Abstract
Augmenting live cells with novel signal transduction capabilities is a key objective in genetic engineering and synthetic biology. We showed earlier that two-component signaling pathways could function in mammalian cells, albeit while losing their ligand sensitivity. Here we show how to transduce small molecule ligands in a dose-dependent fashion into gene expression in mammalian cells using two-component signaling machinery. First, we engineer mutually complementing truncated mutants of a histidine kinase unable to dimerize and phosphorylate the response regulator. Next, we fuse these mutants to protein domains capable of ligand-induced dimerization, which restores the phosphoryl transfer in a ligand-dependent manner. Cytoplasmic ligands are transduced by facilitating mutant dimerization in the cytoplasm, while extracellular ligands trigger dimerization at the inner side of a plasma membrane. These findings point to the potential of two-component regulatory systems as enabling tools for orthogonal signaling pathways in mammalian cells.
Introduction
The engineering of novel signaling modalities in cells has already led to breakthroughs in basic research, biotechnology, and medicine1–5. The approaches encompass protease-triggered transduction6–9, rewired10 or chimeric11 receptors, redesigned scaffolds1, intein splicing12, 13, reengineered notch receptors14, and multi-input logic circuits15, 16. Some approaches rely on native signaling and thus are not fully decoupled from the host cell, while others utilize a single-turnover proteolysis of the receptor and an irreversible release of a transcriptional activator. Creating multiple-turnover yet sufficiently orthogonal signaling pathways still represents a major challenge. Earlier we showed that components of prokaryotic two-component systems (TCSs) were able to perform phosphoryl transfer and elicit phosphorylation-dependent gene regulation in mammalian cells17. TCSs' histidine/aspartate phosphorelay, different from vertebrate serine/tyrosine/threonine relay, pointed to their potential as orthogonal signaling modality in mammalian cells. Yet, dose-dependent response to a stimulus has remained elusive.
A typical TCS comprises a membrane-spanning histidine kinase sensor protein (HK) and its cognate cytoplasmic partner called a response regulator (RR). With a few exceptions18, an HK sensor forms a homodimer. A signal is transduced via HK autophosphorylation that relies on two distinct dimer conformations. In the unstimulated conformation, the catalytic ATP-binding (CA) domain is distant from the histidine in the dimerization and histidine-containing phosphotransfer (DHp) domain (Fig. 1a). Ligand binding to the extracellular HK domain induces conformational change and places the bound ATP closer to the histidine, resulting in phosphotransfer19. The phosphorylated histidine in the DHp domain transfers the phosphate to the aspartate in the receiver domain of the cognate RR20. The phosphorylated RR binds to its target promoters and regulates gene expression. In addition, an unphosphorylated HK can dephosphorylate an RR, actively shutting down the signaling21. The balance between the kinase and phosphatase activities of an HK determines signaling intensity and dynamics22.
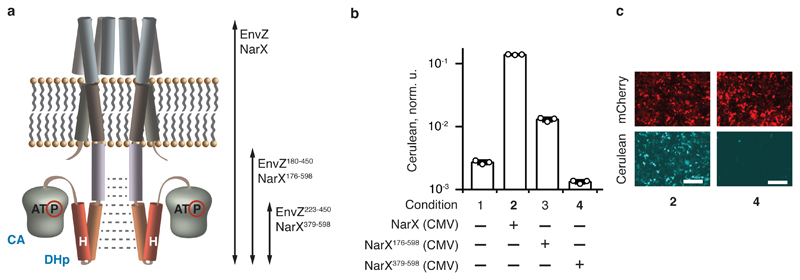
Fig. 1. Identification of HK domains with reduced intrinsic signaling.
a, A representation of an HK receptor in a cell membrane. CA domain with bound ATP and DHp domain containing the phosphorylatable histidine (H) are shown. Dimerization is indicated with dotted gray lines. The arrows span the various tested domains (see also Supplementary Fig. 2). b, Signaling capacity of truncated NarX. The bars show Cerulean levels in cells coexpressing NarL and an indicated NarX variant in the presence of NarL-inducible Cerulean reporter. Cerulean expression is normalized to the transfection control and shown as mean ± SD of independent biological triplicates. The circles indicate individual measurements. c, Microscopy images of HEK293 cells for conditions shown in bold in panel b. The top and the bottom rows show, respectively, the expression of constitutive mCherry transfection control (red), and pathway-induced Cerulean reporter (cyan) in the same transfection. The numbers correspond to the conditions in b. The white scale bar is 200 μm. The DNA constructs are described in Supplementary Fig. 1. The results were reproduced at least once in an independent experiment.
In this study, we show that external signals can be transduced in a dose-dependent fashion via two-component signaling into gene expression in mammalian cells. The underlying engineered cascade utilizes ligand-triggered proximity of mutually-complementing mutants of a histidine kinase cytoplasmic domain. We demonstrate dose-dependent signaling by cytoplasmic and extracellular ligands, the former via ligand-induced protein-protein interactions and the latter via re-engineered G protein-coupled receptors (GPCR)/β-arrestin interaction.
Results
Impact of N-terminal deletion on HK activity
Previously, we transplanted TCS to mammalian cells by (i) human codon optimization of the nucleic acid sequences coding for HK and RR; (ii) fusing a mammalian transactivator domain to an RR; and (iii) creating an RR-responsive promoter by fusing RR binding DNA sequences upstream of a minimal Pol II promoter (YB-TATA15) driving a gene of interest. We observed that the full-length HKs, including EnvZ and NarX, triggered constitutive signaling in mammalian cells when coexpressed with their cognate RRs OmpR-VP48 and NarL-VP48 (labeled for brevity OmpR and NarL) and in the presence of the RR-responsive reporters, suggesting that the ligand-induced conformation change was impaired17. In addition, that study showed that HK fluorescent fusions were excluded from the nucleus. Therefore, an RR had to be phosphorylated outside, and drive gene expression inside, the nucleus, indicating that the RR can move freely between the nucleus and the cytoplasm.
Our observations suggested that in mammalian cells, a full-length HK dimerized in a phosphotransfer-enabling conformation despite its transmembrane domains. We tested whether N-terminal deletions of an HK would reduce dimerization and signaling (Supplementary Figs. 1 and 2a). We cloned complete cytoplasmic domains of envZ and narX (EnvZ180-450 and NarX176-598), as well as reduced-size domains starting about 20 amino acids upstream of the phosphorylatable histidine (EnvZ223-450 and NarX379-598, Supplementary Fig. 1). A similar deletion in the HK RstB led to homodimer dissociation23. Truncated EnvZ mutants trigger constitutive signaling similar to wild-type EnvZ (Supplementary Fig. 2bc), agreeing with the earlier finding that DHp domain of EnvZ is able to homodimerise24. However, NarX deletions show size-dependent reduction in signaling, dropping to background levels with NarX379-598 (Fig. 1bc). Possible explanations include (1) a reduction in protein stability; (2) a decrease in the kinase activity and/or an increase in the phosphatase activity of the truncated mutant and (3) the inability of the mutant to dimerize on its own. The latter explanation would support the establishment of a synthetic signaling process with NarX379-598 via restoring its dimerization in a ligand-dependent fashion. Constitutive dimerization despite truncation, as observed with EnvZ, would exclude the possibility of establishing ligand-induced signaling.
Fusion to dimerizing proteins reactivates short NarX
To determine if forced dimerization could restore signaling, SYNZIP1 and SYNZIP2 proteins, known to form strong heterodimers in mammalian cells25, were fused to NarX379-598. We reasoned that if the dimerization alone was impaired due to truncation, these fusions would regain the capacity to dimerize and transduce the signal downstream. Increased reporter expression in the presence of SYNZIP1 and SYNZIP2 N-terminal (but not C-terminal) fusion pair, consistent with the position of the sensor domain relative to the cytoplasmic domain in the full-length HK (Supplementary Figs. 1 and 3), hinted at the possibility of forced dimerization as a signaling mechanism.
Our components are expressed from constitutive promoters, and changes in reporter expression in the presence of small molecule ligands, could be an artifact resulting from modulating promoter activity rather than bona fide signaling. To eliminate such artifacts, we evaluated a number of constitutive promoters, including CMV, full-length EF1a and a reduced-size, 5-fold weaker variant EF1a-V1 (Altamura et al, in preparation). Epinephrine and procaterol, known to induce both TCS and mammalian signalling, induce the CMV promoter by a factor of two, while EF1α promoter is not affected (Supplementary Fig. 4a). Further, narX driven by the weaker EF1a-V1 generates as much signalling as the wild-type EF1α, while narL expression from the stronger promoter results in stronger signalling (Supplementary Fig. 4b). In light of these results we opted to use EF1α-V1 to drive narX-based constructs and full-length EF1α to drive narL.
