Significance
Programmed death-1 homolog (PD-1H) is a cell surface molecule that coinhibits T cells, with important roles in regulating immune responses in mouse models of tumor, autoimmune, and inflammatory diseases. Monoclonal antibodies (mAbs) or small-molecule reagents targeting PD-1H are being tested for therapeutic effect in clinical trials. However, efforts to uncover molecular details of PD-1H function have been hampered by the lack of structural information. We have solved the atomic structure of PD-1H to 1.9-Å resolution, revealed unique structural features, and investigated how these features contribute to coinhibitory function. Our results will guide the design of future studies and agents targeting this important immune signaling molecule.
Keywords: PD-1H, VISTA, structure, T cell coinhibition, cancer immunotherapy
Abstract
Programmed death-1 homolog (PD-1H), a CD28/B7 family molecule, coinhibits T cell activation and is an attractive immunotherapeutic target for cancer and inflammatory diseases. The molecular basis of its function, however, is unknown. Bioinformatic analyses indicated that PD-1H has a very long Ig variable region (IgV)-like domain and extraordinarily high histidine content, suggesting that unique structural features may contribute to coinhibitory mechanisms. Here we present the 1.9-Å crystal structure of the human PD-1H extracellular domain. It reveals an elongated CC′ loop and a striking concentration of histidine residues, located in the complementarity-determining region-like proximal half of the molecule. We show that surface-exposed histidine clusters are essential for robust inhibition of T cell activation. PD-1H exhibits a noncanonical IgV-like topology including an extra “H” β-strand and “clamping” disulfide, absent in known IgV-like structures, that likely restricts its orientation on the cell surface differently from other IgV-like domains. These results provide important insight into a molecular basis of T cell coinhibition by PD-1H.
Host immune responses to antigens are tightly regulated in activation and inhibition by the cosignaling network (1, 2) so as to maintain homeostasis. CD28/B7 family molecules, part of the Ig superfamily (IgSF), including those critical for cosignaling modulation, are attractive therapeutic targets for cancer, autoimmune, and inflammatory diseases. Biologics targeting CD28/B7 family members have been developed with notable successes, including US Food and Drug Administration approval of drugs targeting the programmed death-1 homolog (PD-1)/PD-L1 coinhibitory pathway to treat a broad spectrum of human solid tumors (3) and blocking CD80/CD86/CD28 costimulatory pathway by using CTLA-4–Ig fusion protein to treat rheumatoid arthritis or transplantation rejection (4, 5). Although these breakthrough drugs have had remarkable clinical outcomes, only a portion of patients responds to treatment, motivating efforts to discover new costimulatory and coinhibitory pathways and explore their therapeutic potentials, alone or in combination with other immunotherapies (6, 7).
PD-1H (also called V domain Ig suppressor of T cell activation [VISTA], Gi24, Dies-1, or DD1α) is a more recently identified cell surface coinhibitory molecule of the CD28/B7 gene family (8, 9). We and others have reported that PD-1H can function as an inhibitory ligand on antigen-presenting cells and regulate T cell responses, and ablation of PD-1H by either genetic knockout or antagonist antibody can boost T cell immune responses against tumors in mouse models (10, 11). PD-1H may also play critical roles in the regulation of inflammation and autoimmune diseases, as shown in mouse models of graft-versus-host disease (GVHD) (9), acute hepatitis (10), encephalitis (8, 12), lupus (13, 14), asthma (15), and psoriasis (16). We showed that PD-1H could also function as a coinhibitory receptor on T cells. PD-1H agonistic mAb dramatically regulates antigen-specific CD4 T cell responses and protects mice from GVHD, acute hepatitis, and asthma (9, 10, 15). Up-regulated PD-1H in prostate cancer patients was also shown to associate with resistance to ipilimumab (CTLA-4 mAb) (17). Furthermore, it has been shown that targeting PD-1H may synergize with other nonredundant pathways, like PD-1 blockade, to achieve optimal tumor clearance efficacy in experimental mouse models (18). Therefore, PD-1H may be an important molecule in the regulation of immune responses and a potential target for immunotherapy.
While PD-1H is highly conserved across species, even between mouse and zebrafish (19), it appears to be distantly related to other CD28 or B7 family members (9). PD-1H is considered homologous to both the T cell coinhibitory receptor PD-1 as well as the IgV-like domain of B7-H1 (PD-L1) (8, 9). These disparate homologies to both counter receptors may be connected to the reported capacity of PD-1H to act as both a ligand and a receptor in T cell responses. Our bioinformatic and sequence analyses reveal several unique structural features, such as an unusual large IgV domain and a strikingly high percentage of histidine residues. These initial observations prompted us to determine the crystal structure of human PD-1H. The structure, together with our functional studies, provide insight into the molecular basis of PD-1H function, and inform the design of therapeutics targeting this important immune signaling molecule.
Results
PD-1H Has a Very Long IgV-Like Domain with a Unique CC′ Loop and Exceptional Histidine Content.
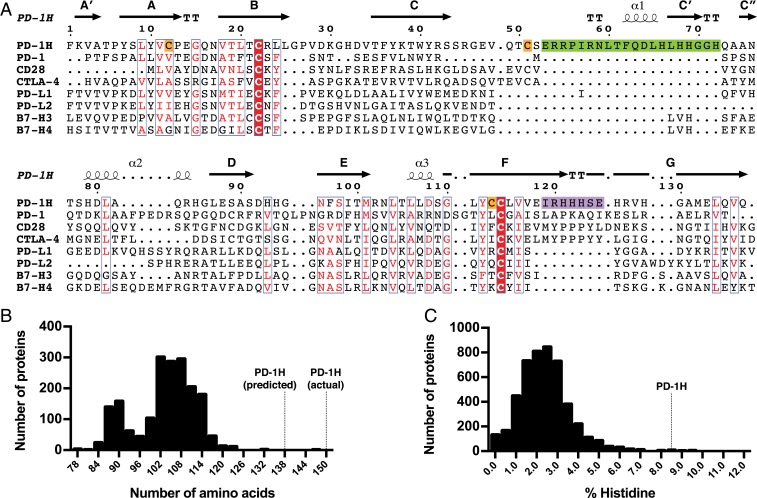
We performed bioinformatic analysis to reveal a number of conspicuous features of the PD-1H amino acid sequence that are unique among the CD28/B7 family members. First, alignment of the human PD-1H amino acid sequence with related CD28 and B7 family members showed that PD-1H contains ∼20 contiguous amino acids in the CC′ region that do not align with other family members (Fig. 1A). This inserted sequence represents an increase of a sizeable fraction of the typical length of an Ig domain (70 to 110 amino acids), markedly differentiating PD-1H from related proteins. Given the large size (161 amino acids) of the PD-1H extracellular domain (ECD) despite only having a single IgV-like domain, we sought to quantitatively assess how the length of PD-1H compares to other IgV-like domains. We plotted all IgV-like domain lengths as defined by the domain boundaries from the Pfam database’s V-set (PF07686) family (20), derived from 1,873 experimentally determined structures of V-set domains (Fig. 1B). This confirmed that PD-1H—with a predicted IgV-like domain length of 137 amino acids—is indeed a clear outlier, significantly longer than every other V-set Ig domain in this database. In fact, our structural study revealed that the actual IgV-like domain of PD-1H is even larger than predicted (149 amino acids), imparted by the inclusion of an unexpected “H” β-strand (discussed later).
