Abstract
Sub-cortical volumetric differences were associated with attention-deficit/hyperactivity disorder (ADHD) in a recent multi-site, mega-analysis of 1713 ADHD persons and 1529 controls. Because there was a wide range of effect sizes among the sub-cortical volumes, it is possible that selective neuronal vulnerability plays a role in these volumetric losses. To address this possibility, we used data from Allen Brain Atlas to investigate variability in gene expression profiles between subcortical regions of typically developing brains. We tested the hypothesis that the expression of genes in a set of curated ADHD candidate genes and five a priori selected, biological pathways would be associated with the ENIGMA findings. Across the subcortical regions studied by ENIGMA, gene expression profiles for three pathways were significantly correlated with ADHD-associated volumetric reductions: apoptosis, oxidative stress, and autophagy. These correlations were strong and significant for children with ADHD, but not for adults. Although preliminary, these data suggest that variability of structural brain anomalies in ADHD can be explained, in part, by the differential vulnerability of these regions to mechanisms mediating apoptosis, oxidative stress, and autophagy.
Keywords: ADHD, autophagy, apoptosis, oxidative stress, neurodevelopment, brain volumes, transcript expression, imaging genomics, microarray
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is a common neurodevelopmental disorder characterized by impairing symptoms of hyperactivity, impulsivity, and inattention. The prevalence is estimated at 5–7% of school-age children and 2.5–5% of adults.1 Decades of research have documented environmental2 and genetic1 risk factors along with both structural and functional brain abnormalities.3 These data suggest that ADHD is a multifactorial condition caused by the confluence of many environmental and genetic risks that affect neurodevelopment and, ultimately, lead to symptoms of the disorder.1 It is unknown, however, why some brain regions are impacted by environmental and/or genetic risks and others are not.
Structural magnetic resonance imaging (MRI) studies revealed small but consistent volumetric reductions in ADHD brains compared to those of typically developing subjects. Meta-analyses of structural MRI studies have reported area and volume reductions of cerebellar regions, the splenium of the corpus callosum, total and right cerebral volume, and the right caudate in ADHD children e.g., 4. Whole-brain voxel-based morphometry meta-analyses have consistently demonstrated gray matter reductions of the caudate, globus pallidus, and putamen in both children and adults with ADHD. There is also evidence for effects of pharmacological treatment and age: volumetric changes are more pronounced in people with ADHD who have not been treated with medications and these changes tend to diminish with increasing age.5
The ADHD subgroup of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) Consortium investigated subcortical brain volumes in data from 23 cohorts with a total sample size of over 3,200 subjects, including children and adults (age range: 4 – 63 years, median = 14, 66% males).6 ENIGMA ADHD used both meta-analyses (relying on summary statistics) and mega-analyses (relying on raw data) to test for group differences in subcortical volumes. Mega-analysis demonstrated that ADHD probands had significantly smaller volumes for accumbens (Cohen’s D = 0.15), amygdala (D = 0.19), caudate (D = 0.11), hippocampus (D = 0.11), and putamen (D = 0.14) when compared to typically developing individuals. Meta-analysis yielded similar results and same ordering of effect sizes, however there was no significant difference detected for the volumes of the hippocampus or accumbens between ADHD cases and unaffected comparison subjects in the meta-analysis. In both the mega- and meta-analyses, no significant differences were observed for volumes of the pallidum or thalamus. Subcortical volume differences were strongest for children, with no significant findings for adults. The subcortical volumes were not influenced by lifetime stimulant medication, presence of comorbid psychiatric disorders, or ADHD symptom severity.6
The ENIGMA ADHD results raise intriguing questions: Why are the different subcortical regions affected to different degrees? Is this evidence of selective neuronal vulnerability (SNV), i.e., the selective sensitivity of particular neuronal populations to stressors?7 Neurons present a wide variety of distinct morphological and functional characteristics reflecting the unique gene expression profile of each neuronal population. As a consequence, different neurons respond differently to damaging stimuli regardless of whether these stimuli derive from genetic variants or environmental insults. There is evidence of SNV for neurodegenerative disorders. For example, neurons from the entorhinal cortex, hippocampus CA1 region, frontal cortex, and amygdala are the most sensitive to neurodegeneration in Alzheimer’s disease. These neurons are selectively vulnerable to conditions such as energy deprivation and oxidative stress. Activating cell stress pathways accumulates beta-amyloid protein and tau-based tangles in these vulnerable neurons. In Parkinson’s disease, dopaminergic neurons from the substantia nigra pars compacta are particularly vulnerable to mitochondrial dysfunction and elevated levels of reactive oxygen species. In Huntington’s, striatal medium spiny GABAergic neurons (MSNs) are the main neuronal population affected by the mutant Huntingtin, possibly leading to mitochondrial dysfunction. The selective loss of the neuronal populations cited above are responsible in large part for the clinical manifestations observed in these neurodegenerative disorders.8, 9
SNV could explain why the ENIGMA study found some subcortical structures to show greater ADHD-associated volumetric loss than others. If so, understanding SNV in ADHD could point to biological pathways involved in the disorder. To assess this possibility, we tested hypotheses about the expression profiles of candidate genes and pathways in the subcortical structures evaluated by the ENIGMA ADHD Working Group. We, a priori, selected five pathways previously implicated in ADHD, which we hypothesized would be associated with SNV: neurotransmission regulation, oxidative stress, neurodevelopment, apoptosis, and autophagy.10–13We also selected a set of ADHD candidate genes that had been proposed by a panel of ADHD experts and further implicated via GWAS.14
Materials and Methods
Identification of ADHD Candidate Genes and Pathways
For each pathway, we identified relevant genes by surveying gene sets made available through data sets curated by Ingenuity Pathway Analysis (IPA) (version 26127183, November-30–2015, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) and the Molecular Signature Database (MSigDB) (version 5, August-15–2015, http://software.broadinstitute.org/gsea/msigdb). We downloaded the following biological pathways from IPA: (1) axon guidance signaling, (2) dopamine degradation, (3) dopamine receptor signaling, (4) GABA receptor signaling, (5) glutamate receptor signaling, (6) glutamate degradation, (7) noradernergic/adrenaline degradation, (8) neuron signaling, (9) NRF2-guided oxidative stress response, (10) production of reactive oxygen species, (11) serotonin degradation, and (12) serotonin receptor signaling. In addition, we extracted similar gene sets and pathways available through MSigDB.
