ABSTRACT
Sexual reproduction is the most common form of reproduction among eukaryotes, which is characterized by a series of massive cellular or tissue renovations. Recent studies have revealed novel functions of autophagy during sexual reproductive processes, ranging from yeast to mammals. In mammals, autophagy is indispensable for spermatogenesis and oogenesis, and it participates in early embryonic development and maternal-fetus crosstalk to ensure the development of embryos or fetuses. Thus, autophagy provides the molecular basis for resource allocation among parents and their offspring, providing an important way to benefit the next generation.
Abbreviations: ATG: autophagy-related; Becn1: beclin 1, autophagy related; CMA: chaperone-mediated autophagy; epg: ectopic PGL granules; ES: ectoplasmic specialization; EVTs: extravillous trophoblasts; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; PCD: programmed cell death; PTB: preterm birth; STB: syncytiotrophoblast
KEYWORDS: Autophagy, embryogenesis, gametogenesis, resource allocation, sexual reproduction
Introduction
Sexual reproduction is the most common form of reproduction in eukaryotes, it involves the combination of genetic information from 2 types of specialized reproductive cells called gametes, and creates a new organism who is the blend of both parents. Sexual reproduction is a very complex process, it is characterized by 2 major processes: meiosis, by which the diploid germ cells reduce their chromosome number to produce the haploid gametes; and fertilization, by which 2 gametes fuse with each other to restore the original number of chromosomes. Sexual reproduction processes are even more complex in mammals, for instance after meiosis, drastic morphological and physiological transformations are needed to form the mature gametes, a female’s large ovum (or egg) or a male’s smaller sperm. After fertilization, embryogenesis starts, accompanying maternal-fetal crosstalk, the fetus subsequently develops into a mature baby in the womb [1].
Almost all these stages are characterized by massive cellular/tissue renovations. During sexual reproduction, a large amount of resources is switched to reproductive processes instead of somatic cell proliferation or maintenance. According to the life history theory [2], a tradeoff exists between somatic cells and sex cells. For instance, in Caenorhabditis elegans and Drosophila melanogaster, depletion of progenitor germ cells delays senescence and increases the lifespan of the organism [3–5]. Similarly, castration in rats and some humans prolongs the lifespan [6,7]. The opposite of these circumstances is also true. Once organisms choose to reproduce, resource utilization to sustain the organism is reduced [8,9]. Some animals may invest resources to develop ‘secondary sex characteristics’ to get the right to mate with their spouse [10], even though these conspicuous phenotypes are a handicap to their survival [11]. Other factors that affect lifespan include the number of offspring, the timing of birth, and the initiation of menopause in females [9,12,13]. Therefore, resource allocation between somatic cells and germ cells is not only very important for successful sexual reproduction but also for the long-term survival of an organism or even a species. The mechanisms underlying resource allocation during sexual reproduction have been uncovered recently, and autophagy was found to be an important regulator for resource allocation during these processes.
As an evolutionarily conserved process, autophagy delivers cytoplasmic material such as long-lived proteins and organelles to the lysosome for degradation to provide raw materials for the survival of cells under stress conditions [14–16]. Recent findings have revealed novel functional roles of autophagy in the reproductive process, with an emphasis on resource allocation. Therefore, autophagy might provide a molecular basis for resource allocation during sexual reproduction, thus providing an important way to benefit our offspring.
The pivotal role of autophagy in resource allocation
Autophagy is an intracellular catabolic process that delivers cell components, including proteins, lipids and whole organelles, to lysosomes for degradation and recycling. It includes microautophagy, chaperone-mediated autophagy (CMA), and macroautophagy. Microautophagy occurs by uptake directly at the lysosomal membrane and degrades small molecules in the cytoplasm [17]. CMA requires protein unfolding by chaperone proteins to directly transport substrate proteins across the lysosomal membrane [18,19]. Macroautophagy is mediated by double-membrane vesicles to engulf cytoplasmic cargos that then fuse with lysosomes for degradation [20,21]. Because macroautophagy is the major and extensively studied type of autophagy in sexual reproduction, we will refer to macroautophagy simply as “autophagy” in the following parts of this review. Autophagy can be further divided into bulk and selective autophagy. Upon nutrient deprivation or other relevant stimulations, bulk autophagy leads to the random uptake of cytoplasm into the phagophore, the precursor to the autophagosome. Selective autophagy is responsible for clearing certain components such as protein aggregates, and damaged or superfluous organelles, and they are named after the particular organelle cleared, such as mitophagy (mitochondria), reticulophagy (endoplasmic reticulum), lipophagy (lipid droplets), pexophagy (peroxisome), and so on [22] (Figure 1).
Figure 1.
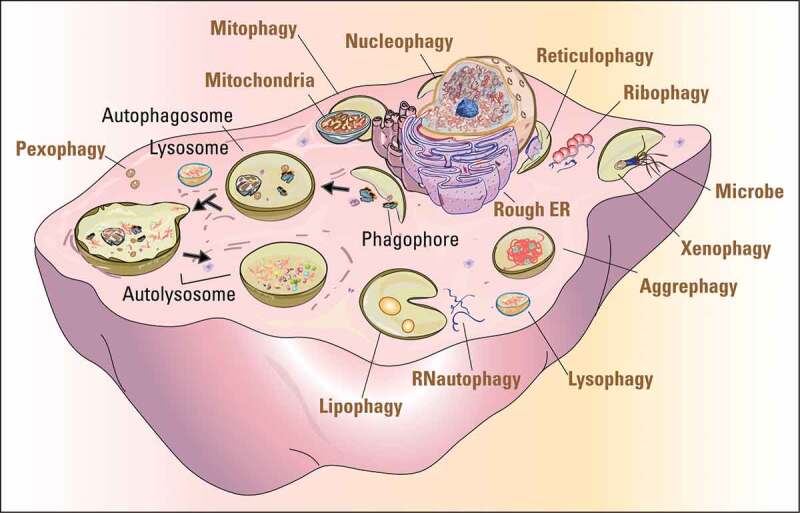
The pivotal role of autophagy in protein and organelle recycling. The cartoon shows the processes of autophagy (black fonts) and the selective degradation of various organelles and other materials (ocher fonts). The dark arrows indicate the sequential processes of autophagy from initiation to final degradation.
The molecular machinery of autophagy is evolutionarily conserved from yeast to mammals, and more than 40 autophagy-related (ATG) proteins have been characterized [23]. In the initiation stage of autophagosome formation, proautophagic signals activate an ULK1/Atg1-ATG13 complex, which triggers the formation of a phagophore [24]. Subsequently, a ubiquitin-activating E1 like enzyme, ATG7, activates ATG12 and then facilitates ATG12–ATG5-ATG16L1 complex formation [25,26]. This complex can function as an E3 ligase for the second ubiquitin-like conjugation system (ATG7 and ATG3), conjugating MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) to the lipid-containing membrane. The LC3-lipid-containing membrane drives membrane expansion to form an autophagosome [27]. The mature autophagosomes fuse with lysosomes to form autolysosomes, and finally the inner contents are digested by lysosomal hydrolases and are further recycled [28].
