Abstract
Spinal and bulbar muscular atrophy (SBMA) is a neuromuscular disease caused by a polyglutamine (polyQ) expansion in the androgen receptor (AR). Despite the fact that the monogenic cause of SBMA has been known for nearly 3 decades, there is no effective treatment for this disease, underscoring the complexity of the pathogenic mechanisms that lead to a loss of motor neurons and muscle in SBMA patients. In the current review, we provide an overview of the system-wide clinical features of SBMA, summarize the structure and function of the AR, discuss both gain-of-function and loss-of-function mechanisms of toxicity caused by polyQ-expanded AR, and describe the cell and animal models utilized in the study of SBMA. Additionally, we summarize previously conducted clinical trials which, despite being based on positive results from preclinical studies, proved to be largely ineffective in the treatment of SBMA; nonetheless, these studies provide important insights as researchers develop the next generation of therapies.
Key Words: Polyglutamine, androgen receptor, neurodegenerative disease, motor neuron, spinal and bulbar muscular atrophy.
History and Background
Progressive proximal spinal and bulbar muscular atrophy (SBMA) was first described by Dr. William R. Kennedy et al. in 1968 following the examination of 2 families in which 11 members, all male, presented with a late onset, slowly progressive neuromuscular disorder [1]. Decades later, the genetic basis for SBMA was identified as the expansion of a polymorphic tandem CAG repeat within the first exon of the androgen receptor (AR) gene [2]. SBMA, the first known CAG trinucleotide repeat disorder, now exists within a disease family that includes Huntington’s disease (HD), dentatorubropallidoluysian atrophy (DRPLA), and spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7, and 17.
Clinical Manifestations
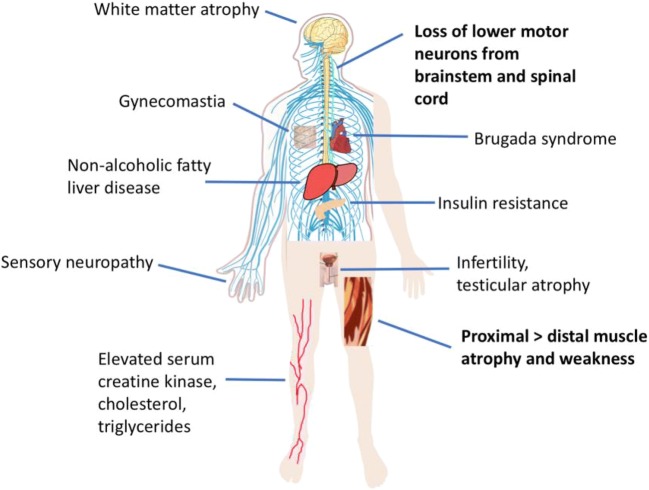
The clinical manifestations of SBMA are summarized in Fig. 1. The majority of SBMA patients initially present with proximal lower limb weakness, with symptom onset typically occurring in men between 30 and 50 years of age [1, 3]. Muscle strength declines slowly, at a rate of approximately 2% per year [4], with additional symptoms appearing over time, including tremor, muscle cramps, fasciculations, dysarthria, and dysphagia [3]. Degeneration of the bulbar musculature predisposes patients to potentially fatal aspiration-induced pneumonia, which was identified as the leading cause of death in a natural history study of SBMA [3]. The predominance of neuromuscular symptoms in SBMA patients corresponds with disease pathology, which is characterized by a loss of lower motor neurons in the anterior horn of the spinal cord and in the brainstem [5, 6], as well as by signs of cell-autonomous toxicity in muscle [7–9]. Evidence of both neurogenic and myogenic abnormalities are observed in patient muscle, including atrophic and morphologically abnormal muscle fibers, fiber-type grouping, and centralized nuclei [5, 8, 10]. Indeed, the best-characterized metabolic indicator of SBMA is serum creatine kinase (CK), which is elevated to approximately 3 to 4 times the normal range in the majority of SBMA patients [11, 12]. This is higher than would be expected for a purely neurogenic disease, underscoring the fact that primary myopathy contributes to SBMA [9].
Fig. 1.
Clinical manifestations of SBMA. Although neuromuscular dysfunction (bold) is the primary clinical feature of SBMA, system-wide disturbances have been reported. Symptoms related to androgen insensitivity, altered metabolism, and sensory neuropathy also affect the quality of life of SBMA patients
In addition to neuromuscular symptoms, SBMA patients also present with signs of androgen insensitivity, such as gynecomastia, infertility, and testicular atrophy [3, 13–15]; however, the neurologic symptoms of SBMA are not caused by a loss of AR function, as these are not observed in patients with complete androgen insensitivity syndrome [16]. Androgen insensitivity is also the presumed cause of the urinary tract symptoms experienced by some SBMA patients, as low levels of androgens have been associated with bladder outlet obstruction [12, 17] and the motor neurons that innervate the bulbospongiosus muscle do not undergo neurodegeneration [18]. It should be noted that this does not rule out cell-autonomous dysfunction of pelvic floor muscles as a contributing factor in urinary tract symptoms [12].
Numerous studies have also noted electrophysiological dysfunction and axon loss in the sensory neurons of SBMA patients, highlighting the fact that disease pathology extends beyond the neuromuscular system [5, 19–27]. Sensory neuron disruption causes a number of symptoms in patients, including vibratory sensation [28], neuropathic pain [28, 29], and numbness [30, 31]. Because these sensory symptoms do not affect quality of life to the same extent as motor symptoms in most patients, the involvement of sensory neurons in disease has not received focused attention despite being a common feature in the manifestation of SBMA.
Metabolic disturbances have also been reported in SBMA patients, with a number of recent studies greatly expanding our understanding of and appreciation for the system-wide effects of mutant AR. For example, impaired glucose homeostasis is now understood to be a common feature of SBMA [12, 32, 33]. Although the results of these studies offer conflicting evidence for whether fasting blood glucose levels are normal [14], low [33], or high [12, 32, 34] in SBMA patients, there seems to be consensus regarding an increase in insulin resistance in SBMA patients, as measured by the homeostasis model assessment of insulin resistance (HOMO-IR) [32–34]. In fact, a recent study found a significant correlation between insulin resistance and motor dysfunction in a cohort of SBMA patients [33]. Although reduced insulin receptor protein levels in skeletal muscle appear to be one contributing factor, the mechanism regarding insulin resistance in SBMA patients requires further elucidation [33]. Regardless, these results suggest that signs of metabolic disruption in SBMA patients may become a useful tool in predicting motor function impairment. Dysregulation of other metabolic indicators have also been reported in a subset of SBMA patients, including low body mass index, bone density, and HDL, and high blood pressure, total cholesterol, and triglycerides [12, 14, 33]. Moreover, SBMA patients are at a high risk for nonalcoholic fatty liver disease [32]. Interestingly, hepatic AR-knockout mice similarly develop insulin resistance, indicating that loss of AR function in SBMA patients may contribute to this phenotype [35].
As the number of known clinical presentations of SBMA continuously expands, it is worth noting that even rarely reported symptoms, when taken together, provide clues for the fundamental mechanisms of cellular toxicity that contribute to SBMA. Although not considered a primary aspect of disease manifestation, some studies have noted subtle cognitive impairment in SBMA patients, finding diminished working memory and executive function [36, 37]. Moreover, a neuropsychological analysis of 20 SBMA patients found a mentalizing deficiency, as determined by performance on the Faux Pas Test [38]. These findings suggest the involvement of the frontal lobe in SBMA pathogenesis, which is further supported by the fact that SBMA patients have glucose hypometabolism [39] and extensive atrophy of white matter in frontal areas of the brain [40]. More recently, however, a study in a larger cohort of SBMA patients (64 patients) found no impairment in neuropsychological performance [41]. Indeed, SBMA patients scored significantly higher in the Prose Memory test than controls, suggesting that earlier findings are not reproduced in a larger cohort and, thus, likely do not represent substantive changes [41].
Additionally, although there is no evidence of cardiomyopathy in SBMA patients [42], Brugada syndrome and other electrocardiogram (ECG) abnormalities have been identified, indicating myocardial involvement in disease which, if undetected, can lead to sudden death [12, 43]. Other rarely reported symptoms include poor sleep quality [44], jaw drop [45], selective bulbar dysfunction [46], and myotonia [43]. The heterogeneity in the clinical presentation of SBMA, a monogenic disease, suggests that environmental factors and genetic modifiers greatly shape patient phenotype.
Unique symptoms are also associated with a highly expanded CAG repeat. In a patient with 68 CAGs (currently the longest repeat identified in an SBMA patient), disease onset occurred at the age of 18, and by age 29, symptoms included autonomic nervous system dysfunction and abnormal sexual development [47]. Taken together, it is clear that SBMA affects a wide range of cell types and tissues and that, although motor neuron degeneration and neuromuscular disruption are considered hallmarks of the disease, a wider view of SBMA pathogenesis may yield new therapeutic opportunities and mechanistic insights.
Although a highly expanded CAG repeat length may be associated with additional symptoms [47], there is extensive evidence demonstrating that in SBMA, CAG repeat length inversely correlates with the age of symptom onset but not with the rate of disease progression [3, 30, 31, 48–50]. Although one study posited that a longer CAG repeat (greater than 47) is associated with a motor-dominant phenotype and a shorter CAG repeat (less than or equal to 47) is associated with a sensory-dominant phenotype (as determined by measuring compound muscle and sensory nerve action potentials) [28], a follow-up study that controlled for patient age at examination reproduced only the negative correlation between CAG repeat length and compound muscle action potential, finding no correlation between CAG repeat length and sensory nerve action potential [51]. In patient tissue, CAG repeat length directly correlates with the frequency of nuclear aggregates of polyQ-expanded AR in motor neurons [28]. Taken together, it appears that in the typical patient population, CAG repeat length correlates with age of symptom onset and AR aggregation, but not with the rate of disease progression nor with any particular symptoms of the disease. However, the characterization of additional, unique symptoms in an SBMA patient with a CAG repeat length of 68 leaves open the possibility that CAG repeat length can influence the severity of disease.
Androgen Receptor
Androgen Receptor Structure and Functional Domains
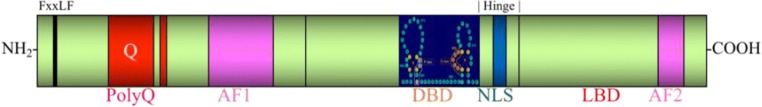
The androgen receptor gene (GenBank: M20132.1) is located on the X chromosome at position q11-12 and contains 8 exons spanning more than 90 kb, encoding a 919 amino acid protein (Fig. 2). Along with the estrogen receptor (α and β), the progesterone receptor, the mineralocorticoid receptor, and the glucocorticoid receptor, the AR is a member of the steroid hormone receptor subfamily of the nuclear receptor superfamily. Like other nuclear receptors, of which there are at least 49 in the human genome [52], the androgen receptor consists of 4 defined domains: an amino-terminal domain (NTD), a highly conserved DNA-binding domain (DBD), a hinge domain, and a carboxyl-terminal ligand-binding domain (LBD) [53].
Fig. 2.
Schematic of the androgen receptor. Schematic representation of the androgen receptor (AR), depicting key functional domains of the protein. Shown here are the size and location of the DNA-binding domain (DBD) from amino acids 539 to 627, the hinge domain from amino acids 628 to 670, and the ligand-binding domain (LBD) from amino acids 671 to 919. Additionally, the polymorphic polyglutamine tract (polyQ) is depicted in the NTD along with the FxxLF motif, which participates in AR intra- and intermolecular amino-carboxyl (N/C) terminal interactions. The nuclear localization signal (NLS) is shown spanning amino acids 617 to 634 in the DBD and hinge regions. Also depicted are the two transactivation domains of the AR: the activation function 1 (AF-1) domain in the NTD and the activation function 2 (AF-2) domain in the LBD
The length, sequence, and structure of the NTD vary between the steroid hormone receptors but typically contain at least one transactivation domain, referred to as the activation function 1 (AF-1) domain. The largest domain of the AR, the NTD spans amino acids 1 to 538 and contains a polymorphic polyglutamine tract as well as polyproline and polyglycine tracts. Although the AR NTD is intrinsically disordered in the absence of binding partners [54], over 150 coregulators are known to bind the AR NTD and coactivator binding can promote structure in the NTD [55, 56]. Additionally, the AR NTD contains an FxxLF motif (23FQNLF27) that participates in intra- and intermolecular amino-carboxyl (N/C) terminal interactions via charge-clamp hydrogen bonding with residues within the LBD [57, 58]. Recently, the first 36 amino acids of the AR NTD were identified as a putative mitochondrial localization sequence (MLS) [59]. The discovery of a bona fide MLS in the AR NTD adds to prior evidence that both wild-type and mutant AR localize to mitochondria [59–61].
The AR DNA-binding domain extends from amino acids 539 to 627 and contains two zinc finger domains composed of 3 α-helices [62]. Within the first zinc finger domain, a P-box comprised of 5 amino acids (residues 577-581) interacts with the major groove of DNA at androgen response elements (AREs) [63]. Consensus AREs consist of 2 hexameric, palindromic motifs spaced by 3 base pairs (AGAACAnnnTGTTCT) [64, 65]. The second zinc finger in the AR DBD contains a 5-amino-acid sequence referred to as the dimerization box (D-box) (residues 596-600), which, in part, mediates AR homodimerization [66]. AR binds to DNA as a dimer, with each member of the dimer binding half of the ARE [62, 67]. Like the progesterone receptor and the glucocorticoid receptor, the AR binds to canonical AREs in a ‘head-to-head’ orientation, although it may bind to noncanonical AREs as a head-to-tail homodimer [62]. Although AR homodimerization precedes DNA binding, populations of both dimerized and monomeric AR can be detected in the nucleus [66]. Intriguingly, FRET analysis demonstrated that the AR N/C interaction occurs predominantly between AR molecules of the mobile nuclear population, suggesting that DNA binding abolishes the N/C interaction, possibly allowing coactivator binding to the AF-2 domain in the AR LBD [68].