We combined gene sets from IPA and MSigDB and eliminated duplicate gene identifiers after merging discrete annotations into five biological pathways: (1) autophagy, (2) apoptosis, (3) neurodevelopment (e.g., axon guidance, axonogenesis), (4) neurotransmitter regulation (e.g., neurotransmitter release, receptor activity, and metabolism), and (5) oxidative stress (e.g., production of reactive oxygen species, oxidative stress responses). The following 43 ADHD candidate genes comprised a gene set for a separate analysis: ADRA1A, ADRA1B, ADRA2A, ADRA2C, ADRB2, ADRBK2, ARRB1, BDNF, CDH13, CHRNA4, COMT, CSNK1E, DBH, DDC, DRD1, DRD2, DRD3, DRD4, FADS1, FADS2, HES1, HTR1B, HTR1E, HTR2A, HTR2C, HTR3B, MAOA, MAOB, NFIL3, NR4A2, PER1, PER2, SLC18A2, SLC6A1, SLC6A2, SLC6A3, SLC6A4, SLC9A9, SNAP25, STX1A, SYT1, TPH1, and TPH2. These genes had been previously nominated as ADHD risk genes by the International Multisite ADHD Genetics (IMAGE) group.15 As a group, these genes were associated with ADHD in a meta-analysis of genome-wide association studies.14 These 43 ADHD candidate genes overlap with pathways for neurodevelopment, neurotransmitter regulation, and apoptosis, and provide additional genes that are not implicated by the remaining pathways.
Microarray Data Import and Management
We downloaded publicly available phenotypic and transcriptome-wide molecular data for typically developing human subjects released through the Allen Brain Atlas website (http://www.brain-map.org).16 We targeted microarray data of postmortem brain samples from six adult donors (24 years to 55 years) whose brain tissues were eligible for inclusion in the Allen Brain Atlas microarray survey as these subjects had no known psychiatric or neurological disorder, brain injury or disease, history of epilepsy, drug or alcohol dependence, prion disease, infectious disease, cancer deaths, chronic renal failure, on a ventilator for > 1 hour, or had a time of death > 30 hours. Microarray data were acquired with existing normalization and adjustment for batch effects included. Expression data conformed to the log2-scale. We applied median summarization to collapse many-to-one mappings of probe sets into single estimates of gene expression within samples.
High resolution sub-cortical gene expression data were targeted for the following brain regions: (1) nucleus accumbens, (2) amygdalohippocampal transition zone, (3) body, head, and tail of caudate nucleus, (4) internal and external segments of globus pallidus, (5) bank and lateral bank of parahippocampal gyrus, (6) putamen, and (7) reticular nucleus of thalamus. Each of the sub-cortical regions that were dissected from adult samples included left and right hemispheres. The median expression of genes was derived across hemispheres and within micro-dissected sub-cortical structures to obtain an average spatial measure of gene expression across 7 regions (Supplementary Figure 1). Sample sizes for each brain region and the total number of genes detected in the Allen Brain cohort are provided in Supplementary Table 1. Note that we omitted samples from a separate Allen Brain Atlas cohort used for RNA-sequencing experiments with coverage of pre- and postnatal (perinatal to adulthood) ages, because those data had only a limited number of brain regions examined by ENIGMA. The number of genes overlapping between pathways is shown in Supplementary Table 2. We found that the normalized pathway expression levels within brain regions were highly correlated between the Allen Brain Atlas and two additional cohorts (Genotype Tissue Expression Project and Human Brain Transcriptome, Supplementary Figure 2).
Statistical Analyses
We compared the average effect sizes between the eight brain regions included in ENIGMA’s meta-analysis6 using two-tailed T-tests. Results from our analysis are provided in Supplementary Table 3. The largest difference between regions was amygdala and pallidum (P = 1.72e-04). Despite there being small effect sizes overall, statistically significant regional differences observed in the average ADHD brain suggests that this variability is meaningful and warrants further investigation.
