The objective was to determine the in vitro antimicrobial susceptibility of Pseudomonas aeruginosa isolates cultured from cystic fibrosis (CF) patients and explore associations between strain sequence type and susceptibility. Fourteen antibiotics and antibiotic combinations, including the novel antibacterial peptide murepavadin, were tested for activity against 414 Pseudomonas aeruginosa isolates cultured from respiratory samples of CF patients.
KEYWORDS: Pseudomonas aeruginosa, cystic fibrosis, murepavadin, sequence type, susceptibility testing
ABSTRACT
The objective was to determine the in vitro antimicrobial susceptibility of Pseudomonas aeruginosa isolates cultured from cystic fibrosis (CF) patients and explore associations between strain sequence type and susceptibility. Fourteen antibiotics and antibiotic combinations, including the novel antibacterial peptide murepavadin, were tested for activity against 414 Pseudomonas aeruginosa isolates cultured from respiratory samples of CF patients. The complete genomes of the isolates were sequenced, and minimum spanning trees were constructed based on the sequence types (STs). Percentages of resistance according to CLSI 2019 breakpoints were as follows: cefepime, 14%; ceftazidime, 11%; ceftazidime-avibactam, 7%; ceftolozane-tazobactam, 3%; piperacillin-tazobactam, 12%; meropenem, 18%; imipenem, 32%; aztreonam, 23%; ciprofloxacin, 30%; gentamicin, 30%; tobramycin, 12%; amikacin, 18%; and colistin, 4%. Murepavadin MIC50 and MIC90 were 0.12 mg/liter and 2 mg/liter, respectively. There were no apparent clonal clusters associated with resistance, but higher MICs did appear to occur more often in STs with multiple isolates than in single ST isolates. In general, the CF isolates showed a wide genetic distribution. P. aeruginosa CF isolates exhibited the lowest resistance rates against ceftolozane-tazobactam, ceftazidime-avibactam, and colistin. Murepavadin demonstrated the highest activity on a per-weight basis and may therefore become a valuable addition to the currently available antibiotics for treatment of respiratory infection in people with CF.
INTRODUCTION
Cystic fibrosis (CF) is a life-limiting inherited disease with a frequency of approximately 1 in 2,500 live births. Chronic bacterial pulmonary infection leads to irreversible damage of the lung structure and to a decline in lung function, which is the main cause of mortality and morbidity (1). Pseudomonas aeruginosa is the most frequently isolated pathogen, chronically infecting up to 80% of adult CF patients (1). Pulmonary exacerbations occur frequently in people with CF chronically infected with P. aeruginosa and are associated with reduced survival and quality of life. Inhaled suppressive antibiotic therapy has been fundamental in improving quality of life, preserving lung function, and reducing exacerbation frequency in CF patients chronically infected with P. aeruginosa (1, 2). As the median predicted survival of CF birth cohorts now exceeds 40 years and as the number of surviving adults is increasing, treatment with inhaled antibiotics may be required for decades (3, 4). Unfortunately, CF pathogens are progressively more resistant to available antibiotics, and up to 45% of CF patients are colonized with multidrug-resistant (MDR) isolates (5). Novel antibiotic agents for intravenous treatment of exacerbation and for inhalation therapy are therefore urgently needed.
Murepavadin, a 14-amino-acid cyclic peptide antibiotic, represents the first member of a novel class of antibacterials targeting the outer membrane protein. The drug binds to the lipopolysaccharide transport protein D in the outer membrane of P. aeruginosa, causing lipopolysaccharide alterations and ultimately killing the bacterium (6). The drug exhibits specific bactericidal activity against P. aeruginosa, and it has activity against isolates resistant to all, or virtually all, other commercially available antibiotics (6). Murepavadin has little to no effect on other bacterial species, which could possibly lead to a lower risk of eliciting cross-resistance with other antibiotics. Its development as an intravenous formulation was recently halted due to unexpected kidney injury findings. However, the development of a formulation for inhaled therapy is ongoing and may prove a valuable addition to the therapeutic options for treating P. aeruginosa lung infection in people with CF or other patients with chronic bronchial colonization with this organism, such as those with bronchiectasis.
