ABSTRACT
Evolution is replete with reuse of genes in different contexts, leading to multifunctional roles of signaling factors during development. Here, we explore osteoclast regulation during skeletal development through analysis of colony-stimulating factor 1 receptor (csf1r) function in the zebrafish. A primary role of Csf1r signaling is to regulate the proliferation, differentiation and function of myelomonocytic cells, including osteoclasts. We demonstrate the retention of two functional paralogues of csf1r in zebrafish. Mutant analysis indicates that the paralogues have shared, non-redundant roles in regulating osteoclast activity during the formation of the adult skeleton. csf1ra, however, has adopted unique roles in pigment cell patterning not seen in the second paralogue. We identify a unique noncoding element within csf1ra of fishes that is sufficient for controlling gene expression in pigment cells during development. As a role for Csf1r signaling in pigmentation is not observed in mammals or birds, it is likely that the overlapping roles of the two paralogues released functional constraints on csf1ra, allowing the signaling capacity of Csf1r to serve a novel function in the evolution of pigment pattern in fishes.
KEY WORDS: Zebrafish, csf1r, Osteoclast, Bone, Pigment, Evolution
Summary: The sub-functionalization of csf1r paralogous genes in teleost fish coincides with the evolution of a novel function in regulating pigmentation.
INTRODUCTION
Paralogous genes, or cases of close gene copies due to duplication, represent a unique case in which to study changes in gene function throughout evolution (Ohno, 1970). Related genes, arisen from a whole-genome duplication, or ohnologues, are often lost due to drift unless these genes evolve new functions (neo-functionalization) or, alternatively, partition previously performed roles (sub-functionalization) (Kassahn et al., 2009). Instances of gene duplication can relax constraint on individual gene function and allow compartmentalization as well as evolution of novel functions. However, overlap in function of retained genes can also lead to a bias in the expression of phenotypic variability due to buffering/redundancy and inter-dependent phenotypes. It is unclear how such dependent mechanisms shape the phenotypic response of organisms and contribute to diversification in development and evolution.
Ray-finned fishes represent the most specious lineage of vertebrates, harboring great diversity in skeletal shape, size and function. Resorption of bone during skeletal development as well as homeostasis is an essential aspect to the formation of shape, structure and function of the skeleton. The form of bones of all vertebrates arises through the interactions between two main cell types: osteoblasts, which lay down new bone, and osteoclasts, which serve to remove bone (Apschner et al., 2011; Weigele and Franz-Odendaal, 2016). The role of osteoclasts in bone remodeling is quite complex, depending on the local cellular environment as well as mechanical forces on the tissue. These interactions are not yet well defined, but it is clear that they are important in the response and adaptability of the skeleton, including the dentition, to functional demands. Osteoclast differentiation and function are dependent on signaling through colony-stimulating factor1 (csf1) (Felix et al., 1990; Hakeda et al., 1998; Mellis et al., 2011) and its receptor, colony-stimulating factor 1 receptor (csf1r), a receptor tyrosine kinase expressed by cells of the monocyte/macrophage lineage and necessary for their differentiation and function (Barreda et al., 2004; Chitu and Stanley, 2017; Dai et al., 2002; Herbomel et al., 2001; Oosterhof et al., 2018; Ryan et al., 2001). In addition, in teleost fishes, csf1ra has been found to be expressed in neural crest progenitors and to act to regulate pigmentation cell differentiation and pattern (Parichy et al., 2000), a trait not related with Csf1 signaling in other vertebrates. It has been argued that selection on pigmentation phenotypes in fishes have contributed to their diversification (Irion et al., 2016; Salis et al., 2019). However, the additional role of csf1r function in regulating skeletal development may constrain the phenotypic spectrum of morphological variation in evolution.
Teleost fishes, which include the zebrafish, share an evolutionary whole-genome duplication event. Whereas previous analyses of csf1r gene evolution in marine fishes have uncovered two csf1r paralogues in many lineages (Braasch et al., 2006), in zebrafish or carp, members of a common teleost lineage Ostariophysi, a functional second paralogue could not be identified. Rather, phylogenetic analysis supported the loss of this paralogue due to drift (Braasch et al., 2006). Recently, a homologue of csf1ra, csf1rb, was identified in zebrafish regulating microglia density and distribution (Oosterhof et al., 2018). This csf1r paralogue showed an additive effect in the regulation of microglial density, suggesting a shared role in microglial regulation (Oosterhof et al., 2018). It remains unclear whether both genes are functionally redundant or whether a sub-functionalization occurred as effects in other tissues were not analyzed.
Here, we describe phylogenetic and functional evidence for retention of a functional csf1rb paralogue in zebrafish that has overlapping and non-overlapping roles with csf1ra in skeletal remodeling. However, csf1ra has adopted a unique role in regulating pigmentation not seen in the retained paralogue. The functional buffering provided by the two paralogues may have provided context for the development of a novel function of csf1ra in development and important in the evolution of diversity.
RESULTS
Recent work established a potential paralogue of csf1r that is maintained in zebrafish (Oosterhof et al., 2018). In order to understand patterns of csf1r gene evolution, we assessed the phylogenetic relationship of the existing, annotated csf1r orthologues in vertebrates (Fig. S1). The ‘b’ orthologues did not strongly differentiate as a single clade (>70 bootstrap probability). However, following Braasch et al. (2006), analysis of synteny among csf1r in the genomes of teleost fishes supports the conclusions that the ‘b’ orthologues share a common genomic structure on the chromosome and are differentiated from ‘a’ orthologues (specifically loss of hmgxb3 and retention of camk2a). We further assessed clustering of the putative csf1rb paralogue with closely related Kit and Pdgf genes from zebrafish. An unrooted maximum likelihood tree shows strong support for the newly identified csf1r paralogue, csf1rb, as being in the csf1r gene family (Fig. S2A).
csf1r paralogues are differentially regulated during early development
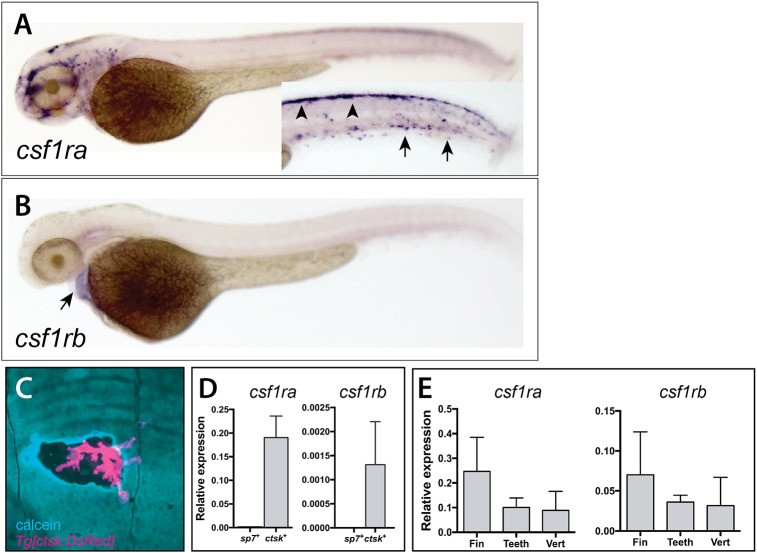
To assess potential different roles of csf1r paralogues in development, we analyzed expression of both csf1ra and csf1rb during early development and in scales of the adult zebrafish. Consistent with what has been previously reported for csf1ra (Herbomel et al., 2001; Parichy et al., 2000; Thisse et al., 2008), we detected expression in migrating neural crest (dorsal) and in early macrophages at 2 dpf (arrowheads and arrows, Fig. 1A). We saw no csf1rb expression in either neural crest or early macrophages. Rather, csf1rb is expressed in the developing heart at 2 dpf (Fig. 1B).
Fig. 1.
Divergent expression of csf1ra and csf1rb during zebrafish early development and in scales of the adult zebrafish. Whole-mount in situ hybridization of csf1ra (A) and csf1rb (B). (A) csf1ra is expressed in cells of the pigment and hematopoietic lineages at 2 dpf; inset shows the trunk in different focal plane with pigment cells precursors (arrowheads) and hematopoietic cells (arrows). (B) In contrast, csf1rb is expressed at only low levels in the heart (arrow) at 2 dpf. (C) Expression of Tg[cstk:DsRed] in osteoclasts localizes at areas of remodeling, as shown by increased calcein staining (blue) at the periphery of the remodeled pit. (D,E) Analysis of expression of csf1r paralogues in isolated cells and from adult skeletal tissues. (D) Expression of csfr1a and csf1rb in isolated adult osteoblasts (sp7+ cells) and osteoclasts (ctsk+ cells) from wild-type zebrafish. (E) Detection of both paralogues in isolated tissues [fin, teeth and vertebra (vert)].
