Abstract
The sebaceous gland (SG) is an essential component of the skin, and SG dysfunction is debilitating1, 2. Yet, the cellular bases for its origin, development and subsequent maintenance remain poorly understood. Here, we apply large-scale quantitative fate mapping to define the patterns of cell fate behaviour during SG development and maintenance. We show that the SG develops from a defined number of lineage-restricted progenitors that undergo a programme of independent and stochastic cell fate decisions. Following an expansion phase, equipotent progenitors transition into a phase of homeostatic turnover, which is correlated with changes in the mechanical properties of the stroma and spatial restrictions on gland size. Expression of the oncogene KrasG12D results in a release from these constraints and unbridled gland expansion. Quantitative clonal fate analysis reveals that, during this phase, the primary effect of the Kras-oncogene is to drive a constant fate bias with little effect on cell division rates. These findings provide insight into the developmental programme of the SG, as well as the mechanisms that drive tumour progression and gland dysfunction.
The complexity of the skin epidermis, comprising the interfollicular epidermis (IFE) and hair follicles, is established during development. Dermal cues promote hair follicle formation, and as primitive follicles elongate into the underlying dermis, specialised epidermal cells forming the SG and infundibular (IFN) lineages appear3, 4. In homeostatic epidermis, terminally differentiated sebocytes within the SG release sebum to lubricate and water proof the epithelium1. Fate mapping studies have revealed that SGs, like the IFE and IFN, are maintained as autonomous compartments during tissue homeostasis5. Multiple models have been proposed for SG maintenance, including models where specialised cells located either within or outside the gland are the source of replenishment5–9. Moreover, developmental studies suggest that SGs are derived from more than 1 Lrig1 expressing cells10–12. Yet, a general consensus is still lacking for how the SG forms, how it is subsequently maintained and how oncogene activation affects cell behaviour.
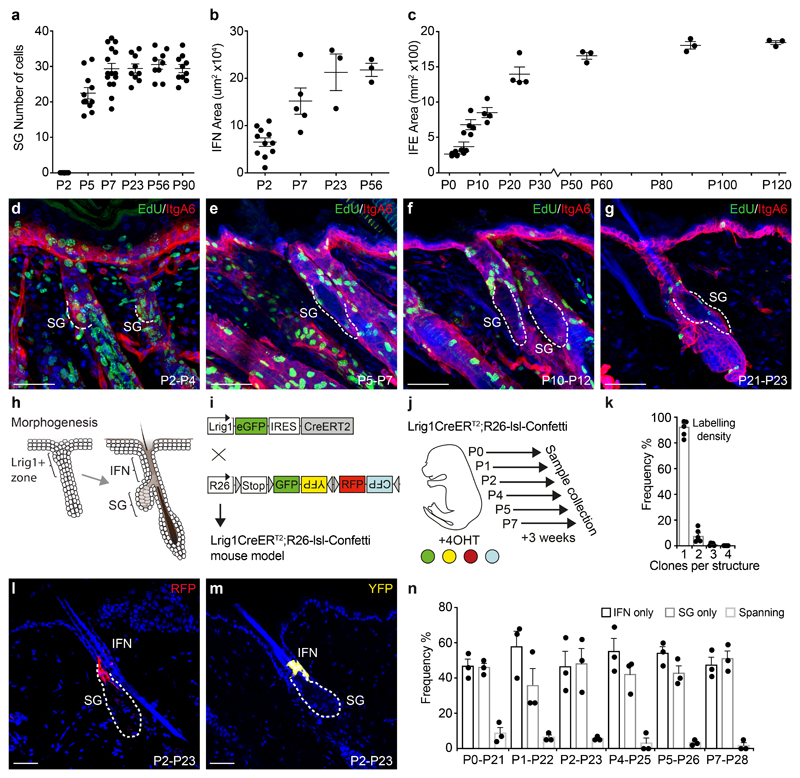
Like many epithelia, SGs consist of proliferative basal cells anchored to a basement membrane, and a differentiated suprabasal compartment. To understand cell behaviour leading to SG formation, we first investigated the patterns of growth during development. Interestingly, distinct parts of the epidermis reach full size in an asynchronous manner, with growth of the SG in awl/auchene and zig-zag hair types arresting at P7 (Sox2+ and Sox2-, respectively; 30±7 cells)13, the IFN by P23, while the IFE reached its adult size around P56 (Figure 1a-c, S1a,b). Surprisingly, SGs associated with awl/auchene- and zig-zag-hair types appear to develop synchronously and the observed variability in SG size was preserved into adulthood (Figure S1c,d). Furthermore, the transition from morphogenesis to homeostasis correlated with a decline in division rate from P7 (Figure 1d-g, S1e-h).
Figure 1. Epidermal morphogenesis characterised by tissue-specific growth and rapid sebaceous gland formation.
(a) Individual SG sizes (cell numbers), at P2: n=7 glands, P5: n=11, P7: n=15, P23: n=9, P56: n=9, P90: n=10. Infundibulum (IFN) area at P2: n=11 follicles, P7: n=5, P23: n=3, P56: n=3. Data pooled from 3-5 animals/time point. Interfollicular epidermis (IFE) defined as total back skin area at P2: n=4 mice, P5: n=4, P7: n=4, P12: n=4, P23: n=4, P56: n=3, P90: n=3, P120: n=3. Data are means±S.E.M. (d-g) IntegrinA6 (ItgA6, red) and EdU (green) detected in rendered confocal z-stacks of back skin. Representative images reflect 48-hour EdU pulse chase experiments from day P2, P5, P10 or P21 (h) Schematic illustration of the premature and adult hair follicle with an Lrig1+ area indicating the prospective SG and IFN. (i) Strategy for reporter induction in Lrig1+ cells. (j) Induction of different cohorts to delineate SG morphogenesis. (k) Fraction of clones found in upper hair follicle structures (IFN or SG) at P7 following 4OHT induction at P2. Data are means±S.E.M. (n=5 animals). (l, m) Typical RFP (red) and YFP (yellow) clones detected in rendered confocal z-stacks of back skin following a 3-week chase (Images representative of 3 mice). (n) Clone location within the SG, IFN or both (spanning) following a 3-week trace from P0, P1, P2, P4, P5 or P7. Data are means±S.E.M (n=3 animals/group). Demarcated lines represent the boundary between dermis and epidermis at the site of the prospective and adult SG. Nuclei are counterstained with DAPI (blue). Scale bars, 50μm.
To define the cellular dynamics of these cells in the upper hair follicle, which express Lrig1, during tissue morphogenesis we labelled these cells using an Lrig1eGFPiresCreERT2 mouse model (Figure 1h,i S1i-l)5, 14, 15. Postnatal clonal-level labelling at multiple timepoints from P0 to P7, followed by a 3-week chase revealed a decreasing frequency of clones that extended between the SG (most often the duct region) and the IFN (Figure 1j-n). This finding was substantiated in complementary experiments with much higher labelling frequencies using both K14 and Lrig1 promoter-driven Cre lines (Figure S1m-p). Altogether, this demonstrated that the majority of upper follicle cells were lineage allocated at the onset of morphogenesis. Similar early developmental lineage allocation and progenitor specification has been observed in other tissue arising from pools of equipotent fetal progenitors16–18.
