Abstract
The basal ganglia and limbic system, particularly the thalamus, putamen, internal and external globus pallidus, substantia nigra, and sub-thalamic nucleus, comprise a clinically relevant signal network for Parkinson’s disease. In order to manually trace these structures, a combination of high-resolution and specialized sequences at 7T are used, but it is not feasible to routinely scan clinical patients in those scanners. Targeted imaging sequences at 3T have been presented to enhance contrast in a select group of these structures. In this work, we show that a series of atlases generated at 7T can be used to accurately segment these structures at 3T using a combination of standard and optimized imaging sequences, though no one approach provided the best result across all structures. In the thalamus and putamen, a median Dice Similarity Coefficient (DSC) over 0.88 and a mean surface distance less than 1.0mm were achieved using a combination of T1 and an optimized inversion recovery imaging sequences. In the internal and external globus pallidus a DSC over 0.75 and a mean surface distance less than 1.2mm were achieved using a combination of T1 and inversion recovery imaging sequences. In the substantia nigra and subthalamic nucleus a DSC of over 0.6 and a mean surface distance of less than 1.0mm were achieved using the inversion recovery imaging sequence. On average, using T1 and optimized inversion recovery together significantly improved segmentation results than over individual modality (p<0.05 Wilcoxon sign-rank test).
Keywords: subcortical grey matter, segmentation, multi-atlas, multi-modal
1. Introduction
The subcortical grey matter is a collection of nuclei situated near the forebrain [1]. These nuclei are primarily involved in connecting distinct portions of the brain, to serve as major functional systems within the brain [1]. For instance, the globus pallidus internal receives GABAergic signaling from the putamen and relays that to the sub-thalamic nucleus. Many subcortical structures have been implicated in one or more diseases [2]. In addiction, dopamine is dysregulated in the putamen and adjacent structures causing dependence phenotypes [3]. In Parkinson’s disease, several subcortical structures undergo Lewy body growth and that growth plays a significant role in the motor phenotypes associated with the disease [4].
Recently, specialized imaging sequences have been developed for studying subcortical grey matter using clinical magnetic resonance (MR) scanners. In particular, the Fast Grey Matter Acquisition T1 Inversion Recovery (FGATIR) sequence was developed to improve subcortical grey matter contrast with the surrounding tissue [5]. The FGATIR sequence uses a longer inversion time than standard T1-weighted approaches, such as Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE), to null the white matter and accentuate the deep brain structures. FGATIR images accentuate the sub-thalamic nucleus, lamina separating the internal and external globus pallidus, and the thalamus amongst other important structures. On the other hand, many important subcortical structures are still difficult to parcellate using the FGATIR [6]. Higher field strength scanners are needed, but these scans at higher field strengths are not clinically feasible in most contexts [7].
In this work, we investigate the efficacy of sequences acquirable in a clinically tolerable setting, namely standard T1-weighted MPRAGE scans and T1-weighted FGATIR scans acquired at 3T. The structures considered in this work are the substantia nigra (SN), subthalamic nucleus (STN), internal globus pallidus (GPI), external globus pallidus (GPE), putamen, and thalamus. These subcortical structures were manually delineated using a combination of scans acquired at 3T and 7T. Furthermore, we compare segmentation using only one modality, either the MPRAGE or FGATIR, to multi-modal segmentation using the enhanced and complimentary contrast patterns present to improve the overall segmentation results. The primary aspect of novelty of this work is the investigation of multi-modal MRI in the context of subcortical segmentation.
2. Methods
We propose a multi-atlas segmentation algorithm for automated segmentation of the subcortical grey matter. This approach uses multi-modal atlases derived using imaging acquired at 3T and 7T. The segmentation uses the imaging sequences acquired at 3T to assess the effectiveness of segmenting subcortical structures using clinically feasible acquisitions.