We engineered a transgenic line to label mature osteoclasts that drives the expression of DsRed under the control of the medaka cathepsin K promoter to visualize osteoclast presence and behavior in zebrafish, (Fig. S3A). This promoter was previously defined as being specific for osteoclasts in medaka and zebrafish (Chatani et al., 2011; To et al., 2012). The cathepsin K (ctsk) transgenic line (Tg[ctsk:DsRed]) labeled cells in developing bones in areas where bone remodeling normally occurs (Fig. 1C; Fig. S3B-D). Importantly, DsRed expression colocalized with TRAP staining in scales, consistent with the ctsk transgenic line labeling active mature osteoclasts (Fig. S3D). As detection of csf1r paralogues in many tissues was difficult to achieve using standard in situ methods, we capitalized on the specificity of this line to experimentally isolate osteoclast cell populations using fluorescence-activated cell sorting (FACS) to assess expression of csf1r paralogues in skeletal cells (Fig. S4B). As a comparison, we used a second transgenic line Tg[sp7:EGFP] (DeLaurier et al., 2010) to isolate pre-osteoblasts from comparable tissue sources (Fig. S4A). We confirmed isolation of osteoclasts and osteoblasts through analysis of the expression of known differentiation markers for these cell populations (Fig. 1D; Fig. S4C). Consistent with the known role of Csf1r in osteoclast maturation (e.g. Chitu and Stanley, 2017), we were able to detect expression of both csf1ra and csf1rb in mature osteoclasts isolated from adult zebrafish (Fig. 1D). This expression was specific to osteoclasts, as sp7+ osteoblasts showed no expression of either csf1r paralogue. After this, we isolated RNA from individual skeletal elements (fin, teeth and vertebra) with detectable populations of Tg[ctsk:DsRed]-positive cells and we were able to determine that both csf1r paralogues were expressed in these tissues (Fig. 1E), complementing the detection of osteoclasts associating with the skeletal structures.
Isolation of csf1ra and csf1rb zebrafish mutants
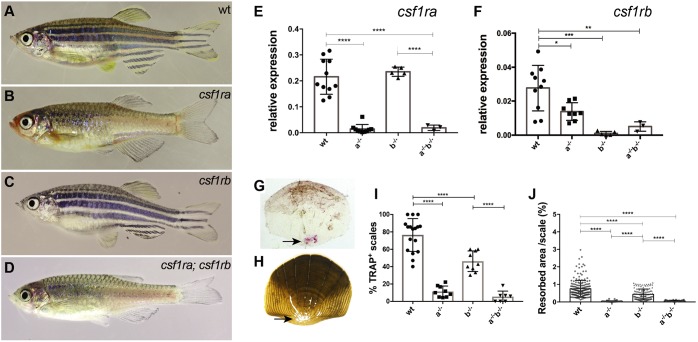
To test function of Csf1r receptors in zebrafish, we isolated mutants in both csf1r paralogues. In a mutagenesis screen, we isolated a mutant allele of csf1ra through expression of its recessive phenotype of altered adult pigmentation (Henke et al., 2017) (Fig. 2B). The loss of xanthophores and reduction of melanophores seen in mh5 mirrors the phenotype previously observed in the panther and salz und pfeffer mutants due to loss of function of csf1ra (Parichy et al., 2000). Sequencing of the coding region of csf1ra in the mh5 mutant revealed a G to T substitution at bp 1466, predicted to cause early truncation of the protein at position 454 (E454X; Fig. S2B). The mutation is tightly linked to the pigmentation phenotype and has not been observed in wild-type zebrafish populations. Given the phenotypic similarity to known alleles of csf1ra, and the severity of the predicted change, the G to T substitution is thought to be the causative mutation underlying the mutant phenotype.
Fig. 2.
Adult phenotypes of csf1ra and csf1rb mutants. (A-D) Representative photomicrographs of adult (A) wild-type zebrafish (wt), (B) homozygous csf1ramh5, (C) csf1rbmh108 and (D) csf1ramh5;csf1rbmh112 mutant zebrafish. (E,F) Quantitative RT-PCR of csf1ra and csf1rb in isolated scales of wild type, or mutants singly homozygous for csf1ra (a−/−) or csf1rb (b−/−), and of double mutants (a−/−b−/−). (G,H) Representative photomicrographs of scales collected from 10 wpf wild-type fish stained using TRAP (G, arrow) and (H) modified Von Kossa staining (arrow). (I,J) Quantitation of TRAP and Von Kossa staining in wild-type and mutant scales. For TRAP activity, each data point represents the percentage of scales stained by TRAP obtained by evaluating 10 scales per fish. For Von Kossa analysis, each data point represents the percentage of resorbed area per individual scale. Data are mean±s.d. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Second, we used CRISPR/Cas9 genome editing to generate loss-of-function alleles of csf1rb. We induced small INDELs at the csf1rb locus by targeting exon 3 of the csf1rb gene with a pool of 5 guide RNAs. We isolated two mutant zebrafish lines, csf1rbmh108 and csf1rbmh112. csf1rbmh108 leads to an induced frame shift in exon 3 (c.479-482delinsTGAATTAT) predicted to cause an early truncation of the protein. The csf1rbmh112 deletion is also predicted to lead to early truncation (c.476-488del; Fig. S2C). In contrast to the csf1ra mutants, which are easily identifiable by their aberrant pigmentation, homozygous csf1rb mutants do not show a detectable morphological phenotype as adults (Fig. 2C). Double csf1ra;csf1rb mutants exhibit a pigmentation phenotype indistinguishable from the csf1ra single mutants (Fig. 2D).
To determine the effect of these mutations on Csf1r activity, the expression of csf1ra (Fig. 2E) and csf1rb (Fig. 2F) was analyzed by qRT-PCR on scales of mutants and wild-type siblings. In each csf1ra or csf1rb single mutant, we were unable to detect significant expression of the affected gene; thus, it is likely that both mutants represent null alleles. It is unlikely that the effect on expression is solely due to reduction in the number of osteoclasts, as both csf1ra and csf1rb are seen expressed in the mutant of the other paralogue. csf1rb expression is significantly reduced in csf1ra mutants, however, suggesting that the loss of csf1ra may limit the population of cells normally expressing csf1rb or, alternatively, that signaling through csf1ra is necessary to maintain csf1rb expression. In csf1ra;csf1rb double mutants, there was no detectable expression of either gene (Fig. 2E,F).
csf1r paralogues and remodeling of the post-embryonic zebrafish skeleton
Maturation and remodeling of the zebrafish skeleton occurs during juvenile development and throughout adulthood. Zebrafish have both mononucleated and multinucleated osteoclasts that participate in normal bone remodeling (Witten et al., 2001; Witten and Huysseune, 2009). Mononucleated osteoclasts are numerous and participate in laminar bone resorption without the presence of lacunar pits, whereas multinucleated osteoclasts are large and fewer in number but have more differentiated resorption pits. We confirmed the broad spectrum of nucleation in zebrafish osteoclasts in adult tissue. Hoechst staining of isolated ctsk+ cells in adult zebrafish skeletal tissue demonstrated the recovery of a wide diversity of single and multinucleated cells (Fig. S4E,F). The presence and activity of both populations of cells in tissues in situ can be detected through analysis of the activity of tartrate-resistant acid phosphatase (TRAP) using histochemical methods. We addressed the role of each csf1r paralogue in regulating the number and function of osteoclasts in late development.
We took advantage of the dynamic homeostasis of scale mineralization in fish to assess osteoclast activity (de Vrieze et al., 2014). The scales that form across the flank of a fish are uniquely suited for analysis of matrix resorption and provide individual test cases for osteoclast function. As an indirect measure of the presence as well as the localization of osteoclasts, we performed an enzymatic assay for TRAP on scales from csf1ra and csf1rb mutants, csf1ra;csf1rb double mutants, and their respective wild-type siblings (Fig. 2G,I). On average, 75% of scales from wild-type fish had TRAP staining, with localized areas of staining flanking the center and in broad rings at the more distal aspect of the scale (Fig. 2G). In contrast, in homozygous csf1ra mutant fish, on average ∼10% of scales have TRAP staining, suggesting that some osteoclasts can still be generated in the absence of csf1ra (Fig. 2I). TRAP staining was not drastically affected in scales from csf1rb mutants (∼50%); however, double mutants show little TRAP activity (<5% Fig. 2I). Scales containing lateral line canals were not selected, as these have been shown to have high levels of reworking and osteoclast numbers (Wada et al., 2014).
