Abstract
Phosphoinositides play crucial roles in intracellular membrane dynamics and cell signaling, with phosphatidylinositol (PI) 3-phosphates being the predominant phosphoinositide lipids at endosomes and lysosomes, whereas PI 4-phosphates, such as phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), are enriched at the cell surface including sites of endocytosis. How PI 4-phosphates and PI 3-phosphates are dynamically interconverted within the endocytic pathway and how this is controlled in space and time remains poorly understood. Here, combining live imaging, genome engineering, and acute chemical and genetic manipulations, we found that local synthesis of PI(3,4)P2 by phosphatidylinositol 3-kinase C2α at plasma membrane clathrin-coated pits is spatially segregated from its hydrolysis by the PI(3,4)P2-specific inositol polyphosphate 4-phosphatase 4A (INPP4A). We observed that INPP4A is dispensable for clathrin-mediated endocytosis and is undetectable in endocytic clathrin-coated pits. Instead, we found that INPP4A partially localizes to endosomes and that loss of INPP4A in HAP1 cancer cells perturbs signaling via AKT kinase and mTOR complex 1. These results reveal a function for INPP4-mediated PI(3,4)P2 hydrolysis in local regulation of growth factor and nutrient signals at endosomes in cancer cells. They further suggest a model whereby synthesis and turnover of PI(3,4)P2 are spatially segregated within the endocytic pathway to couple endocytic membrane traffic to growth factor and nutrient signaling.
Keywords: clathrin, endocytosis, phosphoinositide, phosphatidylinositol kinase (PI kinase), phosphatidylinositol phosphatase, endosome, lysosome, mTOR complex (mTORC), imaging, nutrient signaling
Introduction
Phosphoinositides are a minor class of comparably short-lived membrane phospholipids that mediate crucial cellular and organismal functions, including cell signaling and regulation of intracellular membrane traffic (1, 2). The different PI3 species differ regarding their subcellular localization, with PI 4-phosphates being concentrated in the exocytic pathway and at the plasma membrane and PI 3-phosphates, such as phosphatidylinositol 3-phosphate (PI(3)P), predominantly found at endosomes and lysosomes (1, 2). Hence, as membranes are internalized from the plasma membrane, they have to undergo conversion of their phosphoinositide content from PI 4-phosphates, in particular PI(4,5)P2, to PI 3-phosphates, e.g. PI(3)P, the major signature lipid of endosomes and lysosomes (3).
How this phosphoinositide conversion occurs and how it is spatiotemporally controlled is unclear. Prior studies have established that PI(4,5)P2, a lipid required for endocytic clathrin-coated pit (CCP) formation (4), is hydrolyzed during late stages of CME by PI(4,5)P2 5-phosphatases (5–7). Work by several laboratories has shown that loss of PI(4,5)P2 as CCPs mature is accompanied by clathrin-mediated recruitment of phosphatidylinositol 3-kinase C2α (PI3KC2α) (8, 9), a class II PI 3-kinase family member implicated in human disease (10) that may synthesize PI(3,4)P2 (9) and/or PI(3)P, possibly depending on cell type and its nanoscale localization (11, 12). Depletion of PI3KC2α stalls CCP dynamics and results in cell surface accumulation of nonconstricted U-shaped intermediates of CME (i.e. prior to membrane scission by dynamin), a phenotype rescued by active WT but not kinase-inactive mutant PI3KC2α (9), indicating that PI 3-phosphate synthesis by PI3KC2α is required for CCP maturation and membrane constriction. Conflicting models have been postulated regarding the exact nature of the lipid product synthesized by PI3KC2α during CME. Based on the activity of native PI3KC2α immunoprecipitated from mammalian cells in combination with knockdown/rescue experiments and analysis of the cellular PI 3-phosphate content, it has been postulated that PI3KC2α specifically synthesizes PI(3,4)P2 at endocytic CCPs (9) to facilitate CCP membrane constriction via lipid-mediated recruitment of the dynamin-associated Phox homology-Bin-amphiphysin-rvs domain protein sorting nexin 9 (SNX9 and its close homolog SNX18) to the invagination neck (13). Further support for this model (Fig. 1A, model I) derives from the fact that loss of SNX9/18 or mutation of its PI 3-phosphate–binding PX domain phenocopies loss of PI3KC2α with respect to CME (13, 14). Moreover, re-expression of hyperactive PI3KC2α in cells depleted of the endogenous enzyme results in elevated plasma membrane PI(3,4)P2 levels and facilitates CME in living cells (15), suggesting a tight relationship between local PI(3,4)P2 synthesis by PI3KC2α and the efficiency of CME.
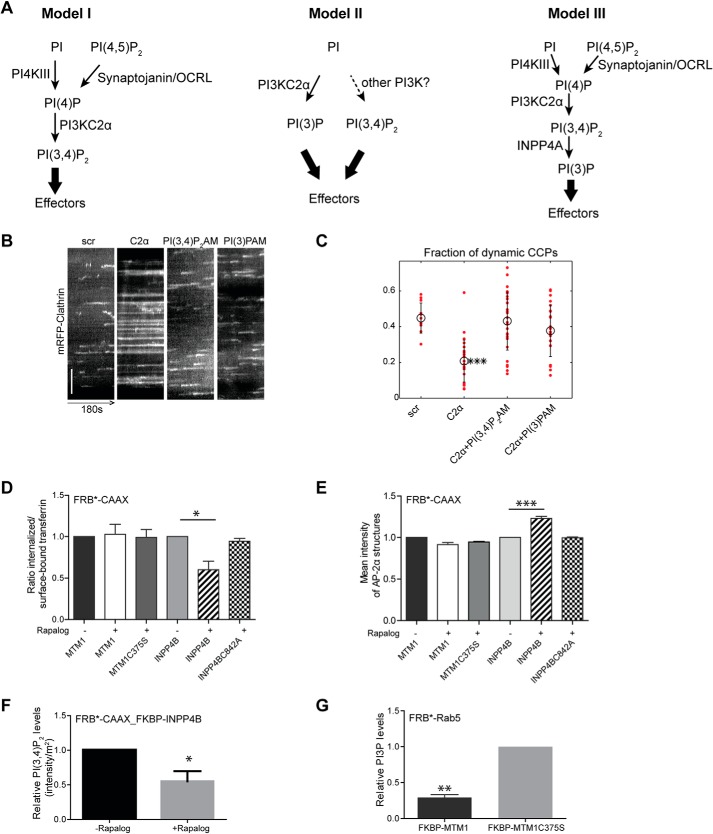
Figure 1.
Acute manipulation of PI 3-phosphates reveals an endocytic function of PI(3,4)P2. A, proposed models of phosphoinositide conversion in CME. Model I, CCP maturation is accompanied by lipid conversion from PI(4,5)P2 to PI(4)P PI(3,4)P2 generated by PI3KC2α. PI(3,4)P2 can recruit effector proteins (e.g. SNX9/18) to facilitate CCP maturation. Model II, PI3KC2α synthesizes PI(3)P at the plasma membrane, alleviating the need for PI(3,4)P2 4-phosphatases. Effector proteins may bind to PI(3)P to facilitate CCP maturation. The kinase responsible for PI(3,4)P2 synthesis has not been identified in this study (16). Model III, PI(3,4)P2 synthesized locally by PI3KC2α is converted rapidly to PI(3)P at CCPs by the PI(3,4)P2-specific 4-phosphatase INPP4A, resulting in generation of PI(3)P. Effector proteins may bind to PI(3)P to facilitate CCP maturation. B and C, supply of exogenous PI(3,4)P2/AM or PI(3)P/AM rescues stalled CCP dynamics in PI3KC2α-depleted cells. Kymographs of COS7 cells expressing mRFP-clathrin fluorescence over 180 s by TIRF imaging are shown in B. y axis scale bar = 5 μm. Scr, scrambled. C, fraction of dynamic CCPs analyzed by quantitative automated 2D tracking. Mean ± S.E., *** p < 0.001. D, acute depletion of PI(3,4)P2 by rapalog-induced recruitment of INPP4B impairs transferrin CME. Depletion of a postulated plasma membrane PI(3)P pool by rapalog-induced recruitment of the PI(3)P 3-phosphatase MTM1 had no effect. Phosphatase inactive variants of INPP4B (C842A) or MTM1 (C375S) did not impair transferrin-CME. Bar diagrams represent the ratio of internalized (10 min, 37 °C) to surface transferrin (45 min, 4 °C). Mean ± S.E., n = 3 independent experiments. 790, 890, 916, 444, 332, and 1097 cells from three independent experiments were analyzed. *, p < 0.05; one-way ANOVA. E, accumulation of AP-2α–containing CCPs in PI(3,4)P2-depleted cells. Bar diagrams represent the mean intensity of endocytic AP-2α–containing CCPs. Mean ± S.E.; n = 3 independent experiments. 179, 158, 119, 109, 119, and 116 cells from three independent experiments were analyzed. ***, p < 0.001; one-way ANOVA. F, rapalog-induced recruitment of INPP4B to the plasma membrane results in decreased PI(3,4)P2 levels. Mean ± S.E, n = 3 independent experiments. 69 cells were analyzed from three independent experiments for each condition. *, p < 0.05; unpaired t test. G, rapalog-induced endosomal recruitment of WT but not inactive (C375S) MTM1 depletes PI(3)P from endosomes. Mean ± S.E., n = 3 independent experiments. 54 and 64 cells were analyzed. **, p < 0.01; unpaired t test.