NarX mutants in CA and DHp domains complement each other
Following up on the preliminary indications, we attempted to improve the dynamic range of induction. HKs fall into two families with respect to the autophosphorylation mechanism26. In the “cis”-family, the phosphoryl group of an CA-domain bound ATP is transferred to the phosphorylatable histidine of the same monomer. In the “trans”-family, phosphoryl transfer takes place between the two monomers (Fig. 2a). For all HKs, phosphotransfer can be prevented by either mutating the ATP binding site or the histidine. However, a heterodimer between complementing ATP binding site and histidine mutants of a “trans”-family HK would still be able to signal via unidirectional phosphate transfer from the histidine mutant to the ATP-binding site mutant23 (Fig. 2a).
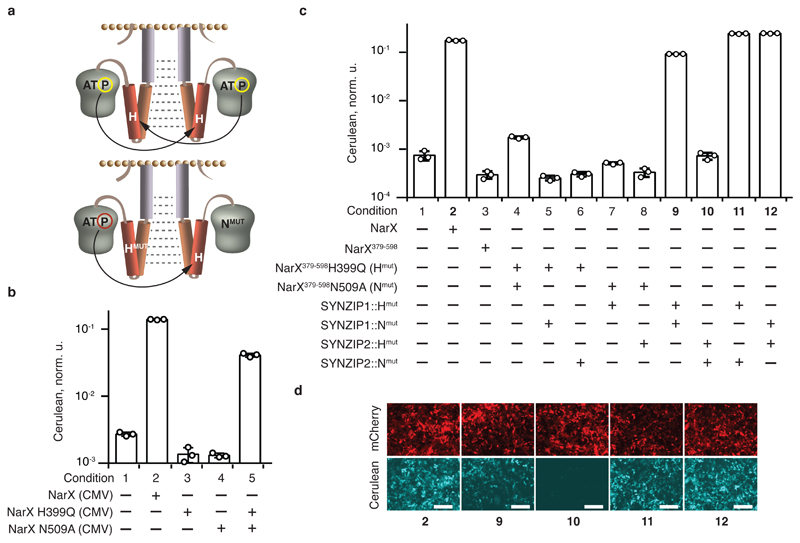
Fig. 2. Restoration of two-component signaling via forced dimerization.
a, Schematics of trans-autophosphorylation. The top and bottom schemes illustrate, respectively, the phosphorylation in a homodimer of a wild-type HK, and in a heterodimer of complementing ATP binding site and histidine mutants. b, Complementation activity assays for full-length NarX. The bars show Cerulean levels in cells coexpressing NarL and an indicated NarX variant, in the presence of NarL-inducible Cerulean reporter. c, Reestablishment of signaling via fusions to SYNZIP1 and SYNZIP2. The bars show Cerulean levels in cells coexpressing NarL and the indicated combinations of NarX- and SYNZIP-derived constructs, in the presence of NarL-inducible Cerulean reporter. See Supplementary Fig. 5 for schematic illustration of the protein fusions. d, Microscopy images of HEK293 cells for conditions shown in bold in panel c. The top and the bottom rows show, respectively, the expression of constitutive mCherry transfection reporter (red), and pathway-induced Cerulean reporter (cyan) in the same transfection. The numbers correspond to the conditions in panel c. Cerulean expression is normalized to the transfection control and shown as mean ± SD of independent biological triplicates. The circles indicate individual measurements. The white scale bars in d correspond to 200 μm. The DNA constructs are described in Supplementary Fig. 1. The results were reproduced at least once in an independent experiment.
We hypothesized that dimerization between complementing mutants could improve the signaling due to reduced phosphatase activity of the mutant HK towards its cognate response regulator21. Complementation had been shown in vitro with purified EnvZ, PhoR and RstB23, 27, and we tested it in mammalian cells with NarX. We mutated asparagine N509 likely important for ATP binding (see Methods), and the phosphorylatable histidine of NarX. Complementation assay with the full-length NarX mutants (Fig. 2b) shows that the asparagine or the histidine mutants are unable to signal on their own, as expected, while co-expression of the complementing mutants partially restores the signaling, suggesting that NarX belongs to the “trans” HK family.
Next, the sequences coding for NarX379-598 mutated at the histidine or the asparagine were fused at their 5'-terminus to the sequences coding for SYNZIP1 and SYNZIP2, creating, respectively, SYNZIP1::NarX379-598H399Q (SYNZIP1::Hmut) and SYNZIP2::NarX379-598N509A (SYNZIP2::Nmut) (Supplementary Figs. 1 and 5ab) and vice versa (SYNZIP2::Hmut and SYNZIP1::Nmut). These pairs restore signaling to the level obtained with full-length, wild-type NarX, generating a much stronger signal compared to SYNZIP fusions to NarX379-598(Figs. 2c, compare conditions 2,3,11 and 12, and Fig. 2d). When one or both of the fused SYNZIP domains are missing, the signaling stops (Fig. 2c, conditions 4-8, and Supplementary Fig. 5c), indicating that the dimerization of the fusion domains is required to restore the signaling. Interestingly, the pair of complementing NarX mutants, both fused to SYNZIP1 (SYNZIP1::Hmut and SYNZIP1::Nmut), also results in elevated signaling activity (Fig. 2c, condition 9), consistent with the (weaker) effect observed under similar conditions with the wild type domain (Supplementary Fig. 5d, condition 9). We hypothesize that SYNZIP1 domain is capable of homodimerizing, albeit with a reduced affinity compared to SYNZIP1-SYNZIP2 interaction. Indeed, original literature suggests that SYNZIP1, and to much lesser extent SYNZIP2, exist as both a monomer and a dimer (Fig. 1a in Thompson et al25).
The contribution of the individual mutants to signaling efficiency was assessed by coexpressing SYNZIP fusions to mutant and wild-type NarX domains (Supplementary Fig. 5d, conditions 7 and 8). The inclusion of at least one mutant improves the induction compared to the wild-type sequence. The major contribution is due to the histidine mutant SYNZIP1::Hmut but the asparagine mutant SYNZIP1::Nmut is also beneficial, with their combination resulting in superior performance.
The dose-response was determined by varying plasmid dosage of the NarX-derived constructs (Supplementary Fig. 5e). Signaling activity increases with plasmid dosage. The full-length wild-type NarX exhibits the highest dosage sensitivity, likely reflecting the strongest dimerization constant, the SYNZIP1-SYNZIP2 pair shows a reduced but comparable dimerization behavior, and SYNZIP1 clearly shows inferior dimerization. Thus, reducing the plasmid amount reduces SYNZIP1-SYNZIP1 signaling to background while still generating strong signaling via SYNZIP1-SYNZIP2 dimerization. NarX acts catalytically, with NarX:NarL ratio of about 1:10 (based on relative promoter strength and plasmid dosage) fully activating the system. This is consistent with two-component signaling stoichiometry of about 1:30 HK:RR ratio in E. coli28.