As osteoclasts function to resorb mineralized tissue, we quantified matrix resorption in fish scales by staining mineralized tissue with von Kossa staining and measuring the area of unstained material at the focus of the scale (Fig. 2H). Unstained pits or tracks in the scale, which correspond to common sites of TRAP staining in wild-type fish, are assumed to represent sites of osteoclast activity (de Vrieze et al., 2014). Evidence of resorption was detected in 99.7% of wild-type scales [coefficient of variation (CV) 5.5%]. Relative to each scale's overall size, an average of 1.2% of the area was resorbed in wild-type scales (Fig. 2J). Using this method, we detected significantly reduced resorption in csf1ra and csf1ra;csf1rb double mutant scales to less than 0.2%; csf1rb mutants also showed a significant, but more subtle, decrease in reworking (Fig. 2J).
csf1r paralogues differentially affect form of the vertebral skeleton
Previous work on csf1ra-deficient mutant zebrafish noted alterations in the formation of the neural and hemal arches in adult vertebrae (Charles et al., 2017; Chatani et al., 2011). The analysis of osteoclast distribution with our Tg[ctsk:DsRed] transgenic line shows localization of mature, resorbing osteoclasts to the forming vertebral neural and hemal arches (Fig. S5). Isolated vertebrae indicate that both csf1r paralogues are expressed in these structures (Fig. 1D,E). The proportion of DsRed-expressing cells in csf1ra mutant vertebrae was reduced in contrast to the csf1rb mutant vertebrae which still retained wild-type levels of ctsk+ cells. This is easiest seen in juveniles (2 weeks post-fertilization, Fig. S5A-C), however in 4-month-old individuals the number of osteoclasts on the formed centra and arches was extremely variable (Fig. S5D-F). This is consistent with what has been found in Csf1r knock-out mice, where residual macrophage populations were found, indicating that other growth factors can partially compensate for loss of Csf1r (Dai et al., 2002).
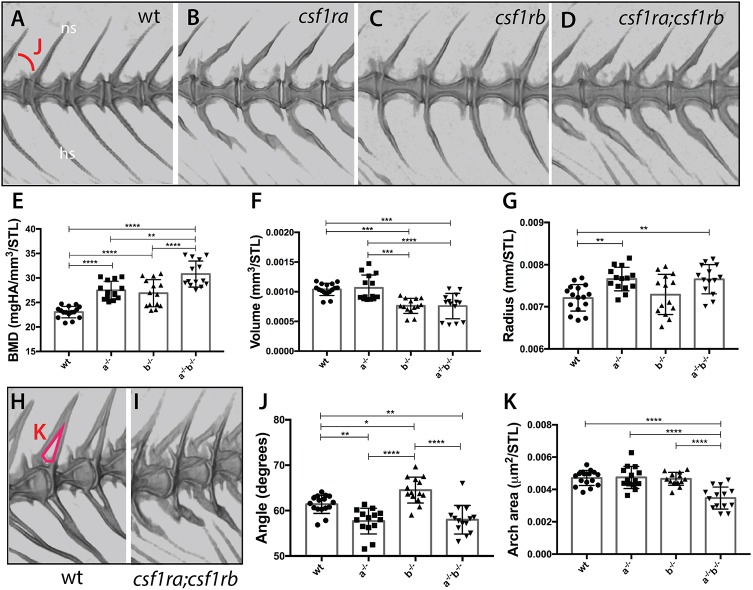
Using microcomputed tomography (µCT), we quantified metrics of vertebral shape and bone density in adult mutant fish (Charles et al., 2017) (Fig. 3). As shown previously (Charles et al., 2017; Chatani et al., 2011), csf1ra mutants show an observable alteration in the shape of the vertebrae and the pitch of the arches (Fig. 3B). In contrast, csf1rb mutants show vertebrae and arches that are morphologically similar to those of wild-type siblings (Fig. 3A,C). Further analysis showed that the angles of the arches in csf1rb mutants were significantly increased (Fig. 3J). In contrast, the arch angles in csf1ra and double mutant fish were decreased. Although arch angle and area of the arches are not completely independent measures, the area of arches remains unaffected in either single mutant (Fig. 3H, red, K). However, the combined action of csf1ra and csf1rb has a significant effect on the arch area, suggesting synergistic effects of deficiencies of csf1r paralogues on this phenotype (Fig. 3I,K). These data suggest that csfr1 paralogues may have nuanced effects in regulating vertebral morphology.
Fig. 3.
Loss of csf1r paralogue function leads to unique and shared phenotypes in the skeleton. MicroCT imaging and quantification of skeletal metrics of the spine of age-matched csf1r mutants and wild-type adult siblings. (A-D) MicroCT renderings of the lateral aspect of the spine showing overall morphology and integration of serial vertebrae. The neural spines (ns) and hemal spines (hs) are indicated. The measure used for the quantification of angle of the arch (J) is noted in A. (E-G,J,K) Quantification of vertebral shape and density in csf1r single and double mutants: (E) bone mineral density (BMD), (F) bone volume, (G) vertebral radius, (J) angle and (K) area of the neural arch. All measurements, with the exception of angle, were standardized to standard length (STL). (H,I) visualization of neural arch area in wild-type (H) and csf1ra; csf1rb mutant (I) zebrafish; measured area used for quantification of the arch area (K) is indicated in red in H. Data are mean±s.d. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Increased bone density can occur in the context of decreased osteoclastic activity. In this regard, we found that csf1ra and csf1rb deficiency leads to an increase in bone density of the vertebral centra in an apparent additive manner (Fig. 3E). In parallel, csf1rb deficiency results in a reduction in bone volume, not seen in csf1ra mutants (Fig. 3F). Furthermore, loss of csf1ra leads to an increase in the radius of the vertebral body, while csf1rb mutants are unaffected (Fig. 3G). Thus, both paralogues are required for the regulation of shape and structure of the skeleton, with shared and unique structural components that are sensitive to alterations in their function.
Synergistic effects of csf1r paralogues on zebrafish dentition
Teeth are a component of the vertebrate dermal skeleton. Zebrafish develop teeth in a predictable pattern along the ventral/peripheral aspect of the last ceratobranchial arch (Fig. S6A) (Huysseune and Witten, 2006). With growth, the arch increases in size and teeth are replaced as new teeth are formed in succession (Fig. S6B). In zebrafish, resorption by osteoclasts at the tooth base is necessary for tooth loss (Huysseune et al., 1998; Witten et al., 2001; Witten and Huysseune, 2009). We reasoned that the dentition might be a sensitive skeletal structure with which to assess the role of csf1r in osteoclastic bone remodeling.
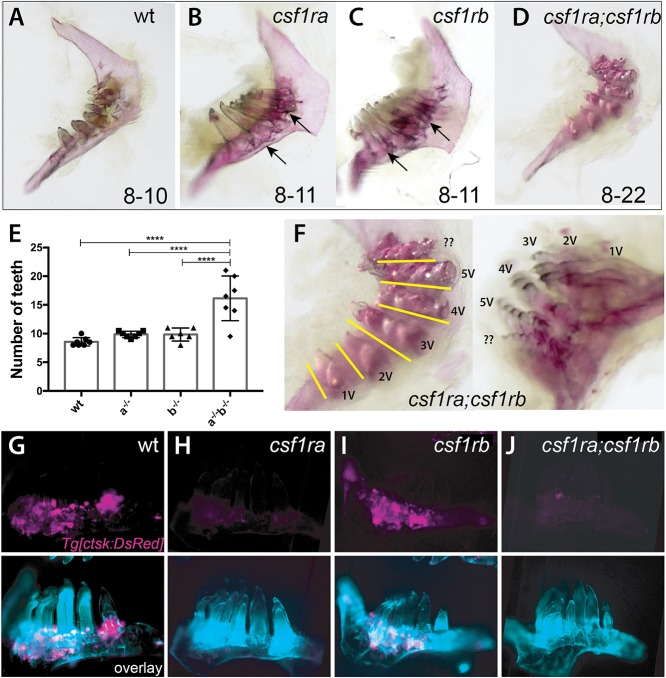
In staining for osteoclasts present in the mature dentition of adult fish, we observed intense TRAP staining in the arch up to the base of individual teeth (Fig. S6C). The presence of osteoclasts in this region was confirmed using the Tg[ctsk:DsRed] transgenic line, where osteoclasts were localized to areas of high bone deposition, as shown by calcein staining (Fig. S6D-F). Despite the abundance of osteoclasts in this region and variation in numbers in single mutants (Fig. 4G-J), whole-mount skeletal staining showed little change in the number or shape of teeth in csf1ra or csf1rb single mutants (Fig. 4B,C,E). However, changes in the retention of attachment bone can be seen at the base of the forming teeth (Fig. 4B,C, arrows). Quite strikingly, csf1ra;csf1rb double mutants show extreme polyodontia, with up to 20 teeth on a single arch (Fig. 4D-F). The teeth within these extended dentitions remain normal in shape and pattern, forming bouquets of teeth aligned in ventral-dorsal tracts (Fig. 4F, line). Thus, these findings suggest that the normal homeostasis of the zebrafish dentition is quite sensitive to loss of csf1r receptor activity, showing shared function of both receptors in remodeling of bone surrounding and ankylosing to the formed teeth, having synergistic functions in regulating tooth number in mature fish.