2.1. Atlas Imaging
Nine healthy subjects were scanned at 3T and 7T. At 7T, a series of 0.7mm isotropic T1-weighted MPRAGE (Inversion Time (TI)/ Repetition Time (TR)/Echo Time (TE) =[400,640,960,1120]/4.74/2.09ms) was acquired and a susceptibility weighted image slab through the midbrain acquired at 0.2x0.2x1.1mm was acquired sagitally, coronally, and axially (TR/TE/Flip Angle (FA)=1952/23ms/45° for all orientations). At 3T, a 1.0mm isotropic resolution T1-weighted MP-RAGE (TI/TR/TE=925/8.1/2.7ms) and an FGATIR scan was acquired for additional mid-brain contrast (TI/TR/TE=400/7.39/3.43).
2.2. Manual Segmentation
For each subject, the 7T T1-weighted MP-RAGE with the inversion time of 960ms was used as the reference space. The other 7T MP-RAGE scans, 3T MP-RAGE, 7T high-resolution susceptibility weighted slabs, and 3T FGATIR were co-registered to the reference space. The following structures were manually labeled on the left hemisphere for one subject: GPI, GPE, STN, SN, thalamus, and the putamen. The labeled atlas was then registered to each of the other eight subjects and the labels were deformed to the target space using the Reg Aladin rigid affine transformation algorithm in NiftyReg [8]. The deformed labels were then manually corrected. Finally, each subject was flipped laterally and the flipped image was registered to the standard space image. Each subject’s labels were deformed in the laterally flipped space and the results were manually corrected. The final result was nine subjects with left and right labels for the GPI, GPE, STN, SN, thalamus, and putamen. All manual segmentations were done using CranialVault and the CRAVE Tools [9] (by Pierre F. D’Haese).
2.3. Segmentation Algorithm
First, each subject, the 3T T1-weighted MRI was automatically segmented with the BrainCOLOR protocol [10] (www.neuromorphometrics.com) following a standard multi-atlas whole brain segmentation (WBS) approach [11]. Briefly, the target image was affinely registerd to MNI space. From a population of 45 atlases (45 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) dataset [12] with BrainCOLOR labeling protocol are considered as the 45 atlases), the 15 atlases geodesically most similar to the target are then empirically selected [13]. These 15 atlases are non-rigidly registered to the target image using the Advance Normalization Tools (ANTs) Symmetric Normalization algorithm (SyN) [14]. Finally, the registered atlas images and labels are fused to the target image using Hierarchical Non-Local Spatial STAPLE [15].
Thus, WBS was used to localize the particular regions of interest. In particular, the thalamus label from the WBS was used to localize the thalamus, the globus pallidus and putamen labels from the WBS were used to localize the GPI, GPE, and putamen, and the diencephalon label from the WBS was used to localize the SN and STN. The bounding box of each of these regions of interest was identified and dilated by 5mm. Finally, the labels and T1 and FGATIR intensities were extracted from these bounding boxes and saved as reduced field of view (RFOV) atlases.
For a given target, the target was segmented with the BrainCOLOR protocol. A series of targets (RFOV) were created following the protocol defined above. The RFOV atlases were co-registered to the RFOV targets. All registrations were performed using with ANTs and the SyN algorithm [16]. After registration, joint label fusion (JLF) was used. In all cases, the same collection of imaging modalities was used for the segmentation [17]. Finally, each structure’s segmentation was reinserted into the standard image space. All operations for creating and manipulating using RFOV atlases and images used custom MATLAB (www.mathworks.org) code.
3. Results
Each of the nine healthy subjects was segmented in a leave-one-out cross validation scheme. First, each subject was segmented using the T1-weighted MRI, the FGATIR, and multi-modally with the T1 and FGATIR. Second, each subject’s scans were flipped left-right to produce a second set of atlases. Each subject was then segmented with the 16 atlases, leaving out the atlas and the flipped version of the atlas. As a result, each subject was segmented six times, twice with each combination of modalities. The results are divided into three pieces for ease of visualization: diencephalon (STN and SN), GPI and GPE, and thalamus and putamen. For each segmentation result the Dice Similarity Coefficient (DSC), mean surface distance (MSD), and Hausdorff distance (HD) were calculated.
