Abstract
Single-domain VHH antibodies are promising reagents for medical therapy. A conserved disulfide bond within the VHH framework region is known to be critical for thermal stability, however, no prior studies have investigated its influence on the stability of VHH antibody–antigen complexes under mechanical load. Here, we used single-molecule force spectroscopy to test the influence of a VHH domain’s conserved disulfide bond on the mechanical strength of the interaction with its antigen mCherry. We found that although removal of the disulfide bond through cysteine-to-alanine mutagenesis significantly lowered VHH domain denaturation temperature, it had no significant impact on the mechanical strength of the VHH:mCherry interaction with complex rupture occurring at ∼60 pN at 103–104 pN/sec regardless of disulfide bond state. These results demonstrate that mechanostable binding interactions can be built on molecular scaffolds that may be thermodynamically compromised at equilibrium.
Keywords: Antibodies, biophysics, disulfide bond, nanobody, single-molecule force spectroscopy
Single-domain antibodies are affinity proteins derived from the variable domains of heavy chain-only antibodies found in the immune systems of Camelidae.1−4 These N-terminal variable domains (VHH domains), also referred to as “nanobodies”, are being developed for medical therapy, and their biophysical properties are of high interest. VHHs possess several advantageous features as compared with conventional full-size IgG antibodies, for example, their relative ease of production, small size (∼13 kDa), and high stability. The VHH framework excluding complementarity determining regions (CDRs) 1–3 contains a conserved disulfide bond common to Ig folds found between residues Cys22 and Cys92 following the Kabat numbering scheme. This disulfide is essential for stability, but is not strictly required for maintenance of antigen binding ability,5 and significant efforts have been expended to understand its precise role on binding affinity and thermal stability of VHH domains.
Akazawa-Ogawa and colleagues reported that removal of the canonical disulfide through mutagenesis lowered thermal denaturation temperatures, however, the mutants retained binding affinity.6 Similarly, Pleiner et al. reported that production of VHH domains in an E. coli strain with a reducing cytoplasm exhibited no loss of binding activity.7 Saerens et al. identified a universal VHH framework lacking the conserved disulfide bond that was used as a recipient scaffold for loop grafting,8 and several other reports of VHH frameworks lacking the conserved disulfide bond are readily found.9−11 Removal of the conserved disulfide typically decreases the thermal denaturation temperature of VHHs.9,12 Furthermore, groups have reported that introduction of an additional disulfide bond linking CDRs 1 and 3 strongly stabilizes the VHH folded structure13−16 and increases thermal stability.
Despite this knowledge on VHH binding affinity and folding stability following disulfide bond removal, there is nothing known about its effects on VHH antibody/antigen complexes under load. The response of antibody/antigen complexes to mechanical force is likely to be therapeutically relevant in drug delivery systems where shear stress is present, for example, during the delivery of nanoparticles to cell surfaces under flow. Single-molecule force spectroscopy (SMFS) with the atomic force microscope (AFM) has been used to characterize the mechanical stability of folded domains17−21 and receptor/ligand interactions,22 for example, in studies on biotin/avidin systems,23−25 antibody/antigen complexes,26−29 pathogenic adhesin proteins,30−32 as well as cellulose adhesion domains (Cohesin/Dockerin).33−41 When receptor/ligand complexes are separated under mechanical load, they can dissociate through energetic pathways that differ from those achieved at equilibrium. These pathways are specific to the direction in which force is applied to the complex, which depends on the amino acid positions used to attach the molecules to the surface/cantilever tip.
When formed within folded protein structures, disulfide bonds create rigid staples that pin nonconsecutive residues together, modulating protein mechanical properties by enhancing mechanostability of folded domains,42−44 increasing rupture forces of receptor/ligand complexes,45,46 or in some cases decreasing unfolding forces.44,47,48 To the best of our knowledge, only one prior report examined the mechanical response of a VHH antibody/antigen complex.49 In that system, an anti-GFP VHH was attached to a cantilever tip while GFP was tethered to a surface either through its C- or N-terminus. However, different disulfide bond states were not considered. The role of disulfide bond state on VHH antibody/antigen mechanics therefore remains unknown.
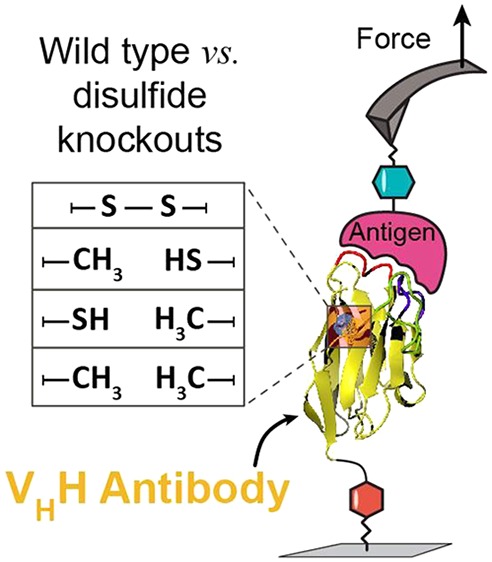
Here, we addressed this question by characterizing the biophysical properties and unbinding energy landscape of a VHH/mCherry receptor/ligand complex using AFM-SMFS while perturbing the disulfide bond. We prepared wild type (WT) and three mutant VHH domains which converted one or both of the conserved cysteines to alanine. We then characterized this 4-member VHH library using thermal denaturation differential scanning fluorescence (DSF), isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and AFM-SMFS to understand the effects of disulfide bond removal on the biophysical performance of VHH.