Fig. 4.
csf1r genes show combinatorial effects in regulating patterning of the dentition and tooth number. (A-D,F) Alizarin Red stained pharyngeal arches from 4 month post-fertilization (mpf) csf1r mutants and wild-type siblings. Numbers on the bottom represent the range of tooth numbers on each arch (n=7). Arrows indicate buildup of bone surrounding base of teeth in csf1ra and csf1rb mutants (B,C). (E) Quantification of the number of teeth represented in csf1r mutants, double mutants and wild-type siblings (****P<0.0001). (F) Higher magnification images of dentitions from csf1ra;csf1rb double mutants showing organization of supernumerary teeth into distinct tracts (yellow line) associated with a row of normal tooth germ patterning in wild-type dentitions (Fig. S6). (G-J) Localization of ctsk-positive cells (magenta) on the dentary of zebrafish csf1r mutants. Bottom, overlay with calcein stain (cyan).
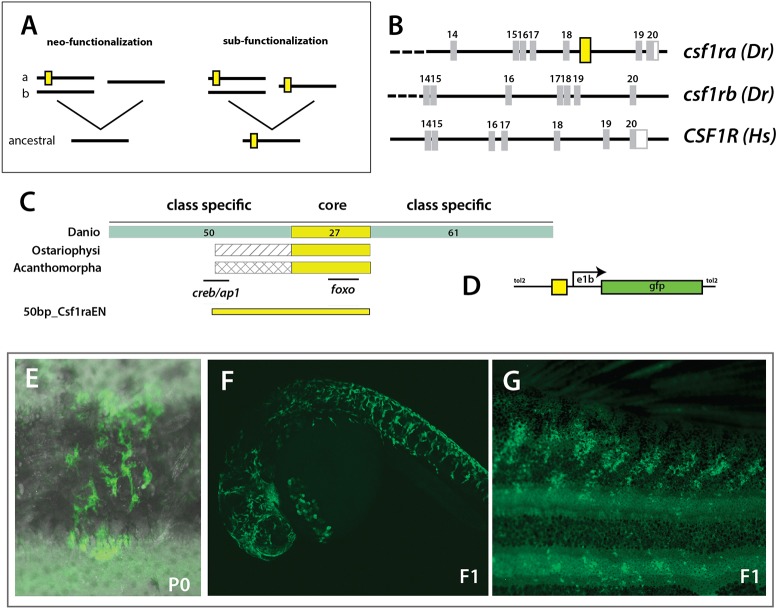
Identification of a unique regulatory enhancer of csf1ra driving novel gene function
The shared activity observed in regulation of the skeleton is in contrast to the early differences in expression observed, as well as to the overall pigmentation phenotype between the mutants analyzed (Fig. 2B,C). Given the required function for csf1ra in osteoclast formation as well as microglia distribution (Oosterhof et al., 2018), we hypothesized that there would be a specific and evolutionary unique gain of cis-regulatory control in csf1ra, shared among all teleost fishes, but unique from other vertebrates (Fig. 5A). Making use of the 11-way genomic evolutionary rate profiling (GERP) track in Ensembl, we identified a 51 bp conserved non-coding element (CNE) within csf1ra that is not seen in csf1rb (Fig. 5B; Fig. S7). We did not find evidence of this sequence in human or other amniote genomes, suggesting it is unique to teleosts (Fig. 5B); this is consistent with a gain of regulation in csf1ra (neo-functionalization, Fig. 5A). The identified CNE is conserved throughout evolution of teleost fishes with a broad order-specific region in Danio species flanking a highly conserved core region shared by all teleosts (Fig. 5C, Fig. S7). When the 50 bp CNE is placed upstream of an E1b minimal promoter (Fig. 5D), we find that it is sufficient to drive reporter expression within stellate pigment cells in clones of injected wild-type fish (P0; Fig. 5E) as well as in identified transgenic lines (F1, Fig. 5F,G).
Fig. 5.
Evolution of a unique enhancer controlling csf1ra expression in pigmentation and its variation. (A) Schematic of different mechanistic hypotheses of evolution of csf1r regulation. Duplication of an ancestral csf1r gene has led to two lineages in vertebrates within teleost fishes having two copies, ‘a’ and ‘b’, and tetrapods having one copy. The functional difference between ‘a’ and ‘b’ could be a consequence of neo-functionalization, where ‘a’ gained functionality in pigment cells; or sub-functionalization, where ‘a’ and ‘b’ retained partial functionality of the ancestral gene; yellow box represents a hypothetical enhancer regulating expression in pigment precursors. (B) Identification of a conserved non-coding element (CNE) among fishes within csf1ra, not found in csf1rb or tetrapod csf1r sequences; Dr, Danio rerio; Hs, Homo sapiens; yellow box represents the position of the CNE. (C) Schematic and alignment of putative enhancer. Within Danios a broad 150 bp sequence is conserved with a ∼27 bp core sequence showing a 100% identity with all other teleost fishes (ostariophysans and acanthomorphs). The numbers of bp in each region are indicated. (D) Schematic of expression construct containing a 50 bp conserved region (50bp_Csf1raEN). (E-G) Representative clone of pigment cells observed in injected adult animals and in 1 dpf and adult individuals in the F1 generation; overlay of brightfield and fluorescence (E).
DISCUSSION
Diversity in skeletal shape among vertebrates provides an opportunity to define shared and dissimilar mechanisms through which skeletal remodeling occurs. Previous genetic analyses in the zebrafish and medaka have shown an essential role for csf1ra in regulating osteoclast development and activity (Chatani et al., 2011; Mantoku et al., 2016). In zebrafish, however, the effect on skeletal development and remodeling was slight (Chatani et al., 2011). Such a limited skeletal phenotype raises questions about the importance of Csfr1 signaling in regulating skeletal form of the zebrafish, as well as the applicability of teleost fish as a model for studying osteoclast-mediated skeletal remodeling in mammals. Here, we identify broad retention of a second csf1r gene within teleost lineages. We identify mutants in both csf1r paralogues in the zebrafish, and define specific and shared functions of these genes in regulating post-embryonic development.
Functional analysis of a csf1rb paralogue and the broader role of csf1r function in skeletal remodeling
We confirmed the known phenotype of csf1ra-deficient zebrafish: having reduced numbers of osteoclasts resulting in altered vertebral arch shape and increased bone mass (Charles et al., 2017; Chatani et al., 2011). However, for the most part, the skeletal shape does not seem overly affected in this mutant in comparison with the osteopetrotic phenotypes observed in rodent models of csf1 or csf1r deficiencies (Cotton and Gaines, 1974; Dobbins et al., 2002; Van Wesenbeeck et al., 2002). On finding a potential csf1rb paralogue in zebrafish, we asked whether loss of both copies of csf1r could better reflect shared roles in osteoclast function in vertebrate post-embryonic development. Each paralogue has quite distinct expression patterns early in development, suggesting diversification of regulation (Fig. 1). Although we do not find evidence for csf1rb being expressed during early development of hematopoietic cells, functionally both paralogues are essential for osteoclast function and expression is seen in mature osteoclasts, suggesting csf1rb is active in these lineages at later time points in juvenile development.
Overall, csf1rb shares functions with csf1ra in remodeling of the skeleton. Often, their activities are combinatorial with additive and synergistic effects. Patterning of the spine is particularly interesting as the effects of each paralogue can be detailed by variation in vertebrae characteristics. These phenotypes are consistent with the localization of osteoclasts on the arches during development (Fig. S5). We notice a difference in effect of csf1ra in maintaining resting numbers of osteoclasts compared with csf1rb. However, we find that the number of osteoclasts is highly variable in wild-type adults; thus, it is difficult to assign specific roles to each paralogue in terms of effect on number or localization of osteoclast populations on bone (Fig. S5). An analysis of the differential role of csf1r paralogues on microglia during development suggest regulation of cell migration may play a large role in the observed phenotype (Herbomel et al., 2001; Oosterhof et al., 2018); however, this is not specifically tested here. The genetic dissociation between neural arch and vertebral phenotypes has also been observed in a medaka osteopetrosis model (To et al., 2015). The modular effect of csf1r genes is intriguing as it would allow different regulation of these areas during development through functional variation of specific Csf1r receptors.
