Abstract
Jaw bones and teeth originate from the first pharyngeal arch and develop in closely related ways. Reciprocal epithelial-mesenchymal interactions are required for the early patterning and morphogenesis of both tissues. Here we review the cellular contribution during the development of the jaw bones and teeth. We also highlight signaling networks as well as transcription factors mediating tissue-tissue interactions that are essential for jaw bone and tooth development. Finally, we discuss the potential for stem cell mediated regenerative therapies to mitigate disorders and injuries that affect these organs.
1. An overview of jaw bone and tooth development
The maxilla and mandible together form the lower part of the facial skeleton, which performs important functions in our daily life. The jaw bones serve as anchors for the teeth, which are critical for mastication and speech.
In vertebrates, the maxilla and mandible, like most of the other craniofacial bones, are derived from cranial neural crest cells (CNCCs). These cells are known for their multipotency and their extensive migration through the embryo (Chai et al., 2000; Le Douarin, Creuzet, Couly, & Dupin, 2004; Noden, 1975; Thiery, Duband, & Delouvee, 1982). During early development, CNCCs migrate out from the hindbrain (rhombomere segments r1–r7), traveling along the dorsal-ventral axis as loosely connected streams that ultimately come to populate the pharyngeal arches. Shortly after first pharyngeal arch (PA1) patterning, a group of mesenchymal cells condenses and develops into Meckel’s cartilage (MC). The MC in each half of the mandible lengthens ventromedially and dorsolaterally, until the two eventually come together to fuse at the distal tip of the mandibular arch (Richany, Bast, & Anson, 1956). Meanwhile, lateral to the MC, mandibular bone starts to form. In the maxilla, the ossification process begins slightly later than in the mandible. At the cellular level, condensed mesenchymal cells undergo differentiation into osteoblasts with the guidance of a series of osteogenic transcriptional regulators, such as Dlx5, Runx2, and Osterix (Baek, Kim, de Crombrugghe, & Kim, 2013; Zhang, 2010). As mandibular ossification progresses, the bony tissue approaches and ultimately wraps around the MC, while the cartilaginous tissue of the MC becomes hypertrophic and degenerates in a process similar to endochondral ossification. Eventually, multinuclear phagocytotic cells called chondroclasts resorb the calcified cartilaginous matrix. In the most distal and proximal regions, the symphysis, condyle, and mandibular angle are formed through endochondral ossification. The rest of the posterior portion of MC may contribute to the formation of the sphenomandibular ligament (Moore, Persaud, & Samperio, 1999). Jaw bone development continues postnatally and ceases around 20 years of age (Love, Murray, & Mamandras, 1990).
Tooth development can be roughly divided into two major events: crown formation, which happens mainly at the embryonic stages, and root development, which begins around postnatal day 3 in the mouse. The first morphological sign of tooth initiation in the mouse is evident at around embryonic day (E)11.5 (Theiler stage 19), with the thickening of epithelial tissue called the dental placode. This tissue continues to proliferate and form the tooth bud. Meanwhile, the mesenchymal tissue around the tooth bud condenses and forms the tooth germ. With the proliferation and in-folding of the epithelium, the tooth bud progresses through the cap and bell stages. During these stages, stem cells residing in the dental mesenchyme and dental epithelium become committed and form odontoblasts and ameloblasts, respectively. Odontoblasts form dentin whereas ameloblasts contribute to enamel formation.
After the crown has formed, the dental epithelium elongates and grows apically to form a bilayered epithelial structure between the dental papilla and dental follicle called Hertwig’s epithelial root sheath (HERS), which functions as a signaling center to guide root formation. In mammals, HERS is a transient structure. After its movement to the cervical loop of the enamel organ, it undergoes perforation and eventually apoptosis, leaving a mesh-like matrix on the root surface. CNC-derived dental mesenchyme is also critical for this developmental event. It gives rise to multiple tissue types including odontoblasts, dental pulp cells, cementoblasts and periodontal ligament (PDL) cells. Traditionally, researchers believed that mesenchymal cells receive signals from the HERS for tooth root elongation (Cate, 1996). Recently, using an inducible Cre line, researchers began to uncover important cell populations as well as signaling within mesenchymal tissue that also play essential roles during tooth root development (Feng et al., 2017; Li, Parada, & Chai, 2017).
In the following sections, we will take a closer look at the different cellular components and molecular networks that regulate different stages of jaw bone development, then turn to tooth development. We will also discuss the potential for stem cell mediated regenerative therapies to mitigate disorders and injuries that affect these organs.
2. Early development of the first pharyngeal arch
2.1. Cellular contributions to mandible and maxilla development
The neural crest is a fascinating and extensively studied cell population largely due to its unique properties. Neural crest cells (NCCs) originate at the ectodermal border of the neural plate. As the neural tube closes, the NCCs undergo epithelial to mesenchymal transition (EMT) and migrate into the mesodermal mesenchyme ventrolaterally; therefore, they are referred to as ectomesenchymal cells (Loring & Erickson, 1987; Teillet, Kalcheim, & Le Douarin, 1987). Based on their original location along the rostral-caudal axis, NCCs can be further divided into four populations: cranial, cardiac, vagal, and trunk (Gilbert, 2000). CNCCs contribute to most of the craniofacial bones, including the maxilla and mandible, as well as cartilage, nerves, and connective tissue in the face. They migrate out of the dorsal neural tube and soon divide into streams which will later enter the pharyngeal arches. This striking pattern of NCC migration is closely related to the rhombomeric organization of the hindbrain. Two gene families are particularly critical for establishing unique segmental identities for hindbrain: the Hox genes and Ephrin/Eph receptors. Extensive studies have shown that Hox genes account for the antero-posterior identity of rhombomeric segments (Barrow, Stadler, & Capecchi, 2000; Krumlauf, 1994; Lumsden & Krumlauf, 1996; McGinnis & Krumlauf, 1992; Studer et al., 1998). During the period of craniofacial morphogenesis in the mouse embryo, Hox gene expression is not detectable in NCCs derived from rhombomeres 1 and 2, which later form the entire facial skeleton. Targeted inactivation of Hoxa2 results in homeotic transformation of skeletal elements derived from the second branchial arch into more anterior structures, leading to a duplication of MC adjacent to the otic capsule (Gendron-Maguire, Mallo, Zhang, & Gridley, 1993; Rijli et al., 1993). Interestingly, NCCs derived from both Hox-negative and Hox-positive regions can further differentiate into cartilage and bone; however, intramembranous ossification only takes place in the Hox-negative region. Higher osteogenic capacity and more robust in vivo bone regeneration have also been observed in progenitor cells derived from Hox-negative CNCCs compared to skeletal progenitor cells from the mesoderm (Chung et al., 2009; Leucht et al., 2008).
The Ephrins and Eph receptors are expressed in the different rhombomeres in a non-overlapping pattern that mediates cell sorting at the boundaries of odd- and even-numbered rhombomeres. This cell sorting process is essential for the segmental streams of migrating NCCs and for preventing the intermingling of cells between adjacent rhombomeres (Mellitzer, Xu, & Wilkinson, 1999; Xu, Mellitzer, Robinson, & Wilkinson, 1999). Inactivation of this signaling pathway using truncated Eph receptors disturbs the boundaries and leads to abnormal migration of third arch NCCs into the second and fourth arch territories (Smith, Robinson, Patel, & Wilkinson, 1997).
Other mechanisms that govern CNCC migration include contact inhibition of locomotion and cell repolarization, co-attraction, and chemotaxis controlled by multiple signaling molecules, such as Complement3a (C3a) and the C3a receptor, stromal cell-derived factor (SDF), VEGF, GDNF, and endothelin (Carmona-Fontaine et al., 2008; Olesnicky Killian, Birkholz, & Artinger, 2009). Simoes-Costa and Bronner identified a hierarchical gene regulatory network composed of a series of transcriptional factors that are specifically expressed in CNCCs in a spatially and temporally restricted manner during CNCC induction and early migration (Simoes-Costa & Bronner, 2016).
Cells from paraxial mesoderm give rise to the muscle component and some skeletal tissue in the posterior part of the head. In the mandibular arch, mesodermal tissue in the center is surrounded by CNCCs, with a clear cell-cell boundary between the two; together they form the mesenchymal core of the pharyngeal arch (Chai & Maxson, 2006). Even though mesodermal tissue does not directly contribute to jaw bone formation, it can still affect the development of the maxilla and mandible through tissue-tissue interactions. Ablation of Tbx1, which is exclusively expressed in the mesoderm, leads to defects in the formation of the proximal mandible (Aggarwal et al., 2010). Pharyngeal ectoderm and endoderm together cover the mesenchymal core. The ectoderm is essential for regulating the fate of CNCCs during mandibular morphogenesis, whereas the establishment of ectodermal identity is independent of CNCCs (Veitch, Begbie, Schilling, Smith, & Graham, 1999). The pharyngeal endoderm makes a limited contribution to craniofacial development. However, the pharyngeal pouch, which is formed by endodermal tissue, serves as a signaling center for tissue-tissue interaction. Using lineage tracing techniques, cells from different origins can be visualized in whole embryos and in cross-sections of the first pharyngeal arch. Post-migratory CNCCs, mesoderm- and ectoderm-derived cells are detectable in Wnt1Cre;R26R, Myf5Cre;R26R, and K14Cre;R26R embryos, respectively, which provide valuable information that can help us gain a better understanding of dynamic cell-cell interactions and identify the regulatory mechanisms that are active during craniofacial development (Chai & Maxson, 2006).
