Abstract
A critical issue facing humanity is the rise in antimicrobial resistance. Although increasing research effort is spent on developing novel drugs, few alternatives reach the clinic. Phage therapy is an old idea, long practiced in eastern European countries and gaining serious attention in the western world. In this Opinion piece, we outline a strategy that harnesses the ability of bacteriophages (phages) to impose strong selection on their bacterial hosts: instead of avoiding resistance, this therapy relies on the target organism resisting the phage. By using phages to both kill bacterial cells and ‘steer’ survivors towards resistant but more compromised phenotypes, we can potentially improve treatment outcomes.
Antimicrobial treatments work by reducing the number of viable pathogens, and their persistent application in a population of infected individuals invariably selects for resistant strains [1-3]. Antimicrobial resistance is a growing clinical challenge, reducing the ability of physicians to care adequately for patients [4]. Compounding the problem, the discovery of novel antimicrobial compounds has decreased over recent decades [5]. These two interrelated challenges have led to the emergence of untreatable multi-drug resistant microbes and associated increases in morbidity and mortality [6-8]. In light of this crisis, there are growing calls to revitalize research into alternative and complementary treatments such as anti-virulence drugs [9], vaccines [10], anti-resistance adjuvants [11], diagnostic-informed treatments [12], and the topic of this article – phage therapy [13].
Phage therapy has a long history of great successes and perplexing failures [13, 14]. Although the exact causes of failure are often unclear and barring inappropriate choice, the most likely mechanism is the evolution of resistance to phage [15]. Given the pervasive risk of resistance evolution and the possible worsening conditions should a therapy fail, phages and the strategies used to apply them need to be chosen very carefully. Several candidates have been proposed to combat resistance evolution, including phage cocktails and combinations [16], phage-antibiotic combinations [15], phage training [17, 18], phage engineering [19], and approaches targeted at structural resistance such as biofilms [20].
Another approach that we advocate here is to deliberately use the evolution of resistance to attain therapeutic objectives. The basis of this idea relies on trade-offs between resistance to phage and other bacterial traits, namely virulence factors and antibiotic resistance. Such approaches have been previously discussed [21, 22] [13] [23], and it is our aim to unite these into a single concept that we call “phage steering”. Phage steering is similar to other strategies by its immediate aim of killing phage-sensitive bacteria. The novelty of phage steering is that remaining bacterial cells will be phage resistant, but somehow more vulnerable to other antimicrobials or have reduced virulence. In either case, target bacteria not eliminated by the phage persist but either cause far less damage to the host or are more sensitive to antibiotics. The notion of ‘steering’ has previously been discussed in the context of preventing or guiding the emergence of drug resistance in microbial pathogens [24, 25] and cancers [26]. Phage steering differs by a focus on actively selecting for specific phage resistances that are correlated with improved infection outcomes. It is thus arguably the phage that steers the infection and not directly the practitioner.
Phage steering
Bacterial surface factors are integral to disease phenotypes. These factors have diverse functions in different environments, mediating attachment, secretion, or nutrient uptake [27-29]. In a pathogen context, surface factors are often described as virulence factors or antibiotic resistance mechanisms, as they can mediate attachment and damage to hosts, and antibiotic efflux, respectively. For example, the opportunistic pathogen Pseudomonas aeruginosa has 306 described virulence factors of which 45% are predicted by PSORTb to be localized to the cell membrane [30].
Given the critical involvement of surface factors in disease phenotypes, we argue that phages binding to these factors would reduce the number of bacteria and, importantly, select against these factors – leading to reductions in virulence or antibiotic resistance among surviving bacterial populations. Accomplishing this requires accounting for the evolution of surface factor interactions between phages and bacteria over short evolutionary time scales, i.e., during treatment of a patient.
The main idea promoted here is that bacterial evolution of phage resistance can generate negative feedbacks for bacterial fitness (i.e., trade-offs) on key traits associated with disease. Several in vitro studies indicate these trade-offs (see [23] for examples of virulence reduction driven by phage and [31] for reduction in antibiotic resistance). Moreover, in a promising recent clinical case, an appropriate phage was identified by screening for the desired effect of steering a bacterial phenotype and successfully administered to a patient [32]. We propose that a range of future therapies can be designed that capitalize on phage steering.
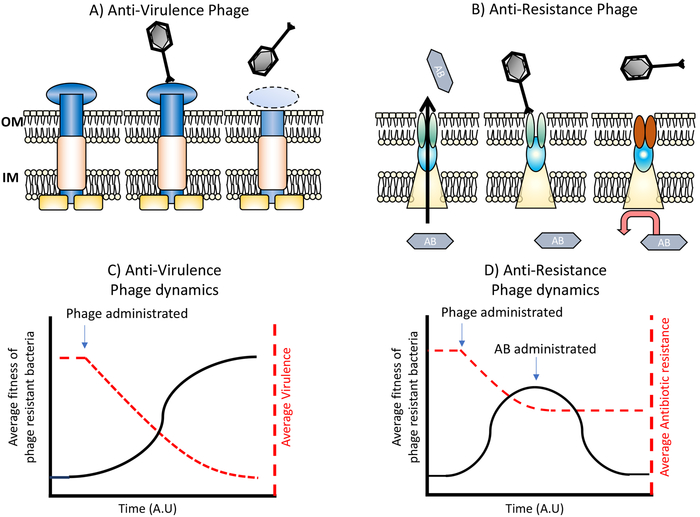
The current prospects for phage therapy are limited by a number of factors: improvements or investments in; regulation, supply chains, and basic biology need to occur before any routine use in the clinic (see Box 1 for discussion of Practice and Safety). It is with this in mind that we describe the scenarios under which we believe phage steering can contribute and hope that our discussion will help illuminate the field and spark further interest and research into these ideas. Although there are many ways in which steering can be employed, we focus on the two most understood and most likely to be put into practice: reduced expression / activity of bacterial surface factors contributing to virulence (Figure 1 A & C) and reduced expression / activity of bacterial surface factors contributing to antimicrobial resistance (Figure 1 B & D).
Box 1. Practicality and Safety.
Phage therapy (in the West) is typically performed as a compassionate care/release study, where phages that can target the focal pathogen are either selected from library stock or isolated de novo from the environment, normally within a limited time window. Choice of candidates for phage steering would be similar to other phage therapy strategies [62], with the notable exception that the former would require identification of targets that correspond to trade-offs, that when selected, would somehow compromise (e.g., either reduced virulence or increased antibiotic sensitivity) the population of bacteria remaining after the lysis of sensitive cells.