3.1. Diencephalon
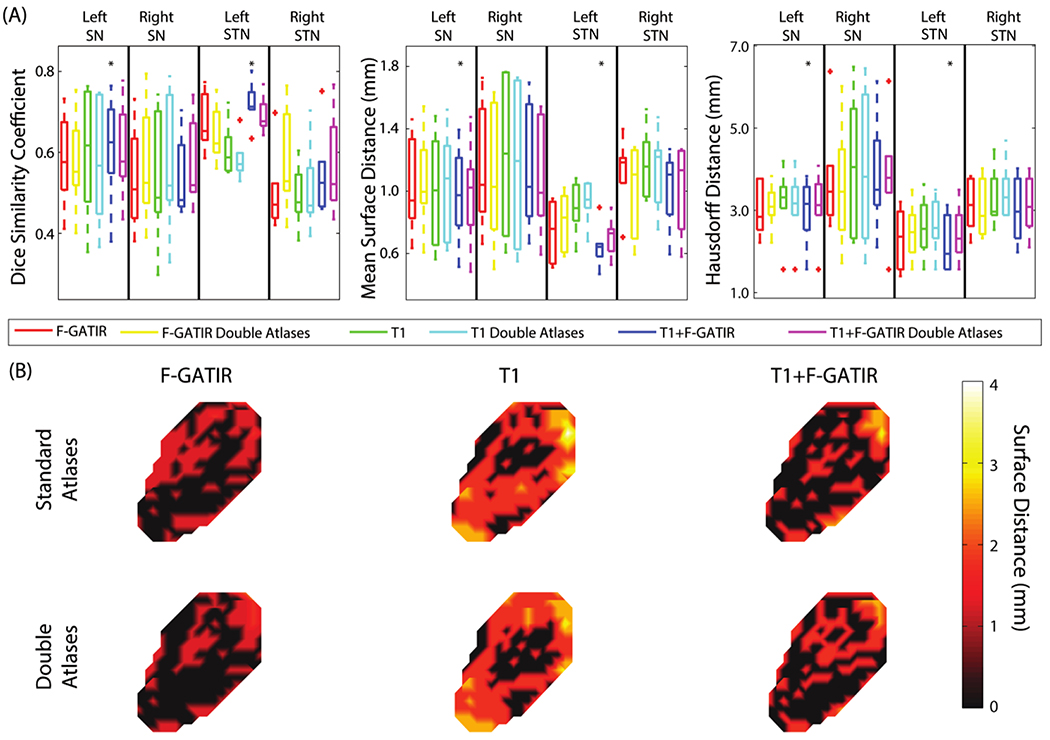
Four structures were segmented in the diencephalon: the left STN, right STN, left SN, and right SN (figure 1). For the left SN, the segmentation with T1 and FGATIR outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.65, median MSD of 0.98 mm, and a median HD of 3.11 mm. For the right SN, no approach significantly outperformed other approaches. For the left STN, segmentation with T1 and FGATIR outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.70, median MSD of 0.61 mm, and a median HD of 2.06 mm. For the right STN, no approach outperformed other approaches (Wilcoxon sign-rank test).
Figure 1:
Segmentation results for structures in the dienchephalon. Quantitative segmentation results are shown in (A). For the left SN, multi-modal segmentation with T1 and FGATIR outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the right SN no segmentation approach outperformed other approaches. For the left STN, multi-modal segmentation with T1 and FGATIR outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the right STN no segmentation approach outperformed other approaches. In (B), surface distances between the true and estimated segmentations for the left SN are shown for the six proposed segmentation approaches.