2.2. Molecular identity of the developing mandible and maxilla
Multiple heterotopic graft experiments in birds and amphibians have clearly suggested that jaw patterning information is passively carried by the NCCs and maintained throughout subsequent development (Noden, 1978a, 1978b, 1983). However, other experiments also demonstrated the importance of environmental cues from other tissues, including cephalic ectoderm, neuroectoderm, and pharyngeal endoderm. For example, tissue-specific loss-of-function of Fgf8 in the first arch ectoderm of murine embryos results in a severe mandible phenotype with loss of the majority of the bone structure (Trumpp, Depew, Rubenstein, Bishop, & Martin, 1999). The fact that numerous genes involved in jaw development are turned on only after CNCCs reach PA1 suggests that patterning of CNCCs within PA1 also relies on environmental cues.
Taking these findings into account, the next step is to address how CNCCs are patterned within PA1. Depew and Compagnucci proposed a very interesting predictive model, known as the “hinge and caps” model, to explain the jaw patterning process (Depew & Compagnucci, 2008). Genes such as Satb2, expressed in the “caps” located near the distal midline of the mandibular process of the first arch and the lambdoidal junctions where the frontonasal prominence meets the maxillary process, are important for the coordination and evolution of the jaws (Depew & Compagnucci, 2008). This model explains some of the similarities between the maxilla and mandible during development. However, how the distinct identities of the upper and lower jaws are established at this early patterning stage is a question still needing to be answered. Endothelin signaling-mediated expression of distal-less genes is critical for establishing the difference between the maxilla and mandible. During PA1 patterning, Endothelin 1 is expressed in the ectoderm at the distal end of the mandibular process whereas Endothelin receptor A (Ednra) is expressed exclusively in the mesenchyme with an intensity gradient from the distal to the proximal region, suggesting that endothelin signaling mediates mandible patterning through epithelial-mesenchymal interaction. Loss of Ednra results in a homeotic transformation of mandible to maxilla, supporting its important function in mandible identity establishment (Sato et al., 2008). Molecularly, Dlx5/6 expression is downregulated in the mandibular processes of Ednra mutant mice (Ruest, Xiang, Lim, Levi, & Clouthier, 2004). Dlx5/6−/− mice show a similar phenotype, suggesting that Dlx5/6 are downstream transcriptional regulators of endothelin signaling (Depew, Lufkin, & Rubenstein, 2002). Six1, which is expressed on the oral side of both the maxillary and mandibular processes, negatively regulates endothelin signaling to maintain maxillary identity. In Six1−/− mice, ectopic endothelin signaling is found in the proximal end of first pharyngeal arch, which leads to the formation of a cartilage-capped, rod-shaped bone at the zygomatic arch (Tavares, Cox, Maxson, Ford, & Clouthier, 2017). Recently, using a zebrafish model Barske and colleagues found that Nr2f nuclear receptor plays a crucial role independent of endothelin signaling in the patterning of the upper jaw (Barske et al., 2018). This finding broadened the general consensus regarding patterning of the maxilla and mandible and strongly suggests that multiple signaling pathways contribute to inter-pharyngeal arch patterning.
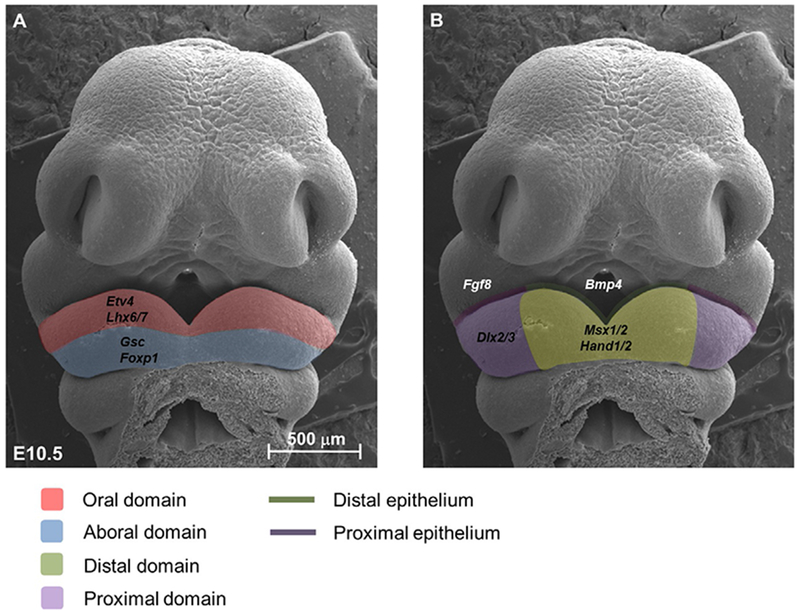
Within the mandibular process of the first pharyngeal arch, the CNCC-derived mesenchyme is patterned along the proximal-distal and oral-aboral axes (Chai & Maxson, 2006). During this process, mesenchymal cells receive signals secreted from mandibular ectoderm, activating downstream patterning genes. Major signaling pathways such as Bmp, Fgf, Shh and Wnt form a complex regulatory network to control the establishment of different domains (Fig. 1). Fgf8 is expressed at the proximal end of the mandibular process epithelium, activating multiple patterning genes expressed in the oral mesenchyme, including two specific Lim-homeobox domain genes, Lhx6 and Lhx7. Meanwhile, Bmp4 expressed in the distal epithelium antagonizes Fgf signaling to establish the distal domain (Fig. 1B). The genes Msx1 and Alx, which are expressed in the distal mesenchyme, have also been identified as downstream targets of Bmp signaling. Wnt signaling overlaps with Bmp signaling, and loss of Wnt signaling by knocking out R-spondin2 (Rspo2), a canonical Wnt signaling activator, results in down-regulation of both Bmp and Fgf signaling, suggesting a tight signaling interaction between the proximal and distal domains (Jin, Turcotte, Crocker, Han, & Yoon, 2011). Shh ligand, which is expressed in the pharyngeal endoderm during patterning, is also a survival factor for CNC-derived mesenchymal cells. Proximal Shh signaling is downstream of Wnt signaling in the distal domain, whereas both Wnt and Shh signaling are mediated by epithelial Islet expression (Li, Fu, et al., 2017). Tucker and colleagues have demonstrated the biological significance of proximal-distal domain establishment by blocking Bmp signaling at the distal end of the mandibular process. They found a transformation of tooth identity from incisor to molar with ectopic expression of Barx1 in the distal mesenchyme (Tucker, Matthews, & Sharpe, 1998).
Fig. 1.
Patterning of the first branchial arch. Frontal view of a scanning electron microscopic image of an embryonic day (E) 10.5 mouse embryo shows that the first branchial arch can be divided into oral/aboral domains (A) and proximal/distal domains (B). In the oral domain, Fgf signaling from the proximal oral epithelium regulates the expression of Etv4 and Lhx6/7, which prevent the expansion of Gsc expression in the aboral domain. Bmp4 is expressed in the distal oral epithelium, regulating the expression of Msx1 and Msx2.
Unlike the proximal-distal axis, the regulatory mechanism of oral-aboral patterning remains elusive. Fgf signaling is certainly critical for multiple genes expressed in the oral domain of the CNC-derived mesenchyme. Specifically, Fgf8 is expressed in the proximal oral ectoderm of both the maxillary and mandibular processes, and its downstream read-out, Etv4, is expressed in the oral half of the mandibular mesenchyme. Lhx6/7, which is directly regulated by Fgf8, is also expressed in the oral mesenchyme (Fig. 1A). Interestingly, Fgf8 has also been reported to positively regulate goosecoid (Gsc) expression, which is present in the Lhx6/7-negative aboral region (Tucker, Yamada, Grigoriou, Pachnis, & Sharpe, 1999). Endothelin 1 (Et-1) is another signaling molecule that might control oral-aboral axis establishment. Et-1 is expressed in the aboral ectoderm of the mandibular process. Multiple studies have shown that endothelin signaling is important for restricting oral mesenchyme gene expansion into the aboral side. For example, endothelin can directly induce Gsc expression and negatively regulate Six1 expression (Tavares et al., 2017; Tucker et al., 1999). The biological function of oral-aboral axis patterning is still not very clear. Interestingly, both Gsc and Lhx6/7 knockout mice have shortened mandibles; however, Gsc mutant mice show a defect at the proximal end whereas Lhx6/7 mutant mice have a defect at the distal end (Denaxa, Sharpe, & Pachnis, 2009; Rivera-Perez, Wakamiya, & Behringer, 1999). This suggests that there might be a dynamic interaction between proximal-distal and oral-aboral axis patterning.
