [Google Scholar]
- 78).Brzosko Z., Mierau S.B., Paulsen O. (2019) Review neuromodulation of spike-timing-dependent plasticity: Past, present, and future. Neuron 103, 563–581. [DOI] [PubMed] [Google Scholar]
- 79).Hendricks J.C., Sehgal A., Pack A.I. (2000) The need for a simple animal model to understand sleep. Prog. Neurobiol. 61, 339–351. [DOI] [PubMed] [Google Scholar]
- 80).Shaw P.J., Cirelli C., Greenspan R.J., Tononi G., Campbell S.S., Tobler I., et al. (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- 81).Andretic R., Shaw P.J. (2005) Essentials of sleep recordings in Drosophila: Moving beyond sleep time. Methods Enzymol. 393, 759–772. [DOI] [PubMed] [Google Scholar]
- 82).Huber R., Hill S.L., Holladay C., Biesiadecki M., Tononi G., Cirelli C. (2004) Sleep homeostasis in Drosophila melanogaster. Sleep 27, 628–639. [DOI] [PubMed] [Google Scholar]
- 83).Cirelli C., Bushey D., Hill S., Huber R., Kreber R., Ganetzky B., et al. (2005) Reduced sleep in Drosophila shaker mutants. Nature 434, 1087–1092. [DOI] [PubMed] [Google Scholar]
- 84).Kamb A., Iverson L.E., Tanouye M.A. (1987) Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell 50, 405–413. [DOI] [PubMed] [Google Scholar]
- 85).Papazian D.M., Schwarz T.L., Tempel B.L., Jan Y.N., Jan L.Y. (1987) Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science 237, 749–753. [DOI] [PubMed] [Google Scholar]
- 86).Tempel B.L., Papazian D.M., Schwarz T.L., Jan Y.N., Jan L.Y. (1987) Sequence of a probable potassium channel component encoded at Shaker locus of Drosophila. Science 237, 770–775. [DOI] [PubMed] [Google Scholar]
- 87).Bushey D., Huber R., Tononi G., Cirelli C. (2007) Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J. Neurosci. 27, 5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Koh K., Joiner W.J., Wu M.N., Yue Z., Smith C.J., Sehgal A. (2008) Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Wu M.N., Joiner W.J., Dean T., Yue Z., Smith C.J., Chen D., et al. (2010) SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat. Neurosci. 13, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Wu M., Robinson J.E., Joiner W.J. (2014) SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr. Biol. 24, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Shi M., Yue Z., Kuryatov A., Lindstrom J.M., Sehgal A. (2014) Identification of Redeye, a new sleepregulating protein whose expression is modulated by sleep amount. eLife 2014, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Pimentel D., Donlea J.M., Talbot C.B., Song S.M., Thurston A.J.F., Miesenböck G., et al. (2016) Operation of a homeostatic sleep switch. Nature 536, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Ueno T., Tomita J., Tanimoto H., Endo K., Ito K., Kume S., et al. (2012) Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15, 1516–1523. [DOI] [PubMed] [Google Scholar]
- 94).Kume K., Kume S., Park S.K., Hirsh J., Jackson F.R. (2005) Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25, 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Rogulja D., Young M.W. (2012) Control of sleep by cyclin A and its regulator. Science 335, 1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Afonso D.J.S., Liu D., Machado D.R., Pan H., Jepson J.E.C., Rogulja D., et al. (2015) TARANIS functions with Cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr. Biol. 25, 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Stavropoulos N., Young M.W. (2011) insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72, 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Pfeiffenberger C., Allada R. (2012) Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a Dopamine arousal pathway in Drosophila. PLoS Genet. 8, e1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99).Toda H., Williams J.A., Gulledge M., Sehgal A. (2019) A sleep-inducing gene, nemuri, links sleep and immune function in Drosophila. Science 363, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100).Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101).Hodgkin J. (1983) Male phenotypes and mating efficiency in CAENORHABDITIS ELEGANS. Genetics 103, 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Klass M.R. (1983) A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 22, 279–286. [DOI] [PubMed] [Google Scholar]
- 103).Friedman D.B., Johnson T.E. (1988) Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J. Gerontol. 43, B102–B109. [DOI] [PubMed] [Google Scholar]