Our primary analyses tested for an association between the average gene expression levels in each pathway and the magnitude of sub-cortical volumetric changes in ADHD participants as measured by the standardized mean difference (Cohen’s D) as computed by the ENIGMA Consortium, correcting for age, sex, and total intracranial volume.6 Using microarray data from the Allen Brain Atlas and pathway-based annotations acquired from IPA and MSigD, we arrived at a single estimate of pathway expression per targeted brain region by calculating the mean expression of genes within pathways (± 95% confidence intervals). Pearson’s correlation coefficients tested the association of pathway expression levels with volumetric changes in ADHD across sub-cortical regions. We adjusted p-values using the Bonferroni procedure to control for testing multiple pathways.
Gene set analysis with GWAS data for ADHD
We supplied summary statistics from the largest available GWAS of ADHD17 to the software MAGMA (v1.06)18 to perform a gene set enrichment analysis focusing on the six pathways evaluated in our study. The NCBI hg19 reference genome was used to map SNPs to genes with a ±20 kilobase window added to the start and end position of every gene. Gene significance scores were calculated by MAGMA based on the mean SNP significance. A linear regression model was then used to test for a difference in mean gene significance scores between genes inside and outside of a defined pathway. The regression models included covariates for minor allele frequency, SNP density, and linkage disequilibrium between genes.
Code availability
The code (R script) used to run the analysis is available from the authors upon request.
Results
Pathways Associated with Sub-Cortical Volume Changes in ADHD
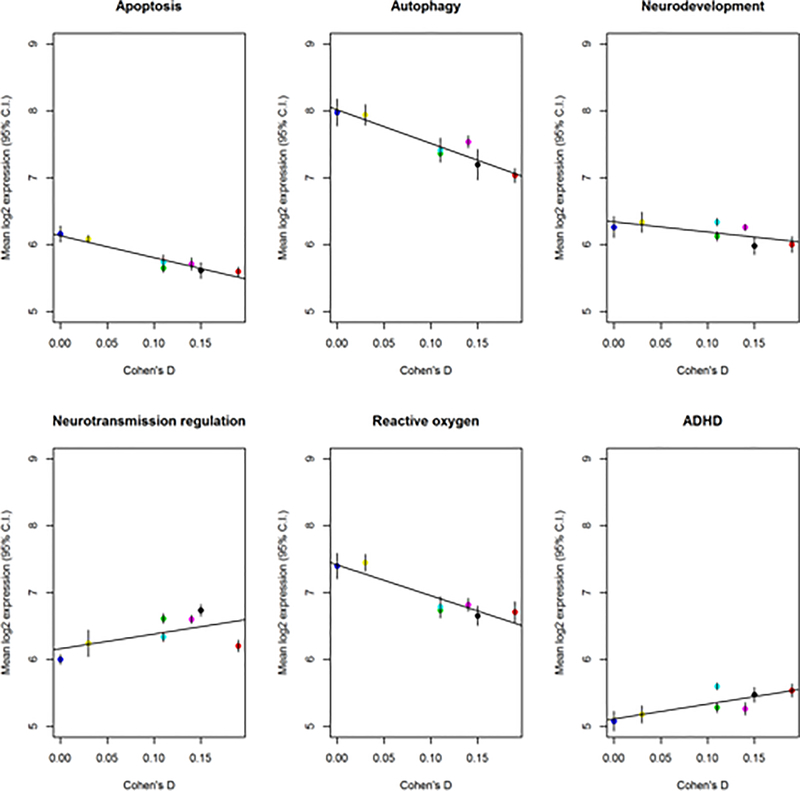
For each pathway, Figure 1 plots the mean and 95% confidence intervals of the mean expression level against Cohen’s D for each pathway along with the best fitting linear regression. We found large and significant correlations between Cohen’s D (children and adults combined) and expression of three pathways: apoptosis, autophagy, and oxidative stress (Table 1A). The significance was strongest for the apoptosis pathway (r = −0.953, uncorrected p-value = 9.0E-04, Bonferroni p-value = 5.4e-03). These findings were the same when analyses were limited to the child subset (Table 1B). In contrast, we found no significant correlations between pathway expression levels and Cohen’s D effect sizes for adults with ADHD (Table 1C). We performed a post hoc analysis by combining expression values for genes in apoptosis and autophagy pathways, in addition to apoptosis, autophagy, and oxidative stress pathways. The combined pathways showed significant correlations with Cohen’s D effect sizes for adults or children with ADHD (apoptosis + autophagy: r = −0.942, p-value = 1.5e-03; apoptosis + autophagy + oxidative stress: r = −0.942, p-value = 1.5e-03).
Figure 1:
Comparison of pathway expression levels and the Cohen’s d effect sizes from ENIGMA’s ADHD vs. control comparisons of subcortical brain regions. Points correspond to mean expression of pathway with 95% confidence intervals indicated by vertical bars. The best fitting regression is denoted by a red line. The Cohen’s d for each brain region are as follows: (1) pallidum = 0.00, (2) thalamus = 0.03, (3) hippocampal formation = 0.11, (4) caudate = 0.11, (5) accumbens = 0.15, (6) putamen = 0.14, and (7) amygdala = 0.19.
