Abstract
Background
Prior colonization by Klebsiella pneumoniae and vancomycin-resistant Enterococci (VRE) is associated with subsequent infection, particularly in intensive care unit (ICU) populations. Screening for VRE colonization, but not K. pneumoniae, is routinely performed in some health care systems. Identification of patient factors associated with K. pneumoniae colonization could enable infection prevention.
Methods
ICU patients were screened for VRE and K. pneumoniae by rectal swab culture over 2 time periods: July–October 2014 (n = 1209) and January–May 2016 (n = 1243). Patient demographics, baseline laboratory data, comorbidities, and outcomes were analyzed. 16S rRNA gene-based analysis was performed on a subset of patients (n = 248) to identify microbiota characteristics associated with VRE and K. pneumoniae colonization.
Results
K. pneumoniae colonization (17.3% of patients in the 2014 cohort, 7.3% in 2016) was significantly associated with VRE colonization in multivariable analysis (P = .03 in 2016; P = .08 in 2014). VRE colonization was associated with poor underlying health, whereas K. pneumoniae colonization was associated with advanced age. The most prevalent operational taxonomic units were Escherichia coli/Shigella spp., Klebsiella, and Enterococcus, consistent with high rates of detectable K. pneumoniae and VRE by culture. Microbial community structure in noncolonized patients was significantly different from those with VRE, K. pneumoniae, or both, attributable to differences in the relative abundance of Klebsiella and Enterococcus.
Conclusions
K. pneumoniae co-colonizes with VRE and is a predominant taxon in ICU patients, but colonization was not associated with significant comorbidities. Screening for K. pneumoniae and VRE simultaneously could be an efficient approach for novel infection prevention strategies.
Keywords: Klebsiella, vancomycin-resistant Enterococci, colonization, infection, microbiome
Klebsiella pneumoniae is a gram-negative bacillus that causes pneumonia, bloodstream infections, and urinary tract infections in hospitalized or immunocompromised patients [1]. It is a leading cause of health care–associated infections (HAIs), causing around 10% of HAIs annually [2]. Additionally, K. pneumoniae is becoming increasingly resistant to antibiotics, including carbapenems, complicating treatment of infections and leading the Centers for Disease Control and Prevention to label Klebsiella an urgent public health threat [3]. Prior colonization with K. pneumoniae is significantly associated with subsequent infection, and 80% of infections in colonized patients are caused by their colonizing strain [4, 5]. The acquisition of Carbapenem-resistant K. pneumoniae (CRKP) in the hospital is strongly associated with colonization pressure, and cohorting patients colonized with CRKP is more effective at preventing transmission than any other infection control measure alone [6–8]. Therefore, understanding risk factors for K. pneumoniae colonization could direct screening measures to vulnerable patient populations, preventing the spread of colonization throughout the hospital and identifying patients at risk for later infection.
Vancomycin-resistant Enterococci (VRE) is a gram-positive coccus that causes around 20 000 HAIs annually. Like K. pneumoniae, VRE colonization is a risk factor for later infection [9, 10], with a sensitivity of 94%, specificity of 78%, and relative risk of 24.15 for bacteremia [11]. To prevent patient-to-patient spread of VRE, many hospitals perform routine surveillance cultures of rectal swabs and place detectably colonized patients in contact precautions [12–14]. K. pneumoniae and VRE infect a similar patient population of hospitalized, immunocompromised patients, and previous antibiotic exposure is a risk factor for colonization with antibiotic-resistant strains of either organism [1, 13, 15]. We have previously shown high K. pneumoniae colonization rates in patients screened for VRE [4]. Despite colonizing similar patients, the biological and epidemiological links between VRE and K. pneumoniae are unclear.
Culturing VRE surveillance samples for K. pneumoniae and analyzing patient medical records could identify factors associated with colonization in patients susceptible to infection. Upon patient identification, 1 potential intervention strategy might involve manipulating the microbiota to better resist colonization by exogenous pathogens. Previous studies have begun to identify intestinal microbiota signatures associated with acquisition of VRE and other multidrug-resistant organisms [16–18]. For example, CRKP colonization has been associated with the presence of 2 specific operational taxonomic units (OTUs; Desulfovibrio and Ruminococcaceae) [19], but this finding may be specific to the specific Klebsiella clone. Identifying species found in the microbiota that are broadly associated with K. pneumoniae colonization could enable widely applicable approaches to infection prevention.
The objective of this study was to identify patient factors associated with colonization with K. pneumoniae in ICU patients. We screened samples from 2 patient cohorts, 1 from 2014 and 1 from 2016, for K. pneumoniae colonization and analyzed the associations between colonization and patient demographics, baseline laboratory data, and comorbidities. In addition, 16S rRNA sequencing of the microbiome was performed to determine if there was a microbiota signature associated with K. pneumoniae colonization. Understanding risk factors for K. pneumoniae colonization might inform surveillance practices for K. pneumoniae colonization in the hospital setting.
METHODS
Sample Collection
This study was conducted at Michigan Medicine, a tertiary care center with 1000 beds, and approved by the Institutional Review Board. Rectal swabs were obtained through Michigan Medicine’s VRE surveillance program, wherein adult patients admitted to the intensive care unit (ICU) or hematology/oncology wards have a rectal swab taken upon admission and then weekly until a positive VRE culture is obtained or the patient is discharged. We included swabs collected from adults during 2 time periods: July 31, 2014, to October 31, 2014 (n = 1800), and January 10, 2016, to May 25, 2016 (n = 1824).