3. Jaw bone development
3.1. Meckel’s cartilage
After PA1 is patterned and before any ossification center starts to form, a group of CNC-derived mesenchymal cells condense and differentiate into chondrocytes to form a pair of symmetric, rod-shaped cartilages named Meckel’s cartilage (MC). The MCs elongate along the dorsal-ventral axis and fuse at their distal ends to form the mandibular symphysis. Most mammalian cartilages ossify and become bony structures, although some retain their cartilaginous character, such as the tracheal, nasal, and articular cartilages. However, the MC has a more complex fate. Both its distal and proximal extremities undergo ossification to form part of the anterior portion of the mandible bone as well as the incus and malleus bones of the middle ear. The intermediate part of MC undergoes dedifferentiation to become fibrous tissue.
Concerning the functional significance of MC, the consensus is that the presence of MC is indispensable for mandibular development, because it serves as a template for mandibular formation. Mutant mouse models that have abnormal MC formation often show later mandibular defects as well (Li, Fu, et al., 2017; Matsui & Klingensmith, 2014; Yahiro, Higashihori, & Moriyama, 2017). However, the molecular mechanisms and interactions that regulate MC are still largely unknown. Numerous genes and signaling pathways have been reported to relate to the formation and degradation of the MC. Like other cartilaginous tissues, MC formation is also mediated by Sox9, which is a well-known chondrogenic transcriptional regulator expressed in chondroblasts and mature chondrocytes. Loss of Sox9 in the CNCC lineage inevitably affects the formation of MC, causing complete absence of MC during the entire course of mandibular development (Mori-Akiyama, Akiyama, Rowitch, & de Crombrugghe, 2003). Interestingly, although the mandibular bone is severely defective in these mutant mice, early osteogenic markers are found ectopically expressed in the craniofacial region at E15.5. This study suggests that the presence of MC is necessary but not sufficient for mandibular bone development.
Shh signaling is also critical for MC induction. Targeted deletion of Shh expression in these tissues leads to an increase of cell death in the mandibular arch and complete absence of MC (Billmyre & Klingensmith, 2015). This phenotype is also associated with severe mandibular defects. The importance of Shh signaling for MC formation is also revealed by other experiments. Ablation of Islet expression in oral epithelium results in micrognathia with defective MC morphology. Hh signaling is downregulated in these mutants and the mandible phenotype can be partially rescued using Isl1Shh-Cre; Tg-pmes-Ihh compound mice, which overexpress transgenic Ihh (Li, Fu, et al., 2017).
During degradation of the MC, autophagy and chondrocyte apoptosis play crucial roles. Starting from E15, Beclin1, a central regulator of autophagy, can be detected in prehypertrophic and hypertrophic chondrocytes located in the central portion facing the proximal end of the incisor teeth. LC3 and Caspase 3 expression is detectable at a slightly later stage in the same location, which suggests that autophagy occurs prior to hypertrophic chondrocyte cell death; this is the final fate of the majority of MC (Yang, Zhang, Liu, Zhou, & Li, 2012). Bmp signaling is involved in the degeneration of MC. In Noggin−/− mutant mice, in which Bmp signaling is over-activated, MC is significantly thickened due to elevated cell proliferation and remains in an unossified state at the caudal end at E18.5. With sustained Bmp signaling, the middle portion of MC fails to degrade and undergoes endochondral ossification to form mandibular bone (Wang, Zheng, Chen, & Chen, 2013). Recently, epigenetic regulation, specifically histone methylation, has been revealed as another factor that affects MC degradation. In the absence of Setdb1, an enzyme that methylates the lysine 9 residue of the histone H3 protein (H3K9), the MC develops a similar phenotype to that of Noggin−/− mutants. Enlargement and persistence of MC are found with over-activated Bmp signaling (Yahiro et al., 2017). Because methylation of the lysine residues of histone H3 negatively regulates gene expression, this study indicates that the withdrawal of Bmp signaling from MC during degeneration is controlled by Setdb1-mediated histone methylation and is required for normal mandibular development.
3.2. Mandibular bone osteogenesis
Both intramembranous and endochondral ossification contribute to the formation of mandibular bone (Lee et al., 2001). The majority of the mandible (the intermediate portion) is ossified in an intramembranous fashion, which is characterized by mesenchymal stem cells initially proliferating and forming a small, dense cluster. These stem cells then undergo differentiation into osteoblasts with an associated morphological change from spindle-shaped to columnar. Meanwhile, the osteoblasts create an extracellular matrix called osteoid tissue, which contains Type-I collagen fibrils and is able to bind calcium salts (Amano et al., 2010). Finally, the osteoid tissue mineralizes to form rudimentary bone tissue with mature osteocytes in the middle and active osteoblasts at the osteogenic front. During this process, a series of well-defined osteogenic markers are expressed at each stage of differentiation. Dlx5 has been identified as one of the earliest markers to be expressed in committed osteoprogenitor cells. Dlx5 also induces Runx2, which is a master regulator for activating the program of osteoblastogenesis and is also expressed in early committed osteoprogenitor cells (Kawane et al., 2014). Runx2-deficient mice completely lack bone tissue due to the arrested differentiation of osteoblasts. Interestingly, cartilage formation in these mutants is only mildly affected, notably including MC, again suggesting that the presence of MC is not sufficient for mandibular bone development (Shibata et al., 2004). Alp and Osterix (Osx) are two other factors that are involved in the later stages of osteogenesis. They are expressed in the differentiated osteoblasts and are downstream ofRunx2. Loss of Osx in CNCC derivatives leads to the absence of almost all craniofacial skeletal structures, suggesting that Osx is required for craniofacial bone formation by CNC-derived cells (Baek et al., 2013).
The distal-most region of the mandibular bone is ossified through endochondral ossification, the other essential bone-forming process that occurs during embryonic development. The onset of this process is similar to chondrogenesis, in which a group of mesenchymal cells condense and differentiate into Sox9-positive chondrocytes to secrete collagen types II, IX, and XI and aggrecan. Then these chondrocytes undergo maturation from a proliferative stage to a hypertrophic stage and eventually undergo apoptosis due to a drastic change in the micro-environment, leaving the cartilaginous remnants as the scaffold for the osteoblasts laying down bone matrix. Recently, lineage tracing studies have enabled us to observe the fate of these hypertrophic chondrocytes (HCs) directly, and there is some recent evidence showing that these HCs may undergo transdifferentiation and continue with a new role in the osteoblast lineage. Col10a1-Cre can specifically target HCs, including late HCs that express Mmp13 and Osx. By crossing these mice with a reporter line, it has been found that HC-derived cells become Col1a1-expressing osteoblasts and sclerostin (Sost)-expressing osteocytes during bone tissue development as well as in bone injury repair (Yang, Tsang, Tang, Chan, & Cheah, 2014). Many major signaling pathways are associated with endochondral ossification, including Bmp, Hh, Wnt, Notch, and retinoic acid signaling. In the distal mandible, Ihh-null mice show reduced chondroprogenitor cell proliferation that results in altered endochondral ossification and an abnormal mandibular symphysis. This phenotype is partially rescued by ablation of Gli3 expression, which is a negative regulator of symphyseal development (Sugito et al., 2011). Golgi-associated N-sulfotransferase 1 (Ndst1) also regulates mandibular symphysis development. Ndst1 catalyzes sulfation of heparan sulfate proteoglycan (HS-PG) glycosaminoglycan chains, which are found on the cell surface as well as in the extracellular matrix and mediate numerous developmental processes. Ndst1-null mice have severe craniofacial skeletal defects including a fused mandibular symphysis due to ectopic osteogenesis at the newborn stage (Yasuda et al., 2010). Ihh signaling was also found to be expanded during the ossification of the mandible in these mice, which might contribute to the up-regulation of Osterix and collagen I expression seen in these mutants.
The proximal end of the mandible is composed of three eminences, namely the coronoid, condyle, and angular processes. At the osteogenic front, unlike in the primary cartilage, progenitor cells express both osteogenic and chondrogenic markers, such as Runx2, Osterix, and Sox9. Since these cells have the potential to differentiate into either osteoblasts or chondrocytes, they are called osteochondroprogenitor cells. During the fate determination of these progenitor cells, Tgf-β signaling is critical. Conditional inactivation of Tgfbr2 in CNCCs leads to increased osteoprogenitor differentiation and disrupted chondrogenesis in the proximal region of the mandible. Enhanced Col I expression and weakened Sox9 expression are found in the same region, suggesting that osteochondroprogenitor cells lean toward the osteogenic rather than chondrogenic lineage. Moreover, by ablating Dlx5, which is an early osteogenic regulator, the mandibular phenotype of these mutants can be partially rescued (Oka et al., 2007).
