Abstract
Segmentation of lungs with acute respiratory distress syndrome (ARDS) is a challenging task due to diffuse opacification in dependent regions which results in little to no contrast at the lung boundary. For segmentation of severely injured lungs, local intensity and texture information, as well as global contextual information, are important factors for consistent inclusion of intrapulmonary structures. In this study, we propose a deep learning framework which uses a novel multi-resolution convolutional neural network (ConvNet) for automated segmentation of lungs in multiple mammalian species with injury models similar to ARDS. The multi-resolution model eliminates the need to tradeoff between high-resolution and global context by using a cascade of low-resolution to high-resolution networks. Transfer learning is used to accommodate the limited number of training datasets. The model was initially pre-trained on human CT images, and subsequently fine-tuned on canine, porcine, and ovine CT images with lung injuries similar to ARDS. The multi-resolution model was compared to both high-resolution and low-resolution networks alone. The multi-resolution model outperformed both the low- and high-resolution models, achieving an overall mean Jacaard index of 0.963±0.025 compared to 0.919±0.027 and 0.950±0.036, respectively, for the animal dataset (N = 287). The multi-resolution model achieves an overall average symmetric surface distance of 0.438 ± 0.315 mm, compared to 0.971 ± 0.368 mm and 0.657 ± 0.519 mm for the low-resolution and high-resolution models, respectively. We conclude that the multi-resolution model produces accurate segmentations in severely injured lungs, which is attributed to the inclusion of both local and global features.
Keywords: Computed tomography, Segmentation, Deep learning
Graphical Abstract

1. Introduction
X-ray computed tomography (CT) produces high-resolution volumetric reconstructions of anatomy. The intensity values in a CT image reflect the density of the tissue, producing high contrast between low-density lungs and the surrounding soft tissue. High-resolution CT images allow for intricate visualization of lung texture, vasculature, and airway segments. CT imaging is routinely utilized for diagnosing lung pathologies, guiding treatment, monitoring progression, and characterizing lung diseases.
The acute respiratory distress syndrome (ARDS) is associated with severe respiratory failure in the presence of diffuse inflammation, increased pulmonary vasculature permeability, and loss of lung tissue aeration (Coppola et al., 2017). Radiographically, this condition presents with diffuse bilateral opacification in the dependent lung (Gattinoni et al., 2001; Cereda et al., 2019). While a plain chest x-ray image can confirm the diagnosis of ARDS, it may not provide specific information on the locus of injury or its spatial heterogeneity. CT thus has more clinical utility for diagnostic imaging in ARDS, since it can differentiate pathological phenotypes and provide information regarding treatment response (Gattinoni et al., 2006), as well as characterize spatial heterogeneity of injury and regional mechanical properties (Kaczka et al., 2011; Perchiazzi et al., 2014; Fernandez-Bustamante et al., 2009; Gattinoni et al., 2006; Paula et al., 2016; Carvalho et al., 2008). Quantitative CT (qCT) of the lung enables objective assessment of injury, in contrast to the subjective interpretation of a radiologist. QCT has also been used for evaluating response to mechanical ventilation (Black et al., 2008; Godet et al., 2018; Perchiazzi et al., 2011) and monitoring injury progression (Cereda et al., 2016, 2017). Spatial and temporal heterogeneity of parenchymal tissue strain in ARDS can also be measured through registration of dynamically imaged lungs (Herrmann et al., 2017) or lungs imaged at distending pressures (Kaczka et al., 2011; Perchiazzi et al., 2014; Cereda et al., 2017).
In general, CT imaging is not easily nor often performed on humans with ARDS. Patients with ARDS are often critically ill and it is too risky to transfer the patient to a CT scanner. CT imaging is not required to diagnose ARDS, but rather is used most often clinically to assess complications such as infection, pneumothorax, pulmonary embolism, etc. Therefore, use of quantitative CT in ARDS is currently more prevalent in research settings with animal models of ARDS. We anticipate that quantitative CT may become a more valuable clinical tool as automated image analysis software is developed and validated experimentally. For example, there is growing support for use of CT scans to facilitate differentiation of ARDS phenotypes, or guide patient-specific optimal ventilation settings (Cereda et al., 2019; Zompatori et al., 2014; Sheard et al., 2012).
A necessary precursor to the application of qCT in ARDS is lung segmentation, which distinguishes intrapulmonary tissues and structures from the surrounding chest wall and mediastinum. Intensity-based segmentation methods are widely used for CT images of the thorax, since there is normally high contrast between the air-filled lungs and surrounding tissues (Brown et al., 1997; Kemerink et al., 1998; Hu et al., 2001; Ukil & Reinhardt, 2004). However these methods fail to include dense pathologies, such as non-aerated edematous and atelectatic regions commonly observed in ARDS. Lungs with ARDS are particularly challenging to segment, since the injury pattern is often diffuse and predominates in posterior dependent regions in supine subjects. Lungs with peripheral injury patterns are particularly more challenging to segment compared to those with more interior patterns, because there is little to no contrast between the injured parenchyma and non-pulmonary structures. Furthermore, consolidated regions have no textural features that make it distinguishable from the surrounding soft tissue.
Several studies have investigated different segmentation techniques for injured lungs. Shape prior methods rely on modeling the variations in lung shape and are better suited for segmentation of injured lungs compared to intensity-based methods. Sun et al. (2012) proposed a 3D robust active shape model to approximate a lung segmentation in lung cancer subjects. The model is initialized using rib cage information and refined used an optimal surface finding graph search (Li et al., 2006). Similarly Sofka et al. (2011) use a statistical shape model which is initialized using automatically detected landmarks on the carina, ribs, and spine. Soliman et al. (2016) proposed a joint 3D Markov-Gibbs random field (MGRF) model which combines an active shape model with first and second order appearance models to segment normal and pathological lungs. Segmentation-by-registration is another shape-prior-based approach which uses image registration to find the mapping between the image to be segmented and one or more images, i.e. an atlas, with known segmentations. A semiautomatic approach using segmentation-by-registration in longitudinal images of rats with surfactant depletion was proposed by Xin et al. (2014). A limitation of this approach is the requirement of manual segmentation of baseline (i.e., pre-injury) CT scan. Moreover their method relies on a relatively time-consuming image registration algorithm, requiring 4 to 6 hours for completion. Similar segmentation-by-registration approaches have also been proposed (Sluimer et al., 2005; Zhang et al., 2006; van Rikxoort et al., 2009; Pinzón et al., 2014; Pinzón, 2016). Anatomic information from the airways and rib cage has been used to identify the boundaries between injured parenchyma and surrounding soft tissue in injured lungs (Cuevas et al., 2009). A wavelet-based approach was proposed by Talakoub et al. (2007). However their method may fail for severely injured lungs, for which no discernible parenchymal boundary information is present.
Manual segmentation is still widely used for segmentation of lungs with acute injury or ARDS, since current automated methods are not reliable. However, manual segmentation is tedious, time-consuming, and subject to high intra- and inter-observer variability. Furthermore, manual lung segmentations are typically performed on 2D transverse slices of the thorax, which limits global context and produces segmentations that may not appear smooth in sagittal or coronal sections. For large datasets, such as dynamic (i.e., 4D) CT images with multiple phases or time points, manual segmentation is not practical. Thus, accurate and efficient segmentation of injured lungs remains a major obstacle limiting the clinical use of qCT in ARDS and other heterogeneous lung pathologies.
