Abstract
MicroRNAs (miRNAs or miRs) are small noncoding RNAs derived from genome to control target gene expression. Recently we have developed a novel platform permitting high-yield production of bioengineered miRNA agents (BERA). This study is to produce and utilize novel fully-humanized BERA/miR-328-3p molecule (hBERA/miR-328) to delineate the role of miR-328-3p in controlling nutrient uptake essential for cell metabolism. We first demonstrated successful high-level expression of hBERA/miR-328 in bacteria and purification to high degree of homogeneity (>98%). Biologic miR-328-3p prodrug was selectively processed to miR-328-3p to suppress the growth of highly-proliferative human osteosarcoma (OS) cells. Besides glucose transporter protein type 1, gene symbol solute carrier family 2 member 1 (GLUT1/SLC2A1), we identified and verified large neutral amino acid transporter 1, gene symbol solute carrier family 7 member 5 (LAT1/SLC7A5) as a direct target for miR-328-3p. While reduction of LAT1 protein levels by miR-328-3p did not alter homeostasis of amino acids within OS cells, suppression of GLUT1 led to a significantly lower glucose uptake and decline in intracellular levels of glucose and glycolytic metabolite lactate. Moreover, combination treatment with hBERA/miR-328 and cisplatin or doxorubicin exerted a strong synergism in the inhibition of OS cell proliferation. These findings support the utility of novel bioengineered RNA molecules and establish an important role of miR-328-3p in the control of nutrient transport and homeostasis behind cancer metabolism.
Key words: Bioengineered RNA, MiR-328, LAT1, GLUT1, Chemosensitivity, Cancer
Abbreviations: 2-NBDG, 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxyglucose; ABCG2, ATP-binding cassette subfamily G member 2; ACN, acetonitrile; Au/Uv, absorbance unit of ultraviolet-visible spectroscopy; BCRP, breast cancer resistant protein; BERA, bioengineered miRNA agent; CI, combination index; CPT, cisplatin; DOX, doxorubicin; E. coli, Escherichia coli; ESI, electrospray ionization; Fa, fraction affected; FPLC, fast protein liquid chromatography; GLUT1, glucose transporter protein type 1; hBERA, humanized bioengineered miRNA agent; HCC, hepatocellular carcinoma; HPLC, high-performance liquid chromatography; hsa, Homo sapiens; htRNASer, human seryl-tRNA; IS, internal standard; KRB, Krebs–Ringer bicarbonate; LAT1, large neutral amino acid transporter 1; LC–MS/MS, liquid chromatography–tandem mass spectroscopy; MCT4, monocarboxylate transporter 4; miR or miRNA, microRNA; MRE, miRNA response elements; MRM, multiple reaction monitoring; mTOR, mammalian target of rapamycin; ncRNA, noncoding RNAs; nt, nucleotide; OS, osteosarcoma; PAGE, polyacrylamide gel electrophoresis; PTEN, phosphatase and tensin homolog; PVDF, Polyvinylidene fluoride; RAGE, receptor for advanced glycosylation end products; RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction; SLC2A1, 7A5, 16A3, solute carrier family 2 member 1, family 7 member 5, family 16 member 3; WT, wild type
Graphical abstract
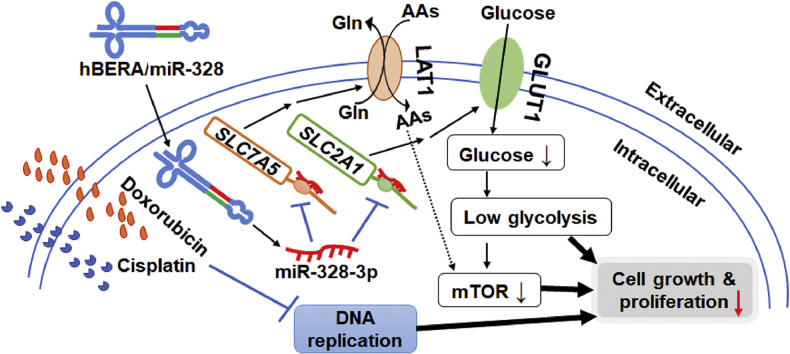
Fully-humanized, bioengineered miR-328-3p agent reduces glycolysis through the suppression of protein levels of targeted transporters GLUT1 and LAT1 in human osteosarcoma cells, leading to synergistic antiproliferative effects when combined with chemotherapeutic drugs.
1. Introduction
MicroRNAs (miRs or miRNAs) are a superfamily of small (usually 18–25 nucleotides in length) noncoding RNAs (ncRNAs) derived from the genome1, 2, 3. Through interactions with corresponding miRNA response element (MRE) within the 3′-untranslated region (3′UTR) of targeted transcript to achieve the regulation of target gene expression at the posttranscriptional level1, 2, 3, 4, miRNAs are involved in the control of various cellular processes in development, such as cell cycle, proliferation, differentiation, metabolism, migration, apoptosis, autophagy, and senescence. Biogenic miRNAs may promote or suppress disease initiation and progression including tumorigenesis and metastasis, while the biogenesis and final expression levels of certain miRNAs are significantly altered in diseased cells1, 2, 3, 5. With an improved understanding of the roles of miRNAs in human diseases as well as development of means to manipulate miRNA-controlled processes, large efforts have been made to develop novel miRNA-based therapeutic strategies such as those for the treatment of lethal cancer6, 7, 8, 9.
Being the most common type of primary bone malignancy, osteosarcoma (OS) occurs predominantly among children and adolescents while it is rarely diagnosed in adults. OS exhibits a high rate of metastasis especially pulmonary metastasis, in addition to local and regional destruction to the bone and surrounding tissues10, 11. Although the development of neoadjuvant chemotherapy and refinement of surgical techniques have markedly improved the survival rate of patients with localized OS, pulmonary metastasis complicates the treatment and results in a sharp drop of overall survival rate down to 20%–30%12, 13. Moreover, the incidence of recurrences is very high in pediatric OS patients, e.g., 30%–50% for patients with initial localized OS and 80% of patients with metastasis at diagnosis14. Therefore, it is important to investigate novel therapeutic strategies to combat against aggressive or recurrent OS that would help extend the lifespans of pediatric patients.
Among a number of miRNAs strongly implicated in cancer, miR-328-3p is commonly downregulated in many types of tumors, including OS, lung, colon, breast, and hepatocellular carcinoma (HCC)15, 16, 17, 18, 19, 20. However, there are some discrepancies in the roles of miR-328 in carcinogenesis which was found to exhibit antiproliferative activities by most studies15, 16, 17, 18 amid reports of pro-proliferative effects19, 20. This could be due to the variation in cell-specific actions of a miRNA or simply the intrinsic difference between the dominant form miR-328-3p and trivial miR-328-5p, which the latter might have been overlooked. Our previous research has revealed that miR-328-3p negatively regulates the expression of breast cancer resistance protein, gene symbol ATP-binding cassette subfamily G member 2 (BCRP/ABCG2) behind multidrug resistance, and subsequently elevates intracellular drug accumulation and sensitizes cancer cells to chemotherapeutics21. A recent study has also showed that miR-328-3p, downregulated in colon cancer patient samples, is able to modulate the expression of glucose transporter protein type 1, gene symbol solute carrier family 2 member 1 (GLUT1/SLC2A1)17, which might have implications in cancer metabolism. Another latest study have demonstrated a significant downregulation of miR-328-3p levels in OS tissues and cell lines, and identified matrix metalloproteinase-16 (MMP-16) as a direct target for miR-328-3p showing antiproliferative activity against OS cells15. Further studies have also indicated that intratumoral administration of miR-328-3p is effective to improve the sensitivity of OS xenograft tumor to radiotherapy in mouse models22. Rather, the mechanistic actions of antiproliferative miR-328-3p remain elusive, especially in the control of nutrient disposition and homeostasis critical for cell metabolism.
