Abstract
Mediterranean spotted fever is a reemerging acute tick-borne infection produced by the α-proteobacterium, Rickettsia conorii. Rickettsia conorii infects vascular endothelial cells producing disseminated plasma leakage, manifesting as nonspecific fever, headache, and maculopapular rash. Because there are no available tests of early infection, Mediterranean spotted fever is often undiagnosed and untreated, resulting in significant mortality. To address this critical need, we have applied a quantitative proteomics pipeline for analyzing the secretome of primary human umbilical vein endothelial cells. Of the 104 proteins whose abundance changed significantly in the R. conorii–infected human umbilical vein endothelial cells’ secretome, 46 proteins were up-regulated: 45 were host secreted proteins (including cytokines), and 1 was a rickettsial protein, the putative N-acetylmuramoyl-l-alanine amidase RC0497. Proteins with sequence highly homologous to RC0497 were found to be shared by many species of the spotted fever group rickettsiae, but not typhus group rickettsiae. Quantitative targeted proteomics studies of plasma from a mouse model of sublethal and lethal R. conorii identified RC0497 in the blood, and its circulating levels were proportionally associated with infection outcome. Finally, the presence of RC0497 in the serum samples from a cohort of humans presenting with acute rickettsioses was confirmed. The detection of RC0497 has the potential to be a sensitive and specific marker for acute rickettsial spotted rickettsioses.
The genus Rickettsia, a group of nonmotile, Gram-negative, intracellular α-proteobacteria, has been increasing in global medical and veterinary health importance because of their endemicity and reemergence. From the clinical and antigenic perspective, rickettsial diseases are classified into two groups, spotted fever and typhus. Rickettsia conorii, the agent of Mediterranean spotted fever (MSF; boutonneuse fever), is one of the most virulent strains. MSF is prevalent throughout the Mediterranean basin, Africa, the Middle East, and India.1 In humans, the MSF presents as an eschar, acute fever, headache, maculopapular rash, and vascular leakage; and the patients may eventually develop pulmonary and cerebral edema without proper treatment.
The clinical manifestations of acute MSF are a consequence of the tropism of rickettsiae for endothelial cells.2,3 Rickettsial organisms bind Ku70 on cholesterol-rich microdomains and enter endothelial cells through a calcium-dependent zipper-like entry mechanism.4, 5, 6, 7, 8 Viable organisms subsequently lyse intracellular phagosomes via phospholipase D and hemolysin activities,9,10 and they replicate in the cytoplasm. A characteristic of the spotted fever group is that R. conorii organisms spread intercellularly as a consequence of directional actin polymerization without producing early cellular injury.11, 12, 13 Oxidative stress produced by replication of R. conorii results in peroxidation of endothelial membranes and disruption of the endothelial barrier function, producing vascular leakage.2,14
The pulmonary and cerebral edema in patients infected with rickettsiales can cause significant morbidity and mortality if there are delays in diagnosis and treatment.2 The reference standard for diagnosis of rickettsial diseases is a fourfold increase in antibody titer by the indirect immunofluorescence antibody assay using paired serum samples 2 to 4 weeks after onset of illness.15 Patients usually do not have diagnostic serum antibody titers during the first week of illness, and a negative result by immunofluorescence antibody does not exclude the diagnosis. Therefore, many cases are initially misdiagnosed, accounting for adverse outcomes,16 and consequently, the epidemiology of the disease is underreported. A highly specific and sensitive biomarker for early diagnosis of rickettsia infection is greatly needed.
A major challenge in the development of rickettsial diagnostics is that the infection is localized to the vascular endothelium, and bacteremia is not typically present.2 To circumvent this limitation, we described herein a pipeline for identifying the markers for early R. conorii infection (Figure 1). First, the dynamic changes in the endothelial secretome in response to R. conorii infection were analyzed. The secretome of primary human umbilical vein endothelial cells (HUVECs) infected with R. conorii was profiled. With this approach, the putative N-acetylmuramoyl-l-alanine amidase RC0497 was identified as the most abundant rickettsial protein in R. conorii–infected HUVEC-conditioned medium and a highly specific candidate for a diagnosis of rickettsial infection. Next, proteomics studies of plasma from mice infected with either sublethal or lethal doses of R. conorii were conducted, and a cluster of increased amounts of acute phase reactants and coagulation factors was identified. As expected, no rickettsial proteins were detected in this discovery study, confirming the low abundance of bacterial proteins in the circulation. To increase the sensitivity of detection, a stable isotope dilution (SID)–parallel reaction monitoring (PRM) assay was developed to detect RC0497 in plasma of infected animals, and its circulating levels were found to be proportional to the bacterial burden. Finally, an immunoprecipitation (IP) enrichment–selected reaction monitoring (SRM) assay was developed to detect the low abundance of circulating RC0497, and this assay was validated as a diagnostic approach in a cohort of humans presenting with acute rickettsial infections. Because RC0497 is conserved in >10 species of the rickettsial spotted fever group, we propose that its detection is a specific marker for diagnosis of acute infection. This assay will also advance the epidemiologic studies of this infection.
Figure 1.
Workflow of identification of the biomarkers for rickettsial infection and the severity of infection. The biomarker development workflow has three steps, each indicated in the arrows. In step 1, the potential protein markers for Rickettsia conorii infection were identified via proteomics profiling of the secretome of human umbilical vein endothelial cells (HUVECs) infected with R. conorii. In step 2, the presence of the secretome markers in the plasma was verified using a mouse model of rickettsial infection, and the potential markers for the severity of rickettsia infection were identified via proteomics profiling of the plasma from the animals with different doses of R. conorii. In step 3, the protein markers identified in steps 1 and 2 were verified using the plasma from patients with rickettsia infection. LD, lethal dose; SLD, sublethal dose. RC0497, putative N-acetylmuramoyl-L-alanine amidase RC0497.
Materials and Methods
Ethical Approval
Animal experiments were performed according to the NIH Guide for Care and Use of Laboratory Animals17 and approved by the University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee (number 90-07082). This study of human subjects was conducted under approval from the UTMB Institutional Review Board (numbers 08 to 258) and was compliant with all applicable federal regulations governing the protection of human subjects.
Materials
All reagents and solvents in liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses were at least high-performance LC grade (Fisher Scientific, Fair Lawn, NJ). Ammonium bicarbonate and acetic acid were purchased from Sigma-Aldrich (St. Louis, MO). Iodoacetamide, acetonitrile, dithiothreitol, methanol, and formic acid were purchased from Thermo Scientific (Waltham, MA). Urea ultra was from MP Biomedicals (Santa Ana, CA). Sequencing-grade modified trypsin and endoproteinase Lys-C were from Promega (Madison, WI). Recombinant rickettsial putative N-acetylmuramoyl-l-alanine amidase RC0497 was purchased from MyBiosource (San Diego, CA).
Rickettsia
Rickettsia conorii (Israeli spotted fever strain) was obtained from ATCC (Manassas, VA). For in vitro cell infection, R. conorii Israeli spotted fever strain was cultivated in Vero cells. To purify rickettsiae, cells were homogenized and rickettsiae were diluted in a 10% suspension of sucrose-phosphate-glutamate buffer (0.218 mmol/L sucrose, 3.8 mmol/L KH2PO4, 7.2 mmol/L K2HPO4, 4.9 mmol/L monosodium glutamic acid, pH 7.0) and stored at −80°C. For mouse inoculation, R. conorii (Malish 7 strain) was obtained from ATCC (catalog number VR-613). Rickettsiae were propagated in the yolk sacs of specific pathogen-free embryonated chicken eggs, as described previously.18 The concentrations of stock rickettsiae cultured in both yolk sacs and cell culture were determined by plaque assay, as described previously.19 The rickettsial stock was stored at −80°C until used, and all of the experiments described in this study were performed in a certified biosafety level 3 laboratory at UTMB.
Cell Cultures and Infection with R. conorii
The culture of HUVECs and R. conorii infection were conducted, as described previously.20 The culture medium was supplemented EGM-Plus endothelial cell growth medium (Lonza, Walkersville, MD; catalog number CC-5035) without the addition of gentamicin sulfate and amphotericin-B or fetal bovine serum. For infection, 15 × 106 primary HUVECs were infected with R. conorii in biosafety level 3 containment, and subsequently the conditioned medium was collected 24 hours after infection. The conditioned medium was filtered through a 0.22-μm membrane filter to remove infectious rickettsiae in accordance with University of Texas Medical Branch Institutional Biosafety Committee–approved protocols.
Trypsin Digestion of the Secretome of HUVECs Infected with R. conorii
The secretome was isolated, as described previously.21 Briefly, the conditioned medium was collected and centrifuged at 2000 × g at 4°C for 20 minutes to remove any dead cells. The supernatant was centrifuged at 10,000 × g at 4°C for 10 minutes to remove cell debris. The supernatant was further concentrated using Amicon ultra-4 centrifugal filters-3K (Millipore, Billerica, MA). Then, 200 μL of 8 mol/L urea was added into centrifugal filters to denature the proteins. Next, the proteins were reduced with 10 mmol/L dithiothreitol for 30 minutes, followed by alkylation with 30 mmol/L iodoacetamide for 60 minutes at room temperature in the dark. The centrifugal filters were centrifuged at 14,000 Χ g at 4°C for 15 minutes, and 200 μL of 50 mmol/L of ammonium bicarbonate (pH 8.0) was added into the sample. The centrifugal filters were centrifuged at 14,000 × g at 4°C for 15 minutes again. The sample that remained in the filter was transferred into a 0.5-mL microcentrifugation tube. The proteins were digested first with 1.0 μg endoproteinase Lys-C–trypsin (Promega) for 12 hours at 37°C, then diluted two times with 50 mmol/L of ammonium bicarbonate, and further digested with 1.0 μg trypsin (Promega) for 16 hours at 37°C. The digestion was terminated with 0.5% trifluoroacetic acid. The peptides were desalted on a reversed-phase SepPak C18 cartridge (Waters, Milford, MA) before LC-MS/MS analysis.
Label-Free Quantification of Protein Expression
The desalted peptides were reconstituted in 30 μL 4% acetonitrile/0.1% formic acid and analyzed on an Easy nLC1000 UHPLC-Q Exactive Orbitrap LC-MS system (Thermo Scientific, San Jose, CA), as previously described.22 A 2-hour linear gradient from 2% solvent A (0.1% formic acid in water) to 35% solvent B (0.1% formic acid in acetonitrile) was used for each LC-MS/MS run. Data-dependent acquisition in positive ion mode was used. The spray voltage was 2.1 kV. The resolution of full scan range from 400 to 1400 m/z is 70,000, the maximum injection time is 80 milliseconds, and the automatic gain control was set to 1 × 106. The top 15 ions in each survey scan were selected for MS2 with a resolution of 17,500. The collision energy for higher-energy collisional dissociation was set to 30, the maximum injection time was 60 milliseconds, and the automatic gain control was 1 × 105. Ions selected for MS/MS were dynamically excluded for 30 seconds after fragmentation.