TCS rewires FKBP/FRB interaction to gene expression
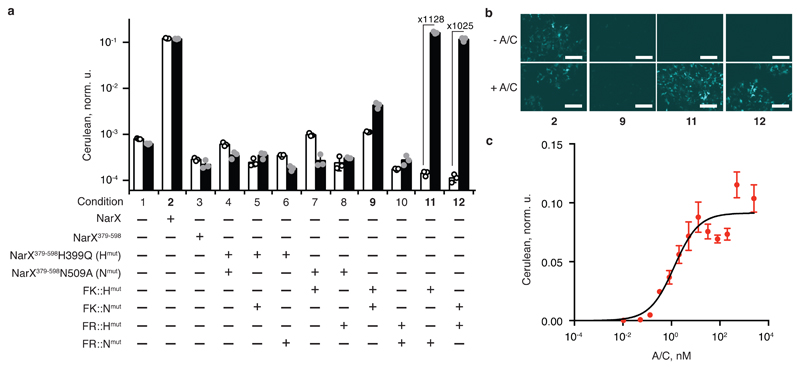
To enable bona fide signaling, the dimerization of the NarX domains should be controlled by an external stimulus. A well characterized ligand-induced heterodimerization takes place between the proteins FK506-binding protein 12 (FKBP) and FKBP12-rapamycin binding domain (FRB) mutant FRBT2098L in the presence of A/C heterodimerizer (a rapamycin analog C16-(S)-7-methylindolerapamycin, known also as AP21967)29, 30. To find out if NarX domains are capable of transducing this interaction (Supplementary Fig. 6a), complementing NarX mutants were fused at their N-terminus to the FKBP (FK) and the FRBT2098L (FR) proteins, respectively, resulting in FK::NarX379-598H399Q (FK::Hmut) and FR::NarX379-598N509A (FR::Nmut), and the inverse pair FR::Hmut and FK::Nmut (Supplementary Fig. 1). First, we confirmed that A/C did not affect the wild-type NarX/NarL signaling (Fig. 3a, conditions 2 and 3). Next, we probed the ligand-induced signaling by expressing different fusion variants and control constructs in HEK293 cells in the presence of the response regulator NarL and NarL-inducible reporter with and without the ligand (Fig. 3ab and Supplementary Fig. 6bc). In all the cases except with wild-type NarX, there is no signaling in the absence of the ligand. High ligand concentration induces strong signaling with a dynamic range above 3 orders of magnitude (Fig. 3a, conditions 11 and 12) only when (i) the NarX-derived domains contain complementary histidine and asparagine mutations and (ii) they are fused, respectively, to FKBP and FRB interaction partners. The dose-response (Fig. 3c) shows an expected Hill function dependency (Supplementary Table 1), with the EC50 of 1.3 nM, similar to the published values of 10 nM31 and 36 nM29.
Fig. 3. Transduction of cytoplasmic ligand to gene expression.
a, Ligand-induced signaling with the help of FKBP/FRB fusions. The bars show Cerulean levels in cells coexpressing NarL and the indicated combination of NarX- and FKBP- or FRB- derived constructs, in the presence of NarL-inducible Cerulean reporter. The white and the black bars show Cerulean expression in the absence and in the presence of 100 nM of A/C ligand, respectively. The fold change of A/C induction is shown for conditions 11 and 12. See Supplementary Fig. 6 for schematic illustrations of the process. b, Microscopy images of HEK293 cells for conditions shown in bold in panel a. The top and the bottom rows show, respectively, the reporter expression in the absence and in the presence of A/C. The numbers correspond to the conditions in panel a. c, Dose response to the amount of A/C dimerizer. NarL-regulated Cerulean reporter expression is shown for different A/C concentrations in the presence of NarL and the pair FR::Hmut and FK::Nmut. In this figure, Cerulean levels are normalized to the transfection control and averaged over a biological triplicate, shown as mean ± SD. The circles indicate individual measurements. The white scale bar in b corresponds to 200 μm. The DNA constructs are described in Supplementary Fig. 1. The results were reproduced at least once in an independent experiment.
Dimerization is the driving force behind TCS signaling
The above observations begged the question whether the dimerization was the only reason behind restored signaling, or if some of the data could also be explained by NarX truncated mutant stabilization due to fused domains. Indeed, in our preliminary experiments (Supplementary Fig. 3), fusing SYNZIP2 alone to NarX379-598 resulted in a small increase over the background, SYNZIP1 alone in a slightly stronger increase, and the coexpression of SYNZIP1 and SYNZIP2 pair had the strongest effect. However, as we discuss above, both SYNZIP1 and SYNZIP2 are able to homodimerize, with the former creating a medium-strength and the latter a very weak homodimer; SYNZIP1-SYNZIP2 form a strong heterodimer. The ranking of the effects observed with NarX fusions is therefore fully consistent with the dimerization strength of the fused domains; HK mutant stabilization would result in similar output increase for all the variants irrespective of the dimerization constant of the fused domains.
Another evidence comes from the ligand-controlled FKBP-FRB system (Fig. 3), which allows direct observation of the effect triggered by the fused domain dimerization as opposed to a putative dimerization of the stabilized NarX domains. We mirrored the experiments described in the previous paragraph, this time using FK and FR pair (Supplementary Fig. 7). The FK and FR domains were fused at the N-terminus of NarX379-598 resulting in FK::NarX379-598 and FR::NarX379-598, respectively. The fusion of the FK or FR domain alone to NarX379-598 slightly increases the reporter expression compared to "naked" NarX379-598 (Supplementary Fig. 7, conditions 3-5); the presence of the A/C heterodimerizer does not affect the output in this case. However, coexpressing FK::NarX379-598 and FR::NarX379-598 (Supplementary Fig. 7, condition 6) results in strong ligand-dependent induction. Accordingly, the mere fusion of a protein sequence at the N-terminus of NarX379-598, which might potentially stabilize the truncated NarX, has a very weak effect of its own, if any. In summary, our dataset shows that the dimerization is the main factor that restores NarX-enabled signaling.
Rewiring the GPCR/β-arrestin interaction via TCS
Many transmembrane signaling pathways involve protein-protein interactions at the cytoplasmic surface of the lipid bilayer, including an important class of signaling pathways initiated by GPCR, a family of a few hundred proteins32, 33. A key step of GPCR signal transduction is a complex formation between the GPCR itself and the β-arrestin protein34, followed by GPCR internalization, recycling, and signaling35, 36. This interaction was used to rewire GPCR signaling by specific proteolytic cleavage of a fused transcriptional activator6, 8, for example, in a Tango assay6, which employs transcriptional activator tTA fused to a GPCR via a TEV protease cleavage site. In that assay, tTA is released when β-arrestin-2, fused to TEV protease, is recruited to the GPCR following stimulation with a GPCR ligand (Supplementary Fig. 8a). One of the GPCRs engineered in that work6 to increase β-arrestin affinity is a fusion between the ligand binding domain of procaterol-activated Beta-2 adrenergic receptor (β2AR), and the cytoplasmic fragment of Vasopressin V2 receptor (V2R)6, or β2AR∆C::V2R∆N. To examine whether GPCR stimulation could be rewired via TCS machinery (Fig. 4a), we fused this chimeric GPCR to the sequence coding for NarX379-598H399Q (β2AR∆C::V2R∆N::Hmut) and β-arrestin-2 to NarX379-598N509A (β-arrestin::Nmut). Inverse fusions were constructed as well (Supplementary Fig. 1).
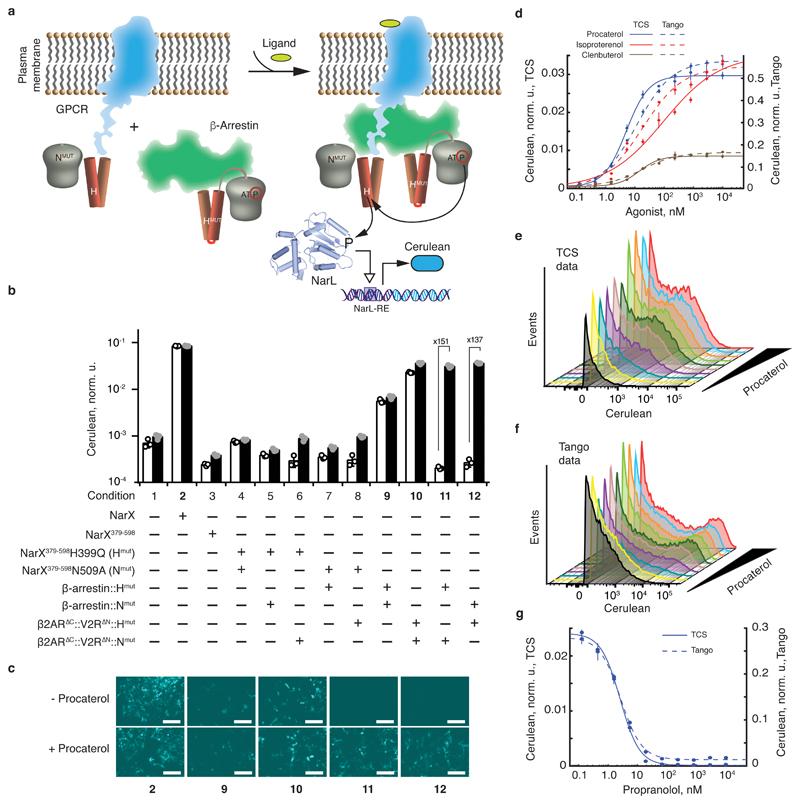
Fig. 4. Rewiring the GPCR activity to the expression of a reporter gene.