In this study, we determined the activity of murepavadin and 13 licensed antipseudomonal antibiotics used in CF care against clinical CF P. aeruginosa isolates. To ensure that a sufficiently diverse and representative population was tested, the isolates were typed using a multilocus sequence typing (MLST) approach with whole-genome sequencing (WGS) data.
RESULTS
Susceptibility testing.
The MIC50, MIC90, and susceptibility percentages for the antibiotics tested are listed in Table 1. Of all drugs tested, murepavadin expressed the highest activity on a per-weight basis, with a MIC50 of 0.12 mg/liter and a MIC90 of 2 mg/liter (see Fig. S1A and B in the supplemental material). Eleven strains (2.7%) had MICs exceeding 16 mg/liter, but this was not specifically associated with elevated MICs for the other antipseudomonal antibiotics. The second lowest MIC50 value was that of meropenem (0.25 mg/liter); however, the MIC90 of meropenem was higher, at 16 mg/liter. Applying both EUCAST and CLSI breakpoints, 76% of the strains were susceptible to meropenem. Ceftolozane-tazobactam and colistin had MIC90 values identical to those of murepavadin (2 mg/liter) but higher MIC50 values.
TABLE 1.
MICs and susceptibility of 414 Pseudomonas aeruginosa isolates from cystic fibrosis patients
| Drug | Value for the isolate group or type (mg/liter) |
Susceptibility (%) by standard (n = 414)a
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All strains (n = 414) |
Pediatric (n = 111) |
Adult (n = 288) |
Mucoid (n = 112) |
Small-colony variant (n = 63) |
EUCAST |
CLSI |
||||||||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | S | I | R | S | I | R | |
| Murepavadin | 0.12 | 2 | 0.12 | 1 | 0.25 | 4 | 0.12 | 2 | 0.25 | 8 | ND | ND | ND | ND | ND | ND |
| Cefepime | 4 | 32 | 4 | 16 | 8 | 64 | 8 | 32 | 8 | >128 | 75 | 25 | 75 | 11 | 14 | |
| Ceftazidime | 2 | 64 | 2 | 8 | 2 | 128 | 2 | 8 | 2 | 128 | 80 | 20 | 80 | 4 | 11 | |
| Ceftazidime-avibactam | 2 | 8 | 2 | 4 | 2 | 8 | 1 | 8 | 2 | 8 | 93 | 7 | 93 | 7 | ||
| Ceftolozane-tazobactam | 1 | 2 | 0.5 | 2 | 1 | 4 | 1 | 2 | 1 | 4 | 95 | 5 | 95 | 1 | 3 | |
| Piperacillin-tazobactam | 4 | 128 | 4 | 32 | 4 | >256 | 2 | 128 | 2 | >256 | 81 | 19 | 81 | 7 | 12 | |
| Meropenem | 0.25 | 16 | 0.25 | 4 | 0.5 | 16 | 0.5 | 16 | 0.5 | 16 | 76 | 12 | 12 | 76 | 6 | 18 |
| Imipenem | 2 | 32 | 2 | 16 | 2 | 32 | 2 | 32 | 1 | 32 | 68 | 32 | 59 | 8 | 32 | |
| Aztreonam | 8 | 128 | 8 | 32 | 4 | 256 | 2 | 64 | 2 | >256 | 77 | 23 | 62 | 14 | 23 | |
| Ciprofloxacin | 1 | 8 | 0.5 | 4 | 2 | 8 | 1 | 8 | 2 | 16 | 39 | 61 | 53 | 17 | 30 | |
| Gentamicin | 4 | 64 | 4 | 32 | 8 | 64 | 4 | 16 | 8 | >128 | 53 | 47 | 53 | 17 | 30 | |
| Tobramycin | 1 | 16 | 0.5 | 8 | 2 | 16 | 1 | 4 | 2 | 64 | 84 | 16 | 84 | 5 | 12 | |
| Amikacin | 16 | 64 | 8 | 64 | 16 | 128 | 16 | 64 | 16 | >128 | 47 | 21 | 32 | 68 | 14 | 18 |
| Colistin | 1 | 2 | 1 | 2 | 1 | 2 | 0.5 | 1 | 1 | 4 | 93 | 7 | 93 | 3 | 4 | |
S, susceptible; I, intermediately susceptible; R, resistant; ND, no breakpoint defined.
