Abstract
Ubiquinone 8 (coenzyme Q8 or Q8) mediates electron transfer within the aerobic respiratory chain, mitigates oxidative stress, and contributes to gene expression in Escherichia coli. In addition, Q8 was proposed to confer bacterial osmotolerance by accumulating during growth at high osmotic pressure and altering membrane stability. The osmolyte trehalose and membrane lipid cardiolipin accumulate in E. coli cells cultivated at high osmotic pressure. Here, Q8 deficiency impaired E. coli growth at low osmotic pressure and rendered growth osmotically sensitive. The Q8 deficiency impeded cellular O2 uptake and also inhibited the activities of two proton symporters, the osmosensing transporter ProP and the lactose transporter LacY. Q8 supplementation decreased membrane fluidity in liposomes, but did not affect ProP activity in proteoliposomes, which is respiration-independent. Liposomes and proteoliposomes prepared with E. coli lipids were used for these experiments. Similar oxygen uptake rates were observed for bacteria cultivated at low and high osmotic pressures. In contrast, respiration was dramatically inhibited when bacteria grown at the same low osmotic pressure were shifted to high osmotic pressure. Thus, respiration was restored during prolonged growth of E. coli at high osmotic pressure. Of note, bacteria cultivated at low and high osmotic pressures had similar Q8 concentrations. The protection of respiration was neither diminished by cardiolipin deficiency nor conferred by trehalose overproduction during growth at low osmotic pressure, but rather might be achieved by Q8-independent respiratory chain remodeling. We conclude that osmotolerance is conferred through Q8-independent protection of respiration, not by altering physical properties of the membrane.
Keywords: respiration, cardiolipin, trehalose, ubiquinone, stress, coenzyme Q, LacY, osmotolerance, ProP, proteoliposomes
Introduction
Phospholipid membranes are highly permeable to water but not to polar solutes. Thus, abrupt changes in external solute concentration cause water to rapidly leave or enter cells. In Escherichia coli, osmotically induced dehydration is associated with inhibition of energy-linked functions that include respiration and active transport (1–3). Well-defined osmoregulatory systems respond by mediating solute synthesis or the uptake of exogenous osmolytes to restore cellular hydration (4). For example, osmosensing transporters activate to mediate osmolyte uptake as other transporters inactivate at high osmotic pressure. Evidence suggests that osmotically induced changes to physical properties of the cytoplasmic membrane contribute to the activation of osmosensing transporters (5–7). However, the biochemical basis for the inhibition and subsequent restoration of other energy linked functions is not understood.
Recent reports indicate that cultivation in a high-osmotic pressure medium increases the concentrations of two E. coli membrane lipids: cardiolipin (CL)3 (8) and ubiquinone 8 (also known as coenzyme Q8 or Q8) (9). CL associates with particular membrane proteins, including respiratory enzymes (10). In addition, the osmotic pressure required to activate osmosensing transporter ProP is a direct function of the proportion of anionic phospholipid in E. coli (CL plus phosphatidylglycerol (PG)) (11). Q8 is a redox-active lipid that plays three well-established physiological roles in E. coli: it mediates electron transfer from dehydrogenases to terminal oxidases within the respiratory chain, its reduced form (ubiquinol 8) mitigates oxidative stress by serving as an antioxidant, and it is implicated in the regulation of gene expression (12). Other ubiquinones and isoprenoid lipids influence physical properties of phospholipid membranes (13–18). However, in most cases, the physiological significance of these effects is unclear.
Sévin and Sauer (9) reported that the Q8 content of E. coli increased 110-fold during growth in a high-osmotic pressure medium, that Q8 deficiency impaired the osmotolerance of E. coli although it did not exacerbate oxidative stress, and that exogenous Q10 (the mammalian ubiquinone variant) restored osmotolerance to Q8-deficient E. coli. Reasoning that Q10 could not substitute for Q8 as a respiratory electron carrier in E. coli, they concluded that the elevation of Q8 concentration contributed to the osmotic stress tolerance of E. coli by affecting physical properties of the cytoplasmic membrane. In fact, evidence indicates that Q10 can substitute for Q8 as a respiratory electron carrier in E. coli (19).
We further examined the impact of Q8 on respiration, osmotolerance, and membrane properties in E. coli. Proton-solute symporters ProP and LacY served as indicators of those impacts: ProP activates as lactose transporter LacY inactivates in response to osmotic upshifts (3). Here we confirm that a Q8 biosynthetic lesion impairs the osmotolerance of E. coli during growth in minimal salts media without exogenous osmolytes. We further show that Q8 deficiency impairs respiration and the activities of ProP and LacY in E. coli cells. We report that Q8 supplementation decreases the membrane fluidity of liposomes prepared from an E. coli polar lipid extract but does not alter the osmotic activation of ProP in proteoliposomes (ProP-supplemented liposomes in which a protonmotive force can be imposed without respiration). Thus, Q8 influences the osmoregulatory action of ProP by supporting respiration, not by altering physical properties of the membrane.
During these studies, we observed similar oxygen uptake rates for E. coli cells cultivated in low- and high-osmotic pressure media. This was surprising, because comparable osmotic upshifts dramatically inhibit oxygen uptake (2). Such data suggest that respiration is protected by cellular changes that occur during growth at high osmotic pressure. Remarkably, our data show no difference in Q8 content between E. coli cells cultivated at low and high osmotic pressure. Thus, respiration is not protected by elevating Q8. Here, we show that respiration is also not protected by elevating CL or trehalose. Thus, other changes, such as Q8-independent remodeling of the respiratory chain, may protect respiration in E. coli cells grown at high osmotic pressure.
Results
Ubiquinone is required for the growth of E. coli at high osmolality
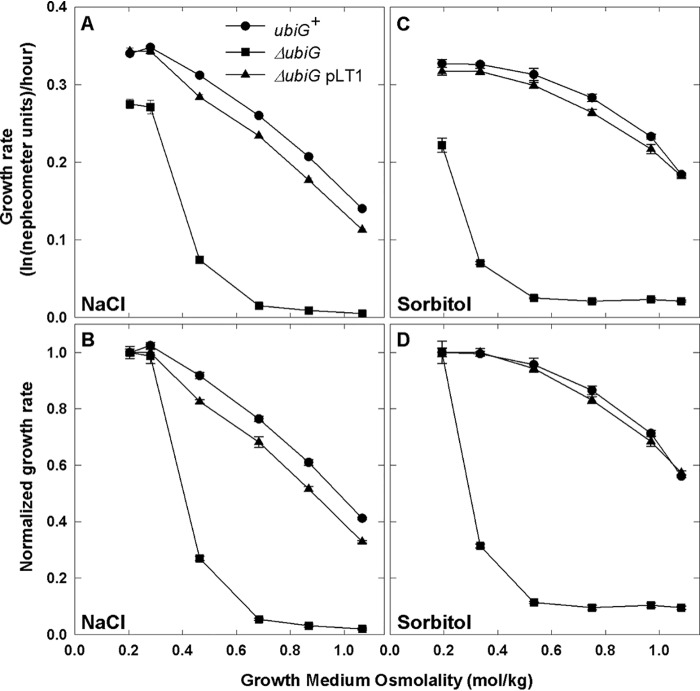
Sévin and Sauer (9) reported that the salinity tolerance of E. coli BW25113 was impaired by mutation ΔubiG785::kan, which blocks Q8 synthesis (20), in Keio Collection strain JW2226. Key characteristics of Keio Collection strains such as growth rate, ribosomal content, and protein expression may be affected by other mutations present in BW25113 (21). However, the ΔubiG785::kan mutation also impaired salinity tolerance in the background of WT E. coli (MG1655) during cultivation in the M9 minimal salts medium used by Sévin and Sauer (9) (Fig. S1A) or in MOPS, a standard minimal medium for physiological studies of E. coli (22) (Fig. 1A). Lesion ΔubiG785::kan affected the growth and osmotolerance of E. coli strains MG1655 (Fig. 1 and Fig. S1), BW25113 (Fig. S2), and WG350 (data not shown) similarly. Complementation with plasmid-borne ubiG restored growth to the ΔubiG785::kan derivative of strain MG1655 (ΔubiG pLT1 in Fig. 1A), confirming that the ubiG lesion was directly responsible for the decreased salinity tolerance.
Figure 1.
Ubiquinone deficiency impairs the growth and osmotolerance of E. coli. E. coli strains MG1655 (WT, circles), WG1533 (MG1655 ΔubiG785::kan, squares) and WG1591 (WG1533 pLT1, triangles) were cultivated in NaCl-free MOPS medium, and growth was monitored nephelometrically after subculturing in media supplemented with NaCl (50–500 mm) (A and B) or sorbitol (100–900 mm) (C and D), as described under “Experimental procedures.” Similar results were obtained when the bacteria were cultivated in MOPS medium (which contains 50 mm NaCl) before subculturing (data not shown), and plasmid vector pBAD24 did not restore growth to strain WG1533 (Fig. S4). Means ± S.E. (error bars) of four replicate measurements from one of two replicate experiments are shown. In B and D, each growth rate is normalized with respect to the mean growth rate observed at the lowest osmolality.
Observing similar effects of the ubiG mutation and anaerobiosis, Sévin and Sauer (9) attributed the impact of the ubiG defect on growth at low salinity to impaired respiration. This effect can be masked and the impact of Q8 deficiency on salinity tolerance highlighted by normalizing each growth rate with that obtained at the lowest salinity tested (e.g. Fig. 1 (compare A and B) and Fig. S2 (compare A and B)). The growth of the ΔubiG785::kan mutant appeared to be inhibited more than that of WT E. coli as the medium osmolality increased (Fig. 1). This was true when the osmolality was adjusted with NaCl (Fig. 1, A and B) or with sorbitol, which is not metabolized by E. coli K-12 (Fig. 1, C and D). Thus, the effects of these agents were osmotic, justifying presentation of the data in terms of measured medium osmolalities, where the osmolality (Π/RT, units of mol/kg) is the osmotic pressure (Π) at a particular temperature (T) and R is the gas constant.
The ubiG lesion impairs the activity of osmosensing transporter ProP by impairing respiration
Redundant osmolyte accumulation mechanisms promote the growth of E. coli K-12 in high-osmolality media (4). They include the trehalose biosynthetic system (OtsAB) as well as broad specificity osmolyte transporters ProP and ProU. In addition, transporter BetT mediates choline uptake, whereas BetB and BetA mediate the oxidation of choline to glycine betaine. Respiration generates the protonmotive force that powers proton-osmolyte symport via transporter ProP (23) and lactose transport via its paralogue, LacY (24), in aerobic E. coli. LacY served as a control in these experiments as it is not osmotically activated, nor does it contribute to osmotolerance.