The hinge domain of the AR (residues 628-670) is so-called because it serves as a flexible bridge between the DBD and LBD. Although this region is poorly conserved, it nonetheless serves a number of important functions. A bipartite nuclear localization signal (NLS) (617RKCYEAGMTLGARKLKKL634), encompassing the end of the DBD and the first 8 residues of the hinge region, regulates AR nuclear import [69]. Binding of the AR to the importin α–importin β complex is mediated by the basic residues within 629RKLKKL634, as mutation of K630, K632, or K633 reduces this interaction and mutation of all 3 lysines completely prevents the interaction [70]. Beyond their role in AR nuclear import, the first 12 amino acids of the hinge region, referred to as the carboxyl-terminal extension of the DBD, are also involved in DNA binding, specifically in mediating the selectivity of AR transactivation at canonical versus noncanonical AREs [71]. Importantly, the role of residues 627 to 634 in nuclear import and DNA binding are not mutually exclusive, as the interaction between the AR and importin α dissociates in the nucleus prior to DNA binding [72]. Given their role in mediating AR nuclear localization, it is perhaps surprising that deletion of amino acids 629 to 634 results in an increase in AR transcriptional activity on some reporters despite a dramatic reduction in AR nuclear import [73]. Although coactivator recruitment is not significantly affected by this deletion, the N/C interaction was markedly enhanced [73]. Subsequent studies have further elucidated the mechanism by which deletion of 629 to 634 increases AR transactivation based on the understanding that, in the nucleus, AR molecules exist in both a mobile fraction and an immobile fraction, which is believed to be DNA-bound (this is supported by the fact that DNA-binding mutants exist in a mobile fraction) [68, 74]. Mutation of residues 629 to 630 as well as 632 to 633 greatly reduces the immobile fraction of the AR and correspondingly decreases its residence time on DNA by ~10-fold [75]. Thus, despite reducing AR nuclear import, deletion of 629 to 634 may increase AR transcriptional activity by increasing the rate at which AR cycles on DNA (a mechanism demonstrated for the estrogen receptor) [72, 76]. Moreover, the fact that mutations in the hinge region are associated with prostate cancer and increase AR transcriptional activity supports the notion that the hinge region plays a key regulatory (predominantly inhibitory) role in AR activity [73]. There is also evidence that the hinge domain plays a role in facilitating the intramolecular N/C interaction, as overexpression of sirtuin 1 (SIRT1), which deacetylates the AR at K630, K632, and K633, reduces the intramolecular N/C interaction [77].
The C-terminal LBD of the AR, spanning amino acids 671 to 919, is modestly conserved in terms of sequence, but its 12 α-helix secondary structure is well-conserved [78]. Upon hormone binding, the position of helix 12 shifts, covering the ligand binding pocket (composed of the other 11 α-helices) and exposing the AF-2 domain to coactivator binding [58, 79]. Antagonist binding to steroid hormone receptors causes helix 12 to reposition differently, blocking the binding of some coactivators and/or recruiting the binding of corepressors [80]. As previously described, the AF-2 domain of the AR LBD binds to the 23FQNLF27 motif in the NTD via both intra- and intermolecular interactions. Temporally, hormone binding in the cytoplasm induces an intramolecular N/C interaction, which is followed by intermolecular N/C interactions in the nucleus [66]. This interaction is essential for the transactivation of certain genes (one notable exception being the mouse mammary tumor virus (MMTV)) [81]. Additionally, recent work revealed that the AR LBD can homodimerize in a head-to-head conformation, with the interface of AR monomers centered around helix 5 [82]. Perhaps because the primary interactor with the AF-2 domain is the FxxLF motif in the NTD, interactions between coactivators and the AR AF-2 domain are typically weaker than those formed with the AF-1 domain [83]. Thus, the AF-2 domain plays a diminished role in AR transcriptional activity compared with other steroid hormone receptors [84]. Mutagenesis experiments have identified an additional role for the LBD in AR nuclear export, as a 75-amino-acid region (residues 743-817) is both necessary and sufficient for AR nuclear export in a PC3 prostate cancer cell model [85]. The putative nuclear export signal (NES) contained within this region is leptomycin B-insensitive, suggesting that this regulatory motif is not a candidate for chromosome region maintenance 1 (CRM-1)–mediated nuclear export upon hormone withdrawal [85–87].
Cellular Trafficking
Prior to ligand binding, the AR resides in the cytoplasm in an inactive aporeceptor complex that contains chaperones (Hsc70, Hsp40, Hsp90, HIP, HOP), p23, and immunophilins (Cyp40, FKBP51, FKBP52) [88, 89]. The aporeceptor complex maintains the AR in a high affinity state for hormone binding [90], whereas an association with importin 7 retains AR in the cytoplasm by blocking the interaction between importin α and the bipartite NLS of the AR [91]. It has been proposed that karyopherins such as importin α act as chaperones by binding exposed basic residues on their client proteins [92, 93]. Unlike some other steroid hormone receptors, the AR is rapidly degraded in its unliganded state [94].
Testosterone is primarily produced by Leydig cells in the testes in response to secretion of luteinizing hormone (LH) from the pituitary gland [95]. Extracellularly, most testosterone is bound by sex hormone-binding globulin (SHBG) and albumin, but the free form can passively diffuse across cell membranes [95]. In cells expressing the enzyme 5α-reductase, testosterone is converted intracellularly into the more potent androgen, dihydrotestosterone (DHT) [95, 96]. Notably, there are 3 isoenzymes of the steroid 5α-reductase (SRD5A) family, characterized by different expression patterns and kinetic properties [97, 98]. Upon binding of testosterone or DHT, the AR undergoes a conformational change, dissociating from the aporeceptor complex and importin 7 [91]. This conformational change also induces the intramolecular N/C interaction and exposes the NLS to binding by importin α, facilitating nuclear import [66, 70].
Following nuclear translocation, the AR binds to DNA, canonically as a homodimer [66], but can also be tethered to DNA through interactions with other transcriptional regulators. Coactivator binding to the AR primarily via the AF-1 domain of the NTD leads to the recruitment of RNA polymerase II at the promoter, thus enabling transcription [99].
There is evidence that the AR is exported from the nucleus and subsequently degraded in the cytoplasm by the ubiquitin–proteasome system [100]. In addition to the NES identified in the AR LBD, residues in both the DBD and the hinge region also appear to regulate AR nuclear export. A double mutation in the DBD (F582, 583A) is sufficient to block nuclear export; however, DNA binding itself is not required for AR nuclear export, as the DNA-binding mutant V581F does not affect export [101, 102]. Additionally, phosphorylation of the AR at serine 650 has been shown to regulate nuclear export of the AR [103–105]. In COS-7 and LNCaP cells, phosphorylation of S650 is mediated by the MAPK kinase 4/JNK and MAPK kinase 6/p38 stress kinase signaling pathways. Inhibition of JNK and p38 reduced the nuclear export of wild-type AR in COS-7 cells to an equivalent extent as a phospho-null (S650A) mutation but had no effect on AR with a phospho-mimic (S650D) mutation [103]. Consistent with these findings, knockdown of MAP3K11, an upstream regulator of JNK activity, decreased S650 phosphorylation in LNCaP and C2-4B cells [106]. Additionally, inhibition of protein phosphatase 1 alpha (PP1α) by tautomycin led to an increase in S650 phosphorylation and a decrease in AR nuclear accumulation in LNCaP cells [104]. Notably, increasing AR nuclear export is associated with a decrease in its transcriptional activity, whereas blocking export increases AR transcriptional activity [101, 103, 104, 107].
Polyglutamine Expanded AR
Structural Changes
The length of the polyQ tract in the AR NTD causes structural changes with important implications for AR function. Although the NTD is relatively unstructured in the absence of coregulator binding [56], expansion of the polyQ tract leads to an increase in α-helical structure, as determined by circular dichroism and nuclear magnetic resonance (NMR) spectroscopy [108, 109]. Correspondingly, shorter polyQ tracts exhibit reduced α-helical structure [108, 110]. As polyQ tract length increases, unconventional hydrogen bonds accumulate between glutamine side chains and main chain carbonyl groups, stabilizing α-helices [109]. Other studies have found that expanded polyQ tracts increase the propensity of a protein (including the AR) to form antiparallel β sheets [111–114]. The critical threshold for disease is approximately 40 glutamines in several of the polyQ repeat disorders, suggesting that a fundamental structural change may occur at this length. At about 40Q, the polyQ tracts of different proteins may form β-pleated sheet structures capable of self-associating into polar zippers (so-named because they resemble the leucine zipper interaction that mediates the binding of c-Jun to c-Fos) [115]. Additionally, a cylindrical β-sheet structure has been shown to be unstable at 20Q, but stable and capable of nucleating further helical growth at 40Q [116]. Relative to AR20Q, AR45Q also exhibits increased binding to the hydrophobic probe 8-anilinonaphthalene-1-sulphonic acid and increased sensitivity to urea-induced unfolding [108]. This correlates with data suggesting that the stability of AR is inversely proportional to its polyQ tract length [117]. Although an expanded polyQ tract has not been shown to alter the AR N/C interaction, blocking the N/C interaction of polyQ-expanded AR is protective in SBMA cell and animal models [118, 119]. Disruption of the N/C interaction substantially increases phosphorylation of the AR at S16, and its protective effect is dependent on S16 phosphorylation [118]. Whether the polyQ expansion affects the N/C interaction or S16 phosphorylation remains an open question; however, given that the intermolecular N/C interaction stabilizes the AR homodimer [120, 121], it may be that disrupting the N/C interaction exerts a protective effect irrespective of whether or not the polyQ expansion disrupts these processes.
Altered Posttranslational Modifications
The AR is known to be regulated by phosphorylation, ubiquitination, methylation, SUMOylation, palmitoylation, and acetylation, with some of these posttranslational modifications altered by the polyQ expansion. For example, reduced phosphorylation of the mutant AR at S650 may contribute to its impaired nuclear export [105]. Additionally, hyperacetylation of the AR at K630, K632, and K633 correlates with enhanced toxicity of AR with acetyl-mimic K630, K632, and K633Q/T mutations and with reduced toxicity of AR with K630, K632, and K633R mutations in cells [122]. PolyQ-expanded AR also exhibits increased arginine methylation relative to wild-type AR, due to an enhanced interaction with protein arginine methyltransferase 6 (PRMT6) [123]. Correspondingly, knockdown of PRMT6 was protective in SBMA MN-1 cells and knockdown of the Drosophila homolog of PRMT6 (DART8) suppressed the degenerative eye phenotype of flies expressing AR52Q [123]. Other studies have demonstrated that modulating the posttranslational modifications of the mutant AR, even in the absence of a baseline difference with wild-type AR, can be protective. For example, increasing AR phosphorylation at S215 and S792 (Akt consensus sites) reduces AR toxicity in cell and animal models of SBMA [124, 125]. Similarly, both wild-type and polyQ-expanded AR are phosphorylated at S96 by cyclin-dependent kinase 2 (CDK2), and reducing the phosphorylation of the mutant AR S96 by activation of the adenylyl cyclase (AC)/protein kinase A (PKA) signaling pathway is protective in cell and animal models of SBMA [126]. Furthermore, phosphorylation of the AR at S514 (a MAPK consensus site) blocks mutant AR-induced toxicity and caspase-3 cleavage of the AR in HEK293T and MN-hybrid cells [127]. Blocking the SUMOylation of mutant AR attenuates muscle atrophy and behavioral deficits and extends survival in a mouse model of SBMA, highlighting a protective effect of increasing AR transcriptional activity in vivo [128]. Taken together, these data indicate both that polyQ-expanded AR exhibits an altered posttranslational modification profile and that modulating the posttranslational modifications of the mutant AR can reduce its toxicity in SBMA cell and animal models.
Altered Trafficking and Function
Considering that the AR interacts with approximately 250 different proteins [129], it is unsurprising that structural changes in the AR NTD caused by a polyQ expansion can alter this interactome. For example, Pax Transactivation-domain-interaction Protein (PTIP) has been shown to interact with mutant AR, but not with wild-type AR [130]. Given the role of PTIP in DNA damage repair, it was hypothesized that this aberrant interaction could impair the ability of PTIP to perform its normal function [130]. In a similar way, retinoblastoma protein (Rb) interacts more strongly with mutant AR than with wild-type AR, inappropriately inducing E2F1 transcriptional activation in a Drosophila model of SBMA [131]. Cytochrome c oxidase subunit Vb (COXVb) interacts more strongly with soluble wild-type AR than with polyQ-expanded AR, but importantly, also colocalizes with AR aggregates [61]. This altered interaction (perhaps especially the sequestration of COXVb into AR aggregates) could contribute to the mitochondrial dysfunction observed in cell and animal models of SBMA [60, 132], as well as in SBMA patient tissue [133]. Moreover, it may be that the potential for mitochondrial localization of AR, driven in part by its mitochondrial localization signal [59], contributes to both COXVb sequestration and mitochondrial dysfunction. The extent to which the sequestration of proteins into AR inclusions contributes to proteotoxicity will be more fully addressed below.
Although polyQ-expanded AR translocates to the nucleus with comparable kinetics as wild-type AR [128, 134], both its intranuclear mobility and nuclear export rate are reduced [105]. Additionally, a number of studies have demonstrated that the length of the AR polyQ tract is inversely proportional to its transcriptional activity [135–137]. Indeed, there is evidence that increased transcriptional activity of AR with shorter polyQ tracts corresponds to an increased risk of prostate cancer [138]. The effect of polyQ tract length on AR transcriptional activity may be cell-type dependent, however, as one study found a positive correlation between polyQ length and AR transcriptional activity in C2C12 skeletal muscle cells [139]. There are several potential mechanisms by which polyQ tract length and AR transcriptional activity may be related. As discussed above, changes in the structure of the AR NTD can change the AR interactome, including interactions with coactivators [135, 140]. Additionally, it has been shown that N-terminal fragments of polyQ-expanded AR can interact with and disrupt the transcriptional activity of full-length polyQ-expanded AR [141]. Along similar lines, it is possible that accumulation of the mutant AR into small, potentially difficult-to-detect oligomers may contribute to its reduced transcriptional activity. Intriguingly, a more recent study found increased binding of AR64Q to DNA (specifically, to the MMTV promoter) despite reduced transactivation [134]. This could reflect a reduced capacity for the mutant AR to appropriately cycle on/off DNA, perhaps due to altered cofactor recruitment. Indeed, the cyclical assembly/disassembly of transcription cofactors with the estrogen receptor directly relates to its transcriptional activity [76]. The pathogenic consequences of enhanced DNA binding by polyQ-expanded AR have not been studied.