Dictyostelium discoideum
For AFM-SMFS studies, we cloned the VHH(WT) domain containing two cysteines in frame with the fourth domain of F-actin cross-linking filamin (ddFLN4).50 We cloned the gene for the target antigen (mCherry) in frame with a “found in various architectures” (FIVAR) domain.31 The ddFLN4 and FIVAR domains were located at the C-termini of their respective fusion proteins and contained C-terminal ybbR and hexahistidine tags. The ybbR tag was used to covalently and site-specifically immobilize the proteins onto coenzyme A (CoA)-functionalized AFM cantilever tips and coverglass surfaces, respectively, via 4′-phosphopantetheinyl transferase (Sfp)-mediated ligation51 (Supporting Information). The nomenclature for these proteins is VHH(WT)-ddFLN4 and mCherry-FIVAR where the yBBR and hexahistidine tags are omitted for brevity.
In addition to VHH(WT)-ddFLN4, we produced three mutant VHHs where either one or both cysteines was mutated to alanine. These mutants, denoted VHH(C24A), VHH(C98A), and VHH(C24A, C98A) lacked the ability to form the conserved disulfide bond. All proteins were produced both as fusions with ddFLN4 and FIVAR which served as marker/fingerprint domains for AFM-SMFS, as well as in isolated form for SPR, ITC, and DSF measurements. Protein sequence are provided in the Supporting Information.
Proteins were expressed and purified from E. coli SHuffle cells using metal ion affinity and size exclusion chromatography. E. coli SHuffle cells are commonly used for cytoplasmic production of VHH domains and facilitate easier purification and higher yields as compared with periplasmic expression.52−56 Protein molecular weight and purity were confirmed via denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry (Supplementary Figure 1). We confirmed disulfide bonding in VHH(WT) using Ellman’s reagent (5,5-dithio-bis(2-nitrobenzoic acid)) to measure the presence of free cysteines under native and denaturing conditions. The semiquantitative results shown in Supplementary Table 1 indicated that >75% of VHH(WT) molecules contained oxidized disulfides.
To understand the effect of disulfide bond removal on equilibrium affinity, kinetic off rates and thermal denaturation temperatures, we tested VHH domains as isolated proteins lacking the marker domains using ITC, SPR, and DSF (Table 1). ITC indicated that VHH(WT) had the highest affinity (KD = 68.97 nM) and VHH(C24A, C98A) had the lowest affinity (KD = 169.92 nM) whereas VHH(C24A) and VHH(C98A) exhibited intermediate affinities (KD = 86.96 and 121.8 nM, respectively). These differences in bulk affinity suggested that under equilibrium conditions, removal of the disulfide bond slightly destabilized the bound complex. SPR results (Table 1) demonstrated the same trend as ITC, albeit with overall higher KD values. We attributed the lower affinities measured by SPR to the surface immobilization protocol which relied on N-hydroxysuccinimide coupling to lysine groups on the surfaces of the proteins, which can lower apparent affinity. Thermal denaturation analysis by DSF indicated that removal of the canonical disulfide bond in the 3 mutant VHH domains decreased the first thermal melting peak by >5 °C. VHH(WT) furthermore exhibited an additional second thermal melting transition at 70.2 °C that was not observed for any of the mutants.
Table 1. Quantification of Binding Affinity and Thermal Stability of WT and Mutant VHH Domainsa.
| ITC |
SPR |
DSF |
||||||
|---|---|---|---|---|---|---|---|---|
| sample | ΔH (cal·mol–1) | ΔS (cal·mol–1·deg–1) | KD (nM) | koff (10–3 s–1) | kon (103 M–1 s–1) | KD (nM) | Tm1 (°C) | Tm2 (°C) |
| VHH(WT) | –5187 | 15.4 | 68.97 ± 12.70 | 8.53 ± 0.04 | 52.34 ± 15.4 | 162.97 ± 47.53 | 52.35 ± 0.04 | 70.2 ± 0.14 |
| C24A | –4691 | 16.6 | 86.96 ± 9.81 | 10.67 ± 0.04 | 55.25 ± 17.91 | 193.12 ± 61.18 | 47.25 ± 0.04 | ND |
| C98A | –4318 | 17.2 | 121.80 ± 17.36 | 10.82 ± 0.04 | 55.82 ± 9.98 | 193.84 ± 31.38 | 46.80 ± 0.10 | ND |
| C24A/C98A | –4120 | 17.2 | 168.92 ± 31.66 | 11.55 ± 0.06 | 54.70 ± 19.30 | 211.15 ± 85.04 | 47.02 ± 0.06 | ND |
ND, not detected; errors indicate 95% confidence intervals (see respective methods in Supporting Information).