Ceftolozane-tazobactam was the drug with highest susceptibility rate: 95% of the isolates had MICs of ≤4 mg/liter, the breakpoint for susceptibility of both EUCAST and CLSI guidelines. Colistin and ceftazidime-avibactam demonstrated susceptibility greater than 90%. Susceptibility to ceftazidime-avibactam was higher than that to ceftazidime alone (Table 1 and Fig. S2), indicating a role for β-lactamases in the resistance of P. aeruginosa isolates to this cephalosporin. High resistance rates were found, in particular, for ciprofloxacin, gentamicin, and aztreonam. Of the aminoglycosides tested, tobramycin was the most active, with MIC50 and MIC90 values 4-fold lower than those of gentamicin and amikacin (Table 1 and Fig. S3).
MIC50 and MIC90 values were, in general, equal or higher for isolates from the 288 adult patients than for isolates from the 111 pediatric patients; the difference was significant for 10 of the 14 antibiotics (P < 0.05, by a Mann Whitney U test). Sixty-three isolates were typed as small-colony variants, and 112 isolates were mucoid. The MICs of the mucoid isolates were generally comparable to or lower than those of the total population (P < 0.05 for ceftazidime-avibactam, piperacillin-tazobactam, aztreonam, and colistin, by a Mann Whitney U test); small-colony variants displayed higher MIC values, in particular higher MIC90 values (P < 0.05 for murepavadin and the three aminoglycosides). No differences in MIC50 and MIC90 values were observed between the different countries.
Whole-genome sequencing.
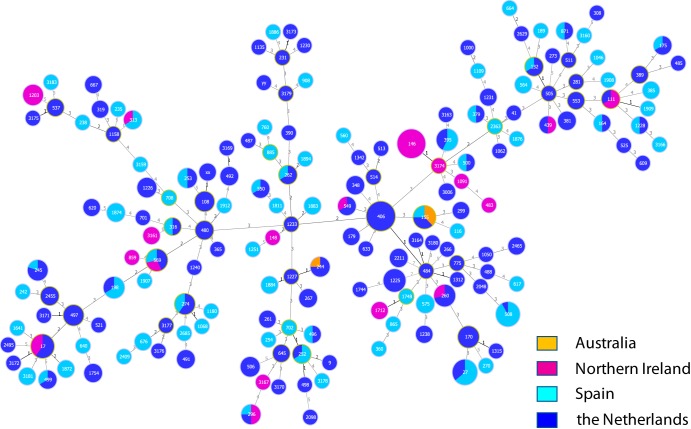
A whole-genome sequence was obtained for 412 of the 414 tested isolates, which fell into 165 different sequence types (STs). Figure 1 shows the relationship between the numbers of isolates with different STs, the genetic relation between the STs, and the countries where the strains were isolated. Isolates from The Netherlands, Spain, and Northern Ireland largely overlapped although some small clusters could be observed that differed between the three countries, such as clusters of ST406 isolates in The Netherlands (the Dutch epidemic strain) (7), of ST508 isolates in Spain, and of ST146 isolates in Northern Ireland (Liverpool epidemic strain) (7). A limited clustering of MICs of ≥MIC90 could be observed in certain STs with a higher number of isolates; small clusters of up to three isolates and single isolates appeared to have a higher likelihood of a MIC of ≤MIC50 (P < 0.05, by Mann Whitney U test for 8 of the 14 antibiotics) (see also Fig. S4A to N in the supplemental material).