3.2. Globus Pallidus
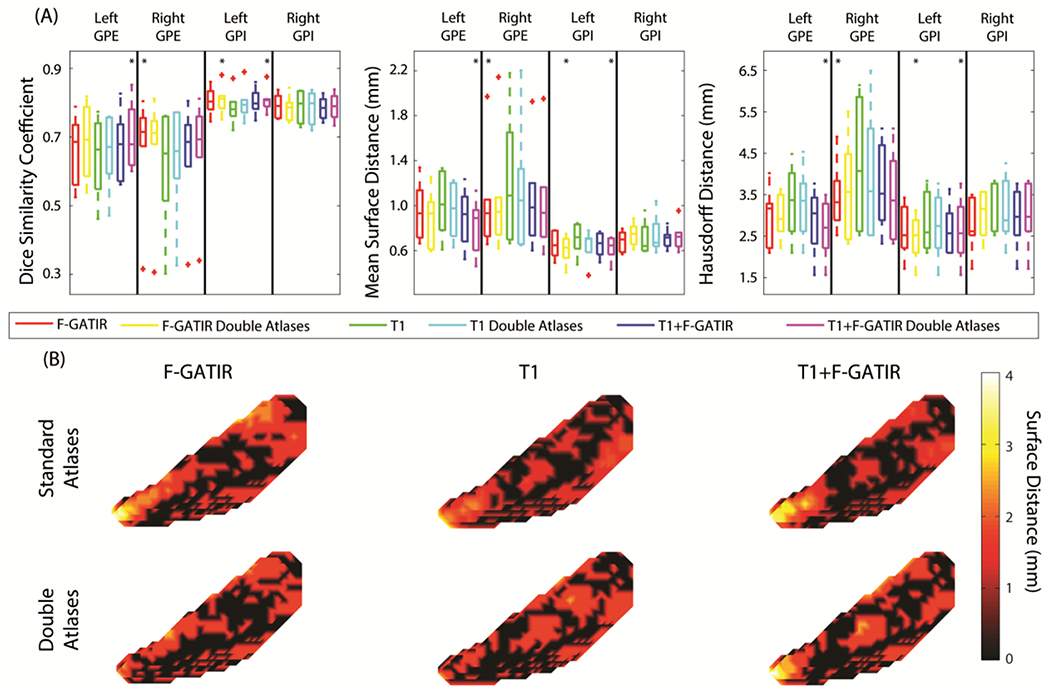
Two structures were segmented in the globus pallidus: the GPI and GPE. These structures were segmented bilaterally and resulted in four total structures segmented (figure 2). For the left GPE, the segmentation with T1 and FGATIR including flipped atlases outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.68, median MSD of 0.94 mm, and a median HD of 2.70 mm. For the right GPE, segmentation with FGATIR outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.71, a median MSD of 0.96 mm, and a median HD of 3.42 mm. For the left GPI, segmentation with FGATIR with flipped atlases and multi-modal segmentation with T1 and FGATIR and flipped atlases outperformed other approaches but were not statistically distinguishable from each other (p<0.05 Wilcoxon sign-rank test), with median DSC values of 0.80 and 0.81, median MSD values of 0.68 and 0.69 mm, and median HD values of 2.50 and 2.52 mm respectively. For the right GPI, no approach significantly outperformed another.
Figure 2:
Segmentation results for structures in the globus pallidus. Quantitative segmentation results are shown in (A). For the left GPE, multi-modal segmentation with T1 and FGATIR with double atlases outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the right GPE segmentation with FGATIR outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the left GPI, multi-modal segmentation with T1 and FGATIR with doubled atlases and segmentation with FGATIR with doubled atlases outperformed other approaches but were not distinguishable amongst each other (*; p<0.05; Wilcoxon sign-rank test). For the right GPI no segmentation approach outperformed other approaches. In (B), surface distances between the true and estimated segmentations for the left GPI are shown for the six proposed segmentation approaches.