Table 1A.
Correlation of Sub-cortical Volumetric Changes in ADHD with Pathway Expression Levels across Seven Brain Regions in Adult Microarray Samples.
| Pathway label | Pearson’s r | p-value | Bonferroni p-value |
|---|---|---|---|
| ADHD candidate genes | 0.771 | 4.22E-02 | 0.253 |
| Apoptosis | −0.953 | 9.00E-04 | 0.0054 |
| Autophagy | −0.948 | 1.20E-03 | 0.007 |
| Neurodevelopment | −0.676 | 9.52E-02 | 0.57 |
| Neurotransmitter regulation | 0.557 | 1.94E-01 | 1.0 |
| Oxidative Stress | −0.915 | 3.90E-03 | 0.0243 |
Table 1B.
Correlation of Sub-cortical Volumetric Changes in Children with ADHD with Pathway Expression Levels across Seven Brain Regions in Adult Microarray Samples.
| Pathway label | Pearson’s r | p-value | Bonferroni p-value |
|---|---|---|---|
| ADHD candidate genes | 0.688 | 0.87 | 0.52 |
| Apoptosis | −0.953 | 0.00086 | 0.0052 |
| Autophagy | −0.899 | 0.0059 | 0.036 |
| Neurodevelopment | −0.662 | 0.11 | 0.63 |
| Neurotransmitter regulation | 0.699 | 0.08 | 0.48 |
| Oxidative stress | −0.949 | 0.0011 | 0.0066 |
Table 1C.
Correlation of Sub-cortical Volumetric Changes in Adults with ADHD with Pathway Expression Levels across Seven Brain Regions in Adult Microarray Samples.
| Pathway label | Pearson’s r | p-value | Bonferroni p-value |
|---|---|---|---|
| ADHD candidate genes | 0.584 | 0.17 | 1.00 |
| Apoptosis | −0.676 | 0.095 | 0.54 |
| Autophagy | −0.792 | 0.033 | 0.20 |
| Neurodevelopment | −0.761 | 0.047 | 0.28 |
| Neurotransmitter regulation | 0.226 | 0.626 | 1.00 |
| Oxidative stress | −0.579 | 0.173 | 1.00 |
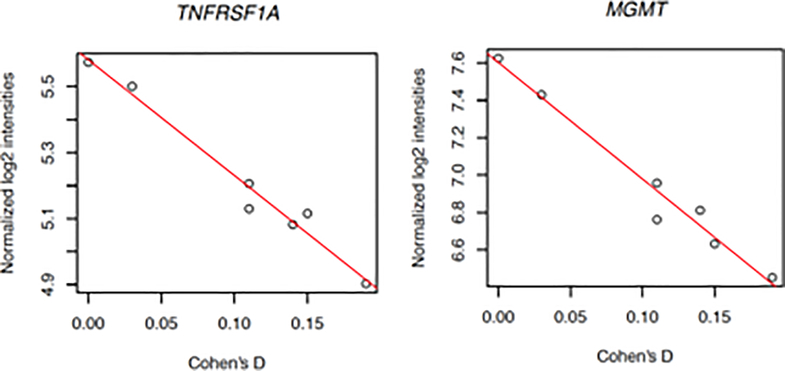
Given these findings, we then performed a post hoc test for each of 704 genes constituting the statistically significant pathways for the combined Cohen’s D effect sizes for subcortical volume differences in children and adults with ADHD. We identified two genes that remained statistically significant after correcting for testing 704 genes (Figure 2): TNFRSF1A (Pearson’s r = −0.986, p-value = 3.84e-05, Bonferroni p-value = 0.027) and MGMT (Pearson’s r = −0.983, p-value = 6.67e-05, Bonferroni p-value = 0.046). None of the 704 genes examined were significantly correlated with subcortical volume changes in children with ADHD after multiple testing correction. The correlation of TNFRSF1A with Cohen’s D estimates for children with ADHD was consistent in direction and nominally significant before correction (r = −0.945, uncorrected p-value = 0.0014, Bonferroni p-value = 0.95). However, the top ranking gene by correlation with subcortical volume differences in children with ADHD was HMOX1 (r = −0.975, uncorrected p-value = 1.9e-04, Bonferroni p-value = 0.13).
Figure 2:
A strongly negative correlation of TNFRSF1A expression and Cohen’s d was identified in adult brain samples (Pearson’s r = −0.986, p-value < 3.84e-05, Bonferroni p-value < 0.027). A similar correlation was found for MGMT with the same samples (Pearson’s r = −0.983, p-value = 6.67e-05, Bonferroni p-value = 0.046). A total of 704 genes were examined across pathways for apoptosis, autophagy, and oxidative stress.
Pathways Not Enriched with GWAS Signals for ADHD
We observed no significant enrichment of GWAS associations among genes contained in the apoptosis gene set from our analysis with MAGMA (Beta = 0.198, SE = 0.164, uncorrected p-value = 0.56). In total, none of the pathways we examined showed a significant enrichment of GWAS signals for ADHD (Supplementary Table 4).