FIG 1.
Minimum spanning tree of 412 Pseudomonas aeruginosa respiratory isolates recovered from cystic fibrosis patients. The plot indicates the distribution of isolates over geographic regions, as indicated on the figure. Sequence types (STs) were based on seven housekeeping genes as determined by whole-genome sequencing. The numbers in the circles indicate the STs assigned by PubMLST (www.pubmlst.org). XX and YY are isolates with partially deleted housekeeping genes which could not be assigned an ST. The circle size indicates the number of isolates with the same ST. The numbers on the lines between the circles indicate the number of allele differences between two STs.
DISCUSSION
In this study, the novel antimicrobial cyclic peptide, murepavadin, was highly active against P. aeruginosa recovered from CF patients, displaying a higher activity on a per-weight basis than comparator antibiotics. Ceftolozane-tazobactam displayed the lowest percent resistance of the tested antibiotics; this may be due in part to the higher intrinsic activity of this combination in P. aeruginosa and in part to the lack of exposure of CF patients to this relatively novel drug combination (8, 9).
Susceptibility of the CF isolates to murepavadin was lower than previously reported for non-CF clinical P. aeruginosa isolates, including MDR P. aeruginosa; the MIC90 value (2 mg/liter) was higher than the previously reported 0.12 to 0.25 mg/liter (6, 10) and even higher in isolates recovered from adult patients. The driving mechanism behind the increased MIC values cannot be explained by exposure of the bacteria to the drug, nor were there indications for cross-resistance with other antibiotics. Further analysis of the genetic data of these CF isolates may yield an explanation for this phenomenon.
Pharmacokinetic/pharmacodynamic (PK/PD) studies in neutropenic mouse models found that the efficacy of murepavadin correlated best with the area under the concentration-time curve for the unbound fraction of the drug (fAUC)/MIC. The mean fAUC/MIC required for stasis was 27.78 mg/liter, and the mean fAUC/MIC required for a 1-log reduction was 39.85 mg/liter. The corresponding values for the epithelial lining fluid (ELF) were 34.5 and 49.4, respectively (11). For non-CF clinical isolates, these PK/PD targets were readily attained with the applied dosage regimens used in previous clinical studies (12). The development of murepavadin as an intravenous formulation was halted due to adverse findings in clinical trials. However, the development of murepavadin as an inhalant is continuing and is supported by recent in vivo studies which investigated the pharmacokinetics, tolerability, and efficacy of murepavadin administered by intratracheal application in P. aeruginosa lung infection models (13).
CF patients generally have chronic polymicrobial respiratory infection with an array of bacterial species, which may be symbiotic or which may compete with one another. It has been reported that 40 to 51% of CF patients colonized with P. aeruginosa harbor multiple phenotypes of the microorganism but that these mostly constitute different growth forms of the same strain (14). In the same report, only 8 to 11% of colonized patients carried more than a single P. aeruginosa ST. A number of epidemic strains have been described that have been associated with lower susceptibility to antibiotics and, in some cases, with increased virulence (7). However, up to half of CF patients are colonized with unique STs, and this fraction may increase even more as segregation policies in CF centers continue to have an effect.
In order to provide a relevant insight into susceptibility of P. aeruginosa in CF patients, a diverse collection of isolates from different countries, some with previously determined STs, was selected. The wide variety of isolates and the fact that the distribution appears to be similar in The Netherlands, Spain, and Northern Ireland suggest that this sample is representative for P. aeruginosa isolates from CF patients, at least for those in Western Europe.
Analysis of the distribution of the MICs for the different antipseudomonal antibiotics did not yield clear patterns of clonal clusters associated with resistance. This finding and the fact that isolates from pediatric patients were more susceptible than those from adults are consistent with the idea that CF patients mostly acquire unique P. aeruginosa isolates from the environment, isolates which during chronic infection gradually develop resistance. Furthermore, it appeared that STs with only a single isolate more frequently had MICs lower than or equal to the MIC50, while STs with multiple isolates may represent epidemic CF isolates that express more antibiotic resistance.
