Abstract
Whereas a wide variety of in vitro models have been developed and validated to assess the effect of specific food ingredients on the human gut microbiome, such models have only been developed and applied to a limited extent for companion animals. Since the use of pre- and probiotics to improve gut health is an emerging research topic in the field of companion animals and as dogs are often used as laboratory animals in developing and testing of pharmaceuticals, the current study aimed to establish an adequate canine in vitro model. This consisted of a four-stage reactor composed of a stomach and small intestinal compartment followed by a proximal and distal colon. This semi-continuous gastrointestinal tract model allowed a long-term, region-dependent, and pH-controlled simulation of the colon-associated microbial community of dogs. Upon reaching a functional steady state, the simulated canine microbial community composition proved to be representative of the in vivo situation. Indeed, the predominant bacterial phyla present in the in vitro proximal and distal colon corresponded with the main bacterial phyla detected in the fecal material of the dogs, resulting in an average community composition along the simulated canine gastrointestinal tract of 50.5% Firmicutes, 34.5% Bacteroidetes, 7.4% Fusobacteria, 4.9% Actinobacteria, and 2.7% Proteobacteria. A parallel in vivo–in vitro comparison assessing the effects of fructooligosaccharides (FOS) on the canine microbial community composition showed a consistent stimulation of Lactobacillus concentrations in the in vivo fecal samples as well as in the in vitro canine gut model. Furthermore, the in vitro platform provided additional insights about the prebiotic effect of FOS supplementation of dogs, such as a reduced abundance of Megamonas spp. which are only present in very low abundance in in vivo fecal samples, indicating an interesting application potential of the developed canine in vitro model in research related to gastrointestinal health of dogs.
Keywords: dog, FOS, in vitro, SCIME, validation
Introduction
The canine gastrointestinal tract harbors a complex community of bacteria, fungi, archaea, and viruses, in the same order of magnitude as the host cells (Sender et al., 2016). With 109 to 1010 CFU/g, the amount of bacterial cells present in the large intestine of dogs outnumbers the other parts of the canine gastrointestinal tract (Honneffer et al., 2014). Similar to humans, the composition of the intestinal microbial community of dogs mainly consists of the bacterial phyla Firmicutes, Bacteroides, Proteobacteria, Fusobacteria, and Actinobacteria in a similar distribution, thereby covering 99% of all bacteria in the canine intestine (Middelbos et al., 2007; Turnbaugh et al., 2009; Suchodolski, 2011; Schmitz and Suchodolski, 2016). It has been shown that the colon-associated canine microbiome plays a crucial role in food digestion, prevention from pathogenic infection, bioconversion of endogenous and exogenous compounds and immunomodulation (Garcia-Mazcorro et al., 2012). Therefore, much attention has recently been given to strategies that modulate the composition and metabolism of the canine intestinal microbial population in order to improve canine health (Pinna and Biagi, 2014). The most extensively studied approach for microbial community modulation in dogs is the use of prebiotics. Prebiotic compounds are classified as nondigestible substrates that are selectively used by the gut microbial community, thereby conferring a health benefit for the host (Gibson et al., 2017). Although the use of novel next-generation sequencing techniques gave rise to new insights regarding effects of prebiotics on microbial structure and function, final conclusions remain conflicting (Sunvold et al., 1995; Swanson et al., 2002a, 2002b; Flickinger et al., 2003; Hesta et al., 2003; Suchodolski, 2011; Beloshapka et al., 2013; Pinna and Biagi, 2014), emphasizing the need of additional studies to unravel the effect of prebiotics on the canine gastrointestinal community.
As in vivo studies of dietary effects on the intestinal microbiota frequently encounter technical and ethical constraints, much attention has been given to the development of in vitro models. Whereas a wide variety of in vitro gut models to study the effect of test products, including prebiotics, on the gut microbiome have been developed and validated for human applications (Van den Abbeele et al., 2010), the use of such models for companion animals such as dogs remains limited. Moreover, since dogs are often used as laboratory animals in the development of pharmaceutical compounds, the establishment of an adequate canine in vitro gut model is of high interest.
