Significance
The gold-standard model for structural analysis of F1-ATPase has been bovine mitochondrial F1 (bMF1), but its rotational dynamics remain elusive. This study analyzes rotational characteristics of bMF1. bMF1 showed 3 distinct dwells in rotation, “binding dwell,” “catalytic dwell,” and “short dwell,” in each 120° step of rotation. While the positions of binding and catalytic dwell are similar to those of human mitochondrial F1 (hMF1), bMF1 shows short dwell at a distinctively different position from the corresponding dwell of hMF1, implying variety in the timing of the putative reaction at short dwell, phosphate release or ADP release. Single-molecule manipulation experiments revealed that the affinity change of ATP is a major torque-generating step.
Keywords: F1-ATPase, bovine mitochondrial F1, single-molecule analysis, molecular motor
Abstract
The reaction scheme of rotary catalysis and the torque generation mechanism of bovine mitochondrial F1 (bMF1) were studied in single-molecule experiments. Under ATP-saturated concentrations, high-speed imaging of a single 40-nm gold bead attached to the γ subunit of bMF1 showed 2 types of intervening pauses during the rotation that were discriminated by short dwell and long dwell. Using ATPγS as a slowly hydrolyzing ATP derivative as well as using a functional mutant βE188D with slowed ATP hydrolysis, the 2 pausing events were distinctively identified. Buffer-exchange experiments with a nonhydrolyzable analog (AMP-PNP) revealed that the long dwell corresponds to the catalytic dwell, that is, the waiting state for hydrolysis, while it remains elusive which catalytic state short pause represents. The angular position of catalytic dwell was determined to be at +80° from the ATP-binding angle, mostly consistent with other F1s. The position of short dwell was found at 50 to 60° from catalytic dwell, that is, +10 to 20° from the ATP-binding angle. This is a distinct difference from human mitochondrial F1, which also shows intervening dwell that probably corresponds to the short dwell of bMF1, at +65° from the binding pause. Furthermore, we conducted “stall-and-release” experiments with magnetic tweezers to reveal how the binding affinity and hydrolysis equilibrium are modulated by the γ rotation. Similar to thermophilic F1, bMF1 showed a strong exponential increase in ATP affinity, while the hydrolysis equilibrium did not change significantly. This indicates that the ATP binding process generates larger torque than the hydrolysis process.
FoF1-ATP synthase (or ATP synthase) is one of the most ubiquitous enzymes found in the mitochondrial inner membrane, chloroplast thylakoid membrane, and bacterial plasma membranes (1, 2). FoF1-ATP synthase catalyzes the ATP synthesis reaction coupled with H+ (or sodium in some bacteria) translocation, which is driven by proton motive force (pmf) across membranes. This enzyme is structurally and functionally separated into 2 components, F1 and Fo, both of which are rotary molecular motors. F1 is the protruding portion from the membrane and possesses catalytic reaction centers for ATP synthesis. Fo is the membrane-embedded portion and conducts H+ translocation across the membrane. In the whole ATP synthase complex, F1 and Fo interconvert the free energy of ATP hydrolysis and pmf, via the mechanical rotation of the rotor complex. When the free energy of ATP hydrolysis per turn of the rotor complex exceeds the pmf per turn, FoF1-ATP synthase catalyzes the reverse reaction and hydrolyzes ATP, pumping H+ to generate pmf.
F1-ATPase (F1), when isolated from Fo, hydrolyzes ATP to ADP and inorganic phosphate (Pi). Upon catalysis, F1 rotates the rotor complex against the surrounding stator ring, on which the catalytic reaction centers are located. The subunit composition of F1 is α3β3γδε in both bacterial and mammalian types. However, the δ and the ε subunits are not equivalent in bacterial and mammalian types. The minimum complex as a rotary motor is the α3β3γ subcomplex.
The atomic structures of F1 have been intensively studied by X-ray crystallography since the first report on bovine mitochondrial F1, bMF1, in 1994 (3). The first crystal structure revealed most of the basic structural features of F1, which were repeatedly confirmed in later structural studies on bMF1 and other F1s (4–6). F1 is composed of the α3β3 stator ring and the central rotor complex of the γε in bacterial types and the γδε in mammalian types. In the α3β3 stator ring, the α and the β subunits are arranged alternately. The catalytic sites reside on one side of the αβ interface, while the other side of the αβ interface binds to ATP; however, it is catalytically impotent and thereby termed the noncatalytic site. The catalytic residues are mostly located on the β subunit, except for the catalytically critical arginine residue termed the “arginine finger” on the α subunit (3, 7–9). Among the 3 β subunits, 2 β subunits bind to nucleotides: one β binds to the ATP analog AMP-PNP and the other binds to ADP [later revealed to also bind to azide (5)]. These β subunits, termed βTP and βDP, respectively, adopt so-called closed conformation, in which the C-terminal helical domain rotates inwardly to the rotor γ subunit. The third subunit, βempty, has no bound nucleotide and adopts an open conformation, swinging the C-terminal domain outwardly. From the structural features, it has been proposed that ATP binding triggers the open-to-closed conformational transition of the β subunit, which is a major power-stroking motion. The conformational transition of the β subunit was later visualized using the single-fluorescence polarization technique (10), Förster resonance energy transfer (11), and high-speed atomic force microscopy (12).
Later crystallographic studies showed that βDP can bind to AMP-PNP (4) or a transition-state analog (13), while βTP predominantly binds to AMP-PNP unless AMP-PNP is omitted from the crystallization medium. Therefore, βDP is thought to represent the catalytically active conformational state for cleavage of bound ATP. This contention is supported by several studies (7, 14, 15). In some crystal structures, βempty has a small anionic ligand: phosphate (6), thiophosphate (16), with the implication that βempty represents the pre-/post-Pi-release state, as supported in a theoretical study (17).
Since its first visualization, the rotary dynamics of F1 have been well studied by single-molecule rotation assay (18). In a typical rotation assay, the α3β3 ring is immobilized on the glass surface, and a probe for rotational imaging is attached onto the protruding component of the γ subunit. For high-speed imaging of rotation practically not affected with viscous friction against a rotating probe, nanoprobes are used such as 40-nm gold colloid (19, 20) or nanorods (21). For the torque measurement or rotation manipulation, 100- to 500-nm-diameter polystyrene beads or magnetic beads are used. Single-molecule rotation analysis has elucidated the basic features of F1 rotation and the chemomechanical coupling scheme of rotation.
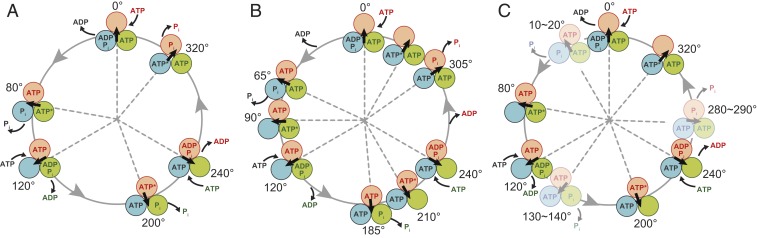
Among F1s characterized by the rotation assay thus far, thermophilic Bacillus PS3 (TF1) is the best characterized due to its high conformational stability and clear stepping behaviors. The rotation analysis of TF1 established the reference reaction scheme, although some variations for different F1s have been observed as described later. The unitary step size of the rotation is 120°, each coupled with a single turnover of ATP hydrolysis, reflecting the pseudo 3-fold symmetry of the structure. The 120° step rotation is divided into 2 substeps of 80° and 40° (19), each intervened by ATP-waiting dwell (binding dwell) or catalysis-waiting dwell (catalytic dwell), respectively (22). During binding dwell, another β releases ADP (23, 24), and Pi release is suggested to occur during catalytic dwell (24, 25). Considering that each β exerts a single turnover of ATP hydrolysis upon a single turn of the rotor, and the reaction phase is different by 120° among 3 βs, the reaction scheme is proposed as shown in Fig. 1A (25), although another scheme has also been proposed (26).
Fig. 1.
