Abstract
We have previously shown that the retinoid-related orphan receptor alpha (RORα) phosphorylation plays a pivotal role in sulfotransferase 1E1 gene regulation within mouse liver. Here, we found serine 100–phosphorylated RORα orchestrates constitutive androstane receptor (CAR) and hepatocyte nuclear factor 4 alpha (HNF4α) to induce CYP2B6 by phenobarbital (PB) in human primary hepatocytes (HPHs). RORα knockdown using small interfering RNAs suppressed CYP2B6 mRNAs in HPH, whereas transient expression of RORα in COS-1 cells activated CYP2B6 promoter activity in reporter assays. Through chromatin immunoprecipitation (IP) and gel shift assays, we found that RORα in the form of phosphorylated (p-) S100 directly bound to a newly identified RORα response element (RORα response element on CYP2B6 promoter, −660/−649) within the CYP2B6 promoter in untreated or treated HPH. In PB-treated HPH, p-Ser100 RORα was both enriched in the distal phenobarbital response element module (PBREM) and the proximal okadaic acid response element (OARE), a known HNF4α binding site. Chromatin conformation capture assay revealed direct contact between the PBREM and OARE only in PB-treated HPH. Moreover, CAR preferably interacted with phosphomimetically mutated RORα at Ser100 residue in co-IP assay. A gel shift assay with a radiolabeled OARE module and nuclear extracts prepared from PB-treated mouse liver confirmed that HNF4α formed a complex with Ser 100–phosphorylated RORα, as shown by supershifted complexes with anti–p-Ser100 RORα and anti-HNF4α antibodies. Altogether, the results established that p-Ser100 RORα bridging the PBREM and OARE orchestrates CAR and HNF4α to form active chromatin complex during PB-induced CYP2B6 expression in human primary hepatocytes.
SIGNIFICANCE STATEMENT
CYP2B6 is a vital enzyme for the metabolic elimination of xenobiotics, and it is prone to induction by xenobiotics, including phenobarbital via constitutive androstane receptor (CAR) and hepatocyte nuclear factor 4 alpha (HNF4α). Here, we show that retinoid-related orphan receptor alpha (RORα), through phosphorylated S100 residue, orchestrated CAR-HNF4α interaction on the CYP2B6 promoter in human primary hepatocyte cultures. These results signify not only the role of RORα in the molecular process of CYP2B6 induction, but it also reveals the importance of conserved phosphorylation sites within the DNA-binding domain of the receptor.
Introduction
Cytochrome P450 2B6, or CYP2B6, is a drug-metabolizing enzyme that is mainly expressed in the liver. The enzyme is responsible for the metabolism of about 2%–10% therapeutic drugs (Hedrich et al., 2016). Several of its drug substrates, including antimalarial artemisinin (Simonsson et al., 2003), efavirenz (Robertson et al., 2008) and carbamazepine (Oscarson et al., 2006), and nonsubstrates, such as phenobarbital (PB) and phenytoin (Wang et al., 2004), can induce the enzyme posing a potential drug-drug interaction. In fact, regulatory agencies recommend a routine clinical risk assessment for CYP2B6 induction by a new therapeutic product (Zhang et al., 2009; Fahmi et al., 2016). Thus, CYP2B6 induction as well as its polymorphism remain important parameters determining therapeutic or toxic outcomes of certain pharmaceuticals, including efavirenz and cyclophosphamide (Desta et al., 2007; Wang et al., 2013; Zanger and Klein, 2013; Hedrich et al., 2016).
Over the past few decades, the molecular mechanism of CYP2B6 induction has been extensively studied and it is well understood that the constitutive androstane receptor (CAR) [nuclear receptor (NR) 1I3] mediates CYP2B6 induction by phenobarbital, a prototype CYP2B6 inducer (Sueyoshi et al., 1999; Wang and Negishi, 2003; Wang et al., 2004; Negishi, 2017). Studies revealed that two DNA elements, the distal phenobarbital response element module (PBREM) and the proximal okadaic acid response element (OARE), on the CYP2B6 regulatory region are crucial for the induction of the gene by CAR activators (Swales et al., 2005; Inoue and Negishi, 2008). In the human or rodent liver primary cells, PB induces dephosphorylation of CAR at Threonine 38, which, in turn, initiates nuclear localization and occupation of PBREM motif and subsequent gene induction (Mutoh et al., 2009). Studies have shown that hepatocyte nuclear factor 4 alpha (HNF4α), which constitutively occupies a direct repeat protein-binding motif separated by a single nucleotide (DR1) unit within the OARE motif in the CYP2B6 proximal promoter and is also a key factor for the induction of the CYP2B6 gene (Inoue and Negishi, 2009).
Ser100 of retinoid-related orphan receptor alpha (RORα) represents a highly conserved phosphorylation site between the two zinc fingers in the nuclear receptor superfamily DNA-binding domain. Amino acid sequence comparison among the receptors suggests the site is conserved in 41 out of 46 total human nuclear receptors (Negishi, 2017). In a recent publication, we found RORα was phosphorylated at Ser100 in mouse liver and that phosphorylation reversed RORα from suppressor of mouse sulfotransferase (Sult) 1e1 gene to coactivator (Fashe et al., 2018), showing that RORα can regulate its transcriptional activity through phosphorylation/dephosphorylation and dramatically affect the DNA binding as well as protein-protein interaction properties of the receptor (Hashiguchi et al., 2016; Fashe et al., 2018). Phosphorylation of this conserved site in other nuclear receptors has also been shown (Sun et al., 2007; Sueyoshi et al., 2019). For example, phosphorylation of CAR Thr38 was shown to be a key element for its functions in gene induction mechanism by phenobarbital (Mutoh et al., 2009, 2013). Furthermore, we have observed that equivalent phosphorylation in farnesoid X receptor (FXR) Ser154 (Hashiguchi et al., 2016), retinoid X receptor α (RXRα) Thr167 (Sueyoshi et al., 2019), and estrogen receptor α (ERα) Ser216 (Shindo et al., 2013) contributes to regulation of their respective biologic activities. Thus, the conserved site of phosphorylation may endow each nuclear receptor with unidentified phases of functional regulation.
In this report, we have identified a role of RORα in CYP2B6 gene induction by phenobarbital in human primary hepatocytes through phosphorylation of conserved Ser100, which enhanced RORα to orchestrate CAR and HNF4α interactions and CYP2B6 transcriptional activation. Previously, RORα had been implicated in the regulation of several CAR target genes, including Cyp2b10, Sult1e1, and Sult2a1 in mouse liver cells (Kang et al., 2007; Fashe et al., 2018) and CYP7B1, CYP2C8, and SULT2A1 gene in human liver cells (Echchgadda et al., 2007; Wada et al., 2008; Chen et al., 2009; Ou et al., 2013). However, the modulation of RORα transcriptional activity via Ser100 phosphorylation and its role in regulation of CYP2B6 expression has not been reported. Here, utilizing gene reporter assays, chromatin immunoprecipitation (ChIP), coimmunoprecipitation (co-IP), chromatin conformation capture (3C), and electromobility shift assay (EMSA), we report that Ser100–phosphorylated RORα binds to a newly identified response element, RORα response element (RORE) on CYP2B6 promoter, to activate gene transcription in cultures of human primary hepatocytes and synergized CAR activation of CYP2B6 gene expression. Situated between the PBREM and OARE motifs, where CAR and HNF4α bind, respectively, phosphorylated (p-) Ser100 RORα enhanced CAR and HNF4α to form an active chromatin complex on the CYP2B6 promoter in response to PB to induce gene expression.
Materials and Methods
Antibodies, Chemicals, Reagents, and Animal Treatments.