Ihh signaling is also required in temporomandibular joint (TMJ) formation and condyle growth. In Ihh-null mice, TMJ development is severely compromised. Condylar cartilage growth, polymorphic cell proliferation, and PTHrP expression are all inhibited in these mice. This phenotype can be partially reversed by ablation of Gli3, a natural inhibitor of Hh signaling (Shibukawa et al., 2007).
3.3. Hemifacial microsomia
Hemifacial microsomia (HFM) is a common congenital defect that has an incidence ranging from 1:3500 to 1: 5600 (Hartsfield, 2007). It is primarily characterized by unilateral hypoplasia of the mandible and ear; other craniofacial malformations sometimes associated with HFM include facial palsy, cleft lip/palate and orbital defects. Three possible hypotheses have been raised to explain HFM: vascular abnormality and hemorrhage, disrupted development of MC, and abnormal development of CNCCs (Chen, Zhao, Shen, & Dai, 2018). Small molecule drugs such as thalidomide can cause local hemorrhage or vascular abnormalities, and accordingly, Poswillo established an animal model that mimics the phenotypes of HFM using triazene and thalidomide treatment (Poswillo, 1973). However, the exact mechanism that leads to the HFM phenotypes is still unclear. MC is closely associated with the formation of the mandible and middle ear. Disruption of the development of MC often leads to mandibular hypoplasia. A recent study showed that loss of VEGF expression in the CNCCs impairs blood vessel growth, leading to insufficient blood supply to MC and ultimately causing mandibular hypoplasia (Wiszniak et al., 2015). This study revealed the internal connections among these three possible pathogenic mechanism models and provides insight into potential prevention and treatment strategies for HFM.
3.4. Quantitative analysis using dynamic imaging and anatomical landmarks
Traditionally, researchers have relied on regular histology and whole-mount skeletal staining to document normal and abnormal craniofacial development. Recently, microCT imaging utilizing defined anatomical landmarks has made it possible to analyze defects quantitatively in the mandible and maxilla. Based on the landmarks that have been established with morphometrics, we are able to identify nuanced differences between normal and abnormal skeletal growth, which provides a basis for understanding the localized and overall influence of mutations associated with disease (Ho et al., 2015). Percival and colleagues systematically analyzed the craniofacial skeletal structures of control and Fgfr2+/P253R mice at multiple embryonic stages. They found that certain bones are significantly reduced in volume in the mutants, while others are not. Interestingly, they also found that the density of bone tissues formed through intramembranous and endochondral ossification differ during development (Percival, Huang, Jabs, Li, & Richtsmeier, 2014).
To uncover maxillary and mandibular phenotypes that are uniquely associated with specific mutant models, precise measurement and comparison between microCT data of different samples are required. FaceBase, which is a collaborative NIDCR-funded consortium, has developed a web-based platform (available at facebase.org) that allows users to rotate and view each facial bone in any position. One can select any facial bone and view its anatomical landmarks based on Mouse Development (Rossant & Tam, 2002), and also calculate the distance between any two anatomical landmarks. These measurements can serve as the basis for evaluating normal/abnormal facial bone development.
4. Tooth development
4.1. Early interaction between odontogenic ectoderm and ectomesenchyme
Similar to mandible patterning, odontogenic signaling for tooth initiation also relies on epithelial-mesenchymal interactions. As early as E10, Bmp4 expression is restricted to the oral ectoderm where the incisors will form. By the time of tooth initiation, Bmp4 expression switches from the epithelium to the underlying mesenchyme corresponding to the condensation beneath the epithelial thickening. In the later bud and cap stages, Bmp4 expression is restricted to the tooth germ. Msx1, one of the downstream targets of Bmp4, is also expressed in the CNC-derived ectomesenchyme (Vainio, Karavanova, Jowett, & Thesleff, 1993). Interestingly, Msx1 can also positively regulate Bmp4 expression, and this feedback loop is critical for tooth initiation and morphogenesis (Chen, Bei, Woo, Satokata, & Maas, 1996). Msx1 null mice show an arrest of molar tooth development at the bud stage with reduced Bmp4 expression (Satokata & Maas, 1994). This data suggests that Msx1 is not only expressed in response to signals from the dental epithelium, but also regulates downstream target genes to provide feedback to the dental epithelium.
Pax9 is another transcription factor that is essential for tooth initiation. At E11, it is also expressed in the ectomesenchyme corresponding to the future dental mesenchyme condensation. Pax9-deficient mice experience arrest of tooth development at the bud stage with reduced mesenchymal cell condensation. Bmp4, Msx1 and Lef1 all are downregulated in these mutant mice (Peters, Neubuser, Kratochwil, & Balling, 1998). Pax9 expression can be induced by Fgf8 secreted from the oral epithelium, and Pax9 expression is downregulated in Fgf8−/− mice (Neubuser, Peters, Balling, & Martin, 1997). Interestingly, Fgf8 cannot induce Bmp4 expression or vice versa. Instead, they work antagonistically to establish incisor and molar formation domains (Chai & Maxson, 2006).
Shh expression in the tooth-forming regions of PA1 starts at E11.5. Shh expression is highly restricted to the epithelial thickening of the future tooth germ (Bitgood & McMahon, 1995). Shh signaling is responsible for the localized proliferation of the epithelial tissue, which in turns invaginates into the underlying mesenchyme to form a tooth bud. The localization of Shh expression is accomplished through antagonism of Wnt7b expression in the non-tooth-forming epithelium (Sarkar et al., 2000). Ectopic application of Shh protein to the non-dental oral ectoderm of E10.5 mandible explants leads to formation of multiple ectopic epithelial invaginations after 3 days in culture. Moreover, inhibition of Shh signaling in a mandible culture with blocking antibody results in failed tooth bud formation with reduced cell proliferation and increased apoptosis after 3 days of culture, again suggesting that Shh regulates epithelial cell proliferation and survival in the developing tooth germ (Cobourne, Hardcastle, & Sharpe, 2001). In vivo studies using transgenic mouse models have also demonstrated the importance of Shh for tooth development. Loss of Shh in the oral epithelium (K14-Cre;Shhfl/fl) severely affects tooth development, including retardation of tooth growth, abnormal placement of teeth in the jaw, and disrupted tooth morphogenesis. Interestingly, Shh can also regulate the pattern of developing cusps; the lingual side of the tooth is more severely affected than the buccal side when it is disrupted (Dassule, Lewis, Bei, Maas, & McMahon, 2000).
4.2. Signaling regulating dentin and enamel formation
In the late bell stage, mineralized dentin and enamel tissues are formed by odontoblasts from the dental mesenchyme and ameloblasts from the dental epithelium. The formation of dentin and enamel takes place at the interface between the mesenchyme and epithelium and is regulated by multiple signaling pathways through tissue-tissue interactions. The enamel knot, which lies at the tip of the future cusp, serves as a signaling center that mediates the differentiation of odontoblasts and ameloblasts (Fig. 2). More than 10 signaling molecules belonging to the BMP, FGF, Hh, and Wnt families are expressed in the primary enamel knot to guide not only dentin and enamel formation, but also cusp patterning during tooth morphogenesis (Jussila & Thesleff, 2012).
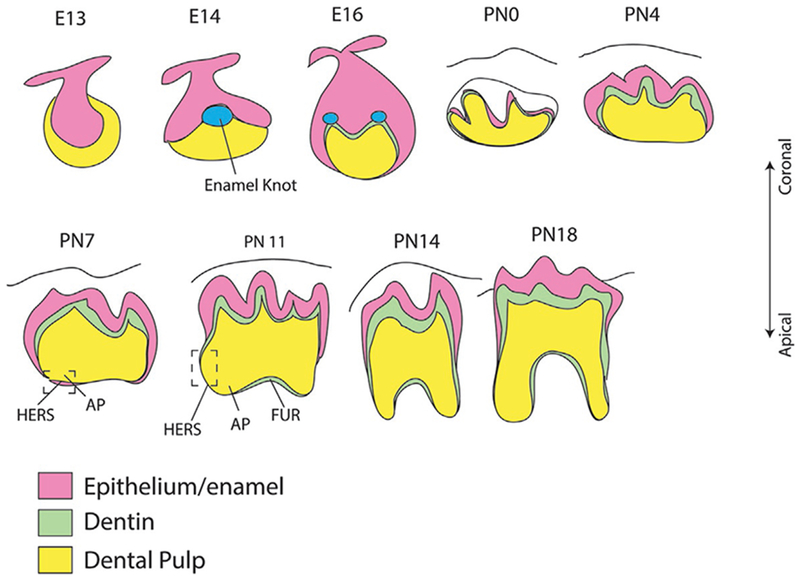
Fig. 2.