Chemomechanical coupling rotation schemes of TF1 (A), hMF1 (B), and bMF1 (C). Each circle and arrow represents the catalytic state of the β subunit and the angular positions of the γ subunit, respectively. 0° is defined as the position of γ subunit where a β subunit (orange) binds to ATP. The asterisks following “ATP” represent the catalytically active state to undergo hydrolysis of bound ATP.
Recent statistical analysis (27) revealed that TF1 makes a small rotation upon catalysis during catalytic dwell that is too small to be detected in conventional image analysis, suggesting that the catalytic dwell is split into hydrolysis and Pi-release dwells. The split of the catalytic dwell was also proposed in studies on the rotation of yeast mitochondrial F1 (28) and human mitochondrial F1 (hMF1) (29). The work on hMF1 showed that the 120° rotation was resolved into 3 substeps: 65°, 25°, and 30°. Each step was initiated by ATP binding, presumably Pi release and hydrolysis. Therefore, the dwells before the 65°, 25°, and 30° substeps are referred to as the binding dwell, Pi-release dwell, and catalytic dwell, respectively. The reaction scheme of hMF1 was proposed as shown in Fig. 1B. Due to the close sequence homology of hMF1 and bMF1 [∼99% in the α and β subunits and ∼93% in the γ subunit (16)], it is expected that the reported rotation behavior of hMF1 is similar to bMF1. From the viewpoint of the structure–function relationship of F1, the correlation between dwells and conformational states found in crystal structures is important to determine. Assays with inhibitors suggest that the Pi-release dwell corresponds to the state found in the majority of bMF1 crystal structures, including the first crystal structure (3), ground-state structure (4), and thiophosphate-bound structure (16). However, there are still differences in amino acid sequences between hMF1 and bMF1, and the investigation of the exact correlation between rotary dynamics and atomic structure of F1 requires a rotation assay with F1 from the same species used in the crystal structure analysis. Although a preliminary study on bMF1 was reported (30), basic characteristics of bMF1 have not been analyzed.
A single-molecule rotation assay of F1 enabled not only detailed kinetic analysis of stepping rotation but also manipulation experiments when combined with a magnetic tweezers system. The manipulation experiment of TF1 was first conducted for the direct demonstration of ATP synthesis upon the reverse rotation of the γ subunit (31, 32). After that, it has become a major focus how F1 modulates the rate and equilibrium constants of elementary reaction steps: binding, hydrolysis, and product releases. To assess this issue, a stall-and-release experiment was conducted to determine the rate constant and equilibrium constant of ATP binding or hydrolysis of ATP bound on the catalytic site as a function of rotary angle that formed a basis for following theoretical studies (17, 33–38). Significantly larger angle dependence of ATP binding than hydrolysis revealed that TF1 generates larger torque in the ATP binding step than in the hydrolysis step. However, the stall-and-release experiments have been conducted only for TF1 (39) and the generality of these findings remains unclear.
In this study, we investigated the γ rotation of bMF1 and found several differences in rotation dynamics between bMF1 and hMF1, from which we propose the reaction scheme for bMF1 shown in Fig. 1C. Based on the reaction scheme, we also analyzed the angle dependence of ATP affinity change as well as the modulation of the equilibrium constant of ATP hydrolysis by conducting a stall-and-release experiment. The single-molecule manipulation analysis revealed the general features of angle dependence of binding and catalysis are well-conserved across the species, suggesting that the torque generation mechanism is common among F1s, although the stepping behaviors have some variations.
Results
Construct of bMF1 for Rotation Assay.
Recombinant bovine mitochondrial F1 composed of α, β, γ, δ, and ε subunits was coexpressed with assembly factors, AF1 and AF2, and purified according to a previous report (30) with slight modifications (Materials and Methods). Two cysteines were introduced in the protruding part of the γ subunit at A99 and S191. They were specifically biotinylated to attach 40-nm gold nanoparticles or magnetic beads with ∼200-nm diameter as an optical probe. For immobilization, 9 histidine residues (His-tag) were introduced at the N terminus of the β subunit. The resultant bMF1 (His-tag, γA99C, γS191C) showed normal ATPase as shown below and was referred to as the wild type. In order to ensure the complex stability of bMF1, the bMF1 solution was diluted down to 10 nM and concentrated with a 100-kDa ultrafiltration filter to remove dissociated small subunits such as the δ and ε subunits. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed the genuine subunit composition was retained after dilution, showing the complex of bMF1 is stable at the nanomolar condition where the single-molecule rotation assay is to be conducted (SI Appendix, Fig. S1).
ATP-Driven Rotation.
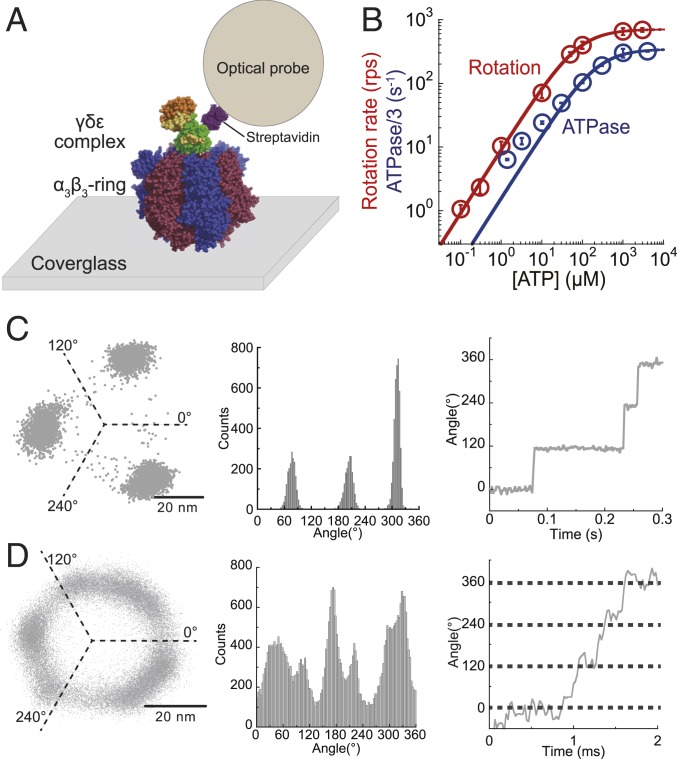
To observe genuine rotation of bMF1, 40-nm gold colloid attached to the γ subunit of bMF1 was observed under various [ATP]s at 23 ± 2 °C (Fig. 2A) with a dark-field microscope (20). Images were recorded at 125 to 45,000 frames per second (fps) (22 to 8,000 μs per frame), dependent on [ATP]. Red data points in Fig. 2B show the Michaelis–Menten curve of the rotation rate, in which the maximum rotation rate (Vmax) and Michaelis constant (Km) are 707 revolutions per second (rps) and 77 μM, respectively. The maximum rotation rate of 707 rps was comparable with that of human mitochondrial F1 (hMF1), 741 rps, and remarkably faster than those of bacterial F1s: 129 rps for thermophilic Bacillus PS3 (TF1) (19) and 449 rps for E. coli F1 (EF1) (40). Considering the coupling ratio of 3 ATPs per turn, the maximum rotation rate corresponded to the ATP hydrolysis rate of 2,121 per s. We also measured the ATP hydrolysis rate of bMF1 in solution with ATP-regeneration system (blue data points in Fig. 2B) and determined and as 1,037 per s and 218 μM, respectively. The ATPase rates measured in solution were lower than the estimated catalytic rate from the rotation rate at all [ATP]s. Significantly lower catalytic rates than expected from the rotation rate were often reported in other F1s (19, 28, 41, 42). This is due to ADP inhibition, which is an inactive state of F1 transiently halting catalysis and rotation. The ADP-inhibited state lowers the time-averaged rotation rate in the single-molecule rotation assay and ATP hydrolytic activity, determined as an ensemble average of molecules in solution.
Fig. 2.