Recently, deep learning with convolutional neural networks (ConvNets) has dominated a wide range of applications in computer vision, with the ability to perform image classification, localization, segmentation, and registration that at times surpass human-level accuracy. Deep learning enables computers to learn robust and predictive features directly from raw data, rather than relying on explicit rule-based algorithms or learning from human-engineered features. Deep learning systems are also more robust and computationally efficient. Within the specialized field of medical imaging, ConvNets successfully detect skin cancer (Esteva et al., 2017), classify lung nodules (Shen et al., 2015), and segment various anatomic and pathological structures (Prasoon et al., 2013; Li et al., 2014; Ronneberger et al., 2015; Anthimopoulos et al., 2016; Shin et al., 2016). A recent survey on deep learning applied to medical imaging is given by Litjens et al. (2017).
A major challenge limiting the use of ConvNets for medical image segmentation is computing hardware. Graphical processing units (GPUs) are essential for efficient ConvNet training, but current GPUs have limited memory. Training ConvNet models using high-resolution volumetric images requires prohibitively large amounts of GPU memory. As a compromise, most methods extract 2D slices or 3D patches with local extent. Figure 1 illustrates several approaches to low-memory image representation using various degrees of downsampling and/or cropping. Approaches relying on aggressive cropping will sacrifice global information and 3D smoothness, in favor of high anatomic resolution. By contrast, downsampling approaches preserve global context at the expense of small-scale features. Another challenge for deep learning in medical imaging is the limited availability of labeled training data. Deep learning methods require large training datasets to fit millions of free model parameters. Expert annotation of training data is time-consuming and laborious, especially for volumetric medical images, which typically have upwards of 500 2D slices for a single thoracic CT scan. Furthermore, the pathological derangements associated with rare diseases yield very small cohorts of subjects.
Figure 1:
Given an image with size and resolution as illustrated in (a), where the grid represents the voxel lattice, (b)-(e) illustrate techniques for subsampling the image in order to reduce the memory requirement for training a ConvNet on a GPU. (b) downsampling reduces the number of voxels by combining intensity values of multiple voxels, thereby decreasing the image resolution. In this example, a downsampling factor of four is used for each dimension, i.e., the size of the image is reduced from 163 to 43 voxels. If the downsampling factor sufficiently large, the memory requirement can be reduced enough to utilize the full image extent during training on a GPU. (c)-(e) cropping uses the original image resolution, however, only a portion of the image voxels are extracted as denoted by the regions that have gridlines in (c)-(e). (c) the slab has full extent in two dimensions, but limited extent in the third dimension. (d) the 3D patch has limited extent in all three spatial dimensions. (e) the 2D slice has full extent in two spatial dimensions, however, no 3D context is available. Note, (b) and (d) representations are both 43 voxels, and thus require the same memory, however, (b) has global extent with low resolution, whereas (d) has local extent with high resolution. The text below each figure describes the relative image resolution, spatial extent, and required memory for each scenario.
These challenges need to be addressed for successful application of ConvNets for segmentation of ARDS lungs. Global contextual information, such as the surrounding anatomic features, is necessary for segmentation of injured lungs, since local intensity is non-distinguishable from surrounding tissue. Current methods that use 2D slices or 3D patches are not ideal, as these do not consider global features. Furthermore, limited annotated training data of ARDS lungs is available due to the time necessary to produce manual segmentations. The main contributions of this work are:
A multi-resolution ConvNet model which has the capacity to learn both local and global features in large volumetric medical images.
Fully automated and computationally efficient segmentation of injured lungs obtained from CT imaging.
Segmentation of lungs across multiple mammalian species using limited annotated training data.
The multi-resolution ConvNet cascade makes use of both low-resolution and high-resolution models to enable multi-scale learning. In a pilot study, we proposed a multi-resolution ConvNet model and evaluated the model on a small dataset of porcine subjects with acute lung injury (Gerard et al., 2018). In this work, we further evaluate the multi-resolution model using an extensive dataset consisting of porcine, canine, and ovine subjects. Given the wide variability of animal species and large imaging datasets used in experimental ARDS research, the ability of an automated lung segmentation algorithm to generalize across species is critical for widespread utility. Furthermore, we show the utility of the multi-resolution model through comparison with two other ConvNet-based models. We hypothesized that a model incorporating both global and local information would be superior to models that use only local or global information for this type of injury. The importance of global and local information is explored by comparing the proposed model to a conventional high-resolution model which uses image slabs, and a low-resolution model which uses aggressive downsampling. The high-resolution model sacrifices global contextual information in favor of high-resolution information whereas the low-resolution model sacrifices high-resolution detail in favor of global context. A transfer learning approach is used which allows training the model using a limited amount of mammalian ARDS training data, after first pre-training the model using an extensive image database of human lungs without ARDS.
2. Datasets and Reference Standards
We utilized a dataset consisting of CT scans from four different species: human, canine, porcine, and ovine. Hereinafter the collection of human images is referred to as the human dataset, while the collection of canine, porcine, and ovine images is referred to as the animal dataset. The human dataset was used to pre-train the model, while the animal dataset was used for fine-tuning. The human dataset consisted of 3418 CT scans, including 3113 subjects with chronic obstructive pulmonary disease (COPD) and 305 subjects with idiopathic pulmonary fibrosis (IPF). The animal dataset consisted of 301 scans of subjects with various experimental models of ARDS: 76 scans of canine subjects with an oleic acid injury, 152 scans of porcine subjects with an oleic acid injury, 27 scans of ovine subjects with a saline lavage injury, and 46 scans of ovine subjects with a lipopolysaccharide (LPS) injury.
2.1. Human Dataset
The human dataset consisted of CT scans acquired from three large-scale clinical trials: COPDGene, SPIROMICS, and PANTHER-IPF.. COPDGene is a large multi-institutional clinical trial studying genetics and imaging biomarkers of COPD subjects (Regan et al., 2011). The subset of COPDGene subjects used in this study had images acquired at total lung capacity (TLC), functional residual capacity (FRC), and residual volume (RV). TLC scans were acquired at 120 kVp and 200 mAs. FRC and RV scans were acquired at 120 kVp and 50 mAs. SPIROMICS is also a multi-institutional clinical trial studying subpopulations and intermediate outcomes in COPD subjects (Couper et al., 2014; Sieren et al., 2016; Woodruff et al., 2016). The SPIROMICS subjects used in this study had images acquired at TLC and RV (Sieren et al., 2016). The IPF dataset was obtained from an ancillary study of PANTHER-IPF (Idiopathic Pulmonary Fibrosis Clinical Research Network et al., 2012, 2014). This study used high-resolution CT images to identify IPF textural features and their relations to disease progression (Salisbury et al., 2017). This dataset consisted of scans acquired at TLC (Salisbury et al., 2017).
2.2. Animal Dataset
2.2.1. Porcine Dataset
The porcine dataset was obtained from a study of alternative mechanical ventilation modalities to treat ARDS, approved by the University of Iowa Institutional Animal Care and Use Committee. Pigs approximately 10 to 15 kg in size were scanned under baseline conditions and following maturation of acute lung injury induced by infusion of oleic acid into the superior vena cava. 3DCT images were acquired during breath-hold maneuvers at constant airway pressures of 0, 5, 10, 15, 20, 25, and 30 cmH2O. 4DCT images were acquired during mechanical ventilation using three ventilator modalities: conventional pressure-controlled ventilation, high-frequency oscillatory ventilation, and multi-frequency oscillatory ventilation (Kaczka et al., 2015; Herrmann et al., 2017). All images were acquired using a Siemens Somatom Force scanner, with 120 kVp, 90 mA s, and 0.5 mm slice thickness for 3DCT, or 80 kVp, 150 mAs, and 0.6 mm slice thickness for 4DCT. The 4DCT images have a limited axial coverage of 5.76 cm, which excludes the apices and bases of the lungs.