Scheme of tooth development. Tooth development begins with the evagination of the epithelium into the underling mesenchyme to form the tooth bud. During crown formation, enamel knots serve as the signaling center to mediate the differentiation of odontoblasts and ameloblasts, and the patterning of the cusps. Starting from postnastal day 3.5, bilayered HERS grows apically and guides the tooth root development. HERS, Hertwig’s epithelial root sheath; AP, apical papilla; FUR, furcation.
Even though the search for master transcriptional regulators for odontogenesis is still ongoing, the importance of Bmp signaling has been well characterized. Multiple growth factors in the Bmp family have been shown to induce terminal differentiation of odontoblasts in vitro (Nakashima, 1994; Tasli, Aydin, Yalvac, & Sahin, 2014; Zhu et al., 2018). Loss of Smad4, a common mediator of Bmp and Tgf-β signaling, in the CNC-derived mesenchyme leads to defects in odontoblast differentiation and formation of ectopic bone-like structure in the dentin-forming region. More interestingly, despite the defect in dentin formation, enamel formation appears unaffected in these mutant mice, suggesting that functional odontoblast differentiation is not required for ameloblast differentiation (Li et al., 2011).
There is also evidence showing that Wnt signaling is associated with dentinogenesis. Wnt10a is specifically expressed in the primary and secondary enamel knots. In addition, Axin2, the canonical Wnt signaling transducer and read-out, is expressed in developing odontoblasts and dental pulp cells. Both loss- and gain-of-function of Wnt signaling in early odontoblasts have been studied using OC-Cre;WlsCO/CO and OC-Cre;Catnblox(ex3)/+ mutant mice, respectively. Loss of Wnt signaling through inactivation of Wntless (Wls) leads to compromised odontoblast maturation, down-regulation of the terminal differentiated odontoblast marker Dspp, and reduced dentin thickness, whereas over-activation of Wnt signaling through overexpression of β-catenin results in excessive dentin and cementum formation, which may be due to prematurely differentiated odontoblasts (Bae et al., 2015; Kim et al., 2011). These studies collectively indicate that temporo-spatial regulation of Wnt/beta-catenin signaling is essential for normal odontoblast differentiation and dentin formation.
Bmp signaling, which is one of the few signaling pathways that transmits bidirectional signals between epithelial and mesenchymal tissues, also regulates ameloblast differentiation and enamel formation. Ablation of Bmp2 in odontoblasts and dental pulp cells using Osx-Cre;Bmp2fl/fl results in a severe phenotype of both incisors and molars with a thin, hypomineralized enamel layer (Feng, Yang, et al., 2011). Two factors downstream of Bmp signaling, Runx2 and Osterix, are also related to ameloblast differentiation. In vitro culture of mouse ameloblast lineage cells (mALCs) revealed that Runx2 physically interacts with Fam50a to increase its binding affinity to the ameloblastin (Ambn) promoter (Kim et al., 2018). Tooth morphogenesis initially progresses normally in Osterix null mice; however, markers of mature ameloblasts (Enam, Amelx, Mmp20, Amtn, Klk4) have limited expression in incisors and molar tissues of these mice, and they lack enamel matrix (Bae et al., 2018).
Proper ion exchange is critical for the demineralization and remineralization processes during dentin and enamel formation because it regulates and maintains the required calcium and pH homeostasis. Multiple human syndromes and diseases including Timothy syndrome, Olmsted syndrome and osteopetrosis are associated with defects in enamel and dentin formation with impaired biomineralization. Patients with these conditions have mutations in genes encoding various ion channel-related proteins, such as CACNA1, TRPV3, CLCN7 and AE2 (Duan, 2014). ClC-5 functions as a Cl−/H+ exchanger and plays an important role in pH regulation. ClC-5 knockout mice have abnormal dentin, similar to the characteristics of dentinogenesis imperfecta in humans, possibly due to an overexpression of Tgf-β signaling (Duan et al., 2009). The cystic fibrosis transmembrane conductance regulator (CFTR), which is a transporter-class ion channel, also regulates Cl− exchange. CFTR is expressed in ameloblasts during amelogenesis. CFTR-ΔF508 pigs have hypomineralized and visibly disorganized enamel tissue, and a similar phenotype is observed in Cftr-deficient mice, with soft, chalky white incisor enamel that degenerates shortly after completion of the secretory phase of amelogenesis (Lacruz et al., 2012).
4.3. Tooth root development
Tooth root development is mainly driven by two cell populations: the dental epithelium-derived HERS and the CNC-derived dental pulp stem cells. The HERS contains a transient cell population that serves as a signaling center and provides different types of factors that trigger tooth root elongation (Fig. 2). Dental pulp stem cells can be identified using Gli1 in the apical portion of the dental papilla at postnatal day 3.5 (P3.5), and then later take up permanent residence in the apical region of the elongated tooth root (Feng et al., 2017; Liu et al., 2015). Lineage tracing studies showed that these cells proliferate and populate the entire tooth root mesenchyme during development. A large body of research has shown that disruption of the HERS or dental pulp stem cells through targeted genetic modification disturbs the development of the tooth root. An array of growth and transcription factors has been uncovered based on their expression patterns, and multiple mutant animal models with tooth root defects have been generated (Li, Parada, & Chai, 2017).
During HERS formation, Bmp signaling is activated in both the dental epithelium and mesenchyme. Recent studies have shown that a Bmp-Smad4-Shh-Gli1 signaling network regulates the fate of the transient dental epithelial stem cells, which are Sox2 + , in the mouse molar (Li et al., 2015). Specifically, loss of Bmp signaling in the dental epithelium leads to an expansion of Shh signaling, which in turn causes the maintenance of the cervical loop structure and retention of Sox2 + dental epithelial stem cells postnatally. As a result, HERS formation is delayed and tooth root development is arrested. Interestingly, loss of Shh ligand expression in the dental epithelium rescues this tooth root phenotype, suggesting that the Bmp/Shh signaling cascade is critical for HERS formation and root development. Consistent with this, ablation of Msx2, which is a direct downstream target of Smad-mediated Bmp signaling, also leads to shortened molar roots (Aioub et al., 2007). As mentioned above, multiple Bmp ligands including Bmp2, 3, 4 and 7 as well as phosphorylated Smad 1/5/8, which indicates activation of Bmp signaling, are expressed in the dental mesenchyme, implying that mesenchymal Bmp signaling plays a role in tooth root development. Indeed, using Gli1-CreERT;Bmpr1afl/fl mice to specifically knock out Bmp type I receptor in dental pulp stem cells results in impaired tooth root formation. An odontoblast differentiation defect is also observed in these mice, as indicated by loss of Klf4 expression in the pre-odontoblast region; Klf4 may serve as a switch for regulating odontoblast differentiation (Feng et al., 2017).
Evidence shows that an Nfic/Hh signaling cascade also regulates tooth root development. Nfic−/− mice show a tooth root defect and down-regulation of Hh signaling attenuator Hhip. Therefore, these mice have an expansion of Hh signaling similar to that of KRT14-rtTA;tetO-Cre;Smad4fl/fl mutant mice. Treatment of Nfic−/− mice with Hh inhibitor partially rescues cell proliferation and root morphology (Liu et al., 2015). These data suggest that the proper regulation of Hh signaling and activation through the Bmp/Nfic/Hh network is critical for tooth root development.
4.4. Tooth and jaw bone interaction
Teeth and jaw bones have a common origin: they both arise from the first pharyngeal arch and they develop in closely related ways. Multiple mouse models show that mutations affecting early mandible development also have an impact on tooth formation (Denaxa et al., 2009; Peters et al., 1998; Satokata & Maas, 1994). Msx1 is a critical patterning gene expressed in the distal half of the mandibular arch during early stages of tooth development. Later, Msx1 is strongly expressed in the dental mesenchyme. Msx1−/− mice exhibit both mandibular abnormalities and tooth defects. The overall length of the mandible is slightly shorter and the alveolar ridge is absent in these Msx1 mutant mice, and tooth development fails to progress past the bud stage (Satokata & Maas, 1994). This phenotype suggests that Msx1 is not only needed for the differentiation of dental follicle cells into alveolar bone osteoblasts, but also required as feedback to the epithelium for progression of the tooth bud to the cap and bell stages. Similarly, Pax9 is also expressed in the mandibular arch at the early patterning stage and in the dental mesenchyme. In Pax9-deficient mice, alveolar bones and coronoid processes are missing and tooth development stalls at the bud stage. Interestingly, Msx1 and Pax9 not only show closely overlapping expression patterns, but also physically interact with each other to guide tooth formation (Ogawa, Kapadia, Wang, & D’Souza, 2005).
The teeth and jaw bones also interact through mechanical force. After tooth extraction, a reduction of the alveolar ridge is commonly observed. One explanation for this bone resorption is that the forces on the bone are reduced after tooth loss so that less bone is needed (Hansson & Halldin, 2012). Clinically, a bone allograft in combination with a membrane is used to improve the ridge dimensions in these patients who will require dental implants to restore their dentition (Iasella et al., 2003).