To test the effects of disulfide removal on VHH/mCherry complex stability from a mechanical perspective, we used AFM-SMFS (Figure 1a). A cantilever tip bearing covalently linked mCherry-FIVAR was brought into contact with a surface bearing covalently linked VHH(WT)-ddFLN4, forming a molecular complex. The cantilever was retracted at constant speed and in cases when a molecular complex had successfully formed, the force–distance traces exhibited sawtooth like features characteristic of marker domain unfolding and VHH/mCherry complex rupture (Figure 1b). The approach-retract cycle was repeated tens of thousands of times while actuating the XY stage between each measurement to sample a different surface-bound VHH domain in each trace. Large data sets consisting of typically ∼20 000 force versus extension curves were filtered using contour length transformation to identify specific interactions.57
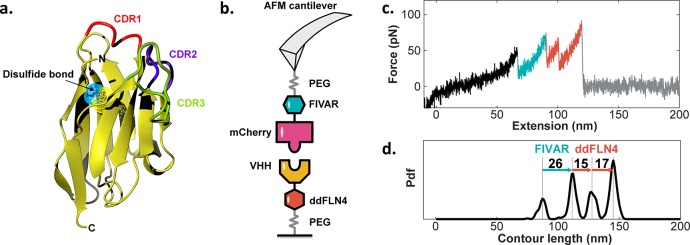
Figure 1.
AFM-SMFS on VHH/mCherry complexes. (a) Homology model of VHH domain. The disulfide bond is shown as surface mesh in blue and yellow. Modeling was carried out using the ABodyBuilder server.58 (b) Measurement setup with mCherry-FIVAR attached to the cantilever and VHH-ddFLN4 attached to the substrate. (c) Typical force versus extension trace showing unfolding of FIVAR (cyan) and ddFLN4 (orange) marker domains, followed by complex rupture. (d) Contour length histogram of the data in panel (c). Increments between peaks were used to make domain assignments to unfolding events.
Following unfolding and stretching of FIVAR (Figure 1b, cyan) and ddFLN4 (Figure 1b, orange) via an intermediate state, the final peak represented rupture of a single VHH/mCherry complex. Following rupture, the cantilever lost its tether to the surface and the force dropped to zero (Figure 1b, gray). Domain assignments were made using wormlike chain polymer transformation and contour length analysis as previously described.57 A typical contour length histogram (Figure 1c) showed peak-to-peak length increments corresponding to the amino acid sequence lengths of ddFLN4 and FIVAR minus the folded domain lengths.
Because the forces required to unfold the ddFLN4 and FIVAR marker domains and the force required to rupture the VHH/mCherry complex all lie in a similar range, we observed traces where the order of marker domain unfolding was reversed or where only one of the marker domains unfolded prior to complex rupture. In order to avoid biases59 during energy landscape reconstruction, we included traces showing one and even zero marker domains in the analysis while applying a minimal contour length filter to remove nonspecific adhesion events close to the surface (<60 nm = 2× PEG spacer length). Supplementary Figure 2 in the Supporting Information shows example traces containing zero, one, and two marker domain unfolding events prior to complex rupture and their overall prevalence in a representative data set.
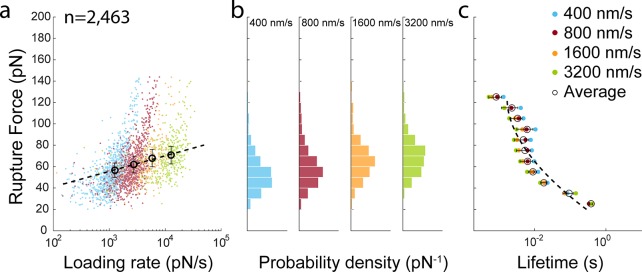
To obtain energy landscape parameters, we performed AFM-SMFS measurements at pulling speeds of 400, 800, 1600, and 3200 nm/s and detected 2,463 specific rupture events from analysis of individual force–extension traces (Figure 2a). The complex rupture force distributions (Figure 2b) show that rupture of the VHH(WT)/mCherry occurs most probably at ∼60–70 pN at loading rates of ∼103–104 pN/s. Force-dependent bound lifetimes (Figure 2c) were generated using histogram transformation and fitting as described by Dudko, Hummer, and Szabo (DHS).60 We assumed a harmonic well with a cusplike energy barrier (v = 0.5) to fit the lifetime data and obtain values for the activation barrier height (ΔG‡), distance to the transition state (Δx) and intrinsic lifetime (t0), yielding: ΔG‡ = 9.04 kBT; Δx = 0.46 nm; and t0 = 2.07 s from this measurement.