Given the above, identifying steering phages will be challenging, but certainly possible given the enormous diversity of phages in the environment. Moreover, an unexplored but plausible alternative to phage discovery is to engineer them. Because engineering constructs can be patented whereas natural phages cannot easily, this may promote regulation and spur investment.
Safety is a major consideration in phage therapy [63], and given its relatively recent history, considerations for phage steering have not yet been discussed in any detail. Because lysogenic phages may carry antibiotic resistance genes, transfer toxins such as those of the vibrio cholera phage (CTX) [64], and are likely to be less effective at selecting for resistance than lytic phages – outside of being engineered – they will not be candidates for steering.
Likewise, safety considerations for other phage therapies also apply to steering phages. These include avoiding phages that increase virulence [36], induce biofilm formation [65] or prophage activity [66], or select for the emergence of mutators [67]. For example, chronic infections of P. aeruginosa in cystic fibrosis patients may be associated with hyper-mutators [68] and with increased antibiotic resistance [69]. Although selection for compensatory mutations is a potential concern in phage steering, this is unlikely, both due to the sparse cell numbers remaining after phage introduction [70], and (as discussed in Box 3) the screening for phages that limit the ascension of phenotypically resistant bacteria.
Figure 1. Phage steering of bacterial virulence and antibiotic resistance.
Selection for bacterial resistance to phages at specific targets reduces bacterial virulence (A and C) or antibiotic resistance (B and D). As seen in C, the fitness of resistant bacteria (black line) increases due to phage pressure, while virulence (red line) declines. D shows a similar interaction, but this time focusing on antibiotic resistant mechanisms. The fitness of phage-resistant bacteria (black line) increases as phage replicate, and there is coetaneous decline in average antibiotic resistance (red line). Subsequent administration of antibiotics would reduce the fitness of these now antibiotic sensitive cells.
Attenuating virulence
Virulence is a widely used term with different definitions in different fields. Here we employ it to mean the in vivo expression of bacterial factors that damage the host [9], and bacterial life-history traits such as in vivo growth rate and competitiveness with commensals in the microbiome [33]. We focus below on virulence factors required for phage attachment and outline evidence of reduced virulence following phage exposure (Table 1.).
Table 1.
Bacterial virulence factors with defined phage that can potentially act as steering agents
| Target | Description |
|---|---|
| Porins. | A clear example in a natural system was shown in recent work by Seed and colleagues [83], where phage predation on the pathogen V. cholerae led to a reduction in virulence in humans. Resistance to the phage ICP2 can occur in V. cholerae by modification of a major outer membrane porin OmpU. OmpU is linked with resistance to bile and anionic detergents [84]. The receptor OmpU has previously been demonstrated to be important for bacterial virulence in infections [85-87] and reduction in the receptor expression level leads to reduced virulence and colonization of V. cholerae in an animal model. The authors findings for cases of cholera where the phage ICP2 was present were consistent with dampened virulence benefitting the patient. |
| Type IV pili. | Phages can select for less virulent bacteria via alteration in Type IV pili [88]. Changes in the pili reduce phage attachment, but may also impair other functions, namely those involved in initiating bacterial infection [89]. Thus, trade-offs involved in selection for phage resistance via alteration of pili could result in reduced virulence [90]. Consistent with this idea, Betts et al. 2016 showed that exposure to Type IV pili phage selected for altered bacterial pilF gene [91]. However, it remains to be determined whether this resistance leads to lower virulence in a host. Among the virulence genes identified by Turner et al. in P. aeruginosa, the pilF gene was not essential in a cystic fibrosis sputum medium or CF sputum [41], implying that loss of this factor will not be strongly counter-selected. This suggests that phages capable of attaching and initiating an infection via Type IV pili are strong candidates for anti-virulence therapies in cystic fibrosis patients infected with P. aeruginosa, provided that pili facilitate virulence in the CF lung environment (see Figure 1 A for a schematic). If this is the case, we predict that bacteria evolving resistance to such phage will lose the ability to fully express Type IV pili and exhibit attenuated virulence. |
| Lipopolysaccharides. | Bacterial surface lipopolysaccharides (LPS) are used by a wide range of phages to initiate infection [92-94]. LPS is also associated with virulence as an endotoxin in many Gram-negative bacteria [95-97]. It can stimulate the Toll-Like-Receptor 4 immune response, which leads to increased cellular damage in infections [98]. Modification of LPS can achieve phage resistance and reductions in virulence [99, 100]. Chart and colleagues were among the first to show a link between phage and LPS virulence reduction [99]. Using a range of Salmonella enteritidis phages (40 phages in total), they demonstrated that LPS modifications following phage infection lead to reduced bacterial virulence in mice [99]. Santander and Robeson similarly showed that phage resistance induced the loss of the O-polysaccharide in the LPS structure, resulting in reduced virulence of S. enteritidis in Caenorhabditis elegans [100]. In most Gram-negative bacteria, the LPS is an essential structure, but modifications are common [101]. For example, isolates of P. aeruginosa from cystic fibrosis patients commonly have altered LPS structures [97, 102], but it is unclear whether this is due to increased fitness within the lung environment, phage predation, or some other interaction with the human host [44, 97, 102]. |
| Siderophores. | Bacteria require iron for metabolism and many bacterial pathogens are adapted to scavenge for iron when the latter is limiting. These bacteria produce siderophore molecules, which strip iron from other ligands. Some phages use iron binding receptors (e.g., TonB) on bacterial membranes as attachment sites [103] (see [23] [13] for list of phage targets identified as virulence factors). In chronic infections like CF, where available iron is abundant but typically in the Fe3+ form, siderophores can chelate the iron to the usable Fe2+; however, over time (during the long-term course of disease) the majority of iron in the lung becomes ferrous which is readily available without siderophores. During persistent infections siderophores might still contribute to virulence by further disrupting iron homeostasis but provide little to no fitness benefit for the pathogen [104]. Phages that select against these receptors could reduce harm to patients, corresponding to the category where bacteria resist by losing or modifying TonB suffer little or no fitness consequences as iron is readily available. |
As early as 1931, D’Herelle noted that reduction in virulence was a hallmark of successful phage therapy, stating that bacterial modifications following phage treatment could lead to attenuated virulence if the bacteria are not completely eradicated. He included reduction in virulence as one of the three principles of phage therapy: first, the action of the phages in reducing the number of bacteria; second, the reduction in virulence; and third, an increase in phagocytosis due to lysins acting to opsonize pathogens [34]. Almost a full century later we are still reporting virulence reduction as an interesting yet unexploited phenomenon.