ATP-driven rotation of bMF1. (A) A schematic image of the single-molecule rotation assay of bMF1. The α3β3-ring is immobilized on a glass surface, and a detection probe is attached to the γ subunit via biotin–streptavidin interaction. (B) [ATP] versus the rate of rotation (red) or ATPase/3 (blue). The mean value and the SD for each data point are shown with circles and error bars, respectively (n = 20 to 25 for measurement of rotation rate, n = 3 for measurement of ATPase). Solid lines represent Michaelis–Menten fittings; Vmax: 707 ± 5 rps, Km: 77 ± 2 μM for rotation rate; : 346 ± 11 s−1, : 218 ± 26 μM for ATPase/3 (fitted parameter ± fitting error). (C and D) x–y plot, angular histogram, and time course of rotation found at 300 nM ATP (C) and at 3 mM ATP (D). The recording rate was 500 and 45,000 fps, respectively.
Rotation trajectories projected on the x–y plane showed stepping rotation of bMF1 (Fig. 2 C and D). At low [ATP]s well below Km, bMF1 showed 3 distinctive pauses with 120° intervals (Fig. 2C) that should correspond to ATP-binding dwell. The histograms of the dwell time for ATP binding showed a single exponential decay function (SI Appendix, Fig. S2A). The rate constants determined from the dwell-time histograms were proportional to [ATP] as expected, giving the rate constant of ATP binding (), 3.4 × 107 M−1⋅s−1. This is mostly comparable to that of TF1 (3.0 × 107 M−1⋅s−1) (19) and hMF1 (2.7 × 107 M−1⋅s−1) (29). At high [ATP]s over Km, several bMF1 molecules showed 6 pauses as found in the rotation of hMF1 (29), although many of the molecules did not show 6 clear pauses. Subpauses were detected in the angle histograms by eye in 4 of 23 molecules. Fig. 2D showed x–y projections of a trajectory and the corresponding histogram of angular position observed at 3 mM ATP. The time course also shows multiple pauses within one revolution. Three of the 6 pauses should correspond to catalytic dwell as found in TF1 and hMF1. The estimated time constant of ATP binding at 3 mM should be less than 10 μs, too short to be detected. Thus, the intervening pause is not binding dwell. These suggest that bMF1 makes an intervening pause in addition to catalytic dwell. Note that the response time of the 40-nm gold nanoparticle was ∼0.1 ms, and thereby submillisecond events are principally blurred and difficult to resolve. Therefore, the dwell-time analysis at high [ATP] was impractical.
ATPγS-Driven Rotation.
To resolve the rotation and dwells more clearly, we observed rotation in the presence of ATPγS, which is a slowly hydrolyzable ATP analog. The previous rotation assays showed that ATPγS slows the ATP hydrolysis on TF1 (22) and also presumably release of thiophosphate on hMF1 (29). Rotation rates of bMF1 were determined at various [ATPγS]s to draw the Michaelis–Menten curve (SI Appendix, Fig. S3A). Vmax and Km were determined to be 20.3 rps and 2.2 μM, respectively. As expected, Vmax was largely suppressed at about 35 times slower than that of the ATP-driven rotation. The binding constant of ATPγS, , was estimated from to be 3.0 × 107 M−1⋅s−1, which was almost identical to .
At high [ATPγS]s over Km, bMF1 showed distinct pauses separated by 120° steps, corresponding to 3 dense clusters in the x–y plot of rotation (Fig. 3 A, Inset). A closer look at the time courses showed additional short pauses during 120° steps (Fig. 3A), showing that bMF1 makes 2 types of dwells, hereafter referred to as long dwell (blue in Fig. 3A) and short dwell (orange in Fig. 3A) in a 120° rotation. Short dwell was not always observed in each 120° step due to the short lifetime. To identify the dwells and steps of rotation objectively, we employed a nonparametric change-point (CP) analysis to detect angular changes in the rotary traces based on the permutation test (27). Unlike other parametric-level detection methods in single-molecule time series analysis, such as hidden Markov modeling (43), our nonparametric CP analysis does not require any a priori assumption of the noise model and the number of pauses/steps in the rotary trace. CP analyses have also been shown to outperform the commonly used method of binning and thresholding the time series that may introduce artifacts to the waiting time kinetics in low signal-to-noise cases (44). Details of our CP detection method are given in SI Appendix, Supplementary Information Text and Fig. S4.
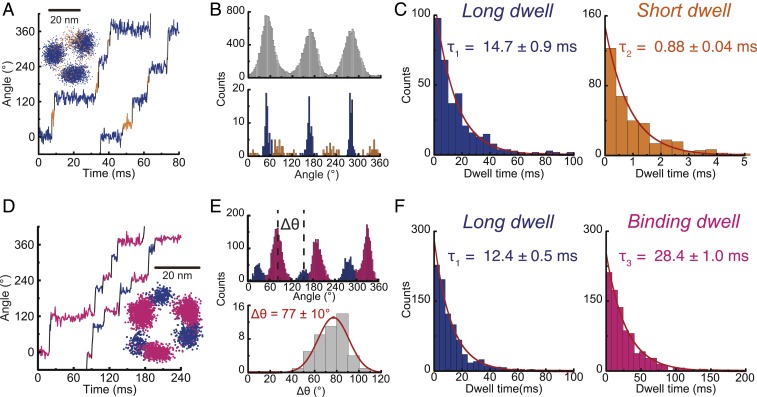
Fig. 3.
ATPγS-driven rotation of bMF1. From Michaelis–Menten fitting, Vmax and Km were estimated to be 20.3 ± 1.0 rps and 2.2 ± 0.5 μM (fitted parameter ± fitting error), respectively (SI Appendix, Fig. S3A). (A) Time course of rotation at 1 mM ATPγS recorded at 10,000 fps. CP analysis detects long (blue) and short (orange) dwell. (Inset) x–y plot. (B) Histograms for dwell position analysis. (Upper) The conventional histogram of angle positions from all data points from the time course. (Lower) The histogram of angle positions of dwells detected by CP analysis. (C) Dwell-time analysis of long and short dwells at 1 mM (n = 6). Values are fitted parameter ± fitting error. (D) Time course of rotation at 1 μM ATPγS recorded at 1,000 fps. (Inset) x–y plot. (E) Histograms of angle positions. (Upper) The conventional histogram from all data points. (Lower) The histogram of the angle positions of long dwell from binding dwell (Δθ), defined in the upper panel. Values are mean ± SD (n = 45, 15 molecules). (F) Dwell time analysis of long and binding dwell at 1 μM (n = 4). Values are fitted parameter ± fitting error.
The presence of short dwells was confirmed in the CP analysis. Fig. 3B shows angular histograms of a representative molecule. Fig. 3 B, Upper shows a conventional angular histogram prepared from all data points of a time course trajectory. Fig. 3 B, Lower shows the histogram constructed from the CP intervals, denoted as a histogram of CP intervals, in which the angular positions between 2 successive CPs are represented by the median angle of the interval. Each CP interval gives a single count in the histogram regardless of the dwell length, contrary to the conventional angle histogram where a longer dwell provides more counts. It is evident from Fig. 3B that the histogram of CP intervals clearly showed 3 clusters corresponding to the short dwells (orange) between distinctly high peaks of long dwells (blue). The angular position of short dwell was 60° from a long dwell at the left side (clockwise side) (SI Appendix, Fig. S5A). The dwell time was analyzed for long and short dwells, both of which showed single exponential decay functions, giving time constants of 14 to 15 ms for long dwell and 0.7 to 0.9 ms for short dwell (Fig. 3C and SI Appendix, Fig. S6). It should be noted that in addition to long and short dwells, distinctively long pauses over 1 s were also observed occasionally (SI Appendix, Fig. S7 A–C). We attributed the occasional long pause to ADP inhibition, considering that all characterized F1s in the rotation assay showed long dwells during rotation by ADP inhibition on the order of seconds (40, 41, 45, 46). The position of the ADP-inhibition dwell coincided with the position of long dwell (SI Appendix, Fig. S7C), suggesting that long dwells correspond to catalytic dwell where F1 executes the ATP cleavage reaction (22).