Figure 2.
Dynamic force spectroscopy on VHH(WT)/mCherry complexes. (a) VHH(WT)/mCherry complex rupture events detected at four pulling speeds from 400 to 3 200 nm/s. Colored points represent rupture events of individual molecular complexes. Open circle markers represent the most probable rupture force and average loading rate at each pulling speed. The dashed line represents the Bell-Evans fit to the open circle. (b) Rupture force histograms at each pulling speed. (c) Lifetime plot obtained using histogram transformation based on the DHS model with v = 0.5. The dashed line represents the fit to the DHS model.
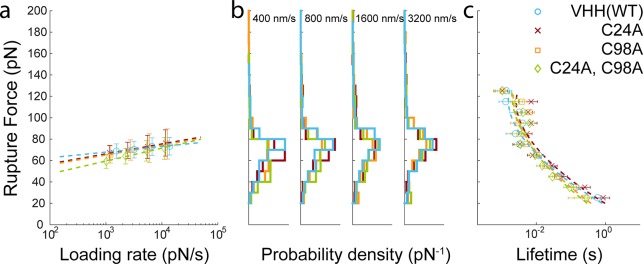
When protein–protein interactions are mechanically dissociated, it is difficult to determine if dissociation is driven by unfolding of one of the binding partners and loss of the binding epitope structure, or by global loss of contact between two well-folded proteins.61 Given that disulfide bonding supports overall VHH fold stability, if VHH/mCherry mechanical dissociation were driven by loss of VHH tertiary structure, then removal of the disulfide bond could potentially lower the forces at which the VHH/mCherry complex dissociates. We performed AFM-SMFS on VHH(C24A), VHH(C98A), and VHH(C24A, C98A) using identical expression, purification and surface chemical procedures as for VHH(WT). We obtained several thousand specific single-molecule interaction traces for each mutant and analyzed the rupture force behavior (Figure 3). As shown in Figure 3a, the dynamic force spectra for all three mutants look remarkably similar to that of VHH(WT) with VHH/mCherry rupture events occurring at ∼60 pN at loading rates between 103–104 pN/s. Rupture force histograms (Figure 3b) and lifetime analyses (Figure 3c) reflect the fact that no significant differences were observed upon mutagenesis of one or both cysteines. The fitted parameters based on least-squares fitting of the force versus lifetime plot to the DHS model with a cusp-like barrier are shown in Table 2.
Figure 3.
SMFS experiments on VHH(WT) and disulfide bond knockout mutants C24A, C98A, and C24A/C98A. (a) Dynamic force spectra of VHH library members obtained from four pulling speeds from 400 to 3, 200 nm/s. Points shown represent the most probable rupture force versus average loading rate at a given pulling speed. The dashed line represents the Bell-Evans model fit. (b) Rupture force histograms obtained at each pulling speed. (c) Data fitting with the DHS model was used to extract energy landscape parameters shown in Table 2.
Table 2. Most Probable Rupture Forces (Errors Are Full Width at Half-Maximum for Each Rupture Force Distribution) Obtained Different Pulling Speeds and Energy Landscape Parameters (±SD) Based on the DHS Model (v = 0.5).
| most
probable rupture force (pN) |
Dudko–Hummer–Szabo
model |
||||||
|---|---|---|---|---|---|---|---|
| sample | 400 nm/s | 800 nm/s | 1600 nm/s | 3200 nm/s | ΔG (kBT) | Δx (nm) | t0 (s) |
| VHH_WT | 68.78 ± 9.85 | 70.21 ± 10.15 | 73.04 ± 12.29 | 73.42 ± 15.65 | 10.91 ± 1.57 | 0.61 ± 0.16 | 11.02 ± 7.93 |
| C24A | 67.18 ± 14.12 | 69.42 ± 15.33 | 72.72 ± 19.63 | 76.44 ± 22.55 | 10.47 ± 3.01 | 0.59 ± 0.20 | 12.16 ± 8.91 |
| C98A | 65.67 ± 16.07 | 69.82 ± 19.01 | 71.6 ± 22.71 | 75.46 ± 25.16 | 9.22 ± 1.96 | 0.49 ± 0.21 | 3.06 ± 2.87 |
| C24A/C98A | 59.11 ± 12.76 | 64.07 ± 14.65 | 69.06 ± 19.13 | 71.42 ± 20.54 | 8.93 ± 1.93 | 0.46 ± 0.20 | 2.12 ± 2.01 |
The similarity of the SMFS results from all four members of the VHH library demonstrates that the mechanostability of the VHH/mCherry interaction is independent of the disulfide bond state and supports an interpretation that VHH remains folded at the time of bond rupture. If complex rupture was driven by VHH domain unfolding, one would expect that removal of the stabilizing disulfide bond would result in lower rupture forces. The limiting factor in VHH/mCherry mechanostability is therefore likely to be noncovalent interactions at the binding interface, not overall mechanical stability of VHH. This effect could be geometry/linkage dependent, and anchoring VHH at a different position in its sequence could result in a case where disulfide bonding stabilizes the interaction under force. It is known that CDR3 is the most crucial CDR for binding affinity. A superficial inspection of the structure (Figure 1a) indicates that the C-terminal anchor geometry for VHH could easily direct tensile forces directly to CDR3 and across the binding interface, and indeed our results are consistent with such a scenario. Despite disulfide bonding being an important contributor to VHH thermal stability, our results indicate it plays a relatively minor role in antigen binding both under equilibrium conditions where the differences measured by ITC and SPR were small, as well as under mechanical load as tested by AFM-SMFS. As a burgeoning class of immunotherapeutic molecules, VHH domains represent an interesting case study in the interplay between thermal stability, antigen binding affinity, and molecular mechanical properties.
