Macrolides are the cornerstone of Mycobacterium abscessus multidrug therapy, despite that most patients respond poorly to this class of antibiotics due to the inducible resistance phenotype that occurs during drug treatment. This mechanism is driven by the macrolide-inducible ribosomal methylase encoded by erm(41), whose expression is activated by the transcriptional regulator WhiB7.
KEYWORDS: Mycobacterium abscessus, WhiB7, beta-galactosidase, erm(41), inducible resistance, macrolides, therapeutic activity, zebrafish
ABSTRACT
Macrolides are the cornerstone of Mycobacterium abscessus multidrug therapy, despite that most patients respond poorly to this class of antibiotics due to the inducible resistance phenotype that occurs during drug treatment. This mechanism is driven by the macrolide-inducible ribosomal methylase encoded by erm(41), whose expression is activated by the transcriptional regulator WhiB7. However, it has been debated whether clarithromycin and azithromycin differ in the extent to which they induce erm(41)-mediated macrolide resistance. Herein, we show that macrolide resistance is induced more rapidly in various M. abscessus isolates upon exposure to azithromycin than to clarithromycin, based on MIC determination. Macrolide-induced expression of erm(41) was assessed in vivo using a strain carrying tdTomato placed under the control of the erm(41) promoter. Visualization of fluorescent bacilli in infected zebrafish demonstrates that azithromycin and clarithromycin activate erm(41) expression in vivo. That azithromycin induces a more rapid expression of erm(41) was confirmed by measuring the β-galactosidase activity of a reporter strain in which lacZ was placed under the control of the erm(41) promoter. Shortening the promoter region in the lacZ reporter plasmid identified DNA elements involved in the regulation of erm(41) expression, particularly an AT-rich motif sharing partial conservation with the WhiB7-binding site. Mutation of this motif abrogated the macrolide-induced and WhiB7-dependent expression of erm(41). This study provides new mechanistic information on the adaptive response to macrolide treatment in M. abscessus.
INTRODUCTION
Members of the Mycobacterium abscessus complex are rapidly growing nontuberculous mycobacteria (NTM) of increasing clinical significance due to the rising burden of associated pulmonary disease, particularly in cystic fibrosis (CF) patients (1). In CF patients, infection with M. abscessus correlates with a more rapid decline in lung function and can represent an obstacle to subsequent lung transplantation (2–4). The possible person-to-person transmission of virulent clones among the CF population makes this situation even more worrisome (5, 6). From a taxonomic view, the complex currently comprises three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense (7). These subspecies exhibit different clinical outcomes and drug susceptibility profiles with respect to antibiotic treatments. M. abscessus subsp. abscessus is notoriously acknowledged as the most drug-resistant mycobacterial species, characterized by a wide panel of acquired and innate drug-resistance mechanisms against nearly all antitubercular drugs, as well as to most classes of antibiotics (8). These mechanisms rely largely on the expression of enzymes able to modify and inactivate rifamycins (9), aminoglycosides (10), tetracyclines (11), or β-lactams (12) or on the presence of multiple efflux pumps of the MmpL family, which afford low-level resistance to clofazimine and bedaquiline (13, 14). Therapeutic treatments require prolonged courses of multiple antibiotics, usually combining a macrolide, a β-lactam (cefoxitin or imipenem), and an aminoglycoside (15). Macrolides represent the cornerstone of M. abscessus subsp. abscessus multidrug therapy (15), but treatment success rates are poor, particularly in macrolide-resistant strains. Acquired resistance to macrolides usually occurs at low rates through point mutations at positions 2058 or 2059 of the 23S rRNA rrl gene (16). In addition, M. abscessus subsp. abscessus harbors an inducible ribosomal methylase encoded by erm(41), which represents the primary mechanism of intrinsic resistance against macrolides (17). This adaptive resistance mechanism can be manifested by prolonged incubation of the bacilli up to 14 days in drug-susceptible testing assays, rather than the typical 3 to 4 days of exposure to other drugs. A single nucleotide exchange (of T28C, resulting in a Trp10Arg replacement) in erm(41) is associated with a loss of methylase activity in M. abscessus subsp. abscessus, abrogating the inducible resistance mechanism and thereby resulting in a macrolide-susceptible phenotype (18, 19). This T28C polymorphism is found in 15 to 20% of clinical isolates (20, 21). In contrast to M. abscessus subsp. abscessus or M. abscessus subsp. bolletii, M. abscessus subsp. massiliense has been associated with clarithromycin (CLR) susceptibility due to deletions (bases 64 to 65 and 159 to 432) within erm(41) (17, 22), explaining the more favorable clinical outcomes upon CLR treatment (23). However, the role and contribution of the Erm(41)-dependent mechanism in azithromycin (AZM) resistance is less clear and has been the subject of discrepancies between studies. In accordance with an early report of Brown et al. (24), it was subsequently reported that MICs of AZM for the M. abscessus complex tend to be higher than those of CLR (18). In contrast to these studies, Choi et al. reported that AZM is a poor inducer of erm(41) gene expression compared to CLR and, therefore, should be preferred in antibiotic therapy for M. abscessus infections (25). However, this work was inconsistent with another study in which both molecules provoked a comparable resistance phenotype in M. abscessus (19). A recent study also showed that erm(41) expression increased after exposure to either macrolide for 7 days and it was proposed that this should not be an argument for choosing between CLR and AZM for therapy (26). However, most studies are based on the in vitro determination of the MIC to cultures exposed to macrolides and data reporting the Erm(41)-dependent inducible mechanisms in the infected host are scarce. Except for a study reporting the increased expression of erm(41) transcripts in M. abscessus-infected mice treated with CLR, but not with AZM (25), it remains to be fully established whether macrolide-inducible resistance occurs in vivo, and whether induction occurs rapidly upon drug treatment in the infected host.
In the context of this long debate, the present study aimed at revisiting the erm(41) inducible mechanisms through the following means: (i) evaluation of the in vitro drug susceptibility of clinical isolates belonging to the M. abscessus complex to CLR and AZM; (ii) visualization of induced expression of erm(41) in zebrafish infected with an M. abscessus reporter strain in which a fluorescent marker has been placed under the control of the erm(41) promoter region; and (iii) dissection of the erm(41) promoter to uncover important DNA regions that participate directly in this regulatory process.
RESULTS
Macrolide-inducible resistance of smooth and rough M. abscessus CIP104536T is medium independent.
The current debate regarding the level of resistance induced by CLR or AZM has been hypothesized to be dependent on the composition of the medium used for the determination of the MICs of the two drugs. In particular, Choi et al. (25) used Middlebrook 7H9, which contains a large amount of inorganic salts and has a slightly acidic pH (pH 6.8), whereas Bastian et al. (18), following the Clinical and Laboratory Standards Institute (CLSI) recommendations, used cation-adjusted Mueller-Hinton broth (CaMHB), which has a higher pH (pH 7.4) than 7H9. Because macrolide activity is sensitive to pH (27), it was hypothesized that the different media used in various studies may have influenced the MIC values. Herein, we evaluated and compared macrolide-induced resistance to both drugs at various time points (3, 5, 7, 10, and 14 days posttreatment [dpt]) in three different media (CaMHB, 7H9, and Sauton’s). When assaying the smooth (S) morphotype of the reference strain CIP104536T, we consistently found that AZM induced a more rapid upshift of the MIC than CLR in all three media (Fig. S1A in the supplemental material). The increase in MIC of AZM started after 3 dpt and rose progressively to reach a MIC of 256 μg/ml at 10 dpt. The same MIC value was reached at 14 dpt with CLR, indicating that, although the intensity of induced resistance is comparable for both macrolides, the kinetics of resistance was delayed with CLR compared to AZM in vitro. As expected, amikacin (AMK), used as a nonrelevant control drug, failed to show a similar drug-induced phenotype upon a prolonged exposure time. As observed with the S variant, the kinetics of resistance to AZM in the rough (R) variant was more rapid than that with CLR and this macrolide-induced resistance was independent of the medium used (Fig. S1B). Overall, these results indicate that both S and R morphotypes respond similarly to macrolide treatment regardless of the medium.
