Abstract
ARFGEF1 encodes a guanine exchange factor involved in intracellular vesicle trafficking, and is a candidate gene for childhood genetic epilepsies. To model ARFGEF1 haploinsufficiency observed in a recent Lennox Gastaut Syndrome patient, we studied a frameshift mutation (Arfgef1fs) in mice. Arfgef1fs/+ pups exhibit signs of developmental delay, and Arfgef1fs/+ adults have a significantly decreased threshold to induced seizures but do not experience spontaneous seizures. Histologically, the Arfgef1fs/+ brain exhibits a disruption in the apical lining of the dentate gyrus and altered spine morphology of deep layer neurons. In primary hippocampal neuron culture, dendritic surface and synaptic but not total GABAA receptors (GABAAR) are reduced in Arfgef1fs/+ neurons with an accompanying decrease in the number of GABAAR-containing recycling endosomes in cell body. Arfgef1fs/+ neurons also display differences in the relative ratio of Arf6+:Rab11+:TrfR+ recycling endosomes. Although the GABAAR-containing early endosomes in Arfgef1fs/+ neurons are comparable to wildtype, Arfgef1fs/+ neurons show an increase in the number of GABAAR-containing lysosomes in dendrite and cell body. Together, the altered endosome composition and decreased neuronal surface GABAAR results suggests a mechanism whereby impaired neuronal inhibition leads to seizure susceptibility.
Keywords: Lennox-Gastaut Syndrome, Epilepsy, GABAA receptor, endocytic cycle
Introduction
Epileptic encephalopathy (EE) is a group of brain disorders often with childhood onset and accompanied by serious neurocognitive consequences (Jain et al., 2013). While linkage and association studies have suggested that ARFGEF1 is involved in genetic epilepsy (Wallace et al., 1996; Piro et al., 2011; Addis et al., 2018), it was one of several candidate genes and no specific causal ARFGEF1 variants were indicted previously.
ARFGEF1 encodes Brefeldin A (BFA) inhibited guanine-nucleotide-exchange protein-1, also known as BIG1 (Addis et al., 2018); and is highly conserved across mammals and eukaryotes (Wright et al., 2014). Arfgef1 belongs to the GBF/BIG family based on the relatively large molecular size compared to other ARF-GEFs and a distinctive feature – sensitivity to Brefeldin A (BFA) (Yamaji et al., 2000). Among multiple domains, the BFA-sensitive Sec7 catalytic domain is the most studied (Le et al., 2013; Lin et al., 2013; Zhou et al., 2013). Arfgef1 selectively activates Class I ARFs to initiate conversion of ARF-GDP to ARF-GTP during the process of intracellular vesicle formation and trafficking (Zhao et al., 2002). Earlier studies also showed that the N-terminal DCB domain interacts with Arl1, targeting Arfgef1 to the trans-Golgi network (TGN) membrane (Galindo et al., 2016); the C-terminus interacts with kinesin and myosin (Saeki et al., 2005; Shen et al., 2008) as well as regulating cell surface localization of ABCA1 or GABAAR (Lin et al., 2013; Li et al., 2014). The role of Arfgef1 in the TGN has been studied in detail (Manolea et al., 2008; Boal and Stephens, 2010; Lowery et al., 2013), where it activates Arf1-GTP to recruit clathrin coats, AP1 and Arf binding proteins, also known as GGAs (Lowery et al., 2013). These vesicle-forming components with cargo-sorting capability form new vesicles and traffic cargo from TGN to endosomes or plasma membrane (Shen et al., 2006).
An earlier study suggested that BIG2 (Arfgef2) is functionally associated with endosomal integrity whereas Arfgef1 is associated with the Golgi (Shen et al., 2006). However, newer evidence suggests that Arfgef1 is also localized on endosomes (D'Souza et al., 2014). Since the main Arfgef1 target, Arf1, localizes on recycling endosomes and regulates trafficking from recycling endosome to plasma membrane (Nakai et al., 2013) and GGAs are known to facilitate trafficking between TGN and lysosomes (Hirst et al., 2000), it is reasonable to suggest that Arfgef1 may have a role in post-TGN endosomal trafficking, similar to that of BIG2. However, this aspect of Arfgef1 is understudied.
Arfgef1 regulates neurite development in vitro (Zhou et al., 2013). Recently, different groups demonstrated that Arfgef1 is required both for the survival of cortical deep layer neurons and for the initiation of neuronal myelination (Teoh et al., 2017; Miyamoto et al., 2018). Another in vitro study showed that decreased Arfgef1 abundance is coupled with decrease of plasma membrane GABAAR without affecting total GABAAR, resulting in impaired chloride ion influx (Li et al., 2014). However, it is not yet known directly whether Arfgef1 has a role in phenotypes that rely on maintaining inhibition:excitation balance in vivo.
Understanding the underlying disease etiology and mechanisms may provide insight for new therapies, especially needed for severe childhood epilepsies. Here, we examine the effect of Arfgef1 haploinsufficiency in a new mouse line based on a Lennox-Gastaut patient that carries a de novo loss of function mutation in ARFGEF1.
Results
ARFGEF1 de novo nonsense mutation in a Lennox-Gastaut Syndrome patient.
ARFGEF1 was detected as a candidate gene in a boy diagnosed with Lennox-Gastaut Syndrome. Seizures began at 6 months of age, and were subsequently diagnosed as infantile spasms. By 3 years of age, the patient exhibited generalized convulsions as well as myoclonic, tonic, and atonic seizures. Neurological exams showed global developmental delay and axial hypotonia. He had no language and no meaningful vocalizations. He was unable to pull to stand or support his own weight. General physical exam results were normal and no dysmorphic features was observed. The electroencephalogram (EEG) initially showed hypsarrhythmia and infantile spasms. Subsequent EEGs showed the emergence of generalized slow spike-and-wave activity along with paroxysmal fast activity and persistence of multifocal epileptiform discharges, superimposed on a high amplitude, disorganized background. He was treated initially with ACTH, followed by multiple medications including vigabatrin, levetiracetam, clonazepam, zonisamide, phenobarbital, lamotrigine, valproic acid, and rufinamide. He was also treated with the ketogenic diet and underwent a corpus callosotomy. Despite these interventions the seizures remained severely refractory.
Exome sequencing performed as part of the Epi4k Collaborative (Epi4K Consortium et al., 2013) revealed a single nucleotide variant resulting in a premature translational stop codon in exon 30 of the ARFGEF1 gene (ENST00000262215.3:c.4365C>A; p.Cys1455Ter). The patient also has a missense mutation in the BMP2 gene (ENST00000378827.4:c.328C>A; p.Arg110Ser), classified as “probably damaging” based on a PolyPhen-2 score of 0.968. However, since BMP2 variants have not yet been associated with neurological disease in human or mouse, and because Arfgef1 null mouse mutants have abnormal brain development (Teoh et al., 2017), we considered ARFGEF1 the more likely candidate.
Genetic analysis of Arfgef1 haploinsufficiency
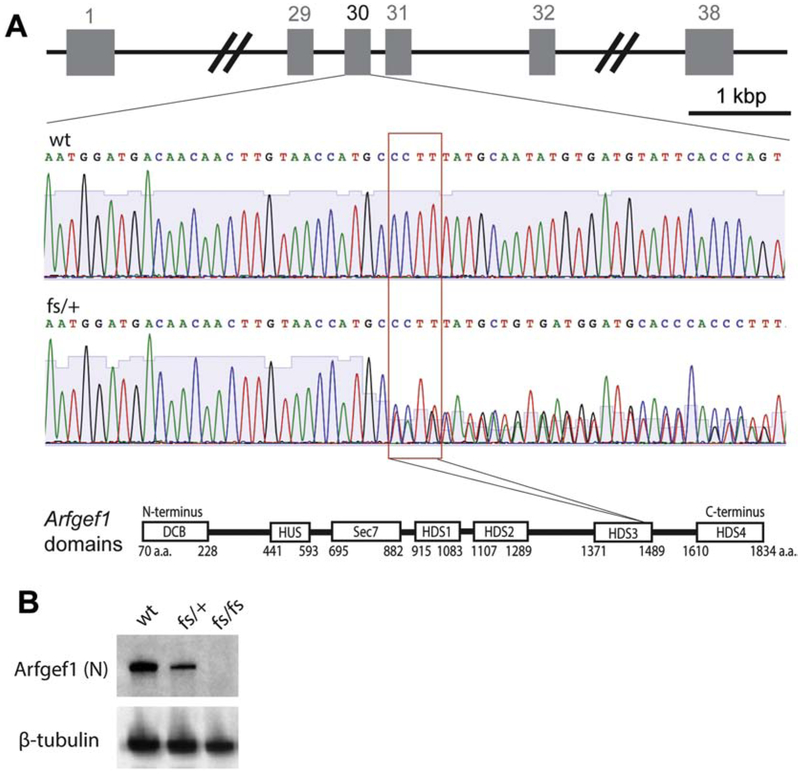
We generated an Arfgef1 knockout mouse line on the C57BL/6NJ strain background by using CRISPR/Cas9 to target the orthologous region on mouse Chromosome 1 (Fig. 1 A; Methods). Line Arfgef1em3Frk (shown hereafter as Arfgef1fs) has a 4-nt frameshift mutation in exon 30, resulting in a premature termination codon (MQYVMYSPSI*) closely mimicking the patient’s mutation. Although heterozygous mice are viable and fertile, all homozygotes died within one day of birth, similar to a previous report of the Arfgef1 knockout mouse (Teoh et al., 2017). The presence of cDNA and protein in whole brain at embryonic day 17.5 was examined using primer pairs flanking N-terminal DCB domain, Sec7 domain, exon 29-31 (mutation in exon 30) and C-terminal HDS4 domain (Fig. S1). The Arfgef1 protein was not detected in Arfgef1fs/fs whole brain lysate using Arfgef1 antibodies with N-terminal epitopes (Fig. 1B). In Arfgef1fs/+ brain, Arfgef1 protein was lower in abundance than wildtype, consistent with haploinsufficiency. The brain size of E17.5 embryos from Arfgef1fs/+ were comparable to wildtype except that the Arfgef1fs/fs forebrain was notably smaller.