Acknowledgments
This work was supported by the University of Basel, ETH Zurich, an ERC Starting Grant (MMA- 715207), the NCCR in Molecular Systems Engineering, and the SNSF (Project 200021_175478). We thank Prof. Florian Seebeck and Dr. Duy Tien Ta for helpful discussions. We thank Mariia Beliaeva for assistance with ITC.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.9b02062.
Supplementary figures (mass spectrometry, representative force–extension curves, Ellman’s assay), experimental materials and methods, and protein amino acid sequences(PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Goldman E. R.; Liu J. L.; Zabetakis D.; Anderson G. P. Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview. Front. Immunol. 2017, 8, 865. 10.3389/fimmu.2017.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher D.; Helma J.; Schneider A. F. L.; Leonhardt H.; Hackenberger C. P. R. Nanobodies: Chemical Functionalization Strategies and Intracellular Applications. Angew. Chem., Int. Ed. 2018, 57 (9), 2314–2333. 10.1002/anie.201708459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helma J.; Cardoso M. C.; Muyldermans S.; Leonhardt H. Nanobodies and Recombinant Binders in Cell Biology. J. Cell Biol. 2015, 209 (5), 633–644. 10.1083/jcb.201409074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudikoff S.; Pumphrey J. G. Functional Antibody Lacking a Variable-Region Disulfide Bridge. Proc. Natl. Acad. Sci. U. S. A. 1986, 83 (20), 7875–7878. 10.1073/pnas.83.20.7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa-Ogawa Y.; Uegaki K.; Hagihara Y. The Role of Intra-Domain Disulfide Bonds in Heat-Induced Irreversible Denaturation of Camelid Single Domain VHH Antibodies. J. Biochem. 2016, 159 (1), 111–121. 10.1093/jb/mvv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiner T.; Bates M.; Trakhanov S.; Lee C.-T.; Schliep J. E.; Chug H.; Böhning M.; Stark H.; Urlaub H.; Görlich D.. Correction: Nanobodies: Site-Specific Labeling for Super-Resolution Imaging, Rapid Epitope-Mapping and Native Protein Complex Isolation. eLife 2016, 5. 10.7554/eLife.15597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens D.; Pellis M.; Loris R.; Pardon E.; Dumoulin M.; Matagne A.; Wyns L.; Muyldermans S.; Conrath K. Identification of a Universal VHH Framework to Graft Non-Canonical Antigen-Binding Loops of Camel Single-Domain Antibodies. J. Mol. Biol. 2005, 352 (3), 597–607. 10.1016/j.jmb.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Hagihara Y.; Saerens D. Engineering Disulfide Bonds within an Antibody. Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844 (11), 2016–2023. 10.1016/j.bbapap.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Li T.; Bourgeois J.-P.; Celli S.; Glacial F.; Le Sourd A.-M.; Mecheri S.; Weksler B.; Romero I.; Couraud P.-O.; Rougeon F.; et al. Cell-Penetrating Anti-GFAP VHH and Corresponding Fluorescent Fusion Protein VHH-GFP Spontaneously Cross the Blood-Brain Barrier and Specifically Recognize Astrocytes: Application to Brain Imaging. FASEB J. 2012, 26 (10), 3969–3979. 10.1096/fj.11-201384. [DOI] [PubMed] [Google Scholar]
- Hagihara Y.; Matsuda T.; Yumoto N. Cellular Quality Control Screening to Identify Amino Acid Pairs for Substituting the Disulfide Bonds in Immunoglobulin Fold Domains. J. Biol. Chem. 2005, 280 (26), 24752–24758. 10.1074/jbc.M503963200. [DOI] [PubMed] [Google Scholar]
- Hagihara Y.; Mine S.; Uegaki K. Stabilization of an Immunoglobulin Fold Domain by an Engineered Disulfide Bond at the Buried Hydrophobic Region. J. Biol. Chem. 2007, 282 (50), 36489–36495. 10.1074/jbc.M707078200. [DOI] [PubMed] [Google Scholar]
- Kunz P.; Zinner K.; Mücke N.; Bartoschik T.; Muyldermans S.; Hoheisel J. D. The Structural Basis of Nanobody Unfolding Reversibility and Thermoresistance. Sci. Rep. 2018, 8 (1), 7934. 10.1038/s41598-018-26338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabetakis D.; Olson M. A.; Anderson G. P.; Legler P. M.; Goldman E. R. Evaluation of Disulfide Bond Position to Enhance the Thermal Stability of a Highly Stable Single Domain Antibody. PLoS One 2014, 9 (12), e115405. 10.1371/journal.pone.0115405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y.; Kandalaft H.; Ding W.; Ryan S.; van Faassen H.; Hirama T.; Foote S. J.; MacKenzie R.; Tanha J. Disulfide Linkage Engineering for Improving Biophysical Properties of Human VH Domains. Protein Eng., Des. Sel. 2012, 25 (10), 581–589. 10.1093/protein/gzs055. [DOI] [PubMed] [Google Scholar]