Rotation was observed at [ATPγS]s below Km, which was expected to resolve the rotation into binding dwell and long dwell. The recording rate was set at 1,000 fps to preserve image data storage of the high-speed camera. This allowed long-time observations. As expected, 2 types of dwells were found during 120° rotations in both the angle histogram and time course (Fig. 3 D and E). In the region below Km, the binding pause limited the overall rotation rate, showing longer dwells than long dwells. In the time course and angle histogram, the longer pauses were identified as binding dwells (pink). Relatively shorter pauses were assigned as long dwells. The angle distance between long dwell and binding dwell was 77° (Fig. 3 E, Lower). This is mostly consistent with hMF1 (29) and TF1 (22). As expected, when [ATPγS] was decreased, the duration time of binding dwell was lengthened. Fig. 3F and SI Appendix, Fig. S6 show the dwell-time histogram for binding dwell and long dwell. The time constant of long dwell was constant at 11 to 13 ms, consistent with the abovementioned value (14 ms). The length of binding dwell depended on [ATPγS]s as expected, giving the rate constants of ATPγS binding (), 2.9 × 107 M−1⋅s−1, well consistent with (3.0 × 107 M−1⋅s−1) determined from the Michaelis–Menten analysis. In the rotation assay at low [ATPγS]s, a distinctively long pause attributable to ADP inhibition was again observed at the angle of long dwell (SI Appendix, Fig. S7 D–F), suggesting that long dwell corresponds to catalytic dwell. Short dwells were not detected throughout the rotation assay with low [ATPγS]s, probably because short dwells of ATPγS rotation are too short to be detected with the recoding frame rate (1 ms per frame).
Rotation of bMF1(βE188D).
To confirm that long dwell is +80° from binding angle, we tested a mutant F1 (βE188D) in the rotation assay. This glutamic acid is highly conserved in primary sequences among all F1s. In crystal structures, this glutamic acid interacts with the γ phosphate via a coordinated water molecule. A quantum mechanics/molecular mechanics study revealed that this glutamic acid accelerated the ATP cleavage reaction, promoting the rate-limiting proton relay (7). When βE190 of TF1 (equivalent to βE188 of bMF1) was replaced with aspartic acid (D), the rate constant of ATP cleavage step was greatly slowed over 320-fold (22).
SI Appendix, Fig. S3B shows the rotation rates of bMF1(βE188D) at various [ATP]s. The data points were well fitted with the Michaelis–Menten curve with Vmax and Km of 1.2 rps and 1.2 μM, respectively. As expected, Vmax was largely suppressed, which was about 600 times lower than that of wild-type bMF1. The ATP binding constant, , was estimated to be 3.2 × 106 M−1⋅s−1, which was 10 times lower than of the wild type.
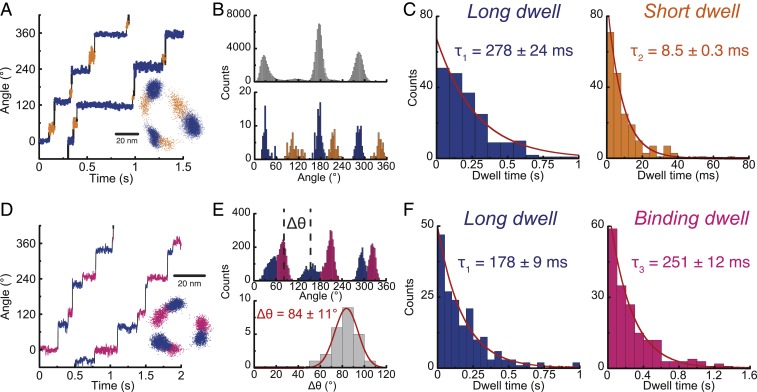
The stepping behaviors of bMF1(βE188D) were well consistent with those found in the ATPγS-driven rotation of the wild-type bMF1. At high [ATP]s, we again observed long and short dwells during 120° steps (Fig. 4 A–C). The dwell position histogram based on CP analysis showed that short dwell was located at 48° between long dwells (Fig. 4 B, Lower and SI Appendix, Fig. S5B). Histograms of durations of long and short dwells showed a single exponential decay function with time constants of 220 to 280 ms and 6 to 12 ms, respectively (Fig. 4F and SI Appendix, Fig. S8). At low [ATP]s, bMF1(βE188D) showed that long dwell occurred at +84° from binding dwell (Fig. 4 D–F). Dwell-time histograms determined the time constants of long dwell to be 180 to 230 ms and binding dwell to be 251 ms for 1 μM ATP, 75 ms for 3 μM ATP, and 27 ms for 10 μM ATP. The rate constant of ATP binding was determined to be 4.3 × 106 M−1⋅s−1, which is mostly consistent with that estimated from the abovementioned Michaelis–Menten analysis. Thus, rotation assay of ATPγS and bMF1(βE188D) confirmed that bMF1 makes long dwell at +80° from binding dwell and short dwell at +50° to 60° from long dwell, that is, +10° to 20° from binding dwell.
Fig. 4.
ATP-driven rotation of bMF1(βE188D). From Michaelis–Menten fitting, Vmax and Km were estimated to be 1.24 ± 0.03 rps and 1.15 ± 0.13 μM (fitted parameter ± fitting error), respectively (SI Appendix, Fig. S3B). (A) Time course of rotation at 1 mM ATP recorded at 2,000 fps. CP analysis detects long (blue) and short (orange) dwell. (Inset) x–y plot. (B) Histograms for dwell position analysis. (Upper) The conventional histogram of angle positions from all data points from the time course. (Lower) The histogram of angle positions of dwells detected by CP analysis. (C) Dwell-time analysis of long and short dwells at 1 mM (n = 5). Values are fitted parameter ± fitting error. (D) Time course of rotation at 1 μM ATP recorded at 500 fps. (Inset) x–y plot. (E) Histograms of angle positions. (Upper) The conventional histogram from all data points. (Lower) The histogram of the angle positions of long dwell from binding dwell (Δθ), defined in the upper panel. Values are mean ± SD (n = 24, 8 molecules). (F) Dwell time analysis of long and binding dwell at 1 μM (n = 5). Values are fitted parameter ± fitting error.
Stall by AMP-PNP.
The rotation assays with ATPγS or bMF1(βE188D) showed that long dwell occurred at +80° from binding angle. In addition, the coincidence of long-dwell angle with the angle of the ADP-inhibited state suggested that long dwell represented catalytic dwell where F1 executes the cleavage reaction. To further confirm these findings, we investigated the pause positions of bMF1 by blocking rotation with AMP-PNP, a nonhydrolyzable ATP analog to stall rotation at the angle of cleavage.
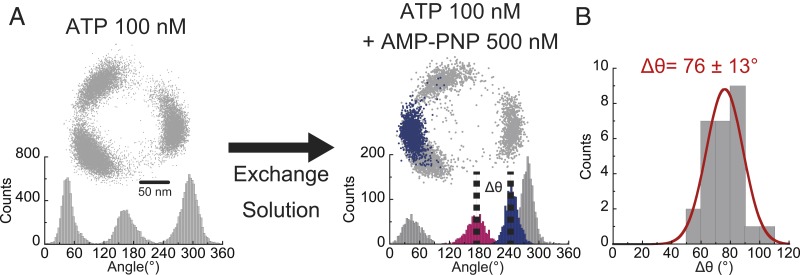
The rotation of the γ subunit of bMF1 was visualized with magnetic beads as a rotation probe because AMP-PNP–inhibited bMF1 could be reactivatable with magnetic tweezers, which allowed repeated experiments for the molecules. Rotation was observed at 100 nM ATP, where clear pauses at the ATP binding dwells were observed at 3 positions (Fig. 5A). Recording rate was 30 fps. After confirming the 3 pauses as binding dwell in a turn, the solution of 100 nM ATP plus 500 nM AMP-PNP was gently introduced into a flow cell to minimize interference of rotation by buffer flow. Typically, molecules stopped rotation within 3 min after buffer exchange. Once lapsed into AMP-PNP inhibition, bMF1 molecules never resumed rotation unless forcibly rotated over +360° with magnetic tweezers. It should be noted that ADP inhibition is rarely observed at 100 nM ATP (SI Appendix, Fig. S9A). The mean duration time of ADP inhibition observed at 2 mM ATP was ∼25 s, which is evidently shorter than the duration time of AMP-PNP inhibition, which is over 4 min (SI Appendix, Fig. S9B). In this experiment, after confirming that the pause lasted over 4 min, we defined the pause as an AMP-PNP stall. The pause angle of AMP-PNP inhibition was evidently different from the angles for binding dwell. The angular distance of the AMP-PNP stall from the nearest binding dwell on the left side was +76° (Fig. 5B), which is consistent with the position of long dwell. Thus, the angular position of ATP hydrolysis was confirmed at +80° from the angle of binding dwell, which is the same position as long dwell.