Figure 1. Generation and genotyping of Arfgef1fs mice.
A, CRISPR/Cas9 targeting Arfgef1 exon 30 on mouse Chromosome 1 resulted in a 4-nt frameshift mutation (Arfgef1fs). The heterozygotes (Arfgef1fs/+) were verified with Sanger sequencing. The red box shows the sites of mutation for Arfgef1fs correspond to HDS3 domain in Arfgef1 protein. B, Western blot was performed using E17.5 brain lysate (n=3 animals per genotype) and detected using antibody targeting Arfgef1 based on an N-terminal epitope, with ß-tubulin as loading control.
Developmental delay in Arfgef1fs/+ mouse pups
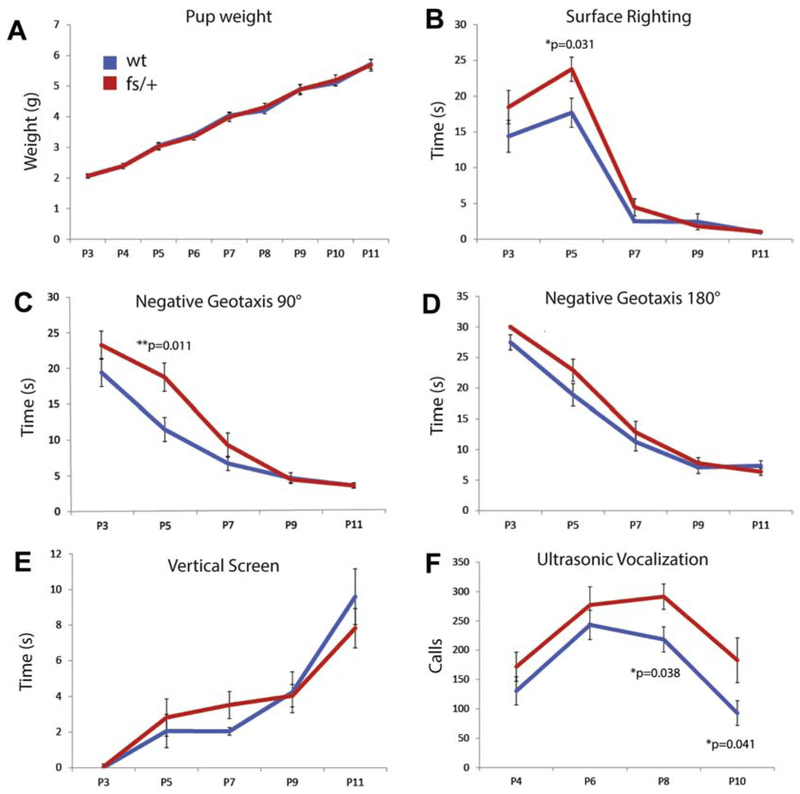
We studied the impact of the Arfgef1fs/+ genotype in mouse pups, to explore parallels with the Lennox-Gastaut Syndrome patient. Newborn pups are known to perform typical behavioral activities that serve as benchmarks for neurodevelopmental delay during postnatal development (Hill et al., 2008). Arfgef1fs/+ pup body weight did not change significantly from postnatal day (PND) 3 to PND11 (Fig. 2A), suggesting that the feeding behavior was normal. However, in the first week several milestones showed modest deficit, including latencies to surface righting reflex and negative geotaxis (Fig. 2B, C), while vertical screen holding was similar to wildtype (Fig. 2D, E). Pups younger than PND12 emit ultrasonic vocalization (USV) when separated from dam and littermates (Ferhat et al., 2016). The USV calls in wildtype pups peaked at PND6 and then gradually diminished. In contrast, the peak incidence and diminishment of USV calls was significantly delayed in Arfgef1fs/+ (Fig. 2F), suggestive of developmental delay. Representative samples of USV calls from wildtype and Arfgef1fs/+ pups are shown in Fig.S2.
Figure 2. Developmental milestone and ultrasonic vocalization of Arfgef1fs/+ pups.
A, There was no difference in body weight of Arfgef1fs/+ and wildtype pups from PND3 to PND11. B, C, The latency to surface righting or negative geotaxis (90° turn) of Arfgef1fs/+ pups is longer than wildtype at PND5. D, E, There is no difference in latency between genotypes in negative geotaxis (180° turn) or vertical screen holding. F, The incidence of maternal separation-associated ultrasonic vocalizations is higher at PND8 and PND10 for Arfgef1fs/+ compared to wildtype. Data were collected from pups of both sexes; wildtype, n=22 animals; Arfgef1fs/+, n=23 animals from 7 litters. Permutation p-values shown were determined using the Wilcoxon rank-sum test.
Morphological defects in Arfgef1fs/+ brains and neurons
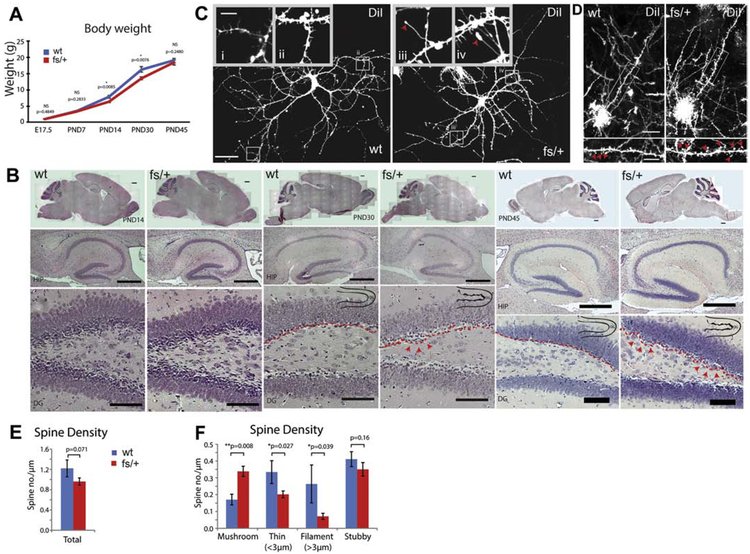
An earlier study showed that the Arfgef1 knockout heterozygotes have normal brain size during embryogenesis (Teoh et al., 2017), but the postnatal brain was not examined. First, we observed that Arfgef1fs/+ mice showed transient weight loss by PND14, and catch-up to wildtype by PND45 (Fig. 3A). Using this as a guide for histological examination, we then compared Arfgef1fs/+ and wildtype brain at PND14, PND30 and PND45 (Fig. 3B). The overall brain size of Arfgef1fs/+ was similar to wildtype, but dysregulated growth in the dentate gyrus lining of Arfgef1fs/+ hippocampus was apparent from PND30 (Fig. 3B, red arrowheads). We then examined the morphology of single neuron by diolistic labeling using DiI, a lipophilic dye that diffuses through the membrane lipid bilayer and stains the whole cell structure, first in primary neuron culture and then in brain slices. At 14 days in vitro (DIV), we observed long, filamentous and tertiary branch-like structures with larger growth cones on the dendrite of Arfgef1fs/+ primary hippocampal neurons (Fig. 3C, red arrowheads). These tertiary structures did not adhere to the culture dish surface compared to the respective primary dendrite. Larger growth cone size has been shown to correlate with slower neurite growth rate (Ren and Suter, 2016), suggesting a delay in dendritic branch maturation. In Arfgef1fs/+ brain slices, although the total number of dendritic spines (per unit length) was similar in deep layer neurons of both genotypes (Fig. 3D, 3E), the distribution of spine morphological subtypes was altered. In Arfgef1fs/+ dendrites, the fraction of mushroom-shaped spines was significantly higher than observed in wildtype (Fig. 3D, bottom inset, red arrowheads), whereas there were fewer thin spines and filopodia (Fig. 3F). Since mushroom spines on cortical deep layer neurons are typically more persistent than the immature thin spines or filopodia (Trachtenberg et al., 2002), this evidence suggested changes in dendritic spine plasticity.