Aggregation
PolyQ-expanded AR exhibits not only a partial loss of its normal transcriptional function (as described above), but also a gain of new, toxic properties. Indeed, it is clear that the neurologic symptoms of SBMA are not caused by a loss of AR function, as these are not observed in patients with complete androgen insensitivity syndrome [16]. Instead, SBMA is caused by the acquisition of a toxic property or properties of the mutant AR, with aberrant aggregation likely conferring some of these toxic effects. In SBMA patients, nuclear, and to a lesser extent cytoplasmic, AR aggregates are found throughout the CNS as well as in peripheral tissues [18, 28, 142, 143]. These aggregates are morphologically granular and do not associate with membranes [143]. Mitochondria are found in proximity to AR aggregates, suggesting that the process of aggregation (and/or the maturation of aggregates) requires energy [144]. By atomic force microscopy (AFM), AR65Q was reported to form fibrils 300 to 600 nm in length, whereas AR22Q formed only annular oligomers 120 to 180 nm in diameter [145]. AR aggregation species have been further evaluated by SDS-agarose gel electrophoresis (SDS-AGE), an established method for resolving large aggregates of polyQ proteins [146–148]. By SDS-AGE, polyQ-expanded AR forms 2 distinct aggregation species. Slow-migrating aggregates contain full-length AR, are SDS-soluble, and are larger and more heterogeneous by AFM. Fast-migrating aggregates, on the other hand, contain both full-length AR and N-terminal AR fragments, are largely SDS-insoluble, and are smaller and more homogeneous by AFM [149, 150]. Additionally, slow-migrating aggregation species are detected by the 3B5H10 monoclonal antibody, which recognizes expanded polyQ tracts in low molecular weight oligomers, but not in higher molecular weight inclusion bodies [151].
Early studies of AR aggregation in SBMA patient tissue failed to detect AR aggregates with antibodies directed to the C terminus of the AR [18, 143], which was determined to be due to proteolytic cleavage of the mutant AR in a subsequent analysis in an SBMA mouse model [152]. This process was further elucidated in vitro, with both biochemical and imaging assays revealing that mutant AR aggregates first as a full-length protein, becoming proteolyzed over the course of inclusion maturation [150]. Moreover, soluble AR aggregates were shown to contain full-length protein [150]. Although there is no correlation between the number of AR inclusion bodies and neurodegeneration in a Drosophila model of SBMA [153]—in agreement with the finding that fully formed inclusion bodies of polyQ-expanded huntingtin are associated with a reduced risk of death in a cell model of HD [154]—it is clear that AR aggregation disrupts normal cellular function at least partially via the sequestration of other proteins. Proteins integrally important for normal cell function and survival have been found in AR inclusions, such as the transcriptional coactivators CREB-binding protein (CBP) and steroid receptor coactivator-1(SRC-1) [155]; the chaperones Hsp40, Hsp70, and Hsp90 [144, 156–158]; and components of the ubiquitin–proteasome system: ubiquitin, REG-gamma, NEDD8, and PA700 and 26S proteasome caps [18, 144, 159, 160]. Thus, although the extent to which soluble and/or insoluble AR aggregation species contribute to SBMA pathogenesis is uncertain, it seems likely that at least some forms of AR aggregates contribute to toxicity in SBMA.
Modeling SBMA
In Vitro Models
In the decades since the genetic cause of SBMA was identified, a number of cell models have been developed to recapitulate aspects of the disease (Table 1). Both polyQ length- and hormone-dependent toxicity are considered essential phenotypes for an SBMA model system, but the various SBMA cell lines have distinct advantages and disadvantages in mirroring other aspects of the human disease.
Table 1.
Modeling SBMA—in vitro models
| Cell model | Species | Cell type | PolyQ length | Aggregation | Hormone-dependent cell death | Reference |
|---|---|---|---|---|---|---|
| PC12 | Rat | Pheochromocytoma-derived | 10Q/112Q | Yes | Yes | [161] |
| NSC-34 | Mouse | Motor neuron-derived | 0Q/23Q/46Q | Yes | No | [165] |
| MN-1 | Mouse | Motor neuron-derived | 24Q/65Q/100Q | No | No | [123, 163] |
| C2C12 | Mouse | Immortalized myoblasts | 24Q/100Q | No | No | [170] |
| iPSC-derived NPC | Human | Neuronal precursor cells | Varied | Yes | No | [168] |
| Mesenchymal stem cells | Human | Adipose-derived | Varied | Yes* | No | [173] |
SBMA cell models are further reviewed by Pennuto and Basso [225]
*Mesenchymal stem cells from SBMA patients form aggregates upon proteasomal inhibition via treatment with MG132
A PC12 cell model of SBMA, in which rat pheochromocytoma-derived cells express full-length human AR under the control of a tetracycline-inducible promoter, is unique among SBMA cell models in that PC12 cells readily form intranuclear inclusions of aggregated AR in a polyQ length- and hormone-dependent manner [161]. Although the role of intranuclear inclusions in SBMA pathogenesis is still debated, the inclusions formed in PC12 cells reproduce the aberrant AR proteolysis observed in SBMA patient autopsy material [18]. PC12 cells expressing AR112Q also exhibit DHT-dependent cell death, and importantly, several studies have demonstrated that a change in toxicity and/or AR aggregation in the PC12 cell model correlates with altered phenotypes in mouse models of SBMA [118, 122, 128, 162].
Another widely studied immortalized cell model of SBMA is the motor neuron-derived 1 (MN-1) model, in which AR24Q, AR65Q, or AR100Q is expressed under the control of the CMV promoter [123, 163]. These cells, originally generated by fusing embryonic mouse spinal cord motor neurons with mouse neuroblastoma cells [164], express motor neuron markers, making them a useful tool for studying the effect of polyQ-expanded AR expression in a disease-relevant cell type, as the use of primary motor neuron cultures and iPSC-derived motor neurons requires considerably more time and resources. Although these cells recapitulate the mitochondrial dysfunction that has been observed in patient tissue [60], AR65Q and AR100Q do not aggregate or cause hormone-dependent cell death in MN-1 cells.
Similarly, mouse neuroblastoma-spinal cord (NSC)-34 cells have been utilized as a motor neuron-like cell line in the study of SBMA [165]. NSC-34 cells stably expressing AR46Q exhibit cytoplasmic and perinuclear aggregates [165], potentially reflective of the cytoplasmic aggregates observed in some patient tissues [28, 142]. In the absence of hormone, AR46Q expression in NSC-34 cells decreases flux through the proteasome [166]. Testosterone treatment and subsequent AR aggregation restores proteasomal function, suggesting that sequestration of the mutant, misfolded AR into cytoplasmic aggregates preserves protein quality control in the cell [166]. These cytoplasmic AR aggregates activate autophagy in NSC-34 cells but are not properly recruited to newly formed autophagosomes, demonstrating that autophagic flux is also impaired by misfolded AR [167] (a finding replicated in other SBMA models [168]). Thus, these cells have served as a useful model in the study of protein quality control pathways in SBMA.
Corresponding with a growing recognition in the SBMA research community that muscle plays a primary role in disease pathogenesis [8, 9, 169], a muscle cell model of SBMA was developed in which C2C12 myoblasts transduced by lentivirus stably express AR24Q or AR100Q [170]. As in MN-1 cells, AR100Q does not aggregate or cause hormone-dependent cell death in C2C12 cells, although morphological analysis of these cells revealed that AR100Q expression blocks the trophic effect of DHT [170]. Treatment of C2C12 myoblasts with the β-agonist clenbuterol restored the trophic effect of DHT, which was predictive of improved muscle pathology and motor function in a mouse model of SBMA [170]. Moreover, C2C12 cells expressing AR97Q display reduced expression of the creatine transporter SLC6A8, a finding that was mirrored in SBMA patients [171].
It is important to note that in the aforementioned immortalized SBMA cell models, AR is overexpressed and contains a polyQ tract that far exceeds what is typically found in peripheral blood lymphocytes of patients (the longest patient polyQ tract length reported to date being 68Q) [47]. In order to study endogenously expressed AR in human cells, several groups have recently generated and characterized iPSC-derived neuronal cells and mesenchymal cells from SBMA patients [168, 172–174]. The study of both iPSC-derived motor neurons and iPSC-derived neuronal precursor cells (NPCs) has revealed novel SBMA phenotypes, including reduced histone deacetylase 6 activity [172] and autophagic flux defects [168], both of which were subsequently observed in patient tissue. Additionally, iPSC-derived motor neurons from SBMA patients were recently found to have defects in neurite morphology, decreased protein translation, reduced survival in prolonged culture, and dysregulation of neuronal-related signaling pathways [174]. Adipose-derived mesenchymal stem cells isolated from SBMA patients provide researchers with an additional tool, as aggregation of mutant AR can be induced in these cells by treatment with the proteasome inhibitor MG132 [173].
In Vivo Models
Transgenic Drosophila models allow for high-throughput genetic and pharmacologic screening in vivo. Using the GAL4/UAS system to drive cell-type–specific expression of mutant AR, Drosophila models of SBMA have been shown to display hormone-dependent toxicity in photoreceptor neurons, motor neurons, and pan-neuronally [153, 175, 176]. Upon exposure to DHT, flies expressing polyQ-expanded AR exhibit both motor function deficits as well as neuromuscular junction pathology that mirrors what is observed in some mouse models [153, 175, 176]. It should be noted that Drosophila expressing wild-type AR also exhibit toxicity in response to DHT, although to a lesser extent than those expressing polyQ-expanded AR [123]. This is likely due to overexpression of the AR, but it nonetheless raises an important caveat in the interpretation of data derived from this model.
A number of SBMA mouse models have been generated using a variety of genetic strategies, each with a unique set of strengths and weaknesses in their ability to recapitulate the human disease (Table 2). Transgenic mice expressing AR97Q under the control of the chicken β-actin promoter with a cytomegalovirus (CMV) enhancer express high levels of the mutant AR throughout all tissues [177]. Male mice lose weight by about 6 weeks of age with onset of motor impairment by 8 weeks of age, as determined by latency to fall from an accelerating rotarod [177]. Additionally, chicken β-actin-AR97Q transgenic mice are hypoactive and develop gait abnormalities [177]. Both neuronal and muscle pathology are present in these mice, as AR aggregates are detectable throughout the CNS as well as in the muscle and in the heart [177]. Additionally, histological analyses have revealed extensive muscle fiber abnormalities in this mouse model, including fiber-type grouping and atrophic fibers as well as hypertrophic fibers with centralized nuclei [125]. The 50% survival rate of these mice is approximately 3 months [177]. Female mice display a markedly less severe phenotype, but it is notable that they do develop weight loss and motor function deficits at later time points [177].
Table 2.
Modeling SBMA—mouse models
| Mouse model | Promoter | Motor dysfunction | Decreased lifespan | Motor neuron pathology | Muscle pathology | PolyQ length dependent | Reference |
|---|---|---|---|---|---|---|---|
| Transgenic AR97Q | Chicken β-actin | Yes (8 weeks) | Yes | Yes | Yes | Yes | [177] |
| Transgenic AR121Q* | Chicken β-actin | Yes (4 weeks) | Yes | Yes | Yes | Yes | [176] |
| Transgenic AR112Q | Prion protein | Yes (8 weeks) | No | Yes | No | Yes | [178] |
| YAC transgenic AR100Q | Endogenous human | Yes (11 months) | Yes | Yes | Yes | Yes | [180] |
| BAC transgenic AR121Q† | Endogenous human | Yes (13 weeks) | Yes | Yes | Yes | Yes | [8] |
| Transgenic AR22Q | Human skeletal α-actin | Yes | Yes | Yes | Yes | No | [181] |
| Knock-in AR113Q | Endogenous mouse | Yes (8 weeks) | No‡ | No | Yes | Yes | [185] |
SBMA animal models are further reviewed by Pennuto and Basso [225]
*Chicken β-actin-AR121Q mice express a polyQ tract encoded by alternating CAG/CAA repeats
†BAC AR121Q mice contain loxP sites flanking exon 1
‡Urinary tract obstruction leads to early death in a subset of AR113Q knock-in mice. Mice that survive this bottleneck live a normal lifespan
Recently, a new transgenic mouse model was generated in which expression of AR121Q (encoded by alternating CAG/CAA repeats) is similarly driven by a CMV enhancer and chicken β-actin promoter, but with expression levels in the muscle comparable to endogenous mouse AR (notably, the expression level in the spinal cord is still higher than endogenous) [176]. These mice exhibit a rapid decline in body weight, rotarod performance, and grip strength beginning at 4 weeks of age and a drastically shortened lifespan, with a 50% survival rate of about 7 weeks [176]. Ubiquitin-positive aggregates of the mutant AR are detectable in the CNS as well as in skeletal muscle, and histological analyses of the muscle revealed type 1 to type 2 fiber-type switching and atrophied myofibers [176]. Disease in male mice is hormone dependent; however, female mice were not evaluated [176]. Given that these mice express 3-fold less AR in the muscle than the previously described chicken β-actin-AR97Q model [176], it is likely that the longer polyQ tract of the AR121Q (CAG/CAA) model enhances the severity of the phenotype.
A more slowly progressive phenotype is observed in the prion protein (PrP) promoter-AR112Q mouse model of SBMA [178]. In this model, AR is predominantly expressed in the CNS with some, but substantially less, expression in peripheral tissues, including muscle [118, 178]. Male PrP-AR112Q mice first display a rotarod deficit at 8 weeks of age [118] and, similar to SBMA patients, develop slowly progressive motor dysfunction but not a significantly shortened lifespan [118, 122, 162, 178]. As transgenic AR expression is low in the peripheral tissues of these mice, motor dysfunction is likely due to disrupted neuronal function in this model [178]. Additionally, intranuclear inclusions of aggregated AR can be detected in the brain and spinal cord of PrP-AR112Q mice [178]. PrP-AR112Q females display only mild behavioral deficits at later time points [178].