As reviewed by Suchodolski (Suchodolski, 2011), prebiotic in vitro studies in the field of companion animals are restricted to short-term batch experiments (Sunvold et al., 1995; Barry et al., 2011). Such experiments are often not representative of the complex colonic environment as they lack pH control, repeated feeding cycles and refreshment of the media, and therefore do not simulate accurately the in vivo situation. This indicates a clear need for better designed in vitro gut models simulating the gastrointestinal tract of companion animals.
In the current study, a dynamic in vitro gut model simulating the canine gastrointestinal tract was developed with focus on the colon-associated microbial community. To validate the developed canine gut model, a simultaneous in vivo–in vitro comparison was performed in which beagle dogs were subjected to common prebiotic treatment in vivo, whereas the fecal microbiota of the dogs was used as inoculum for a parallel in vitro prebiotic supplementation study.
Materials and Methods
Simulator of the Canine Intestinal Microbial Ecosystem
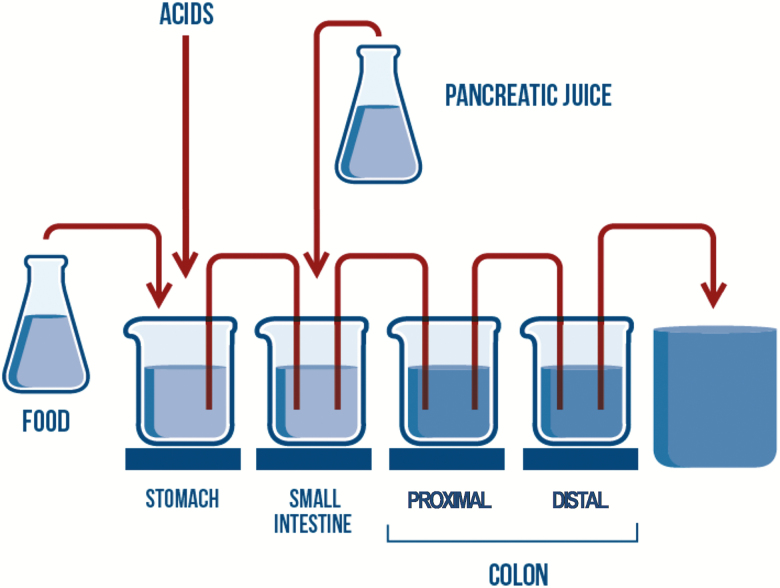
Considering the high similarity at the compositional and functional levels between the dog and human gut microbiota, the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) as described by Molly et al. (1993) was adapted to simulate the canine gastrointestinal tract. The Simulator of the Canine Intestinal Microbial Ecosystem (SCIME) set-up included four temperature-controlled reactors representing the different parts of the canine gastrointestinal tract, i.e., stomach, small intestine, proximal colon (PC), and distal colon (DC), as shown in Figure 1. The system was operated at 39 °C and kept anaerobic by daily flushing with N2 gas. The first two reactors were of the fill-and-draw principle to simulate food uptake and digestion. In the stomach compartment, a defined amount of simulated nutritional medium, composed of 9 g/L dog food (Hill’s Science Plan Adult Advance Fitness Lamb and Rice), 4 g/L mucin, 0.5 g/L cystein (Sigma-Aldrich, Bornem, Belgium), 4 g/L special peptone, and 1.5 g/L yeast extract (Oxoid, Aalst, Belgium), was added twice a day. Similarly, simulated pancreatic juice, composed of 2 g/L bile extract (Oxgall, Difco, Bierbeek, Belgium), 0.9 g/L pancreatin (Applichem, Zedelgem, Belgium), and 12.5 g/L NaHCO3 (VWR, Heverlee, Belgium), was added to the small intestinal compartment upon entering of the gastric suspension into this reactor. The last two reactor vessels, representing the canine large intestine, were continuously stirred with constant volume and pH control, and received the digested suspension from the small intestinal compartment at predefined intervals, with 12 h in-between each feeding cycle. The pH controllers, peristaltic pumps for liquid transfer, and flushing equipment were incorporated in an automated setup controlled by LabVIEW software (TWINSHIME, ProDigest, Zwijnaarde, Belgium). Residence times and pH were controlled to resemble in vivo conditions in the different parts of the canine gastrointestinal tract. Table 1 shows an overview of the parameters implemented in the SCIME model when compared with the SHIME setup.