Resistance to macrolides is rapidly induced in the M. abscessus complex strains upon exposure to CLR and AZM.
We next investigated the kinetics of macrolide-inducible resistance in clinical strains isolated from cystic fibrosis (CF) and non-CF patients in CaMHB. Clinical M. abscessus subsp. abscessus strains 2069 and 3022, from non-CF patients, were characterized by a more rapid increase in the MIC values for AZM than for CLR, similar to the reference strain CIP104536T (Fig. S2A and S2B). However, in the case of strain 2069, the resistance to CLR reached a plateau at 128 μg/ml at 10 dpt, whereas for strain 3022 the MIC continued to rise up to 256 μg/ml at 14 dpt, sharing a profile similar to the CF strain 2648. In contrast, strain 1298 failed to show inducible resistance to either macrolide (Fig. S2A). Due to the important contribution of the polymorphism at the 28th nucleotide of erm(41), where the T28 sequevar (Trp10) shows inducible resistance while the C28 sequevar (Arg10) remains susceptible to macrolides (18), we sequenced erm(41) in M. abscessus subsp. abscessus strains 1298, 2069, 2648, 3022, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense. All M. abscessus subsp. abscessus strains possess an erm(41) gene identical to the M. abscessus CIP104536T reference strain, with a Trp in position 10, except for strain 1298 carrying a Leu residue instead of a Phe in position 34 (not shown). To our knowledge, this point mutation, presumably inactivating Erm(41) and conferring susceptibility to macrolides, has not been yet reported (20). The M. abscessus subsp. bolletii S strain carrying the T28 sequevar also showed inducible resistance to macrolides, similarly to M. abscessus subsp. abscessus (Fig. S2C). As anticipated, M. abscessus subsp. massiliense was found to be intrinsically susceptible to CLR and AZM due to a genetic lesions in erm(41) that renders the enzyme nonfunctional (18) (Fig. S2D). We next constructed an unmarked deletion mutant of erm(41) using a recently developed strategy based on a new suicide vector, pUX1-katG, which allows the easy and rapid generation of genetic deletions in the M. abscessus chromosome, facilitated by the presence of the brightly red fluorescent tdTomato positive selectable marker and the katG counter selectable marker (10, 13, 28) (Fig. S3). In agreement with a previous report (25), this mutant remained fully susceptible to CLR and AZM, confirming the direct involvement of Erm(41) in macrolide-inducible resistance (Fig. S2E).
Together, these results confirm that M. abscessus subsp. abscessus clinical isolates and M. abscessus subsp. bolletii carry a functional Erm(41) methylase undergoing a faster increase in resistance to AZM than to CLR, and that the newly discovered F34L substitution in Erm(41) abrogates the macrolide-induced resistance.
Inducible resistance to macrolides occurs in infected zebrafish.
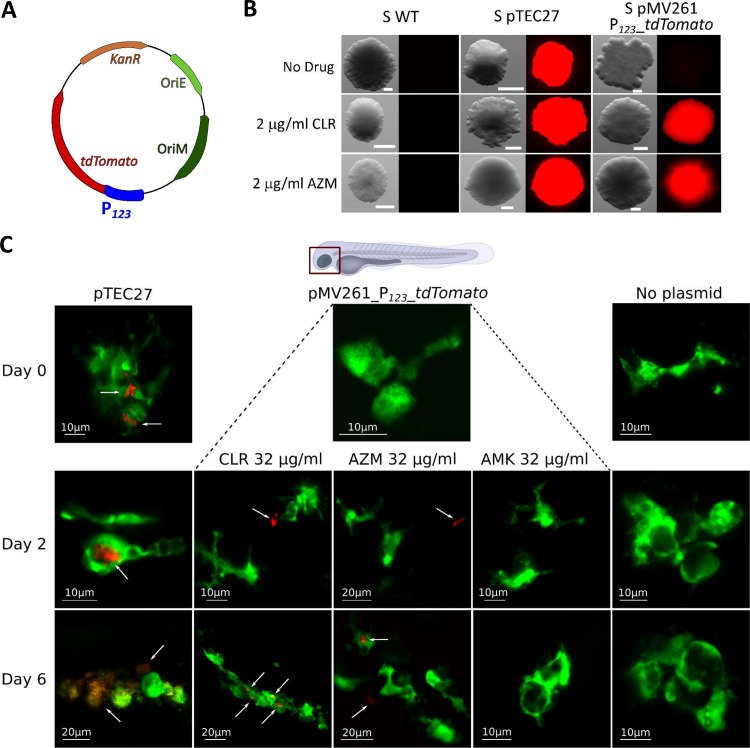
The vast majority of studies related to macrolide-inducible resistance are based on MIC determination and/or determination by quantitative reverse transcriptase PCR (qRT-PCR) of the level of erm(41) transcripts in vitro (18, 19, 26). Despite the clinical importance, reports describing whether macrolide-inducible resistance occurs during patient therapy are scarce. However, one study reported increased expression of erm(41) transcripts in M. abscessus-infected mice treated with CLR but not with AZM (25). We took advantage of the transparency of zebrafish embryos to visualize and monitor the induced expression of erm(41) during macrolide treatment of larvae infected with M. abscessus. Indeed, this recently developed animal model allows easy assessment of the suitability and sensitivity of clinically relevant drugs, along with the ability to visualize, in a dose- and time-dependent manner, the dynamics of infection in the presence of an active compound (12, 29–31). To do so, a fluorescent reporter M. abscessus strain was designed, carrying the plasmid pMV261_P123_tdTomato, in which the tdTomato gene was cloned under the control of the 123 bp promoter region of erm(41) (Fig. 1A). To verify the usefulness of this construct, the recombinant strain was first plated onto Middlebrook 7H10 medium containing 100 μg/ml kanamycin supplemented or not with 2 μg/ml CLR or AZM. Examination of the plates under a fluorescence microscope revealed the presence of red fluorescent colonies in the presence of either macrolide, while no fluorescence was detected on plates lacking these drugs (Fig. 1B). As a positive control for fluorescence, a strain harboring pTEC27, which allows constitutive expression of tdTomato in M. abscessus (32), was also included in these experiments (Fig. 1B). These results clearly highlight the potential of this reporter strain to monitor macrolide-inducible erm(41) expression in an infected host. Whether red fluorescent bacilli can be visualized directly in M. abscessus-infected embryos upon treatment with macrolides was next investigated. Around 1000 CFU of the reporter strain were microinjected into the caudal vein of the mpeg:GFP-CAAX transgenic embryos harboring green fluorescent macrophages (33) which, after 96 h, were exposed to 32 μg/ml CLR or AZM (directly added to the water). Fish were observed under the fluorescence microscope at 0, 2, and 6 dpt. Red fluorescent bacilli could be detected at 2 dpt with either macrolide, while no signal was detected in control embryos (M. abscessus without plasmid) or embryos treated with AMK as a nonrelevant drug, indicating that expression of tdTomato occurred very rapidly during infection in a living animal and was specifically induced by macrolides (Fig. 1C). At later time points (6 dpt), red fluorescent bacilli were detected inside individualized macrophages or inside aggregated macrophages forming granulomas. Red bacilli could also be observed outside macrophages, although they may reside inside neutrophils (which cannot be visualized in this transgenic zebrafish line harboring only fluorescent macrophages), shown to rapidly engulf M. abscessus (34). It remains, however, difficult to explain at this stage whether already fluorescent bacteria are phagocytosed by macrophages or whether macrolides penetrate macrophages/granulomas to induce tdTomato expression in intracellular bacilli.
FIG 1.