Fig. 1.
PD-1H has unique sequence features in the CC′ and FG loops, a very long IgV domain, and a large number of conserved histidines. (A) Multiple-sequence alignment of PD-1H and CD28/B7 family member IgV domains. Secondary structural annotations are based on our crystal structure of PD-1H. Similar residues are colored red, and invariant residues have red backgrounds. A long insertion in PD-1H between the C- and C′-strands is highlighted in green, the PD-1H sequence corresponding to the CDR3-like region in the FG loop is highlighted in purple, and additional cysteines that are invariant among PD-1H orthologs are highlighted in orange. (B) Histogram of all IgV domain lengths as defined by Pfam. (C) Histogram of histidine content in all annotated type I transmembrane ECDs.
Another unique feature of PD-1H is a striking enrichment of histidines in its ECD. In particular, the FG loop region of PD-1H, corresponding to the critical MYPPPY motifs of CD28 and CTLA-4 and the complementarity-determining region (CDR)3 region of T cell receptors (TCRs) and antibodies, consists of a triplet of conserved histidines (Fig. 1A). To assess how PD-1H compares to other proteins in histidines, we obtained sequences of all type 1 transmembrane ECDs longer than 20 amino acids from UniProt (21) and plotted their histidine content (Fig. 1C). We found that PD-1H is indeed drastically higher in histidine content than almost any other type 1 transmembrane ECD, with histidine making up 8.6% of its extracellular residues; in contrast, the mean histidine content among all type 1 transmembrane ECDs is only 2.4%. Comparing PD-1H orthologs showed that most of these histidines are conserved, suggesting that they may confer unique functions to PD-1H (SI Appendix, Fig. S1).
General Features of the PD-1H Crystal Structure.
We carried out structural studies of PD-1H to elucidate the unique features suggested by our bioinformatic investigation. PD-1H is an extensively glycosylated protein, which results in an apparent molecular mass of ∼35 to 45 kDa as assayed by size-exclusion chromatography (SEC) and SDS/PAGE, despite only having a predicted molecular mass of ∼20 kDa (SI Appendix, Fig. S2). Although we were able to obtain crystals with protein partially deglycosylated with Peptide:N-glycosidase F (PNGase F), these crystals did not diffract, possibly due to defects caused by remaining glycans. We then inhibited glycan remodeling during protein expression with swainsonine to maintain glycans in their high-mannose forms, rendering them susceptible to enzymatic digestion with Endoglycosidase H (22). This method allowed more complete deglycosylation and led to crystals that diffracted to high resolution (1.9 Å). The structure was solved by molecular replacement using the IgV-like domain of PD-L1 (23) as a search model (Table 1). The final refined structure includes residues 1 to 26 and 33 to 153, with residues 27 to 32 (which corresponds to the CDR1-like loop) disordered and not seen. Electron density connected to Asn17 could be unambiguously interpreted as the remaining N-acetylglucosamine residue (SI Appendix, Fig. S3).
Table 1.
Data collection and refinement statistics
| Data collection and refinement | PD-1H (Protein Data Bank ID 6U6V) |
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 28.82, 55.83, 42.27 |
| α, β, γ (°) | 90, 97.69, 90 |
| Resolution (Å) | 41.9–1.9 (1.95–1.90) |
| Rsym or Rmerge | 0.097 (0.697) |
| Mean I/σI | 10.4 (1.3) |
| CC1/2 | (0.530) |
| Completeness (%) | 95.2 (75.5) |
| Redundancy | 3.4 (2.8) |
| Refinement | |
| Resolution (Å) | 41.9–1.9 |
| No. reflections | 9,503 |
| Rwork/Rfree | 0.209/0.245 (0.323/0.373) |
| No. atoms | |
| Protein | 1,163 |
| Ligand | 14 |
| Water | 35 |
| Average B-factors (Å2) | |
| Protein | 42.9 |
| Ligand | 48.6 |
| Water | 41.8 |
| rmsd | |
| Bond lengths (Å) | 0.016 |
| Bond angles (°) | 2.1 |
| Ramachandran favored (%) | 95.1 |
| Ramachandran allowed (%) | 4.9 |
| Ramachandran outliers (%) | 0 |
Statistics in parentheses indicate those for the highest resolution shell.
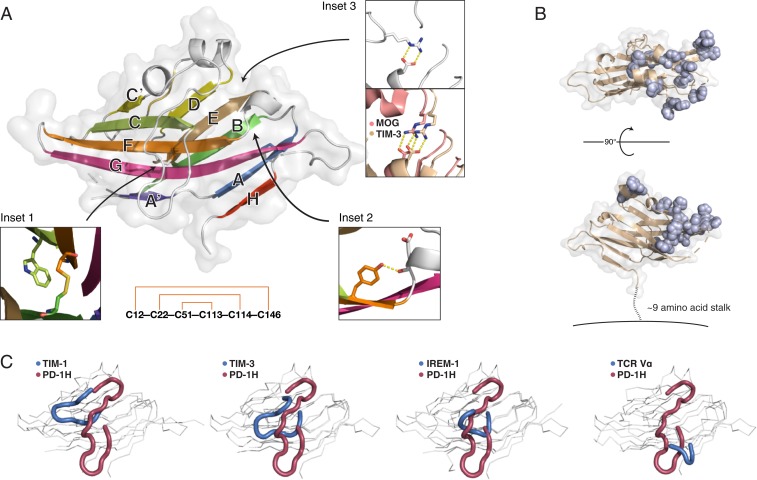
The overall structure of PD-1H adopts a β-sandwich conformation with the H-, A-, G-, F-, C-, and C′-strands comprising the front face and the A′-, B-, E-, D-, and C″-strands forming the back face (Fig. 2A). In common with other IgSF members, PD-1H includes the canonical disulfide (Cys22–Cys114) bridging the B- and F-strands, thus locking the front sheet to the back sheet, as well as the conserved tryptophan (Trp40) that packs with this disulfide and helps form the hydrophobic core (Fig. 2 A, Inset 1). In addition, the hydrogen bond between the main-chain carbonyl oxygen of Asp-108 prior to the F-strand and Tyr112 in the F-strand comprise the “Tyr-corner” that is common among IgSF domains (24) (Fig. 2 A, Inset 2). However, PD-1H lacks the conserved salt bridge between the arginine ahead of the D-strand and the aspartate prior to the B-strand [e.g., Arg68–Asp92 in MOG (25), Arg82–Asp105 in TIM-3 (26)]. In its place, instead, an alternative salt bridge (Arg58–Asp108) with a unique orientation is made that involves the unusually long CC′ loop (Fig. 2 A, Inset 3). The crystal structure also clarified the role of the invariant cysteine residues that are not conserved with other B7 family members. In addition to the canonical B–F disulfide, an additional disulfide clamps the N-terminal A′-strand to the C-terminal H-strand, and another disulfide connects the CC′ loop to the F-strand. These disulfides follow an “outside-in” pattern of connectivity (Fig. 2 A, Lower).