Antibodies against RORα (no. PP-H3910-00) and HNF4α (no. PP-H1415-00) were purchased from Perseus Proteomics Inc (Tokyo, Japan). Anti–p-Ser100 RORα peptide antibody (αP-Ser100 RORα) and anti-RORα were custom-generated by Genescript (Piscataway, NJ). Anti-GFP–horseradish peroxidase (HRP) (no. ab6663) and anti-FLAG-HRP (no. S8592) were obtained from Abcam (Cambridge, MA) or Sigma-Aldrich (St. Louis, MO), respectively. High Capacity Archive kit, lipofectamine RNAiMAX Reagent, Power SYBR Green Master Mix (no. A25741), TaqMan Universal Master Polymerase Chain Reaction (PCR) Mix (no. 4304437), TaqMan PCR probes [no. Hs00231959_1 for human CAR (hCAR), no. Hs00536545_1 for human RORα, no. Hs00167937_1 for CYP2B6, no. Hs01060665_g1 for β-actin, and no. Hs00609178_1 for human G6PC] were obtained from Life Technologies Corporation (Grand Island, NY). Dual luciferase assay system and TNT Coupled Reticulocyte Lysate Systems were from Promega (Madison, WI), and phosphatase inhibitor cocktail-2 and -3 were from Sigma-Aldrich. ChIP IT Express kit was from active motif (Carlsbad, CA). Polynucleotide T4 kinase, T4 DNA ligase, and AluI were from New England Biolabs (Ipswich, MA). Small interfering RNAs (siRNAs) targeting RORα or scrambled control siRNAs were from Dharmacon (Lafayette, CO). All animals were housed in a room maintained at 22°C with a 12:12-hour light/dark cycle (7:00 AM to 7:00 PM). Mice were fed ad libitum with National Institutes of Health–31 Open Formula Autoclavable diet (Zeigler). Phenobarbital (10 mg/kg) or vehicle (PBS) was administered intraperitoneally and kept for 24 hours for liver nuclear extraction and immunohistochemic analyses. All animal procedures were approved by the Animal Care and Use Committee at National Institute of Environmental Health Sciences at the National Institutes of Health and performed humanly in accordance with the Public Health Service Policy.
Plasmids.
All reporter constructs harboring CYP2B6-promoter DNA fragments used in this study were prepared in pGL3 vector and were described in previous research works (Swales et al., 2005). RORE was deleted within −1.8 kb CYP2B6 promoter and placed in front of luciferase gene in PGL3 vector. RORα plasmids were described in our previous publication (Fashe et al., 2018).
Cell Culture and Transfection.
Human primary hepatocytes were obtained from Life Technologies Corporation and cultured in William’s E Medium without phenol red (no. A1217601) supplemented with 100 nM dexamethasone and L-glutamine. COS-1 and Huh7 cells were in Dulbecco’s modified Eagle’s medium. All media were supplemented with fetal bovine serum and penicillin (100 U/ml) and streptomycin (100 µ/ml), and the cells were cultured at 5% CO2 and 37°C. Transfections with a given plasmid were performed with FUGENE 6 per the manufacturer’s instructions, whereas lipofectamine RNAiMAX Reagent was used for the transfection of siRNAs into the human primary hepatocytes.
ChIP Assays.
The ChIP assays were carried out using active motif’s ChIP IT EXPRESS kit per the manufacturer’s instructions. Briefly, human primary hepatocytes were seeded in 10-cm culture dish overnight before phosphate buffer (PBS) or 1 mM PB treatment of 6 hours. Then the culture medium was washed off, and the cells were incubated in 1% formaldehyde in PBS at room temperature for 10 minutes followed by a 5-minute incubation in glycine. The cells were lysed, nuclear material was collected, and the chromatins were sheared. Overnight immunoprecipitations were performed with anti-RORα, p-Ser100 RORα, HNF4α, or anti-RXRα antibodies, mouse normal IgG, and G protein–conjugated magnetic beads. After washing the beads, the formalin cross-linking was reversed, and PCR amplification was achieved by using primers targeting selected motifs (PBREM, RORE, and OARE) of CYP2B6 gene regulatory region (Table 1).
TABLE 1.
Primers and oligos used in gel mobility shift (EMSA), ChIP, and 3C assays
| Application/Motif | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| EMSA | ||
| NR1/PBREM | TCAGGGTCAGGAAAGTACAGTT | AACTGTACTTTCCTGACCCTGA |
| 2B6-RORE | TCTTGACCTCAATTGATC | GATCAATTGAGGTCAAGA |
| DR1/OARE | ACCTGGACTTTGATATCTC | GAGATATCAAAGTCCAGGT |
| ChIP | ||
| PBREM | TTACTGTGTGTAAAGCACTTC | GACAAACAGTCCTATTTGTAAG |
| 2B6-RORE | CCGAGTAGCTGGGATTAAAAGTACCCA | CTTCCCAACGTGCTGGGATTACAG |
| OARE | GGACAAATGCATGCAAGCAC | TGACCTGATGCCTATCCCTTTAC |
| 3C | ||
| PBREM/OARE | CTGTCAGAGGATGTGTGGGTG | GGCAGGGATACAGTGGTAGAG |
3C Assays.
3C assays were performed in human primary hepatocytes (HPHs). The cells were seeded in a 10-cm culture dish and treated with PBS or 1 mM PB for 6 hours. The cells were incubated in 2% formaldehyde in PBS for 10 minutes before a 5-minute treatment with glycine. Then the cells were harvested in a lysis buffer made of 50 mM Tris (pH 7.5), 150 mM NaCl, 5 mM EDTA, 0.5% NP40, and 1% Triton X in the presence of protease inhibitor cocktail and placed on ice for 10 minutes. The lysates were centrifuged, and the pellet was taken up in 440 µl distilled H2O, 60 µl Cutsmart buffer and SDS (0.3%) were added, and the mixture was incubated at 37°C and 225 rpm for 1 hour followed by addition of 2.5% Triton X-100 in the same incubation conditions for another hour. Restriction enzyme digestion and ligation were achieved by AluI and T4 DNA ligase, respectively. The formalin fixations were reversed at 65°C overnight in the presence of proteinase K (20 µg/ml), which was followed by an RNase treatment of 45 minutes at 37°C, and DNAs were extracted by QIAquick spin column per manufacturer’s instructions. Primers were designed and amplifications were made using Green Taq, and amplicons were separated in a 2% agarose gel from which the expected bands were excised, and gel was extracted to confirm the identity of the amplicon by sequencing.
Gel Shift Assays.
Gel mobility shift assays were described in our previous study (Fashe et al., 2018). Shortly, radioactive probes were generated by end-labeling double-stranded oligonucleotides (NR1/PBREM, 2B6-RORE, or DR1/OARE, Table 1) with [γ-32P]ATP in the presence of T4 polynucleotide kinase. Mock (pcDNA3.1), CAR, RXRα, RORα, and HNF4α proteins were in vitro translated using TNT Coupled Reticulocyte Lysate Systems per the manufacturer’s instructions. One microliter of the protein mixture was mixed with the radioactive probes and incubated at room temperature for 10 minutes before electrophoresis in acrylamide gel.
Reporter Assay.
COS-1 cells in 24-well plates were cotransfected with expression vectors for CAR, HNF4α, RORα, CAR, and RORα or HNF4α and RORα for reporter assays. Five reporter constructs harboring CYP2B6 regulatory region, termed in this study as −1.8 kb, −1.8 kb ∆PBREM, −1.8 kb ∆RORE, or −1.8 kb ∆OARE, were used in this study. Renilla luciferase plasmids were also included in the transfection mixture to normalize the luminescent activity. The relative luciferase activity was measured after 48 hours using the Dual Luciferase Assay System (Promega).
Real-Time PCR.
Total RNAs were extracted from human primary hepatocytes treated with PBS/PB in Trizol reagent, which were reverse transcribed using a High Capacity Archive kit. The mRNAs were quantified by real-time PCR using TaqMan universal PCR master mix and probes for RORα and CYP2B6. mRNA expression of hCAR, human G6PC, CYP7B1, SULT2A1, and human β-actin was also measured as a positive or negative control for the expression of RORα in human primary hepatocytes treated with siRNAs.
Co-IP and Western Blot Assays.