A recent imaging study based on composite protein/lipid-binding probes derived from the clathrin uncoating factor auxilin, a protein that acts post-scission in CME, proposed a different model for endocytic phosphoinositide conversion. According to this model (Fig. 1A, model II), short bursts of PI(3)P and PI(3,4)P2 are observed only after vesicle scission, with PI3KC2α responsible for the majority of the late PI(3)P signal (16), whereas PI(3,4)P2 may be synthesized by another enzyme, conceivably a class I PI 3-kinase. Although PI3KC2α, in principle, is capable of synthesizing PI(3)P, e.g. on endosomes (17, 18), this model conflicts with the accumulation of nonconstricted CCPs, i.e. a step prior to scission, in PI3KC2α-depleted cells or cells from PI3KC2α knockout mice (9). Finally, based on in vitro studies using liposomes, it has been proposed that PI3KC2α indeed produces PI(3,4)P2 at CCPs, but this lipid may be rapidly converted to PI(3)P by the PI(3,4)P2-specific inositolpolyphosphate 4-phosphatase INPP4A, an enzyme previously localized to endosomes (19). This PI(3)P pool then serves to recruit SNX9 to late-stage CCPs (Fig. 1A, model III), where it may facilitate assembly of F-actin (20).
Whether PI(3,4)P2-to-PI(3)P conversion can occur at the plasma membrane or at endosomes in vivo is unclear but of key physiological importance, as distinct PI 3-phosphates differentially control endolysosomal sorting and signaling functions in cells. For example, PI(3)P has been shown to activate mTORC1 (21), promote kinesin 1–mediated dispersion of late endosomes or lysosomes to the cell periphery (22), and foster endosomal membrane recruitment of the PI(3,4,5)P3/PI(3,4)P2 3-phosphatase PTEN (23). Conversely, local synthesis of PI(3,4)P2 at late endosomes or lysosomes by PI3KC2β, a close relative of PI3KC2α activated under conditions of growth factor deprivation (24), facilitates the perinuclear clustering of lysosomes and represses mTORC1 signaling (24, 25).
In this study, we combine live imaging, genome engineering, and acute chemical and genetic manipulations to reveal that local synthesis of PI(3,4)P2 by PI3KC2α at CCPs is spatially segregated from its hydrolysis at endosomes by INPP4A to locally regulate cell signaling via Akt and mTORC1.
Results
Acute manipulation of PI 3-phosphates reveals an endocytic function for PI(3,4)P2
A distinguishing feature of the various proposed models of PI3KC2α-mediated PI 3-phosphate generation in CME is their dependence on PI(3,4)P2 (Fig. 1A, model I) versus PI(3)P (Fig. 1A, models II and III). We first probed whether PI(3,4)P2 and/or PI(3)P are sufficient to restore defective CME in the absence of endogenous PI3KC2α. We generated a novel set of membrane-permeant lipid tools comprising acetoxymethylester (AM)–protected variants of PI(3)P and PI(3,4)P2 carrying long-chain fatty acids that, in addition, also harbored a cross-linkable diazirine and an alkyne group not used here (the synthesis of membrane-permeant diazirine- and alkyne-containing PI(3,4)P2 and PI(3)P lipids will be described in detail elsewhere)4 (Fig. S1A). Upon cell entry, AM-derivatized PI(3)P and PI(3,4)P2 are deprotected by cellular esterases, resulting in release of the unprotected lipid (26, 27). When we analyzed the lifetimes of CCPs by total internal reflection (TIRF) microscopy combined with automated particle tracking (28), we observed a striking accumulation of long-lived static endocytic structures and a corresponding reduction in productive dynamic CCPs in COS7 cells depleted of endogenous PI3KC2α, in agreement with previous data (9, 15). Membrane-permeant PI(3)P and PI(3,4)P2 were equally effective in restoring defective CCP dynamics in PI3KC2α-depleted cells (Fig. 1, B and C). Hence, CCP maturation, in principle, can be driven by either PI(3)P or PI(3,4)P2, consistent with the fact that the PI3KC2α effector SNX9/18 binds to PI(3)P and PI(3,4)P2 with equal affinity and via the same site (13, 14, 29). To probe whether PI(3)P and/or PI(3,4)P2 are also required for CME, we acutely depleted either of these PI 3-phosphates from the plasma membrane via rapalog-induced recruitment of the PI(3)P 3-phosphatase MTM1 (1, 30, 31) or the PI(3,4)P2-specific 4-phosphatase INPP4B (1, 9, 32) to the cell surface. Acute depletion of plasma membrane PI(3,4)P2 by active WT INPP4B (Fig. 1F and Fig. S1B) impaired CME, as evidenced by accumulation of the endocytic clathrin adaptor AP-2 in enlarged static CCPs at the cell surface (Fig. 1D) and a corresponding reduction in CME of transferrin (Fig. 1E). Catalytically inactive INPP4B affected neither PI(3,4)P2 levels nor CME. Depletion of a postulated (16) cell surface pool of PI(3)P (not detectable by conventional PI(3)P probes such as 2× FYVE [Fab 1 (yeast orthologue of PIKfyve), YOTB, Vac 1 (vesicle transport protein), and EEA1] in our hands) by rapalog-induced plasma membrane recruitment of active MTM1 did not affect CME (Fig. 1, D and E), although endosomal PI(3)P was effectively eliminated by MTM1 targeting to Rab5-positive endosomes (Fig. 1G and Fig. S1C) (30), where most intracellular PI(3)P resides. Similar observations were made previously upon constitutive targeting of INPP4B or MTM1 to the plasma membrane (9). These experiments thus suggest that CME is facilitated by a plasma membrane–localized endocytic pool of PI(3,4)P2.
CCP dynamics are controlled by synthesis of PI(3,4)P2 mediated by PI3KC2α
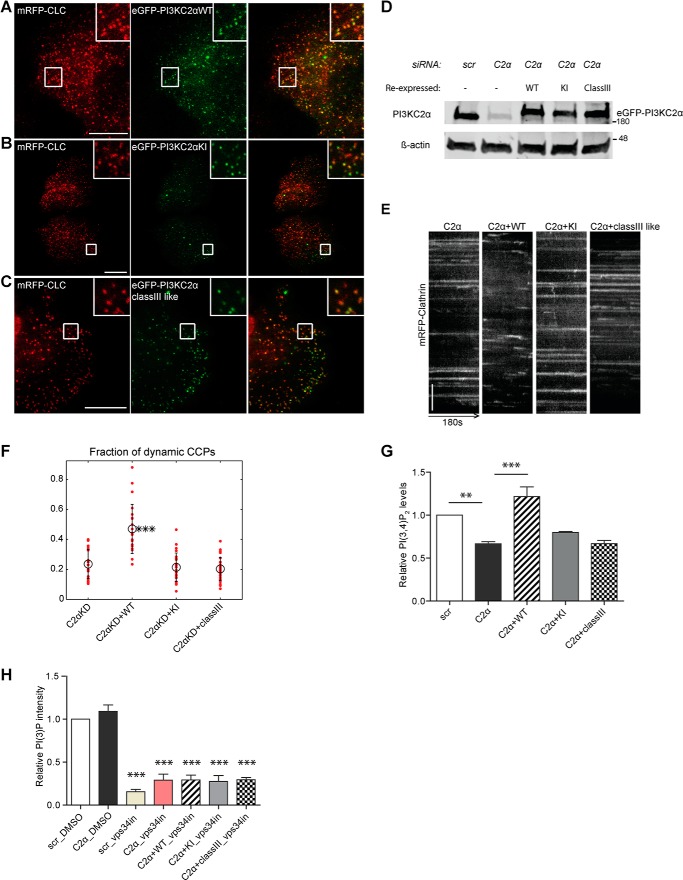
To challenge these data using an independent approach, we analyzed the ability of WT and different mutants forms of PI3KC2α to rescue defective CME in cells lacking the endogenous enzyme. PI3KC2α-depleted COS7 cells were transfected with plasmids encoding eGFP-tagged WT PI3KC2α, a kinase-inactive mutant of PI3KC2α, or a re-engineered class III–like variant carrying the substrate-binding loop of the PI(3)P-selective PI 3-kinase Vps34 that synthesizes PI(3)P but is unable to generate PI(3,4)P2, as demonstrated earlier (9). As PI3KC2α, in addition to its role in endocytosis, also regulates endosomal recycling of transferrin (17), the effects of PI3KC2α manipulation on CME cannot simply be monitored by measuring the efficiency of transferrin uptake in genetic knockdown/rescue experiments. We therefore analyzed the effect of the different PI3KC2α variants on CCP dynamics as a direct readout of its role in regulating CME. The WT and kinase-inactive and class III–like mutants were correctly targeted to endocytic CCPs (Fig. 2, A–C) and expressed at similar levels (Fig. 2D). The accumulation of long-lived static endocytic CCPs in COS7 cells depleted of endogenous PI3KC2α was effectively rescued by re-expression of siRNA-resistant WT PI3KC2α but not by kinase-inactive or class III-like mutant PI3KC2α (Fig. 2, E and F). Moreover, WT but not kinase-inactive or class III–like mutant PI3KC2α restored the partial depletion of plasma membrane PI(3,4)P2 in PI3KC2α knockdown COS7 cells (Fig. 2G), suggesting a tight correlation between PI(3,4)P2 synthesis by PI3KC2α and CCP dynamics. Importantly, class III–like mutant PI3KC2α was perfectly able to restore defects in transferrin recycling (17) or sonic hedgehog signaling from Rab8-positive endosomes (18) via endosomal synthesis of PI(3)P. This pool of PI(3)P is likely small and/or spatiotemporally restricted (at least in COS7 cells), as we failed to detect PI3KC2α-mediated PI(3)P synthesis in living COS7 cells either under control conditions or upon selective inhibition of Vps34 (Fig. 2H and Fig. S2), the major PI(3)P-synthesizing enzyme in eukaryotic cells. Collectively, these data indicate that CCP dynamics and, therefore, CME are regulated by local synthesis of PI(3,4)P2 by PI3KC2α.