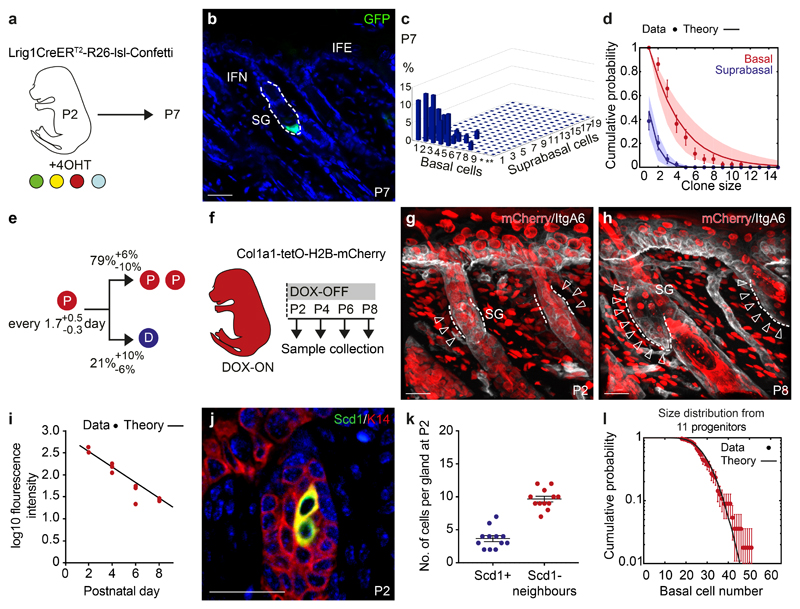
Based on markers for early lineage commitment we identified P2 as a key stage, where emerging hair follicles were largely homogenous (Figure S2a,b). To probe further the cell dynamics of precursors from P2 onwards, we performed quantitative clonal lineage tracing (Figure 2a, S2c)19. Three-dimensional clone composition analysis at P7 revealed that, over the 5-day chase, clones acquired a broad exponential-like distribution of basal and suprabasal cell numbers, independent of their position in the SG, suggesting that all cells contributed equally to SG morphogenesis (Figure 2b,c, S2d,e). To test this quantitatively, we turned to biophysical modelling20, 21 and found that the basal and suprabasal cell distributions could be recapitulated with high accuracy by an effective model in which, every 1.7 days on average, progenitors choose stochastically between symmetric cell division or differentiation/transfer into the suprabasal compartment, with a heavy bias towards division (Figure 2d,e, S2f; For details, and comparisons with other types of models, see Supplementary Note). To challenge the model, we measured cell division rates based on the fluorescence decay of doxycycline-inducible H2B-mCherry (Figure 2f-h, S2g,h). The reduction in average H2B-mCherry intensity within the basal SG cell compartment followed an exponential-like decay that fit the division rate inferred from the clonal data (Figure 2i).
Figure 2. Lineage restriction and cellular dynamics of sebaceous gland morphogenesis.
(a) Strategy for tissue collection at P7 following 4OHT administration at P2. (b) Detection of a representative GFP (green) clone at P7 in rendered confocal z stacks. (c) Frequency of individual clones scored in 3D composed of a given number of basal cells and subrabasal cells (d) Biophysical analysis based on an effective model with a single equipotent progenitor pool provides a good fit to the clonal fate data for both the basal (red) and suprabasal (blue) clone size distribution at P7. The model prediction is illustrated by a line and shaded areas represent the 95% confidence interval. Data are means±S.D. (c,d) n=44 clones from a total of five mice, *=10-19 and **=20-39 basal cells. (e) Biophysical modelling predicts the indicated probabilities for either the symmetrical division of a progenitor (P, red) or differentiation (D, blue) every 1.5 days. (f) Strategy for cell cycle analysis using Col1a1-tetO-H2BmCherry mouse model following a 2-week doxycycline (DOX) pulse and chase starting at P2. (g, h) H2BmCherry (red) and Integrin A6 (ItgA6, white) detected in rendered confocal z stacks in back skin at indicated time points. Arrows indicate label-diluted areas lining the Integrin alpha 6+ basal layer. Images representative of 3 animals. (i) Intensity of fluorescence in the basal compartment of individual SGs at indicated time points. Line represents fluorescent decay rate as predicted in (e). Lines indicate the medians. Demarcated lines represent the boundary between dermis and epidermis at the prospective or adult SG. n=3 animals per group with 17-26 glands counted per animal. (j) Detection of K14 (red) and Scd1 (green) in the upper hair follicle at P2. Images representative of 3 experiments. (k) Number of Scd1- cells surrounding Scd1+ cells. Data are means±S.E.M (n=12 SGs in 3 animals). (l) Experimental gland size distribution (number of basal cells, dots), and prediction (thick line) based on the independent behaviour of 11 SG precursors following the stochastic rules of panel (e) (n=44 clones in 5 mice. Data in graphs displayed as mean±S.E.M. Nuclei are counterstained with DAPI (blue). Scale bars, 50μm.
Following the assessment during morphogenesis, we noted that, at P7, individual clones constituted 9±1% (mean±S.E.M.) of the SG basal compartment, suggesting that, at P2, 11±1 (mean±S.E.M.) progenitors contribute to the formation of the SG. This estimate fitted well with the average number of cells surrounding the first lipid-synthesising cells at P2 (Figure 2j,k). Given the relatively large variance in SG size itself (Figure 1a, S1a), we questioned whether individual fate choices of precursors were uncorrelated, or whether coordination between precursors regulate the eventual SG size. Strikingly, using the stochastic rules inferred from the clonal lineage tracing data, numerical simulations of SG growth and homeostatic size, based on the independent expansion of 11 progenitors resulted in size distributions that matched the experimental data very well (Figure 2l). This suggests that the observed heterogeneity in SG sizes may simply arise from independent and stochastic fate choices of its founder precursors, rather than from coordinated fate decisions during morphogenesis. Moreover, the bulk of the clone size data analysed was well explained by the model, further supporting the approximate synchrony observed for SG initiation; although small asynchronies in the timing of SG specification and variations in the initial number of cells at P2 cannot be excluded. Together our analysis demonstrated that the SG is formed by lineage segregated progenitors, which undergo imbalanced stochastic cell fate decisions fuelling growth.
