Abstract
Background
Efficacious therapy for triple negative breast cancer (TNBC) continues to be a profound clinical challenge, but the key driven genes and convoluted signaling pathways are still unknown.
Material/Methods
A total of 223 samples (163 TNBC and 60 healthy breast tissues) were taken and deeply integrated analyzed by R software from 4 expression profiles in the study, including GSE53752, GSE45827, GSE65194, and GSE38959. We examined differentially expressed genes (DEGs) and screen for critical genes and pathways enrichment. The protein-protein interaction (PPI) network of DEGs-associated was built through the STRING Version: 11.0 database and Cytoscape software to filter the hub gene. Then, we verified hug gene expression levels through the Oncomine database. Also, we analyzed the prognostic value of TNBC patient’s hub genes using the Kaplan-Meier plotter database.
Results
In our study, we filter out 365 DEGs, including 212 upregulated genes and 153 downregulated genes. Then, 10 hub genes were picked out by the intersection of 12 algorithms. At the same time, we discovered that CXCR4 and CXCL10 overexpression are favorable prognostic factors for recurrence-free survival of TNBC through the Kaplan-Meier plotter database.
Conclusions
Our research found that CXCR4 and CXCL10 overexpressed, and they were a favorable prognostic factor in patients with TNBC. CXCR4 and CXCL10 might be effective targets for TNBC therapy.
MeSH Keywords: Chemokine CXCL10; Genes, abl; Receptors, CXCR4
Background
Triple negative breast cancer (TNBC) is a common type of breast cancer (BC), which lacks expression of the estrogen, progesterone and human epidermal growth factor receptor 2 [1]. This subtype represents 12% to 17% of the breast cancers [2]. It often occurs in young women [3], and it is more likely to recurrence and metastasize [4]. As this subtype lacks molecular targets, patients with TNBC cannot be treated with human epidermal growth factor receptor 2 (HER2)-targeted therapy or endocrine therapy. Currently, chemotherapy is the primary treatment for patients with TNBC [5]. Unfortunately, many tumors have significant drug resistance, and rapid recurrence and metastasis after neoadjuvant chemotherapy [6]. As of now, TNBC still has very limited treatment options and poor prognosis [7]. Therefore, it is necessary to further explore the potential therapeutic genes and novel targets of TNBC.
For the TNBC, we re-analyzed the gene expression profiles of GSE45827 [8], GSE65194 [9–11], GSE53752 [12] and GSE38959 [13] and determined the differentially expressed genes (DEGs) in 163 TNBC and 60 normal breast tissues samples. Bioinformatics analysis was used for functional enrichment analysis of DEGs, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Next, we constructed a protein-protein interaction (PPI) network to filter hub genes related to TNBC and validation of hub gene expression levels by the Oncomine database. We performed a survival analysis of the hub gene by the Kaplan-Meier plotter database. We defined the DEGs, and enriched the biological functions and candidate genes, to provide new biomarkers of TNBC patients for early diagnosis and treatment.
Material and Methods
Data source
Data sets of our study were all from the Gene Expression Omnibus (GEO) public database (https://www.ncbi.nlm.nih.gov/geo/) and 4 sets of gene expression profiling chips (GEPC) are selected, including GSE38959 (GPL4133; Agilent-014850 Whole Human Genome Microarray 4 × 44 K G4112F), GSE53752 (GPL7264; Agilent-012097 Human 1A Microarray (V2) G4110B), GSE45827 and GSE65194 (GPL570; [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array).
Differentially expressed genes (DEGs)
The robust multi-array average (RMA) approach performed for background correction and normalization. The original GEO data were then converted into expression measures using the affy R package. Limma R package was subsequently employed for identifying differentially expressed genes (DEGs). We use P<0.05 and |logFC| >1 as data processing standards for analysis. We analyzed each data set and cross-screened differentially expressed genes using Venn 2.1 webtool (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Enrichment analysis
GO function enrichment analysis consists of 3 parts, namely the biological process (BP), molecular function (MF) and cellular component (CC). KEGG is a database for storing genomic and biological pathways. In this study, we performed GO function and KEGG pathway enrichment analysis of DEGs by DAVID 6.7 database tool (https://david.ncifcrf.gov/). The P-value of <0.05 to be considered statistically significant.
PPI and hub gene
We analyze the PPI network of DEGs through the STRING v11.0 database (https://string-db.org/cgi/input.pl?sessionId=JUci4htYBrbs and input_page_active_form=multiple_identifiers), which select score >0.4 to extract PPIs for DEGs. Then, visualize the PPI network with Cytoscape v3.7.1 software. Nodes with higher connectivity degree are more likely to maintain overall network stability. Next, we use the cytoHubba to calculate the degree of each protein node. In this study, we identified 10 hub genes by 12 algorithms (Maximal Clique Centrality (MCC), Density of Maximum Neighborhood Component (DMNC), Maximum Neighborhood Component (MNC), Degree, Edge Percolated Component (EPC), Bottleneck (BN), EcCentricity, Closeness, Radiality, Betweenness, Stress, Clustering Coefficient). Then, Oncomine was utilized to investigate hub gene expression of breast cancer in multiple datasets.
ONCOMINE analysis
ONCOMINE is a publicly available genome-wide online cancer microarray database (https://www.oncomine.org/resource/login.html). In our study, we generated a P-value by Student’s t-test in the TNBC group compared with the normal control group, which fold change was 2 and P-value was 0.01.