Figure 2.
CCP dynamics are controlled by synthesis of PI(3,4)P2 mediated by PI3KC2α. A–C, TIRF microscopy snapshots of living COS7 cells, illustrating recruitment of WT or mutant variants of PI3KC2α to endocytic CCPs labeled by mRFP-clathrin light chain (CLC). COS7 cells depleted of endogenous PI3KC2α were transfected with plasmids encoding siRNA-resistant WT (A), kinase-inactive (KI, B), or class III–like mutant (class III–like, C; i.e. PI3KC2α that can only produce PI(3)P but not PI(3,4)P2) eGFP-PI3KC2α. Scale bar = 10 μm. D, immunoblot illustrating equal expression of WT and mutant variants of PI3KC2α in COS7 cells depleted of endogenous PI3KC2α. β-Actin was analyzed as a loading control. Numbers indicate the molecular mass of the immunoblotted proteins in kilodaltons. Scr, scrambled. E and F, rescue of defective CCP dynamics by WT but not kinase-inactive or class III–like mutant eGFP-PI3KC2α expressed in PI3KC2α-depleted cells. E, representative kymographs of mRFP-clathrin puncta (180 s) in PI3KC2α-depleted COS7 cells expressing WT or mutant variants of PI3KC2α. y axis scale bar = 5 μm. F, fraction of dynamic CCPs analyzed by quantitative automated 2D tracking. G and H, PI(3,4)P2 and PI(3)P levels in PI3KC2α-depleted COS7 cells re-expressing siRNA-resistant WT, kinase-inactive, or class III–like mutant eGFP-PI3KC2α. Vps-34 inhibitor (10 μm) was added to the indicated samples to inhibit PI(3)P synthesis by Vps34 to reveal possible pools of PI(3)P generated by re-expressed eGFP-PI3KC2α variants. Mean ± S.E., n = 3 independent experiments. More than 66 cells for each condition from three independent experiments were analyzed. **, p < 0.01; ***, p < 0.0001; one-way ANOVA.
The PI(3,4)P2-specific 4-phosphatase INPP4 is absent from CCPs and dispensable for CME
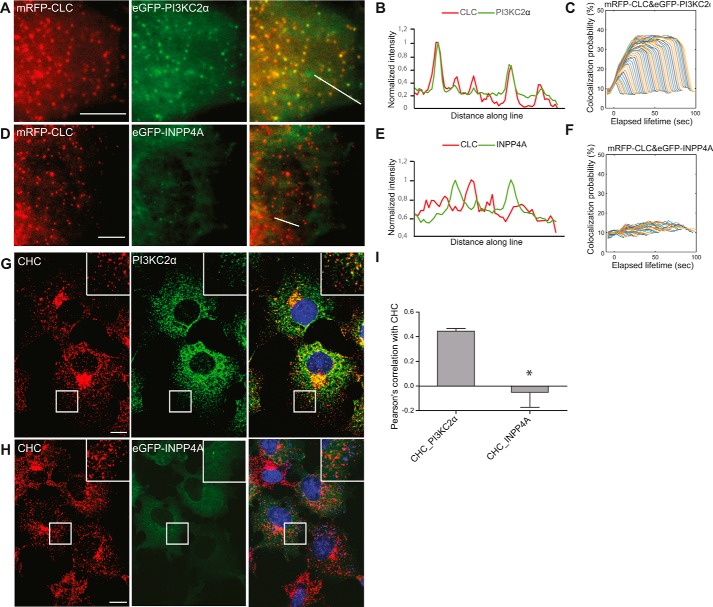
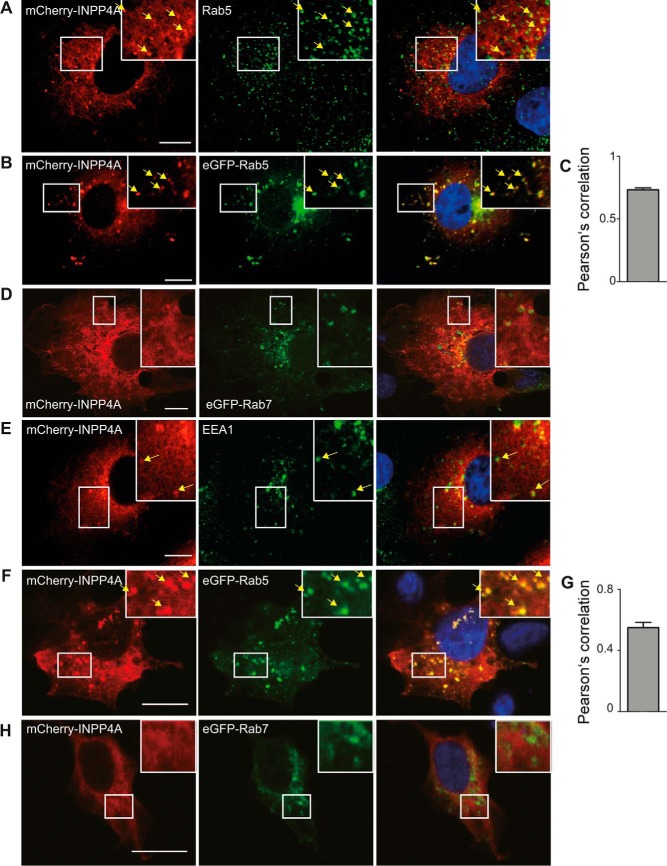
Our data are incompatible with PI3KC2α-mediated PI(3)P synthesis (Fig. 1A, model II) at CCPs, in agreement with a recent study (33), indicating that the phosphoinositide sensors generated by He et al. (16) may be nonspecific. However, they do not distinguish between a direct role of PI(3,4)P2 in recruitment of endocytic effector proteins such as SNX9/18 (Fig. 1A, model I) and an indirect role of PI(3,4)P2 as a substrate for the PI(3,4)P2-specific 4-phosphatase INPP4A to locally produce PI(3)P at CCPs, which then recruits SNX9/18 (Fig. 1A, model III). The latter mechanism requires INPP4A to act locally at CCPs to generate PI(3)P from PI(3,4)P2. It further predicts that loss of INPP4A should phenocopy endocytic defects seen in cells lacking either PI3KC2α or SNX9/18. To test these predictions, we monitored the localization and dynamics of eGFP-INPP4A or eGFP-PI3KC2α in living COS7 cells coexpressing monomeric red fluorescent protein-clathrin light chain by two-color TIRF imaging with automated particle tracking. PI3KC2α displayed striking colocalization with mRFP-clathrin light chain across all lifetime cohorts, in agreement with its key regulatory role in CME (Fig. 3, A–C). In striking contrast, INPP4A was largely cytoplasmic and absent from CCPs, with no correlation to clathrin recruitment (Fig. 3, D–F). eGFP-PI3KC2α also strongly colocalized with endogenous clathrin at endocytic CCPs in fixed cells (Fig. 3, G and I), whereas eGFP-INPP4A was absent from endocytic clathrin puncta (Fig. 3, H and I). Instead, we found that, in COS7 cells, a fraction of fluorescent protein–tagged INPP4A colocalized with markers of early endosomes, such as endogenous Rab5 or EEA1 (Fig. 4, A and E), whereas colocalization with late endosomal LAMP1 or endogenous Rab7 (Fig. S3) was less overt. Overexpression of eGFP-Rab5 facilitated recruitment of INPP4A to early endosomes in transfected COS7 (Fig. 4, B and C) or HAP1 cancer cells (Fig. 4, F and G). In contrast, overexpression of eGFP-Rab7 did not affect INPP4A localization in either COS7 (Fig. 4D) or HAP1 cells (Fig. 4H). These data indicate that INPP4A is absent from CCPs and instead localizes, at least in part, to early endosomes, consistent with earlier data (19).