Recently we have established a novel tRNA/pre-miRNA-based platform to achieve high-yield and large-scale production of bioengineered miRNA agents (BERAs) through microbial fermentation, which represent a new class of ncRNA molecules for research on miRNA biology and development of miRNA therapeutics23, 24, 25, 26. Distinguished from chemically-synthesized miRNA mimics carrying extensive and various forms of chemical modifications, BERAs are produced and folded within living cells and comprised of a minimal level of posttranscriptional modifications that should better capture the structures, physicochemical properties, biological functions, and safety profiles of cellular RNAs9, 27. Our studies have proved the concept that recombinant BERA/miR-34a molecule is active in the control of miR-34a target gene expression in human cells, and thus effective to inhibit cancer cell proliferation, xenograft tumor growth, and pulmonary metastasis23, 24, 28, 29. Further studies have also demonstrated that BERA/miR-1291 prodrug is able to improve chemosensitivity through the downregulation of efflux transporters and other cancer-related genes30, 31. This approach is also employed for successful production of BERA/miR-328-3p molecule that indeed suppresses the expression of transporter BCRP/ABCG2 to sensitize drug-resistant cells32. Nevertheless, all these tRNA/pre-miRNA carriers consist of a bacterial tRNA. To make recombinant RNAs more compatible to human cells, we have committed to creating fully-humanized BERAs (hBERAs) by replacing the bacterial tRNA with a human tRNA.
Herein, we presented the design of a novel hBERA/miR-328 molecule by using human seryl-tRNA (htRNASer)/pre-miR-34a carrier (Fig. 1A). After successful expression and purification of hBERA/miR-328, we demonstrated that target miR-328-3p was selectively released from biologic ncRNA molecule in human OS cells to exert antiproliferative activity. Furthermore, we verified for the first time that the large neutral amino acid transporter 1, gene symbol solute carrier family 7 member 5 (LAT1/SLC7A5) was a direct target of miR-328-3p, besides glucose transporter GLUT1/SLC2A1. As a result, hBERA/miR-328 was effective to reduce both LAT1 and GLUT1 protein levels in human OS cells. In addition, downregulation of GLUT1 by miR-328-3p led to the disruption of glucose uptake and homeostasis, and subsequently a strong synergistic antiproliferation activity against OS cells with chemotherapeutic drug cisplatin or doxorubicin.
Figure 1.
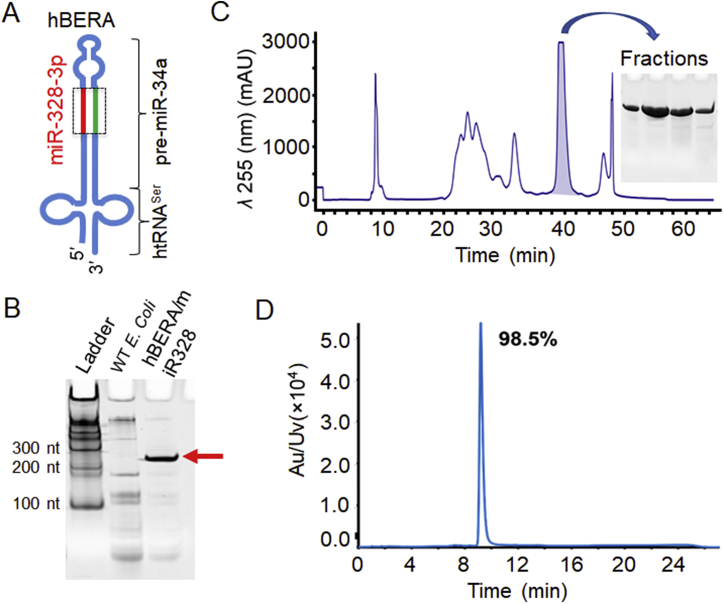
Production of fully-humanized biologic miR-328-3p molecule. (A) Schematic illustration of the fully-humanized bioengineered RNA agent (hBERA) bearing the target miR-328-3p (hBERA/miR-328, 192 nucleotides in length). The carrier consists of a human serine tRNA (htRNASer) and a modified pre-miR-34a23, in which miR-34a duplexes are replaced by target miR-328 sequences. (B) High-level heterogeneous expression of hBERA/miR-328 through bacterial fermentation, as demonstrated by urea-PAGE analysis. Total RNA from wild type (WT) Escherichia coli was used for comparison. (C) FPLC trace during the purification of hBERA/miR-328. The insert shows urea-PAGE analysis of target RNA fractions collected during FPLC separation. (D) The purity of isolated hBERA/miR-328-3p (98.5%) was determined by HPLC analysis. Au/Uv, absorbance unit of ultraviolet-visible spectroscopy; nt, nucleotide.
2. Materials and methods
2.1. Chemicals and materials
Trizol reagent, BCA protein assay kit, RPMI 1640 medium, fetal bovine serum, Lipofectamine 3000, 0.05% trypsin-EDTA, and RIPA lysis buffer were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Protease inhibitor cocktail was bought from Sigma–Aldrich (St. Louis, MO, USA). Bovine serum albumin and dimethyl sulfoxide were bought from VWR (Radnor, PA, USA). Polyvinylidene fluoride (PVDF) membrane, Western blot ECL substrate kit, and blotting-grade blocker were purchased from Bio-Rad (Hercules, CA, USA). Direct-zol RNA MiniPrep kit was bought from Zymo Research (Irvine, CA, USA). All other solvents and chemicals of analytical grade were purchased from either Thermo Fisher Scientific or Sigma–Aldrich.
2.2. Cell culture and transfection
HEK293 cells and human OS MG63 and 143B cells were purchased from the American Type Culture Collection (Manassas, VA, USA), and maintained in DMEM or RPMI 1640 medium supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air. Cells were transfected with target plasmids or RNAs by using Lipofectamine 3000 reagent, according to the manufacturer's instructions.
2.3. Production of recombinant RNA agents
Expression and purification of fully-humanized bioengineered RNA agents (hBERA) bearing the target miR-328-3p (hBERA/miR-328; Fig. 1A) and corresponding sephadex aptamer tagged htRNASer (control RNA) were conducted as we described recently9, 23, 25, 32 with some modifications (unpublished data), in particular, by replacing bacterial tRNA with human tRNASer (Fig. 1A). Briefly, the recombinant hBERA/miR-328 expression plasmid was constructed by using htRNASer/pre-miR-34a as the carrier and substituting the miR-34a duplexes with target miR-328 sequences with In-Fusion® HD cloning kit (Takara, Mountain View, CA, USA), and further confirmed by DNA sequencing (Genscript, Piscataway, NJ, USA). Recombinant ncRNAs were overexpressed in Escherichia coli HST08 on small scale (15 mL) for initial assessment and then large scale (0.5 L) for mass production. Purification of target RNA molecules was achieved by using the anion exchange fast protein liquid chromatography (FPLC) methods23, 32 on an NGC QUEST 10PLUS FPLC system (Bio-Rad). The purity of isolated RNAs were estimated by denaturing urea (8 mol/L) polyacrylamide (8%) gel electrophoresis (PAGE) and quantitated accurately by the high-performance liquid chromatography (HPLC) method25 on a Prominence Ultra-Fast Liquid Chromatography system (Shimadzu Corporation, Kyoto, Japan) . Endotoxin levels were measured by using Limulus Amebocyte Lysate Pyrogent-5000 kinetic assay (Lonza, Walkersville, MD, USA). Recombinant RNAs with high homogeneity (≥ 98%) and low endotoxin activity (≤ 5 EU/μg RNA) were used in the following studies.