The essential role of osteoclasts in shaping the dentition and tooth replacement
Of the skeletal defects observed in csf1ra;csf1rb deficient zebrafish, the effects on tooth number are quite striking. Either csf1ra or csf1rb function is sufficient to complement the loss of the other paralogue to permit adequate removal of older teeth as new teeth are formed. The bouquet formation of the dentition in the double knockout is suggestive that osteoclasts are essential for clearance of successive teeth in a series. Similarly in medaka, a broad inhibition of osteoclast function leads to the formation of supernumerary teeth (Mantoku et al., 2016; To et al., 2015). In teleost fishes, this manifestation appears to be a failure of tooth shedding. Mice do not show tooth replacement, therefore comparable effects on dental patterning cannot be addressed. However, an important manifestation of osteoclast deficiency in mice is the failure of tooth eruption (Cotton and Gaines, 1974; Dobbins et al., 2002; Tiffee et al., 1999; Van Wesenbeeck et al., 2002). Our data detail the essential role of osteoclast function for normal tooth replacement mechanisms in the zebrafish. The synergistic effects of csf1r paralogue functions on the retention of teeth reveal an intricate skeletal patterning process yet to be uncovered in analyses of natural variants or in previously identified experimental mutants.
The evolution of a key role for csf1ra in pigment pattern diversification in teleost fishes
Teleost fishes express a broad array of pigmented varieties and pigmentation patterns are often under intense selection and are key components of fish speciation (Irion et al., 2016; Salis et al., 2019). Csf1ra function in the zebrafish was first identified through its action in regulating pigment pattern of the adult zebrafish (Haffter et al., 1996; Odenthal et al., 1996; Parichy et al., 2000). However, alteration of Csf1r activity in mammals does not lead to pigmentation defects. Whereas we find both csf1r paralogues are necessary for skeletal remodeling with a combined phenotype comparable with that seen in mammals, csf1ra has a unique role in xanthophore differentiation and stripe formation (Parichy et al., 2000). We find that csf1rb mutants do not show comparable pigmentation phenotypes to csf1ra mutants and do not show any increased effect in combination with csf1ra deficiency. In support of a separate role for csf1ra in regulating pigmentation, we find that csf1ra is uniquely expressed in early neural crest lineages, whereas csf1rb is not detected in these cells (Fig. 1). Thus, the function of csf1ra in neural crest differentiation and pigment pattern formation in fishes is a derived function for this gene. Concordantly, we identify a CNE within zebrafish csf1ra that is unique to that paralogue and highly conserved among teleosts. The CNE is not found among non-teleost vertebrates, suggesting it arose after the whole-genome duplication. The conserved CNE is sufficient to regulate expression in pigment cell during zebrafish development, suggesting this is a component of the specialization of csf1ra.
In diverse teleost fishes, csf1ra has been implicated in functional variation in pigmentation (Parichy and Johnson, 2001; Quigley et al., 2005). The pivotal role during pigment pattern formation is thought to drive diversification of the csf1ra gene in these groups (Salzburger et al., 2007). However, the essential role of csf1ra in regulating skeletogenesis would be expected to constrain the capacity of this gene in driving pigment diversity in evolution. Our finding of an essential shared role for csf1rb in skeletal remodeling in zebrafish suggests that sub-functionalization of csf1r paralogues provides buffering of function and thus the means for csf1ra to evolve new roles in neural crest and pigment pattern differentiation (Fig. 6), consistent with the broad retention of pigment-related gene paralogues in evolution of teleosts (Braasch et al., 2009; Lorin et al., 2018). We provide here the first molecular and functional evidence of this neo-functionalization.
Fig. 6.
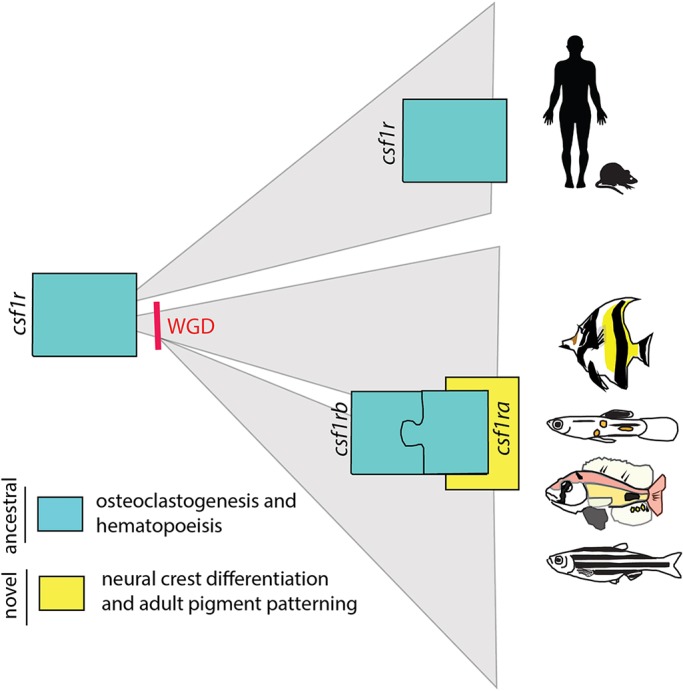
Model of the evolutionary diversification of Csf1r function. Csf1r signaling is conserved in all vertebrates. CSF1R plays an important and shared role in hematopoiesis, including osteoclast differentiation. In teleost fishes, the csf1r locus was duplicated after the teleost-specific whole-genome duplication (WGD) event and existing paralogues are retained in most lineages. From mutational analysis, it is clear that, in teleost fish, csf1r genes combine to regulate osteoclast activity similar to mammals (ancestral function, blue). However, each csf1r paralogue has combined and singular roles in regulating remodeling. We proposed a model in which, in teleosts, csf1ra acquired a novel function in regulating pigment pattern specification (new function, yellow). This role would be a derived function for this gene and has led to differential selection of csf1ra in regulating the pigment pattern of the adult fish.
MATERIALS AND METHODS
Phylogenetic analysis
The phylogenetic relationship between zebrafish csf1ra, csf1rb, pdgfra, pdgfrb, kita and kitb was established by retrieving the amino acid sequences from NCBI (O'Leary et al., 2016) and ENSEMBL (Yates et al., 2016) databases. Protein sequences were aligned using MUSCLE (Edgar, 2004) multiple sequence alignment tool. A phylogenetic tree was constructed based on amino acid difference with the maximum likelihood algorithm in MEGA version 7.0.18 (Kumar et al., 2016). The reliability of the tree was assessed by bootstrapping, using 500 replicates, and bootstrap support ≥70 was considered significant. The resulting tree was imported to iTOL version 3.2.4 (Letunic and Bork, 2016) for final editing.
To examine the phylogenetic relationship between zebrafish csf1ra and csf1rb paralogues and their corresponding vertebrate orthologues, the amino acid sequences of Astyanax mexicanus, Clupea harengus, Ctenopharyngodon idellus, Cyprinodon variegatus, Danio rerio, Esox lucius, Homo sapiens, Latimeria chalumnae, Mus musculus, Oreochromis niloticus, Oryzias latipes, Poecilia reticulata, Pygocentrus nattereri, Salmo salar, Sinocyclocheilus anshuiensis, Sinocyclocheilus grahami, Sinocyclocheilus rhinocerous, Takifugu rubripes and Xenopus tropicalis were retrieved from NCBI and ENSEMBL databases. The same strategy described before was used to construct a phylogenetic tree. Accession numbers of protein sequences used in the analysis are listed in Table S1.
Zebrafish husbandry and identification of mutant lines
Zebrafish strains were maintained under standard conditions (Westerfield, 1993) in compliance with internal regulatory review at Boston Children's Hospital. At the indicated age, fish were euthanized with 20% MS-222 and scales were collected from the anterior area of either side of the trunk.
The csf1ramh5 mutant line was isolated in the Harris lab in an ENU mutagenesis screen. The mutation in csf1ra was identified by a candidate gene approach due to similarity to panther and salz und pfeffer mutants. A missense mutation in csf1ra was detected in cDNA from homozygous 4 dpf csf1ramh5 mutant larvae amplified using two primer combinations [csf1ra-f1 (ACGGTCCAGTGACGTTTCTT) with csf1ra-r2 (CTCTTTGCCCAGACCGTAAG); and csf1ra-f2 (GGCAGTGACACCTTCTCCAT) with csf1ra-r1 (GTCTGCAGCTGTTGTGCTGT)] to cover a large component of the csf1ra-coding region (NM_131672.1). For genotyping of individuals from genomic DNA, we used primers csf1ra_g_f1 (CGGCCTTCTGTGAATTTTGT) and csf1ra_g_r1 (TGCCTAATTACCCAAACTTGC). The csf1ra mutation is consistently linked to the pigment phenotype.