In vivo induction of macrolide resistance in M. abscessus-infected zebrafish embryos. (A) Schematic representation of the plasmid pMV261_P123_tdTomato used to transform M. abscessus smooth (S) variant CIP104536T, where P123 represents the full-length promoter region of erm(41). (B) Bacteria with no plasmid (WT), with the pTEC27 plasmid which constitutively expresses the red fluorescent protein tdTomato, or with the pMV261_P123_tdTomato plasmid for monitoring macrolide-inducible expression of tdTomato were plated on 7H10OADC supplemented or not with 2 μg/ml CLR or AZM. Plates were incubated at 37°C for 5 days and observed under a fluorescence microscope. Scale bars represent 0.200 μm. (C) Macrolide-inducible resistance by CLR and AZM occurs in vivo. Tg(mpeg1:GFP-CAAX) zebrafish embryos, harboring green fluorescent macrophages, were injected in the caudal vein with 900 to 1,000 CFU of the three M. abscessus strains described in panel B. At 24 hpi, embryos were exposed to osmotic water supplemented with 32 μg/ml CLR, AZM, or AMK (day 0 of treatment) and fluorescence was monitored at 2 and 6 days posttreatment. Shown is representative confocal live imaging. White arrows indicate red fluorescent bacteria.
Overall, these results indicate that macrolide-inducible resistance occurs in the zebrafish model of infection.
Low macrolide concentrations induce a more rapid expression of erm(41) than high concentrations in M. abscessus.
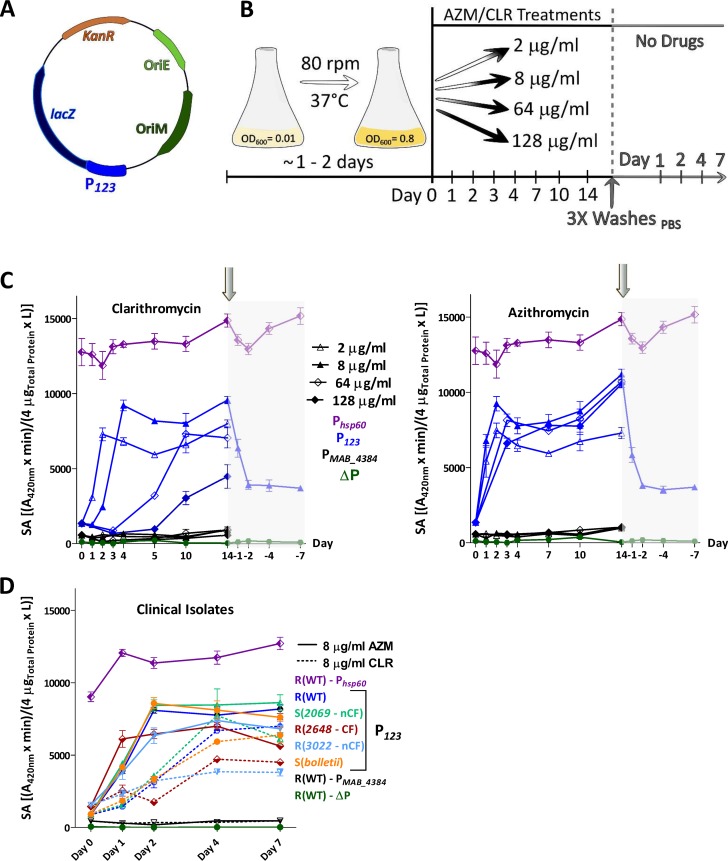
The pMV261_P123_lacZ plasmid containing the lacZ gene encoding the β-galactosidase (β-Gal) gene under the control of the erm(41) promoter was designed to tackle the fluctuations in erm(41) gene expression during exposure of the culture to various drug treatments (Fig. 2A). This strain was exposed to 2, 8, 64, and 128 μg/ml of either CLR or AZM and lacZ expression was determined by measuring β-Gal activity using 2-nitrophenyl β-d-galactopyranoside (ONPG) at different time points over a 14-day period (Fig. 2B). Exposure to 2 μg/ml of CLR induced rapid β-Gal activity during the first 2 days of treatment, which plateaued thereafter. Increasing the drug concentration resulted in similar but delayed specific activity curves. At the highest concentration tested, the lag phase lasted for 7 days prior to the induction of lacZ expression. This is unlikely to be linked to the impaired growth of the cultures treated with high concentrations of macrolide as the specific activity is expressed in relation to the total amount of soluble proteins. The maximum specific activity was, however, lower than the β-Gal activity produced when lacZ was placed under the control of the strong constitutive hsp60 promoter (Fig. 2C, left). As expected, when the promoterless construct was introduced in M. abscessus, this resulted in the loss of β-Gal activity. Moreover, in another strain in which the erm(41) promoter region was substituted by the promoter region of MAB_4384, encoding a TetR protein previously characterized (35), no activity was detected. This further supports that CLR-dependent expression of lacZ was specific to the erm(41) promoter region. Treatment with increasing concentrations of AZM showed slightly different trends, characterized by a faster induction of lacZ expression at low concentrations (2 and 8 μg/ml) than at high concentrations (64 and 128 μg/ml) and a significantly shorter lag phase with AZM than with CLR (Fig. 2C, right). Cultures treated with 64 μg/ml of AZM reached a β-Gal activity of approximately 8,000 units in only 3 days, whereas 10 days were needed for cultures treated with the same CLR concentration to reach this level of activity. Together, these results suggest that erm(41) transcriptional expression is more rapidly induced by AZM than by CLR, in agreement with the MIC results (Fig. S1 and S2).
FIG 2.
Kinetics of macrolide-induced expression of erm(41) using lacZ reporter strains. (A) Schematic representation of the pMV261_P123_lacZ used to transform the M. abscessus CIP104536T rough variant. (B) Schematic protocol of the β-Gal assay to evaluate the kinetics of macrolide-induced resistance. (C) The M. abscessus strain carrying the pMV261_P123_lacZ was first exposed to various concentrations of CLR and AZM for 14 days. Antibiotics were then removed by washing the bacterial pellets several times with PBS prior to β-Gal activity measurements for the next 7 days. (C) The β-Gal activity profile of M. abscessus CIP104536T rough variant exposed to 2, 8, 64, and 128 μg/ml CLR (left) or AZM (right). A pMV261 derivative containing lacZ cloned under the control of the constitutive hsp60 promoter (Phsp60) was used as a positive control. A promoterless construct (ΔP) and a pMV261_lacZ construct carrying the promoter of an unrelated gene (PMAB_4384) were included as negative controls. (D) Similar experiments as in panel C were carried out using different M. abscessus subsp. abscessus clinical strains (2069, 2648, and 3022) as well as M. abscessus subsp. bolletii transformed with pMV261_P123_lacZ and exposed to either CLR or AZM (8 μg/ml) for 7 days. All experiments were performed on 2 separate occasions with a total of 6 technical replicates. The specific activity of β-Gal is given in (A420nm × min)/(4 μgTotal protein × liters) as described in the Materials and Methods section ± standard deviation.
To ask whether the rapid increase in AZM-dependent expression of lacZ was restricted to the reference strain, the influence of both macrolides on lacZ expression was assessed in clinical strains (M. abscessus subsp. abscessus strains 2069, 2648, 3022 and M. abscessus subsp. bolletii) transformed with pMV261_P123_lacZ. In all cases, treatment with 8 μg/ml AZM induced a more rapid induction of lacZ expression than treatment with 8 μg/ml CLR, although the intensity of the response varied from one strain to another and was always below the activity associated with the hsp60 promoter (Fig. 2D). For instance, lacZ induction in strain 3022 was significantly slower than in strain 2069.
Overall, these results confirm that, although both macrolides induce a potent transcriptional expression of erm(41), AZM induces a more rapid response than does CLR.
Both CLR and AZM reduce the efficacy of AMK in M. abscessus.