All data were analyzed with the MaxQuant software version 1.5.2.823,24 with the Andromeda search engine.25 The false discovery rate (FDR) was set to 1% for both proteins and peptides, and a minimum length of seven amino acids was specified. The Andromeda search engine was used for the MS/MS spectra search against a combined SwissProt human and rickettsial database (downloaded on December 2015, containing 20,193 human protein entries, 4476 rickettsial protein entries, and 247 contaminants). Carbamidomethylation of cysteine was set as fixed modification, and methionine oxidation was set as variable modification. A maximum of two missed cleavages had enzyme specificity as C-terminal to Arg and Lys. The match between runs feature of MaxQuant was used to transfer identifications to other LC-MS/MS runs based on their masses and retention time (maximum deviation, 0.7 minutes) in quantification experiments. Quantifications were performed with the label-free algorithms in MaxQuant.23 At least one razor peptide was required for quantification. Perseus version 1.5.5.326 was used to analyze the MaxQuant output, including statistics, hierarchical clustering, and principal component analysis (PCA). Reversed identifications and proteins identified only by site modification were strictly excluded from further analysis. After filtering (two valid values in at least one group), remaining missing values were imputed from a normal distribution (width, 0.3 of SD; down shift, 1.8 of SD). The t-test was performed to identify the significantly differentially expressed proteins with a permutation-based FDR <0.01. The log2 label-free quantification intensity of protein expression was z-score normalized for each row. Hierarchical clustering of the z-normalized log2 label-free quantification intensity was performed using euclidean distances between means. The number of clusters was set as 300. Genome ontology enrichment analysis of molecular functions and biological function in differentially expressed proteins was performed using Panther version 13.0 (http://pantherdb.org, last accessed March 30, 2018).27 This classification uses an evolutionary framework to infer protein functions in a species-independent manner.27 The phylogenomics analysis of RC04975 was employing PhyML 3.128 (http://www.atgc-montpellier.fr/phyml, last accessed June 5, 2019). The alignment of the protein sequences of RC0497 from different rickettsial strains was done using T-COFFE 11.00.829 (http://tcoffee.crg.cat, last accessed June 16, 2019).
SID–SRM-MS Validation of Differentially Expressed Secreted Proteins
The SID–SRM-MS assays of selected proteins were developed, as described previously.30 For each peptide, three to five SRM transitions were monitored. The signature peptides and SRM parameters are listed in Supplemental Table S1. The proteins were trypsin digested, as described above. An aliquot of 10 μL of 50 fmol/μL diluted stable isotope-labeled (SIS) peptides (Thermo Scientific, San Jose, CA) was added to each tryptic digest. These samples were desalted with a ZipTip C18 cartridge (Millipore, Burlington, MA). LC-SRM-MS analysis was performed, as described previously,30,31 with a TSQ Vantage triple quadrupole coupled with Eksigent NanoLC-2D HPLC system (AB SCIEX, Dublin, CA). The peak areas in the extract ion chromatography of the native and SIS version of each signature peptide were integrated using Xcalibur 2.1 (Thermo Scientific). The SRM data were manually inspected to ensure peak detection and accurate integration.
In Vivo Challenge with R. conorii
Age- and sex-matched C3H/HeN mice were purchased from Charles River Laboratories (Wilmington, MA). Mice were inoculated intravenously through the tail vein with R. conorii Malish 7 strain with either a sublethal dose (SLD) 0.1 LD50 (5 × 103 plaque-forming units) or a lethal dose (LD) 3 LD50 (1.5 × 105 plaque-forming units) of rickettsiae. Negative controls were inoculated with 300 μL of sucrose-phosphate-glutamate buffer alone. Mice were monitored daily for signs of illness, including ruffled fur, hunched posture, and decreased activity. Mice were sacrificed on day 4 after infection. Mice were anesthetized by inhalational isoflurane (Isoflurane USP; Piramal Healthcare Limited, Digwal, India) and euthanized by carbon dioxide inhalation, followed by cervical dislocation. Whole mouse blood was recovered by cardiac puncture immediately after euthanasia. All animal experiments were conducted in a certified animal biosafety level 3 laboratory at the UTMB. The UTMB Animal Care and Use Committee approved all experiments and procedures, and experiments in mice were performed according to NIH's Guide for the Care and Use of Laboratory Animals.17
Trypsin Digestion of Mouse Plasma or Human Serum and Label-Free Quantification
Plasma (10 μL) was aliquoted from each sample, and 50 μL of 9 mol/L urea was added. The protein concentration was measured by bicinchoninic acid assay. Approximately 200 μg of proteins from each sample were used for protein digestion. The proteins were digested first with endoproteinase Lys-C/Trypsin (Promega) and then trypsin, as described above. The tryptic peptides were desalted, as described above.
PRM Analysis of Rickettsia Protein RC0497
An aliquot of SIS peptides of rickettsia protein RC0497 [LLLSLDSTGEK(13C615N2)] was added into each sample. For PRM analyses, the peptides were analyzed with Easy nLC1000 UHPLC-Q Exactive Orbitrap LC-MS system (Thermo Scientific, San Jose, CA). The resolution of full scan was 70,000 (@m/z 200), the target automatic gain control value was set to 3 × 106, and maximum fill time was 200 milliseconds for full scan; and 17,500 (@m/z 200), a target automatic gain control value of 2 × 105, and maximum fill times of 100 milliseconds for MS2 scan. PRM targeted the pair of peptides of rickettsia protein RC0497 (native and SIS peptides). The assessment of the detection of peptides was performed after acquisition using Skyline version 3.6.0.9321.32,33 For each peptide evaluated, the signals of the five most intense fragment ions (as defined in spectra of SIS peptides of RC0497) were extracted from each corresponding MS/MS spectrum. The MS/MS spectra with five fragment ions detected were submitted to spectral matching. The comparison of the relative intensities of these fragments with those defined in the reference composite MS/MS spectrum was performed on the basis of the dot product (dotp) value. In addition, the retention time of the native and SIS peptides was used as an additional acceptance criterion. The variation of the retention time between the analyte peptides and their SIS counterparts should be within 0.05 minutes.
Generation of Anti-RC0497 Antibody
Protein RC0497 was expressed in Escherichia coli using the recombinant plasmid gifted by Dr. Sanjeev Sahni (University of Texas Medical Branch, Galveston, TX). After expression, the protein was purified using Ni-resin and dialyzed to remove imidazole and benzamidine to optimize thrombin activity, followed by removal of His-tag. Rabbit polyclonal antibodies against RC0497 were generated at Rockland Immunochemicals, Inc. (Limerick, PA). The antibodies then underwent either protein A or affinity purification. The reactivity of polyclonal antibody against RC0497 was confirmed with immunoblotting and IP-SRM.
IP of RC0497 and Trypsin Digestion
Approximately 100 μL of serum was suspended in 1 mL of low ionic strength immunoprecipitation buffer (50 mmol/L NaCl; 25 mmol/L HEPES, pH 7.4; 1% IGEPAL CA-630; 10% glycerol; 1 mmol/L fresh dithiothreitol; and protease inhibitor cocktail). An aliquot of 4 μg of anti-RC0497 antibody or control IgG (rabbit polyclonal; Santa Cruz Biotechnology, Dallas, TX) was used in each IP. The mixture was incubated overnight at 4°C; then, 30 μL of protein A magnetic beads (Dynabeads; Invitrogen, Carlsbad, CA) was added. After incubation at 4°C for 4 hours, the beads were separated from the supernatant with a magnetic stand. The beads were washed with phosphate-buffered saline five times before trypsin digestion. The trypsin digestion was performed as previously described.34
Results
Quantitative Proteomics Analysis of Secretome of HUVECs Infected with R. conorii
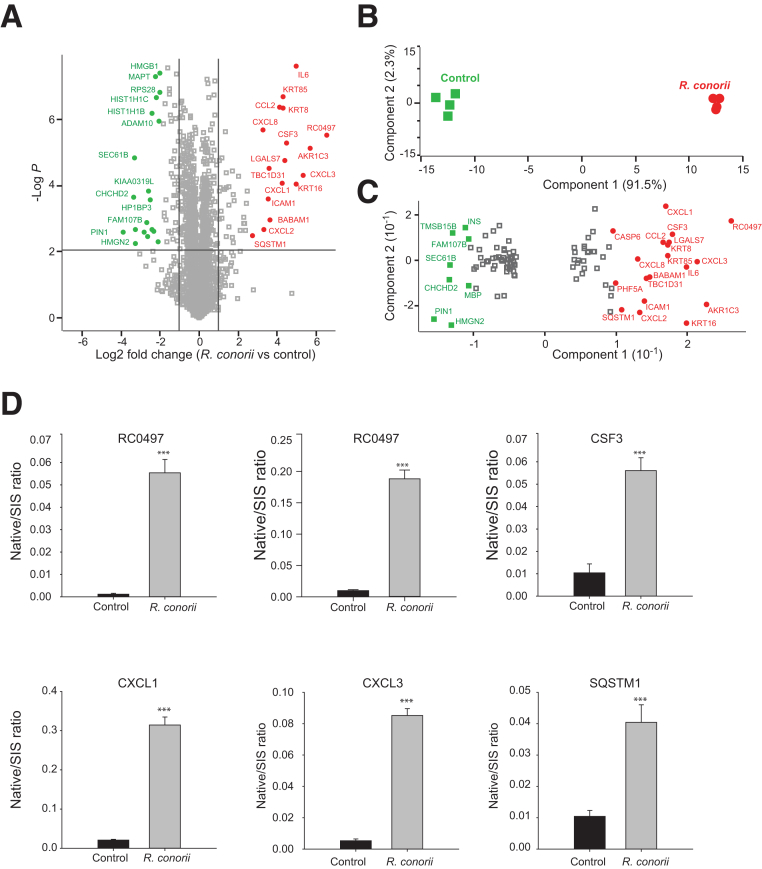
HUVECs are a standardized model for early responses to rickettsial vascular infection.35 To understand the spectrum of proteins secreted/released by endothelial cells in response to R. conorii infection, a quantitative proteomics analysis of the proteins in the conditioned medium of HUVECs with and without R. conorii infection was conducted (Figure 1). The mass spectrometry raw data and proteomics have been deposited in the ProteomeXchange Consortium via the PRIDE36 partner repository (https://www.ebi.ac.uk/pride/archive; data set identifier PXD013548). In this study, 2655 proteins were identified with 1% FDR. Among them, 2620 proteins were quantified in all the replicates (Supplemental Table S2). The intensity of each protein in biological and technical replicates had an excellent agreement (Pearson correlation r > 0.95) (Supplemental Figure S1), confirming that the MS quantification of proteins was robust and reproducible. Proteins satisfying two criteria (t-test with permutation-based FDR <0.01 and a twofold change in the abundance) were considered to be significantly changed by rickettsial infection. With this approach, 104 proteins were identified whose abundance in the conditioned medium was significantly changed by R. conorii infection, including 45 up-regulated and 59 down-regulated proteins. The volcano plot of the logarithm-transformed fold changes in the secretome of HUVECs plotted versus the t-test P values is shown in Figure 2A. Some most significantly up-regulated or down-regulated proteins were highlighted in red (up-regulated by R. conorii infection) and green (down-regulated). A PCA of the 104 significant proteins confirmed the successful quantitative separation between the rickettsia-infected versus control HUVECs (Figure 2B), indicating that the protein abundances in the conditioned medium were sufficiently informative to discriminate the two groups. Next, the proteins driving the separation (the loadings of the multidimensional PCA) were examined, and the proteins with the highest power to separate the rickettsia-infected HUVECs from the control HUVECs were identified. These proteins are highlighted in red and green in Figure 2C and tabulated in Table 1. The proteins on the far right of Figure 2C include the rickettsial protein—putative N-acetylmuramoyl-l-alanine amidase RC0497, human inflammatory cytokines and chemokines [IL-6, CXCL1, CXCL2, CXCL3, CXLC8, and granulocyte colony-stimulating factor 3 (CSF3)], and the autophagy protein sequestosome-1 (SQSTM1/p62), a protein that has been reported to be potentially involved in interactions of rickettsiae with mammalian host cells.37,38
Figure 2.
