Abstract
Both the timing and molecular determinants of metastasis are unknown, hindering treatment and prevention efforts. Here we characterize the evolutionary dynamics of this lethal process by analyzing exome sequencing data from 118 biopsies from 23 colorectal cancer (CRC) patients with metastases to the liver or brain. The data show low primary tumor-metastasis genomic divergence, where canonical driver genes were acquired early. Analysis within a spatial tumor growth model and statistical inference framework indicates that early disseminated cells commonly (81%, 17/21 evaluable patients) seed metastases while the carcinoma is clinically undetectable (typically <0.01 cm3). We validated the association between early drivers and metastasis in an independent cohort of 2,751 CRCs, demonstrating their utility as biomarkers of metastasis. This new conceptual and analytical framework provides quantitative in vivo evidence that systemic spread can occur early in CRC and illuminates strategies for patient stratification and therapeutic targeting of the canonical drivers of tumorigenesis.
Editorial summary:
Analysis of evolutionary dynamics of colorectal cancers and paired distant brain or liver metastases provides evidence that early disseminated cancer cells seed metastases before the carcinoma is clinically undetectable.
Introduction
Metastasis is the primary cause of cancer patient death, but the timing and molecular determinants of this process are largely uncharacterized1–3. In particular, when and how metastatic competence is specified is of clinical significance. The prevailing linear progression model posits that metastatic capacity is acquired late following the gradual accumulation of somatic alterations, such that only a subset of cells evolve the capacity to disseminate and seed metastases4–7. At odds with this view, gene expression signatures from primary tumors are predictive of distant recurrence indicating that metastatic cells constitute a dominant subpopulation in primary tumor8,9. In addition, disseminated tumor cells have been identified in patients with early breast lesions10 and in mouse models of early breast and pancreatic cancers11–13. However, the timing of metastatic dissemination has not been evaluated in human cancers due to the challenge in obtaining paired primary tumors and distant metastases and the limitations of phylogenetic approaches.
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and leading cause of cancer death14, as well as an excellent model for studying tumor progression given that the initiating driver alterations are well characterized4. The site and resectability of CRC metastases dictate treatment options and prognosis15,16 with liver being the most common site presumably due to venous drainage, and one third of metastatic CRC (mCRC) patients exhibiting liver-exclusive metastasis16. In contrast, brain metastasis is a rare (<4% of mCRCs), but devastating diagnosis with limited therapeutic options and median survival of 3 to 6 months17. In CRC, metastasis is assumed to be seeded by genetically advanced cancer cells that have evolved through a series of sequential clonal expansions4,18. However, CRC progression is not necessarily linear. Rather, we described a Big Bang model of tumor evolution, whereby after transformation some CRCs grow as a single expansion populated by heterogeneous and effectively equally fit subclones, and where most detectable intra-tumor heterogeneity arises early19. These data suggest that some CRCs may be born to be bad, wherein invasive and even metastatic potential is specified early19,20. Effectively neutral evolution has since been reported in other primary tumors21–24, but the ‘mode’ of evolution (effective neutrality versus subclonal selection) has not been evaluated in paired primary tumors and metastases.
Although the metastatic process is largely occult, spatio-temporal patterns of genomic variation in paired primary tumors and metastases embed their evolutionary histories. Here we analyze exome sequencing data from 118 biopsies from 23 mCRC patients with paired distant metastases to the liver or brain to delineate the timing and routes of metastasis and to define metastasis competent clones (Fig. 1). The data show low primary tumor-metastasis genomic divergence (PMGD), where genomic drivers were acquired early. Moreover, through simulation studies, we establish that low PMGD in bulk-sample sequencing data is indicative of early dissemination, contrary to current assumptions2. Phylogeny reconstruction and analysis of the mutational cancer cell fraction (CCF) revealed the early divergence of metastatic lineages and their monoclonal origin. To overcome the limitations of phylogenetic approaches, which cannot resolve the timing of dissemination2,25–28, we developed a spatial computational model of tumor progression and Bayesian statistical inference framework to ‘time’ dissemination in a patient-specific fashion. Further, we validated the association between combinations of early driver genes and metastasis in an independent cohort of 2,751 CRCs, demonstrating their utility as biomarkers of aggressive disease. These results provide quantitative in vivo evidence for early metastatic seeding in mCRC with implications for systemic therapy and earlier detection.
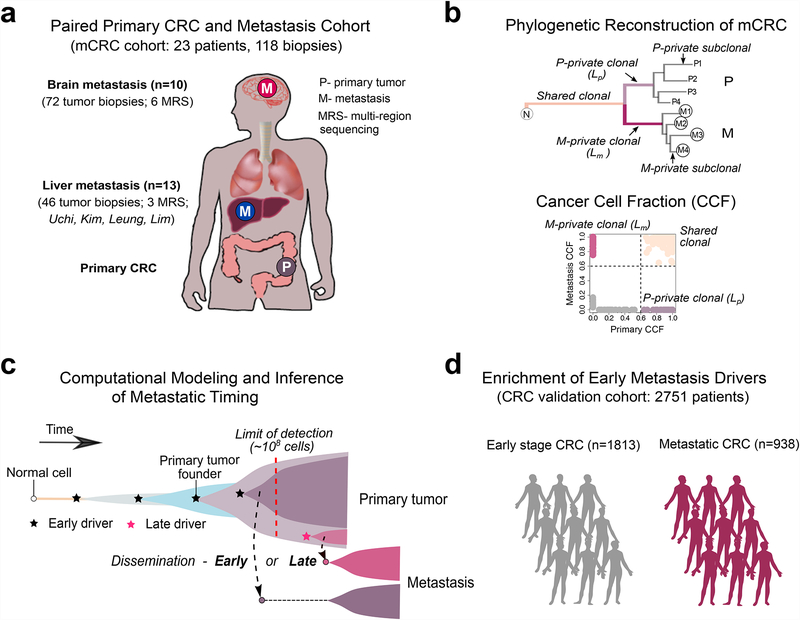
Figure 1. Study overview.
(a) The metastatic colorectal cancer (mCRC) patient cohort includes 118 tumor biopsies from 23 patients. Paired CRCs with metastases to the brain and other sites (liver, lung, lymph nodes) from 10 patients and 72 tumor biopsies were whole-exome sequenced, including 6 cases with multi-region sequencing (MRS) of 3–5 regions each from the primary CRC and metastasis. Additionally, four publicly available cohorts with paired CRCs and liver metastases from 13 patients and 46 tumor biopsies were reanalyzed within the same bioinformatics framework, including 3 cases with MRS. (b) Tumor phylogenies were reconstructed from somatic alterations (sSNVs+indels). The mutational cancer cell fraction (CCF) was computed and compared for each primary CRCs and metastasis pair. (c) Schematic illustration of tumor evolution starting from a normal cell that acquires mutations leading to malignant transformation, growth of the primary tumor, metastatic dissemination, seeding and outgrowth. It is unknown whether dissemination occurs early from a dominant subclone when the size of the primary tumor is below the limits of clinical detection (108 cells or 1 cm3) (early dissemination) or later from a minor subclone after the acquisition of additional driver alterations (late dissemination). To address this question, we developed a 3-D model of tumor growth and statistical inference framework to time metastasis from patient genomic data. (d) We further leveraged a large collection of metastatic (n=938) and non-metastatic (n=1,813) CRCs with targeted sequencing data to evaluate the association between specific combinations of early driver genes (modules) identified in the mCRC cohort.
Results
Overview of clinical cohorts
mCRC patients exhibit varied progression paths where liver-exclusive metastasis and brain metastasis represent extreme scenarios with distinct prognoses15,16. We therefore sought to characterize the genomic landscape, routes and timing of metastasis in mCRC by analyzing exome sequencing data from 118 biopsies from 23 patients with paired distant metastases to the liver or brain (referred to as the mCRC cohort, Fig. 1a, Supplementary Fig. 1, Supplementary Table 1, Methods). To investigate these patterns, we sequenced 72 samples from a unique cohort of 10 mCRC patients with paired brain metastases some of whom had additional metastases to the liver (n=1), lung (n=1) and lymph nodes (n=4). Five patients had brain-exclusive distant metastasis (V402, V514, V855, V953 and V974) estimated to occur in a mere 2–10% of patients with brain metastasis16. For six patients, multi-region sequencing (MRS) of the paired primary and metastasis (P/M pairs) was performed (3–5 regions each), enabling the detailed reconstruction of tumor phylogenies (Fig. 1b). Additionally, we included 46 tumor biopsies from 13 mCRC patients with paired liver metastases after excluding cases with low tumor cell purity (<0.4) (Supplementary Fig. 2) from four published datasets (Uchi21, Kim29, Leung30 and Lim31), analyzed using the same unified bioinformatics framework (Methods). No other sites of metastasis were reported for these patients and MRS was available for 3 P/M pairs (n=2–9 regions each). As we have previously shown, MRS enables more accurate estimation of the cancer cell fraction (CCF) and discrimination between clonal and subclonal mutations relative to single sample sequencing (Fig. 1b, Supplementary Fig. 3)23. Additionally, we leveraged an independent collection of 2,751 CRC patients, including 938 with metastatic disease (stage IV) and 1,813 early stage (stage I-III) patients for whom targeted sequencing data from the MSK-Impact32 and GENIE33 studies were available in order to evaluate the association between specific combinations of early driver genes (modules) defined in the mCRC cohort and metastatic propensity (Fig. 1d, Methods).