a, Schematics of transducing ligand-induced GPCR/β-arrestin interaction34 into gene expression via TCS machinery. b, Ligand-induced reporter activation via GPCR rewiring. NarL-inducible Cerulean expression is shown in the presence of NarL and the indicated NarX-, β2AR∆C::V2R∆N, and/or β-arrestin-derived constructs. The white and the black bars correspond to the absence and the presence of 2 μM procaterol, respectively. The fold-change of procaterol-triggered induction is shown for the conditions 11 and 12. c, Microscopy images of HEK293 cells for selected conditions shown in bold in panel a. The top and the bottom rows show reporter expression without and with 2 μM of procaterol, respectively. The numbers correspond to the conditions in a. d, Reporter expression as a function of different β2AR agonists' concentration. Full lines represent the results obtained with β2AR∆C::V2R∆N::Hmut and β-arrestin::Nmut in the presence of NarL and NarL-regulated Cerulean reporter (TCS). Dotted lines represent the results obtained with the Tango assay using chimeric GPCR βAR∆C::V2R∆N::tTA. The primary Y axis is used for the TCS data and the secondary Y axis is used for Tango data. d, e, Fluorescence histograms in Cerulean-positive cells obtained for increasing amounts of procaterol with the TCS-based transduction (d) and Tango (e). f, Dose response to the antagonist propranolol in the presence of 100 nM of procaterol. The full line shows data obtained with TCS-based transduction and the dashed line summarizes data obtained with Tango assay. Primary and secondary Y-axes are as in (d). All values in the bar chart and the graph are Cerulean levels normalized to transfection control and averaged over a biological triplicate, shown as mean ± SD. The circles indicate individual measurements. The white scale bar in c corresponds to 200 μm. The DNA constructs are described in Supplementary Fig. 1. The results were reproduced at least once in an independent experiment.
First, we confirmed that procaterol did not affect the signaling via NarX/NarL system (Fig. 4b, conditions 2 and 3). Coexpression of the complementary NarX mutants fused, respectively, to the GPCR receptor, β-arrestin, or both, shows a certain amount of procaterol-independent signaling with β-arrestin and the GPCR receptor alone (Fig. 4bc and Supplementary Fig. 8bc), suggesting that these proteins dimerize to some extent in a ligand-independent fashion. Importantly, strong procaterol-triggered signaling takes place when the complementary NarX mutants are fused, respectively, to the GPCR receptor and the β-arrestin (Fig. 4b). The dynamic range obtained with TCS is higher than the one obtained with the comparable Tango assay, even after attempted optimization of the latter using EF1α promoter combination (Supplementary Fig. 8d).
We characterized the dose response of the pair β2AR∆C::V2R∆N::Hmut and β-arrestin::Nmut in the presence of two agonists (procaterol, isoproterenol) and a partial agonist (clenbuterol), which induces lower maximum response compared to the agonists (Fig. 4d). Procaterol and isoproterenol induce strong downstream gene expression in a dose-dependent manner and reach the same maximum response at saturating doses. Partial agonist clenbuterol induces about 3.5 times lower expression, as expected. EC50 values of procaterol, isoproterenol and clenbuterol were determined to be 5 nM, 30 nM and 14 nM, respectively, similar to published values37 and to the values determined using Tango assay (Fig. 4d, Supplementary Table 1). The Tango assay results in higher absolute reporter expression yet higher leakage compared to TCS-based signaling (Fig. 4ef). We also found that the antagonist propranolol inhibited the effect of procaterol with IC50 of about 3 nM in both the TCS and the Tango assays (Fig. 4g). These results demonstrate that TCS-based signaling can faithfully recapitulate a variety of known effects of agonists and antagonists on the GPCR activity, and it can generate quantitative data on interaction parameters.
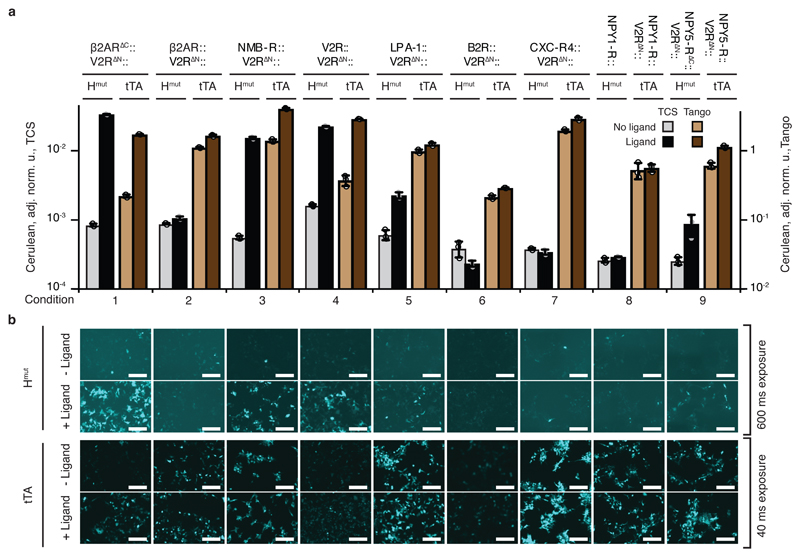
We tested our approach with additional GPCRs and their ligands, inspired by the PRESTO-Tango approach in which full length GPCRs are fused to the V2R∆N, TEV protease cleavage site, and tTa38. We fused six full-length GPCRs, including β2AR, and one truncated GPCR NPY5-R∆C, to V2R∆N followed by NarX379-598H399Q (Hmut). One full-length GPCR NPY1-R was fused directly to Hmut without V2R∆N. This resulted in the total of eight new constructs (Supplementary Fig. 1). TCS-based signal rewiring was examined by coexpressing GPCR::Hmut fusions, β-arrestin::Nmut, and NarL in HEK293 cells in the presence of the NarL-activated reporter. Reporter expression was quantified in the absence or presence of the cognate GPCR ligands at saturating concentrations8, 38. The corresponding Tango cascades utilizing GPCR-tTA fusions, β-arrestin::TEV and tTA-inducible reporter were set up alongside and tested in a similar manner (Fig. 5ab and Supplementary Fig. 9). Using TCS approach, five of the eight GPCR::Hmut fusions triggered more than two-fold reporter induction by their cognate ligands (Fig. 5, conditions 1, 3- 5, 9 and Supplementary Table 2). With the Tango method, only three GPCR::tTA fusions show more than two-fold reporter induction (Fig. 5 and Supplementary Table 2). Thus, the dynamic range of the rewired TCS-based GPCR signaling outperformed the reference benchmark of Tango assay in all cases.
Fig. 5. Multiple GPCRs are rewired to induce gene expression.
a, Characterization of multiple GPCRs rewired via TCS and Tango cascades. Every experimental condition (numbered on X axis) represents a particular GPRC (indicated above the top horizontal line) fused either to Hmut (TCS) or tTA (Tango), as shown. GPCR Hmut fusions are coexpressed with β-arrestin::Nmut and NarL in the presence of NarL-inducible Cerulean reporter. GPCR tTA fusion is coexpressed with β-arrestin-TEV and tTA-inducible TRE-driven Cerulean reporter. Gray and light brown bars correspond to Cerulean expression in the absence of a cognate GPCR ligand, and the black and dark brown bars show Cerulean expression in the presence of a ligand. The primary Y axis is used for the TCS experiments and the secondary Y axis for Tango experiments. b, Representative microscopy images of HEK293 cells show Cerulean reporter expression with (+Ligand) and without the ligand (-Ligand) for TCS-based GPCR induction (two upper rows) and for Tango-based induction (two bottom rows). See Supplementary Fig. 9 for transfection control images. The bar charts display Cerulean level normalized to the transfection control and averaged over an independent biological triplicate as mean ± SD. Circles show individual measurements. The constructs are described in Supplementary Fig. 1. The white scale bars in panel b correspond to 200 μm. The results were reproduced at least once in an independent experiment.