The fact that disease progression in the previously described transgenic mouse models is hormone dependent [177, 178] supports the notion that they are reflective of at least some aspects of the human disease. Nevertheless, it is impossible to predict the extent to which toxicity in these models is amplified (or changed) by the fact that the mutant AR is overexpressed and/or that expression is regulated by an exogenous promoter. To generate an SBMA mouse model in which expression of polyQ-expanded AR is controlled by its endogenous regulatory elements, Sopher et al. utilized a yeast artificial chromosome (YAC) containing 450 kb DNA (with the 180-kb AR gene centrally located) [179, 180]. Two YAC-SBMA lines were characterized, each expressing slightly less AR100Q than endogenous mouse AR [180]. Disease onset in the higher-expressing YAC-SBMA line (80% AR100Q expression compared with endogenous mouse AR) is marked by deficits on the hanging wire test at 11 months of age [180]. Gait abnormalities first appear at 13 months, progressing to severe hindlimb atrophy and paralysis by 17 months [180]. Although aggregated AR was not detected in the neurons of these mice, inclusions were found in astrocytes within the spinal cord and the number of motor neurons was decreased in 16-month-old mice [180]. Extensive muscle pathology is also observed in this mouse model, including atrophied and hypertrophied muscle fibers, abnormal muscle fiber morphology, centralized nuclei, and fiber-type grouping [180]. A similar mouse model was subsequently generated by Cortes et al., in which mice express AR121Q with a floxed first exon on a bacterial artificial chromosome (BAC) containing 50 kb of DNA upstream and 30 kb of DNA downstream of the AR gene [8]. These mice express AR levels similar to the YAC-SBMA model but display a substantially more severe disease phenotype, likely due to a longer AR polyQ tract length. Motor dysfunction onset is at 13 weeks and the 50% survival rate is approximately 18 weeks [8]. The primary advantage of the BAC-SBMA model is that AR expression can be inactivated by Cre-mediated excision in a cell-type–specific manner. Using this approach, it has been shown that knockdown of the AR121Q transgene from skeletal muscle alone is sufficient to cause a dramatic rescue of the disease phenotype [8].
The importance of AR expression level in generating a phenotype in mice is highlighted by the ‘myogenic model’ of SBMA. In these mice, wild-type rat AR (22Q) is highly overexpressed in skeletal muscle, with expression driven by the human skeletal α-actin promoter [181]. Myogenic SBMA mice develop progressive, hormone-dependent motor dysfunction as well as neuromuscular pathology with similarities to what is observed in other SBMA mouse models, albeit with much more rapid demise [181]. This raises important questions as to which phenotypes observed in other transgenic SBMA mouse models are related to a polyQ expansion versus overexpression of the AR. Indeed, overexpression of the wild-type form of ataxin-1, another protein implicated in polyQ disease, also causes disease in mice [182].
Although many aspects of SBMA are recapitulated in transgenic models of SBMA, the knock-in model is the only tool available to SBMA researchers in which AR is expressed at precisely endogenous levels and regulated by endogenous regulatory elements. A knock-in model of SBMA was created in which a large part of exon 1 of the mouse AR gene was replaced with corresponding exon 1 sequences of the human AR containing 21Q or 113Q [183, 184]. Mice expressing AR21Q and female mice do not develop a disease phenotype, whereas male mice expressing AR113Q exhibit decreased grip strength beginning at 8 weeks and subsequently display a progressive motor phenotype [185]. High expression of the AR in the levator ani/bulbocavernosus muscles leads to urinary tract obstruction and early death in a subset of knock-in SBMA mice, but mice that survive this bottleneck live a normal lifespan [185]. Muscle pathology is apparent in these mice and ubiquitin-positive AR aggregates are present in spinal cord motor neurons [185]. Importantly, AR113Q knock-in mice display signs of androgen insensitivity, a key aspect of disease manifestation in SBMA patients that is not observed in other mouse models [185].
Altogether, there are a number of SBMA mouse models available to researchers, each with a unique set of advantages and disadvantages. However, important caveats remain in the use of any mouse model. With particular focus on the study of SBMA, it should be noted that the AR polyQ tract length in each of these models far exceeds the longest found in patients [47]. The length of the polyQ tract can have important implications in the pathology mediated by a polyQ-disease–causing protein, as has been demonstrated in HD [186]. Because polyQ tract length can affect which tissues are affected (and to what degree), caution is warranted in the interpretation of tissue-specific effects in mice expressing AR with extraphysiological polyQ tract lengths. Furthermore, although motor neuron loss was reported in the YAC-SBMA model, other models exhibit no significant motor neuron loss [8, 177, 178, 185]. This is particularly relevant in the testing of therapeutics. Postsymptomatic SBMA patients have substantial motor neuron loss [5]; thus, therapeutics that reverse motor dysfunction in mouse models may not be relevant for patients, as lost motor neurons cannot be recovered.
Clinical Trials and Outcomes
Leuprorelin [187–189]
The requirement for hormone in the pathogenesis of SBMA has been demonstrated in multiple animal models of the disease [175, 177, 178, 185] and is supported by the fact that females homozygous for polyQ-expanded AR display only a mild SBMA phenotype [190]. The therapeutic potential of androgen reduction was first examined in a transgenic mouse model of SBMA and 2 approaches were evaluated. Flutamide, a competitive antagonist of the AR, promoted AR nuclear localization and was unable to rescue the SBMA phenotype in mice, whereas the gonadotropin-releasing hormone (GnRH) agonist leuprorelin acetate led to a substantial rescue of motor function and reduced nuclear AR [191]. Initially, agonism of GnRH receptors by leuprorelin acetate stimulates production of LH and follicle-stimulating hormone (FSH) by the pituitary gland, which increases testosterone production [192]. However, GnRH receptors become desensitized with continuous treatment of leuprorelin, ultimately leading to reduced secretion of LH and FSH by the pituitary gland and decreased production of testosterone in the testes [192]. Treatment of SBMA patients with leuprorelin acetate yielded promising results in a phase 2 clinical trial in which 48 weeks of randomized, placebo-controlled treatment followed by an additional 96 weeks of open-label treatment significantly improved swallowing function [189]. Moreover, autopsy of 1 leuprorelin-treated patient suggested that leuprorelin may decrease the nuclear accumulation of mutant AR in motor neurons of the brainstem and spinal cord [189]. A larger, phase 3 clinical study was then conducted in which 100 SBMA patients (99 placebo controls) were treated with leuprorelin for 48 weeks and evaluated for swallowing function [187]. Contrary to the results of the smaller phase 2 study, no significant improvement in swallowing function was observed [187]. More recently, a follow-up analysis of 36 patients treated with leuprorelin for 84 months (open-label) found a slower decline in motor function than in nontreated controls, as determined by the amyotrophic lateral sclerosis (ALS) Functional Rating Scale (ALSFRS-R), the Limb Norris Score, and the Norris Bulbar Score [188]. Event-free survival was also increased in the leuprorelin-treated group [188]. Although the interpretation of these results is complicated by the lack of a placebo control, taken together, there may be a modest benefit for SBMA patients treated with leuprorelin acetate. Early intervention is likely key, as leuprorelin treatment led to a greater improvement in swallowing function in patients with a disease duration of less than 10 years [187].
Dutasteride [4]
Although the neurologic phenotypes associated with SBMA are caused by a gain of toxic function of the mutant AR, loss of AR function causes a number of deleterious symptoms in patients [15]. Thus, there is rationale for selectively blocking the activation and nuclear translocation of the mutant AR although retaining some of the trophic effects of testosterone [193]. The type 1 and type 2 5α-reductase inhibitor dutasteride blocks the conversion of testosterone to the more potent androgen DHT and is approved for use in the USA for the treatment of benign prostatic hyperplasia (enlarged prostate) [194]. A randomized, double-blind clinical trial was conducted to assess the therapeutic potential of dutasteride in SBMA, in which 25 patients were treated with dutasteride for 24 months and compared with 25 placebo controls [4]. Neither the primary outcome measure, an assessment of muscle strength, nor any quantitative secondary measures improved as a result of Dutasteride treatment, despite the fact that, as predicted, DHT levels decreased approximately 90% in treated patients, whereas testosterone levels were unaffected [4]. These results suggest that testosterone is sufficient to activate and maintain the pathogenic state of mutant AR.
Exercise [195]
Exercise is well-known to be beneficial to overall health, but there is rationale for caution in the use of exercise as a therapy for neuromuscular disorders, as it has been hypothesized that exercise may increase the rate of muscle atrophy in ALS. This is primarily based on 2 observations: first, that high-level athletes are at a greater risk for developing ALS [196]; and second, that when symptom onset is asymmetrical in ALS patients, it occurs disproportionately on the dominant side [197]. This finding is mirrored in SBMA patients who, when symptom onset is asymmetrical, complain of muscle weakness on their dominant side ~70% of the time [14, 30]. Having noted these concerns, thus far, clinical trials studying the effect of exercise on ALS patients have not reported negative outcomes. In fact, studies have shown that exercise significantly improves muscle strength [198, 199] and quality of life scores [199] in ALS patients. There is an additional rationale for the therapeutic potential of exercise in the treatment of SBMA, as exercise is known to increase circulating levels of insulin-like growth factor 1 (IGF-1) [200], which is protective in animal models of SBMA [125, 201]. Based on these data, a clinical study was performed in which 50 SBMA patients participated in either ‘functional exercise’ (24 patients) or stretching (control, 26 patients) for 12 weeks [195]. Overall, exercise did not cause a significant improvement in either the primary outcome measure (Adult Myopathy Assessment Tool (AMAT) score), or on any secondary measures, which included evaluations of muscle strength, balance, quality of life score, and IGF-1 levels [195]. However, when patients were subdivided into high- or low-functioning groups based on their initial AMAT scores, exercise was found to have significantly improved the AMAT scores of low-functioning patients [202]. These findings may suggest that the functional exercise routine performed by patients in this study was too light, particularly for those subdivided into the high-functioning group. The effect of high-intensity training on disease progression in SBMA is currently being investigated (clinicaltrials.gov—NCT02156141).
BVS857 [203]
IGF-1 is known to promote growth and regeneration of skeletal muscle [204] and to protect against motor neuron death in mouse models of ALS [205, 206]. Additionally, IGF-1 signaling activates Akt which, via phosphorylation of the mutant AR at 2 Akt consensus sites (S215 and S792), decreases its ligand binding, nuclear translocation, transcriptional activation, and toxicity in cell and animal models of SBMA [124, 125, 201]. Based on the protective effects of both genetic overexpression of IGF-1 as well as injection of IGF-1 in SBMA mice, a double-blind, placebo-controlled clinical trial was conducted in which BVS857, an IGF-1 mimetic with improved pharmacological properties, was administered to 18 SBMA patients (9 placebo controls) for 12 weeks [203]. Although no serious adverse side effects were reported in the 18 patients receiving BVS857, 11 developed an immune response against BVS857 and 5 patients produced cross-reactive antibodies capable of neutralizing endogenous IGF-1 [203]. Although this immune response did not cause any observable symptoms in these patients, the detection of IGF-1 neutralizing antibodies in patients treated with BVS857 poses a serious challenge to its long-term use as a therapy for SBMA. Despite a relatively small sample size and the short treatment duration, a significant improvement in thigh muscle volume and a trend toward improved lean body mass were reported in BVS857-treated patients [203]. These outcomes indicate that there may be therapeutic potential for IGF-1 in the treatment of SBMA, although activation of the IGF-1 signaling pathway by other means may be necessary to avoid an adverse immune response.
Creatine Monohydrate [207]
It has been previously shown that SBMA patients exhibit reduced serum creatinine levels and that there is an inverse relationship between serum creatinine levels and motor dysfunction [208]. Creatinine is synthesized intracellularly from its precursor phosphocreatine, which is taken into the cell via the creatine transporter SLC6A8 [209]. In both skeletal muscle from SBMA patients and in cultured C2C12 muscle cells expressing mutant AR, SLC6A8 protein expression is decreased, providing a possible mechanism for the observed reduction in serum creatinine in SBMA patients [171]. Additionally, it is known that slow-twitch, type 1 muscle fibers store less phosphocreatine than fast-twitch, type 2 muscle fibers [210], and that type 2 to type 1 fiber switching occurs in SBMA patients and mouse models of SBMA [132, 211, 212]. These data provided the rationale to conduct a randomized, double-blind, placebo-controlled clinical trial to explore the potential of creatine monohydrate supplementation to attenuate muscle weakening in SBMA patients. The study enrolled 45 SBMA patients who were divided evenly between 3 groups: placebo, 10 g creatine monohydrate/day, and 15 g creatine monohydrate/day [207]. Creatine monohydrate was taken orally for 8 weeks, with the primary endpoint at the conclusion of the trial being handgrip strength [207]. Secondary endpoints included respiratory and swallowing function, skeletal muscle mass, and quality of life score [207]. Although the results of this study are not currently available, there is reason for optimism as creatine supplementation has had a beneficial effect in Duchenne muscular dystrophy patients [213–215].
SBMA—Open Questions
Given the lack of an effective treatment for SBMA following multiple clinical studies, fundamental aspects of the disease are continuing to be studied in an effort to more carefully design the next clinical trials. Indeed, new discussions regarding the role of hormone in SBMA have been driven by the fact that clinical trials aimed at suppressing hormone production in SBMA patients have had modest effects on disease progression [4, 187], despite the fact that chemical or surgical castration completely abolishes (and even reverses) disease in mouse models [177, 178, 185, 191]. It has recently been suggested that some subclinical phenotypes in SBMA mouse models may be hormone independent [216], although the authors of this study note that prenatal androgen exposure during development [217] represents an important caveat. Nevertheless, a conversation regarding the role of hormone in disease is worthwhile as efforts to knock down mutant AR mRNA in SBMA patients gain traction based on promising preclinical studies [169, 218]. The question arises: if hormone suppression has little effect on disease progression in postsymptomatic SBMA patients, why will an AR knockdown succeed?
The requirement for androgens in SBMA pathogenesis is exemplified by the fact that expression of mutant AR in heterozygous females primarily results in mild or subclinical symptoms [9, 31]. Although some heterozygous women have been reported to experience muscle weakness (possibly correlated with skewed X-chromosome inactivation of the normal AR gene [219]), evaluation of 2 homozygous women revealed only mild muscle cramping and tremor [190]. This case study clearly indicates that hormone is required for manifestation of the human disease: although it is notable that homozygous females are spared from SBMA, removing hormone from male patients has little effect on disease progression, albeit with treatment starting well after disease onset.