Genomic heterogeneity in CRCs and paired metastases
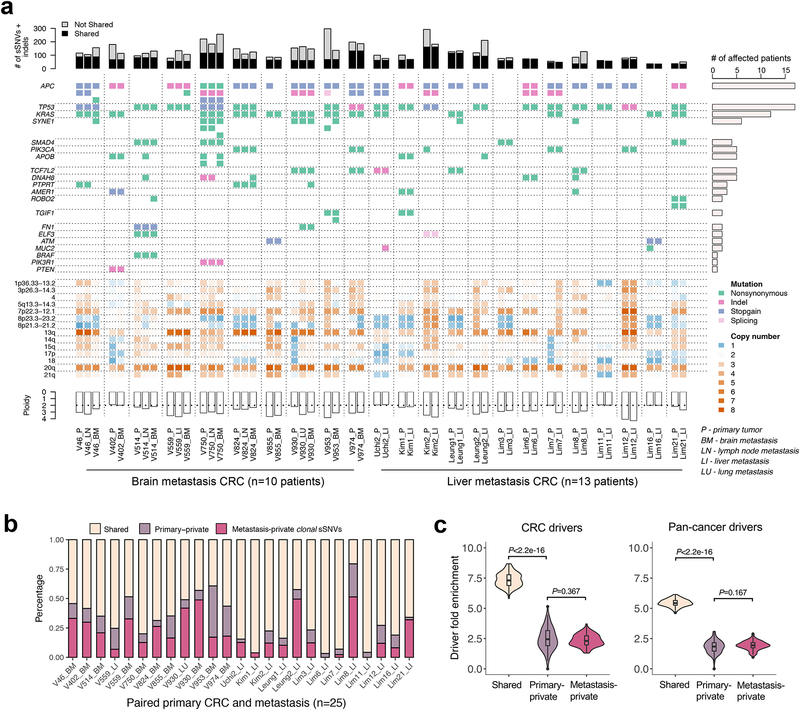
High concordance amongst putative driver genes was observed in the mCRC cohort (Fig. 2a) consistent with previous studies21,29,34–37. For instance, KRAS, TP53, SMAD4, TCF7L2, FN1, ELF3 and ATM mutations were completely concordant between P/M pairs (Fig. 2a, Supplementary Table 2). On average, 70% of high-frequency somatic single nucleotide variants (sSNVs) with CCF>60% (Methods) in any primary tumor or metastasis were shared by both lesions (Fig. 2b). Amongst genes that were mutated in more than five patients, SYNE1 (4/6 patients) and APOB (3/5 patients) tended to be primary or metastasis private and thus likely arose after transformation. Although metastases usually had more private high-frequency sSNVs than the primary tumor (P=0.020, Wilcoxon Rank-Sum Test, Fig. 2b), they were not enriched for CRC drivers (defined based on IntOGen38 and TCGA39) or a published list of pan-cancer drivers40 (Fig. 2c, Supplementary Table 3, Methods). Similar results were obtained when stratifying by brain or liver metastases (Supplementary Fig. 4). These data reflect limited driver gene heterogeneity between P/M pairs and suggest that few additional private genomic drivers were required for metastasis. Somatic copy number alterations (CNAs) were also generally concordant, with chromosomes 7p22.3–12.1, 13 and 20q11–13 exhibiting recurrent amplification and chromosomes 8p23.3–23.2, 8p21.3–21.2, 18 exhibiting recurrent deletion in P/M pairs (Fig. 2a, Supplementary Fig. 5)41. Several putative oncogenes, including PIK3CA, GNAS, SRC, FXR1, MUC4, GPC6, MECOM were recurrently (≥4 patients) amplified in metastases relative to paired primary tumors. Intriguingly, HTR2A (5-hydroxytryptamine receptor 2A), which encodes a receptor for the neurotransmitter serotonin that dually functions as a regulatory factor in the gastrointestinal tract42, was amplified more frequently in brain (4/10) than liver (1/13) metastases (Supplementary Fig. 5).
Figure 2. The mutational landscape and patterns of genetic divergence in paired primary CRCs and metastases.
(a) Concordance amongst somatic alterations (sSNVs, indels and CNAs) in known CRC ‘driver’ genes between paired primary CRCs and metastases. Stacked barplots illustrate the total number of sSNVs and indels in exonic regions with a lower cutoff of variant allele frequency (VAF)=0.1 in the corresponding site (primary or metastasis). (b) The percentage of clonal sSNVs that are shared, primary-private, or metastasis-private out of all clonal sSNVs with CCF>60% in any of paired primaries and distant metastases. (c) Violin plots illustrate the probability density of driver gene fold enrichment amongst shared, primary-private, and metastasis-private clonal non-silent sSNVs based on known CRC or pan-cancer ‘drivers’. The inset box corresponds to the 25th to 75th percentile (interquartile range, IQR); the horizontal line indicates the median; and the vertical line includes data within 1.5 times the IQR. A test statistic was computed based on n=100 down-samplings amongst patients (Methods). P-value, Wilcoxon Rank-Sum Test (two-sided).
We defined the number of metastasis-private (M-private) clonal sSNVs as Lm (merged CCF>60% in the metastasis samples and <1% in the primary tumor samples) and the number of primary tumor-private (P-private) clonal sSNVs as Lp (merged CCF>60% in the primary and <1% in the metastasis), where a cutoff of 60% accurately distinguished clonal and subclonal sSNVs (Fig. 1b, Supplementary Fig. 6, Supplementary Fig. 7a–b). Therefore, we used a merged CCF value of 60% as the cutoff to distinguish clonal and subclonal mutations throughout. Brain metastases exhibited higher Lm than liver metastases (median=24.5 vs 9.5, P=0.01, Wilcoxon Rank-Sum Test), whereas no difference was noted for Lp (median=8.5 vs. 6.0, P=0.70, Wilcoxon Rank-Sum Test) (Supplementary Fig. 7c), potentially reflecting longer progression times (and more cell divisions). Neither Lm (P=0.68, Wilcoxon Rank-Sum Test) nor Lp (P=0.95, Wilcoxon Rank-Sum Test) differed significantly in chemo-naïve versus treated cases despite a slight shift in mutational spectra (A|T->C|G) after chemotherapy (Supplementary Fig. 8).
Gene-ontology analysis showed enrichment for cellular adhesion terms amongst both brain and liver metastasis-private non-silent clonal mutations, but not primary-private clonal or subclonal mutations (Supplementary Table 4). Nervous system development and neuronal differentiation terms were enriched amongst brain and liver metastasis-private clonal mutations and primary tumor-private mutations, consistent with hijacking of the enteric nervous system in gastrointestinal malignancies43. In contrast, primary tumor-private non-silent clonal mutations were enriched for metabolic processes, DNA repair and damage, suggestive of more general deregulation and resource constraints during tumor expansion.
Phylogenetic reconstruction of metastatic CRC
The MRS data revealed extensive intra-tumor heterogeneity (ITH) both within tumors and between P/M pairs (Fig. 3a–b, Supplementary Fig. 9, Supplementary Table 2) and ample mutations for phylogeny reconstruction. We employed FST44 to quantify ITH within tumors (primary tumor or metastasis) in the mCRC cohort based on subclonal sSNVs23 (Methods). Clonal mutations present in all samples don’t contribute to ITH and were excluded in FST calculations. Both the primary tumor (median FST=0.180, range 0.150–0.430) and paired metastases (median FST=0.178, range 0.123–0.271) exhibited high FST values, consistent with rapid genetic diversification (Supplementary Fig. 10a). Proliferative indices based on Ki67 staining were also similar between paired CRCs and metastases (P=0.765, Wilcoxon Signed-Rank Test, Supplementary Fig. 10b).
Figure 3. Within and between lesion heterogeneity in paired primary CRCs and metastases.
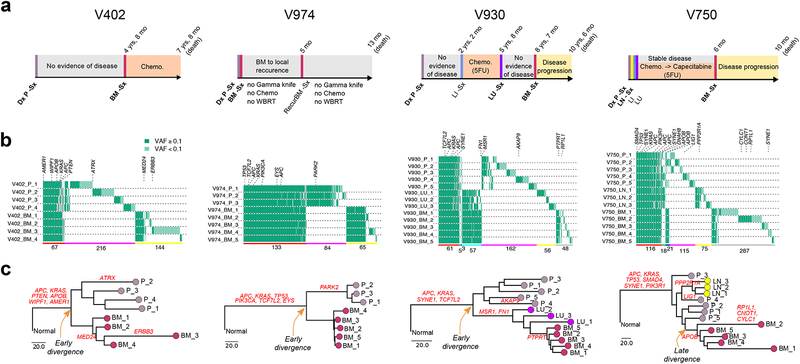
(a) Clinical and treatment history for four representative CRC patients with brain metastases. Dx: diagnosis; Sx: surgical resection. (b) Patterns of within and between lesion heterogeneity amongst sSNVs and indels based on multi-region sequencing of paired primary CRCs and metastases, where canonical CRC driver genes are labeled. The number of mutations shared or private amongst different lesions is indicated below the corresponding colored horizontal bars: ubiquitously P-M shared (red), partially P-M shared (green-M1 or blue-M2), P-private (pink) or M-private (yellow-M1 or gray-M2 or cyan-M1 and M2). P corresponds to primary. M1 and M2 correspond to different metastatic sites in the same patient. (c) Phylogeny reconstruction via maximum parsimony (PHYLIP) based on mutational presence/absence, where canonical CRC drivers genes are labeled. VAF, variant allele frequency.