Survival analysis
In this study, overall survival and relapse-free survival rate was analyzed using samples from 1402 breast cancer patients and 3955 TNBC patients. We used the Kaplan-Meier plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=breast) to assess the prognostic value of BC hub genes, especially in TNBC with estrogen (ER), progesterone (PR) and HER2 negative expression patients. Based on the only Jetset best probe set and the probe IDs for each gene are shown in Table 1. Then, we divided the median values of each gene in each patient of TNBC into the 2 groups. P-value of <0.05 was considered statistically significant.
Table 1.
Top 10 hub genes by intersection of 12 algorithms.
| Gene symbol | Gene description | Algorithms | Probe ID |
|---|---|---|---|
| MKI67 | Marker of Proliferation Ki-67 | MCC, MNC, Degree, EPC, BottleNeck, EcCentricity, Closeness, Radiality, Betweenness, Stress | 212023_at |
| FOXM1 | Forkhead Box M1 | MCC, MNC, Degree, EPC, EcCentricity, Closeness, Radiality, Betweenness, Stress | 202580_at |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase | MNC, Degree, EPC, BottleNeck, EcCentricity, Closeness, Radiality, Betweenness, Stress | 212581_at |
| KIT | KIT Proto-Oncogene Receptor Tyrosine Kinase | MNC, Degree, EPC, BottleNeck, Closeness, Radiality, Betweenness, Stress | 205051_at |
| IGF1 | Insulin Like Growth Factor 1 | MNC, Degree, EPC, Closeness, Radiality, Betweenness, Stress | 209541_at |
| CCNB2 | Cyclin B2 | MCC, Degree, EPC, BottleNeck, EcCentricity, Stress | 202705_at |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A | BottleNeck, EcCentricity, Closeness, Radiality, Betweenness, Stress | 207039_at |
| CXCR4 | C-X-C Motif Chemokine Receptor 4 | MNC, Degree, EPC, Closeness, Radiality, Stress | 217028_at |
| ICAM1 | Intercellular Adhesion Molecule 1 | MNC, Degree, EPC, BottleNeck, Closeness, Radiality | 202638_at |
| CXCL10 | C-X-C Motif Chemokine Ligand 10 | MNC, Degree, EPC, Betweenness | 204533_at |
Results
Differentially expressed genes
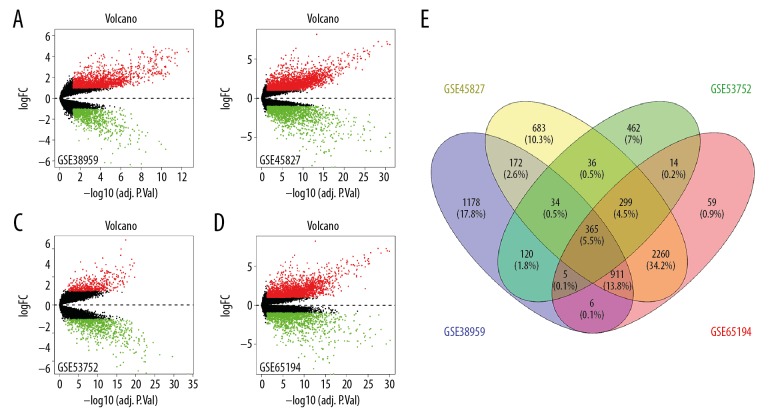
In this study, we selected 4 GEPC (GSE45827, GSE53752, GSE38959, and GSE65194) from the GEO database. GSE45827 and GSE65194 included 41 TNBC and 11 healthy breast samples, GSE53752 contained 51 TNBC samples and 25 healthy breast samples and GSE38959 contained 30 TNBC and 13 healthy breast samples (Table 2). Volcano plots were generated to visualize the distribution of expressed genes between cancer and normal controls from different studies. Red or green dots in the plots represented significantly upregulated or downregulated genes respectively. Using the P<0.05 and |logFC| >1 as cutoff criterion, we recovered 4760, 3919, 1335, and 2791 the DEGs from GEO database GSE45827, GSE65194, GSE53752, and GSE38959 (Figure 1A–1D). Then, we screened 365 differentially expressed genes from 4 data sets through bioinformatics compared to normal breast tissues in the TNBC tissues (Figure 1E), including 153 downregulated genes and 212 upregulated genes (Table 3).
Table 2.
Statistical analysis of the 4 microarray databases.
| Reference (PMID) | Tissue | GEO | Platform | Normal | Tumor | Total number |
|---|---|---|---|---|---|---|
| 27006338 | TNBC | GSE45827 | GPL570 | 11 | 41 | 52 |
| 23144294 | TNBC | GSE65194 | GPL570 | 11 | 41 | 52 |
| 23049873 | TNBC | GSE53752 | GPL7264 | 25 | 51 | 76 |
| 23254957 | TNBC | GSE38959 | GPL4133 | 13 | 30 | 43 |
GEO – Gene Expression Omnibus; TNBC – triple-negative breast cancer.
Figure 1.
Volcano plot and Venn diagram of DEGs in mRNA expression profiling datasets. Volcano plots of DEGs in TNBC and normal tissue in (A) GSE38959, (B) GSE45827, (C) GSE53752 and (D) GSE65194 datasets. The green points represent downregulation of the expression of genes screened of |fold change| <1.0 and a corrected P<0.05. Black points indicate no statistical difference. FC is the fold change. (E) Identification of 365 DEGs from 4 data sets (GSE38959, GSE45827, GSE53752 and GSE65194) using Venny 2.1.0 (available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html). DEGs were identified with classical t-test, statistically significant DEGs were defined with P<0.05 and |log FC| >1 as the cutoff criterion. DEGs – differentially expressed genes; TNBC – triple-negative breast cancer.