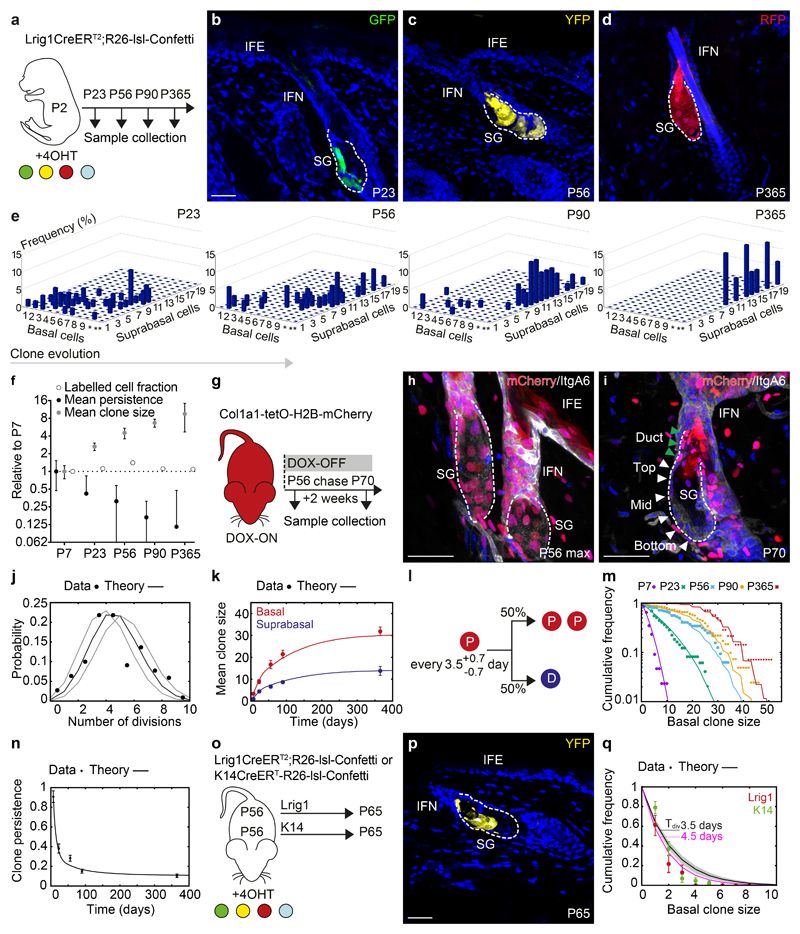
Having determined the cellular dynamics during SG morphogenesis, we turned to consider the homeostatic state. Clonal analysis revealed a steady increase in clone size and a progressive monoclonal conversion of SGs, indicative of clonal competition (Figure 3a-e, S3a). Interestingly, we found a pronounced bimodal distribution of clone sizes, most evident at P90 with small clones primarily located at the SG duct (Figure S3b-d), a structure proposed to be maintained by Gata6 expressing duct-cells22. Duct clones were, however, lost between P90 and P365, and clones encompassing the duct were subsequently connected to the infundibulum, whereas clonally converted SGs consistently lacked contributions to the SG duct (Figure S3b,d-f). This demonstrates that the duct is an integral part of the infundibulum rather than the SG (Figure S3g,h), and suggests that clonal competition in the SG drives a progressive drift towards monoclonality, as observed in other closed niche environments such as the intestinal crypt15, 23. To quantify this process, we turned to a detailed analysis of the non-ductal clones at P23, P56, P90 and P365 (Figure 3e).
Figure 3. Neutral drift dynamics in the context of spatial restriction.
(a) Strategy for clonal labelling of SG-precursors at P2 and tissue collection. (b-d) Representative clones in rendered confocal z-stacks. Images representative of 4-7 animals per timepoint. (e) Composition of clones analysed in 3D projections. Axis indicate clone frequency with a given number of basal and subrabasal cells *=10-19 and **=20-39 basal cells. (f) Labelled cell fraction, clone persistence and clone size (white, black, grey circles respectively) at indicated time points relative to P7. (e,f) P7: n=44 clones from 5 mice, P23: n=49 clones from 4 mice; P56: n=32 clones from 5 mice; P90: n=68 clones from 7 mice; P365: n=15 clones from 3 mice. (g) Strategy for cell cycle analysis using the Col1a1-tetO-H2BmCherry mouse model. (h, i) Detection of H2BmCherry at indicated time points following DOX removal. White and green arrowheads indicate cells in basal layer and SG duct. Images representative of 3 animals. (j) Experimental probability distribution of cell divisions within a 2-week period and stochastic theory based on a fitted proliferation time of 3.5 days (black line) ±0.5 (grey lines, 95% confidence interval). 204 measurements in n=3 animals (k) Biophysical modelling of mean clone size fits the evolution of both basal (red) and suprabasal (blue) compartments. n-numbers as in (e-f) (l) Model-prediction of fate probabilities for equipotent progenitors in the adult SG. P and D refer to progenitor (basal) and differentiated (suprabasal) cells. (m) Experimental basal clone size distributions (dots, same dataset as (b-f) vs. theoretical distribution (lines, using parameters shown in (l)) at all timepoints. (n) Experimental (dots) vs. theoretical (lines, using parameters in (l)) clone persistence in the SG. n-numbers as in (e-f). (o) Strategy for clonal labelling in adulthood using Lrig1CreERT2 and K14CreERT mouse models. (p) Typical clone from Lrig1CreERT2. Image representative of 3 animals. (q) Basal clone size distribution for Lrig1CreERT2 (red) and K14CreERT (green) derived clones (mean±S.D., n=3 mice/group). Black line = model-prediction from panel (l). Best-fit value of 3.5d: line, shaded area: 95% confidence interval. Best-fit division rate (4.5d)=pink line. Data are mean±S.E.M. Nuclei stained with DAPI. Scale bars, 50μm.
A first important finding was that the expansion of the average clone size was perfectly compensated by clone loss, leading to a constant labelled cell fraction (Figure 3f). This demonstrated that Lrig1 marked a representative population of SG cells responsible for long-term SG maintenance as an independent compartment. Before attempting to model the cellular dynamics, we wished to constrain the parameter set by measuring independently the basal cell division rate during homeostasis. The decay in H2B-mCherry showed that cells divide on average every 3.5±0.5 days (Figure 3g-j). With this division rate, we tested whether the most parsimonious model, involving equipotent progenitors choosing stochastically between symmetric division and differentiation with equal probability, could explain the dataset.
We found that the model provided a good quantitative prediction of the evolution of the basal clone size over time. Here, the average clone size first increased linearly from P7, as expected from neutral drift dynamics, followed by a slower convergence towards a plateau, as expected from a finite compartment drifting towards monoclonality (Figure 3k). Considering the measured division rates, the fast speed of monoclonal conversion suggested that few, if any, divisions resulted in asymmetric fate outcome (P->P+D, Supplementary Note; Figure 3l). To provide a more quantitative test of the model, we examined basal clone size distributions prior to monoclonal conversion (P7 and P23). The clonal distributions converged towards a characteristic exponential scaling form, as predicted by the model (Figure 3m). Importantly, this behaviour is a hallmark of neutral drift dynamics of an equipotent population24. At P56, P90 and P365, this scaling behaviour disappeared, as expected in a process of monoclonal conversion. The proposed model for SG maintenance in the presence of constrained expansion could further be confirmed quantitatively by the initial drop in clone persistence and subsequent fixation of labelled glands (Figure 3n). One should note that the observed exponential character of the clone size distribution cannot discriminate between intrinsic and extrinsic modes of fate regulation at the level of individual cell divisions. To resolve such behaviour, a detailed examination of temporal lineage dependences would be required by e.g. live-imaging25. We conclude that the population-wide dynamics of the SG lineage followed a model of long-term stochastic cell fate behaviour in which monoclonal conversion arose through spatial restrictions in the total size of the progenitor niche.