- 104).Kenyon C., Jean C., Grensch E., Adam R., Ramon T. (1993) A C. elegans mutant that twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- 105).Morris J.Z., Tissenbaum H.A., Ruvkun G. (1996) A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536–539. [DOI] [PubMed] [Google Scholar]
- 106).Kimura K.D., Tissenbaum H.A., Liu Y., Ruvkun G. (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946. [DOI] [PubMed] [Google Scholar]
- 107).Kenyon C.J. (2010) The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- 108).Uno M., Nishida E. (2016) Lifespan-regulating genes in C. elegans. NPJ Aging Mech. Dis. 2, 16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109).Sedensky M.M., Meneely P.M. (1987) Genetic analysis of halothane sensitivity in Caenorhabditis elegans. Science 236, 952–954. [DOI] [PubMed] [Google Scholar]
- 110).Humphrey J.A., Hamming K.S., Thacker C.M., Scott R.L., Sedensky M.M., Snutch T.P., et al. (2007) A putative cation channel and its novel regulator: Cross-species conservation of effects on general anesthesia. Curr. Biol. 17, 624–629. [DOI] [PubMed] [Google Scholar]
- 111).Jospin M., Watanabe S., Joshi D., Young S., Hamming K., Thacker C., et al. (2007) UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr. Biol. 17, 1595–1600. [DOI] [PubMed] [Google Scholar]
- 112).Yeh E., Ng S., Zhang M., Bouhours M., Wang Y., Wang M., et al. (2008) A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 6, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Pierce-Shimomura J.T., Chen B.L., Mun J.J., Ho R., Sarkis R., McIntire S.L. (2008) Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 20982–20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Kippert F., Saunders D.S., Blaxter M.L. (2002) Caenorhabditis elegans has a circadian clock. Curr. Biol. 12, R47–R49. [DOI] [PubMed] [Google Scholar]
- 115).Saigusa T., Ishizaki S., Watabiki S., Ishii N., Tanakadate A., Tamai Y., et al. (2002) Circadian behavioural rhythm in Caenorhabditis elegans. Curr. Biol. 12, R46–R47. [DOI] [PubMed] [Google Scholar]
- 116).Simonetta S.H., Migliori M.L., Romanowski A., Golombek D.A. (2009) Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS One 4, e7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117).Simonetta S.H., Golombek D.A. (2007) An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J. Neurosci. Methods 161, 273–280. [DOI] [PubMed] [Google Scholar]
- 118).Raizen D.M., Zimmerman J.E., Maycock M.H., Ta U.D., You Y., Sundaram M.V., et al. (2008) Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572. [DOI] [PubMed] [Google Scholar]
- 119).Nagy S., Tramm N., Sanders J., Iwanir S., Shirley I.A., Levine E., et al. (2014) Homeostasis in C. elegans sleep is characterized by two behaviorally and genetically distinct mechanisms. eLife 3, e04380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Driver R.J., Lamb A.L., Wyner A.J., Raizen D.M. (2013) DAF-16/FOXO regulates homeostasis of essential sleep-like behavior during larval transitions in C. elegans. Curr. Biol. 23, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121).Monsalve G.C., Van Buskirk C., Frand A.R. (2011) LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21, 2033–2045. [DOI] [PubMed] [Google Scholar]
- 122).Davis K.C., Raizen D.M. (2017) A mechanism for sickness sleep: Lessons from invertebrates. J. Physiol. 595, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Trojanowski N.F., Nelson M.D., Flavell S.W., Fang-yen C., Raizen D.M., Trojanowski X.N.F., et al. (2015) Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J. Neurosci. 35, 14571–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Nelson M.D., Lee K.H., Churgin M.A., Hill A.J., Van Buskirk C., Fang-Yen C., et al. (2014) FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr. Biol. 24, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Iannacone M.J., Beets I., Lopes L.E., Churgin M.A., Fang-Yen C., Nelson M.D., et al. (2017) The RFamide receptor DMSR-1 regulates stress-induced sleep in C. elegans. eLife 6, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Streisinger G., Walker C., Dower N., Knauber D., Singer F. (1981) Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291, 293–296. [DOI] [PubMed] [Google Scholar]