2.4. RNA isolation and reverse transcription quantitative real-time PCR (RT-qPCR)
MG63, 143B and HEK293 cells were seeded in 24-well plates at a density of 4 × 104 cells per well and incubated overnight. The cells were then transfected with 15 nmol/L hBERA/miR-328, control RNA, or lipofectamine 3000 reagent only (vehicle). Forty-eight hours later, total RNAs were isolated with the Direct-zol RNA MiniPrep kit (Zymo Research) and quantified with a NanoDrop spectrophotometer (Thermo Scientific, Rockford, IL, USA). Reverse transcription was conducted by NxGen M-MuLV reverse transcriptase (Lucigen, Middleton, WI, USA) and random hexamers (for U6) or stem-loop primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACGGAA-3′ (for miR-328-3p). Then qPCR analyses were performed with iTaq™ Universal SYBR® Green Supermix (Bio-Rad) and the following gene specific primers, forward 5′-ATATCTGGCCCTCTCTGCCC-3′, reverse 5′-GTGCAGGGTCCGAGGT-3′ for miR-328-3p, and forward 5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-AACGCTTCACGAATTTGCGT-3′ for U6, on a CFX96 Touch real-time PCR system (Bio-Rad). The relative level of mature miR-328-3p over internal standard (U6) was calculated as 2ΔΔCT, in which ΔΔCT = ΔCT treatment group (analyte–internal standard) − ΔCT control group (analyte–internal standard).
2.5. Protein isolation and Western blot analyses
MG-63 and 143B cells were seeded in 6-well plates at a density of 3 × 105 cells/well and treated with 15 nmol/L hBERA/miR-328 or control RNA and then harvested at 48 h post-transfection. Total protein of the harvested cells was isolated with RIPA lysis buffer supplemented with complete protease inhibitors. Protein concentrations were quantitated using a BCA protein assay kit (Thermo Fisher Scientific). Proteins (30 μg per lane) were separated on a 10% SDS-PAGE gel and electrophoretically transferred onto PVDF membranes. After a 2-h incubation with 5% nonfat milk, membranes were incubated with primary antibodies against GLUT1 (1:2000 dilution, ab32551, Abcam, Cambridge, MA, USA), LAT1 (1:500 dilution, SC374232, Santa Cruz Biotechnology, Dallas, TX, USA), mammalian target of rapamycin (mTOR, 1:500 dilution, 2983S, Cell Signaling Technology, Danvers, MA, USA), p-mTOR (1:500 dilution, 2971S, Cell Signaling Technology) or β-actin (1:5000 dilution, A5441, Sigma–Aldrich) overnight at 4 °C. The membranes were then incubated with horseradish peroxidase-labeled secondary antibodies (anti-mouse, 1:3000 dilution, Cell Signaling Technology; anti-rabbit, 1:10,000 dilution, Jackson ImmunoResearch, West Grove, PA, USA) at room temperature for 2 h, followed by incubation in Clarity Western ECL substrates. Images were obtained with a ChemiDoc MP Imaging System (Bio-Rad). Protein band intensity values were determined by Image Laboratory software (Bio-Rad) and then normalized to the β-actin levels in corresponding samples for comparison.
2.6. Identification of MRE sites and luciferase reporter assay
Putative MREs for miR-328-3p within the 3′UTR of SLC7A5/LAT1 and SLC2A1/GLUT1 were identified by using TargetScan (http://www.targetscan.org/). Full length of human LAT1 3′ UTR and GLUT1 3′ UTR was cloned into downstream of humanized firefly luciferase gene within pEZX-MT06 plasmid (Genecopoeia, Rockville, MD, USA), and were named as pEZX-MT06-LAT1-3′ UTR and pEZX-MT06-GLUT1-3′ UTR, respectively. Luciferase reporter assays were performed as previously reported with minor modifications21, 30, 33. Briefly, HEK293 cells were seeded in 96-well plates at a density of 1.5 × 104 cells/well and incubated overnight. Cells were transfected with pEZX-MT06-LAT1-3′ UTR or pEZX-MT06-GLUT1-3′ UTR alone, or along with bioengineered miR-328 or control RNA. Forty-eight hours post-transfection, both the firefly luciferase activity and Renilla luciferase activity (internal control) were measured with a dual-luciferase reporter assay kit (Promega, Madison, WI, USA) on a SpectraMax M3 Microplate Reader (Molecular Devices, LLC., San Jose, CA, USA), according to the manufacturer's protocols. The firefly luciferase activity was normalized to Renilla luciferase activity and then the corresponding 3′ UTR-expressing plasmid alone treatment for comparison. Each treatment was performed in five replicates.
2.7. Cell viability assay
Cells were seeded in 96-well plates at 5 × 103 cells per well and transfected with 15 nmol/L hBERA/miR-328 or control RNA. Cell viability was determined at designated time points (0, 24, 48, 72, and 96 h post-transfection) by using a CellTiter-Glo Luminescent cell viability assay kit (Promega) according to manufacturer's protocol. The relative cell viability values were calculated by normalizing the luminescence intensities at particular time points to untreated cells (0 h). All experiments were conducted in five replicates. For drug combination study, cells were treated with various concentrations of hBERA/miR-328 alone (0–50 nmol/L), chemotherapeutic drug alone (doxorubicin 0–500 nmol/L; cisplatin 0–50,000 nmol/L), or their combinations (hBERA/miR-328 plus doxorubicin at a fixed ratio 1:10; hBERA/miR-328 plus cisplatin at a fixed ratio 1:1000) for 48 h. Cell viability was determined by MTT assays31, and the data were normalized to vehicle control (0%) to calculate the degrees of inhibition.
To estimate the EC50 of each drug, cell viability data were fitted to an inhibitory, normalized response model with variable slop, Y = 100/{1 + 10^[(Log IC50−X) × HillSlope]} (GraphPad Prism, San Diego, CA, USA). To define the interaction of hBERA/miR-328 and chemotherapeutics in the inhibition of OS cell growth, combination index (CI) at each fraction affected (Fa) was calculated by CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA)34. CI > 1 indicates an antagonistic interaction, CI = 1 indicates additive interaction, and CI < 1 indicates synergism between drugs.
2.8. LC–MS/MS quantification of amino acids, glucose and lactate
Intracellular and extracellular levels of amino acids, glucose and lactate were quantitated with a Shimadzu Prominence ultra-fast liquid chromatography system coupled to an AB Sciex 4000 QTRAP tandem mass spectrometry (AB Sciex, Framingham, MA, USA). All the samples were separated on an Intrada Amino Acid column (50 mm × 3 mm, 3 μm; ImtaktUSA, Portland, OR, USA). Data were analyzed and processed by the Analyst software (Version 1.6.2, AB Sciex).
To determine extra- and intra-cellular amino acids, cells were seeded in 6-well plates at a density of 2.5 × 105 cells/well (143B) or 3 × 105 cells/well (MG63), and then treated with 15 nmol/L hBERA/miR-328 or control RNA. Forty-eight hours later, the culture medium was diluted with acetonitrile (ACN)/water (v/v = 4:1) supplemented with internal standard (IS, 4-chloro-phenylalanine, 0.2 μmol/L) and 1% formic acid to 1000 folds, vortexed for 5 min, and centrifuged at 16,000×g for 15 min at 4 °C. The supernatant was transferred to a new vial and subjected to LC–MS/MS analysis. Meanwhile, cells were harvested and frozen with liquid nitrogen and subsequently thawed with ultrasonication in water, followed by two more freeze–thaw cycles to get cell lysates. A 100 μL of cell lysates with a protein concentration of 200 ng/μL was added to 300 μL ACN consisting of 1% formic acid and IS (final concentration is 0.2 μmol/L), and then the mixture was vortexed for 5 min and centrifuged at 16,000×g for 15 min. The supernatants were transferred to new vials and injected for LC–MS/MS analysis. Amino acids were separated through gradient elution with an aqueous solution A (100 mmol/L ammonium formate) and solvent B (5% water + 95% ACN + 0.3% formic acid) at a flow rate of 0.6 mL/min, as we reported recently35. All compounds (amino acids, IS) were detected and quantified in positive electrospray ionization (ESI) mode and multiple reaction monitoring (MRM) mode under optimized conditions35.