3.3. Thalamus and Putamen
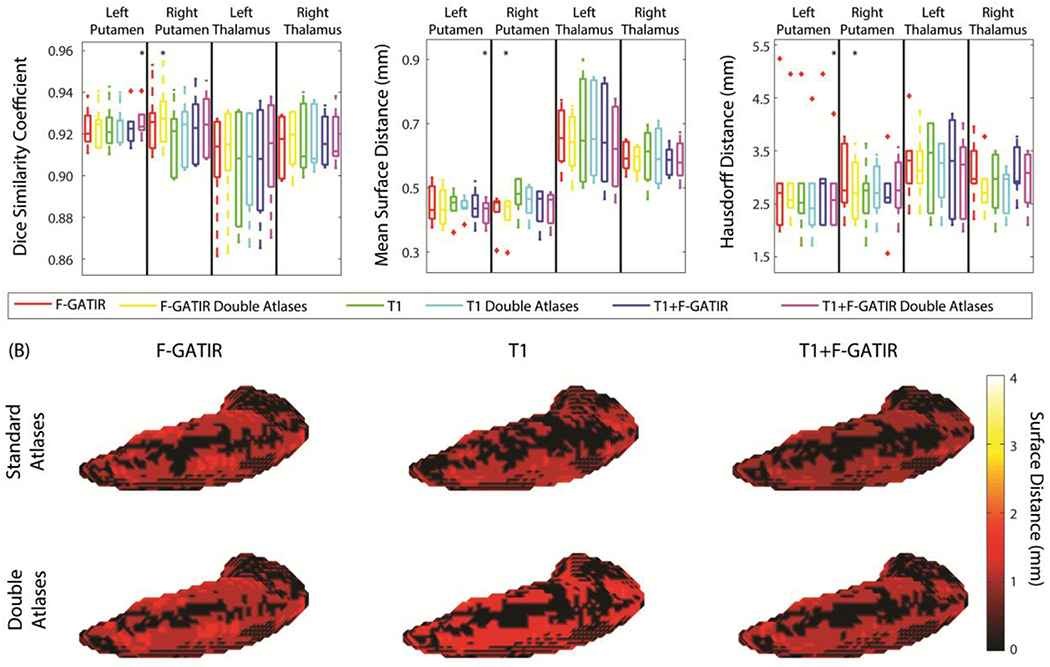
The left and right thalamus and putamen were segmented resulting in four total structures (figure 3). For the left putamen the segmentation with T1 and FGATIR including flipped atlases outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.93, a median MSD of 0.42 mm, and a median HD of 2.50 mm. For the right putamen, segmentation with FGATIR including flipped atlases outperformed other approaches (p<0.05 Wilcoxon sign-rank test) with a median DSC of 0.93, a median MSD of 0.45 mm, and a median HD of 2.61 mm. For the left thalamus, no approach significantly outperformed the others. Finally, for the right thalamus, no approach significantly outperformed the others.
Figure 3:
Segmentation results for the putamen and thalamus. Quantitative segmentation results are shown in (A). For the left putamen, multi-modal segmentation with T1 and FGATIR with double atlases outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the right putamen segmentation with FGATIR with doubled atlases outperformed other approaches (*; p<0.05; Wilcoxon sign-rank test). For the left thalamus, no segmentation approach outperformed other approaches For the right thalamus, no segmentation approach outperformed other approaches. In (B), surface distances between the true and estimated segmentations for the left putamen are shown for the six proposed segmentation approaches.
4. Discussion
In this work, we presented segmentation approaches for segmenting six subcortical structures bilaterally. These segmentation approaches considered the effect of imaging modality on segmentation results. Two distinct imaging modalities were considered. First, a standard T1-weighted MPRAGE, a sequence commonly acquired in clinical and research settings, was acquired for nine subjects. Second, a T1-weighted FGATIR, a specialized sequence with enhanced contrast in subcortical structures, was acquired for the same nine subjects. A series of 7T T1-weighted MPRAGE scans with varying inversion times and high-resolution susceptibility weighted slabs were acquired on the nine subjects. Then, an expert in subcortical anatomy manually delineated the thalamus, putamen, internal and external globus pallidus, sub-thalamic nucleus, and substantia nigra bilaterally.