Table 3.
The 365 Differentially Expressed Genes (DEGs) were identified from 4 profile datasets, including 212 upregulated genes and 153 downregulated genes in the Triple-Negative Breast Cancer (TNBC) tissues compared to normal breast tissues.
| DEGs | Genes name |
|---|---|
| Upregulated | CXCR4,IRAK1,GPSM2,CCNB1,CAPS,CCNE1,FOXM1,TOR3A,EN1,TTLL4,GNB4, KIF14,MAD2L1, TTC13, KIF4A, SQLE, MCM10, TYMS, MELK, HN1, NDC80, OIP5, CCNA2, HRASLS, MMP1, STMN1, CKS2, DEPDC1, BRI3BP, IDO1, LMNB1, SPAG5, CCNB2, PRC1, LRP8, CCNE2, MCM4, WISP1, CXCL10, CKAP2L, SULF1, CDCA2, RPP25, EXO1, HPSE, KDELR2,COL11A1, HIST1H1C, HORMAD1, NEK2,TMEM206, BAIAP2L1, C1orf106, ATP6V0B, EZH2, CHEK1, KIF11, TAP1, KIFC1, MASTL, VGLL1, ARNTL2, HIST2H2AA4, RRS1, PYCR1, KCNK1, CENPA, RNFT2, CDCA7, CENPN, BUB1, TNFRSF12A, OASL, TRIP13, CDC7, UBE2S, DDIT4, LYN, IFI6, MND1, SDC1, RRM2, HNRNPAB, TOP2A, FEN1, RAD54L, PKMYT1, LRRC59, KIF18A, FAM83D, CHAF1B, PPP1R14C, HDGF, COL5A1, PRAME, CTHRC1, UHRF1, CDKN2A, RECQL4, CDKN3, CENPM, FAM64A, CENPF, RANGAP1, CDCA8, TPX2, ANP32E, KPNA2, ANLN, BIRC5, LAMP3, KLRG2, RACGAP1, EZR, CDC6, APOBEC3B, AURKA, FN1, AURKB, CTSB, FANCA, TMEM79, SPP1, IL4I1, IMPA2, USP18, DHCR7, ZWINT, GTSE1, NUF2, PTTG1, CDCA5, UBE2T, BCL2A1, RGS1, TEAD4, PPP1CA, ECT2, MMP11, MYBL1, GGH, HMOX1, PARP12, PPP1R14B, TACC3, CLEC7A, DCBLD1, CFL1, CEP55, GZMB, MFAP2, DLGAP5, RAD51AP1, TBC1D7, MKI67, TMEM123, IL32, LYZ, NMU, GAPDH, SLC39A1, KIAA0101, HSD17B6,GINS2, DONSON, E2F8, TSTA3, S100P, HMGB3, HOMER3, FLVCR1, SLC39A7, C16orf59, ADAMDEC1, TMEM45A, IDH2, CXCL11, STK38L, OAS3, ZIC1, BLM, KIF2C, CCNYL1, TUBB3, PBK, S100A11, PHF5A, TK1, SHCBP1, ASPM, INHBA, ATAD2, BRIP1, IFI30, GJB2, ICAM1, CXCL9, SPC25, KIF15, HJURP, ATP6V1C2, RSAD2, RAD51, HMMR, ART3, KIF20A, SOX11, P4HB, CHAC2, TTK, NCAPG, NUSAP1 |
| Downregulated | KLHL13, IGF1, PPP1R14A, NAP1L2, FGFR1, NOSTRIN,NFIA, ARL4A, CNN1, MAOA, BCL2, MYBPC1, IRS1, BDH2, OXTR, TFAP2B, ZBTB20, HLF, DCX, LAMB3, PLSCR4, VIT, CMYA5, SEMA3G, PDGFD, FAM3B, NTN4, KIT, SCN4B, C1orf115, SH3BGRL2, HOXA5, RUNX1T1, WIF1, FMOD, SETBP1, MME, SCGB1D2, GPD1L, FOXO1, PDK4, CA12, BEND5, RERGL, CTTNBP2, GRAMD1C, NR3C2, SASH1,NTRK2, FTO, OLFM4, TCEAL7, PLAT, NBEA, ABCA6, C4orf32, PDGFA, MUCL1, APOD, CBX7, RAI2, SPATA18, LHFP, NME5, GNG11, RHOJ PGR, SRPX, NAV3, SSPN, AK5, ANKRD30A, GFRA1, CPE