The products of this autophagic degradation are transported back to the cytoplasm and are used to sustain cell homeostasis in response to nutrient deprivation or other stress conditions [29]. A broad requirement for autophagy-related genes in lifespan extension has been elegantly demonstrated in yeasts, worms, flies, and mice [19]. Therefore, autophagy regulates intracellular resources, optimizes their distribution, and plays a pivotal role in resource allocation in many physiological conditions.
Conserved roles of autophagy in resource allocation during sexual reproduction
Autophagy acts as a governor to regulate resource allocation during sexual reproduction in different taxonomic categories. In yeast, nitrogen starvation is an efficient way to induce the sexual reproductive process of sporulation, which suggests that resource limitation is essential to initiate sexual reproduction [30]. During this process, autophagy is highly activated and any defect in autophagy prevents yeast from initiating meiosis [31]. In Schizosaccharomyces pombe (fission yeast), autophagy is also required for the integrity of the meiotic nuclear structure, efficient meiosis progression, and accurate meiotic chromosome segregation [16]. Recently, we have found that autophagy contributes to premeiotic DNA replication by selective degradation of a meiotic negative regulator [32]. Thus, in unicellular eukaryotes, autophagy might orchestrate resource limitation, by degrading some unnecessary components and negative regulators to induce the sexual reproductive process.
In C. elegans, autophagy critically regulates reproduction at various stages. P granules are specialized protein-RNA aggregates that are found exclusively in germ cells and work as regulators of germ cell development [33]. P granules need to be cleared to ensure proper differentiation of the germline and provide the necessary resources for embryogenesis [34]. Disruption of ATG homologs epg-2, epg-3, epg-4 and epg-5 leads to P granule accumulation [35–37], and SEPA-1 (Suppressor of Ectopic P granules in Autophagy mutant 1) directly binds to a key P granule element, PGL-3, and also to the autophagy protein LGG-1/Atg8. This ‘bridging’ mediates the specific recognition and degradation of P granule components via autophagy [35]. Moreover, a large proportion of germline cells at the meiotic pachytene stage undergo programmed cell death (PCD), and the dead cells might provide cytoplasmic components to support the development of oocytes [38]. In addition, autophagy removes apoptotic cell corpses for resource allocation [39,40]. After fertilization, autophagy is essential for removing paternal mitochondria and mitochondrial DNA before the 64-cell embryonic stage, and autophagy mutant zygotes, such as atg-7, atg-13, bec-1, lgg-1 or lgg-3 fail to remove them in late embryos and even in larvae [41–43]. During nematode zygotic stages, membrane organelles in spermatids, which are specialized vesicular structures, are engulfed by phagophores [44]. Thus, autophagy extensively participates in ‘self-eating’ and resource allocation to promote pre-existing resource recycling to ensure the nematode’s reproduction.
A potential role of autophagic vacuoles in the nurse cell of D. melanogaster has been proposed for nearly 40 years [45]. During oogenesis, PCD primarily takes place at 3 stages of egg chamber development, including the germarium before the follicle cell layer form, the pre-vitellogenic stages, and the egg nears maturation stages [46]. Cell death in the germarium and pre-vitellogenic stages occurs sporadically in well-fed flies and increases dramatically in response to developmental abnormalities or poor environmental conditions. In contrast, late-stage cell death occurs during the development of the egg, and nurse cells undergoing PCD transport their cytoplasm through ring canals into the oocytes to support their normal development [46]. Autophagy takes place in the germarium, during midoogenesis, and in dying nurse cells. Germline-specific knockdown of autophagy results in stage 14 egg chambers with some persisting nurse cell nuclei and further cell death [47,48]. In follicle cells, autophagy is essential for proper oogenesis and might contribute to communication between the follicle cells and germline cells [49]. Thus, autophagy participates in Drosophila’s reproduction by primarily mediating appropriate cell death to provide resources for oogenesis.
Autophagy also plays important roles in resource allocation in plant sexual reproduction. The tapetum is the innermost layer of the anther and provides both nutrients and lipid components to developing microspores, pollen grains, and pollen coat. Similar to nematodes and flies, PCD of the tapetum is indispensable for fertility in plants. In rice, autophagy mutants showed complete sporophytic male sterility and fail to accumulate lipid and starch components in pollen grains at the flowering stage, indicating autophagy within tapetum cells is required for the metabolic regulation and nutrient supply in anthers [50]. In Arabidopsis thaliana, ATG6 has been reported to be essential for pollen germination [51]. Thus, although the functional roles of autophagy in reproduction are diverse in different model organisms, its core function in resource allocation is well conserved (Table 1).
Table 1.
The functional roles of autophagy in the sexual reproduction of different species.
| Organisms | Functions | Autophagy-related genes | Refs |
|---|---|---|---|
| Yeast | Protein or organelle recycling | ATG7 | [14,15,52] |
| Chromosome segregation | ATG1, ATG7, ATG14 | [16] | |
| Premeiotic DNA replication | ATG12 | [32] | |
| C. elegans | P granule degradation | epg-2, epg-3, epg-4, epg-5, sepa-1 | [35–37] |
| Apoptotic cell corpse clearance | bec-1 | [39] | |
| Physiological germline cell death | atg-2, atg-3, atg-5, lgg-1, epg-1, epg-4, | [40] | |
| Paternal mitochondria removal | atg-7, bec-1, lgg-1, lgg-3, epg-1 | [41–43] | |
| Drosophila | Cell death in the ovary | Atg1, Atg7, Atg13, Vps34 | [47,48] |
| Follicle cell development | Atg1, Atg13 | [49] | |
| Arabidopsis thaliana | Pollen germination | ATG6 | [50] |
| Oryza sativa | Nutrient supply in tapetum cells | OsATG7 | [51] |
| Mouse | Testosterone biosynthesis | Atg5, Atg7 | [62] |
| Ectoplasmic specialization assembly | Atg5, Atg7 | [64] | |
| Acrosome biogenesis | Atg7, Sirt1, Tbc1d20 | [69,72,73] | |
| Spermatid differentiation | Atg7 | [74] | |
| Prevent excessive loss of oocytes | Becn1, Atg7 | [76,77] | |
| Promote progesterone synthesis | Becn1 | [83] | |
| Early embryogenesis | Atg5, Becn1 | [84,85] | |
| Organ remodeling | Rb1cc1, Ambra1 | [91,92] | |
| Prevent placental infection | Atg16l1 | [98] | |
| Labor | Atg4c, Atg7 | [101] | |
| Prevent neonatal lethality |
Atg3, Atg5, Atg7, Atg9, Atg16l1 |
[27,53–56] |
Autophagy-dependent resource allocation during mammalian reproduction
Mammals have far more complex reproductive systems than others. Autophagy participates in 4 major reproductive processes in mammals: spermatogenesis, oogenesis, embryogenesis, and maternal-fetal crosstalk. Autophagy might work as a resource governor to regulate resource allocation during these processes.