Another intriguing case study may hold additional insights. This report describes a male-to-female transgender SBMA patient who developed full disease phenotype despite 15 years of treatment with an anti-androgen (leading to undetectable levels of androgens) [220]. Although this patient noted gynecomastia before starting therapy, she did not develop muscle weakness until 4 years after anti-androgen treatment and was not diagnosed with SBMA until 6 years after beginning treatment [220]. In this case, the anti-androgen used by the patient, spironolactone, caused some AR nuclear localization, aggregation, and transcriptional activation in an SBMA cell model as well as slight, but significant toxicity in a Drosophila model [220]. Although the effect of spironolactone in causing these phenotypes was substantially reduced compared to DHT, these findings do provide 1 explanation for the development of SBMA in this patient. Nonetheless, it is notable that disease in this patient was not slowed relative to her brother despite presymptomatic anti-androgen treatment. Muscle weakness was noted by the patient (AR49Q) at age 29 and her brother (AR50Q) at age 34 [220].
These two case studies could indicate that there is a developmental aspect to SBMA, wherein androgen exposure that occurs in males prenatally or during puberty sets the disease in motion before symptoms appear. Additional changes to the cellular environment that occur with aging may ultimately lead to motor neuron death later in life, even if circulating androgen levels are reduced. Hormone is clearly a requirement for disease, as even homozygous females exhibit only mild symptoms; however, it is also clear that reducing androgen levels in postsymptomatic patients has limited effects on disease progression. Although 14 years of androgen deprivation slowed disease progression of 1 SBMA patient relative to historical data [221], it should be noted that this patient began androgen deprivation (by treatment with leuprorelin acetate) only 6 months after symptom onset [221]. Thus, the slow disease progression of this patient highlights the importance of early intervention as much as the potential benefits of long-term androgen deprivation.
Additionally, the question of which cell type(s) drive disease is of vital importance for the next generation of therapies. Some studies have suggested that peripheral knockdown of the AR is sufficient to rescue disease phenotypes in SBMA mouse models [8, 169]. Should these findings translate to patients, it would simplify drug delivery and spare androgen receptor function in the CNS [222]. However, there is also evidence that expression of the AR in the CNS drives disease [178, 218]. Additionally, SBMA patients suffer from sensory neuropathies, further suggesting a primary role for CNS AR in disease.
Thus far, all of the studies conducted in SBMA mouse models suffer from the caveat that, in order to drive an observable behavioral phenotype, AR with a supraphysiological polyQ tract length is expressed. In HD, long polyQ tracts are associated with juvenile onset HD, which is characterized by dysfunction of different brain regions (notably, the cerebellum) than adult onset HD [186]. Given that polyQ tract length can affect which cell types are disrupted by the mutant protein (and to what degree), caution is therefore warranted in the interpretation of tissue-specific effects in these SBMA mouse models. It is well-established that toxicity in motor neurons can drive muscle pathology and that toxicity in muscle can drive motor neuron pathology. Consequently, similar phenotypes could be achieved in SBMA mice regardless of whether muscle or motor neurons drive disease. The goal, however, is to determine which cell types are critical for disease in human SBMA patients. This question was addressed in HD by utilizing an allelic series of knock-in mice to gather transcriptomics data on various tissues in response to huntingtin with a range of polyQ tract lengths [223]. Perhaps a similar approach in an SBMA mouse model could shed light on how polyQ tract length impacts the cell types affected by the mutant AR. Given that polyQ tract length inversely correlates with disease onset, identifying the genes that change most in response to a longer polyQ tract could be important, as these genes must contribute to the disease. Moreover, determining if transcriptomic/proteomic phenotypes at a physiologically relevant polyQ tract length occur first in motor neurons or muscle could guide efforts to effectively deliver SBMA therapies moving forward.
Related to the question of the tissue involved in disease initiation, the extent to which tissue-specific somatic instability of the expanded CAG tract of mutant AR contributes to disease remains to be determined. Somatic mosaicism is primarily observed in cardiac and skeletal muscle of SBMA patients—tissues with high levels of AR expression [224]. Additionally, a higher CAG mosaicism index in SBMA patient blood inversely correlated with age of onset in a study of 46 SBMA patients, although statistical significance was not reached for other clinical features of disease [30]. Clarifying the role of somatic instability in SBMA pathogenesis could add to our understanding of why specific cells/cell types are vulnerable to polyQ-expanded AR.
It remains to be determined which direction(s) of study will ultimately lead to a treatment for SBMA, but until effective treatments exist, further exploration in all directions is imperative. The studies summarized here reflect broad efforts to advance our understanding of SBMA on multiple fronts, with the ultimate goal of achieving a clinically meaningful outcome in patients.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18(7):671–80. doi: 10.1212/wnl.18.7.671. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. [DOI] [PubMed]
- 3.Atsuta N, Watanabe H, Ito M, Banno H, Suzuki K, Katsuno M, et al. Natural history of spinal and bulbar muscular atrophy (SBMA): a study of 223 Japanese patients. Brain. 2006;129(Pt 6):1446–55. [DOI] [PubMed]
- 4.Fernández-Rhodes LE, Kokkinis AD, White MJ, Watts CA, Auh S, Jeffries NO, et al. Efficacy and safety of dutasteride in patients with spinal and bulbar muscular atrophy: a randomised placebo-controlled trial. Lancet Neurol. 2011;10(2):140–7. doi: 10.1016/S1474-4422(10)70321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobue G, Hashizume Y, Mukai E, Hirayama M, Mitsuma T, Takahashi A. X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain. 1989;112(Pt 1):209–32. doi: 10.1093/brain/112.1.209. [DOI] [PubMed] [Google Scholar]
- 6.Amato AA, Prior TW, Barohn RJ, Snyder P, Papp A, Mendell JR. Kennedy’s disease: a clinicopathologic correlation with mutations in the androgen receptor gene. Neurology. 1993;43(4):791–4. doi: 10.1212/wnl.43.4.791. [DOI] [PubMed] [Google Scholar]
- 7.Banno H, Katsuno M, Suzuki K, Tanaka F, Sobue G. Pathogenesis and molecular targeted therapy of spinal and bulbar muscular atrophy (SBMA) Cell Tissue Res. 2012;349(1):313–20. doi: 10.1007/s00441-012-1377-9. [DOI] [PubMed] [Google Scholar]
- 8.Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, Ly L, et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 2014;82(2):295–307. doi: 10.1016/j.neuron.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorarù G, D'Ascenzo C, Polo A, Palmieri A, Baggio L, Vergani L, et al. Spinal and bulbar muscular atrophy: skeletal muscle pathology in male patients and heterozygous females. J Neurol Sci. 2008;264(1–2):100–5. doi: 10.1016/j.jns.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Katsuno M, Adachi H, Waza M, Banno H, Suzuki K, Tanaka F, et al. Pathogenesis, animal models and therapeutics in spinal and bulbar muscular atrophy (SBMA) Exp Neurol. 2006;200(1):8–18. doi: 10.1016/j.expneurol.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Chahin N, Sorenson EJ. Serum creatine kinase levels in spinobulbar muscular atrophy and amyotrophic lateral sclerosis. Muscle Nerve. 2009;40(1):126–9. doi: 10.1002/mus.21310. [DOI] [PubMed] [Google Scholar]
- 12.Querin G, Bertolin C, Da Re E, Volpe M, Zara G, Pegoraro E, et al. Non-neural phenotype of spinal and bulbar muscular atrophy: results from a large cohort of Italian patients. J Neurol Neurosurg Psychiatry. 2016;87(8):810–6. doi: 10.1136/jnnp-2015-311305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner CL, Griffin JE, Wilson JD, Jacobs LD, Murray KR, Fischbeck KH, et al. X-linked spinomuscular atrophy: a kindred with associated abnormal androgen receptor binding. Neurology. 1992;42(11):2181–4. doi: 10.1212/wnl.42.11.2181. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes LE, Freeman BK, Auh S, Kokkinis AD, La Pean A, Chen C, et al. Clinical features of spinal and bulbar muscular atrophy. Brain. 2009;132(Pt 12):3242–51. doi: 10.1093/brain/awp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejager S, Bry-Gauillard H, Bruckert E, Eymard B, Salachas F, LeGuern E, et al. A comprehensive endocrine description of Kennedy’s disease revealing androgen insensitivity linked to CAG repeat length. J Clin Endocrinol Metab. 2002;87(8):3893–901. doi: 10.1210/jcem.87.8.8780. [DOI] [PubMed] [Google Scholar]
- 16.Brinkmann AO. Molecular basis of androgen insensitivity. Mol Cell Endocrinol. 2001;179(1–2):105–9. doi: 10.1016/s0303-7207(01)00466-x. [DOI] [PubMed] [Google Scholar]
- 17.Koritsiadis G, Stravodimos K, Mitropoulos D, Doumanis G, Fokitis I, Koritsiadis S, et al. Androgens and bladder outlet obstruction: a correlation with pressure-flow variables in a preliminary study. BJU Int. 2008;101(12):1542–6. doi: 10.1111/j.1464-410X.2008.07521.x. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Miwa S, Kobayashi Y, Merry DE, Yamamoto M, Tanaka F, et al. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann Neurol. 1998;44(2):249–54. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Sobue G, Doyu M, Mukai E, Hashizume Y, Mitsuma T. Primary sensory neurons in X-linked recessive bulbospinal neuropathy: histopathology and androgen receptor gene expression. Muscle Nerve. 1995;18(3):301–8. doi: 10.1002/mus.880180306. [DOI] [PubMed] [Google Scholar]
- 20.Harding AE, Thomas PK, Baraitser M, Bradbury PG, Morgan-Hughes JA, Ponsford JR. X-linked recessive bulbospinal neuronopathy: a report of ten cases. J Neurol Neurosurg Psychiatry. 1982;45(11):1012–9. doi: 10.1136/jnnp.45.11.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olney RK, Aminoff MJ, So YT. Clinical and electrodiagnostic features of X-linked recessive bulbospinal neuronopathy. Neurology. 1991;41(6):823–8. doi: 10.1212/wnl.41.6.823. [DOI] [PubMed] [Google Scholar]
- 22.Polo A, Teatini F, D'Anna S, Manganotti P, Salviati A, Dallapiccola B, et al. Sensory involvement in X-linked spino-bulbar muscular atrophy (Kennedy’s syndrome): an electrophysiological study. J Neurol. 1996;243(5):388–92. doi: 10.1007/BF00868997. [DOI] [PubMed] [Google Scholar]
- 23.Kachi T, Sobue G, Sobue I. Central motor and sensory conduction in X-linked recessive bulbospinal neuronopathy. J Neurol Neurosurg Psychiatry. 1992;55(5):394–7. doi: 10.1136/jnnp.55.5.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sperfeld AD, Karitzky J, Brummer D, Schreiber H, Häussler J, Ludolph AC, et al. X-linked bulbospinal neuronopathy: Kennedy disease. Arch Neurol. 2002;59(12):1921–6. doi: 10.1001/archneur.59.12.1921. [DOI] [PubMed] [Google Scholar]
- 25.Antonini G, Gragnani F, Romaniello A, Pennisi EM, Morino S, Ceschin V, et al. Sensory involvement in spinal-bulbar muscular atrophy (Kennedy’s disease) Muscle Nerve. 2000;23(2):252–8. doi: 10.1002/(sici)1097-4598(200002)23:2<252::aid-mus17>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Ferrante MA, Wilbourn AJ. The characteristic electrodiagnostic features of Kennedy’s disease. Muscle Nerve. 1997;20(3):323–9. doi: 10.1002/(SICI)1097-4598(199703)20:3<323::AID-MUS9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.Guidetti D, Vescovini E, Motti L, Ghidoni E, Gemignani F, Marbini A, et al. X-linked bulbar and spinal muscular atrophy, or Kennedy disease: clinical, neurophysiological, neuropathological, neuropsychological and molecular study of a large family. J Neurol Sci. 1996;135(2):140–8. doi: 10.1016/0022-510x(95)00283-8. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Katsuno M, Banno H, Takeuchi Y, Atsuta N, Ito M, et al. CAG repeat size correlates to electrophysiological motor and sensory phenotypes in SBMA. Brain. 2008;131(Pt 1):229–39. doi: 10.1093/brain/awm289. [DOI] [PubMed] [Google Scholar]
- 29.Manganelli F, Iodice V, Provitera V, Pisciotta C, Nolano M, Perretti A, et al. Small-fiber involvement in spinobulbar muscular atrophy (Kennedy’s disease) Muscle Nerve. 2007;36(6):816–20. doi: 10.1002/mus.20872. [DOI] [PubMed] [Google Scholar]
- 30.Fratta P, Nirmalananthan N, Masset L, Skorupinska I, Collins T, Cortese A, et al. Correlation of clinical and molecular features in spinal bulbar muscular atrophy. Neurology. 2014;82(23):2077–84. doi: 10.1212/WNL.0000000000000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariotti C, Castellotti B, Pareyson D, Testa D, Eoli M, Antozzi C, et al. Phenotypic manifestations associated with CAG-repeat expansion in the androgen receptor gene in male patients and heterozygous females: a clinical and molecular study of 30 families. Neuromuscul Disord. 2000;10(6):391–7. doi: 10.1016/s0960-8966(99)00132-7. [DOI] [PubMed] [Google Scholar]
- 32.Guber RD, Takyar V, Kokkinis A, Fox DA, Alao H, Kats I, et al. Nonalcoholic fatty liver disease in spinal and bulbar muscular atrophy. Neurology. 2017;89(24):2481–90. doi: 10.1212/WNL.0000000000004748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatsuji H, Araki A, Hashizume A, Hijikata Y, Yamada S, Inagaki T, et al. Correlation of insulin resistance and motor function in spinal and bulbar muscular atrophy. J Neurol. 2017;264(5):839–47. doi: 10.1007/s00415-017-8405-3. [DOI] [PubMed] [Google Scholar]
- 34.Rosenbohm A, Hirsch S, Volk AE, Grehl T, Grosskreutz J, Hanisch F, et al. The metabolic and endocrine characteristics in spinal and bulbar muscular atrophy. J Neurol. 2018;265(5):1026–36. doi: 10.1007/s00415-018-8790-2. [DOI] [PubMed] [Google Scholar]
- 35.Lin HY, Yu IC, Wang RS, Chen YT, Liu NC, Altuwaijri S, et al. Increased hepatic steatosis and insulin resistance in mice lacking hepatic androgen receptor. Hepatology. 2008;47(6):1924–35. doi: 10.1002/hep.22252. [DOI] [PubMed] [Google Scholar]