Figure 3.
The PI(3,4)P2 4-phosphatase INPP4A is absent from CCPs. A and D, representative TIRF images of living COS7 cells expressing mRFP-CLC together with eGFP-PI3KC2α (A) or eGFP-INPP4A (D). In contrast to eGFP-PI3KC2α, eGFP-INPP4A did not detectably colocalize with clathrin. Scale bars = 5 μm. B and E, fluorescence intensity profiles of mRFP-CLC (red) and eGFP-PI3KC2α or eGFP-INPP4A (green), measured along the lines drawn in A and D, respectively. C and F, intensity profiles of PI3KC2α (C) and INPP4A (F) at CCPs analyzed by automated 2D tracking across as a function of CCP lifetime. G and H, confocal images of COS7 cells immunostained for clathrin heavy chain (CHC) and endogenous PI3KC2α (G) or expressed eGFP-INPP4A (H). Note the strong colocalization of PI3KC2α but not INPP4A with clathrin. Scale bars = 10 μm. I, Pearson's correlation coefficients determined from two-color confocal microscopy imaging of clathrin with PI3KC2α or INPP4A. Mean ± S.E., n = 3 independent experiments. 72 and 71 cells from three independent experiments were analyzed. *, p < 0.05, unpaired t test.
Figure 4.
INPP4A partially localizes to EEA1- and Rab5-containing endosomes. A, B, D, and E, confocal images of COS7 cells transiently expressing mCherry-INPP4A immunostained for endogenous Rab5 (A), overexpressed eGFP-Rab5 (B), overexpressed eGFP-Rab7 (D), or endogenous EEA1 (E). Note the partial colocalization of mCherry-INPP4A with the early endosome markers Rab5 and EEA1. Scale bars = 10 μm. C and G, Pearson's correlation coefficients determined from two-color confocal microscopy imaging of COS7 (C) and HAP1 (G) cells expressing mCherry-INPP4A and eGFP-Rab5. Mean ± S.E.; 61 and 46 cells were analyzed. F and H, confocal images of HAP1 cells transiently expressing mCherry-INPP4A and eGFP-Rab5 (F) or eGFP-Rab7 (H). Scale bars = 10 μm.
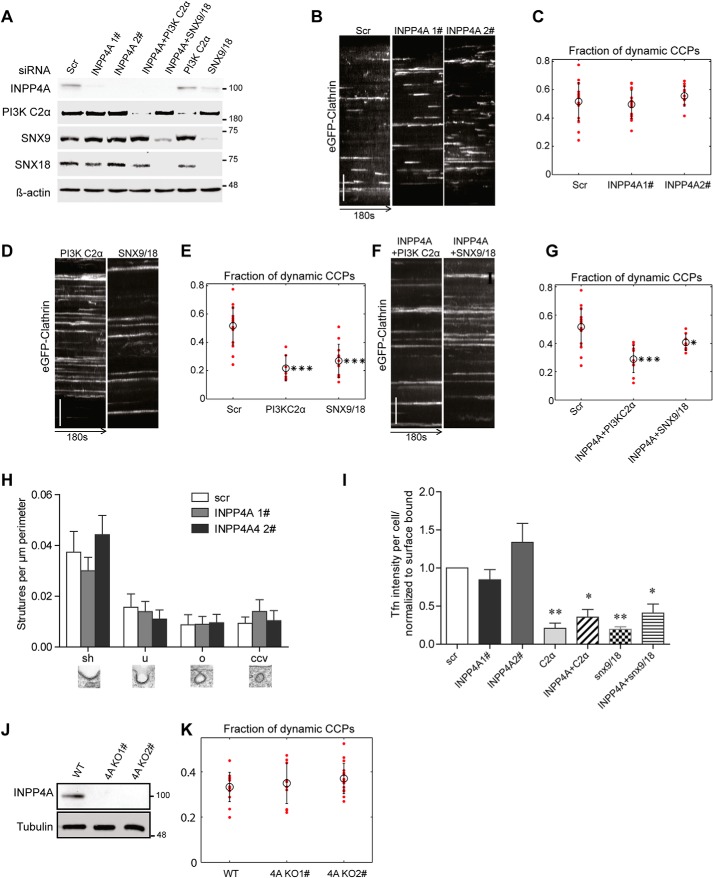
It is possible that INPP4A functionally regulates CME without being enriched at CCPs. Hence, we analyzed the consequences of INPP4A depletion on CCP dynamics and ultrastructure in COS7 cells, which lack expression of INPP4B (Fig. S4E), the only other INPP4 family member in mammals. Depletion of INPP4A by either of two distinct siRNAs (Fig. 5A) had no effect on the dynamics of CCPs (Fig. 5, B and C), in contrast to the accumulation of static AP-2–enriched CCPs observed in COS7 cells depleted of either PI3KC2α or SNX9/18 (Fig. 5, D and E, and Fig. S4A). Double loss of INPP4A and either PI3KC2α or SNX9/18 impaired CCP dynamics (Fig. 5, F and G, and S4A), akin to single loss of PI3KC2α or SNX9/18. These data suggest that PI3KC2α and SNX9/18 act upstream of INPP4A within the endocytic pathway. Consistent with our results regarding CCP dynamics, depletion of INPP4A also did not affect the ultrastructure or number of endocytic intermediates (Fig. 5H) or the efficiency of transferrin-CME (Fig. 5I). To corroborate these data from siRNA knockdown experiments, we generated INPP4A KO clones using CRISPR/Cas9 (Fig. 5J). INPP4A KO COS7 cells were indistinguishable from WT cells with respect to CCP dynamics (Fig. 5, J and K) or plasma membrane AP-2 levels (Fig. S4B).
Figure 5.
INPP4A is dispensable for CME. A, immunoblot analysis of COS7 cells transfected with the indicated siRNAs confirms efficient depletion of their targets. β-Actin was analyzed as a loading control (INPP4A 2 was used for co-knockdown with snx9 and PI3KC2α). Scr, scrambled. B, representative kymographs of eGFP-clathrin puncta over 180 s in control cells (treated with scrambled siRNA) or cells depleted of INPP4A by two different siRNAs. y axis scale bar = 5 μm. C, fraction of dynamic CCPs in control cells (Scr) or cells depleted of INPP4A by two different siRNAs, analyzed by quantitative automated 2D tracking. D and F, kymographs of eGFP-clathrin puncta over 180 s in control cells (Scr) or cells depleted of PI3KC2α or SNX9/SNX18 either alone (D) or together with INPP4A (F). y axis scale bars = 5 μm. E and G, fraction of dynamic CCPs in control cells (Scr) or cells depleted of PI3KC2α or SNX9/SNX18 either alone (E) or together with INPP4A (G), analyzed by quantitative automated 2D tracking. H, ultrastructural analysis of endocytic intermediates in control cells (Scr) or cells depleted of INPP4A by two different siRNAs by morphometric quantification of thin-section electron microscopy images. The different morphological groups are indicated below the bar diagram. I, depletion of INPP4A by two different siRNAs does not impair CME of transferrin. Bar diagrams represent the ratio of internalized (10 min, 37 °C) to surface-bound transferrin (45 min, 4 °C). Mean ± S.E., n = 3 independent experiments. 1202, 633, 761, 559, 645, 764, and 464 cells from three independent experiments were analyzed. *, p < 0.05; **, p < 0.01; one-way ANOVA. J, loss of INPP4A expression in INPP4A KO COS7 cells. Tubulin was analyzed as a loading control. Numbers in A and J indicate the molecular mass of the immunoblotted proteins in kilodaltons. K, fraction of dynamic CCPs in control or INPP4A KO COS7 cells, analyzed by quantitative automated 2D tracking.
In summary, our data show that INPP4A is probably absent from CCPs and dispensable for CME. Our combined results are therefore most compatible with a model according to which PI(3,4)P2 is synthesized locally at endocytic CCPs by PI3KC2α and is hydrolyzed downstream in the endosomal pathway by INPP4 (Fig. 1A, model I) and possibly other phosphatases, e.g. PTEN (43).