Discussion
Our analyses support our hypothesis that pathways mediating apoptosis, autophagy, and oxidative stress might play a role in the variability of volumetric differences between people with and without ADHD. These findings, albeit preliminary, suggest that normal variability in these pathways may regulate sensitivity to the genetic and environmental events that lead to ADHD. Because we lack gene expression and brain volumetric data from the same samples, we cannot infer whether pathways are up- or down-regulated between people with and without ADHD. Because the significant correlations we reported cannot be interpreted as causal relationships, experimental work in animal models investigating the functional role of genetic variants linked with brain volume regulation is needed. Ideally, studies of ADHD brain tissue would be used to confirm these results but such data are not possible to obtain because tissue resource centers have not made ADHD a focus of collection efforts. Therefore, we caution against extrapolating our findings from “normal” brain tissue to the ADHD brain.
Pathways mediating oxidative stress, autophagy, and apoptosis showed the strongest correlations with the magnitude of volumetric anomalies associated with ADHD. We had selected these pathways a priori based on prior studies of ADHD (see Introduction). These findings suggest that the expression of genes and environmental factors regulating programmed cell death may mediate risk for ADHD and that their differential effects on subcortical regions are due to regional differences in susceptibility to these effects.
The gene expression results cannot parse genetic and environmental sources of SNV. To get some leverage on this issue, we tested for significant enrichment of common variant GWAS signals for ADHD among the gene sets examined in this study. These analyses were not significant, which suggests that genetic sources of SNV may be limited or lie in rare variants. Consistent with this, according to the LD Hub database, the genetic correlations between ADHD the subcortical volumes for brain regions examined in this study are low and not significant.19 Estimates of genetic correlations produced by LD Hub are based on effect sizes of common variation from GWAS. Taken together these finding suggest that future work should investigate environmental risk factors as a source of SNV in subcortical structures. Consistent with this, a study of monozygotic twins discordant for ADHD found exhibit epigenetic differences among genes highly expressed in the developing cerebellum, striatum, and thalamus, in addition to genes functionally related to neurodevelopment and neurotransmitter regulation.20
We found lower expression of oxidative stress pathway genes in regions with greater ADHD-associated volumetric loss. This may indicate that regions which are a priori less able to process reactive oxygen species (ROS) are more susceptible to their effects. ROS cause cellular damage through oxidation of critical cellular components, such as enzymes, membrane lipids, and DNA.21, 22 To avoid such damage, the enzymatic antioxidant system within cells maintains a low concentration of ROS by generating reducing agents and ROS-scavengers, such as glutathione and ascorbic acid.22, 23 Because the brain metabolizes over 20% of the body’s oxygen and does not contain a robust antioxidant system, it is disproportionately susceptible to ROS damage.21, 24
Our hypothesis about oxidative stress had been motivated by a meta-analysis that found a significant association between ADHD and peripheral measures of oxidative stress that could not be accounted for by publication biases.10 Because the association between ADHD and peripheral antioxidant levels was not significant, the authors concluded that patients with ADHD have normal levels of anti-oxidant production but that their response to oxidative stress is insufficient, leading to oxidative damage. Indirect evidence for oxidative stress in ADHD also comes from studies showing treatment efficacy for anti-oxidant compounds e.g., 25, 26, 27. Copy number variants associated with ADHD are also significantly enriched for pathways relevant for ROS regulation.12 Increased oxidative stress has been found in the cortex, striatum, and hippocampus of established animal models of ADHD (spontaneously hyperactive rats).28
Cellular stressors, such as ischemia, excitotoxicity, ROS, reduced cell nutrient availability and decreased growth factor signaling, cause macromolecular and organelle damage and increase the production and aggregation of misfolded proteins. These stressors initiate autophagy to remove damaging molecules and provide cellular nutrients.29 Autophagy (“self-eating”) is a bulk degradative, largely pro-survival pathway 30. It degrades cellular components, such as proteins and mitochondria, through the endo-lysosomal system. Through this mechanism it protects cells from nutrient and growth factor deprivation, protein aggregation, apoptotic signaling and damaged cellular organelles.29 Its proper function is critical and considered a necessary homeostatic mechanism for neuronal development and function.31, 32
The rate of autophagy during development modulates the proliferation and differentiation of neural progenitor cells33 and is instrumental in regulating neuronal size.7 Thus, neurons with decreased autophagic function are more susceptible to cellular stressors34, 35 and are likely to have perturbations in size and morphology.33 Consistent with this, our finding suggest that low expression of autophagic genes is associated with greater ADHD-associated loss in regional sub-cortical brain volumes.