2.2.2. Canine Dataset
The canine dataset was obtained from a study of respiratory mechanics in subjects with acute lung injury (Kaczka et al., 2011), approved by the Johns Hopkins University Institutional Animal Care and Use Committee. Dogs approximately 22 to 33 kg in size were scanned under baseline conditions and following maturation of acute lung injury induced by infusion of oleic acid into the pulmonary artery. 3DCT images were acquired during breath-hold maneuvers at constant airway pressures of 0, 5, 10, 15, and 20 cmH2O. Images were acquired using a Siemens Somatom Sensation 16-slice scanner, with 137 kVp, 165 mA s, and 2.5 mm slice thickness.
2.2.3. Ovine Dataset 1
The first ovine dataset was obtained from a study of prone vs. supine positioning to treat subjects with ARDS, approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. Sheep approximately 20 to 30 kg in size were scanned following acute lung injury induced by saline lavage. 3DCT images were acquired during breath hold maneuvers at inflation levels corresponding to end-expiration (PEEP 5 cmH20), end-inspiration (tidal volume 8 mL/kg), and mean airway pressure during mechanical ventilation. Images of prone sheep were rotated 180 degrees to align anatomical features in the corresponding supine orientation. Images were acquired using a Siemens Biograph combined PET-CT scanner, with 120 kVp, 80 mA s, and 0.5 mm slice thickness.
2.2.4. Ovine Dataset 2
The second ovine dataset was obtained from a study of subjects with ARDS, approved by the Johns Hopkins University and University of Iowa Institutional Animal Care and Use Committees (Fernandez-Bustamante et al., 2012). Subjects approximately 25 to 45 kg in size were scanned under baseline conditions and following acute lung injury induced by intravenous infusion of lipopolysaccharide (LPS). 3DCT images were acquired using respiratory-gated CT imaging at inflation levels corresponding to end-expiration and end-inspiration during mechanical ventilation. Images were acquired using a Siemens Somatom Sensation 16- or 64-slice scanner, with 120 kVp, 250 or 180 mA s, and 1.5 or 1.2 mm slice thickness.
2.2.5. Ground Truth
Manual segmentations were generated semi-automatically using Pulmonary Analysis Software Suite (PASS, University of Iowa Advanced Pulmonary Physiomic Imaging Laboratory (Guo et al., 2008)), and then manually corrected by an expert image analyst. The PASS software uses a conventional intensity-based lung segmentation followed by large airway removal and hole-filling using morphological operations. Diffuse injured regions of the lung were not included in the automated segmentation and thus required manual inclusion. Manual correction was performed using 3D Slicer software (Fedorov et al., 2012). The expert image analysts were instructed to include injured regions using the rib cage for guidance and narrow HU windowing for enhanced boundary visualization when necessary. The analysts were instructed to include pulmonary vasculature once it enters the lungs to create a smooth boundary at the mediastinum. The vena cava was excluded from the segmentation as it only passes through the lung and is not involved in pulmonary circulation, although it is fully surrounded by the parenchyma of the accessory lobe in porcine, canine, and ovine species. Tracings were primarily done on axial slices, however, sagittal and coronal views were also used to provide 3D context and consistent boundary identification across axial slices. Five expert image analysts were involved with manual corrections. Depending on the injury severity manual correction took anywhere from 4 to 6 hours per case.
3. Methods
3.1. Overview
A multi-resolution ConvNet model was proposed for the lung segmentation of the CT images (Section 3.3.3), designed to handle severely injured lungs across multiple mammalian species. The multi-resolution model was compared to the performances of the low-resolution (Section 3.3.1) and high-resolution (Section 3.3.2) models alone. All models used the same underlying ConvNet architecture, referred to as Seg3DNet (Section 3.2). However, the spatial resolution of the training data was varied. Training images with different spatial resolutions result in different ranges of feature scales learned by each model. Due to the limited number of scans for each species in the animal dataset, transfer learning from the human dataset was used for training all models (Section 3.4).
3.2. Convolutional Neural Network
The underlying ConvNet architecture used in each of the three models was a fully convolutional network (FCN) called Seg3DNet (Gerard et al., 2019), see Figure 2. The network has an encoder and decoder module, similar to the popular U-Net architecture (Ronneberger et al., 2015). However, Seg3DNet was extended to three spatial dimensions and the decoder was designed to use less GPU memory. The input and output to the network were both images with three spatial dimensions of the same size. The input image was transformed to increasingly abstract image representations using a hierarchy of network layers. Each intermediate image representation has three spatial dimensions and a fourth dimension representing different feature types. Henceforth, we refer to the fourth dimension as the channel dimension, analogous to that of RGB images. The output of Seg3DNet is an image with |Y| channels, where Y is the class set. The task of lung segmentation is treated as a binary segmentation process (i.e., |Y| = 2), where the classes correspond to lung tissue and background.
Figure 2:
Seg3DNet architecture consists of an encoder and decoder network. The encoder network learns a multi-scale image representation by transforming the input image using convolutional and max pooling (i.e. downsampling) layers. The decoder network combines the image representations at each scale to produce the output prediction of voxel-wise classification probability using deconvolution (i.e. upsampling) and convolutional layers. Each image representation (illustrated as cubes) is a 3D image with N channels (denoted in lower left of each cube). This can also be thought of as N 3D images, or N activation maps, where each activation map represents a different feature type. Different layer operations are represented as arrows, e.g. convolutional layers are represented as thick black arrows.
The encoder network consists of L resolution levels, where each resolution level l ∈ {0, …, L−1} consists of two convolutional layers followed by a maximum pooling layer. The number of filters used in each convolutional layer at level l is defined as Nl = 2l+5 such that the number of channels, or activation maps, increases by a factor of two at each level. The decoder network upsampled the image representation at the end of each level, back to the input image resolution using deconvolution layers, and combined the multi-scale features using two subsequent convolutional layers. Each voxel in the output image was a floating point number corresponding to the probability that the voxel was part of the lung field.
Convolutional layers use kernels with spatial extent of 3 × 3 × 3 voxels. Zero-padding was used such that the spatial size of the image representation remained unchanged. Maximum pooling with a kernel size of 2 × 2 × 2 voxels and stride of 2 × 2 × 2 voxels was used which effectively spatially downsampled the image representation by a factor of two along each spatial dimension, with the number of feature maps remaining unchanged. Batch normalization and a rectified linear unit (ReLu) activation function was used after each convolutional layer, with the exception of the last layer. The last layer used a softmax vector nonlinearity (Equation 1). The output of the softmax function yielded the conditional probability distribution that a voxel x belongs to each class y ∈ Y.
| (1) |
For a binary classification, the predicted probability distribution can unambiguously be represented as a single floating-point number. The predicted probability that x belonged to the lung was denoted as . The predicted probability that x belongs to the background is therefore .
3.3. Models
3.3.1. Low-Resolution Model
The low-resolution model consisted of a single Seg3DNet, which was trained using aggressively downsampled CT images and lung segmentations. All training images were downsampled to constant image size of 64 × 64 × 64 voxels, regardless of image size. This corresponded to a downsampling factor of roughly eight along each spatial dimension. At this size, the entire image can be entered as an input to the network, which allowed for global features to be learned. However, exact boundary information is lost with downsampling. Images from the 4D porcine dataset, which were reconstructed as slabs, were padded with axial slices prior to downsampling to avoid large deformation of the lung due to large differences in the axial and transverse image extents. By comparison, the other datasets exhibited less differences between axial and transverse image extents, resulting in varying degrees of lung deformation and differences in proportion of lung to non-lung when resizing all images to a fixed image size. These variations were allowed in the training data to make the model robust to different scanning and reconstruction parameters and thereby avoid initial preprocessing steps. Gaussian smoothing was performed prior to downsampling to avoid aliasing. The intensity range was clipped to obtain values between −1024 and 1024 HU and then normalized to have a mean of zero and standard deviation of one. The output of the low-resolution model was then upsampled to the original resolution using b-spline interpolation.