The host and microbe interspecies interactions are an area of much interest, where the response to antibiotics is likely due to factors additional to bacterial killing (1). For example, studies in which sputa from CF patients were analyzed by deep sequencing of 16S rRNA genes have failed to establish differences between the microbiota of CF patients during chronic infection, during acute exacerbations, and after systemic antibiotic treatment (15). The correlation between susceptibility test results and outcome of antibiotic treatment is also not clearly established for pulmonary infection in CF (16). Nevertheless, the use of antipseudomonal antibiotics for early eradication of P. aeruginosa, for treatment of exacerbations, and for chronic suppressive inhalation therapy have significantly contributed to improving the quality of life and life expectancy of CF patients (17). Furthermore, in practice, the choice of antibiotic regimens is guided by susceptibility testing results, particularly when patients fail to respond to first-line therapy. Therefore, the results from susceptibility tests in this study suggest that drugs such as ceftolozane-tazobactam and ceftazidime-avibactam may be useful for the treatment of CF pulmonary infections. In addition, murepavadin may be added to this list if an inhaled formulation is developed.
MATERIALS AND METHODS
Isolates.
Pulmonary P. aeruginosa isolates from CF patients were selected as follows. Isolates from The Netherlands (n = 238) consisted of 130 isolates with unique sequence types (STs) that were collected in 2007 (14) and 2013, 20 isolates from 10 patients that were collected with intervals of 5 to 10 years between 2007 and 2014, and 88 prospectively collected isolates from 2015 to 2016, which could include multiple isolates with different morphotypes from one specimen. Isolates from Spain (n = 114) consisted of 100 isolates collected in a multicenter national CF study in 2013 and 2014 (18) and 14 isolates from three patients with changing morphotypes over a 4-year period. Isolates from Northern Ireland (n = 58) consisted of 38 isolates collected in previous multicenter CF studies (19, 20), 19 clinical isolates collected in 2015 to 2016, and one clinical strain commonly used in animal models of infection (Q502). Isolates from Australia (n = 4) consisted of clinical isolates from 2015 to 2016. Mucoid and small-colony morphotypes were recorded. Age group (pediatric versus adult) was available for 399/414 isolates. Identification of all isolates was confirmed by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) (Bruker Daltonics, Germany) and by whole-genome sequencing.
Susceptibility testing.
MICs were determined in cation-adjusted Mueller-Hinton broth by standard ISO broth microdilution with two frozen panels, one of which was supplied by Trek Diagnostic Systems (Westlake, Ohio) with ceftazidime, aztreonam, meropenem, imipenem, ciprofloxacin, tobramycin, and colistin and one of which was made in-house by Polyphor AG (Basel, Switzerland) with ceftazidime-avibactam, cefepime, piperacillin-tazobactam, gentamicin, amikacin, and murepavadin. The decision to produce the second set of frozen panels was prompted by the prolonged time required for commercial production. The antibiotics and their ranges tested were as follows: cefepime, 0.12 to 128 mg/liter; ceftazidime, 0.25 to 256 mg/liter; ceftazidime-avibactam, 0.25 to 256 mg/liter; piperacillin-tazobactam, 0.25 to 256 mg/liter; meropenem, 0.06 to 64 mg/liter; imipenem, 0.125 to 128 mg/liter; aztreonam, 0.25 to 256 mg/liter; gentamicin, 0.125 to 128 mg/liter; tobramycin, 0.125 to 128 mg/liter; amikacin, 0.125 to 128 mg/liter; ciprofloxacin, 0.03 to 32 mg/liter; colistin, 0.25 to 16 mg/liter; and murepavadin, 0.016 to 16 mg/liter. Tazobactam and avibactam were tested at fixed doses of 4 mg/liter. Susceptibility to ceftolozane-tazobactam (range, 0.016 to 256 mg/liter) was determined by gradient diffusion testing using a Liofilchem MIC test strip (Liofilchem, Abruzzi, Italy) as per the manufacturer’s instructions. (At the time of the study this antibiotic combination was not available as a pure compound for use in the frozen plates). Escherichia coli ATCC strain 25922, P. aeruginosa ATCC strain 27853, and MDR P. aeruginosa strain PA3140 (with a murepavadin MIC of 0.25 mg/liter) were included as run controls. MIC50 and MIC90 values were determined, and percentages of susceptible, intermediate, and resistant isolates were calculated using CLSI and EUCAST breakpoints (21, 22).