Tumor phylogenies were reconstructed using sSNVs and small insertions and deletions (indels) across multiple regions of each P/M pair using the maximum-parsimony method45. Distant metastases corresponded to monophyletic clades in all but one (Kim1) case (8/9 with MRS) (Fig. 3c, Supplementary Fig. 9, Methods), consistent with the unique origin of the metastatic lineage. Inspection of the phylogeny for Kim1 indicated that the liver metastasis preceded the primary tumor, which is improbable and likely due to metastasis-specific loss of heterozygosity (LOH) spanning multiple mutations. In most patients, the metastatic lineage diverged prior to genetic diversification of the primary tumor (V402, V930, V953, V974, Uchi2; early divergence), whereas divergence occurred during diversification of the primary tumor in patients V750, V824 and Kim2 (late divergence). All brain metastases and most liver metastases harbored many private clonal sSNVs, but lacked shared subclonal sSNVs with the primary tumors, consistent with monoclonal seeding (Supplementary Fig. 11–12), as demonstrated by simulation studies (Supplementary Fig. 13). Two liver metastases (Lim6 and Lim11), exhibited enrichment for shared subclonal mutations, but lacked metastasis-private clonal mutations, consistent with polyclonal seeding (Supplementary Fig. 12–13). These data suggest that distant metastases are often seeded by a single clone (a single cell or a group of genetically similar cells). Notably, the phylogenetic tree for case V930 indicates that the brain metastasis derived from the lung metastasis, in-line with the patient’s clinical history (Fig. 3). Brain metastases and regional lymph node (LN) metastases formed separate clades in the two cases in which they were profiled (V750, V824), indicative of their independent clonal origin from primary tumor (Fig. 3c, Supplementary Fig. 9), consistent with polyguanine-repeat analysis46.
The finding that paired CRCs and metastases formed separate phylogenetic clades in most patients suggests that metastatic dissemination may occur early such that the primary tumor has sufficient time to accumulate many unique clonal mutations after dissemination. However, phylogenetic divergence may occur much earlier than dissemination (Supplementary Fig. 14) and phylogenetics cannot resolve the timing of dissemination2,25–28. As such, we next sought to investigate the determinants of PMGD and to quantify the timing of metastasis.
The timing of dissemination and P-M genomic divergence
To model the evolutionary dynamics of metastasis, we developed a 3-D agent-based computational model to simulate the spatial growth, progression and lineage relationships of realistically sized patient tumors under varied parameters19,23 (Fig. 4a, Supplementary Fig. 15, Supplementary Table 5, Methods). We model the growth of a primary CRC starting from a single founder cell and assume that the metastasis is seeded by a random single cell on periphery of primary tumor, yielding primary and metastatic tumors composed of ~109 cells (~10 cm3). To account for distinct modes of tumor evolution, we simulate effective neutrality and stringent subclonal selection19,23, resulting in four evolutionary scenarios for P/M pairs: Neutral/Neutral (N/N), Neutral/Selection (N/S), Selection/Neutral (S/N) and Selection/Selection (S/S) (Fig. 4a, Supplementary Fig. 15–16, Methods). Using this simulation framework, where ground-truth values are known, we evaluated the relationship between the number of M-private clonal sSNVs (Lm) and primary CRC size at the time of dissemination (Nd) in hundreds of virtual paired P/M tumors, where size is a surrogate for time since cell division rates are unknown (Methods).
Figure 4. Correlation between the Lp, Lm and H and primary carcinoma size at the time of dissemination (Nd).
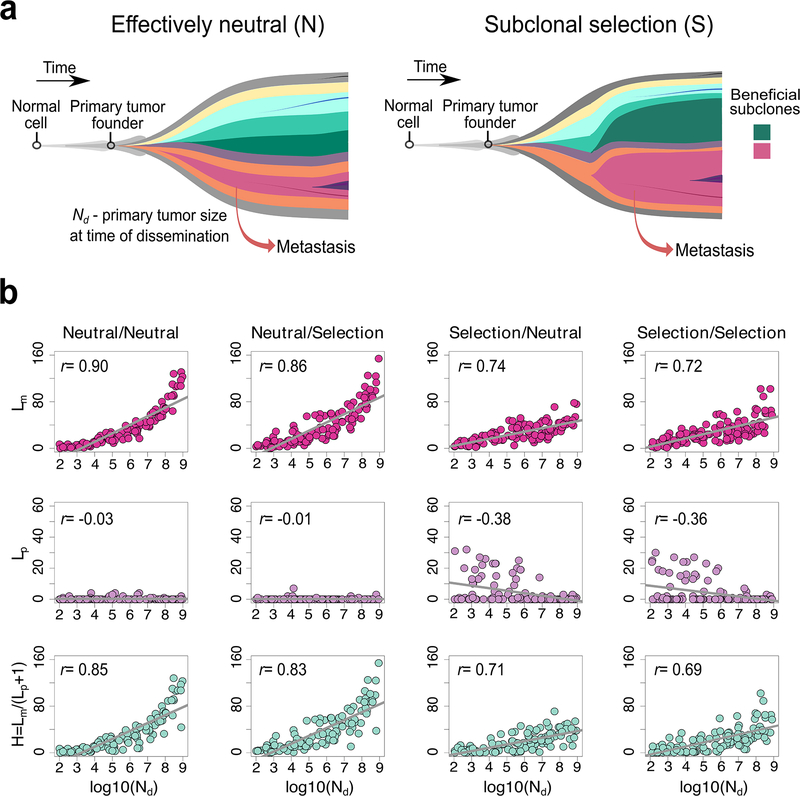
(a) Schematic illustration of effectively neutral (N) evolution and stringent subclonal selection (S), two distinct evolutionary modes that can occur during growth of the primary tumor or metastasis. It is assumed that metastatic dissemination occurs during expansion of the primary CRC where Nd corresponds to the size of the primary carcinoma at the time of dissemination. (b) The correlation between the timing of dissemination, Nd, and Lm, Lp or H, based on the spatial simulation of tumor growth (n=100 tumors for each scenario; Pearson’s r is reported). Lp and Lm correspond to the number of private clonal sSNVs (CCF>60% in one site and CCF<1% on the other site) in the whole primary carcinoma and metastasis, respectively and H=Lm/(Lp+1).
To define Lm, we first evaluated M-private clonal sSNVs with respect to relatively high-frequency sSNVs in the whole primary tumor (CCF>1%). Thus any clonal sSNV in the metastasis will be M-private if the CCF<1% in the primary tumor. We found that Lm is positively correlated with Nd under all four evolutionary scenarios (Fig. 4b). The positive relationship between Lm and Nd remains significant when accounting for variation in mutation rate, cell birth/death rate and selection intensity during tumor growth (Supplementary Fig. 17). We next evaluated Lm by simulating sequencing reads from variable numbers of primary tumor regions (n=1, 10, 50 or 100) while considering the whole metastasis as a bulk sample within our computational model. The positive correlation between Lm and Nd was highly significant under all sampling scenarios, pointing to the robustness of this observation (Supplementary Fig. 18). As expected, smaller Lm was observed when a greater number of primary tumor regions were sequenced because fewer mutations were M-private (Supplementary Fig. 18). Mathematical analysis of the special case of neutral evolution and exponential growth further demonstrates the positive relationship between Lm and Nd (Supplementary Note, Eq. S6). These data suggest that later dissemination results in more clonal mutations in the metastasis, many of which are at low frequency in the primary tumor and often undetectable in bulk sequencing. Accordingly, later dissemination will give rise to more metastasis-private clonal mutations in real sequencing data, leading to higher PMGD. It should be noted that if sampling of the primary tumor was exhaustive or if the metastasis-founder (M-founder) clone could be traced, neither of which are generally practical for studies of human tumors, one would expect very small Lm values and no correlation between Lm and Nd since all mutations in the M-founder cell that accumulated during primary tumor growth would be captured. In contrast, the number of P-private clonal sSNVs (Lp) exhibited slightly negative correlation with Nd when CRCs grew under stringent selection (S/N or S/S), whereas under neutral evolution (N/N or N/S) Lp≈0, regardless of the timing of dissemination (Fig. 4b, Supplementary Fig. 17).
We defined early dissemination as Nd <108 cells (~1 cm3 in volume), the size at which CRCs are generally clinically detectable, and late dissemination as Nd≥108 cells. To establish intuition for the relationship between PMGD and Nd, we defined H=Lm/(Lp+1). In the simulation studies, H was positively correlated with Nd (Fig. 4b, Supplementary Fig. 17), indicating that larger H values are associated with later dissemination. Indeed, late dissemination typically results in large H (>20). The observation that most patients in the mCRC cohort exhibited small H values (median=2.4, range: 0.5–23.5) suggests that early dissemination may be relatively common. While H is strongly associated with the timing of dissemination, it does not capture all components of PMGD, including the mutation rate as this is cancelled out in the division of Lm over Lp. Additionally, variation in Lp due to differences in the mode of evolution and sampling bias contribute to noise in H. To account for these sources of variability while estimating the timing of dissemination in individual patients we turned to a powerful statistical inference framework grounded in population genetic theory.