RSPO3, CX3CR1, ABCA5, TMTC1, CYBRD1, TFF1, SYNM,C2orf40,AMIGO2, AGR2, PTN, CXCL14, ITIH5, LGR6, AGR3, KIAA1683, DACH1, PTHLH, SLC40A1, CD36, DCLK1, TSHZ2, NR3C1, DNALI1, EDN3, EDNRB, MYH11, EGFLAM, SALL2, NRG2, RUNDC3B,GLI3,INHBB,SCGB2A1, HOXA7, DYNLRB2, PDE2A, ATP1A2, ACADSB, IGSF10, CDO1, SEPP1, SDPR, FOXA1, CXCL12,CX3CL1, COL14A1, SCGB2A2, ABAT, CAV1, TNS4, CCL28, JAM2, IRX2, HOXA4, DST, ITM2A, GPRASP1, C1orf226, FMO2, C3orf18, IL33, ME3, SCUBE2, PDZK1, CIRBP, CLDN8, PPP1R3C, STC2, THSD4, MAMDC2, TESC, ADAMTS5, AR, TFF3, SFRP1, SEMA3C, GRAMD3, NDN |
Functional enrichment analyses
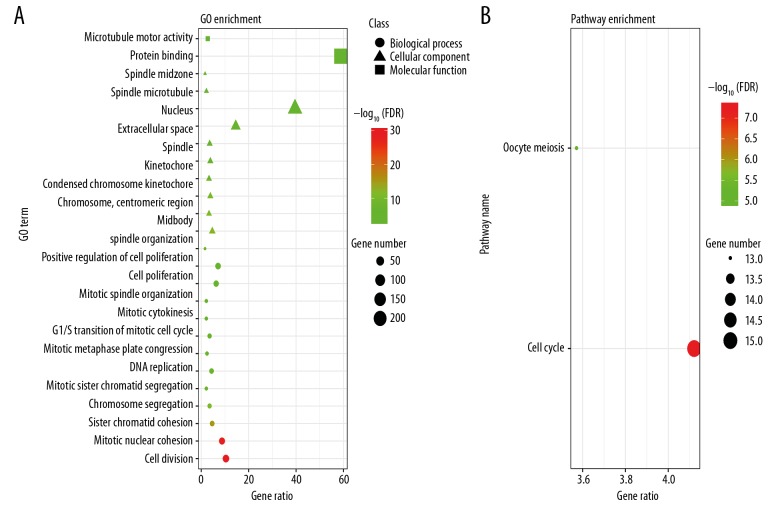
We used the DAVID online tool to perform GO and KEGG functional analysis of DEGs (Table 4). Functional enrichments of genes with FDR <0.05 were obtained. GO enrichment analysis is divided into BP, CC, and MF, including mitotic nuclear division, cell division, sister chromatid cohesion, chromosome segregation, mitotic sister chromatid segregation, DNA replication, mitotic metaphase plate congression, mitotic cytokinesis, G1/S transition of mitotic cell cycle, mitotic spindle organization, cell proliferation, positive regulation of cell proliferation, spindle organization; midbody, condensed chromosome kinetochore, chromosome centromeric region, kinetochore, spindle, spindle pole, extracellular space, nucleus, spindle microtubule, spindle midzone; microtubule motor activity, protein binding. In addition, we found that KEGG pathway analysis of DEGs is mainly enriched in oocyte meiosis and cell cycle. The results are shown in Figure 2.
Table 4.
Significantly enriched GO terms and KEGG pathways of DEGs.
| Category | Term | Description | Count | FDR |
|---|---|---|---|---|
| BP Term | GO: 0051301 | Cell division | 38 | 9.09348E-14 |
| BP Term | GO: 0007067 | Mitotic nuclear division | 32 | 3.88578E-13 |
| BP Term | GO: 0007062 | Sister chromatid cohesion | 17 | 2.98826E-07 |
| BP Term | GO: 0007059 | Chromosome segregation | 13 | 1.32402E-05 |
| BP Term | GO: 0000070 | Mitotic sister chromatid segregation | 8 | 0.000666199 |
| BP Term | GO: 0006260 | DNA replication | 16 | 0.000750953 |
| BP Term | GO: 0007080 | Mitotic metaphase plate congression | 9 | 0.000851431 |
| BP Term | GO: 0000082 | G1/S transition of mitotic cell cycle | 13 | 0.001349094 |
| BP Term | GO: 0000281 | Mitotic cytokinesis | 8 | 0.002021202 |