Whole-genome sequencing (WGS) and analysis.
Bacterial DNA was purified using a Qiacube with a DNeasy Blood and Tissue kit with the enzymatic lysis protocol (Qiagen, Carlsbad, CA) and used to prepare a library for sequencing with the MiSeq or Nextseq (Illumina, San Diego, CA) platforms, using a NexteraXT library prep kit (Illumina). Contigs were assembled with the SPAdes genome assembler, version 3.6.2. The assembled contigs were used to determine the STs with the multilocus sequence typing (MLST) module (version 2.0) from the Center for Genomic Epidemiology (Technical University of Denmark, Copenhagen, Denmark; accessed 28 October 2018) (23) and PubMLST (accessed 11 January 2019 [https://pubmlst.org/]) (24).
Supplementary Material
ACKNOWLEDGMENTS
Tim Kidd kindly provided the Australian isolates for this study. Judith Vlooswijk and Sjoukje van Gorkum performed susceptibility testing, and Barry Benaissa-Trouw performed WGS.
This research project received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement number 115721, resources of which are composed of financial contributions from the European Union Seventh Framework Program (FP7/2007-2013) and in-kind contributions from the European Federation of Pharmaceutical Industries.
Francesca Bernardini and Glenn Dale were employed by Polyphor AG. No other authors have conflicts of interest to report.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Elborn JS. 2016. Cystic fibrosis. Lancet 388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 2.Ryan G, Singh M, Dwan K. 2011. Inhaled antibiotics for long-term therapy in cystic fibrosis. Cochrane Database Syst Rev 3:CD001021. doi: 10.1002/14651858.CD001021.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Burgel PR, Bellis G, Elborn JS. 2017. Modelling future trends in cystic fibrosis demography using the French Cystic Fibrosis Registry: update and sensitivity analysis. Eur Respir J 50:1700763. doi: 10.1183/13993003.00763-2017. [DOI] [PubMed] [Google Scholar]
- 4.Dodge JA, Lewis PA, Stanton M, Wilsher J. 2007. Cystic fibrosis mortality and survival in the UK: 1947–2003. Eur Respir J 29:522–526. doi: 10.1183/09031936.00099506. [DOI] [PubMed] [Google Scholar]
- 5.Sherrard LJ, Tunney MM, Elborn JS. 2014. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384:703–713. doi: 10.1016/S0140-6736(14)61137-5. [DOI] [PubMed] [Google Scholar]
- 6.Sader HS, Dale GE, Rhomberg PR, Flamm RK. 2018. Antimicrobial activity of murepavadin tested against clinical isolates of Pseudomonas aeruginosa from the United States, Europe, and China. Antimicrob Agents Chemother 62:e00311-18. doi: 10.1128/AAC.00311-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkins MD, Somayaji R, Waters VJ. 2018. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev 31:e00019-18. doi: 10.1128/CMR.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho JC, Fiorenza MA, Estrada SJ. 2015. Ceftolozane/tazobactam: a novel cephalosporin/β-lactamase inhibitor combination. Pharmacotherapy 35:701–715. doi: 10.1002/phar.1609. [DOI] [PubMed] [Google Scholar]
- 9.Vickery SB, McClain D, Wargo KA. 2016. Successful use of ceftolozane-tazobactam to treat a pulmonary exacerbation of cystic fibrosis caused by multidrug-resistant Pseudomonas aeruginosa. Pharmacotherapy 36:e154–e159. doi: 10.1002/phar.1825. [DOI] [PubMed] [Google Scholar]
- 10.Sader HS, Flamm RK, Dale GE, Rhomberg PR, Castanheira M. 2018. Murepavadin activity tested against contemporary (2016–17) clinical isolates of XDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:2400–2404. doi: 10.1093/jac/dky227. [DOI] [PubMed] [Google Scholar]