Quantitative evidence for early metastatic seeding in CRC
In order to infer the timing of dissemination Nd, mutation rate u (per cell division in exonic regions) and mode of tumor evolution in P/M pairs, we developed SCIMET (Spatial Computational Inference of MEtastatic Timing), which couples our spatial (3D) agent-based model of tumor evolution with a statistical inference framework based on Approximate Bayesian Computation (ABC)47,48 (Fig. 4a, Supplementary Fig. 15–16, Supplementary Fig. 19, Supplementary Table 6–7, Methods). ABC is well established in population genetics and has been utilized to infer the parameters of tumor evolution19,49. Since the patient genomic data were generally consistent with monoclonal seeding, we assumed that a single cell seeds the metastasis (Lim6 and Lim11 were therefore excluded from this analysis). Evaluation of SCIMET on virtual tumors demonstrates the accurate recovery of the mutation rate and timing of dissemination (Supplementary Fig. 20).
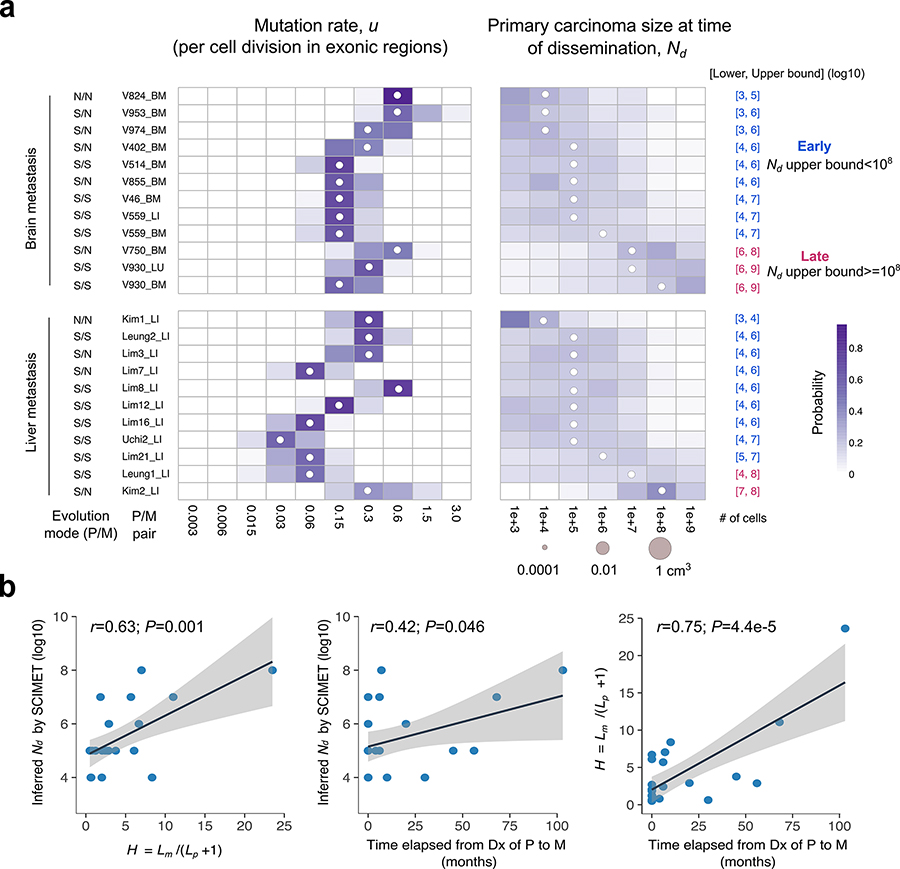
The majority (90%) of CRCs and metastases (57%) exhibited patterns consistent with subclonal selection (Fig. 5a). Inference of patient-specific mutation rates via SCIMET showed an order of magnitude variation across patients (inferred u or , corresponding to 10−9-10−8 mutations per base pair per cell division). Strikingly, in 83% (19/23) P/M pairs from 17/21 patients, dissemination was estimated to occur early when the primary CRC was below the limits of clinical detection (inferred Nd or cells) and typically when the primary tumor was composed of fewer than 106 cells using conservative estimates (Fig. 5a, Supplementary Table 1, Methods). The inferred Nd values were also significantly smaller than the tumor size documented at the time of diagnosis in this cohort. Of note, early dissemination was common irrespective of the site of distant metastasis (8/10 brain, 10/12 liver, 1/1 lung). Congruent results were also obtained when accounting for higher ratios of cell birth/death rates in the primary CRC and metastasis (Supplementary Fig. 21), the collective dissemination of small clusters of cells (n=10 cells) (Supplementary Fig. 22) or single-region sampling (Supplementary Fig. 23). Amongst the four cases where late dissemination was inferred, three had MRS data, enabling comparison with their phylogenies. For two patients (V750 brain metastasis and Kim2 liver metastasis) late dissemination was consistent with the tumor phylogeny (Fig. 3c, Supplementary Fig. 9). For patient V930, late dissemination was inferred for both the lung and brain metastases, consistent with the large H values (brain: H=23.5; lung: H=11). However, the tumor phylogeny indicates early divergence of the metastatic lineage (Fig. 3c). This case illustrates that phylogenetic divergence can occur before dissemination (Supplementary Fig. 14), emphasizing the need for a quantitative evolutionary framework to ‘time’ metastasis.
Figure 5. Patient-specific inference of the timing of metastasis in CRC.
(a) Heatmap of the posterior probability distributions inferred by SCIMET for the mutation rate u (per cell division in exonic regions) and Nd (timing of metastatic dissemination relative to primary carcinoma size) in individual P/M pairs (n=23) from 21 mCRC patients. The median of the inferred posterior distribution (referred to as the inferred Nd or) is indicated by a white circle at the corresponding value. For patients with more than one distant metastasis, each was analyzed independently. The mode of tumor evolution in each P/M pair was determined based on model selection within the statistical inference framework (Methods). We define early dissemination as Nd (upper bound) <108 cells (~1 cm3 in volume) and use the 3rd quartile of the posterior distribution as the upper bound to be conservative. Late dissemination is defined as Nd (upper bound) ≥108. P/M pairs where dissemination and seeding are inferred to have occurred early are denoted in blue, whereas those inferred to have disseminated late are denoted in magenta. (b) Correlations between the inferred timing of dissemination () based on SCIMET and the H metric as well as the time elapsed from diagnosis (Dx) of the primary to diagnosis of the metastasis (n=23). The Pearson’s r and P values are reported. Shading corresponds to 95% confidence interval of the linear regression.
The inferred values based on SCIMET were positively correlated with H (Pearson’s r=0.63, P=0.001, Fig. 5b), consistent with the observation that the H metric reflects the timing of dissemination. Additionally, both and H were positively correlated with the time elapsed between diagnosis of the primary CRC and distant metastasis (Fig. 5b), implying that metastases that are diagnosed later likely disseminated later. Further, we estimated the time span between metastatic dissemination and surgical resection of the primary tumor by employing an approximate analytical function for our spatial tumor growth model and find that dissemination often occurred more than 3 years before surgery (Supplementary Fig. 24, Supplementary Note).
Metastasis-associated early driver gene modules
As noted above, most canonical drivers were clonal and shared between paired primaries and metastases (Fig. 2), indicative of their early acquisition before transformation. Taken this together with the finding that cancer cells seed metastases early in the majority of mCRCs in this cohort, specific combinations of early driver genes (modules) may confer metastatic competence. In support of this view, oncogene engineering of four canonical early driver genes (APC, KRAS, TP53, SMAD4) in wild-type primary colon organoids yielded metastases upon xenotransplantation50. Similarly, in a mouse model of CRC, oncogenic Kras in combination with Apc and Trp53 deficiency was sufficient to drive metastasis51.
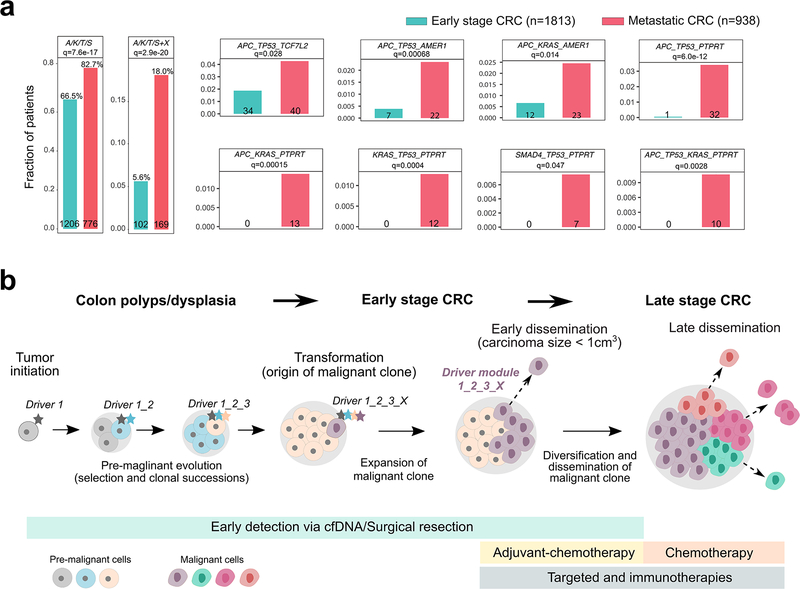
We therefore evaluated the association between the early driver modules defined in the mCRC cohort and metastatic proclivity by analyzing a collection of 2,751 CRC patients, including 938 with metastatic disease (stage IV) and 1,813 early-stage (stage I-III) CRC patients that were prospectively sequenced as part of the MSK-Impact32 and GENIE33 studies (Methods). Strikingly, we find that numerous early driver gene modules were significantly enriched in metastatic relative to early stage CRCs in this independent dataset after correction for multiple hypothesis testing (Fig. 6a, Supplementary Fig. 25, Supplementary Table 8, Methods). These modules consist of a backbone of canonical ‘core’ CRC drivers (combinations of APC, KRAS, TP53 or SMAD4, abbreviated A/K/T/S) with one additional candidate metastasis driver (TCF7L2, AMER1 or PTPRT). Collectively, the ‘core’ modules plus an additional candidate metastasis driver shows a statistically significant enrichment in metastatic versus early stage CRCs (18% vs. 5.6%, respectively, q=2.9 × 10−20). Examination of the prevalence and enrichment of individual modules indicates that PTPRT mutations in combination with canonical drivers were almost exclusively observed in metastatic patients (Fig. 6a, Supplementary Fig. 25). Thus, PTPRT appears to be a highly specific driver of metastasis. PTPRT mutations were previously reported in 26% of colorectal cancers52 and loss of PTPRT in CRC and in head and neck squamous cell cancers results in increased STAT3 activation and cellular survival53,54. It has therefore been proposed that PTPRT mutations may be predictive biomarkers for STAT3 pathway inhibitors, illuminating new therapeutic opportunities54. Other modules involving AMER1 and TCF7L2 were also significantly enriched in metastatic cases, but were less specific perhaps because an additional driver defines the module. Thus we identify a compendium of metastasis driver modules that can inform the stratification and therapeutic targeting of patients with aggressive disease.