Previous studies have proposed that SG maintenance involved a directional bias, where progenitors at the top of the gland replenished cells lost through differentiation at the bottom6, 26. Such a model would imply that persisting “top”-clones remain anchored to the basal layer, expanding anisotropically towards the bottom. Moreover, loss of differentiated cells should occur preferentially in clones localised towards the bottom. However, such behaviour was not supported by the data, where irrespective of location, clones included differentiated progeny without statistical biases in size or composition (Figure S2d-e), and no correlation between clone size and isotropy (Figure S3k-m). If anything, expansion of bottom-clones post-P23 was slightly larger than those in the top, which could be explained by higher proliferation rates (±12%) in cells at the bottom (Figure 3i, S3n,o, Supplementary Note). Importantly, the introduction of this small spatial gradient in cell division rate, within the same framework of stochastic division/differentiation choices, did not markedly change the model predictions in relation to clone size or survival (Figure S3p,q, Supplementary Note).
To further test the model’s predictive power, we performed two short-term lineage tracing experiments initiated during homeostasis, targeting K14 and Lrig1 expressing cells (Figure 3o). In both cases, the model predicted faithfully the clone size distributions and persistence with a proliferation rate that was broadly consistent with that inferred from the H2B-mCherry dilution assay (Figure 3p,q). We conclude that the observed drift of labelled glands towards monoclonality during homeostasis resulted from stochastic and neutral competition between equipotent progenitor cells confined in a spatially-restricted niche.
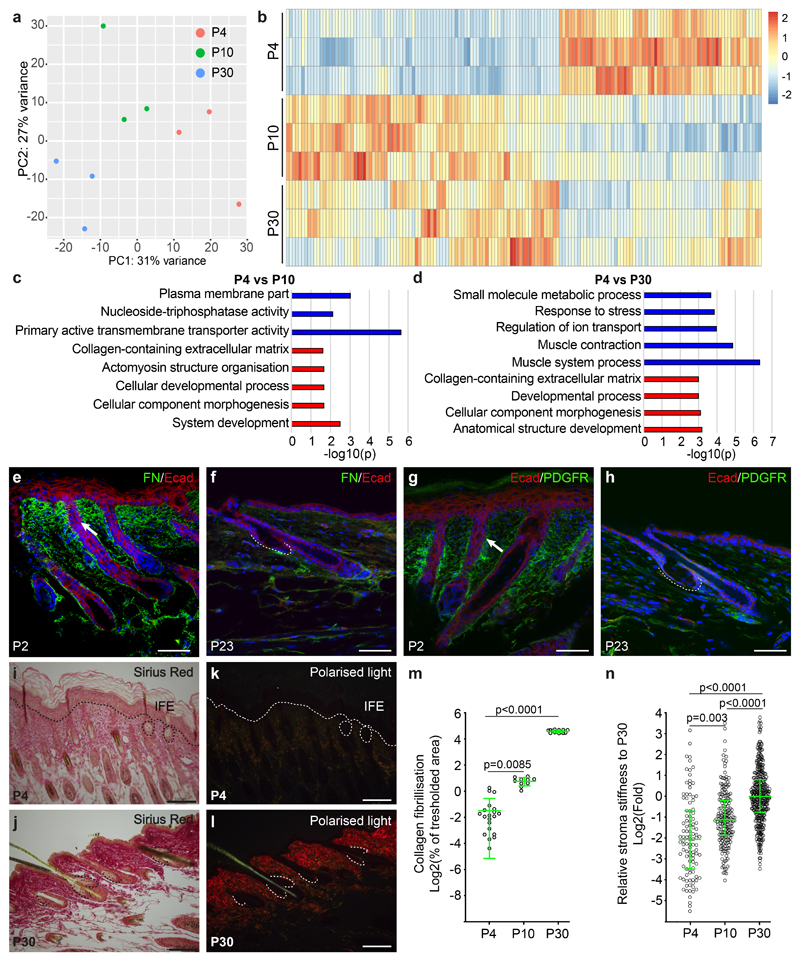
To address how the sudden transition in cell behaviour between morphogenesis and homeostasis is orchestrated at the molecular level, we isolated RNA from SGs during morphogenesis (P4), immediately after the transition into homeostasis (P10) and homeostasis (P30). The temporal identities were evident in the principal component analysis (Figure 4a), with an enrichment of genes associated with developmental processes and extracellular matrix at P4 when compared to the other two homeostatic time points (Figure 4b-d; Table S1). At the protein level, there was a profound decrease in Fibronectin that surrounds the epithelium during homeostasis and decreased levels of Pdgfra+ mesenchymal cells, suggesting a reduced fibroblast density (Figure 4e-h). Stromal collagen deposition, visualised by Sirius red staining and polarised light microscopy showed enhanced fibrillisation in homeostatic samples, and Atomic Force Microscopy (AFM) measurements revealed an increase over time in the elastic modulus of the SG stroma (Figure 4i-n). This suggested that SGs expanded within a pliable environment during morphogenesis, and that changes in spatial constraints from the mechanical microenvironment could promote the transition into homeostasis.
Figure 4. Dynamic extracellular matrix remodelling and sebaceous gland development.
(a) Principal component analysis of RNA-seq samples from morphogenesis and homeostasis. (b) Heatmap of differentially expressed genes (Log2fold change > 0.5; FDR < 0.05 based on Benjamini-Hochberg multiple correction method following a Wald test for statistical significance), comparing the expression profile of epithelial SG progenitors isolated from P4 to that of P10 and P30. (c-d) GO-term enrichment analysis highlighting the biological processes enriched in the P4 samples compared to P10 and P30 time points. (a-d) The overrepresentation test was performed using a Benjamini-Hochberg False Discovery Rate for multiple correction following a Fisher exact test for statistical significance. (e, f) Detection of fibronectin and (g, h) PDGFRa during morphogenesis (P2) and homeostasis (P23). (i-l) Sirius red staining of back skin sections for collagen deposition in brightfield (i, j) and fibrillisation visualized using polarized light (k, l) during morphogenesis and homeostasis (a-l) 3 mice were analysed for each time point. (m) Quantification of progressive fibrillisation at the indicated time points using the thresholded area method in ImageJ software on histological Sirius red stained sections (P4: n=21, P10: n=10, P30: n=14 individual measurements pooled from 3 animals per timepoint. Axis indicates the percentage of a given area positive for fibrillar collagen visualized in polarized contrast illumination. Data displayed as a scattered dot plot with line indicating means±S.D. (n) Atomic force microscopy measurement of stroma Elastic modulus in areas surrounding the SG using a spherical probe at time points indicated shown as fold change when compared to P30. Data displayed as a scattered dot plot with line indicating median with interquartile range. Number of individual measurements P4: n=103, P10: n=115, P30: n=395 pooled from 3 individual mice per timepoint. (m, n) Significance was estimated based on a Kruskal-Wallis test. Nuclei are counterstained with DAPI (blue). Scale bars are 100 μm.