The male reproductive system
High-quality sperm and mating rights are the key issues for males’ success in reproduction [57], and all of them are dependent on testis in mammals. The testis consists of some somatic cells (such as Leydig and Sertoli cells) and germ cells. The Leydig cells localize in the testicular interstitium and produce androgen hormones, such as testosterone, androstenedione, and dehydroepiandrosterone (DHEA), to promote the development of secondary sexual characteristics and drive sexual behavior under the control of pituitary hormones [58]. The Sertoli cells are the major somatic cells in the seminiferous tubules that support germ cells and construct the microenvironment for spermatogenesis. In the seminiferous tubules, diploid spermatocytes undergo a series of successive cellular renovation and differentiation, comprising spermatogonial mitosis, spermatocytic meiosis and spermiogenesis to produce haploid sperms [59]. Autophagy participates in almost all of the above-mentioned processes to ensure the successful male reproductive function.
Autophagy is extremely active in Leydig cells [60]. We, together with other groups, found that low levels of autophagic flux in Leydig cells are associated with the decline of serum testosterone [61,62]. By generating 2 Leydig cell-specific autophagy-deficient mouse models, we found that the disruption of autophagy influences the sexual behavior of aging male mice, due to a defect in cholesterol uptake and further reduced serum testosterone, which is similar to the symptoms of late-onset hypogonadism. Further investigation revealed that once autophagic flux is disrupted, SLC9A3R2/NHERF2 (solute carrier family 9 [sodium/hydrogen exchanger], member 3 regulator 2) accumulates in Leydig cells, which results in the downregulation of SCARB1/SR-BI (scavenger receptor class B, member 1) and eventually leads to insufficient cholesterol supply [62]. Thus, autophagy in Leydig cells might regulate cholesterol allocation to produce androgen hormones and support spermatogenesis.
Sertoli cells are nurse cells that provide structural support and nourishment to developing germ cells during spermatogenesis. Functional cell interconnections in the seminiferous tubules are maintained through both Sertoli-Sertoli cell and Sertoli-germ cell junctions [63]. Furthermore, ectoplasmic specialization (ES) is essential for Sertoli-Sertoli and Sertoli-germ cell communication to support all phases of germ cell development and maturity. We recently revealed a novel role for autophagy in Sertoli-germ cell communication where it regulates the cytoskeleton to maintain the proper organization of the ES. The well-organized cytoskeleton structure was disturbed in both autophagy-deficient testis and Sertoli cells. A negative regulator of cytoskeleton organization, PDLIM1 (PDZ and LIM domain 1 [elfin]), was found to interact with LC3 and be degraded via the autophagy pathway. In autophagy-deficient Sertoli cells, PDLIM1 accumulation leads to cytoskeletal disorganization, disrupting both apical and basal ES, and finally affects spermatogenesis [64]. Thus, autophagy plays a very important role in spermiogenesis by regulating cytoskeletal organization and Sertoli-germ cell communication.
In male germ cells, autophagy is involved in spermatid differentiation, including acrosome biogenesis, spermatozoa flagella biogenesis and cytoplasm removal. The acrosome is a special membranous structure located at the anterior part of the sperm head [65], which is essential for sperm-egg fusion and male fertility [66]. The malformation or loss of the acrosome results in globozoospermia, which is a rare but severe human infertility syndrome [67]. Acrosome biogenesis starts when proacrosomic granules derived from trans Golgi stacks are transported to and accumulated in the concave region of the nuclear surface, followed by fusion with each other to form a single large acrosomic granule that associates with the nuclear membrane [68]. We found that germ cell-specific atg7 knockout results in a globozoospermia-like phenotype due to a malformed acrosome. Moreover, autophagy is involved in proacrosomic vesicle transport or fusion in the acrosome [69]. Because both acrosomes and lysosomes share low internal acidic pH, and they contain many lysosomal-related enzymes [70], the acrosome is thought to be a form of modified lysosome or a lysosome-related organelle [71]. Together with our new results, we propose that the acrosome might originate from an autolysosome rather than lysosome alone. In support of the above hypothesis, Sirt1 (sirtuin 1) and Tbc1d20 (TBC1 domain family, member 20) were found to participate in acrosome biogenesis by regulating autophagic flux [72,73].
Except for the acrosome biogenesis defect in atg7 null mice, we found that the spermatozoa motility also dropped significantly with some extra cytoplasm retained on the mature sperm head. This defect is also due to the accumulation of PDLIM1, which needs to be degraded by autophagic pathway to facilitate the proper organization of the cytoskeleton in spermatids [74]. Therefore, autophagy participates in not only the transport of proacrosomic granules but also cytoplasm removal during spermiogenesis, and both of them are essential for the maturation of sperm.
In summary, at least 3 novel functions of autophagy have been uncovered in the male reproductive system. Autophagy mediates the crosstalk between spermatids and Sertoli or Leydig cells (Figure 2) and plays an important role in resource allocation to ensure proper spermatogenesis.
Figure 2.
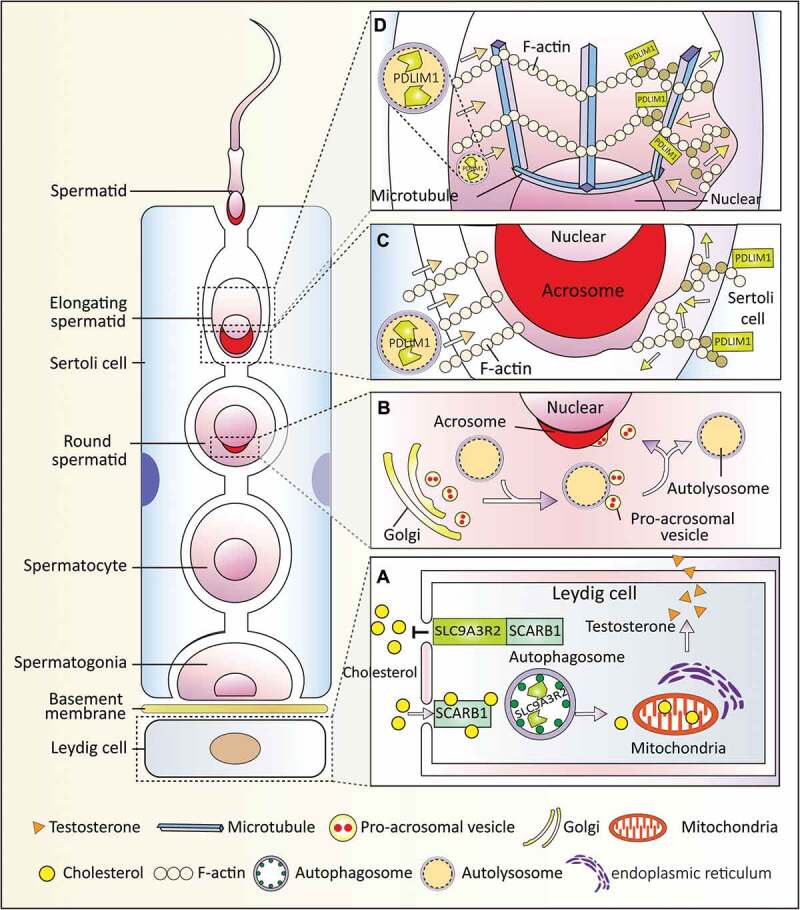
Autophagy-dependent resource allocation in the male reproductive system. Autophagy participates in somatic-germ cell interactions and spermiogenesis in the male reproductive systems. (a) Autophagy participates in testosterone synthesis by regulating cholesterol uptake. In the Leydig cells, SCARB1 functions as a receptor for high-density lipoprotein, and autophagy promotes cholesterol uptake by eliminating its negative regulator SLC9A3R2. (b) Autophagy is involved in acrosome biogenesis. Autophagy participates in proacrosomic vesicle transport and further fusion into a single acrosome on the nuclear membrane. (c and d) Autophagy regulates cytoplasm removal and sperm head shaping by selective degradation of a negative cytoskeleton assembly regulator (PDLIM1) in both Sertoli cells (c) and spermatids (d). PDLIM1 is degraded by autophagy to ensure the proper organization of the cytoskeleton in the apical ES of the Sertoli cell and in the manchette of spermatids.