Bacterial Identification and Patient Population
Rectal swabs (ESwab Collection and Transport System, Becton Dickinson, Franklin Lakes, NJ, USA) were sent to Michigan Medicine’s Clinical Microbiology laboratory and cultured to identify VRE using VRESelect chromogenic media (Bio-Rad, Hercules, CA, USA). Swabs were stored in glycerol at –80ºC. To identify Klebsiella pneumoniae in the samples, swabs were either cultured on MacConkey agar plates before freezing (2014 cohort) or after thawing (2016 cohort), and 10 μL of sample was streaked. Three mucoid lactose-fermenting colonies were isolated and subcultured onto MacConkey agar plates. If <3 mucoid lactose-fermenting colonies were present from the sample, then all colonies were subcultured. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis was used for bacterial identification from the subcultures. Only patients with rectal swabs screened for both K. pneumoniae and VRE that were collected during patients’ first encounter with Michigan Medicine during the study periods were included: 1209 for the 2014 cohort and 1243 for the 2016 cohort.
Demographic Definitions
Patient electronic medical records (EMRs) were reviewed for patient variables associated with colonization. Comorbidity data were collected from the patient’s first encounter during the study. Comorbidities were defined using the Elixhauser Comorbidity Index [20]. The Elixhauser Comorbidity Score was calculated as previously described [21, 22]. Antibiotic usage was defined as antibiotic administration that occurred within 30 days preceding rectal swab collection and that started at least 48 hours before rectal swab collection. Patients were considered to have a K. pneumoniae clinical culture if their EMRs indicated a positive clinical blood, urine, or respiratory culture for K. pneumoniae within 90 days after their first rectal swab. Patients were considered to have a K. pneumoniae infection if the clinical culture met case definitions for a bloodstream infection, pneumonia, or a urinary tract infection within 90 days after their first rectal swab. Bloodstream infection was defined as any blood culture positive for K. pneumoniae. Pneumonia was defined based on new or progressive radiographic infiltrate plus at least 2 of 3 clinical features (fever >38ºC, leukocytosis or leukopenia, and purulent secretions) [23]. Urinary tract infection was defined based on National Healthcare Safety Network (NHSN) case definitions [24].
Antibiotic susceptibility results (Sensititre, ThermoFisher Scientific) were extracted for Klebsiella-colonized patients with subsequent positive clinical cultures and reviewed for interpretations of extended-spectrum beta-lactamase and carbapenem resistance in the specimen report.
DNA Purification and Microbiota Analysis
The MagAttract PowerMicrobiome DNA/RNA Kit (Qiagen, Inc., Hilden, Germany) optimized for the EpMotion 5075 (Eppendorf, Hamburg, Germany) was used to isolate DNA from swabs. The V4 region of the bacterial 16S rRNA genes was polymerase chain reaction–amplified [25] from 1 μL of the sample DNA, as previously described [26]. The 500 cycle MiSeq Reagent Kit v.2 (Illumina, catalog No. MS-102-2003) was used to prepare amplicons for sequencing on a MiSeq (Illumina, San Diego, CA, USA) by the University of Michigan Microbial Systems Molecular Biology Laboratory using the manufacturer’s protocols [26].
Sequences were quality-trimmed and processed using mothur v.1.39.5 [27] as described previously [19]; chimeras were removed using UCHIME [28] and reads aligned to the SILVA 16S rRNA sequence database [29]. Samples with <2500 sequences were excluded from further analysis. Using mothur’s opticlust algorithm [30], sequences were clustered into OTUs at 97% similarity. The Ribosomal Database Project was used to classify OTUs [31]. Inverse Simpson’s index, shared OTUs, and the Yue and Clayton dissimilarity index were calculated from unfiltered OTU data. Principal components analysis and analysis for molecular variance (AMOVA) were calculated in mothur. Basic R packages were used to visualize data. We created Dirichlet multinomial mixture models using R v.3.3.2 and used the DirichletMultinomial v.1.6.0 package [32] to assign all samples to community types, as proposed by Holmes et al. [33]. We determined the number of community types by comparing the Laplace approximation with the negative log posterior likelihood and identifying the point at which an increase in Dirichlet components results in minor improvements in model fit.
Statistical Analysis
We compared the distributions of continuous variables among K. pneumoniae–colonized patients with those of noncolonized patients using the Student t test; for categorical variables, we used the chi-square or Fisher exact test. Variables significantly associated with K. pneumoniae colonization in either cohort at α = 0.05 were entered into a multivariate logistic model predicting K. pneumoniae colonization. Univariate analysis comparing co-colonized, K. pneumoniae–colonized, VRE-colonized, and noncolonized patients was conducted using 1-way analysis of variance for continuous variables and the chi-square or Fisher exact test for categorical variables. Additional analyses comparing the colonized groups with the noncolonized group were performed using Tukey’s multiple comparisons test for continuous variables and the chi-square or Fisher exact test for categorical variables with the hybrid Wilson/Brown method of estimating confidence intervals (Prism 7, Graph Pad). AMOVA was used to distinguish the microbiota structure between patient groups. The nonparametric Kruskal-Wallis test was used for multigroup microbiota comparisons.
RESULTS
VRE Colonization Is Associated With K. pneumoniae Colonization
Of the 1209 rectal swabs collected from eligible patients between July 31 to October 31, 2014 (2014 cohort), 209 (17.3%) were colonized with K. pneumoniae. In the 2016 cohort, collected from 1243 eligible patients between January 10 and May 25, 2016, 91 (7.3%) were colonized with K. pneumoniae (Supplementary Figure 1). In addition to increased prevalence of K. pneumoniae colonization in 2014 (P < .0001), women represented a higher percentage and white patients represented a smaller percentage of patients. Conversely, length of stay was significantly longer in the 2016 cohort (Table 1). Based on significant differences between the cohorts, the results were stratified based on cohort year, with the goal of finding consistent associations across 2 cohorts that differ by date and K. pneumoniae colonization rates.