CRISPR-Cas9 technology and specific guide RNAs (gRNA) designed against csf1rb were used for the generation of mutants. Specifically, five gRNAs were designed using CHOPCHOP (Montague et al., 2014) (chopchop.cbu.uib.no) targeting csf1rb exon 3 (gRNA1-GGTCATCCCAGGAGGAACAG, gRNA2-GCAGTATAATTCATCCCAGG, gRNA3-GCCTGGGATGAATTATACTG, gRNA4-GATACTGCGGATCCCAGACG and gRNA5-GTCTGTATGGTGAACATGAG). Guide RNAs were synthesized as described by Gagnon et al. (2014). Briefly, templates for gRNA transcription were generated by annealing gene-specific oligonucleotides containing the T7 promoter sequence (5′-TAATACGACTCACTATA-3′), the target site without the protospacer adjacent motif (PAM) sequence and a complementary region to a constant oligonucleotide encoding the reverse complement of the tracrRNA tail. Guide RNAs were transcribed using the MEGAshortscript T7 kit (Ambion). Cas9 mRNA was synthesized from a linearized pCS2-nCas9n plasmid (Jao et al., 2013) using the mMESSAGE mMACHINE SP6 kit (Ambion). To generate mutant fish, a pool of the five gRNAs (100 ng/μl) and Cas9 mRNA (500 ng/μl) were injected into one-cell stage zebrafish AB wild-type embryos.
CRISPR-induced mutations were identified though amplification of a 743 bp genomic region containing csf1rb exon 3 using primers 5′-AAACAGATCCCGACAAAGCC-3′ and 5′-CGTCCGATTTTGACCGCG-3′ followed by T7 endonuclease digestion. To establish mutant lines, injected fish were grown to adulthood and outcrossed to AB wild-type fish. The nature of induced INDELs created by CRISPR/Cas9 was identified in the F1 progeny by sequencing of subcloned PCR products.
Generation of cathepsin K osteoclast reporter line
We used a 3 kb upstream region of the medaka (Oryzias latipes) cathepsin K gene, previously shown to drive expression in osteoclasts in zebrafish (Chatani et al., 2011), and cloned it into the p5E-MCS plasmid (228, Tol2kit; Kwan et al., 2007) in front of the DsRed sequence flanked by FRT sites. To enable simultaneous generation of an osteoclast reporter and an osteoclast-specific line driving Cre expression, sequence encoding Cre (from the plasmid pOG231; O'Gorman et al., 1997) was cloned in a middle entry gateway plasmid (pENTR/D-TOPO, Life Technologies). For generation of an expression reporter construct, we used the multisite Gateway system in which the 3′ entry plasmid (302, p3E-polyA) and the destination vector (395, pDestTol2CG2) were obtained from the Tol2kit (Kwan et al., 2007). As a marker for transgenesis, the pDestTol2CG2 destination vector contains a cmlc2 promoter driving GFP expression in the heart. The construct was made via recombination from gateway-based clones using LR clonase II (Life Technologies).
To generate transgenic fish, plasmid DNA (10 ng/μl) and transposase RNA (10 ng/μl) were injected into one-cell stage zebrafish embryos. These animals were raised to adulthood and outcrossed to Tuebingen (Tue) wild-type fish. Progeny were screened for cardiac GFP expression at 48 hpf and raised to establish the Tg[Ola.ctsk:FRT-DsRed-STOP-FRT,Cre, cmlc2:GFP] transgenic line (referred to here as Tg[ctsk:DsRed]). Tg[ctsk:DsRed] transgenic fish were crossed with csf1ramh5 and csf1rbmh108 to visualize the cathepsin K-expressing cells in mutant lines. Progeny were stained overnight with 100 μg/l calcein (Sigma) solution in water as counterstain for mineralized matrix.
In situ hybridization
csf1ra and csf1rb in situ hybridization was performed in scales using a modified version of the Thisse et al. protocol (Thisse and Thisse, 2008). At 24 hpf, 30 mg/l of N-Phenylthiourea (PTU, Sigma) was added to the developing embryos to prevent pigment development. For in situ hybridization, 700-800 bp of unique sequence for each gene were amplified from cDNA [csf1ra, (f) ACTGGACTTCCTCGCTGCTA, (r) GTCTGCAGCTGTTGTGCTGT; csf1rb, (f) CGACCTCCTCAACTTCTTGC, (r) TCAGGTGCCTTCGAAGTTCT] and used to transcribe sense and antisense digoxigenin (DIG)-labeled riboprobes with DIG RNA labeling kit SP6/T7 (Roche). Probes were hybridized at 60°C against 24, 48 and 96 hpf wild-type larva and in adult scales, and the hybridization was visualized using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP) staining.
Cell isolation and sorting
The transgenic lines Tg[ctsk:DsRed] and Tg[sp7:EGFP] (DeLaurier et al., 2010) were used to isolate osteoclasts and osteoblasts, respectively. Protocol was based on methodologies outlined by Madel et al. (2018) and Buettner et al. (2015). Fish were euthanized and tissue samples harvested for isolation of cells. Tissues were digested by vigorous shaking at 30°C in dissociation solution [0.21% trypsin-EDTA (Invitrogen), 16 mg/ml collagenase from Clostridium histolyticum, type IA (Sigma)] for 15 min. The cell suspension was washed with Dulbecco's Modified Eagle Media (DMEM) supplemented with 10% of fetal bovine serum (FBS, Invitrogen) and filtered on a 40 µm nylon mesh. The filtered cell suspension was layered on Histopaque-1077 (Sigma) at a 2:1 ratio and centrifuged at 400 g, without acceleration or break, for 30 min in a swinging bucket rotor. Both the ring and the pellet were collected in PBS+2%FBS. Immediately prior to sorting, cells were stained with 1X SYTOX Blue (Invitrogen). After doublet exclusion, live cells were identified by the absence of SYTOX Blue staining. EGFP or DsRed-positive cells were separated on an FACSAria II cell sorter (BD Biosciences) using an 85 µm nozzle at a flow rate of ∼2000 events/s. For RNA studies, sorted cells were collected directly in RNA lysis buffer for subsequent RNA extraction.
For analysis of DNA content, after isolation cells were counted and stained using Hoechst 33342 (Sigma) 10 µg/ml per 106 cells per 500 µl at 37°C for 30 min (method adapted from Madel et al., 2018; Schmid and Sakamoto, 2001). Cells were washed in PBS+2% FBS prior to FACS analysis. After removal of doublets, DsRed+ or EGFP+ cells we identified and analyzed according to Hoechst 33342 expression. A Hoechst 33342 histogram was plotted and sp7:EGFP+ cells were used to determine gating strategy for mononuclear cells.
RNA isolation and quantitative RT-PCR
Scales from adult fish were collected in Trizol reagent (Life Technology), the tissue was mechanically homogenized and RNA was extracted using the Direct-zol RNA kit (Zymo Research). Sorted cells were collected directly in RNeasy plus micro kit (Qiagen) lysis buffer and RNA was extracted according to the manufacturer's recommendation.
Reverse transcription was performed with the Protoscript first strand cDNA synthesis kit (New England Biolabs). Quantitative real-time PCR (qRT-PCR) was performed using Sybr green reagent (Life Technologies) in duplicate. Expression levels were normalized to tubulin. RT-PCR primers used in the analysis are: csf1ra (f) AATCAGAACCACCTGTCCCG; csf1ra (r) CGACCAGGTTAAAGGCCACA; csf1rb (f) TAAAAGGCAATGCACGGCTG, csf1rb (r) CGCAAAATCTGGTTGTGGCA; ctsk (f) CGCCTTCAGATACGTCAGCA, ctsk (r) GGTCAGTGCCCTCTCGTTAC; sp7 (f) TCGATTCTGGAGGAGGAAACAC, sp7 (r) CGTCCGTTTTTAGCGCTCTG; tubulin (f) GTACGTGGGTGAGGGTATGG, tubulin (r) ACACAGCAGGCAGCATTTCTA.
Tartrate resistant acid phosphatase staining
Tartrate resistant acid phosphatase (TRAP) staining solution was made from equal volumes of solution A and solution B. Solution A contained naphtol AS-MX phosphate (100 μg/ml), N,N-dimethylformamide (1% v/v) in acetate buffer (0.1 M); solution B contained Fast Red LB Violet Salt (600 μg/ml) in acetate buffer (0.1 M). After mixing solutions A and B, sodium tartrate (50 mM) was added and the pH adjusted to 5.0. The solution was then filtered to remove large particles.
Freshly collected scales from 10 weeks post-fertilization (wpf) fish were washed in PBS and fixed in 4% paraformaldehyde at room temperature for 1 min. Samples were washed twice with distilled H2O and stained with pre-warmed TRAP staining solution at 37°C for 15 min. The number of scales with TRAP staining was counted and calculated as percentage of total scales (minimum of 10 scales per fish, 10 fish per group was used).
