Abstract
Kestose, a fructooligosaccharide (FOS) with one fructose monomer linked to sucrose, is a key component of the prebiotic activity of FOS. This study aimed to evaluate the prebiotic potential of Kestose in terms of the impact on population change in the intestinal microbiota and fecal short-chain fatty acid (SCFA) concentration in dogs. Kestose 2 g per dog was administered daily with conventional diet to 6 healthy, adult beagle dogs for 8 weeks followed by 4 weeks of follow-up period without Kestose supplementation. Fresh fecal samples were obtained before and every 4 weeks until the end of the follow-up period. Genomic DNA extracted from the fecal samples was subjected to 16S rRNA gene analysis using next generation sequencer and to quantitative polymerase chain reaction (qPCR). Fecal acetate, propionate, butyrate, lactate and ethanol concentrations were measured by high-performance liquid chromatography. 16S rRNA gene analysis and qPCR showed increasing trend of genus Bifidobacterium after Kestose supplementation while genera Bacteroides and Sutterella decreased. Clostridium perfringens decreased below the detection limit within first 4 weeks after starting Kestose supplementation. Fecal butyrate concentration was significantly increased at week 8 and returned to the base level after 4 weeks of the washing period. To the best of our knowledge, this is the first study to reveal effect of Kestose on the populational changes in fecal microbiota and fecal butyrate concentration in dogs.
Keywords: butyrate, dog, intestinal microbiota, kestose, prebiotics
It is well-known that the intestinal microbiota has direct and indirect impact on the health of the multiple organ systems of the host [6, 30, 31]. Growing interests in relationships between gut microbiota and health or pathological status are also growing in the field of veterinary science, and for example, altered microbiome populations in canine inflammatory bowel diseases [54, 62] and lymphoma [17], has been reported in dogs. Probiotics, prebiotics, and synbiotics are the most popular approaches aiming to normalize or improve the gastrointestinal environment using microbiota. Prebiotics are a non-viable food component that confers a health benefit on the host and is associated with the modulation of the microbiota [41]. The major representative of prebiotics is oligosaccharides, which are metabolized by the intestinal microbiota, resulting in the accumulation of short-chain fatty acids (SCFAs) that have various beneficial effects on the host [8]. Bifidogenic activity of prebiotics is one of the most important effects on host health [46]. Fructooligosaccharides (FOSs), which are oligosaccharides with one or more fructose monomers, are well-characterized prebiotics in humans, and studies have suggested their beneficial properties in rodents; for example, treatment with FOS prevented the development of preneoplastic lesions [14] and the delayed onset of senescence [36]. FOS consist of different ratios of 1-kestose (Kestose), nystose, and fructofranosylnystose, which have 1 to 3 fructose monomers linked with sucrose [8]. Of the FOS components, recent in vitro study using human intestinal lactobacilli and bifidobacteria suggested that Kestose content was crucial for the prebiotic activity of FOS [13, 38]. In addition, marked proliferation of bifidobacteria and increase in the cecal level of SCFA was observed in rats fed a Kestose-supplemented diet [59].
Several studies have reported that dietary prebiotic fibers and/or FOS modulates gastrointestinal microbiota in dogs [5, 15, 16, 26, 33, 39, 40, 42, 51] and cats [3, 4, 16, 50]. On the other hand, prebiotic property of Kestose has not been evaluated for its influence on the intestinal microbiota in dogs. The purpose of this study was to elucidate the prebiotic potential of Kestose in terms of changes in the intestinal microbiota population and fecal SCFA concentration.
MATERIALS AND METHODS
Animals
All procedures for the animal experiment in this study were approved by the animal research committee at the Tokyo University of Agriculture and Technology (approval number, #28-58). Healthy adult beagles (n=6) maintained in laboratory were used in this study. Of these, three neutered females were 6 years old and three intact males were 2 years old. Their body weight was 9.3–11.4 kg.
Study design
The whole study period was 12 weeks; the dogs were fed the same regular maintenance diet (Vita-one®, Nippon Petfood Co., Ltd., Tokyo, Japan; 200 g/dog once a day) during the study period. Ingredients of this maintenance diet include: corn, bran, chicken meal, powdered beef, powdered pork, defatted soybean, gamma-linolenic acid, brewing yeast, dried cabbage, oligosaccharide, casein phosphopeptide, minerals, vitamins, and amino acids. Each dog was fed a tablet form of Kestose (2 g once a day) along with the regular diet, for 8 weeks. Fresh feces of approximately 2 g/dog were sampled beginning from day 1, just before starting Kestose supplementation, and on the first days of week 4, 8 and 12. Fresh feces were gently taken directly from the dogs’ rectum using a gloved finger to minimize contamination and hemorrhage. The fecal samples taken before starting Kestose (week 0) were immediately stored at −20°C for a month then subsequently stored at −80°C until further use. The fecal samples taken at week 4, 8 and 12 were immediately stored at −80°C until further use.
DNA extraction from fecal samples
Genomic DNA was extracted from the feces according to the method reported by Takahashi et al. [56]. Frozen fecal samples were thawed on ice, then 100 mg of each sample was suspended in 4 M guanidium thiocyanate, 100 mM Tris-HCl (pH 9.0), and 40 mM EDTA and then disrupted with zirconia beads using a FastPrep FP100A instrument (MP Biomedicals, Santa Ana, CA, USA). DNA was extracted from the bead-treated suspensions using a Magtration System 12GC and GC series MagDEA DNA 200 (Precision System Science, Chiba, Japan). Following the estimation of DNA concentrations of each sample by spectrophotometry, ND-1000 (NanoDrop Technologies, Wilmington, DE, USA), the final concentration of the DNA samples was adjusted to 10 ng/µl.