Table 1.
Demographic Information and Colonization Rates for the 2014 and 2016 Cohort
| Year | |||
|---|---|---|---|
| Patient Variable, No. (%) | 2014 (n = 1209) | 2016 (n = 1243) | P Value |
| Sex–female | 591 (48.9) | 548 (44.1) | .017 |
| Age, mean ± SD, y | 57.9 ± 16.4 | 58.9 ± 16.4 | .119 |
| Race–white | 990 (81.9) | 1059 (85.2) | .028 |
| Black | 128 (10.6) | 121 (9.7) | |
| Other | 91 (7.5) | 63 (5.1) | |
| Admits from nursing home | 3 (0.2) | 2 (0.2) | .683 |
| Length of stay, mean ± SD, d | 11.5 ± 17.0 | 13.2 ± 17.4 | .017 |
| Death within 30 d | 110 (9.1) | 120 (9.7) | .637 |
| Elixhauser comorbidity score, mean ± SD | 7.22 ± 8.7 | 6.92 + 7.5 | .363 |
| Klebsiella pneumoniae– colonized | 209 (17.3) | 91 (7.3) | <.0001 |
| VRE-colonized | 118 (9.8) | 96 (7.7) | .074 |
| Primary service–surgery | 216 (17.9) | 224 (18.0) | .094 |
| GynOnc | 117 (9.7) | 87 (7.0) | |
| Cardiac | 119 (9.8) | 100 (8.1) | |
| HemOnc | 181 (15.0) | 194 (15.6) | |
| Medical critical care | 78 (6.5) | 69 (5.6) | |
| Medicine | 229 (19.0) | 249 (20.0) | |
| Burn | 17 (1.4) | 13 (1.0) | |
| Cardiothoracic surgery | 184 (15.2) | 222 (17.9) | |
| Other | 68 (5.6) | 85 (6.8) | |
Differences between the cohorts were calculated using the Student t test for continuous variables and the chi-square or Fisher exact test for categorical variables.
Abbreviation: VRE, vancomycin-resistant Enterococci.
To identify variables consistently associated with K. pneumoniae rectal colonization, univariate analysis was performed on demographic, comorbidity, and baseline laboratory data from each cohort (Table 2). The Elixhauser comorbidity score was not significantly associated with colonization in either cohort (2014: 7.57 ± 8.47 vs 7.14 ± 8.80; P = .52; 2016: 7.55 ± 6.37 vs 6.87 ± 7.57; P = .40). A positive clinical culture for K. pneumoniae was associated with rectal colonization in both the 2014 (odds ratio [OR], 5.0; 95% confidence interval [CI], 2.82–9.2; P < .0001) and 2016 (OR, 3.5; 95% CI, 1.38–9.07; P = .0263) cohorts. The sensitivity of detectable colonization for a subsequent positive culture was 48.9% (95% CI, 35.3%–62.8%) and 20.8% (95% CI, 9.2%–40.5%), and the specificity was 83.9% (95% CI, 81.8%–86.0%) and 92.9% (95% CI, 91.4%–94.3%) in the 2014 and 2016 cohorts, respectively. The negative predictive value for subsequent infection was consistently high in both cohorts (2014: 97.6%; 95% CI, 96.5%–98.4%; 2016: 98.4%; 95% CI, 97.4%–98.9%). We reviewed antibiotic resistance among clinical cultures from Klebsiella-colonized patients. Consistent with the low institutional resistance rate for K. pneumoniae, 1 extended-spectrum beta-lactamase isolate was found in a 2016 clinical culture, and no CRKP were detected. Clinical infection was significantly associated with K. pneumoniae colonization in the 2014 cohort (OR, 5.5; 95% CI, 2.51–12.20; P < .0001). Infections were higher among patients who were colonized with K. pneumoniae in the 2016 cohort as well (OR, 2.85; 95% CI, 0.61–11.0); however, this did not reach statistical significance. In the cohort from 2014, patients colonized with K. pneumoniae were more likely to be female, have hypertension, have lower baseline albumin levels, and be older than noncolonized patients. In the 2016 cohort, K. pneumoniae–colonized patients were more likely to die within 30 days, have a neurologic disorder, and have underlying pulmonary circulation comorbidities. VRE colonization (2014: OR, 1.57; 95% CI, 0.95–2.57; 2016: OR, 2.20; 95% CI, 1.258–3.877) and low albumin baseline (2014: 3.44 ± 0.62 colonized vs 3.56 ± 0.65 not colonized; P = .027; 2016: 3.34 ± 0.58 vs 3.47 ± 0.62; P = .072) were potential shared risk factors for K. pneumoniae colonization between the cohorts.
Table 2.