The examples in Table 1 of virulence-associated receptors targeted by specific phages demonstrate that, in principle, phages have the potential to reduce a bacterial pathogen’s per capita virulence in situ. We suggest that with appropriate development, phages could be used therapeutically to both reduce bacterial population size and the virulence of surviving phage-resistant cells [35]. However, care should be taken with generalizations, since not all phage-induced changes in the targeted bacterial receptors will affect virulence. Practitioners of phage therapies targeting LPS and other surface factors will have to be mindful of whether any virulence attenuation is sufficient to meet therapeutic objectives. Moreover, increases in virulence after phage exposure have also been reported [36]. Key examples can be found in work using the filamentous phage Phi-RSS. Phi-RSS can increase the virulence of its host Ralstonia solanacearum (other phages have been shown to reduce virulence [37]). However, the alteration of virulence in these examples appears to be due to changes in the phage life cycle and not from selection against receptors to which the phage attach [38]. Obviously, such phages would be omitted from therapeutic consideration.
Co-opting bacterial resistance to antibiotics
Another approach to improving therapeutic outcomes is for phages to also select against antibiotic resistance mechanisms. For example, Salmonella enterica requires a TolC efflux pump for effective resistance against Ciprofloxacin [39]. In screening for phages unable to bind to tolC knockout strains, Ricci & Piddock isolated a phage that uses this resistance mechanism as its attachment receptor [40]. Such cases where phages target structures providing resistance to antibiotics are of particular interest since they could extend the life of antibiotics that otherwise would have been discarded due to intrinsic or evolved resistance.
Phages that attach and infect via efflux pumps potentially alter the balance between selection for resistance to the phage and resistance to an antibiotic. Chan et al isolated an outer membrane porin-dependent lytic phage of P. aeruginosa, designated Omko1 [31]. Omko1 binds to the oprM subunit of the mexXY efflux pump responsible for resistance to a wide range of antibiotics in P. aeruginosa (see Figure 1B for a schematic). After selection with this phage, a significant reduction in in vitro resistance to a range of antibiotics was described by Chan et al. It is yet to be determined if phage-mediated selection can repeatedly produce the same effect in vivo. However, Omko1 was recently used successfully in two compassionate release studies, one of a long-term P. aeruginosa pericardial infection [32] and the second in a CF patient with a high abundance of P. aeruginosa (B. Chan, pers. comm.). In the latter case, the post-phage treated isolates of P. aeruginosa showed a significant decrease in antibiotic resistance even though the patient continued to receive antibiotic treatment. This suggests that phage-mediated selection can be strong and sufficiently sustained to produce bacterial evolutionary responses during the course of patient treatment.
Interestingly, the orpM gene is non-essential in a range of clinical settings [41]. This might facilitate the evolution of phage resistance through modification of the efflux pump, and at the same time impose a substantial fitness cost to bacteria when they come into contact with antibiotics. Thus, a phage treatment may be a robust therapeutic approach even in the presence of drugs, with the desirable side-effect of selecting against antibiotic resistance. In explicit terms, used appropriately this phage has the potential to revert existing antibiotic resistance in patients, thus an infection may be rendered treatable with antibiotics again.
Considerations of phage steering as a therapeutic
Current approaches to treating bacterial pathogens with antibiotics select for resistance [1, 3, 42, 43]. Employing phages as stand-alone agents or in combination with other antimicrobials shows great promise, but discussion and study have been limited to only a few of the many possible strategies. Our argument is that phages can be employed to do more than their primary objective of reducing bacterial density. Anti-virulence phages can select for a range of bacterial phenotypes, resulting in reductions in disease severity on both an individual and public health scale – thereby reducing the need for antibiotic applications and conserving antibiotic sensitivity. Anti-resistance phages can drive antibiotic sensitivity in otherwise antibiotic resistant bacterial pathogens, and when used in combination with antibiotics, result in treatment success. However, given the complexity of phage-bacteria interactions, several considerations are important in developing effective phage steering therapies for clinical use.
First, the availability and phenotypic impact of bacterial surface factor mutants requires consideration. Not all mutations in surface factors will lead to desirable patient outcomes, and, similarly, not all pathogenic phenotypes involve surface factors. For example, a mutation in a surface factor that reduces phage binding may have little or no effect on virulence. Although LPS endotoxins are virulence factors, structures like lipid A and O antigens within LPS can be changed (e.g., sugar modification [44]), reducing the absorption of phages, but have little effect on disease progression or severity [45].
Second, the phages need to impose and maintain selection on the target surface factor for steering to be most effective. Maintaining this selection might be disrupted if the phage and bacteria co-evolve [46-48]; thus selection on a target surface factor may change through time. Moreover, resistance to phage can be generated by a number of mechanisms other than receptor modification. For example, under certain conditions receptor modification is predicted to maintain phages in the presence of active CRISPR-Cas immunity (see Box 2) [49]. Even less appreciated is how bacterial virulence and antibiotic resistance are impacted over short evolutionary timescales – i.e., the duration of treatment. Bacterial escape mutants, such as those that form biofilms, may maintain unmodified surface factors and still contribute to virulence [50]. This could be compounded by phages switching surface factors, i.e., evolving to use novel receptors [51]. This suggests more generally that for steering to be successful the range of resistance mechanisms employed by the bacterium need to be understood in order to minimize the risk of unanticipated treatment failures.
Box 2. Alternative resistance mechanisms and indirect targets.
Adaptation of the surface receptor is not the only mechanism by which bacteria can evolve phage resistance. Considerable recent research has investigated how bacteria resist phage attack (for a review, see [71]). This has yielded very promising biotechnology applications, such as CRISPR-Cas [72], and multiple other resistance mechanisms, such as restriction modification systems [73]. It will be important to know how these mechanisms contribute to the short-term evolution of resistance to phage selection, and whether they are involved in coevolutionary arms races [49]. What little in vitro evidence exists suggests that coevolution gradually shifts from ever-escalating arms race dynamics to fluctuating selection dynamics [74, 75], indicating increasing costs of accumulating attack and defence mechanisms. Such costs (of bacterial defence) are of particular interest in the context of phage therapy since they could reduce the ability of the bacterial pathogen to respond to demanding environments, or to actively modify their environments. Recent work on community dynamics of phage therapy employing a virulent lysogenic phage in vitro has shown that the form of resistance could change depending on the local bacterial community [76]. We discuss the impacts of phage steering on community dynamics in Box 3.