The impact of mutation ΔubiG785::kan on ProP function was first explored by determining the growth rates of E. coli strains WG1230 (proP+ ubiG+) and WG1535 (proP+ ubiG−) as a function of the osmolality in the presence and absence of osmolyte glycine betaine (GB) (Fig. 2). Growth stimulation by glycine betaine indicates ProP activity in these MG1655 derivatives, which lack all other osmolyte accumulation mechanisms (Table 1). GB stimulated the growth of both strains, but it appeared to be more effective in the ubiG+ strain (WG1230) than in its ΔubiG785::kan derivative (WG1535).
Figure 2.
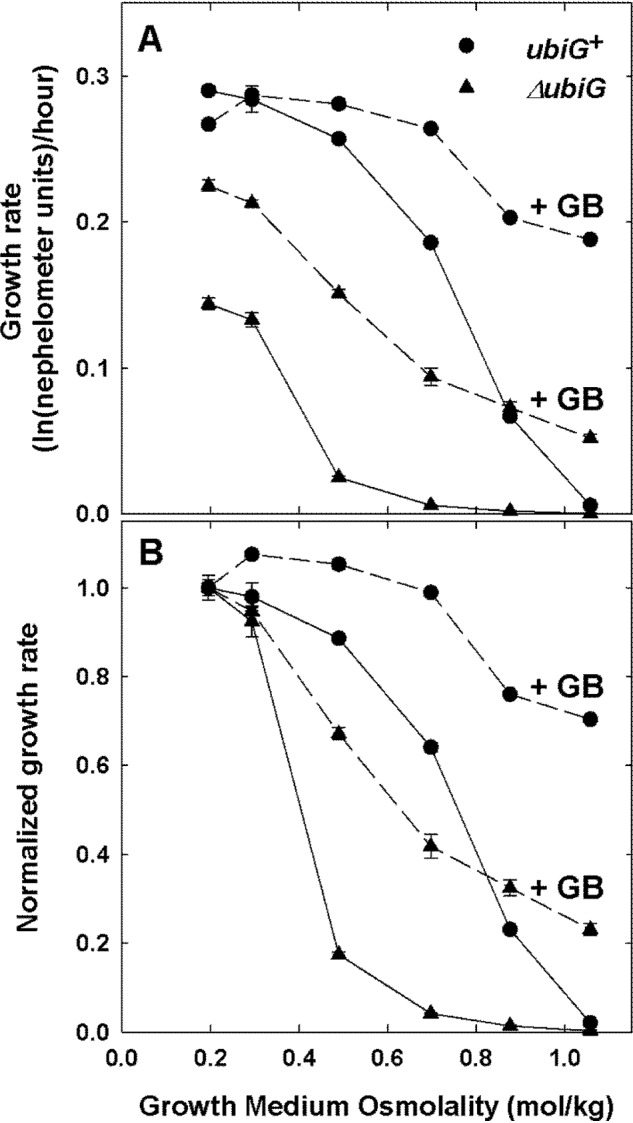
Ubiquinone deficiency impairs osmoprotection mediated by transporter ProP. E. coli strains WG1230 (MG1655 ΔproW859::FRT ΔbetT856::FRT ΔotsA847::FRT proP+ ubiG+, circles) and WG1535 (WG1230 proP+ ΔubiG785::kan, triangles) were cultivated as described in the legend to Fig. 1 and subcultured with (dashed lines) or without (solid lines) osmolyte GB (1 mm) in NaCl-supplemented MOPS medium. Growth was monitored nephelometrically, and data are presented as described under “Experimental procedures” and in the legend to Fig. 1. Error bars, S.E.
Table 1.
E. coli strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| DH5α | F− ϕ80 dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk− mk−) supE44 λ− thi-1 gyrA relA1 | Ref. 59 |
| BKT12 | W3110 ΔclsA856::FRT ΔclsB861::FRT ΔclsC788::kan | Ref. 39 |
| BW25113 | F− λ− rph-1 Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) Δ(rhaD-rhaB)568 hsdR514 | Ref. 71 |
| JP20 | W3110 ΔotsBA::FRT ΔtreA::FRT ΔtreC::FRT ΔtreF::FRT ΦampH::lacI-Ptac-otsBA::FRT | Ref. 43 |
| JW2226 | BW25113 ΔubiG785::kan | Ref. 60 |
| MG1655 | WT (F− λ− rph-1) | Refs. 72 and 73 |
| W3110 | F− λ− IN(rrnD-rrnE)1 rph-1 | Ref. 73 |
| WG1230 | MG1655 ΔproW859::FRT ΔbetT856::FRT ΔotsA847::FRT | Ref. 41 |
| WG1533 | MG1655 ΔubiG785::kan | This work |
| WG1535 | WG1230 ΔubiG785::kan | This work |
| WG350 | F−lacZ trp rpsL thi Δ(putPA)101 Δ(proU)600 Δ(proP-melAB)212 | Ref. 74 |
| WG708 | WG350 pBAD24 | Ref. 27 |
| WG709 | WG350 pDC79 | Ref. 75 |
| WG1541 | WG350 ΔubiG785::kan | This work |
| WG1542 | WG1541 pDC79 | This work |
| WG1590 | WG1533 pBAD24 | This work |
| WG1591 | WG1533 pLT1 | This work |
| WG1610 | MG1655 pDC347 | |
| pBAD24 | Expression vector | Ref. 76 |
| pDC79 | Encodes proP under the control of pBAD and AraC in pBAD24 | Ref. 75 |
| pDC347 | Encodes full-length clsA under the control of pBAD and AraC in pBAD24 | Ref. 77 |
| pLT1 | Encodes full-length ubiG under the control of pBAD and AraC in pBAD24 | This work |
ProP activity is a sigmoid function of the assay medium osmolality in cells cultivated at low osmolality (25). Such direct measurement revealed that mutation ΔubiG785::kan decreased ProP activity ∼10-fold (Fig. 3A). Proline was the ProP substrate for these measurements, as proline and glycine betaine are essentially equivalent as ProP substrates (26), and proline is more readily available in radiolabeled form than glycine betaine. The osmoregulation of transporter activity was retained, as there was no significant change in the osmolality at which transporter activity was half-maximal (Fig. 3A (inset) and Table S1 (parameters Π½/RT and B)). The ubiG defect could affect ProP activity by impairing generation of the protonmotive force and by altering membrane properties. A similar, dramatic impairment of LacY activity (Fig. 3B and Table S1) suggested that respiration, and hence generation of the protonmotive force, was impaired in the Q8-deficient bacteria. Osmotic inhibition of LacY activity was not observed, because these experiments involved a narrower osmolality range than was employed for previous work (1, 3). In addition, this work was done with intact cells, whereas an earlier report was based on cytoplasmic membrane vesicles (3).
Figure 3.
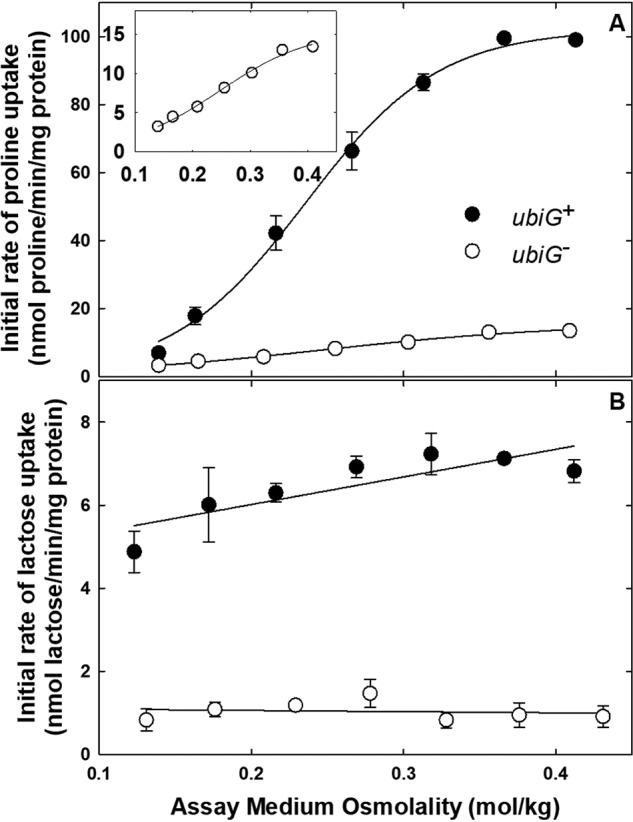
Q8 deficiency impairs the activities of E. coli transporters ProP and LacY. E. coli strains WG709 (ubiG+, closed circles) and WG1542 (ΔubiG785::kan, open circles) were cultivated in NaCl-free MOPS medium, initial rates of proline (A) and lactose (B) uptake were determined, and data were analyzed as described under “Experimental procedures.” Assay medium osmolalities were adjusted with NaCl at concentrations in the range 0–0.15 m. Means ± S.E. (error bars) of triplicate measurements from one of two replicate experiments are shown. The osmotic activation of ProP in the ubiG− E. coli strain is shown in the inset to A. The regression lines are shown in the figure, and the osmolality response parameters for the individual experiments are shown in Table S1.
Oxygen uptake measurements directly demonstrated the inhibition of respiration by the ubiG lesion and an osmotic upshift (Fig. 4A). As expected, mutation ΔubiG785::kan dramatically decreased the rate of oxygen uptake by bacteria cultivated and incubated at low osmolality (compare circles and squares, Low - Low). That effect was reversed when the ubiG defect was complemented with plasmid pLT1 (compare circles, squares, and triangles, Low - Low). As reported previously (1), respiration was also dramatically inhibited when bacteria cultivated at low osmolality were introduced to a high-osmolality medium (Fig. 4A, Low - Low versus Low - High). Respiration at high osmolality could be restored to ubiG+ bacteria by long-term cultivation in a high-osmolality medium (Fig. 4A, High - High). Similar patterns were seen when the osmolalities of the media were adjusted with NaCl (Fig. 4A) or sorbitol (Fig. 4B).
Figure 4.