- Govaert J.; Pellis M.; Deschacht N.; Vincke C.; Conrath K.; Muyldermans S.; Saerens D. Dual Beneficial Effect of Interloop Disulfide Bond for Single Domain Antibody Fragments. J. Biol. Chem. 2012, 287 (3), 1970–1979. 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Cao Y. Protein Mechanics: From Single Molecules to Functional Biomaterials. Acc. Chem. Res. 2010, 43 (10), 1331–1341. 10.1021/ar100057a. [DOI] [PubMed] [Google Scholar]
- Yu H.; Siewny M. G. W.; Edwards D. T.; Sanders A. W.; Perkins T. T. Hidden Dynamics in the Unfolding of Individual Bacteriorhodopsin Proteins. Science 2017, 355 (6328), 945–950. 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa I.; Fernández J. M.; Garcia-Manyes S. Direct Quantification of the Attempt Frequency Determining the Mechanical Unfolding of Ubiquitin Protein. J. Biol. Chem. 2011, 286 (36), 31072–31079. 10.1074/jbc.M111.264093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl Z. N.; Yang W.; Marszalek P. E. Direct Observation of Multimer Stabilization in the Mechanical Unfolding Pathway of a Protein Undergoing Oligomerization. ACS Nano 2015, 9 (2), 1189–1197. 10.1021/nn504686f. [DOI] [PubMed] [Google Scholar]
- Hughes M. L.; Dougan L. The Physics of Pulling Polyproteins: A Review of Single Molecule Force Spectroscopy Using the AFM to Study Protein Unfolding. Rep. Prog. Phys. 2016, 79 (7), 076601. 10.1088/0034-4885/79/7/076601. [DOI] [PubMed] [Google Scholar]
- Johnson K. C.; Thomas W. E. How Do We Know When Single-Molecule Force Spectroscopy Really Tests Single Bonds?. Biophys. J. 2018, 114 (9), 2032–2039. 10.1016/j.bpj.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers C. E.; Crozat E.; Chu C.; Moy V. T.; Sherratt D. J.; Howarth M. A Streptavidin Variant with Slower Biotin Dissociation and Increased Mechanostability. Nat. Methods 2010, 7 (5), 391–393. 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico F.; Moy V. T. Energy Landscape Roughness of the Streptavidin-Biotin Interaction. J. Mol. Recognit. 2007, 20 (6), 495–501. 10.1002/jmr.841. [DOI] [PubMed] [Google Scholar]
- Sedlak S. M.; Bauer M. S.; Kluger C.; Schendel L. C.; Milles L. F.; Pippig D. A.; Gaub H. E. Monodisperse Measurement of the Biotin-Streptavidin Interaction Strength in a Well-Defined Pulling Geometry. PLoS One 2017, 12 (12), e0188722. 10.1371/journal.pone.0188722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienberger F.; Kada G.; Mueller H.; Hinterdorfer P. Single Molecule Studies of Antibody-Antigen Interaction Strength versus Intra-Molecular Antigen Stability. J. Mol. Biol. 2005, 347 (3), 597–606. 10.1016/j.jmb.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Morfill J.; Blank K.; Zahnd C.; Luginbühl B.; Kühner F.; Gottschalk K.-E.; Plückthun A.; Gaub H. E. Affinity-Matured Recombinant Antibody Fragments Analyzed by Single-Molecule Force Spectroscopy. Biophys. J. 2007, 93 (10), 3583–3590. 10.1529/biophysj.107.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwesinger F.; Ros R.; Strunz T.; Anselmetti D.; Güntherodt H. J.; Honegger A.; Jermutus L.; Tiefenauer L.; Pluckthun A. Unbinding Forces of Single Antibody-Antigen Complexes Correlate with Their Thermal Dissociation Rates. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (18), 9972–9977. 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulchek T. A.; Friddle R. W.; Langry K.; Lau E. Y.; Albrecht H.; Ratto T. V.; DeNardo S. J.; Colvin M. E.; Noy A. Dynamic Force Spectroscopy of Parallel Individual Mucin1-Antibody Bonds. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (46), 16638–16643. 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bausier P.; Labate C.; Towell A. M.; Derclaye S.; Geoghegan J. A.; Dufrêne Y. F. Staphylococcus Aureus Clumping Factor A Is a Force-Sensitive Molecular Switch That Activates Bacterial Adhesion. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (21), 5564–5569. 10.1073/pnas.1718104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles L. F.; Bayer E. A.; Nash M. A.; Gaub H. E. Mechanical Stability of a High-Affinity Toxin Anchor from the Pathogen Clostridium Perfringens. J. Phys. Chem. B 2017, 121, 3620. 10.1021/acs.jpcb.6b09593. [DOI] [PubMed] [Google Scholar]