Clinical and Demographic Characteristics of Patients With and Without Klebsiella pneumoniae Colonization
| 2014 Cohort | 2016 Cohort | |||||
|---|---|---|---|---|---|---|
| Variable | No. | Kp-Colonized | P Value | No. | Kp-Colonized | P Value |
| Female | 591 | 116 (19.63) | .035 | 548 | 37 (6.75) | .494 |
| Male | 618 | 93 (15.05) | 695 | 54 (7.77) | ||
| Age | .024 | .254 | ||||
| <50 y | 347 | 42 (12.10) | 344 | 21 (6.10) | ||
| 51–60 y | 275 | 51 (18.55) | 249 | 14 (5.62) | ||
| 61–70 y | 306 | 59 (19.28) | 333 | 26 (7.81) | ||
| 71+ y | 281 | 57 (20.28) | 317 | 30 (9.46) | ||
| Race | .386 | .119 | ||||
| White | 990 | 169 (17.1) | 1059 | 71 (6.7) | ||
| Black | 128 | 27 (21.1) | 121 | 14 (11.5) | ||
| Other | 91 | 13 (14.2) | 63 | 6 (9.5) | ||
| Length of stay | .954 | .877 | ||||
| <4 d | 357 | 63 (17.65) | 333 | 22 (6.61) | ||
| 4–7 d | 267 | 48 (17.98) | 277 | 20 (7.22) | ||
| 7–14 d | 302 | 52 (17.22) | 267 | 19 (7.12) | ||
| 14+ d | 283 | 46 (16.25) | 366 | 30 (8.20) | ||
| Death in 30 d | 110 | 23 (20.91) | .292 | 120 | 15 (12.50) | .022 |
| No death in 30 d | 1099 | 186 (16.92) | 1123 | 76 (6.77) | ||
| Later K. pneumoniae clinical culture | 47 | 23 (48.9) | <.0001 | 24 | 5 (20.8) | .0263 |
| No later K. pneumoniae clinical culture | 1162 | 186 (16.0) | 1219 | 86 (7.1) | ||
| Later K. pneumoniae infection | 25 | 13 (52.0) | <.0001 | 11 | 2 (18.1) | .1897 |
| No later K. pneumoniae infection | 1184 | 196 (16.6) | 1232 | 89 (7.2) | ||
| VRE-colonized | 96 | 23 (23.96) | .072 | 118 | 16 (13.56) | .006 |
| Not VRE-colonized | 1113 | 186 (16.71) | 1125 | 75 (6.67) | ||
| Neurologic disorder | 95 | 16 (16.84) | >.999 | 65 | 11 (16.92) | .005 |
| No neurologic disorder | 1114 | 193 (17.32) | 1178 | 80 (6.79) | ||
| Congestive heart failure | 127 | 25 (19.69) | .457 | 123 | 14 (11.38) | .097 |
| No congestive heart failure | 1082 | 184 (17.01) | 1120 | 77 (6.88) | ||
| Pulmonary circulation disorder | 72 | 13 (18.06) | .872 | 67 | 10 (14.93) | .026 |
| No pulmonary circulation disorder | 1137 | 196 (17.24) | 1176 | 81 (6.89) | ||
| Peripheral vascular disease | 90 | 20 (22.22) | .194 | 99 | 7 (7.07) | >.999 |
| No peripheral vascular disease | 1119 | 189 (16.89) | 1144 | 84 (7.34) | ||
| Hypertension | 449 | 92 (20.49) | .027 | 198 | 10 (5.05) | .233 |
| No hypertension | 760 | 117 (15.40) | 1045 | 81 (7.75) | ||
| Admitted overnight 180 d prior | 701 | 124 (17.69) | .664 | 676 | 55 (8.14) | .907 |
| Not admitted overnight 180 d prior | 508 | 85 (16.73) | 567 | 36 (6.35) | ||
| Antibiotic exposure | 110 | 12 (10.91) | .064 | 170 | 10 (5.88) | .438 |
| No antibiotic exposure | 1099 | 197 (17.93) | 1073 | 81 (7.55) | ||
| Central line | 92 | 12 (13.04) | .263 | 147 | 7 (4.76) | .205 |
| No central line | 1117 | 197 (17.93) | 1096 | 84 (7.66) | ||
| Urinary catheter | 87 | 17 (19.54) | .564 | 136 | 5 (3.68) | .084 |
| No urinary catheter | 1122 | 192 (17.11) | 1107 | 86 (7.77) | ||
| Hemoglobin baseline | .204 | .554 | ||||
| <9.9 | 313 | 58 (18.53) | 319 | 29 (9.09) | ||
| 10–11.7 | 303 | 58 (19.14) | 316 | 22 (6.96) | ||
| 11.8–13.4 | 303 | 55 (18.15) | 303 | 19 (6.27) | ||
| 13.5+ | 288 | 38 (13.19) | 303 | 21 (6.93) | ||
Differences between colonized and not-colonized patients were calculated using the Student t test for continuous variables and the chi-square or Fisher exact test for categorical variables.
Abbreviations: Kp, Klebsiella pneumoniae; VRE, vancomycin-resistant Enterococci.
In the final multivariable models, VRE colonization was associated with K. pneumoniae colonization with similar point estimates in both cohorts (2014: OR, 1.58; 95% CI, 0.92–2.62; 2016: OR, 2.07; 95% CI, 1.06–3.83), as was age >70 (OR, 1.73; 95% CI, 1.09–2.78; OR, 1.8; 95% CI, 0.96–3.47) (Table 3). Black race (OR, 1.38; 95% CI, 0.85–2.2; OR, 1.91; 95% CI, 0.94–3.63) had similar point estimates in both but did not reach statistical significance in either cohort. In the 2014 cohort, male gender was inversely associated with K. pneumoniae colonization (OR, 0.7; 95% CI, 0.51–0.96), but this was not observed in the 2016 cohort (OR, 1.14; 95% CI, 0.71–1.85). Similarly, hypertension was associated with colonization in the 2016 cohort (OR, 0.49; 95% CI, 0.22–0.97), but the point estimate was reversed in the 2014 cohort (OR, 1.33; 95% CI, 0.97–1.83).