The female reproductive system
In mammals, the reproductive processes in females include folliculogenesis, ovulation, luteinization, fertilization, embryogenesis and implantation, all involving dramatic physiological changes and resource allocation from mother to offspring. Autophagy has been found to play multiple roles during all these processes.
During the establishment of the primordial follicle pool in most mammalian ovaries, significant germ cell loss occurs around the time of birth. Autophagy could protect the oocytes from excessive loss in the neonatal ovaries [75], which is highly associated with the quantity of oocyte during oogenesis. In Becn1+/- ovaries, perturbed autophagy leads to a 56% reduction of germ cells at postnatal day 1 [76]. Recently, we found that germ cell-specific atg7 knockout results in more than 50% oocyte cell loss during the neonatal transition period [77]. In contrast, the induced autophagy in cultured ovaries treated with rapamycin for 5 days results in a 25%-30% increase in oocyte number. Actually, fetuses always obtain nutrients from the mother before their births; once born, the transplacental nutrient supply is suddenly interrupted, and the babies have to start the arterial supply system, thus facing severe starvation until nutrient supply is restored. Germ cells are sensitive to nutrient deprivation, and autophagy might be induced to protect the excessive loss of the germ cells [77] likely by mobilizing prestored materials. Male germ cells have a self-renewal ability, whereas female germ cells cannot proliferate after birth; thus only the oocytes show a massive loss. These results suggest that autophagy may act as a protective or survival mechanism during germ cell cyst breakdown and the formation of the primordial follicle pool in mammalian ovaries [75].
During folliculogenesis, only 1% of all follicles develop up to the maturation stage and eventually the fertilization stage, whereas others undergo atresia at different developmental stages, indicating that a tradeoff is involved to optimize resource allocation in this process [78]. Traditionally, follicle atresia was thought to be triggered by massive granulosa cell apoptosis, but some researchers found that autophagy is involved in atresia in different species, such as rat [79], human [80], goose and quail [81]. Some recent investigations suggest that BCL2 (B cell leukemia/lymphoma 2) family members might serve as the key regulator in balancing apoptosis and autophagy during follicle atresia [82].
The corpus luteum is the main endocrine structure for progesterone synthesis in the female reproductive system. After ovulation, the remains of the ovulated follicle are transformed into the corpus luteum, which maintains an appropriate level of progesterone secretion to ensure offspring survival throughout the whole gestation period. Autophagy promotes lipid droplet formation and progesterone synthesis in luteal cells, which prevents the fetus from preterm birth (PTB) [83]. Because progesterone is the key mediator of reproductive costs, such as preparing the uterus for embryonic implantation and supporting placenta development, autophagy might further participate in hormone-regulated maternal behavior.
The studies described above have highlighted the indispensable roles of autophagy in oogenesis, fertilization, and gestation (Figure 3). Autophagy not only plays an important role in metabolism and homeostasis but also serves as a global organizer of the resources in these processes.
Figure 3.
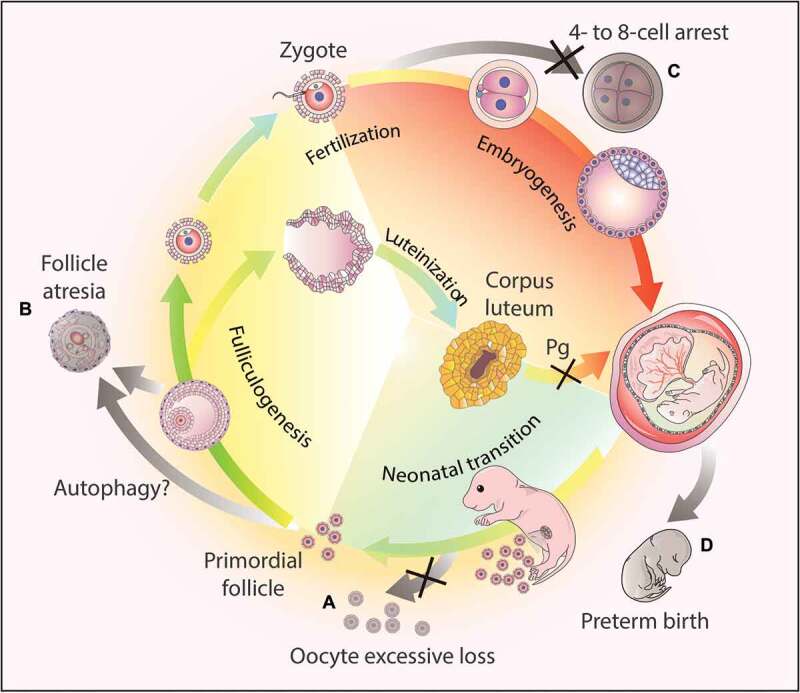
Autophagy-dependent resource allocation in the female reproductive processes. (a) Autophagy might protect the oocytes from excesive loss in the neonatal ovaries. (b) Autophagy participates in follicle atresia, albeit the exact mechanism remains to be elucidated. (c) Autophagy actively participates in early embryogenesis, and the disruption of the autophagic flux results in 4- to 8-cell arrest. (d) Hormone synthesis. After ovulation, the corpus luteum forms from the remains of the ovulated follicles to secrete progesterone (Pg). Once autophagy is disrupted in this process, the synthesis of progesterone decreases due to insufficient lipid droplets in the luteal cells, and finally leads to preterm birth.