Staining of bone mineralized matrix
Whole-mount silver nitrate staining was defined after a modified von Kossa staining method (de Vrieze et al., 2014). Scales from 10 wpf fish were stained immediately after collection with freshly prepared silver nitrate (2.5% AgNO3 in distilled H2O, Sigma) for 30 min at room temperature. Scales were washed in distilled H2O before being observed under a microscope. Analysis of the resorbed area was performed by applying a threshold to the image in Image J (Schneider et al., 2012). At least ten fish per group, with ten scales per fish were used for each experiment.
Teeth from adult (4mpf) csf1ramh5, csf1rbmh108, csf1ramh5;csf1rbmh112 and wild-type siblings were stained overnight in an 100 μg/ml Alizarin Red solution (Sigma) and destained in 0.5% KOH solution.
Micro-computed tomography
Adult (4 mpf) csf1ramh5, csf1rbmh108, csf1ramh5;csf1rbmh112 and wild-type siblings were euthanized and fixed in 10% formalin for 24 h and kept in 70% ethanol. Caudal vertebrae and teeth were imaged using micro-computed tomography (μCT) with a voxel size of 6 μm, using an X-ray tube potential of 55 kVp, an X-ray intensity of 0.145 mA and an integration time of 600 ms (Scanco µCT35). Vertebral analysis was performed using custom software to determine neural arch area and angle, vertebral volume, radius, and bone mineral density, as performed previously (Charles et al., 2017). Two sets of three seed points were placed manually on the anterior and posterior faces of the vertebral body. The radius was determined from fitting each set to a circle and calculating the average. The length was defined as the distance between the centers of the fitted circles. The volume was total number of voxels defined as bone by the thresholding algorithm and the angle was measured between the ray connecting the averaged circumference and a second line defined by seed points placed on the arch. With the exception of the angle, all parameters were normalized with the standard length (STL) of each fish. Thresholding was performed equally through all samples. Analysis of variant measures at different thresholds showed comparable data between wild-type and mutant fish. 3D reconstruction was made in Amira 6.0 (FEI) software.
Generation of csf1ra-CNE overexpression reporter lines
Using the 11-way genomic evolutionary rate profiling (GERP) track in ENSEMBL, we identified a conserved non-coding element (CNE) within csf1ra. To examine sequence similarities between Danios, Gadus morhua, Takifugu rubripes, Tetraodon nigroviridis, Oreochromis niloticus, Gasterosteus aculeatus, Oryzias latipes, Poecilia formosa, Xiphophorus maculatus and Astyanax mexicanus, genomic csf1ra sequences were retrieved from NCBI (O'Leary et al., 2016) and ENSEMBL (Yates et al., 2016) databases. CNE sequences were aligned using Clustal Omega multiple sequence alignment tool (Madeira et al., 2019). TFbind was used for transcription factors binding site prediction (Tsunoda and Takagi, 1999). Accession numbers of genomic DNA sequences used in the analysis are listed in Table S2.
Enhancer sequences (50 bp) from D. rerio were cloned into the E1b-GFP-Tol2 plasmid upstream of the E1b minimal promoter (Birnbaum et al., 2012). To generate transgenic fish, plasmid DNA (10 ng/μl) and transposase RNA (10 ng/μl) were injected into one-cell stage Tue wild-type zebrafish embryos. These animals were raised to adulthood and outcrossed to Tue wild-type fish. Progeny were screened for GFP expression at 24 hpf and raised to establish the transgenic line.
Statistical analysis
Statistical analysis was performed using SPSS Statistics 23.0 (IBM). Continuous variables were expressed as mean±s.d. Groups were compared using analysis of variance (ANOVA) test followed by Bonferroni's multiple comparison test. Statistical significance was set at P<0.05.
Supplementary Material
Acknowledgements
The authors thank Mrs. Althea James for assistance in zebrafish husbandry, the Flow Cytometry Research Facility at Boston Children's Hospital, and David Parichy in sharing unpublished whole genome data from select Danio species.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.C.-L., K.H., K.U., J.F.C., M.P.H.; Methodology: J.C.-L., K.H., J.D., J.F.C., M.P.H.; Validation: J.C.-L., K.H., J.F.C., M.P.H.; Formal analysis: J.C.-L., K.H., K.U., J.D., J.F.C., M.P.H.; Investigation: J.C.-L., K.H., K.U., J.D., J.F.C., M.P.H.; Resources: M.L.W., M.P.H.; Data curation: J.C.-L., K.H., J.D., J.F.C., M.P.H.; Writing - original draft: M.P.H.; Writing - review & editing: J.C.-L., K.H., K.U., J.F.C., M.L.W., M.P.H.; Visualization: J.C.-L., K.H., J.D., J.F.C., M.P.H.; Supervision: J.F.C., M.L.W., M.P.H.; Project administration: J.F.C., M.L.W., M.P.H.; Funding acquisition: J.F.C., M.L.W., M.P.H.
Funding
This work was partially supported by the National Institutes of Health (U01 DE024434 to M.P.H., K08 AR062590 to J.F.C. and GM122471 to D.P.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.181834.supplemental
References
- Apschner A., Schulte-Merker S. and Witten P. E. (2011). Not all bones are created equal - using zebrafish and other teleost species in osteogenesis research. Methods Cell Biol. 105, 239-255. 10.1016/B978-0-12-381320-6.00010-2 [DOI] [PubMed] [Google Scholar]
- Barreda D. R., Hanington P. C. and Belosevic M. (2004). Regulation of myeloid development and function by colony stimulating factors. Dev. Comp. Immunol. 28, 509-554. 10.1016/j.dci.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Birnbaum R. Y., Clowney E. J., Agamy O., Kim M. J., Zhao J., Yamanaka T., Pappalardo Z., Clarke S. L., Wenger A. M., Nguyen L. et al. (2012). Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 22, 1059-1068. 10.1101/gr.133546.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I., Salzburger W. and Meyer A. (2006). Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol. Biol. Evol. 23, 1192-1202. 10.1093/molbev/msk003 [DOI] [PubMed] [Google Scholar]
- Braasch I., Brunet F., Volff J.-N. and Schartl M. (2009). Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol. Evol. 1, 479-493. 10.1093/gbe/evp050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner A., Sundaram S., Vyas H., Yu T., Mathavan S. and Winkler C. (2015). Fluorescence-activated cell sorting (FACS) of osteoblasts and osteoclasts for RNA sequencing in a medaka, Oryzias latipes (Temming & Schlegel, 1846), osteoporosis model. J. Appl. Ichthyol. 34, 481-488. 10.1111/jai.13660 [DOI] [Google Scholar]
- Charles J. F., Sury M., Tsang K., Urso K., Henke K., Huang Y., Russell R., Duryea J. and Harris M. P. (2017). Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis. Bone 101, 162-171. 10.1016/j.bone.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatani M., Takano Y. and Kudo A. (2011). Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish. Dev. Biol. 360, 96-109. 10.1016/j.ydbio.2011.09.013 [DOI] [PubMed] [Google Scholar]
- Chitu V. and Stanley E. R. (2017). Regulation of embryonic and postnatal development by the CSF-1 receptor. Curr. Top. Dev. Biol. 123, 229-275. 10.1016/bs.ctdb.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton W. R. and Gaines J. F. (1974). Unerupted dentition secondary to congenital osteopetrosis in the Osborne-Mendel rat. Proc. Soc. Exp. Biol. Med. 146, 554-561. 10.3181/00379727-146-38146 [DOI] [PubMed] [Google Scholar]
- Dai X.-M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., Sylvestre V. and Stanley E. R. (2002). Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111-120. 10.1182/blood.V99.1.111 [DOI] [PubMed] [Google Scholar]
- de Vrieze E., Moren M., Metz J. R., Flik G. and Lie K. K. (2014). Arachidonic acid enhances turnover of the dermal skeleton: studies on zebrafish scales. PLoS ONE 9, e89347 10.1371/journal.pone.0089347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaurier A., Eames B. F., Blanco-Sánchez B., Peng G., He X., Swartz M. E., Ullmann B., Westerfield M. and Kimmel C. B. (2010). Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 48, 505-511. 10.1002/dvg.20639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins D. E., Sood R., Hashiramoto A., Hansen C. T., Wilder R. L. and Remmers E. F. (2002). Mutation of macrophage colony stimulating factor (Csf1) causes osteopetrosis in the tl rat. Biochem. Biophys. Res. Commun. 294, 1114-1120. 10.1016/S0006-291X(02)00598-3 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792-1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix R., Cecchini M. G. and Fleisch H. (1990). Macrophage colony stimulating factor restores in vivo bone resorption in the op/op osteopetrotic mouse. Endocrinology 127, 2592-2594. 10.1210/endo-127-5-2592 [DOI] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Odenthal J., Mullins M. C., Lin S., Farrell M. J., Vogelsang E., Haas F., Brand M., van Eeden F. J. M., Furutani-Seiki M. et al. (1996). Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206, 260-276. 10.1007/s004270050051 [DOI] [PubMed] [Google Scholar]