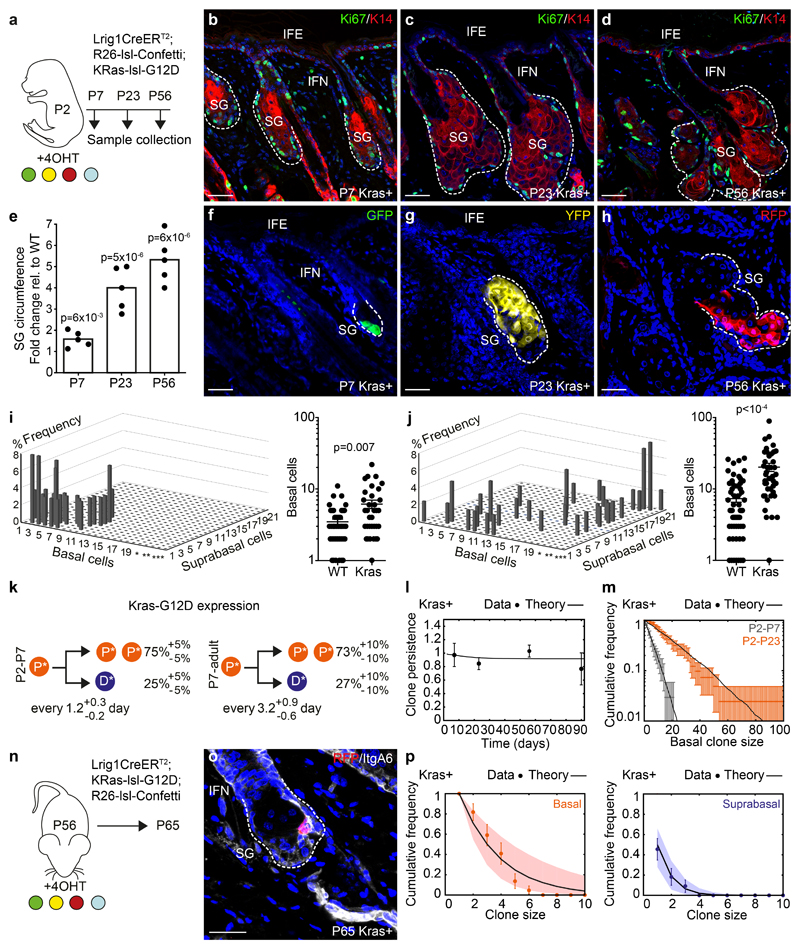
To further develop the regulatory basis of SG development and maintenance, we questioned how cell dynamics changed under perturbation. Introduction of the constitutively active G12D mutation into the endogenous allele of Kras in SG progenitors resulted in persistent gland expansion, with proliferation confined to the basal layer and differentiated cells to suprabasal layers (Figure 5a-h, S4a-m)27. Quantitative clonal analysis showed that, although the inferred cell division rates from P2 to P7 was increased, the most dramatic change occurred after P7 (Figure 5i,j). Detailed analysis showed that mutant cell fate dynamics post-P7 could be captured by a model with a persistent imbalance towards symmetric renewal (P->P+P: 73% versus 50% during homeostasis; Figure 5k, S4n-p, Supplementary Note). This was consistent with the observed SG growth, indicating that KrasG12D released cells from the spatial confinement that operated during homeostasis and the attendant cellular competition. Infrequent Confetti allele recombination implied that clonally labelled KrasG12D mutant cells were in competition with unlabelled cells bearing the same mutation. This hypothesis was supported by further analysis of the model, which showed that a heterogeneous scenario, with mixed competition between KrasG12D cells and wild type cells would translate to a statistically significant perturbation of the clonal dynamics, not observed in the data (Figure 5i,j, S4q, Supplementary Note), although we cannot exclude small fractions of wild-type cells remanining. Interestingly, although ECM deposition and cellular composition of the dermis resembled that observed in homeostasis, the release from spatial confinement of SGs upon Kras activation was aligned with a reduction in matrix stiffness (as measured by AFM) to similar levels as observed during morphogenesis (Figure S5a-g). Given that Lrig1 during early morphogenesis is expressed by mesenchymal cells, it remains a possibility that cell fate changes are partially driven by KrasG12D expression in fibroblasts, although the number of proliferating cells and cellular density in the mesenchyme surrounding the SG is not perturbed (Figure S4e-h, S5c-d).
Figure 5. Cellular dynamics under reduced spatial constraints.
(a) Strategy for clonal labelling of precursors with KrasG12D mutation. (b-d) Detection of Ki67 (green) and K14 (red) at indicated timepoints (Images representative of 3 animals/timepoint). (e) Relative SG size as fold change between WT and KrasG12D-mutants. Kras-n=5 glands/timepoint (from 2 animals), WT-n=8 glands/timepoint (P7, P23) and 6 glands (P56)/timepoint (from 3 animals). Statistical significance was assessed using a two-tailed unpaired Student’s t-test (f-h) Detection of typical clones in rendered confocal z-stacks. Images representative of 3 animals/timepoint. (i, j) Left plots: clone-compositions at P7 and P23 in KrasG12D mutants. Here, Y-axis indicate clone frequency, x-axis basal cells and z-axis number of subrabasal cells. *=21-40, **=41-60 and ***=61-100 basal cells. P7-n=34 clones from 3 animals; P23-n=41 clones from 4 animals. Right plots display basal footprint of same KrasG12D clones but compared to WT clones. WT: P7-n=44 clones in 3 animals, P23-n=49 clones from 4 mice. (k) Cell fate probabilities (from l-p) of progenitors between P2-P7 and after P7 in KrasG12D-mutants (*). P: basal progenitors and D: suprabasal differentiated cells. (l) Experimental vs theoretical (line, from parameters in (k) clone persistence (P7: n=426 follicles in 7 animals; P23: n=253 follicles in 4 animals; P56: n=253 follicles in 5 animals; P90: n=160 follicles in 3 animals). (m) Basal clone size distribution at P7 (grey) and P23 (orange) (n as in i, j). Lines represent the best-fit prediction (used to extract P2-P7 parameters in (k)). Data displayed as means±S.D. (n) Strategy for clonal labelling during adulthood in KrasG12D-mutants. (o) Representative clones (RFP) and integrinA6 (grey) in rendered z-stacks in SG with activated KrasG12D. Image representative of 3 animals. (p) Clone size distributions according to basal (red) and suprabasal (blue) cells in KrasG12D-mutants are well-fit by a single exponential, consistent once again with a single equipotent progenitor population. Lines represent the best-fit prediction (used to extract P7-adult parameters in (k), while dots represent the average. (o-p) n=23 clones/3 animals; WT data from Figure 3o). Statistical significance was assessed using a non-paired Mann-Whitney test. Data in graphs displayed as mean±S.E.M. Nuclei stained with DAPI. Scale bars, 50μm.