Early embryogenesis
Drastic protein/mRNA turnover and organelle remodeling take place after fertilization, suggesting that nutrient distribution must be altered to adapt for the next generation. Autophagy is highly induced within 4 h after fertilization, and autophagy-dependent protein/RNA and organelle degradation are involved in this process [84]. Autophagy is also essential for early embryonic development [84]. A complete autophagy deficiency leads to mouse embryo arrest at 4- to 8-cell stages and ultimately results in embryonic lethality. However, when fertilizing atg5-/- oocytes containing maternally inherited ATG5 proteins with atg5-/- sperm, embryos develop normally, but then experience postnatal death. This result suggests that autophagy is vital in pre-implantation development, but not later development of the embryo. Further analysis showed that the protein synthesis rates in autophagy-deficient embryos were decreased by 30% compared with wild-type embryos [84], indicating autophagy promotes metabolic resource allocation during the pre-implantation development stage.
During early embryogenesis, a gradual decrease in Atg gene transcription takes place during the morula to blastocyst stages [85], which is proposed to prevent the destruction of crucial factors needed for further embryonic development [86] or for implantation and invasion into the maternal uterus [87]. When implantation is delayed, the mouse blastocysts go into a dormant state [88], and autophagy can be induced to extend the longevity of the dormant blastocysts in the uterus during the delayed implantation [89]. During this dormant state, autophagy might be activated to degrade unnecessary components to sustain the survival of these embryos at the cost of developmental fitness, such as impaired fetal growth.
After implantation, gastrulation begins, and autophagy is very important for this stage to progress normally. Severe developmental delay is observed in Becn1-defective embryos, and they die at E7.5 [85]. Remarkably, autophagy may prevent the accumulation of dead cells, contributing to dead cell corpse clearance resulting from PCD during cavitation [90]. Furthermore, autophagy is also associated with organ remodeling during embryonic development. RB1CC1/FIP200 (RB1-inducible coiled-coil 1; a potential mammalian counterpart of yeast Atg17) is essential for the growth of liver and heart, and its absence in mice leads to embryonic death at mid/late gestation [91]. AMBRA1 (autophagy/beclin 1 regulator 1) regulates neural tube cell proliferation and maintains their survival during the development of the mouse nervous system [92]. Taken together, autophagy not only promotes the removal of ‘outdated’ cells but also facilitates the development of ‘updated’ cells and organs, highlighting the essential role of autophagy in resource allocation during early embryogenesis.
Maternal-fetal crosstalk
During pre-implantation, the development of the early embryo relies primarily on the resources that are stored in the egg, whereas after implantation, the ectoderm of the embryo develops into the placenta to support fetal growth and development via various maternal resources. Autophagy also plays essential roles in maintaining the maternal-fetal crosstalk.
After implantation, the trophoblast cells differentiate into villous trophoblasts and extravillous trophoblasts (EVTs). The EVTs invade the maternal decidua and aggregate in the lumen of spiral arteries, producing a trophoblastic plug to support embryo and placental growth. Autophagy is highly activated in the EVTs, and its disruption suppresses the invasion and vascular remodeling process [93]. Moreover, altered autophagy in villous trophoblasts of the placenta is observed in women experiencing pre-eclampsia and intrauterine growth restriction [94], suggesting a possible target to treat complicated pregnancies in the future.
The syncytiotrophoblast (STB), a multinucleated continuous cell layer, is another important component of the placenta and comes from the fused cytotrophoblast, which covers the surface of the placenta. The STB not only promotes resource allocation [95,96] but also secretes a variety of hormones and factors to contribute to embryonic development and homeostasis [97]. In normal-term human placenta, increased autophagic activity is found during trophoblast syncytialization, and a high level of ATG16L1 is associated with the resistance to infection in STB. The loss of ATG16L1 makes the placenta more susceptible to infections, which can lead to PTB in mice [98]. Thus, autophagy promotes resistance to pathogen invasion in STB.
Autophagy is also involved in the labor process. Granulosa cell-specific becn1 knockout significantly reduces progesterone synthesis in mice, which leads to PTB [83]. Additionally, induced autophagy through rapamycin treatment rescues PTB in mice with uterine Trp53 deletion [99]. The labor process also matters a lot. Compared with a spontaneous vaginal delivery placenta, autophagy in a cesarean section placenta is highly activated, probably in response to nutrition restriction due to the fasting prior to surgery [100]. Moreover, a relationship between altered autophagic flux and inflammation-induced preterm labor has been reported, as ATG4C and ATG7 decrease significantly in the uterus and the placenta during inflammation-induced preterm labor [101]. Altered autophagic flux enhances inflammatory responses by stimulating the activation of NFKB/NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B cells) and proinflammatory cytokines/chemokines in both the uterus and the placenta. This evidence suggests that autophagy-dependent resource allocation occurs at the eve of birth, and autophagy contributes to the resource allocation from mother to fetus, eventually contributing to maternal-fetal crosstalk.
Conclusions and perspective
Sexual reproduction is an important part of an individual’s life, and autophagy plays important roles in resource allocation, including prestored resource mobilization and the clearance of unnecessary subcellular components to ensure the normal execution of this process. Subsequently, recycled materials can support further cellular renovation during various stages of sexual reproduction including spermatogenesis, folliculogenesis, fertilization, embryogenesis, placental formation and labor. Therefore, autophagy regulates resource allocation and provides a way to ‘eat ourselves’ to benefit our offspring.
Although several novel functions of autophagy have been uncovered during the reproductive processes, many open questions remain to be addressed in the future. For example, the ubiquitin-proteasome system (UPS) also participates in resource allocation by promoting the degradation of short-lived proteins [102]. The relationship between these 2 degradation systems during the reproductive process remains to be elucidated. Do microautophagy and CMA also play a role in the reproductive process? What are the functional roles of different selective autophagy types during reproduction? Do these autophagy types have their own substrates or do they share some common roles during cellular component recycling? Although the interface between some nursing cells and germ cells has been uncovered, whether autophagy is involved in nutrient supply to the germ cells is unclear. Moreover, both the tapetum of the plant anther and the STB of the human placenta share a common feature: they are multinuclear structures that mediate resource exchange between 2 different cell types or tissues. Autophagy in tapetum cells is required for metabolic regulation and nutrient supply in anthers [103], does autophagy also play a similar role for the nutrient supply in the STB? To address these questions, researchers need to generate more specialized animal models and develop more cutting edge methods. In combination with high-resolution imaging and high-throughput omics, the above-mentioned systems may provide great avenues to comprehensively understand the functional roles of autophagy in the future [104,105].
In addition to the previously mentioned experimental challenges, autophagy-related clinical studies in reproductive medicine have their own set of challenges. Many of them are far from well conducted. More efforts are needed to establish the causal relationship between autophagic defects and reproduction-related diseases. Autophagy-specific activators or inhibitors are urgently needed to treat reproduction-related diseases such as late-onset hypogonadism or primary ovarian insufficiency. Therefore, we think that the following decades will witness the vigorous development and clinical application of autophagy-based investigations.