Fig. 2.
PD-1H crystal structure reveals a striking surface organization of histidines and a long, circuitous CC′ loop. (A) The human PD-1H extracellular domain structure in cartoon and surface representation. β-Strands are labeled according to standard IgV nomenclature, but including an additional unique H-strand. (Inset 1) Zoomed view of the canonical B–F disulfide packing against the conserved tryptophan to form the hydrophobic core between β-sheets. (Inset 2) Zoomed view of the “Tyrosine-corner” that is present in PD-1H and is common in IgV domains. (Inset 3) PD-1H (Left) lacks the conserved arginine-aspartate salt bridge in IgV proteins, such as MOG (salmon, Right) or TIM-3 (wheat, Right), instead using an arginine from the CC′ loop to form an alternative salt bridge at the same location. (Lower): “Outside–in” pattern of disulfide bonding connectivity in the PD-1H extracellular domain. (B) The structure of PD-1H in 2 views as a cartoon (wheat) with a transparent surface. All histidine residues are displayed in sphere representation (light blue) to emphasize their spatial clustering within one-half of the PD-1H ECD, proximal to the CDR-equivalent region. A short stalk of ∼9 amino acids (dashed line) connects the ECD to the membrane (solid line). (C) Comparison of CC′ loop regions from PD-1H (red) and other IgV-like domains (blue), including those with long CC′ loops.
To better understand the PD-1H structure in the context of other known structures, we performed automated structural comparison using the DALI server (27). Nearly all of the hits with highest similarity were various TCR Vα domains. Overall, the topology of PD-1H is most similar to that of TCR Vα and CTLA-4, with an N-terminal β-strand (A′) that originates on the back face of the β-sandwich structure before crossing over and continuing in the front face (as the A-strand) (SI Appendix, Fig. S4). However, PD-1H differs from these structures in having an additional α-helix between its C″- and D-strands, and even more strikingly in having an additional β-strand after the final G-strand that redirects the peptide chain such that it emerges from an unusual location closer to the N-terminal (CDR-equivalent) end of the ectodomain.
PD-1H Has a Striking Spatial Concentration of Conserved Histidines and a Tethered CC′ Protrusion.
One of the key bioinformatics observations was the curiously high number of histidine residues within the ECD. Surprisingly, the PD-1H structure revealed that the spatial density of histidine residues is even more striking than would be predicted by the high percentage of histidines alone, as they are locally concentrated in the CDR-proximal half of the molecular surface but are totally absent from the CDR-distal half (Fig. 2B). With this distinctive organization, they are likely oriented away from the membrane and readily available for interaction with putative counterreceptors.
Certain IgSF proteins, such as those of the TIM family (28) and the IREM family (29), contain long extensions in their CC′ loops. Often, these protrusions are tethered to the Ig-like domain via 1 or multiple disulfide bonds, and may serve functional roles. PD-1H represents a remarkable example of this feature, with an unusually extended CC′ loop whose position is reinforced by an invariant disulfide bond (Cys51–Cys113) as well as a salt bridge (Arg58–Asp108) that substitutes for the conserved interaction between the arginine before the D-strand and the aspartate prior to the B-strand in other IgSF family members (Fig. 2C). While extensions in the CC′ loop often reach out toward the FG loop, the PD-1H CC′ loop instead reaches toward the stalk (C terminal to the IgV-like domain). Furthermore, the long CC′ loop of PD-1H follows an unusually winding path to pack against the body of the Ig domain, compared to other CC′ loops.
Conserved Histidine Clusters on PD-1H Are Functionally Important.
Due to the high concentration of clustered histidine residues, and their conservation and notable position at the CDR-equivalent region of PD-1H, we hypothesized that these histidines may contribute to a coinhibitory mechanism by PD-1H. Similarly, we speculated that the distinctive CC′ loop may also be functionally important, as in proteins such as TIM family members (28). We generated recombinant PD-1H–hFc fusion protein variants including alanine substitutions of surface-exposed histidine residues (HA123-hFc), or deletion of the CC′ loop (ΔCC′-hFc). To rule out the possibility of differences in the overall protein structures upon mutation, all protein variants were compared by SEC (SI Appendix, Fig. S5). All mutant variants eluted at approximately the same position as the wild-type (WT) PD-1H–hFc, suggesting no gross differences in the size, shape, or oligomeric state of the protein due to mutation.
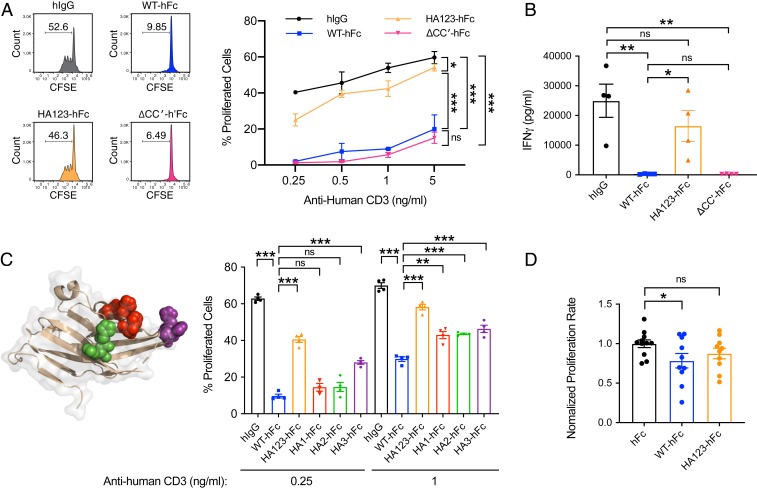
Using human T cell activation assays, we compared these mutants against WT PD-1H (WT-hFc) or human immunoglobulin G (hIgG) for their ability to suppress anti-CD3–induced T cell activation (Fig. 3A and SI Appendix, Fig. S6A). While WT-hFc significantly inhibited T cell proliferation compared to hIgG, the mutation of histidines to alanines largely abrogated this inhibition. In contrast, ΔCC′-hFc remained inhibitory. We further quantified the cytokine IFN-γ, which is associated with T cell effector function, from the cell culture supernatants after T cell activation in the presence of WT, mutant, or control proteins (Fig. 3B and SI Appendix, Fig. S6B). Consistent with T cell proliferation, IFN-γ was strikingly inhibited by WT PD-1H–hFc as well as ΔCC′-hFc; in contrast, inhibition was largely abrogated by the histidine→alanine mutations.
Fig. 3.