Loss of INPP4A in HAP1 cells perturbs signaling via Akt and mTORC1
Apart from its function in endocytosis, PI(3,4)P2 has well-known roles in cell signaling. Plasma membrane PI(3,4)P2, generated by the concerted action of class I PI 3-kinase and the 5-phosphatase SHIP2/INPP5 (33, 34), activates Akt, a gatekeeper for cell survival and an inducer of nutrient signaling via the late endosomal/lysosomal mTORC1 complex. Conversely, local synthesis of a late endosomal/lysosomal pool of PI(3,4)P2 by PI3KC2β, an enzyme activated when growth factor signaling ceases (25), represses mTORC1 activity via an Akt-independent mechanism in serum-deprived cells (24). Hence, PI(3,4)P2 differentially regulates cell signaling via Akt and mTORC1, depending on its subcellular location and cellular nutrient conditions.
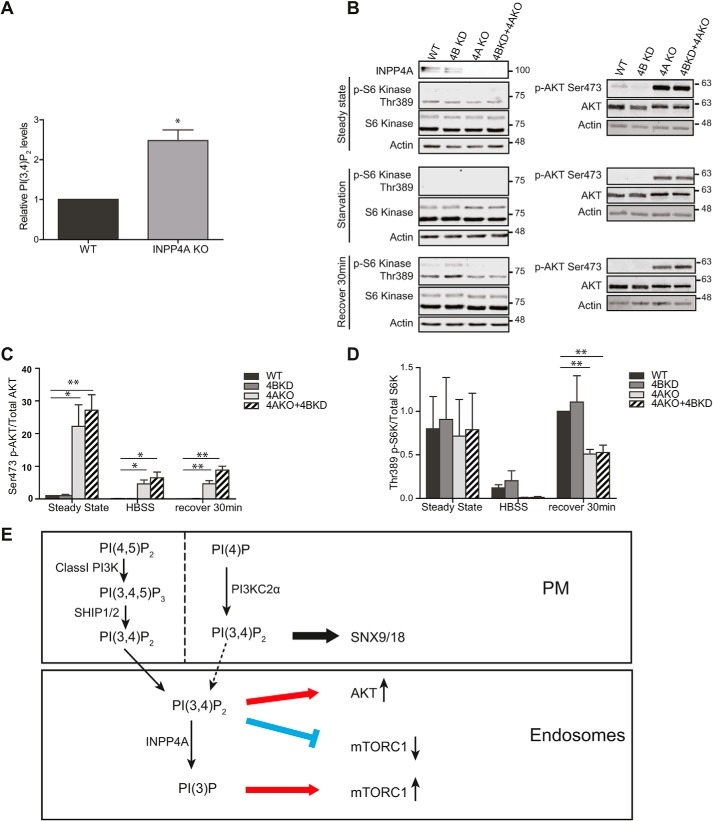
As a fraction of INPP4A is localized to endosomes (19) (Fig. 4), where it may hydrolyze PI(3,4)P2, we hypothesized that INPP4A might control cell signaling via Akt and/or mTORC1. The levels of INPP4A and its close relative INPP4B vary greatly between different cell types and tissues. For example, loss of INPP4B-mediated PI(3,4)P2 hydrolysis is associated with overactive Akt signaling in breast cancer cells (32, 35, 36). Among the cell types studied, we found INPP4A to be highly expressed in HAP1 cells, a near-haploid cancer cell line characterized by elevated PI(3,4,5)P3/PI(3,4)P2 production via hyperactive class I PI 3-kinase signaling (37). COS7 cells, used for most of our studies described above, expressed very low levels of INPP4A, whereas INPP4B was not detectable in either of these cell lines (Fig. S4E). To test whether loss of INPP4A might alter PI(3,4)P2-dependent signaling cascades, we analyzed INPP4A KO HAP1 cells. These cells showed normal CME (Fig. S5F; akin to INPP4A KO in COS7 cells (Fig. 5)) but displayed elevated levels of PI(3,4)P2 (Fig. 6A) that correlated with overactive Akt signaling (Fig. 6, B and C). Additional knockdown of INPP4B in INPP4A KO cells did not result in any further elevation of pAkt signaling (Fig. 6, B and C), indicating that INPP4A is the major, if not the only, physiologically relevant INPP4 family member in HAP1 cells. Interestingly, INPP4A KO cells also displayed delayed recovery of mTORC1 activity upon refeeding post-starvation, monitored by the levels of the mTORC1-specific substrate pS6 kinase 1 (Fig. 6, B and D). This is consistent with a local role of endolysosomal PI(3,4)P2 in repressing mTORC1 signaling in serum-starved cells (24). These phenotypes were not observed in INPP4A KO COS7 cells, which express the enzyme at very low levels (Fig. S4, C and D, and S5, A–D). Collectively, these results show that INPP4A, by controlling the levels of endosomal PI(3,4)P2, differentially regulates growth and nutrient signaling via Akt and mTORC1 in HAP1 cancer cells.
Figure 6.
Loss of INPP4A in HAP1 cells perturbs signaling via Akt and mTORC1. A, relative levels of PI(3,4)P2 in HAP1 WT and INPP4A KO cells. Mean ± S.E., n = 3 independent experiments. 126 and 128 cells from three independent experiments were analyzed. *, p < 0.05, unpaired t test. B, levels of phospho-Thr389-p70 S6K, total p70 S6K, phospho-Ser473-AKT, and total AKT at steady state, during starvation, or recovery (30 min) from starvation in HAP1 WT, INPP4B-depleted (KD), INPP4AKO, and INPP4BKD+INPP4AKO HAP1 cells. Note the elevated pAKT signal and delayed recovery of pS6K1 levels in INPP4AKO cells. C and D, quantification. Mean ± S.E., n = 3 independent experiments. *, p < 0.05; **, p < 0.01; unpaired t test. Numbers indicate the molecular mass of the immunoblotted proteins in kilodaltons. E, proposed model for the spatial segregation of PI(3,4)P2 synthesis and turnover in the endocytic pathway.
Discussion
In this study, we provide evidence that synthesis and turnover of PI(3,4)P2 are spatially segregated within the endocytic pathway to couple endocytic membrane traffic to growth factor and nutrient signaling. We show that, although an exogenous supply of either PI(3)P or PI(3,4)P2 can, in principle, rescue functional endocytic defects elicited by loss of PI3KC2α, CCP dynamics in living cells are mainly controlled by PI(3,4)P2 under physiological conditions, in contrast to the model suggested by He et al. (16) on the basis of auxilin-derived lipid/protein sensors; see also the study by Hammond and co-workers (33) regarding their specificity. Although PI3KC2α is able to synthesize PI(3)P in vitro and at endosomes in vivo (17, 18), the high PI(4)P-to-PI substrate ratio at the plasma membrane (38) would render PI(3)P synthesis at cell surface CCPs inefficient. Instead, we propose that PI3KC2α uses the abundant plasma membrane lipid PI(4)P, whose local levels may be further elevated by clathrin-associated PI(4,5)P2 5-phosphatases, to synthesize an endocytic pool of PI(3,4)P2 that serves to recruit effector proteins such as SNX9/18 (Fig. 6E) and the flat BAR domain protein FCHSD2 (39) to promote CCP maturation prior to dynamin-mediated membrane fission. Unlike SNX9/18, FCHSD2 binds to PI(3,4)P2 but not to PI(3)P and has been suggested to be responsible for the majority of local actin polymerization at CCPs (39).
Our results also do not support a function of the PI(3,4)P2-specific 4-phosphatase INPP4A/B in CME, proposed from in vitro experiments (20). We find INPP4A to be absent from CCPs, and loss of INPP4A in COS7 or HAP1 cells neither perturbs CCP dynamics nor causes defects in CME at the ultrastructural or functional levels (Figs. 3–5). Our findings, together with earlier data by Shin et al. (19), indicate instead that INPP4A operates at the level of early endosomes to convert PI(3,4)P2, either derived from endocytic pools (e.g. PI3KC2α) or from class I PI 3-kinase/SHIP2 signaling downstream of receptor activation, to PI(3)P (Fig. 6E). INPP4-mediated generation of a local pool of PI(3)P after endocytic vesicle fission has occurred (i.e. at the level of early endosomes) is consistent with the late timing of PI(3)P appearance post-scission, monitored by auxillin–2× FYVE (16) and confirmed by us (data not shown), and may contribute to the multiple roles of PI(3)P in the endolysosomal system and in autophagy (1, 40). Conceivably, it could also aid endosomal recruitment of SNX9 (20) or other PI(3)P-binding actin-polymerizing factors to promote endosomal F-actin polymerization.