4.5. Dental stem cells
Unlike mouse molars and all human teeth, mouse incisors grow continuously throughout the lifetime of the animal due to adult stem cells residing within the tissue. These stem cells undergo self-renewal and maintain the homeostasis of the dentin and enamel. Harada and colleagues first identified the stellate reticulum, which is located in the cervical loop area, as the putative site of dental epithelial stem cells (Harada et al., 1999). Starting from E14.5, Sox2 expression is found in the labial cervical loop of the mouse incisor, and from E16.5 to E18.5, Sox2 expression is more restricted to the proximal tip of the cervical loop, which corresponds to the location of the putative epithelial stem cell population. Lineage tracing showed that Sox2 positive cells contribute to all epithelial lineages of the tooth (Juuri et al., 2012). This differentiation process involves a group of multipotent progenitor cell progeny called transit-amplifying (TA) cells, which are adjacent to the stem cell population. Notch signaling has been reported to be critical for stem cell maintenance as well as fate determination. Notch1, Notch2, and lunatic fringe, a Notch homolog found in Drosophila, are all expressed by cervical loop epithelial cells (Harada et al., 1999). In explant culture, inhibition of Notch signaling using DAPT leads to a reduction of cell proliferation and increased apoptosis in the epithelial stem cell niche (Felszeghy, Suomalainen, & Thesleff, 2010). Signals from the mesenchyme play a role in the self-renewal and differentiation of the stem cell population in the incisor epithelium. Fgf3 is expressed in the mesenchyme underlying the cervical loop region. Fgf10-expressing cells partially overlap with the population that expresses Fgf3 and surround the whole cervical loop epithelium, while Fgfr1b is strongly expressed in the basal epithelial cells and stratum intermedium, suggesting that Fgf signaling might regulate the continuous growth of the incisor epithelium. Interestingly, Fgf3-deficient mice have relatively normal tooth development, which may be due to compensation by Fgf10. In Fgf3−/− ;Fgf10+/− compound mutant mice, the lower incisors are shorter and frequently broken. They also have hypoplastic morphology of the cervical loop and either very thin or missing enamel (Wang et al., 2007). In addition, even though Fgf3 is not required for early cervical loop morphogenesis, its asymmetric expression pattern may be important for the difference in size between the labial and lingual portions of the cervical loop. Fgf3 induces cell proliferation in the incisor epithelium. Consistent with this, Follistatin−/− mice exhibit an enlarged lingual cervical loop along with ectopic expression of Fgf3 in the lingual dental mesenchyme underlying the epithelium (Wang et al., 2004).
Recent studies have revealed the identity of mesenchymal stem cells (MSCs) in mouse incisors. When dental pulp-derived cells are cultured, a group of cells with MSC characteristics such as clonogenic, multi-lineage differentiation and expression of defining markers such as CD90, CD73, and CD105 can be rapidly isolated. Despite this, it took more than 10 years to identify the in vivo location of this heterogeneous cell population. Feng and colleagues first showed that NG2-labeled pericytes contribute to odontoblasts during growth as well as after damage to the dental pulp (Fig. 3) (Feng, Mantesso, De Bari, Nishiyama, & Sharpe, 2011). However, in both cases, pericyte-derived odontoblasts only account for 15% of the whole population, suggesting that another source of MSCs may contribute the majority of odontoblasts and dental pulp. In addition, NG2+ pericytes do not contribute to incisor homeostasis, suggesting that different stem cell populations are primarily responsible for tissue homeostasis and injury repair.
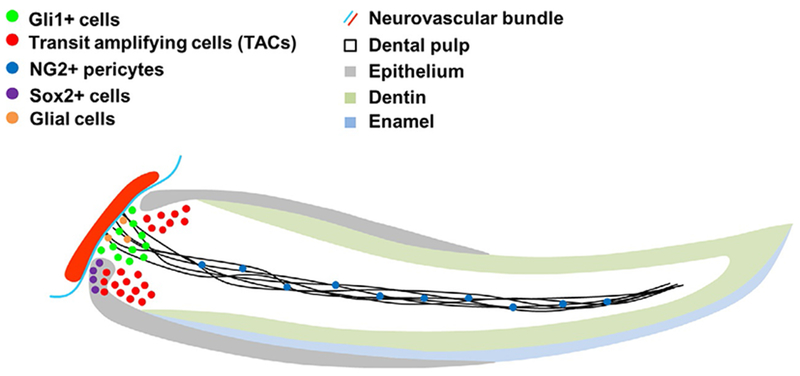
Fig. 3.
Stem cell population in the mouse incisor. In the labial cervical loop, Sox2+ epithelial stem cells are responsible for replenishing the enamel and epithelial tissue. Gli1+ and glial cells residing in the mesenchyme between the labial and lingual cervical loop regions close to the neurovascular bundle represent slow-cycling stem cells. Transit amplifying cells located either in the mesenchyme close to the labial and lingual cervical loop region or in the labial cervical loop are derived from self-renewing stem cells. NG2+ pericytes located on the abluminal surface of endothelial cells contribute to mesenchymal tissue repair after injury.
Since this initial finding, peripheral nerve-associated glia were identified as another source of dental MSCs (Fig. 3). Using two different ERT2-Cre drivers targeting glial cells (Plp1 and Sox10) combined with reporters, lineage tracing studies showed that these cells can contribute to the odontoblast population during incisor growth, homeostasis, and injury repair. However, depending on the dosage of tamoxifen injection given to these mice, the amount of Schwann-cell-derived progeny varies from 4% to 47%, again suggesting the existence of another source of MSCs (Kaukua et al., 2014).
General consensus holds that the most proximal end of the mouse incisor, namely the mesenchyme between the lingual and labial epithelial cervical loops, serves as a stem cell niche that supplies new cells for replacement of tissue loss due to occlusion and abrasion. This model was confirmed through identification of a small group of label-retaining cells detectable in this location after a 4-week chase period. These Gli1 + cells surround the neurovascular bundle and receive Shh signal from the sensory nerve (Fig. 3) (Zhao et al., 2014). After 4 weeks of lineage tracing, the typical turn over time for odontoblasts and ameloblasts in mouse incisors, almost 100% of odontoblasts and pulp cells are derived from these Gli1 + cells. Interestingly, these Gli1 + cells do not express surface markers that define MSCs in vitro, such as CD105, CD146, and Sca1, which are highly expressed in the NG2 + pericytes that are derived from these Gli1 + cells. This finding will have an important impact on the definition and identification of MSCs in vivo (Zhao et al., 2014).
5. Stem cells and regenerative therapies
5.1. Mandibular distraction osteogenesis, growth factors, and stem cell treatment
Mandibular hypoplasia is one of the most common congenital malformations, and can be either non-syndromic or associated with other anomalies, as in Pierre Robin sequence or Marfan syndrome. Surgical intervention is required in many cases due to the breathing and swallowing difficulties caused by posterior tongue displacement and the resulting airway compromise. Mandibular distraction osteogenesis (MDO) is the current standard treatment for micrognathia and proceeds in three stages: (i) the initial latency stage, which starts after an osteotomy is created, allowing the initial healing and callus formation; (ii) the activation stage, during which the ends of the bone are gradually moved apart, allowing new bone to form in the gap; and (iii) the consolidation stage, once the mandible reaches the optimal length, allowing for the final maturation of the newly formed bone.
Different growth factors, stem cells, and adjuvant procedures have been tested in an attempt to promote the healing process during distraction osteogenesis in several animal models. Multiple bone morphogenetic proteins including Bmp2, Bmp4, and Bmp7 enhance bone volume, remodeling, and mature bone formation in both rat and rabbit models (Mizumoto, Moseley, Drews, Cooper, & Reddi, 2003; Yonezawa, Harada, Ikebe, Shinohara, & Enomoto, 2006). Nerve growth factor induces bone formation around the regenerating axons and improves mechanical strength and histomorphometric outcomes in a rabbit model (Cao et al., 2012). Insulin-like growth factor appears to increase the mineral deposition rate, suggesting a positive anabolic effect (Stewart et al., 1999).
Bone marrow-derived mesenchymal stem cells (BMMSCs) undergo osteogenic differentiation following stimulation from certain biological signals and are relatively easy to harvest and amplify in culture. In several MDO models, BMMSC-treated groups showed significant higher radio-density and bone volume and thickness in the early consolidation stage (Aykan et al., 2013; Kim, Cho, Lee, & Hwang, 2013; Ma et al., 2013). In another study, stromal cell-derived factor-1 (SDF-1) was shown to facilitate migration of MSCs in vitro and in vivo. In a rat model of MDO, the recruitment of endogenous MSCs to the injury site was significantly enhanced by SDF-1 treatment, which provides a new insight into how manipulation of endogenous MSCs could be used to enhance the bone healing process (Cao et al., 2013).