- 127).Holtzman N.G., Kathryn Iovine M., Liang J.O., Morris J. (2016) Learning to fish with genetics: A primer on the vertebrate model Danio rerio. Genetics 203, 1069–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128).Chakrabarti S., Streisinger G., Singer F., Walker C. (1983) Frequency of γ-ray induced specific locus and recessive lethal mutations in mature germ cells of the zebrafish, BRACHYDANIO RERIO. Genetics 103, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Grunwald D.J., Streisinger G. (1992) Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet. Res. 59, 103–116. [DOI] [PubMed] [Google Scholar]
- 130).Solnica-Krezel L., Schier A.F., Driever W. (1994) Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136, 1401–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131).Mullins M.C., Hammerschmidt M., Haffter P., Nüsslein-Volhard C. (1994) Large-scale mutagenesis in the zebrafish: In search of genes controlling development in a vertebrate. Curr. Biol. 4, 189–202. [DOI] [PubMed] [Google Scholar]
- 132).Driever W., Fishman M.C. (1996) The zebrafish: Heritable disorders in transparent embryos. J. Clin. Invest. 97, 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133).Eisen J.S. (1996) Zebrafish make a big splash. Cell 87, 969–977. [DOI] [PubMed] [Google Scholar]
- 134).Driever W., Solnica-Krezel L., Schier A.F., Neuhauss S.C., Malicki J., Stemple D.L., et al. (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46. [DOI] [PubMed] [Google Scholar]
- 135).Haffter P., Granato M., Brand M., Mullins M.C., Hammerschmidt M., Kane D.A., et al. (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36. [DOI] [PubMed] [Google Scholar]
- 136).Henke K., Daane J.M., Hawkins M.B., Dooley C.M., Busch-Nentwich E.M., Stemple D.L., et al. (2017) Genetic screen for postembryonic development in the Zebrafish (Danio rerio): Dominant mutations affecting adult form. Genetics 207, 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Amatruda J.F., Shepard J.L., Stern H.M., Zon L.I. (2002) Zebrafish as a cancer model system. Cancer Cell 1, 229–231. [DOI] [PubMed] [Google Scholar]
- 138).Shepard J.L., Amatruda J.F., Stern H.M., Subramanian A., Finkelstein D., Ziai J., et al. (2005) A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl. Acad. Sci. U.S.A. 102, 13194–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139).Stern H.M., Zon L.I. (2003) Cancer genetics and drug discovery in the zebrafish. Nat. Rev. Cancer 3, 533–539. [DOI] [PubMed] [Google Scholar]
- 140).Gaiano N., Amsterdam A., Kawakami K., Allende M., Becker T., Hopkins N. (1996) Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 383, 829–832. [DOI] [PubMed] [Google Scholar]
- 141).Nagayoshi S., Hayashi E., Abe G., Osato N., Asakawa K., Urasaki A., et al. (2008) Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like. Development 135, 159–169. [DOI] [PubMed] [Google Scholar]
- 142).Sivasubbu S., Balciunas D., Davidson A.E., Pickart M.A., Hermanson S.B., Wangensteen K.J., et al. (2006) Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 123, 513–529. [DOI] [PubMed] [Google Scholar]
- 143).Lawson N.D., Wolfe S.A. (2011) Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev. Cell 21, 48–64. [DOI] [PubMed] [Google Scholar]
- 144).Granato M., van Eeden F.J., Schach U., Trowe T., Brand M., Furutani-Seiki M., et al. (1996) Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 123, 399–413. [DOI] [PubMed] [Google Scholar]
- 145).Darland T., Dowling J.E. (2001) Behavioral screening for cocaine sensitivity in mutagenized zebrafish. Proc. Natl. Acad. Sci. U.S.A. 98, 11691–11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146).Petzold A.M., Balciunas D., Sivasubbu S., Clark K.J., Bedell V.M., Westcot S.E., et al. (2009) Nicotine response genetics in the zebrafish. Proc. Natl. Acad. Sci. U.S.A. 106, 18662–18667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147).Orger M.B., de Polavieja G.G. (2017) Zebrafish behavior: Opportunities and challenges. Annu. Rev. Neurosci. 40, 125–147. [DOI] [PubMed] [Google Scholar]
- 148).Leung L.C., Wang G.X., Madelaine R., Skariah G., Kawakami K., Deisseroth K., et al. (2019) Neural signatures of sleep in zebrafish. Nature 571, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149).Frøland Steindal I.A., Whitmore D. (2019) Circadian clocks in fish. What have we learned so far? Biology (Basel) 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150).Vatine G., Vallone D., Gothilf Y., Foulkes N.S. (2011) It’s time to swim! Zebrafish and the circadian clock. FEBS Lett. 585, 1485–1494. [DOI] [PubMed] [Google Scholar]