3.3.2. High-Resolution Model
The high-resolution model consisted of a single Seg3DNet, which was trained using high-resolution CT images. The CT images were resampled to isotropic voxel sizes: 1 mm voxels for humans, dogs, and sheep and 0.6 mm for pigs. Resampling was performed to achieve consistent voxel sizes and relative anatomical size scales between scans and species. This corresponded to a downsampling factor of less than two along each dimension. At this resolution, the entire CT image was too large for GPU memory. Therefore, axial slabs of size 256×256×32 were sampled at multiple axial positions for training the model. This limited the amount of global context that could be learned by the high-resolution model, since specific anatomic features from the entire lung field could not be learned. The intensity range was clipped to obtain values between −1024 and 1024 HU and then normalized to have a mean of zero and standard deviation of one.
3.3.3. Multi-Resolution Model
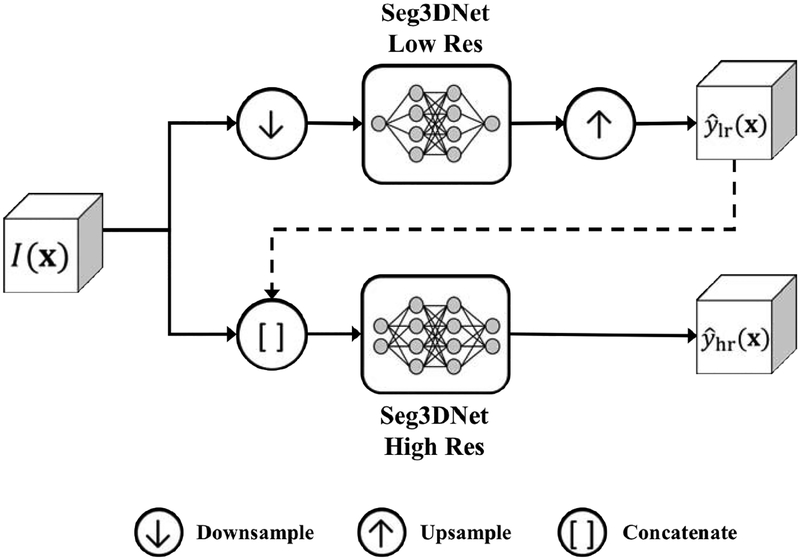
The multi-resolution model consisted of two Seg3DNets, utilizing both low-resolution and high-resolution models, which were linked to allow information learned by the low-resolution model to be exploited by the high-resolution model. The two Seg3DNets were trained sequentially. In the first stage, the low-resolution model was trained on aggressively downsampled images as described in Section 3.3.1. In the second stage, the high-resolution model was trained, similarly to the model described in Section 3.3.2. However, the low-resolution model prediction was included in the input, in addition to the high-resolution image. Combining the low-resolution and high-resolution models eliminated the necessity of choosing between global contextual information and precise boundary detail when training with limited GPU memory. The multi-resolution model is illustrated in Figure 3.
Figure 3:
Multi-resolution model. The upper pipeline corresponds to the low-resolution model and the lower pipeline corresponds to the high-resolution model. Global information learned in the low-resolution model was used in the high-resolution model, denoted by the dashed line.
3.4. Training
Transfer learning (Oquab et al., 2014) was used for training all models. A model was first pre-trained using the human training dataset. The model learned from the human dataset was then used to initialize the animal model. Fine-tuning of the network was then performed using the animal dataset.
Due to the limited number of animal scans with lung injury, a five-fold cross validation was performed for training the animal model. The animal dataset was split into five groups (approximately 60 images per group), four of which were used for training. The model performance was evaluated on the remaining group. This sequence was performed five times, allowing all images in the animal dataset to be used for both training and evaluation. Each of the five groups had the same number of images and approximately equal representation for each species. Five-fold cross validation was performed for training all models, using identical splitting to obtain fair comparisons.
A binary cross entropy loss function was used for training. The loss for each voxel x was given by
| (2) |
where y(x) is the true class label for voxel x, y(x) = 1 for lung and y(x) = 0 for background, and is the predicted probability that voxel x belongs to the lung class. The total loss for each image was obtained as the average loss over all voxels in the image. The loss function was optimized with respect to the free parameters (the convolution kernels), using standard backpropagation. Adam optimization (Kingma & Ba, 2014) was used for training, with a learning rate of 5 × 10−4 for pre-training and 5 × 10−5 for fine-tuning. Prior to pre-training, all free parameters were initialized using Xavier normal initialization (Glorot & Bengio, 2010). The networks were trained using a P40 NVIDIA GPU with 24 GB RAM. Total training time was approximately 48 hours for each model.
3.5. Post-processing
The output predicted by each model was a lung probability image - that is, an image with floating point values between 0 and 1, representing the probability that a given voxel belonged to the lung. A simple post-processing step was used to obtain a final binary lung segmentation, by applying a probability threshold of 0.5. Subsequently, a 3D connected component analysis was performed on the thresholded image. The two largest connected components were retained, corresponding to the left and right lungs, and any remaining components were discarded. In some cases, the left and right lungs form one connected component due to adjacent boundaries. These cases were automatically identified based on the ratio of volumes for the two largest connected components, and only the largest connected component was retained. In all cases, the result was a binary segmentation which did not distinguish between left and right lungs.
3.6. Quantitative Evaluation
The proposed multi-resolution, low-resolution, and high-resolution models were quantitatively evaluated by comparison to manual segmentations. Three metrics were used to assess agreement: the Jacaard index, average symmetric surface distance (ASSD), and maximum surface distance (MSD). The Jacaard index is a measure of volume overlap given by
| (3) |
where |·| is the set cardinality, and P ∩ M and P ∪ M are the intersection and union, respectively, of the set of voxels predicted to be lung in the automated segmentation P and the set of voxels defined as lung in the manual segmentation M. The Jacaard index has values ranging from zero to one, with one indicating perfect agreement. ASSD was used to measure the distance between the predicted lung boundary BP and manually generated lung boundary BM. The distance between a voxel x and a set of voxels on boundary B was defined as
| (4) |
where d(x, y) is the Euclidean distance between voxels x and y. The ASSD between BP and BM was defined as
| (5) |
The MSD between BP and BM was defined as
| (6) |
Both ASSD and MSD are greater or equal to zero, with zero being perfect agreement. A Friedman test was used to evaluate effect of the model on segmentation performance, as measured by Jacaard index, ASSD, and MSD.