Apoptosis is a programmed cellular death pathway that indiscriminately degrades cellular proteins and causes cell shrinkage and pyknosis.36 This orderly removal of cells does not cause inflammation to nearby tissue37 and is critical during development.36 In neurons, apoptotic molecules play an important role in synaptic scaling and pruning.38–41 Apoptotic dysregulation has been associated with attention disorders41, 42 and is regulated by methylphenidate.11 Lo et al.41 hypothesized that lower apoptotic signals in attention disorders cause a failure in synaptic scaling and thus information processing. They further suggested that decreased apoptotic pathway expression leads to decreased neuronal downscaling and thus overstimulation of neuronal networks when processing novel stimuli. Consistent with this, we found gene expression in apoptotic pathways to correlate with ADHD-associated decreases in regional sub-cortical brain volumes.
Autophagy and apoptosis act together to determine the fate of cells.43–46 However, the biological context is important, as they can activate, antagonize or assist one another in their activities.30, 45, 46 Thus, it is not surprising that these pathways are co-regulated. Since autophagy acts to regulate apoptotic signals,47 decreased expression of critical autophagic proteins, such as Beclin-1 and other autophagy genes, would make cells more sensitive to apoptosis.30, 46 Moreover, autophagy is upregulated by ROS and protects against its cytotoxic effects.24, 48–51
Lower autophagic gene expression could predispose neurons to apoptotic cell death in critical brain regions and lead to the region-specific volumetric reductions we found in the ADHD-ENIGMA study. Lower autophagic expression would make neurons more susceptible to ROS, due to the lack of appropriate defective protein degradation and mitochondrial turnover.21, 24 In addition, decreased autophagy would disinhibit apoptotic signaling, further adding to selective neuronal vulnerability.46 Because ROS, autophagy and apoptosis are co-regulated in a compensatory fashion, it is especially notable that all were implicated in our study. In addition, significant overlaps between apoptosis and autophagy, and apoptosis and ROS gene sets were found, which might explain why all three pathways were significant in our analysis.
Additional analysis of individual genes in the apoptosis, autophagy, and ROS pathways identified the TNFRSF1A gene as significant after correcting for multiple comparisons. TNFRSF1A encodes the tumor necrosis factor receptor 1 (TNFR1), one of the major receptors for the pro-inflammatory cytokine TNF. The activation of the TNFR1 receptor results in the formation of two opposing signaling complexes depending on the circumstances upon which it occurs. While Complex I regulates the expression of anti-apoptotic proteins, Complex II triggers cell death processes.52 As a result of complex downstream signaling processes, TNF presents both neuroprotective and neurodegenerative effects that varies according to brain regions.53 Consistent with our findings, a neuroimaging study on typically developing adults reported significant associations between TNFRSF1A polymorphisms and volume of the caudate and putamen. However, there was no evidence of association with hippocampal volume. TNFRSF1A is a genome-wide significant locus for multiple sclerosis 54, mothers with multiple sclerosis are at risk for having ADHD children55 and diffusion tensor imaging suggests that aberrant myelination is associated with ADHD.56 Thus, TNFRSF1A should be considered for further study in ADHD.
The MGMT gene, which encodes O-6-Methylguanine-DNA methyltransferase, was the only other gene that showed a strong correlation with ADHD brain volumetric differences after multiple testing correction. The enzyme encoded by MGMT protects cells from DNA damage caused by alkylation, which is significant for maintaining cell integrity.57 No literature on MGMT relationship to ADHD has been found, however, it would be intriguing to explore the relationship in the context of DNA-damage repair and ADHD susceptibility.
The correlations supporting our conclusions were strong and significant for children with ADHD, but not for adults. This pattern is consistent with the original ENIGMA report which found significant case-control volumetric differences for children but not adults. 6 One explanation for these findings is that environmental risk factors early in development (e.g., anoxia, maternal smoking) lead to increased oxidative stress, autophagy and apoptosis which, in turn, lead to reduced brain volumes. If the molecular defenses against these risk factors are upregulated and persist through development, it is possible that they slowly lead to a reduction in oxidative stress and, in some patients, a normalization of brain volumes. Consistent with this hypothesis, a meta-analysis found that the association between ADHD and peripheral measures of oxidative stress was significant for children but not adults.10
Our results, albeit intriguing, are limited in several ways. We cannot make causal statements about the relationship of expression levels and volumetric changes as the two sources of data were from different samples. In addition, gene expression studies do not provide evidence about protein functioning. For example, a gene with low transcript number does not necessarily have decreased function, as post-translational modification or other pathway modifiers can increase activity. The Allen Brain data are based on a very small sample of typically developing adults that will not generalize to all populations of interest. Also, expression data for the pallidum was not available in the Allen Brain Atlas. Instead, we used data for the globus pallidus which, with the ventral pallidum, forms the larger pallidum examined in the ENIGMA ADHD study.
Despite these limitations, our work is the first to propose a mechanism to explain the differential vulnerability of brain regions to the as yet unknown mechanisms that cause ADHD. This leads us to further hypothesize that future etiologic studies of ADHD will implicate genetic and environmental risk factors implicating pathways regulating oxidative stress, autophagy and apoptosis.
Supplementary Material
Acknowledgements
Dr. Faraone is supported by the K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway, the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no 602805, the European Union’s Horizon 2020 research and innovation programme under grant agreement No 667302 and NIMH grants 5R01MH101519 and U01 MH109536-01. Dr. Akutagava-Martins’ contribution was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil.