Functional role of PD-1H histidine clusters in T cell coinhibition. (A) T cell proliferation after activation with α-CD3 in the presence of control hIgG, WT PD-1H, or mutant PD-1H where either all 3 histidine clusters were mutated to alanine (HA123) or the CC′ loop was deleted (ΔCC′). The numbers in each of 4 CFSE profiles indicate percentages of proliferated cells. (B) Cytometric bead array staining of IFN-γ in supernatants from T cells activated in the presence of control or PD-1H protein variants. (C) Similar experiment as A but with histidine clusters (in different colors) mutated individually. For A–C, representative results of at least 3 independent experiments were shown. (D) T cell proliferation after activation by xeno-antigen in vivo in NSG mice in the presence of control (n = 11), WT (n = 10), or mutant PD-1H with histidine clusters mutated to alanine (n = 10). Proliferation percentages were normalized as folds of mean proliferation percentage of WT-hFc group. Results were pooled from 2 independent experiments. Statistical analyses were carried out with a 2-tailed Student’s t test, and all error bars reflect SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
To further map which specific histidines contribute to coinhibition, we subdivided the exposed histidine residues into spatial clusters and tested alanine mutations of individual clusters (HA1-hFc, HA2-hFc, or HA3-hFc) (Fig. 3C and SI Appendix, Fig. S6C). When individual histidine clusters were mutated to alanine, PD-1H–hFc lost some ability to inhibit T cell proliferation, but not as much as the triple-cluster mutant (HA123-hFc), suggesting that separate surface-exposed histidine clusters may all contribute to T cell coinhibition. The histidine cluster corresponding to CDR3 (HA3-hFc) appeared to have the greatest effect, especially at submaximal activating signals. In addition to the in vitro assay, we also evaluated the inhibitory function of WT PD-1H–hFc and the triple-cluster histidine mutant using an in vivo xenogeneic human T cell activation model (Fig. 3D). Human peripheral blood mononuclear cells (PBMCs) were injected into the peritoneal cavity of immune-deficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice, in which human T cells would be activated by the xeno-antigen presented by mouse antigen-presenting cells. As expected, human T cell proliferation in mice preinjected with WT PD-1H–hFc–expressing plasmid was significantly inhibited compared to those in mice given the control hFc plasmid. However, inhibition was not significant in mice injected with the histidine mutant (HA123-hFc) plasmid. Together with the results of the in vitro functional assays, our findings indicate that the histidine clusters play an important role in the inhibitory function of PD-1H as a ligand.
We performed in vitro assays in the presence of MIH65, a human PD-1H recombinant monoclonal antibody (BD Biosciences) to gain additional insight into the inhibition mechanism of PD-1H. MIH65 was able to partially block WT PD-1H–mediated inhibition of T cell proliferation (SI Appendix, Fig. S7). Interestingly, it seems that histidine mutation would not impair the antagonistic activities of MIH65. This result indicates that the mechanism of antagonism by MIH65 is likely independent of the blockage of these histidines. This suggests that other important molecular determinants of PD-1H function may exist, and this is in fact consistent with our finding that mutating histidines does not completely restore T cell proliferation to an equivalent level as control-treated cells.
PD-1H Deviates Profoundly from IgV-Like Topologies by a Tethered H β-Strand and a “Clamping” Disulfide That Enforce a Unique Point of Emergence for Its Stalk.
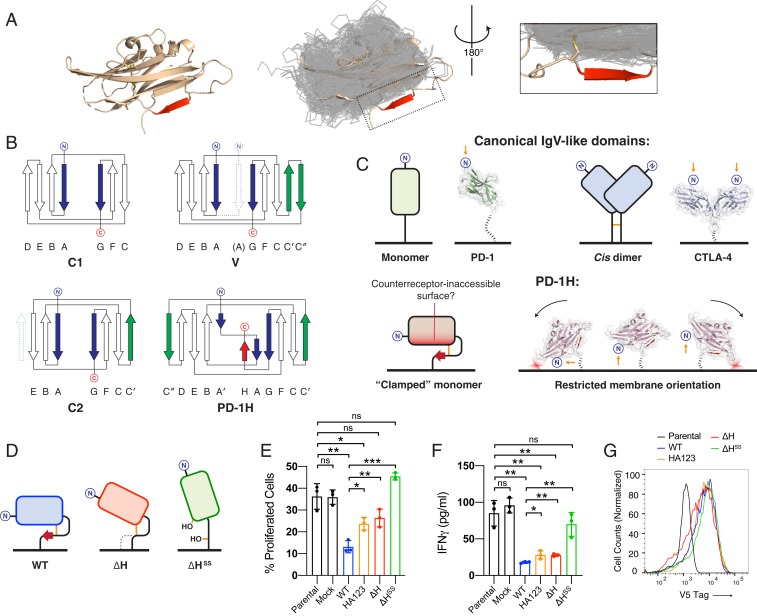
The PD-1H IgV-like domain is larger than those of all other known structures of IgV-like domains in the Pfam database’s V-set Ig family. In addition to the long CC′ loop, the unusual length is also imparted by an unexpected extra β-strand after the G-strand, which would normally be the last strand in the domain. This strand is “clamped” to the IgV-like domain by a disulfide bond connecting it to the A′-strand. To assess whether this H-strand has any correlates in other V-set Ig domain-containing structures, all 1,873 V-set domains from the Pfam database were extracted and superposed on the PD-1H structure (Fig. 4A). PD-1H clearly stands out as the only structure among these V-set domains that has this extra H-strand. Comparing the PD-1H sequence to other CD28 family members reveals that the H-strand is located in a region that corresponds to where the stalk or transmembrane regions are found in other family members, highlighting its novelty (SI Appendix, Fig. S8).
Fig. 4.