Drug-inducible resistance is a mechanism of adaptive response to antibiotic treatment that is not genetically programed. As such, this reversible phenomenon is expected to disappear after stopping the drug treatment. Our reporter strain allowed us to ask whether erm(41) expression is reversed after removal of the drugs. In the experimental protocol used, bacteria were treated for 14 days with 8 μg/ml CLR (Fig. 2C, left) or with 8 μg/ml AZM (Fig. 2C, right), after which the cultures were extensively washed with phosphate-buffered saline (PBS) and the β-Gal activity further monitored for 7 days in the absence of drugs (Fig. 2B). Rapidly after washing, lacZ expression dropped during the next 2 days and then remained unchanged until the end of the experiment (7 days post washing) at significantly higher levels (around 4,000 units) than prior to drug addition. To assess the possibility that macrolide-pretreated cultures are more tolerant than unexposed cultures, bacteria containing pMV261_P123_lacZ were first preexposed or not with 8 μg/ml of CLR or AZM for 4 days. Once the optimal β-Gal activity was reached, the cultures were carefully washed with 1× PBS and bacterial pellets were resuspended and incubated in macrolide-free Sauton’s medium for two more days, a time point at which a β-Gal activity of 4,000 units was measured (not shown). From this stage (considered day 0), the MIC of CLR, AZM, and AMK were determined in Sauton’s medium at 3, 5, 7, 10, and 14 days (Table 1). At day 3, the untreated cultures exhibited MIC values of 1, 4, and 16 μg/ml for CLR, AZM, and AMK, respectively, while the macrolide-pretreated cultures exhibited higher MIC values. The CLR-treated culture displayed MICs of 64, 128, and 16 μg/ml against CLR, AZM, and AMK, respectively. This effect was even more pronounced with the AZM-treated culture (128, 256, and 32 for CLR, AZM, and AMK, respectively) and further increased at day 5 and day 7, with MIC values exceeding 256 μg/ml. While the MIC of AMK remained unchanged for the untreated culture (16 to 32 μg/ml), pretreatment with CLR induced cross-resistance to AMK (64 g/ml), as previously reported (36). We found this was also true with AZM (Table 1), indicating that, like CLR, exposure to AZM reduces the activity of AMK.
TABLE 1.
Enduring resistance levels to macrolides and amikacin following preexposure of M. abscessus to CLR or AZM
| Growth conditionsa | MIC μg/ml |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 3 |
Day 5 |
Day 7 |
Day 10 |
Day 14 |
|||||||||||
| CLR | AZM | AMK | CLR | AZM | AMK | CLR | AZM | AMK | CLR | AZM | AMK | CLR | AZM | AMK | |
| Untreated | 1 | 4 | 16 | 16 | 64 | 16 | 64 | 128 | 16 | 128 | 256 | 32 | 256 | >256 | 32 |
| CLR pretreated | 64 | 128 | 16 | 256 | >256 | 32 | >256 | >256 | 32 | >256 | >256 | 64 | >256 | >256 | 64 |
| AZM pretreated | 128 | 256 | 32 | >256 | >256 | 64 | >256 | >256 | 64 | >256 | >256 | 64 | >256 | >256 | 64 |
M. abscessus CIP104536T (rough variant) was pretreated or not with 8 μg/ml of either CLR or AZM for 4 days, washed, and incubated in macrolide-free medium for 2 additional days. MIC of CLR, AZM, and AMK were determined according to the CLSI guidelines at different time points. The MICs are given in μg/ml. Results are representative from three individual experiments with similar values.
Identification of a whiB7 box upstream of erm(41) required for macrolide-induced expression.
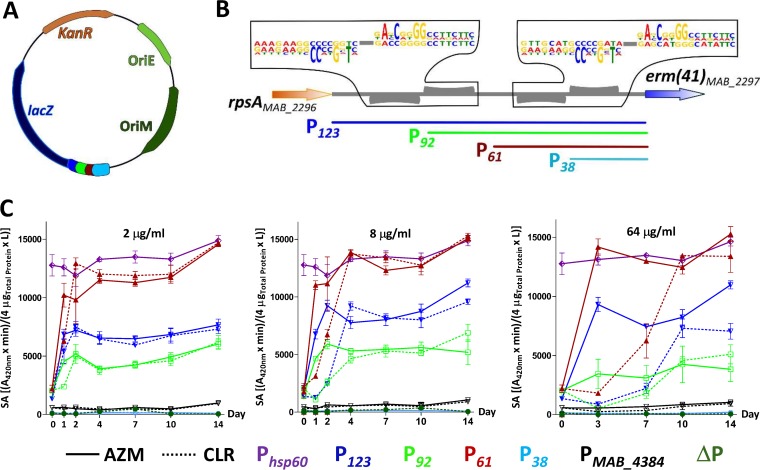
Careful examination of the promoter region using the MEME software identified two pairs of conserved repetitive sequences. To gain further insights into the regulatory DNA elements responsible for macrolide induction, the P123 promoter region in pMV261_ P123_lacZ was replaced by DNA fragments of 92 bp, 61 bp, and 38 bp derived from the original P123. The segment sizes were chosen to progressively shorten the degenerated inverted repeats (Fig. 3A and B). These DNA motifs may eventually represent the target of transcriptional factors that contribute to erm(41) transcriptional regulation. Cultures were exposed to various concentrations of AZM or CLR and lacZ expression was monitored over time by measuring the β-Gal activity. In the presence of 2 μg/ml of either macrolide, the optimal specific activity of the full-length construct (123 bp) plateaued at around 7,000 units, while the 92-bp construct was associated with a reduction in activity, to around 5,000 units (Fig. 3C, left). This indicates that the construct lacks an important regulatory element that is required for normal transcriptional levels. Unexpectedly, further shortening the promoter region down to 61 bp led to an opposite effect, with a much more pronounced β-Gal activity than with P123, reaching the levels of the strong hsp60 promoter. As shown earlier, these effects were always more rapid when the various strains were exposed to AZM than to CLR. Similar profiles were obtained by treating the various strains with 8 μg/ml (Fig. 3C, middle) or 64 μg/ml (Fig. 3C, right) of macrolides. In sharp contrast, the strain carrying pMV261_P38_lacZ failed to express any β-Gal activity, indicating that P38 is missing important regulatory information. As expected, when either the promoterless construct (ΔP) or the pMV261_PMAB_4384_lacZ were introduced in M. abscessus, no β-Gal activity was detected (Fig. 3C). This suggests the presence of two important regulatory sequences in P123: one located between the 5′ end of the P61 construct and the 5′ end of the P38 construct that positively regulates the expression and one located at the 5′ end of the 92 bp segment that possibly participates in the downregulation of erm(41) expression.
FIG 3.
Dissection of the erm(41) promoter region and impact on lacZ expression. (A) Schematic representation of the intergenic region between rpsA and erm(41). Two pairs of weakly conserved palindromes were identified using the MEME Suite 4.20.0 online tool. (B) Derivatives of pMV261_lacZ containing various truncated intergenic regions (where P123, P92, P61, and P38 correspond to the size of the promoter region in base pairs) were generated according to this bioinformatics analysis and shown using a color code. (B) Schematic representation of the intergenic region between rpsA and erm(41). Two pairs of weakly conserved palindromes were identified using the MEME Suite 4.20.0 online tool. (C) β-Gal activity profile of the M. abscessus strains transformed with the various lacZ reporter constructs monitored over a period of 14 days and exposed to 2 μg/ml (left), 8 μg/ml (middle), or 64 μg/ml (right) of either CLR (dotted lines) or AZM (normal lines). A pMV261 derivative containing lacZ cloned under the control of the constitutive hsp60 promoter was used as a positive control (Phsp60). A promoterless construct (ΔP) and a pMV261_lacZ construct carrying the promoter of an unrelated gene (PMAB_4384) were included as negative controls. The results were obtained from two separate experiments with a total of 6 technical replicates. The specific activity of β-Gal is given in (A420nm × min)/(4 μgTotal protein × liters) as described in the Materials and Methods section ± standard deviation.