Given their importance to bacterial fitness, virulence and antibiotic resistance expression are expected to be tightly regulated. Phage steering could apply selection to a wide range of the pathogenic mechanisms by causing down- and/or dysregulation of master regulators. An example is the interaction between the bacteria Bordetella spp and the phage BPP-1. BPP-1 utilizes the pertactin autotransporter (Prn) for secretion of peractin, which is used in adhesion to epithelial cells [77]. Prn is under the genetic control of the Bvg two-component system [78]. Bvg also controls the expression of multiple virulence determinants, such as the Type III secretion system [79]. Thus, if the evolution of resistance to phage BPP-1 involves mutations in the Bvg cassette, this may cause negative pleiotropic effects on the expression of one or more of the many virulence factors under its control. This example illustrates how virulence can be managed indirectly through phage-induced selection on genes that do not directly interact with the phage but control the expression of the phage attachment site.
A third consideration is the impact that phage steering has on the fitness of the selected, resistant bacterial pathogen. Changes in receptors or structures with essential functions (such as LPS in most Gram-negative bacteria) are unlikely to involve gene deletions, and those that do will likely have higher fitness costs when compared to mutants that make modifications to motifs or reduce the expression of the receptor [52]. This difference in fitness would lead to the mutants with slight modifications outcompeting the full gene deletion strains. In contrast, receptors with non-essential functions may be lost or down-regulated due to phage selection without lowering bacterial fitness, and in fact as seen in Tn-seq experimentation [41, 53] could increase bacterial fitness, yet still have the desired outcome of lowering either virulence or antibiotic resistance [54]. Box 3 provides an overview of what makes a ‘good’ phage target.
Box 3. What is a good target for phage steering?
Depending on the environment, a number of outer membrane structures may actively contribute to virulence or antibiotic resistance [9, 21, 22]. For example, work by Turner et al. identified several virulence factors in P. aeruginosa that are potentially directly costly to the pathogen in a clinically relevant environment (i.e., the mutants were enriched in vitro) [41, 53]. These genes would make ideal candidates for phage steering due to the apparent lack of counter-selection to maintain functionality. It is thus in the bacteria’s ‘best interest’ to not express these genes if they are not under positive selection. If this is bolstered by the addition of phage that apply selection on these same genes, then resistant bacteria are expected to reduce the associated phenotype (i.e., virulence or antibiotic resistance) [23, 31, 80]. Given encouraging findings that certain phages may select for antibiotic sensitivity [31, 80], future research should consider longer term effects where there is a coevolutionary response between the antagonists [48].
Chronic infections are often polymicrobial in nature and are receiving increasing attention [81]. Phage have been shown to affect the types of mutation that occur when bacteria compete in complex communities [76]. Surface factors that mediate interactions between the different species or with the environment are potential targets for phage steering. Given the complexity of even the simplest in situ microbial communities, while it might be possible to reduce pathogen burden by selecting against another species in the microbiome that facilitates the pathogen [82], in practice we currently lack the required understanding of complex community dynamics to make this a viable therapeutic option.
Finally, as suggested above, phage steering is unlikely to be used as a standalone antimicrobial therapy. Steering phages work by killing sensitive bacteria and selecting for resistant strains that are otherwise vulnerable. To be a viable strategy, either the resistant strains need to be compromised so as to be avirulent, or need to be eliminated directly by additional treatments and/or by the immune system. These objectives are most readily achieved through combinations with other antimicrobial agents in the form of phage-phage or phage-antibiotic cocktails. In this context, phage steering plays the role of both primary treatment (bacterial killing) and an adjuvant (compromised survivors) to more inclusive therapies.
Concluding remarks
Ecological interactions govern life and how it adapts and responds to changing environments. This applies to classic ecosystems as well as to disease ecosystems, and it would be short-sighted to not use this knowledge to its full potential [33]. Our understanding of phage ecology in an infection context is still in its infancy [55, 56]. The classic view of infection as a simple interaction between the host and the pathogen is being replaced with an appreciation that there exists a complex community ecology [33, 57, 58]. Some pathogens actively alter their environments to foster growth, reproduction, and colonization to the detriment of the host [59, 60]. Understanding how these interactions can be disrupted will open the way for novel and effective therapeutics [33] (see also Outstanding Questions).
Outstanding Questions.
What is the range of bacterial surface factors that steering phage can target?
To what extent is phage steering evolution proof, is it robust to compensatory mutations?
Can phage steering work on a public health scale?
How does phage steering drive coevolution?
What is the impact of steering phage on integrated lysogenic phage?
How well would phage steering work under constant antibiotic pressure?
Can these principles be applied to other selective agents such as porins or lysins?
How does phage target choice evolve under therapeutic conditions?
Phages will likely not completely supplant antibiotics. There is increasing effort in the search for novel antimicrobials [61], especially how antibiotic combinations increase both the efficacy of treatments and minimize the emergence and spread of antibiotic resistance. Nevertheless, phages augment the arsenal available to successfully treat certain bacterial pathogens [33]. Even if phages are not sufficient as a standalone alternative treatment to antibiotics, their ability to compromise bacterial virulence or alter bacterial vulnerability to antibiotics merits attention. If used in an evolutionarily rational way, phages can both improve treatment success and extend the useful lifespans of antibiotics [15]. Thus, counterintuitively, the selection for resistance may actually form an important part of the antimicrobial arsenal.
Highlights.
Antibiotic resistance is a global issue that currently causes increased morbidity and mortality. It is crucial to expand and explore alternative strategies to combat the crisis.
The use of bacteriophages, the viruses of bacteria, is one of the many alternative strategies being considered and developed.
Bacteriophage therapy conventionally works by reducing the number of bacteria at a given site in a manner analogous to antimicrobials.
Bacteriophage therapy also shares the same problem as conventional antimicrobials: the rapid emergence of resistance.