| BP Term | GO: 0007052 | Mitotic spindle organization | 8 | 0.002592039 |
| BP Term | GO: 0008283 | Cell proliferation | 23 | 0.006602397 |
| BP Term | GO: 0008284 | Positive regulation of cell proliferation | 26 | 0.010896295 |
| BP Term | GO: 0007051 | Spindle organization | 6 | 0.018277854 |
| CC Term | GO: 0030496 | Midbody | 17 | 6.28264E-06 |
| CC Term | GO: 0000775 | Chromosome, centromeric region | 12 | 1.52009E-05 |
| CC Term | GO: 0000777 | Condensed chromosome kinetochore | 14 | 1.83106E-05 |
| CC Term | GO: 0000776 | Kinetochore | 12 | 0.000657356 |
| CC Term | GO: 0005819 | Spindle | 14 | 0.000966442 |
| CC Term | GO: 0000922 | Spindle pole | 13 | 0.001994984 |
| CC Term | GO: 0005615 | Extracellular space | 53 | 0.002367057 |
| CC Term | GO: 0005634 | Nucleus | 144 | 0.018757183 |
| CC Term | GO: 0005876 | Spindle microtubule | 8 | 0.028575601 |
| CC Term | GO: 0051233 | Spindle midzone | 6 | 0.034840011 |
| MF Term | GO: 0005515 | Protein binding | 214 | 0.015739274 |
| MF Term | GO: 0003777 | Microtubule motor activity | 10 | 0.041584748 |
| KEGG PATHWAY | hsa04110 | Cell cycle | 15 | 0.000666844 |
| KEGG PATHWAY | hsa04114 | Oocyte meiosis | 13 | 0.00711138 |
BP – biological process; CC – cellular component; DEG – differentially expressed gene; ECM – extracellular matrix; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
Functional analyses of DEGs in TNBC. (A) GO enrichment significance items of DEGs in different functional groups (top 10 biological processes, cellular components and molecular functions). (B) KEGG pathway enrichment analysis of DEGs. DEGs – differentially expressed genes; TNBC – triple-negative breast cancer; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
PPI and hub gene
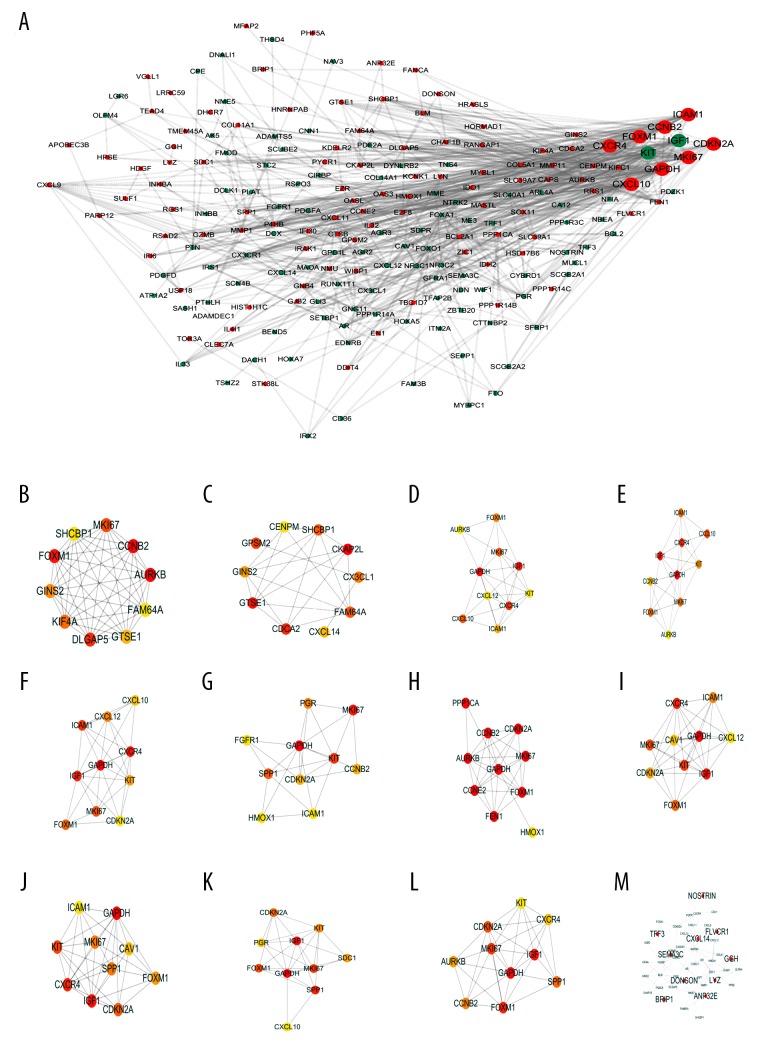
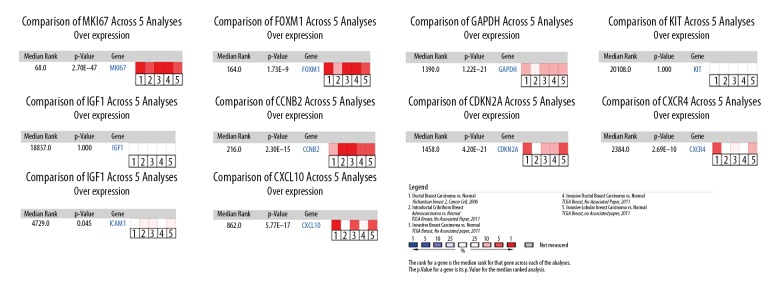
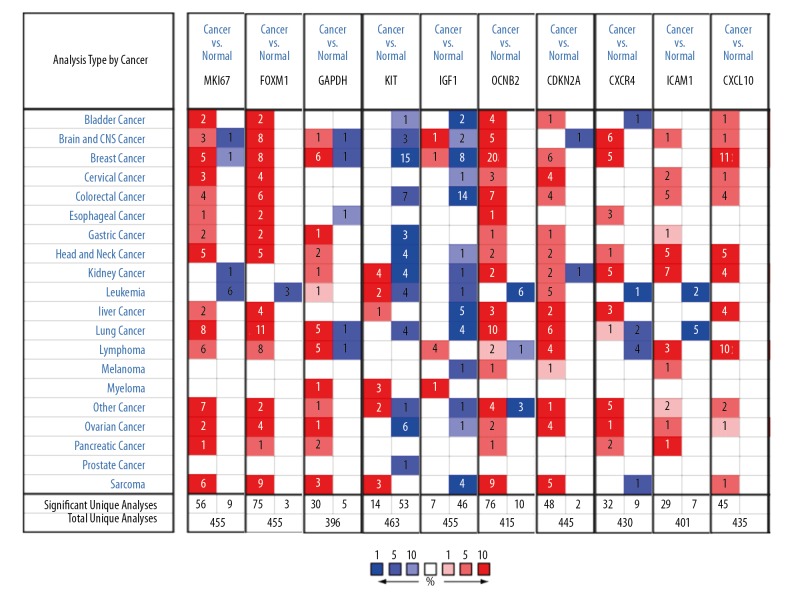
We enter the DEGs into STRING Version: 11.0 database to forecast PPI, and then the date of PPI network was refined using Cytoscape. After removing the isolated and partially connected nodes, the complex network of DEGs was established in Figure 3A. Evaluation of the top 10 genes by intersection of 12 algorithms with PPI networks (Table 1, Figure 3B–3M). The results showed that the expression of MKI67, FOXM1, GAPDH, CCNB2, CDKN2A, CXCR4, ICAM1, and CXCL10 had marked differences among different datasets in BC (Figure 4).