Fig. 5.
Pause positions stalled by AMP-PNP. (A) Experimental procedure. After observing binding dwell at 100 nM [ATP] (Left), 500 nM AMP-PNP and 100 nM ATP was introduced into the reaction mixture (Right). Blue data points represent the positions when rotation was blocked with AMP-PNP. (B) The angular distance (Δθ) of AMP-PNP inhibition from the nearest binding dwell (pink) (n = 27). Values are mean ± SD.
Angle-Dependent Modulation of Reaction Rates and Equilibriums.
Identification of rotation angles for ATP binding and ATP hydrolysis is fundamental to elucidate how F1 interconverts chemical energy of ATP hydrolysis into mechanical rotation. One of the most distinctive features that discriminate F1 from other molecular motor proteins is that F1 largely modulates chemical equilibriums of catalytic reaction steps depending on rotary angle to achieve ATP synthesis upon reversed rotation (31, 32). In a previous study (39), we established a “stall-and-release” experiment with magnetic tweezers, which allows for measurements of the rate constant and equilibrium constant of ATP binding or ATP hydrolysis as a function of rotary angle. This experiment revealed quantitative aspects of the “binding-change mechanism.” It was shown that TF1 exponentially increased affinity to ATP by 235-fold upon rotation by 60°, while it increased the equilibrium constant of ATP hydrolysis/synthesis only by 3-fold. From these results, the contribution of affinity change for torque generation was estimated to be 21 to 54 pN nm, while that of hydrolysis was only 4 to 17 pN nm (2).
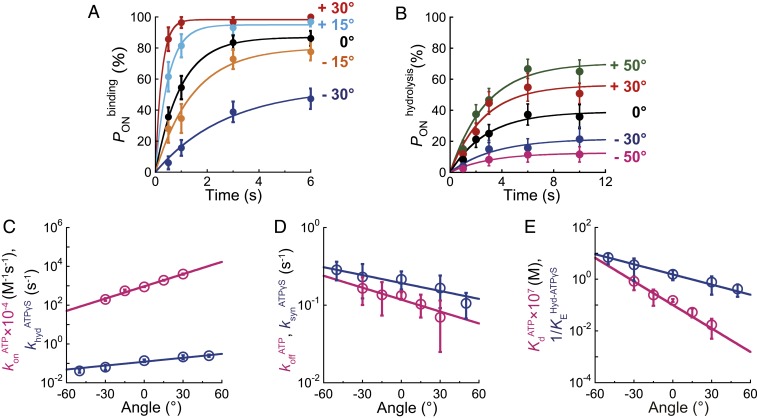
To investigate the angle-dependent modulation of affinity change and hydrolysis equilibrium of bMF1, we conducted a “stall-and-release” experiment. The experimental procedure was as follows. Rotation was observed under conditions where the target reaction, ATP binding or hydrolysis, was the rate-limiting step in the overall rotation rate. For ATP-binding, [ATP] was lowered to 100 nM, in which the mean waiting time for ATP binding was 0.9 s, while other reaction steps should be completed within 1 ms. For ATP-hydrolysis measurement, the intrinsic time constant for ATP hydrolysis, less than 0.5 ms, is too short for manipulation. Therefore, we observed rotation of bMF1(βE188D) in the presence of ATPγS, in which the catalytic dwell was prolonged to 4.0 s. When F1 paused to wait for the target reaction to occur, we stalled the rotation of bMF1 at the targeted angle with magnetic tweezers. After the set time period lapsed, bMF1 was released from the magnetic tweezers. Principally, bMF1 showed 2 behaviors: returning to the original waiting angle or stepping to the next waiting angle. Returning indicated that F1 had not executed the waiting reaction during the stall. We refer to that case as “OFF.” Stepping indicated that F1 had already executed the reaction and torque had been generated on the magnetic beads. That is referred to as an “ON” case. By determining the probability of ON cases (PON), we measured the probability of reaction as a function of rotary angle.
Fig. 6A shows time courses of the probability of ATP binding, , measured at the stall angles in the presence of 100 nM [ATP]. increased with the stall time and reached a plateau level, suggesting that the ATP binding reaction is reversible and in equilibrium with ATP release into solution. Assuming the reversible reaction scheme, F1 + ATP ⇄ F1-ATP, the time courses were fitted with the equation to determine and as functions of rotary angle, where and represent the rate constants of binding and release, respectively. Fig. 6B shows time courses of the probability of ATP hydrolysis, , that also shows typical saturation curves, reaching equilibrium levels. The time courses were fitted with , where and represent the rate constants of hydrolysis and synthesis, respectively.
Fig. 6.
Angle dependence of ATP binding and ATP hydrolysis in bMF1. (A) Time course of at 100 nM ATP in bMF1(WT). Each data point was obtained from 21 to 67 trials using 5 to 13 molecules. (B) Time course of at 1 mM ATPγS in bMF1(βE188D). Each data point was obtained from 28 to 64 trials using 3 to 8 molecules. (C–E) Angle dependence of ATP binding and ATP hydrolysis. 0° represents the initial position of ATP binding or hydrolysis before manipulation. The directions for “forward” and “reverse” reactions are defined as that for ATP hydrolysis (counterclockwise) and ATP synthesis (clockwise), respectively. Pink and blue represent ATP binding/ATP release and ATP hydrolysis/ATP synthesis. (C) Modulation of forward reactions upon rotation ( in pink and in blue). (D) Modulation of reverse reactions upon rotation ( in red and in blue). (E) Modulation of equilibrium constants upon rotation ( in pink and in blue). The circles and error bars in each data point represent the fitted parameter and fitting error determined in A and B.
Fig. 6 C–E show the determined rate constants and equilibrium constants plotted against the rotary angle, where “0” degree is defined as the mean angle for ATP binding or ATP hydrolysis. The directions for “forward” and “reverse” reactions are defined as that for ATP hydrolysis (counterclockwise) and ATP synthesis (clockwise), respectively. In both ATP binding and ATP hydrolysis, forward reactions are exponentially accelerated upon forward rotation, while the reverse reactions are exponentially decelerated. As a result, equilibriums are both exponentially changed to stabilize binding and hydrolysis states upon forward rotation. All rate constants and equilibrium constants were fitted with simple exponential functions as summarized in SI Appendix, Table S1. The determined functions show that the angle dependence is more remarkable in the binding process than hydrolysis: and increase/decrease upon rotation from −30° to +30°, by a factor of 20 and 2.4, respectively, and and are increased/decreased by a factor of 3.4 and 1.4, respectively. Binding affinity to ATP is largely modulated compared with the hydrolysis equilibrium. changed from 8.3 × 10−8 to 1.7 × 10−9 upon rotation from −30° to +30°, while changed from 3.6 to 0.8. The deviation of or against the rotary angle represents the free energy change upon rotation and thus is a good barometer of the contribution to torque generation. The free energy change upon binding affinity change and equilibrium shift to hydrolysis state is 17 to 49 and 7 to 16 pN nm, respectively (2, 39). This strongly suggests that binding affinity change contributes to torque generation more than the hydrolysis step. Although the degree of free energy change found in bMF1 is slightly lower than TF1, the observations are consistent.