Quantitative proteomics analysis of the secretome of primary human umbilical vein endothelial cells (HUVECs) infected with Rickettsia conorii. The HUVECs were infected with R. conorii for 24 hours. The cell culture media were collected, and the secretory proteins from HUVECs were analyzed by label-free liquid chromatography–tandem mass spectrometry. A: Volcano plot of protein expression in the secretome of HUVECs infected with R. conorii. The dots above the horizontal line are proteins whose levels in the secretome of HUVECs were significantly changed in response to R. conorii infection (t-test; permutation-based FDR, 1%). Vertical lines indicate the twofold change cutoff. Some most significantly up-regulated or down-regulated proteins are labeled. B: Projection of principal component analysis of 104 proteins whose levels in the secretome of HUVECs were significantly changed in response to R. conorii infection (t-test with permutation-based FDR of 1% and twofold change cutoff). The first two components of data variability of 104 proteins, from four replicates of control HUVECs (green squares) and four replicates of R. conorii–infected HUVECs, are shown. C: Scatter plot depicts the protein feature loadings of component 1 and component 2 of the PCA in Figure 2B. Protein features showing significant importance in dividing control and rickettsial groups were labeled. D: Stable isotope dilution–selected reaction monitoring (SID-SRM)–MS validation of differentially expressed proteins. Five proteins that were up-regulated in the HUVEC secretome by R. conorii were selected for further validation with quantitative SID-SRM-MS. RC0497 was measured with two signature peptides. Error bars are the SEM of SRM measurements. ***P < 0.001 versus control (t-test). ADAM10, disintegrin and metalloproteinase domain-containing protein 10; AKR1C3, aldo-keto reductase family 1 member C3; BABAM1, BRISC and BRCA1-A complex member 1; CCL2, C-C motif chemokine 2; CHCHD2, coiled-coil-helix-coiled-coil-helix domain-containing protein 2; CSF, granulocyte colony-stimulating factor; CXCL1, growth-regulated alpha protein; CXCL2, C-X-C motif chemokine 2; CXCL3, C-X-C motif chemokine 3; CXCL8, interleukin-8; FAM107B, protein FAM107B; HIST1H1B, histone H1.5; HIST1H1C, histone H1.2; HMGB1, high mobility group protein B1; HMGN2, non-histone chromosomal protein HMG-17; HP1BP3, heterochromatin protein 1-binding protein 3; ICAM1, intercellular adhesion molecule 1; IL6, interleukin-6; INS, insulin; KIAA0319L, dyslexia-associated protein KIAA0319-like protein; KRT16, keratin, type I cytoskeletal 16; KRT8, keratin, type II cytoskeletal 8; KRT85, keratin, type II cuticular Hb5; LGALS7, galectin-7; MAPT, microtubule-associated protein tau; MBP, myelin basic protein; PHF5A, PHD finger-like domain-containing protein 5A; PIN1, peptidyl-prolyl cis-trans isomerase NIMA-interacting 1; RC0497, Putative N-acetylmuramoyl-L-alanine amidase RC0497; RPS28, 40S ribosomal protein S28; SEC61B, protein transport protein Sec61 subunit beta; SIS, stable isotope labeled; SQSTM1, sequestosome-1; SQSTM1, sequestosome-1; TBC1D31, TBC1 domain family member 31; TMSB15B, thymosin beta-15B.
Table 1.
The Proteins with the Highest Discrimination Power between Rickettsia conorii–Infected HUVECs and Control HUVECs
| Protein names | Accession no. | Genes | Organism | Log2 FC (R. conorii versus Ctrl) | −log10 P value |
|---|---|---|---|---|---|
| Putative N-acetylmuramoyl-l-alanine amidase RC0497 | Q92IC3 | RC0497 | R. conorii | 6.54 | 5.51 |
| Aldo-keto reductase family 1 member C3 | P42330 | AKR1C3 | Homo sapiens | 5.69 | 5.14 |
| C-X-C motif chemokine 3 | P19876 | CXCL3 | H. sapiens | 5.34 | 4.30 |
| Keratin, type I cytoskeletal 16 | P08779 | KRT16 | H. sapiens | 4.99 | 4.05 |
| IL-6 | P05231 | IL6 | H. sapiens | 4.99 | 7.61 |
| Granulocyte colony-stimulating factor | P09919 | CSF3 | H. sapiens | 4.47 | 5.29 |
| Galectin-7 | P47929 | LGALS7 | H. sapiens | 4.38 | 4.76 |
| Keratin, type II cuticular Hb5 | P78386 | KRT85 | H. sapiens | 4.33 | 6.68 |
| Keratin, type II cytoskeletal 8 | P05787 | KRT8 | H. sapiens | 4.33 | 6.33 |
| Growth-regulated α protein | P09341 | CXCL1 | H. sapiens | 4.26 | 4.06 |
| C-C motif chemokine 2 | P13500 | CCL2 | H. sapiens | 4.15 | 6.38 |
| BRISC and BRCA1-A complex member 1 | Q9NWV8 | BABAM1 | H. sapiens | 3.65 | 2.97 |
| TBC1 domain family member 31 | Q96DN5 | TBC1D31 | H. sapiens | 3.60 | 4.52 |
| Intercellular adhesion molecule 1 | P05362 | ICAM1 | H. sapiens | 3.54 | 3.58 |
| C-X-C motif chemokine 2 | P19875 | CXCL2 | H. sapiens | 3.31 | 2.67 |
| IL-8 | P10145 | CXCL8 | H. sapiens | 3.26 | 5.68 |
| Sequestosome-1 | Q13501 | SQSTM1 | H. sapiens | 2.72 | 2.48 |
| PHD finger-like domain-containing protein 5A | Q7RTV0 | PHF5A | H. sapiens | 2.47 | 3.84 |
| Caspase-6 | P55212 | CASP6 | H. sapiens | 2.40 | 3.47 |
| Myelin basic protein | P02686 | MBP | H. sapiens | −2.65 | 2.44 |
| Protein FAM107B | Q9H098 | FAM107B | H. sapiens | −2.70 | 2.88 |
| Insulin | P01308 | INS | H. sapiens | −2.81 | 2.58 |
| Thymosin β-15B | P0CG35 | TMSB15B | H. sapiens | −3.27 | 2.65 |
| Nonhistone chromosomal protein HMG-17 | P05204 | HMGN2 | H. sapiens | −3.29 | 2.25 |
| Protein transport protein Sec61 subunit β | P60468 | SEC61B | H. sapiens | −3.34 | 4.85 |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial | Q9Y6H1 | CHCHD2 | H. sapiens | −3.36 | 3.65 |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Q13526 | PIN1 | H. sapiens | −3.89 | 2.60 |
Accession numbers of the proteins in the table are from the Uniprot protein database (http://www.uniprot.org).
Ctrl, control; FC, fold change; HUVEC, human umbilical vein endothelial cell.
The changes in the abundance of these proteins in the secretome of HUVECs were further validated with independent quantitative SID-SRM-MS, a targeted MS approach for the detection and accurate quantification of proteins in a complex background.30,39 SID-SRM-MS provides structural specificity and, therefore, is the most accurate approach available for direct quantification of target proteins in a complex mixture. SRM assays were developed for the measurement of five proteins: RC0497, CSF3, CXCL1, CXCL3, and SQSTM1. Using two proteotypic peptides (SDFPAEQIGK and LLLSLDSTGEK), a marked increase of RC0497 abundance was observed in the conditioned medium of R. conorii–infected HUVECs (Figure 2D). Similarly, SID-SRM-MS assays confirmed the elevated secretion of CSF3, CXCL1/3, and SQSTM1 in response to R. conorii infection (Figure 2D).
Host Responses to R. conorii Infection
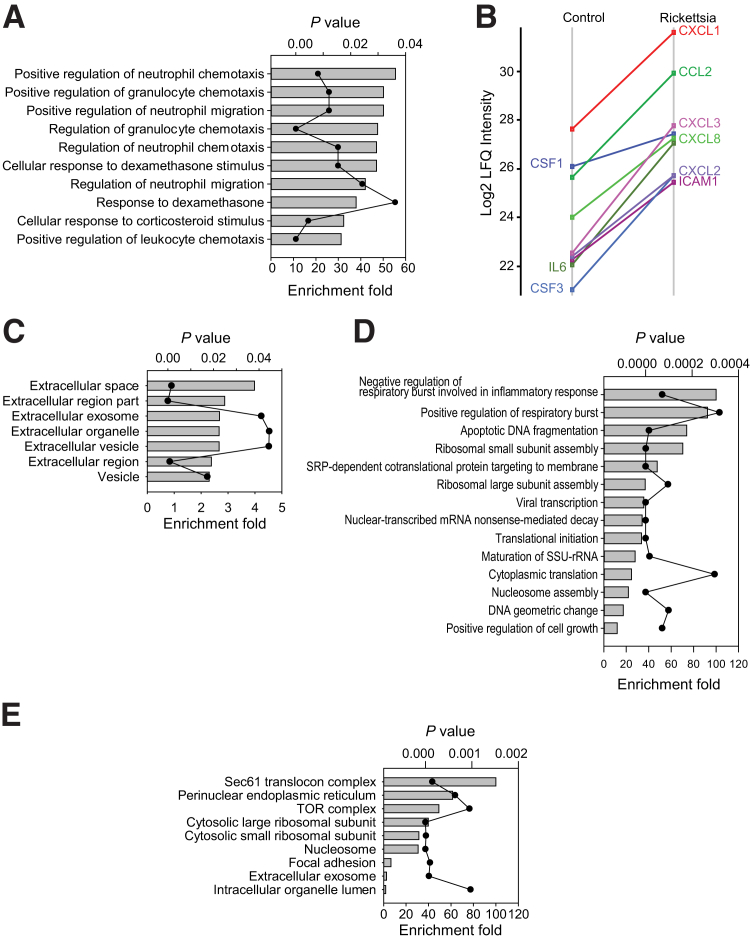
The Genome Ontology (GO) biological process enrichment analysis of the 45 up-regulated human proteins found that these proteins are involved in positive regulation of neutrophil chemotaxis, chemokine-mediated signaling pathways, and inflammation response (Figure 3A). Within this group, the secretion of IL-6, CXCL1/2/3/8, chemokine (C-C motif) ligand 2, CSF1/3, and intercellular adhesion molecule 1 was dramatically induced by R. conorii infection (Figure 3B). The GO cellular component analysis of these 45 proteins showed that the extracellular space proteins were highly enriched, suggesting that these proteins were within the secretory pathway (Figure 3C). More than half of the 45 proteins were identified as extracellular space proteins. Most cytokines and chemokines have NH2-terminal signal peptides and are secreted through classic secretory pathways. By contrast, some proteins, such as SQSTM1 and aldo-keto reductase family 1 member C3, lack signal peptides and are known to be secreted via exosomes, a typical cellular response to stress-autophagy.40, 41, 42 In contrast, the GO biological process enrichment analysis of the 59 down-regulated proteins found that these proteins were highly enriched for negative regulation of respiratory burst involved in inflammatory response, apoptotic DNA fragmentation, and ribosomal small subunit assembly (Figure 3D). Most down-regulated proteins (50 of 59) are organelle proteins. GO cellular component analysis of these 59 proteins showed that Sec61 translocon complex, perinuclear endoplasmic reticulum, and target of rapamycin complex were highly enriched (Figure 3E). According to ExoCarte,43 a web-based database of exosomal proteins, 19 of the 59 proteins are extracellular exosome proteins.
Figure 3.