- Milles L. F.; Schulten K.; Gaub H. E.; Bernardi R. C. Molecular Mechanism of Extreme Mechanostability in a Pathogen Adhesin. Science 2018, 359 (6383), 1527–1533. 10.1126/science.aar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Malinowska K. H.; Bernardi R. C.; Milles L. F.; Jobst M. A.; Durner E.; Ott W.; Fried D. B.; Bayer E. A.; Schulten K.; et al. Ultrastable Cellulosome-Adhesion Complex Tightens under Load. Nat. Commun. 2014, 5, 1–35. 10.1038/ncomms6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Bernardi R. C.; Malinowska K. H.; Durner E.; Ott W.; Bayer E. A.; Schulten K.; Nash M. A.; Gaub H. E. Mapping Mechanical Force Propagation through Biomolecular Complexes. Nano Lett. 2015, 15 (11), 7370–7376. 10.1021/acs.nanolett.5b02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobst M. A.; Schoeler C.; Malinowska K.; Nash M. A. Investigating Receptor-Ligand Systems of the Cellulosome with AFM-Based Single-Molecule Force Spectroscopy. J. Vis. Exp. 2013, 82, e50950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott W.; Jobst M. A.; Schoeler C.; Gaub H. E.; Nash M. A. Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes: The Current Toolbox. J. Struct. Biol. 2017, 197 (1), 3–12. 10.1016/j.jsb.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Durner E.; Ott W.; Nash M. A.; Gaub H. E. Post-Translational Sortase-Mediated Attachment of High-Strength Force Spectroscopy Handles. ACS Omega 2017, 2 (6), 3064–3069. 10.1021/acsomega.7b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S. W.; Nash M. A.; Fried D. B.; Slutzki M.; Barak Y.; Bayer E. A.; Gaub H. E. Single-Molecule Dissection of the High-Affinity Cohesin-Dockerin Complex. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (50), 20431–20436. 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdorfer T.; Bernardi R. C.; Meinhold A.; Ott W.; Luthey-Schulten Z.; Nash M. A.; Gaub H. E. Combining in Vitro and in Silico Single-Molecule Force Spectroscopy to Characterize and Tune Cellulosomal Scaffoldin Mechanics. J. Am. Chem. Soc. 2017, 139, 17841. 10.1021/jacs.7b07574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M. A.; Smith S. P.; Fontes C. M.; Bayer E. A. Single versus Dual-Binding Conformations in Cellulosomal Cohesin–dockerin Complexes. Curr. Opin. Struct. Biol. 2016, 40, 89–96. 10.1016/j.sbi.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Vera A. M.; Carrión-Vázquez M. Direct Identification of Protein-Protein Interactions by Single-Molecule Force Spectroscopy. Angew. Chem., Int. Ed. 2016, 55 (45), 13970–13973. 10.1002/anie.201605284. [DOI] [PubMed] [Google Scholar]
- Beedle A. E. M.; Mora M.; Lynham S.; Stirnemann G.; Garcia-Manyes S. Tailoring Protein Nanomechanics with Chemical Reactivity. Nat. Commun. 2017, 8, 15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D.; Perisic O.; Peng Q.; Cao Y.; Lam C.; Lu H.; Li H. Single-Molecule Force Spectroscopy Reveals a Mechanically Stable Protein Fold and the Rational Tuning of Its Mechanical Stability. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (22), 9278–9283. 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A.; Alonso-Caballero Á.; Fertin M.; Poly S.; De Sancho D.; Perez-Jimenez R. The Influence of Disulfide Bonds on the Mechanical Stability of Proteins Is Context Dependent. J. Biol. Chem. 2017, 292 (32), 13374–13380. 10.1074/jbc.M117.784934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrance O. E.; Hann E.; Kaminska R.; Housden N. G.; Derrington S. R.; Kleanthous C.; Radford S. E.; Brockwell D. J. A Force-Activated Trip Switch Triggers Rapid Dissociation of a Colicin from Its Immunity Protein. PLoS Biol. 2013, 11 (2), e1001489. 10.1371/journal.pbio.1001489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Caballero A.; Schönfelder J.; Poly S.; Corsetti F.; De Sancho D.; Artacho E.; Perez-Jimenez R. Mechanical Architecture and Folding of E. Coli Type 1 Pilus Domains. Nat. Commun. 