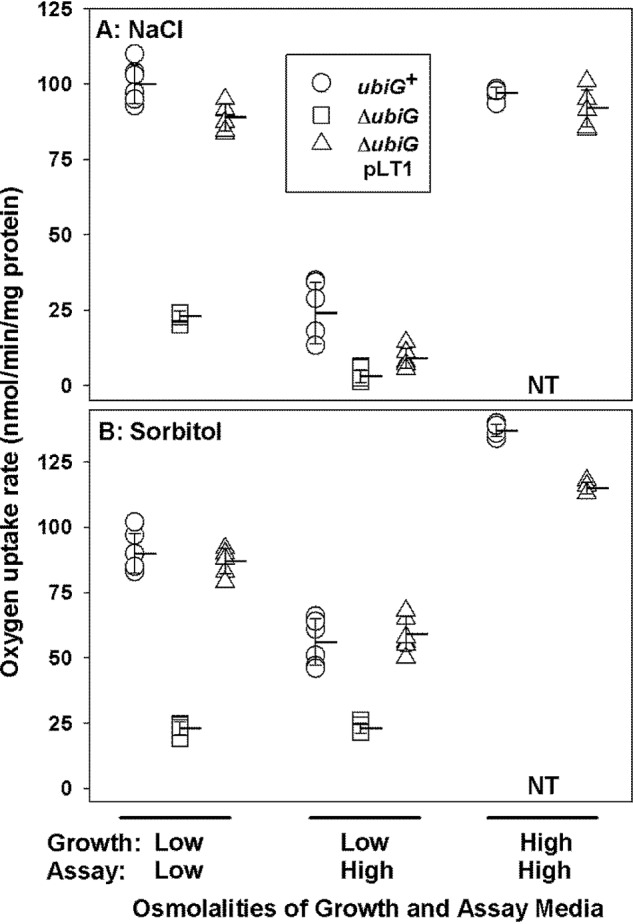
The role of ubiG in aerobic respiration by E. coli. E. coli strains MG1655 (ubiG+, circles), WG1533 (MG1655 ΔubiG785::kan, squares), and WG1591 (MG1655 ΔubiG785::kan pLT1, triangles) were cultivated in NaCl-free MOPS medium as described under “Experimental procedures.” That medium contained no NaCl (the low-osmolality medium, 0.12 mol/kg) or 450 mm NaCl (the high-osmolality medium, 0.97 mol/kg) (A) or no added sorbitol (the low-osmolality medium, 0.12 mol/kg), or 800 mm sorbitol (the high-osmolality medium, 0.99 mol/kg) (B). NT, not tested (strain WG1533 does not grow in the high-osmolality media). Oxygen uptake was measured in media with the same specifications, as indicated. Triplicate measurements from each of two experiments were combined. Horizontal lines with error bars represent means ± S.D.
The above results show that respiration (Fig. 4) and the activities of protonmotive force–dependent transporters ProP and LacY (Figs. 2 and 3 and Table S1) are inhibited by a ubiG lesion (i.e. Q8 deficiency) or osmotic upshifts in E. coli cells. Also, the restoration of respiration afforded by cultivation of E. coli in a high-osmolality medium is independent of the osmolyte transporters (ProP, ProU, and BetT), as it does not require provision of their substrates (e.g. GB, proline, or choline) (Fig. 4). However, these data do not reveal whether changes to physical properties of the membrane contribute to the observed effects.
Elevation of Q8 alters membrane fluidity but does not affect ProP function
Histidine-tagged ProP (ProP-His6) can be purified and reconstituted in liposomes comprised of an E. coli polar lipid extract, creating proteoliposomes (PRLs) (27) in which the protein retains its physiological membrane orientation (3). These PRLs lack the respiratory chain, but osmoregulated ProP activity can be supported by an artificially imposed protonmotive force (27, 28). The osmolality at which ProP becomes active in cells and PRLs is directly proportional to the anionic phospholipid content, and particularly the CL content, of the host membrane (8, 11). Liposomes and PRLs were therefore employed to assess the impact of Q8 on physical properties of the membrane and, in turn, on ProP function.
The limited water solubilities of ubiquinones (29) render the creation of ubiquinone (Q)-supplemented liposomes and PRLs challenging. The extent of Q incorporation was therefore determined for liposomes prepared in three ways: lipid film hydration alone (30), lipid film hydration with detergent dialysis (27), and lipid film hydration with freeze-thaw (14). In each case, monodisperse, unilamellar liposomes were generated by subsequent extrusion through microporous membranes (30). Supplementary Q was not well-incorporated into the lipid bilayer by lipid film hydration alone or with detergent dialysis. This was indicated by low Q recovery upon extrusion of such preparations (Table 2) and by the presence of a yellow residue on the extrusion membrane. Successful Q8 incorporation was achieved by lipid film hydration with freeze-thaw, as assessed by high Q recovery and no yellow residue. As for Q10 (14), only ∼2% Q8 (weight of Q8/weight of lipid) was recovered from liposomes prepared after supplementation of phospholipid with 3% Q8 (Table 2). Based on these results, data based on other liposome preparation procedures and preparations designed to contain significantly larger proportions of Q should be considered with care (9, 16, 31).
Table 2.
Ubiquinone (Q) content of ubiquinone-supplemented liposomes
Liposomes were prepared, lipid was extracted from the aqueous dispersions, and lipid contents were measured as described under “Experimental procedures.”
| Liposome preparation |
Lipid analysis |
||||||
|---|---|---|---|---|---|---|---|
| Q/lipid % (w/w) | Film hydration | Detergent dialysis | Freeze-thaw | Extrusion | Lipida recovery % (w/w) | Qb recovery % (w/w) | Measured Q/lipid % (w/w) |
| 2.0 | + | − | − | − | 100 ± 16 | 86 ± 1 | 1.7 ± 0.3 |
| + | − | − | + | 82 ± 17 | 32 ± 3 | 0.6 ± 0.1 | |
| 2.0 | + | + | − | − | 79 ± 11 | 95 ± 2 | 2.4 ± 0.3 |
| + | + | − | + | 80 ± 18 | 25 ± 1 | 0.8 ± 0.1 | |
| 1.0 | + | − | + | − | 95 ± 5 | 92 ± 4 | 1.0 ± 0.1 |
| + | − | + | + | 89 ± 7 | 84 ± 6 | 0.9 ± 0.1 | |
| 3.0 | + | − | + | − | 95 ± 5 | 98 ± 2 | 3.1 ± 0.2 |
| + | − | + | + | 89 ± 7 | 57 ± 2 | 2.0 ± 0.1 | |
a The mean weight ± S.E. of total lipid (as determined by gravimetric analysis for four independent liposome extracts) relative to the quantity of phospholipid added to the preparation (as specified by Avanti Polar Lipids, Inc.).
b The mean weight of Q ± S.E. (as determined by HPLC analysis for four independent liposome extracts) relative to the weight of Q added to the preparation. The liposomes prepared by lipid film hydration, alone, were supplemented with Q10. Those prepared by lipid film hydration with detergent dialysis or freeze-thaw were supplemented with Q8.
The steady-state anisotropy of diphenylhexatriene (DPH) fluorescence was used to indicate the impact of Q8 on membrane fluidity (or perhaps, more correctly, membrane order), a representative physical property (32) (Fig. 5). Liposomes containing only Q8 present in the E. coli polar lipid extract (0.11 ± 0.05% (Q8 relative to total lipid, w/w)) were compared with liposomes supplemented with 1% Q8 (w/w). Sévin and Sauer (9) reported that 1% Q8 (w/w) was present in the membrane of E. coli cultivated at high osmolality. DPH fluorescence was measured as a function of temperature in the range 20–45 °C (Fig. 5). As expected, the fluorescence anisotropy of DPH in both preparations decreased with increasing temperature, indicating an increase in membrane fluidity. In addition, the fluorescence anisotropy of DPH in the Q8-supplemented liposomes was higher than that in the unsupplemented liposomes at every temperature (Fig. 5). The decrease in membrane fluidity due to Q8 supplementation corresponded to the impact of approximately a 5 °C decrease in temperature. Thus, Q8 affected a physical property of the membrane. Similar analyses have shown that other ubiquinones decrease the fluidities of membranes with other phospholipid compositions (14, 15, 31, 33, 34).
Figure 5.
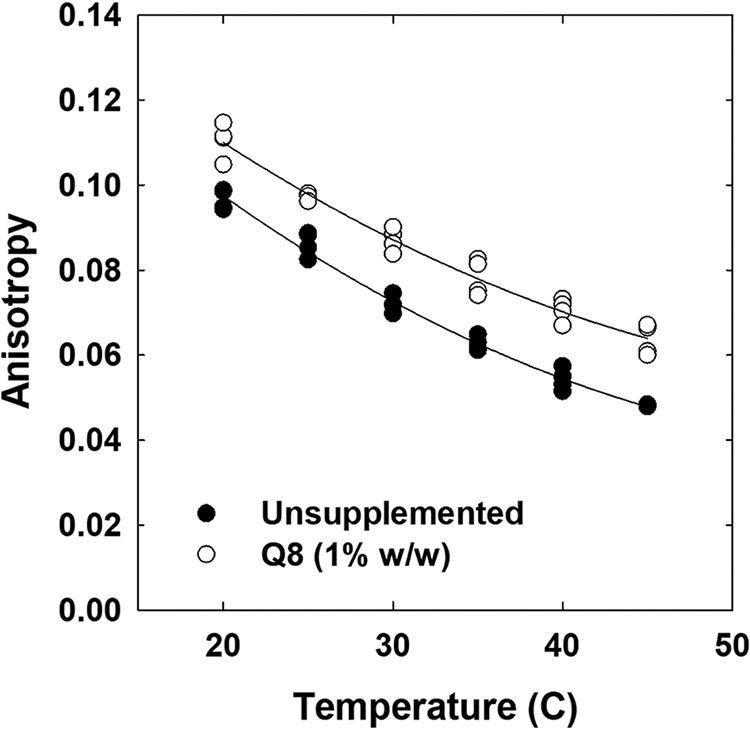
Q8 increases the anisotropy of DPH fluorescence in liposomes. Liposomes comprised of a polar lipid extract from E. coli, with or without supplementary Q8, were prepared in 0.1 m potassium phosphate, pH 7.4, 0.1 mm K+-EDTA and labeled with DPH (0.2 mol % relative to phospholipid) before the anisotropy of DPH fluorescence was measured as described under “Experimental procedures.” Q8 was omitted (closed circles) or added to attain 1% (open circles) Q8 relative to lipid (w/w). Measurements were made at 5 °C intervals over a 20–45 °C temperature range. Duplicate measurements from each of two experiments are shown.
Next, the impact of Q8 supplementation on ProP activity was analyzed using PRLs. Unsupplemented or Q8-supplemented liposomes, the latter with a Q8 proportion of 2% (w/w), were fused with PRLs containing ProP-His6. The resulting preparations contained only endogenous Q8 (0.11 ± 0.05% (w/w) or 1% Q8 (w/w)). Liposome fusion is an established technique for the adjustment of lipid composition (7, 11). Transport assays revealed no effect of Q8 supplementation on the osmolality dependence of ProP activity (Fig. 6 and Table S2).
Figure 6.
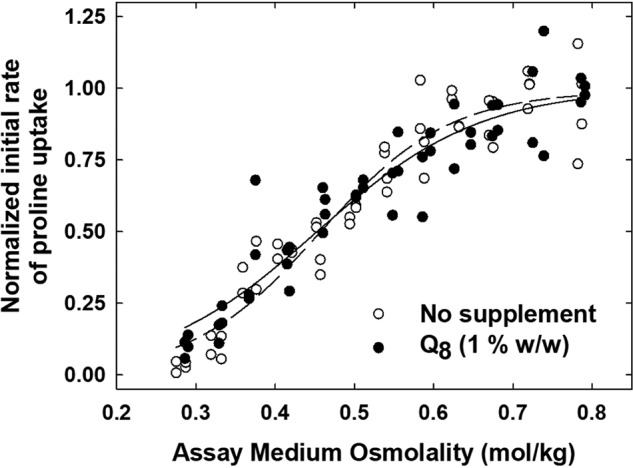
Q8 does not affect ProP activity in proteoliposomes. ProP-His6–containing PRLs were fused with E. coli lipid liposomes (open circles) or Q8-supplemented E. coli lipid liposomes (for a final Q8 concentration of 1% (w/w)) (closed circles), initial rates of proline uptake were measured, and data were analyzed as described under “Experimental procedures.” The osmolalities of the assay media were adjusted with sorbitol (0–0.48 m). For each preparation, data from two replicate experiments were fit to Equation 1, the uptake activities were normalized on the resulting Amax values, and the data were combined and again fit to Equation 1. The resulting regression lines are shown in the figure, and the osmolality response parameters for the individual experiments are shown in Table S2.