4. Results
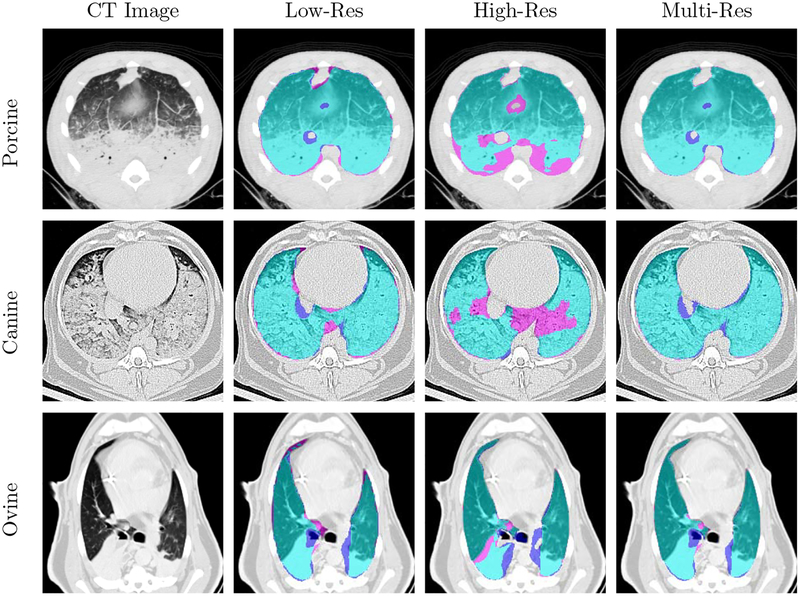
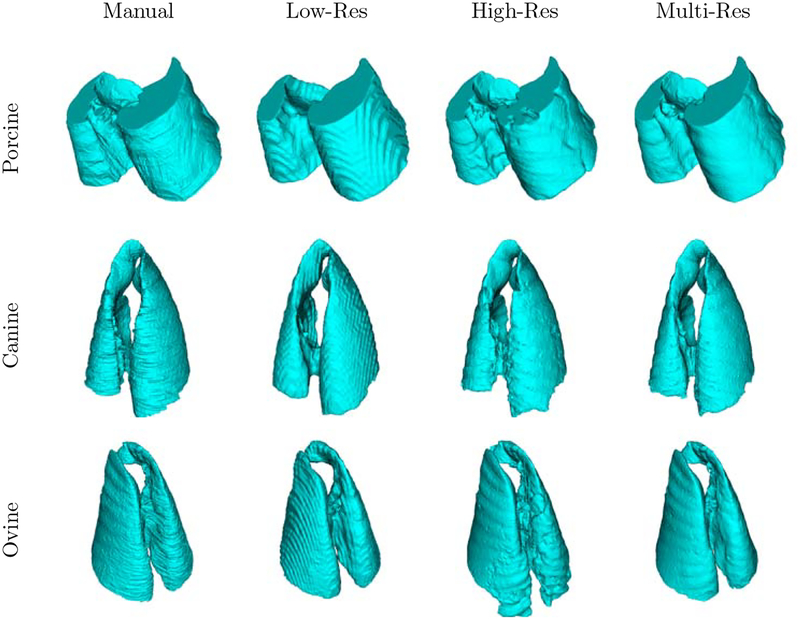
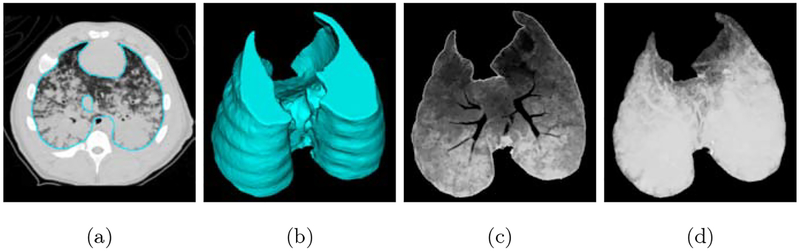
Axial slice views and surface renderings of segmentations produced by the different models are shown in Figure 4 and Figure 5, respectively. Surface renderings, minimum intensity projections, and maximum intensity projections are illustrated in Figure 6, to emphasize extent of injury and the inclusion of this injury in the predicted segmentation. Multi-resolution segmentation results for ten subjects are displayed in Figure 7 to emphasize the large variations in lung shape and size, as well as variations in injury severity, appearance, and spatial distribution.
Figure 4:
Lung segmentation results for porcine, canine, and ovine subjects in the top, middle, and bottom rows, respectively. True positives, false negatives, and false positives are denoted in cyan, magenta, and purple, respectively.
Figure 5:
Surface rendering of lung segmentations of porcine, canine, and ovine subjects in the top, middle, and bottom rows, respectively.
Figure 6:
Multi-resolution model results. (a) contour of predicted segmentation overlaid on CT image, (b) surface rendering of predicted segmentation, (c) minimum intensity projection of voxels included in predicted segmentation, and (d) maximum intensity projection of voxels included in predicted segmentation.
Figure 7:
Multi-resolution model segmentation results for representative cases from five porcine subjects (first column), three ovine subjects (second column, rows 1–3), and two canine subjects (second column, rows 4–5). Each segmentation result is displayed to the right of the corresponding CT image. True positives, false negatives, and false positives are denoted in cyan, magenta, and purple, respectively.
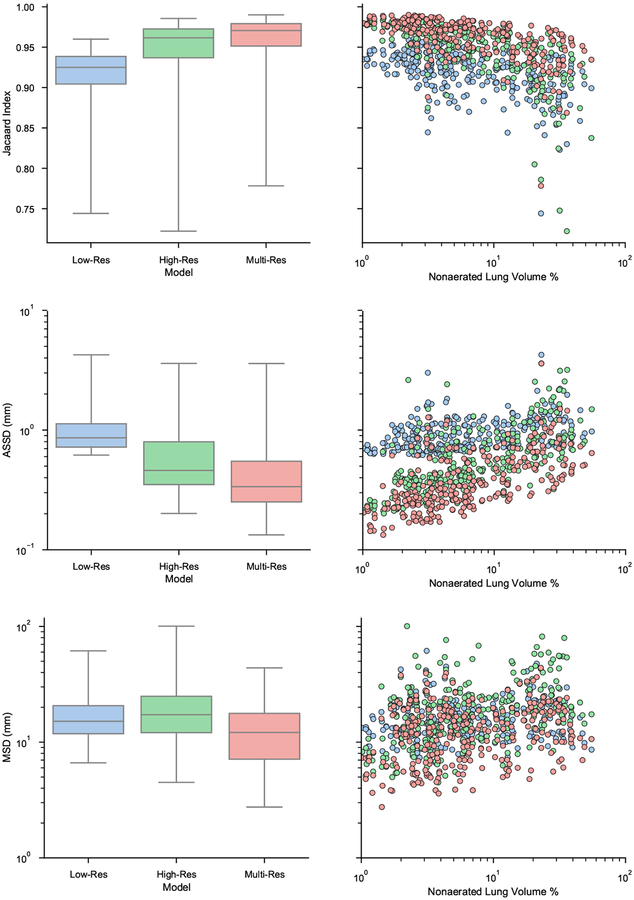
Quantitative results for each model are shown in Figure 8. Each row illustrates a different metric. The left column shows overall boxplot distributions for each model and the right column shows a scatter plot of each result stratified by the percent of lung volume that is non-aerated. The percent of lung volume that is non-aerated is used as a surrogate for the severity of lung injury. Voxels with HU greater than −100 were considered non-aerated.
Figure 8:
Quantitative comparison of the three models. Each row depicts a different metric, from top to bottom: Jacaard index, ASSD, and MSD. Left column shows overall boxplots for each model. Right column shows a scatter plot depicting result for each lung image, stratified by the percent of non-aerated volume.
The Friedman teat revealed that there was a statistically significant effect of the model on segmentation performance as measured by Jacaard index, ASSD, and MSD (p < 0.001). A Conover post hoc test revealed that there were a significant differences between each pair of models. The multi-resolution model outperformed both low-resolution and high-resolution models (p < 0.001) and the high-resolution model outperformed the low-resolution model (p < 0.001). Linear regression was used to model the correlation between injury severity and segmentation performance. The linear regression slope and coefficient of determination (R2) for each model are shown in Table 1.