A well reported phenomena of bacteriophage interactions is the reduction of virulence of phage resistant bacteria and recent studies have shown that resistance to phage can also reduce resistance to antimicrobials.
Phage therapy could be used as a selective force against pathogenic surface factors, manipulating the evolution of the pathogen in favor of the infected patient.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108 Suppl 2:10871–7. Epub 2011/06/22. doi: 10.1073/pnas.1100299108. PubMed PMID: 21690376; PubMed Central PMCID: PMCPMC3131826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith RA, M'Ikanatha NM, Read AF. Antibiotic resistance: a primer and call to action. Health Commun. 2015;30(3):309–14. Epub 2014/08/14. doi: 10.1080/10410236.2014.943634. PubMed PMID: 25121990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samore MH, Lipsitch M, Alder SC, Haddadin B, Stoddard G, Williamson J, et al. Mechanisms by Which Antibiotics Promote Dissemination of Resistant Pneumococci in Human Populations. American Journal of Epidemiology. 2005;163(2):160–70. doi: 10.1093/aje/kwj021. [DOI] [PubMed] [Google Scholar]
- 4.Schaberle TF, Hack IM. Overcoming the current deadlock in antibiotic research. Trends in microbiology. 2014;22(4):165–7. Epub 2014/04/05. doi: 10.1016/j.tim.2013.12.007. PubMed PMID: 24698433. [DOI] [PubMed] [Google Scholar]
- 5.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24(1):71–109. Epub 2011/01/15. doi: 10.1128/cmr.00030-10. PubMed PMID: 21233508; PubMed Central PMCID: PMCPMC3021209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. PubMed PMID: 25232278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanwar J, Das S, Fatima Z, Hameed S. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;2014:541340-. Epub 2014/07/16. doi: 10.1155/2014/541340. PubMed PMID: 25140175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–83. PubMed PMID: 25859123. [PMC free article] [PubMed] [Google Scholar]
- 9.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nature Reviews Microbiology. 2014;12(4):300. [DOI] [PubMed] [Google Scholar]
- 10.Jansen KU, Anderson AS. The role of vaccines in fighting antimicrobial resistance (AMR). Human vaccines & immunotherapeutics. 2018;14(9):2142–9. Epub 2018/05/23. doi: 10.1080/21645515.2018.1476814. PubMed PMID: 29787323; PubMed Central PMCID: PMCPMC6183139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright GD. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends in microbiology. 2016:1–10. doi: papers3://publication/doi/ 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 12.McAdams D, Wollein Waldetoft K, Tedijanto C, Lipsitch M, Brown SP. Resistance diagnostics as a public health tool to combat antibiotic resistance: A model-based evaluation. PLoS biology. 2019;17(5):e3000250–e. doi: 10.1371/journal.pbio.3000250. PubMed PMID: 31095567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kortright KE, Chan BK, Koff JL, Turner PE. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell host & microbe. 2019;25(2):219–32. Epub 2019/02/15. doi: 10.1016/j.chom.2019.01.014. PubMed PMID: 30763536. [DOI] [PubMed] [Google Scholar]
- 14.Lin DM, Koskella B, Lin HC. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8(3):162–73. doi: 10.4292/wjgpt.v8.i3.162. PubMed PMID: 28828194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Barcelo C, Hochberg ME. Evolutionary Rationale for Phages as Complements of Antibiotics. Trends in microbiology. 2016;24(4):249–56. Epub 2016/01/21. doi: 10.1016/j.tim.2015.12.011. PubMed PMID: 26786863. [DOI] [PubMed] [Google Scholar]
- 16.Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future microbiology. 2013;8(6):769–83. Epub 2013/05/25. doi: 10.2217/fmb.13.47. PubMed PMID: 23701332. [DOI] [PubMed] [Google Scholar]
- 17.Betts A, Vasse M, Kaltz O, Hochberg ME. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol Appl. 2013;6(7):1054–63. Epub 2013/07/15. doi: 10.1111/eva.12085. PubMed PMID: 24187587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friman VP, Soanes-Brown D, Sierocinski P, Molin S, Johansen HK, Merabishvili M, et al. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. Journal of evolutionary biology. 2016;29(1):188–98. Epub 2015/10/18. doi: 10.1111/jeb.12774. PubMed PMID: 26476097. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Batra H, Dong J, Chen C, Rao VB, Tao P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front Microbiol. 2019;10:954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhry WN, Concepcion-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PloS one. 2017;12(1):e0168615. Epub 2017/01/12. doi: 10.1371/journal.pone.0168615. PubMed PMID: 28076361; PubMed Central PMCID: PMCPMC5226664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vale PF, Fenton A, Brown SP. Limiting damage during infection: lessons from infection tolerance for novel therapeutics. PLoS Biol. 2014;12(1):e1001769. Epub 2014/01/28. doi: 10.1371/journal.pbio.1001769. PubMed PMID: 24465177; PubMed Central PMCID: PMCPMC3897360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vale PF, McNally L, Doeschl-Wilson A, King KC, Popat R, Domingo-Sananes MR, et al. Beyond killing: Can we find new ways to manage infection? Evolution, Medicine, and Public Health. 2016;2016(1):148–57. doi: 10.1093/emph/eow012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.León M, Bastías R. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol. 2015;6:343-. doi: 10.3389/fmicb.2015.00343. PubMed PMID: 25954266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith M, Cobey S. Fitness Landscapes Reveal Simple Strategies for Steering Evolution to Minimize Antibiotic Resistance. bioRxiv. 2016:093153. doi: 10.1101/093153. [DOI] [Google Scholar]