PD-1H deviates drastically from known IgV-like topologies by a tethered, H β-strand and “clamping” disulfide that enforce an unusual orientation on membrane. (A) Exhaustive structural alignment of all IgV-like domains (transparent gray ribbons) in the Pfam V-set Ig set of solved crystal structures onto the PD-1H structure (Left, wheat cartoon). PD-1H clearly stands out as having an extra β-strand (red) with no correlate in other known IgV-like domains. (B) PD-1H breaks topological patterns of IgSF subsets. In C1 domains, the first (“A”) and last (“G”) strands are parallel (blue), which is the basis of the separation of the N- and C-terminal ends of this domain to opposite sides. In C2 domains, an additional C′-strand (green) and a lack of a D-strand (dashed green) are compensatory, maintaining the relative orientation of the N- and C-terminal ends. Similarly, in IgV-like domains, C′- and C″-strands (green) also preserve this relative orientation. However, PD-1H breaks this pattern, with its unique last strand (red) antiparallel to the first (A/A′) strand (blue), leading to the proximal positioning of the C- and the N-terminal ends. (C) Model of differential display from the membrane based on PD-1H’s unusual stalk location. (D) Design of membrane-bound, full-length mPD-1H constructs expressed on 293T-KbOVA artificial antigen-presenting cells for cell-based T cell activation experiments. (E) CFSE labeled OT-I T cells were stimulated with UV-irradiated 293T-KbOVA cell lines expressing membrane-bound, full-length WT or mutant PD-1H for 3 d. CFSE profiles of OT-I T cells are shown. (F) IFN-γ levels produced by OT-I cells in culture supernatants are compared. For E and F, representative results of 2 independent experiments are shown. Statistical analyses were carried out with a 2-tailed Student’s t test, and all error bars reflect SEM. (G) Expression levels of membrane-bound, full-length mPD-1H constructs, determined by FACS staining of V5 expression tag. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
The additional H-strand bestows on PD-1H a unique topology that restricts its orientation on the cell surface. Ig domains, which are composed of 7 to 9 antiparallel β-strands, can be further divided into topological types (e.g., V-set, C1-set, C2-set) based on different 3D orientation of secondary structural elements. Importantly, despite variations in topology, the N- and C-terminal ends are located in the opposite sides of the canonical IgV-like domains (Fig. 4 B and C). For example, compared to the C1-set, the C2-set possesses 1 fewer strand in the front face but 1 additional strand in the back face. This combination of strand addition and loss is compensatory, so that the relative orientation of the 2 strands at the N- and C-terminal ends remains antiparallel, and the peptide chain consequently emerges from opposite ends of the domain. Similarly, in V-set domains, the addition of 2 antiparallel β-strands also preserves the opposite locations of N- and C-terminal ends. In notable contrast, PD-1H represents an apparent exception to this rule, with its C-terminal stalk emerging proximally to its N terminus. The structural basis of this unusual architecture is imparted by its H β-strand. While this region had been predicted to be part of a long-disordered stalk (8), the disulfide tethering of the H-strand as part of the Ig domain results in a substantially shorter stalk of ∼9 amino acids. While other IgSF molecules, such as CD45 may be rigidly displayed with the stalk emerging from a point distal to the N terminus (30), the short stalk and its position at the midpoint of the PD-1H Ig domain is likely to change the angle from which the IgV-like domain is displayed from the cell surface, given the restricted rotations allowed without clashing with the membrane (Fig. 4C). Why PD-1H adopts this unique topology is unclear, but it may play an important role in limiting the surface of the protein that can be presented for interaction with partner molecules including the counterreceptors for PD-1H.
We performed cell-based assays to validate that this molecular feature plays a functional role. We initially tried to generate Fc-fusion proteins with H-strand deletion or mutation in order to test them in the human T cell activation assay. However, well-behaved proteins for these mutants could not be obtained with extracellular domains alone, rendering them unsuitable for this functional assay. Therefore, we explored a cell-based mouse T cell activation assay established previously in our laboratory (31, 32) that would be compatible with membrane-bound, full-length proteins. In this assay, an HEK293T-Kb-ovalbumen (OVA) cell line (293T-KbOVA) stably expressing a mouse H-2Kb molecule and a chicken OVA 257-264 peptide (OVA257-264) is used as antigen-presenting cells, to activate mouse CD8+ OT-I TCR transgenic T cells (OT-I). To test the function of PD-1H and its mutants, we generated 293T-KbOVA cell lines that stably express membrane-bound, full-length WT murine PD-1H (mPD-1H, WT), or mPD-1H mutants with the H-strand deleted (ΔH), or with the H-strand deleted and additionally the 2 clamping cysteines (Cys12 and Cys145) mutated to serine (ΔHSS) (Fig. 4D). We activated purified OT-I cells by incubating them with UV-irradiated, mPD-1H–expressing 293T-KbOVA cells (Fig. 4E). Compared to parental or mock, WT mPD-1H–transduced 293T-KbOVA cells significantly inhibited T cell proliferation. Deletion of the H-strand significantly reduced the inhibitory function of mPD-1H, suggesting that this strand may play a role in PD-1H function.
Interestingly, mutating the clamping disulfide in addition to deleting the H-strand led to a complete loss of mPD-1H inhibitory function. We also quantified the IFN-γ produced by OT-I cells in the cell culture supernatants after T cell activation (Fig. 4F). Consistent with the T cell proliferation, more IFN-γ was produced when the H-strand was deleted, especially when the clamping disulfide was also lost. In this cell-based assay, we also included HA123 histidine cluster mutant; consistent with our results from the fusion protein-based assay, this HA123-transduced 293T-KbOVA cell also partially lost the inhibitory function compared to WT or mock cell. In order to confirm WT PD-1H and mutants are expressed at similar level on the 293T-KbOVA cell lines, we stained the V5 expression tag which is fused with PD-1H at the C terminus. We found that all of the PD-1H constructs were expressed at similar levels (Fig. 4G), so the loss-of-function by mutation should not be caused by different expression level of PD-1H.
Discussion
PD-1H is a newly identified regulator of T cell function (8, 9), with outstanding questions about its molecular form and functions. By extensive bioinformatics and sequence analyses, we have uncovered several unique structural features of PD-1H, including a strikingly high percentage of histidine residues, most of which are highly conserved. In comparison with related CD28/B7 family members, PD-1H also has 4 invariant cysteine residues absent from other IgSF members, which are predicted to be exposed on the surface of the protein and may be available for intermolecular hetero- or homophilic bridges (8). The native oligomeric state of PD-1H has been uncertain previously, with a report of homophilic binding capacity (33). By determining the crystal structure of human PD-1H to a resolution of 1.9 Å together with functional studies, our results provide insight into the unresolved features of PD-1H, illuminate a mechanistic basis of PD-1H function, and establish a structural framework to guide the design of future studies and agents targeting PD-1H.
Several intriguing features of PD-1H can be seen from its sequence. Chief among these is a conspicuous quantity of histidine residues within its ECD. Many of these histidines are highly conserved, which further suggests their potential functional importance. Not only is the raw percentage of histidine residues significantly higher than almost any other type I transmembrane ECD, but their local concentration on the PD-1H molecular surface is even higher, as they are clustered in the CDR-proximal half of PD-1H. Obtaining a high-resolution structure allowed us to separate these clusters and test their individual importance in T cell coinhibition. T cell activation assays revealed that all 3 histidine clusters appear to contribute to T cell coinhibition, with the histidine triplet that comprises the FG loop apparently the most important. Notably, this is the region that corresponds to the CDR3 region of TCRs and antibodies, where it is the most important player in determining the specificities of binding to antigen (34). CDR3-equivalent regions are also often used by other CD28/B7 family members in receptor–ligand interactions, such as CTLA-4 (35, 36), CD28 (37), ICOS (38), and CD8 (39). It is conceivable that, in a similar manner, the histidine triplet in the CDR3-like loop of PD-1H may be involved in a direct interaction with counterreceptors.
With the counterreceptors and their functions for PD-1H yet to be fully elucidated, the exact function of these histidines remains unclear. Histidine is a versatile amino acid, with the ability to make 5 types of interactions, and with 2 protonation states (40). Compared to other ionizable amino acids, its pKa (∼6.5) uniquely positions histidine as a potential “pH sensor,” as it is largely unprotonated at physiological pH (∼7.4) but largely protonated in acidic environments such as tumors or areas of inflammation (41). It is tempting to speculate that PD-1H may have a particular sensitivity to pH, conferred by this density of histidines, thereby regulating its function as an important immune modulator in those environments. For example, PD-1H may be differentially regulated at the cell surface or may make different interactions with counterreceptors or cofactors (e.g., metals or membrane lipids), depending on the pH of the local environment. Perhaps unsurprisingly (given the high surface density of histidines), even PD-1H ECD constructs that lack any His6-tag are strongly captured by Ni-NTA resin, and require up to 500 to 1,000 mM imidazole to elute. Given this observation, one possibility is that interactions of PD-1H with its counterreceptors involve metal coordination, although this remains speculative.