- 36.Soukup GR, Sperfeld AD, Uttner I, Karitzky J, Ludolph AC, Kassubek J, et al. Frontotemporal cognitive function in X-linked spinal and bulbar muscular atrophy (SBMA): a controlled neuropsychological study of 20 patients. J Neurol. 2009;256(11):1869–75. doi: 10.1007/s00415-009-5212-5. [DOI] [PubMed] [Google Scholar]
- 37.Kasper E, Wegrzyn M, Marx I, Korp C, Kress W, Benecke R, et al. Minor cognitive disturbances in X-linked spinal and bulbar muscular atrophy, Kennedy’s disease. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(1–2):15–20. doi: 10.3109/21678421.2013.837927. [DOI] [PubMed] [Google Scholar]
- 38.Di Rosa E, Sorarù G, Kleinbub JR, Calvo V, Vallesi A, Querin G, et al. Theory of mind, empathy and neuropsychological functioning in X-linked spinal and bulbar muscular atrophy: a controlled study of 20 patients. J Neurol. 2015;262(2):394–401. doi: 10.1007/s00415-014-7567-5. [DOI] [PubMed] [Google Scholar]
- 39.Lai TH, Liu RS, Yang BH, Wang PS, Lin KP, Lee YC, et al. Cerebral involvement in spinal and bulbar muscular atrophy (Kennedy’s disease): a pilot study of PET. J Neurol Sci. 2013;335(1–2):139–44. doi: 10.1016/j.jns.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Kassubek J, Juengling FD, Sperfeld AD. Widespread white matter changes in Kennedy disease: a voxel based morphometry study. J Neurol Neurosurg Psychiatry. 2007;78(11):1209–12. doi: 10.1136/jnnp.2006.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcato S, Kleinbub JR, Querin G, Pick E, Martinelli I, Bertolin C, et al. Unimpaired neuropsychological performance and enhanced memory recall in patients with SBMA: a large sample comparative study. Sci Rep. 2018;8(1):13627. doi: 10.1038/s41598-018-32062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Querin G, Melacini P, D'Ascenzo C, Morandi L, Mazzini L, Silani V, et al. No evidence of cardiomyopathy in spinal and bulbar muscular atrophy. Acta Neurol Scand. 2013;128(6):e30–2. doi: 10.1111/ane.12140. [DOI] [PubMed] [Google Scholar]
- 43.Araki K, Nakanishi H, Nakamura T, Atsuta N, Yamada S, Hijikata Y, et al. Myotonia-like symptoms in a patient with spinal and bulbar muscular atrophy. Neuromuscul Disord. 2015;25(11):913–5. doi: 10.1016/j.nmd.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Romigi A, Liguori C, Placidi F, Albanese M, Izzi F, Uasone E, et al. Sleep disorders in spinal and bulbar muscular atrophy (Kennedy’s disease): a controlled polysomnographic and self-reported questionnaires study. J Neurol. 2014;261(5):889–93. doi: 10.1007/s00415-014-7293-z. [DOI] [PubMed] [Google Scholar]
- 45.Sumner CJ, Fischbeck KH. Jaw drop in Kennedy’s disease. Neurology. 2002;59(9):1471–2. doi: 10.1212/01.wnl.0000033325.01878.13. [DOI] [PubMed] [Google Scholar]
- 46.Praline J, Guennoc AM, Malinge MC, de Toffol B, Corcia P. Pure bulbar motor neuron involvement linked to an abnormal CAG repeat expansion in the androgen receptor gene. Amyotroph Lateral Scler. 2008;9(1):40–2. doi: 10.1080/17482960701553915. [DOI] [PubMed] [Google Scholar]
- 47.Grunseich C, Kats IR, Bott LC, Rinaldi C, Kokkinis A, Fox D, et al. Early onset and novel features in a spinal and bulbar muscular atrophy patient with a 68 CAG repeat. Neuromuscul Disord. 2014;24(11):978–81. doi: 10.1016/j.nmd.2014.06.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni W, Chen S, Qiao K, Wang N, Wu ZY. Genotype-phenotype correlation in Chinese patients with spinal and bulbar muscular atrophy. PLoS One. 2015;10(3):e0122279. doi: 10.1371/journal.pone.0122279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinnreich M, Sorenson EJ, Klein CJ. Neurologic course, endocrine dysfunction and triplet repeat size in spinal bulbar muscular atrophy. Can J Neurol Sci. 2004;31(3):378–82. doi: 10.1017/s0317167100003486. [DOI] [PubMed] [Google Scholar]
- 50.Igarashi S, Tanno Y, Onodera O, Yamazaki M, Sato S, Ishikawa A, et al. Strong correlation between the number of CAG repeats in androgen receptor genes and the clinical onset of features of spinal and bulbar muscular atrophy. Neurology. 1992;42(12):2300–2. doi: 10.1212/wnl.42.12.2300. [DOI] [PubMed] [Google Scholar]
- 51.Kim H, Lim YM, Lee EJ, Oh YJ, Kim KK. Correlation between the CAG repeat size and electrophysiological findings in patients with spinal and bulbar muscular atrophy. Muscle Nerve. 2018;57(4):683–6. doi: 10.1002/mus.25977. [DOI] [PubMed] [Google Scholar]
- 52.Robinson-Rechavi M, Carpentier AS, Duffraisse M, Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17(10):554–6. doi: 10.1016/s0168-9525(01)02417-9. [DOI] [PubMed] [Google Scholar]
- 53.Brinkmann AO, Faber PW, van Rooij HC, Kuiper GG, Ris C, Klaassen P, et al. The human androgen receptor: domain structure, genomic organization and regulation of expression. J Steroid Biochem. 1989;34(1–6):307–10. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 54.Lavery DN, McEwan IJ. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry. 2008;47(11):3360–9. doi: 10.1021/bi702221e. [DOI] [PubMed] [Google Scholar]
- 55.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 56.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43(11):3008–13. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 57.Langley E, Zhou ZX, Wilson EM. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270(50):29983–90. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 58.He B, Gampe RT, Kole AJ, Hnat AT, Stanley TB, An G, et al. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell. 2004;16(3):425–38. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 59.Bajpai P, Koc E, Sonpavde G, Singh R, Singh KK. Mitochondrial localization, import, and mitochondrial function of the androgen receptor. J Biol Chem. 2019;294(16):6621–34. doi: 10.1074/jbc.RA118.006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranganathan S, Harmison GG, Meyertholen K, Pennuto M, Burnett BG, Fischbeck KH. Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18(1):27–42. doi: 10.1093/hmg/ddn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beauchemin AM, Gottlieb B, Beitel LK, Elhaji YA, Pinsky L, Trifiro MA. Cytochrome c oxidase subunit Vb interacts with human androgen receptor: a potential mechanism for neurotoxicity in spinobulbar muscular atrophy. Brain Res Bull. 2001;56(3–4):285–97. doi: 10.1016/s0361-9230(01)00583-4. [DOI] [PubMed] [Google Scholar]
- 62.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A. 2004;101(14):4758–63. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lallous N, Dalal K, Cherkasov A, Rennie PS. Targeting alternative sites on the androgen receptor to treat castration-resistant prostate cancer. Int J Mol Sci. 2013;14(6):12496–519. doi: 10.3390/ijms140612496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helsen C, Dubois V, Verfaillie A, Young J, Trekels M, Vancraenenbroeck R, et al. Evidence for DNA-binding domain–ligand-binding domain communications in the androgen receptor. Mol Cell Biol. 2012;32(15):3033–43. doi: 10.1128/MCB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verrijdt G, Haelens A, Claessens F. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol Genet Metab. 2003;78(3):175–85. doi: 10.1016/s1096-7192(03)00003-9. [DOI] [PubMed] [Google Scholar]
- 66.van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J Cell Sci. 2012;125(Pt 8):1970–9. [DOI] [PubMed]
- 67.Wong CI, Zhou ZX, Sar M, Wilson EM. Steroid requirement for androgen receptor dimerization and DNA binding. Modulation by intramolecular interactions between the NH2-terminal and steroid-binding domains. J Biol Chem. 1993;268(25):19004–12. [PubMed] [Google Scholar]
- 68.van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, et al. Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol. 2007;177(1):63–72. doi: 10.1083/jcb.200609178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem. 1994;269(18):13115–23. [PubMed] [Google Scholar]
- 70.Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci. 2008;121(Pt 7):957–68. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- 71.Schoenmakers E, Alen P, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, et al. Differential DNA binding by the androgen and glucocorticoid receptors involves the second Zn-finger and a C-terminal extension of the DNA-binding domains. Biochem J. 1999;341(Pt 3):515–21. [PMC free article] [PubMed] [Google Scholar]
- 72.Clinckemalie L, Vanderschueren D, Boonen S, Claessens F. The hinge region in androgen receptor control. Mol Cell Endocrinol. 2012;358(1):1–8. doi: 10.1016/j.mce.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 73.Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67(9):4514–23. doi: 10.1158/0008-5472.CAN-06-1701. [DOI] [PubMed] [Google Scholar]
- 74.Farla P, Hersmus R, Geverts B, Mari PO, Nigg AL, Dubbink HJ, et al. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J Struct Biol. 2004;147(1):50–61. doi: 10.1016/j.jsb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 75.Tanner TM, Denayer S, Geverts B, Van Tilborgh N, Kerkhofs S, Helsen C, et al. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci. 2010;67(11):1919–27. doi: 10.1007/s00018-010-0302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 77.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, et al. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26(21):8122–35. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3(1):87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 79.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 80.Heldring N, Pawson T, McDonnell D, Treuter E, Gustafsson JA, Pike AC. Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J Biol Chem. 2007;282(14):10449–55. doi: 10.1074/jbc.M611424200. [DOI] [PubMed] [Google Scholar]
- 81.He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem. 2002;277(28):25631–9. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 82.Nadal M, Prekovic S, Gallastegui N, Helsen C, Abella M, Zielinska K, et al. Structure of the homodimeric androgen receptor ligand-binding domain. Nat Commun. 2017;8:14388. doi: 10.1038/ncomms14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christiaens V, Bevan CL, Callewaert L, Haelens A, Verrijdt G, Rombauts W, et al. Characterization of the two coactivator-interacting surfaces of the androgen receptor and their relative role in transcriptional control. J Biol Chem. 2002;277(51):49230–7. doi: 10.1074/jbc.M209322200. [DOI] [PubMed] [Google Scholar]
- 84.Callewaert L, Verrijdt G, Christiaens V, Haelens A, Claessens F. Dual function of an amino-terminal amphipatic helix in androgen receptor-mediated transactivation through specific and nonspecific response elements. J Biol Chem. 2003;278(10):8212–8. doi: 10.1074/jbc.M210744200. [DOI] [PubMed] [Google Scholar]
- 85.Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278(43):41998–2005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- 86.Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Jänne OA. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. Journal of Cell Science. 2000;113(Pt 17):2991–3001. doi: 10.1242/jcs.113.17.2991. [DOI] [PubMed] [Google Scholar]
- 87.Tyagi RK, Lavrovsky Y, Ahn SC, Song CS, Chatterjee B, Roy AK. Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol Endocrinol. 2000;14(8):1162–74. doi: 10.1210/mend.14.8.0497. [DOI] [PubMed] [Google Scholar]
- 88.Pratt WB, Welsh MJ. Chaperone functions of the heat shock proteins associated with steroid receptors. Semin Cell Biol. 1994;5(2):83–93. doi: 10.1006/scel.1994.1012. [DOI] [PubMed] [Google Scholar]
- 89.Cano LQ, Lavery DN, Bevan CL. Mini-review: Foldosome regulation of androgen receptor action in prostate cancer. Mol Cell Endocrinol. 2013;369(1–2):52–62. doi: 10.1016/j.mce.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 90.Fang Y, Fliss AE, Robins DM, Caplan AJ. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271(45):28697–702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- 91.Ni L, Llewellyn R, Kesler CT, Kelley JB, Spencer A, Snow CJ, et al. Androgen induces a switch from cytoplasmic retention to nuclear import of the androgen receptor. Mol Cell Biol. 2013;33(24):4766–78. doi: 10.1128/MCB.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jäkel S, Mingot JM, Schwarzmaier P, Hartmann E, Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21(3):377–86. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173(3):677–92.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kemppainen JA, Lane MV, Sar M, Wilson EM. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J Biol Chem. 1992;267(2):968–74. [PubMed] [Google Scholar]
- 95.Ramaswamy S, Weinbauer GF. Endocrine control of spermatogenesis: role of FSH and LH/ testosterone. Spermatogenesis. 2014;4(2):e996025. doi: 10.1080/21565562.2014.996025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grino PB, Griffin JE, Wilson JD. Testosterone at high concentrations interacts with the human androgen receptor similarly to dihydrotestosterone. Endocrinology. 1990;126(2):1165–72. doi: 10.1210/endo-126-2-1165. [DOI] [PubMed] [Google Scholar]
- 97.Azzouni F, Godoy A, Li Y, Mohler J. The 5 alpha-reductase isozyme family: a review of basic biology and their role in human diseases. Adv Urol. 2012;2012:530121. doi: 10.1155/2012/530121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamana K, Labrie F, Luu-The V. Human type 3 5α-reductase is expressed in peripheral tissues at higher levels than types 1 and 2 and its activity is potently inhibited by finasteride and dutasteride. Horm Mol Biol Clin Investig. 2010;2(3):293–9. doi: 10.1515/HMBCI.2010.035. [DOI] [PubMed] [Google Scholar]
- 99.Chang CY, McDonnell DP. Androgen receptor-cofactor interactions as targets for new drug discovery. Trends Pharmacol Sci. 2005;26(5):225–8. doi: 10.1016/j.tips.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 100.Gong Y, Wang D, Dar JA, Singh P, Graham L, Liu W, et al. Nuclear export signal of androgen receptor (NESAR) regulation of androgen receptor level in human prostate cell lines via ubiquitination and proteasome-dependent degradation. Endocrinology. 2012;153(12):5716–25. doi: 10.1210/en.2012-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Black BE, Holaska JM, Rastinejad F, Paschal BM. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr Biol. 2001;11(22):1749–58. doi: 10.1016/s0960-9822(01)00537-1. [DOI] [PubMed] [Google Scholar]