2018, 9 (1), 2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giganti D.; Yan K.; Badilla C. L.; Fernandez J. M.; Alegre-Cebollada J. Disulfide Isomerization Reactions in Titin Immunoglobulin Domains Enable a Mode of Protein Elasticity. Nat. Commun. 2018, 9 (1), 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl P.; Kwok C. H.; Manderson G.; Speicher D. W.; Discher D. E. Forced Unfolding Modulated by Disulfide Bonds in the Ig Domains of a Cell Adhesion Molecule. Proc. Natl. Acad. Sci. U. S. A. 2001, 98 (4), 1565–1570. 10.1073/pnas.98.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamecka K.; Severin P. M.; Milles L. F.; Gaub H. E.; Leonhardt H. Energy Profile of Nanobody-GFP Complex under Force. Phys. Biol. 2015, 12 (5), 056009. 10.1088/1478-3975/12/5/056009. [DOI] [PubMed] [Google Scholar]
- Schwaiger I.; Kardinal A.; Schleicher M.; Noegel A. A.; Rief M. A Mechanical Unfolding Intermediate in an Actin-Crosslinking Protein. Nat. Struct. Mol. Biol. 2004, 11 (1), 81–85. 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- Yin J.; Straight P. D.; McLoughlin S. M.; Zhou Z.; Lin A. J.; Golan D. E.; Kelleher N. L.; Kolter R.; Walsh C. T. Genetically Encoded Short Peptide Tag for Versatile Protein Labeling by Sfp Phosphopantetheinyl Transferase. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (44), 15815–15820. 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib I.; Smolarek D.; Hattab C.; Grodecka M.; Hassanzadeh-Ghassabeh G.; Muyldermans S.; Sagan S.; Gutiérrez C.; Laperche S.; Le-Van-Kim C.; et al. V(H)H (nanobody) Directed against Human Glycophorin A: A Tool for Autologous Red Cell Agglutination Assays. Anal. Biochem. 2013, 438 (1), 82–89. 10.1016/j.ab.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Zarschler K.; Witecy S.; Kapplusch F.; Foerster C.; Stephan H. High-Yield Production of Functional Soluble Single-Domain Antibodies in the Cytoplasm of Escherichia Coli. Microb. Cell Fact. 2013, 12, 97. 10.1186/1475-2859-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Huang H. Expression of Single-Domain Antibody in Different Systems. Appl. Microbiol. Biotechnol. 2018, 102 (2), 539–551. 10.1007/s00253-017-8644-3. [DOI] [PubMed] [Google Scholar]
- Ta D. T.; Redeker E. S.; Billen B.; Reekmans G.; Sikulu J.; Noben J.-P.; Guedens W.; Adriaensens P. An Efficient Protocol towards Site-Specifically Clickable Nanobodies in High Yield: Cytoplasmic Expression in Escherichia Coli Combined with Intein-Mediated Protein Ligation. Protein Eng. Des. Sel. 2015, 28 (10), 351–363. [DOI] [PubMed] [Google Scholar]
- Billen B.; Vincke C.; Hansen R.; Devoogdt N.; Muyldermans S.; Adriaensens P.; Guedens W. Cytoplasmic versus Periplasmic Expression of Site-Specifically and Bioorthogonally Functionalized Nanobodies Using Expressed Protein Ligation. Protein Expression Purif. 2017, 133, 25–34. 10.1016/j.pep.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Liu H.; Ta D. T.; Nash M. A. Mechanical Polyprotein Assembly Using Sfp and Sortase-Mediated Domain Oligomerization for Single-Molecule Studies. Small Methods 2018, 2, 1800039. 10.1002/smtd.201800039. [DOI] [Google Scholar]
- Leem J.; Dunbar J.; Georges G.; Shi J.; Deane C. M. ABodyBuilder: Automated Antibody Structure Prediction with Data–driven Accuracy Estimation. MAbs 2016, 8 (7), 1259–1268. 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Verdorfer T.; Gaub H. E.; Nash M. A. Biasing Effects of Receptor-Ligand Complexes on Protein-Unfolding Statistics. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2016, 94 (4–1), 042412. 10.1103/PhysRevE.94.042412. [DOI] [PubMed] [Google Scholar]
- Dudko O. K.; Hummer G.; Szabo A. Theory, Analysis, and Interpretation of Single-Molecule Force Spectroscopy Experiments. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (41), 15755–15760. 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles L. F.; Gaub H. E. Is Mechanical Receptor Ligand Dissociation Driven by Unfolding or Unbinding?. bioRxiv 2019, 593335. 10.1101/593335. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.