- 25.Allen RC, Brown SP. Modified Antibiotic Adjuvant Ratios Can Slow and Steer the Evolution of Resistance: Co-amoxiclav as a Case Study. mBio. 2019;10(5):e01831–19. doi: 10.1128/mBio.01831-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallaher JA, Enriquez-Navas PM, Luddy KA, Gatenby RA, Anderson ARA. Spatial Heterogeneity and Evolutionary Dynamics Modulate Time to Recurrence in Continuous and Adaptive Cancer Therapies. Cancer Research. 2018;78(8):2127. doi: 10.1158/0008-5472.CAN-17-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrova OE, Sauer K. Sticky Situations: Key Components That Control Bacterial Surface Attachment. Journal of Bacteriology. 2012;194(10):2413. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green ER, Mecsas J. Bacterial Secretion Systems: An Overview. Microbiol Spectr. 2016;4(1): 10.1128/microbiolspec.VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015. PubMed PMID: 26999395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS pathogens. 2010;6(8):e1000949. Epub 2010/08/17. doi: 10.1371/journal.ppat.1000949. PubMed PMID: 20711357; PubMed Central PMCID: PMCPMC2920840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–15. Epub 2010/05/13. doi: 10.1093/bioinformatics/btq249. PubMed PMID: 20472543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Scientific reports. 2016;6:26717. Epub 2016/05/27. doi: 10.1038/srep26717. PubMed PMID: 27225966; PubMed Central PMCID: PMCPMC4880932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan BK, Turner PE, Narayan D, Kim S, Mojibian HR, Elefteriades JA. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evolution, Medicine, and Public Health. 2018;2018(1):60–6. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochberg ME. An ecosystem framework for understanding and treating disease. Evolution, Medicine, and Public Health. 2018;2018(1):270–86. doi: 10.1093/emph/eoy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.d'Herelle F Bacteriophage as a Treatment in Acute Medical and Surgical Infections. Bulletin of the New York Academy of Medicine. 1931;7(5):329–48. Epub 1931/05/01. PubMed PMID: 19311785; PubMed Central PMCID: PMCPMC2095997. [PMC free article] [PubMed] [Google Scholar]
- 35.Smith HW, Huggins MB. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. Journal of general microbiology. 1982;128(2):307–18. Epub 1982/02/01. doi: 10.1099/00221287-128-2-307. PubMed PMID: 7042903. [DOI] [PubMed] [Google Scholar]
- 36.Hosseinidoust Z, van de Ven TG, Tufenkji N. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol. 2013;79(19):6110–6. Epub 2013/07/31. doi: 10.1128/aem.01421-13. PubMed PMID: 23892756; PubMed Central PMCID: PMCPMC3811365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Álvarez B, Biosca EG. Bacteriophage-Based Bacterial Wilt Biocontrol for an Environmentally Sustainable Agriculture. Front Plant Sci. 2017;8:1218-. doi: 10.3389/fpls.2017.01218. PubMed PMID: 28769942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Askora A, Yamada T. Two different evolutionary lines of filamentous phages in Ralstonia solanacearum: their effects on bacterial virulence. Frontiers in Genetics. 2015;6(217). doi: 10.3389/fgene.2015.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricci V, Tzakas P, Buckley A, Coldham NC, Piddock LJV. Ciprofloxacin-Resistant <em>Salmonella enterica</em> Serovar Typhimurium Strains Are Difficult To Select in the Absence of AcrB and TolC. Antimicrobial Agents and Chemotherapy. 2006;50(1):38. doi: 10.1128/AAC.50.1.38-42.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricci V, Piddock LJV. Exploiting the Role of TolC in Pathogenicity: Identification of a Bacteriophage for Eradication of <em>Salmonella</em> Serovars from Poultry. Applied and Environmental Microbiology. 2010;76(5):1704. doi: 10.1128/AEM.02681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of <em>Pseudomonas aeruginosa</em> in cystic fibrosis sputum. Proceedings of the National Academy of Sciences. 2015;112(13):4110. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipsitch M, Singer RS, Levin BR. Antibiotics in agriculture: when is it time to close the barn door? Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):5752–4. Epub 2002/05/02. doi: 10.1073/pnas.092142499. PubMed PMID: 11983874; PubMed Central PMCID: PMCPMC122845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipsitch M, Samore MH. Antimicrobial use and antimicrobial resistance: a population perspective. Emerging infectious diseases. 2002;8(4):347–54. Epub 2002/04/25. doi: 10.3201/eid0804.010312. PubMed PMID: 11971765; PubMed Central PMCID: PMCPMC2730242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King JD, Kocincova D, Westman EL, Lam JS. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate immunity. 2009;15(5):261–312. Epub 2009/08/28. doi: 10.1177/1753425909106436. PubMed PMID: 19710102. [DOI] [PubMed] [Google Scholar]
- 45.Ernst RK, Moskowitz SM, Emerson JC, Kraig GM, Adams KN, Harvey MD, et al. Unique lipid a modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. The Journal of infectious diseases. 2007;196(7):1088–92. Epub 2007/09/01. doi: 10.1086/521367. PubMed PMID: 17763333; PubMed Central PMCID: PMCPMC2723782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maura D, Morello E, du Merle L, Bomme P, Le Bouguénec C, Debarbieux L. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environmental microbiology. 2012;14(8):1844–54. doi: 10.1111/j.1462-2920.2011.02644.x. [DOI] [PubMed] [Google Scholar]
- 47.Torres-Barceló C Phage Therapy Faces Evolutionary Challenges. Viruses. 2018;10(6). doi: 10.3390/v10060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Betts A, Kaltz O, Hochberg ME. Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proceedings of the National Academy of Sciences of the United States of America. 2014; 111(30):11109–14. Epub 2014/07/16. doi: 10.1073/pnas.1406763111. PubMed PMID: 25024215; PubMed Central PMCID: PMCPMC4121802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurney J, Pleska M, Levin BR. Why put up with immunity when there is resistance: an excursion into the population and evolutionary dynamics of restriction-modification and CRISPR-Cas. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2019;374(1772):20180096. Epub 2019/03/25. doi: 10.1098/rstb.2018.0096. PubMed PMID: 30905282; PubMed Central PMCID: PMCPMC6452257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darch SE, Kragh KN, Abbott EA, Bjarnsholt T, Bull JJ, Whiteley M. Phage Inhibit Pathogen Dissemination by Targeting Bacterial Migrants in a Chronic Infection Model. mBio. 2017;8(2). Epub 2017/04/06. doi: 10.1128/mBio.00240-17. PubMed PMID: 28377527; PubMed Central PMCID: PMCPMC5380840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science (New York, NY). 2002;295(5562):2091–4. Epub 2002/03/16. doi: 10.1126/science.1067467. PubMed PMID: 11896279. [DOI] [PubMed] [Google Scholar]