Figure 3. Morphological characterization of Arfgef1fs/+ mice.
A, Body weight of Arfgef1fs/+ mice is lower at PND14 and PND30 compared to wildtype mice. E17.5, wildtype, n=16; Arfgef1fs/+, n=12 from 4 litters; PND14, 30 and 45, wildtype, n=7 mice; Arfgef1fs/+, n=8 mice from 5 litters. B, H&E-stained sagittal sections shows irregular bumps (red arrowheads) along Arfgef1fs/+ dentate gyrus lining (red dotted lines) at PND30 and PND45. n=3 animals per genotype for each time-point. Scale bars, sagittal sections, 500 μm; HIP, 500 μm; DG, 100 μm. C, In DIV14 primary hippocampal neuron culture, filamentous tertiary structures with large growth cones (red arrowheads) appeared on Arfgef1fs/+ neurons; wildtype: 4 neurons from 1 animal; Arfgef1fs/+, n=11 neurons from 3 animals. Scale bars, 50 μm; inset, 10 μm. D, Single deep layer neuron labeled using diolistic method in PND14 deep layer neurons. Mushroom spines are marked by red arrowheads. E & F, The total spine density is similar between Arfgef1fs/+ neurons and wildtype neurons. However, spine subtype density (mushroom, thin and filament) are significantly different; n=3 animals each genotype. Scale bars, 20 μm; enlarged box, 10 μm. Data were collected from mice of both sexes. HIP, hippocampus; DG, dentate gyrus.
Seizure susceptibility of Arfgef1fs/+ adult mice
In four years of breeding, Arfgef1fs/+ mice show no signs of spontaneous convulsive seizure throughout their lifetime, nor was there evidence of seizure or epileptiform activity in 48 hr video-EEG recordings of heterozygous adult mice (Fig. S3). However, both male and female heterozygotes have a significantly reduced electroconvulsive threshold to the minimal forebrain clonic seizure endpoint, compared with wildtype littermates (Wilcoxon test with permutation p=0.003 and p=0.001, respectively; Fig. 4A). They are also susceptible to tonic-clonic seizures induced by subcutaneous GABAAR antagonist pentylenetetrazol (PTZ) at a dose (40 mg/kg) near the threshold for wildtype C57BL/6NJ mice. Thus, 92.7% of Arfgef1fs/+ heterozygotes reached the tonic-clonic seizure or worse endpoint, in contrast to 18.2% of wildtype mice (p<0.0001, Fisher’s Exact Test). The average latency to the first tonic-clonic seizure was also significantly shorter in Arfgef1fs/+ mice, by an average of 788 sec (Wilcoxon test with permutation p<0.001; Fig. 4B).
Figure 4. Seizure susceptibility of Arfgef1fs/+ mice.
A, Electroconvulsive threshold to the minimal clonic seizure endpoint is significantly lower in Arfgef1fs/+ mice of both sexes (male, wildtype, n=11; Arfgef1fs/+, n= 18; female, wildtype, n=11; Arfgef1fs/+, n= 15). B, The latency to PTZ-induced tonic-clonic seizure is significantly shorter for Arfgef1fs/+ mice; wildtype, n=4; Arfgef1fs/+ n= 11. Permutation p-values shown were determined using the Wilcoxon rank-sum test.
Decreased dendritic surface GABAAR in Arfgef1fs/+ neurons
Li and colleagues (2014) previously demonstrated that siRNA-mediated depletion of Arfgef1 could reduce functional GABAAR at the membrane surface (Li et al., 2014). Given the morphological features and seizure susceptibility of Arfgef1fs/+ mice, we examined GABAAR localization in primary hippocampal neuron culture. Through immunofluorescence staining of permeabilized neurons, we determined that the total number of GABAAR puncta in the Arfgef1fs/+ proximal dendrite was similar to that of wildtype (Fig. 5A, D). However, on non-permeabilized neurons, dendritic surface-bound GABAAR puncta count decreased in Arfgef1fs/+ (Fig. 5B, E). GABAAR puncta colocalized with synaptophysin, a synaptic marker, also decreased (Fig. 5C, F), together suggesting a decrease in synaptic GABAAR density along the dendritic surface. Initial assessment of synaptic properties of hippocampal pyramidal neurons from these cultures at DIV14 showed that Arfgef1fs/+ neurons had a nominal increase in mean mEPSC frequency and no difference in mIPSC frequency or in mEPSC or mIPSC amplitude (Figure S4A, B, C).
Figure 5. Membrane surface GABAAR and synaptic GABAAR distribution in Arfgef1fs/+ neurons.
A, There is no difference in the localization of total GABAAR puncta in proximal dendrite. B, GABAAR localize on dendritic surface (representative punctum pointed by red arrowheads) at DIV14. Regions without GABAAR puncta on dendritic surface are marked by white arrowheads. Surface GABAAR, wildtype, n=19:4 (# neurons:# mice); Arfgef1fs/+ n=23:7. Total GABAAR, wildtype, n=19:4; Arfgef1fs/+, n=23:7. All scale bars, 10 μm. C, At DIV14, synaptic GABAAR puncta (white arrowheads) on the dendritic surface are fewer on Arfgef1fs/+ dendrite. wildtype, n=19:4; Arfgef1fs/+, n=12:4. All scale bars, 10 μm. D, E, Graphs show quantification of the dendritic surface GABAAR density and total GABAAR density in dendrite. F, Graph shows the synaptic GABAAR density and total synapse density on dendritic surface. Density is defined as punctum number per dendritic length in μm.
Disrupted distribution of GABAAR-containing trafficking vesicles in Arfgef1fs/+ neurons
Because the total GABAAR puncta count remained unchanged, we reasoned that the decrease in synaptic GABAAR may result from malfunction either in receptor trafficking or in recycling to the plasma membrane. That is, GABAAR might either be endocytosed but cannot be recycled to the plasma membrane (Li et al., 2014), or there is slowdown in trafficking of GABAAR from Golgi to plasma membrane.
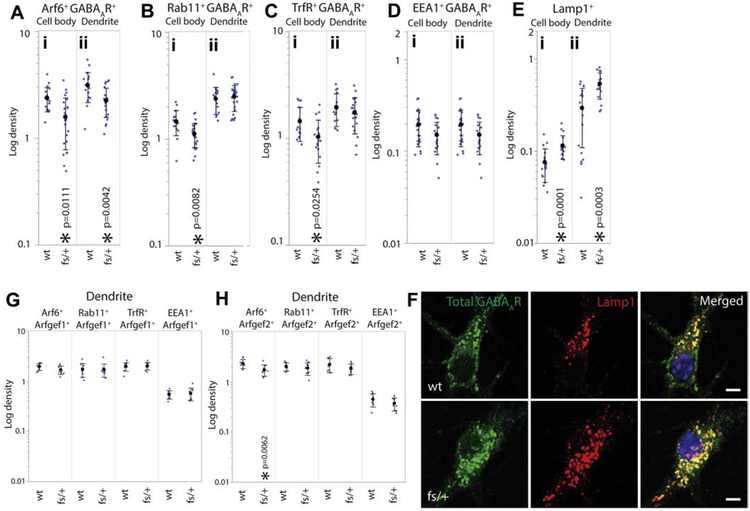
There is significant heterogeneity in the type and subcellular localization of recycling endosomes in neurons, and this heterogeneity is in part represented by Arf6+, Rab11+, TrfR+ markers. For example, Arf6+ but not Rab11+ and TrfR+ recycling endosomes respond to nerve growth factor (NGF) stimulation, suggesting functional differences in recycling endosome species (Kobayashi and Fukuda, 2013). Immunofluorescent staining of primary hippocampal neurons showed that Arf6+, Rab11+ and Trf+ puncta colocalized with GABAAR+ puncta in dendrite and cell body (Fig.S5A, B, C). Vesicle-GABAAR colocalization was quantified using object-based colocalization (see Methods). We found that the number of Arf6+, Rab11+ and Trf+ vesicles each decreased in the Arfgef1fs/+ neuronal cell body (Fig. S6Ai, Bi, Ci), as were those that colocalized with GABAAR (Fig. 6Ai, Bi, Ci). These results suggest either an increase in the dendritic GABAAR recycling, leading to reduction in retrograde trafficking into the cell body, or increase in the recycling endosome that mature into late endosomes. The former is less likely because we observed less dendritic surface-bound GABAAR. Findings in dendrites further reinforced this idea: in Arfgef1fs/+ dendrites, Arf6+ recycling endosomes decreased in number compared to wildtype (Fig. S6Aii), including those that colocalized with GABAAR (Fig. 6Aii). In contrast, the number of Rab11+ and TrfR+ recycling endosomes (Fig. S6Bii, Cii) including those colocalized with GABAAR (Fig. 6Bii, Cii) in Arfgef1fs/+ dendrite were comparable to wildtype. These results suggest that Arfgef1 haploinsufficiency not only reduces the number of Arf6+, Rab11+, TrfR+ recycling endosomes in cell body, but also specifically affects Arf6-dependent dendritic GABAAR recycling.