5.2. Dentin repair and regeneration
Following an injury or lesion in a tooth, odontoblasts in and around the injury site are damaged and the tooth is at risk of infection. In this case, isolated pericytes in the dental pulp and glial cells close to the injury site undergo rapid proliferation and differentiate into odontoblast-like cells that generate reparative dentin to protect the exposed pulp (Pang Yvonne et al., 2015). At the same time, stem cells residing in the proximal incisor niche also contribute to replenishing the odontoblasts. The signaling mechanism that guides this reparative process remains unclear. Wnt signaling, which is activated in TA cells as well as in the region of newly differentiated odontoblasts, is critical for the transition from slow-cycling stem cells to rapidly proliferating progenitor cells. Moreover, mice with an activated Wnt pathway due to loss of Axin2, which represses Wnt signaling in a ligand-dependent manner, show a much stronger repair response than controls (Hunter Daniel et al., 2015). Tgf-β signaling has also been shown to play a role in dental stem cell differentiation. Non-ionizing, low-power laser treatment activates latent, endogenous Tgf-β1 via a specific methionine residue, thereby promoting stem cell differentiation, which subsequently significantly increases the amount of dentin regeneration in rat injury model (Arany et al., 2014).
6. Conclusion and future directions
The development of the jaws and teeth are closely related, not only due to their common origins involving cranial neural crest cells, but also the regulatory mechanisms and factors that they share. Many transgenic mouse models exhibit malformations in both the teeth and jaws, suggesting a close relationship between them. Despite the significant progress we have made in understanding the regulatory mechanisms behind jaw and tooth development over the past few decades, there are still several unanswered questions, especially regarding the early stages of their development, including how patterning processes contribute to mesenchymal cell fate determination as well as bone and cartilage formation. Future studies, with the help of emerging techniques such as single-cell RNA sequencing, will enable us to identify and trace heterogeneous cell populations within the mandibular process, which could shed light on the regulatory mechanism of cell fate determination. Understanding the molecular regulation of each stage of the development of the jaws and teeth will not only improve our knowledge of the etiology of developmental defects, but also facilitate the treatment of related diseases using stem cells and tissue regeneration.
Acknowledgments
We would like to thank Julie Mayo and Bridget Samuels for critical reading of the manuscript and discussion, and funding support from the National Institute of Dental and Craniofacial Research, NIH (R37 DE012711 and U01 DE024421) to Yang Chai.
References
- Aggarwal VS, Carpenter C, Freyer L, Liao J, Petti M, & Morrow BE (2010). Developmental Biology, 344, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aioub M, Lezot F, Molla M, Castaneda B, Robert B, Goubin G, et al. (2007). Bone, 41, 851–859. [DOI] [PubMed] [Google Scholar]
- Amano O, Doi T, Yamada T, Sasaki A, Sakiyama K, Kanegae H, et al. (2010).Journal of Oral Biosciences, 52, 125–135. [Google Scholar]
- Arany PR, Cho A, Hunt TD, Sidhu G, Shin K, Hahm E, et al. (2014). Science Translational Medicine, 6, 238ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykan A, Ozturk S, Sahin I, Gurses S, Ural AU, Oren NC, et al. (2013). The Journal of Craniofacial Surgery, 24, e169–e175. [DOI] [PubMed] [Google Scholar]
- Bae JM, Clarke JC, Rashid H, Adhami MD, McCullough K, Scott JS, et al. (2018). Journal of Bone and Mineral Research, 33, 1126–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae CH, Kim TH, Ko SO, Lee JC, Yang X, & Cho ES (2015). Journal of Dental Research, 94, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek WY, Kim YJ, de Crombrugghe B,& Kim JE (2013). Biochemical and Biophysical Research Communications, 432, 188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow JR, Stadler HS, & Capecchi MR (2000). Development, 127, 933–944. [DOI] [PubMed] [Google Scholar]
- Barske L, Rataud P, Behizad K, Del Rio L, Cox SG, & Crump JG (2018). Developmental Cell, 44, 337–347.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre KK, & Klingensmith J (2015). Developmental Dynamics, 244, 564–576. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, & McMahon AP (1995). Developmental Biology, 172, 126–138. [DOI] [PubMed] [Google Scholar]
- Cao J,Wang L, Du ZJ, Liu P, Zhang YB, Sui JF, et al. (2013). The British Journal of Oral & Maxillofacial Surgery, 51, 937–941. [DOI] [PubMed] [Google Scholar]
- Cao J, Wang L, Lei DL, Liu YP, Du ZJ, & Cui FZ (2012). Oral Surgery, Oral Medicine, Oral Pathology, and Oral Radiology, 113, 48–53. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, et al. (2008). Nature, 456, 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate AT (1996). Oral Diseases, 2, 55–62. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P Jr., Han J, Rowitch DH, et al. (2000). Development, 127, 1671–1679. [DOI] [PubMed] [Google Scholar]
- Chai Y, & Maxson RE Jr. (2006). Developmental Dynamics, 235, 2353–2375. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, & Maas R (1996). Development, 122, 3035–3044. [DOI] [PubMed] [Google Scholar]
- Chen Q, Zhao Y, Shen G, & Dai J (2018). Journal of Dental Research, 97, 1297–1305. [DOI] [PubMed] [Google Scholar]
- Chung IH, Yamaza T, Zhao H, Choung PH, Shi S, & Chai Y (2009). Stem Cells, 27, 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobourne MT, Hardcastle Z, & Sharpe PT (2001). Journal of Dental Research, 80, 1974–1979. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, & McMahon AP (2000). Development, 127, 4775–4785. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Sharpe PT, & Pachnis V (2009). Developmental Biology, 333, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, & Compagnucci C (2008). Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 310, 315–335. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, & Rubenstein JL (2002). Science, 298, 381–385. [DOI] [PubMed] [Google Scholar]
- Duan X (2014). Journal of Dental Research, 93, 117–125. [DOI] [PubMed] [Google Scholar]
- Duan X, Mao Y, Yang T, Wen X, Wang H, Hou J, et al. (2009). Archives of Oral Biology, 54, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Felszeghy S, Suomalainen M, & Thesleff I (2010). Differentiation, 80, 241–248. [DOI] [PubMed] [Google Scholar]
- Feng J, Jing J, Li J, Zhao H, Punj V, Zhang T, et al. (2017). Development, 144, 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Mantesso A, De Bari C, Nishiyama A, & Sharpe PT (2011). Proceedings of the National Academy of Sciences of the United States of America, 108, 6503–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, Gluhak-Heinrich J, Yang W, Wang L, et al. (2011). Cells, Tissues, Organs, 194, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron-Maguire M, Mallo M, Zhang M, & Gridley T (1993). Cell, 75, 1317–1331. [DOI] [PubMed] [Google Scholar]
- Gilbert SF (2000). Developmental biology (6th ed.). Sunderland, MA: Sinauer Associates. [Google Scholar]
- Hansson S, & Halldin A (2012). Journal of Dental Biomechanics, 3 1758736012456543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P,Jung H-S, Mustonen T, Wang YA,& Thesleff I (1999). The Journal of Cell Biology, 147, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsfield JK (2007). Orthodontics & Craniofacial Research, 10, 121–128. [DOI] [PubMed] [Google Scholar]
- Ho TV, Iwata J, Ho HA, Grimes WC, Park S, Sanchez-Lara PA, et al. (2015). Developmental Biology, 400, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter Daniel J, Bardet C, Mouraret S, Liu B, Singh G, Sadoine J, et al. (2015). Journal of Bone and Mineral Research, 30, 1150–1159. [DOI] [PubMed] [Google Scholar]
- Iasella JM, Greenwell H, Miller RL, Hill M, Drisko C, Bohra AA, et al. (2003). Journal of Periodontology, 74, 990–999. [DOI] [PubMed] [Google Scholar]
- Jin YR, Turcotte TJ, Crocker AL, Han XH, & Yoon JK (2011). Developmental Biology, 352, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M, & Thesleff I (2012). Cold Spring Harbor Perspectives in Biology, 4, a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, et al. (2012). Developmental Cell, 23, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukua N, Shahidi MK, Konstantinidou C, Dyachuk V, Kaucka M, Furlan A, et al. (2014). Nature, 513, 551–554. [DOI] [PubMed] [Google Scholar]
- Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M, et al. (2014).Journal of Bone and Mineral Research, 29, 1960–1969. [DOI] [PubMed] [Google Scholar]
- Kim IS, Cho TH, Lee ZH, & Hwang SJ (2013). Tissue Engineering. Part A, 19, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Hur SW,Jeong BC, Oh SH, Hwang YC, Kim SH, et al. (2018).Journal of Cellular Physiology, 233, 1512–1522. [DOI] [PubMed] [Google Scholar]