Collectively, these data suggest that Q8 deficiency altered ProP activity by impairing respiration-dependent generation of the protonmotive force (observed in cells; Figs. 3 and 4), not by altering physical properties of the membrane (observed in liposomes (Fig. 5) and PRLs (Fig. 6 and Table S2). By extension, they also suggest that the osmotolerance exhibited by the growth of E. coli in high-osmolality media results from changes that protect respiration.
Does increased Q8 synthesis protect respiration in E. coli cultivated at high osmolality?
Respiration is dramatically inhibited by osmotic upshifts, but E. coli can adapt to restore respiration during long-term cultivation in a high-osmolality medium. For example, the rate of oxygen uptake by WT bacteria cultivated and maintained at a high osmolality in NaCl-supplemented medium (0.97 mol/kg) is similar to that of bacteria cultivated and maintained at low osmolality (0.12 mol/kg), even though there is a 76% decrease in the oxygen uptake rate by bacteria cultivated at the same low osmolality, and then shifted to the same high osmolality with NaCl (Fig. 4A). The impact of an osmotic upshift imposed with sorbitol was less dramatic (Fig. 4B), but respiration was again restored during long-term cultivation in sorbitol-supplemented medium. The protection of respiration attained in the steady state is consistent with the more rapid growth of ubiG+ than of ubiG− bacteria at high osmolality.
Sévin and Sauer (9) attributed the growth of E. coli at high osmolality to the accumulation of Q8 and concluded that Q8 conferred osmotolerance by affecting physical properties of the cytoplasmic membrane. In their work, an E. coli metabolite extract containing 13C-labeled Q10 was added to chloroform-methanol extracts from bacteria cultivated at low and high osmolality. The resulting samples were analyzed by flow-injection MS. Summed intensities of signals attributed to 12C-Q8 were divided by summed intensities of signals attributed to 13C-Q10, and these ratios were then used to generate calibration curves and to quantify endogenous Q8. Q8 was reported to be below the limit of detection in bacteria cultivated at low osmolality, increasing 110-fold to constitute 1% (w/w) relative to phosphatidylethanolamine (PE) plus PG in bacteria cultivated at high osmolality (9). The impact of Q on physical properties of the membrane was assessed by imaging morphological changes in liposomes (either unsupplemented or supplemented with Q10 (5% (w/w)) before and after osmotic upshocks.
In the present work, bacteria were cultivated as for the experiments reported in Figs. 3 and 4, and total Q8 (Q8 plus Q8H2) was determined by LC-MS/MS as described under “Experimental procedures.” As expected, total Q8 was below the limit of detection in ΔubiG785::kan bacteria, and complementation with plasmid-borne ubiG restored total Q8 to the level observed in the WT bacteria (Table 3). As for the complementation of bacterial growth at high osmolality (Fig. S3), restoration of Q8 did not require arabinose induction of transcription of the plasmid-borne ubiG gene (Table 3).
Table 3.
Cultivation of E. coli under osmotic stress does not elevate total Q8 concentration
Bacteria were cultivated and harvested, and Q8 was extracted and measured as described under “Experimental procedures.”
| Strain | Relevant genotype | Growth medium supplement | Mean total Q8a ± S.D. |
|---|---|---|---|
| nmol/mg protein | |||
| BW25113 | ubiG+ | Nil | 1.1 ± 0.5 |
| NaCl | 1.9 ± 1.0 | ||
| JW2226 | ΔubiG785::kan | Nil | ND |
| MG1655 | ubiG+ | Nil | 1.7 ± 0.9 |
| NaCl | 2.4 ± 0.7 | ||
| Sorbitol | 2.1 ± 0.5 | ||
| WG1533 | ΔubiG785::kan | Nil | ND |
| WG1591 | ΔubiG785::kan | Nil | 2.7 ± 1.2 |
| NaCl | 1.6 ± 0.2 | ||
| pLT1 (ubiG+) | Sorbitol | 2.1 ± 0.9 |
a The mean weight ± standard deviation (S.D.) of Total Q8 for 5 independent extracts relative to the quantity of protein in the cell suspension from which each was derived. ND = Not Detected.
The following observations and conclusions are inconsistent with those of Sévin and Sauer (9). Remarkably, no change in total Q8 content was detected upon cultivation of ubiG+ bacteria in high-osmolality, NaCl- or sorbitol-supplemented media, as compared with those cultivated in low-osmolality medium (Table 3). PE and PG constitute ∼95% of the phospholipid in E. coli (35), phospholipids are the predominant lipid species (36), and the proportion of protein to lipid in E. coli is ∼6 (37). On this basis and given that the molecular weight of Q8 is 727, 2 nmol of Q8 per mg of protein (Table 3) would correspond with ∼1% Q8 relative to PE plus PG (w/w). Thus, the concentration of Q8 in bacteria cultivated at low or high osmolality (Table 3) was comparable with that reported previously, but only at high osmolality (9).
Q8 at the concentration that we observe did affect a physical property of the membrane (fluidity) (Fig. 5). However, Q8 concentrations are not elevated during cultivation at high osmolality and hence cannot account for the observed osmotolerance or protection of respiration.
What protects respiration during bacterial growth at high osmolality?
The proportion of CL among E. coli phospholipids increases more than 2-fold during growth in a high-osmolality medium (8) or in stationary growth phase (38). Three cardiolipin synthases are encoded by the genome of E. coli K-12: ClsA mediates CL synthesis during exponential-phase growth at low or high osmolality, whereas ClsB and ClsC mediate CL synthesis during stationary phase (39). In contrast to the strong effect of a ubiG deletion (Fig. 1), deletion of clsA, clsB, and clsC had no effect on the osmotolerance of WT E. coli (Fig. S4A, compare CL synthase–deficient strain BKT12 (triangles) with its WT parent, W3110 (circles)). In addition, overexpression of clsA during bacterial cultivation at low osmolality did not protect respiration from an osmotic upshift (Fig. S4B). Thus, CL accumulation during bacterial growth at high osmolality is not the change that protects respiration.
Meury (2) showed that osmotic upshifts dramatically impair respiration in E. coli. The restoration of cell volume and respiration corresponded with the osmotically induced uptake of exogenous K+ or osmolyte glycine betaine or with trehalose synthesis (2, 40). We therefore tested the hypothesis that trehalose accumulation protects respiration.
Data in Figs. 1 and 2 illustrate the contribution of trehalose synthesis to the osmotolerance of E. coli cells that lack osmolyte uptake systems and/or exogenous osmolytes (compare the osmolality dependence of the growth of WT E. coli (MG1655, Fig. 1A) and E. coli WG1230, which cannot synthesize trehalose (Fig. 2A)) (41, 42). The effect of the trehalose biosynthetic lesion is less profound than that of the ubiG deletion (compare WT E. coli (ubiG+) and its ΔubiG785::kan derivative (Fig. 1A)), but trehalose may at least partially protect respiration from cellular dehydration at high osmolality.
In E. coli JP20, trehalose synthesis is conferred by isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible, LacI and Ptac-mediated expression of an otsAB operon integrated in ampH (the native otsAB operon is deleted from this strain) (43). With IPTG induction, the trehalose content in strain JP20 cultivated at low osmolality is similar to that in the WT parent strain (WG3110) cultivated at high osmolality (Table 4 and Fig. S5) (43). If trehalose accumulation were sufficient to protect respiration, E. coli JP20 cultivated in IPTG-supplemented, low-osmolality medium would respire at the same rate when introduced to low- and high-osmolality assay media. Oxygen uptake assays revealed no such protection of respiration (Fig. 7).
Table 4.
The trehalose content of E. coli cells cultivated at low and high osmolalities
Bacteria were cultivated in low-osmolality (LOM) or high-osmolality (HOM) NaCl-supplemented MOPS medium, with or without IPTG (0.1 mm), as for the oxygen uptake assays. Trehalose was extracted and estimated by TLC as described under “Experimental procedures.” A representative chromatogram is shown in Fig. S5.
| Strain | Growth condition | Repa | Trehalose contentb |
|---|---|---|---|
| nmol/OD600 unit | |||
| W3110 | LOM | 1 | NDc |
| 2 | ND | ||
| W3110 | HOM | 1 | 13.0 ± 0.7 |
| 2 | 9.5 ± 1.5 | ||
| JP20 | LOM, −IPTG | 1 | ND |
| 2 | ND | ||
| JP20 | LOM, +IPTG | 1 | 16.3 ± 1.6 |
| 2 | 11.5 ± 1.1 |
a Replicate. Two independent experiments were performed for each strain under each growth condition.
b The quantity of trehalose detected by TLC as described under “Experimental procedures” is normalized to the optical density (600 nm) of the culture from which the sample was derived. Trehalose contents are shown as the means ± S.E. of three replicates for each experiment.
c ND, not detected.
Figure 7.