Table 3.
Multivariable Logistic Regression Model for Klebsiella pneumoniae Colonization in 2014 and 2016
| 2014 Cohort | 2016 Cohort | |||
|---|---|---|---|---|
| Variable | Odds Ratio (95% CI) | P Value | Odds Ratio (95% CI) | P Value |
| VRE colonization | 1.58 (0.92–2.62) | .08 | 2.07 (1.06–3.83) | .03 |
| Male gender | 0.7 (0.51–0.96) | .03 | 1.14 (0.71–1.85) | .6 |
| Race (ref. white) | ||||
| Black | 1.38 (0.85–2.2) | .18 | 1.91 (0.94–3.63) | .06 |
| Other | 0.71 (0.27–1.59) | .45 | 1.29 (0.3–3.88) | .68 |
| Unknown | 1.13 (0.44–2.55) | .78 | 2.94 (0.44–11.63) | .17 |
| Days in hospital | 0.99 (0.98–1) | .22 | 1 (0.98–1.01) | .6 |
| Albumin ≤3 | 0.95 (0.69–1.31) | .77 | 0.87 (0.47–1.53) | .64 |
| Age (ref. <50 y) | ||||
| 50–60 y | 1.66 (1.04–2.66) | .03 | 0.84 (0.37–1.84) | .66 |
| 60–70 y | 1.72 (1.1–2.71) | .02 | 1.17 (0.6–2.31) | .65 |
| Over 70 y | 1.73 (1.09–2.78) | .02 | 1.8 (0.96–3.47) | .07 |
| Death within 30 d | 1.25 (0.74–2.05) | .39 | 1.68 (0.85–3.15) | .12 |
| Neurologic disorder | 1.07 (0.58–1.88) | .81 | 2.07 (0.9–4.4) | .07 |
| Pulmonary circulation disorder | 0.96 (0.47–1.82) | .91 | 2.18 (0.95–4.57) | .05 |
| Hypertension | 1.33 (0.97–1.83) | .08 | 0.49 (0.22–0.97) | .05 |
Abbreviations: CI, confidence interval; VRE, vancomycin-resistant Enterococci.
VRE Colonization Is Associated With Comorbidities and Poor Outcomes
After establishing the association between K. pneumoniae and VRE colonization in ICU patients, we tested the hypothesis that co-colonized patients have more comorbidities and worse health outcomes. We grouped the cohorts into patients colonized with both K. pneumoniae and VRE, patients colonized with K. pneumoniae or VRE, and patients colonized with neither organism and performed univariate analysis with postanalysis, comparing each colonized group with the noncolonized group (Supplementary Table 1). Across both cohorts, the Elixhauser score was higher and albumin baseline was lower in the both-positive and VRE-positive colonization groups compared with the both-negative group. Also in both cohorts, death within 30 days, central lines, and urinary catheters were significantly associated with VRE colonization only, compared with the both-negative group. In the 2014 cohort but not the 2016 cohort, patients colonized with both organisms and only K. pneumoniae were more likely to have a later K. pneumoniae infection compared with patients colonized by neither. Overall, VRE and dual-colonized patients appeared to have poorer overall health compared with noncolonized patients, whereas K. pneumoniae–colonized patients did not.
Klebsiella and Enterococcus OTUs Dominate the Gut Microbiota in Colonized Patients
The positive association between VRE and K. pneumoniae colonization suggested that the microbiota may differ between colonized (either alone or with both) and noncolonized patients. To examine these differences and better understand potential mechanisms of colonization, we performed 16S rRNA gene-based analysis on a total of 248 available rectal swabs from the 2014 cohort. Of those swabs, 7 were from patients co-colonized with VRE and K. pneumoniae, 20 were from patients colonized with VRE, 31 were from patients colonized with K. pneumoniae, and 190 swabs were from patients not colonized with either organism at baseline.