Temperature Dependence of Maximum Rotation Rate.
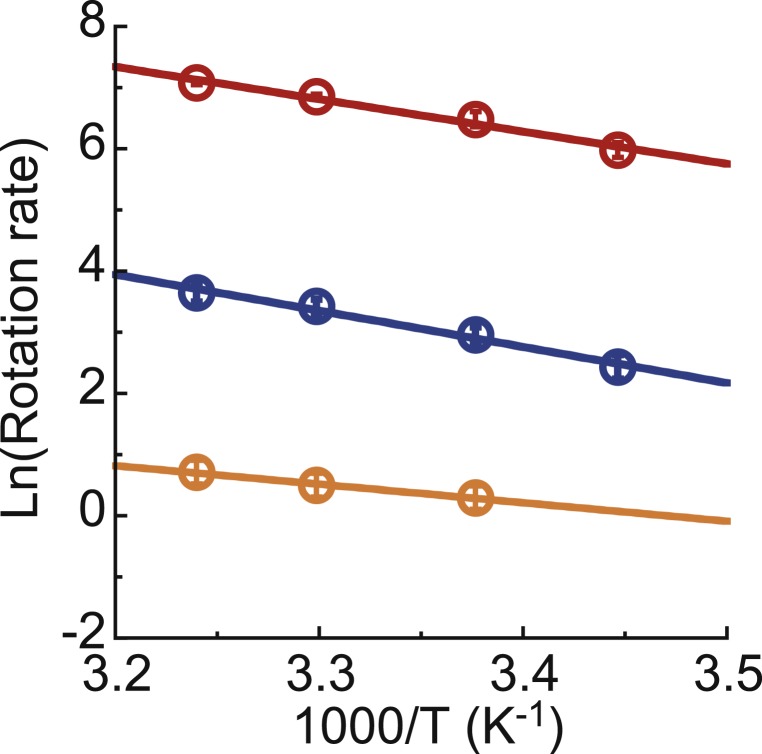
All of the abovementioned experiments were conducted at 23 ± 2 °C. In order to confirm that the reaction scheme is essentially not different in a wide range of temperatures including those near the physiological temperature of bMF1, the rotation rate at the ATP-saturated condition (1 mM) was observed at temperatures ranging from 17 °C to 35.5 °C. The rotation of bMF1(βE188D) or with ATPγS was also analyzed. The resulting Arrhenius plot showed the clear linearity in the range of temperature examined in all conditions (Fig. 7), indicating that the catalytic dwell is the kinetically bottlenecked reaction determining the overall reaction rate from 17 °C to 35.5 °C. This suggests that the reaction scheme found at room temperature is valid at a wide range of temperatures.
Fig. 7.
Arrhenius plot of rotation rate at substrate-saturated condition: (red) 1 mM ATP of bMF1(WT), (blue) 1 mM ATPγS of bMF1(WT), and (orange) 1 mM ATP of bMF1(βE188D). The mean value and the SD for each data point are shown with circles and error bars, respectively (n = 6 to 33).
The temperature dependence of the rotation rate of bMF1 is essentially the same as those of TF1. The Q10 factor of the rotation rate of bMF1 (1.3 to 1.9) is almost the same as the Q10 factors of ATP hydrolysis (1.9) and Pi release (1.6) of TF1 (47). As a result, the activation free energy, calculated from , was 56 to 72 kJ/mol, also well consistent with the values obtained previously for TF1 (47), and EF1 (48, 49). These results indicate that the transition states of the catalytic dwell of bMF1 are the same as those for TF1.
Discussion
Catalytic Event in Long Dwell and Binding Dwell.
This study investigated the fundamental features of bMF1 rotation under 3 conditions: in the presence of ATP, in the presence of ATPγS, and, by using a mutant F1, bMF1(βE188D). The latter 2 conditions were employed to slow down the cleavage step for resolving rotation into clear substeps. In all conditions, we observed long and short dwells under substrate-saturated conditions and long dwell and binding dwell in the region below Km. Although the short dwell in the ATP-driven rotation was too short for analysis, the rotation assays with ATPγS or with bMF1(βE188D) showed coincident angle assignments for short and long dwells: When the angular position for binding dwell was defined as 0°, long dwell was at +80° and short dwell at +10 to 20°.
ATPγS and the βE188D mutation are known to specifically slow down the hydrolysis step, although several studies suggested that the release step of thiophosphate or phosphate was also slowed down to some degree (7, 22). Therefore, it is reasonable to identify long pause as the hydrolysis waiting state. The combination of ATPγS and the βE188D mutation supports this assertion: In the presence of ATPγS, bMF1(βE188D) showed long dwell of 4.0 s. This is extended from the original long pause (0.5 ms) by a factor of 8,000, which is very close to the expected value (12,000) from multiplication of individual extension factors: 30 by ATPγS and 400 by βE188D. The agreement well supports that both ATPγS and βE188D slowed down the same reaction step at which the bound ATP is hydrolyzed. Another support is the observation of ADP-inhibited pause: The rotating bMF1 transiently stopped at the angle of long dwell for a few seconds. To date, all F1s characterized in the rotation assay have shown the transient pause by ADP inhibition at catalytic angle. This was confirmed in the inhibition experiment with AMP-PNP, which halts F1 rotation at the hydrolysis step. Unlike the case of ADP inhibition, once bMF1 stopped rotation it never resumed rotation even after forcible rotation with magnetic tweezers. The pause angle of bMF1 inhibited by AMP-PNP was found at the angle for long dwell. All experimental results show that long dwell represents the hydrolysis waiting state of bMF1. This is consistent with the findings of TF1 and hMF1, in which the catalytic pause is at +80 to 90° from the binding angle.
The remaining uncertainty is the short pause. Considering the high similarity of amino acid sequences between bMF1 and hMF1, it was expected that the short pause would correspond to the intermitting pause found in the rotation assay of hMF1 [referred to as the “1st dwell” or the “Pi dwell” in a previous study (29)]). Actually, the short pause of bMF1 and the 1st dwell of hMF1 were both found between ATP-binding angle and catalytic angle. This suggests that short pause of bMF1 is also the pause for phosphate release, as considered for the 1st dwell of hMF1. However, the addition of phosphate or thiophosphate at concentrations from 10 μM to 100 mM in the assay solution did not show a clear impact on the rotation behavior of bMF1 in current conditions (SI Appendix, Fig. S10). Thus, the chemical state of short pause of bMF1 remains to be elucidated.
Rotation Scheme of bMF1.
Fig. 1C shows the proposed reaction scheme of bMF1. Considering the findings of the present study as well as the crystal structures of bMF1 in which 2 of 3 catalytic sites are always occupied with bound nucleotides, ATP hydrolysis is assigned to be at 200° when the angle for ATP binding on the catalytic site is defined as 0°. This is also along the reaction schemes of TF1 and hMF1 (Fig. 1 A and B). One prominent difference in the reaction scheme among species is the number of substeps: TF1 shows 2 distinctive substeps. A clear difference between mammalian F1s is the position of the pause between binding dwell and catalytic dwell: +65° from binding dwell in hMF1 and +10 to 20° in bMF1. As a result, substep size is also different: 65°, 25°, and 30° substeps for hMF1, while bMF1 makes 10 to 20°, 50 to 60°, and 40° substeps. Note while experimentally this had not been determined, careful data analysis based on a data-mining method found that TF1 also makes small substeps during catalytic dwell (27).
There are also some distinctive differences in the kinetics of rotation between bMF1 and hMF1, although overall kinetic parameters such as Vmax and Km are mostly the same. In the rotation of bMF1, the duration time of catalytic dwell was always longer than short dwell, although the reverse is true in the rotation of hMF1: The 1st dwell was longer than catalytic dwell in hMF1. The source of these differences found in substeps and kinetics between bMF1 and hMF1 is unknown. The amino acid sequences are overall quite similar between bMF1 and hMF1. The α and β subunits share mostly identical sequences (99%), whereas the γ subunit shows relatively lower homology, 93% (16). Therefore, the most probable explanation is that the structural difference of the γ subunit causes differences in the kinetics and stepping behavior.