Gene Ontology (GO) annotation enrichment analysis of differentially expressed secretory proteins. A: GO biological process enrichment analysis of 45 up-regulated secretory proteins. Bars represent fold enrichment of GO annotation; scatter-line plot represents the P value of enrichment analysis. B: Expression of proteins that are involved in the inflammatory response. C: GO cellular component enrichment analysis of 45 up-regulated secretory proteins. D: GO biological function enrichment analysis of 59 down-regulated secretory proteins. E: GO cellular component enrichment analysis of 59 down-regulated secretory proteins. CSF3, granulocyte colony-stimulating factor 3; CCL, chemokine (C-C motif) ligand; ICAM1, intercellular adhesion molecule 1; LFQ, label-free quantification; SSU-rRNA, small subunit ribosomal ribonucleic acid; TOR, target of rapamycin.
Quantitative Proteomics Analysis of the Plasma from Mice Infected with R. conorii
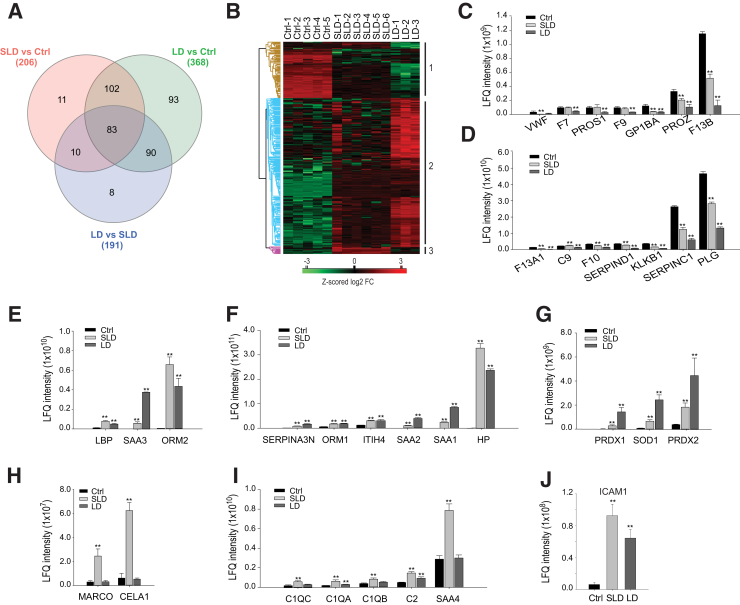
To identify proteins induced in a small animal model of rickettsial infection, mice were infected with an SLD (n = 6) or LD (n = 3) of R. conorii; uninfected animals were used as controls (Ctrls; n = 5). Label-free LC-MS/MS analysis of the mouse plasma identified 608 proteins with 1% FDR. Of these, 534 proteins were quantified in all the samples (Supplemental Table S3). The mass spectrometry raw data have been deposited in the ProteomeXchange Consortium via the PRIDE36 partner repository (https://www.ebi.ac.uk/pride/archive; data set identifier PXD013548). Comparing the intensity of each protein in biological and technical replicates showed excellent agreement (Pearson correlation r = 0.90 to 0.96) (Supplemental Figure S2A), confirming that the reproducibility of the quantification was excellent and robust. To identify the plasma proteins that indicate rickettsial infection or the severity of the disease, three pairwise comparisons were performed: SLD versus Ctrl, LD versus Ctrl, and SLD versus LD. Two criteria, t-test with permutation-based FDR 0.01 and twofold change in the abundance, were used to determine the significant hits in each pairwise comparison (Supplemental Figure S2, B–D). The experiments identified 206, 368, and 191 proteins that met both criteria from each pairwise comparison, respectively. The overlaps of the significant proteins identified from three pairwise comparisons are shown in the Venn diagram (Figure 4A). Overall, a total of 397 significant proteins were identified in the three pairwise comparisons.
Figure 4.
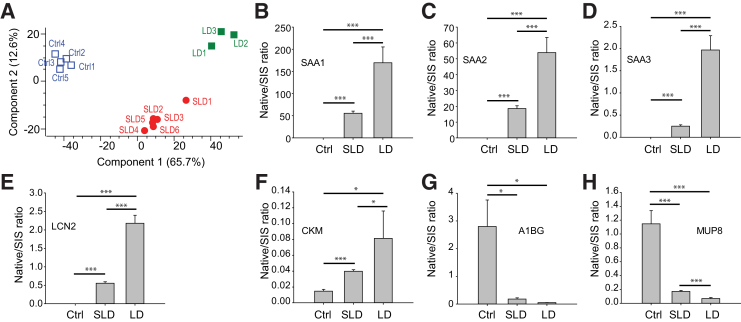
Differentially expressed serum proteins in response to Rickettsia conorii infection. A: Venn diagram of the overlap of the differentially expressed serum proteins from three pairwise comparisons. Data are given for sublethal dose (SLD), lethal dose (LD), and control (Ctrl). B: Heat map of the expression of proteins that are differentially expressed in at least one pairwise comparison. Brown cluster, the proteins were down-regulated after R. conorii infection; blue cluster, the proteins were up-regulated by R. conorii and more so in mice with lethal dose; purple cluster, the proteins were up-regulated by sublethal dose of R. conorii. C and D: The expression of proteins involved in blood coagulation. E and F: The expression of proteins involved in the acute response. G: The expression of proteins involved in the removal of reactive oxygen species. H and I: The expression of proteins involved in the defense response. J: The abundance of intercellular adhesion molecule 1 in the plasma. n = 6 SLD (A); n = LD 3 (A); n = 5 Ctrl (A). **P < 0.01 versus control (t-test with permutation-based false discovery rate of <0.01 in the pairwise comparison). C2, complement C2; C9, complement component C9; C1QA, complement C1q subcomponent subunit A; C1QB, complement C1q subcomponent subunit B; C1QC, complement C1q subcomponent subunit C; CELA1, chymotrypsin-like elastase family member 1; HP, haptoglobin; ITIH4, inter-α-trypsin inhibitor heavy chain H4; KLKB1, plasma kallikrein; LBP, lipopolysaccharide-binding protein; LFQ, label-free quantification; MARCO, macrophage receptor MARCO; ORM, α-1-acid glycoprotein; PLG, plasminogen; PRDX, peroxiredoxin; PROS1, vitamin K-dependent protein S; PROZ, vitamin K-dependent protein Z; SAA, serum amyloid A; SERPINA3N, serine protease inhibitor A3N; SERPINC1, antithrombin-III; SERPIND1, heparin cofactor 2; SOD1, superoxide dismutase; VWF, von Willebrand factor.
Unsupervised hierarchical clustering of these 397 proteins identified three clusters (Figure 4B). The first cluster (Figure 4B) represents proteins whose abundance in plasma was reduced by the rickettsial infection. The GO biological process enrichment analysis of the proteins in this cluster over the reference human proteome found that the proteins are involved in the removal of high-density lipoprotein particle, alternative complement pathway, fibrinolysis, and blood coagulation (Supplemental Table S4). Figure 4, C and D, show the abundances of the proteins that regulate blood coagulation, including plasminogen, coagulation factors (F7, F9, F10, F13A1, and F13B), and von Willebrand factor. These proteins were depleted from the plasma in response to the R. conorii infection, and the magnitude of depletion correlated with infection severity. The second cluster (Figure 4B) identified plasma proteins up-regulated by R. conorii infection, more significantly in the animals treated with an LD of R. conorii. The GO biological process enrichment analysis of the proteins in this cluster determined that proteins involved in the removal of superoxide radicals, gluconeogenesis, and acute-phase response were highly enriched. Figure 4, E and F, show the abundances of the proteins that mediate the acute-phase response. Many of these acute-phase proteins, such as haptoglobin, serum amyloid A-1,2,3 (SAA1/2/3), α-1-acid glycoprotein 1 and 2, and inter-α-trypsin inhibitor heavy chain H4, are expressed by the liver and secreted into plasma,44 suggesting that R. conorii infection induced a robust hepatic acute-phase response. Superoxide dismutase (Cu-Zn) and peroxiredoxin 1 and 2 were up-regulated in the plasma of infected animals, indicating that the host response to R. conorii infection involved antioxidant pathways (Figure 4G). The third cluster (Figure 4B) has plasma proteins that were up-regulated more significantly by the SLD of R. conorii in comparison to the LD infection. The GO biological function enrichment analysis showed that host defense proteins were highly enriched in this cluster. The proteins belong to this category, including macrophage receptor Marco, chymotrypsin-like elastase family member 1 (Figure 4H), and components of the classic pathway of complement systems (Figure 4I). Intercellular adhesion molecule 1, a marker identified in the secretome of R. conorii–infected HUVECs, was elevated in the plasma of R. conorii–infected mice (Figure 4J), indicating the activation of endothelial cells in response to R. conorii infection.45 Other cytokines identified in the secretome of R. conorii–infected HUVECs were undetected in the label-free mass spectrometric analysis of mouse plasma, probably because the presence of high abundance of classic plasma proteins, such as albumin and hepatic acute-phase proteins, hindered the detection of low-abundance cytokines by mass spectrometry.
A PCA of the 397 significantly changed proteins yielded quantitative separation among the animals in the SLD, LD, and uninfected control groups (Figure 5A), indicating that the protein abundances in plasma were sufficiently informative to discriminate between these groups. To identify the proteins with the highest power to separate the SLD group from the control group, a PCA analysis of the 206 significant proteins in the pairwise comparison between SLD and Ctrl was performed. The SLD group was separated entirely from the control group (Supplemental Figure S3A). Next, the proteins driving the separation (the loadings of the multidimensional PCA) were examined to identify the proteins with the highest power to separate these two groups (Supplemental Figure S3B and Table 2). The proteins that have the highest discriminating power between LD and Ctrl groups were also identified by PCA analysis (Supplemental Figure S3, C and D, and Table 2). Many proteins that differentiated LD from controls overlapped with the proteins that separated SLD group from the control group. PCA analysis identified 17 proteins that can differentiate the LD group from SLD group, including uteroglobin, creatine kinase M-type, and plasminogen activator inhibitor 1 (Supplemental Figure S3, E and F, and Table 2).
Figure 5.
Identification of plasma protein markers for Rickettsia conorii infection. A: Principal component analysis of plasma proteins that were significantly changed by sublethal dose (SLD) and lethal dose (LD) of R. conorii infection. Blue open squares are control animals (Ctrl), red solid circles are individual animals with SLD R. conorii infection, and green solid squares are animals with lethal dose R. conorii infection. B–H: Stable isotope dilution–selected reaction monitoring–mass spectrometry validation of plasma protein markers for R. conorii infection and the severity of the infection. n = 6 SLD (A); n = 3 LD (A); n = 5 Ctrl (A). *P < 0.05, ***P < 0.001 (t-test). A1BG, alpha-1B-glycoprotein; CKM, creatine kinase M-type; LCN2, neutrophil gelatinase-associated lipocalin; MUP8, major urinary protein 8; SAA, serum amyloid A; SIS, stable isotope labeled.
Table 2.