Table 1:
Slope and coefficient of determination results for model performance vs. injury severity.
| Model | Jacaard index | ASSD | MSD | |||
|---|---|---|---|---|---|---|
| Slope | R2 | Slope | R2 | Slope | R2 | |
| Multi-Res | −1.20 × 10−3 | 0.316** | 1.33 × 10−2 | 0.206** | 9.96 × 10−2 | 0.021* |
| Low-Res | −1.13 × 10−3 | 0.201** | 1.06 × 10−2 | 0.096** | 4.70 × 10−2 | 0.004 |
| High-Res | −2.23 × 10−3 | 0.437** | 2.62 × 10−2 | 0.298** | 3.94 × 10−1 | 0.086** |
p < 0.001,
p < 0.05
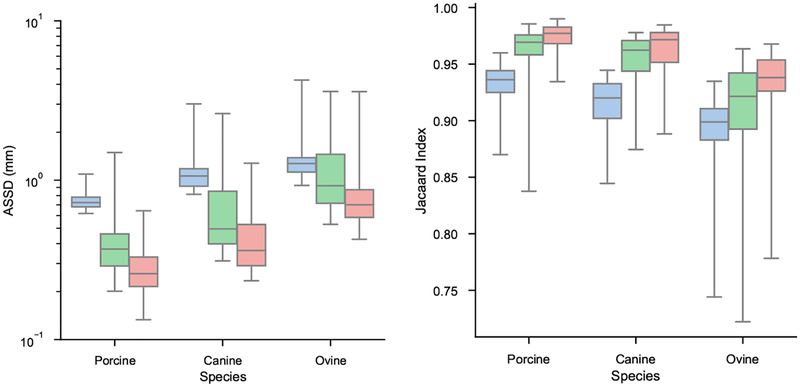
Results stratified by species and cross validation fold are displayed in Figures 9 and 10, respectively. The results show all models performed best on the porcine datasets, and worst on the ovine datasets. All folds of the cross validation performed equally well.
Figure 9:
ASSD and Jacaard index distributions stratified by species. Blue is low-resolution model, green is high-resolution model, and red is multi-resolution model.
Figure 10:
ASSD and Jacaard index distribution stratified by cross validation fold. Blue is low-resolution model, green is high-resolution model, and red is multi-resolution model.
5. Discussion
In this study, we developed a method for cross-species fully automatic lung segmentation, with emphasis on subjects with acute injury patterns similar to ARDS. This injury is particularly challenging to segment due to its diffuse anatomic appearance in dependent lung regions, resulting in little to no contrast between the posterior lung boundary and the surrounding soft tissue. Thus, a novel multi-resolution ConvNet model was proposed to incorporate both high-resolution local features and low-resolution global contextual information.
The multi-resolution model was compared to each of its components - a high-resolution model and a low resolution-model. All models showed high performance in terms of ASSD and Jacaard index, with the multi-resolution model out-performing the others. The superiority of the multi-resolution model is a direct consequence of access to both local detail and global context. The use of the low-resolution model’s prediction as an additional input channel to enhance the high-resolution model resulted in improved prediction beyond the capabilities of either individual component. Furthermore, qualitatively the high-resolution model produced segmentations that were more smooth reflecting the true lung topology. Whereas the low-resolution model produced segmentations with “staircasing” artifacts resulting from upsampling the low-resolution segmentation.
The high-resolution model has less global context compared to the low-resolution and multi-resolution models. However, this model is still trained on large 3D slabs. These 3D slabs were larger than the 2D slices and small 3D patches used in current ConvNet approaches (see Figure 1). This is a direct advantage of the Seg3DNet architecture’s reduced demand on GPU memory (Gerard et al., 2019). Thus we may expect models trained on 2D slices or 3D patches to perform worse compared to either the high-resolution or multi-resolution models in this study, due to further reductions in global context.
Despite aggressive downsampling, the low-resolution model demonstrated robust performance across all levels of injury severity. The same degree of segmentation accuracy was obtained in both healthy and severely non-aerated lungs, indicating that the low-resolution model relies on global context and anatomic features which are consistent despite variable lung aeration. Nevertheless, there may be an upper bound of the low-resolution model’s performance imposed by the inability to predict precise boundary location without local edge information. This is evidence by the comparatively poor performance of the low-resolution model in well-aerated lungs, for which boundaries are easily detected and global context does not provide much additional advantage. Other advantages of the low-resolution model include reduced training time, inference time, and GPU memory requirement. Therefore this model may be preferred over a 2D slice model, which has very limited global context and sacrifices 3D smoothness.
All segmentation models were evaluated on three different mammalian species, consistently performing best on the porcine dataset and worst on the ovine dataset. The lower performance on the ovine dataset can likely be attributed to the lower amount of training data for this species. The proposed method nonetheless showed high performance on all species. Rule-based systems often fail to generalize across species, requiring such methods to be specially tuned for each dataset. More advanced methods such as statistical shape models also cannot seamlessly handle multiple species, since an underlying assumption is that the data comes from a Gaussian distribution. By contrast, the ConvNet models used in this study be be able to exploit features consistent across mammalian species, such as the relative position of various thoracic organs and skeletal structures. The use of multiple species in training data may improve the ability of the model to robustly generalize mammalian lung segmentation in CT images.
Subjects were stratified by injury severity using percent of non-aerated lung volume as a surrogate for injury severity. All models showed a decrease in segmentation performance with increasing injury severity for both Jacaard index and ASSD. The high-resolution model showed the largest decline in performance with increasing injury severity, indicating that global information becomes more important for cases exhibiting a greater extent of injury. Although there were small decrements in performance with increasing injury severity, the multi-resolution model still demonstrated acceptable performance for subjects with severe injury, achieving voxel-level ASSD error for lungs with 30% non-aeration. Another factor to consider is the validity of the manual segmentations in these severely diseased cases. Injury results in little or no contrast at lung boundaries, and therefore greater subjectivity in manual segmentation, which could contribute to an apparent decrease in performance.
Although the proposed method generates lung segmentations that have a smooth surface in 3D, manual segmentation can vary by several millimeters between slices. Manual segmentation was performed on 2D slices, and when the lung boundary is poorly defined manually choosing a consistent boundary across slices is very difficult. Defining the boundary in lungs with diffuse consolidation is very subjective. Thus the intra- and inter-observer variation would likely be quite high on these cases, although it was not assessed in this study due to the availability of only a single manual segmentation per image.
The proposed method utilizes transfer learning since there is a limited number of annotated images of ARDS subjects. It is assumed that some features learned in the source domain (human subjects without ARDS) are useful in the target domain (animal subjects with ARDS). However, we did not investigate how much the transfer learning in particular improved the segmentation in the target domain, as opposed to simple training from scratch on the target domain. In future work, it would be interesting to quantify how much the use of transfer learning contributes to reduction of training time, retention of pre-learned features, and improved accuracy of predictions. It seems likely transfer learning is most valuable for very small training datasets, with diminishing returns as the training dataset size increases.
Several of the datasets used in this study include multiple 3D images of the same subject, e.g., 4DCT images or static images acquired at different airway pressures. The proposed method is limited to processing a single 3D image and therefore cannot learn patterns between multiple images. In future work, we plan to explore a 4D algorithm to exploit temporal patterns in image sequences, for example a recurrent neural network. A 4D algorithm would be especially useful for cases that have improved aeration resulting in higher boundary contrast in some images compares to others in the same sequence.
The run-time for executing the proposed multi-resolution model on a single 3D image was 40 seconds using a GPU. If a GPU is unavailable, the model could also be executed on a CPU with a run-time of approximately 2.5 minutes. This is fully automated and requires no user intervention. For comparison, manual segmentation of these images takes anywhere from 4 to 6 hours per case, depending on the experience of the analyst or the severity of lung injury. Our proposed method mitigates the prohibitive time and labor costs of injured lung segmentation, enabling rapid and accurate quantitative CT analysis for ARDS in both clinical and experimental settings.