Correlation with Crystal Structures.
The present work reveals that bMF1 has at least 3 distinctive conformational states: binding dwell state, short dwell state, and catalytic dwell state. Obviously, the catalytic dwell state principally corresponds to the bMF1 crystal structures. It has been suggested that the current resolved crystal structures of bMF1 represent the catalytically active state, as supported by several studies, including the crystal structure with transient state analog, beryllium fluoride, and theoretical analysis (13). However, variations among crystal structures have been reported to date. They differ in bound nucleotides, inhibitors, inorganic ligands, and conformational states of subunits. Particularly, there is variety in rotational orientation of the γ subunit in crystal structures. Although it should depend on methods for structural alignment and analysis, the maximum difference in the angular orientation of the γ subunit has been reported to be over 30°. Particularly, when βE binds to thiophosphate the γ subunit is positioned at −30° (16) from that found in crystal structures with mitochondrial inhibitor proteins (50, 51). This feature is almost consistent with the rotation scheme proposed for hMF1, where phosphate release triggers rotation from the phosphate-releasing state at +65° from the binding site to the hydrolysis waiting state at +90°. However, the actual bMF1 does not show a dwell at around −30° from the catalytic dwell position. Although it is possible to assign short pause as the phosphate-releasing state, the angular distance between short and catalytic dwells, 50 to 60°, is too large. Thus, it is still unclear which crystal structure(s) exactly correspond to the catalytic dwell state.
Angle Dependence of Catalytic Power of bMF1.
One of the most remarkable features of F1-ATPase that discriminates it from other molecular motors is that F1-ATPase can reverse the catalytic reaction to synthesize ATP from ADP and phosphate when the rotation is reversed. This means that all catalytic reaction steps should be modulated with the rotation angle. To investigate this characteristic feature in detail, we developed a “stall-and-release” experiment. In the previous study on TF1, we found that the affinity to ATP was exponentially increased with forward rotation, while the equilibrium constant of hydrolysis was only slightly increased (39). The estimated free energy release upon the progressive affinity change of ATP was 21 to 54 pN nm. In contrast, upon equilibrium shift of hydrolysis, it was only 4 to 17 pN nm, where the lower-limit values represent the estimation directly from the angle dependence and the upper-limit values represent the corrected values with consideration of the torsional elasticity of the γ subunit (52). In this study, we confirmed the generality of the angle-dependent catalysis modulation: bMF1 progressively and exponentially increased affinity to ATP while the hydrolysis equilibrium shifted slightly upon forward rotation. The free energy change upon rotation was 17 to 49 pN nm for binding change and 7 to 16 pN nm for equilibrium shift of hydrolysis. Although the estimated free energy change was lower in both reactions compared to TF1, similar angle dependence was confirmed. Considering that thermophilic V1-ATPase (TV1) showed much weaker angle dependence of binding affinity (53), the strong angle-dependent affinity change is a conserved characteristic feature of F1-ATPase among species.
Materials and Methods
Preparation of bMF1.
The bMF1 plasmid, a gift from T. Suzuki, Tokyo Institute of Technology, Tokyo, was introduced into the FoF1-deficient Escherichia coli strain BL21. The recombinant E. coli strain was cultured in Super Broth medium containing 100 μg/mL carbenicillin and 25 μg/mL tetracycline for 24 h at 27 °C. To avoid dissociation of the bMF1 complex, purification was performed throughout at room temperature, 23 ± 2 °C. Harvested cells were suspended in 50 mM imidazole-HCl (pH 7.2) containing wash buffer A (40 mM potassium phosphate buffer, pH 7.5, 100 mM K2SO4, 10% glycerol, and 0.2 mM ATP), disrupted using an ultrasound disintegrator, and subjected to ultracentrifugation (81,000 × g, 20 min, 25 °C). The supernatant was introduced into Ni-Sepharose FF resin (GE Healthcare). After binding of bMF1 to the resin, it was washed with more than 10 volumes of wash buffers A and B (100 mM imidazole containing wash buffer A) to remove contaminant proteins. Subsequently, bMF1 was eluted with 50 mL of elution buffer (400 mM imidazole containing wash buffer A). The eluted fraction was applied to a gel-filtration column (Superdex 200; GE Healthcare) that had been previously equilibrated with gel-filtration buffer (40 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5 mM EDTA, pH 8.0, 10% glycerol, and 0.2 mM ATP). The fractions were collected and concentrated using a centrifugal concentrator (30-kDa, Centricon50; Millipore Corp.). The concentration of bMF1 was determined using bovine serum albumin (BSA) as a standard.
Measurement of ATPase Activity.
ATPase activity was measured at 25 °C in 50 mM Hepes-KOH (pH 7.5) containing 50 mM KCl, various concentrations of MgCl2, and ATP-regenerating system supplemented with 0.2 mM NADH and 0.1 mg/mL lactate dehydrogenase. The ATPase activity was calculated from the maximum slope of the absorbance of NADH during 5 s after the start of measurement.
Rotation Assay.
To visualize the rotation of bMF1, 2 cysteines on a rotor γ subunit (γA99C, γS191C) were biotinylated to attach 40-nm gold nanoparticles or magnetic beads as an optical probe. The bMF1 rotation assay was performed in the same manner as previously described (25), except for slight modifications. The procedures were as follows. The flow cell was constructed from 2 cover glasses (18 × 18 mm2 and 24 × 32 mm2; Matsunami Glass) using double-sided tape as a spacer. The bMF1 of ∼1 nM in the basal buffer was infused into the flow cell and incubated for 5 to 10 min. After that, unbound bMF1 molecules were washed out with the basal buffer containing 10 mg/mL BSA. Then, 40-nm gold nanoparticles or magnetic beads were infused and incubated for 5 to 10 min. Unbound beads were washed out with the basal buffer containing indicated concentrations of substrate. The basal buffer for bMF1 assay contained 50 mM Hepes-KOH (pH 7.5), 50 mM KCl, and various concentrations of MgCl2. When ATP was used, an ATP-regenerating system (1 mM phosphoenolpyruvate and 50 μg/mL pyruvate kinase) was added to the reaction mixture.
In the rotation assays with the 40-nm gold colloid, the rotating colloid particle that was attached to the γ subunit of bMF1 was observed using a dark-field microscope with a 60× objective lens at the recording rate of 125 to 45,000 fps (FASTCAM-1024PCI; Photron). The localization precision was 1 to 2 nm with signal-to-noise ratio ranging from 60 to 100 (20). For observation of the magnetic beads, a phase-contrast microscope (IX-70 or IX-71; Olympus) with a 100× objective lens at 30 fps (FC300M; Takex) was used.
The rotation assay was performed at 23 ± 2 °C, room temperature, except for 17 °C, 30 °C, and 35.5 °C in the temperature-dependence experiment (Fig. 7). For assay at 17 °C, the microscope room was cooled with an equipped air conditioner, and temperature was monitored with a thermometer attached on the flow cell on the microscopic stage. For assay at 30 °C or 35.5 °C, an objective lens heater (MATS-75R;, Tokai Hit Corp.) was used. Actual temperature of the sample was monitored the same as in the assay at 17 °C.
Data Analysis.
To suppress the effect of focus drifts on analysis, we have corrected data using nonspecific binding molecules on a glass surface before analysis. To avoid undesired fluctuation, the median filter (±2 frames) was applied to the time course in Figs. 3A and 4A. To estimate time constants, the histograms of the dwell time were fitted by a single exponential decay function. Pause angles were determined by fitting the angle distribution with a Gaussian function in Figs. 3E, 4E, and 5B. For the visualization and estimation of short pause between long pauses, CP analysis was applied to the time traces shown in Figs. 3A and 4A, as described in SI Appendix, Supplementary Information Text.
Manipulation with Magnetic Tweezers.