To analyze extra- and intra-cellular glucose and lactate levels, cell culture medium was diluted to 5000-fold with water. Likewise, cell lysate samples were diluted with water to achieve the same protein concentration (200 ng/μL). Then 40 μL cell lysates or diluted medium were mixed with 120 μL ACN consisting of 0.4% formic acid and IS 5-fluorouracil (final concentration 0.5 μmol/L) to precipitate the protein. The mixture was vortexed for 5 min and centrifuged at 16,000× g for 15 min. The supernatant was transferred to a new vial and injected for LC–MS/MS analysis. Analytes were separated by using the following mobile phase, solution C (96% water + 4% ACN + 0.2% formic acid) and solution D (5% water + 95% ACN + 0.2% formic acid) at a flow rate of 0.4 mL/min. Column was eluted with 85% solution D for 1.5 min, which was decreased to 60% solution D over 2.5 min, maintained at 100% for 2.9 min, and then returned to initial condition and maintained for another 2.9 min. The mass spectrometric analysis was conducted by using ESI and negative-ion MRM, specifically, m/z 178.9–88.9 for glucose, m/z 88.8–43.0 for lactate, and m/z 129–42 for the IS 5-fluorouracil.
2.9. Glucose uptake assay
MG63 or 143B cells were seeded in 96-well plates at 5 × 103 cells per well and treated with 15 nmol/L hBERA/miR-328 or control RNA for 48 h. Cells were washed twice with 37 °C glucose-free Krebs–Ringer bicarbonate (KRB) buffer (pH 7.4), and pre-incubated with glucose-free KRB buffer in a 5% CO2 incubator at 37 °C for 15 min. Then the incubation buffer was replaced with 100 μL of fresh KRB buffer supplemented with 600 μmol/L 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxyglucose (2-NBDG, Thermo Scientific) and incubated with the cells for another 1 h, followed by three washes with ice-cold PBS. After adding another 100 μL ice-cold PBS, fluorescence intensity was determined immediately at excitation/emission wavelength of 465/540 nm on a SpectraMax Microplate Reader for the assessment of glucose uptake capacity. Experiments were conducted in five replicates, and data were normalized to vehicle control group (set as 100%) for comparison.
2.10. Statistical analysis
All values are expressed as mean ± SD, and different treatment groups were compared by one-way or two-way ANOVA (Prism, GraphPad Software Inc., San Diego, CA, USA). Difference was considered as statistically significant when P value was less than 0.05 (P < 0.05).
3. Results
3.1. Fermentation production of novel hBERA/miR-328 agent and purification to high degree of homogeneity
Our recent studies have established a tRNA/pre-miRNA-based platform that offers high-level, large-scale, and cost-effective production of biologic miRNA agents through microbial fermentation23, 26. In this study, a new fully-humanized BERA carrying the warhead miR-328-3p, namely hBERA/miR-328, was designed by using the htRNASer/pre-miR-34a as a carrier and replacing miR-34a duplexes with target miR-328-3p sequences (Fig. 1A). After successful cloning of target ncRNA-expressing plasmid and transformation of the common E. coli strain HST08, total bacterial RNA was extracted and subjected to urea-PAGE analysis. The appearance of a new and strong band at expected size in bacteria transformed with hBERA/miR-328-expressing plasmid (Fig. 1B), as compared to wild type bacteria, demonstrated a successful, high-level heterogeneous expression of target hBERA/miR-328 molecule.
Fully-humanized biologic miR-328-3p agent was thus purified from bacterial RNA by using an anion exchange FPLC method, as reported23, 25. Recombinant hBERA/miR-328 was clearly separated from bacterial RNAs, and target fractions were collected (Fig. 1C). Fractions showing a high degree of homogeneity, as verified by urea-PAGE analysis (Fig. 1C), were combined and desalted to offer final hBERA/miR-328 product. Further analyses by HPLC and Limulus Amebocyte Lysate Pyrogent-5000 kinetic assays confirmed a high purity (>98%; Fig. 1D) and low endotoxin level (≤ 5 EU/μg RNA), respectively, for the FPLC-isolated hBERA/miR-328 molecule. Likewise, pure control RNA namely htRNASer was obtained and utilized in the following studies. Given the high-level expression of target RNAs, we readily obtained ∼3 and ∼8 mg of pure control RNA and hBERA/miR-328, respectively, from half liter bacterial culture.
3.2. Bioengineered hBERA/miR-328 agent is processed to target miR-328-3p in human carcinoma cells to inhibit cell proliferation
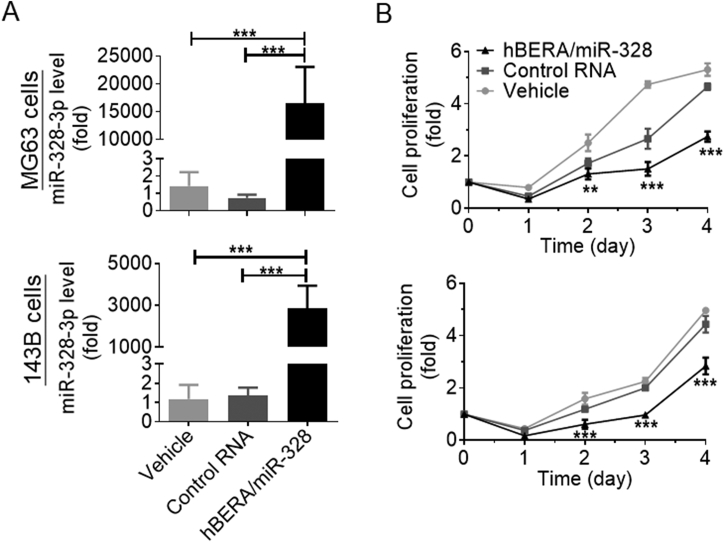
To determine if biologic hBERA/miR-328 molecule could be processed to mature miR-328-3p in human OS 143B and MG63 cells, selective stem-loop RT-qPCR assay was conducted to determine mature miR-328-3p levels in cells after 48 h treatment with recombinant ncRNAs. The data showed that miR-328-3p levels were remarkably higher in both 143B and MG63 cells transfected with hBERA/miR-328 than vehicle and control RNA treatment (***P < 0.001; Fig. 2A). To further assess the impact of miR-328-3p on the growth of OS cells, cell viability was monitored over time. The results showed that growth of both 143B and MG63 cells was significantly suppressed by hBERA/miR-328, as compared to vehicle and control RNA (**P < 0.01, ***P < 0.001; Fig. 2B). Together, these findings demonstrated that high-levels of mature miR-328-3p were successfully released from recombinant hBERA/miR-328 agent to exert antiproliferative activity against human OS cells.
Figure 2.
Recombinant hBERA/miR-328 is processed to target miR-328-3p in human carcinoma cells to inhibit cell proliferation. MG63 and 143B cells were transfected with 15 nmol/L hBERA/miR-328, control RNA or vehicle for 48 h. Levels of miR-328-3p (A) in hBERA/miR-328-treated cells were remarkably higher than the controls. ***P < 0.001, n = 4/group (one-way ANOVA with Bonferroni's post-hoc test). As a result, MG63 and 143B cell growth (B) were greatly inhibited by miR-328-3p, as compared with either control RNA or vehicle treatment. Values are the mean ± SD; **P < 0.01, ***P < 0.001, n = 5/group (two-way ANOVA with Bonferroni's post-hoc test).