Figure 6. Enrichment of early driver gene modules in mCRC and clinical implications of early dissemination.
(a) The enrichment of canonical ‘core’ CRC driver genes (APC, KRAS, TP53 or SMAD4; A, K, T or S) plus recurrent mutations in candidate drivers (AMER1, ATM, BRAF, PIK3CA, PTPRT or TCF7L2) identified in the mCRC cohort was evaluated in an independent cohort of 2,751 CRC patients. The combined barplots (left) illustrate the overall frequency of the ‘core’ module alone or with an additional candidate driver (‘X’) in early stage versus metastatic CRCs. Individual barplots indicate the frequency of specific ‘modules’. Q-values are based on two-sided Fisher’s exact tests with Benjamini–Hochberg adjustment. (b) Three stages of CRC progression are outlined: pre-malignancy (between initiation and transformation), early-stage (between transformation and dissemination) and late-stage (after dissemination). A set of potential interventions to prevent cancer mortality targets each stage: for pre-malignant lesions, resection (after detection via colonoscopy or possibly cell free DNA; cfDNA); for early-stage CRC, surgical resection and possibly adjuvant chemotherapy; and for late-stage CRC, chemotherapy and/or targeted/immune therapies. Given the high rate (80% here) of early dissemination, prior to clinical detectability of the early-stage CRC, detection and resection of pre-malignant lesions will have the greatest impact on preventing cancer mortality. For tumors that undergo dissemination prior to clinical detectability, surgical resection alone, even of a small tumor, cannot prevent metastasis. Once the early-stage tumor is discovered, newly defined metastatic modules (panel a) may inform patient stratification to aid the directed use of adjuvant chemotherapy.
Discussion
We describe a novel theoretical and analytical framework that yields quantitative in vivo measurement of the dynamics of metastasis in a patient-specific manner, while accounting for confounding factors, including the founder event, the mode of tumor evolution, mutation rate variation and tissue sampling bias. By analyzing genomic data from paired primary CRCs and distant metastases to the liver and brain from five patient cohorts within this evolutionary framework, we demonstrate that metastatic seeding often occurs early (17/21 patients), when the carcinoma is clinically undetectable (~104-108 cells or 0.0001–1 cm3) and years before diagnosis and surgery (Fig. 5, Supplementary Fig. 21–24). The observation that early metastatic seeding was prevalent irrespective of the site of distant metastasis, suggests the generalizability of these results. Moreover, dissemination was early even when considering liver-exclusive and brain-exclusive metastases, which represent extremes in terms of their prevalence and prognosis. Collectively, these finding indicate that CRCs can be born to be bad, wherein invasive and metastatic potential is specified early19,20,55, illuminating the need to target the canonical drivers of tumorigenesis. However, not all tumors will metastasize and there is an urgent need to identify biomarkers associated with aggressive disease.
Towards this end, we validated metastasis-associated driver modules in an independent cohort, thereby defining the molecular features of metastasizing clones. The overlap with drivers of initiation and combinatorial structure of these modules may explain why few drivers of metastasis have been identified to date. While the canonical driver landscape is relatively sparse, there are nonetheless many possible combinations of mutations that collectively disrupt key signaling pathways (WNT, TP53, TGFB, EGFR and cellular adhesion) enabling niche independence and outgrowth at foreign sites50.
Of note, the vast majority (90%) of primary tumors in the mCRC cohort exhibited subclonal selection consistent with the metastatic clone having a selective growth advantage (Fig. 5a). In contrast, a smaller proportion of early stage (I-III) CRCs (33%) exhibited patterns consistent with subclonal selection23, suggesting that the mode of tumor evolution may correlate with disease stage or aggressiveness, although larger studies are needed to investigate this trend. Whereas drivers were not enriched in metastases when all cases were considered (Fig. 2c), stratifying by the mode of tumor evolution revealed the enrichment of private high-frequency (CCF>20%) driver mutations in metastases evolving under stringent selection compared to those evolving neutrally (Supplementary Fig. 26), implying that further subclonal driver mutations may occur during the growth of some metastases. Nonetheless, a sizeable proportion (43%) of distant metastases evolved neutrally, potentially reflecting the high fitness of the metastatic clone, consistent with a fitness plateau56.
The finding that early dissemination resulting in successful metastatic seeding can occur before the primary tumor is clinically detectable in the majority (80%) of mCRC patients in this cohort underscores the importance of detecting malignancy at the earliest possible stage (Fig. 6b). Such small tumors fall below the detection limits for current imaging modalities, but advances in profiling circulating cell-free tumor DNA may ultimately enable earlier non-invasive detection57,58. Importantly, a considerable number of mCRC patients did not exhibit early systemic spread, suggesting that colonoscopy can be beneficial in this subgroup. Our data also raise the possibility that early-stage patients harboring combinations of driver genes that confer a high risk of metastasis may particularly benefit from adjuvant chemotherapy to target micro-metastatic disease59. While the clinical utility of this approach needs to be prospectively evaluated, our findings provide a rationale for patient stratification and therapeutic targeting.
Methods
Clinical specimens, pathology review and sequencing studies
Briefly, archived formalin-fixed paraffin-embedded (FFPE) tissue specimens from 10 patients with metastatic CRC, including primary tumor, matched metastases and adjacent normal colon tissue, were obtained from the Medical University of Vienna brain metastasis bio-bank, which was established in accordance with ethical guidelines (approval 078/2004). Tissue specimens were collected during the course of routine clinical care and clinical data were retrieved by retrospective chart review. All samples were de-identified and patients in the brain metastasis cohort were deceased prior to initiating this study. Brain metastases were available for all patients (BM, n=10) and for several patients metastases to the liver (LI, n=1), lung (LU, n=1), and regional lymph nodes (LN, n=4) were also available (Supplementary Table 1). For 6 of the 10 patients, multiple specimens (n=3–5) from both the primary and metastasis were sampled and sequenced (Supplementary Table 1). Histological sections were independently reviewed by expert pathologists (A.B, P.B, C.J.S). The Ki67 proliferative index was determined via immunohistochemical staining, as previously described60. Consistent with the growth of CRC brain metastases in an expansive rather than infiltrating fashion61, no normal brain parenchyma was observed within the main brain metastasis lesion.
For all patients regions of high-cellularity (>60%) were selected for DNA isolation using the QIAamp DNA FFPE Tissue Kit (Qiagen). Libraries were prepared using the Agilent SureSelect Human All Exon kit or Ilumina Nextera Rapid Capture Exome (NCRE) kit for sequencing on the Illumina Hiseq 2000/2500 or Nextseq 500. Paired sequencing reads were aligned to human reference genome build hg19 with BWA (v.0.7.10)62. Duplicate reads were flagged with Picard Tools (v.1.111). Aligned reads were further processed with GATK 3.4.0 for local re-alignment around insertions and deletions and base quality recalibration.
We also analyzed de-identified exome sequencing data from metastatic colorectal cancer patients in four published datasets (Uchi21, Kim29, Leung30 and Lim31) using the same unified bioinformatics framework detailed below. After excluding tumors with low purity (<0.4), we retained 46 tumor specimens from 13 mCRC patients with paired liver metastases and refer to this as the liver metastasis cohort.