Figure 6. Distribution of intracellular vesicles colocalized with GABAAR in wildtype and Arfgef1fs/+ DIV14 neurons.
A, B, C, D, E, Graphs show the density of GABAAR colocalized with Arf6+, Rab11+, TrfR+ recycling endosomes, EEA1+ early endosomes and Lamp1+ lysosomes in cell body and dendrite. Arf6+ GABAAR+, wildtype, n=19:4 (# neurons:# mice); Arfgef1fs/+, n=23:7. Rab11+ GABAAR+, wildtype, n=16:4; Arfgef1fs/+, n=27:7; TrfR+ GABAAR+, wildtype, n=15 :4; Arfgef1fs/+, n=18:3; EEA1+ GABAAR+, wildtype, n=21:6; Arfgef1fs/+, n=19:5; Lamp1+ GABAAR+, wildtype, n=18:6; Arfgef1fs/+, n=18:5. F & G, Graphs show the density of Arf6+, Rab11+, TrfR+ and EEA1+ vesicles colocalized with Arfgef1 or Arfgef2 in dendrite. For each vesicle type plus Arfgef1 or Arfgef2 data set, wildtype, n=3:3; Arfgef1fs/+, n=3:3. H, The number of Lamp+ lysosomes and GABAAR-containing lysosomes increase in Arfgef1fs/+ neurons. All scale bars, 5 μm. Density is shown in log scale.
It is useful to consider the impact of Arfgef1 genotype on the relative ratio of Arf6+, Rab11+, and TrfR+ recycling endosomes, because of the interelationship between these microorganelles. Pairwise comparisons of mutant to wildtype showed that the recycling endosome ratio was significantly skewed in Arfgef1fs/+ dendrite (p=0.006) but not in cell body (p=0.252). The imbalance in recycling endosome populations in dendrite could be the reason for the decrease of dendritic surface GABAAR.
The early endosome is an intermediate organelle, which can either turn into recycling endosomes or mature into lysosomes (Naslavsky and Caplan, 2018). Earlier evidence showed that Arfgef1 also localizes on early endosomes and functions similarly as Arfgef1 localizes in the trans-Golgi network (D'Souza et al., 2014). Reduction in the number of early endosomes in the Arfgef1fs/+ dendrite could explain the decrease in recycling endosomes. Immunofluorescent staining of primary hippocampal neurons showed that EEA1+ puncta colocalized with GABAAR+ puncta in cell body and dendrite (Fig. S5D). However, when compared to wildtype, there were no changes in the number of EEA1+ early endosomes in dendrite or cell body of Arfgef1fs/+ neurons (Fig. S6Di, ii), or those that colocalize with GABAAR (Fig. 6Di, ii).
Multiple studies in non-neuronal cells demonstrated that Arfgef2 but not Arfgef1 colocalizes with Arf6+, Rab11+, TrfR+ or EEA1+ endosomes (Togawa et al., 1999; Ishizaki et al., 2008; D'Souza et al., 2014). In DIV 14 primary hippocampal neurons, we found that both Arfgef1 (Fig. S7A, B, C, D) and Arfgef2 (Fig.S7E, F, G, H) puncta overlapped with Arf6+, Rab11+, TrfR+ or EEA1+ puncta. The number of Arfgef1+ puncta colocalized with Arf6+, Rab11+, TrfR+ or EEA1+ puncta were not different between Arfgef1 genotypes (Fig. 6F). However, the number of Arfgef2+ puncta that colocalized with Arf6+, but not Rab11+, TrfR+ and EEA1+, is significantly lower than wt in Arfgef1fs/+ dendrites (Fig. 6G). This suggests that Arfgef2 might compensate for the reduction of Arfgef1 at Golgi in the cell body.
Accumulation of GABAAR in lysosomes in Arfgef1fs/+ neurons
The decrease of surface GABAAR accompanied by decrease in recycling endosomes suggest that the internalized receptors might have been targeted elsewhere. It has been shown that internalized GABAAR that fail to recycle to the plasma membrane are directed to lysosomes for degradation (Kittler et al., 2004). This led us to ascertain whether reduction in surface GABAAR was accompanied by lysosomal accumulation of GABAAR. Indeed, in cell body and dendrite of Arfgef1fs/+ neurons, Lamp1+ lysosome number (Fig. S6Ei,ii), and those colocalized with GABAAR (Fig.6Ei, ii, H) increased compared to wildtype. This showed that GABAAR in Arfgef1fs/+ neurons were redirected to lysosomes instead of being recycled to the plasma membrane.
Discussion
Arfgef1fs/+ mice were generated to model a Lennox-Gastaut Syndrome patient heterozygous for a de novo mutation that creates a premature translational stop in the Arfgef1 C-terminus, and thus a haploinsufficient state via presumed nonsense-mediated decay. Arfgef1fs/+ mice do not have spontaneous convulsions, as is sometimes the case with genotypically accurate mouse models of childhood epilepsy that either do not have spontaneous seizures (Frankel et al., 2009; Amendola et al., 2014) or have seizures of a different type than described in the respective patients (Warner et al., 2016; Kovacevic et al., 2018). However, Arfgef1fs/+ do have a low threshold to induced seizures, indicating clear seizure susceptibility. Their abnormal performance in developmental milestone tasks, modest but significant, also suggests global developmental delay and possibly hypotonia, like the patient. The predicted maturation rate for mouse is 150 times faster than human in the first month. Thus, the age at which the Arfgef1fs/+ developmental features are most apparent, between ages PND5 and PND7, is equivalent to a 2-3 year child (Flurkey et al., 2007). This very rapid postnatal maturation in mouse may also partly account for compensatory neurodevelopmental features that prevent a more severe outcome, such as spontaneous seizures.
Arfgef1fs/+ animals have a normal lifespan, whereas Arfgef1fs/fs die perinatally; in agreement with the earlier report of Arfgef1 knockout mice (Teoh et al., 2017), except the forebrain defect evident at E17.5 in Arfgef1fs/fs mice, is more severe. This difference could be due merely to genetic background: the earlier study did not use an inbred strain but rather a mixed 129/SvJ;C57BL/6J background in which hybrid vigor may have mitigated severity.
The GABAAR is a membrane-bound ligand-gated ion channel selective for chloride ions (Moroni et al., 2011). Accumulation of GABAAR at the synaptic site is required for inhibitory synapse formation and for fast synaptic inhibition (Bogdanov et al., 2006). Other than being trafficked from Golgi to plasma membrane (Luscher et al., 2011), surface GABAA receptors also undergo constitutive endocytosis and then are recycled back to the membrane surface or delivered to lysosome for degradation (Kittler et al., 2000). Our results suggest that both copies of Arfgef1 are required to maintain normal endosome composition, which is in turn critical for receptor recycling.
Arl1 recruits Arfgef1 to early endosomes (D'Souza et al., 2014). Internalized membrane surface receptors fed into early endosomes have several fates. Surface receptors stay in early endosomes if they are not quickly recycled to plasma membrane from early endosomes or delivered into recycling endosomes for slow recycling. The early endosomes with surface receptors that are not destinied for recycling then mature into lysosomes for degradation (Kittler et al., 2004; Naslavsky and Caplan, 2018). In Arfgef1fs/+ neurons, there was no change in dendritic early endosome number but Arf6+ recycling endosomes were available albeit fewer in number, suggesting that less early endosomes are destined for recycling – as supported by our observation of decreased GABAAR on the Arfgef1fs/+ dendrite surface; these vesicles mature into lysosomes, which are more abundant in the Arfgef1fs/+ cell body.