16S rRNA gene sequence analysis using next generation sequencing (NGS)
Canine fecal bacterial 16S rRNA gene (16S rDNA) was analyzed by NGS using the MiSeq system (Illumina, San Diego, CA, USA) as previously described [56]. The V3-V4 hypervariable regions of 16S rDNA were amplified using polymerase chain reaction (PCR) from microbial genomic DNA using the universal primers for bacteria (341f and R806, Table 1) [10, 35] and the dual-index method [24]. Barcoded amplicons were sequenced using the paired-end method and were modified to 2 × 284-bp cycle run on the MiSeq system using MiSeq Reagent Kit version 3 (600 Cycle) (Illumina). After the alignment, the overlapping regions within the paired-end reads were merged and primer regions were omitted, which resulted in a 430 bp sequence. Only the reads with >99% of their sequence with quality value scores of ≥20 were extracted for further analysis [56]. The chimeric sequence detected by Usearch6.1.544_i86 was excluded [12]. Based on the sequences, taxonomic position of the sequences was identified at 97% similarity using the Metagenome@KIN analysis software (World Fusion, Tokyo, Japan) and the TechnoSuruga Lab Microbial Identification database DB-BA 10 (TechnoSuruga Laboratory, Shizuoka, Japan) [24, 27].
Table 1. List of primers and thermal cycling profiles used in this study.
| Target | Primer name | Oligonucleotide sequence | Reference strains for standard curves | PCR profile |
|---|---|---|---|---|
| All Bacteria | 341f | CCTACGGGAGGCAGCAG | Escherichia coli JCM 1649T | 95°C (5 sec)–60°C (20 sec)–72°C (20 sec)/35 cycles |
| 534r | ATTACCGCGGCTGCTGG | |||
| Actinobacteria | Act920F3 | TACGGCCGCAAGGCTA | Bifidobacterium longum subsp. Longum | 95°C (20 sec)–54°C (20 sec) –72°C (50 sec)/35 cycles |
| Act1200R | TCRTCCCCACCTTCCTCCG | JCM 1217T | ||
| Bacteroidetes | Bact934F | GGARCATGTGGTTTAATTCGATGAT | Bacteroides fragilis DSM 2151T | 95°C (5 sec)–55°C (30 sec) /35cycles |
| Bact1060R | AGCTGACGACAACCATGCAG | |||
| Fusobacteia | Fusobacteria F | GATCCAGCAATTCTGTGTGC | Fusobacterium nucleatum subsp. Nucleatum JCM 8532T | 95°C (5 sec)–55°C (20 sec)–72°C (50 sec)/35 cycles |
| Fusobacteria R | CGAATTTCACCTCTACACTTGT | |||
| Clostridium cluster XIV | CXIV-F1 | GAWGAAGTATYTCGGTATGT | Clostridium clostridioforme JCM 1291T | 95°C (5 sec)–52°C (30 sec)–72°C (30 sec)/35 cycles |
| CXIV-R2 | CTACGCWCCCTTTACAC | |||
| Bacteroides spp. | HuBac594Bhqf m | GTTGTGAAAGTTTGCGGCTCAACC | Bacteroides fragilis DSM 2151T | 95°C (5 sec)–60°C (30 sec)/35 cycles |
| HuBac692r | CTACACCACGAATTCCGCCT | |||
| Bifidobacterium spp. | BifiLM26F | GATTCTGGCTCAGGATGAACGC | Bifidobacterium longum subsp. JCM1217T | 95°C (5 sec)–60°C (20 sec)–72°C (20 sec)/35 cycles |
| Bif228R | CTGATAGGACGCGACCCCAT | |||
| Fusobacterium spp. | * | * | * | 95°C (5 sec)–64°C (30 sec) /35cycles |
| Lactobacillus spp. | LactoR’F | CACAATGGACGMAAGTCTGATG | Lactobacillus casei JCM 1134T | 95°C (20 sec)–56°C (20 sec) –72°C (50 sec)/35 cycles |
| LBFR | CGCCACTGGTGTTCTTCCAT | |||
| Sutterella spp. | ** | ** | ** | 95°C (5 sec)–57°C (20 sec)–72°C (20 sec)/35 cycles |
| Clostridium perfringens | Cperf 165F | CGCATAACGTTGAAAGATGG | Clostridium perfringens JCM 1290T | 95°C (5 sec)–60°C (30 sec) /35cycles |
| Cperf269R | CCTTGGTAGGCCGTTACCC | |||
| Faecalibacterium prausntzii | FPR-2F | GGAGGAAGAAGGTCTTCGG | Faecalibacterium prausnitzii ATCC 27768T | 95°C (5 sec)–57°C (20 sec)–72°C (50 sec)/35 cycles |
| Fprau645R | AATTCCGCCTACCTCTGCACT | |||
*Fusobacterium Detection Kit (TechnoSuruga Laboratory, Shizuoka, Japan). **Sutterella Detection Kit (TechnoSuruga Laboratory).
Quantitative analysis of intestinal microorganisms in dog feces using real-time PCR (qPCR)
Using the extracted DNA sample used in NGS analysis, quantitative analysis of the following intestinal organisms was performed using real-time PCR detecting each 16S rDNA: all bacteria [35], phylum Actinobacteria [2], phylum Bacteroidetes [20], phylum Fusobacteria [22], Clostridium cluster XIV [48], Bacteroides spp. [28], Bifidobacterium spp. [19], Fusobacterium spp., Lactobacillus spp. [49], Sutterella spp., Clostridium perfringens [37], and Faecalibacterium prausnitzii [43]. The list of primers and qPCR cycle conditions are indicated in Table 1. Each 16S rDNA of strains described in Table 1 was used for generating standard curves.