The Host Proteins with the Highest Discrimination Power between Controls and SLD and LD Rickettsia conorii Infection
| Protein names | Accession no. | Genes | Log2 FC | −log10 P value |
|---|---|---|---|---|
| SLD versus control | ||||
| Serum amyloid A-2 protein | P05367 | SAA2 | 11.26 | 13.76 |
| Serum amyloid A-1 protein | P05366 | SAA1 | 10.02 | 9.34 |
| Proteoglycan 4 | Q9JM99 | PRG4 | 9.35 | 9.77 |
| Haptoglobin | Q61646 | HP | 8.97 | 10.12 |
| Neutrophil gelatinase-associated lipocalin | P11672 | LCN2 | 8.70 | 7.38 |
| Band 3 anion transport protein | P04919 | SLC4A1 | 8.42 | 6.59 |
| Serum amyloid A-3 protein | P04918 | SAA3 | 8.22 | 7.00 |
| Monocyte differentiation antigen CD14 | P10810 | CD14 | 8.04 | 8.00 |
| α-1-Acid glycoprotein 2 | P07361 | ORM2 | 7.75 | 11.05 |
| Chymotrypsinogen B | Q9CR35 | CTRB1 | 7.05 | 4.76 |
| Glycosylation-dependent cell adhesion molecule 1 | Q02596 | GLYCAM1 | −3.97 | 3.31 |
| Major urinary protein 2 | P11589 | MUP2 | −4.33 | 7.12 |
| Thrombospondin-1 | P35441 | THBS1 | −4.41 | 2.99 |
| α-1B-glycoprotein | Q19LI2 | A1BG | −4.43 | 6.46 |
| Receptor-type tyrosine-protein phosphatase N2 | P80560 | PTPRN2 | −4.77 | 5.47 |
| Major urinary protein 4 | P11590 | MUP4 | −5.20 | 4.17 |
| Major urinary proteins 11 and 8 | P04938 | MUP8 | −6.70 | 4.49 |
| LD versus control | ||||
| Serum amyloid A-2 protein | P05367 | SAA2 | 13.28 | 9.65 |
| Serum amyloid A-1 protein | P05366 | SAA1 | 11.79 | 6.29 |
| Serum amyloid A-3 protein | P04918 | SAA3 | 10.91 | 5.15 |
| Neutrophil gelatinase-associated lipocalin | P11672 | LCN2 | 10.56 | 5.05 |
| Proteoglycan 4 | Q9JM99 | PRG4 | 9.29 | 6.11 |
| Argininosuccinate synthase | P16460 | ASS1 | 9.29 | 5.88 |
| Band 3 anion transport protein | P04919 | SLC4A1 | 9.25 | 5.40 |
| Heat shock protein HSP 90-α | P07901 | HSP90AA1 | 9.15 | 5.51 |
| Triosephosphate isomerase | P17751 | TPI1 | 8.60 | 3.71 |
| Major urinary proteins 11 and 8 | P04938 | MUP8 | −6.42 | 3.93 |
| α-1B-glycoprotein | Q19LI2 | A1BG | −6.63 | 5.65 |
| Major urinary protein 2 | P11589 | MUP2 | −7.00 | 5.52 |
| Insulin-like growth factor–binding protein complex acid labile subunit | P70389 | IGFALS | −7.12 | 3.47 |
| LD versus SLD | ||||
| Uteroglobin | Q06318 | SCGB1A1 | 7.19 | 3.38 |
| Creatine kinase M-type | P07310 | CKM | 5.91 | 3.43 |
| Plasminogen activator inhibitor 1 | P22777 | SERPINE1 | 5.43 | 4.84 |
| Cytosolic 10-formyltetrahydrofolate dehydrogenase | Q8R0Y6 | ALDH1L1 | 5.37 | 4.03 |
| Histone H4 | P62806 | HIST1H4A | 5.26 | 3.80 |
| Argininosuccinate lyase | Q91YI0 | ASL | 5.22 | 3.28 |
| Inorganic pyrophosphatase | Q9D819 | PPA1 | 5.18 | 4.12 |
| Adenylyl cyclase-associated protein 1 | P40124 | CAP1 | 5.17 | 2.56 |
| Estradiol 17 β-dehydrogenase 5 | P70694 | AKR1C6 | 5.03 | 2.23 |
| Nicotinate-nucleotide pyrophosphorylase (carboxylating) | Q91X91 | QPRT | 5.01 | 3.24 |
| Ribonuclease UK114 | P52760 | HRSP12 | 5.01 | 2.10 |
| Tubulin β-4B chain | P68372 | TUBB4B | 4.93 | 4.91 |
| Chymotrypsin-like elastase family member 3B | Q9CQ52 | CELA3B | −4.30 | 3.67 |
| Anionic trypsin-2 | P07146 | PRSS2 | −4.55 | 2.33 |
| Insulin-like growth factor–binding protein complex acid labile subunit | P70389 | IGFALS | −4.68 | 2.96 |
| Carboxypeptidase A1 | Q7TPZ8 | CPA1 | −5.38 | 3.35 |
| BPI fold-containing family A member 2 | P07743 | BPIFA2 | −5.67 | 6.69 |
There were three experimental groups: SLD (n = 6), LD (n = 3), and control (n = 5). Accession numbers of the proteins in the table are from the Uniprot protein database (http://www.uniprot.org).
FC, fold change; LD, lethal dose; SLD, sublethal dose.
Several protein markers, including SAA1/2/3, neutrophil gelatinase-associated lipocalin (LCN2), creatine kinase M-type, alpha-1B-glycoprotein (A1BG), and major urinary protein 8 (MUP8), were selected for further validation with quantitative SID-SRM-MS assays. SID-SRM-MS analysis confirmed the result from the label-free proteomics analysis (Figure 5, B–H). For example, the abundance of SAA1/2/3, LCN2, and creatine kinase M-type was significantly elevated during R. conorii infection (Figure 5, B–F); and the magnitude of up-regulation increased with the severity of infection. Meanwhile, SID-SRM-MS confirmed the down-regulation of A1BG and MUP8 during R. conorii infection (Figure 5, G and H).
SID-PRM-MS Analysis of Rickettsial RC0497 Protein in Mouse Plasma
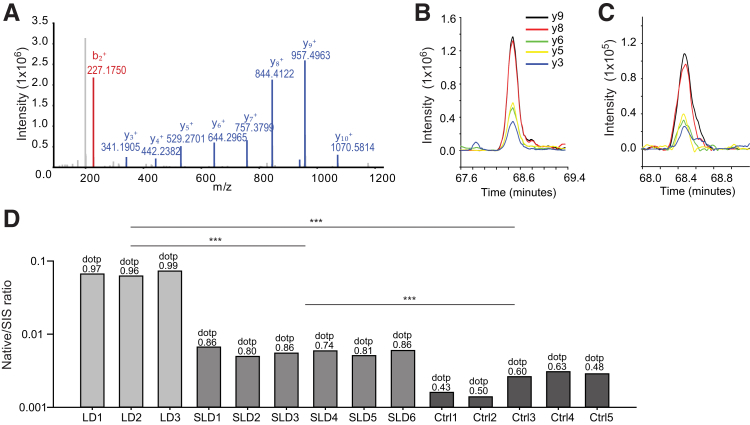
In the analysis of the plasma proteome of the animals infected with R. conorii, no rickettsial protein was identified, including RC0497, probably because of their low abundance in the mouse plasma. To increase the sensitivity of detection, a targeted SID-PRM assay was developed for RC0497. The stable isotope-labeled RC0497 signature peptide LLLSLDSTGE[K(13C6,15N2)] of RC0497 was used to determine the best PRM transitions (Figure 6A). Extracted ion chromatograms for transitions of LLLSLDSTGE[K(13C6,15N2)] and their unlabeled counterparts, yielded by tryptic digest from the plasma of mice infected with LD of R. conorii, are shown in Figure 6, B and C. The similarity in peak shape and retention time of native RC0497 peptide and the stable isotope-labeled standard peptides was examined. The extracted ion chromatogram of the native RC0497 peptide in the plasma of the animal with lethal R. conorii infection (Figure 6C) was remarkably similar to that of the standard peptide (Figure 6B). The dotp value was used to quantitatively measure the degree of the match between spectral library MS/MS and the extracted ion chromatogram of the corresponding transitions of peptide LLLSLDSTGEK.46 A high dotp value (ranging from a value of 0 to 1) indicates a better match and the absence of interfering signals.47 The PRM measurement of RC0497 peptides from LD samples had superior dotp values (>0.95), indicating the unambiguous identification of RC0497 peptide; five of six SLD samples had dotp values >0.8; dotp values of uninfected samples were 0.3 to 0.63, which indicates the absence of the peptides in the sample (Figure 6D). Together, the PRM data confirmed the presence of RC0497 in the plasma of R. conorii–infected animals. Moreover, RC0497 abundance was positively correlated with the load of R. conorii and severity of the infection. Because RC0497 is unique to Rickettsia, and the most highly abundant rickettsial protein in the endothelial secretome, it is an ideal biomarker candidate for the diagnosis of acute rickettsial infections.
Figure 6.
Stable isotope dilution–parallel reaction monitoring–mass spectrometry (SID-PRM-MS) validation of rickettsial protein RC0497. A: Annotated MS/MS spectrum of stable isotope-labeled RC0497 peptide LLLSLDSTGE[K(13C6,15N2)]. B: Liquid chromatography (LC) chromatogram for transitions of stable isotope-labeled standard peptide LLLSLDSTGE[K(13C6,15N2)]. C: LC chromatogram for transitions of native peptide LLLSLDSTGEK from the serum of mice with lethal dose (LD) rickettsial infection. D: SID-PRM-MS quantification of RC0497 peptide LLLSLDSTGEK in the mouse serum. The y axis uses a log10 scale. Dot-product (dotp) values show the expected similarity in peak shape of native peptides to the library spectra. ***P < 0.001 (t-test). Ctrl, control mice; SIS, stable isotope labeled; SLD, sublethal dose.
Conservation of RC0497 in Major Spotted Fever Group Rickettsiae
In this study, RC0497 was identified in the conditioned medium of HUVECs infected with R. conorii with 10 peptides. To determine whether this protein was conserved in other rickettsial strains, a Blast and sequence alignment analysis of RC0497 protein was conducted against the UniProt protein database. This analysis identified 29 rickettsial sequences that resemble RC0497. Proteins with sequences highly homologous to RC0497 are present in Rickettsia rickettsii, Rickettsia monacensis, Rickettsia amblyommaties, Rickettsia parkeri, Rickettsia sibirica, Rickettsia africae, Rickettsia philipii, Rickettsia japonica, and others (Supplemental Table S5). The phylogenomic analysis of protein RC0497 suggests that this protein is highly conserved across diverse rickettsial strains, including R. rickettsii, the causative agent of Rocky Mountain spotted fever (Supplemental Figure S4). Notably, no typhus group rickettsiae were identified. Detection of RC0497 has potential for diagnosis of a wide variety of rickettsial spotted fever infections, and our targeted SRM-MS or PRM-MS assays could be directly transferrable to studies in these other infections. This hypothesis will require direct confirmation measuring RC0497 in the biofluids of mice or humans infected with different spotted fever group rickettsial species.