Huh7 cells seeded in 10-cm culture dishes were transfected with expression plasmids of GFP-RORα or its S100A or S100D mutants in the presence or absence of hCAR-V5 or HNF4α-V5. The cells were lysed in a buffer composed of 20 mM Tris-HCl (pH, 7.5), 100 mM NaCl, 0.5 mM EDTA, and 1% Triton -X in 10% glycerol supplemented with protease inhibitor cocktail. The cell debris was removed by centrifugation, and 250 μg protein of the lysate was mixed with anti-GFP agarose overnight, washed (3 times) with the lysis buffer, and subjected to 1× SDS/PAGE sample buffer for Western blot assay. The input (5%) and the IP samples were electrophoresed in 10% acrylamide gel at 150 V and transferred onto polyvinylidene difluoride membranes at 15 V for 1 hour. The membranes were probed with anti-V5 (HRP) or anti-GFP (HRP) antibodies for 1 hour at room temperature and visualized by C-DiGit Chemiluminescent Western Blot Scanner (LI-COR, Inc., Lincoln, NE) using WesternBright Sirius HRP substrate (Advansta Inc, CA).
Statistical Analysis.
Multiple groups were analyzed by two-way analysis of variance with Tukey’s multiple comparison test to obtain P values for the observed differences and 95% confidence interval (CI) for activation ratios. Two groups were compared by unpaired Student’s t test. These statistical analyses were conducted using the software GraphPad Prism.
Results
RORα Regulates the Basal Gene Expression of CYP2B6 in Human Primary Hepatocyte Cultures.
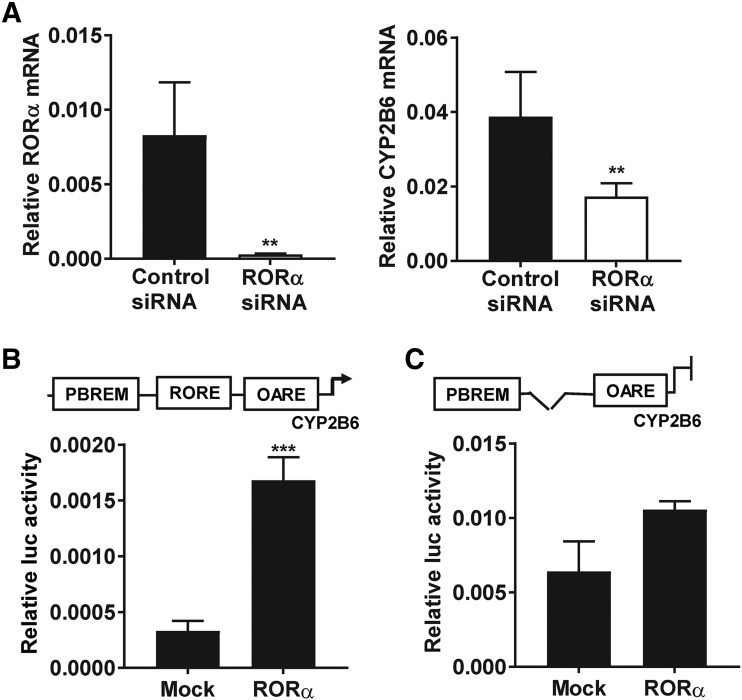
Expression of CYP2B6 mRNAs was studied in human primary hepatocytes treated with control siRNAs or siRNAs targeting RORα for 3 days. Expression RORα mRNAs were suppressed by 96% in the cells treated with siRNAs targeting RORα compared with the control ones. In the same cultures, the basal expression of CYP2B6 was suppressed (P < 0.05) along with RORα mRNAs (Fig. 1A). To confirm the effect of RORα suppression in human primary hepatocytes, we measured the expression of known RORα target genes, such as SULT2A1, G6PC, and CYP7B1. As expected, the change in the expression of these genes was also statistically significant, whereas the expression of CAR mRNA, a non-RORα target gene, was not affected (Supplemental Fig. 1). This clearly suggests that RORα regulates the basal gene expression of CYP2B6 in human primary hepatocytes.
Fig. 1.
RORα regulates the basal expression of CYP2B6 in the liver cells. (A) The expression of CYP2B6 mRNAs was suppressed when RORα expression was attenuated by treatment with siRNAs targeting RORα (n = 5). (B and C) RORα activated CYP2B6 reporter gene. Cell-based reporter assays were performed in COS-1 cells transiently expressing RORα and a luciferase reporter gene harboring −1.8 kb CYP2B6 promoter (n = 3). RORα activated this −1.8 kb CYP2B6 reporter but was unable to do so when the RORα response element was deleted as shown in (C). Luc on vertical axes stands for luciferase. The asterisk (*) between the groups represents statistically significant difference (**P < 0.01; ***P < 0.001). Mean and S.D. are shown for each group of treatment.
To support these observations, we performed a cell-based reporter assay. For this purpose, RORα and a −1.8 kb CYP2B6 promoter placed in front of luciferase gene in PGL3 vector were cotransfected into COS-1 cells. Supporting the above observations, RORα activated a −1.8 kb CYP2B6 promoter in COS-1 cells (Fig. 1B). It has been reported that RORα regulates its target genes by directly binding a DNA core motif AGGTCA preceded by A/T rich sequence (Giguère et al., 1995; Jetten et al., 2001). Thus, we analyzed this −1.8 kb CYP2B6 gene promoter for an RORE sequence and identified an element (5′... TGACCTCAATTG…3′), termed as 2B6-RORE, located −660 and −649 upstream of the CYP2B6 transcription start site. To confirm whether the transcriptional activity of RORα was attributed to its binding to 2B6-RORE motif, we deleted this putative RORE sequence within the −1.8 kb CYP2B6 promoter (1.8 kbΔRORE) and performed a cell-based reporter assay. RORα could not activate the −1.8 kbΔRORE reporter, which suggests the 2B6-RORE is the key element for the CYP2B6 gene activation by RORα (Fig. 1C). Moreover, we performed a gel shift assay using a radioactive probe prepared by labeling the 2B6-RORE with [γ-32P]ATP, and an In Vitro translated RORα proteins. The results indicated that RORα could strongly bind to this sequence (Fig. 2A).
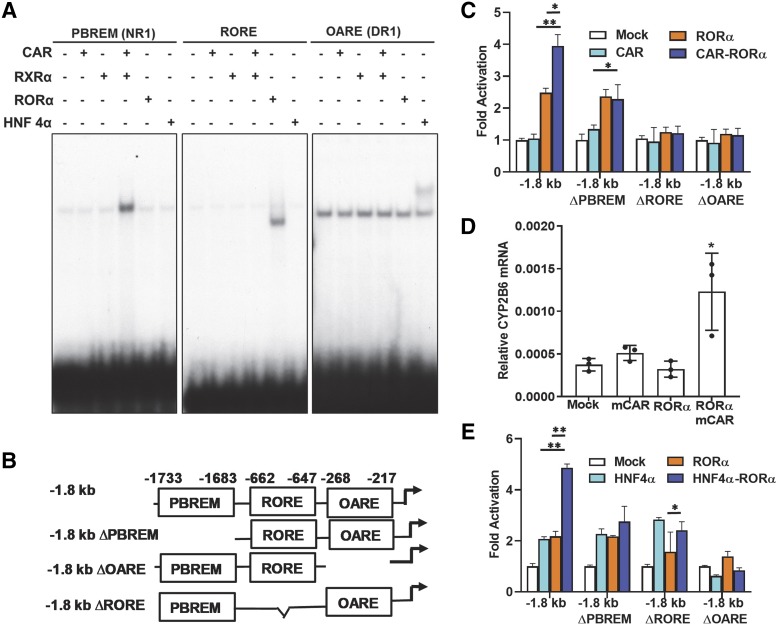
Fig. 2.
RORα coactivated CYP2B6 promoter with CAR or HNF4α. (A) RORα binding DNA element in CYP2B6 promoter was identified by gel shift analyses. Gel shift assay was performed as in Materials and Methods using 32P labeled DNA probes shown in Table 1. The assay revealed RORα bound to its putative RORE in CYP2B6 promoter. CAR and HNF4α bound NR1 and DR1, respectively, as previously reported (Sueyoshi et al., 1999; Inoue and Negishi, 2008). (B) Luciferase reporter constructs used in this study. Three elements (PBREM, 2B6-RORE, and OARE) in CYP2B6 gene promoter and their locations relative to transcriptional start site are shown. (C) RORα and CAR coactivated CYP2B6 reporter genes in COS-1 cells. RORα and CAR coactivation was observed only with the intact −1.8 kb CYP2B6 promoter. Statistical analysis was performed with values between single-factor and two-factor cotransfection. (D) RORα and CAR synergistic activation of CYP2B6 gene. CAR, RORα, or both were transfected into Huh7 cells, and CYP2B6 mRNA expression was measured from total RNAs. (E) RORα and HNF4α coactivated CYP2B6 reporter genes in COS-1 cells. HNF4α and CAR coactivation was observed only with the intact −1.8 kb CYP2B6 promoter. (C and E) Statistical analysis results were shown between single-factor and two-factor cotransfection. (C–E) The asterisk (*) between the groups represents statistically significant difference (*P < 0.05; **P < 0.01). Mean and S.D. are shown for each transfected group (n = 3).