Measurement of SCFA, lactate and ethanol concentrations in feces
Fecal SCFA (acetate, propionate, and butyrate), lactate, and ethanol concentrations were measured using high-performance liquid chromatography (HPLC) (Shimadzu corporation, Kyoto, Japan) using the HPX-87H column (Bio-Rad Laboratories, Hercules, CA, USA). As for sample preparation, 100 mg of each fecal sample was suspended in 150 µl ultrapure water. Following centrifugation at 15,000 ×g at 4°C for 5 min, 80 µl of the supernatant and 120 µl of 8 mM H2SO4 were mixed and filtered using a low protein-binding hydrophilic PTFE membrane with a pore size of 0.45 µm (Merck Millipore, Darmstadt, Germany). This filtrate was used for HPLC analysis.
Statistical analysis
All of the results were compared according to the following two phases of this study: week 0 vs weeks 4 and 8, indicating effects caused by Kestose consumption (phase 1), and week 8 vs week 12, indicating effects caused by ceasing Kestose (phase 2). Shapiro-Wilk testing revealed that data of metagenomic analysis and quantitative PCR analysis were not normally distributed; hence Wilcoxon signed rank test was used for statistical analysis of both phase 1 and 2. In order to correct significance level in comparing data in phase 1, false discovery rate control was done using Benjamini–Hochberg procedure. Shapiro-Wilk testing revealed that data of SCFA concentration was normally distributed; hence Tukey’s multiple comparison test and paired t-test in phase 1 and 2 respectively, was used for statistical analyses. All of the statistical analyses were performed using the software IBM SPSS Statistics version 25 (IBM Japan, Ltd., Tokyo, Japan).
RESULTS
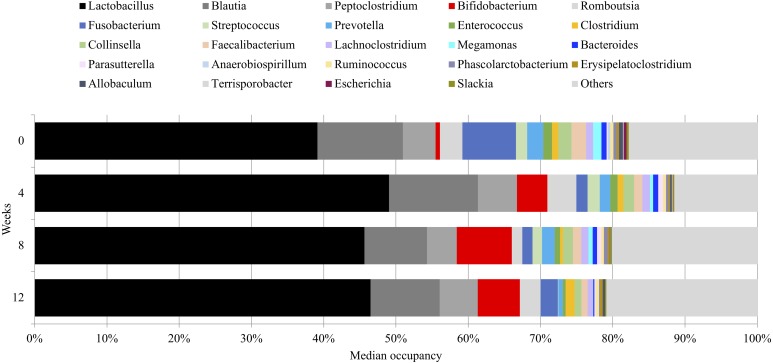
All dogs completed the 12-week study period without any clinical complications. After removal of chimeric sequences, 23,692 ± 651, 20,042 ± 574, 17,593 ± 687 and 17,753 ± 539 (mean ± SE) reads were obtained from samples collected at weeks 0, 4, 8 and 12, respectively, by 16S rRNA gene sequence analysis. The evenness and richness of gut microbiota of the tested dogs were analyzed by the Shannon, Chao and Simpson indices, which no statistical differences were observed. The ranges and median occupancies of the bacterial taxa are provided in Table S1. 16S rRNA gene sequence analysis revealed that the phylum Firmicutes was present in the largest population, accounting for 67.3% before starting Kestose supplementation and then 78.1, 66.6, and 70.5% at 4, 8 and 12 weeks, respectively (Table S1). Within the phylum Firmicutes, the genus Lactobacillus was present in the highest population, accounting for a median of 39.1% before starting Kestose supplementation. Daily Kestose supplementation resulted in a slight increase, yet not statistically significant, in the population of the genus Lactobacillus: 49.0, 45.6, and 46.5% at 4, 8, and 12 weeks, respectively (Fig. 1). Within phylum Actinobacteria, the occupancy of genus Bifidobacterium showed increasing trend after Kestose supplementation, at weeks 4 (median 4.19%) and 8 (median 7.64%) compared to week 0 (median 0.63%), then it slightly declined at week 12 (median 5.83%) which was a month after seceding Kestose (Fig. 1, Table S1). On the other hand, occupancy of phylum Fusobacteria genus Fusobacterium showed decreasing trend after Kestose supplementation, at weeks 4 (median 1.53%) and 8 (median 1.44%), compared to week 0 (median 7.42%) (Fig. 1, Table S1). Significant decrease in occupancies between week 8 and 12 were observed in phylum Bacteroidetes, genus Prevotella, genus Megamonas, genus Bacteroides, genus Butyricicoccus (P=0.043, respectively; Fig. 1, Table S1).
Fig. 1.
The median occupancy of each genus of canine fecal microbiome detected by 16S rRNA sequence analysis before (week 0) and after daily Kestose supplementation (weeks 4 and 8) and then after the cessation of daily Kestose for 4 weeks (week 12).