Somatic SNV detection and filtering
sSNVs were called by MuTect (v.1.1.7)63 with paired tumor and normal sequencing data. sSNVs failing MuTect’s internal filters, having fewer than 10 total reads or 3 variant reads in the tumor sample, fewer than 10 reads in the normal sample, or mapping to paralogous genomic regions were removed. Additional Varscan (v.2.3.9)64 filters were applied to remove sSNVs with low average variant base qualities, low average mapping qualities among variant supporting reads, strand bias among variant supporting reads and high average mismatch base quality sums among variant supporting reads, either within each tumor sample or across all tumor samples from the same patient. Additional filtering removed sSNVs detected in a panel of normals (PON) by running MuTect in single-sample mode with less stringent filtering criteria (artifact detection mode). sSNVs called in at least two normal samples were included in the PON sSNV list. For FFPE samples, sSNVs called in samples from one patient were checked against samples from all other patients to flag those that might be artifactual. The maximal observed variant allele frequencies (VAF) across all samples from each patient were calculated based on raw output files from MuTect. sSNVs with maximal observed VAFs between 0.01 and 0.05 in at least two other patients were removed. Small insertions and deletions (indels) were called with Strelka (v.1.0.14) and annotated by Annovar (v.20150617)65. sSNVs and small insertions and deletions (indels) in protein coding regions were retained for downstream analyses. Additional filters were applied to exclude possible artifactual sSNVs due to the processing of FFPE specimens. Specifically, artifacts among C>T/G>A sSNVs with bias in read pair orientation were filtered in each individual FFPE sample, similar to the approach of Costello et al.66.
For patients with MRS data, we sought to exploit this information by retrieving read counts for sSNVs across samples from the same patient. To obtain depth and VAF information across all samples from the same patient, for each sSNV and in each tumor sample that an sSNV was not originally called in, the total reads and variant supporting reads were counted using the mpileup command in SAMtools (v.1.2)67. Only reads with mapping quality ≥ 40 and base quality at the sSNV locus ≥ 20 were counted and used to calculate VAF values for that sSNV.
Copy-number analysis, tumor purity and CCF estimation
Copy number analysis was performed using TitanCNA (v.1.5.7)68. Briefly, TitanCNA uses depth ratio and B-allele frequency information to estimate allele-specific absolute copy numbers with a hidden Markov model, and estimates tumor purity and clonal frequencies. Only autosomes were used in copy number analysis. First, for each patient, germline heterozygous SNP at dbSNP 138 loci were identified using SAMtools and SnpEff (v.3.6) in the normal sample. HMMcopy (v.0.99.0)69 was used to generate read counts for 1000-bp bins across the genome for all tumor samples. Whole-exome sequences (WES) from multiple normal samples per patient were pooled separately for the purpose of calculating read counts in the bins and the pooled normal read depth data were used as controls for the calculation of depth ratios only. TitanCNA was used to calculate allelic ratios at the germline heterozygous SNP loci in the tumor sample and depth ratios between the tumor sample and the pooled normal data in bins containing those SNP loci. Only SNP loci within WES covered regions were then used to estimate allele-specific absolute copy number profiles. TitanCNA was run with different numbers of clones (n=1–3). One run was chosen for each tumor sample based on visual inspection of fitted results, with preference given to the results with a single clone unless results with multiple clones had visibly better fit to the data. Results from tumor samples from the same patient were inspected together to ensure consistency. Overall ploidy and purity for each tumor sample was calculated from the TitanCNA results. For the public datasets including liver-exclusive mCRCs, cases with estimated purity >0.4 in both the primary tumor and paired metastases (Supplementary Fig. 2) were included since low purity hinders accurate SNV/CNA calling.
Mutational cancer cell fractions (CCFs) were estimated with CHAT (v 1.0)70. CHAT includes a function to estimate the CCF of each sSNV by adjusting its variant allele frequency (VAF) based on local allele-specific copy numbers at the sSNV locus. sSNV frequencies and copy number profiles estimated from previous steps were used to calculate CCFs for all sSNVs in autosomes (using a modified function). The CCFs were also adjusted for tumor purity. The merged CCF of each sSNV is computed by integrating CCFs from multiple regions when MRS data is available:
| Eq. (1) |
where di and CCFi are the sequencing depth and cancer cell fraction estimation in region i, respectively. Of note, the vast majority (99%) of P-M shared sSNVs have CCF (or merged CCF) > 60%, a cutoff that also optimally distinguishes the site-private clonal and subclonal sSNV clusters (Supplementary Fig. 6). We thus use 60% as the CCF cutoff to define clonal versus subclonal sSNVs in the primary-metastasis genomic divergence (PMGD) analysis.
Data processing for downstream analyses
For each tumor site (primary or metastasis) in a patient, the average CCF estimate of a sSNV is set to 0 if neither of these two criteria are met: a) VAF ≥ 0.03 and variant read count ≥ 3; b) VAF ≥ 0.1 in any of the regions. The following additional filters were applied to summarize the MRS P/M data in a given patient:
Filter out sSNVs without VAF ≥ 0.05 and variant read count ≥ 3 or VAF ≥ 0.1 in any samples from this pair of sites
Filter out sSNVs with total read depth < 20 from either of the two tumor sites
Filter out all sSNVs in chromosome regions with LOH in all specimens from one tumor site but not in all samples from the other tumor site.
For sSNVs not present in any specimens with LOH, filter out sSNVs satisfying the following criteria in specimens from at least one of the two tumor sites: a) absent in some samples with LOH; b) not absent in any samples without LOH.
Driver enrichment analysis
Driver fold enrichment was determined based on colorectal adenocarcinoma (COAD) driver genes (defined by combining IntOGen v.2016.538 and TCGA39 including 221 genes, Supplementary Table 3) or all pan-cancer drivers, including 369 high-confidence genes40 harboring non-silent coding sSNVs out of the total number of genes with non-silent coding sSNVs. The resulting metric was normalized by the fraction of driver genes out of all genes in the human genome. Clonal mutations (CCF>60% in P or M; merged CCF was used for MRS data) were divided into three sets representing shared, primary-private and metastasis-private mutations, where only distant metastases were considered. Driver gene fold enrichment was calculated for each set of mutations by randomly sampling 15 of 25 P/M pairs from the whole cohort, aggregating them to calculate one driver enrichment score, and repeating this 100 times (n=100 down-samplings) to derive a test statistic. For each down-sampling, the driver enrichment score was calculated as:
| Eq. (2) |
where n(all non-silent clonal) and n(driver non-silent clonal) correspond to the total number of non-silent clonal mutations and the number of non-silent clonal mutations in driver genes, respectively. Here n(driver genes) and n(total genes) correspond to the total number of drivers reported for CRC (n=221) or pan-cancer (n=369) and the number of coding genes in the genome (n=22,000), respectively.
Orthogonal validation of early metastasis driver gene modules
Clinical annotations and targeted sequencing data were obtained for the GENIE33 (v.3.0) and MSK-Impact32 CRC cohorts from Synapse (http://synapse.org/genie) and cBioPortal (http://www.cbioportal.org/study?id=crc_msk_2018), respectively. The MSK-Impact cohort includes early-stage primary CRCs, primary CRCs that are known to have metastasized and the metastatic lesion (predominantly liver) from 1,099 mCRC patients and a total of 1,134 samples with available sequencing and clinical covariates including stage, microsatellite status, and time to metastasis. Since the mCRC “discovery” cohort did not include microsatellite unstable (MSI+) cases, these were removed as were cases with POLE mutations. Microsatellite stable (MSS) samples were divided into early-stage non-metastatic samples (n=57), metastatic primary tumors (n = 440) and metastatic samples (n = 498).
The GENIE cohort is composed of 39,600 samples profiled with different targeted sequencing panels from which CRC samples were selected (oncotree codes: COADREAD, COAD, CAIS, MACR, READ and SRCCR). In order to avoid duplicated samples, all MSK-Impact samples from the GENIE cohort were removed, as were duplicated samples from the same patient, resulting in 2,666 samples, 1,756 of which were from primary tumors. As the GENIE cohort does not currently include stage or outcome information, all primaries are assumed to be non-metastatic, although some may be stage IV or diagnosed as metastatic in the future.
All possible combinations of recurrent putative M-driver genes (APC, TP53, KRAS, SMAD4, PIK3R1, BRAF, AMER1, TCF7L2, PIK3CA, PTPRT and ATM) identified in the mCRC cohort were evaluated in metastatic relative to early stage cases using a two-sided Fisher’s exact test (Benjamini–Hochberg adjustment for multiple testing). The enrichment analysis was calculated for the combined MSK-Impact and GENIE primary CRC cohort, as well as for the MSK-Impact cohort alone (Supplementary Table 8). Importantly, as the number of genes in a module increases, the specificity of the association with metastasis increases, but the frequency of the module and in turn power to detect an association decreases (Supplementary Fig. 25). While combining datasets may potentially introduce some biases, because we assume that all GENIE primary samples are non-metastatic and MSS, this will render our analyses conservative. Indeed, it is worth noting that while these results are already highly significant, they are likely conservative for several reasons: i) due to the short follow-up time, some early-stage cases may develop metastases, ii) imbalanced sample size with nearly twice as many early stage versus metastatic cases, iii) several putative M-drivers identified in the mCRC cohort are not represented on the targeted sequencing panel and hence cannot be evaluated.
Phylogenetic tree reconstruction and FST computation
We ran PHYLIP71 (http://www.trex.uqam.ca/index.php?action=phylip&app=dnapars) and applied the Maximum Parsimony method to reconstruct the phylogeny of multiple specimens from individual patients based on the presence or absence of SNVs and indels. When multiple maximum parsimony trees were reported, we chose the top ranked solution. FigTree (http://tree.bio.ed.ac.uk/software/Figtree/) was employed to visualize the reconstructed trees. We computed the FST statistic for each primary tumor or metastasis using the Weir and Cockerham method44 based on the adjusted frequency of subclonal sSNVs (merged CCF<60%) identified in MRS data. Clonal mutations (merged CCF>60%) don’t contribute to ITH and were excluded in FST calculations.