Funding Statement
This work was supported by the National Natural Science Foundation of China [91649202]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDA16020701]; National Key R&D Program of China [2016YFA0500901] .
Acknowledgments
We thank Li Yu and Tracey Baas for critical reading of the manuscript. This work was funded by the National Natural Science Foundation of China (grants 91649202), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA16020701), and the National Key R&D Program of China (grant 2016YFA0500901).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Smith JM, Szathmary E.. The major transitions in evolution. New York: Oxford University Press; 1997. [Google Scholar]
- [2].Stearns SC The evolution of life histories. 1992. (575 S81). [Google Scholar]
- [3].Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010. August;1204:156–162. PubMed PMID: 20738286. [DOI] [PubMed] [Google Scholar]
- [4].Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999. May 27;399(6734):362–366. PubMed PMID: 10360574. [DOI] [PubMed] [Google Scholar]
- [5].Flatt T. Survival costs of reproduction in Drosophila. Exp Gerontol. 2011. May;46(5):369–375. PubMed PMID: 20970491. [DOI] [PubMed] [Google Scholar]
- [6].Drori D, Folman Y. Environmental effects on longevity in the male rat: exercise, mating, castration and restricted feeding. Exp Gerontol. 1976;11(1–2):25–32. PubMed PMID: 1278267. [DOI] [PubMed] [Google Scholar]
- [7].Min KJ, Lee CK, Park HN. The lifespan of Korean eunuchs. Curr Biol. 2012. September 25;22(18):R792–R793. PubMed PMID: 23017989. [DOI] [PubMed] [Google Scholar]
- [8].Gavrilov LA, Gavrilova NS. Evolutionary theories of aging and longevity. ScientificWorldJournal. 2002. February 7;2:339–356. PubMed PMID: 12806021; PubMed Central PMCID: PMCPMC6009642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wainer-Katsir K, Zou JY, Linial M. Extended fertility and longevity: the genetic and epigenetic link. Fertil Steril. 2015. May;103(5):1117–1124. PubMed PMID: 25796320. [DOI] [PubMed] [Google Scholar]
- [10].Starr C, Taggart R, Evers C, et al. Biology: the unity and diversity of life. Toronto: Nelson Education; 2014. [Google Scholar]
- [11].Gould SJ. The origin and function of “bizarre” structures: antler size and skull size in the “Irish elk,” Megaloceros giganteus. Evolution. 1974;28(2):191–220. [DOI] [PubMed] [Google Scholar]
- [12].Ehrlich S. Effect of fertility and infertility on longevity. Fertil Steril. 2015. May;103(5):1129–1135. PubMed PMID: 25934598. [DOI] [PubMed] [Google Scholar]
- [13].Korpelainen H. Fitness, reproduction and longevity among European aristocratic and rural Finnish families in the 1700s and 1800s. Proc Biol Sci. 2000. September 7;267(1454):1765–1770. PubMed PMID: 12233775; PubMed Central PMCID: PMCPMC1690744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005. September 9;280(36):31582–31586. PubMed PMID: 16027116. [DOI] [PubMed] [Google Scholar]
- [15].Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 2011. February 25;6(2):e17412. PubMed PMID: 21364763; PubMed Central PMCID: PMCPMC3045454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsuhara H, Yamamoto A. Autophagy is required for efficient meiosis progression and proper meiotic chromosome segregation in fission yeast. Genes Cells. 2016. January;21(1):65–87. PubMed PMID: 26696398. [DOI] [PubMed] [Google Scholar]
- [17].Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012. April;69(7):1125–1136. PubMed PMID: 22080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014. January;24(1):92–104. PubMed PMID: 24281265; PubMed Central PMCID: PMC3879702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018. September;19(9):579–593. PubMed PMID: 30006559. DOI: 10.1038/s41580-018-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013. March 21;495(7441):389–393. PubMed PMID: 23455425. [DOI] [PubMed] [Google Scholar]
- [21].Nascimbeni AC, Giordano F, Dupont N, et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. Embo J. 2017. July 14;36(14):2018–2033. PubMed PMID: 28550152; PubMed Central PMCID: PMC5509996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell. 2017. April 10;41(1):10–22. PubMed PMID: 28399394; PubMed Central PMCID: PMC5395098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018. May;20(5):521–527. 10.1038/s41556-018-0092-5. PubMed PMID: 29686264. [DOI] [PubMed] [Google Scholar]
- [24].Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000. September 18;150(6):1507–1513. PubMed PMID: 10995454; PubMed Central PMCID: PMCPMC2150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007. December 28;282(52):37298–37302. PubMed PMID: 17986448. [DOI] [PubMed] [Google Scholar]
- [26].Fujita N, Itoh T, Omori H, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008. May;19(5):2092–2100. PubMed PMID: 18321988; PubMed Central PMCID: PMCPMC2366860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sou YS, Waguri S, Iwata J, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008. November;19(11):4762–4775. PubMed PMID: 18768753; PubMed Central PMCID: PMCPMC2575156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. PubMed PMID: 21801009. [DOI] [PubMed] [Google Scholar]
- [29].Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008. August;4(6):740–743. PubMed PMID: 18567941. [DOI] [PubMed] [Google Scholar]
- [30].Mata J, Lyne R, Burns G, et al. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002. September;32(1):143–147. PubMed PMID: 12161753. [DOI] [PubMed] [Google Scholar]
- [31].Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993. October 25;333(1–2):169–174. PubMed PMID: 8224160. [DOI] [PubMed] [Google Scholar]
- [32].Wen FP, Guo YS, Hu Y, et al. Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy. 2016;12(4):671–688. PubMed PMID: 27050457; PubMed Central PMCID: PMCPMC4835973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Voronina E. The diverse functions of germline P-granules in Caenorhabditis elegans. Mol Reprod Dev. 2013. August;80(8):624–631. PubMed PMID: 23150384. [DOI] [PubMed] [Google Scholar]
- [34].Yang P, Zhang H. You are what you eat: multifaceted functions of autophagy during C. elegans development. Cell Res. 2014. January;24(1):80–91. PubMed PMID: 24296782; PubMed Central PMCID: PMCPMC3879703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y, Yan L, Zhou Z, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009. January 23;136(2):308–321. PubMed PMID: 19167332. [DOI] [PubMed] [Google Scholar]
- [36].Tian Y, Li Z, Hu W, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010. June 11;141(6):1042–1055. PubMed PMID: 20550938. [DOI] [PubMed] [Google Scholar]
- [37].Zhang G, Lin L, Qi D, et al. The composition of a protein aggregate modulates the specificity and efficiency of its autophagic degradation. Autophagy. 2017. September 2;13(9):1487–1495. PubMed PMID: 28806108; PubMed Central PMCID: PMCPMC5612515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gumienny TL, Lambie E, Hartwieg E, et al. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999. February;126(5):1011–1022. PubMed PMID: 9927601. [DOI] [PubMed] [Google Scholar]