- 151).DeBruyne J., Hurd M.W., Gutiérrez L., Kaneko M., Tan Y., Wells D.E., et al. (2004) Isolation and phenogenetics of a novel circadian rhythm mutant in zebrafish. J. Neurogenet. 18, 403–428. [DOI] [PubMed] [Google Scholar]
- 152).Tan Y., DeBruyne J., Cahill G.M., Wells D.E. (2008) Identification of a mutation in the Clock1 gene affecting zebrafish circadian rhythms. J. Neurogenet. 22, 149–166. [DOI] [PubMed] [Google Scholar]
- 153).Chiu C.N., Prober D.A. (2013) Regulation of zebrafish sleep and arousal states: Current and prospective approaches. Front. Neural Circuits 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154).Chiu C.N., Rihel J., Lee D.A., Singh C., Mosser E.A., Chen S., et al. (2016) A zebrafish genetic screen identifies neuromedin U as a regulator of sleep/wake states. Neuron 89, 842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Paigen K. (2003) One hundred years of mouse genetics: An intellectual history. I. The classical period (1902–1980). Genetics 163, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156).Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., et al. (2000) Genealogies of mouse inbred strains. Nat. Genet. 24, 23–25. [DOI] [PubMed] [Google Scholar]
- 157).Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158).Siepka S.M., Takahashi J.S. (2005) Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol. 393, 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159).Ralph M.R., Menaker M. (1988) A mutation of the circadian system in golden hamsters. Science 241, 1225–1227. [DOI] [PubMed] [Google Scholar]
- 160).Lowrey P.L., Shimomura K., Antoch M.P., Yamazaki S., Zemenides P.D., Ralph M.R., et al. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161).Russell W.L., Kelly E.M., Hunsicker P.R., Bangham J.W., Maddux S.C., Phipps E.L. (1979) Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. U.S.A. 76, 5818–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Hitotsumachi S., Carpenter D.A., Russell W.L. (1985) Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc. Natl. Acad. Sci. U.S.A. 82, 6619–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163).McDonald J.D., Bode V.C., Dove W.F., Shedlovsky A. (1990) Pahhph-5: A mouse mutant deficient in phenylalanine hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 87, 1965–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164).Pittendrigh C.S., Daan S. (1976) A functional analysis of circadian pacemakers in nocturnal rodents I. The stability and lability of spontaneous frequency. J. Comp. Physiol. A 106, 223–252. [Google Scholar]
- 165).Vitaterna M.H., King D.P., Chang A.M., Kornhauser J.M., Lowrey P.L., McDonald J.D., et al. (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166).Reijmers L.G., Coats J.K., Pletcher M.T., Wiltshire T., Tarantino L.M., Mayford M. (2006) A mutant mouse with a highly specific contextual fear-conditioning deficit found in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen. Learn. Mem. 13, 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167).Kumar V., Kim K., Joseph C., Thomas L.C., Hong H., Takahashi J.S. (2011) Second-generation high-throughput forward genetic screen in mice to isolate subtle behavioral mutants. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl.), 15557–15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168).Hrabé de Angelis M.H., Flaswinkel H., Fuchs H., Rathkolb B., Soewarto D., Marschall S., et al. (2000) Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25, 444–447. [DOI] [PubMed] [Google Scholar]
- 169).Hrabé de Angelis M., Balling R. (1998) Large scale ENU screens in the mouse: Genetics meets genomics. Mutat. Res. 400, 25–32. [DOI] [PubMed] [Google Scholar]
- 170).Nolan P.M., Kapfhamer D., Bućan M. (1997) Random mutagenesis screen for dominant behavioral mutations in mice. Methods 13, 379–395. [DOI] [PubMed] [Google Scholar]
- 171).Nolan P.M., Peters J., Strivens M., Rogers D., Hagan J., Spurr N., et al. (2000) A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 25, 440–443. [DOI] [PubMed] [Google Scholar]
- 172).Potter P.K., Bowl M.R., Jeyarajan P., Wisby L., Blease A., Goldsworthy M.E., et al. (2016) Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat. Commun. 7, 12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173).Tei H., Okamura H., Shigeyoshi Y., Fukuhara C., Ozawa R., Hirose M., et al. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512–516. [DOI] [PubMed] [Google Scholar]