The imbalance of recycling and early endosome populations may also explain why there are more lysosomes in Arfgef1fs/+. AP1 and GGAs, recruited by the Arfgef to the trans-Golgi network, are required to regulate lysosomal enzyme trafficking from the trans-Golgi to lysosomes (Le Borgne and Hoflack, 1998; Hirst et al., 2000; Lowery et al., 2013). Depletion of Arfgef1 alone does not affect the level of GGAs in the trans-Golgi network (Boal and Stephens, 2010; Miyamoto et al., 2018). It has also been demonstrated that the number of synaptic GABAARs increase in GGA-deficient mice (Walker et al., 2016). However, depletion of Arfgef1 reduces the formation of AP-1 complex. Notably, such depletion does not entirely eliminate AP-1 (Miyamoto et al., 2018). Since Arfgef2 can compensate for Arfgef1 in the trans-Golgi network (Ramaen et al., 2007), AP-1 and GGAs levels are required for maintaining the network through to lysosomal trafficking, thus the degradation rate would be be maintained in Arfgef1fs/+ neurons. In our study, although there is significantly less dendritic surface-bound GABAAR, some GABAAR can still be transported to the membrane surface. Thus, the reduction of surface GABAAR is most likely due to reduced recycling, leading to increased GABAAR accumulation in lysosomes.
We can only speculate why Arfgef1 haploinsufficiency in mouse is less severe than the human disease it models, or why more ARFGEF1 variants have not been found in epilepsy. Lennox-Gastaut Syndrome is a genetically heterogeneous complex childhood epilepsy, if a primary gene is known at all. Arfgef1 homozygous knockouts are very severely impaired. One possibility is that a very delicate balance of gene dosage, combined with a limited window of late gestational or early postnatal vulnerability, allows the more rapid mouse postnatal development to push through. A more discrete possibility is that ARFGEF1 is actually a modifier mutation, requiring another predisposing variant for severe disease. In multiple prior studies, ARFGEF1 was reported as a candidate together with at least one other gene. For example, in a large Australian family spanning three generations in which CRH had been proposed as a EE candidate gene (Wallace et al., 1996), Piro and colleagues later suggested that ARFGEF1 was also plausible (Piro et al., 2011). In another study of copy number variants (CNV), the authors reported an EE patient with onset of 10 years with ARFGEF1 and CSPP1 as candidate genes (Addis et al., 2018). In our study, the Lennox-Gastaut patient also carried a nonsynonymous BMP2 variant; while BMP2 is not yet associated with epilepsy at least one study has shown that BMP2 signaling promotes the differentiation of some GABAergic neurons (Yao et al., 2010). Regardless, the specific impact of Arfgef1 haploinsufficiency on endosomal balance and dendritic vesicle fate is an attractive pathogenic mechanism and therapeutic target opportunity that may apply to other forms of childhood epilepsy.
Materials and Methods
Generation of Arfgef1 mice
All mice breeding and husbandry were done within the Institute of Comparative Medicine in Columbia University. The mice were kept under a 12-hr day-night cycle with unlimited access to water and food. Mice at two to six-months-old were used for mating. All animal-related procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Columbia University. The germline mutation mice were generated in the Genetic Engineering and Technology Core at The Jackson Laboratory (Bar Harbor, ME). In mouse, the Arfgef1 gene is on Chromosome 1 and consists of 38 exons. Briefly, a guide RNA directing Cas9 endonuclease together with a single-stranded repair template targeting exon 30 was injected into single-cell mouse embryos of C57BL/6NJ line. Founder line were genotyped using PCR (primer F3, 5’-GTCTGAAGTGAAGCACGTTGG-3’ and primer R3, 5’-CAGTGGGGTCAACGTGTTATG-3’), restriction enzyme digestion (MwoI; New England Biolabs, R0573L) and DNA sequencing (primer F2, 5’-GTGGCTAGAGAGGCTCGTTTT-3’). Identified founder mice were crossed with wildtype C57BL/6NJ, resulting Arfgef1fs line (4-nt deletion in exon 30).
RT-PCR
Total RNA was collected from wildtype and Arfgef1fs/+ brains at embryonic day 17.5 (E17.5) using TRIzol reagent (Ambion, 15596026). Reverse transcription was performed using Superscript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, 11752-050) according to the manufacturer's protocol. PCR was then performed using primers flanking Arfgef1 cDNA corresponding to the exons 3-6 (N-terminal DCB domain), exons 13-17 (Sec7 domain), exons 29-31 (mutation site) and exons 31-38 (C-terminal HDS4 domain) with primerd flaking actin as control. The PCR products were subjected to electrophoresis on 2% agarose gels. The primers used in this experiment were listed below.
DCB domain
Forward: 5'- CAC CCT TCC ACC AGT GAA ATC A -3'
Reverse: 5'- TGC TGC CGA TGT CTT TCT CTT TC -3'
Amplicon size: 495bp
Sec7 domain
Forward: 5'- GGA ATT GGC AGC TAC AGT ACA CAG -3'
Reverse: 5'- CCT GTG GAC TGT GAA GGT CTG -3'
Amplicon size: 505bp
Exon 29-31
Forward: 5'- GAC AAC ATG AAA TTG CCA GAA CAG C -3'
Reverse: 5'- GAT ATC CAG GGT GCA GTT ACA CG -3'
Amplicon size: 285bp
HSD4 domain
Forward: 5'- GCT GGA ACT CAT CCA GAC CAT C -3'
Reverse: 5'- TGA GCT TTA AAC CTG CTG TCA CTG -3'
Amplicon size: 524bp
Actin
Forward: 5'- ACG ATA TCG CTG CGC TGG -3'
Reverse: 5'- GAG CAT CGT CGC CCG C -3'
Amplicon size: 72bp
Antibodies
The following primary antibodies were used: Anti-Arfgef1 N-terminal epitope (ThermoFisher Scientific, PA5-54894; Santa Cruz, sc-376790; sc-50391), anti-Arfgef1 C-terminal epitope (Bethyl, A300-998A), anti-Arfgef2 C-terminal epitope (Sigma-Aldrich, MABS1246), Anti-GABAA Receptor with epitope for extracellular domain of the β2,3 subunits (Millipore Sigma, MAB341), anti-synaptophysin (ThermoFisher Scientific, PA1-1043), anti-lamp1 (DSHB, 1D4B), anti-eea1 (R&D Systems, AF8047), anti-transferrin receptor (Abcam, ab84036), anti-Rab11 (ThermoFisher Scientific, 71-5300), anti-Arf6 (ThermoFisher Scientific, PA1-093), anti-map2 (Millipore Sigma, Ab5622), anti-ankG (Santa Cruz, sc-28561), anti-ß-tubulin (Proteintech, 10094-1-AP and 66240-1-Ig), anti-GAPDH (Proteintech, 60004-1-Ig), anti-lamin B1 (Proteintech, 66095-1-lg). The following secondary antibodies were used: Goat anti-mouse HRP-conjugated IgG (Proteintech, SA00001-1), goat anti-rabbit HRP-conjugated IgG (Proteintech, SA00001-2), Goat anti-mouse Alexa Fluor 488-conjugated IgG (ThermoFisher Scientific, A11017), goat anti-rabbit Alexa Fluor 594-conjugated IgG (ThermoFisher Scientific, A11072), goat anti-rat Alexa Fluor 594-conjugated IgG (ThermoFisher Scientific, A11007), donkey anti-sheep NL557-conjugated IgG (R&D Systems, NL010).
Western blotting
Brain lysates were collected using the trichloroacetic acid precipitation method. Briefly, the lysates were separated by SDS-PAGE and transferred to Immobilon-PSQ PVDF Membrane (Millipore Sigma, ISEQ00010). The membranes were incubated with primary antibodies overnight at 4°C, followed by incubation with the appropriate HRP-conjugated secondary antibody for 1 hr at room temperature. Samples were washed three times in PBS with 0.05% Tween-20 (PBST) after each step. Signals were developed using Amersham ECL Western Blotting Detection Reagent (GE Healthcare, RPN2106) and visualized using western blot imaging system (Azure Biosystems, Azure C400).
Video-EEG
Mice of both sexes, aged 5 weeks and 8 weeks were anesthetized through intraperitoneal injection of 400mg/kg 2,2,2-Tribromoethanol (Sigma Aldrich, T48402). Surgery was performed to drill four burr holes. The first two holes located 1 mm anterior to bregma on both sides. The third hole located 2 mm posterior to bregma on the left side. The fourth hole located over the cerebellum as reference. Four teflon-coated silver wires soldered on a microconnector (Mouser electronics, 575-501101) were placed between dura mater and pia mater and a dental cap was applied. Post-operative mice were given 5mg/kg carprofen (Zoetis, Rimadyl injectable) subcutaneously and a recovery time of 48 hours before electroencephalography (EEG) recording. The recovered mice were then connected to Natus Quantum programmable amplifier (Natus Neurology, 014160) for 24 hours over one day-night cycle. The mice activity was video-recorded simultaneously using a night-vision-enabled Sony IPELA EP500 camera. Differential amplification recordings were recorded pair-wise between the three electrodes and a referential electrode, resulted in a montage of 6 channels for each mouse. The EEG data was analyzed using Natus Database v8.5.1.