RORα and CAR Coactivate CYP2B6 Promoter.
Since the CYP2B6 gene is an archetypal xenobiotic-inducible gene and the nuclear receptor CAR generally spearheads the molecular process of induction of this gene, we would like to define the role of RORα in the molecular mechanism of CYP2B6 gene induction. For this purpose, we performed gene reporter analysis using CYP2B6 −1.8 kb and its deletion mutant reporters shown in Fig. 2B. As a first experiment, RORα, CAR, or both were cotransfected with −1.8 kb 2B6 reporter into COS-1 cells. The results in Fig. 2C clearly showed RORα and CAR synergistically activated the reporter, since CAR + RORα cotransfection showed 3.97- (95% CI ratio; 3.12–4.84), 3.93- (95% CI ratio; 3.08–4.78), and 2.48-fold (95% CI ratio; 1.63–3.33) activation over mock, CAR, or RORα cotransfected reporters, respectively. We analyzed the role of each of the three DNA elements (i.e., PBREM, 2B6-RORE, and OARE) in the activation of CYP2B6 promoter by preparing reporter constructs by deleting each element within the −1.8 kb CYP2B6 promoter (Fig. 2B). Interestingly, deletion of PBREM, 2B6-RORE, or OARE abolished the synergy between CAR and RORα, suggesting that the three motifs were all important for the synergistic activation of the CYP2B6 gene by CAR and RORα (Fig. 2C). To further confirm the coactivation of the CYP2B6 gene by CAR and RORα, we transfected Huh7 cells with expression plasmids of CAR, RORα, or both and treated the cells with DMSO or 1,4-Bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-Tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) for 24 hours. We could observe that strong induction of the CYP2B6 gene by TCPOBOP only when CAR and RORα were cotransfected, confirming the synergy between the two nuclear receptors (Fig. 2D). Moreover, the binding of RORα onto the OARE motif was further enhanced by coexpression of CAR in Huh7 cell per ChIP assay data (Supplemental Fig. 2).
Similarly, since HNF4α binds the OARE motif that was crucial for the activation of CYP2B6 promoter, we performed a similar reporter assay using the reporter constructs discussed above in COS-1 cells. The results showed RORα and HNF4α coactivated the CYP2B6 promoter. Deletion of PBREM did not affect the activity of either receptor, but deletion of any of the three motifs (i.e., PBREM, 2B6-RORE, and OARE) abolished the observed coactivation (Fig. 2E). In an effort to investigate the actual interaction between HNF4α and RORα on the CYP2B6 promoter, we also performed a ChIP assay using anti-HNF4α antibody in Huh7 cells transiently expressing RORα. It was observed that the binding of HNF4α to the OARE motif was enhanced in cells transiently expressing RORα but not CAR (Supplemental Fig. 2).
RORα Mediated the Contact between PBREM and OARE in PB-Treated Human Primary Hepatocytes.
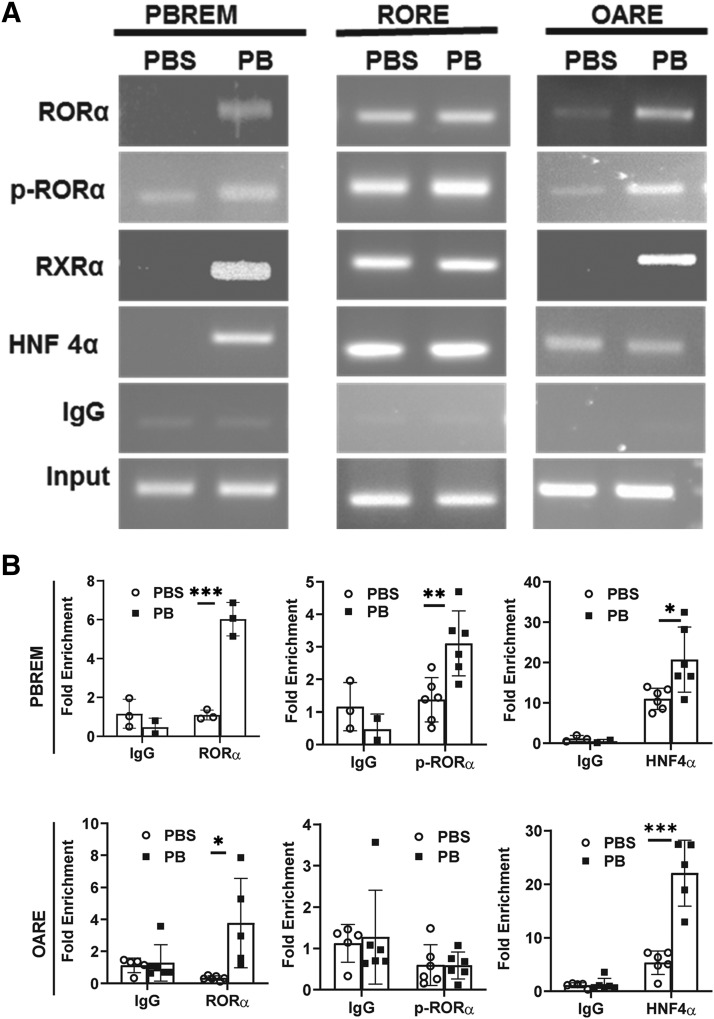
Having established that CAR and RORα coactivate the CYP2B6 promoter and also knowing that the three motifs, PBREM, 2B6-RORE, and OARE, are critical for the activation of the CYP2B6 gene promoter, we were interested to examine the dynamics of CAR, RORα, and HNF4α interactions with the aforementioned DNA motifs on the CYP2B6 promoter in untreated and PB-treated human primary hepatocyte cultures. For this purpose, we performed ChIP assays using anti-RXRα, anti-RORα, anti–p-Ser100-RORα, and anti-HNF4α antibodies. As reported in the previous studies, anti-RXRα, antibody against a CAR partner, was used here as an indicator of CAR binding to PBREM (Saito et al., 2013; Hori et al., 2016). All of the evaluated antibodies immunoprecipitated 2B6-RORE chromatin region in both untreated and treated conditions. In contrast, all of the antibodies precipitated more PBREM and OARE chromatin region in PB-treated than nontreated human primary hepatocytes (Fig. 3A). We also showed quantitative ChIP results for PBREM and OARE in Fig. 3B, and the results suggested essentially the same with Fig. 3A, excepting the marginal increase of p-Ser100 RORα–precipitated band for OARE. Nonetheless, substantial amount of p-Ser100 RORα bound on PBREM, RORE, and OARE in PB-treated cells. These observations suggest that p-Ser100 RORα exists on 2B6-RORE, and after PB treatment, drastic DNA-protein interaction changes were introduced in the CYP2B6 promoter.
Fig. 3.
Phenobarbital induced p-Ser100 RORα onto the PBREM and OARE motifs in human primary hepatocytes. (A) ChIP-PCR assays were performed in human primary hepatocytes using antibodies against RORα, p-Ser100 RORα, HNF4α, and RXRα to find interaction of these factors with PBREM, 2B6-RORE, and OARE. Though both RORα and p-Ser100-RORα were detected at RORE before and after PB treatment, PB initiated increase of DNA bands corresponding to RORα and p-RORα interaction with PBREM and OARE. (B) The same set of antibodies and PCR primers was used for quantitative ChIP-PCR analysis for OARE and PBREM. The asterisk (*) between the groups represents statistically significant difference (*P < 0.05; **P < 0.01, ***P < 0.001). Mean and S.D. are shown for each transfected group (n = 6).