3.3. MiR-328-3p directly acts on the 3′ UTR of SLC7A5 and SLC2A1 to regulate LAT1 and GLUT1 protein levels, respectively, and thus modulate mTOR expression in human OS cells
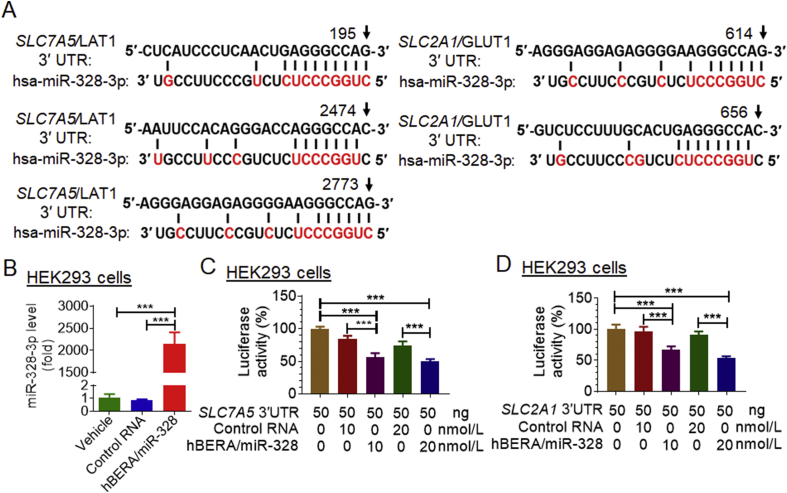
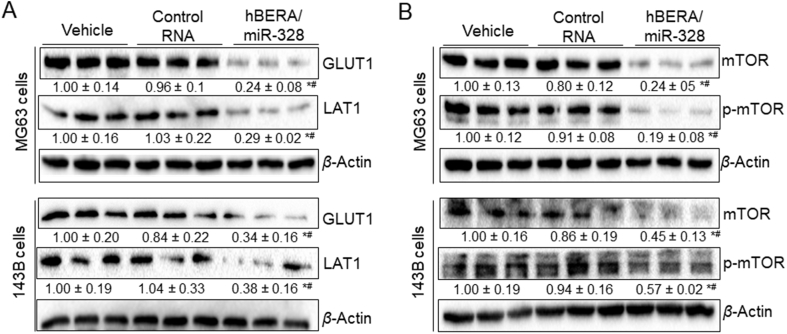
Transporters LAT1/SLC7A5 and GLUT1/SLC2A1 are critical for nutrient supply for cancer cell metabolism and proliferation, which are both upregulated in malignant tumor cells36, 37. After identifying computationally that miR-328-3p might directly target the 3′ UTR of SLC7A5/LAT1 (Fig. 3A), in addition to SLC2A1/GLUT1 reported very recently17, we aimed to investigate the regulation of both LAT1 and GLUT1 by bioengineered miR-328-3p in this study. Firstly, we conducted dual luciferase report assays to examine the interactions of miR-328-3p with 3′ UTRs of SLC7A5 and SLC2A1. Our data showed that, following the release of target mature miR-328-3p from hBERA/miR-328 in HEK293 cells (Fig. 3B), both SLC7A5 (Fig. 3C) and SLC2A1 (Fig. 3D) 3′ UTR-luciferase reporter activities were reduced by 30%–50%, supporting the actions of miR-328-3p on SLC7A5 and SLC2A1 3′ UTRs. Secondly, immunoblot analyses were performed to directly define the influence of miR-328-3p on the protein levels of LAT1 and GLUT1 in OS cells. As shown in Fig. 4A, both LAT1 and GLUT1 protein levels were significantly (P < 0.05) suppressed by hBERA/miR-328 in human OS 143B and MG63 cells, compared to either vehicle or control RNA treatment, suggesting a role of miR-328-3p in the regulation of LAT1 and GLUT1.
Figure 3.
Both SLC7A5/LAT1 and SLC2A1/GLUT1 are the direct targets of miR-328-3p. (A) Computational analyses identified multiple putative MREs for miR-328-3p within the 3′UTR of both SLC7A5 and SLC2A1 transcripts. (B) High levels of miR-328-3p were generated from hBERA/miR-328 in HEK293 cells, as determined by qPCR analyses. (C) and (D) Dual luciferase reporter assay revealed that miR-328-3p significantly inhibited SLC7A5 and SLC2A1 3′ UTR-luciferase reporter activities in HEK293 cells, as compared to controls. Values are the mean ± SD (n = 5–6/group); ***P < 0.001, one-way ANOVA with Bonferroni's post-hoc test. hsa, Homo sapiens.
Figure 4.
Bioengineered miR-328-3p reduces the protein levels of GLUT1 and LAT1 in human osteosarcoma cells, and subsequently decreases mTOR and p-mTOR protein levels. Cells were treated with hBERA/miR-328, control RNA or vehicle for 48 h, and protein levels were determined by Western blot analyses with selective antibodies. β-Actin was used as a loading control. Protein levels were normalized to vehicle group for comparison. Values are the mean ± SD (n = 3/group); *P < 0.05 and #P < 0.05, compared to control RNA and vehicle, respectively (one-way ANOVA with Bonferroni's post-hoc test).
We further evaluated the consequent effects of downregulation of LAT1 and GLUT1 by miR-328-3p on mTOR levels, a central regulator of mammalian metabolism, given the facts that nutrients including glucose and amino acids are sensed to regulate mTOR signaling38 while LAT1 and GLUT1 are major transporters for the transport of amino acids and glucose, respectively35, 36. The results showed that both mTOR and p-mTOR protein levels were significantly lower (P < 0.05) in human OS 143B and MG63 cells treated with hBERA/miR-328, as compared to either vehicle or control RNA treatment (Fig. 4B). Taken together, these findings indicated that miR-328-3p regulated the expression of both LAT1 and GLUT1 transporters through targeting of their 3′ UTRs, and subsequently modulated mTOR pathway.
3.4. Bioengineered miR-328-3p shows no or minimal effects on the homeostasis of amino acids in human OS cells
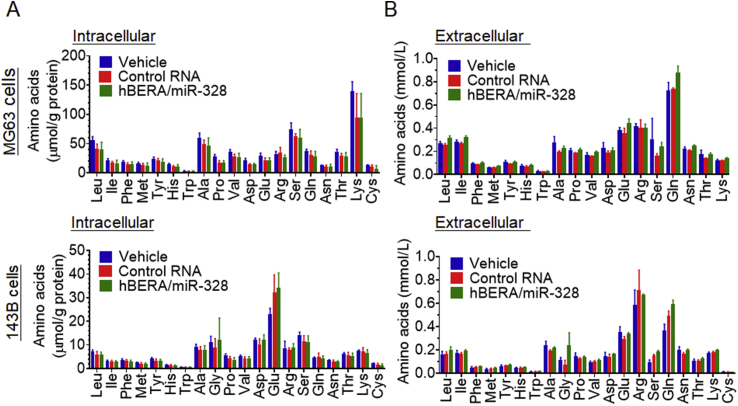
To determine to what degree the homeostasis of proteinogenic amino acids would be altered following the downregulation of LAT1 by biologic miR-328-3p (Fig. 4A), an accurate and selective LC–MS/MS method35 was utilized to measure both intracellular and extracellular concentrations of individual amino acids at 48 h post-treatment. The data showed that intracellular (Fig. 5A) and extracellular (Fig. 5B) concentrations were largely variable among these amino acids, whereas there was no statistical difference in any specific amino acid levels between 143B or MG63 cells treated with hBERA/miR-328 and vehicle or control RNA. The results suggest that miR-328-3p-mediated downregulation of LAT1 had no or limited impact on the homeostasis of amino acids in human OS cells.
Figure 5.
Intracellular and extracellular amino acid levels are not significantly altered by miR-328-3p in human osteosarcoma cells. Intracellular (A) and extracellular (B) amino acid concentrations were quantitated with an accurate and selective LC–MS/MS method at 48 h post-treatment of MG63 and 143B cells with hBERA/miR-328, control RNA and vehicle. Amino acid levels in cell lysates were normalized to corresponding protein concentrations. Values are the mean ± SD (n = 3/group).