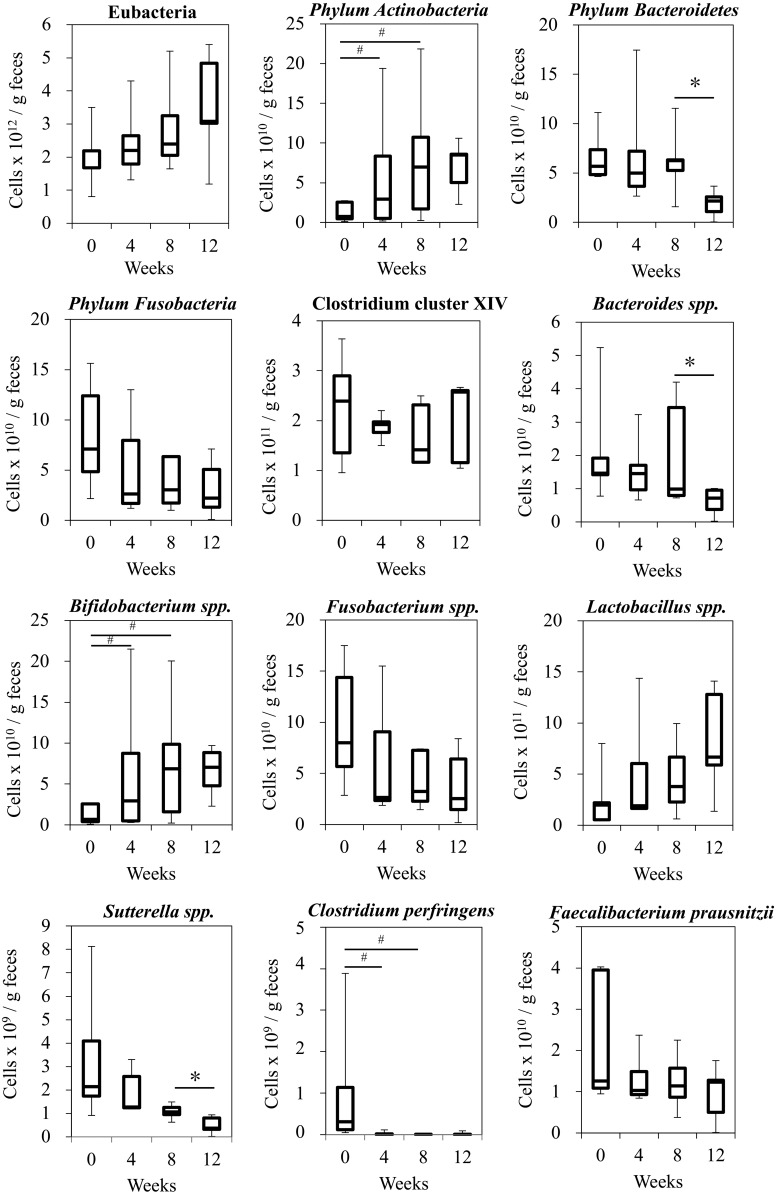
Quantitative analysis of the selected populations using real-time PCR revealed how the selected populations changed after Kestose intake. All members of eubacteria slightly increased after starting Kestose supplementation, yet not statistically significant. Similarly, increasing trends were observed in phylum Actinobacteria and genus Bifidobacterium during Kestose supplementation, and their numbers at week 12 stayed about the same level as those at week 8. The phylum Bacteroidetes, genus Bacteroides, and genus Sutterella showed no change up to 8 weeks during Kestose supplementation, but their numbers were significantly decreased at week 12 (P=0.043). Clostridium perfringens decreased to below the detection limit as early as 4 weeks after starting Kestose supplementation and remained below the detection limit during the study period. Phylum Fusobacteria and genus Fusobacterium showed decreasing trend during and after Kestose supplementation. Faecalibacterium prausnitzii and Clostridium cluster XIV showed no change in number during the study period (Fig. 2).
Fig. 2.
The bacterial count of representative species before (week 0), during (week 4 and 8) and after (week 12) Kestose supplementation, detected using qPCR of 16S rDNA from genomic DNA extracted from fecal samples. Median ± interquartile range and complete range, *P<0.05, #P<0.1.
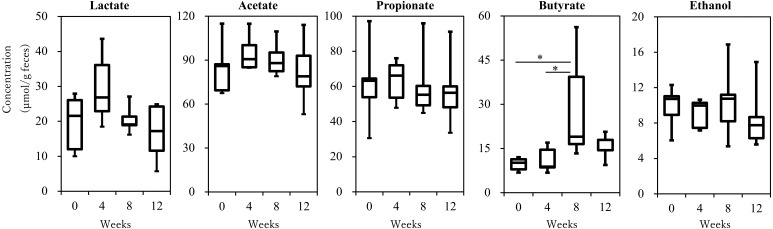
The fecal concentrations of SCFAs, lactate, and ethanol (µmol/g feces, in wet weight) were analyzed. Although lactate, acetate, and propionate did not show significant changes during the study period, butyrate was significantly increased at week 8 after starting Kestose supplementation (28.89 ± 8.21 µmol/g) compared with that at week 0 (9.65 ± 1.00 µmol/g, P=0.03) and week 4 (11.16 ± 1.96 µmol/g, P=0.048). Its concentration decreased to the basal level by week 12, which is after Kestose had been discontinued for 4 weeks (Fig. 3).
Fig. 3.
The fecal concentration of short-chain fatty acids, lactate, and ethanol analyzed using high-performance liquid chromatography before (week 0), during (week 4 and 8) and after (week 12) Kestose supplementation. Median ± interquartile range and complete range, *P<0.05.