Verification of RC0497 and Host Proteins in the Sera of Patients with Acute Rickettsiosis
The presence of RC0497 in the serum of patients with rickettsial infection was first verified with targeted MS analysis, but the level of RC0497 in the serum of patients with rickettsial infection was found to be much lower than in the in vivo model, and the sensitivity of PRM or SRM was not enough for detecting RC0497 in patients' sera. To increase the sensitivity of the assay, an IP-SRM assay was developed in which the RC0497 protein was captured from serum with an RC0497-specific antibody and followed by SRM analysis of the enriched protein. Because there is no commercial RC0497 antibody, a polyclonal antibody directed against RC0497 was developed. The reactivity and specificity of the antibody was first examined against RC0497 using immunoblotting. The culture medium and cell lysate of HUVECs infected with R. conorii were collected. The presence of RC0497 in the medium and cell lysate was probed with the anti-RC0497 antibody. The culture medium and cell lysate collected from uninfected cells were used as controls. RC0497 was detected in the culture medium and cell lysate of HUVECs infected with R. conorii; there was no significant detection signal in the negative controls, suggesting that the anti-RC0497 antibody has excellent reactivity and specificity against RC0497 (Supplemental Figure S5A). Next, it was tested whether this antibody was feasible for immunoprecipitation. Recombinant RC0497 was spiked into human serum at the concentrations of 10 and 100 ng/mL. Then, RC0497 protein was exposed to the anti-RC0497 antibody or control IgG, respectively, and pulled down with protein A/G beads. The proteins captured on the beads were digested with trypsin, as described previously.34 The abundance of RC0497 pull down was measured by anti-RC0497 antibody or IgG with SID-SRM-MS. The RC0497 was significantly enriched by anti-RC0497 antibody relative to negative control IgG (Supplemental Figure S5B). These data suggest that anti-RC0497 antibody efficiently pulls down RC0497 and can be used for IP-SRM-MS analysis.
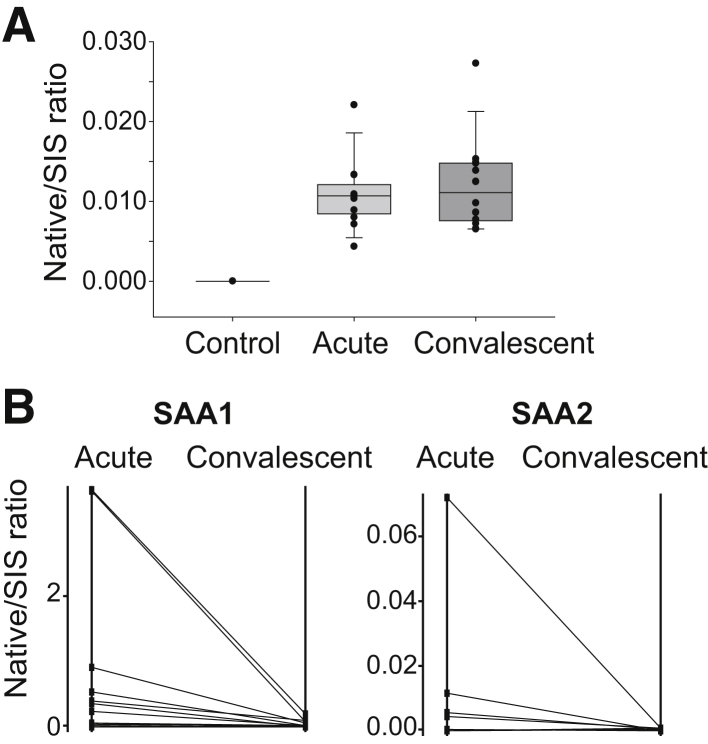
Next, 13 deidentified paired serum samples (acute and convalescent) from confirmed cases of Mediterranean spotted fever collected from discarded diagnostic material were used. Acute serum samples were taken when the patients presented in the hospital during the acute phase of the disease. The convalescent samples were taken 10 to 126 days after the acute samples were taken (Supplemental Table S6). MSF infection was diagnosed by immunofluorescence antibody. The demographic information of participants and the diagnosis of rickettsial infection are tabulated in Supplemental Table S6. Serum samples from six healthy individuals free of rickettsial infection were used as controls (Supplemental Table S7). The IP-SRM-MS assay described above was used to measure the abundance of RC0497 in these samples. The level of RC0497 was elevated in the serum samples from patients with acute MSF relative to healthy controls (Figure 7A). However, a cohort with samples collected at different stages of the acute phase of infection will be needed to determine how soon after onset RC0497 can be detected in the blood. The level of RC0497 in the serum samples from convalescent patients was also elevated relative to healthy controls; and in some cases, the level of RC0497 was even higher than that in the patients' acute samples. The convalescent RC0497 levels were binned into early (10 to 30 days) and late (70 to 126 days) after the acute samples were taken. There is a trend, not statistically significant, for lower levels in the later samples (Supplemental Figure S6). Further experimentation will determine the time course for clearing the antigen after resolution of the infection, which is now enabled by the findings and assays developed by this study.
Figure 7.
Quantification of RC0497 and host proteins in serum from patients infected with Rickettsia conorii. A: Stable isotope dilution–selected reaction monitoring–mass spectrometry (SID-SRM-MS) analysis of RC0497. B: SID-SRM-MS analysis of serum amyloid A-1 protein (SAA1) and serum amyloid A-2 protein (SAA2). SIS, stable isotope labeled.
Also, SRM assays were developed for human host proteins SAA1 and SAA2. Both proteins were significantly up-regulated in the serum of mice infected with R. conorii. The levels of these two proteins in the paired human serum collected during the acute phase of R. conorii infection and convalescence were measured. Compared with convalescence, the levels of SAA1 and SAA2 were markedly elevated, suggesting that the acute-phase response reactants were activated during the early stage of infection and were cleared during convalescence (Figure 7B).
Discussion
Rickettsia conorii is the arthropod-transmitted etiological agent of MSF. After entry, rickettsiae replicate intracellularly, triggering an innate response, resulting in the secretion of cytokines, interferons, cell surface adhesion molecules (including E-selectin, vascular cell adhesion molecule 1, intercellular adhesion molecule 1,48, 49, 50 and αVβ3 integrin51), and procoagulants (tissue factor and von Willebrand factor).52, 53, 54 These endothelial cellular responses mediate essential aspects of the pathobiology of natural human infections, including microvascular hemorrhage, endothelial leakage, and multiorgan failure.2,55 The absence of sensitive clinical assays for accurate diagnosis during the initial clinical presentation at a time when antibiotic treatment would be efficacious has been a major problem.56
Rickettsia conorii replicates primarily in the human endothelium and spreads via intercellular transfer without producing rapid endothelial lysis, resulting in a low abundance of extracellular bacterial proteins in the blood during acute infection. The detection of these markers in the blood with mass spectrometry is hindered by the high abundance of host plasma proteins, such as albumin and hepatic acute-phase response proteins. Therefore, despite significant advances in the development of quantitative proteomics, identification of diagnostic rickettsial antigens in human infections has been a considerable challenge. In addition, because the endothelial infection induces limited cell lysis, PCR-based approaches lack sufficient sensitivity for early detection of disease. To circumvent these limitations, a quantitative proteomics pipeline was applied for analysis of the secretome of primary HUVECs infected with R. conorii to identify 45 host-derived proteins and 1 rickettsia protein, RC0497. RC0497 was identified with 11 peptides and is the only rickettsial protein that was found in the conditioned medium supernatant. In an established mouse model of R. conorii infection, 206 differentially expressed host acute-phase reactants and coagulation factors were identified, but no rickettsial proteins were identified. Therefore, a SID-PRM-MS assay was developed to detect RC0497 present in low abundance in plasma, and it was demonstrated that its circulating levels are proportional to the bacterial burden. Finally, using an IP-enrichment-SRM assay, RC0497 was validated as a diagnostic target in a cohort of humans presenting with acute rickettsial infections versus healthy controls. RC0497 was detectable in convalescence, which is consistent with the previous finding that R. conorii RNA is detectable in the mouse liver and lungs on day 13 after infection.57 Informatics analysis showed that RC0497 is shared with >10 species of the spotted fever group, including R. rickettsii, the causative agent of Rocky Mountain spotted fever. We propose that detection of RC0497 is a sensitive and specific marker for acute spotted fever rickettsial infections.
Our proteomics profiling takes advantage of the fact that all species of rickettsiae infect endothelial cells as a primary target3 and that the secretome is dynamically changed in response to intracellular replication of pathogens.22 This approach provides deep insight into the dynamic changes in the secreted proteins induced by intracellular rickettsial replication. Despite the large numbers of innate cytokines and adhesion factors produced by endothelial cells, surprisingly few rickettsial proteins are found in the conditioned medium, consistent with its nonlytic behavior. Using this approach, it was determined that putative N-acetylmuramoyl-l-alanine amidase, RC0497, is secreted from infected endothelial cells. Another advantage of focusing on the endothelial secretome is that it lends itself to transparent transition to diagnostic assays using blood. Although blood is a complex biofluid populated with highly abundant carrier proteins,58 that does not lend itself to straightforward biomarker discovery approaches, targeted proteomics in blood yield clinically useful diagnostics because this fluid is readily accessible.15,59 Herein, we demonstrate translating discovery proteins to targeted proteomics using parallel reaction monitoring techniques. Although PRMs improve the sensitivity of protein detection, significant increases in sensitivity of detection can be achieved using a preenrichment step. The preenrichment using affinity purification significantly enhances the signal/noise ratio of SRM by reducing background noise.30,31
Rickettsia conorii infection activates the Janus kinase–STAT1–ubiquitin-like protein ISG15 pathway and induces the reprogramming of plasma membrane integrin/cadherin signaling.20 Analysis of the endothelial secretome identified enhanced secretion of cytokines and chemokines, IL-6, CXCL8, CXCL1, and CXCL3 in a manner consistent with previous studies.60 In vivo, CXCL8 secretion is responsible for neutrophil recruitment, and IL-6 is involved in monocyte activation and stimulation of the hepatic acute-phase response.44,60 The earlier finding that rickettsiae up-regulate the intercellular adhesion molecule 1 adhesion factor responsible for promoting the transmigration of activated neutrophils was reproduced.61 The studies herein identify the release of SQSTM1, suggesting the potential involvement of autophagy pathway in the interactions of rickettsiae with human endothelial cells.
In this study, we extended our previously characterized mouse model that has been useful for investigating the immune response to spotted fever group rickettsial infections.62,63 The label-free proteomics studies enabled identification of pathways involved in sublethal versus lethal infections. Of interest, the complement cascade and the macrophage protein Marco were uniquely expressed in sublethal disease (Figure 4H). Activation of complement C3 and the terminal complex has been observed in human plasma during R. conorii infection.64 The complement pathway may play an important role in bacterial lysis and inflammation. Marco is a member of the class A macrophage scavenger receptor family that binds to bacteria,65 playing a role in the antimicrobial innate response. The elevation of these factors may be related to mechanisms involved in the resolution of infection.
The R. conorii genome is predicted to contain approximately 1374 proteins. Of these proteins, our study uniquely identified RC0497 as the most highly abundant protein secreted during rickettsial endovascular infection. RC0497 is a putative N-acetylmuramoyl-l-alanine amidase, an enzymatic activity important in peptidoglycan catabolism. As a protein involved in cell wall metabolism and biogenesis, RC0497 may be secreted during endothelial replication, accounting for its high abundance. In informatics comparisons, 10 proteotypic peptides of RC0497 were identified that are conserved with other members of the spotted fever group rickettsiae. Although this protein has not yet been measured in other spotted fever rickettsial infections, RC0497 is likely to be a diagnostic marker of acute spotted fever infections not only in MSF but also in Rocky Mountain spotted fever in the United States and Latin America. The epidemiology of the spotted fever group of rickettsial infections is muddled by misdiagnosis, underdiagnosis, and underreporting. Our identification of RC0497 and development of an assay that detects the antigen in acute infections will enable early diagnosis, treatment, and accurate public health notification of spotted fever rickettsial infections.
Conclusions
Using an integrated secretome-animal model for diagnostic and biomarker identification, we identified a putative N-acetylmuramoyl-l-alanine amidase, RC0497, as a specific marker of acute R. conorii infections. This study will advance the detection, treatment, and epidemiologic study of spotted fever rickettsial infections.