Spatial agent-based modeling of tumor progression
We extended our previously described three-dimensional agent-based tumor evolution framework19,23 to model tumor growth, mutation accumulation and metastatic dissemination after malignant transformation under different evolutionary scenarios in P/M pairs, namely Neutral/Neutral (N/N), Neutral/Selection (N/S), Selection/Neutral (S/N) or Selection/Selection (S/S). Pre-malignant clonal expansions prior to transformation do not alter the genetic heterogeneity within a tumor thus were not modeled (Fig. 1c, Fig. 4a and Supplementary Fig. 15) and we assume that dissemination occurs after malignant transformation of the founding carcinoma cell since invasion (a cardinal feature of carcinomas) is a requirement for metastasis. We have previously employed this framework to model primary tumor evolution23. In this model, spatial tumor growth is simulated via the expansion of deme subpopulations (composed of ~5k cells with diploid genome), mimicking the glandular structures often found in colorectal tumors and metastases and consistent with the number of cells found in individual colorectal cancer glands (~2,000–10,000 cells)72. Model assumptions are detailed in Supplementary Table 5. The deme subpopulations expand within a defined 3D cubic lattice (Moore neighborhood, 26 neighbors), via peripheral growth while cells within each deme are well-mixed without spatial constraints and grow via a random birth-and-death process (division probability p and death probability q=1-p at each generation). The notion of peripheral growth is supported by recent studies indicating that cancer cells at the periphery of the tumor proliferate much faster than those at the center73. Moreover, peripheral growth results in a power law model of net tumor growth (Supplementary Fig. 15b), and is supported by data in colorectal cancer74. The first deme is generated via the same birth-and-death process, beginning with a single transformed founding tumor cell. Here we employ the following parameters: p=0.55 and q=0.45 for the deme expansion in both the primary tumor and metastasis. Thus the cell birth/death probability ratio for the founding lineage is p/q=0.55/0.45≈1.2. This is supported by the observation that there is no significant difference in proliferation rates based on Ki67 staining of paired CRCs and brain metastases (Supplementary Fig. 10b), as previously reported in liver metastases75. Based on these values of p and q, approximately 3 years are required from transformation to the diagnosis of primary carcinoma (~109 cells) (Supplementary Fig. 15b). Once a deme exceeds the maximum size (10,000 cells), it splits into two offspring demes via random sampling of cells from a binomial distribution [Nc, p=0.5], where Nc is the current deme size.
During the growth of the primary CRC, a single cell from a random deme at the tumor periphery is randomly chosen to seed the metastasis supported by mounting pathological evidence of invasive cells in tumor front and that blood vessels are also mostly distributed in the invasive front in CRC76. The total cell number at the time of metastatic dissemination is denoted by Nd. The metastasis grows via the same model as the primary tumor, starting from the disseminated tumor cell(s).
During each cell division, the number of neutral passenger mutations acquired in the coding portion of the genome follows a Poisson distribution with mean u. Thus, the probability that k mutations occurred in each cell division is as follows:
| Eq. (3) |
where an infinite sites model and constant mutation rate are assumed during tumor progression. For simplicity, we do not simulate CNAs, LOH, or aneuploidy, and all mutations are heterozygous. Under the neutral model, all somatic mutations are assumed to be neutral passenger events and do not confer a fitness advantage, whereas in the subclonal selection model, beneficial mutations (or advantageous mutations) arise stochastically via a Poisson process with mean us during each cell division. We assume us=10−5 per cell division in the genome23,77. We investigated a relatively strong positive selection coefficient (s=0.1), where s specifies the increase in cell division probability per cell division when a beneficial mutation occurs in the neutral cell lineage. The cell birth and death probabilities for a selectively beneficial clone are ps=p×(1+s) and qs=1-ps=1-p×(1+s), respectively, thus the selective advantage is defined as s=ps/p-1. We selected s=0.1 since we have previously shown that the resultant patterns of between-region genetic divergence can be clearly distinguished from those arising under effectively neutral growth23.
During simulation of primary and metastatic growth, each mutation is assigned a unique index that is recorded with respect to its genealogy and host cells, enabling analysis of the mutational frequency in a sample of tumor cells or the whole tumor during different stages of growth. We simulate growth until the primary and metastasis reach a size of ~109 cells (or ~10 cm3) comparable to the size of the clinical samples studied here which ranged from 4–15 cm in maximum diameter. To simulate each of the four scenarios of P/M growth, namely N/N, N/S, S/N or SS, we employed a mutation rate u=0.3 per cell division in the exonic region (corresponding to 5×10−9 per site per cell division in the 60Mb diploid coding regions) and selection coefficients s=0 and s=0.1 when modeling neutral evolution and subclonal selection, respectively, during growth of the primary tumor or metastasis. Under each of the four scenarios of P/M growth, 100 time points (representing the primary tumor size at the time of dissemination, Nd) were sampled at random from a uniform distribution, log10(Nd)~U(2,9), each giving rise to independent P/M pairs. The CCF from the whole tumor in both the P and M lesions were obtained for each sSNV (site). CCFs > 60% in one site and CCFs < 1% in the other site were used to count the number of P-private and M-private clonal sSNVs (Lp and Lm, respectively), consistent with the strategy employed for patient samples.
Spatial Computational Inference of MEtastatic Timing (SCIMET)
We sought to infer two parameters that govern the dynamics of metastasis, namely u, the mutation rate per cell division in the exonic region and Nd, primary tumor size at the time of dissemination based on our spatial tumor simulation framework. The two parameters of interest (u and Nd) were randomly sampled from a prior discrete uniform distribution, namely 10 values from 0.003 to 3 for u; and 7 values from 103 to 109 cells (on log10 scale) for Nd (Supplementary Table 6–7, Supplementary Fig. 19). Discrete prior distributions for u and Nd were used to estimate the order of magnitude rather the precise values of these two parameters. We simulated 70,000 paired primary tumors and metastases (composed of 109 cells each) under each of the four evolutionary scenarios (N/N, N/S, S/N or S/S). After generating the virtual P/M tumors, multiple regions (n=4) each composed of ~106 cells are sampled from an octant of tumor sphere, as was done for our clinical samples (Supplementary Fig. 19). The VAF of all sSNVs in the sampled bulk subpopulation is considered the true VAF (denoted by fT), whereas the observed allele frequency is obtained via a statistical model that mimics the random sampling of alleles during sequencing. Specifically, we employ a Binomial distribution (n, fT) to generate the observed VAF at each site given its true frequency fT and number of covered reads n. The number of covered reads at each site is assumed to follow a negative-binomial distribution (negative binomial(size, depth)) where depth is the mean sequencing depth and size corresponds to the variation parameter78. We assume depth=80 and size=2 for the sequencing data in each tumor region. A mutation is called when the number of variant reads is ≥3, thereby applying the same criteria as for the patient tumors. The observed VAF for each mutation is converted to CCF and the merged CCF from four regions were computed (Eq.(1)) to mimic the patient genomic data. The nine summary statistics used to fit the CCF data are described in Supplementary Fig. 19 and Supplementary Table 6. The median values of the posterior probability distributions obtained from SCIMET are referred to as the inferred parameter values ( and ). To be conservative, we define early dissemination as Nd (upper bound)<108 cells (~1 cm3 in volume) using the 3rd quartile of the posterior distribution as the upper bound, whereas late dissemination is defined as Nd (upper bound) ≥108 cells (Fig. 5a). We also evaluated the robustness of SCIMET to a higher birth/death rate ratio (Supplementary Fig. 21), collective dissemination by a cell cluster (n=10 cells, Supplementary Fig. 22) or single-region sequencing data (Supplementary Fig. 23). Of note, both a higher birth/death rate ratio and single-region sequencing data would result in overestimation of the timing of metastatic dissemination. A higher birth/death rate ratio yields a higher tumor growth rate thus the primary tumor size at the time of dissemination would be larger than for a lower birth/death rate ratio. Single-region sampling results in a larger number of metastasis-private clonal mutations (larger Lm and larger H) compared with multi-region sequencing, thus the timing of dissemination would be overestimated in accordance with the positive correlation between Lm or H and Nd. Overall, these comparisons demonstrate the robustness of SCIMET to different model assumptions.
We employ a version of ABC based on the Acceptance-Rejection Algorithm79 to estimate posterior probability distributions for the parameters of interest θ(u, Nd). The ABC version of rejection sampling is as follows:
For i=1 to K under model M(N/N, N/S, S/N or S/S):
Sample parameters θ′ from the prior distribution π(θ)
Simulate data D′ using model M with the sampled parameters θ′, and summarize D′ as summary statistics S′
Accept θ′ if d(S′, S)<ε, for a given tolerance rate ε, where d(S′, S) is a measure of Euclidean distance between S′ and S
Go to 1
Using this scheme, we are able to approximate the posterior distribution by: P(θ|d(S′, S)<ε). We use a common variation of ABC47,80 where rather than using a fixed threshold, ε, we sort all K distances calculated in by d(S′, S) (Step 3), and accept the θ′ that generated the smallest 100×η percent distances. We use η=0.01 so that the posterior is composed of 70,000×0.01=700 data points. The ABC procedure is performed using the R package abc81. To determine the most probable model of tumor evolution (N/N, N/S, S/N or S/S) in P/M pairs, we run the postpr method implemented in the R package abc, which integrates all simulation data from the four models to run the ABC procedures (steps 1–4) and outputs the probability of each model based on the posterior distribution. The model (N/N, N/S, S/N or S/S) with the highest probability was selected.