Figure 3.
(A–M) Protein-protein interaction network and top 10 hub genes. Red node indicates the upregulated gene and green node indicates the downregulated gene.
Figure 4.
Meta-analysis of differentially expressed hub genes. Meta-analysis of MKI67, FOXM1, GAPDH, KIT, IGF1, CCNB2, CDKN2A, CXCR4, ICAM1, and CXCL10 gene expressions in TNBC and normal tissue shows that they are significantly differentially expressed. Statistical analysis was performed using the Oncomine gene expression database. The heat maps represent the relative expression in patients with the indicated TNBC compared with normal tissue. Red indicates overexpression in TNBC patients and blue indicates under expression. The reported median ranks and P values consider all indicated studies simultaneously. TNBC – triple-negative breast cancer.
ONCOMINE analysis of hub genes
To detect the expression of hub genes, we researched that revealed hub genes mRNA expression levels of BC were significantly higher than healthy tissue samples by Oncomine analysis datasets of different cancer types (Figure 5).
Figure 5.
The mRNA expression pattern of hub genes in different tumor types. mRNA expression upregulated (red), downregulated (blue). The p value threshold is 0.01.
Survival analysis
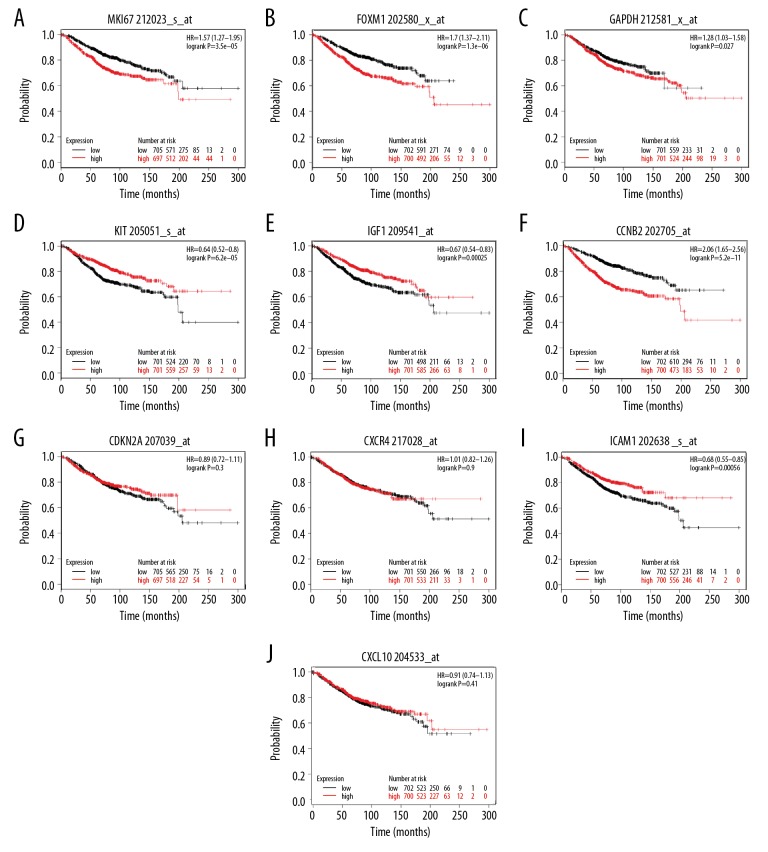
We evaluated hub genes using the Kaplan-Meier plotter tool for analysis of overall survival and relapse-free survival in 1402 BC patients. Simultaneously, we detected that high expression of KIT, IGF1 ICAM1, low expression of MKI67, FOXM1, GADPH, and CCNB2 hub gene contributes to overall survival in BC patients. These hub genes (MKI67, FOXM1, GAPDH, KIT, IGF1, CCNB2, and ICAM1) were closely related to overall survival of BC patients. However, no significant difference was found in the CDKN2A, CXCR4, and CXCL10 of hub genes for overall survival analysis (Figure 6).
Figure 6.
(A–J) Kaplan-Meier plotter online tool for detecting overall survival of 10 hub genes in triple-negative breast cancer.
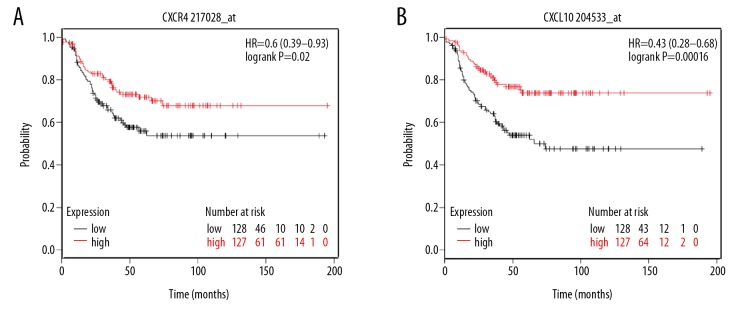
However, only overexpression of CXCR4 (hazard ration [HR]=0.6; 95% confidence interval [CI]: 0.39–0.93; P=0.02; n=386) and CXCL10 (HR=0.43; 95% CI: 0.28–0.68; P=0.00016; n=386) were the favorable prognostic factors related to relapse-free survival in TNBC patients. The results are shown in Figure 7.