While other IgV-like domains are generally 70 to 110 amino acids, the PD-1H IgV-like domain (∼149 amino acids) is drastically longer than every other one in the Pfam V-set Ig structure family. This additional length is largely explained by the presence of an H β-strand, as well as a long CC′ loop. Both of these features involve disulfide bonds conserved among PD-1H orthologs, suggesting that these features may play an important role in PD-1H function or regulation. The CC′ loop is unusual in both its length and the circuitous path it takes as it packs against the rest of PD-1H. Combined with our identification of the FG loop histidines as functionally important, this raises the intriguing possibility that the CC′ loop may have a capacity to interact with the FG loop in a manner similar to that of TIM family members (e.g., by extending toward the FG loop and chelating a metal cation) (42). However, our in vitro assays were not able to detect a functional contribution of the CC′ loop. Still, it is important to note that histidine mutation did not completely block T cell inhibition, and an antagonist mAb may inhibit PD-1H function through an independent mechanism, suggesting that there are likely other molecular determinants that contribute to PD-1H function.
The H-strand appears to be completely unique to PD-1H. Its inclusion, with no other compensatory strand addition or deletion, breaks the canonical topological patterns among IgSF sets. This has the effect of forcing the stalk to emerge from an unusual position closer to the N-terminal end of the domain and CDR-like regions. Because the stalk is only ∼9 amino acids, this likely limits the membrane orientation and surface regions of PD-1H that are available for interactions with counterreceptors. Indeed, we found that deleting the H-strand caused diminished inhibitory function of membrane-bound full-length mPD-1H, and further mutation of the potentially orientation-restricting clamping disulfide led to a complete loss of function. These results suggest that the unusual membrane orientation, likely imparted by the H-strand and the clamping disulfide may be an important determinant in PD-1H function. This may occur through limiting which surfaces of PD-1H are accessible to counterreceptors, or through positioning of important features of PD-1H such as histidine clusters so that they can interact with proximal partners [e.g., membrane components, such as lipids, as has been proposed (42) for TIM-4]. We cannot rule out the possibility that our crystal structure represents one of multiple possible conformations of PD-1H; it is tempting to speculate that reduction and oxidation of this clamping disulfide, or alternative folding pathways involving other pairings of cysteines, may allow the structure of PD-1H to be regulated according to certain cues.
Given its extensive glycosylation, it is possible that glycans may also contribute to PD-1H function. However, our in vitro experiments in which glycans were unaltered, maintained in high-mannose form, or enzymatically removed did not show a notable contribution of glycans. In all 3 cases, T cell proliferation was dampened to similar levels, with only a small increase in proliferation after glycan removal at the highest dose of anti-human CD3 (SI Appendix, Fig. S9). Nonetheless, given PD-1H’s broad expression pattern, multiple reported potential ligands, and ability to function as both a receptor and ligand, it is possible that certain molecular determinants are important in some contexts and not others; therefore, we cannot rule out the functional importance of the glycans (as well as the CC′ loop) in other settings.
Previous predictions left uncertain the role of the invariant cysteine residues that are unique to PD-1H, with the suggestion that they may be exposed and available for intermolecular disulfides (8). However, our crystal structure revealed that the ECD cysteines are in fact all involved in intramolecular disulfides. Interestingly, the connectivity of these cysteines follows an “outside–in” pattern, which suggests that the first 3 cysteines (in the A-strand, B-strand, and CC′ loop, respectively) likely remain unpaired during translation until they are eventually oxidized in the reverse order, raising questions of how incorrect pairings are prevented, such as those between PD-1H molecules. While there is a report of a potential homophilic, noncovalent dimer of PD-1H (33), our crystal structure did not support a dimeric form, with crystal contacts too small to represent dimeric interfaces and a space group inconsistent with a simple homodimer. Furthermore, both SEC and reducing/nonreducing SDS/PAGE of deglycosylated PD-1H exhibited a molecular mass consistent with that predicted for a monomer.
With new therapeutic agents sought to target PD-1H, some of which are undergoing clinical trials, we expect our structure to provide a valuable structural framework in the design of potential therapies. In particular, we have identified histidine clusters that appear to be important for T cell coinhibitory function. An orally bioavailable small-molecule inhibitor of PD-1H (as well as PD-L1 and PD-L2) called CA-170 is currently undergoing a clinical trial (NCT02812875) in advanced solid tumors and lymphomas. Interestingly, CA-170 is a 1,3,4‐oxadiazole that has some structural similarity to histidine (43). It is possible that its mechanism of PD-1H antagonism may be connected to this structural similarity, given the importance of histidines in PD-1H function. For example, CA-170 may compete for a binding site of PD-1H’s histidines, thereby blocking PD-1H from making functional interactions.
In this study, we evaluated the inhibitory function of WT and mutant PD-1H in T cell activation only as a ligand; however, studies have suggested that PD-1H also works as a receptor on either T cells or other cell types (9, 10, 44). The specific residues used by PD-1H to make these interactions may be different when functioning as a ligand or as a receptor. Future studies aimed at uncovering the molecular determinants of PD-1H function as a receptor, and whether the key features revealed by the crystal structure perform similar functions when acting as a receptor, should prove invaluable in furthering our understanding the molecular mechanisms of coinhibition by PD-1H.
Materials and Methods
Bioinformatic Analysis of PD-1H Amino Acid Sequence.
Multiple sequence alignments of PD-1H and other CD28/B7 family receptors or other PD-1H orthologs were generated using Clustal Omega with the Kalign algorithm and processed with ESPript (45) for presentation. To generate the histogram of IgV-like domain lengths, individual IgV-like domains were extracted from the Pfam database’s V-set Ig (PF07686) containing solved crystal structures and their lengths plotted using GraphPad Prism 7.0 (GraphPad Software). To generate the histogram of histidine content, type 1 transmembrane sequences were retrieved from UniProt (21), their ECDs were extracted, and the percent of histidine in the ECDs were plotted using GraphPad Prism 7.0.
Protein Expression and Purification.
The extracellular domain of human PD-1H (residues 1 to 161) was cloned into a pcDNA3.4 vector (Thermo Fisher) with an N-terminal signal peptide from CRYPα and a C-terminal Mpro protease site and His6-tag. Plasmids were transfected into Expi293 cells (Thermo Fisher) using standard manufacturer protocols but with the addition of 20 µM swainsonine (Cayman Chemical) at the time of transfection. Secreted protein in the supernatant was harvested, dialyzed into Ni-NTA binding buffer (50 mM Tris pH 8.0, 300 mM NaCl), and purified using Ni-NTA affinity chromatography (Qiagen) and SEC using a Superdex 200 column (GE Life Sciences) equilibrated with HBS. Peak fractions were concentrated and digested using Endoglycosidase Hf (New England Biolabs) at pH 5.2 and 37 °C for 48 h using standard manufacturer protocols. Digested protein was repurified using SEC, with fractions corresponding to deglycosylated protein pooled and concentrated to ∼7.4 mg mL−1 in HBS for crystallization.