- [39].Huang S, Jia K, Wang Y, et al. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy. 2013. February 1;9(2):138–149. PubMed PMID: 23108454; PubMed Central PMCID: PMCPMC3552879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang H, Lu Q, Cheng S, et al. Autophagy activity contributes to programmed cell death in Caenorhabditis elegans. Autophagy. 2013. December;9(12):1975–1982. PubMed PMID: 24185352. [DOI] [PubMed] [Google Scholar]
- [41].Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 2011. December;21(12):1662–1669. PubMed PMID: 22105480; PubMed Central PMCID: PMCPMC3234996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Al Rawi S, Louvet-Vallee S, Djeddi A, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011. November 25;334(6059):1144–1147. PubMed PMID: 22033522. [DOI] [PubMed] [Google Scholar]
- [43].Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011. November 25;334(6059):1141–1144. PubMed PMID: 21998252. [DOI] [PubMed] [Google Scholar]
- [44].Merlet J, Rubio-Pena K, Al Rawi S, et al. Autophagosomal sperm organelle clearance and mtDNA inheritance in C. elegans. Adv Anat Embryol Cell Biol. 2018. November 23. DOI: 10.1007/102_2018_1. [PubMed PMID: 30467692]. [DOI] [PubMed] [Google Scholar]
- [45].Witkus ER, Altman LG, Taparowsky JA. An investigation of the presence of smooth endoplasmic reticulum and GERL during vitellogenesis in the ovary of Drosophila melanogaster. Exp Cell Biol. 1980;48(6):373–383. PubMed PMID: 6773830. [PubMed] [Google Scholar]
- [46].Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol. 2013. November;23(11):567–574. PubMed PMID: 23968895; PubMed Central PMCID: PMCPMC3839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nezis IP, Shravage BV, Sagona AP, et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010. August 23;190(4):523–531. PubMed PMID: 20713604; PubMed Central PMCID: PMCPMC2928014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hou YC, Chittaranjan S, Barbosa SG, et al. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008. September 22;182(6):1127–1139. PubMed PMID: 18794330; PubMed Central PMCID: PMCPMC2542474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Barth JM, Szabad J, Hafen E, et al. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011. June;18(6):915–924. PubMed PMID: 21151027; PubMed Central PMCID: PMCPMC3131947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kurusu T, Koyano T, Hanamata S, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014. May;10(5):878–888. PubMed PMID: 24674921; PubMed Central PMCID: PMCPMC5119067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Harrison-Lowe NJ, Olsen LJ. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy. 2008. 4;Apr(3):339–348. PubMed PMID: 18227644. [DOI] [PubMed] [Google Scholar]
- [52].Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992. October;119(2):301–311. PubMed PMID: 1400575; PubMed Central PMCID: PMCPMC2289660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004. December 23;432(7020):1032–1036. PubMed PMID: 15525940. [DOI] [PubMed] [Google Scholar]
- [54].Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005. May 9;169(3):425–434. PubMed PMID: 15866887; PubMed Central PMCID: PMCPMC2171928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saitoh T, Fujita N, Hayashi T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009. December 8;106(49):20842–20846. PubMed PMID: 19926846; PubMed Central PMCID: PMCPMC2791563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008. November 13;456(7219):264–268. PubMed PMID: 18849965. [DOI] [PubMed] [Google Scholar]
- [57].Robert T. Paternal investment and sexual selection. Sexual Selection & the Descent of Man. New York: Aldine de Gruyter; 1972. p. 136-179. [Google Scholar]
- [58].Purvis K, Cusan L, Hansson V. Regulation of steroidogenesis and steroid action in Leydig cells. J Steroid Biochem. 1981. December;15:77–86. PubMed PMID: 7040817. [DOI] [PubMed] [Google Scholar]
- [59].Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1–15. PubMed PMID: 19856159. [DOI] [PubMed] [Google Scholar]
- [60].Tang XM [The autophagic activity of Leydig cells in normal rat testes]. Shi Yan Sheng Wu Xue Bao. 1988. March;21(1):119–129. PubMed PMID: 3201844. [PubMed] [Google Scholar]
- [61].Li WR, Chen L, Chang ZJ, et al. Autophagic deficiency is related to steroidogenic decline in aged rat Leydig cells. Asian J Androl. 2011. November;13(6):881–888. PubMed PMID: 21822295; PubMed Central PMCID: PMCPMC3739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gao F, Li G, Liu C, et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J Cell Biol. 2018. June 4;217(6):2103–2119. PubMed PMID: 29618492; PubMed Central PMCID: PMCPMC5987723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998. August;9(4):411–416. PubMed PMID: 9813187. [DOI] [PubMed] [Google Scholar]
- [64].Liu C, Wang H, Shang Y, et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy. 2016. May 3;12(5):814–832. PubMed PMID: 26986811; PubMed Central PMCID: PMCPMC4854559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kang-Decker N, Mantchev GT, Juneja SC, et al. Lack of acrosome formation in Hrb-deficient mice. Science. 2001. November 16;294(5546):1531–1533. PubMed PMID: 11711676. [DOI] [PubMed] [Google Scholar]
- [66].Jin M, Fujiwara E, Kakiuchi Y, et al. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc Nat Acad Sci. 2011;108(12):4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu C, Li W. The molecular mechanism underlying acrosome biogenesis. spermatogenesis: molecular mechanisms, regulation and biological perspectives. New York: Nova Science Publishers; 2016. [Google Scholar]
- [68].Mortimer D, Menkveld R. Sperm morphology assessment–historical perspectives and current opinions. J Androl. 2001. Mar–Apr;22(2):192–205. PubMed PMID: 11229793 [PubMed] [Google Scholar]
- [69].Wang H, Wan H, Li X, et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014. July;24(7):852–869. PubMed PMID: 24853953; PubMed Central PMCID: PMCPMC4085765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Moreno RD, Alvarado CP. The mammalian acrosome as a secretory lysosome: new and old evidence. Mol Reprod Dev. 2006. November;73(11):1430–1434. PubMed PMID: 16894549. [DOI] [PubMed] [Google Scholar]
- [71].Hartree EF. The acrosome-lysosome relationship. J Reprod Fertil. 1975. July;44(1):125–126. PubMed PMID: 1171229. [DOI] [PubMed] [Google Scholar]
- [72].Liu C, Song Z, Wang L, et al. Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development. 2017. February 1;144(3):441–451. PubMed PMID: 28003215. [DOI] [PubMed] [Google Scholar]