3.5. Bioengineered miR-328-3p modulates glucose uptake and homeostasis in human OS cells
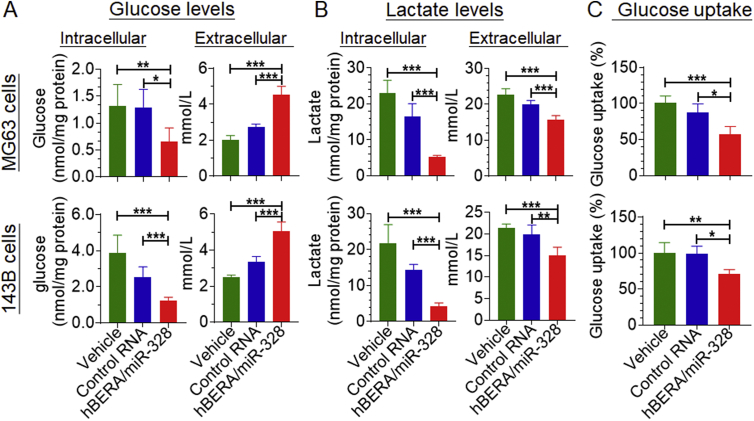
We thus examined whether the reduction of GLUT1 expression by miR-328-3p (Fig. 4A) would change glucose metabolism. Levels of glucose and lactate, the latter of which is an end metabolite of glycolysis, within human OS cells (intracellular) as well as cell culture medium (extracellular) were measured with a selective and accurate LC–MS/MS assay. The data revealed that intracellular glucose concentrations decreased around 50% by bioengineered miR-328-3p in both MG63 and 143B cells, as compared with control RNA (Fig. 6A). In contrast, extracellular glucose levels were 30%–40% higher in MG63 and 143B cells treated with biologic miR-328-3p than corresponding cells transfected with control RNA (Fig. 6A). Meanwhile, both intracellular and extracellular lactate concentrations were significantly reduced by miR-328-3p (Fig. 6B).
Figure 6.
Bioengineered miR-328-3p modulates the glycolytic profiles of human osteosarcoma cells via the suppression of glucose uptake capacity. Cells were treated with 15 nmol/L hBERA/miR-328, control RNA or vehicle. Glucose (A) and lactate (B) levels in cell lysate (intracellular) and medium (extracellular) were measured by LC–MS/MS method at 48 h post-treatment. Intracellular glucose and lactate levels were normalized to total protein levels in corresponding samples. Values are mean ± SD (n = 6/group). (C) Glucose uptake capacity was reduced by 30%–50% by miR-328-3p in cells, as compared with control RNA or vehicle treatments, which was determined by using 2-NBDG-based assay at 48 h post-treatment. Values are mean ± SD (n = 4-5/group); *P < 0.05, **P < 0.01, and ***P < 0.001; one-way ANOVA with Bonferroni's post-hoc test.
Since the RPMI 1640 for the culture of OS cells is a lactate-free medium, the lower intracellular and extracellular lactate levels in miR-328-3p-treated OS cells were likely caused by the decline of intracellular glucose concentrations that is presumably due to the change in glucose uptake, as indicated by higher extracellular glucose levels in miR-328-3p-treated cells. To test this hypothesis, we further employed the 2-NBDG-based glucose uptake assay to directly determine the impact of hBERA/miR-328 on cellular glucose uptake capacity. The data showed that glucose uptake capacity was indeed reduced by 30%–40% in both MG63 and 143B cells following the transfection with miR-328-3p, when compared with control RNA treatment (Fig. 6C). Together, these findings revealed that bioengineered miR-328-3p was effective to suppress intracellular glucose levels through the reduction of glucose uptake (Fig. 6), following the downregulation of GLUT1 protein levels in human OS cells (Fig. 4A).
3.6. Combination treatment with bioengineered miR-328-3p and chemotherapeutic drug produces synergistic and potent antiproliferative effects against human OS cells
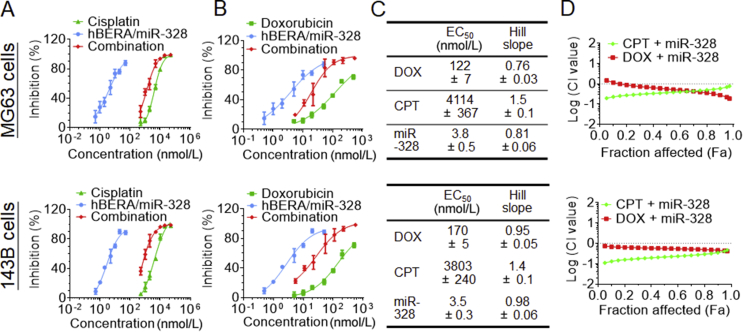
As monotherapy usually offers a limited degree of efficacy and the above data showed that bioengineered hBERA/miR-328 molecule itself exhibited antiproliferative activity against human OS cells, at least partially due to the inhibition of nutrient supply via modulation of SLC transporter expression, we evaluated whether co-administration of hBERA/miR-328 with chemotherapeutics would result in a synergism for the control of OS cell proliferation. Our results revealed that miR-328, doxorubicin or cisplatin monotherapy exerted a dose-dependent inhibitory effect on the proliferation of human OS cells (Fig. 7A and B), whose EC50 and Hill slope values were readily determined (Fig. 7C). As expected, co-administration of hBERA/miR-328 largely enhanced the chemosensitivity of both 143B and MG63 cells, as manifested by a left shift of the dose-response curve of doxorubicin and cisplatin (Fig. 7A and B). Using Chou-Talalay approach34 and demonstrated by the CI-Fa plots (Fig. 7D), synergism was clearly identified for the combined use of hBERA/miR-328 and chemotherapeutics. Compared with doxorubicin, cisplatin showed a greater degree of synergistic effects, as indicated by relatively lower CI values for cisplatin plus miR-328 at most corresponding Fa points than doxorubicin plus miR-328. These results demonstrated the benefits of miR-328-3p-based combination therapy for the control of human OS cell proliferation.
Figure 7.
Combination of biologic miR-328-3p with doxorubicin (DOX) or cisplatin (CPT) exhibits synergistic effects in the inhibition of osteosarcoma cell proliferation. (A) Cisplatin and (B) doxorubicin, alone or combined with miR-328-3p, exerted a dose-dependent response in the inhibition of MG63 and 143B cell viability, as determined by MTT assay at 48 h post-treatment. (C) The estimated EC50 and Hill slop values for the antiproliferative activities of miR-328-3p, DOX and CPT. Values are mean ± SD (n = 4/group). (D) Chou-Talalay (CI-Fa) plots revealed a synergism for combined use of miR-328 and DOX or CPT in the control of human osteosarcoma cell proliferation.
4. Discussion
Our recent studies have established a robust ncRNA bioengineering technology using a bacterial tRNA-fused human pre-miRNA as a carrier23, 26, 31, 39 that permits high-yield and large-scale production of novel BERAs. To minimize the potential risk of bacterial tRNA to human cells, we intended to develop hBERA production system by utilizing human tRNA instead of bacterial tRNA (unpublished data). The present study exemplified successful heterogenous expression and purification of target hBERA/miR-328 molecule by using htRNASer/pre-miR-34a as the carrier, which was selectively processed to mature miR-328-3p in human OS cells, and subsequently suppressed cell proliferation. Further studies verified two direct targets for miR-328-3p, SLC2A1/GLUT1 and SLC7A5/LAT1, critical for the transport of nutrients essential for cell metabolism, proliferation and growth. Indeed, suppression of GLUT1 expression by hBERA/miR-328 led to a significantly lower glucose uptake and glycolysis capacity, whereas the perturbation of LAT1 did not alter the homeostasis of amino acids in OS cells. Additionally, our study showed that coadministration of hBERA/miR-328 with cisplatin or doxorubicin synergistically inhibited the proliferation of OS cells.
Accumulating evidences have demonstrated that many miRNAs are involved in the regulation of drug metabolism and disposition via targeting of enzymes, transporters or their regulatory factors9, 40, 41, offering new insights into the interindividual variations in pharmacotherapy as well as development of new therapeutics. These findings are also in line with the discoveries on the roles of miRNAs in the control of development and progression of various diseases including lethal cancer. This is particularly notable for some miRNAs such as miR-129130, 33, 42 and miR-32815, 17, 21 that are able to target genes directly involved in both drug disposition and disease progression. Nevertheless, miRNA research and drug development are dominated by the use of chemo-engineered miRNA mimics and virus- or plasmid-based miRNA-expressing materials. By contrast, BERAs produced and folded in living cells are a novel class of ncRNA molecules that should better resemble the structural, physicochemical and biological properties of genome-transcribed ncRNAs for basic and translational research9, 27, 43.