Our results show that multiple ligand-GPCR interactions can be transduced by the TCS. Successfully-rewired GPCRs recognize different types of ligands (Supplementary Fig. 10a), suggesting that the ligand class is not a success factor. A phylogenetic tree of all the crystalized class A GPCRs and the eight tested GPCRs (Supplementary Fig. 10a) shows that two of the three non-responsive GPCRs, CXC-R4 and B2R, cluster together, suggesting that a subclass of the GPCR might be more challenging to rewire. Aligning the C-terminal domains of the successfully-rewired GPCRs, we found a conserved sequence SLLCCxR ~9 amino acids downstream of the C-terminus of 7th transmembrane helix (Supplementary Fig. 10b). This sequence is absent in NPY1-R, CXC-R4, and B2R. The cysteine in the consensus sequence was described as the palmitoylation site of β2AR39 and V2R40, suggesting that the ability to rewire might depend on this sequence or on GPCR palmitoylation state. Some of the fusions that failed to induce in our hands using either the TCS or Tango approach, e.g., β2AR::V2R∆N, had been shown to respond better in a cell line with stably integrated signaling components and a luciferase reporter38. Lastly, the same GPCR can respond differently depending on the fusion structure. For example, β2AR∆C::V2R∆N::Hmut, but not β2AR::V2R∆N::Hmut, responds to procaterol (Fig. 5, conditions 1 and 2), highlighting the importance of a linker between the GPCR and the truncated NarX domain. These observations point toward possible future fine-tuning of our approach.
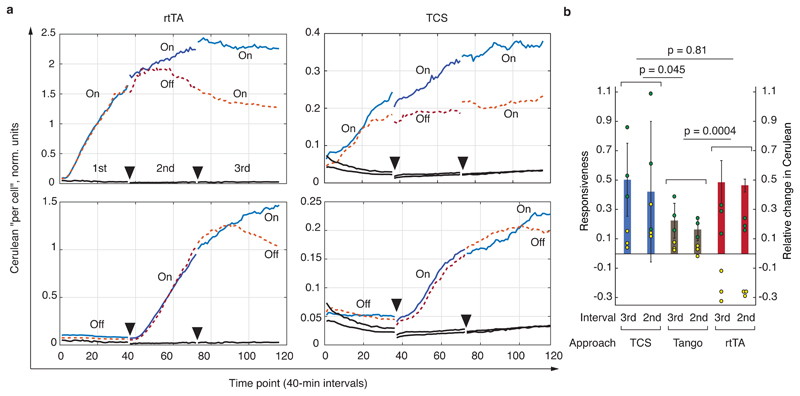
Next, we compared the dynamic response of TCS-enabled signaling to the canonical Doxycycline-inducible rtTA-regulated expression and the Tango approach. We tracked Cerulean expression in HEK293 cell expressing the components of GPCR signaling rewired via either the TCS,the Tango, or the rtTA approach (Fig. 6 and Supplementary Figs. 11 and 12). The rtTA system represents the "best-case" scenario in which DNA binding is modulated instantaneously by the small molecule Doxycycline. The time course was followed over three time intervals during which the inducer (procaterol for GPCR sensors, Doxycycline for rtTA) was either added (On) or withheld (Off) (Methods, Fig. 6a, Supplementary Fig. 11). Despite noisy raw data, a number of conclusions can be made. The Tango system is the leakiest among the three. It has the fastest response when initially activated in the first (but not the second) incubation interval, however, the response slows down quickly and becomes slower compared to the TCS and the rtTA. We speculate that the membrane-bound receptors, accumulated before the first induction, are rapidly processed by the TEV protease, and after this phase, the signal is generated by newly synthesized receptors, which results in slower response. Moreover, membrane-bound GPCRs whose tTA domain has been cleaved can still compete for ligand and β-arrestin binding, reducing the response even further. On the other hand, the induction dynamics of both TCS fusion and rtTA are qualitatively very similar. Because the modulation of rtTA binding to DNA with Doxycycline is very fast, we conclude that the induction dynamics of TCS fusion, in our assay, is on par with the benchmark. It is much more robust compared to Tango, which slows down very quickly after the first burst.
Fig. 6. Dynamic characterization of the various signaling approaches.
a, Selected time lapse traces of normalized output expression. Charts show, from left to right, the response of rtTA-Doxycycline (rtTA) system, and GPCR signaling via TCS pathway (Hmut fusions). Each trace is labeled either On or Off, with On indicating the presence of a ligand and Off means absence of a ligand during the measurement interval. Dotted lines indicate time course that include an Off condition during the 2nd or the 3rd interval, and solid lines are time courses with On conditions during both the 2nd and the 3rd interval. Full triangles indicate the endpoints of the measurement intervals. Solid black lines are background readouts. See Supplementary Fig. 11 for the entire dataset including the time traces obtained with the Tango assay. b, Responsiveness compared for the three approaches. The interval (3rd or 2nd, see also top left chart in panel a) indicated on the X axis is the interval during which the On and the Off responses were compared. The yellow and the green dots, respectively, represent the relative change in the expression of the Cerulean reporter (see Methods and Supplementary Fig. 12) during the On (green) and Off (yellow) intervals. The bars indicate the responsiveness during each interval, i.e., the difference between the relative Cerulean change during an On and Off interval (Methods), and the error bars are calculated from the raw data (circles) using appropriate error propagation. The p-values are the products of the p-values calculated for the responsiveness comparison between matched intervals (2nd vs 2nd and 3rd vs 3rd, two-sided t-test, 8 degrees of freedom, also see Methods). The primary Y axis shows responsiveness and the secondary Y axis shows relative change in the Cerulean expression. The entire experiment was performed once, every condition was measured as a biological triplicate, and the conclusions are drawn upon analysis of six independent measurements (three for the 2nd and three for the 3rd interval comparison).
In order to assess the inactivation dynamics of the various sensor systems, we developed a robust measure we call "responsiveness" (Supplementary Fig. 12, Methods). In this analysis, the Tango system shows the worst responsiveness, consistent with qualitative examination of the time course data and with the hypothesis of rapid depletion of functional sensors following irreversible cleavage by the TEV fused to β-arrestin. Both the TCS fusion and the rtTA have superior responsiveness and are statistically indistinguishable from one another (Fig. 6b). Thus, also from the aspect of signaling shutdown, the TCS fusion behaves in our assay on par with rtTA.
Discussion
Implementing non-native signaling modalities in cells in general and mammalian cells in particular is highly desirable for rational control of cell behavior and ultimately, engineering novel cellular functions for basic research, biotechnology and medicine. Two-component histidine/aspartate signaling has apparently co-evolved independently of the serine/threonine/tyrosine signaling, and to the best of our knowledge, not a single instance of histidine to aspartate phosphoryl transfer has been described in vertebrate cells41. Native mechanism of TCS signal transduction in prokaryotes relies on ligand-induced conformation change of the HK dimer in the membrane, but direct implementation of this mechanism in mammalian cells has been elusive. Instead, here we pursued a different strategy to achieve the same end result by controlling the switch between dissociated and associated states of the HK cytoplasmic domains. In the cases shown here, the switching was accomplished by ligand-induced dimerization of proteins fused, respectively, to histidine and asparagine mutants of a truncated cytoplasmic domain of NarX. A similar qualitative effect is observed when wild-type truncated domain is used instead of the mutants. However, the quantitative performance is inferior and more importantly, the resulting effect does not distinguish between ligand-induced dimerization of the two interaction partners and homo-dimerization of one of the partners.
Our approach retains many of the features of the original prokaryotic signaling. It is an amplifying, multiple turnover process with a single NarX dimer capable of phosphorylating multiple copies of the response regulator NarL, which in turn can induce multiple transcription initiation events. Our time-lapse data suggest that single-turnover Tango approach results in rapid response desensitization due to the quick depletion of the GPCR molecules fused to the tTA moiety, and possible competition between newly synthesized and cleaved GPCR receptors for ligand and β-arrestin binding. In contrast, the TCS-based rewired GPCR signaling dynamics are comparable to posttranslational regulation of rtTA-driven gene expression both during induction and shutdown. These characteristics will allow the development of sensor cells with more robust and sustainable responsiveness, compared to the ones utilizing irreversible protein turnover.
Multiplexing is a potential advantage of our approach. In principle, multiplexing can be achieved by building a library of HK cytoplasmic domains belonging to the “trans” family that fail to dimerize on their own, similar to NarX short cytoplasmic domain. Multiple sets of mutually-complementary pairs and their cognate response regulators and inducible promoters can in principle operate in parallel, to probe either cytoplasmic or transmembrane interactions. Systematic screening of multiple HK candidates is a promising strategy to uncover such pairs in an unbiased fashion.