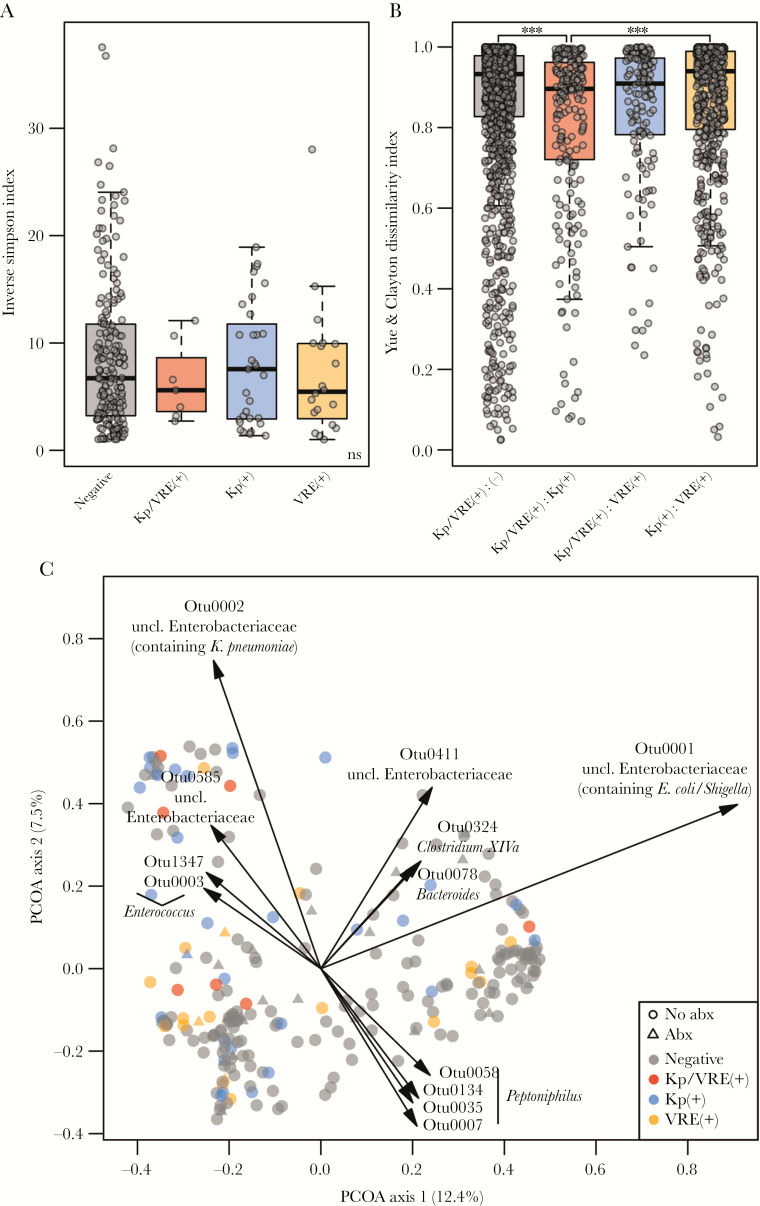
Overall diversity of the microbiota communities (inverse Simpson’s diversity) did not significantly differ across the 4 groups (Figure 1A). However, beta-diversity indices, reflective of microbial community similarity between the groups, demonstrated that co-colonized individuals exhibited a more similar microbiota to K. pneumoniae–colonized patients than to VRE-colonized or noncolonized patients (Figure 1B). Further, co-colonized patients and K. pneumoniae patients shared more operational taxonomic units (OTUs) than co-colonized patients and VRE-colonized patients (Supplementary Figure 2). AMOVA based on the Yue and Clayton theta dissimilarity index [34] indicated significant differences between groups overall (P < .001) (Supplementary Table 2) and between the noncolonized group compared with the co-colonized group (P = .048), the VRE-positive group (P < .001), or the K. pneumoniae–positive group (P < .001). The community composition also differed significantly between the K. pneumoniae– and VRE-colonized groups (P = .003) (Figure 1C). Community modeling by Dirichlet multinomial mixtures (DMMs) separated the samples into 4 community state types (CSTs) (Supplementary Figure 3) [32, 33]. Although no single CST was correlated with colonization, patients colonized with any combination of K. pneumoniae and VRE were significantly more likely to have CST1 or 2 compared with CST 3 and 4 combined (P = .05). Principal coordinate analysis with overlayed biplots of OTUs indicated that 2 OTUs of Enterobacteriaceae (OTU1 and OTU2) and 1 Enterococcus OTU (OTU3) were strong components in driving these influences (Figure 1C). Further comparisons of the sequences clustered into each of the OTUs revealed that OTU1 contained sequences classified as Escherichia coli/Shigella, whereas OTU2 contained K. pneumoniae, K. variicola, and Enterobacter species. OTU3 contained sequences classifying as Enterococcus species. Consistent with numbering convention, these were the most predominant OTUs in these samples.
Figure 1.
A, Alpha diversity (inverse Simpson index) and (B) beta diversity, or community structure similarity (Yue & Clayton dissimilarity) in the microbiota of noncolonized (“negative”), co-colonized (“Kp/VRE(+)”), K. pneumoniae–colonized (“Kp(+)”), and VRE-colonized (“VRE(+)”) patients (Kruskal-Wallis adjusted with Benjamini-Hochberg method and Dunn’s post hoc; *P < .01; **P < .001; ***P < .0001). C, Principal components analysis of the fecal microbiota community in all 4 patient groups, based on Yue & Clayton dissimilarity (AMOVA, P < .05). Abbreviations: AMOVA, analysis for molecular variance; Kp, Klebsiella pneumoniae; VRE, vancomycin-resistant Enterococci.
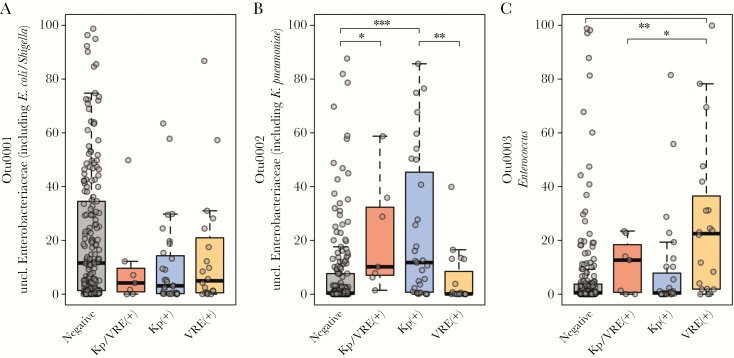
We compared the relative abundance of these predominant OTUs in each of the patient groups (Figure 2). The OTU2 containing Klebsiella was significantly more abundant in samples from patients colonized with K. pneumoniae alone (P < .0001) or co-colonized with VRE (P = .013) compared with patients colonized with neither organism. Similarly, Enterococcus (OTU3) was higher in samples from patients colonized by VRE (P = .0002). OTU1 (containing E.coli/Shigella sequences) was less abundant in patients colonized with K. pneumoniae compared with no colonization but did not reach statistical significance (P = .08). To determine if these dominant OTUs were driving the overall differences observed in the microbiomes based on colonization state, we removed Klebsiella OTU2 and Enterococcus OTU3 and repeated our analyses. In this modified data set, AMOVA did not detect significant differences between the 4 colonization states (P = .182).