- 174).Sun Z.S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., Lee C.C. (1997) RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003–1011. [DOI] [PubMed] [Google Scholar]
- 175).Shearman L.P., Zylka M.J., Weaver D.R., Kolakowski L.F., Reppert S.M. (1997) Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269. [DOI] [PubMed] [Google Scholar]
- 176).Albrecht U., Sun Z.S., Eichele G., Lee C.C. (1997) A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 177).Takumi T., Taguchi K., Miyake S., Sakakida Y., Takashima N., Matsubara C., et al. (1998) A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 17, 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178).Zylka M.J., Shearman L.P., Weaver D.R., Reppert S.M. (1998) Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110. [DOI] [PubMed] [Google Scholar]
- 179).Hogenesch J.B., Chan W.K., Jackiw V.H., Brown R.C., Gu Y.Z., Pray-Grant M., et al. (1997) Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 272, 8581–8593. [DOI] [PubMed] [Google Scholar]
- 180).Ikeda M., Nomura M. (1997) cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem. Biophys. Res. Commun. 233, 258–264. [DOI] [PubMed] [Google Scholar]
- 181).Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., et al. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- 182).Hogenesch J.B., Gu Y.Z., Jain S., Bradfield C.A. (1998) The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183).van der Spek P.J., Kobayashi K., Bootsma D., Takao M., Eker A.P., Yasui A. (1996) Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics 37, 177–182. [DOI] [PubMed] [Google Scholar]
- 184).Hsu D.S., Zhao X., Zhao S., Kazantsev A., Wang R.P., Todo T., et al. (1996) Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry 35, 13871–13877. [DOI] [PubMed] [Google Scholar]
- 185).Todo T., Ryo H., Yamamoto K., Toh H., Inui T., Ayaki H., et al. (1996) Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 272, 109–112. [DOI] [PubMed] [Google Scholar]
- 186).Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., et al. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- 187).Griffin E.A., Staknis D., Weitz C.J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771. [DOI] [PubMed] [Google Scholar]
- 188).van der Horst G.T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630. [DOI] [PubMed] [Google Scholar]
- 189).Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., et al. (2002) Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844. [DOI] [PubMed] [Google Scholar]
- 190).King D.P., Takahashi J.S. (2000) Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 23, 713–742. [DOI] [PubMed] [Google Scholar]
- 191).Siepka S.M., Yoo S.-H.H., Park J., Song W., Kumar V., Hu Y., et al. (2007) Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192).Yoo S., Mohawk J.A., Siepka S.M., Shan Y., Huh S.K., Hong H.-K., et al. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193).Godinho S.I.H., Maywood E.S., Shaw L., Tucci V., Barnard A.R., Busino L., et al. (2007) The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316, 897–900. [DOI] [PubMed] [Google Scholar]
- 194).Keesler G.A., Camacho F., Guo Y., Virshup D., Mondadori C., Yao Z. (2000) Phosphorylation and destabilization of human period I clock protein by human casein kinase Iε. Neuroreport 11, 951–955. [DOI] [PubMed] [Google Scholar]
- 195).Busino L., Bassermann F., Maiolica A., Lee C., Nolan P.M., Godinho S.I.H., et al. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904. [DOI] [PubMed] [Google Scholar]
- 196).Hirano A., Yumimoto K., Tsunematsu R., Matsumoto M., Oyama M., Kozuka-Hata H., et al. (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118. [DOI] [PubMed] [Google Scholar]
- 197).Lee C., Etchegaray J.P., Cagampang F.R., Loudon A.S., Reppert S.M. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867. [DOI] [PubMed] [Google Scholar]
- 198).Lee H., Chen R., Lee Y., Yoo S., Lee C. (2009) Essential roles of CKIδ and CKIε in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199).Shimomura K., Kumar V., Koike N., Kim T.-K., Chong J., Buhr E.D., et al. (2013) Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. eLife 2, e00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200).Debruyne J.P., Noton E., Lambert C.M., Maywood E.S., Weaver D.R., Reppert S.M. (2006) A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477. [DOI] [PubMed] [Google Scholar]