DISCUSSION
The first part of this study showed population changes within the fecal microbiota before, during and after Kestose supplementation using 16S rRNA gene sequencing and quantitative PCR analysis in dogs. Lactobacillus spp. was the most dominant group, showing an additional slight increase after daily Kestose supplementation. As lactobacilli is known as a common inhabitant of all parts of the canine intestine [18, 52, 57], results obtained in this study are consistent with the previous reports. Similarities in the transition of the quantitative analysis results between eubacteria and Lactobacillus spp. may come from the large population impact of lactobacilli on the entire eubacteria. Median occupancy and number of the genus Bifidobacterium, respectively, where increased after Kestose supplementation, and the values remained higher than the basal level even after four-week period without Kestose supplementation; however, these changes remained as a trend and were not statistically significant. Bifidobacterium spp. is known to consume Kestose, resulting in strong bifidogenic activity of Kestose which has been reported in both in vitro and in vivo studies in humans and rodents [38, 55, 59]. Yet not specifically Kestose, previous studies have shown synbiotic and FOS to cause Bifidobacterium spp to increase in feces in dogs [5, 15, 40]. Although it was speculated that small number of dogs and considerable inter-dog difference resulted in less statistical power, it was suggested that fecal occupancy and cell numbers of Bifidobacterium spp. increased in dogs, possibly due to their preferential consumption of Kestose. Bifidobacteria are usually regarded as beneficial intestinal microbes in various animals and their activities are linked with the health of the host [25], the possibility of positive effects on health is expected in dogs as well; however, the evidence of such specific effects has not been elucidated. Populations which decreased during or after Kestose supplementation were Phylum Bacteroidetes, Bacteroides spp., Sutterella spp, and Clostridium perfringens. In one previous study in dogs, Bacteroides spp. decreased with addition of inulin [5]. Another study in dogs reported no change in fecal Bacteroides spp. with or without synbiotics [15]. No previous reports showing quantitative change of Sutterella spp. with regard to prebiotics in dogs could be found. In terms of Clostridium perfringens, Rinkinen et al. reported that canine-origin Lactobacilli reduced adhesion of C. perfringens ex vivo [45], while a metanalysis showed that number of C. perfringens was not affected by prebiotics in dogs [40]. In human reports, decrease in C. perfringens along with FOS uptake has been shown [23]. Although the decrease of C. perfringens observed in this study was statistically a trend, immediate reduction below the detection limit after starting Kestose supplementation suggested an effect of Kestose either directly or indirectly. Decreased populations may have lost in competition or by having been overwhelmed by other species which increased, possibly Bifidobacterium. Another possible reason for the decrease in these populations may have been change in fecal pH associated with the observed increase in the butyrate concentration. Butyrate is known to lower luminal pH, which leads to a less ideal environment for some species to proliferate. However, fecal pH was not evaluated in this study. It was an important finding that decreased populations after Kestose supplementation are known to be associated with gastrointestinal disorders. Clostridium perfringens and genus Sutterella were shown to increase in the feces of dogs with acute diarrhea compared with that in feces of healthy dogs using pyrosequencing and qPCR assays [53]. Association of Bacteroides spp. and host health is somewhat unclear. Certain article reported an increase of the microbe in the feces of dogs with chronic diarrhea [26], but opposite findings were also reported [34, 53]. Questions of what are “good” or “bad” populations and what is the ideal healthy balance in canine intestinal microbiota is still far from being fully understood; however, considering the general information from human and rodents, the population changes observed in this study may be a part of the positive prebiotic effect exerted by Kestose supplementation. Gut microbial diversity seems to be a beneficial marker of a “healthy gut”, and dietary fiber intake has been linked to decrease temporal microbial diversity in gut without any unwelcome outcomes [61]. In this study, Kestose supplementation did not affect the alpha-diversity.
The latter part of this study showed changes in SCFA, lactate, and ethanol concentrations in association with Kestose supplementation in dogs. Although fecal lactate, acetate, and propionate concentrations showed no change during the study period, butyrate concentration significantly increased at 8 weeks after starting daily Kestose supplementation. In humans, most of SCFAs are rapidly absorbed and metabolized by the host [21, 60], therefore the whole production of SCFA is difficult to be precisely determined [21]. However, since rate of its absorption is not likely to fluctuate depending on how much it is produced, higher concentration of fecal SCFA, representing remnant after absorption, was considered to reflect its voluminous production within large intestine, and fecal SCFAs are analyzed in other studies with similar concept [8, 15, 40, 59]. Increased production of SCFA within large intestine is known to have various beneficial effect to the host [1, 9, 15, 21, 47, 63]. The supplementation of prebiotic oligosaccharide has been previously shown to increase the fecal SCFA concentration in dogs [40, 44]. Supplementation with Kestose in particular, has been shown to result in a ten-fold increase of the butyrate concertation in rat cecal content [59], and similar effect was suggested to have taken place in dogs in this study. The results of fecal SCFA concentrations in this study cannot be well explained in relation with the results of microbiota analysis. As described, the major microbial change by Kestose supplementation is the proliferation of bifidobacteria; however, their major end-products are usually lactate and acetate, and not butyrate [7, 11]. Although major butyrate producers in canine intestines have not yet been characterized, certain species in the Clostridium cluster XIV and F. prausnitzii are major butyrate producers in human intestines [29]. These organisms were not significantly increased after Kestose supplementation. A possible reason for this discrepancy may have been due to the presence of unidentified butyrate producers in canine intestines. Butyrate is known to have beneficial effects in other animals; it is a major energy source for epithelial cells [21], induces regulatory T cells [1], promotes apoptosis in human colonic carcinoma cells [63], regulates intestinal inflammation [47], and is associated with the improvement of metabolic syndrome [9]. As dogs also suffer from many homologous disorders seen in humans, such as various tumors and inflammatory diseases, the possibility of beneficial prebiotic effects of Kestose supplementation, by the stimulation of butyrate production should be further elucidated.
The largest limitation of this study is the small number of dogs available to include in the study, due to the limited capacity for giving good animal care within the laboratory. In addition, considerable inter-dog difference, which is a common difficulty in any study using large animals, resulted in less statistical power. Differences in storage temperatures between samples collected at week 0 (stored at −20°C) and weeks 4, 8 and 12 (stored at −80°C) might be also a possible concern. Sample storage conditions, in general, may have an impact on results of microbiota analysis, however, with regard to the differences between −20°C and −80°C, immediate freezing of fecal samples in either temperature for a short period of time resulted in similar relative abundances of bacterial taxon and alpha-diversity estimations [32, 58]. It is also previously reported that immediate freezing at −20°C or below has been considered the gold standard for microbiome preservation [44]. As for the current result, the microbiota in week 12, which is after four-week washing period, showed a pattern returning or approaching to that of week 0, different from weeks 4 and 8 which was under the influence of Kestose ingestion. Although, to speak strictly, it remains unknown whether the sample storage temperature in this study did affect the results, authors concluded that differences in storage temperatures did not have significant impact in our studies, from above background.