- 52.Wright RCT, Friman V-P, Smith MCM, Brockhurst MA. Cross-resistance is modular in bacteria–phage interactions. PLOS Biology. 2018;16(10):e2006057. doi: 10.1371/journal.pbio.2006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa Acute Burn and Chronic Surgical Wound Infection. PLOS Genetics. 2014;10(7):e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends in microbiology. 2012;20(7):336–42. doi: papers3://publication/doi/ 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abedon ST. Phage evolution and ecology. Advances in applied microbiology. 2009;67:1–45. Epub 2009/02/28. doi: 10.1016/s0065-2164(08)01001-0. PubMed PMID: 19245935. [DOI] [PubMed] [Google Scholar]
- 56.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. Epub 2011/06/21. doi: 10.4161/bact.1.1.14942. PubMed PMID: 21687533; PubMed Central PMCID: PMCPMC3109452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conrad D, Haynes M, Salamon P, Rainey PB, Youle M, Rohwer F. Cystic fibrosis therapy: a community ecology perspective. American journal of respiratory cell and molecular biology. 2013;48(2):150–6. Epub 2012/10/30. doi: 10.1165/rcmb.2012-0059PS. PubMed PMID: 23103995; PubMed Central PMCID: PMCPMC3604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends in ecology & evolution. 2007;22(3):133–9. Epub 2006/12/02. doi: 10.1016/j.tree.2006.11.005. PubMed PMID: 17137676. [DOI] [PubMed] [Google Scholar]
- 59.Popat R, Harrison F, da Silva AC, Easton SA, McNally L, Williams P, et al. Environmental modification via a quorum sensing molecule influences the social landscape of siderophore production. Proc Biol Sci. 2017;284(1852). Epub 2017/04/14. doi: 10.1098/rspb.2017.0200. PubMed PMID: 28404780; PubMed Central PMCID: PMCPMC5394672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollein Waldetoft K, Raberg L. To harm or not to harm? On the evolution and expression of virulence in group A streptococci. Trends in microbiology. 2014;22(1):7–13. Epub 2013/11/19. doi: 10.1016/j.tim.2013.10.006. PubMed PMID: 24238777. [DOI] [PubMed] [Google Scholar]
- 61.Martens E, Demain AL. The antibiotic resistance crisis, with a focus on the United States. The Journal of antibiotics. 2017;70(5):520–6. Epub 2017/03/02. doi: 10.1038/ja.2017.30. PubMed PMID: 28246379. [DOI] [PubMed] [Google Scholar]
- 62.Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Current pharmaceutical biotechnology. 2010;11(1):2–14. Epub 2009/01/01. doi: 10.2174/138920110790725311. PubMed PMID: 20214604. [DOI] [PubMed] [Google Scholar]
- 63.Rohde C, Resch G, Pirnay J-P, Blasdel BG, Debarbieux L, Gelman D, et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial Phage Resistance, Phage Training and Prophages in Bacterial Production Strains. Viruses. 2018;10(4):178. doi: 10.3390/v10040178. PubMed PMID: 29621199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waldor MK, Mekalanos JJ. Lysogenic Conversion by a Filamentous Phage Encoding Cholera Toxin. Science (New York, NY). 1996;272(5270):1910. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 65.Moulton-Brown CE, Friman V-P. Rapid evolution of generalized resistance mechanisms can constrain the efficacy of phage–antibiotic treatments. Evol Appl. 2018;11(9):1630–41. doi: 10.1111/eva.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blasdel BG, Ceyssens P-J, Chevallereau A, Debarbieux L, Lavigne R. Comparative transcriptomics reveals a conserved Bacterial Adaptive Phage Response (BAPR) to viral predation. bioRxiv. 2018:248849. doi: 10.1101/248849. [DOI] [Google Scholar]
- 67.Pal C, Macia MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450(7172):1079–81. Epub 2007/12/07. doi: 10.1038/nature06350. PubMed PMID: 18059461. [DOI] [PubMed] [Google Scholar]
- 68.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science (New York, NY). 2000;288(5469):1251–4. Epub 2000/05/20. doi: 10.1126/science.288.5469.1251. PubMed PMID: 10818002. [DOI] [PubMed] [Google Scholar]
- 69.Macia MD, Blanquer D, Togores B, Sauleda J, Perez JL, Oliver A. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrobial agents and chemotherapy. 2005;49(8):3382–6. doi: 10.1128/AAC.49.8.3382-3386.2005. PubMed PMID: 16048951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacLean RC, Vogwill T. Limits to compensatory adaptation and the persistence of antibiotic resistance in pathogenic bacteria. Evol Med Public Health. 2014;2015(1):4–12. Epub 2014/12/24. doi: 10.1093/emph/eou032. PubMed PMID: 25535278; PubMed Central PMCID: PMCPMC4323496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–27. doi: papers3://publication/doi/ 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 72.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY). 2013;339(6121):819–23. Epub 2013/01/05. doi: 10.1126/science.1231143. PubMed PMID: 23287718; PubMed Central PMCID: PMCPMC3795411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1972;69(10):2904–9. Epub 1972/10/01. doi: 10.1073/pnas.69.10.2904. PubMed PMID: 4342968; PubMed Central PMCID: PMCPMC389671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host–parasite coevolutionary arms races give way to fluctuating selection. Ecology letters. 2011;14(7):635–42. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 75.Gurney J, Aldakak L, Betts A, Gougat-Barbera C, Poisot T, Kaltz O, et al. Network structure and local adaptation in co-evolving bacteria–phage interactions. Molecular Ecology. 2017;26(7):1764–77. doi: 10.1111/mec.14008. [DOI] [PubMed] [Google Scholar]
- 76.Alseth EO, Pursey E, Lujan AM, McLeod I, Rollie C, Westra ER. Bacterial biodiversity drives the evolution of CRISPR-based phage resistance in <em>Pseudomonas aeruginosa</em>. bioRxiv. 2019:586115. doi: 10.1101/586115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diavatopoulos DA, Hijnen M, Mooi FR. Adaptive evolution of the Bordetella autotransporter pertactin. Journal of evolutionary biology. 2006;19(6):1931–8. Epub 2006/10/17. doi: 10.1111/j.1420-9101.2006.01154.x. PubMed PMID: 17040390. [DOI] [PubMed] [Google Scholar]