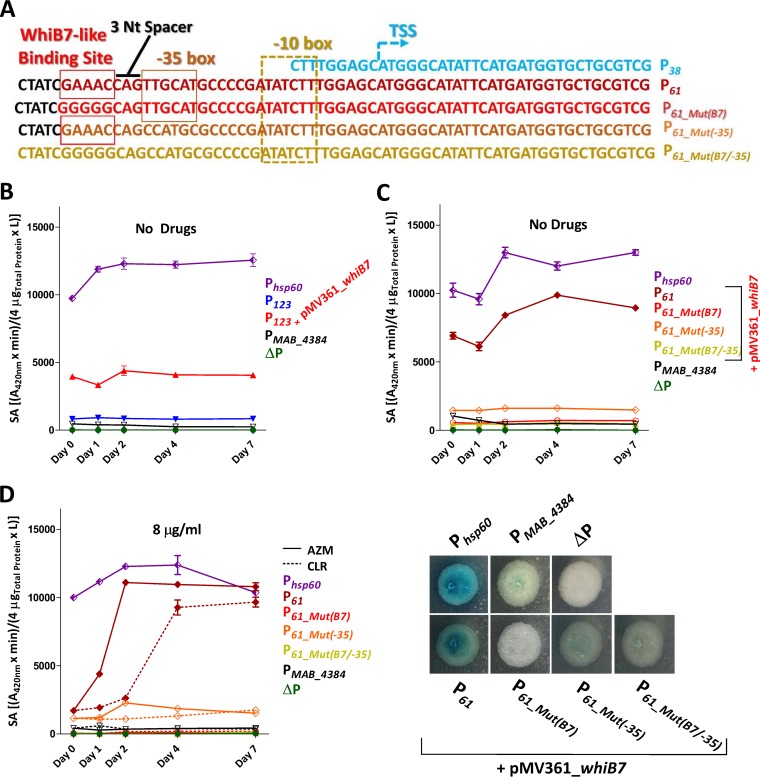
By analyzing the sequence of P61 missing in P38, we identified a putative −35 box (Fig. 4A). In addition, WhiB7 is known as a master regulator that activates transcription of many different genes involved in resistance to macrolides and amikacin (36, 37), but its direct implication in this process remains to be defined. We therefore scrutinized more thoroughly the DNA sequence of P61 and found a sequence that resembles the typical AT-rich DNA sequence recognized by WhiB7. This sequence displays partial conservation with a motif located in the region upstream of whiB7 in various mycobacterial species (38) and which is missing in P38 (Fig. 4A). This putative 5 bp WhiB7-binding site (GAAAC) is located 3 nucleotides upstream of a potential −35 box, similarly to the sites and sequences defining the whiB7 promoter (38, 39). However, the optimal distance of 16 to 19 nucleotides between the −35 and −10 hexamers is not conserved, as they are separated by only 7 nucleotides (Fig. 4A). Mycobacterial promoters have been identified with hexamer distances of between 6 and 35 bp (40). To test whether WhiB7 influences erm(41) expression by binding to this motif, the integrative plasmid pMV361_whiB7 was introduced into M. abscessus carrying pMV261_P123_lacZ and β-Gal activity was followed for 7 days in the absence of drug treatment. Fig. 4B clearly shows that lacZ was constitutively expressed at much higher levels in the strain containing pMV361_whiB7, indicating that WhiB7 is a positive regulator of erm(41) expression, in accordance with previous findings (36, 37). To further illuminate the contribution of the GAAAC motif as a potential WhiB7-binding site, this sequence was mutated in pMV261_P61_lacZ, (designated P61_Mut(B7)), as illustrated in Fig. 4A. Introduction of the mutation abrogated lacZ expression both in liquid cultures (Fig. 4C, upper) and on agar plates (Fig. 4C, lower). In addition, mutating the putative −35 box was accompanied by a sharp drop in β-Gal activity and mutations in both the whiB7 and the −35 boxes abrogated lacZ expression, suggesting that both regions are required for regulation and the transcriptional expression of erm(41) (Fig. 4C). To conclusively demonstrate the importance of the GAAAC motif for macrolide-inducible resistance, we treated the three mutated strains (pMV261_P61_Mut(B7)_lacZ, pMV261_P61_Mut(−35)_lacZ, and pMV261_P61_Mut(B7/-35)_lacZ) with either 8 μg/ml CLR or AZM for 7 days. This almost completely abolished the β-Gal activity upon treatment with either drug, compared to the pMV261_P61_lacZ control strain carrying the intact motifs (Fig. 4D).
FIG 4.
Implications of a WhiB7-binding site required for macrolide-inducible expression of erm(41). (A) DNA sequences of P38 and P61 as well as three mutated constructs of the P61 promoter region were introduced into the lacZ reporter strains. These nucleotide changes were introduced in the putative WhiB7-binding site (P61_Mut(B7)) and the erm(41) putative −35 box (P61_Mut(−35)), either individually or together (P61_Mut(B7/-35)). All constructs were used to transform M. abscessus CIP104536T rough variant. The transcriptional starting site (TSS) has been determined previously (17). (B) In the absence of macrolides, constitutive expression of whiB7 (provided by the integrative pMV361_whiB7, red) in M. abscessus pMV261_P123_lacZ correlates with a significantly stronger β-Gal activity than in a strain carrying pMV261_P123_lacZ alone, implicating WhiB7 as a positive regulator of P123. (C) M. abscessus rough variant containing the P61, P61_Mut(B7), P61_Mut(−35), or P61_Mut(B7/-35) derivatives fused to lacZ were transformed with pMV361_whiB7. All these mutations abrogated the β-Gal activity in liquid cultures (upper) and on solid medium (lower). Exponentially growing M. abscessus cultures were spotted onto 7H10 plates containing 50 μg/ml X-Gal. The plates were subsequently incubated for 3 days at 37°C and bacteria visualized for their blue/white coloration. (D) The presence of macrolides failed to induce β-Gal activity in the mutated promoter constructs as evidenced by monitoring the lacZ activity in the presence of 8 μg/ml of CLR or AZM. A pMV261 derivative containing lacZ cloned under the control of the constitutive hsp60 promoter was used as positive control (Phsp60). A promoterless construct (ΔP) and a pMV261_lacZ construct carrying the promoter of an unrelated gene (PMAB_4384) were included as negative controls. The results were obtained from two separate experiments with a total of 6 technical replicates. The specific activity of β-Gal is given in (A420nm × min)/(4 μgTotal protein × liters) as described in the Materials and Methods section ± standard deviation.
Collectively, these results indicate that the 61 bp promoter region comprises a critical WhiB7 binding motif that is necessary for macrolide-inducible erm(41) expression.
DISCUSSION
M. abscessus is a challenging pathogen that causes severe respiratory infections in patients with underlying lung disorders and these infections have become more common recently. This is of particular concern due to the natural resistance of M. abscessus to most conventional antibiotics (8). Macrolides are important components of the multidrug therapy against M. abscessus sensu lato and the American Thoracic Society/Infectious Diseases Society of America guidelines for the treatment of nontuberculous mycobacteria (NTM) recommend use of one of these agents as part of a multidrug regimen (15). It was previously shown that clarithromycin (CLR) is a stronger inducer of Erm(41) than azithromycin (AZM), with higher mRNA expression and a more rapid increase in MICs during prolonged incubation (25), leading to the conclusion that AZM may be preferred to CLR for the treatment of M. abscessus infections. It was then reported that the median MICs of AZM for M. abscessus sensu lato were markedly higher than those of CLR (41), while both macrolides induce resistance similarly (26). The present results confirm that M. abscessus subsp. bolletii and M. abscessus subsp. abscessus clinical isolates carrying a functional Erm(41) methylase undergo a more rapid increase in the MIC for AZM than for CLR, which was linked neither to the composition of the medium nor to the morphotype of the strains. Based on these results, induction of resistance cannot be a criterion for choosing between CLR and AZM for M. abscessus subsp. abscessus and M. abscessus subsp. bolletii drug treatment.