These nine subjects were then used in a leave-one-out cross-validation to assess the segmentation accuracy using only T1-weighted MPRAGE, only T1-weighted FGATIR, and multi-modally with the MPRAGE and FGATIR. In general, the multi-modal segmentation outperformed the other approaches and furthermore including atlases flipped laterally tended to improve segmentation results, but there was no single approach that outperformed in all cases. Fortunately, the proposed segmentation approach does not require all segmentations to be performed with the same modalities. This allows flexibility in which sequences are used to segment each structure. From a practical perspective, the lack of a globally best method is disappointing as it would seem to necessitate that different methods would be needed for different structures. We choose to view this result in a more positive light in that pragmatic segmentations are possible with a variety of different imaging modalities. Hence, it is possible to consider tradeoffs in imaging time, sequence availability, and segmentation approaches when designing a study to use multi-modal MRI for segmentation of subcortical structures.
Several recent works have used multi-modal MRI to deal with subcortical structure segmentation. Traynor et al. showed that adding T1 with T2 MRI improve thalamus segmentation based on a priori anatomical hypothesis and T1/T2 values[18]. Hasan et al. utilized T1, T2 and DTI maps to identify lesions appearance frequency in cerebral subcortical, lobar white and cortical gray matter subdivisions [19]. Haegelen et al. evaluated the effectiveness of using T1 and T2 MRI on different registration approaches and the patch-based method to identify deep brain structures [20]. Similarly, Xiao et al. proposed patch based label fusion methods to segment the STN and its adjacent structures using T1 and T2 MRIs [21]. Forstmann et al. presented a data set that includes whole-brain and reduced field-of-view T1 MP2RAGE and T2-W scans of the subcortex and brainstem with an ultra-high resolution at a sub-millimeter scale, and the data can be used to develop new algorithms that help to build high-resolution atlases [22]. For instance, Visser et al. use Forstmann’s dataset multimodal (T1, T2, and DTI) method for subcortical segmentation and apply it to the striatum and globus pallidus, substantia nigra, subthalamic nucleus and red nucleus [23, 24]. D’Albis et al. developed PyDBS, which is an automated image processing workflow for deep brain stimulation surgery that integrates with a T1-T2 multi-modality input pipeline [25]. Liu et al. showed a multi-modal (T1 and T2) learning-based method using regression forests to automatically localize the target in pre-operative MR brain scans [26]. Ewert et al. used T1, T2, proton density, T2 relaxometry to parcellate STN and GPi functional zones using structural connectivity to the cortex and the thalamus respectively [27]. Kim et al. demonstrated that Susceptibility Weighted Image (SWI) and T2 approach significantly improves volumetric segmentation subcortical structures such as the basal ganglia and thalamus at 7T [28]. Coron et al. employed T1-, T2 -weighted and quantitative susceptibility mapping (QSM) information to automatically segment major thalamic subnuclear groups [29]. Li et al. proposed a QSM/T1 multi-atlas approach to segment subcortical on QSM images [30]. We note that the Dice accuracy is imperfect on the smaller structure (e.g., ~0.6 for the substantia nigra and STN). For population studies, this may be sufficient to detect group differences. However, careful consideration of the spatial accuracy would be needed before interventional applications would be advised.
These sequences that we proposed in this work are of interest because they are all acquirable on a clinical population. As a result, the proposed segmentation approaches can be translated to clinical populations and thus aid in the clinical workflow. In particular, the STN and GPI are common targets for deep brain stimulation surgery (DBS) [31]. DBS is a surgery commonly used in Parkinson’s disease to mitigate the motor symptoms of the disease. Generalization for pathological anatomies will require additional evaluation to ensure that the registration procedures can adapt between controls and patients. Alternatively, patient atlases could be acquired. Overall, this work is a meaningful step toward understanding the effects of imaging sequence on segmentation of subcortical grey matter structures and optimizing the algorithms for segmentation of these data.