The TCS-based signaling is intrinsically orthogonal to the endogenous mammalian signaling because HKs are unable to phosphorylate human proteins, including those involved in signaling. The interface between the TCS-based pathway and a mammalian cell is the output protein induced by the RR. This can be an arbitrary gene product, including a component of a mammalian signaling pathway, be it a cell surface receptor or an intermediate. Building interfaces between artificial and endogenous signaling holds great promise in the field of cell engineering and reprogramming.
Methods
Plasmid Construction
Plasmids were constructed using standard cloning techniques. All restriction enzymes used in this work were purchased from New England Biolabs (NEB). Q5 High-Fidelity DNA Polymerase (NEB) was used for fragment amplification. Single-stranded oligonucleotides were synthesized by Sigma-Aldrich. Digestion products or PCR fragments were purified using GenElute Gel Extraction Kit or Gen Elute PCR Clean Up Kit (Sigma-Aldrich). Ligations were performed using T4 DNA Ligase (NEB) by temperature cycle ligation with 140 cycles between 30 s at 10°C and 30 s at 30°C. Gibson assembly was done as described below. 5 μl of the ligation product or the Gibson assembly product were transformed into chemically competent E.Coli DH5α or E. coli TOP10 that were plated on LB Agar with Ampicillin at 100 μg/ml. The resulting clones where screened directly by colony-PCR (Dream Taq Green PCR Master Mix, Thermo Scientific). We expanded single clones in LB Broth Miller Difco (BD) supplemented with ampicillin and purified plasmid DNA using GenElute Plasmid Miniprep Kit (Sigma-Aldrich). All the resulting plasmids were sequence-verified by Microsynth using Sanger sequencing method. The DNA for mammalian transfection was obtained from 100 ml of liquid culture using the Promega PureYield™ Plasmid Midiprep System (A2495). The recovered DNA was further purified using the Norgen Endotoxin Removal Kit Mini (Cat.# 27700) or Midi (Cat.# 52200). A short cloning procedure for each construct used in this work is described in Supplementary Note 1.
Gibson assembly protocol
The Gibson assembly42 was performed in 10 μl final volume by mixing vectors (0.018 pmol) and inserts (0.09 pmol) in 1X Gibson assembly buffer (0.1 M Tris-HCl, pH 7.5, 0.01 M MgCl2, 0.2 mM dGTP, 0.2 mM dATP, 0.2 mM dTTP, 0.2 mM dCTP, 0.01 M DTT, 5% (w/v) PEG-8000, 1 mM NAD), 0.04 units of T5 exonuclease (NEB), 0.25 units of Phusion DNA polymerase (NEB) and 40 units of Taq DNA ligase (NEB). Negative controls for Gibson assemblies included vectors alone. Gibson assembly reactions were incubated at 50°C for 1 h.
Identification of CA mutants of NarX
Mutational analysis of the amino acids present in the CA domain of EnvZ allowed to identify the asparagine in the position 347 as a critical residue for kinase activity of EnvZ27. The CA domains of the histidine kinase belongs to the large family of the ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase protein (Superfamily 55874, http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?sunid=55874). To identify if the N347 of EnvZ is conserved in NarX, we aligned EnvZ and NarX with proteins containing the ATPase domain. The alignment identified the asparagine N509 for NarX as a conserved residue that is potentially important for ATP binding.
Cell culture
The experiments in this work are performed with HEK293 purchased from Life Technologies (Cat # 11631-017). Cells were cultured at 37 ºC, 5% CO2 in DMEM (Gibco, Life Technologies; Cat # 41966-052), supplemented with 10% FBS (Sigma-Aldrich; Cat # F9665) and with 1% Penicillin/Streptogamine Solution (Sigma-Aldrich, Cat #P4333). Splitting was performed every 3-4 days using 0.25% Trypsin- EDTA (Gibco, Life technologies; Cat # 25200-072). Cultures were propagated for at most two months before being replaced by fresh cell stock.
Transfections
All transfections were performed using Lipofectamine 2000 Transfection Reagent (Life Technologies; Cat#11668027). All transfections except the time lapse experiment were performed in 24-well plates (Thermo Scientific Nunc; NC-142475) and 400 ng of DNA was transfected. The cells were seeded 24 h before transfection at 50 000 cells per well in 500 μl of DMEM. The plasmids for each sample were mixed as indicated in Supplementary Tables 7-15 and 17-27 and completed with Opti-MEM I Reduced Serum (Gibco, Life Technologies Cat # 31985-962) to a final volume of 50 μl. 1.5 μl of lipofectamine 2000 was diluted in 50 μl Opti-MEM I per sample to a final ratio of 3.75 μl per 1 μg DNA. After an incubation of at least 5 minutes the diluted Lipofectamine was added to the mixed DNA sample. The resulting mixture was briefly mixed by gentle vortexing and incubated for 20 minutes at room temperature before being added to the cells. Four hours after the DNA was added to the cells, the medium was removed and replaced with 500 μl of fresh medium with or without the chemical at the desired concentration. The reagents used were A/C Heterodimerizer (Clontech; Cat# 635057), Procaterol (Sigma; Cat# P9180-10MG), Bradykinin acetate salt (Sigma; Cat#B3259) Lysophosphatidic Acid (Santa cruz Biotech; Cat#SC201053), [Arg8]-Vasopressin (Tocris; Cat#2935), NMB (Tocris; Cat#1908), Recombinant Human CXCL12 (SDF-1α, Lys22 to lys89) (Biolegend; Cat#581202), NPY (Sigma; Cat#N5017), Isoproterenol (Sigma; Cat# I6504), Clenbuterol (Sigma; Cat# C5423) and Propranolol (Sigma; Cat# P0884). The preparation of the different chemicals is described in Supplementary Note 2.
Microscopy
Microscopy images were taken about 48 hours after transfection. We used a Nikon Eclipse Ti microscope equipped with a mechanized stage and temperature control chamber held at 37 °C during the image acquisition. The excitation light was generated by a Nikon IntensiLight C-HGFI mercury lamp and filtered through a set of optimized Semrock filter cubes. The resulting images were collected by an Hamamatsu, ORCA R2 camera using a 10X or 4X objective. Each Semrock cube is assembled from an excitation filter, a dichroic mirror and an emission filter. In order to minimize the crosstalk between the different fluorescent proteins we used the following setup: Cerulean: CFP HC (HC 438/24, BS 458, HC 483/32), mCherry: TxRed HC (HC 624/40, BS 593, HC 562/40). The images were acquired with an exposure of 40 ms or 600 ms for Cerulean and 40 ms for mCherry. The acquired images were processed by ImageJ software performing uniform contrast-enhancement to improve visualization. In Figs. 1, 2 and Supplementary Fig. 2, the parameters used for the Cerulean images are 40 ms exposure, LUT values 0-7500 and the parameters used for the mCherry image are 40 ms exposure, LUT values 0-25000. In Fig. 3 and Supplementary Fig. 6, the parameters used for the Cerulean images are 40 ms exposure, LUT values 0-7500 and the parameters used for the mCherry are 40 ms exposure, LUT values 0-12500. In Fig. 4 and Supplementary Fig. 8, the parameters used for the Cerulean images are 40 ms exposure, LUT values 0-5000 and the parameters used for the mCherry are 40 ms exposure, LUT values 0-20000. In Fig. 5, the parameters used for the Cerulean images of cell transfected with GPCR fused to mutated NarX (Hmut) are 600 ms exposure, LUT values 1200-8500, and the parameters used for the Cerulean images of cells transfected with GPCR fused to tTA are 40 ms exposure, LUT values 0-20000. In Supplementary Fig. 9, the parameters used for the Cerulean images are the same as the ones used for Fig. 5; the parameters used for the mCherry images of cells transfected with GPCR fused to mutated NarX (Hmut) are 40 ms exposure, LUT values 0-20000 and the parameters used for the mCherry images of cells transfected with GPCR fused to tTA are 40 ms exposure, LUT values 0-12500.