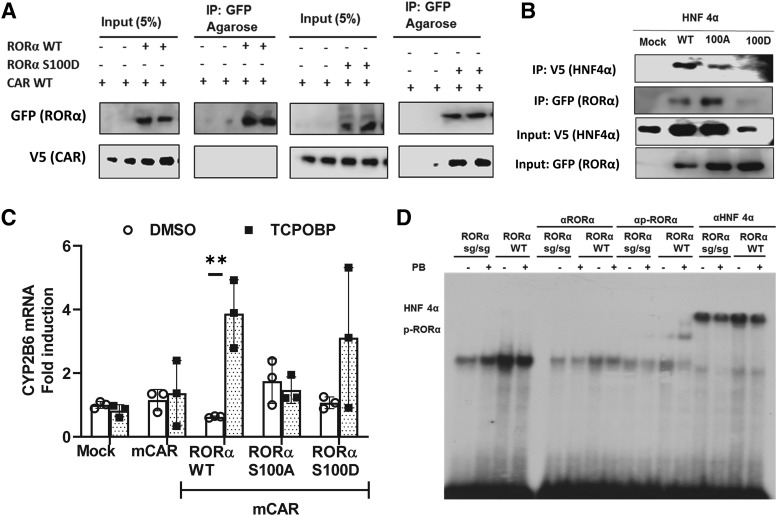
To find roles of RORα phosphorylation at Ser100 in the molecular interaction between RORα with CAR or HNF4α, we performed co-IP assay as shown in Fig. 4, A and B. We used phosphomimetic mutant RORα with Asp100 instead of Ser (RORα S100D) in these co-IP experiments. CAR preferably interacted with RORα S100D in Huh7 cells transiently expressing CAR and RORα wild type (WT) or RORα S100D (Fig. 4A). Unexpectedly, coexpression of RORα mutants with HNF4α in Fig. 4B suggested RORα S100D destabilized HNF4α protein. Although the mechanism for this HNF4α destabilization was not clear at this point, this suggested some molecular communication between these factors existed. Even with consideration of different amount of HNF4α in the input samples, HNF4α and RORα interaction was clearly shown in this co-IP assay (Fig. 4B). In HepG2 cells that were treated with TCPOBOP, which is a mouse CAR (mCAR)-specific ligand activator, quantitative mRNAs (real-time–quantitative PCR) analysis revealed that CYP2B6 mRNAs were induced in cells transiently expressing mCAR and RORα WT or RORα S00D but not RORα S100A, suggesting that p-Ser100 RORα was crucial for the CAR-RORα synergy (Fig. 4C).
Fig. 4.
p-Ser100 RORα functional interaction with CAR and HNF4α. (A) RORα S100D mutant interacted with CAR. Coimmunoprecipitation assay was performed in Huh7 cells transiently expressing RORα WT or its phosphomimetic mutant, RORα S100D, in the presence or absence of CAR. CAR preferably interacted with RORα S100D. (B) HNF4α protein was destabilized by coexpression with RORα S100D. RORα WT, RORα S100A, or RORα S100D were cotransfected into Huh7 cells for a co-IP assay. (C) CYP2B6 gene coactivation by CAR and RORα S100D mutant. Expression plasmids of mCAR were transfected into HepG2 cells in the presence of RORα WT, RORα S100A, or RORα S100D, and CYP2B6 mRNA expression was measured. The asterisk (*) between the groups represents statistically significant difference (**P < 0.01). Mean and S.D. are shown for each group (n = 3). (D) HNF4α and p-Ser100 RORα binding to OARE. Gel shift analysis was performed as in Materials and Methods section using DR1 radioactive probe shown in Table 1 and mouse liver nuclear extracts. Stronger binding was observed with nuclear extracts from RORα WT mice liver than with RORα sg/sg, which lacks one exon in lateral organ boundaries domain of RORα gene. Though anti-RORα antibody blocked the gel shift, phospho-specific anti-Ser100 RORα antibody produced a super shift with only WT PB-treated mice liver nuclear extracts. Anti-HNF4α antibody supershifted the bands.
To find S100-phosphorylated RORα in 2B6-RORE binding-protein complex, we performed EMSA assay as shown in Fig. 4D. A radioactive 2B6-OARE was incubated with nuclear extracts prepared from PBS- or PB-treated RORα WT or RORα staggerer mice (mutant mice with one exon deletion that codes a part of ligand-biding domain of RORα) liver as described in the Materials and Methods section. A single shifted band was observed, and the band intensity was stronger with the nuclear extracts from RORα WT than with the nuclear extracts from RORα staggerer mice livers. Anti-HNF4α antibody supershifted the complex, and anti-RORα blocked the formation of the complex, suggesting the multiple components, including RORα and HNF4α, existed in the shifted bands. Interestingly, anti–p-Ser100 RORα supershifted the complex formed with nuclear extracts from RORα WT mice livers, suggesting that p-Ser100 RORα was also a member of the complex, and phosphorylation of this site increased by PB (Fig. 4D).
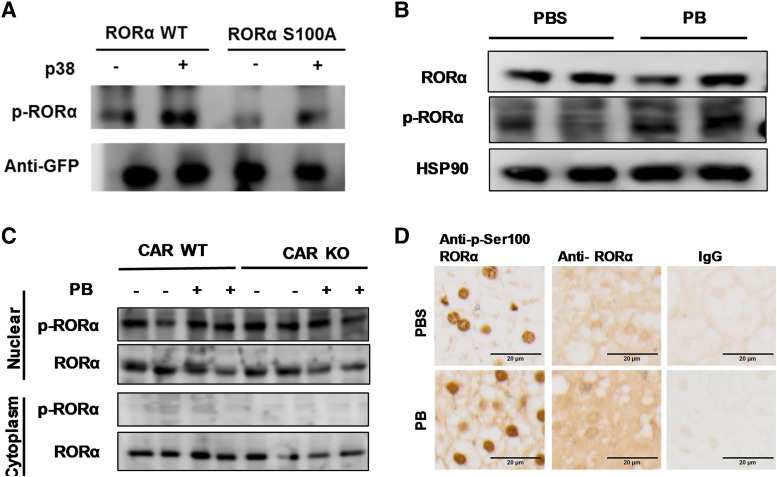
Since the data suggested that p-Ser100 RORα was an essential component of PB induction of the CYP2B6 gene in human primary hepatocytes, we aimed to confirm expression as well as the subcellular localization of p-Ser100 RORα in liver cells. First, we analyzed phosphorylation of RORα with P38 mitogen-activated protein (MAP) kinase in HepG2 cells ectopically expressing the kinase and WT RORα or RORα S100A. The results in Fig. 5A indicated protein band reacting with anti–p-Ser 100 RORα antibody clearly increased when WT RORα was cotransfected in HepG2 cells. Slight increase of the bands with same migration in RORα S100A–transfected HepG2 may represent endogenous RORα in HepG2 extracts. Thus, this Western blot analysis of the cell lysates suggested that RORα S100 residue was phosphorylated by p38 (Fig. 5A).
Fig. 5.
p-Ser100 RORα exists in nucleus in mouse liver. (A) RORα phosphorylation by p38 MAP kinase in HepG2 cell. HepG2 cells were transfected with RORα WT or RORα S100A with p38 MAP kinase expression vectors. Western blot assay was employed to determine phosphorylation of RORα S100 residue. (B) Expression of RORα and p-Ser100 RORα in human primary hepatocytes. The cells were treated with PBS or PB for 24 hours and the Western blot assay was determined in whole-cell lysate by anti-RORα or p-Ser100 RORα antibodies. HSP90, heat shock protein 90. (C and D) CAR WT and CAR KO mice treated with PB for 24 hours, and livers were collected for Western blot (C) or immunohistochemic staining (D) with anti-RORα or anti–p-Ser-RORα antibodies. Both immunohistochemic staining and Western blot assay findings suggested that though RORα was expressed both in the cytoplasm and nuclear fractions of the mouse liver, p-Ser100 RORα expression was limited in the nucleus.