5. Acknowledgements
This research was supported by NSF CAREER 1452485 (Landman), R01NS095291 (Dawant), and T32LM012412 (Malin). This study was also supported by NIH 5R01NS056307, 5R21NS082891 and in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors have no conflicts of interests with this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6 References
- [1].Alexander GE and Crutcher MD, “Functional architecture of basal ganglia circuits: neural substrates of parallel processing,” Trends in neurosciences, vol. 13, no. 7, pp. 266–271, 1990. [DOI] [PubMed] [Google Scholar]
- [2].Albin RL, Young AB, and Penney JB, “The functional anatomy of basal ganglia disorders,” Trends in neurosciences, vol. 12, no. 10, pp. 366–375, 1989. [DOI] [PubMed] [Google Scholar]
- [3].Yin HH and Knowlton BJ, “The role of the basal ganglia in habit formation,” Nature Reviews Neuroscience, vol. 7, no. 6, pp. 464–476, 2006. [DOI] [PubMed] [Google Scholar]
- [4].Brown P, “Oscillatory nature of human basal ganglia activity: relationship to the pathophysiology of Parkinson’s disease,” Movement Disorders, vol. 18, no. 4, pp. 357–363, 2003. [DOI] [PubMed] [Google Scholar]
- [5].Sudhyadhom A, Haq IU, Foote KD, Okun MS, and Bova FJ, “A high resolution and high contrast MRI for differentiation of subcortical structures for DBS targeting: the Fast Gray Matter Acquisition T1 Inversion Recovery (FGATIR),” Neuroimage, vol. 47, pp. T44–T52, 2009. [DOI] [PubMed] [Google Scholar]
- [6].Xiao Y, Beriault S, Pike GB, and Collins DL, “Multicontrast multiecho FLASH MRI for targeting the subthalamic nucleus,” Magnetic resonance imaging, vol. 30, no. 5, pp. 627–640, 2012. [DOI] [PubMed] [Google Scholar]
- [7].Jovicich J et al. , “MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths,” Neuroimage, vol. 46, no. 1, pp. 177–192, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Addington J et al. , “North American prodrome longitudinal study (NAPLS 2): overview and recruitment,” Schizophrenia research, vol. 142, no. 1, pp. 77–82, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].D’Haese P-F et al. , “CranialVault and its CRAVE tools: A clinical computer assistance system for deep brain stimulation (DBS) therapy,” Medical image analysis, vol. 16, no. 3, pp. 744–753, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Klein A, Dal Canton T, Ghosh SS, Landman B, Lee J, and Worth A, “Open labels: online feedback for a public resource of manually labeled brain images,” in 16th Annual Meeting for the Organization of Human Brain Mapping, 2010. [Google Scholar]
- [11].Iglesias JE and Sabuncu MR, “Multi-atlas segmentation of biomedical images: A survey,” (in eng), Med Image Anal, vol. 24, no. 1, pp. 205–19, August 2015, doi: 10.1016/j.media.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marcus DS, Wang TH, Parker J, Csemansky JG, Morris JC, and Buckner RL, “Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults,” Journal of cognitive neuroscience, vol. 19, no. 9, pp. 1498–1507, 2007. [DOI] [PubMed] [Google Scholar]
- [13].Asman AJ, Huo Y, Plassard AJ, and Landman BA, “Multi-atlas learner fusion: An efficient segmentation approach for large-scale data,” Medical image analysis, vol. 26, no. 1, pp. 82–91,2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Avants BB, Tustison NJ, Song G, Cook PA, Klein A, and Gee JC, “A reproducible evaluation of ANTs similarity metric performance in brain image registration,” (in eng), Neuroimage, vol. 54, no. 3, pp. 2033–44, February 2011, doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Asman AJ and Landman BA, “Hierarchical performance estimation in the statistical label fusion framework,” Med Image Anal, vol. 18, no. 7, pp. 1070–81, October 2014, doi: 10.1016/j.media.