Acknowledgment
We thank Dr. Sanjeev Sahni (University of Texas Medical Branch, Galveston, TX) for kindly providing the recombinant plasmid for expressing RC0497 in Escherichia coli and SCMM Selected Reaction Monitoring facility for the use of the facility.
Footnotes
Supported by National Center for Advancing Translational Sciences /NIH grants UL1TR001439 (A.R.B.) and UL1TR002373 (A.R.B.), University of Texas Medical Branch Sealy Center for Molecular Sciences (SCMM) pilot funding, and the Institute for Translational Sciences.
Disclosures: A.R.B., Y.Z., D.H.W., and R.F. have submitted a patent provisional application related to this project entitled Methods and Compositions for Diagnosis of Rickettsia Infection (provisional patent application 62/892,048).
Current address of J.B., Department of Biological Sciences, Sam Houston State University, Houston, TX; of C.S., Laboratory of Tumor Immunology and Biology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD; of G.V., School of Public Health, University of California, Berkeley, Berkeley, CA; of A.R.B., Institute for Clinical and Translational Research, University of Wisconsin–Madison, Madison, WI.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2019.10.007.
Contributor Information
Yingxin Zhao, Email: yizhao@utmb.edu.
Allan R. Brasier, Email: abrasier@wisc.edu.
Supplemental Data
Multi–scatter plots of the protein intensities between the samples and replicates. The Pearson correlation of the pairwise comparison of protein intensities between samples and replicates is shown in each scatter plot.
Quantitative proteomics analysis of the plasma of mice infected with Rickettsia conorii. The mice were infected with R. conorii at two different doses [sublethal dose (SLD) and lethal dose (LD)]. The uninfected mice (Ctrl) were used as controls. The mouse plasma proteins were analyzed by label-free liquid chromatography–tandem mass spectrometry. A: Multi–scatter plots of the protein intensities between the samples and replicates. The Pearson correlation of the pairwise comparison of protein intensities between samples and replicates is shown in each scatter plot. B–D: Volcano plot of the comparison of protein expression between SLD and Ctrl (B), LD and Ctrl (C), and LD and SLD (D). The dots above the horizon line are proteins for which the abundance was significantly changed in response to R. conorii infection (t-test; permutation-based false discovery rate, 1%). Vertical lines indicate the twofold change cutoff horizontal lines, the FDR-corrected P value cutoff. Some most significantly up-regulated or down-regulated proteins are labeled. n = 6 SLD (A, B, and D); n = 3 (A, C, and D); n = 5 Ctrl (A, B, and C).
Principal component analysis (PCA). A: PCA of plasma proteins that were significantly changed by sublethal dose (SLD) Rickettsia conorii infection. Red solid circles are individual animals with SLD R. conorii infection. Uninfected animals [controls (Ctrls)] are in blue. B: Loading plot of the variables (proteins) that led to the group clustering shown in A. Red proteins are increased by R. conorii infection, and green proteins are decreased. C: PCA of plasma proteins that were significantly changed by lethal dose (LD) R. conorii infection. Green squares are individual animals with LD R. conorii infection. Uninfected animals (controls) are in blue. D: Loading plot of the variables (proteins) that lead to the group clustering shown in C. Green proteins are increased by LD R. conorii infection, and blue proteins are decreased. E: PCA of plasma proteins that were significantly changed by different doses of R. conorii infection. Green squares are individual animals with LD R. conorii infection. Red solid circles are animals with SLD R. conorii infection. F: Loading plot of the variables (proteins) that led to the group clustering shown in E. Green proteins are increased by LD R. conorii infection, and red proteins are decreased.
Phylogenomics analysis of protein N-acetylmuramoyl-l-alanine amidase from different rickettsia species. The percentage branch support values (red text) derived from maximum likelihood analysis in PhyML 3.1 are indicated on branches. The nodes of the phylogenomic tree are the accession number of RC0497 in each rickettsia species. The RC0497 from Rickettsia conorii identified in this study is in bold. The accession numbers of RC0497 proteins are from the Uniprot database (http://www.uniprot.org).
The evaluation of the antibody (Ab) against Rickettsia RC0497 protein. A: Immunoblotting of RC0497 with a polyclonal antibody against RC0497. B: Immunoprecipitation–selected reaction monitoring (SRM) analysis of RC0497 using a polyclonal anti-RC0497 antibody. IgG was used as the negative control. The RC0497 was pulled down by the polyclonal anti-RC0497 antibody or IgG, and then three RC0497 peptides were measured with stable isotope dilution–SRM–mass spectrometry. *P < 0.05, ***P < 0.001 (t-test). Lys, cell lysate; SIS, stable isotope labeled; SN, supernatant of cell culture medium.
Box plot of the level of serum RC0497 after acute phase of Rickettsia conorii infection. The convalescent samples were taken 10 to 126 days after the acute-phase samples were taken from patients infected with R. conorii. The data were binned into two groups: early phase of convalescence (10 to 30 days) and late phase (70 to 126 days). SIS, stable isotope labeled.
References
- 1.Rovery C., Brouqui P., Raoult D. Questions on Mediterranean spotted fever a century after its discovery. Emerg Infect Dis. 2008;14:1360–1367. doi: 10.3201/eid1409.071133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker D.H., Valbuena G.A., Olano J.P. Pathogenic mechanisms of diseases caused by Rickettsia. Ann N Y Acad Sci. 2003;990:1–11. doi: 10.1111/j.1749-6632.2003.tb07331.x. [DOI] [PubMed] [Google Scholar]
- 3.Valbuena G., Walker D.H. Infection of the endothelium by members of the order Rickettsiales. Thromb Haemost. 2009;102:1071–1079. doi: 10.1160/TH09-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker T.S., Winkler H.H. Penetration of cultured mouse fibroblasts (L cells) by Rickettsia prowazeki. Infect Immun. 1978;22:200–208. doi: 10.1128/iai.22.1.200-208.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez J.J., Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117:5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- 6.Chan Y.G., Cardwell M.M., Hermanas T.M., Uchiyama T., Martinez J.J. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 2009;11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeng R.L., Goley E.D., D'Alessio J.A., Chaga O.Y., Svitkina T.M., Borisy G.G., Heinzen R.A., Welch M.D. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 8.Martinez J.J., Seveau S., Veiga E., Matsuyama S., Cossart P. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123:1013–1023. doi: 10.1016/j.cell.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 9.Renesto P., Dehoux P., Gouin E., Touqui L., Cossart P., Raoult D. Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J Infect Dis. 2003;188:1276–1283. doi: 10.1086/379080. [DOI] [PubMed] [Google Scholar]
- 10.Whitworth T., Popov V.L., Yu X.J., Walker D.H., Bouyer D.H. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect Immun. 2005;73:6668–6673. doi: 10.1128/IAI.73.10.6668-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaechter M., Bozeman F.M., Smadel J.E. Study on the growth of Rickettsiae, II: morphologic observations of living Rickettsiae in tissue culture cells. Virology. 1957;3:160–172. doi: 10.1016/0042-6822(57)90030-2. [DOI] [PubMed] [Google Scholar]
- 12.Teysseire N., Chiche-Portiche C., Raoult D. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res Microbiol. 1992;143:821–829. doi: 10.1016/0923-2508(92)90069-z. [DOI] [PubMed] [Google Scholar]
- 13.Heinzen R.A., Hayes S.F., Peacock M.G., Hackstadt T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993;61:1926–1935. doi: 10.1128/iai.61.5.1926-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silverman D.J., Santucci L.A. Potential for free radical-induced lipid peroxidation as a cause of endothelial cell injury in Rocky Mountain spotted fever. Infect Immun. 1988;56:3110–3115. doi: 10.1128/iai.56.12.3110-3115.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broadhurst L.E., Kelly D.J., Chan C.T., Smoak B.L., Brundage J.F., McClain J.B., Miller R.N. Laboratory evaluation of a dot-blot enzyme immunoassay for serologic confirmation of illness due to Rickettsia conorii. Am J Trop Med Hyg. 1998;58:786–789. doi: 10.4269/ajtmh.1998.58.786. [DOI] [PubMed] [Google Scholar]
- 16.Paddock C.D., Greer P.W., Ferebee T.L., Singleton J., Jr., McKechnie D.B., Treadwell T.A., Krebs J.W., Clarke M.J., Holman R.C., Olson J.G., Childs J.E., Zaki S.R. Hidden mortality attributable to Rocky Mountain spotted fever: immunohistochemical detection of fatal, serologically unconfirmed disease. J Infect Dis. 1999;179:1469–1476. doi: 10.1086/314776. [DOI] [PubMed] [Google Scholar]
- 17.Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council . Eighth Edition. National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 18.Bechelli J., Smalley C., Zhao X., Judy B., Valdes P., Walker D.H., Fang R. MyD88 mediates instructive signaling in dendritic cells and protective inflammatory response during Rickettsial infection. Infect Immun. 2016;84:883–893. doi: 10.1128/IAI.01361-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker D.H., Popov V.L., Wen J., Feng H.M. Rickettsia conorii infection of C3H/HeN mice: a model of endothelial-target rickettsiosis. Lab Invest. 1994;70:358–368. [PubMed] [Google Scholar]
- 20.Zhao Y., Valbuena G., Walker D.H., Gazi M., Hidalgo M., De Sousa R., Oteo J.A., Goez Y., Brasier A.R. Endothelial cell proteomic response to Rickettsia conorii infection reveals activation of the Jak-STAT-ISG15 pathway and reprogramming plasma membrane integrin/cadherin signaling. Mol Cell Proteomics. 2016;15:289–304. doi: 10.1074/mcp.M115.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maroto R., Zhao Y., Jamaluddin M., Popov V.L., Wang H., Kalubowilage M., Zhang Y., Luisi J., Sun H., Culbertson C.T., Bossmann S.H., Motamedi M., Brasier A.R. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J Extracell Vesicles. 2017;6:1359478. doi: 10.1080/20013078.2017.1359478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Jamaluddin M., Zhang Y., Sun H., Ivanciuc T., Garofalo R.P., Brasier A.R. Systematic analysis of cell-type differences in the epithelial secretome reveals insights into the pathogenesis of respiratory syncytial virus-induced lower respiratory tract infections. J Immunol. 2017;198:3345–3364. doi: 10.4049/jimmunol.1601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 26.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 27.Gaudet P., Livstone M.S., Lewis S.E., Thomas P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 29.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., Brasier A.R. Applications of selected reaction monitoring (SRM)-mass spectrometry (MS) for quantitative measurement of signaling pathways. Methods. 2013;61:313–322. doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y.X., Widen S.G., Jamaluddin M., Tian B., Wood T.G., Edeh C.B., Brasier A.R. Quantification of activated NF-kappa B/RelA complexes using ssDNA aptamer affinity-stable isotope dilution-selected reaction monitoring-mass spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.008771. M111.008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egertson J.D., MacLean B., Johnson R., Xuan Y., MacCoss M.J. Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat Protoc. 2015;10:887–903. doi: 10.1038/nprot.2015.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Sun H., Zhang J., Brasier A.R., Zhao Y. Quantitative assessment of the effects of trypsin digestion methods on affinity purification-mass spectrometry-based protein-protein interaction analysis. J Proteome Res. 2017;16:3068–3082. doi: 10.1021/acs.jproteome.7b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker D.H., Firth W.T., Edgell C.J. Human endothelial cell culture plaques induced by Rickettsia rickettsii. Infect Immun. 1982;37:301–306. doi: 10.1128/iai.37.1.301-306.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A.F., Ternent T., Brazma A., Vizcaino J.A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchiyama T., Kishi M., Ogawa M. Restriction of the growth of a nonpathogenic spotted fever group rickettsia. FEMS Immunol Med Microbiol. 2012;64:42–47. doi: 10.1111/j.1574-695X.2011.00879.x. [DOI] [PubMed] [Google Scholar]