Comparing GPCR rewiring via TCS and Tango methods
See Supplementary Note 2 for the exact formulations of the GPCR ligands. NMB-R is induced with NMB (10 μM); V2R is induced with [Arg8]-Vasopressin(10 μM); LPA-1 is induced with Lysophosphatidic Acid (10 μM); B2R is induced with Bradykinin acetate salt (10 μM), CXC-R4 is induced with recombinant human CXCL12 (0.2 μM). NPY (0.5 μM) is used as ligand for the modified NPY1-R and NPY5-R. Procaterol (2 μM) is used as a ligand for the two variants of the modified β2AR. Note that the architecture of NPY1-R and NPY5-R fused to NarX379-598 H399Q (NPY1-R::Hmut and NPY5-R∆C::V2R∆N::Hmut) is slightly different from the architecture of these GPCRs fused to tTA (NPY1-R::V2R∆N::tTA and NPY5-R::V2R∆N::tTA).
Time lapse experiment
2500 cells per well in 125 μl of DMEM were seeded in a 96 well plate (Grenier bio-one 655090). 24 hours after the seeding, 100 ng of DNA was transfected. The plasmids for each sample were mixed as indicated in Supplementary Table 16 and completed with Opti-MEM I Reduced Serum (Gibco, Life technologies Cat # 31985-962) to a final volume of 12.5 μl. 0.375 μl of lipofectamine 2000 was diluted in 12.5 μl Opti-MEM I per sample to have a final ratio of 3.75 μl per 1 μg of DNA. After an incubation of at least 5 minutes, the diluted Lipofectamine was added to the mixed DNA sample. The resulting mixture was briefly mixed by gentle vortexing and incubated for 20 minutes at room temperature before being added to the cells. 4 hours after the DNA was added to the cells the medium was removed and replaced with 125 μl of fresh DMEM. The plate was placed on the stage of a Nikon Eclipse Ti microscope equipped with a mechanized stage and temperature/CO2 control chamber held at 37 °C/5%. 14 hours after the medium has been changed, 25 μl of DMEM with or without the chemical at the desired concentration were added to the cells. The procaterol solution was prepared as indicated in Supplementary note 2. Doxycycline hyclate (Sigma; Cat#D9891) solution at 100 μg/ml was prepared in water (Invitrogen; Cat#10977-035) and them diluted in DMEM to obtain the solution with the appropriate concentration. The plate was put back in the incubator chamber of the microscope for 23.3 hours, which constituted the first incubation interval. At the end of the first incubation interval, the cells were washed by replacing the medium with 200 μl of fresh DMEM. The cells were incubated at 37°C for 5 minutes and the medium was replaced with 150 μl of DMEM with or without the inducer at the desired concentration. The plate was put back in the incubator chamber of the microscope for 23.3 hours, constituting the second incubation interval. At the end of the second incubation time the medium was replaced as described previously. The plate was put back in the incubator chamber of the microscope for 29.3 hours, constituting the third incubation period. During all the incubation intervals microscopy images were taken every 40 minutes. Cells were imaged with Nikon Eclipse Ti microscope using a 10x PlanFluor (NA 0.3) objective, using a SPECTRA X LED light source (Lumencor). The resulting images were collected by an Hamamatsu, ORCA R2 camera using. The following setup was used for each image Cerulean 1: Excitation 438/24, Dichroic 458, Emission 480/17, exposure time 30 ms; Cerulean 2: Excitation 438/24, Dichroic 458, Emission 480/17, exposure time 100 ms; mCherry: Excitation 549/15, Dichroic 562, Emission 593/40, exposure time 50 ms.
Image analysis was based on the approach developed earlier: the individual time course sequences were extracted using ImageJ and a custom script, and saved as multiframe tiff files. The signals were quantified following previously described procedure43. Briefly, the images were uploaded into Matlab workspace and rows of pixels (in the X direction) were processed one by one. 10 pixels next to the image edge were discarded to avoid stitching artefacts. A row of pixels was considered as a 1D intensity profile, which was smoothened using a Matlab function smooth(row,0.015,'loess'); and then the background was subtracted using the function msbackadj(xvec,Yout,'WindowSize',30). Pixels above the designated threshold (75 units for Cerulean signal, 500 units for Cherry signal) were considered as being robustly above background. The intensities of individual pixels were added up to give the integrated signal of a given field of view, and the number of pixels above the threshold was recorded. The average "above-background" pixel intensity was computed by dividing the total signal by the number of above-threshold pixels. Provided that on average, all cells have similar pixel footprint, this value correlates with the average signal intensity per cell. The "per cell" intensity of the transfection control mCherry was calculated in a similar fashion. Lastly, the "per cell" intensity of the Cerulean signal was divided by the "per cell" intensity of the mCherry control to generate the time course shown in the figures of this article. Cerulean 1 channel was used to analyze rtTA and Tango data, and Cerulean 2 to analyze TCS data.
Responsiveness analysis
We introduce the measure we call responsiveness to compare time course experiments in a robust fashion. We consider time courses that are identical to each other during two out of three intervals and differ in one interval, during which the sensor is induced in one experimental arm and uninduced in another arm. For example, we compare the sequence On-On-On and On-Off-On and focus on the second interval in which the induction differs. The relative change in the Cerulean reporter is calculated as follows (see also Supplementary Fig. 12). First, a linear fit is performed using the raw dataset collected during the interval in question. For TCS and Tango methods, all datapoints are used while for the rtTA method, the first 18 timepoints are discarded for both On and Off intervals due to biphasic behavior. Next, Cerulean expression is re-calculated for the first and the last timepoints of the interval using the fitted parameters. For an On interval, these values are denoted YI,ON and YF,ON (I stands for initial and F for final), while for the Off interval the values are denoted YI,OFF and YF,OFF. The relative change during the On interval is calculated using the formula
and the relative change during the Off interval as
Lastly, the responsiveness is the difference between the two relative changes, and it is always calculated for the same interval, e.g., between the On and Off changes during the second time interval or during the third one:
For the biological triplicate time course dataset (Fig. 6b, Supplementary Fig. 11), the responsiveness was calculated by averaging the relative change during the On interval and subtracting the average relative change during the Off interval, with appropriate error propagation applied:
where Q = a-b and δX is the standard deviation of X.
The p-values in Fig. 6b are calculated as follows. A two-sided t-test comparing the responsiveness between pairs of signaling approaches in the same interval (i.e., second or third) is performed according to the following formula
where and represent, respectively, the averaged relative change during the On and the Off intervals for the first approach, and and represent the same for the second approach. The values represent their standard deviation estimates calculated from the biological triplicates corresponding to these means. The p-value is then estimated from the t-value given 8 degrees of freedom (12 individual measurements in total less 4 mean values). The p-values in Fig. 6b are the products of the individual p-values corresponding to the comparison in the second and the third intervals, and they represent the overall probability that there is no difference in responsiveness between two compared approaches.
Flow Cytometry
The cells were prepared for flow cytometry analysis 48 h after transfection by removing the medium and incubating the cells with 200 μl StemPro™ Accutase™ Cell Dissociation Reagent (Gibco, cat # A11105-01) at 37°C for 5 minutes. After incubation, the plates were transferred on ice. To avoid potential cell damage the samples were prepared in successive batches so that no single sample was kept on ice for more than 1 h. The prepared samples were measured using a BD LSR Fortessa II Cell Analyzer with a combination of excitation and emission that minimizes the crosstalk between different fluorescent reporters. Cerulean and AmCyan were measured with a 445 nm excitation laser coupled to a 473/10 nm emission filter. mCherry was measured with a 561 nm excitation laser coupled to a 600 nm longpass filter and 610/20 emission filter. The Cerulean and the mCherry were measured, respectively, at PMT voltage of 330 and 310 in all the experiments except for the experiment presented in Supplementary Fig. 4a, in which mCherry was measured at PMT voltage of 325. The data presented in Supplementary Fig. 6 were obtained by measuring the AmCyan and mCherry, respectively, at PMT voltage of 350 and 315. SPHERO RainBow Calibration particles (Spherotech; Cat # RCP-30-5A, BD) were used to ensure constant device performance.