Figure 2.
Relative abundance of OTU0001, OTU0002, and OTU0003 in noncolonized (“negative”), co-colonized (“Kp/VRE(+)”), K. pneumoniae–colonized (“Kp(+)”), and VRE-colonized patients (“VRE(+)”; Kruskal-Wallis adjusted with Benjamini-Hochberg method and Dunn’s post hoc; *P < .01; **P < .001; ***P < .0001). Abbreviations: Kp, Klebsiella pneumoniae; VRE, vancomycin-resistant Enterococci.
DISCUSSION
We and others have shown previously that K. pneumoniae colonization is associated with subsequent infection [4, 5]. This study sought to identify patient factors associated with K. pneumoniae colonization in ICU patients, including VRE colonization, which might be used to target screening programs to patients most likely to benefit from infection prevention interventions. In 2 separate cohorts of 1209 and 1243 patients, the only consistent patient factor associated with K. pneumoniae colonization was advanced age. By contrast, colonization with VRE alone or VRE plus K. pneumoniae was associated with worse baseline comorbidities than K. pneumoniae alone. However, there was an important microbiological factor: VRE colonization was associated with K. pneumoniae colonization. Importantly, the differences in the microbiome between groups seem to be driven by Klebsiella and Enterococcus themselves, with a higher abundance of a Klebsiella OTU in Klebsiella and co-colonized groups and a higher abundance of an Enterococcus OTU in VRE-colonized groups compared with the noncolonized group.
Outcomes of colonization include potential transmission to other patients and increased risk of infection in the colonized patient. This study was not designed to detect transmission, although there are reports of transmission from a colonizing strain from ICU settings across the globe [4, 5]. Further, K. pneumoniae infections occur frequently in colonized patients, as shown in a previous analysis of the 2014 cohort, where a significant association between infection and colonization was observed [4]. The current analysis using the first collected rectal swabs from each patient’s first hospital encounter and their fecal microbiome demonstrated the same association with subsequent Klebsiella clinical cultures.
The consistent association between VRE and K. pneumoniae colonization has several implications. Because VRE status is routinely determined in units with an active surveillance program, detectable VRE colonization could be used to identify patients who are potentially colonized with K. pneumoniae. As a biological implication, this association suggests that VRE and K. pneumoniae interact in the fecal microbiome. Unlike infections caused by other health care–associated pathogens, such as Clostridium (Clostridioides) difficile [35], we did not observe differences in the overall diversity of the microbiota across the patient groups. This suggests that K. pneumoniae does not require a depauperate microbiome, lacking in number and variety of species, to persist and may be found in the range of intestinal microbial communities seen in this hospital. VRE has similar dynamics, although it tends to colonize patients with more comorbidities than K. pneumoniae.
Our microbiota analysis also indicates that K. pneumoniae and Enterococcus are predominant taxa in the microbiome of ICU patients. Intestinal domination with Enterococcus and CRKP is associated with subsequent infections [36, 37]. Data from our study indicate that detectable colonization may represent dominance of that taxa in the microbiome, and not simply its presence, perhaps explaining why detectable colonization with VRE or K. pneumoniae is also associated with subsequent infection.
A previous study demonstrated that CRKP and VRE neither compete nor synergize in the murine model, and the 2 organisms reside within the same intestinal regions but occupy distinct metabolic niches [38]. Further, different indigenous OTUs classified as Enterobacteriaceae in mice impact susceptibility to another enteric pathogen, Salmonella [39]. It is possible that similar mechanisms in humans exist, where 1 endogenous, nonpathogenic organism can confer protection against other similar organisms.
A strength of this study was the use of 2 large cohorts of patients separated over time with extensive available chart data and known VRE and K. pneumoniae colonization status. VRE colonization was associated with K. pneumoniae colonization in each cohort despite significant differences in overall K. pneumoniae colonization rates between cohorts. An intriguing possibility is that these differences in colonization rates may be related to differences in ambient temperature between the summer of 2014 and winter of 2016 (Supplementary Figure 1). K. pneumoniae bacteremia rates are highly correlated with temperature [40]. Perhaps this fluctuation is due to changing colonization rates that in turn affect infection rates. The significant differences in demographics and overall comorbidities between cohorts were both a strength and a limitation, highlighting the robust association between K. pneumoniae and VRE but perhaps confounding identification of other variables associated with colonization.
In summary, this study demonstrates a reproducible association between detectable VRE and K. pneumoniae colonization and suggests that each can colonize varied intestinal microbial communities found in hospitalized patients.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This project was funded by a University of Michigan MCubed 2.0 award to C.E.W., B.F., and M.A.B., by the Centers for Disease Control and Prevention (contract number 200-2016-91966) to M.A.B. and A.M.S., and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number K08 AI119182) to R.J.W.
Disclaimer. The views expressed in written conference materials or publications and by speakers and moderators at Department of Health and Human Services–sponsored conferences do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention of trade names, commercial practices, or organizations imply endorsement by the US Government.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data availability. Raw sequences are available in SRA (BioProjectID: PRJNA556249, BioSampleIDs: SAMN12685404-SAMN12685651).