By using a new carrier htRNASer/pre-miR-34a, we achieved a high-yield production of fully-humanized biologic miR-328-3p agent in current study that offered us unique tools to investigate the mechanistic actions of miR-328-3p in the regulation of nutrient transport and homeostasis in cancer metabolism. In addition to GLUT1/SLC2A1 reported recently17, we identified and verified another nutrient transporter LAT1/SLC7A5 as a direct target of miR-328-3p. LAT1 is responsible for the uptake of essential large neutral amino acids such as leucine, isoleucine, asparagine, and the efflux of glutamine36, 44. Cancerous cells are addicted to nutrients such as glucose and amino acids to meet the demand for rapid proliferation and growth, and LAT1 is usually upregulated in carcinoma cells including osteosarcoma15, 17, 44 to encounter an increased and continuous supply of proteinogenic amino acids. Although miR-328-3p was able to reduce LAT1 protein levels, amino acid levels were not altered significantly in OS cells, suggesting that other pathways such as uptake/export by other transporters, biosynthesis, and biodegradation may compensate to maintain the overall homeostasis of amino acids when LAT1 expression is downregulated. Indeed, a particular amino acid (e.g., arginine) may be transported by multiple SLC transporters, synthesized from and converted to other metabolites by various enzymes, highlighting the complexity in the regulatory network underlying amino acid homeostasis and the need for identification of critical factors for the control of specific amino acids. In addition, the ways how amino acid homeostasis is governed could be variable among different types of carcinoma cells. Therefore, the effects of miR-328-3p and other candidate miRNAs on amino acid homeostasis in other types of cancer cells warrant further investigations.
Cancer cells preferentially metabolize glucose via aerobic glycolysis, which is a less efficient energy supply approach than mitochondrial oxidative phosphorylation, the main metabolic approach used by normal cells45, 46. This phenomenon, characterized by elevated glucose uptake, enhanced glycolysis and lactate production, is termed as Warburg effect47, 48, and it is recognized not only as one of the main hallmarks of cancer but also a viable target for cancer therapy. It has been reported that removal of glucose from cell culture media decreases glycolysis and promotes cell death in cancer cells49, 50. In this study, we demonstrated that hBERA/miR-328 was selectively processed to target miR-328-3p to suppress GLUT1 protein levels in human OS cells, which led to a lower glucose uptake rate, and subsequently a decrease of intracellular glucose and lactate concentrations as well as glycolysis capacity. We cannot exclude possible effects of miR-328-3p on other transporters or enzymes involved in glycolysis or more generally cancer cell metabolism and proliferation because miR-328-3p is indeed predicted to target other factors such as monocarboxylate transporter 4, gene symbol solute carrier family 16 member 3 (MCT4/SLC16A3) by computational analysis despite that it is not verified yet by any experiments. These findings on the downregulation of GLUT1 expression and reduction of glycolysis provide insights into mechanisms underlying the antiproliferative activity of miR-328-3p.
Because of the presence of multiple and compensatory mechanisms behind complex cancer cell signaling pathways and networks, monotherapy acting on single target or pathway is usually effective in only a small fraction of patients or may not be effective at all. Therefore, combination therapy targeting multiple pathways or processes is a general practice in the clinic which indeed has been proved as much more effective. Cisplatin and doxorubicin are two chemotherapeutic drugs commonly used for the treatment of OS, which suppress tumor cell growth via the interference of DNA replication12, 13. To explore the utility of hBERA/miR-328 in the control OS cell growth and achieve optimal outcomes, we assessed the antiproliferative activity when combined with cisplatin or doxorubicin in this study. Our results revealed a strong synergism for combination therapy with hBEAR/miR-328 and either cisplatin or doxorubicin in the inhibition of OS cell proliferation. These findings are consistent with previous studies showing that inhibition of GLUT1 is able to enhance the anticancer effects of cisplatin and doxorubicin51, 52, and supporting novel strategy to combat cancer through targeting multiple metabolic circuses.
5. Conclusions
A novel fully-humanized bioengineered RNA agent bearing miR-328-3p warhead was heterogeneously expressed in bacteria at high yield and purified to high homogeneity by using our newly-established, fermentation-based ncRNA bioengineering technology. The bioengineered miR-328-3p prodrug was specifically processed to target miR-328-3p to inhibit human OS cell growth. LAT1, an important amino acid transporter, was verified as a new direct target of miR-328-3p, whereas levels of extra- and intra-cellular amino acid levels were not altered by miR-328-3p in OS cells. Furthermore, biologic miR-328-3p reduced GLUT1 protein levels in OS cells, leading to a lower glucose uptake and glycolysis rate. In addition, combination of biologic miR-328-3p with chemotherapy, cisplatin or doxorubicin, synergistically inhibited the proliferation of OS cells. These findings exemplify the development of biologic ncRNA agents and demonstrate a new role for miR-328 in the regulation of xenobiotic/nutrient transport and homeostasis underlying cancer metabolism.
Acknowledgments
Ai-Xi Yu is supported by Hubei Province Scientific and Technological Innovation Key Project (No. 2019ACA136, China). Ai-Ming Yu is supported by National Institute of General Medical Sciences grant (No. R01GM113888) and National Cancer Institute grant (No. R01CA225958), National Institutes of Health (USA). Wanrong Yi is supported by a fellowship from the Chinese Scholarship Council (No. 201706270162, China). The authors also appreciate the access to the Molecular Pharmacology Shared Resources funded by the UC Davis Comprehensive Cancer Center Support Grant awarded by the National Cancer Institute grant (P30CA093373, USA).
Footnotes
Peer review under responsibility of Institute of Materia Medica,Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Ai-Xi Yu, Email: yuaixi@whu.edu.cn.
Ai-Ming Yu, Email: aimyu@ucdavis.edu.
Author contributions
All authors participated in research design, performed data analysis, wrote or contributed to the writing of the manuscript. Wanrong Yi, Meijuan Tu, Zhenzhen Liu, Chao Zhang, and Neelu Batra conducted experiments. Ai-Ming Yu and Ai-Xi Yu contributed to new reagents or analytical tools.