Seizure susceptibility tests
Electroconvulsive threshold testing was performed in mice 7-10 weeks of age as described previously with minor modification(Frankel et al., 2001)s (Frankel et al., 2001). Briefly, a drop of a topical anesthetic (0.5% tetracaine in 0.9% NaCl) was placed on each eye of a restrained mouse, and current was applied using silver transcorneal electrodes connected to an electroconvulsive stimulator (Ugo Basile model 7801) using the following parameters: 299 Hz frequency 1.6 ms pulse width, 0.2 s pulse duration, variable current. Mice were tested approximately daily in 0.5 mA increments until the minimal clonic forebrain seizure endpoint was reached. For analysis the mean integrated root mean square (iRMS) current is reported for each genotype-sex group.
Chemical seizure susceptibility was performed using subcutaneous injection of mice between 9 and 10 weeks of age using pentylenetetrazol (PTZ), a GABAA receptor antagonist. Mice were observed for 30 min and incidence of and latency to the first tonic-clonic seizure endpoint was recorded. The PTZ dose of 40 mg/kg was predetermined to be near the threshold for the tonic-clonic seizure endpoint in C57BL/6NJ mice of this age.
The Wilcoxon non-parametric rank-sum test with 1000 permutations was used in R to determine permutation p-values for elecroconvulsive threshold and for latency to PTZ-induced tonic-clonic seizure.
Developmental milestones and ultrasonic vocalization
All pups from the same litter were kept with respective mother throughout the experimental period. They were numbered with tattoo on the soles at PND2 and evaluated for neurological reflexes on PND 3, 5, 7, 9 and 11. Each subject was tested at the same three-hour time window of a day. The pups were weighted each day during the test. For righting reflex test, the pup was placed on its back on a flat and hard surface. The time a pup took to right itself on all four paws was recorded. For negative geotaxis test, the pup was placed with head facing downward on a 20 × 20 cm flat wire mesh screen slanted at 45° angle. The time a pup took to turn around and face the angle of 90° and 180° from the starting downward direction was recorded. For vertical screen grasping test, the pups were placed on a 20 × 20 cm flat wire mesh standing vertically at an angle of 90° to a flat surface. The time a pup was able to maintain itself on the mesh wire was recorded. All the times were recorded using a stopwatch for a maximum of 30 seconds (s). All test were carried out under room temperature. The pups were returned to the mother immediately after the tests.
Ultrasonic vocalization from a pup being isolated from its mother and littermates was recorded on PND 4, 6, 8 and 10. Briefly, a random pup was removed gently from its nest into an isolation container filled with clean bedding. The container was then placed into a sound attenuating chamber equipped with an UltraSoundGate Condenser Microphone CM 16 (Avisoft Bioacoustics, 40011). The microphone connected to an UltraSoundGate 116 USB audio device (Avisoft Bioacoustics) was linked to a computer installed with Avisoft RECORDER v2.97, where the acoustic data was recorded at a sampling rate of 250,000 Hz in 16 bit format. The software was programmed to filtered away acoustic frequency lower than 15 kHz to reduce background noise interference. Each pup was recorded for 3 min, weighted, and then returned to nest with mother immediately. The number of calls emitted by the pups was counted by an independent operator blinded from the genotypes. The Wilcoxon non-parametric rank-sum test with 1000 permutations was used in R to determine permutation p-values.
Hematoxylin and eosin staining
Mice of respective ages were perfused. The brains were carefully extracted and cut along the lambda-bregma points. Pieces of brains were fixed in 4% (w/v) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB) at pH 7.4 overnight at 4°C. After serial dehydration steps, the brains were embedded in paraffin (Sigma, P3808). The brains were sectioned at 5 μm thickness using Leica RM2125 microtome. Every tenth section were collected on a glass slide (Matsunami Glass, SUMGP14) and subjected to HE staining. Briefly, the slices were rehydrated and stained with hematoxylin then counterstained with eosin. These slices were subsequently dehydrated using ethanol and xylene and mounted with coverslip using Permount (Fisher Chemical, SP15-500). Histological images were acquired using a Nikon Eclispse E800M light microscope and NIS-elements v4.51. Images were merged using automated function in Photoshop CC 2015.
Ex vivo single neuron diolistic labeling and spine analysis
The postnatal day (PND) 14 brains were collected and fixed as described above. These brains were transferred into fresh 2% PFA in 0.1 M PB at pH 7.4 and stored at 4°C for up to two months before use. The brains were embedded in 4% agarose and shot with a gene gun (Biorad, 1652451) loaded with homemade bullets consisting tungsten microbeads (Biorad, 1652267) coated with DiI (Invitrogen, D282) at 80 PSI. After gentle washing, the brains were immersed in fresh 2% PFA in 0.1 M PB at pH 7.4 and stored in a dark moisten chamber for 3 days at room temperature to allow dye diffusion. Finally, the brains were sliced at 300uM using a vibrotome (Leica, VT1200) and mounted with coverslip using glycerol. Z-stacks images were acquired immediately using Zeiss LSM-800 confocal microscope and Zen v2.3. The quantification and morphological analysis of spines were done using NeuronStudio v0.9.92. In agreement with literature, the spine shapes were categorized as filopodia, thin, mushroom and stubby (Qiao et al., 2016). Spines with a neck are classified as thin or mushroom based on the head diameter. Spines longer than 3 mm were classified as filopodia. Spines without a neck are classified as stubby.
Primary hippocampal neuron culture
Primary hippocampal neurons were collected according to Fath’s method with little modification (Fath et al., 2009). Briefly, hippocampi were dissected from embryonic day (E) 17.5 embryos, dissociated through trituration with the aid of papain (Worthington, LK003178) pre-incubation. A total of 1 × 105 cells was plated in 35 mm culture dish containing acid-treated coverslips coated with poly-D-lysine (Sigma-Aldrich, P7886) and laminin (Gibco, 23017015). The primary hippocampal neurons were kept in culture medium containing Neurobasal-A medium (Gibco, 10888022) supplemented with B27 (Gibco, 17504044) and Glutamax (Gibco, 35050061). Cytosine β-D-arabinofuranoside (Sigma-Aldrich, C1768) was added once to a final concentration of 3 μM on the next day after 50% medium change. The cultures were maintained at 37°C with 5% CO2 in a humidified incubator. Every three days, 50% expensed medium were replaced with fresh culture medium.
Immunofluorescence
DIV14 neurons were fixed for 10 min at 4°C with 4% (w/v) PFA in 0.1 M PB at pH 7.4. The fixed neurons were incubated in blocking buffer containing 5% goat serum (Gibco, 16210064) and 0.1% saponin (Sigma-Aldrich, 47036) in PBS pH 7.4 (Gibco, 10010031) at room temperature for 30 min. Blocked neurons were subsequently incubated with primary antibodies in blocking buffer at 4°C overnight followed by incubation with appropriate secondary antibodies and DAPI in blocking buffer at room temperature for 2 hrs. Samples were washed three times in PBS after each step. The stained neurons on coverslips were mounted on glass slides using Fluoromount-G (Southern Biotech, 0100-01) and sealed with nail polish after drying at room temperature overnight. Z-stacks images were acquired using Zeiss LSM-800 confocal microscope and Zen v2.3. The quantification and analysis of labeled vesicle and GABAaR puncta were done using Imaris v9.2.1 software. Due to the limitation of the point spread function (PSF) for tiny objects in confocal microscopy, colocalization is quantified using an object-based method (Dunn et al., 2011). Briefly, vesicle and GABAaR puncta stained with antibodies in cell body or dendrite are recognized and segmented in different channels using Imaris v9.2.1. If a punctum in green channel and a punctum in red channel localize within 1 μm proximity (center to center) the signal pair will be counted as colocalized. There were no significant differences found in the dendrite length and the cell body area where the data were collected. Therefore, the number of puncta in dendrite was normalized using dendrite length (μm) and the number of puncta in cell body was normalized using cell body area (μm2). The normalized numbers were presented as density (in dendrite, vesicle number per μm; in cell body, vesicle number per μm2).