6. Conclusion
In this study, a multi-resolution ConvNet cascade was proposed to enable learning of both local and global features in large 3D and 4D CT image sets despite limitations in GPU memory. This method was applied to automatic segmentation of lungs with acute injury in three different mammalian species. We confirmed that both global and local features are important for segmentation of injured lungs by comparing a multi-resolution model to its isolated low-resolution and high-resolution components. The proposed multi-resolution model performed best in terms of Jacaard Index, ASSD, MSD, as well as segmentation boundary smoothness, demonstrating the importance of both global and local features for the task of injured lung segmentation. Furthermore, we have demonstrated that the proposed method is able to generalize across canine, porcine, and ovine subjects despite a limited number of training datasets, due to transfer learning from a large human dataset without ARDS and learning anatomic features consistent across species.
Acknowledgment
This work was supported in part by the following grants: NIH RO1 HL142625, NIH R01 CA166703, NIH STTR R41 HL140640-01, NIH R01 HL094639, NIH/NHLBI 1UG3HL140177-01A1, and Merck-sponsored Investigator-initiated Study MISP 58429.
S.E. Gerard received support from a Presidential Fellowship through the University of Iowa Graduate College and from a NASA Iowa Space Grant Consortium Fellowship.
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Peer-Reviewed Medical Research Program under Award No. W81XWH-16-1-0434. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
We thank the COPDGene, SPIROMICS, and PANTHER-IPF investigators for providing the human image datasets used in this study. The COPDGene study is supported by NIH grants R01 HL089897 and R01 HL089856. SPIROMICS was supported by contracts from the NIH/NHLBI (HHSN268200900013C, HHSN268200900014C, HHSN268200900015C, HHSN268200900016C, HHSN268200900017C, HHSN268200900018C, HHSN268200900019C, HHSN268200900020C). The PANTHER-IPF study is supported by NIH grant R01 HL091743.
We gratefully acknowledge Eric A. Hoffman and Junfeng Guo for providing ground truth lung segmentations for the human dataset.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, & Mougiakakou S (2016). Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Medical Imaging, 35, 1207–1216. [DOI] [PubMed] [Google Scholar]
- Black CB, Hoffman A, Tsai L, Ingenito E, Suki B, Kaczka D, Simon B, & Lutchen K (2008). Impact of positive end-expiratory pressure during heterogeneous lung injury: insights from computed tomographic image functional modeling. Annals of Biomedical Engineering, 36, 980–991. [DOI] [PubMed] [Google Scholar]
- Brown MS, Mcnitt-Gray MF, Mankovich NJ, Goldin JG, Hiller J, Wilson LS, & Aberie D (1997). Method for segmenting chest CT image data using an anatomical model: Preliminary results. IEEETMI, 16, 828–839. [DOI] [PubMed] [Google Scholar]
- Carvalho AR, Spieth PM, Pelosi P, Melo MFV, Koch T, Jandre FC, Giannella-Neto A, & de Abreu MG (2008). Ability of dynamic airway pressure curve profile and elastance for positive end-expiratory pressure titration. Intensive Care Medicine, 34, 2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereda M, Xin Y, Goffi A, Herrmann J, Kaczka DW, Kavanagh BP, Perchiazzi G, Yoshida T, & Rizi RR (2019). Imaging the injured lung: Mechanisms of action and clinical use. Anesthesiology: The Journal of the American Society of Anesthesiologists,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereda M, Xin Y, Hamedani H, Bellani G, Kadlecek S, Clapp J, Guerra L, Meeder N, Rajaei J, Tustison NJ et al. (2017). Tidal changes on CT and progression of ARDS. Thorax, 72, 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereda M, Xin Y, Meeder N, Zeng J, Jiang Y, Hamedani H, Profka H, Kadlecek S, Clapp J, Deshpande CG et al. (2016). Visualizing the propagation of acute lung injury. Anesthesiology: The Journal of the American Society of Anesthesiologists, 124, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola S, Froio S, & Chiumello D (2017). Lung imaging in ARDS In Acute Respiratory Distress Syndrome (pp. 155–171). Springer. [Google Scholar]
- Couper D, LaVange LM, Han M, Barr RG, Bleecker E, Hoffman EA, Kanner R, Kleerup E, Martinez FJ, Woodruff PG et al. (2014). Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS). Thorax, 69, 492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas L, Spieth P, Carvalho A, de Abreu M, & Koch E (2009). Automatic lung segmentation of helical-ct scans in experimental induced lung injury. In 4th European Conference of the International Federation for Medical and Biological Engineering (pp. 764–767). Springer. [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, & Thrun S (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature, 542, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M et al. (2012). 3D Slicer as an image computing platform for the quantitative imaging network. Magnetic Resonance Imaging, 30, 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bustamante A, Easley R, Fuld M, Mulreany D, Chon D, Lewis J, & Simon B (2012). Regional pulmonary inflammation in an endotoxemic ovine acute lung injury model. Respiratory Physiology & Neurobiology, 183, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bustamante A, Easley RB, Fuld M, Mulreany D, Hoffman EA, & Simon BA (2009). Regional aeration and perfusion distribution in a sheep model of endotoxemic acute lung injury characterized by functional computed tomography imaging. Critical Care Medicine, 37, 2402–2411. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Caironi P, Cressoni M, Chiumello D, Ranieri VM, Quintel M, Russo S, Patroniti N, Cornejo R, & Bugedo G (2006). Lung recruitment in patients with the acute respiratory distress syndrome. New England Journal of Medicine, 354, 1775–1786. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Caironi P, Pelosi P, & Goodman LR (2001). What has computed tomography taught us about the acute respiratory distress syndrome? American journal of respiratory and critical care medicine, 164, 1701–1711. [DOI] [PubMed] [Google Scholar]
- Gerard SE, Herrmann J, Kaczka DW, & Reinhardt JM (2018). Transfer learning for segmentation of injured lungs using coarse-to-fine convolutional neural networks In Image Analysis for Moving Organ, Breast, and Thoracic Images (pp. 191–201). Springer. [Google Scholar]
- Gerard SE, Patton TJ, Christensen GE, Bayouth JE, & Reinhardt JM (2019). FissureNet: A deep learning approach for pulmonary fissure detection in CT images. IEEE Trans. Medical Imaging, 38, 156–166. doi: 10.1109/TMI.2018.2858202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorot X, & Bengio Y (2010). Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics (pp. 249–256). [Google Scholar]
- Godet T, Jabaudon M, Blondonnet R, Tremblay A, Audard J, Rieu B, Pereira B, Garcier J-M, Futier E, & Constantin J-M (2018). High frequency percussive ventilation increases alveolar recruitment in early acute respiratory distress syndrome: an experimental, physiological and CT scan study. Critical Care, 22, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Fuld MK, Alford SK, Reinhardt JM, & Hoffman EA (2008). Pulmonary Analysis Software Suite 9.0: Integrating quantitative measures of function with structural analyses In Brown M, de Bruijne M, van Ginneken B, Kiraly A, Kuhnigk JM, Lorenz C, Mori K, & Reinhardt J (Eds.), First International Workshop on Pulmonary Image Analysis (pp. 283–292). [Google Scholar]
- Herrmann J, Hoffman EA, & Kaczka DW (2017). Frequency-selective computed tomography: Applications during periodic thoracic motion. IEEE Transactions on Medical Imaging, 36, 1722–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Hoffman EA, & Reinhardt JM (2001). Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans. Medical Imaging, 20, 490–498. [DOI] [PubMed] [Google Scholar]