References
- 1. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Magill SS, Edwards JR, Bamberg W, et al. Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. Atlanta: Centers for Disease Control and Prevention; 2014. [Google Scholar]
- 4. Martin RM, Cao J, Brisse S, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 2016; 1:e00261–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorrie CL, Mirceta M, Wick RR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 2017; 65:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clement L, Valerie TS, Helene P, et al. Is cohorting the only solution to control carbapenemase producing Enterobacteriaceae outbreaks? A single centre experience. J Hosp Infect. 2018; 99:390–395. [DOI] [PubMed] [Google Scholar]
- 7. Fournier S, Monteil C, Lepainteur M, et al. Long-term control of carbapenemase-producing Enterobacteriaceae at the scale of a large French multihospital institution: a nine-year experience, France, 2004 to 2012. Euro Surveill 2014; 19:20802. [DOI] [PubMed] [Google Scholar]
- 8. Okamoto K, Lin MY, Haverkate M, et al. Modifiable risk factors for the spread of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae among long-term acute-care hospital patients. Infect Control Hosp Epidemiol 2017; 38:670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hendrix CW, Hammond JM, Swoboda SM, et al. Surveillance strategies and impact of vancomycin-resistant enterococcal colonization and infection in critically ill patients. Ann Surg 2001; 233:259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pettigrew MM, Johnson JK, Harris AD. The human microbiota: novel targets for hospital-acquired infections and antibiotic resistance. Ann Epidemiol 2016; 26:342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alevizakos M, Gaitanidis A, Nasioudis D, et al. Colonization with vancomycin-resistant Enterococci and risk for bloodstream infection among patients with malignancy: a systematic review and meta-analysis. Open Forum Infect Dis 2017; 4:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Montecalvo MA, Shay DK, Gedris C, et al. A semiquantitative analysis of the fecal flora of patients with vancomycin-resistant Enterococci: colonized patients pose an infection control risk. Clin Infect Dis 1997; 25:929–30. [DOI] [PubMed] [Google Scholar]
- 13. Bonten MJ, Slaughter S, Ambergen AW, et al. The role of “colonization pressure” in the spread of vancomycin-resistant Enterococci: an important infection control variable. Arch Intern Med 1998; 158:1127–32. [DOI] [PubMed] [Google Scholar]
- 14. Calfee DP, Giannetta ET, Durbin LJ, et al. Control of endemic vancomycin-resistant Enterococcus among inpatients at a university hospital. Clin Infect Dis 2003; 37:326–32. [DOI] [PubMed] [Google Scholar]
- 15. Slaughter S, Hayden MK, Nathan C, et al. A comparison of the effect of universal use of gloves and gowns with that of glove use alone on acquisition of vancomycin-resistant Enterococci in a medical intensive care unit. Ann Intern Med 1996; 125:448–56. [DOI] [PubMed] [Google Scholar]
- 16. Halpin AL, de Man TJ, Kraft CS, et al. Intestinal microbiome disruption in patients in a long-term acute care hospital: a case for development of microbiome disruption indices to improve infection prevention. Am J Infect Control 2016; 44:830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Araos R, Tai AK, Snyder GM, et al. Predominance of Lactobacillus spp. among patients who do not acquire multidrug-resistant organisms. Clin Infect Dis 2016; 63:937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ubeda C, Bucci V, Caballero S, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seekatz AM, Bassis CM, Fogg L, et al. Centers for Disease Control and Prevention Epicenters Program Gut microbiota and clinical features distinguish colonization with Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae at the time of admission to a long-term acute care hospital. Open Forum Infect Dis 2018; 5(X):XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elixhauser A, Steiner C, Harris DR, Coffey RM. Comorbidity measures for use with administrative data. Med Care 1998; 36:8–27. [DOI] [PubMed] [Google Scholar]
- 21. van Walraven C, Austin PC, Jennings A, et al. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care 2009; 47:626–33. [DOI] [PubMed] [Google Scholar]
- 22. Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol 1992; 45:613–9. [DOI] [PubMed] [Google Scholar]
- 23. American Thoracic Society. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005; 171:388–416. [DOI] [PubMed] [Google Scholar]
- 24. Centers for Disease Control and Prevention. Urinary Tract Infection (Catheter-Associated Urinary Tract Infection [CAUTI] and Non-Catheter-Associated Urinary Tract Infection [UTI]) and Other Urinary System Infection [USI]) Events. Atlanta: Centers for Disease Control and Prevention; 2016. [Google Scholar]
- 25. Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seekatz AM, Theriot CM, Molloy CT, et al. Fecal microbiota transplantation eliminates Clostridium difficile in a murine model of relapsing disease. Infect Immun 2015; 83:3838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007; 35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2017; 2:e00073–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37:D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morgan M. DirichletMultinomial: Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data. R package version 1.22.0. 2018. [Google Scholar]
- 33. Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 2012; 7:e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yue JC, Clayton MK. A similarity measure based on species proportions. Commun Stat-Theor Met 2005; 34:2123–31. [Google Scholar]
- 35. Rea MC, O’Sullivan O, Shanahan F, et al. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J Clin Microbiol 2012; 50:867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimasaki T, Seekatz A, Bassis C, et al. Centers for Disease Control and Prevention Epicenters Program Increased relative abundance of Klebsiella pneumoniae Carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin Infect Dis 2019; 68:2053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caballero S, Carter R, Ke X, et al. Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and Carbapenem-resistant Klebsiella pneumoniae. PLoS Pathog 2015; 11:e1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Velazquez EM, Nguyen H, Heasley KT, et al. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat Microbiol 2019; 4:1057–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anderson DJ, Richet H, Chen LF, et al. Seasonal variation in Klebsiella pneumoniae bloodstream infection on 4 continents. J Infect Dis 2008; 197:752–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.