Figure 1.
SCIME reactor setup. Schematic overview of the SCIME (Simulator of the Canine Intestinal Microbial Ecosystem) simulating the full canine gastrointestinal tract.
Table 1.
SCIME parameters
| SCIME | SHIME | |||||
|---|---|---|---|---|---|---|
| Volume, mL | Residence time, h | pH | Volume, mL | Residence time, h | pH | |
| Stomach | 140 | 1 | 2.00 | 200 | 2 | 2.00 |
| Small intestine | 200 | 4 | 6.80 | 200 | 4 | 6.80 |
| Proximal colon | 100 | 6 | 5.60 to 5.90 | 500 | 20 | 5.60 to 5.90 |
| Transverse colon | — | — | — | 800 | 32 | 6.10 to 6.40 |
| Distal colon | 167 | 10 | 6.65 to 6.90 | 600 | 24 | 6.60 to 6.40 |
| Feeding regimen | 2×/day | 3×/day |
Reactor setup of the Simulator of the Canine Intestinal Microbial Ecosystem (SCIME) including reactor volumes (mL), residence times (h), and pH compared with the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) (Possemiers et al., 2004).
In Vivo Validation Study
The animals were housed at the Faculty of Veterinary Medicine (Ghent, Belgium) according to the European animal welfare conditions. Prior to the study, dogs were adapted to the standard diet (Hill’s Science plan adult advance Fitness Lamb and Rice) during 14 d. During the in vivo trial, 10 beagle dogs (five male, five female) were fed the standard diet during a 10-d control period. This was followed by a 20-d treatment period, during which the standard diet was supplemented with 3% (w:w) fructooligosaccharides (FOS) from chicory root (Fibrulose, Cosucra Groupe Warcoing S.A., Belgium). Four out of the 10 dogs were randomly selected for validation of the in vitro SCIME model. During the control period, fresh fecal material from these four dogs was collected in an anaerobic container using an anaerobic bag (Anaerogen, Oxoid, Aalst, Belgium) for transport. Fecal material was processed within the hour after defecation as previously reported by De Boever et al. (2000) and used for inoculation of four parallel in vitro SCIME platforms. After inoculation, the canine microbial community was allowed to stabilize during 14 d to the in vitro reactor conditions. Following this stabilization period, the in vitro experiment consisted of a 2-wk control period in which the standard nutritional medium was administered, followed by a 2-wk treatment period where the standard nutritional medium was supplemented with 3% FOS (Fibrulose, Cosucra Groupe Warcoing S.A., Belgium), corresponding to a daily dose of 7.5 g/d.
Sample Collection
Sampling of each reactor vessel of the SCIME was performed every 2 d during the stabilization, control, and treatment periods. Liquid samples for subsequent analysis of microbial metabolic activity were immediately frozen at −20 °C, while pelleted cells (5 min, 9,000 × g) originating from 1 mL liquid sample were frozen at −20 °C for subsequent molecular analysis.
During the in vivo trial, fecal material was collected 30 min after defecation every 2 d during the control and treatment periods and stored at −20 °C prior to analysis. Fecal material for metabolic analysis was diluted in distilled water 12.5% (w:v) and analyzed immediately. DNA extraction was performed directly on 0.5 g fecal material.
Microbial Metabolic Activity
Short-chain C2-C6 fatty acids (SCFA), including isoforms C4-C6, were measured by gas chromatography as described by Andersen et al. (2014). Ammonium concentrations were determined by steam distillation as described by Possemiers et al. (2004).
Microbial Community Composition
Total DNA was extracted using the method described previously by Van den Abbeele et al. (2018). The extracted DNA was dissolved in DNAse-free water and stored at −20 °C for subsequent analysis. Firstly, the microbial community composition was determined through Illumina sequencing. The V1-2 region of the 16S rRNA gene was amplified as previously described (Camarinha-Silva et al., 2014) with some minor modifications. In the first 20 cycles of the PCR, the 16S rDNA target was enriched using the well-documented 27F and 338R primers (Lane, 1991; Etchebehere and Tiedje, 2005) as specified by Chaves-Moreno et al. (2015). Libraries were sequenced on an Illumina MiSeq platform. Furthermore, quantitative PCR (qPCR) for Lactobacillus spp. was performed as previously reported by Furet et al. (2009).