- [73].Sidjanin DJ, Park AK, Ronchetti A, et al. TBC1D20 mediates autophagy as a key regulator of autophagosome maturation. Autophagy. 2016. October 2;12(10):1759–1775. PubMed PMID: 27487390; PubMed Central PMCID: PMCPMC5079675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shang Y, Wang H, Jia P, et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy. 2016. September;12(9):1575–1592. PubMed PMID: 27310465; PubMed Central PMCID: PMCPMC5082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sun YC, Wang YY, Sun XF, et al. The role of autophagy during murine primordial follicle assembly. Aging (Albany NY). 2018. February 5;10(2):197–211. PubMed PMID: 29410391; PubMed Central PMCID: PMCPMC5842841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gawriluk TR, Hale AN, Flaws JA, et al. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction. 2011. June;141(6):759–765. PubMed PMID: 21464117. [DOI] [PubMed] [Google Scholar]
- [77].Song ZH, Yu HY, Wang P, et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis. 2015. January;15(6):e1589. PubMed PMID: 25590799; PubMed Central PMCID: PMCPMC4669757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang J, Liu Y, Yao W, et al. Initiation of follicular atresia: gene networks during early atresia in pig ovaries. Reproduction. 2018. July;156(1):23–33. PubMed PMID: 29743261. [DOI] [PubMed] [Google Scholar]
- [79].Choi J, Jo M, Lee E, et al. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertil Steril. 2011. March 15;95(4):1482–1486. PubMed PMID: 20630503. [DOI] [PubMed] [Google Scholar]
- [80].Serke H, Vilser C, Nowicki M, et al. Granulosa cell subtypes respond by autophagy or cell death to oxLDL-dependent activation of the oxidized lipoprotein receptor 1 and toll-like 4 receptor. Autophagy. 2009. October;5(7):991–1003. PubMed PMID: 19730000. [DOI] [PubMed] [Google Scholar]
- [81].Markstrom E, Svensson E, Shao R, et al. Survival factors regulating ovarian apoptosis – dependence on follicle differentiation. Reproduction. 2002. January;123(1):23–30. PubMed PMID: 11869183. [DOI] [PubMed] [Google Scholar]
- [82].Yuan J, Zhang Y, Sheng Y, et al. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11(7):1081–1098. PubMed PMID: 26060891; PubMed Central PMCID: PMCPMC4590641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gawriluk TR, Ko C, Hong X, et al. Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proc Natl Acad Sci U S A. 2014. October 7;111(40):E4194–203. PubMed PMID: 25246579; PubMed Central PMCID: PMCPMC4210046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tsukamoto S, Kuma A, Murakami M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008. July 4;321(5885):117–120. PubMed PMID: 18599786. [DOI] [PubMed] [Google Scholar]
- [85].Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003. December 9;100(25):15077–15082. PubMed PMID: 14657337; PubMed Central PMCID: PMCPMC299911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Egli D, Rosains J, Birkhoff G, et al. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447(7145):679. [DOI] [PubMed] [Google Scholar]
- [87].Oh SY, Roh CR. Autophagy in the placenta. Obstet Gynecol Sci. 2017. May;60(3):241–259. PubMed PMID: 28534010; PubMed Central PMCID: PMCPMC5439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Weitlauf HM. Metabolic changes in the blastocysts of mice and rats during delayed implantation. J Reprod Fertil. 1974. July;39(1):213–224. PubMed PMID: 4604758. [DOI] [PubMed] [Google Scholar]
- [89].Lee JE, Oh HA, Song H, et al. Autophagy regulates embryonic survival during delayed implantation. Endocrinology. 2011. May;152(5):2067–2075. PubMed PMID: 21363932. [DOI] [PubMed] [Google Scholar]
- [90].Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007. March 9;128(5):931–946. PubMed PMID: 17350577. [DOI] [PubMed] [Google Scholar]
- [91]. Gan B, Peng X, Nagy T, et al. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006. October 9;175(1):121–133. PubMed PMID: 17015619; PubMed Central PMCID: PMCPMC2064504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007. June 28;447(7148):1121–1125. PubMed PMID: 17589504. [DOI] [PubMed] [Google Scholar]
- [93].Saito S, Nakashima A. Review: the role of autophagy in extravillous trophoblast function under hypoxia. Placenta. 2013. March;34 Suppl:S79–84. PubMed PMID: 23306070. [DOI] [PubMed] [Google Scholar]
- [94].Nakashima A, Aoki A, Kusabiraki T, et al. Autophagy regulation in preeclampsia: pros and cons. J Reprod Immunol. 2017. September;123:17–23. PubMed PMID: 28869810. [DOI] [PubMed] [Google Scholar]
- [95].Goldman-Wohl D, Yagel S. United we stand not dividing: the syncytiotrophoblast and cell senescence. Placenta. 2014. June;35(6):341–344. PubMed PMID: 24709558. [DOI] [PubMed] [Google Scholar]
- [96].Huppertz B. IFPA award in placentology lecture: biology of the placental syncytiotrophoblast–myths and facts. Placenta. 2010. March;31 Suppl:S75–81. PubMed PMID: 20042237. [DOI] [PubMed] [Google Scholar]
- [97]. Lash GE, Robson SC, Bulmer JN. Review: functional role of uterine natural killer (uNK) cells in human early pregnancy decidua. Placenta. 2010. March;31 Suppl:S87–S92. PubMed PMID: 20061017. [DOI] [PubMed] [Google Scholar]
- [98].Cao B, Macones C, Mysorekar IU. ATG16L1 governs placental infection risk and preterm birth in mice and women. JCI Insight. 2016. December 22;1(21):e86654. PubMed PMID: 28018968; PubMed Central PMCID: PMCPMC5161251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hirota Y, Cha J, Yoshie M, et al. Heightened uterine mammalian target of rapamycin complex 1 (mTORC1) signaling provokes preterm birth in mice. Proc Natl Acad Sci U S A. 2011. November 1;108(44):18073–18078. PubMed PMID: 22025690; PubMed Central PMCID: PMCPMC3207648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Signorelli P, Avagliano L, Virgili E, et al. Autophagy in term normal human placentas. Placenta. 2011. June;32(6):482–485. PubMed PMID: 21459442. [DOI] [PubMed] [Google Scholar]
- [101].Agrawal V, Jaiswal MK, Mallers T, et al. Altered autophagic flux enhances inflammatory responses during inflammation-induced preterm labor. Sci Rep. 2015. March;23(5):9410. PubMed PMID: 25797357; PubMed Central PMCID: PMCPMC4369745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Douglas PM, Dillin A. The disposable soma theory of aging in reverse. Cell Res. 2014. January;24(1):7–8. PubMed PMID: 24189044; PubMed Central PMCID: PMCPMC3879701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Kurusu T, Koyano T, Kitahata N, et al. Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal Behav. 2017. September 2;12(9):e1365211. PubMed PMID: 28873038; PubMed Central PMCID: PMCPMC5640179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104]. Harland RM. A new view of embryo development and regeneration. Science. 2018. June 1;360(6392):967–968. PubMed PMID: 29853675. [DOI] [PubMed] [Google Scholar]
- [105].Filippi-Chiela EC, Bueno E Silva MM, Thome MP, et al. Single-cell analysis challenges the connection between autophagy and senescence induced by DNA damage. Autophagy. 2015;11(7):1099–1113. PubMed PMID: 25701485; PubMed Central PMCID: PMCPMC4590630. [DOI] [PMC free article] [PubMed] [Google Scholar]