Proteins for in vitro human T cell activation experiments were cloned into the same expression vector but with a C-terminal human IgG1 Fc (hFc). HA1-hFc includes alanine mutations at H66, H68, and H72; HA2-hFc includes alanine mutations at H93 and H94; HA3-hFc includes alanine mutations at H121, H122, and H123; HA123-hFc contains alanines at all of these sites. ΔCC′-hFc has residues S44 through R55 deleted, and an additional C113S mutation so that the C113 (which would normally be S-S bonded to C51 in this deleted region) would not be unpaired after deletion of this loop, in order to maintain good behavior of this mutant. These residues were chosen based on our solved structure, such that the residues immediately before and after the deleted region are proximal in space, so that the overall structure of the protein may be more confidently preserved. Secreted proteins in Expi293 supernatants were harvested, purified using Protein A affinity chromatography (GE Life Sciences) and SEC using a Superdex 200 column equilibrated with PBS, and concentrated to 3 to 13 mg mL−1.
Crystallization and Data Collection.
Crystals were grown at 25 °C using a microbatch under oil method by mixing 0.3 µL protein with 0.3 µL of crystallization buffer consisting of 100 mM Bis-Tris:HCl pH 6.0 and 28% (vol/vol) polyethylene glycol monomethyl ether (MW 2000) (Sigma-Aldrich) under a drop of paraffin oil and silicon oil in a 2:1 ratio. Crystals suitable for diffraction took 3 to 4 wk to grow. Crystals were cryoprotected with crystallization buffer supplemented with 30% (vol/vol) glycerol, flash-frozen in liquid nitrogen, and diffraction data were collected at the Advanced Photon Source (beamline 24-ID-E). Data were processed using HKL2000 (46).
Structure Determination and Refinement.
The PD-1H crystal structure was solved using molecular replacement with the program Phaser (47), with the PD-L1 IgV-like domain (23) lacking interstrand loops as a search model. The initial model was refined using Refmac5 (48), autorebuilt with Buccaneer (49), and manually rebuilt with Coot (50). Iterative cycles of refinement and manual rebuilding were carried out to improve the model. During refinement, clear electron density was seen connected to Asn-17 in an Fo – Fc difference Fourier map, allowing unambiguous assignment of an N-acetylglucosamine residue. The structure was refined to a final R-factor of 20.9% and an Rfree of 24.5%, using data in the resolution range 41.9 to 1.9 Å. The final model includes 1,163 atoms in PD-1H, 14 atoms in the attached N-acetylglucosamine, and 35 water molecules. The data collection and refinement statistics are summarized in Table 1.
Structural Comparisons.
For comparisons of PD-1H and structurally homologous proteins, we performed automated structural comparison using the DALI server (27) using our PD-1H structure as a search query. Structures were superimposed and displayed with PyMOL (The PyMOL Molecular Graphics System, v2.0 Schrödinger, LLC). To compare the H-strand region of PD-1H with known IgV-like domain structures, individual IgV-like domains were extracted from the crystal structures of the Pfam database’s V-set Ig domain (PF07686) and superimposed on the PD-1H structure with PyMOL’s super or align commands. For figure generation, 5 structures that exhibited strand swapping were omitted for clarity (like all others, these structures also lacked any residues in the location corresponding to the H-strand of PD-1H).
Mice and Cells.
NSG mice were purchased from the Jackson Laboratory and maintained in our laboratory. Female mice were used for in vivo experiments at 2 mo of age. All mouse procedures were performed in Yale University’s animal facility and all mouse studies were approved by Yale University’s Institutional Animal Care and Use Committee. Buffy coats were purchased from New York Blood Center. PBMCs were isolated by using SepMate PBMC Isolation tubes (Stemcell Technologies) and stored in liquid nitrogen for in vitro and in vivo experiments.
In Vitro Human T Cell Proliferation Assay.
Ninety-six–well plates were coated with 5 µg/mL human IgG, or WT or mutated PD-1H fused with human IgG1 Fc tag at 4 °C overnight. Human PBMCs were labeled with 5 µM 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) and seeded in the plates at 2 × 105 per well. Soluble anti-human CD3ε OKT3 was added in culture in a range of concentrations. After culturing for 3 d, culture supernatants were collected for cytokine detection by human cytometric bead array (CBA). Cells were harvested for flow cytometry staining. CFSE profiles in the human CD45+ human CD3+ gate were analyzed. Antibodies for flow cytometry were purchased from Biolegend. Human Th1/Th2/Th17 CBA kit was purchased from BD Biosciences.
In Vitro Mouse OT-I CD8+ T Cell Activation by HEK293T-Kb-OVA Cell Lines.
Full-length mPD-1H, including its native signal peptide, was inserted into the pLenti7.3/V5-TOPO-GFP lentivector upstream of the C-terminal V5 tag (Thermo Fisher). For the ΔH construct, residues Met146 through Asn149 corresponding to the H-strand seen in our human PD-1H structure were deleted. For the ΔHSS construct, the outermost paired cysteines (Cys12 and Cys145, corresponding to human Cys146) were mutated to serines, in addition to the same H-strand deletion. Lentiviruses were generated with mock lentivector, WT or mutant mPD-1H lentivector and pPACKH1 packaging kit (System Biosciences) in HEK293T cells. HEK293T-KbOVA (293T-KbOVA) cell line was transduced with each lentivirus carrying either mPD-1H WT or mutant genes. Cells were stained by anti-mouse PD-1H monoclonal antibody (mam82 clone, made in our laboratory), and GFP+ mPD-1H+ cells were sorted by BD FACSAriaII. Polyclonal stable cell lines were maintained after sorting. To confirm the expression level of mPD-1H, the C-terminal V5 expression tag was detected by intracellular staining with anti-V5 monoclonal antibody (2F11F7, Thermo Fisher). OT-I T cells were purified from lymph nodes and spleen of C57BL/6-Tg(TcraTcrb)1100Mjb/J mouse (Jackson Laboratories) with EasySep Mouse CD8+ T Cell Isolation Kit (Stemcell) and labeled with 5 µM CFSE. Next, 2 × 105 OT-I cells were cocultured with 4 × 104 UV-radiated parental, mock transduced, mPD-1H WT- or mutant-transduced 293TKbOVA cells in 96 well flat bottom plate (Corning). Three days later, cells were harvested and stained by anti-CD3 and anti-CD8 (BD). CFSE profiles on CD3+CD8+ gate were analyzed on Attune NxT cytometer (Life Technology). IFN-γ in culture supernatant was detected by CBA mouse inflammation kit (BD Biosciences).
In Vivo Human T Cell Activation with Xeno-Antigen.