Overall, findings of the present study demonstrated that daily Kestose supplementation resulted in its bifidogenic activity and increased fecal butyrate concentration, while Bacteroides spp., C. perfringens, and Sutterella spp. decreased in dogs. These findings suggest that Kestose may be a promising prebiotic supplement for dogs. Further studies including clinical trials are warranted to evaluate its potency for clinical application.
Supplementary
Acknowledgments
The project was funded by the B Food Science Co., Ltd. Support for this project was also provided by the Graduate School of Animal Life Science, Tokyo University of Agriculture and Technology. The funder provided support by providing experimental instruments and consumable products.
REFERENCES
- 1.Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., Kim S., Fritz J. V., Wilmes P., Ueha S., Matsushima K., Ohno H., Olle B., Sakaguchi S., Taniguchi T., Morita H., Hattori M., Honda K.2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236. doi: 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 2.Bacchetti De Gregoris T., Aldred N., Clare A. S., Burgess J. G.2011. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 86: 351–356. doi: 10.1016/j.mimet.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 3.Barry K. A., Wojcicki B. J., Middelbos I. S., Vester B. M., Swanson K. S., Fahey G. C., Jr.2010. Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J. Anim. Sci. 88: 2978–2987. doi: 10.2527/jas.2009-2464 [DOI] [PubMed] [Google Scholar]
- 4.Barry K. A., Middelbos I. S., Vester Boler B. M., Dowd S. E., Suchodolski J. S., Henrissat B., Coutinho P. M., White B. A., Fahey G. C., Jr., Swanson K. S.2012. Effects of dietary fiber on the feline gastrointestinal metagenome. J. Proteome Res. 11: 5924–5933. doi: 10.1021/pr3006809 [DOI] [PubMed] [Google Scholar]
- 5.Beloshapka A. N., Dowd S. E., Suchodolski J. S., Steiner J. M., Duclos L., Swanson K. S.2013. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol. Ecol. 84: 532–541. doi: 10.1111/1574-6941.12081 [DOI] [PubMed] [Google Scholar]
- 6.Buford T. W.2017. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome 5: 80. doi: 10.1186/s40168-017-0296-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell J. M., Fahey G. C., Jr., Wolf B. W.1997. Selected indigestible oligosaccharides affect large bowel mass, cecal and fecal short-chain fatty acids, pH and microflora in rats. J. Nutr. 127: 130–136. doi: 10.1093/jn/127.1.130 [DOI] [PubMed] [Google Scholar]
- 8.Campbell J. M., Bauer L. L., Fahey G. C., Hogarth A. J. C. L., Wolf B. W., Hunter D. E.1997. Selected fructooligosaccharide (1-kestose, nystose, and 1 F -β-fructofuranosylnystose) composition of foods and feeds. J. Agric. Food Chem. 45: 3076–3082. doi: 10.1021/jf970087g [DOI] [Google Scholar]
- 9.Canfora E. E., Jocken J. W., Blaak E. E.2015. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 11: 577–591. doi: 10.1038/nrendo.2015.128 [DOI] [PubMed] [Google Scholar]
- 10.Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., Knight R.2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 Suppl 1: 4516–4522. doi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan S. H., Louis P., Flint H. J.2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70: 5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R.2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. doi: 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo A., Nakamura S., Konishi K., Nakagawa J., Tochio T.2016. Variations in prebiotic oligosaccharide fermentation by intestinal lactic acid bacteria. Int. J. Food Sci. Nutr. 67: 125–132. doi: 10.3109/09637486.2016.1147019 [DOI] [PubMed] [Google Scholar]
- 14.Frederico A., Gomides F., Oliveira De Paula S., Gonçalves R. V., Licursi De Oliveira L., Lúcia De Luces C., Ferreira F., Comastri D. S., Do Carmo M., Peluzio G.2014. Prebiotics prevent the appearance of aberrant crypt foci (ACF) in the colon of Balb/C mice for increasing the gene expression of p16 protein. Nutr Hosp. Nutr. Hosp. 3030: 883–890. [DOI] [PubMed] [Google Scholar]
- 15.Gagné J. W., Wakshlag J. J., Simpson K. W., Dowd S. E., Latchman S., Brown D. A., Brown K., Swanson K. S., Fahey G. C., Jr.2013. Effects of a synbiotic on fecal quality, short-chain fatty acid concentrations, and the microbiome of healthy sled dogs. BMC Vet. Res. 9: 246–256. doi: 10.1186/1746-6148-9-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Mazcorro J. F., Barcenas-Walls J. R., Suchodolski J. S., Steiner J. M.2017. Molecular assessment of the fecal microbiota in healthy cats and dogs before and during supplementation with fructo-oligosaccharides (FOS) and inulin using high-throughput 454-pyrosequencing. PeerJ 5: e3184. doi: 10.7717/peerj.3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavazza A., Rossi G., Lubas G., Cerquetella M., Minamoto Y., Suchodolski J. S.2018. Faecal microbiota in dogs with multicentric lymphoma. Vet. Comp. Oncol. 16: E169–E175. doi: 10.1111/vco.12367 [DOI] [PubMed] [Google Scholar]
- 18.Grześkowiak Ł., Endo A., Beasley S., Salminen S.2015. Microbiota and probiotics in canine and feline welfare. Anaerobe 34: 14–23. doi: 10.1016/j.anaerobe.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueimonde M., Tölkkö S., Korpimäki T., Salminen S.2004. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70: 4165–4169. doi: 10.1128/AEM.70.7.4165-4169.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X., Xia X., Tang R., Zhou J., Zhao H., Wang K.2008. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 47: 367–373. doi: 10.1111/j.1472-765X.2008.02408.x [DOI] [PubMed] [Google Scholar]