In human primary hepatocyte whole-cell lysate, Western blot analysis revealed that RORα and p-Ser100 RORα were expressed (Fig. 5B). Among multiple bands recognized with anti–p-Ser100 RORα antibody, the band with strongest intensity indicated by the arrow showed exactly the same migration with the band recognized by anti-RORα antibody in the Fig. 5B upper panel. Consistent with the ChIP assay in Fig 3, PB treatment did not change the p-Ser RORα expression in these cells. Next, we analyzed Ser100-phosphorylated RORα subcellular localization in mouse liver (Fig. 5, C and D). We performed Western blotting analysis with nuclear and cytoplasmic extracts (Fig. 5C) and immunohistochemistry analysis of liver sections (Fig. 5D). Ser100-phosphorylated RORα was clearly observed in mouse liver cells, and it was localized within the nucleus per both Western blot and immunohistochemic staining data.
PB Induced Change in CYP2B6 Chromatin Conformation in Human Primary Hepatocytes.
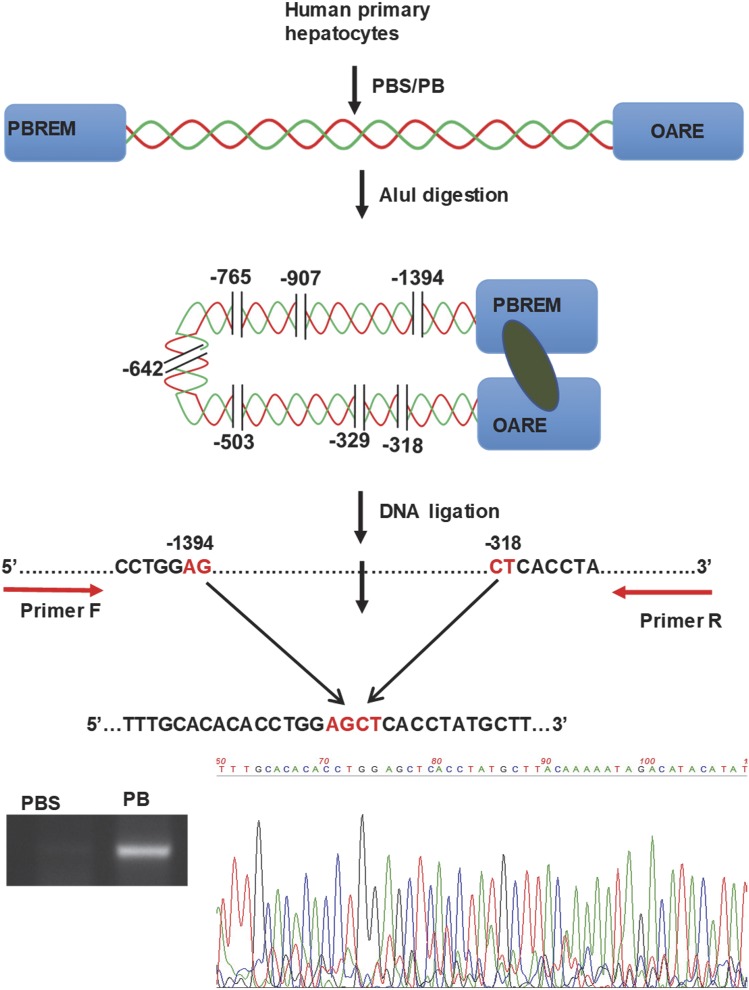
We studied chromatin conformation changes within −1.8 kb CYP2B6 promoter using 3C assay in human primary hepatocytes treated with PBS or 1 mM PB (Fig. 6). Restriction enzyme AluI has seven different restriction sites between the PBREM and OARE motifs within the proximal −1.8 kb of the CYP2B6 promoter. As described in the Materials and Methods section above, after AluI digestion and subsequent ligation, the DNAs were amplified using PCR primers designed to amplify DNA fragments between −1495 and −195 in such a way that without digestion the size of the amplicon would be 1.3 kb and with AluI digestion without ligation there would be no amplicon. However, we detected an amplicon size of about 210 bp in the PB-treated but not PBS-treated primary hepatocytes, and the identity of the amplicon was confirmed by sequencing as shown in Fig. 6. This suggested that PB induced a substantial change in the CYP2B6 chromatin conformation that enabled two DNA fragments consisting of PBREM and OARE, respectively, to coexist in the protein-DNA complex and efficiently ligate the two fragments.
Fig. 6.
Phenobarbital induced change in CYP2B6 chromatin conformations in human primary hepatocytes. 3C assay was performed using a restriction enzyme AluI and forward and reverse primers starting at −1495 and −195 upstream CYP2B6 transcription site, respectively. After digestion and ligation, the primers amplified a 207-bp DNA fragment only in PB-treated samples, as confirmed by agarose gel electrophoresis and subsequent sequencing.
Discussion
We found RORα siRNAs suppressed expression of CYP2B6 mRNA in human primary hepatocyte cultures. Consistent with this observation, RORα activated a −1.8 kb CYP2B6 promoter in a cell-based reporter assay with COS-1 cells. Next, we identified a RORα-responsive DNA element located between −668 and −645 bp upstream of the CYP2B6 transcription start site and named it 2B6-RORE. The assay also revealed that though p-Ser 100-RORα binding to 2B6-RORE is constitutive and no change was observed after PB treatment in these cells, this protein was enriched on the PBREM after PB treatment. Similarly, both HNF4α and CAR were enriched on the PBREM motifs after PB treatment. Although we have no anti-CAR antibody working for ChIP analysis, we consider that RXRα ChIP results in Fig. 3 suggested CAR binding on PBREM, since previous results showed PB treatment of HPH translocate cytoplasmic localized CAR into nucleus (Li et al., 2009), and CAR-RXRα heterodimer strongly binds to the NR1 sequence in PBREM (Sueyoshi et al., 1999). Although it seems unlikely, the possibility of other RXRα partner involved in the PBREM ChIP complex remains. Consistent with the ChIP results, our 3C assay indicated that chromatin containing the CYP2B6 promoter went through dynamic structural modulation by PB treatment, such that both the PBREM and OARE elements existed in the same complex, as evidenced by efficient ligation that occurred between the DNA fragments including these elements. Taking all these observations together, it is inevitable to postulate that through binding their respective response elements, CAR, RORα, and HNF4α comediate the induction of CYP2B6 gene expression in human primary hepatocytes (Fig. 7).
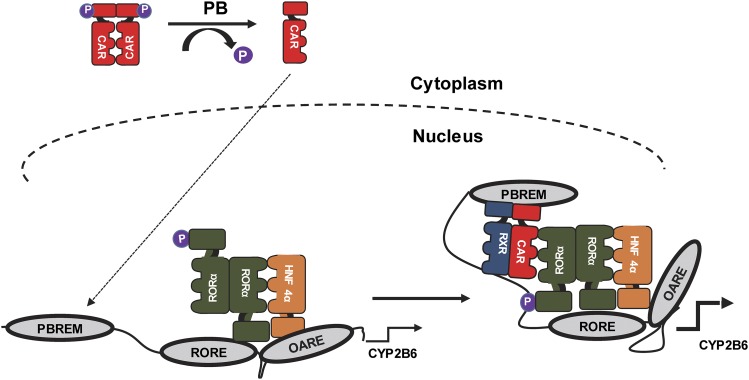
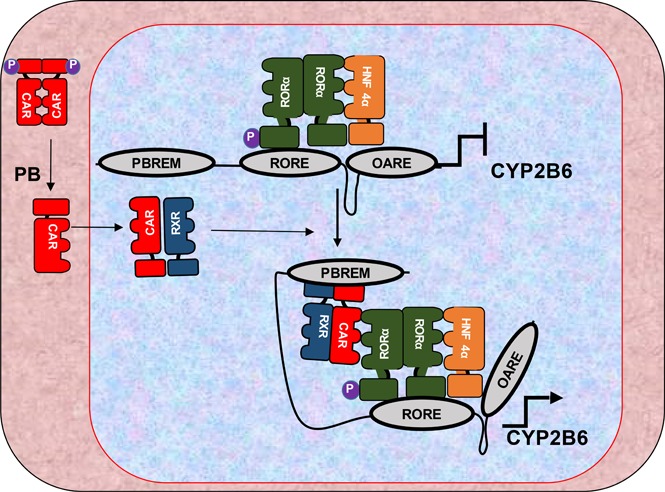
Fig. 7.