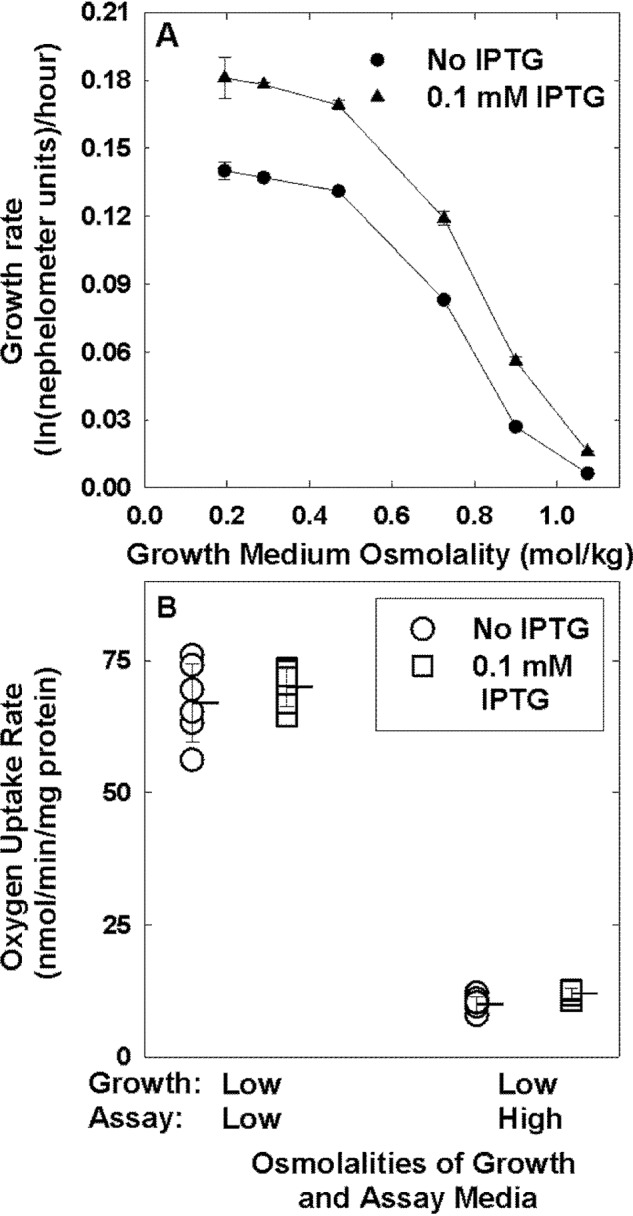
Trehalose accumulation during growth of E. coli at low osmotic pressure does not protect respiration from an osmotic upshift. A, growth rates of E. coli strain JP20 (W3110 ΔotsBA::FRT ΔtreA::FRT ΔtreC::FRT ΔtreF::FRT ΦampH::lacI-Ptac-otsBA::FRT) in NaCl-supplemented MOPS media of the indicated osmolalities without (circles) or with (triangles) IPTG (0.1 mm) were determined, and the data were analyzed as described in the legend to Fig. 1. Error bars, S.E. B, E. coli strain JP20 was cultivated in low osmolality MOPS medium, without (circles) or with (squares) IPTG (0.1 mm), and oxygen uptake was measured in low- and high-osmolality MOPS media as described in the legend to Fig. 4. The high-osmolality medium was supplemented with NaCl. Triplicate measurements from each of two experiments were combined. Horizontal lines with error bars represent means ± S.D.
Discussion
Changes to the osmotic pressure of the cellular environment trigger rapid transmembrane water fluxes. For example, as the osmotic pressure increases, water leaves E. coli on a sub-second time scale (44, 45). Such water efflux is forestalled by the rapid activation of available osmoregulatory systems that include K+ transporter Trk and osmolyte transporter ProP (4). In the absence of exogenous osmolytes, glutamate accumulates due to the inhibition of protein synthesis and serves as the predominant K+ counterion. K+ glutamate accumulation only partially restores cell functions and growth, whereas osmolyte accumulation (e.g. glycine betaine uptake) is fully effective (46).
This rapid and well-characterized osmoregulatory response overlaps slower osmoadaptive mechanisms that require cellular remodeling and have been less fully investigated. This report concerns the protection of respiration afforded by prolonged incubation and growth of E. coli in a high-osmotic pressure medium: the rates of oxygen uptake by bacteria cultivated in low- and high-osmotic pressure media are similar, even though respiration is dramatically inhibited when bacteria cultivated at the same low osmotic pressure are introduced to a medium with the same high osmotic pressure (Fig. 4). Long-term osmoadaptation is known to involve the synthesis of osmoregulatory proteins (e.g. OtsA, OtsB, ProP, ProU, BetT, BetB, and BetA), a phospholipid (CL), and two osmolytes (trehalose and glycine betaine, the latter synthesized through the action of the Bet proteins only when choline is available exogenously) (4). Here, we have shown that the protection of respiration achieved during growth in a minimal salts medium at high osmotic pressure, without exogenous osmolytes, does not result from osmotically induced increases in the cellular content of Q8 (9) (which does not increase; Table 3), CL (Fig. S4), or trehalose (Fig. 7).
The osmoadaptive mechanism that protects respiration may be related to the mechanism by which osmotic upshifts inhibit respiration. By measuring DPH fluorescence anisotropy, Culham et al. (5) showed that osmotic upshifts decreased the membrane fluidity of liposomes comprised of E. coli phospholipid. Such a decrease may render the intramembrane diffusion of electron carriers rate-limiting for respiration, and osmotic adaptation of the respiratory system may be designed to address that limitation.
The hypothesis that intramembrane diffusion of electron carriers may limit the respiratory rate has been extensively tested, primarily with experimental systems that replicate the mitochondrial respiratory chain (e.g. see Ref. 47). Such analyses yielded a random diffusion (or fluid state) model: respiration was proposed to occur via random collisions among independent respiratory chain constituents (48). That model was challenged by mounting evidence for the existence of respiratory supercomplexes, enzyme complexes comprised of respiratory chain components. Such complexes clearly exist in mitochondria, and there is mounting evidence for their existence in bacteria (49, 50). The existence of respiratory supercomplexes was predicted by the “solid-state” model for respiratory chain organization (51). However, recent work suggests that, at least in E. coli, the respiratory chain is highly plastic. Changes in respiratory chain composition, including the synthesis or recruitment of alternative quinones (52), accommodate diverse oxygen levels and terminal electron acceptors. Furthermore, evidence suggests that interactions among respiratory chain constituents are dynamic, not static (49). Ultimately, however, the functional impact of respiratory supercomplexes remains unclear (53, 54).
Sévin and Sauer (9) were the first to propose that a respiratory chain modification would accommodate osmotic stress, but their lipidomics analysis incorrectly identified increased Q8 concentration as that modification. The following two factors may have contributed to the differences between the results reported here and by Sévin and Sauer (9). First, the lipidomics data provided by Sévin and Sauer included only two molecular species specific for Q8 (octaprenyl-ubiquinone and octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinone). Each metabolite was detected in two forms (−H and +F). For each metabolite, the quantities of one of the two forms in samples derived from bacteria grown at low and high osmolality were not statistically significantly different. For the other form, the quantities were only marginally different. Second, the flow-injection TOF MS analysis employed by those authors provides qualitative, not quantitative, data. To overcome this limitation, Sévin and Sauer (9) inferred absolute concentrations from 12C-Q8/13C-Q10 ratios using a calibration curve of 12C-Q8 and 13C-Q10 standards. However, that calibration curve relied on 13C-Q10 rather than 13C-Q8, it showed poor correlation, and the data for samples extracted from E. coli grown at low and high osmolalities were outside and borderline with the standard curve. In combination, these limiting factors could have led to incorrect assignment and quantification of Q8 by Sévin and Sauer. In contrast, we employed quantitative and qualitative LC-MS/MS analysis for the determination of Q8.
Previous reports of the Q8 content of E. coli cells from exponential-phase, aerobic cultures vary widely. They are difficult to compare because analytical methodologies and reporting units also vary. For example, Wallace et al. (55) reported 2.26 nmol of Q8/mg of membrane protein (∼11 nmol/mg of cell protein), and Loiseau et al. (56) reported ∼100 pmol/mg of wet cells (∼0.6 nmol/mg of protein, assuming that dry weight is 30% of cell weight and protein is 55% of cell dry weight (37)). Our data are reasonably consistent with these reports.
This study shows directly that respiration is protected during long-term osmoadaptation. It is possible that osmotic stress induces the synthesis of an alternate quinone electron carrier, related to but distinct from Q8, or the synthesis of other respiratory chain constituents. Alternatively, the organization of the respiratory chain may change during growth at high osmotic pressure: for example, respiratory supercomplexes may form.
The concept that quinones play physiological roles related to their impact on membrane physical properties is attractive (9, 57). However, this study provided no evidence for such a role in E. coli osmoadaptation. The established role of Q8 in respiration fully accounted for its involvement in osmoadaptation.
Experimental procedures
Bacterial strains and plasmids
The E. coli strains and plasmids used for this work are listed in Table 1, and primers are listed in Table S3. Molecular biological techniques were performed as described (58, 59). The ubiG genes of E. coli strains MG1655, WG350, and WG1230 were replaced with ΔubiG::kan by P1 cml crl 100 bacteriophage-mediated transduction with E. coli JW2226 (60) as donor and selection on LB Km (kanamycin, 50 μg/ml). The mutation was confirmed via PCR with the primers listed in Table S3. Reaction mixtures were prepared as described (61), and PCR was performed with a PerkinElmer GeneAmp PCR System 2400 thermal cycler (Woodbridge, Canada) programmed as follows: 5 min at 94 °C; 26 cycles with 45 s at 94 °C, 30 s at 57 °C, and 2 min/kb at 72 °C; 5 min at 72 °C. Taq polymerase, purchased from Invitrogen (Burlington, Canada), was used unless otherwise indicated, and oligonucleotides were purchased from Eurofins MWG Operon (Huntsville, AL).
To construct plasmid pLT1, ubiG was amplified by PCR with chromosomal DNA from E. coli MG1655 as template, primers ubiGus and ubiGds (Table S3), and Pwo DNA polymerase (0.1 units/ml) (Roche Applied Science, Laval, Canada). The small NcoI-HindIII fragment of vector pBAD24 was replaced with the ubiG amplicon. Transformants of E. coli DH5α were selected on LB Amp (50 μg/ml ampicillin), and the identity of the plasmid was confirmed by DNA sequencing (Advanced Analysis Centre, University of Guelph, Guelph, Canada).
Culture media and growth conditions
Bacteria from frozen stocks were introduced into LB medium (62) and incubated in a shaking incubator at 200 rpm and 37 °C. They were subcultured into either M9 minimal salts (9) or NaCl-free MOPS minimal medium and further incubated as described. The latter was MOPS medium (22) from which NaCl was omitted. Minimal media were supplemented with NH4Cl (9.5 mm) as nitrogen source and fructose (0.2%, w/v) as carbon source. l-Tryptophan (0.005%, w/v) and thiamine hydrochloride (0.0001%, w/v) were included to meet auxotrophic requirements. NaCl or sorbitol was used to adjust medium osmolalities, which were measured with a VAPRO Vapor Pressure Osmometer 5520 (Claremont, Canada). Ampicillin (100 μg/ml) was added to maintain plasmids. Culture optical densities at 600 nm (OD600) were determined with a Pharmacia LKB NovaSpec II spectrophotometer.
Growth curves
Bacterial growth was monitored using a Nephelostar microplate nephelometer (BMG Labtech, Ortenberg, Germany) as described (41) with the following modifications. Bacteria were subcultured into M9 or MOPS minimal medium (2%, v/v) and incubated until the ubiG+ strain reached an OD600 of 0.5. They were then subcultured (1% (v/v) for ubiG+ strains, and 2% (v/v) for ubiG− strains) into the same medium supplemented with NaCl (50–500 mm) or sorbitol (100–900 mm), with or without glycine betaine (1 mm), or arabinose (1.33, 13.3, or 133 μm), as indicated. Aliquots (200 μl, four replicates per test) were distributed into sterile 96-well microtiter plates (Thermo Fisher Scientific, Ottawa, Canada) which were covered with a Breathe-Easy® sealing membrane (Sigma-Aldrich, Oakville, Canada). Bacterial growth was monitored for 19 h at 36 °C. The natural logarithms of the nephelometer units measured at each time point were plotted versus time. Growth rates, determined by linear regression, were the slopes of these plots during the exponential phase of growth, as illustrated in Fig. S1. Growth rates were normalized by dividing each growth rate by that obtained at the lowest NaCl or sorbitol concentration tested.