- 11.Melchers MJ, Teague J, Warn P, Hansen J, Bernardini F, Wach A, Obrecht D, Dale GE, Mouton JW. 2019. Pharmacokinetics and pharmacodynamics of murepavadin in neutropenic mouse models. Antimicrob Agents Chemother 63:e01699-18. doi: 10.1128/AAC.01699-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wach A, Dembowsky K, Dale GE. 2018. Pharmacokinetics and safety of intravenous murepavadin infusion in healthy adult subjects administered single and multiple ascending doses. Antimicrob Agents Chemother 62:e02355-17. doi: 10.1128/AAC.02355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardini F, Dale GE, Wach A, Obrecht D. 2019. Pharmacokinetics and pharmacodynamics of murepavadin (POL7080) in neutropenic lung infection models when evaluated by aerosol administration, abstr WS01-4. J Cyst Fibros 18(Suppl 1):S2. doi: 10.1016/S1569-1993(19)30120-1.31679724 [DOI] [Google Scholar]
- 14.van Mansfeld R, de Vrankrijker A, Brimicombe R, Heijerman H, Teding van Berkhout F, Spitoni C, Grave S, van der Ent C, Wolfs T, Willems R, Bonten M. 2016. The effect of strict segregation on Pseudomonas aeruginosa in cystic fibrosis patients. PLoS One 11:e0157189. doi: 10.1371/journal.pone.0157189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgan MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somayaji R, Parkins MD, Shah A, Martiniano SL, Tunney MM, Kahle JS, Water VJ, Elborn JS, Bell SC, Flume PA, VanDevanter DR, Antimicrobial Resistance in Cystic Fibrosis International Working Group. 2019. Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: a systematic review. J Cystic Fibros 18:236–243. doi: 10.1016/j.jcf.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Castellani C, Duff AJ, Bell SC, Heijerman HG, Munck A, Ratjen F, Sermet-Gaudelus I, Southern KW, Barben J, Flume PA, Hodková P, Kashirskaya N, Kirszenbaum MN, Madge S, Oxley H, Plant B, Schwarzenberg SJ, Smyth AR, Taccetti G, Wagner TO, Wolfe SP, Drevinek P. 2018. ECFS best practice guidelines: the 2018 revision. J Cyst Fibros 17:153–178. doi: 10.1016/j.jcf.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 18.de Dios Caballero J, Del Campo R, Royuela A, Solé A, Máiz L, Olveira C, Quintana-Gallego E, de Gracia J, Cobo M, de la Pedrosa EG, Oliver A, Cantón R. 2016. Bronchopulmonary infection-colonization patterns in Spanish cystic fibrosis patients: results from a national multicenter study. J Cyst Fibros 15:357–365. doi: 10.1016/j.jcf.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Muhlebach MS, Hatch JE, Einarsson GG, McGrath SJ, Gilipin DF, Lavelle G, Mirkovic B, Murray MA, McNally P, Gotman N, Davis Thomas S, Wolfgang MC, Gilligan PH, McElvaney NG, Elborn JS, Boucher RC, Tunney MM. 2018. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: a multisite study. Eur Respir J 52:1800242. doi: 10.1183/13993003.00242-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCaughey G, McKevitt M, Elborn JS, Tunney MM. 2012. Antimicrobial activity of fosfomycin and tobramycin in combination against cystic fibrosis pathogens under aerobic and anaerobic conditions. J Cyst Fibros 11:163–172. doi: 10.1016/j.jcf.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 21.European Committee on Antimicrobial Susceptibility testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 22.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley KA, Bray JE, Maiden MC. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.