- 21.Hamer H. M., Jonkers D., Venema K., Vanhoutvin S., Troost F. J., Brummer R. J.2008. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27: 104–119. doi: 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- 22.Hermann-Bank M. L., Skovgaard K., Stockmarr A., Larsen N., Mølbak L.2013. The Gut Microbiotassay: a high-throughput qPCR approach combinable with next generation sequencing to study gut microbial diversity. BMC Genomics 14: 788. doi: 10.1186/1471-2164-14-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hidaka H., Tashiro Y., Eida T.1991. Proliferation of bifidobacteria by oligosaccharides and their useful effect on human health. Bifidobact. Microflora 10: 65–79. doi: 10.12938/bifidus1982.10.1_65 [DOI] [Google Scholar]
- 24.Hisada T., Endoh K., Kuriki K.2015. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch. Microbiol. 197: 919–934. doi: 10.1007/s00203-015-1125-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isolauri E.2012. Development of healthy gut microbiota early in life. J. Paediatr. Child Health 48 Suppl 3: 1–6. doi: 10.1111/j.1440-1754.2012.02489.x [DOI] [PubMed] [Google Scholar]
- 26.Jia J., Frantz N., Khoo C., Gibson G. R., Rastall R. A., McCartney A. L.2010. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol. Ecol. 71: 304–312. doi: 10.1111/j.1574-6941.2009.00812.x [DOI] [PubMed] [Google Scholar]
- 27.Kasai C., Sugimoto K., Moritani I., Tanaka J., Oya Y., Inoue H., Tameda M., Shiraki K., Ito M., Takei Y., Takase K.2015. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 15: 100. doi: 10.1186/s12876-015-0330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Layton A., McKay L., Williams D., Garrett V., Gentry R., Sayler G.2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72: 4214–4224. doi: 10.1128/AEM.01036-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis P., Flint H. J.2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294: 1–8. doi: 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- 30.Lynch S. V., Pedersen O.2016. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 375: 2369–2379. doi: 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 31.Masuoka H., Shimada K., Kiyosue-Yasuda T., Kiyosue M., Oishi Y., Kimura S., Yamada A., Hirayama K.2017. Transition of the intestinal microbiota of dogs with age. Biosci. Microbiota Food Health 36: 27–31. doi: 10.12938/bmfh.BMFH-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy S., Gilliland S. E.2007. Isolation and characterization of Lactobacillus species having potential for use as probiotic cultures for dogs. J. Food Sci. 72: M94–M97. doi: 10.1111/j.1750-3841.2007.00310.x [DOI] [PubMed] [Google Scholar]
- 33.Middelbos I. S., Vester Boler B. M., Qu A., White B. A., Swanson K. S., Fahey G. C., Jr.2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One 5: e9768. doi: 10.1371/journal.pone.0009768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minamoto Y., Otoni C. C., Steelman S. M., Büyükleblebici O., Steiner J. M., Jergens A. E., Suchodolski J. S.2015. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 6: 33–47. doi: 10.1080/19490976.2014.997612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer G., de Waal E. C., Uitterlinden A. G.1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura S., Kondo N., Yamaguchi Y., Hashiguchi M., Tanabe K., Ushiroda C., Kawahashi-Tokuhisa M., Yui K., Miyakoda M., Oku T.2014. Daily feeding of fructooligosaccharide or glucomannan delays onset of senescence in SAMP8 mice. Gastroenterol. Res. Pract. 2014: 303184. doi: 10.1155/2014/303184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolan T., Hands R. E., Bustin S. A.2006. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1: 1559–1582. doi: 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- 38.Ose R., Hirano K., Maeno S., Nakagawa J., Salminen S., Tochio T., Endo A.2018. The ability of human intestinal anaerobes to metabolize different oligosaccharides: Novel means for microbiota modulation? Anaerobe 51: 110–119. doi: 10.1016/j.anaerobe.2018.04.018 [DOI] [PubMed] [Google Scholar]
- 39.Panasevich M. R., Kerr K. R., Dilger R. N., Fahey G. C., Jr., Guérin-Deremaux L., Lynch G. L., Wils D., Suchodolski J. S., Steer J. M., Dowd S. E., Swanson K. S.2015. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br. J. Nutr. 113: 125–133. doi: 10.1017/S0007114514003274 [DOI] [PubMed] [Google Scholar]
- 40.Patra A. K.2011. Responses of feeding prebiotics on nutrient digestibility, faecal microbiota composition and short-chain fatty acid concentrations in dogs: a meta-analysis. Animal 5: 1743–1750. doi: 10.1017/S1751731111000887 [DOI] [PubMed] [Google Scholar]
- 41.Pineiro M., Asp N. G., Reid G., Macfarlane S., Morelli L., Brunser O., Tuohy K.2008. FAO Technical meeting on prebiotics. J. Clin. Gastroenterol. 42 Suppl 3 Pt 2: S156–S159. doi: 10.1097/MCG.0b013e31817f184e [DOI] [PubMed] [Google Scholar]
- 42.Pinna C., Vecchiato C. G., Bolduan C., Grandi M., Stefanelli C., Windisch W., Zaghini G., Biagi G.2018. Influence of dietary protein and fructooligosaccharides on fecal fermentative end-products, fecal bacterial populations and apparent total tract digestibility in dogs. BMC Vet. Res. 14: 106. doi: 10.1186/s12917-018-1436-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez-Farias C., Slezak K., Fuller Z., Duncan A., Holtrop G., Louis P.2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 101: 541–550. doi: 10.1017/S0007114508019880 [DOI] [PubMed] [Google Scholar]