Further support for a role of INPP4A at endosomes derives from our analysis of INPP4A function in signaling-active HAP1 cells. We show that loss of INPP4A results in hyperactivity of the PI(3,4)P2 effector Akt and local repression of mTORC1 signaling by accumulated endolysosomal PI(3,4)P2 pools, consistent with the idea that PI(3,4)P2 differentially regulates cell signaling, depending on its subcellular location and cellular nutrient conditions (24, 25, 34). How such differential regulation, e.g. of Akt and mTORC1, is accomplished at the molecular mechanistic level is unclear. PI(3,4)P2 can directly bind to Akt and has been shown to be required for sustained Akt activation (32, 36, 41, 42), possibly in conjunction with additional factors, e.g. other proteins activated downstream of class I PI 3-kinase signaling cascades. The mechanism of PI(3,4)P2-mediated repression of mTORC1 activity at late endosomes/lysosomes remains incompletely understood but may involve inhibitory association of 14-3-3 proteins with the Raptor subunit of mTORC1 (24), although additional mechanisms may contribute. Future studies will be needed to examine the cross-talk and regulation of different PI(3,4)P2 effectors in cell signaling.
A somewhat surprising finding is the observation that loss of INPP4A in COS7 cells, which do not endogenously express INPP4B, did not result in grossly elevated PI(3,4)P2 levels. Hence, PI(3,4)P2 must be hydrolyzed by other phosphatases in these cells. A promising candidate for this activity is the PI 3-phosphatase PTEN, an enzyme localized to endosomes (23), in addition to its established function in removal of PI(3,4,5)P3 at the plasma membrane (41). Recent work by several laboratories has uncovered a direct role of PTEN in PI(3,4)P2 hydrolysis in several cell types (33, 43). Moreover, consistent with a role in PI(3,4)P2 hydrolysis, PTEN has been found to reduce signaling at nascent CCPs and to slow their maturation (44). It therefore seems likely that loss of function of INPP4A/B is compensated by the 3-phosphatase activity of PTEN, resulting in production of PI(4)P, a lipid found in secretory organelles, including recycling endosomes (31), and at the plasma membrane (45). How PI(3,4)P2 is funneled to these enzymatically distinct pathways of turnover remains enigmatic and will need to be addressed in future studies.
Experimental procedures
Cell culture
COS7 cells obtained from the ATCC were cultured in DMEM (Gibco) with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco). HAP1 WT cells and INPP4A KO cells were obtained from HorizonTM and cultured in Iscove's modified Dulbecco's medium (Gibco) supplied with 10% FBS and 1% penicillin/streptomycin. Cells were not used beyond passage 30. All cell lines were routinely tested for mycoplasma contaminations on a monthly basis.
Antibodies and reagents
For immunoblotting, we used β-actin (Sigma, A-5441, 1:5000, mouse), PI3KC2α (homebrewed polyclonal, 1:2000, rabbit), PI3KC2β(BD Transduction Laboratories, 611342, 1:1000, mouse), Snx9 (Protein Tech, 15721, 1:1000, rabbit), Snx18 (GeneTex, GTX119426, 1:5000, rabbit), INPP4A (Abcam, ab109622, 1:1000, rabbit), INPP4B (Santa Cruz, sc-12318, 1:750, goat), PTEN (Cell Signaling, 9552, 1:1000, rabbit), tubulin (Sigma, T5168, 1:2000, mouse), GFP (Clontech, 632381, 1:2500, mouse), Akt (Cell Signaling, 4691, 1:1000, rabbit), phospho-Akt538 Ser473 (Cell Signaling, 4060, 1:1000, rabbit), p70 S6K (Cell Signaling, 2708, 1:1000, rabbit), and phospho-p70 S6K 542 Thr389 (Cell Signaling, 9234, 1:1000, rabbit). For immunofluorescence, we used PI(3,4)P2 (Echelon Biosciences, Z-P034b, 1:150, mouse), clathrin heavy chain (homebrewed, 1:100, mouse), AP2 (homebrewed, polyclonal, 1:50, mouse), GFP (Abcam, ab6556, 1:500, rabbit), Rab5 (BD Transduction Laboratories, 610724, 1:75, mouse), Rab7 (Cell Signaling, 9367, rabbit, 1:100), and LAMP1 (BD Biosciences, 555798, 1:100, mouse). HRP-conjugated or LI-COR 800CW and 680RD IR secondary antibodies were used. Quantification was performed via IR fluorescence with Odyssey FC detection.
siRNAs
All siRNA sequences were used as 21-mers, including 3′-dTdT overhangs. The sequence of the PI3K C2α-targeting siRNA used in this study was 5′-GCACAAACCCAGGCTATTT-3′. The INPP4A siRNA sequences were as follows: siRNA1, 5′-CAUCAUAGGUUGCAUUUAA-3′; siRNA2, 5′-GAUCGAAAGCCAAAUAGUU-3′ (this siRNA was used in the majority of experiments). The INPP4B siRNA sequences were as follows: siRNA1, 5′-CGAUGAAAUUGGAAUGUUA-3′; siRNA2, 5′-GAAGGAUUGUUAAGUACAU-3′. The Snx9 siRNA sequences was 5′-GGACAGAACGGGCCTTGAA-3′. The Snx18 siRNA sequence was 5′-CACCGACGAGAAAGCCUGGAA-3′. Sigma Mission Universal Negative Control 1 was used as a control.
siRNA and plasmid transfection
Cells were seeded and transfected with siRNA on day 1 using jetPRIME (Polyplus Transfection) according to the manufacturer's instructions by reverse transfection, expanded on day 2 before a second round of knockdown was performed on day 3 using X-tremeGENE (Roche) according to the manufacturer's instructions, and processed on day 4. Rescue transfections were performed on day 3 using X-tremeGENE according to the manufacturer's instructions. For experiments not involving siRNA, plasmid transfections were done using jetPRIME.
Transferrin uptake and surface labeling
Cells treated with siRNA or transfected with different plasmids were seeded on coverslips coated with Matrigel (BD Biosciences). Before the experiment, cells were treated in serum-starved medium for 1 h and used for transferrin uptake or transferrin receptor surface labeling. For the rapalog-induced system (Takara, AP21967), rapalog or DMSO was added during 1-h starvation. For transferrin uptake, cells were incubated with 25 mg/ml transferrin–Alexa 647 (Molecular Probes, Invitrogen) for 10 min at 37 °C. Cells were first washed twice with ice-cold PBS containing 10 mm MgCl2 and then acid-washed at pH 5.3 (0.2 m sodium acetate and 200 mm sodium chloride) on ice for 1 min to remove surface-bound transferrin. Cells were then washed twice more with ice-cold PBS containing 10 mm MgCl2 and fixed with 4% paraformaldehyde (PFA) for 45 min at room temperature. For surface labeling, cells were incubated with 25 mg/ml transferrin–Alexa 647 at 4 °C for 45 min, washed three times with ice-cold PBS (10 mm MgCl2) on ice for 1 min, and fixed with 4% PFA for 45 min at room temperature. Transferrin labeling was analyzed using a Nikon Eclipse Ti microscope and ImageJ software. Internalized transferrin per cell was quantified and normalized to the amount of surface-bound transferrin determined in the same experiment as a measure of the efficiency of internalization.
TIRF microscopy
TIRF imaging was performed using a Zeiss Axiovert200M microscope equipped with an incubation chamber (37 °C and 5% CO2), a 3100 TIRF objective, and a dual-color TIRF setup from Visitron Systems using Slidebook imaging software (3i Inc.). For analysis of CCP dynamics, a time-lapse series of 3 min with a frame rate of 2 s/frame was recorded.
Automated CCPs tracking
CCPs were tracked automatically (46) and statistically analyzed as follows. An instantaneous density was used as a metric for CCP lifetime density. Conceptually, the instantaneous density is the average density of CCPs (of a specific lifetime or lifetime range) in a given frame of the movie, i.e. the number of CCPs that are visible per area, in units of [number of events/square microns], with no time dependence. 150 s was set as a threshold to distinguish dynamic from persistent CCPs (i.e. defined as CCPs with lifetimes of more than 150 s). The instantaneous density is calculated by counting the number of visible CCPs per area in each frame. Then the number of dynamic CCPs of the known lifetime (i.e. visible CCPs with uncensored trajectories below 150 s) is corrected for the corresponding observation bias, where the correction factor in this case depends on both CCP lifetime and the frame number, because more censored trajectories are visible at the beginning and the end of the movie than in the middle. The number of persistent CCPs is the sum of both uncensored and censored CCPs of a known lifetime of 150 s or more (which may include structures that are visible continuously for the entire length of the movie) without bias correction. Following this approach, the fraction of dynamic CCPs of all CCPs represents the unitless ratio of dynamic CCP instantaneous density over total CCP instantaneous density, where total = dynamic + persistent.