Declaration of competing interest
The authors are named inventors of patent applications related to RNA bioengineering technology and utilities that are owned by the UC Davis, and Dr. Yu is a founder of AimRNA, Inc., which has an agreement to license the intellectual property.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Chang T.C., Mendell J.T. MicroRNAs in vertebrate physiology and human disease. Annu Rev Genom Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 4.Yu A.M., Tian Y., Tu M.J., Ho P.Y., Jilek J.L. MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos. 2016;44:308–319. doi: 10.1124/dmd.115.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittmann J., Jäck H.M. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 7.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An X., Sarmiento C., Tan T., Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B. 2017;7:38–51. doi: 10.1016/j.apsb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrek H., Batra N., Ho P.Y., Tu M.J., Yu A.M. Bioengineering of a single long noncoding RNA molecule that carries multiple small RNAs. Appl Microbiol Biotechnol. 2019;103:6107–6117. doi: 10.1007/s00253-019-09934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Valentin R.K., Zhu L., Hughes D.P. Bone sarcomas in pediatrics: progress in our understanding of tumor biology and implications for therapy. Paediatr Drugs. 2015;17:257–271. doi: 10.1007/s40272-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlick R., Janeway K., Lessnick S., Randall R.L., Marina N., COG Bone Tumor Committee Children's oncology group's 2013 blueprint for research: bone tumors. Pediatr Blood Cancer. 2013;60:1009–1015. doi: 10.1002/pbc.24429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ta H.T., Dass C.R., Choong P.F., Dunstan D.E. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 13.Luetke A., Meyers P.A., Lewis I., Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Tabone M.D., Kalifa C., Rodary C., Raquin M., Valteau-Couanet D., Lemerle J. Osteosarcoma recurrences in pediatric patients previously treated with intensive chemotherapy. J Clin Oncol. 1994;12:2614–2620. doi: 10.1200/JCO.1994.12.12.2614. [DOI] [PubMed] [Google Scholar]
- 15.Shi J., An G., Guan Y., Wei T., Peng Z., Liang M. MiR-328-3p mediates the anti-tumor effect in osteosarcoma via directly targeting MMP-16. Cancer Cell Int. 2019;19:104. doi: 10.1186/s12935-019-0829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W., Ma C.N., Zhou N.N., Li X.D., Zhang Y.J. Up-regulation of miR-328-3p sensitizes non-small cell lung cancer to radiotherapy. Sci Rep. 2016;6:31651. doi: 10.1038/srep31651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santasusagna S., Moreno I., Navarro A., Muñoz C., Martinez F., Hernández R. MiR-328 mediates a metabolic shift in colon cancer cells by targeting SLC2A1/GLUT1. Clin Transl Oncol. 2018;20:1161–1167. doi: 10.1007/s12094-018-1836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo T., Yan Y., He Q., Ma X., Wang W. miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by targeting RAGE. Oncol Rep. 2018;39:2906–2914. doi: 10.3892/or.2018.6353. [DOI] [PubMed] [Google Scholar]
- 19.Liang F., Cui Z.J., Liu J.D., Liu K.P., Li L., Chen Y.L. Downregulated miR-328 suppressed cell invasion and growth in hepatocellular carcinoma via targeting PTEN. Eur Rev Med Pharmacol Sci. 2018;22:6324–6332. doi: 10.26355/eurrev_201810_16043. [DOI] [PubMed] [Google Scholar]
- 20.Luo X., Yang S., Zhou C., Pan F., Li Q., Ma S. MicroRNA-328 enhances cellular motility through posttranscriptional regulation of PTPRJ in human hepatocellular carcinoma. OncoTargets Ther. 2015;8:3159–3167. doi: 10.2147/OTT.S93056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Y.Z., Morris M.E., Yu A.M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z., Wa Q.D., Lu C., Pan W., Lu Z., Ao J. miR3283p enhances the radiosensitivity of osteosarcoma and regulates apoptosis and cell viability via H2AX. Oncol Rep. 2018;39:545–553. doi: 10.3892/or.2017.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho P.Y., Duan Z., Batra N., Jilek J.L., Tu M.J., Qiu J.X. Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther. 2018;365:494–506. doi: 10.1124/jpet.118.247775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y., Tu M.J., Wang W.P., Qiu J.X., Yu A.X., Yu A.M. Genetically engineered pre-microRNA-34a prodrug suppresses orthotopic osteosarcoma xenograft tumor growth via the induction of apoptosis and cell cycle arrest. Sci Rep. 2016;6:26611. doi: 10.1038/srep26611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W.P., Ho P.Y., Chen Q.X., Addepalli B., Limbach P.A., Li M.M. Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther. 2015;354:131–141. doi: 10.1124/jpet.115.225631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q.X., Wang W.P., Zeng S., Urayama S., Yu A.M. A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res. 2015;43:3857–3869. doi: 10.1093/nar/gkv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho P.Y., Yu A.M. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA. 2016;7:186–197. doi: 10.1002/wrna.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Tu M.J., Yu Y.F., Wang W.P., Chen Q.X., Qiu J.X. Combination therapy with bioengineered miR-34a prodrug and doxorubicin synergistically suppresses osteosarcoma growth. Biochem Pharmacol. 2015;98:602–613. doi: 10.1016/j.bcp.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jian C., Tu M.J., Ho P.Y., Duan Z., Zhang Q., Qiu J.X. Co-targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget. 2017;8:30742–30755. doi: 10.18632/oncotarget.16372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu M.J., Ho P.Y., Zhang Q.Y., Jian C., Qiu J.X., Kim E.J. Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett. 2019;442:82–90. doi: 10.1016/j.canlet.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M.M., Addepalli B., Tu M.J., Chen Q.X., Wang W.P., Limbach P.A. Chimeric microRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos. 2015;43:1129–1136. doi: 10.1124/dmd.115.064493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Tian Y., Tu M.J., Ho P.Y., Batra N., Yu A.M. Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharm Sin B. 2019;9:639–647. doi: 10.1016/j.apsb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu M.J., Pan Y.Z., Qiu J.X., Kim E.J., Yu A.M. MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic cancer cell proliferation and tumorigenesis. Oncotarget. 2016;7:45547–45561. doi: 10.18632/oncotarget.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., Tu M.J., Zhang C., Jilek J.L., Zhang Q.Y., Yu A.M. A reliable LC–MS/MS method for the quantification of natural amino acids in mouse plasma: method validation and application to a study on amino acid dynamics during hepatocellular carcinoma progression. J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1124:72–81. doi: 10.1016/j.jchromb.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafliger P., Charles R.P. The L-type amino acid transporter LAT1-an emerging target in cancer. Int J Mol Sci. 2019;20:E2428. doi: 10.3390/ijms20102428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szablewski L. Expression of glucose transporters in cancers. Biochim Biophys Acta. 2013;1835:164–169. doi: 10.1016/j.bbcan.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta S., Peterson T.R., Sabatini D.M. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M.M., Wang W.P., Wu W.J., Huang M., Yu A.M. Rapid production of novel pre-microRNA agent hsa-mir-27b in Escherichia coli using recombinant RNA technology for functional studies in mammalian cells. Drug Metab Dispos. 2014;42:1791–1795. doi: 10.1124/dmd.114.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano M., Nakajima M. Current knowledge of microRNA-mediated regulation of drug metabolism in humans. Expert Opin Drug Metab Toxicol. 2018;14:493–504. doi: 10.1080/17425255.2018.1472237. [DOI] [PubMed] [Google Scholar]
- 41.Yu A.M., Pan Y.Z. Noncoding microRNAs: small RNAs play a big role in regulation of ADME?. Acta Pharm Sin B. 2012;2:93–101. doi: 10.1016/j.apsb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y.Z., Zhou A., Hu Z., Yu A.M. Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab Dispos. 2013;41:1744–1751. doi: 10.1124/dmd.113.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Z., Yu A.M. Bioengineered non-coding RNA agent (BERA) in action. Bioengineered. 2016;7:411–417. doi: 10.1080/21655979.2016.1207011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhutia Y.D., Babu E., Ramachandran S., Ganapathy V. Amino acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 45.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asgari Y., Zabihinpour Z., Salehzadeh-Yazdi A., Schreiber F., Masoudi-Nejad A. Alterations in cancer cell metabolism: the Warburg effect and metabolic adaptation. Genomics. 2015;105:275–281. doi: 10.1016/j.ygeno.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y., Zhang W., Cao Y., Liu Y., Bergmeier S., Chen X. Small compound inhibitors of basal glucose transport inhibit cell proliferation and induce apoptosis in cancer cells via glucose-deprivation-like mechanisms. Cancer Lett. 2010;298:176–185. doi: 10.1016/j.canlet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 50.EI Mjiyad N., Caro-Maldonado A., Ramírez-Peinado S., Muñoz-Pinedo C. Sugar-free approaches to cancer cell killing. Oncogene. 2011;30:253–264. doi: 10.1038/onc.2010.466. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Cao Y., Zhang W., Bergmeier S., Qian Y., Akbar H. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther. 2012;11:1672–1682. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto T., Jimi S., Migita K., Takamatsu Y., Hara S. Inhibition of glucose transporter 1 induces apoptosis and sensitizes multiple myeloma cells to conventional chemotherapeutic agents. Leuk Res. 2016;41:103–110. doi: 10.1016/j.leukres.2015.12.008. [DOI] [PubMed] [Google Scholar]