Determination of Q8 and Q8H2 from E. coli by LC/tandem MS
LC/MS/MS was used for the determination of Q8 and its reduced form, Q8H2, in E. coli samples. Briefly, bacteria were cultivated as for transport assays. Aliquots (11 ml) were harvested by centrifugation (12,000 × g, 10 min, 4 °C), washed (11 ml of saline (0.85% (w/v) NaCl), resuspended in saline (1.1 ml), and transferred to microcentrifuge tubes. Samples (0.1 ml) were taken for protein analysis; the remaining samples were lyophilized. Each lyophilized sample was resuspended in 100 μl of PBS (137 mm NaCl, 10 mm Na2HPO4, 2.7 mm KCl, 1.8 mm KH2PO4), and internal standard (100 pmol of Q10; Sigma-Aldrich) was added. Each sample was transferred to a 15-ml screw top tube to which 2 ml of methanol and 10 ml of water-washed hexane were added. The mixture was then mixed vigorously for 1 min and centrifuged (1,430 × g, 5 min, 4 °C), and 9 ml of the top hexane layer was collected then dried using a rotary evaporator. The resulting dried lipids were redissolved in 180 μl of ice-cold ethanol (HPLC grade), and 2 μl was injected onto an Agilent 1290 UHPLC system connected to an Agilent 6495 triple-quadrupole mass spectrometer. Analytes were separated on a Luna 5-μm C18(2) 100-Å column (150 × 4.6 mm; Phenomenex) by gradient elution using mobile phase A (2.5 mm ammonium formate in 95:5 methanol/isopropyl alcohol) and mobile phase B (2.5 mm ammonium formate in 100% isopropyl alcohol) at 0.4 ml/min. The gradient consisted of 50% mobile phase B from 0 to 15 min, 50–70% B from 15 to 17 min, 70–100% B from 17 to 19 min, and then 50% mobile phase B from 19 to 24 min. Flow was then directed into the triple quadrupole mass spectrometer with parameters set as follows: gas temperature = 290 °C; gas flow = 14 liters/min; nebulizer pressure = 25 p.s.i.; sheath gas heater = 400 °C; sheath gas flow = 11 liters/min; capillary voltage = 3,000 V. Detection of Q8, Q8H2, and Q10 was by multiple-reaction monitoring (MRM) in positive ion mode using the above general MS parameters with fragmentor voltage at 380 V and cell accelerator voltage at 5 V. In each case, the fragment ions generated by collision-induced dissociation of the [M + H] were used for quantification. MRM settings for the target analytes were as follows (parent ion → fragment ion); Q8 (m/z 727.1 → 197.1) with collision energy (CE) = 33 V; Q8H2 (m/z 746.1 → 197.1) with CE = 33 V; and Q10 (m/z 863.6 → 197.1) with CE = 37 V. Q8 and Q10 were quantified against authentic commercial standards obtained from Avanti Polar Lipids (Alabaster, AL) and Sigma-Aldrich, respectively. The Q8H2 standard was generated from Q8 by a sodium borohydride–mediated reduction.
Preparation of Q-supplemented liposomes
Liposomes were prepared using an E. coli polar lipid extract (Avanti Polar Lipids), with or without Q8 (Avanti Polar Lipids) or Q10 (Sigma-Aldrich). Samples containing Q were covered to limit light exposure. E. coli polar lipid extract (100 mg of lipid in 4 ml of chloroform) was combined with the appropriate ubiquinone solution (in chloroform), the chloroform was removed by rotary evaporation at 37 °C, and the remaining lipid film was dried in a vacuum desiccator for 1 h. The lipid film was rehydrated in 10 ml of rehydration buffer (0.1 m K+-phosphate, pH 7.4, containing 0.5 mm K+-EDTA) by magnetic stirring for 1 h at room temperature. Aliquots (0.8 ml) of each liposome suspension were stored in liquid nitrogen (−196 °C). Before use, they were thawed at room temperature and extruded 21 times through a Whatman Nucleopore track-etched membrane (0.4-μm pore diameter, Millipore Sigma) using a 1-ml LiposoFast extruder (Avestin, Ottawa, Canada) at room temperature. For liposomes prepared by detergent dialysis, the rehydration buffer was supplemented with n-octyl-β-d-glucopyranoside (51.3 mm). Following rehydration, the detergent was removed by dialysis against detergent-free rehydration buffer (3 × 925 ml over 24 h). For liposomes prepared by freeze-thaw, the lipid suspension was rehydrated at 60 °C, subjected to 15 freeze-thaw cycles (freezing in liquid nitrogen for 5 min, thawing at 60 °C for 10 min), stored in liquid nitrogen, and thawed at 60 °C before extrusion.
Lipid analysis: Liposomes
Lipids were extracted from aqueous lipid dispersions essentially as described (63, 64). Briefly, liposome aliquots (0.16 ml) were transferred to 1.5 ml siliconized, snap-capped microcentrifuge tubes (Thermo Fisher Scientific) and combined with chloroform (0.2 ml) and methanol (0.4 ml). After incubation at room temperature for 15 min, the samples were centrifuged at 13,400 × g (5 min). Chloroform (0.2 ml) and rehydration buffer (0.2 ml) were added to achieve phase separation, the samples were centrifuged at 13,400 × g (5 min), and the two-phase solution was stored at 4 °C overnight.
The total lipid content was determined gravimetrically (65). Four aliquots (50 μl) of the organic phase were applied to tared, microaluminum weigh boats (13-mm diameter × 3.5-mm height, Cole-Parmer Canada Co., Montreal, Canada) with a Hamilton syringe and dried to constant weight at room temperature (they were dried under a gentle stream of air for 2 min and then placed under a vacuum for 2.5 h). Each boat was weighed with a Mettler AT250 analytical microbalance (Mettler Toledo, Mississauga, Canada). Two independent gravimetric analyses were conducted per liposome preparation.
The Q content of the lipid extracts was determined by HPLC on a Zorbax XDB-C18 (4.6 × 75 mm, 3.5 μm) reversed-phase column (Agilent Technologies) maintained at 25 °C with an absorbance detector. Four aliquots (20 μl) of the organic phase were injected and eluted isocratically with acetonitrile/tetrahydrofuran/water (55:40:5) at a flow rate of 1 ml min−1 (66). Oxidized Q8 and Q10 were detected at 275 nm (67) with retention times of 3.3 and 5.9 min, respectively. Reduced coenzyme Q8, which absorbs at 290 nm (67), was not detected in these samples. Q8 and Q10 were quantified by area comparison with authentic standards (31.2 ng to 2.5 μg Q) prepared as described (66). Peak fronting could be avoided by diluting samples containing more than 1 μg of Q, and it did not affect quantitation. Two independent HPLC analyses were conducted for each liposome preparation.
Diphenylhexatriene fluorescence anisotropy
Liposomes were labeled with DPH as described (15), with the following modifications. Following extrusion, liposomes (in rehydration buffer) were supplemented with 0.06% DPH (w/w, relative to lipid; ∼0.2 mol%) (Molecular Probes, Inc., Eugene, OR) and incubated for 18–20 h in the dark at room temperature. Fluorescence measurements were performed at the specified temperature using a PTI QuantaMaster QM-8 steady state fluorimeter as described (5). Duplicate measurements from two independent experiments were combined and analyzed as described (5). The data were analyzed by nonlinear regression with the polynomial quadratic function using SigmaPlot 12.5.
Transport assays
Bacteria were cultivated in NaCl-free MOPS medium, and initial rates of proline or lactose uptake by intact cells were determined as described previously (25). The expression of lacY was induced by adding IPTG (1 mm) to the final growth medium. Initial rates of proline and lactose uptake were measured with l-[14C(U)]proline (0.2 mm) (PerkinElmer Life Sciences) and d-[glucose-1-14C]lactose (0.2 mm) (Amersham Biosciences, Bath, UK), respectively. Each measurement was performed in triplicate with two different cell suspensions. The initial rates of substrate uptake as a function of the osmolality were analyzed by linear (LacY data) or nonlinear regression (ProP data) using SigmaPlot 12.5. The latter data were fit to Equation 1 (25),
| (Eq. 1) |
where Amax is the uptake rate that would be observed at high osmolality, R is the gas constant, T is the temperature, Π½/RT is the medium osmolality yielding half-maximal activity, and B is a constant inversely proportional to the slope of the response curve.
ProP-containing PRLs (0.2 mg/ml protein, 60 mg/ml E. coli lipid in 0.1 m potassium phosphate, pH 7.4, 0.5 mm potassium EDTA) were prepared as described (27). The PRLs were then fused with liposomes that were or were not supplemented with Q8. Extruded liposomes (450 μl, 10 mg/ml) were mixed with a quantity of PRLs containing the same quantity of lipid (75 μl). The mixture was centrifuged at 386,000 × g at 20 °C for 22 min. The pellet was resuspended in 150 μl of the same buffer and extruded as described above (see “Preparation of Q-supplemented liposomes”).
Initial rates of proline uptake by the PRLs were measured as described (25). Each assay was performed in duplicate, and two independent experiments were performed. The data were fit to Equation 1 by nonlinear regression as described above (25).
Oxygen uptake assays
Cell suspensions for oxygen uptake measurements were prepared as for transport assays, with the exception that bacteria from the 24-h LB culture were subcultured in both NaCl-free MOPS medium (the low-osmolality medium) and MOPS medium containing 450 mm NaCl or 800 mm sorbitol (the high-osmolality media). Wash and assay buffers were prepared to match the osmolalities of the growth media as described (25).
Oxygen uptake was measured essentially as described by McManus and Josephy (68). Assay buffer (2 ml) was added to the reaction chamber of a Clark oxygen electrode (Qubit Systems, Kingston, Canada). After an equilibration period (1 min), cell suspension (100 μl) was added to the chamber, and oxygen depletion within the chamber was monitored for 10 min. Measurements were performed at room temperature and in triplicate per treatment per sample. All experiments were repeated at least twice. Linear regression was used to determine the initial rate of oxygen depletion (corresponding to bacterial oxygen uptake) using the slope of the line between 1.5 and 4 min of the 10-min measurement period.
Trehalose assays
The trehalose contents of bacterial cells were determined by TLC as follows. Bacterial cultures were prepared as for oxygen uptake assays, and their optical densities (600 nm) were recorded. Bacteria were sedimented from a 24-ml aliquot of the final culture (10 min, 3,400 × g, room temperature) and resuspended in 1 ml of an unsupplemented MOPS medium adjusted with NaCl to attain the same osmolality as the growth medium. The suspension was transferred to a microcentrifuge tube, the cells were sedimented (2 min, 15,800 × g, room temperature), and they were resuspended in TCA (36 μl, 15% (w/v)) and incubated on ice for 10 min. The suspension was centrifuged (2 min, 15,800 × g), and aliquots (2 μl) of the supernatant were spotted on Silica Gel 60 F254 Micro TLC/AL plates (SiliCycle, Quebec City, Canada) with authentic trehalose standards (5, 10, 15, and 20 nmol). The plates were developed with the solvent system n-butanol/ethanol/water (5:3:2 by volume), dried, dipped in methanol/sulfuric acid (95:5, v/v), and charred at 150 °C until the spots appeared (less than 1 min). The quantities of material comigrating with authentic trehalose were estimated by densitometry using a Bio-Rad ChemiDoc XRS+ system and analyzed with ImageJ.