From a theoretical perspective, a first key consequence of eliminating competition between neighbouring progenitors is that most labelled clones should survive long-term. Indeed, using the inferred parameter set, we found that the model could predict accurately the time-dependence of clonal persistence (Figure 5l). Second, while mutant cells remain equipotent, we still expect stochastic cell fate choice (albeit imbalanced) to translate to scaling behaviour of the clone size distribution. Once again, we found excellent quantitative agreement between the model and the experimental clone size distributions at both P7 and P23 (Figure 5m). Both of these predictions were important, as they also validated the hypothesis that the vast majority of SG cells express the KrasG12D mutation. As a third test of the inferred cell dynamics, we induced labelling and KrasG12D expression in adulthood (P56) followed by a 9-day chase. Importantly, we found that a model with an imbalance towards symmetric renewal (73%), but with a division rate similar to that observed in wild-type, predicted both the average basal/suprabasal clone sizes, and their detailed distributions (Figure 5n-p). This result suggested that the observed imbalance in cell fate resulting from KrasG12D expression is a manifestation of altered clonal competition driven by the changes in matrix stiffness, while proliferation rates were less affected.
In this study, we have shown that morphogenesis of SG is driven by approximately eleven equipotent and lineage-allocated progenitors. These progenitors undergo a defined process of stochastic cell division and differentiation with fate choices that appear uncorrelated from neighbouring cells, leading to SGs of variable sizes. Around P7, SG progenitors transfer collectively into a homeostatic programme, where stochastic cell fate choice drives neutral competition between basal progenitors (Figure S5h). In contrast to previous reports, we find no evidence that specialised stem cells residing either within or outside the SG play a role in its maintenance6–8. Rather, our results demonstrate that all basal cells contribute to long-term SG maintenance similar to the IFE20.
Our observations reveal that spatial confinement of the progenitor niche represents a critical parameter for on-going cellular competition between clones during homeostasis. This raises important questions on how niche size is fixed within different tissues and how deviations may affect long-term tissue function. Our data suggest in particular that the biophysical properties of the stroma are important factors that need to be considered to direct the transition from morphogenesis to homeostasis. This is consistent with recent findings that matrix composition and stiffness promote changes in cell fate decisions via mechanical forces during homeostasis, morphogenesis and regeneration28, 29. Our characterisation of cell behaviour in the SG and biophysical properties of their microenvironment emphasise the potential inductive properties of the stroma imposing restrictions on cell behaviour.
Furthermore, quantitative fate mapping studies in the IFE have revealed that oncogene-dependent effects drive persistent tissue expansion30, 31. In line with this, we showed that the primary effect of KrasG12D activation within the SG is to evoke a development-like cell fate bias, rather than increased cell division, to fuel expansion (Figure S5h). This has widespread implications on our understanding of Ras mutations during tumour development, if Ras-activation similarly to the activation from developmental state, affects matrix stiffness. Interestingly, Ras activation in vitro has been shown to elevate some aspects of cell crowding without directly affecting proliferation32. This is analogous to our observations for KrasG12D, where cells maintain their normal proliferation rates while developmental cell fate outcome drives continuous gland expansion. Interestingly, the effect of Ras activation has little effect on intestinal stem cells33, where self-renewal in contrast to the SG relies on Wnt signalling34–36. This means that progenitors that exhibit neutral drift dynamics exist in both Wnt dependent and independent niches, and our results suggest that it is the outer physical boundaries of that niche, in combination with tissue-specific pathways, that governs tissue homeostasis. Further studies are required to define how the changes in the extracellular matrix composition and biophysical properties control stem cell behaviour.
Methods
Mice
The mouse models have all been described previously Lrig1::eGFPiresCreERT2 (Lrig1CreERT2) 5, K14CreERT 37, Kras-lsl-G12D27; Rosa26-lsl-Confetti (R26Confetti)15; R26-TTA/TTA:Col1a1-tetO-H2B-mCherry mice (JAX Stock No: 014602)38. C57BL/6J mice used for the EdU pulse-chase experiments were purchased from Taconic, Denmark. Mouse colonies were housed in SPF (specific pathogen free) animal facilities, with companion mice, and cages were placed under a 12hr light-dark cycle. Food and water were provided ad libitum. This study is compliant with all relevant ethical regulations regarding animal research. The National animal ethics committee in Denmark reviewed and approved all animal procedures (Permit numbers 2013-15-2934-00756 and 2017-15-0201-01381). Female and male animals were used for all experiments with an equal animal gender ratio in the majority of the analysis.
Induction of lineage marks
For clonal induction Lrig1CreERT2-R26Confetti, Lrig1CreERT2-KRasG12D-R26Confetti, and KrasG12D-R26Confetti mice were treated with a single topical application of 1.43 mg 4-hydroxy tamoxifen (4OHT; Sigma-Aldrich) dissolved in Acetone (99% Sigma-Aldrich), applied topically in a total volume of 100uL on approximately 2 cm2 back skin. Adult animals were shaven before application. For high dose experiments 4OHT was administered to pups on two consecutive days starting at P3 intraperitoneally with 1.43 mg 4-hydroxy tamoxifen in corn oil (Sigma).
Cell proliferation
To measure the kinetics of cell proliferation H2B mCherry mice were fed for 2 weeks prior to label dilution with 2 g/kg sterile DOX food pellets (Ssniff-Specialdiäten GmbH). For 48-hour pulse/chase experiments, 8,3ug/g bodyweight EdU (Thermo-Scientific) was administered i.p. in sterile PBS. The dose was adjusted according to bodyweight at the time of injection (t=0). EdU incorporation was visualized in frozen thick sections using a Click-iT™ EdU Alexa Fluor 647 Imaging Kit (Thermo Scientific) according to manufacturers’ protocol. All sections for quantification where counterstained with Diamidino-2-phenylindole dihydrochloride (DAPI; 1μM; Sigma) and IntegrinA6 as described in the section below.