Data Analysis
Flow cytometry data analysis for generating the bar charts was performed using FlowJo software. In this work, the fluorescence values in the bar charts, shown as normalized expression units (Cerulean, norm. u. or AmCyan, norm. u.) are calculated as follows. Live cells are gated based on their forward and side scatter readouts. From this population single cell are gated based on their forward scatter area and forward scatter height. Within this gate, cells positive for Cerulean (or AmCyan) are gated based on a negative control such that 99.9% of cells in this control sample fall outside of the selected gate (Supplementary Fig. 13a-f). For reporter-positive cell population, the mean value of the fluorescent intensity is calculated and multiplied by the frequency of the positive cells. This value is used as a measure for the total reporter signal in a sample and can be defined as Total Intensity (TI). The TI of the Cerulean or AmCyan is normalized by the TI of mCherry-positive cells (constitutive transfection control). The formula is therefore:
Due to lasers alignment done in the course of the project and the resulting changes in the instrument performance, we adjusted the values of Cerulean and mCherry readouts of data presented in Fig. 5 and Supplementary Fig. 4a, to be comparable with readouts presented in other figures. Specifically, Cerulean and mCherry intensity of the 3 brightest population of the SPHERO RainBow Calibration particles (P1, P2 and P3, Supplementary Fig. 13g-h) analyzed were quantified during sample analysis. The values obtained in each experiment was used to determine the conversion factor using a linear fit, and adjust Cerulean and mCherry value of data in the Fig. 5 and Supplementary Fig. 4a to the level of the other figures. The adjusted values were used to determined Reporter intensity in adj. norm. u. as indicated
Sequence analysis
The phylogenetic tree was built and downloaded from GPCRdb44. The neighbor-joining method was used to calculate the distance between all the crystalized GPCR and the eight GPCR that we used. The multiple alignment analysis was built with the MultAlin45 interface by using blosum62 as comparison table.
Supplementary Material
Acknowledgements
The study was funded by ETH Zurich, NCCR Molecular Systems Engineering and Swiss National Science foundation (Grant 31003A_149802). We thank B. Haefliger, R. Altamura, J. Hansen, T. Littmann, G. Bernhardt and C. Stelzer for plasmids, Benenson lab members for discussions, H. M. Kaltenbach for help with statistical analysis, and the members of the Single Cell Unit for their help with imaging and flow cytometry.
Footnotes
Data availability.
All plasmid sequences and data supporting the findings are available from the corresponding author upon request.
Code availability.
The code used to process the time lapse imaging data is available from the corresponding author upon request.
Author contributions.
A.M. conceived research, performed the experiments, analyzed data and wrote the paper. Y.B. conceived research, analyzed data, and wrote the paper.
Competing interests.
A patent application has been filed describing the result in this study, with A.M. and Y.B. listed as coinventors. Y.B. is a coinventor of a background patent to this filing.
References
- 1.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 2.Jackson HJ, Rafiq S, Brentjens RJ. Driving CAR T-cells forward. Nat Rev Clin Oncol. 2016;13:370–383. doi: 10.1038/nrclinonc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet. 2012;13:455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J, Benenson Y. Synthetic biology of cell signaling. Nat Comput. 2016;15:5–13. [Google Scholar]
- 5.Cho JH, Collins JJ, Wong WW. Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell. 2018;173:1426–1438. doi: 10.1016/j.cell.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnea G, et al. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci USA. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz KA, Daringer NM, Dolberg TB, Leonard JN. Rewiring human cellular input-output using modular extracellular sensors. Nature Chemical Biology. 2017;13:202–209. doi: 10.1038/nchembio.2253. [DOI] [PubMed] [Google Scholar]
- 8.Kipniss NH, et al. Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nature Communications. 2017;8 doi: 10.1038/s41467-017-02075-1. 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baeumler TA, Ahmed AA, Fulga TA. Engineering synthetic signaling pathways with programmable dCas9-Based chimeric receptors. Cell Reports. 2017;20:2639–2653. doi: 10.1016/j.celrep.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie M, et al. Beta-cell-mimetic designer cells provide closed-loop glycemic control. Science. 2016;354:1296–1301. doi: 10.1126/science.aaf4006. [DOI] [PubMed] [Google Scholar]
- 11.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mootz HD, Muir TW. Protein splicing triggered by a small molecule. J Am Chem Soc. 2002;124:9044–9045. doi: 10.1021/ja026769o. [DOI] [PubMed] [Google Scholar]
- 13.Slomovic S, Collins JJ. DNA sense-and-respond protein modules for mammalian cells. Nat Methods. 2015;12:1085–1090. doi: 10.1038/nmeth.3585. [DOI] [PubMed] [Google Scholar]
- 14.Morsut L, et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelici B, Mailand E, Haefliger B, Benenson Y. Synthetic biology platform for sensing and integrating endogenous transcriptional inputs in mammalian cells. Cell Reports. 2016;16:2525–2537. doi: 10.1016/j.celrep.2016.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber J, Arter M, Lapique N, Haefliger B, Benenson Y. Model-guided combinatorial optimization of complex synthetic gene networks. Molecular Systems Biology. 2016;12:899. doi: 10.15252/msb.20167265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen J, et al. Transplantation of prokaryotic two-component signaling pathways into mammalian cells. Proc Natl Acad Sci USA. 2014;111:15705–15710. doi: 10.1073/pnas.1406482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera-Cancel G, Ko WH, Tomchick DR, Correa F, Gardner KH. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci USA. 2014;111:17839–17844. doi: 10.1073/pnas.1413983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dago AE, et al. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc Natl Acad Sci USA. 2012;109:E1733–E1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 21.H TN, Noriega CE, Stewart V. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc Natl Acad Sci USA. 2010;107:21140–21145. doi: 10.1073/pnas.1013081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry BP, Palanki R, Dyulgyarov N, Hartsough LA, Tabor JJ. Phosphatase activity tunes two-component system sensor detection threshold. Nature Communications. 2018;9 doi: 10.1038/s41467-018-03929-y. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashenberg O, Rozen-Gagnon K, Laub MT, Keating AE. Determinants of homodimerization specificity in histidine kinases. J Mol Biol. 2011;413:222–235. doi: 10.1016/j.jmb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomomori C, et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 25.Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth Biol. 2012;1:118–129. doi: 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai SJ, Inouye M. Spontaneous subunit exchange and biochemical evidence for trans-autophosphorylation in a dimer of Escherichia coli histidine kinase (EnvZ) J Mol Biol. 2003;329:495–503. doi: 10.1016/s0022-2836(03)00446-7. [DOI] [PubMed] [Google Scholar]
- 27.Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nature Communications. 2014;5 doi: 10.1038/ncomms4258. 3258. [DOI] [PubMed] [Google Scholar]
- 28.Cai SJ, Inouye M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 29.Bayle JH, et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Schramm A, et al. Establishing a Split Luciferase Assay for Proteinkinase G (PKG) Interaction Studies. Int J Mol Sci. 2018;19:20. doi: 10.3390/ijms19041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards SR, Wandless TJ. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. J Biol Chem. 2007;282:13395–13401. doi: 10.1074/jbc.M700498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobilka B, Schertler GFX. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol Sci. 2008;29:79–83. doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Billerbeck S, et al. A scalable peptide-GPCR language for engineering multicellular communication. Nature Communications. 2018;9 doi: 10.1038/s41467-018-07610-2. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci. 2002;115:455–465. doi: 10.1242/jcs.115.3.455. [DOI] [PubMed] [Google Scholar]
- 36.Cahill TJ, et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci USA. 2017;114:2562–2567. doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker JG. The selectivity of β-adrenoceptor agonists at human β1-, β2-and β3-adrenoceptors. British journal of pharmacology. 2010;160:1048–1061. doi: 10.1111/j.1476-5381.2010.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol. 2015;22:362–369. doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Dowd BF, Hnatowich M, Caron MG, Lefkowitz RJ, Bouvier M. Palmitoylation of the human beta 2-adrenergic receptor. Mutation of Cys341 in the carboxyl tail leads to an uncoupled nonpalmitoylated form of the receptor. J Biol Chem. 1989;264:7564–7569. [PubMed] [Google Scholar]
- 40.Sadeghi HM, Innamorati G, Dagarag M, Birnbaumer M. Palmitoylation of the V2 vasopressin receptor. Mol Pharmacol. 1997;52:21–29. doi: 10.1124/mol.52.1.21. [DOI] [PubMed] [Google Scholar]
- 41.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Science signaling. 2009;2:pe13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 42.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Haefliger B, Prochazka L, Angelici B, Benenson Y. Precision multidimensional assay for high-throughput microRNA drug discovery. Nature Communications. 2016;7 doi: 10.1038/ncomms10709. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandy-Szekeres G, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.