Magnetic tweezers were equipped on the microscope stage and controlled by custom software (Celery) (39). In Fig. 6 A and B, kinetic parameters were determined by using a single exponential function according to the reversible reaction scheme. The SD of in Fig. 6 A and B is given as , where N is the number of trials for each experiment.
Data Availability.
Data are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.pg4f4qrjk).
Supplementary Material
Acknowledgments
We thank T. Suzuki (Tokyo Institute of Technology) for the kind gift of the bMF1 plasmid; M. Hara, R. Watanabe, and S. Mori (University of Tokyo) for technical support; and all members of the H.N. laboratory for valuable comments. This work was supported in part by Grants-in-Aid for Scientific Research (25840053 and 16K14706 to H.U.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and Bilateral Joint Research Projects from the Japan Society for the Promotion of Science (to H.U.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data reported in this paper have been deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.pg4f4qrjk).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909407117/-/DCSupplemental.
References
- 1.Walker J. E., The ATP synthase: The understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Noji H., Ueno H., McMillan D. G. G., Catalytic robustness and torque generation of the F1-ATPase. Biophys. Rev. 9, 103–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahams J. P., Leslie A. G. W., Lutter R., Walker J. E., Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Bowler M. W., Montgomery M. G., Leslie A. G. W., Walker J. E., Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 A resolution. J. Biol. Chem. 282, 14238–14242 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Bowler M. W., Montgomery M. G., Leslie A. G. W., Walker J. E., How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. U.S.A. 103, 8646–8649 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabaleeswaran V., Puri N., Walker J. E., Leslie A. G. W., Mueller D. M., Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 25, 5433–5442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi S., et al. , Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single-molecule observations. J. Am. Chem. Soc. 134, 8447–8454 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Komoriya Y., et al. , Principal role of the arginine finger in rotary catalysis of F1-ATPase. J. Biol. Chem. 287, 15134–15142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W., Gao Y. Q., Cui Q., Ma J., Karplus M., The missing link between thermodynamics and structure in F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 874–879 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masaike T., Koyama-Horibe F., Oiwa K., Yoshida M., Nishizaka T., Cooperative three-step motions in catalytic subunits of F1-ATPase correlate with 80° and 40° substep rotations. Nat. Struct. Mol. Biol. 15, 1326–1333 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Sugawa M., et al. , F1-ATPase conformational cycle from simultaneous single-molecule FRET and rotation measurements. Proc. Natl. Acad. Sci. U.S.A. 113, E2916–E2924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchihashi T., IIno R., Ando T., Noji H., High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–759 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Kagawa R., Montgomery M. G., Braig K., Leslie A. G. W., Walker J. E., The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 23, 2734–2744 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittrich M., Hayashi S., Schulten K., ATP hydrolysis in the βTP and βDP catalytic sites of F1-ATPase. Biophys. J. 87, 2954–2967 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuno D., et al. , Correlation between the conformational states of F1-ATPase as determined from its crystal structure and single-molecule rotation. Proc. Natl. Acad. Sci. U.S.A. 105, 20722–20727 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bason J. V., Montgomery M. G., Leslie A. G. W., Walker J. E., How release of phosphate from mammalian F1-ATPase generates a rotary substep. Proc. Natl. Acad. Sci. U.S.A. 112, 6009–6014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okazaki K., Hummer G., Phosphate release coupled to rotary motion of F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 110, 16468–16473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noji H., Yasuda R., Yoshida M., Kinosita K. Jr, Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Yasuda R., Noji H., Yoshida M., Kinosita K. Jr, Itoh H., Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Ueno H., et al. , Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 98, 2014–2023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spetzler D., et al. , Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 45, 3117–3124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimabukuro K., et al. , Catalysis and rotation of F1 motor: Cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizaka T., et al. , Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Adachi K., et al. , Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Watanabe R., Iino R., Noji H., Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Shimo-Kon R., et al. , Chemo-mechanical coupling in F1-ATPase revealed by catalytic site occupancy during catalysis. Biophys. J. 98, 1227–1236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C. B., Ueno H., Watanabe R., Noji H., Komatsuzaki T., ATP hydrolysis assists phosphate release and promotes reaction ordering in F1-ATPase. Nat. Commun. 6, 10223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steel B. C., et al. , Comparison between single-molecule and X-ray crystallography data on yeast F1-ATPase. Sci. Rep. 5, 8773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T., Tanaka K., Wakabayashi C., Saita E., Yoshida M., Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat. Chem. Biol. 10, 930–936 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T., et al. , Expression of mammalian mitochondrial F1-ATPase in Escherichia coli depends on two chaperone factors, AF1 and AF2. FEBS Open Bio 6, 1267–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh H., et al. , Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Rondelez Y., et al. , Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat. Biotechnol. 23, 361–365 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Volkán-Kacsó S., Marcus R. A., Theory for rates, equilibrium constants, and Brønsted slopes in F1-ATPase single molecule imaging experiments. Proc. Natl. Acad. Sci. U.S.A. 112, 14230–14235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volkán-Kacsó S., Marcus R. A., Theory of single-molecule controlled rotation experiments, predictions, tests, and comparison with stalling experiments in F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 113, 12029–12034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkán-Kacsó S., Marcus R. A., Theory of long binding events in single-molecule-controlled rotation experiments on F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 114, 7272–7277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee S., Warshel A., Dissecting the role of the γ-subunit in the rotary-chemical coupling and torque generation of F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 112, 2746–2751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S., Warshel A., Brønsted slopes based on single-molecule imaging data help to unveil the chemically coupled rotation in F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 112, 14121–14122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam K., Pu J., Karplus M., Trapping the ATP binding state leads to a detailed understanding of the F1-ATPase mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 17851–17856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe R., et al. , Mechanical modulation of catalytic power on F1-ATPase. Nat. Chem. Biol. 8, 86–92 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Bilyard T., et al. , High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMillan D. G. G., Watanabe R., Ueno H., Cook G. M., Noji H., Biophysical characterization of a thermoalkaliphilic molecular motor with a high stepping torque gives insight into evolutionary ATP synthase adaptation. J. Biol. Chem. 291, 23965–23977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spetzler D., et al. , Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry 48, 7979–7985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinney S. A., Joo C., Ha T., Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terentyeva T. G., et al. , Dynamic disorder in single-enzyme experiments: Facts and artifacts. ACS Nano 6, 346–354 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Hirono-Hara Y., et al. , Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hisabori T., Kondoh A., Yoshida M., The γ subunit in chloroplast F1-ATPase can rotate in a unidirectional and counter-clockwise manner. FEBS Lett. 463, 35–38 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Watanabe R., Minagawa Y., Noji H., Thermodynamic analysis of F1-ATPase rotary catalysis using high-speed imaging. Protein Sci. 23, 1773–1779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekiya M., Nakamoto R. K., Al-Shawi M. K., Nakanishi-Matsui M., Futai M., Temperature dependence of single molecule rotation of the Escherichia coli ATP synthase F1 sector reveals the importance of γ-β subunit interactions in the catalytic dwell. J. Biol. Chem. 284, 22401–22410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.al-Shawi M. K., Parsonage D., Senior A. E., Thermodynamic analyses of the catalytic pathway of F1-ATPase from Escherichia coli. Implications regarding the nature of energy coupling by F1-ATPases. J. Biol. Chem. 265, 4402–4410 (1990). [PubMed] [Google Scholar]
- 50.Bason J. V., Montgomery M. G., Leslie A. G. W., Walker J. E., Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 111, 11305–11310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gledhill J. R., Montgomery M. G., Leslie A. G. W., Walker J. E., How the regulatory protein, IF1, inhibits F1-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U.S.A. 104, 15671–15676 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okuno D., Iino R., Noji H., Stiffness of γ subunit of F1-ATPase. Eur. Biophys. J. 39, 1589–1596 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Tirtom N. E., Okuno D., Nakano M., Yokoyama K., Noji H., Mechanical modulation of ATP-binding affinity of V1-ATPase. J. Biol. Chem. 288, 619–623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the Dryad Digital Repository (https://doi.org/10.5061/dryad.pg4f4qrjk).