In one case only, macrolide-induced resistance was abrogated (clinical strain 1298) due to a F34L substitution in Erm(41), a mutation that was not previously reported to our knowledge, that presumably inactivates enzymatic activity. MIC data were also corroborated with transcriptional data through the development of a lacZ reporter strain exposed to various doses of either macrolide and supporting the view that expression of erm(41) is more rapidly induced with AZM than with CLR. This reporter strain provides a means to better understand the differential effect of AZM and CLR on promoter activity when bacteria are exposed to various stress conditions, such as antibiotic combinations and medium composition. Importantly, through the development of a tdTomato reporter strain, we were able to image macrolide-induced resistance in infected animals. Thanks to the optical transparency of zebrafish embryos, individual bacilli expressing tdTomato could be detected very early after exposing the embryos to either CLR or AZM (2 dpt). Although qualitative, these results indicate that CLR- and AZM-inducible resistance occurs very rapidly in the infected host, likely explaining the poor effectiveness of macrolide therapy in patients infected with M. abscessus subsp. abscessus or M. abscessus subsp. bolletii (42, 43). While the zebrafish model appears well adapted to monitor the in vivo induction of erm(41), it presents some limitations to study the reverse phenomenon following removal of the drugs. Drugs are likely to accumulate in the embryos and, even after their removal from the water, their presence in fish will sustain the expression of tdTomato. Hence, this would lead to erm(41) expression kinetics that differ from those observed in vitro. However, this tdTomato reporter strain may be particularly suitable to assay next-generation macrolide antibiotics that do not induce the erm(41)-based resistance mechanism and retain activity against M. abscessus. It is conceivable to develop a microbiological assay based on the M. abscessus strain containing pMV261_P123_tdTomato in a 96-well plate format that couples MIC determination with fluorimetric readings, enabling simultaneous assessment in vitro of the activity of a compound and the degree of macrolide-induced resistance. Such an approach could be adapted to the high-throughput screening of large libraries of macrolide scaffolds (44) in a short time frame.
Penetration of drugs into the bacterial cytoplasmic compartment induces an important transcriptional reprograming that results in changes in growth rate, metabolism, and induction of genes involved in drug resistance, such as enzymes that modify either the antibiotic or its target as well as efflux pumps. Interestingly, erm(41), also known as MAB_2297, is located in close proximity to a gene cluster participating in the intrinsic resistance to clofazimine and bedaquiline in M. abscessus, comprising a TetR transcriptional repressor (MAB_2299c) that regulates the transcription of an MmpS/MmpL efflux pump system encoded by MAB_2300/MAB_2301 (13). This gene cluster also possesses a second MmpS/MmpL pair (MAB_2302/MAB_2303) whose role in resistance to antibiotics has yet to be established. These observations point to a multidrug resistance island that participates in the intrinsic resistance to different classes of antibiotics in M. abscessus.
WhiB7 represents one of the best-studied transcriptional activators of the reprograming circuit that belongs to the WhiB family of regulators conserved in actinomycetes (39, 45). The whiB7 gene is induced upon treatment of mycobacteria with several structurally unrelated antibiotics, as well as compounds that can perturb respiration, redox balance, and iron starvation (38, 39, 46, 47). Recently, MAB_3508c, which is 75% identical to the Mycobacterium smegmatis and Mycobacterium tuberculosis whiB7 genes, has been shown to be involved in intrinsic resistance of M. abscessus to various antibiotics, as deletion of this gene results in hypersusceptibility to erythromycin, clarithromycin, streptomycin, spectinomycin, amikacin, and tetracycline (37). In addition, RNA-seq data to determine the whiB7 regulon of M. abscessus identified 128 genes, including erm(41) and eis2 (39). In another study, it was shown that WhiB7 was induced by CLR and was needed for erm(41) induction in M. abscessus (36). This is supported by our study, where constitutive overexpression of WhiB7 led to increased lacZ expression when it was cloned under the control of the erm(41) promoter. In addition, we identified a putative WhiB7-binding site which, when mutated, prevented macrolide-induced resistance, proving that the direct binding of WhiB7 to the specific AT-rich motif is necessary to induce erm(41) expression. This confirms WhiB7 as a primary determinant in transcriptional reprograming in macrolide-inducible resistance in M. abscessus.
As a nongenetically programed mechanism, macrolide-inducible resistance reflects an adaptive response to antibiotic treatment, thought to be reversible upon drug removal. We found that the β-Gal activity, mirroring erm(41) transcriptional expression, rapidly decreased during the 2 days following removal of CLR or AZM from cultures and then remained constant for the 5 following days at levels that were significantly higher than prior to drug addition. This suggests that induced macrolide resistance is only partially reversible, at least under the conditions used, since the specific activity at 7 days postwashing remained significantly higher than the basal level of lacZ expression prior to drug treatment. This phenomenon implies that, even after several rounds of division, the bacteria remain in an activated metabolic state conferring an enduring resistance phenotype. In this particular physiological stage, M. abscessus exhibits very high resistance levels to CLR and AZM and, to a lesser extent, to AMK, compared to previously unexposed bacteria at days 3, 5, 7, and 10. It can be inferred that pretreatment with CLR or AZM sensitizes the cultures, which will respond more efficiently upon reexposure to the drugs. This may confer the bacilli a protective advantage during subsequent drug therapy. This scenario appears to involve the transfer of signaling events through the subsequent generations and may possibly implicate epigenetic regulation by providing newly formed bacteria the capacity to stay in an “on” state for macrolide-inducible resistance. A simple explanation is that during treatment, the bacilli adapt their metabolism so as to continue producing erm(41) transcripts at higher basal levels. That bacteria previously exposed to either CLR or AZM express higher basal levels of erm(41) transcripts (and show higher resistance levels to macrolides and amikacin) may have important clinical consequences. Indeed, recent surveys have identified M. abscessus as a major threat in many CF centers worldwide and epidemiological studies are documenting the transmission from patient to patient of dominant circulating M. abscessus clones that have spread globally between hospitals (6). If this happens, it is possible that patients infected with previously macrolide-exposed bacilli may be more refractory to subsequent macrolide and AMK treatment, complicating the antibiotic-based therapy.
MATERIALS AND METHODS
Strains and growth conditions.
M. abscessus subsp. abscessus CIP104536T reference strain and clinical isolates are listed in Table S1 in the supplemental material. Strains were grown either in Middlebrook 7H9 broth (BD Difco) supplemented with 0.05% Tween 80 (Sigma-Aldrich) and 10% oleic acid, albumin, dextrose, catalase (OADC enrichment; BD Difco) (7H9T/OADC), Sauton’s medium, or cation-adjusted Mueller-Hinton broth (CaMHB; Sigma-Aldrich) at 30°C. On plates, colonies were selected either on Middlebrook 7H10 agar (BD Difco) supplemented with 10% OADC enrichment (7H10OADC) or on LB agar. Antibiotics were purchased from Sigma-Aldrich.
Drug susceptibility assessment.
The MICs were determined according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (48).
Construction of the lacZ and tdTomato reporter strains.
The full-length intergenic region (IR) (designated P123) as well as its truncated versions (designated P92, P61, and P38) were PCR amplified from the M. abscessus CIP104536T genomic DNA using the Phusion polymerase (Thermo Fisher Scientific) with the primers listed in Table S2. To generate the mutated IRs (P61_Mut(B7), P61_Mut(−35), and P61_Mut(B7/-35)), complementary single-stranded DNA segments containing the XbaI and BamHI sites (Table S2) were first denaturated for 15 min at 95°C and then annealed together. All DNA fragments were restricted with XbaI and BamHI and ligated to the linearized pMV261_lacZ (35), digested with the same restriction enzymes, leading to a panel of lacZ reporter constructs, as listed in Table S3. To generate pM261_P123_tdTomato, pMV261_P123_lacZ was digested with BamHI and HindIII to replace lacZ by a linearized BamHI/HindIII-restricted tdTomato gene which was amplified by PCR from pTEC27 using the primers listed in Table S2. The pMV361 containing an apramycin resistance cassette was linearized with the XmnI blunt cutter and ligated to the double-blunt whiB7 PCR product to generate the pMV361_whiB7 construct, allowing constitutive overexpression of whiB7. Plasmids were electroporated into an electrocompetent M. abscessus S strain for the tdTomato reporter construct and M. abscessus R strains for the lacZ constructs.
β-Galactosidase assay.