- 102.Black BE, Vitto MJ, Gioeli D, Spencer A, Afshar N, Conaway MR, et al. Transient, ligand-dependent arrest of the androgen receptor in subnuclear foci alters phosphorylation and coactivator interactions. Mol Endocrinol. 2004;18(4):834–50. doi: 10.1210/me.2003-0145. [DOI] [PubMed] [Google Scholar]
- 103.Gioeli D, Black BE, Gordon V, Spencer A, Kesler CT, Eblen ST, et al. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol Endocrinol. 2006;20(3):503–15. doi: 10.1210/me.2005-0351. [DOI] [PubMed] [Google Scholar]
- 104.Chen S, Kesler CT, Paschal BM, Balk SP. Androgen receptor phosphorylation and activity are regulated by an association with protein phosphatase 1. J Biol Chem. 2009;284(38):25576–84. doi: 10.1074/jbc.M109.043133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arnold FJ, Pluciennik A, Merry DE. Impaired nuclear export of polyglutamine-expanded androgen receptor in spinal and bulbar muscular atrophy. Sci Rep. 2019;9(1):119. doi: 10.1038/s41598-018-36784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whitworth H, Bhadel S, Ivey M, Conaway M, Spencer A, Hernan R, et al. Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen. PLoS One. 2012;7(6):e38950. doi: 10.1371/journal.pone.0038950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shank LC, Kelley JB, Gioeli D, Yang CS, Spencer A, Allison LA, et al. Activation of the DNA-dependent protein kinase stimulates nuclear export of the androgen receptor in vitro. J Biol Chem. 2008;283(16):10568–80. doi: 10.1074/jbc.M800810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol. 2008;41(5):301–14. doi: 10.1677/JME-08-0042. [DOI] [PubMed] [Google Scholar]
- 109.Escobedo A, Topal B, Kunze MBA, Aranda J, Chiesa G, Mungianu D, et al. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. Nat Commun. 2019;10(1):2034. doi: 10.1038/s41467-019-09923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eftekharzadeh B, Piai A, Chiesa G, Mungianu D, García J, Pierattelli R, et al. Sequence context influences the structure and aggregation behavior of a polyQ tract. Biophys J. 2016;110(11):2361–6. doi: 10.1016/j.bpj.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Masino L, Pastore A. A structural approach to trinucleotide expansion diseases. Brain Res Bull. 2001;56(3–4):183–9. doi: 10.1016/s0361-9230(01)00582-2. [DOI] [PubMed] [Google Scholar]
- 112.Palazzolo I, Gliozzi A, Rusmini P, Sau D, Crippa V, Simonini F, et al. The role of the polyglutamine tract in androgen receptor. J Steroid Biochem Mol Biol. 2008;108(3–5):245–53. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 113.Schneider R, Schumacher MC, Mueller H, Nand D, Klaukien V, Heise H, et al. Structural characterization of polyglutamine fibrils by solid-state NMR spectroscopy. J Mol Biol. 2011;412(1):121–36. doi: 10.1016/j.jmb.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 114.Buchanan LE, Carr JK, Fluitt AM, Hoganson AJ, Moran SD, de Pablo JJ, et al. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc Natl Acad Sci U S A. 2014;111(16):5796–801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91(12):5355–8. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc Natl Acad Sci U S A. 2002;99(8):5596–600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lieberman AP, Harmison G, Strand AD, Olson JM, Fischbeck KH. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum Mol Genet. 2002;11(17):1967–76. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 118.Zboray L, Pluciennik A, Curtis D, Liu Y, Berman-Booty LD, Orr C, et al. Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 2015;13(10):2312–23. doi: 10.1016/j.celrep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Orr CR, Montie HL, Liu Y, Bolzoni E, Jenkins SC, Wilson EM, et al. An interdomain interaction of the androgen receptor is required for its aggregation and toxicity in spinal and bulbar muscular atrophy. J Biol Chem. 2010;285(46):35567–77. doi: 10.1074/jbc.M110.146845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou ZX, Kemppainen JA, Wilson EM. Identification of three proline-directed phosphorylation sites in the human androgen receptor. Mol Endocrinol. 1995;9(5):605–15. doi: 10.1210/mend.9.5.7565807. [DOI] [PubMed] [Google Scholar]
- 121.Langley E, Kemppainen JA, Wilson EM. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem. 1998;273(1):92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 122.Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J Neurosci. 2011;31(48):17425–36. doi: 10.1523/JNEUROSCI.3958-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scaramuzzino C, Casci I, Parodi S, Lievens PMJ, Polanco MJ, Milioto C, et al. Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron. 2015;85(1):88–100. doi: 10.1016/j.neuron.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Palazzolo I, Burnett BG, Young JE, Brenne PL, La Spada AR, Fischbeck KH, et al. Akt blocks ligand binding and protects against expanded polyglutamine androgen receptor toxicity. Hum Mol Genet. 2007;16(13):1593–603. doi: 10.1093/hmg/ddm109. [DOI] [PubMed] [Google Scholar]
- 125.Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63(3):316–28. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Polanco MJ, Parodi S, Piol D, Stack C, Chivet M, Contestabile A, et al. Adenylyl cyclase activating polypeptide reduces phosphorylation and toxicity of the polyglutamine-expanded androgen receptor in spinobulbar muscular atrophy. Sci Transl Med. 2016;8(370):370ra181. doi: 10.1126/scitranslmed.aaf9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.LaFevre-Bernt MA, Ellerby LM. Kennedy’s disease. Phosphorylation of the polyglutamine-expanded form of androgen receptor regulates its cleavage by caspase-3 and enhances cell death. J Biol Chem. 2003;278(37):34918–24. doi: 10.1074/jbc.M302841200. [DOI] [PubMed] [Google Scholar]
- 128.Chua JP, Reddy SL, Yu Z, Giorgetti E, Montie HL, Mukherjee S, et al. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J Clin Invest. 2015;125(2):831–45. doi: 10.1172/JCI73214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M. The androgen receptor gene mutations database: 2012 update. Human mutation. 2012;33(5):887–94. doi: 10.1002/humu.22046. [DOI] [PubMed] [Google Scholar]
- 130.Xiao H, Yu Z, Wu Y, Nan J, Merry DE, Sekiguchi JM, et al. A polyglutamine expansion disease protein sequesters PTIP to attenuate DNA repair and increase genomic instability. Hum Mol Genet. 2012;21(19):4225–36. doi: 10.1093/hmg/dds246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Suzuki E, Zhao Y, Ito S, Sawatsubashi S, Murata T, Furutani T, et al. Aberrant E2F activation by polyglutamine expansion of androgen receptor in SBMA neurotoxicity. Proc Natl Acad Sci U S A. 2009;106(10):3818–22. doi: 10.1073/pnas.0809819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giorgetti E, Yu Z, Chua JP, Shimamura R, Zhao L, Zhu F, et al. Rescue of metabolic alterations in AR113Q skeletal muscle by peripheral androgen receptor gene silencing. Cell Rep. 2016;17(1):125–36. doi: 10.1016/j.celrep.2016.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Borgia D, Malena A, Spinazzi M, Desbats MA, Salviati L, Russell AP, et al. Increased mitophagy in the skeletal muscle of spinal and bulbar muscular atrophy patients. Hum Mol Genet. 2017;26(6):1087–103. doi: 10.1093/hmg/ddx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Belikov S, Bott LC, Fischbeck KH, Wrange Ö. The polyglutamine-expanded androgen receptor has increased DNA binding and reduced transcriptional activity. Biochem Biophys Rep. 2015;3:134–9. doi: 10.1016/j.bbrep.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, Coetzee GA. Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum Mol Genet. 2000;9(2):267–74. doi: 10.1093/hmg/9.2.267. [DOI] [PubMed] [Google Scholar]
- 136.Nakajima H, Kimura F, Nakagawa T, Furutama D, Shinoda K, Shimizu A, et al. Transcriptional activation by the androgen receptor in X-linked spinal and bulbar muscular atrophy. J Neurol Sci. 1996;142(1–2):12–6. doi: 10.1016/0022-510x(96)00142-6. [DOI] [PubMed] [Google Scholar]
- 137.Choong CS, Kemppainen JA, Zhou ZX, Wilson EM. Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol Endocrinol. 1996;10(12):1527–35. doi: 10.1210/mend.10.12.8961263. [DOI] [PubMed] [Google Scholar]
- 138.Stanford JL, Just JJ, Gibbs M, Wicklund KG, Neal CL, Blumenstein BA, et al. Polymorphic repeats in the androgen receptor gene: molecular markers of prostate cancer risk. Cancer Res. 1997;57(6):1194–8. [PubMed] [Google Scholar]
- 139.Sheppard RL, Spangenburg EE, Chin ER, Roth SM. Androgen receptor polyglutamine repeat length affects receptor activity and C2C12 cell development. Physiol Genomics. 2011;43(20):1135–43. doi: 10.1152/physiolgenomics.00049.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hsiao PW, Lin DL, Nakao R, Chang C. The linkage of Kennedy’s neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. J Biol Chem. 1999;274(29):20229–34. doi: 10.1074/jbc.274.29.20229. [DOI] [PubMed] [Google Scholar]
- 141.Schiffer NW, Céraline J, Hartl FU, Broadley SA. N-terminal polyglutamine-containing fragments inhibit androgen receptor transactivation function. Biol Chem. 2008;389(12):1455–66. doi: 10.1515/BC.2008.169. [DOI] [PubMed] [Google Scholar]
- 142.Adachi H, Katsuno M, Minamiyama M, Waza M, Sang C, Nakagomi Y, et al. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain. 2005;128(Pt 3):659–70. doi: 10.1093/brain/awh381. [DOI] [PubMed] [Google Scholar]
- 143.Li M, Nakagomi Y, Kobayashi Y, Merry DE, Tanaka F, Doyu M, et al. Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am J Pathol. 1998;153(3):695–701. doi: 10.1016/S0002-9440(10)65612-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8(5):731–41. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 145.Jochum T, Ritz ME, Schuster C, Funderburk SF, Jehle K, Schmitz K, et al. Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochim Biophys Acta. 2012;1822(6):1070–8. doi: 10.1016/j.bbadis.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 146.Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Régulier E, et al. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104(3):846–58. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 147.Sontag EM, Lotz GP, Yang G, Sontag CJ, Cummings BJ, Glabe CG, et al. Detection of mutant huntingtin aggregation conformers and modulation of SDS-soluble fibrillar oligomers by small molecules. J Huntingtons Dis. 2012;1(1):119–32. doi: 10.3233/JHD-2012-129004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sontag EM, Lotz GP, Agrawal N, Tran A, Aron R, Yang G, et al. Methylene blue modulates huntingtin aggregation intermediates and is protective in Huntington’s disease models. J Neurosci. 2012;32(32):11109–19. doi: 10.1523/JNEUROSCI.0895-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Berger TR, Montie HL, Jain P, Legleiter J, Merry DE. Identification of novel polyglutamine-expanded aggregation species in spinal and bulbar muscular atrophy. Brain Res. 2015;1628(Pt B):254–64. doi: 10.1016/j.brainres.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heine EM, Berger TR, Pluciennik A, Orr CR, Zboray L, Merry DE. Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis. J Biol Chem. 2015;290(20):12572–84. doi: 10.1074/jbc.M114.617894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7(12):925–34. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li M, Chevalier-Larsen ES, Merry DE, Diamond MI. Soluble androgen receptor oligomers underlie pathology in a mouse model of spinobulbar muscular atrophy. J Biol Chem. 2007;282(5):3157–64. doi: 10.1074/jbc.M609972200. [DOI] [PubMed] [Google Scholar]
- 153.Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67(6):936–52. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 155.McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9(14):2197–202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 156.Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, Angelidis C, et al. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J Neurosci. 2003;23(6):2203–11. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kobayashi Y, Kume A, Li M, Doyu M, Hata M, Ohtsuka K, et al. Chaperones Hsp70 and Hsp40 suppress aggregate formation and apoptosis in cultured neuronal cells expressing truncated androgen receptor protein with expanded polyglutamine tract. J Biol Chem. 2000;275(12):8772–8. doi: 10.1074/jbc.275.12.8772. [DOI] [PubMed] [Google Scholar]
- 158.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11(5):515–23. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 159.Yersak JM, Montie HL, Chevalier-Larsen ES, Liu Y, Huang L, Rechsteiner M, et al. The 11S proteasomal activator REGγ impacts polyglutamine-expanded androgen receptor aggregation and motor neuron viability through distinct mechanisms. Front Mol Neurosci. 2017;10:159. doi: 10.3389/fnmol.2017.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abel A, Walcott J, Woods J, Duda J, Merry DE. Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum Mol Genet. 2001;10(2):107–16. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 161.Walcott JL, Merry DE. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J Biol Chem. 2002;277(52):50855–9. doi: 10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- 162.Montie HL, Cho MS, Holder L, Liu Y, Tsvetkov AS, Finkbeiner S, et al. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18(11):1937–50. doi: 10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brooks BP, Paulson HL, Merry DE, Salazar-Grueso EF, Brinkmann AO, Wilson EM, et al. Characterization of an expanded glutamine repeat androgen receptor in a neuronal cell culture system. Neurobiol Dis. 1997;3(4):313–23. doi: 10.1006/nbdi.1997.0126. [DOI] [PubMed] [Google Scholar]
- 164.Salazar-Grueso EF, Kim S, Kim H. Embryonic mouse spinal cord motor neuron hybrid cells. Neuroreport. 1991;2(9):505–8. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]
- 165.Simeoni S, Mancini MA, Stenoien DL, Marcelli M, Weigel NL, Zanisi M, et al. Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum Mol Genet. 2000;9(1):133–44. doi: 10.1093/hmg/9.1.133. [DOI] [PubMed] [Google Scholar]