Colocalization probability
Colocalization probability is calculated in analogy to intensity plots of lifetime cohorts (46, 47), except that, for the purposes of this analysis, the fluorescence intensity is replaced by the colocalization probability. The colocalization probability here is defined as the probability that a spot is detected in the second (dependent) color channel within a threshold distance of the first (reference) color channel, where that threshold distance is defined by the resolution limit (Rayleigh criterion for diffraction-based resolution).
EM
For thin-section EM analysis of clathrin-coated structures, cells were grown in 6-cm Petri dishes, fixed with 2% glutaraldehyde in PBS, mechanically detached, post-fixed with 1% osmium tetroxide in cacodylate buffer, and processed for epoxy resin embedding. Embedded cell pellets were ultrathin-sectioned and imaged by transmission EM using a Zeiss 900 transmission electron microscope. Clathrin-coated structures where quantified along the plasma membrane in cell profile montages and normalized to the length of the plasma membrane perimeter.
CRISPR/Cas9-mediated genome engineering
Primers for guide RNA generation in COS7 cells for INPP4A were designed as follows: forward, 5′-GGAAGCCCGCTGCACCGCAC-3′; reverse, 5′-GTGCGGTGCAGCGGGCTTCC-3′. The primers were annealed and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene, 42230) using the BbsI site. COS7 cells were transfected with the px330-plasmid containing the guide RNA and a fluorescent marker (GFP) for indication of successfully transfected cells for 72 h. GFP-expressing cells were selected by FACS and plated on 96-well plates to obtain single-cell clones. Colonies were expanded and screened by immunoblotting using specific anti-INPP4A antibodies to identify KO clones.
Immunofluorescence
Cells were seeded on coverslips coated with Matrigel (BD Biosciences). For PI(3,4)P2 staining, cells were washed once with PBS containing 10 mm MgCl2 and fixed in 2% PFA either with (COS7 cells) or without (Hap1 cells) 0.5% glutaraldehyde for 20 min at room temperature. Cells were washed three times with PBS containing 50 mm NH4Cl and processed for permeabilization with PBS containing 0.5% saponin, 1% BSA, and 10% normal goat serum for 30 min. Primary and secondary antibodies were incubated for 1–2 h in PBS buffer containing 1% BSA and 10% normal goat serum. Cells were washed three times with PBS after primary and secondary antibody treatment and preceding mounting with 4′,6-diamidino-2-phenylindole. For PI(3)P staining, COS7 cells were washed once with PBS containing 10 mm MgCl2 and fixed in 2% PFA for 15 min at room temperature. Cells were washed three times with PBS containing 50 mm NH4Cl and permeabilized with 20 μm digitonin in buffer A (20 mm Pipes (pH 6.8), 137 mm NaCl, and 2.7 mm KCl) for 5 min. After washing with buffer A three times, cells were incubated for 30 min in blocking buffer (buffer A with 5% normal goat serum and 50 mm NH4Cl) containing 0.055 mg/ml GFP-FYVE. Primary and secondary antibodies were incubated for 1 h in buffer A containing 5% normal goat serum. Cells were post-fixed with 2% PFA for 5 min and washed three times with PBS containing 50 mm NH4Cl and twice with PBS.
For antibody staining, COS7 cells were washed once with PBS containing 10 mm MgCl2 and fixed in ice-cold methanol at −20 °C for 10 min. Cells were then blocked for 30 min in blocking buffer (10% normal goat serum in PBS with 0.02% Triton X-100). Primary and secondary antibodies were incubated for 1–2 h in the same buffer. For fixation in 4% PFA, 0.05% saponin was used instead of Triton X-100 in blocking buffer.
Cells were analyzed using a spinning disk confocal microscope (Ultraview ERS, PerkinElmer Life Sciences) with Volocity imaging software (Improvision, PerkinElmer Life Sciences). PI(3)P and PI(3,4)P2 levels were quantified using custom-written ImageJ macros as described earlier (9).
Rescue of defective CME by membrane-permeant PI 3-phosphates
COS7 cells were washed twice in DMEM with 4.5 g/liter glucose without FBS prior to addition of PIP/AMs. PI(3)P/AM and PI(3,4)P2/AM were dissolved in dry DMSO to a stock concentration of 50 mm and mixed with an equal volume of 10% pluronic F127 to enhance solubility. After thorough mixing, the sample was diluted in DMEM to a final concentration of 100 μm and incubated with COS7 cells for 20 min. An equal volume of DMEM supplemented with 20% FBS was added to the cells prior to TIFR imaging.
Data analysis and statistics
Data are expressed as a means ± S.E. of at least three experiments. The statistical significance between groups was evaluated with one-way ANOVA or unpaired t test. Significant differences were accepted at p < 0.05.
Author contributions
H. W., D. L., R. M., C. S., and V. H. conceptualization; H. W., C. B., and D. P. investigation; H. W., D. L., C. B., R. M., D. P., and C. S. methodology; H. W. and V. H. writing-original draft; H. W. and V. H. writing-review and editing; D. L. software; D. L. formal analysis; P. A. K. data curation; C. S. and V. H. supervision; C. S. and V. H. funding acquisition; V. H. project administration.
Supplementary Material
Supported by Deutsche Forschungsgemeinschaft Grant TRR 186/TP 08 (to V. H. and C. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
R. Müller and C. Schultz, manuscript in preparation.
- PI
- phosphatidylinositol
- PI(3)P
- phosphatidylinositol 3-phosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- CCP
- clathrin-coated pit
- CME
- clathrin-mediated endocytosis
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- AM
- acetoxymethylester
- TIRF
- total internal reflection microscopy
- eGFP
- enhanced GFP
- PFA
- paraformaldehyde
- ANOVA
- analysis of variance
- CLC
- clathrin light chain
- PTEN
- phosphatase and tensin homolog
- mRFP
- monomeric red fluorescent protein.
References
- 1. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Paolo G., and De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 3. Posor Y., Eichhorn-Grünig M., and Haucke V. (2015) Phosphoinositides in endocytosis. Biochim. Biophys. Acta 1851, 794–804 10.1016/j.bbalip.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 4. Zoncu R., Perera R. M., Sebastian R., Nakatsu F., Chen H., Balla T., Ayala G., Toomre D., and De Camilli P. V. (2007) Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. U.S.A. 104, 3793–3798 10.1073/pnas.0611733104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Erdmann K. S., Mao Y., McCrea H. J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., and De Camilli P. (2007) A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell 13, 377–390 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nández R., Balkin D. M., Messa M., Liang L., Paradise S., Czapla H., Hein M. Y., Duncan J. S., Mann M., and De Camilli P. (2014) A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. Elife 3, e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verstreken P., Koh T. W., Schulze K. L., Zhai R. G., Hiesinger P. R., Zhou Y., Mehta S. Q., Cao Y., Roos J., and Bellen H. J. (2003) Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 40, 733–748 10.1016/S0896-6273(03)00644-5 [DOI] [PubMed] [Google Scholar]
- 8. Gaidarov I., Smith M. E., Domin J., and Keen J. H. (2001) The class II phosphoinositide 3-kinase C2α is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell 7, 443–449 10.1016/S1097-2765(01)00191-5 [DOI] [PubMed] [Google Scholar]
- 9. Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E., Krauss M., Schultz C., Schmoranzer J., Noé F., and Haucke V. (2013) Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499, 233–237 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]
- 10. Tiosano D., Baris H. N., Chen A., Hitzert M. M., Schueler M., Gulluni F., Wiesener A., Bergua A., Mory A., Copeland B., Gleeson J. G., Rump P., van Meer H., Sival D. A., Haucke V., et al. (2019) Mutations in PIK3C2A cause syndromic short stature, skeletal abnormalities, and cataracts associated with ciliary dysfunction. PLoS Genet. 15, e1008088 10.1371/journal.pgen.1008088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bilanges B., Posor Y., and Vanhaesebroeck B. (2019) PI3K isoforms in cell signalling and vesicle trafficking. Nat. Rev. Mol. Cell Biol. 20, 515–534 10.1038/s41580-019-0129-z [DOI] [PubMed] [Google Scholar]
- 12. Gulluni F., De Santis M. C., Margaria J. P., Martini M., and Hirsch E. (2019) Class II PI3K functions in cell biology and disease. Trends Cell Biol. 29, 339–359 10.1016/j.tcb.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 13. Schöneberg J., Lehmann M., Ullrich A., Posor Y., Lo W. T., Lichtner G., Schmoranzer J., Haucke V., and Noé F. (2017) Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 8, 15873 10.1038/ncomms15873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lo W. T., Vujičić Žagar A., Gerth F., Lehmann M., Puchkov D., Krylova O., Freund C., Scapozza L., Vadas O., and Haucke V. (2017) A Coincidence detection mechanism controls PX-BAR domain-mediated endocytic membrane remodeling via an allosteric structural switch. Dev. Cell 43, 522–529.e4 e524 10.1016/j.devcel.2017.10.019 [DOI] [PubMed] [Google Scholar]