- Hakeda Y., Kobayashi Y., Yamaguchi K., Yasuda H., Tsuda E., Higashio K., Miyata T. and Kumegawa M. (1998). Osteoclastogenesis inhibitory factor (OCIF) directly inhibits bone-resorbing activity of isolated mature osteoclasts. Biochem. Biophys. Res. Commun. 251, 796-801. 10.1006/bbrc.1998.9523 [DOI] [PubMed] [Google Scholar]
- Henke K., Daane J. M., Hawkins M. B., Dooley C. M., Busch-Nentwich E. M., Stemple D. L. and Harris M. P. (2017). Genetic screen for postembryonic development in the zebrafish (Danio rerio): dominant mutations affecting adult form. Genetics 207, 609-623. 10.1534/genetics.117.300187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Thisse B. and Thisse C. (2001). Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274-288. 10.1006/dbio.2001.0393 [DOI] [PubMed] [Google Scholar]
- Huysseune A. and Witten P. E. (2006). Developmental mechanisms underlying tooth patterning in continuously replacing osteichthyan dentitions. J. Exp. Zool. B Mol. Dev. Evol. 306, 204-215. 10.1002/jez.b.21091 [DOI] [PubMed] [Google Scholar]
- Huysseune A., Van der heyden C. and Sire J.-Y. (1998). Early development of the zebrafish (Danio rerio) pharyngeal dentition (Teleostei, Cyprinidae). Anat. Embryol. 198, 289-305. 10.1007/s004290050185 [DOI] [PubMed] [Google Scholar]
- Irion U., Singh A. P. and Nüsslein-Volhard C. (2016). The developmental genetics of vertebrate color pattern formation: lessons from zebrafish. Curr. Top. Dev. Biol. 117, 141-169. 10.1016/bs.ctdb.2015.12.012 [DOI] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. and Chen W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn K. S., Dang V. T., Wilkins S. J., Perkins A. C. and Ragan M. A. (2009). Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 19, 1404-1418. 10.1101/gr.086827.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G. and Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870-1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Letunic I. and Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242-W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorin T., Brunet F. G., Laudet V. and Volff J.-N. (2018). Teleost fish-specific preferential retention of pigmentation gene-containing families after whole genome duplications in vertebrates. G3 8, 1795-1806. 10.1534/g3.118.200201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn R. D. et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636-W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madel M.-B., Ibáñez L., Rouleau M., Wakkach A. and Blin-Wakkach C. (2018). A novel reliable and efficient procedure for purification of mature osteoclasts allowing functional assays in mouse cells. Front. Immunol. 9, 2567 10.3389/fimmu.2018.02567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantoku A., Chatani M., Aono K., Inohaya K. and Kudo A. (2016). Osteoblast and osteoclast behaviors in the turnover of attachment bones during medaka tooth replacement. Dev. Biol. 409, 370-381. 10.1016/j.ydbio.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Mellis D. J., Itzstein C., Helfrich M. H. and Crockett J. C. (2011). The skeleton: a multi-functional complex organ: the role of key signalling pathways in osteoclast differentiation and in bone resorption. J. Endocrinol. 211, 131-143. 10.1530/JOE-11-0212 [DOI] [PubMed] [Google Scholar]
- Montague T. G., Cruz J. M., Gagnon J. A., Church G. M. and Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401-W407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J., Rossnagel K., Haffter P., Kelsh R. N., Vogelsang E., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Hammerschmidt M. et al. (1996). Mutations affecting xanthophore pigmentation in the zebrafish, Danio rerio. Development 123, 391-398. [DOI] [PubMed] [Google Scholar]
- O'Gorman S., Dagenais N. A., Qian M. and Marchuk Y. (1997). Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94, 14602-14607. 10.1073/pnas.94.26.14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. New York: Springer-Verlag. [Google Scholar]
- O'Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733-D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof N., Kuil L. E., van der Linde H. C., Burm S. M., Berdowski W., van Ijcken W. F. J., van Swieten J. C., Hol E. M., Verheijen M. H. G. and van Ham T. J. (2018). Colony-Stimulating Factor 1 Receptor (CSF1R) regulates microglia density and distribution, but not microglia differentiation in vivo. Cell Rep. 24, 1203-1217.e6. 10.1016/j.celrep.2018.06.113 [DOI] [PubMed] [Google Scholar]
- Parichy D. M. and Johnson S. L. (2001). Zebrafish hybrids suggest genetic mechanisms for pigment pattern diversification in Danio. Dev. Genes Evol. 211, 319-328. 10.1007/s004270100155 [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Ransom D. G., Paw B., Zon L. I. and Johnson S. L. (2000). An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127, 3031-3044. [DOI] [PubMed] [Google Scholar]
- Quigley I. K., Manuel J. L., Roberts R. A., Nuckels R. J., Herrington E. R., MacDonald E. L. and Parichy D. M. (2005). Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development 132, 89-104. 10.1242/dev.01547 [DOI] [PubMed] [Google Scholar]
- Ryan G. R., Dai X.-M., Dominguez M. G., Tong W., Chuan F., Chisholm O., Russell R. G., Pollard J. W. and Stanley E. R. (2001). Rescue of the colony-stimulating factor 1 (CSF-1)-nullizygous mouse (Csf1(op)/Csf1(op)) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood 98, 74-84. 10.1182/blood.V98.1.74 [DOI] [PubMed] [Google Scholar]
- Salis P., Lorin T., Laudet V. and Frédérich B. (2019). Magic traits in magic fish: understanding color pattern evolution using reef fish. Trends Genet. 35, 265-278. 10.1016/j.tig.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Salzburger W., Braasch I. and Meyer A. (2007). Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5, 51 10.1186/1741-7007-5-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid I. and Sakamoto K. M. (2001). Analysis of DNA content and green fluorescent protein expression. Curr. Protoc. Cytom. 16, 7.16.1-7.16.10. 10.1002/0471142956.cy0716s16 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thisse B., Wright G. J. and Thisse C. (2008). Embryonic and larval expression patterns from a large scale screening for novel low affinity extracellular protein interactions. ZFIN Direct Data Submission zfin.org/ZDB-PUB-080227-22
- Tiffee J. C., Xing L., Nilsson S. and Boyce B. F. (1999). Dental abnormalities associated with failure of tooth eruption in src knockout and op/op mice. Calcif. Tissue Int. 65, 53-58. 10.1007/s002239900657 [DOI] [PubMed] [Google Scholar]
- To T. T., Witten P. E., Renn J., Bhattacharya D., Huysseune A. and Winkler C. (2012). Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development 139, 141-150. 10.1242/dev.071035 [DOI] [PubMed] [Google Scholar]
- To T. T., Witten P. E., Huysseune A. and Winkler C. (2015). An adult osteopetrosis model in medaka reveals the importance of osteoclast function for bone remodeling in teleost fish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 178, 68-75. 10.1016/j.cbpc.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Tsunoda T. and Takagi T. (1999). Estimating transcription factor bindability on DNA. Bioinformatics 15, 622-630. 10.1093/bioinformatics/15.7.622 [DOI] [PubMed] [Google Scholar]
- Van Wesenbeeck L., Odgren P. R., MacKay C. A., D'Angelo M., Safadi F. F., Popoff S. N., Van Hul W. and Marks S. C. Jr (2002). The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification. Proc. Natl. Acad. Sci. USA 99, 14303-14308. 10.1073/pnas.202332999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H., Iwasaki M. and Kawakami K. (2014). Development of the lateral line canal system through a bone remodeling process in zebrafish. Dev. Biol. 392, 1-14. 10.1016/j.ydbio.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Weigele J. and Franz-Odendaal T. A. (2016). Functional bone histology of zebrafish reveals two types of endochondral ossification, different types of osteoblast clusters and a new bone type. J. Anat. 229, 92-103. 10.1111/joa.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book; A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio), 2nd edn.: University of Oregon Press. [Google Scholar]
- Witten P. E. and Huysseune A. (2009). A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol. Rev. Camb. Philos. Soc. 84, 315-346. 10.1111/j.1469-185X.2009.00077.x [DOI] [PubMed] [Google Scholar]
- Witten P. E., Hansen A. and Hall B. K. (2001). Features of mono- and multinucleated bone resorbing cells of the zebrafish Danio rerio and their contribution to skeletal development, remodeling, and growth. J. Morphol. 250, 197-207. 10.1002/jmor.1065 [DOI] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M. R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L. et al. (2016). Ensembl 2016. Nucleic Acids Res. 44, D710-D716. 10.1093/nar/gkv1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.