- 166.Rusmini P, Sau D, Crippa V, Palazzolo I, Simonini F, Onesto E, et al. Aggregation and proteasome: the case of elongated polyglutamine aggregation in spinal and bulbar muscular atrophy. Neurobiol Aging. 2007;28(7):1099–111. doi: 10.1016/j.neurobiolaging.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 167.Rusmini P, Crippa V, Giorgetti E, Boncoraglio A, Cristofani R, Carra S, et al. Clearance of the mutant androgen receptor in motoneuronal models of spinal and bulbar muscular atrophy. Neurobiol Aging. 2013;34(11):2585–603. doi: 10.1016/j.neurobiolaging.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17(9):1180–9. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 2014;7(3):774–84. doi: 10.1016/j.celrep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Milioto C, Malena A, Maino E, Polanco MJ, Marchioretti C, Borgia D, et al. Beta-agonist stimulation ameliorates the phenotype of spinal and bulbar muscular atrophy mice and patient-derived myotubes. Sci Rep. 2017;7:41046. doi: 10.1038/srep41046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hijikata Y, Katsuno M, Suzuki K, Hashizume A, Araki A, Yamada S, et al. Impaired muscle uptake of creatine in spinal and bulbar muscular atrophy. Ann Clin Transl Neurol. 2016;3(7):537–46. doi: 10.1002/acn3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Grunseich C, Zukosky K, Kats IR, Ghosh L, Harmison GG, Bott LC, et al. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2014;70:12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dossena M, Bedini G, Rusmini P, Giorgetti E, Canazza A, Tosetti V, et al. Human adipose-derived mesenchymal stem cells as a new model of spinal and bulbar muscular atrophy. PLoS One. 2014;9(11):e112746. doi: 10.1371/journal.pone.0112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sheila M, Narayanan G, Ma S, Tam WL, Chai J, Stanton LW. Phenotypic and molecular features underlying neurodegeneration of motor neurons derived from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2019;124:1–13. doi: 10.1016/j.nbd.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 175.Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, et al. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35(5):855–64. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 176.Badders NM, Korff A, Miranda HC, Vuppala PK, Smith RB, Winborn BJ, et al. Selective modulation of the androgen receptor AF2 domain rescues degeneration in spinal bulbar muscular atrophy. Nat Med. 2018;24(4):427–37. doi: 10.1038/nm.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35(5):843–54. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 178.Chevalier-Larsen ES, O’Brien CJ, Wang H, Jenkins SC, Holder L, Lieberman AP, et al. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J Neurosci. 2004;24(20):4778–86. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.La Spada AR, Peterson KR, Meadows SA, McClain ME, Jeng G, Chmelar RS, et al. Androgen receptor YAC transgenic mice carrying CAG 45 alleles show trinucleotide repeat instability. Hum Mol Genet. 1998;7(6):959–67. doi: 10.1093/hmg/7.6.959. [DOI] [PubMed] [Google Scholar]
- 180.Sopher BL, Thomas PS, LaFevre-Bernt MA, Holm IE, Wilke SA, Ware CB, et al. Androgen receptor YAC transgenic mice recapitulate SBMA motor neuronopathy and implicate VEGF164 in the motor neuron degeneration. Neuron. 2004;41(5):687–99. doi: 10.1016/s0896-6273(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 181.Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, Yu Z, et al. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci U S A. 2007;104(46):18259–64. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408(6808):101–6. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 183.Yu Z, Dadgar N, Albertelli M, Scheller A, Albin RL, Robins DM, et al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am J Pathol. 2006;168(1):195–204. doi: 10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Albertelli MA, Scheller A, Brogley M, Robins DM. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol Endocrinol. 2006;20(6):1248–60. doi: 10.1210/me.2006-0021. [DOI] [PubMed] [Google Scholar]
- 185.Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, Robins DM, et al. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006;116(10):2663–72. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Latimer CS, Flanagan ME, Cimino PJ, Jayadev S, Davis M, Hoffer ZS, et al. Neuropathological comparison of adult onset and juvenile Huntington’s disease with cerebellar atrophy: a report of a father and son. J Huntingtons Dis. 2017;6(4):337–48. doi: 10.3233/JHD-170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Katsuno M, Banno H, Suzuki K, Takeuchi Y, Kawashima M, Yabe I, et al. Efficacy and safety of leuprorelin in patients with spinal and bulbar muscular atrophy (JASMITT study): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2010;9(9):875–84. doi: 10.1016/S1474-4422(10)70182-4. [DOI] [PubMed] [Google Scholar]
- 188.Hashizume A, Katsuno M, Suzuki K, Hirakawa A, Hijikata Y, Yamada S, et al. Long-term treatment with leuprorelin for spinal and bulbar muscular atrophy: natural history-controlled study. J Neurol Neurosurg Psychiatry. 2017;88(12):1026–32. doi: 10.1136/jnnp-2017-316015. [DOI] [PubMed] [Google Scholar]
- 189.Banno H, Katsuno M, Suzuki K, Takeuchi Y, Kawashima M, Suga N, et al. Phase 2 trial of leuprorelin in patients with spinal and bulbar muscular atrophy. Ann Neurol. 2009;65(2):140–50. doi: 10.1002/ana.21540. [DOI] [PubMed] [Google Scholar]
- 190.Schmidt BJ, Greenberg CR, Allingham-Hawkins DJ, Spriggs EL. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59(5):770–2. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- 191.Katsuno M, Adachi H, Doyu M, Minamiyama M, Sang C, Kobayashi Y, et al. Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nat Med. 2003;9(6):768–73. doi: 10.1038/nm878. [DOI] [PubMed] [Google Scholar]
- 192.Wilson AC, Meethal SV, Bowen RL, Atwood CS. Leuprolide acetate: a drug of diverse clinical applications. Expert Opin Investig Drugs. 2007;16(11):1851–63. doi: 10.1517/13543784.16.11.1851. [DOI] [PubMed] [Google Scholar]
- 193.Tindall DJ, Rittmaster RS. The rationale for inhibiting 5alpha-reductase isoenzymes in the prevention and treatment of prostate cancer. J Urol. 2008;179(4):1235–42. doi: 10.1016/j.juro.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Toren P, Margel D, Kulkarni G, Finelli A, Zlotta A, Fleshner N. Effect of dutasteride on clinical progression of benign prostatic hyperplasia in asymptomatic men with enlarged prostate: a post hoc analysis of the REDUCE study. BMJ. 2013;346:f2109. doi: 10.1136/bmj.f2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shrader JA, Kats I, Kokkinis A, Zampieri C, Levy E, Joe GO, et al. A randomized controlled trial of exercise in spinal and bulbar muscular atrophy. Ann Clin Transl Neurol. 2015;2(7):739–47. doi: 10.1002/acn3.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gotkine M, Friedlander Y, Hochner H. Triathletes are over-represented in a population of patients with ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(7–8):534–6. doi: 10.3109/21678421.2014.932383. [DOI] [PubMed] [Google Scholar]
- 197.Turner MR, Wicks P, Brownstein CA, Massagli MP, Toronjo M, Talbot K, et al. Concordance between site of onset and limb dominance in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82(8):853–4. doi: 10.1136/jnnp.2010.208413. [DOI] [PubMed] [Google Scholar]
- 198.Drory VE, Goltsman E, Reznik JG, Mosek A, Korczyn AD. The value of muscle exercise in patients with amyotrophic lateral sclerosis. J Neurol Sci. 2001;191(1–2):133–7. doi: 10.1016/s0022-510x(01)00610-4. [DOI] [PubMed] [Google Scholar]
- 199.Bello-Haas VD, Florence JM, Kloos AD, Scheirbecker J, Lopate G, Hayes SM, et al. A randomized controlled trial of resistance exercise in individuals with ALS. Neurology. 2007;68(23):2003–7. doi: 10.1212/01.wnl.0000264418.92308.a4. [DOI] [PubMed] [Google Scholar]
- 200.Schwarz AJ, Brasel JA, Hintz RL, Mohan S, Cooper DM. Acute effect of brief low- and high-intensity exercise on circulating insulin-like growth factor (IGF) I, II, and IGF-binding protein-3 and its proteolysis in young healthy men. J Clin Endocrinol Metab. 1996;81(10):3492–7. doi: 10.1210/jcem.81.10.8855791. [DOI] [PubMed] [Google Scholar]
- 201.Rinaldi C, Bott LC, Chen KL, Harmison GG, Katsuno M, Sobue G, et al. Insulinlike growth factor (IGF)-1 administration ameliorates disease manifestations in a mouse model of spinal and bulbar muscular atrophy. Mol Med. 2012;18:1261–8. doi: 10.2119/molmed.2012.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dahlqvist JR, Vissing J. Exercise therapy in spinobulbar muscular atrophy and other neuromuscular disorders. J Mol Neurosci. 2016;58(3):388–93. doi: 10.1007/s12031-015-0686-3. [DOI] [PubMed] [Google Scholar]
- 203.Grunseich C, Miller R, Swan T, Glass DJ, Mouelhi ME, Fornaro M, et al. Safety, tolerability, and preliminary efficacy of an IGF-1 mimetic in patients with spinal and bulbar muscular atrophy: a randomised, placebo-controlled trial. Lancet Neurol. 2018. 17:1043-1052. [DOI] [PMC free article] [PubMed]
- 204.Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen Comp Endocrinol. 2010;167(3):344–51. doi: 10.1016/j.ygcen.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 205.Kaspar BK, Lladó J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301(5634):839–42. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- 206.Dobrowolny G, Giacinti C, Pelosi L, Nicoletti C, Winn N, Barberi L, et al. Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J Cell Biol. 2005;168(2):193–9. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hijikata Y, Katsuno M, Suzuki K, Hashizume A, Araki A, Yamada S, et al. Treatment with creatine monohydrate in spinal and bulbar muscular atrophy: protocol for a randomized, double-blind, placebo-controlled trial. JMIR Res Protoc. 2018;7(3):e69. doi: 10.2196/resprot.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hashizume A, Katsuno M, Banno H, Suzuki K, Suga N, Mano T, et al. Longitudinal changes of outcome measures in spinal and bulbar muscular atrophy. Brain. 2012;135(Pt 9):2838–48. doi: 10.1093/brain/aws170. [DOI] [PubMed] [Google Scholar]
- 209.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80(3):1107–213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 210.Sant'Ana Pereira JA, Sargeant AJ, Rademaker AC, de Haan A, van Mechelen W. Myosin heavy chain isoform expression and high energy phosphate content in human muscle fibres at rest and post-exercise. J Physiol. 1996;496(Pt 2):583–8. doi: 10.1113/jphysiol.1996.sp021709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yamada S, Hashizume A, Hijikata Y, Inagaki T, Suzuki K, Kondo N, et al. Decreased peak expiratory flow associated with muscle fiber-type switching in spinal and bulbar muscular atrophy. PLoS One. 2016;11(12):e0168846. doi: 10.1371/journal.pone.0168846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rocchi A, Milioto C, Parodi S, Armirotti A, Borgia D, Pellegrini M, et al. Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modified by high-fat diet. Acta Neuropathol. 2016;132(1):127–44. doi: 10.1007/s00401-016-1550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Banerjee B, Sharma U, Balasubramanian K, Kalaivani M, Kalra V, Jagannathan NR. Effect of creatine monohydrate in improving cellular energetics and muscle strength in ambulatory Duchenne muscular dystrophy patients: a randomized, placebo-controlled 31P MRS study. Magn Reson Imaging. 2010;28(5):698–707. doi: 10.1016/j.mri.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 214.Walter MC, Lochmüller H, Reilich P, Klopstock T, Huber R, Hartard M, et al. Creatine monohydrate in muscular dystrophies: a double-blind, placebo-controlled clinical study. Neurology. 2000;54(9):1848–50. doi: 10.1212/wnl.54.9.1848. [DOI] [PubMed] [Google Scholar]
- 215.Tarnopolsky MA, Mahoney DJ, Vajsar J, Rodriguez C, Doherty TJ, Roy BD, et al. Creatine monohydrate enhances strength and body composition in Duchenne muscular dystrophy. Neurology. 2004;62(10):1771–7. doi: 10.1212/01.wnl.0000125178.18862.9d. [DOI] [PubMed] [Google Scholar]
- 216.Xu Y, Halievski K, Katsuno M, Adachi H, Sobue G, Breedlove SM, et al. Pre-clinical symptoms of SBMA may not be androgen-dependent: implications from two SBMA mouse models. Hum Mol Genet. 2018;27(14):2425–42. doi: 10.1093/hmg/ddy142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Arnold AP, Gorski RA. Gonadal steroid induction of structural sex differences in the central nervous system. Annu Rev Neurosci. 1984;7:413–42. doi: 10.1146/annurev.ne.07.030184.002213. [DOI] [PubMed] [Google Scholar]
- 218.Sahashi K, Katsuno M, Hung G, Adachi H, Kondo N, Nakatsuji H, et al. Silencing neuronal mutant androgen receptor in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2015;24(21):5985–94. doi: 10.1093/hmg/ddv300. [DOI] [PubMed] [Google Scholar]
- 219.Ishihara H, Kanda F, Nishio H, Sumino K, Chihara K. Clinical features and skewed X-chromosome inactivation in female carriers of X-linked recessive spinal and bulbar muscular atrophy. J Neurol. 2001;248(10):856–60. doi: 10.1007/s004150170069. [DOI] [PubMed] [Google Scholar]
- 220.Lanman TA, Bakar D, Badders NM, Burke A, Kokkinis A, Shrader JA, et al. Sexual reassignment fails to prevent Kennedy’s disease. J Neuromuscul Dis. 2016;3(1):121–5. doi: 10.3233/JND-150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hijikata Y, Hashizume A, Yamada S, Ito D, Banno H, Suzuki K, et al. Long-term effects of androgen deprivation in a patient with spinal and bulbar muscular atrophy—a case report with 14 years of follow-up. Intern Med. 2019. [DOI] [PMC free article] [PubMed]
- 222.Mhaouty-Kodja S. Role of the androgen receptor in the central nervous system. Mol Cell Endocrinol. 2018;465:103–12. doi: 10.1016/j.mce.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 223.Langfelder P, Cantle JP, Chatzopoulou D, Wang N, Gao F, Al-Ramahi I, et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat Neurosci. 2016;19(4):623–33. doi: 10.1038/nn.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tanaka F, Reeves MF, Ito Y, Matsumoto M, Li M, Miwa S, et al. Tissue-specific somatic mosaicism in spinal and bulbar muscular atrophy is dependent on CAG-repeat length and androgen receptor–gene expression level. Am J Hum Genet. 1999;65(4):966–73. doi: 10.1086/302578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pennuto M, Basso M. In vitro and in vivo modeling of spinal and bulbar muscular atrophy. J Mol Neurosci. 2016;58(3):365–73. doi: 10.1007/s12031-015-0677-4. [DOI] [PubMed] [Google Scholar]