- 15. Wang H., Lo W. T., Vujicic Zagar A., Gulluni F., Lehmann M., Scapozza L., Haucke V., and Vadas O. (2018) Autoregulation of class II α PI3K activity by its lipid-binding PX-C2 domain module. Mol. Cell 71, 343–351.e4 10.1016/j.molcel.2018.06.042 [DOI] [PubMed] [Google Scholar]
- 16. He K., Marsland R. 3rd, Upadhyayula S., Song E., Dang S., Capraro B. R., Wang W., Skillern W., Gaudin R., Ma M., and Kirchhausen T. (2017) Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 552, 410–414 10.1038/nature25146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Campa C. C., Margaria J. P., Derle A., Del Giudice M., De Santis M. C., Gozzelino L., Copperi F., Bosia C., and Hirsch E. (2018) Rab11 activity and PtdIns(3)P turnover removes recycling cargo from endosomes. Nat. Chem. Biol. 14, 801–810 10.1038/s41589-018-0086-4 [DOI] [PubMed] [Google Scholar]
- 18. Franco I., Gulluni F., Campa C. C., Costa C., Margaria J. P., Ciraolo E., Martini M., Monteyne D., De Luca E., Germena G., Posor Y., Maffucci T., Marengo S., Haucke V., Falasca M., et al. (2014) PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev. Cell 28, 647–658 10.1016/j.devcel.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shin H. W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M. R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., Frankel W. N., Solimena M., De Camilli P., and Zerial M. (2005) An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170, 607–618 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daste F., Walrant A., Holst M. R., Gadsby J. R., Mason J., Lee J. E., Brook D., Mettlen M., Larsson E., Lee S. F., Lundmark R., and Gallop J. L. (2017) Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature. J. Cell Biol. 216, 3745–3765 10.1083/jcb.201704061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong Z., Pedersen N. M., Wang L., Torgersen M. L., Stenmark H., and Raiborg C. (2017) PtdIns3P controls mTORC1 signaling through lysosomal positioning. J. Cell Biol. 216, 4217–4233 10.1083/jcb.201611073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raiborg C., Wenzel E. M., Pedersen N. M., Olsvik H., Schink K. O., Schultz S. W., Vietri M., Nisi V., Bucci C., Brech A., Johansen T., and Stenmark H. (2015) Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234–238 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- 23. Naguib A., Bencze G., Cho H., Zheng W., Tocilj A., Elkayam E., Faehnle C. R., Jaber N., Pratt C. P., Chen M., Zong W. X., Marks M. S., Joshua-Tor L., Pappin D. J., and Trotman L. C. (2015) PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell 58, 255–268 10.1016/j.molcel.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marat A. L., Wallroth A., Lo W. T., Müller R., Norata G. D., Falasca M., Schultz C., and Haucke V. (2017) mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356, 968–972 10.1126/science.aaf8310 [DOI] [PubMed] [Google Scholar]
- 25. Wallroth A., Koch P. A., Marat A. L., Krause E., and Haucke V. (2019) Protein kinase N controls a lysosomal lipid switch to facilitate nutrient signalling via mTORC1. Nat. Cell Biol. 21, 1093–1101 10.1038/s41556-019-0377-3 [DOI] [PubMed] [Google Scholar]
- 26. Laguerre A., and Schultz C. (2018) Novel lipid tools and probes for biological investigations. Curr. Opin. Cell Biol. 53, 97–104 10.1016/j.ceb.2018.06.013 [DOI] [PubMed] [Google Scholar]
- 27. Subramanian D., Laketa V., Müller R., Tischer C., Zarbakhsh S., Pepperkok R., and Schultz C. (2010) Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat. Chem. Biol. 6, 324–326 10.1038/nchembio.348 [DOI] [PubMed] [Google Scholar]
- 28. Lehmann M., Lukonin I., Noé F., Schmoranzer J., Clementi C., Loerke D., and Haucke V. (2019) Nanoscale coupling of endocytic pit growth and stability. Sci. Adv. 5, eaax5775 10.1126/sciadv.aax5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yarar D., Surka M. C., Leonard M. C., and Schmid S. L. (2008) SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic 9, 133–146 10.1111/j.1600-0854.2007.00675.x [DOI] [PubMed] [Google Scholar]
- 30. Fili N., Calleja V., Woscholski R., Parker P. J., and Larijani B. (2006) Compartmental signal modulation: endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc. Natl. Acad. Sci. U.S.A. 103, 15473–15478 10.1073/pnas.0607040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ketel K., Krauss M., Nicot A. S., Puchkov D., Wieffer M., Müller R., Subramanian D., Schultz C., Laporte J., and Haucke V. (2016) A phosphoinositide conversion mechanism for exit from endosomes. Nature 529, 408–412 10.1038/nature16516 [DOI] [PubMed] [Google Scholar]
- 32. Gewinner C., Wang Z. C., Richardson A., Teruya-Feldstein J., Etemadmoghadam D., Bowtell D., Barretina J., Lin W. M., Rameh L., Salmena L., Pandolfi P. P., and Cantley L. C. (2009) Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell 16, 115–125 10.1016/j.ccr.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goulden B. D., Pacheco J., Dull A., Zewe J. P., Deiters A., and Hammond G. R. V. (2018) A high-avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J. Cell Biol. 218, 1066–1079 10.1083/jcb.201809026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu S. L., Wang Z. G., Hu Y., Xin Y., Singaram I., Gorai S., Zhou X., Shim Y., Min J. H., Gong L. W., Hay N., Zhang J., and Cho W. (2018) Quantitative lipid imaging reveals a new signaling function of phosphatidylinositol-3,4-bisphophate: isoform- and site-specific activation of Akt. Mol. Cell 71, 1092–1104.e5 10.1016/j.molcel.2018.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fedele C. G., Ooms L. M., Ho M., Vieusseux J., O'Toole S. A., Millar E. K., Lopez-Knowles E., Sriratana A., Gurung R., Baglietto L., Giles G. G., Bailey C. G., Rasko J. E., Shields B. J., Price J. T., et al. (2010) Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc. Natl. Acad. Sci. U.S.A. 107, 22231–22236 10.1073/pnas.1015245107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gasser J. A., Inuzuka H., Lau A. W., Wei W., Beroukhim R., and Toker A. (2014) SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol. Cell 56, 595–607 10.1016/j.molcel.2014.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B. V., Kralovics R., Brummelkamp T. R., Nijman S. M., and Bürckstümmer T. (2014) Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 24, 2059–2065 10.1101/gr.177220.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saheki Y., Bian X., Schauder C. M., Sawaki Y., Surma M. A., Klose C., Pincet F., Reinisch K. M., and De Camilli P. (2016) Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 18, 504–515 10.1038/ncb3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Almeida-Souza L., Frank R. A. W., García-Nafría J., Colussi A., Gunawardana N., Johnson C. M., Yu M., Howard G., Andrews B., Vallis Y., and McMahon H. T. (2018) A flat BAR protein promotes actin polymerization at the base of clathrin-coated pits. Cell 174, 325–337.e14 10.1016/j.cell.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Devereaux K., Dall'Armi C., Alcazar-Roman A., Ogasawara Y., Zhou X., Wang F., Yamamoto A., De Camilli P., and Di Paolo G. (2013) Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE 8, e76405 10.1371/journal.pone.0076405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., and Bilanges B. (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 42. Franke T. F., Kaplan D. R., Cantley L. C., and Toker A. (1997) Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668 10.1126/science.275.5300.665 [DOI] [PubMed] [Google Scholar]
- 43. Malek M., Kielkowska A., Chessa T., Anderson K. E., Barneda D., Pir P., Nakanishi H., Eguchi S., Koizumi A., Sasaki J., Juvin V., Kiselev V. Y., Niewczas I., Gray A., Valayer A., et al. (2017) PTEN regulates PI(3,4)P2 signaling downstream of class I PI3K. Mol. Cell 68, 566–580.e10 10.1016/j.molcel.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosselli-Murai L. K., Yates J. A., Yoshida S., Bourg J., Ho K. K. Y., White M., Prisby J., Tan X., Altemus M., Bao L., Wu Z. F., Veatch S. L., Swanson J. A., Merajver S. D., and Liu A. P. (2018) Loss of PTEN promotes formation of signaling-capable clathrin-coated pits. J. Cell Sci. 131, jcs208926 10.1242/jcs.208926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hammond G. R., Fischer M. J., Anderson K. E., Holdich J., Koteci A., Balla T., and Irvine R. F. (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aguet F., Antonescu C. N., Mettlen M., Schmid S. L., and Danuser G. (2013) Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Loerke D., Mettlen M., Schmid S. L., and Danuser G. (2011) Measuring the hierarchy of molecular events during clathrin-mediated endocytosis. Traffic 12, 815–825 10.1111/j.1600-0854.2011.01197.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.