Immunohistochemistry
Back skin was fixed with 4% paraformaldehyde (PFA) either for 1 hour at room temperature and embedded in optimal cutting temperature compound (OCT, Cell Path) or overnight at 4C followed by transfer to 70% ethanol prior to paraffin block preparation. For basal and suprabasal cell quantifications thick frozen sections were cut in a 70-90 μm thickness using a CM3050S Leica cryostat (Leica Microsystems). While the fluorescence of Lrig1-expressing (eGFP+) cells and fluorescent labels from the confetti allele were detected directly, immunofluorescence staining was done against selected antigens as listed in Supplementary Table 2. Briefly, thick floating sections were incubated for 1hour in PBS supplemented with 0.5% BSA, 0.5% fish skin gelatin, and 0.1% Triton X-100 before incubation overnight at 4C° with primary antibody diluted in blocking buffer. Sections were washed with PBS supplemented with 0.05% Triton X-100 for 3 × 15 min prior to another round of overnight incubation with appropriate secondary antibodies (Supplementary Table 2) supplemented with DAPI. Finally, sections were washed with PBS for 3 × 15 min at room temperature. Back skin frozen sections were incubated with Oil red O working solutions as described39. Paraffin-embedded thin sections (3-4μm) were stained following a rehydration and antigen retrieval under pressure in fresh citric acid buffer pH 6.0. Hematoxylin staining was performed using Hematoxylin QS (Vector) and mounted using DPX mounting medium (Sigma). For immunostainings the thin sections were blocked for 1hour in blocking buffer before incubation with primary antibody overnight at 4C°, washed with PBS with 0.05% Triton X-100 for 3 × 5 min and left at RT with secondary antibodies and DAPI at room temperature for 1 hour, rinsed in PBS for 3 × 5 min. For fluorescence detection sections were mounted in medium comprising 7 parts glycerol (>99% Sigma-Aldrich) and 3 parts Mowiol (Monsanto). Three-dimensional imaging was done using laser scanning confocal microscope (Leica TSC SP8) while two-dimensional H&E sample images were acquired using NDP Zoomer Digital Pathology scanner (Hamamatsu) and NDPview2 software. Images were processed and analysed following three-dimensional reconstruction of z-stacks covering the full 3D orientation of individual clones using Fiji software or IMARIS version 8.4.
RNA sequencing
Fresh mouse back skin was immediately embedded into OCT and snap frozen. The tissue was sectioned at 10 μm, and placed onto a PEN-membrane slide that had been exposed to UV for 20 min. Tissue sections were then stored at -80C until further use. Sections were quickly thawed at room temperature for 1 min and then submerged in 70% Ethanol for 2 min. Slides were then dipped in RNAse free water until all visible remaining OCT was removed and then immediately dipped into Cryosyl Violet Acetate solution (1%) (Sigma) for 30s. The tissue was dehydrated using ascending ethanol solutions 70%, 96%, 100%. Slides were left to airdry for 2 min and used for RNA isolation of SG progenitors on a laser capture microdissection microscope (Zeiss Axio Observer Z1) collection time did not exceed 20 min in order to avoid RNA degradation. RNA isolation was performed using the Qiagen RNAmicro kit. The concentration and integrity of RNA was measured using the Bioanalyser 6000 Pico kit (Agilent). All extracted RNA was used as input material and libraries were prepared by following the SMARTer Stranded Total RNA-Seq kit v2 Pico Input Mammalian user manual (Takara). In brief, samples were fragmented at 94 °C prior to first-strand synthesis. Illumina adaptors and indexes were added to single-stranded cDNA via 5 cycles of PCR. Libraries were hybridized to R-probes for fragments originating from ribosomal RNA to be cleaved by ZapR. The resulting ribo-depleted library fragments were amplified with 12 cycles of PCR. Fragment length was confirmed with the Bioanalyzer DNA 1000 Kit (Agilent) and concentration was quantified with the Qubit dsDNA BR Assay (Life Technologies. Finally, samples were sequenced on the Illumina NextSeq 500 platform (Illumina).
The quality of the raw single-end sequence reads were evaluated using FastqQC and FASTQ Screen. Raw reads were trimmed with Trimmomatic (v0.32). Trimmed reads were mapped to the canonical chromosomes (mm10 genome assembly) using STAR allignment tool (v2.5.1a) with parameters: '--outFilterMatchNmin 16 --outFilterMismatchNmax 5 -- outFilterMismatchNoverLmax 0.1 --sjdbOverhang 69 --twopassMode Basic'. Aligned reads were assigned genes using featureCounts (v1.5.1) with parameters: '–primary -s 2 -J'. RefSeq gene annotation (downloaded from UCSC 2018/08/05) was used both in the alignment and count assignment steps. The raw gene counts were imported into the statistical software R (v3.4.2) and further processed and differentially expressed genes called using the DESeq2 R-package (v1.18.1). Gene expression levels were considered significantly different across groups if we observed log2 fold changes of ≥ 0.5 associated with a false discovery rate (FDR)-adjusted P ≤ 0.05. Gene expression was obtained in the form of nRPKM as described previously. GO-term enrichment analysis for unbiased gene ontology analysis was performed using PANTHER Version 12.0 (http://pantherdb.org/).
Atomic force microscopy
Back skin fresh frozen sections of 10μm thickness were used to perform measurements. The mechanical properties of the samples were studied using a BioScope Catalyst atomic force microscope (Bruker Nano Surfaces), coupled with the optical microscope Leica DMI6000B (Leica Microsystems). After the thermal stabilization, for each sample, 3 to 7 areas of 140 μm2 were analyzed using the “Point and Shoot” method. On average 120 force-distance curves/area, spaced by at least 20 μm, were collected. The experiments of nanoindentation were performed using a 5 μm Borosilicate Glass spherical tip mounted on a V-shaped silicon nitride cantilever having a nominal spring constant of 0.06 N/m (Novascan Technologies). Indentations were performed with a velocity of 2 μm/s, in relative trigger mode and by setting the trigger threshold to 1 nN. The apparent Young's modulus (YM) was calculated using the NanoScope Analysis 1.80 software (Bruker Nano Surfaces), fitting the approaching force curves with the Hertz indentation model and using a Poisson’s ratio of 0.5. All the force-distant curves having a not clear base line, a maximum above or below 1nN or a change of slope in the region of the fitting (minimum and maximum force fit boundary 5% and 25%, respectively) were rejected and not considered for the analysis. To compare all the calculated apparent YM belonging to different datasets, a data normalization was performed as follows: for each experimental dataset the sample P30 was chosen as control and the median value of the corresponding YM distribution was calculated. Then, for all the samples belonging to the same dataset, P30 included, the ratio between each calculated YM value and the YM median value of P30 was performed. The normalized YM values of similar samples but belonging to different experimental datasets could be then pulled together. The obtained normalized plot expressed in fold change of each sample vs P30 is analyzed using the non-parametric Kruskal-Wallis statistical test followed by the Dunn's Multiple Comparison Test.
Sirius red staining and quantification
Picrosirius red analysis was achieved using paraffin sections of back skin stained with 0.1% picrosirius red (Direct Red80, Sigma) and counterstained with Weigert's iron haematoxylin (Diapath) to reveal fibrillar collagen. Sections were then analysed using a Zeiss microscope fitted with an analyzer and a polarizer oriented parallel and orthogonal to each other and quantified using ImageJ software using the thresholded area method. n=3 animals per group were used. Kruskal-Wallis test was performed to determine statistical differences in between groups.