- 201).Ueda H.R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., et al. (2002) A transcription factor response element for gene expression during circadian night. Nature 418, 534–539. [DOI] [PubMed] [Google Scholar]
- 202).Ueda H.R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., et al. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192. [DOI] [PubMed] [Google Scholar]
- 203).Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., et al. (2002) The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. [DOI] [PubMed] [Google Scholar]
- 204).Takahashi J.S. (2017) Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205).Capecchi M.R. (1989) Altering the genome by homologous recombination. Science 244, 1288–1292. [DOI] [PubMed] [Google Scholar]
- 206).Silva A.J., Stevens C.F., Tonegawa S., Wang Y. (1992) Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science 257, 201–206. [DOI] [PubMed] [Google Scholar]
- 207).Grant S.G., O’Dell T.J., Karl K.A., Stein P.L., Soriano P., Kandel E.R. (1992) Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science 258, 1903–1910. [DOI] [PubMed] [Google Scholar]
- 208).Tsien J.Z., Chen D.F., Gerber D., Tom C., Mercer E.H., Anderson D.J., et al. (1996) Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87, 1317–1326. [DOI] [PubMed] [Google Scholar]
- 209).Tsien J.Z., Huerta P.T., Tonegawa S. (1996) The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell 87, 1327–1338. [DOI] [PubMed] [Google Scholar]
- 210).Poo M., Pignatelli M., Ryan T.J., Tonegawa S., Bonhoeffer T., Martin K.C., et al. (2016) What is memory? The present state of the engram. BMC Biol. 14, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211).Brown S.D.M., Moore M.W. (2012) The International Mouse Phenotyping Consortium: Past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212).Ring N., Meehan T.F., Blake A., Brown J., Chen C.-K., Conte N., et al. (2015) A mouse informatics platform for phenotypic and translational discovery. Mamm. Genome 26, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213).de Angelis M.H., Nicholson G., Selloum M., White J., Morgan H., Ramirez-Solis R., et al. (2015) Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 47, 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214).Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., et al. (2016) High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215).Meehan T.F., Conte N., West D.B., Jacobsen J.O., Mason J., Warren J., et al. (2017) Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat. Genet. 49, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216).Rozman J., Rathkolb B., Oestereicher M.A., Schütt C., Ravindranath A.C., Leuchtenberger S., et al. (2018) Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nat. Commun. 9, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217).Cirelli C. (2009) The genetic and molecular regulation of sleep: From fruit flies to humans. Nat. Rev. Neurosci. 10, 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218).Parmentier R., Ohtsu H., Djebbara-Hannas Z., Valatx J.-L., Watanabe T., Lin J.-S. (2002) Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: Evidence for the role of brain histamine in behavioral and sleep-wake control. J. Neurosci. 22, 7695–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219).Chemelli R.M., Willie J.T., Sinton C.M., Elmquist J.K., Scammell T., Lee C., et al. (1999) Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell 98, 437–451. [DOI] [PubMed] [Google Scholar]
- 220).Sakurai T., Amemiya A., Ishii M., Matsuzaki I., Chemelli R.M., Tanaka H., et al. (1998) Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92, 573–585. [DOI] [PubMed] [Google Scholar]
- 221).Lin L., Faraco J., Li R., Kadotani H., Rogers W., Lin X., et al. (1999) The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98, 365–376. [DOI] [PubMed] [Google Scholar]
- 222).Mahoney C.E., Cogswell A., Koralnik I.J., Scammell T.E. (2019) The neurobiological basis of narcolepsy. Nat. Rev. Neurosci. 20, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223).Naylor E., Bergmann B.M., Krauski K., Zee P.C., Takahashi J.S., Vitaterna M.H., et al. (2000) The circadian clock mutation alters sleep homeostasis in the mouse. J. Neurosci. 20, 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224).Laposky A., Easton A., Dugovic C., Walisser J., Bradfield C., Turek F. (2005) Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 28, 395–409. [DOI] [PubMed] [Google Scholar]