For statistical analysis of puncta counts, a log-Poisson mixed model was run using the lme4 package in R (www.R-project.org), with cell body area or dendrite length as a respective covariate and a random effects term included to correct for significant overdispersion in the count data. To determine whether endosome ratios differed between mutant genotype and wildtype, genotype × vesicle type interaction terms were included in a similar model which also included a fixed term for the individual biological from which ratios examined. Statistical comparisons in this study were considered significant if the p-value is less than 0.05.
Single cell patch-clamp recordings
To assess synaptic activity, electrophysiological recordings were carried out from dissociated hippocampal neurons plated at a density of 50000 cells on 15 mm diameter glass coverslips and cultured for 13-15 days. Recordings were made in voltage clamp whole-cell configuration using a Multiclamp 700B amplifier and a Digidata 1550 digital-to-analogue converter (both from Molecular Devices) at a 10 kHz sample frequency. Patch pipettes were fabricated with a P-97 pipette puller (Sutter Instruments) using 1.5 mm outer diameter, 1.28 mm inner diameter filamented capillary glass (World Precision Instruments). The external recording solution contained (in mM): NaCl 145, KCl 5, HEPES 10, Glucose 10, CaCl2 2, MgCl2 2, 0.001 tetrodotoxin, pH 7.3 with NaOH, and osmolality adjusted to 325 mOsm using sucrose. A cesium-based pipette solution contained (in mM): cesium methanesulfonate 130, sodium methanesulfonate 10, EGTA 10, CaCl2 1, HEPES 10, TEA-Cl 10, MgATP 5, Na2GTP 0.5, QX-314 5, pH 7.2 with CsOH, adjusted to 290 mOsm with sucrose. Pipettes had final resistance of approximately 5 MΩ when filled with internal solution. Uncompensated series resistance was <15 MΩ. Series resistance and membrane capacitance were electrically compensated to approximately 70%. Miniature excitatory postsynaptic currents (mEPSCs) and miniature inhibitory postsynaptic currents (mIPSCs), were recorded from the same cell by holding the cells at −60 and 0 mV, respectively. Recordings were acquired for a period of 5 minutes. All recordings were carried out at room temperature (21 – 23 °C). Miniature events were detected offline with Clampfit 12.7 (Molecular Devices) using the template matching function and a minimum threshold of 5 pA. Each event was manually inspected to determine inclusion or rejection in analysis. Further analysis was carried out using R (www.R-project.org).
Supplementary Material
Expression of Arfgef1 RNA. Representative electrophoresis gel images of PCR products (n=3 animals per genotype). The forward (F) and reverse (R) PCR primers flanking the DCB domain, Sec7 domain, exons 29-31 and HDS4 domain of Arfgef1 cDNA were used for PCR. PCR yielded the expected amplicons as shown.
Ultrasonic vocalization (USV) of Arfgef1fs/+ and wildtype pups. Representative USV recordings containing three discrete calls from Arfgef1fs/+ and wildtype pups at PND4, 8, 10 and 12 are shown. See Fig.2 legend for other details.
Electroencephalogram (EEG) recording. EEG recordings from one wildtype and two Arfgef1fs/+ mice showing no obvious seizure or epileptiform activity. These recordings are representative of mice at 5 weeks of age (n=4 animals each genotype); and 8-weeks, (n=3 each).
Synaptic activity of Arfgef1fs/+ hippocampal neurons. A, Examples of membrane traces showing mEPSCs and mIPSCs recorded from wildtype and Arfgef1fs/+ cultures. B & C, Box plots of frequency and amplitude for mESPC and mIPSC. Each point represents the frequency or amplitude of synaptic currents recording from a single neuron. Box indicates median, first and third quartiles. Whiskers extend to the lowest/highest value within the 1.5 × IQR of hinge. wildtype, n=19 neurons (4 mice); Arfgef1fs/+, n=15 neurons (4 mice).
Colocalization of GABAA receptor and intracellular vesicles. A, B, C, D, Optical slices captured using confocal microscopy show the colocalization (white arrowheads) of GABAAR+ puncta and Arf6+, Rab11+, TrfR+ or EEA1+ puncta in the cell body and dendrite. Scale bars, whole neuron, 10 μm; inset, 2 μm.
Quantification of intracellular vesicles. A, B, C, D, Graphs show the density of Arf6+, Rab11+, TrfR+ and EEA1+ puncta in the cell body or dendrite. Sample sizes are mentioned in Fig. 6.
Colocalization of Arfgef1 or Arfgef2 and intracellular vesicles. A, B, C, D, E, F, G, H, Optical slices captured using confocal microscope show the colocalization (white arrowheads) of Arfgef1+ or Arfgef2+ puncta and Arf6+, Rab11+, TrfR+ or EEA1+ puncta in the dendrite. Scale bars, whole neuron, 10 μm; inset, 2 μm.
Highlights.
Arfgef1fs/+ mice have lower seizure threshold but no spontaneous seizure.
Arfgef1fs/+ neurons show reduced dendritic surface GABAAR expression.
Arfgef1fs/+ neurons have fewer GABAAR-containing recycling endosomes and an increase in GABAAR-containing lysosomes.
Acknowledgements
This research is supported by NIH grant R37 NS031348 to WNF, NIH grant U01HG0009610 to JB and NIH/NIA grant RO1AG050658 to FB. We are grateful to Erin Heinzen-Cox, David Goldstein, Daniel Lowenstein and the EPGP and Epi4k project teams for bringing the ARFGEF1 variant to our attention, to Osasumwen Virginia Aimiuwu for support in RNA reverse transcription and to Ryan Dhindsa for his R script for determining permutation statistics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
WNF consults for Praxis Precision Medicines, Inc.. JB consults for Upsher-Smith and GW Pharmaceuticals.
References
- Addis L, Sproviero W, Thomas SV, Caraballo RH, Newhouse SJ, Gomez K, Hughes E, Kinali M, McCormick D, Hannan S, Cossu S, Taylor J, Akman CI, Wolf SM, Mandelbaum DE, Gupta R, van der Spek RA, Pruna D, Pal DK (2018) Identification of new risk factors for rolandic epilepsy: CNV at Xp22.31 and alterations at cholinergic synapses. J Med Genet 55:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola E, Zhan Y, Mattucci C, Castroflorio E, Calcagno E, Fuchs C, Lonetti G, Silingardi D, Vyssotski AL, Farley D, Ciani E, Pizzorusso T, Giustetto M, Gross CT (2014) Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS One 9:e91613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boal F, Stephens DJ (2010) Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS One 5:e9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov Y, Michels G, Armstrong-Gold C, Haydon PG, Lindstrom J, Pangalos M, Moss SJ (2006) Synaptic GABAA receptors are directly recruited from their extrasynaptic counterparts. EMBO J 25:4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza RS, Semus R, Billings EA, Meyer CB, Conger K, Casanova JE (2014) Rab4 orchestrates a small GTPase cascade for recruitment of adaptor proteins to early endosomes. Curr Biol 24:1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH (2011) A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300:C723–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium et al. (2013) De novo mutations in epileptic encephalopathies. Nature 501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath T, Ke YD, Gunning P, Gotz J, Ittner LM (2009) Primary support cultures of hippocampal and substantia nigra neurons. Nat Protoc 4:78–85. [DOI] [PubMed] [Google Scholar]
- Ferhat AT, Torquet N, Le Sourd AM, de Chaumont F, Olivo-Marin JC, Faure P, Bourgeron T, Ey E (2016) Recording Mouse Ultrasonic Vocalizations to Evaluate Social Communication. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Currer JM, Harrison DE (2007) The Mouse in Aging Research. [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS (2001) Electroconvulsive thresholds of inbred mouse strains. Genomics 74:306–312. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Yang Y, Mahaffey CL, Beyer BJ, O'Brien TP (2009) Szt2, a novel gene for seizure threshold in mice. Genes Brain Behav 8:568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo A, Soler N, McLaughlin SH, Yu M, Williams RL, Munro S (2016) Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi. Cell Rep 16:839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JM, Lim MA, Stone MM (2008) Developmental Milestones in the Newborn Mouse. In: Neuropeptide Techniques (G I, ed), pp 131–149: Humana Press. [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS (2000) A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol 149:67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki R, Shin HW, Mitsuhashi H, Nakayama K (2008) Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol Biol Cell 19:2650–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P, Sharma S, Tripathi M (2013) Diagnosis and management of epileptic encephalopathies in children. Epilepsy Res Treat 2013:501981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler JT, Delmas P, Jovanovic JN, Brown DA, Smart TG, Moss SJ (2000) Constitutive endocytosis of GABAA receptors by an association with the adaptin AP2 complex modulates inhibitory synaptic currents in hippocampal neurons. J Neurosci 20:7972–7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler JT, Thomas P, Tretter V, Bogdanov YD, Haucke V, Smart TG, Moss SJ (2004) Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking. Proc Natl Acad Sci U S A 101:12736–12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Fukuda M (2013) Arf6, Rab11 and transferrin receptor define distinct populations of recycling endosomes. Commun Integr Biol 6:e25036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic J, Maroteaux G, Schut D, Loos M, Dubey M, Pitsch J, Remmelink E, Koopmans B, Crowley J, Cornelisse LN, Sullivan PF, Schoch S, Toonen RF, Stiedl O, Verhage M (2018) Protein instability, haploinsufficiency, and cortical hyper-excitability underlie STXBP1 encephalopathy. Brain 141:1350–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Hoflack B (1998) Mechanisms of protein sorting and coat assembly: insights from the clathrin-coated vesicle pathway. Curr Opin Cell Biol 10:499–503. [DOI] [PubMed] [Google Scholar]