The quantification in liquid medium of the β-Gal activity was performed as previously reported (35). Briefly, bacterial cultures were adjusted to an optical density at 600 nm (OD600) of 0.8 in Sauton’s medium supplemented with 0.025% tyloxapol containing the appropriate antibiotics to maintain the plasmids. The β-Gal activity was determined at various time points after the initial addition of CLR or AZM (t = 0). At each time point, 1 ml cultures were centrifuged (4,000 × g for 10 min at room temperature) and the pellets were resuspended in 700 μl 1× phosphate-buffered saline (PBS). Bacterial suspensions were lysed by bead beating for 3 min at 30 Hz. Lysates were centrifuged (16,000 × g for 10 min at room temperature) and the supernatant collected for measuring the OD280 with a NanoDrop 2000c. Four micrograms of the total protein amount present in the supernatant were coincubated with 100 μl of a reaction buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol in 96-well plates at 37°C for 30 min. Thirty-five microliters of a 4 mg/ml solution of 2-nitrophenyl β-d-galactopyranoside (ONPG, Sigma-Aldrich) were added and the absorbance at 420 nm was measured at 30 to 34°C for 30 min using a multimode microplate reader POLARstar Omega (BMG Labtech). The specific β-Gal activity (SAβ-Gal) was defined as follows: (absorbance420nm × min)/(4 μg of total protein × liter of culture). To discard macrolides from bacterial cultures after 14 days of treatment, bacteria were centrifuged at 4000 × g for 10 min at room temperature and washed three times in PBS. The pellets were finally resuspended in Sauton’s medium supplemented with 0.025% tyloxapol containing the appropriate antibiotics to maintain the plasmids. β-Gal activity on solid medium was directly observed by spotting 2.5 μl of cultures at an OD600 of 0.8 onto 7H10OADC and following incubation of the plates at 37°C for 3 days.
Unmarked deletion of erm(41) in M. abscessus.
The pUX1-katG-erm(41) was generated following the same strategy as previously described (13, 14) using the primers listed in Table S2 and introduced into M. abscessus S. Selection of bacteria having undergone the first homologous recombination was done by screening for red fluorescent colonies on 7H10OADC supplemented with 200 μg/ml kanamycin. To promote the second homologous recombination event, the selected clones were subcultured overnight in 7H9OADC in the absence of kanamycin. Cultures were 10-fold-serially diluted and plated onto 7H10OADC containing 50 μg/ml isoniazid (INH) to select for isoniazid-resistant, kanamycin-sensitive, and nonfluorescent colonies. Following genomic DNA preparation of selected colonies, a PCR screening was set up using primer sets binding upstream of the left arm (LA) and inside the right arm (RA) or binding inside the LA and downstream of the RA. Amplicons were subsequently sequenced to confirm the junction between LA and RA and proper deletion of erm(41).
Zebrafish infections and visualization of macrolide inducible resistance.
All zebrafish experiments were conducted in accordance with the guidelines from the European Union for handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and approved by the Direction Sanitaire et Vétérinaire de l’Hérault et Comité d’Ethique pour l’Expérimentation Animale de la région Languedoc Roussillon under the reference number CEEA-LR-1145. Experiments were conducted using the transgenic Tg(mpeg1:GFP-CAAX) zebrafish line carrying green fluorescent macrophages (33). Ages of embryos are expressed as hours post fertilization (hpf). M. abscessus S left untransformed (as a negative control) or carrying either pTEC27 (with constitutive expression of tdTomato) or pMV261-P123-tdTomato (with macrolide-inducible expression of tdTomato) were injected into zebrafish embryos and monitored for fluorescence expression, according to protocols reported previously (49). Briefly, infections were performed by intravenous microinjection of 900 to 1,000 CFU in dechorionated and anesthetized 30 hpf embryos. At 96 hours postinfection (hpi), infected embryos were exposed to either 32 μg/ml CLR or AZM in osmotic water and this treatment was renewed on a daily basis. Red fluorescence was assessed by immobilizing anesthetized embryos in 1% low-melting-point agarose and performing life imaging by confocal microscopy (LSM 880 Airyscan) at day 0, day 2, and day 6 post treatment (dpt). 2D reconstructions of image stacks were done with the ZEN 2 software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Fondation pour la Recherche Médicale (FRM) (grant number DEQ20150331719 to L.K.; grant number ECO20160736031 to M.R.) and the InfectioPôle Sud for funding the Ph.D. fellowship of A.V.G.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Cowman S, van Ingen J, Griffith DE, Loebinger MR. 2019. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J 54:1900250. doi: 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 2.Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, Welinder-Olsson C. 2007. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol 45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esther CR, Esserman DA, Gilligan P, Kerr A, Noone PG. 2010. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J Cyst Fibros 9:117–123. doi: 10.1016/j.jcf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catherinot E, Roux A-L, Macheras E, Hubert D, Matmar M, Dannhoffer L, Chinet T, Morand P, Poyart C, Heym B, Rottman M, Gaillard J-L, Herrmann J-L. 2009. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J Clin Microbiol 47:271–274. doi: 10.1128/JCM.01478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler IC, Chapman SJ, Clayton A, Cullen M, Daniels T, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, Katzenstein TL, Lillebaek T, MacGregor G, Mayell S, Millar M, Modha D, Nash EF, O'Brien C, O'Brien D, Ohri C, Pao CS, Peckham D, Perrin F, Perry A, Pressler T, Prtak L, Qvist T, Robb A, Rodgers H, Schaffer K, Shafi N, van Ingen J, Walshaw M, Watson D, West N, Whitehouse J, Haworth CS, Harris SR, Ordway D, Parkhill J, Floto RA. 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adekambi T, Sassi M, van Ingen J, Drancourt M. 2017. Reinstating Mycobacterium massiliense and Mycobacterium bolletii as species of the Mycobacterium abscessus complex. Int J Syst Evol Microbiol 67:2726–2730. doi: 10.1099/ijsem.0.002011. [DOI] [PubMed] [Google Scholar]
- 8.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 9.Rominski A, Roditscheff A, Selchow P, Böttger EC, Sander P. 2017. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother 72:376–384. doi: 10.1093/jac/dkw466. [DOI] [PubMed] [Google Scholar]
- 10.Rominski A, Selchow P, Becker K, Brülle JK, Dal Molin M, Sander P. 2017. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J Antimicrob Chemother 72:2191–2200. doi: 10.1093/jac/dkx125. [DOI] [PubMed] [Google Scholar]
- 11.Rudra P, Hurst-Hess K, Lappierre P, Ghosh P. 2018. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob Agents Chemother 62:e00119-18. doi: 10.1128/AAC.00119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubée V, Bernut A, Cortes M, Lesne T, Dorchene D, Lefebvre A-L, Hugonnet J-E, Gutmann L, Mainardi J-L, Herrmann J-L, Gaillard J-L, Kremer L, Arthur M. 2015. β-Lactamase inhibition by avibactam in Mycobacterium abscessus. J Antimicrob Chemother 70:1051–1058. doi: 10.1093/jac/dku510. [DOI] [PubMed] [Google Scholar]
- 13.Richard M, Gutiérrez AV, Viljoen A, Rodriguez-Rincon D, Roquet-Baneres F, Blaise M, Everall I, Parkhill J, Floto RA, Kremer L. 2019. Mutations in the MAB_2299c TetR regulator confer cross-resistance to clofazimine and bedaquiline in Mycobacterium abscessus. Antimicrob Agents Chemother 63:e01316-18. doi: 10.1128/AAC.01316-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez AV, Richard M, Roquet-Banères F, Viljoen A, Kremer L. 2019. The TetR-family transcription factor MAB_2299c regulates the expression of two distinct MmpS-MmpL efflux pumps involved in cross-resistance to clofazimine and bedaquiline in Mycobacterium abscessus. Antimicrob Agents Chemother 63:e01000-19. doi: 10.1128/AAC.01000-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 16.Wallace RJ, Meier A, Brown BA, Zhang Y, Sander P, Onyi GO, Böttger EC. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother 40:1676–1681. doi: 10.1128/AAC.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash KA, Brown-Elliott BA, Wallace RJ. 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother 53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastian S, Veziris N, Roux A-L, Brossier F, Gaillard J-L, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurer FP, Rüegger V, Ritter C, Bloemberg GV, Böttger EC. 2012. Acquisition of clarithromycin resistance mutations in the 23S rRNA gene of Mycobacterium abscessus in the presence of inducible erm(41). J Antimicrob Chemother 67:2606–2611. doi: 10.1093/jac/dks279. [DOI] [PubMed] [Google Scholar]