Disclosures
In the past year, Dr. Faraone received income, potential income, travel expenses and/or research support from Lundbeck, Rhodes, Arbor, Pfizer, Ironshore, Shire, Akili Interactive Labs, CogCubed, Alcobra, VAYA Pharma and NeuroLifeSciences. With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD.
Footnotes
Supplementary Information
Supplementary information is available at Molecular Psychiatry’s website.
Bibliography
- 1.Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA et al. Attention-deficit/hyperactivity disorder. Nature Reviews Disease Primers 2015: 15020. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee TD, Middleton F, Faraone SV. Environmental risk factors for attention-deficit hyperactivity disorder. Acta Pediatrica 2007; 96(9): 1269–1274. [DOI] [PubMed] [Google Scholar]
- 3.Rubia K, Alegria AA, Brinson H. Brain abnormalities in attention-deficit hyperactivity disorder: a review. Rev Neurol 2014; 58 Suppl 1: S3–16. [PubMed] [Google Scholar]
- 4.Valera EM, Faraone SV, Murray KE, Seidman LJ. Meta-analysis of structural imaging findings in attention-deficit/hyperactivity disorder. Biol Psychiatry 2007; 61(12): 1361–1369. [DOI] [PubMed] [Google Scholar]
- 5.Spencer TJ, Brown A, Seidman LJ, Valera EM, Makris N, Lomedico A et al. Effect of psychostimulants on brain structure and function in ADHD: a qualitative literature review of magnetic resonance imaging-based neuroimaging studies. J Clin Psychiatry 2013; 74(9): 902–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoogman M, Bralten J, Hibar DP, Mennes M, Zwiers MP, Schweren LS et al. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Michaelis EK. Selective neuronal vulnerability to oxidative stress in the brain. Front Aging Neurosci 2010; 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 2011; 71(1): 35–48. [DOI] [PubMed] [Google Scholar]
- 9.Roselli F, Caroni P. From intrinsic firing properties to selective neuronal vulnerability in neurodegenerative diseases. Neuron 2015; 85(5): 901–910. [DOI] [PubMed] [Google Scholar]
- 10.Joseph N, Zhang-James Y, Perl A, Faraone SV. Oxidative Stress and Attention Deficit Hyperactivity Disorder: A Meta-Analysis J Atten Disord 2015; 19(11): 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reus GZ, Scaini G, Jeremias GC, Furlanetto CB, Morais MO, Mello-Santos LM et al. Brain apoptosis signaling pathways are regulated by methylphenidate treatment in young and adult rats. Brain Res 2014; 1583: 269–276. [DOI] [PubMed] [Google Scholar]
- 12.Thapar A, Martin J, Mick E, Arias Vasquez A, Langley K, Scherer SW et al. Psychiatric gene discoveries shape evidence on ADHD’s biology. Mol Psychiatry 2016; 21(9): 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry 2011; 168(4): 365–377. [DOI] [PubMed] [Google Scholar]
- 14.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49(9): 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry 2006. [DOI] [PubMed] [Google Scholar]
- 16.Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 2013; 41(Database issue): D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demmontis D, Walters RK, Matrin J, Mattheisen M, Als TD, Agerbro E et al. Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. BiorXiv 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11(4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017; 33(2): 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YC, Sudre G, Sharp W, Donovan F, Chandrasekharappa SC, Hansen N et al. Neuroanatomic, epigenetic and genetic differences in monozygotic twins discordant for attention deficit hyperactivity disorder. Mol Psychiatry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lushchak VI. Free Radicals, Reactive Oxygen Species, Oxidative Stresses and Their Classifications. Ukr Biochem J 2015; 87(6): 11–18. [DOI] [PubMed] [Google Scholar]
- 22.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol 2003; 552(Pt 2): 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halliwell B Reactive oxygen species in living systems: source, biochemistry, and role in human disease. Am J Med 1991; 91(3C): 14S–22S. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J 2012; 441(2): 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloch MH, Qawasmi A. Omega-3 fatty acid supplementation for the treatment of children with attention-deficit/hyperactivity disorder symptomatology: systematic review and meta-analysis. J Am Acad Child Adolesc Psychiatry 2011. 50(10): 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chovanova Z, Muchova J, Sivonova M, Dvorakova M, Zitnanova I, Waczulikova I et al. Effect of polyphenolic extract, Pycnogenol, on the level of 8-oxoguanine in children suffering from attention deficit/hyperactivity disorder. Free Radic Res 2006; 40(9): 1003–1010. [DOI] [PubMed] [Google Scholar]
- 27.Garcia RJ, Francis L, Dawood M, Lai ZW, Faraone SV, Perl A. Attention deficit and hyperactivity disorder scores are elevated and respond to NAC treatment in patients with SLE. Arthritis Rheum 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leffa DT, Bellaver B, de Oliveira C, de Macedo IC, de Freitas JS, Grevet EH et al. Increased Oxidative Parameters and Decreased Cytokine Levels in an Animal Model of Attention-Deficit/Hyperactivity Disorder. Neurochem Res 2017. [DOI] [PubMed] [Google Scholar]