- Idiopathic Pulmonary Fibrosis Clinical Research Network, Martinez FJ, de Andrade JA, Anstrom KJ, King TE Jr, & Raghu G (2014). Randomized trial of acetylcysteine in idiopathic pulmonary fibrosis. New England Journal of Medicine, 370, 2093–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idiopathic Pulmonary Fibrosis Clinical Research Network, Raghu G, Anstrom KJ, King TE Jr, Lasky JA, & Martinez FJ (2012). Prednisone, azathioprine, and n-acetylcysteine for pulmonary fibrosis. New England Journal of Medicine, 366, 1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczka DW, Cao K, Christensen GE, Bates JH, & Simon BA (2011). Analysis of regional mechanics in canine lung injury using forced oscillations and 3D image registration. Annals of Biomedical Engineering, 39, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczka DW, Herrmann J, Zonneveld CE, Tingay DG, Lavizzari A, Noble PB, & Pillow JJ (2015). Multifrequency oscillatory ventilation in the premature lung: Effects on gas exchange, mechanics, and ventilation distribution. Anesthesiology: The Journal of the American Society of Anesthesiologists, 123, 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemerink GJ, Lamers RJ, Pellis BJ, Kruize HH, & Van Engelshoven J (1998). On segmentation of lung parenchyma in quantitative computed tomography of the lung. MEDPHYSICS, 25, 2432–2439. [DOI] [PubMed] [Google Scholar]
- Kingma DP, & Ba J (2014). Adam: A method for stochastic optimization. CoRR, abs/1412.6980. URL:http://arxiv.org/abs/1412.6980. [Google Scholar]
- Li K, Wu X, Chen DZ, & Sonka M (2006). Optimal surface segmentation in volumetric images-a graph-theoretic approach. IEEE Transactions on Pattern Analysis and Machine Intelligence, 28, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cai W, Wang X, Zhou Y, Feng DD, & Chen M (2014). Medical image classification with convolutional neural network. In Control Automation Robotics & Vision (ICARCV), 2014 13th International Conference on (pp. 844–848). IEEE. [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, Van Ginneken B, & Sánchez CI (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. [DOI] [PubMed] [Google Scholar]
- Oquab M, Bottou L, Laptev I, & Sivic J (2014). Learning and transferring mid-level image representations using convolutional neural networks. In The IEEE Conference on Computer Vision and Pattern Recognition (CVPR). [Google Scholar]
- Paula LF, Wellman TJ, Winkler T, Spieth PM, Güldner A, Venegas JG, de Abreu MG, Carvalho AR, & Melo MFV (2016). Regional tidal lung strain in mechanically ventilated normal lungs. Journal of Applied Physiology, 121, 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchiazzi G, Rylander C, Derosa S, Pellegrini M, Pitagora L, Polieri D, Vena A, Tannoia A, Fiore T, & Hedenstierna G (2014). Regional distribution of lung compliance by image analysis of computed tomograms. Respiratory Physiology & Neurobiology, 201, 60–70. [DOI] [PubMed] [Google Scholar]
- Perchiazzi G, Rylander C, Vena A, Derosa S, Polieri D, Fiore T, Giuliani R, & Hedenstierna G (2011). Lung regional stress and strain as a function of posture and ventilatory mode. Journal of Applied Physiology, 110, 1374–1383. [DOI] [PubMed] [Google Scholar]
- Pinzón AM (2016). Lung segmentation and airway tree matching: application to aeration quantification in CT images of subjects with ARDS. Ph.D. thesis Université de Lyon. [Google Scholar]
- Pinzón AM, Orkisz M, Richard J-C, & Hoyos MH (2014). Lung segmentation in 3D CT images from induced acute respiratory distress syndrome. In 11th IEEE International Symposium on Biomedical Imaging. [Google Scholar]
- Prasoon A, Petersen K, Igel C, Lauze F, Dam E, & Nielsen M (2013). Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 246–253). Springer. [DOI] [PubMed] [Google Scholar]
- Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, & Crapo JD (2011). Genetic epidemiology of COPD (COPDGene) study design. COPD: Journal of Chronic Obstructive Pulmonary Disease, 7, 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rikxoort EM, de Hoop B, Viergever MA, Prokop M, & van Ginneken B (2009). Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection. Medical Physics, 36, 2934–2947. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, & Brox T (2015). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 234–241). Springer. [Google Scholar]
- Salisbury ML, Lynch DA, Van Beek EJ, Kazerooni EA, Guo J, Xia M, Murray S, Anstrom KJ, Yow E, Martinez FJ et al. (2017). Idiopathic pulmonary fibrosis: The association between the adaptive multiple features method and fibrosis outcomes. American Journal of Respiratory and Critical Care Medicine, 195, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard S, Rao P, & Devaraj A (2012). Imaging of acute respiratory distress syndrome. Respiratory care, 57, 607–612. [DOI] [PubMed] [Google Scholar]
- Shen W, Zhou M, Yang F, Yang C, & Tian J (2015). Multi-scale convolutional neural networks for lung nodule classification. In International Conference on Information Processing in Medical Imaging (pp. 588–599). Springer. [DOI] [PubMed] [Google Scholar]
- Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, & Summers RM (2016). Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Medical Imaging, 35, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieren JP, Newell JD Jr, Barr RG, Bleecker ER, Burnette N, Carretta EE, Couper D, Goldin J, Guo J, Han MK et al. (2016). SPIROMICS protocol for multicenter quantitative computed tomography to phenotype the lungs. American Journal of Respiratory and Critical Care Medicine, 194, 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer I, Prokop M, & Van Ginneken B (2005). Toward automated segmentation of the pathological lung in CT. IEEE Transactions on Medical Imaging, 24, 1025–1038. [DOI] [PubMed] [Google Scholar]
- Sofka M, Wetzl J, Birkbeck N, Zhang J, Kohlberger T, Kaftan J, Declerck J, & Zhou SK (2011). Multi-stage learning for robust lung segmentation in challenging CT volumes. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 667–674). Springer. [DOI] [PubMed] [Google Scholar]
- Soliman A, Khalifa F, Elnakib A, El-Ghar MA, Dunlap N, Wang B, Gimelfarb G, Keynton R, & El-Baz A (2016). Accurate lungs segmentation on CT chest images by adaptive appearance-guided shape modeling. IEEETMI, 36, 263–276. [DOI] [PubMed] [Google Scholar]
- Sun S, Bauer C, & Beichel R (2012). Automated 3-d segmentation of lungs with lung cancer in CT data using a novel robust active shape model approach. IEEE Transactions on Medical Imaging, 31, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talakoub O, Helm E, Alirezaie J, Babyn P, Kavanagh B, Grasso F, & Engelberts D (2007). An automatic wavelet-based approach for lung segmentation and density analysis in dynamic CT. In Computational Intelligence in Image and Signal Processing, 2007. CIISP 2007. IEEE Symposium on (pp. 369–374). IEEE. [Google Scholar]
- Ukil S, & Reinhardt JM (2004). Smoothing lung segmentation surfaces in 3D X-ray CT images using anatomic guidance. In Proc. SPIE Conf. Medical Imaging (pp. 1066–1075). San Diego, CA volume 5370. [Google Scholar]
- Woodruff PG, Barr RG, Bleecker E, Christenson SA, Couper D, Curtis JL, Gouskova NA, Hansel NN, Hoffman EA, Kanner RE et al. (2016). Clinical significance of symptoms in smokers with preserved pulmonary function. New England Journal of Medicine, 374, 1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Song G, Cereda M, Kadlecek S, Hamedani H, Jiang Y, Rajaei J, Clapp J, Profka H, Meeder N et al. (2014). Semiautomatic segmentation of longitudinal computed tomography images in a rat model of lung injury by surfactant depletion. Journal of Applied Physiology, 118, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hoffman EA, & Reinhardt JM (2006). Atlas-driven lung lobe segmentation in volumetric X-Ray CT images. IEEE Transactions on Medical Imaging, 25, 1–16. [DOI] [PubMed] [Google Scholar]
- Zompatori M, Ciccarese F, & Fasano L (2014). Overview of current lung imaging in acute respiratory distress syndrome. [DOI] [PMC free article] [PubMed]