- 78.Moon K, Bonocora RP, Kim DD, Chen Q, Wade JT, Stibitz S, et al. The BvgAS Regulon of <em>Bordetella pertussis</em>. mBio. 2017;8(5):e01526–17. doi: 10.1128/mBio.01526-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuk MH, Harvill ET, Miller JF. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Molecular microbiology. 1998;28(5):945–59. doi: 10.1046/j.1365-2958.1998.00850.x. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Wei Z, Li M, Wang X, Shan A, Mei X, et al. Parasites and competitors suppress bacterial pathogen synergistically due to evolutionary trade-offs. Evolution. 2017;71(3):733–46. doi: 10.1111/evo.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Filkins LM, O'Toole GA. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS pathogens. 2015;11(12):e1005258. Epub 2016/01/01. doi: 10.1371/journal.ppat.1005258. PubMed PMID: 26719892; PubMed Central PMCID: PMCPMC4700991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Estrela S, Brown SP. Community interactions and spatial structure shape selection on antibiotic resistant lineages. PLoS computational biology. 2018;14(6):e1006179. doi: 10.1371/journal.pcbi.1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, et al. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife. 2014;3:e03497. Epub 2014/08/28. doi: 10.7554/eLife.03497. PubMed PMID: 25161196; PubMed Central PMCID: PMCPMC4141277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wibbenmeyer JA, Provenzano D, Landry CF, Klose KE, Delcour AH. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect Immun. 2002;70(1):121–6. Epub 2001/12/19. doi: 10.1128/iai.70.1.121-126.2002. PubMed PMID: 11748172; PubMed Central PMCID: PMCPMC127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell host & microbe. 2013;14(6):652–63. Epub 2013/12/18. doi: 10.1016/j.chom.2013.11.001. PubMed PMID: 24331463; PubMed Central PMCID: PMCPMC3951154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS pathogens. 2013;9(12):e1003800. Epub 2014/01/05. doi: 10.1371/journal.ppat.1003800. PubMed PMID: 24385900; PubMed Central PMCID: PMCPMC3873450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Zhang W, Dong C. Crystal structure of the outer membrane protein OmpU from Vibrio cholerae at 2.2 A resolution. Acta Crystallographica Section D. 2018;74(1):21–9. doi: doi: 10.1107/S2059798317017697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2(5):363–78. Epub 2004/04/22. doi: 10.1038/nrmicro885. PubMed PMID: 15100690. [DOI] [PubMed] [Google Scholar]
- 89.Giltner CL, Nguyen Y, Burrows LL. Type IV Pilin Proteins: Versatile Molecular Modules. Microbiology and Molecular Biology Reviews. 2012;76(4):740. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brockhurst MA, Buckling A, Rainey PB. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci. 2005;272(1570):1385–91. Epub 2005/06/22. doi: 10.1098/rspb.2005.3086. PubMed PMID: 16006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Betts A, Gifford DR, MacLean RC, King KC. Parasite diversity drives rapid host dynamics and evolution of resistance in a bacteria-phage system. Evolution. 2016;70(5):969–78. Epub 2016/04/19. doi: 10.1111/evo.12909. PubMed PMID: 27005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishiguro EE, Ainsworth T, Shaw DH, Kay WW, Trust TJ. A lipopolysaccharide-specific bacteriophage for Aeromonas salmonicida. Canadian journal of microbiology. 1983;29(10):1458–61. Epub 1983/10/01. PubMed PMID: 6661705. [DOI] [PubMed] [Google Scholar]
- 93.Michel A, Clermont O, Denamur E, Tenaillon O. Bacteriophage PhiX174's Ecological Niche and the Flexibility of Its <em>Escherichia coli</em> Lipopolysaccharide Receptor. Applied and Environmental Microbiology. 2010;76(21):7310. doi: 10.1128/AEM.02721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiljunen S, Datta N, Dentovskaya SV, Anisimov AP, Knirel YA, Bengoechea JA, et al. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage φA1122. J Bacteriol. 2011;193(18):4963–72. Epub 2011/07/19. doi: 10.1128/jb.00339-11. PubMed PMID: 21764935; PubMed Central PMCID: PMCPMC3165662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Molecular microbiology. 2004;52(5):1363–73. Epub 2004/05/29. doi: 10.1111/j.1365-2958.2004.04059.x. PubMed PMID: 15165239. [DOI] [PubMed] [Google Scholar]
- 96.Hoare A, Bittner M, Carter J, Alvarez S, Zaldívar M, Bravo D, et al. The Outer Core Lipopolysaccharide of <em>Salmonella enterica</em> Serovar Typhi Is Required for Bacterial Entry into Epithelial Cells. Infect Immun. 2006;74(3):1555. doi: 10.1128/IAI.74.3.1555-1564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. International journal of medical microbiology : IJMM. 2007;297(5):277–95. Epub 2007/05/01. doi: 10.1016/j.ijmm.2007.03.012. PubMed PMID: 17466590; PubMed Central PMCID: PMCPMC1994162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pålsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113(2):153–62. Epub 2004/09/24. doi: 10.1111/j.1365-2567.2004.01976.x. PubMed PMID: 15379975; PubMed Central PMCID: PMCPMC1782563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chart H, Row B, Threlfall EJ, Ward LR. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS microbiology letters. 1989;51(1):37–40. Epub 1989/07/01. doi: 10.1016/0378-1097(89)90073-6. PubMed PMID: 2676707. [DOI] [PubMed] [Google Scholar]
- 100.Javier S, James R. Phage-resistance of Salmonella enterica serovar Enteritidis and pathogenesis in Caenorhabditis elegans is mediated by the lipopolysaccharide. Electronic Journal of Biotechnology; Vol 10, No 4 (2007). 2007. [Google Scholar]
- 101.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. Journal of endotoxin research. 2001;7(1):57–62. Epub 2001/08/25. PubMed PMID: 11521084. [PubMed] [Google Scholar]
- 102.Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Sub-cellular biochemistry. 2010;53:241–53. Epub 2010/07/02. doi: 10.1007/978-90-481-9078-2_11. PubMed PMID: 20593270; PubMed Central PMCID: PMCPMC2933746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, et al. FepA- and TonB-Dependent Bacteriophage H8: Receptor Binding and Genomic Sequence. Journal of Bacteriology. 2007;189(15):5658. doi: 10.1128/JB.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tyrrell J, Callaghan M. Iron acquisition in the cystic fibrosis lung and potential for novel therapeutic strategies. Microbiology (Reading, England). 2016;162(2):191–205. Epub 2015/12/09. doi: 10.1099/mic.0.000220. PubMed PMID: 26643057; PubMed Central PMCID: PMCPMC4772740. [DOI] [PMC free article] [PubMed] [Google Scholar]