Figure 7.
(A, B) Kaplan-Meier plotter online tool for detecting relapse-free survival of CXCR4 and CXCL10 in triple-negative breast cancer.
Discussion
TNBC is the primary subtype of BC with poor prognosis and lack of valid therapeutic targets. Because of the lack of useful therapeutic targets, TNBC patients cannot benefit from endocrine therapy or HER2 targeted therapy, so chemotherapy is the primary adjunct therapy. However, TNBC patients are more susceptible to drug resistance. So, it is necessary to find new specific targets for TNBC patients.
Based on public databases for gene expression and PPI analysis, we identified potential critical genes associated with TNBC. Then, we screened DEGs with normal breast tissue from the GEO database. Among them, we screened 212 upregulated genes and 153 downregulated DEGs associated with GO terms including cell division, mitotic nuclear division, extracellular space, nucleus, midbody, protein binding, and microtubule motor activity, and mainly enriched in the KEGG terms cell cycle and oocyte meiosis. We constructed the PPI network using differentially expressed genes and screened 10 hub genes by the intersection of 12 algorithms, including MKI67, FOXM1, GAPDH, KIT, IGF1, CCNB2, CDKN2A, CXCR4, ICAM1, and CXCL10. Among MKI67, FOXM1, GAPDH, CCNB2, CDKN2A, CXCR4, ICAM1, and CXCL10 were significantly upregulated in TNBC. Finally, we used the Kaplan-Meier plotter tool to predict the prognosis of hub genes in patients with TNBC.
Through the Kaplan-Meier plotter online public tool, overexpression of MKI67, FOXM1, GAPDH, and CCNB2 were associated with unfavorable prognosis in BC patients, no significant difference was found in the CDKN2A, CXCR4, and CXCL10 of hub genes for overall survival analysis (Figure 6). We found that overexpression of CXCR4 and CXCL10 were related to the favorable prognostic factor of TNBC patients (Figure 7).
CXCR4, CXC motif chemokine receptor type 4, is involved in either normal (maintaining stemness [14] or inducing differentiation [15]) or abnormal (developing cancer [16] and other pathologies [17]) events. CXCL10, CXC motif chemokine ligand 10, may accelerate cancer growth in nonimmune cell types and orchestrate an antitumor response [18]. CXCR4 has been associated with several diseases, including human immunodeficiency virus (HIV) infection [19], cancers [17], and warts, hypogammaglobulinemia, immunodeficiency, myelokathexis (WHIM) syndrome [20]. Some research has demonstrated that CXCR4 can be developed to treat these diseases, especially related to the prognosis of cancers such as colorectal cancer [21,22], thyroid carcinoma [23], head and neck squamous cell carcinomas [24], and breast cancer [25]. It was reported that the inhibitors of CXCR4 could benefit to TNBC patients [26], but some reports showed that it could not benefit to triple-negative breast cancer patients [27]. Thus, the role of CXCR4 needs further study.
It has been reported that CXCL10 might be related to the prognosis in pancreatic adenocarcinoma [28] and TNBC [29]. Our research showed that CXCR4 and CXCL10 overexpression could be related to the prognosis of malignant diseases. For TNBC, our studies demonstrated that CXCR4 and CXCL10 were the chemokines related to prognosis. Therefore, CXCR4 and CXCL10 might be an effective prognostic factor and obviously potential therapeutic target for TNBC treatment.
In addition to CXCR4 and CXCL10, the remaining 8 hub genes were the MKI67, FOXM1, GAPDH, KIT, IGF1, CCNB2, CDKN2A, and ICAM1. We found that these genes had no significant difference for TNBC analysis. This result was consistent with the Oncomine database. However, the mechanism of these hub genes is not fully understood in TNBC and needs further study.
Conclusions
Bioinformatics identified 365 differentially expressed genes from the GEO database.