Protein assays
The protein concentrations of cell and PRL suspensions were determined by the bicinchoninic acid (BCA) assay (69) with the BCA kit from Pierce and by the Shaffner–Weissmann assay (70), respectively, with BSA as the protein standard.
Author contributions
L. T. and J. M. W. conceptualization; L. T., A. A., D. E. C., R. S., and J. M. W. formal analysis; L. T., A. A., D. E. C., R. S., and J. M. W. validation; L. T., A. A., D. E. C., R. S., and J. M. W. investigation; L. T., A. A., D. E. C., R. S., and J. M. W. visualization; L. T., A. A., D. E. C., R. S., and J. M. W. methodology; L. T., R. S., and J. M. W. writing-original draft; L. T., A. A., D. E. C., R. S., and J. M. W. writing-review and editing; R. S. and J. M. W. resources; R. S. and J. M. W. supervision; R. S. and J. M. W. funding acquisition; R. S. and J. M. W. project administration.
Supplementary Material
Acknowledgments
We are grateful to Ziqiang Guan (Duke University) for E. coli strain BKT12, to K. T. Shanmugam (University of Florida) for strains W3110 and JP20, to Katrina Edwards and Victor Agmo-Hernandez (Uppsala University) for sharing insights regarding ubiquinone incorporation into liposomes, to Peter Smith (University of Guelph) for access to equipment and assistance with oxygen uptake measurements, to Kevin Rea and Tariq Ahktar (University of Guelph) for access to equipment and assistance with ubiquinone analysis, and to Tariq Akhtar and Stephen Seah (University of Guelph) and Boris Martinac (Victor Chang Cardiac Research Institute) for helpful discussions of this research topic and our data.
This work was supported by Natural Sciences and Engineering Research Council of Canada Discovery Grant RGPIN-2017-05160 (to J. M. W.), Australian Research Council Grant DP170101453 (to R. S. and A. A.), and a Program and a Senior Principal Research Fellowship from the National Health and Medical Research Council of Australia (Grants 1052616 and 1111632) (to R. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S5.
- CL
- cardiolipin
- Π
- osmotic pressure
- A
- the initial rate of substrate (radiolabeled proline) uptake at a given osmolality
- Amax
- the maximum rate of proline uptake via ProP extrapolated to infinite osmolality
- B
- a constant inversely proportional to the slope of the osmolality response curve
- CE
- collision energy
- DPH
- diphenylhexatriene
- GB
- glycine betaine
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MRM
- multiple-reaction monitoring
- OD600
- optical density measured at a wavelength of 600 nm
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PRL
- proteoliposome
- Q
- ubiquinone
- Q8
- ubiquinone 8 or coenzyme Q8
- Q10
- ubiquinone 10 or coenzyme Q10
- LB
- Luria bertani.
References
- 1. Houssin C., Eynard N., Shechter E., and Ghazi A. (1991) Effect of osmotic pressure on membrane energy-linked functions in Escherichia coli. Biochim. Biophys. Acta 1056, 76–84 10.1016/S0005-2728(05)80075-1 [DOI] [PubMed] [Google Scholar]
- 2. Meury J. (1994) Immediate and transient inhibition of the respiration of Escherichia coli under hyperosmotic shock. FEMS Microbiol. Lett. 121, 281–286 10.1111/j.1574-6968.1994.tb07113.x [DOI] [PubMed] [Google Scholar]
- 3. Culham D. E., Romantsov T., and Wood J. M. (2008) Roles of K+, H+, H2O and ΔΨ in solute transport mediated by major facilitator superfamily members ProP and LacY. Biochemistry 47, 8176–8185 10.1021/bi800794z [DOI] [PubMed] [Google Scholar]
- 4. Wood J. M. (2011) Bacterial Osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65, 215–238 10.1146/annurev-micro-090110-102815 [DOI] [PubMed] [Google Scholar]
- 5. Culham D. E., Marom D., Boutin R., Garner J., Ozturk T. N., Sahtout N., Tempelhagen L., Lamoureux G., and Wood J. M. (2018) Dual role of the C-terminal domain in osmosensing by bacterial osmolyte transporter ProP. Biophys. J. 115, 2152–2166 10.1016/j.bpj.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maximov S., Ott V., Belkoura L., and Krämer R. (2014) Stimulus analysis of BetP activation under in vivo conditions. Biochim. Biophys. Acta 1838, 1288–1295 10.1016/j.bbamem.2013.12.017 [DOI] [PubMed] [Google Scholar]
- 7. van der Heide T., Stuart M. C. A., and Poolman B. (2001) On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 20, 7022–7032 10.1093/emboj/20.24.7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsatskis Y., Khambati J., Dobson M., Bogdanov M., Dowhan W., and Wood J. M. (2005) The osmotic activation of transporter ProP is tuned by both its C-terminal coiled-coil and osmotically induced changes in phospholipid composition. J. Biol. Chem. 280, 41387–41394 10.1074/jbc.M508362200 [DOI] [PubMed] [Google Scholar]
- 9. Sévin D. C., and Sauer U. (2014) Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 10, 266–272 10.1038/nchembio.1437 [DOI] [PubMed] [Google Scholar]
- 10. Arias-Cartin R., Grimaldi S., Arnoux P., Guigliarelli B., and Magalon A. (2012) Cardiolipin binding in bacterial respiratory complexes: structural and functional implications. Biochim. Biophys. Acta 1817, 1937–1949 10.1016/j.bbabio.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 11. Romantsov T., Stalker L., Culham D. E., and Wood J. M. (2008) Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J. Biol. Chem. 283, 12314–12323 10.1074/jbc.M709871200 [DOI] [PubMed] [Google Scholar]
- 12. Søballe B., and Poole R. K. (1999) Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145, 1817–1830 10.1099/13500872-145-8-1817 [DOI] [PubMed] [Google Scholar]
- 13. Eriksson E. K., Edwards K., Grad P., Gedda L., and Agmo Hernández V. (2019) Osmoprotective effect of ubiquinone in lipid vesicles modelling the E. coli plasma membrane. Biochim. Biophys. Acta Biomembr. 1861, 1388–1396 10.1016/j.bbamem.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 14. Eriksson E. K., Agmo Hernández V., and Edwards K. (2018) Effect of ubiquinone-10 on the stability of biomimetic membranes of relevance for the inner mitochondrial membrane. Biochim. Biophys. Acta Biomembr. 1860, 1205–1215 10.1016/j.bbamem.2018.02.015 [DOI] [PubMed] [Google Scholar]
- 15. Agmo Hernández V., Eriksson E. K., and Edwards K. (2015) Ubiquinone-10 alters mechanical properties and increases stability of phospholipid membranes. Biochim. Biophys. Acta 1848, 2233–2243 10.1016/j.bbamem.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 16. Katsikas H., and Quinn P. J. (1982) The polyisoprenoid chain length influences the interaction of ubiquinones with phospholipid bilayers. Biochim. Biophys. Acta 689, 363–369 10.1016/0005-2736(82)90270-X [DOI] [PubMed] [Google Scholar]
- 17. Gruszecki W. I., and Strzałka K. (2005) Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740, 108–115 10.1016/j.bbadis.2004.11.015 [DOI] [PubMed] [Google Scholar]
- 18. Van Gelder K., Rea K. A., Virta L. K. A., Whitnell K. L., Osborn M., Vatta M., Khozin A., Skorupinska-Tudek K., Surmacz L., and Akhtar T. A. (2018) Medium-chain polyprenols influence chloroplast membrane dynamics in Solanum lycopersicum. Plant Cell Physiol. 59, 2350–2365 10.1093/pcp/pcy157 [DOI] [PubMed] [Google Scholar]
- 19. Okada K., Kainou T., Tanaka K., Nakagawa T., Matsuda H., and Kawamukai M. (1998) Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur. J. Biochem. 255, 52–59 10.1046/j.1432-1327.1998.2550052.x [DOI] [PubMed] [Google Scholar]
- 20. Hsu A. Y., Poon W. W., Shepherd J. A., Myles D. C., and Clarke C. (1996) Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806 10.1021/bi9602932 [DOI] [PubMed] [Google Scholar]
- 21. Cardinale S., Joachimiak M. P., and Arkin A. P. (2013) Effects of genetic variation on the E. coli host-circuit interface. Cell Rep. 4, 231–237 10.1016/j.celrep.2013.06.023 [DOI] [PubMed] [Google Scholar]
- 22. Neidhardt F. C., Bloch P. L., and Smith D. F. (1974) Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milner J. L., Grothe S., and Wood J. M. (1988) Proline porter II is activated by a hyperosmotic shift in both whole cells and membrane vesicles of Escherichia coli K12. J. Biol. Chem. 263, 14900–14905 [PubMed] [Google Scholar]
- 24. Kaback H. R., and Guan L. (2019) It takes two to tango: the dance of the permease. J. Gen. Physiol. 151, 878–886 10.1085/jgp.201912377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Culham D. E., Henderson J., Crane R. A., and Wood J. M. (2003) Osmosensor ProP of Escherichia coli responds to the concentration, chemistry and molecular size of osmolytes in the proteoliposome lumen. Biochemistry 42, 410–420 10.1021/bi0264364 [DOI] [PubMed] [Google Scholar]
- 26. MacMillan S. V., Alexander D. A., Culham D. E., Kunte H. J., Marshall E. V., Rochon D., and Wood J. M. (1999) The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim. Biophys. Acta 1420, 30–44 10.1016/s0005-2736(99)00085-1 [DOI] [PubMed] [Google Scholar]
- 27. Racher K. I., Voegele R. T., Marshall E. V., Culham D. E., Wood J. M., Jung H., Bacon M., Cairns M. T., Ferguson S. M., Liang W.-J., Henderson P. J. F., White G., and Hallett F. R. (1999) Purification and reconstitution of an osmosensor: transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 38, 1676–1684 10.1021/bi981279n [DOI] [PubMed] [Google Scholar]
- 28. Racher K. I., Culham D. E., and Wood J. M. (2001) Requirements for osmosensing and osmotic activation of transporter ProP from Escherichia coli. Biochemistry 40, 7324–7333 10.1021/bi002331u [DOI] [PubMed] [Google Scholar]
- 29. Ondarroa M., Sharma S. K., and Quinn P. J. (1986) Solvation properties of ubiquinone-10 in solvents of different polarity. Biosci. Rep. 6, 783–796 10.1007/BF01117101 [DOI] [PubMed] [Google Scholar]
- 30. White G. F., Racher K. I., Lipski A., Hallett F. R., and Wood J. M. (2000) Physical properties of liposomes and proteoliposomes prepared from Escherichia coli polar lipids. Biochim. Biophys. Acta 1468, 175–186 10.1016/S0005-2736(00)00255-8 [DOI] [PubMed] [Google Scholar]