- 225).Douglas C.L., Vyazovskiy V., Southard T., Chiu S.-Y., Messing A., Tononi G., et al. (2007) Sleep in Kcna2 knockout mice. BMC Biol. 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226).Tatsuki F., Sunagawa G.A., Shi S., Susaki E.A., Yukinaga H., Perrin D., et al. (2016) Involvement of Ca2+-dependent hyperpolarization in sleep duration in mammals. Neuron 90, 70–85. [DOI] [PubMed] [Google Scholar]
- 227).Takemori H., Okamoto M. (2008) Regulation of CREB-mediated gene expression by salt inducible kinase. J. Steroid Biochem. Mol. Biol. 108, 287–291. [DOI] [PubMed] [Google Scholar]
- 228).Honda T., Fujiyama T., Miyoshi C., Ikkyu A., Hotta-Hirashima N., Kanno S., et al. (2018) A single phosphorylation site of SIK3 regulates daily sleep amounts and sleep need in mice. Proc. Natl. Acad. Sci. U.S.A. 115, 10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229).Funato H., Miyoshi C., Fujiyama T., Kanda T., Sato M., Wang Z., et al. (2016) Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230).Hayasaka N., Hirano A., Miyoshi Y., Tokuda I.T., Yoshitane H., Matsuda J., et al. (2017) Salt-inducible kinase 3 regulates the mammalian circadian clock by destabilizing PER2 protein. eLife 6, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231).Wang Z., Ma J., Miyoshi C., Li Y., Sato M., Ogawa Y., et al. (2018) Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature 558, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232).Mikhail C., Vaucher A., Jimenez S., Tafti M. (2017) ERK signaling pathway regulates sleep duration through activity-induced gene expression during wakefulness. Sci. Signal 10, eaai9219. [DOI] [PubMed] [Google Scholar]
- 233).Ren D. (2011) Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron 72, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234).Monteil A., Cochet-Bissuel M., Lory P., Monteil A. (2014) The sodium leak channel, NALCN, in health and disease. Front. Cell. Neurosci. 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235).Stephens R.F., Guan W., Zhorov B.S., Spafford J.D. (2015) Selectivity filters and cysteine-rich extracellular loops in voltage-gated sodium, calcium, and NALCN channels. Front. Physiol. 6, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236).Hayashi Y., Kashiwagi M., Yasuda K., Ando R., Kanuka M., Sakai K., et al. (2015) Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science 350, 957–961. [DOI] [PubMed] [Google Scholar]
- 237).Speca D.J., Chihara D., Ashique A.M., Bowers M.S., Pierce-Shimomura J.T., Lee J., et al. (2010) Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 6, e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238).Komor A.C., Badran A.H., Liu D.R. (2017) Review CRISPR-based technologies for the manipulation of Eukaryotic genomes. Cell 168, 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239).Varshney G.K., Carrington B., Pei W., Bishop K., Chen Z., Fan C., et al. (2016) A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 11, 2357–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240).Varshney G.K., Pei W., LaFave M.C., Idol J., Xu L., Gallardo V., et al. (2015) High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241).Shah A.N., Davey C.F., Whitebirch A.C., Miller A.C., Moens C.B. (2015) Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods 12, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242).Meltzer H., Marom E., Alyagor I., Mayseless O., Berkun V., Segal-Gilboa N., et al. (2019) Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat. Commun. 10, 2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243).Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., et al. (2018) Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244).Nishiyama J., Mikuni T., Yasuda R. (2017) Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron 96, 755–768.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245).Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729. [DOI] [PubMed] [Google Scholar]
- 246).Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., et al. (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247).Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X. (2016) Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035. [DOI] [PubMed] [Google Scholar]
- 248).Li Q., Li Y., Yang S., Huang S., Yan M., Ding Y., et al. (2018) CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 20, 1315–1325. [DOI] [PubMed] [Google Scholar]