- Kim TH, Lee JY, Baek JA, Lee JC, Yang X, Taketo MM, et al. (2011). Biochemical and Biophysical Research Communications, 412, 549–555. [DOI] [PubMed] [Google Scholar]
- Krumlauf R (1994). Cell, 78, 191–201. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Moffatt P, Chang EH, Bromage TG, Bringas P Jr., et al. (2012). Journal of Cellular Physiology, 227, 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, & Dupin E (2004). Development, 131, 4637–4650. [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim YS, Oh HS, Yang KH, Kim EC, & Chi JG (2001). The Anatomical Record, 263, 314–325. [DOI] [PubMed] [Google Scholar]
- Leucht P, Kim J-B, Amasha R, James AW, Girod S, & Helms JA (2008). Development, 135, 2845–2854. [DOI] [PubMed] [Google Scholar]
- Li J, Feng J, Liu Y, Ho TV, Grimes W, Ho HA, et al. (2015). Developmental Cell, 33, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fu G, Liu Y, Miao X, Li Y, Yang X, et al. (2017). Molecular and Cellular Biology, 37, e00590–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang X,Xu X, Mayo J, Bringas P Jr., Jiang R, et al. (2011). Development, 138, 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Parada C, & Chai Y (2017). Development, 144, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Feng J, Li J, Zhao H, Ho TV, & Chai Y (2015). Development, 142, 3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, & Erickson CA (1987). Developmental Biology, 121, 220–236. [DOI] [PubMed] [Google Scholar]
- Love RJ, Murray JM, & Mamandras AH (1990). American Journal of Orthodontics and Dentofacial Orthopedics, 97, 200–206. [DOI] [PubMed] [Google Scholar]
- Lumsden A, & Krumlauf R (1996). Science, 274, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Ma D, Ren L, Yao H, Tian W, Chen F, Zhang J, et al. (2013). Journal of Orthopaedic Research, 31, 1082–1088. [DOI] [PubMed] [Google Scholar]
- Matsui M, & Klingensmith J (2014). Developmental Biology, 392, 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, & Krumlauf R (1992). Cell, 68, 283–302. [DOI] [PubMed] [Google Scholar]
- Mellitzer G, Xu Q, & Wilkinson DG (1999). Nature, 400, 77–81. [DOI] [PubMed] [Google Scholar]
- Mizumoto Y, Moseley T, Drews M, Cooper VN 3rd, & Reddi AH (2003). The Journal of Bone and Joint Surgery. American Volume, 85-A(Suppl. 3), 124–130. [DOI] [PubMed] [Google Scholar]
- Moore KL, Persaud TVN, & Samperio JO (1999). Embriología clínica. McGraw-Hill Interamericana. [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, & de Crombrugghe B (2003). Proceedings of the National Academy of Sciences of the United States of America, 100, 9360–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M (1994). Journal of Dental Research, 73, 1515–1522. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, & Martin GR (1997). Cell, 90, 247–255. [DOI] [PubMed] [Google Scholar]
- Noden DM (1975). Developmental Biology, 42, 106–130. [DOI] [PubMed] [Google Scholar]
- Noden DM (1978a). Developmental Biology, 67, 313–329. [DOI] [PubMed] [Google Scholar]
- Noden DM (1978b). Developmental Biology, 67, 296–312. [DOI] [PubMed] [Google Scholar]
- Noden DM (1983). Developmental Biology, 96, 144–165. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kapadia H, Wang B, & D’Souza RN (2005). Archives of Oral Biology, 50, 141–145. [DOI] [PubMed] [Google Scholar]
- Oka K, Oka S, Sasaki T, Ito Y, Bringas P Jr., Nonaka K, et al. (2007). Developmental Biology, 303, 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky Killian EC, Birkholz DA, & Artinger KB (2009). Developmental Biology, 333, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Yvonne WY, Feng J, Daltoe F, Fatscher R, Gentleman E, Gentleman Molly M, et al. (2015). Journal of Bone and Mineral Research, 31, 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival CJ, Huang Y, Jabs EW, Li R, & Richtsmeier JT (2014). Developmental Dynamics, 243, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, & Balling R (1998). Genes & Development, 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poswillo D (1973). Oral Surgery, Oral Medicine, and Oral Pathology, 35, 302–328. [DOI] [PubMed] [Google Scholar]
- Richany SF, Bast TH, & Anson BJ (1956). Quarterly Bulletin of Northwestern University Medical School, 30, 331–355. [PMC free article] [PubMed] [Google Scholar]
- Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, & Chambon P (1993). Cell, 75, 1333–1349. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Wakamiya M, & Behringer RR (1999). Development, 126, 3811–3821. [DOI] [PubMed] [Google Scholar]
- Rossant J, & Tam PT (2002). Mouse development: Patterning, morphogenesis, and organogenesis. Elsevier Science. [Google Scholar]
- Ruest L-B, Xiang X, Lim K-C, Levi G, & Clouthier DE (2004). Development (Cambridge, England), 131, 4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar L, Cobourne M, Naylor S, Smalley M, Dale T, & Sharpe PT (2000). Proceedings of the National Academy of Sciences of the United States of America, 97, 4520–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kurihara Y, Asai R, Kawamura Y, Tonami K, Uchijima Y, et al. (2008). Proceedings of the National Academy of Sciences of the United States of America, 105, 18806–18811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, & Maas R (1994). Nature Genetics, 6, 348–356. [DOI] [PubMed] [Google Scholar]
- Shibata S, Suda N, Yoda S, Fukuoka H, Ohyama K, Yamashita Y, et al. (2004). Anatomy and Embryology (Berlin), 208, 273–280. [DOI] [PubMed] [Google Scholar]
- Shibukawa Y, Young B, Wu C, Yamada S, Long F, Pacifici M, et al. (2007). Developmental Dynamics, 236, 426–434. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M, & Bronner ME (2016). Science, 352, 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Robinson V, Patel K, & Wilkinson DG (1997). Current Biology, 7, 561–570. [DOI] [PubMed] [Google Scholar]
- Stewart KJ, Weyand B, van’t Hof RJ, White SA, Lvoff GO, Maffulli N, et al. (1999). British Journal of Plastic Surgery, 52, 343–350. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, et al. (1998). Development, 125, 1025–1036. [DOI] [PubMed] [Google Scholar]
- Sugito H, Shibukawa Y, Kinumatsu T, Yasuda T, Nagayama M, Yamada S, et al. (2011). Journal of Dental Research, 90, 625–631. [DOI] [PubMed] [Google Scholar]
- Tasli PN, Aydin S, Yalvac ME, & Sahin F (2014). Applied Biochemistry and Biotechnology, 172, 3016–3025. [DOI] [PubMed] [Google Scholar]
- Tavares ALP, Cox TC, Maxson RM, Ford HL, & Clouthier DE (2017). Development, 144, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillet MA, Kalcheim C, & Le Douarin NM (1987). Developmental Biology, 120, 329–347. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Duband JL, & Delouvee A (1982). Developmental Biology, 93, 324–343. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JLR, Bishop JM, & Martin GR (1999). Genes & Development, 13, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, & Sharpe PT (1998). Science, 282, 1136–1138. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Yamada G, Grigoriou M, Pachnis V,& Sharpe PT (1999). Development, 126, 51–61. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, & Thesleff I (1993). Cell, 75, 45–58. [PubMed] [Google Scholar]
- Veitch E, Begbie J, Schilling TF, Smith MM, & Graham A (1999). Current Biology, 9, 1481–1484. [DOI] [PubMed] [Google Scholar]
- Wang X-P, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, et al. (2007). PLoS Biology, 5, e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, & Thesleff I (2004). Developmental Cell, 7, 719–730. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zheng Y, Chen D, & Chen Y (2013). Developmental Biology, 381, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniak S, Mackenzie FE, Anderson P, Kabbara S, Ruhrberg C, & Schwarz Q (2015). Proceedings of the National Academy of Sciences of the United States of America, 112, 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Mellitzer G, Robinson V, & Wilkinson DG (1999). Nature, 399, 267–271. [DOI] [PubMed] [Google Scholar]
- Yahiro K, Higashihori N, & Moriyama K (2017). Biochemical and Biophysical Research Communications, 482, 883–888. [DOI] [PubMed] [Google Scholar]
- Yang L, Tsang KY, Tang HC, Chan D, & Cheah KSE (2014). Proceedings of the National Academy of Sciences of the United States of America, 111, 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RT, Zhang C, Liu Y, Zhou HH, & Li ZB (2012). Anatomical Record (Hoboken), 295, 734–741. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Mundy C, Kinumatsu T, Shibukawa Y, Shibutani T, Grobe K, et al. (2010). Journal of Dental Research, 89, 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Harada K, Ikebe T, Shinohara M, & Enomoto S (2006). Journal of Cranio-Maxillofacial Surgery, 34, 270–276. [DOI] [PubMed] [Google Scholar]
- Zhang C (2010). Journal of Orthopaedic Surgery and Research, 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, et al. (2014). Cell Stem Cell, 14, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Ma J, Mu R, Zhu R, Chen F, Wei X, et al. (2018). Life Sciences, 202, 175–181. [DOI] [PubMed] [Google Scholar]