- 44.Ribeiro R. M., Souza-Basqueira M., Oliveira L. C., Salles F. C., Pereira N. B., Sabino E. C.2018. An alternative storage method for characterization of the intestinal microbiota through next generation sequencing. Rev. Inst. Med. Trop. São Paulo 60: e77. doi: 10.1590/s1678-9946201860077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinkinen M., Jalava K., Westermarck E., Salminen S., Ouwehand A. C.2003. Interaction between probiotic lactic acid bacteria and canine enteric pathogens: a risk factor for intestinal Enterococcus faecium colonization? Vet. Microbiol. 92: 111–119. doi: 10.1016/S0378-1135(02)00356-5 [DOI] [PubMed] [Google Scholar]
- 46.Scott K. P., Martin J. C., Duncan S. H., Flint H. J.2014. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 87: 30–40. doi: 10.1111/1574-6941.12186 [DOI] [PubMed] [Google Scholar]
- 47.Segain J. P., Raingeard de la Blétière D., Bourreille A., Leray V., Gervois N., Rosales C., Ferrier L., Bonnet C., Blottière H. M., Galmiche J. P.2000. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 47: 397–403. doi: 10.1136/gut.47.3.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y., Liu C., Finegold S. M.2004. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 70: 6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Songjinda P., Nakayama J., Tateyama A., Tanaka S., Tsubouchi M., Kiyohara C., Shirakawa T., Sonomoto K.2007. Differences in developing intestinal microbiota between allergic and non-allergic infants: a pilot study in Japan. Biosci. Biotechnol. Biochem. 71: 2338–2342. doi: 10.1271/bbb.70154 [DOI] [PubMed] [Google Scholar]
- 50.Sparkes A. H., Papasouliotis K., Sunvold G., Werrett G., Clarke C., Jones M., Gruffydd-Jones T. J., Reinhart G.1998. Bacterial flora in the duodenum of healthy cats, and effect of dietary supplementation with fructo-oligosaccharides. Am. J. Vet. Res. 59: 431–435. [PubMed] [Google Scholar]
- 51.Spears J. K., Karr-Lilienthal L. K., Fahey G. C., Jr.2005. Influence of supplemental high molecular weight pullulan or γ-cyclodextrin on ileal and total tract nutrient digestibility, fecal characteristics, and microbial populations in the dog. Arch. Anim. Nutr. 59: 257–270. doi: 10.1080/17450390500216993 [DOI] [PubMed] [Google Scholar]
- 52.Suchodolski J. S., Camacho J., Steiner J. M.2008. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol. Ecol. 66: 567–578. doi: 10.1111/j.1574-6941.2008.00521.x [DOI] [PubMed] [Google Scholar]
- 53.Suchodolski J. S., Markel M. E., Garcia-Mazcorro J. F., Unterer S., Heilmann R. M., Dowd S. E., Kachroo P., Ivanov I., Minamoto Y., Dillman E. M., Steiner J. M., Cook A. K., Toresson L.2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7: e51907. doi: 10.1371/journal.pone.0051907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suchodolski J. S., Xenoulis P. G., Paddock C. G., Steiner J. M., Jergens A. E.2010. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet. Microbiol. 142: 394–400. doi: 10.1016/j.vetmic.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 55.Suzuki N., Aiba Y., Takeda H., Fukumori Y., Koga Y.2006. Superiority of 1-kestose, the smallest fructo-oligosaccharide, to a synthetic mixture of fructo-oligosaccharides in the selective stimulating activity on bifidobacteria. Biosci. Microflora 25: 109–116. doi: 10.12938/bifidus.25.109 [DOI] [Google Scholar]
- 56.Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M.2014. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9: e105592. doi: 10.1371/journal.pone.0105592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y., Manninen T. J. K., Saris P. E. J.2012. Dominance of Lactobacillus acidophilus in the facultative jejunal Lactobacillus microbiota of fistulated beagles. Appl. Environ. Microbiol. 78: 7156–7159. doi: 10.1128/AEM.01975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tedjo D. I., Jonkers D. M. A. E., Savelkoul P. H., Masclee A. A., van Best N., Pierik M. J., Penders J.2015. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One 10: e0126685. doi: 10.1371/journal.pone.0126685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tochio T., Kitaura Y., Nakamura S., Sugawa C., Takahashi M., Endo A., Shimomura Y.2016. An alteration in the cecal microbiota composition by feeding of 1-kestose results in a marked increase in the cecal butyrate content in rats. PLoS One 11: e0166850. doi: 10.1371/journal.pone.0166850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Topping D. L., Clifton P. M.2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81: 1031–1064. doi: 10.1152/physrev.2001.81.3.1031 [DOI] [PubMed] [Google Scholar]
- 61.Valdes A. M., Walter J., Segal E., Spector T. D.2018. Role of the gut microbiota in nutrition and health. BMJ 361: k2179. doi: 10.1136/bmj.k2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xenoulis P. G., Palculict B., Allenspach K., Steiner J. M., Van House A. M., Suchodolski J. S.2008. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 66: 579–589. doi: 10.1111/j.1574-6941.2008.00556.x [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y., Zhou L., Bao Y. L., Wu Y., Yu C. L., Huang Y. X., Sun Y., Zheng L. H., Li Y. X.2010. Butyrate induces cell apoptosis through activation of JNK MAP kinase pathway in human colon cancer RKO cells. Chem. Biol. Interact. 185: 174–181. doi: 10.1016/j.cbi.2010.03.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.