Among them, TNBC-related 10 hub genes including MKI67, FOXM1, GAPDH, KIT, IGF1, CCNB2, CDKN2A, CXCR4, ICAM1, and CXCL10. All of them upregulated in TNBC except for KIT and IGF1. We found that overexpression of CXCR4 and CXCL10 were the favorable prognostic factor in relapse-free survival in TNBC. These results show that it is necessary to explore the clinical treatment value of CXCR4 and CXCL10 in TNBC, as they might be the possible target for TNBC treatment.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019;9(2):176–98. doi: 10.1158/2159-8290.CD-18-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J, Kobayashi LC, Grundy A, et al. Lifetime moderate-to-vigorous physical activity and ER/PR/HER-defined post-menopausal breast cancer risk. Breast Cancer Res, Treat. 2017;165(1):201–13. doi: 10.1007/s10549-017-4323-4. [DOI] [PubMed] [Google Scholar]
- 3.Villarreal-Garza C, Bargallo-Rocha JE, Soto-Perez-de-Celis E, et al. Real-world outcomes in young women with breast cancer treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2016;157(2):385–94. doi: 10.1007/s10549-016-3811-2. [DOI] [PubMed] [Google Scholar]
- 4.El Ayachi I, Fatima I, Wend P, et al. The WNT10B network is associated with survival and metastases in chemo-resistant triple-negative breast cancer. Cancer Res. 2019;79(5):982–93. doi: 10.1158/0008-5472.CAN-18-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heiser LM, Mills GB, Gray JW. Therapeutic clues from an integrated OMIC assessment of east Asian triple negative breast cancers. Cancer Cell. 2019;35(3):341–43. doi: 10.1016/j.ccell.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo P, Yang J, Liu D, et al. Dual complementary liposomes inhibit triple-negative breast tumor progression and metastasis. Sci Adv. 2019;5(3) doi: 10.1126/sciadv.aav5010. eaav5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heimes AS, Schmidt M. Atezolizumab for the treatment of triple-negative breast cancer. Expert Opin Investig Drugs. 2019;28(1):1–5. doi: 10.1080/13543784.2019.1552255. [DOI] [PubMed] [Google Scholar]
- 8.Gruosso T, Mieulet V, Cardon M, et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8(5):527–49. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maubant S, Tesson B, Maire V, et al. Transcriptome analysis of Wnt3a-treated triple-negative breast cancer cells. PLoS One. 2015;10(4):e0122333. doi: 10.1371/journal.pone.0122333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maire V, Baldeyron C, Richardson M, et al. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One. 2013;8(5):e63712. doi: 10.1371/journal.pone.0063712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maire V, Némati F, Richardson M, et al. Polo-like kinase 1: A potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73(2):813–23. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- 12.Kuo WH, Chang YY, Lai LC, et al. Molecular characteristics and metastasis predictor genes of triple-negative breast cancer: A clinical study of triple-negative breast carcinomas. PLoS One. 2012;7(9):e45831. doi: 10.1371/journal.pone.0045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu M, Yoshimaru T, Matsuo T, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol. 2013;42(2):478–506. doi: 10.3892/ijo.2012.1744. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi N, Zhang TT, Nakanishi T. Involvement of CXCR4 in normal and abnormal development. Cells. 2019;8(2) doi: 10.3390/cells8020185. pii: E185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MM, Chao N, Chhabra S, et al. Plerixafor (a CXCR4 antagonist) following myeloablative allogeneic hematopoietic stem cell transplantation enhances hematopoietic recovery. J Hematol Oncol. 2016;9(1):71. doi: 10.1186/s13045-016-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tulotta C, Stefanescu C, Chen Q, et al. CXCR4 signaling regulates metastatic onset by controlling neutrophil motility and response to malignant cells. Sci Rep. 2019;9(1):2399. doi: 10.1038/s41598-019-38643-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo YD, Jiang X, Sullivan KM, et al. Mobilization of CD8+ T cells via CXCR4 blockade facilitates PD-1 checkpoint therapy in human pancreatic cancer. Clin Cancer Res. 2019;25(13):3934–45. doi: 10.1158/1078-0432.CCR-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownell J, Polyak SJ. Molecular pathways: Hepatitis C virus, CXCL10, and the inflammatory road to liver cancer. Clin Cancer Res. 2013;19(6):1347–52. doi: 10.1158/1078-0432.CCR-12-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RK, Abdul-Jawad S, McCoy LE, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;68(7751):244–48. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott DH, Pastrana DV, Calvo KR, et al. Plerixafor for the treatment of WHIM syndrome. N Engl J Med. 2019;380(2):163–70. doi: 10.1056/NEJMoa1808575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrero-de LasHeras S, Martínez-Balibrea E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J Gastroenterol. 2018;24(42):4738–49. doi: 10.3748/wjg.v24.i42.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, Zheng L, Li D, et al. CXCR4 overexpression is correlated with poor prognosis in colorectal cancer. Life Sci. 2018;208:333–40. doi: 10.1016/j.lfs.2018.04.050. [DOI] [PubMed] [Google Scholar]
- 23.Werner TA, Forster CM, Dizdar L, et al. CXCR4/CXCR7/CXCL12 axis promotes an invasive phenotype in medullary thyroid carcinoma. Br J Cancer. 2017;117(12):1837–44. doi: 10.1038/bjc.2017.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De-Colle C, Menegakis A, Mönnich D, et al. DKTK-ROG: SDF-1/CXCR4 expression is an independent negative prognostic biomarker in patients with head and neck cancer after primary radiochemotherapy. Radiother Oncol. 2018;126(1):125–31. doi: 10.1016/j.radonc.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Xu L, Cheng L, Yang F, et al. JWA suppresses the invasion of human breast carcinoma cells by downregulating the expression of CXCR4. Mol Med Rep. 2018;17(6):8137–44. doi: 10.3892/mmr.2018.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang J, Hurchla MA, Fontana F, et al. CXCR4 protein epitope mimetic antagonist POL5551 disrupts metastasis and enhances chemotherapy effect in triple-negative breast cancer. Mol Cancer Ther. 2015;14(11):2473–85. doi: 10.1158/1535-7163.MCT-15-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefort S, Thuleau A, Kieffer Y, et al. CXCR4 inhibitors could benefit to HER2 but not to triple-negative breast cancer patients. Oncogene. 2017;36(9):1211–22. doi: 10.1038/onc.2016.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian L, Yu S, Yin C, et al. Plasma IFN-γ-inducible chemokines CXCL9 and CXCL10 correlate with survival and chemotherapeutic efficacy in advanced pancreatic ductal adenocarcinoma. Pancreatology. 2019;19(2):340–45. doi: 10.1016/j.pan.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Adams S, Diamond JR, Hamilton E, et al. Atezolizumab plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: A phase 1b clinical trial. JAMA Oncol. 2019;5(3):334–42. doi: 10.1001/jamaoncol.2018.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]