Schematic representation of molecular mechanism of phenobarbital triggered induction of CYP2B6 in human primary hepatocytes. At noninduced condition, RORα and p-Ser100 RORα occupied the 2B6-RORE motif while HNF4α occupied the OARE, and none of the tested factors were detected on the PBREM. In response to PB treatment, the dynamics of the protein-DNA and protein-protein interactions on the CYP2B6 promoter were changed as a result of CAR recruitment onto the PBREM. As previously reported (Mutoh et al., 2009, 2013), dephosphorylated CAR was translocated into the nucleus and commenced the PB induction process. Then p-Ser100 RORα facilitated the interaction between the factors on the PBREM, RORE, and OARE, thereby enabling the formation of transcriptionally active chromatin complex.
In this hypothetical model (Fig. 7), though RORα and p-Ser100 RORα occupied the 2B6-RORE, HNF4α bound to the OARE in untreated cells. Moreover, since HNF4α was detected on the 2B6-RORE in untreated cells and there is no HNF4α direct binding site in 2B6-RORE, there appears to be an interaction between the RORα on the 2B6-RORE and HNF4α on OARE. This interaction was essential for the modulation of the basal expression of the gene, which was confirmed by the suppression of CYP2B6 mRNAs in RORα-silenced primary hepatocytes. On the other hand, on the basis of our 3C assay and series of cell-based reporter analyses, it was remarkably evident that PB induced changes in CYP2B6 chromatin conformation. Thus, it is plausible to suggest that PB triggers this chromatin remodeling by inducing activation of nuclear receptor CAR through dephosphorylation of its Thr38 residue (Mutoh et al., 2009, 2013), CAR binding to the PBREM motif (Sueyoshi et al., 1999), and its interactions with RORα and HNF4α. For the interactions, p-Ser100 RORα on the 2B6-RORE was vital, which was indicated by the co-IP assay that CAR preferably interacted with RORα S100D mutant (Fig. 4A). In ChIP analysis shown in Fig. 3, we observed RXRα on the 2B6-RORE in untreated and PB-treated hepatocytes. It is not clear what form of RXRα-containing heterodimer exists on 2B6-RORE at this stage of research, and future analysis on the RXRα complex will be necessary.
In our earlier work using CAR WT/knockout (KO) and RORα WT/RORαsg/sg mice models, we reported that though PB induced the mSult1e1 gene in CAR WT or RORα WT mice, it did not induce the gene in either CAR KO or RORαsg/sg mice liver (Fashe et al., 2018). Mechanistically, in untreated mouse liver, RORα directly bound to its RORE on mSult1e1 and suppressed gene expression. In PB-treated mouse liver cells, however, RORα was phosphorylated at its S100 residue, which limited its DNA-binding property. Interestingly, this phenomenon facilitated the CAR binding in PB-treated mouse liver cells as a DR4 unit m1E1-PBREM element and RORα binding site through m1E1-RORE shared half-site, and S100 phosphorylated RORα interaction with CAR as coactivator (Fashe et al., 2018). In this study, we observed a statistically significant change in S100-phosphorylated RORα enrichment on PBREM but not on OARE or 2B6-RORE following PB treatment. Thus, it seemed that PB did not affect the S100 RORα phosphorylation on 2B6-RORE, but it induced change in CYP2B6 chromatin conformation through CAR and p-Ser100 RORα interaction, which in turn resulted in an increased p-RORα enrichment on the PBREM in HPH treated with PB. Kinase(s) involved in the phosphorylation of RORα in CYP2B6 gene regulation is not known at this point. We observed ectopically expressed P38 MAP kinase phosphorylated Ser100 of RORα in this report (Fig. 5A), and protein kinase C phosphorylated the Ser in previous report (Fashe et al., 2018). A recent publication (Hori et al., 2016) by our group found phosphorylated P38 MAP kinase interacted with CAR in nucleus after CAR was activated and translocated into nucleus. P38 MAP kinase–dependent phosphorylation of CAR subsequently causes inactivation of this receptor. Similarly, CAR-P38 MAP kinase complex may affect RORα phosphorylation status after PB activation. However, our ChIP analysis in Fig. 4 and Western blotting in Fig. 5 were not able to detect RORα phosphorylation increases by PB treatment. Furthermore, we could not find any evidence CAR-P38 MAP kinase interaction affects RORα phosphorylation observed in Fig. 5A (unpublished data). Thus, physiologic relevance of our finding in Fig. 5A for CYP2B6 gene regulation may not be clear enough at this point, and future analysis will be necessary for establishing P38 MAP kinase roles in RORα Ser100 phosphorylation.
RORα is known to be a key element for circadian gene regulation, in which competition between RORs and REV-ERBs for the shared response element RORE in circadian gene brain and muscle aryl hydrocarbon receptor nuclear translocator–like 1 (Bmal1) plays pivotal roles (Guillaumond et al., 2005; Solt et al., 2011). Although little is known about CYP2B6 circadian activity changes, hepatic clearance of nevirapine, which is largely metabolized by CYP2B6 (Ward et al., 2003), was reportedly higher during daytime in children (Bienczak et al., 2017). Moreover, in rats treated with PB, the magnitude of CYP2B1/2 mRNA induction showed daytime dependence (Kanno et al., 2004), and it has been long known that circadian clock regulates the expression of CYP enzymes (Froy, 2009). Thus, it is conceivable that the RORα regulation of CYP2B6 can be associated with the differential expression CYP2B6 across the biologic clock and how p-Ser100 RORα involved in this regulation will be interesting research target.
In the series of recent publication by our group, numerous biologic processes were shown to be regulated by the phosphorylation of highly conserved phosphorylation site in DNA-binding domain of these receptors (Mutoh et al., 2009, 2013; Shindo et al., 2012, 2013; Hashiguchi et al., 2016; Negishi, 2017; Sueyoshi et al., 2019). These phosphorylation sites are located between two zinc finger domain and conserved in 41 out of 48 human nuclear receptors. First, we observed CAR Thr38 dephosphorylation by PB through the chemical’s inhibitory effect on epidermal growth factor signaling pathway, and the receptor was translocated into nucleus for activation of target genes (Mutoh et al., 2009, 2013). This phosphorylation was later shown to regulate the intramolecular interaction between DNA-binding domain and ligand binding domain of CAR, causing CAR dimer interphase structure and regulation of active monomer-inactive dimer conversion of the receptor (Shizu et al., 2017, 2018). We also found ERα S212 phosphorylation did not affect its activity as transcription factor, and phospho-mimetic mutant of ERα activated distinct set of genes compared with S212A ERα (Shindo et al., 2012). Ser212-phosphorylated ERα was found in peripheral blood neutrophils, and the cells were found to migrate into the mouse uterus (Shindo et al., 2013). Furthermore, FXR Ser154 phosphorylation made this receptor inactive, and this phosphorylated FXR in the nucleus of centrilobular hepatocytes in FXR ligand treated mice (Hashiguchi et al., 2016). Most recently, RXRα Thr167 phosphorylation was found in fasting mouse white adipose tissue, and genes for key enzymes of energy metabolism in the tissue were differentially regulated in the mutant mice with RXRα Thr167 mutated to Ala (Sueyoshi et al., 2019).
Here, we found ROR Ser100 phosphorylation plays key role in CYP2B6 gene induction, and this represents another example of the roles of conserved phosphorylation sites in regulation of nuclear receptor function. Situated on the newly identified RORE, termed here as 2B6-RORE, positioned between the PBREM and OARE motifs of the CYP2B6 promoter, p-Ser RORα facilitated the looping CAR-RXRα onto the proximal CYP2B6 promoter to initiate the assembly of the transcription machinery on the CYP2B6 promoter prior to the induction of gene transcription. Given the diverse functions and near-ubiquitous expression of RORα, numerous biologic processes may be regulated by this RORα phosphorylation. This report will be a prologue for the future findings of such biologic functions regulated by the RORα Ser 100 phosphorylation.
Acknowledgments
The authors thank for protein expression core the laboratory and histology support group of NIEHS.