- 38.Bechelli J., Vergara L., Smalley C., Buzhdygan T.P., Bender S., Zhang W., Liu Y., Popov V.L., Wang J., Garg N., Hwang S., Walker D.H., Fang R. Atg5 supports Rickettsia australis infection in macrophages in vitro and in vivo. Infect Immun. 2019;87 doi: 10.1128/IAI.00651-18. e00651–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange V., Picotti P., Domon B., Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baixauli F., López-Otín C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demory Beckler M., Higginbotham J.N., Franklin J.L., Ham A.J., Halvey P.J., Imasuen I.E., Whitwell C., Li M., Liebler D.C., Coffey R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prunotto M., Farina A., Lane L., Pernin A., Schifferli J., Hochstrasser D.F., Lescuyer P., Moll S. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J Proteomics. 2013;82:193–229. doi: 10.1016/j.jprot.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., Gangoda L., Mathivanan S. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabay C., Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 45.Videm V., Albrigtsen M. Soluble ICAM-1 and VCAM-1 as markers of endothelial activation. Scand J Immunol. 2008;67:523–531. doi: 10.1111/j.1365-3083.2008.02029.x. [DOI] [PubMed] [Google Scholar]
- 46.Toprak U.H., Gillet L.C., Maiolica A., Navarro P., Leitner A., Aebersold R. Conserved peptide fragmentation as a benchmarking tool for mass spectrometers and a discriminating feature for targeted proteomics. Mol Cell Proteomics. 2014;13:2056–2071. doi: 10.1074/mcp.O113.036475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schilling B., MacLean B., Held J.M., Sahu A.K., Rardin M.J., Sorensen D.J., Peters T., Wolfe A.J., Hunter C.L., MacCoss M.J., Gibson B.W. Multiplexed, scheduled, high-resolution parallel reaction monitoring on a full scan QqTOF instrument with integrated data-dependent and targeted mass spectrometric workflows. Anal Chem. 2015;87:10222–10229. doi: 10.1021/acs.analchem.5b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sporn L.A., Lawrence S.O., Silverman D.J., Marder V.J. E-selectin-dependent neutrophil adhesion to Rickettsia rickettsii-infected endothelial cells. Blood. 1993;81:2406–2412. [PubMed] [Google Scholar]
- 49.Dignat-George F., Teysseire N., Mutin M., Bardin N., Lesaule G., Raoult D., Sampol J. Rickettsia conorii infection enhances vascular cell adhesion molecule-1- and intercellular adhesion molecule-1-dependent mononuclear cell adherence to endothelial cells. J Infect Dis. 1997;175:1142–1152. doi: 10.1086/520353. [DOI] [PubMed] [Google Scholar]
- 50.Damas J.K., Davi G., Jensenius M., Santilli F., Otterdal K., Ueland T., Flo T.H., Lien E., Espevik T., Froland S.S., Vitale G., Raoult D., Aukrust P. Relative chemokine and adhesion molecule expression in Mediterranean spotted fever and African tick bite fever. J Infect. 2009;58:68–75. doi: 10.1016/j.jinf.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Bechah Y., Capo C., Grau G., Raoult D., Mege J.L. Rickettsia prowazekii infection of endothelial cells increases leukocyte adhesion through alphavbeta3 integrin engagement. Clin Microbiol Infect. 2009;15 Suppl 2:249–250. doi: 10.1111/j.1469-0691.2008.02675.x. [DOI] [PubMed] [Google Scholar]
- 52.Clifton D.R., Rydkina E., Huyck H., Pryhuber G., Freeman R.S., Silverman D.J., Sahni S.K. Expression and secretion of chemotactic cytokines IL-8 and MCP-1 by human endothelial cells after Rickettsia rickettsii infection: regulation by nuclear transcription factor NF-kappaB. Int J Med Microbiol. 2005;295:267–278. doi: 10.1016/j.ijmm.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Sporn L.A., Haidaris P.J., Shi R.J., Nemerson Y., Silverman D.J., Marder V.J. Rickettsia rickettsii infection of cultured human endothelial cells induces tissue factor expression. Blood. 1994;83:1527–1534. [PubMed] [Google Scholar]
- 54.Sporn L.A., Marder V.J. Interleukin-1 alpha production during Rickettsia rickettsii infection of cultured endothelial cells: potential role in autocrine cell stimulation. Infect Immun. 1996;64:1609–1613. doi: 10.1128/iai.64.5.1609-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansueto P., Vitale G., Cascio A., Seidita A., Pepe I., Carroccio A., di Rosa S., Rini G.B., Cillari E., Walker D.H. New insight into immunity and immunopathology of Rickettsial diseases. Clin Dev Immunol. 2012;2012:967852. doi: 10.1155/2012/967852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapman A.S., Bakken J.S., Folk S.M., Paddock C.D., Bloch K.C., Krusell A., Sexton D.J., Buckingham S.C., Marshall G.S., Storch G.A., Dasch G.A., McQuiston J.H., Swerdlow D.L., Dumler S.J., Nicholson W.L., Walker D.H., Eremeeva M.E., Ohl C.A., Tickborne Rickettsial Diseases Working Group. CDC Diagnosis and management of tickborne rickettsial diseases: Rocky Mountain spotted fever, ehrlichioses, and anaplasmosis--United States: a practical guide for physicians and other health-care and public health professionals. MMWR Recomm Rep. 2006;55:1–27. [PubMed] [Google Scholar]
- 57.Valbuena G., Bradford W., Walker D.H. Expression analysis of the T-cell-targeting chemokines CXCL9 and CXCL10 in mice and humans with endothelial infections caused by rickettsiae of the spotted fever group. Am J Pathol. 2003;163:1357–1369. doi: 10.1016/S0002-9440(10)63494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson N.L., Anderson N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 59.Makridakis M., Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73:2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Kaplanski G., Teysseire N., Farnarier C., Kaplanski S., Lissitzky J.C., Durand J.M., Soubeyrand J., Dinarello C.A., Bongrand P. IL-6 and IL-8 production from cultured human endothelial cells stimulated by infection with Rickettsia conorii via a cell-associated IL-1 alpha-dependent pathway. J Clin Invest. 1995;96:2839–2844. doi: 10.1172/JCI118354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valbuena G., Walker D.H. Effect of blocking the CXCL9/10-CXCR3 chemokine system in the outcome of endothelial-target rickettsial infections. Am J Trop Med Hyg. 2004;71:393–399. [PubMed] [Google Scholar]
- 62.Walker D.H., Popov V.L., Feng H.M. Establishment of a novel endothelial target mouse model of a typhus group rickettsiosis: evidence for critical roles for gamma interferon and CD8 T lymphocytes. Lab Invest. 2000;80:1361–1372. doi: 10.1038/labinvest.3780144. [DOI] [PubMed] [Google Scholar]
- 63.Osterloh A. Immune response against rickettsiae: lessons from murine infection models. Med Microbiol Immunol. 2017;206:403–417. doi: 10.1007/s00430-017-0514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otterdal K., Portillo A., Astrup E., Ludviksen J.K., Schjalm C., Raoult D., Olano J.P., Halvorsen B., Oteo J.A., Aukrust P., Mollnes T.E., Nilsson P.H. Rickettsia conorii is a potent complement activator in vivo and combined inhibition of complement and CD14 is required for attenuation of the cytokine response ex vivo. Clin Microbiol Infect. 2016;22:734.e1–734.e6. doi: 10.1016/j.cmi.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Elomaa O., Sankala M., Pikkarainen T., Bergmann U., Tuuttila A., Raatikainen-Ahokas A., Sariola H., Tryggvason K. Structure of the human macrophage MARCO receptor and characterization of its bacteria-binding region. J Biol Chem. 1998;273:4530–4538. doi: 10.1074/jbc.273.8.4530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multi–scatter plots of the protein intensities between the samples and replicates. The Pearson correlation of the pairwise comparison of protein intensities between samples and replicates is shown in each scatter plot.
Quantitative proteomics analysis of the plasma of mice infected with Rickettsia conorii. The mice were infected with R. conorii at two different doses [sublethal dose (SLD) and lethal dose (LD)]. The uninfected mice (Ctrl) were used as controls. The mouse plasma proteins were analyzed by label-free liquid chromatography–tandem mass spectrometry. A: Multi–scatter plots of the protein intensities between the samples and replicates. The Pearson correlation of the pairwise comparison of protein intensities between samples and replicates is shown in each scatter plot. B–D: Volcano plot of the comparison of protein expression between SLD and Ctrl (B), LD and Ctrl (C), and LD and SLD (D). The dots above the horizon line are proteins for which the abundance was significantly changed in response to R. conorii infection (t-test; permutation-based false discovery rate, 1%). Vertical lines indicate the twofold change cutoff horizontal lines, the FDR-corrected P value cutoff. Some most significantly up-regulated or down-regulated proteins are labeled. n = 6 SLD (A, B, and D); n = 3 (A, C, and D); n = 5 Ctrl (A, B, and C).
Principal component analysis (PCA). A: PCA of plasma proteins that were significantly changed by sublethal dose (SLD) Rickettsia conorii infection. Red solid circles are individual animals with SLD R. conorii infection. Uninfected animals [controls (Ctrls)] are in blue. B: Loading plot of the variables (proteins) that led to the group clustering shown in A. Red proteins are increased by R. conorii infection, and green proteins are decreased. C: PCA of plasma proteins that were significantly changed by lethal dose (LD) R. conorii infection. Green squares are individual animals with LD R. conorii infection. Uninfected animals (controls) are in blue. D: Loading plot of the variables (proteins) that lead to the group clustering shown in C. Green proteins are increased by LD R. conorii infection, and blue proteins are decreased. E: PCA of plasma proteins that were significantly changed by different doses of R. conorii infection. Green squares are individual animals with LD R. conorii infection. Red solid circles are animals with SLD R. conorii infection. F: Loading plot of the variables (proteins) that led to the group clustering shown in E. Green proteins are increased by LD R. conorii infection, and red proteins are decreased.
Phylogenomics analysis of protein N-acetylmuramoyl-l-alanine amidase from different rickettsia species. The percentage branch support values (red text) derived from maximum likelihood analysis in PhyML 3.1 are indicated on branches. The nodes of the phylogenomic tree are the accession number of RC0497 in each rickettsia species. The RC0497 from Rickettsia conorii identified in this study is in bold. The accession numbers of RC0497 proteins are from the Uniprot database (http://www.uniprot.org).
The evaluation of the antibody (Ab) against Rickettsia RC0497 protein. A: Immunoblotting of RC0497 with a polyclonal antibody against RC0497. B: Immunoprecipitation–selected reaction monitoring (SRM) analysis of RC0497 using a polyclonal anti-RC0497 antibody. IgG was used as the negative control. The RC0497 was pulled down by the polyclonal anti-RC0497 antibody or IgG, and then three RC0497 peptides were measured with stable isotope dilution–SRM–mass spectrometry. *P < 0.05, ***P < 0.001 (t-test). Lys, cell lysate; SIS, stable isotope labeled; SN, supernatant of cell culture medium.
Box plot of the level of serum RC0497 after acute phase of Rickettsia conorii infection. The convalescent samples were taken 10 to 126 days after the acute-phase samples were taken from patients infected with R. conorii. The data were binned into two groups: early phase of convalescence (10 to 30 days) and late phase (70 to 126 days). SIS, stable isotope labeled.