Flow cytometry
Keratinocytes were isolated from mouse back skin as previously described40. Hematopoietic and endothelial cells were excluded by negative selection of CD45+ve and CD31+ve cells (PE-Cy7-conjugated, 30-F11 BD Pharmingen and 390, eBioscience) while the live epithelial cell fraction was selected based on positive selection of EpCam (PE-conjugated, clone G8.8, eBioscience) and a lack of DAPI incorporation. Flow cytometric analysis of H2B-mCherry signal intensity in live EpCam+ cells was carried out using an FACSAriaIII (Beckman Coulter) cell sorter and FlowJo software. Unstained samples, single staining and FMO-1 (mCherry) controls were used to define the gate boundaries.
Statistics and reproducibility
Details on the biophysical modelling approach and statistical tests used to study the cellular dynamics of sebaceous gland morphogenesis, homeostasis, and oncogenic activation are provided in the Supplementary Note. The number of biological and technical replicates and the number of animals are indicated in figure legends and text. Sample size was not predetermined. Experiments were performed without methods of randomization or blinding. For all experiments with error bars, the standard error of the mean (S.E.M.) or standard deviation (S.D.) was calculated to indicate the variation within each experiment or sample. Mann Whitney non-parametric tests were used to assess the significance.
Supplementary Material
Acknowledgement
We thank members of the Jensen and Simons lab for comments and suggestions; Bo Porse and Hans Clevers for animal models and CAB, University of Copenhagen for experimental support. This work was supported by Leo Pharma Research Foundation (to KBJ), Lundbeck Foundation (R105-A9755 to KBJ), the Novo Nordisk Foundation (NNF14OC0012927 to KBJ), EMBO Young Investigator programme (to KBJ) and the Wellcome Trust (grant number 098357/Z/12/Z to BDS). BDS is supported by a Royal Society E P Abraham Professorship (RP\R1\180165). This project has received funding from the European Union’s Horizon 2020 research and innovation programme (grant agreements STEMHEALTH ERCCoG682665 and INTENS 668294 to KBJ). The Novo Nordisk Foundation Center for Stem Cell Biology is supported by a Novo Nordisk Foundation grant number NNF17CC0027852. SE, CCF, SP are supported by la Fondation ARC (R18020AA). The authors acknowledge the IRCAN PICMI core facility which is supported by grants from the Conseil Général 06, the FEDER, the Ministère de l'Enseignement Supérieur, the Région Provence Alpes-Côte d'Azur, the Canceropole PACA and INSERM.
Footnotes
Code availability
Computational source code is accessible in GitHub via https://github.com/ehannezo/SG_stemcelldynamics
Data availability
RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE126491. Raw data for Fig. 1-5 and Supplementary Fig. 1-5 have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Author contribution
Conceptualization, MSA, EH, SU, BDS and KBJ; Methodology, MSA, EH, SU, SE, YA, CCF, BDS and KBJ; Investigation, MSA, EH, SU, SE, SP, KB, SS, MM, MTP, JVJ, DC; Formal Analysis, EH, BDS; Writing – Original Draft, MSA, EH, SU, BDS and KBJ; Funding Acquisition, BDS and KBJ; Supervision, BDS and KBJ
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Zouboulis CC, et al. Beyond acne: Current aspects of sebaceous gland biology and function. Rev Endocr Metab Disord. 2016;17:319–334. doi: 10.1007/s11154-016-9389-5. [DOI] [PubMed] [Google Scholar]
- 2.Ehrmann C, Schneider MR. Genetically modified laboratory mice with sebaceous glands abnormalities. Cell Mol Life Sci. 2016;73:4623–4642. doi: 10.1007/s00018-016-2312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paus R, et al. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J Invest Dermatol. 1999;113:523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E. Skin stem cells: rising to the surface. J Cell Biol. 2008;180:273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell stem cell. 2013;13:471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horsley V, et al. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fullgrabe A, et al. Dynamics of Lgr6(+) Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Reports. 2015;5:843–855. doi: 10.1016/j.stemcr.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersson M, et al. TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J. 2011;30:3004–3018. doi: 10.1038/emboj.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 10.Frances D, Niemann C. Stem cell dynamics in sebaceous gland morphogenesis in mouse skin. Dev Biol. 2012;363:138–146. doi: 10.1016/j.ydbio.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Ghazizadeh S, Taichman LB. Multiple classes of stem cells in cutaneous epithelium: a lineage analysis of adult mouse skin. Embo J. 2001;20:1215–1222. doi: 10.1093/emboj/20.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oules B, et al. Mutant Lef1 controls Gata6 in sebaceous gland development and cancer. EMBO J. 2019;38 doi: 10.15252/embj.2018100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development. 2009;136:2815–2823. doi: 10.1242/dev.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen KB, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell stem cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Guiu J, et al. Tracing the origin of adult intestinal stem cells. Nature. 2019;570:107–111. doi: 10.1038/s41586-019-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Keymeulen A, et al. Lineage-Restricted Mammary Stem Cells Sustain the Development, Homeostasis, and Regeneration of the Estrogen Receptor Positive Lineage. Cell Rep. 2017;20:1525–1532. doi: 10.1016/j.celrep.2017.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wuidart A, et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol. 2018;20:666–676. doi: 10.1038/s41556-018-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanpain C, Simons BD. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 2013;14:489–502. doi: 10.1038/nrm3625. [DOI] [PubMed] [Google Scholar]
- 20.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 21.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 22.Donati G, et al. Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat Cell Biol. 2017;19:603–613. doi: 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponder BA, et al. Derivation of mouse intestinal crypts from single progenitor cells. Nature. 1985;313:689–691. doi: 10.1038/313689a0. [DOI] [PubMed] [Google Scholar]
- 24.Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 25.Mesa KR, et al. Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell stem cell. 2018;23:677–686 e674. doi: 10.1016/j.stem.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cottle DL, et al. c-MYC-induced sebaceous gland differentiation is controlled by an androgen receptor/p53 axis. Cell Rep. 2013;3:427–441. doi: 10.1016/j.celrep.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickstrom SA, Niessen CM. Cell adhesion and mechanics as drivers of tissue organization and differentiation: local cues for large scale organization. Curr Opin Cell Biol. 2018;54:89–97. doi: 10.1016/j.ceb.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Danes A, et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature. 2016;536:298–303. doi: 10.1038/nature19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groth A, Willumsen BM. High-density growth arrest in Ras-transformed cells: low Cdk kinase activities in spite of absence of p27(Kip) Cdk-complexes. Cell Signal. 2005;17:1063–1073. doi: 10.1016/j.cellsig.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 34.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 35.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129:95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- 36.Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes & development. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 39.Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 40.Moestrup KS, Andersen MS, Jensen KB. Isolation and In Vitro Characterization of Epidermal Stem Cells. Methods in molecular biology. 2017;1553:67–83. doi: 10.1007/978-1-4939-6756-8_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.