A Monte Carlo cross-validation scheme was performed to assess the performance of SCIMET. This procedure involves randomly sampling a combination of parameters u’ and Nd’ (true parameters) and sampling 10 simulations of the summary statistics S’ under this parameter set to independently run the ABC scheme. The posterior parameters u and Nd with the maximum probability were used as parameter estimates for one simulation. The mean value of posterior u’s and Nd’s in 10 simulations was taken as the parameter estimate (inferred parameters). The process of Monte Carlo sampling and SCIMET inference was repeated 200 times under each of the four evolutionary scenarios (N/N, N/S, S/N, and S/S). Comparison of the inferred versus true parameter values indicates the robustness of this approach (Supplementary Fig. 20).
Data availability
Data have been deposited at the European Genotype Phenotype Archive (EGA) under accession number EGAS00001003573. Data from previously published studies are available at: DDBJ: JGAS00000000060 (Uchi et al.)21, and the SRA: SRP052609 (Kim et al.)29, SRP074289 (Leung et al.)30, SRP041725 (Lim et al.)31.
Code availability
Code used for genomic data analysis and simulation studies available from:
Supplementary Material
Acknowledgments
C. Curtis is supported by the National Institutes of Health through the NIH Director’s Pioneer Award (DP1-CA238296) and NCI Cancer Target Discovery and Development Network (CA217851). This work was funded in part by grants from the American Cancer Society (IRG-58-007-54), the Emerson Collective Cancer Research Fund, and a gift from the Wunderglo Foundation to C.C. Z.H is supported by an Innovative Genomics Initiative (IGI) Postdoctoral Fellowship. The project was supported in part by Cancer Center Support Grants from the National Cancer Institute to the Stanford Cancer Institute (P30CA124435) and the University of Southern California Norris Comprehensive Cancer Center (P30CA014089). We thank Jennifer Caswell-Jin and Arbel Harpak for critical feedback on the manuscript. This study is dedicated to the memory of Gloria Borges, a tireless cancer warrior.
Competing interests
A.S.B has research support from Daiichi Sankyo and honoraria for lectures, consultation or advisory board participation from Roche Bristol-Myers Squibb, Merck, Daiichi Sankyo as well as travel support from Roche, Amgen and AbbVie. M.P has received honoraria for lectures, consultation or advisory board participation from the following for-profit companies: Bayer, Bristol-Myers Squibb, Novartis, Gerson Lehrman Group (GLG), CMC Contrast, GlaxoSmithKline, Mundipharma, Roche, Astra Zeneca, AbbVie, Lilly, Medahead, Daiichi Sankyo, Merck Sharp & Dome. P.B has received travel support, honoraria for lectures, consultation or advisory board participation from the following for-profit companies: Biocartis, Novartis, Pfizer, Roche, Roche Diagnostics. C.C is a scientific advisor to GRAIL and reports stock options, as well as consulting for GRAIL and Genentech.
References
- 1.Vanharanta S & Massague J Origins of metastatic traits. Cancer Cell 24, 410–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turajlic S & Swanton C Metastasis as an evolutionary process. Science 352, 169–75 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lambert AW, Pattabiraman DR & Weinberg RA Emerging Biological Principles of Metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A 105, 4283–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell PJ et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachida S et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates LR et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 32, 169–184 e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Vijver MJ et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347, 1999–2009 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Ramaswamy S, Ross KN, Lander ES & Golub TR A molecular signature of metastasis in primary solid tumors. Nat Genet 33, 49–54 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Sanger N et al. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int J Cancer 129, 2522–6 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Husemann Y et al. Systemic spread is an early step in breast cancer. Cancer Cell 13, 58–68 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Rhim AD et al. EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosseini H et al. Early dissemination seeds metastasis in breast cancer. Nature (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL et al. Colorectal cancer statistics, 2017. CA Cancer J Clin (2017). [Google Scholar]
- 15.Andres A et al. Surgical management of patients with colorectal cancer and simultaneous liver and lung metastases. Br J Surg 102, 691–9 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Vatandoust S, Price TJ & Karapetis CS Colorectal cancer: Metastases to a single organ. World J Gastroenterol 21, 11767–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen TD, Spindler KL, Palshof JA & Nielsen DL Systematic review: brain metastases from colorectal cancer--Incidence and patient characteristics. BMC Cancer 16, 260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fearon ER & Vogelstein B A genetic model for colorectal tumorigenesis. Cell 61, 759–67 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Sottoriva A et al. A Big Bang model of human colorectal tumor growth. Nat Genet 47, 209–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryser MD, Min BH, Siegmund KD & Shibata D Spatial mutation patterns as markers of early colorectal tumor cell mobility. Proc Natl Acad Sci U S A 115, 5774–5779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchi R et al. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLoS Genet 12, e1005778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki Y et al. Multiregion ultra-deep sequencing reveals early intermixing and variable levels of intratumoral heterogeneity in colorectal cancer. Mol Oncol 11, 124–139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun R et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat Genet 49, 1015–1024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bozic I, Gerold JM & Nowak MA Quantifying Clonal and Subclonal Passenger Mutations in Cancer Evolution. PLoS Comput Biol 12, e1004731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong WS, Shpak M & Townsend JP Inferring the Origin of Metastases from Cancer Phylogenies. Cancer Res 75, 4021–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naxerova K & Jain RK Using tumour phylogenetics to identify the roots of metastasis in humans. Nat Rev Clin Oncol 12, 258–72 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Zhao ZM et al. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci U S A 113, 2140–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz R & Schaffer AA The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet 18, 213–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim TM et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Res 21, 4461–72 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Leung ML et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27, 1287–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim B et al. Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels. Oncotarget 6, 22179–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaeger R et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 33, 125–136 e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium APG AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SY et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One 9, e90459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie T et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics 104, 234–41 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Brannon AR et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 15, 454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan IB et al. High-depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver-limited metastatic colorectal cancer. Genome Biol 16, 32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Perez A et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods 10, 1081–2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Network TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martincorena I et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 171, 1029–1041 e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mamlouk S et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat Commun 8, 14093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa Y et al. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 31, 21–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weir BS & Cockerham CC Estimating F-Statistics for the Analysis of Population-Structure. Evolution 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 45.Fitch WM Toward Defining Course of Evolution - Minimum Change for a Specific Tree Topology. Systematic Zoology 20, 406–& (1971). [Google Scholar]
- 46.Naxerova K et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beaumont MA, Zhang W & Balding DJ Approximate Bayesian computation in population genetics. Genetics 162, 2025–35 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marjoram P & Tavaré S Modern computational approaches for analysing molecular genetic variation data. Nat Rev Genet 7, 759–70 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Sottoriva A, Spiteri I, Shibata D, Curtis C & Tavare S Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res 73, 41–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fumagalli A et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A 114, E2357–E2364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boutin AT et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 31, 370–382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304, 1164–6 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Zhang X et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci U S A 104, 4060–4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lui VW et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci U S A 111, 1114–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turajlic S et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594 e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers ZN et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat Genet 50, 483–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen JD et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tie J et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8, 346ra92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casadaban L et al. Adjuvant chemotherapy is associated with improved survival in patients with stage II colon cancer. Cancer 122, 3277–3287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 60.Berghoff AS et al. Differential role of angiogenesis and tumour cell proliferation in brain metastases according to primary tumour type: analysis of 639 cases. Neuropathol Appl Neurobiol 41, e41–55 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Berghoff AS et al. Invasion patterns in brain metastases of solid cancers. Neuro Oncol 15, 1664–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costello M et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41, e67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ha G et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 24, 1881–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ha G et al. Integrative analysis of genome-wide loss of heterozygosity and mono-allelic expression at nucleotide resolution reveals disrupted pathways in triple negative breast cancer. Genome Res (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B & Li JZ A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol 15, 473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Felsenstein J Phylogeny inference package. Cladistics 5, 164–166 (1989). [Google Scholar]
- 72.Siegmund KD, Marjoram P, Woo YJ, Tavare S & Shibata D Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A 106, 4828–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lloyd MC et al. Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer Res 76, 3136–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarapata EA & de Pillis LG A comparison and catalog of intrinsic tumor growth models. Bull Math Biol 76, 2010–24 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Finlay IG, Meek D, Brunton F & McArdle CS Growth rate of hepatic metastases in colorectal carcinoma. Br J Surg 75, 641–4 (1988). [DOI] [PubMed] [Google Scholar]
- 76.Kather JN et al. Identification of a characteristic vascular belt zone in human colorectal cancer. PLoS One 12, e0171378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bozic I et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A 107, 18545–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tavaré S, Balding DJ, Griffiths RC & Donnelly P Inferring coalescence times from DNA sequence data. Genetics 145, 505–18 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao J, Siegmund KD, Shibata D & Marjoram P Ancestral inference in tumors: how much can we know? J Theor Biol 359, 136–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Csilléry K, François O & Blum MG abc: an R package for approximate Bayesian computation (ABC). Methods in ecology and evolution 3, 475–479 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited at the European Genotype Phenotype Archive (EGA) under accession number EGAS00001003573. Data from previously published studies are available at: DDBJ: JGAS00000000060 (Uchi et al.)21, and the SRA: SRP052609 (Kim et al.)29, SRP074289 (Leung et al.)30, SRP041725 (Lim et al.)31.