Plasmids expressing hIgG1 Fc (hFc), WT PD-1H-hFc or HA123-hFc were administered into NSG mice by hydrodynamic injection. One day later, 8 million CFSE-labeled human PBMCs were injected intraperitoneally into the mice. Six days after the PBMC injection, cells from the peritoneal cavity were isolated with medium (RPMI 1640 with 2% FBS). Cells were harvested for flow cytometry staining. CFSE profiles in the human CD45+ human CD3+ gate were analyzed. Antibodies for flow cytometry were purchased from Biolegend.
Statistics.
All in vitro experiments in this study were repeated at least 3 times independently and data were analyzed with GraphPad Prism 7.01. Statistical analyses were carried out with a 2-tailed Student’s t test. P values <0.05 were considered statistically significant differences between groups. Different asterisks were assigned: *P < 0.05; **P < 0.01; ***P < 0.001. Three or 4 replicated wells were used for in vitro proliferation assays. For the in vivo xeno-activation experiment, results of 2 independent experiments were pooled together.
Data Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6U6V).
Supplementary Material
Acknowledgments
We thank the staff of the Northeastern Collaborative Access Team (NE-CAT) beamline 24-ID-E for their assistance with X-ray data collection; Declan Clarke for discussion on bioinformatic approaches; Ti Badri for helping animal studies; and Beth Cadugan for editing the manuscript. This study was supported partially by sponsored research funding from Boehringer Ingelheim, NIH P50 CA196530, P30 CA016359, and the United Technologies Corporation Endowed Chair. NE-CAT beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E beam line is funded by a NIH-Office of Research Infrastructure Programs High-End Instrumentation grant (S10OD021527). This research used resources of the Advanced Photon Source, a US Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
Competing interest statement: In the past 12 months, L.C. has been a consultant/board member for NextCure, AstraZenca, Pfizer, Junshi, Zai Lab, Tayu, Vcanbio, and GenomiCare; is a scientific founder of NextCure and Tayu; and has sponsored research grants from NextCure, Pfizer, and Boehringer Ingelheim.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6U6V).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908711117/-/DCSupplemental.
References
- 1.Chen L., Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 4, 336–347 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y., Yao S., Chen L., Cell surface signaling molecules in the control of immune responses: A tide model. Immunity 34, 466–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Flies D. B., Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremer J. M., et al. , Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N. Engl. J. Med. 349, 1907–1915 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Vincenti F., et al. ; Belatacept Study Group , Costimulation blockade with belatacept in renal transplantation. N. Engl. J. Med. 353, 770–781 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Park H. J., et al. , PD-1 upregulated on regulatory T cells during chronic virus infection enhances the suppression of CD8+ T cell immune response via the interaction with PD-L1 expressed on CD8+ T cells. J. Immunol. 194, 5801–5811 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Sanmamed M. F., Chen L., A paradigm shift in cancer immunotherapy: From enhancement to normalization. Cell 175, 313–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., et al. , VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 208, 577–592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flies D. B., Wang S., Xu H., Chen L., Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J. Immunol. 187, 1537–1541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flies D. B., et al. , Coinhibitory receptor PD-1H preferentially suppresses CD4+ T cell-mediated immunity. J. Clin. Invest. 124, 1966–1975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Mercier I., et al. , VISTA regulates the development of protective antitumor immunity. Cancer Res. 74, 1933–1944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., et al. , Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 111, 14846–14851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceeraz S., et al. , VISTA deficiency accelerates the development of fatal murine lupus nephritis. Arthritis Rheumatol. 69, 814–825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sergent P. A., et al. , Blocking the VISTA pathway enhances disease progression in (NZB × NZW) F1 female mice. Lupus 27, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H., et al. , A crucial role of the PD-1H coinhibitory receptor in suppressing experimental asthma. Cell. Mol. Immunol. 15, 838–845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N., et al. , Immune-checkpoint protein VISTA critically regulates the IL-23/IL-17 inflammatory axis. Sci. Rep. 7, 1485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J., et al. , VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 23, 551–555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., et al. , Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc. Natl. Acad. Sci. U.S.A. 112, 6682–6687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flajnik M. F., Tlapakova T., Criscitiello M. F., Krylov V., Ohta Y., Evolution of the B7 family: Co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7's historical relationship with the MHC. Immunogenetics 64, 571–590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Gebali S., et al. , The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium T. U.; UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang V. T., Spooner R. A., Crispin M., Davis S. J., Glycan remodeling with processing inhibitors and lectin-resistant eukaryotic cells. Methods Mol. Biol. 1321, 307–322 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lin D. Y., et al. , The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. U.S.A. 105, 3011–3016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemmingsen J. M., Gernert K. M., Richardson J. S., Richardson D. C., The tyrosine corner: A feature of most Greek key beta-barrel proteins. Protein Sci. 3, 1927–1937 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clements C. S., et al. , The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 100, 11059–11064 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandhi A. K., et al. , High resolution X-ray and NMR structural study of human T-cell immunoglobulin and mucin domain containing protein-3. Sci. Rep. 8, 17512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi N., et al. , TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27, 927–940 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Márquez J. A., et al. , The crystal structure of the extracellular domain of the inhibitor receptor expressed on myeloid cells IREM-1. J. Mol. Biol. 367, 310–318 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Chang V. T., et al. , Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17, 574–582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y., et al. , Neuron-specific SALM5 limits inflammation in the CNS via its interaction with HVEM. Sci. Adv. 2, e1500637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., et al. , Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 25, 656–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon K. W., et al. , Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 349, 1261669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J. L., Davis M. M., Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13, 37–45 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Schwartz J. C., Zhang X., Fedorov A. A., Nathenson S. G., Almo S. C., Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature 410, 604–608 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Stamper C. C., et al. , Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature 410, 608–611 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Evans E. J., et al. , Crystal structure of a soluble CD28-Fab complex. Nat. Immunol. 6, 271–279 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyay K., Bhatia S., Fiser A., Almo S. C., Nathenson S. G., Structural basis of inducible costimulator ligand costimulatory function: Determination of the cell surface oligomeric state and functional mapping of the receptor binding site of the protein. J. Immunol. 177, 3920–3929 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Wang R., Natarajan K., Margulies D. H., Structural basis of the CD8 alpha beta/MHC class I interaction: Focused recognition orients CD8 beta to a T cell proximal position. J. Immunol. 183, 2554–2564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao S. M., Du Q. S., Meng J. Z., Pang Z. W., Huang R. B., The multiple roles of histidine in protein interactions. Chem. Cent. J. 7, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato Y., et al. , Acidic extracellular microenvironment and cancer. Cancer Cell Int. 13, 89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santiago C., et al. , Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27, 941–951 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasikumar P. G., Ramachandra M., Small-molecule immune checkpoint inhibitors targeting PD-1/PD-L1 and other emerging checkpoint pathways. BioDrugs 32, 481–497 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Flies D. B., Sandler B. J., Sznol M., Chen L., Blockade of the B7-H1/PD-1 pathway for cancer immunotherapy. Yale J. Biol. Med. 84, 409–421 (2011). [PMC free article] [PubMed] [Google Scholar]
- 45.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Storoni L. C., McCoy A. J., Read R. J., Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Cowtan K., The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6U6V).