- Le K, Li CC, Ye G, Moss J, Vaughan M (2013) Arf guanine nucleotide-exchange factors BIG1 and BIG2 regulate nonmuscle myosin IIA activity by anchoring myosin phosphatase complex. Proc Natl Acad Sci U S A 110:E3162–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen S, Yu Y, Zhou C, Wang Y, Le K, Li D, Shao W, Lu L, You Y, Peng J, Huang H, Liu P, Shen X (2014) BIG1, a brefeldin A-inhibited guanine nucleotide-exchange factor, is required for GABA-gated Cl(−) influx through regulation of GABAA receptor trafficking. Mol Neurobiol 49:808–819. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhou C, Neufeld E, Wang YH, Xu SW, Lu L, Wang Y, Liu ZP, Li D, Li C, Chen S, Le K, Huang H, Liu P, Moss J, Vaughan M, Shen X (2013) BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein modulates ATP-binding cassette transporter A-1 trafficking and function. Arterioscler Thromb Vasc Biol 33:e31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery J, Szul T, Styers M, Holloway Z, Oorschot V, Klumperman J, Sztul E (2013) The Sec7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 guanine nucleotide exchange factors to the trans-Golgi network (TGN). J Biol Chem 288:11532–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, Kilpatrick CL (2011) GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70:385–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolea F, Claude A, Chun J, Rosas J, Melancon P (2008) Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol Biol Cell 19:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Torii T, Tago K, Tanoue A, Takashima S, Yamauchi J (2018) BIG1/Arfgef1 and Arf1 regulate the initiation of myelination by Schwann cells in mice. Sci Adv 4:eaar4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni M, Biro I, Giugliano M, Vijayan R, Biggin PC, Beato M, Sivilotti LG (2011) Chloride ions in the pore of glycine and GABA channels shape the time course and voltage dependence of agonist currents. J Neurosci 31:14095–14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W, Kondo Y, Saitoh A, Naito T, Nakayama K, Shin HW (2013) ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol Biol Cell 24:2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S (2018) The enigmatic endosome - sorting the ins and outs of endocytic trafficking. J Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro RM, Molineris I, Ala U, Di Cunto F (2011) Evaluation of candidate genes from orphan FEB and GEFS+ loci by analysis of human brain gene expression atlases. PLoS One 6:e23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Li MX, Xu C, Chen HB, An SC, Ma XM (2016) Dendritic Spines in Depression: What We Learned from Animal Models. Neural Plast 2016:8056370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaen O, Joubert A, Simister P, Belgareh-Touze N, Olivares-Sanchez MC, Zeeh JC, Chantalat S, Golinelli-Cohen MP, Jackson CL, Biou V, Cherfils J (2007) Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem 282:28834–28842. [DOI] [PubMed] [Google Scholar]
- Ren Y, Suter DM (2016) Increase in Growth Cone Size Correlates with Decrease in Neurite Growth Rate. Neural Plast 2016:3497901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki N, Tokuo H, Ikebe M (2005) BIG1 is a binding partner of myosin IXb and regulates its Rho-GTPase activating protein activity. J Biol Chem 280:10128–10134. [DOI] [PubMed] [Google Scholar]
- Shen X, Xu KF, Fan Q, Pacheco-Rodriguez G, Moss J, Vaughan M (2006) Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci U S A 103:2635–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Meza-Carmen V, Puxeddu E, Wang G, Moss J, Vaughan M (2008) Interaction of brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 and kinesin motor protein KIF21A. Proc Natl Acad Sci U S A 105:18788–18793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HW, Morinaga N, Noda M, Nakayama K (2004) BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell 15:5283–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh JJ, Iwano T, Kunii M, Atik N, Avriyanti E, Yoshimura SI, Moriwaki K, Harada A (2017) BIG1 is required for the survival of deep layer neurons, neuronal polarity, and the formation of axonal tracts between the thalamus and neocortex in developing brain. PLoS One 12:e0175888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M (1999) Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem 274:12308–12315. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K (2002) Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420:788–794. [DOI] [PubMed] [Google Scholar]
- Walker KR, Modgil A, Albrecht D, Lomoio S, Haydon PG, Moss SJ, Tesco G (2016) Genetic Deletion of the Clathrin Adaptor GGA3 Reduces Anxiety and Alters GABAergic Transmission. PLoS One 11:e0155799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Berkovic SF, Howell RA, Sutherland GR, Mulley JC (1996) Suggestion of a major gene for familial febrile convulsions mapping to 8q13-21. J Med Genet 33:308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner TA, Shen W, Huang X, Liu Z, Macdonald RL, Kang JQ (2016) Differential molecular and behavioural alterations in mouse models of GABRG2 haploinsufficiency versus dominant negative mutations associated with human epilepsy. Hum Mol Genet 25:3192–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J, Kahn RA, Sztul E (2014) Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci 71:3419–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji R, Adamik R, Takeda K, Togawa A, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M (2000) Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci U S A 97:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Niu G, Sheng Z, Wang Z, Fei J (2010) Identification of a Smad4/YY1-recognized and BMP2-responsive transcriptional regulatory module in the promoter of mouse GABA transporter subtype I (Gat1) gene. J Neurosci 30:4062–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Lasell TK, Melancon P (2002) Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol Biol Cell 13:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Li C, Li D, Wang Y, Shao W, You Y, Peng J, Zhang X, Lu L, Shen X (2013) BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein regulates neurite development via PI3K-AKT and ERK signaling pathways. Neuroscience 254:361–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of Arfgef1 RNA. Representative electrophoresis gel images of PCR products (n=3 animals per genotype). The forward (F) and reverse (R) PCR primers flanking the DCB domain, Sec7 domain, exons 29-31 and HDS4 domain of Arfgef1 cDNA were used for PCR. PCR yielded the expected amplicons as shown.
Ultrasonic vocalization (USV) of Arfgef1fs/+ and wildtype pups. Representative USV recordings containing three discrete calls from Arfgef1fs/+ and wildtype pups at PND4, 8, 10 and 12 are shown. See Fig.2 legend for other details.
Electroencephalogram (EEG) recording. EEG recordings from one wildtype and two Arfgef1fs/+ mice showing no obvious seizure or epileptiform activity. These recordings are representative of mice at 5 weeks of age (n=4 animals each genotype); and 8-weeks, (n=3 each).
Synaptic activity of Arfgef1fs/+ hippocampal neurons. A, Examples of membrane traces showing mEPSCs and mIPSCs recorded from wildtype and Arfgef1fs/+ cultures. B & C, Box plots of frequency and amplitude for mESPC and mIPSC. Each point represents the frequency or amplitude of synaptic currents recording from a single neuron. Box indicates median, first and third quartiles. Whiskers extend to the lowest/highest value within the 1.5 × IQR of hinge. wildtype, n=19 neurons (4 mice); Arfgef1fs/+, n=15 neurons (4 mice).
Colocalization of GABAA receptor and intracellular vesicles. A, B, C, D, Optical slices captured using confocal microscopy show the colocalization (white arrowheads) of GABAAR+ puncta and Arf6+, Rab11+, TrfR+ or EEA1+ puncta in the cell body and dendrite. Scale bars, whole neuron, 10 μm; inset, 2 μm.
Quantification of intracellular vesicles. A, B, C, D, Graphs show the density of Arf6+, Rab11+, TrfR+ and EEA1+ puncta in the cell body or dendrite. Sample sizes are mentioned in Fig. 6.
Colocalization of Arfgef1 or Arfgef2 and intracellular vesicles. A, B, C, D, E, F, G, H, Optical slices captured using confocal microscope show the colocalization (white arrowheads) of Arfgef1+ or Arfgef2+ puncta and Arf6+, Rab11+, TrfR+ or EEA1+ puncta in the dendrite. Scale bars, whole neuron, 10 μm; inset, 2 μm.