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Avants BB, Epstein CL, Grossman M, and Gee JC, “Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain,” (in eng), Med Image Anal, Research Support, N.I.H., Extramural vol. 12, no. 1, pp. 26–41, February 2008, doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang H, Suh JW, Das SR, Pluta JB, Craige C, and Yushkevich PA, “Multi-atlas segmentation with joint label fusion,” Pattern Analysis and Machine Intelligence, IEEE Transactions on, vol. 35, no. 3, pp. 611–623, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Traynor CR, Barker GJ, Crum WR, Williams SC, and Richardson MP, “Segmentation of the thalamus in MRI based on T1 and T2,” Neuroimage, vol. 56, no. 3, pp. 939–950, 2011. [DOI] [PubMed] [Google Scholar]
- [19].Hasan KM, Walimuni IS, Abid H, Wolinsky JS, and Narayana PA, “Multi-modal quantitative MRI investigation of brain tissue neurodegeneration in multiple sclerosis,” Journal of Magnetic Resonance Imaging, vol. 35, no. 6, pp. 1300–1311, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haegelen C et al. , “Automated segmentation of basal ganglia and deep brain structures in MRI of Parkinson’s disease,” International journal of computer assisted radiology and surgery, vol. 8, no. 1, pp. 99–110, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xiao Y et al. , “Patch-based label fusion segmentation of brainstem structures with dualcontrast MRI for Parkinson’s disease,” International journal of computer assisted radiology and surgery, vol. 10, no. 7, pp. 1029–1041, 2015. [DOI] [PubMed] [Google Scholar]
- [22].Forstmann BU, Keuken MC, Schafer A, Bazin P-L, Alkemade A, and Turner R, “Multi-modal ultra-high resolution structural 7-Tesla MRI data repository,” Scientific data, vol. 1, p. 140050, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Visser E et al. , “Automatic segmentation of the striatum and globus pallidus using MIST: multimodal image segmentation tool,” Neuroimage, vol. 125, pp. 479–497, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Visser E, Keuken MC, Forstmann BU, and Jenkinson M, “Automated segmentation of the substantia nigra, subthalamic nucleus and red nucleus in 7 T data at young and old age,” Neuroimage, vol. 139, pp. 324–336, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].D’Albis T, Haegelen C, Essert C, Fernandez-Vidal S, Lalys F, and Jannin P, “PyDBS: an automated image processing workflow for deep brain stimulation surgery,” International journal of computer assisted radiology and surgery, vol. 10, no. 2, pp. 117–128, 2015. [DOI] [PubMed] [Google Scholar]
- [26].Liu Y and Dawant BM, “Multi-modal learning-based pre-operative targeting in deep brain stimulation procedures,” in 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), 2016: IEEE, pp. 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ewert S et al. , “Toward defining deep brain stimulation targets in MNI space: a subcortical atlas based on multimodal MRI, histology and structural connectivity,” Neuroimage, vol. 170, pp. 271–282, 2018. [DOI] [PubMed] [Google Scholar]
- [28].Kim J, Lenglet C, Duchin Y, Sapiro G, and Harel N, “Semiautomatic segmentation of brain subcortical structures from high-field MRI,” IEEE journal of biomedical and health informatics, vol. 18, no. 5, pp. 1678–1695, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Corona V, Lellmann J, Nestor P, Schoenlieb C-B, and Acosta-Cabronero J, “A multicontrast MRI approach to thalamus segmentation,” arXiv preprint arXiv: 1807.10757, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li X et al. , “Multi-atlas tool for automated segmentation of brain gray matter nuclei and quantification of their magnetic susceptibility,” NeuroImage, vol. 191, pp. 337–349, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kahn E et al. , “Deep brain stimulation in early stage Parkinson’s disease: operative experience from a prospective randomised clinical trial,” Journal of Neurology, Neurosurgery & Psychiatry, pp. jnnp-2011-300008, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]