- 20.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash K, Parodi N, Strong A, Gee M, Smith T, Wallace RJ. 2015. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol 53:1211–1215. doi: 10.1128/JCM.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mougari F, Amarsy R, Veziris N, Bastian S, Brossier F, Berçot B, Raskine L, Cambau E. 2016. Standardized interpretation of antibiotic susceptibility testing and resistance genotyping for Mycobacterium abscessus with regard to subspecies and erm41 sequevar. J Antimicrob Chemother 71:2208–2212. doi: 10.1093/jac/dkw130. [DOI] [PubMed] [Google Scholar]
- 22.Kim H-Y, Kim BJ, Kook Y, Yun Y-J, Shin JH, Kim B-J, Kook Y-H. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol Immunol 54:347–353. doi: 10.1111/j.1348-0421.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 23.Roux A-L, Catherinot E, Soismier N, Heym B, Bellis G, Lemonnier L, Chiron R, Fauroux B, Le Bourgeois M, Munck A, Pin I, Sermet I, Gutierrez C, Véziris N, Jarlier V, Cambau E, Herrmann J-L, Guillemot D, Gaillard J-L, OMA group. 2015. Comparing Mycobacterium massiliense and Mycobacterium abscessus lung infections in cystic fibrosis patients. J Cyst Fibros 14:63–69. doi: 10.1016/j.jcf.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Brown BA, Wallace RJ, Onyi GO, De Rosas V, Wallace RJ. 1992. Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae-like organisms. Antimicrob Agents Chemother 36:180–184. doi: 10.1128/aac.36.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi G-E, Shin SJ, Won C-J, Min K-N, Oh T, Hahn M-Y, Lee K, Lee SH, Daley CL, Kim S, Jeong B-H, Jeon K, Koh W-J. 2012. Macrolide treatment for Mycobacterium abscessus and Mycobacterium massiliense infection and inducible resistance. Am J Respir Crit Care Med 186:917–925. doi: 10.1164/rccm.201111-2005OC. [DOI] [PubMed] [Google Scholar]
- 26.Schildkraut JA, Pennings LJ, Ruth MM, de Brouwer AP, Wertheim HF, Hoefsloot W, de Jong A, van Ingen J. 2019. The differential effect of clarithromycin and azithromycin on induction of macrolide resistance in Mycobacterium abscessus. Future Microbiol 14:749–755. doi: 10.2217/fmb-2018-0310. [DOI] [PubMed] [Google Scholar]
- 27.Peters DH, Friedel HA, McTavish D. 1992. Azithromycin. A review of its antimicrobial activity, pharmacokinetic properties and clinical efficacy. Drugs 44:750–799. doi: 10.2165/00003495-199244050-00007. [DOI] [PubMed] [Google Scholar]
- 28.Viljoen A, Gutiérrez AV, Dupont C, Ghigo E, Kremer L. 2018. A simple and rapid gene disruption strategy in Mycobacterium abscessus: on the design and application of glycopeptidolipid mutants. Front Cell Infect Microbiol 8:69. doi: 10.3389/fcimb.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupont C, Viljoen A, Thomas S, Roquet-Banères F, Herrmann J-L, Pethe K, Kremer L. 2017. Bedaquiline inhibits the ATP synthase in Mycobacterium abscessus and is effective in infected zebrafish. Antimicrob Agents Chemother 61:e01225-17. doi: 10.1128/AAC.01225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupont C, Viljoen A, Dubar F, Blaise M, Bernut A, Pawlik A, Bouchier C, Brosch R, Guérardel Y, Lelièvre J, Ballell L, Herrmann J-L, Biot C, Kremer L. 2016. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol 101:515–529. doi: 10.1111/mmi.13406. [DOI] [PubMed] [Google Scholar]
- 31.Bernut A, Le Moigne V, Lesne T, Lutfalla G, Herrmann J-L, Kremer L. 2014. In vivo assessment of drug efficacy against Mycobacterium abscessus using the embryonic zebrafish test system. Antimicrob Agents Chemother 58:4054–4063. doi: 10.1128/AAC.00142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernut A, Herrmann J-L, Kissa K, Dubremetz J-F, Gaillard J-L, Lutfalla G, Kremer L. 2014. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A 111:E943–952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keatinge M, Bui H, Menke A, Chen Y-C, Sokol AM, Bai Q, Ellett F, Da Costa M, Burke D, Gegg M, Trollope L, Payne T, McTighe A, Mortiboys H, de Jager S, Nuthall H, Kuo M-S, Fleming A, Schapira AHV, Renshaw SA, Highley JR, Chacinska A, Panula P, Burton EA, O'Neill MJ, Bandmann O. 2015. Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum Mol Genet 24:6640–6652. doi: 10.1093/hmg/ddv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernut A, Nguyen-Chi M, Halloum I, Herrmann J-L, Lutfalla G, Kremer L. 2016. Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PLoS Pathog 12:e1005986. doi: 10.1371/journal.ppat.1005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard M, Gutiérrez AV, Viljoen AJ, Ghigo E, Blaise M, Kremer L. 2018. Mechanistic and structural insights into the unique TetR-dependent regulation of a drug efflux pump in Mycobacterium abscessus. Front Microbiol 9:649. doi: 10.3389/fmicb.2018.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pryjma M, Burian J, Kuchinski K, Thompson CJ. 2017. Antagonism between front-line antibiotics clarithromycin and amikacin in the treatment of Mycobacterium abscessus infections is mediated by the whiB7 gene. Antimicrob Agents Chemother 61:e01353-17. doi: 10.1128/AAC.01353-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurst-Hess K, Rudra P, Ghosh P. 2017. Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob Agents Chemother 61:e01347-17. doi: 10.1128/AAC.01347-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burian J, Ramón-García S, Sweet G, Gómez-Velasco A, Av-Gay Y, Thompson CJ. 2012. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J Biol Chem 287:299–310. doi: 10.1074/jbc.M111.302588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burian J, Ramón-García S, Howes CG, Thompson CJ. 2012. WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev Anti Infect Ther 10:1037–1047. doi: 10.1586/eri.12.90. [DOI] [PubMed] [Google Scholar]
- 40.Newton-Foot M, Gey van Pittius NC. 2013. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Maurer FP, Castelberg C, Quiblier C, Böttger EC, Somoskövi A. 2014. Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J Antimicrob Chemother 69:1559–1563. doi: 10.1093/jac/dku007. [DOI] [PubMed] [Google Scholar]
- 42.Rollet-Cohen V, Roux A-L, Le Bourgeois M, Sapriel G, El Bahri M, Jais J-P, Heym B, Mougari F, Raskine L, Véziris N, Gaillard J-L, Sermet-Gaudelus I. 2019. Mycobacterium bolletii lung disease in cystic fibrosis. Chest 156:247–254. doi: 10.1016/j.chest.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Koh W-J, Jeon K, Lee NY, Kim B-J, Kook Y-H, Lee S-H, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med 183:405–410. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 44.Pavlović D, Fajdetić A, Mutak S. 2010. Novel hybrids of 15-membered 8a- and 9a-azahomoerythromycin A ketolides and quinolones as potent antibacterials. Bioorg Med Chem 18:8566–8582. doi: 10.1016/j.bmc.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 45.Soliveri JA, Gomez J, Bishai WR, Chater KF. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology (Reading, Engl) 146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 46.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob Agents Chemother 50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woods GL, Brown-Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, Pfyffer GE, Ridderhof JC, Siddiqi SH, Wallace RJ. 2011. Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes: approved standard 2nd ed M24-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 49.Bernut A, Dupont C, Sahuquet A, Herrmann J-L, Lutfalla G, Kremer L. 2015. Deciphering and imaging pathogenesis and cording of Mycobacterium abscessus in zebrafish embryos. J Vis Exp 103:e53130. doi: 10.3791/53130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.