- 31. Jemiola-Rzeminska M., Kruk J., Skowronek M., and Strzalka K. (1996) Location of ubiquinone homologues in liposome membranes studied by fluorescence anisotropy of diphenyl-hexatriene and trimethylammonium-diphenyl-hexatriene. Chem. Phys. Lipids 79, 55–63 10.1016/0009-3084(95)02507-3 [DOI] [PubMed] [Google Scholar]
- 32. Lentz B. R. (1989) Membrane “fluidity” as detected by diphenylhexatriene probes. Chem. Phys. Lipids 50, 171–190 10.1016/0009-3084(89)90049-2 [DOI] [Google Scholar]
- 33. Galassi V. V., and Arantes G. M. (2015) Partition, orientation and mobility of ubiquinones in a lipid bilayer. Biochim. Biophys. Acta 1847, 1560–1573 10.1016/j.bbabio.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 34. Skowronek M., Jemioła-Rzemińska M., Kruk J., and Strzałka K. (1996) Influence of the redox state of ubiquinones and plastoquinones on the order of lipid bilayers studied by fluorescence anisotropy of diphenylhexatriene and trimethylammonium diphenylhexatriene. Biochim. Biophys. Acta 1280, 115–119 10.1016/0005-2736(95)00264-2 [DOI] [PubMed] [Google Scholar]
- 35. Cronan J. E. (2003) Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57, 203–224 10.1146/annurev.micro.57.030502.090851 [DOI] [PubMed] [Google Scholar]
- 36. Dowhan W. (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66, 199–232 10.1146/annurev.biochem.66.1.199 [DOI] [PubMed] [Google Scholar]
- 37. Neidhardt F. C., and Umbarger H. E. (1996) Chemical composition of Escherichia coli. in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C., ed) 2nd Ed., pp. 13–16, ASM Press, Washington, D. C. [Google Scholar]
- 38. Romantsov T., Helbig S., Culham D. E., Gill C., Stalker L., and Wood J. M. (2007) Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64, 1455–1465 10.1111/j.1365-2958.2007.05727.x [DOI] [PubMed] [Google Scholar]
- 39. Tan B. K., Bogdanov M., Zhao J., Dowhan W., Raetz C. R. H., and Guan Z. (2012) Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 10.1073/pnas.1212797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dinnbier U., Limpinsel E., Schmid R., and Bakker E. P. (1988) Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150, 348–357 10.1007/BF00408306 [DOI] [PubMed] [Google Scholar]
- 41. Murdock L., Burke T., Coumoundouros C., Culham D. E., Deutch C. E., Ellinger J., Kerr C. H., Plater S. M., To E., Wright G., and Wood J. M. (2014) Analysis of strains lacking known osmolyte accumulation mechanisms reveals contributions of osmolytes and transporters to abiotic stress protection. Appl. Environ. Microbiol. 80, 5366–5378 10.1128/AEM.01138-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strøm A. R., and Kaasen I. (1993) Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8, 205–210 10.1111/j.1365-2958.1993.tb01564.x [DOI] [PubMed] [Google Scholar]
- 43. Purvis J. E., Yomano L. P., and Ingram L. O. (2005) Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol. 71, 3761–3769 10.1128/AEM.71.7.3761-3769.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alemohammad M. M., and Knowles C. J. (1974) Osmotically induced volume and turbidity changes of Escherichia coli due to salts, sucrose and glycerol, with particular reference to the rapid permeation of glycerol into the cell. J. Gen. Microbiol. 82, 125–142 10.1099/00221287-82-1-125 [DOI] [PubMed] [Google Scholar]
- 45. Pilizota T., and Shaevitz J. W. (2012) Fast, multiphase volume adaptation to hyperosmotic shock by Escherichia coli. PLoS One 7, e35205 10.1371/journal.pone.0035205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Altendorf K., Booth I. R., Gralla J. D., Greie J.-C., Rosenthal A. Z., and Wood J. M. (2009) Osmotic stress. EcoSal Plus. 3, 10.1128/ecosalplus.5.4.5 [DOI] [PubMed] [Google Scholar]
- 47. Chazotte B., and Hackenbrock C. R. (1989) Lateral diffusion as a rate-limiting step in ubiquinone-mediated mitochondrial electron transport. J. Biol. Chem. 264, 4978–4985 [PubMed] [Google Scholar]
- 48. Hackenbrock C. R., Chazotte B., and Gupte S. S. (1986) The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J. Bioenerg. Biomembr. 18, 331–368 10.1007/BF00743010 [DOI] [PubMed] [Google Scholar]
- 49. Magalon A., and Alberge F. (2016) Distribution and dynamics of OXPHOS complexes in the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 1857, 198–213 10.1016/j.bbabio.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 50. Sousa P. M. F., Videira M. A. M., Bohn A., Hood B. L., Conrads T. P., Goulao L. F., and Melo A. M. P. (2012) The aerobic respiratory chain of Escherichia coli: from genes to supercomplexes. Microbiology 158, 2408–2418 10.1099/mic.0.056531-0 [DOI] [PubMed] [Google Scholar]
- 51. Chance B., and Williams G. R. (1955) A method for the localization of sites for oxidative phosphorylation. Nature 176, 250–254 10.1038/176250a0 [DOI] [PubMed] [Google Scholar]
- 52. Sharma P., Teixeira de Mattos M. J., Hellingwerf K. J., and Bekker M. (2012) On the function of the various quinone species in Escherichia coli. FEBS J. 279, 3364–3373 10.1111/j.1742-4658.2012.08608.x [DOI] [PubMed] [Google Scholar]
- 53. Sousa P. M. F., Silva S. T. N., Hood B. L., Charro N., Carita J. N., Vaz F., Penque D., Conrads T. P., and Melo A. M. P. (2011) Supramolecular organizations in the aerobic respiratory chain of Escherichia coli. Biochimie 93, 418–425 10.1016/j.biochi.2010.10.014 [DOI] [PubMed] [Google Scholar]
- 54. Genova M. L., and Lenaz G. (2014) Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta 1837, 427–443 10.1016/j.bbabio.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 55. Wallace B. J., and Young I. G. (1977) Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA− menA− double quinone mutant. Biochim. Biophys. Acta 461, 84–100 10.1016/0005-2728(77)90071-8 [DOI] [PubMed] [Google Scholar]
- 56. Loiseau L., Fyfe C., Aussel L., Hajj Chehade M., Hernández S. B., Faivre B., Hamdane D., Mellot-Draznieks C., Rascalou B., Pelosi L., Velours C., Cornu D., Lombard M., Casadesús J., Pierrel F., Fontecave M., and Barras F. (2017) The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ. J. Biol. Chem. 292, 11937–11950 10.1074/jbc.M117.789164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Clarke C. F., Rowat A. C., and Gober J. W. (2014) Is CoQ a membrane stabilizer? Nat. Chem. Biol. 10, 242–243 10.1038/nchembio.1478 [DOI] [PubMed] [Google Scholar]
- 58. Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 59. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 60. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown E. D., and Wood J. M. (1992) Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J. Biol. Chem. 267, 13086–13092 [PubMed] [Google Scholar]
- 62. Miller J. H. (1972) Experiments in Molecular Genetics, p. 433, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 63. Kates M., Kushwaha S., and Sprott G. (1982) Lipids of purple membrane from extreme halophiles and of methanogenic bacteria. Methods Enzymol. 88, 98–111 10.1016/0076-6879(82)88016-6 [DOI] [Google Scholar]
- 64. Garrett T. A., Guan Z., and Raetz C. R. (2007) Analysis of ubiquinones, dolichols, and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 432, 117–143 10.1016/S0076-6879(07)32005-3 [DOI] [PubMed] [Google Scholar]
- 65. Caputo G. A., and London E. (2013) Analyzing transmembrane protein and hydrophobic helix topography by dual fluorescence quenching. Methods Mol. Biol. 974, 279–295 10.1007/978-1-62703-275-9_13 [DOI] [PubMed] [Google Scholar]
- 66. Orozco D., Skamarack J., Reins K., Titlow B., Lunetta S., Li F., and Roman M. (2007) Determination of ubidecarenone (coenzyme Q10, ubiquinol-10) in raw materials and dietary supplements by high-performance liquid chromatography with ultraviolet detection: single-laboratory validation. J. AOAC Int. 90, 1227–1236 [PMC free article] [PubMed] [Google Scholar]
- 67. Crane F. L., Lester R. L., Widmer C., and Hatefi Y. (1959) Studies on the electron transport system. XVIII. Isolation of coenzyme Q (Q275) from beef heart and beef heart mitochondria. Biochim. Biophys. Acta 32, 73–79 10.1016/0006-3002(59)90554-2 [DOI] [PubMed] [Google Scholar]
- 68. McManus D. C., and Josephy P. D. (1993) A new approach to measurement of redox-cycling activity in Escherichia coli. Arch. Biochem. Biophys. 304, 367–370 10.1006/abbi.1993.1363 [DOI] [PubMed] [Google Scholar]
- 69. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- 70. Schaffner W., and Weissmann C. (1973) Rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56, 502–514 10.1016/0003-2697(73)90217-0 [DOI] [PubMed] [Google Scholar]
- 71. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guyer M. S., Reed R. R., Steitz J. A., and Low K. B. (1981) Identification of a sex-factor-affinity site in E. coli as γδ. Cold Spring Harb. Symp. Quant. Biol. 45, 135–140 10.1101/SQB.1981.045.01.022 [DOI] [PubMed] [Google Scholar]
- 73. Jensen K. F. (1993) The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175, 3401–3407 10.1128/jb.175.11.3401-3407.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Culham D. E., Lasby B., Marangoni A. G., Milner J. L., Steer B. A., van Nues R. W., and Wood J. M. (1993) Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 229, 268–276 10.1006/jmbi.1993.1030 [DOI] [PubMed] [Google Scholar]
- 75. Culham D. E., Tripet B., Racher K. I., Voegele R. T., Hodges R. S., and Wood J. M. (2000) The role of the carboxyl terminal α-helical coiled-coil domain in osmosensing by transporter ProP of Escherichia coli. J. Mol. Recognit. 13, 309–322 [DOI] [PubMed] [Google Scholar]
- 76. Guzman L.-M., Belin D., Carson M. J., and Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Romantsov T., Gonzalez K., Sahtout N., Culham D. E., Coumoundouros C., Garner J., Kerr C. H., Chang L., Turner R. J., and Wood J. M. (2018) Cardiolipin synthase A colocalizes with cardiolipin and osmosensing transporter ProP at the poles of Escherichia coli cells. Mol. Microbiol. 107, 623–638 10.1111/mmi.13904 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.