- 29.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol 2010; 2010: 214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 2013; 1833(12): 3448–3459. [DOI] [PubMed] [Google Scholar]
- 31.Maday S Mechanisms of neuronal homeostasis: Autophagy in the axon. Brain Res 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JA. Neuronal autophagy: a housekeeper or a fighter in neuronal cell survival? Exp Neurobiol 2012; 21(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv X, Jiang H, Li B, Liang Q, Wang S, Zhao Q et al. The crucial role of Atg5 in cortical neurogenesis during early brain development. Sci Rep 2014; 4: 6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Q, Liu L, Chen Y, Li H, Yang L, Wang Y et al. Synaptosome-related (SNARE) genes and their interactions contribute to the susceptibility and working memory of attention-deficit/hyperactivity disorder in males. Prog Neuropsychopharmacol Biol Psychiatry 2015; 57: 132–139. [DOI] [PubMed] [Google Scholar]
- 35.Fekadu J, Rami A. Beclin-1 Deficiency Alters Autophagosome Formation, Lysosome Biogenesis and Enhances Neuronal Vulnerability of HT22 Hippocampal Cells. Mol Neurobiol 2015. [DOI] [PubMed] [Google Scholar]
- 36.Elmore S Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35(4): 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haanen C, Vermes I. Apoptosis and inflammation. Mediators Inflamm 1995; 4(1): 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 2006; 50(6): 855–867. [DOI] [PubMed] [Google Scholar]
- 39.Snigdha S, Smith ED, Prieto GA, Cotman CW. Caspase-3 activation as a bifurcation point between plasticity and cell death. Neurosci Bull 2012; 28(1): 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilman CP, Mattson MP. Do apoptotic mechanisms regulate synaptic plasticity and growth-cone motility? Neuromolecular Med 2002; 2(2): 197–214. [DOI] [PubMed] [Google Scholar]
- 41.Lo SC, Wang Y, Weber M, Larson JL, Scearce-Levie K, Sheng M. Caspase-3 deficiency results in disrupted synaptic homeostasis and impaired attention control. J Neurosci 2015; 35(5): 2118–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valbonesi S, Magri C, Traversa M, Faraone SV, Cattaneo A, Milanesi E et al. Copy number variants in attention-deficit hyperactive disorder: identification of the 15q13 deletion and its functional role. Psychiatr Genet 2015; 25(2): 59–70. [DOI] [PubMed] [Google Scholar]
- 43.Xu P, Gu R, Broster LS, Wu R, Van Dam NT, Jiang Y et al. Neural basis of emotional decision making in trait anxiety. J Neurosci 2013; 33(47): 18641–18653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 2010; 6(3): 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15(2): 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubinstein AD, Kimchi A. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci 2012; 125(Pt 22): 5259–5268. [DOI] [PubMed] [Google Scholar]
- 47.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 2010; 6(7): 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka K, Whelan KA, Chandramouleeswaran PM, Kagawa S, Rustgi SL, Noguchi C et al. ALDH2 modulates autophagy flux to regulate acetaldehyde-mediated toxicity thresholds. Am J Cancer Res 2016; 6(4): 781–796. [PMC free article] [PubMed] [Google Scholar]
- 49.Rodney GG, Pal R, Abo-Zahrah R. Redox regulation of autophagy in skeletal muscle. Free Radic Biol Med 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Wang K, Lei Y, Li Q, Nice EC, Huang C. Redox signaling: Potential arbitrator of autophagy and apoptosis in therapeutic response. Free Radic Biol Med 2015; 89: 452–465. [DOI] [PubMed] [Google Scholar]
- 51.Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Signal 2011; 14(11): 2215–2231. [DOI] [PubMed] [Google Scholar]
- 52.Cabal-Hierro L, Lazo PS. Signal transduction by tumor necrosis factor receptors. Cell Signal 2012; 24(6): 1297–1305. [DOI] [PubMed] [Google Scholar]
- 53.Sriram K, O’Callaghan JP. Divergent roles for tumor necrosis factor-alpha in the brain. J Neuroimmune Pharmacol 2007; 2(2): 140–153. [DOI] [PubMed] [Google Scholar]
- 54.International Multiple Sclerosis Genetics C, Wellcome Trust Case Control C, Sawcer S, Hellenthal G, Pirinen M, Spencer CC et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011; 476(7359): 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Instanes JT, Halmoy A, Engeland A, Haavik J, Furu K, Klungsoyr K. Attention-Deficit/Hyperactivity Disorder in Offspring of Mothers With Inflammatory and Immune System Diseases. Biol Psychiatry 2017; 81(5): 452–459. [DOI] [PubMed] [Google Scholar]
- 56.Onnink AM, Zwiers MP, Hoogman M, Mostert JC, Dammers J, Kan CC et al. Deviant white matter structure in adults with attention-deficit/hyperactivity disorder points to aberrant myelination and affects neuropsychological performance. Prog Neuropsychopharmacol Biol Psychiatry 2015; 63: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondo N, Takahashi A, Ono K, Ohnishi T. DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids 2010; 2010: 543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.