Abbreviations
- 2B6
CYP2B6
- 3C
chromatin conformation capture
- CAR
constitutive androstane receptor
- ChIP
chromatin IP
- CI
confidence interval
- DR1
direct repeat protein-binding motif separated by a single nucleotide
- EMSA
electro mobility shift assay
- ERα
estrogen receptor α
- FXR
farnesoid X receptor
- hCAR
human CAR
- HNF4α
hepatocyte nuclear factor 4 alpha
- HPH
human primary hepatocyte
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- KO
knockout
- MAP
mitogen-activated protein
- mCAR
mouse CAR
- mSult
mouse Sult
- NR
nuclear receptor
- OARE
okadaic acid response element
- p-
phosphorylated
- PB
phenobarbital
- PBREM
PB response element module
- PCR
polymerase chain reaction
- RORα
retinoid-related orphan receptor alpha
- RORE
RORα response element
- RXRα
retinoid X receptor α
- siRNA
small interfering RNA
- Sult
sulfotransferase
- TCPOBOP
1,4-Bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-Tetrachloro-1,4-bis(pyridyloxy)benzene
- WT
wild type
Authorship Contributions
Participated in research design: Fashe, Hashiguchi, Negishi, Sueyoshi.
Conducted experiments: Fashe, Hashiguchi, Sueyoshi.
Performed data analysis: Fashe, Negishi, Sueyoshi.
Wrote or contributed to the writing of the manuscript: Fashe, Sueyoshi.
Footnotes
This work was supported by National Institutes of Health intramural research program Z01ES1005-01.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Bienczak A, Cook A, Wiesner L, Mulenga V, Kityo C, Kekitiinwa A, Walker AS, Owen A, Gibb DM, Burger D, et al. (2017) Effect of diurnal variation, CYP2B6 genotype and age on the pharmacokinetics of nevirapine in African children. J Antimicrob Chemother 72:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Coulter S, Jetten AM, Goldstein JA. (2009) Identification of human CYP2C8 as a retinoid-related orphan nuclear receptor target gene. J Pharmacol Exp Ther 329:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, Flockhart DA, Zanger UM. (2007) Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8:547–558. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. (2007) The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol 21:2099–2111. [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Shebley M, Palamanda J, Sinz MW, Ramsden D, Einolf HJ, Chen L, Wang H. (2016) Evaluation of CYP2B6 induction and prediction of clinical drug-drug interactions: considerations from the IQ consortium induction working group-an industry perspective. Drug Metab Dispos 44:1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fashe M, Hashiguchi T, Yi M, Moore R, Negishi M. (2018) Phenobarbital-induced phosphorylation converts nuclear receptor RORα from a repressor to an activator of the estrogen sulfotransferase gene Sult1e1 in mouse livers. FEBS Lett 592:2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. (2009) Cytochrome P450 and the biological clock in mammals. Curr Drug Metab 10:104–115. [DOI] [PubMed] [Google Scholar]
- Giguère V, McBroom LD, Flock G. (1995) Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol 15:2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumond F, Dardente H, Giguère V, Cermakian N. (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20:391–403. [DOI] [PubMed] [Google Scholar]
- Hashiguchi T, Arakawa S, Takahashi S, Gonzalez FJ, Sueyoshi T, Negishi M. (2016) Phosphorylation of farnesoid X receptor at serine 154 links ligand activation with degradation. Mol Endocrinol 30:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich WD, Hassan HE, Wang H. (2016) Insights into CYP2B6-mediated drug-drug interactions. Acta Pharm Sin B 6:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Moore R, Negishi M. (2016) p38 MAP kinase links CAR activation and inactivation in the nucleus via phosphorylation at threonine 38. Drug Metab Dispos 44:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Negishi M. (2008) Nuclear receptor CAR requires early growth response 1 to activate the human cytochrome P450 2B6 gene. J Biol Chem 283:10425–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Negishi M. (2009) Early growth response 1 loops the CYP2B6 promoter for synergistic activation by the distal and proximal nuclear receptors CAR and HNF4alpha. FEBS Lett 583:2126–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Kurebayashi S, Ueda E. (2001) The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol 69:205–247. [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. (2007) Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics 31:281–294. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Otsuka S, Hiromasa T, Nakahama T, Inouye Y. (2004) Diurnal difference in CAR mRNA expression. Nucl Recept 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen T, Cottrell J, Wang H. (2009) Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): a novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab Dispos 37:1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. (2009) Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3). J Biol Chem 284:34785–34792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, Negishi M. (2013) Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci Signal 6:ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M. (2017) Phenobarbital meets phosphorylation of nuclear receptors. Drug Metab Dispos 45:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscarson M, Zanger UM, Rifki OF, Klein K, Eichelbaum M, Meyer UA. (2006) Transcriptional profiling of genes induced in the livers of patients treated with carbamazepine. Clin Pharmacol Ther 80:440–456. [DOI] [PubMed] [Google Scholar]
- Ou Z, Shi X, Gilroy RK, Kirisci L, Romkes M, Lynch C, Wang H, Xu M, Jiang M, Ren S, et al. (2013) Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORα and RORγ and its potential relevance to human liver diseases. Mol Endocrinol 27:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SM, Maldarelli F, Natarajan V, Formentini E, Alfaro RM, Penzak SR. (2008) Efavirenz induces CYP2B6-mediated hydroxylation of bupropion in healthy subjects. J Acquir Immune Defic Syndr 49 (5):513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Moore R, Negishi M. (2013) Nuclear receptor CAR specifically activates the two-pore K+ channel Kcnk1 gene in male mouse livers, which attenuates phenobarbital-induced hepatic hyperplasia. Toxicol Sci 132 (1):151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo S, Moore R, Flake G, Negishi M. (2013) Serine 216 phosphorylation of estrogen receptor α in neutrophils: migration and infiltration into the mouse uterus. PLoS One 8:e84462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo S, Sakuma T, Negishi M, Squires J. (2012) Phosphorylation of serine 212 confers novel activity to human estrogen receptor α. Steroids 77:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R, Min J, Sobhany M, Pedersen LC, Mutoh S, Negishi M. (2018) Interaction of the phosphorylated DNA-binding domain in nuclear receptor CAR with its ligand-binding domain regulates CAR activation. J Biol Chem 293:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R, Osabe M, Perera L, Moore R, Sueyoshi T, Negishi M. (2017) Phosphorylated nuclear receptor CAR forms a homodimer to repress its constitutive activity for ligand activation. Mol Cell Biol 37:e00649–e00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson US, Jansson B, Hai TN, Huong DX, Tybring G, Ashton M. (2003) Artemisinin autoinduction is caused by involvement of cytochrome P450 2B6 but not 2C9. Clin Pharmacol Ther 74:32–43. [DOI] [PubMed] [Google Scholar]
- Solt LA, Kojetin DJ, Burris TP. (2011) The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem 3:623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Sakuma T, Shindo S, Fashe M, Kanayama T, Ray M, Moore R, Negishi M. (2019) A phosphorylation-deficient mutant of retinoid X receptor α at Thr 167 alters fasting response and energy metabolism in mice. Lab Invest 99:1470–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM. (2007) Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol 21:1297–1311. [DOI] [PubMed] [Google Scholar]
- Swales K, Kakizaki S, Yamamoto Y, Inoue K, Kobayashi K, Negishi M. (2005) Novel CAR-mediated mechanism for synergistic activation of two distinct elements within the human cytochrome P450 2B6 gene in HepG2 cells. J Biol Chem 280:3458–3466. [DOI] [PubMed] [Google Scholar]
- Wada T, Kang HS, Angers M, Gong H, Bhatia S, Khadem S, Ren S, Ellis E, Strom SC, Jetten AM, et al. (2008) Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3). Mol Pharmacol 73:891–899. [DOI] [PubMed] [Google Scholar]
- Wang D, Li L, Yang H, Ferguson SS, Baer MR, Gartenhaus RB, Wang H. (2013) The constitutive androstane receptor is a novel therapeutic target facilitating cyclophosphamide-based treatment of hematopoietic malignancies. Blood 121:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. (2004) Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem 279:29295–29301. [DOI] [PubMed] [Google Scholar]
- Wang H, Negishi M. (2003) Transcriptional regulation of cytochrome p450 2B genes by nuclear receptors. Curr Drug Metab 4:515–525. [DOI] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K. (2013) Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang YD, Zhao P, Huang SM. (2009) Predicting drug-drug interactions: an FDA perspective. AAPS J 11:300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]