Statistical Analysis
Statistical data analyses for the in vivo–in vitro validation study were performed with the statistical software R, version 3.0.2. for Windows (http://www.r-project.org). Linear mixed model analysis was performed using the linear mixed-effects models (lme) package (Laird and Ware, 1982; Lindstrom and Bates, 1988). The REML approach was used to fit the mixed model (Pinheiro and Bates, 1996). The residuals were plotted against the fitted data. The distribution of the residuals and the normality was visually checked. Additionally, normality of the residuals was tested using the Shapiro–Wilk normality test. In case of non-normality, the data were transformed to reach normality. The optimal transformation was determined using the box–cox transformation function (Box and Cox, 1964) in R. P-values to examine the effect of the prebiotic treatment were obtained using ANOVA of the mixed models. P-value of <0.05 was considered statistically significant.
For the in vivo and in vitro experiment, normality of the data and equality of the variances was assessed using the Kolmogorov–Smirnov test and Levene’s test. Comparison of normally distributed data was performed with Student’s test for pairwise comparisons. Comparison of means of non-normally distributed data was evaluated with nonparametric Kruskal–Wallis test. P-value of <0.05 was considered statistically significant.
Bioinformatic processing of the Illumina sequencing data was conducted as previously described (Camarinha-Silva et al., 2014) with some modifications. Pair-end raw sequences were assembled according to Cole et al. (2014) and subsequently aligned (Gotoh algorithm with the SILVA reference database) and preclustered (diff = 2) using MOTHUR (Schloss et al., 2009). Obtained phylotypes were filtered to include only those present in an average abundance of ≥0.001% in all samples. Rarefaction curves and statistics were generated using the vegan package in R. All phylotypes were assigned a taxonomic affiliation based on RDPs Naïve Bayesian Classifier (RDP classifier) (Wang et al., 2007) applying an 80% of threshold. Overall, a total of 1,113,350 reads were obtained (with an average of 34,792 reads per sample) that clustered into 1,942 phylotypes.
Results
Stability of Canine Microbial Community in SCIME
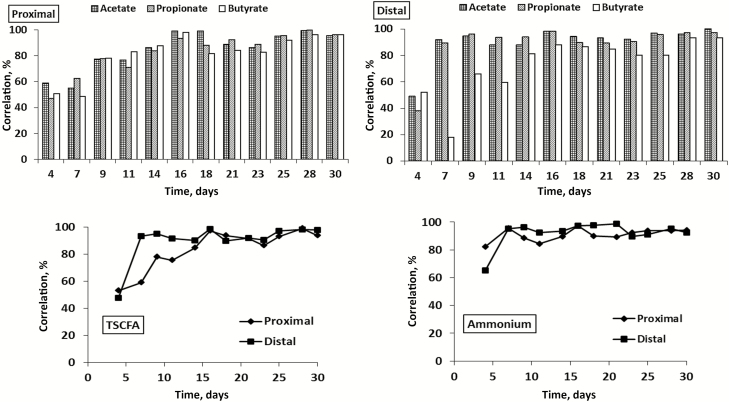
Functional stability of the SCIME reactors was determined by plotting the correlation coefficients of microbial metabolic parameters between a sampling point and its preceding sampling point as a function of time during the stabilization and control periods (Figure 2). It was observed that the calculated correlation coefficients for the measured metabolic parameters, i.e., acetate, propionate, butyrate, total SCFA (sum of acetate, propionate, butyrate, and branched SCFA), and ammonium, exceeded the minimum threshold of 80% within 14 d of reactor operation. In the DC, acetate, propionate, and total SCFA concentrations reached the 80% threshold after 7 d of operation, while a stabilization period of 14 d was needed in the PC. For butyrate, a stabilization period of 14 d was needed in both colon regions in order to reach the stability threshold of 80%. The fastest stability was reached for ammonium concentrations, as the 80% correlation was reached after 4 and 7 d for the PC and DC, respectively.
Figure 2.
Stability of the Canine Microbial Community in SCIME. Variability of microbial metabolic activity obtained in the SCIME during the stabilization (days 1 to 14) and control period (days 15 to 30) expressed as correlation between a sampling point and its preceding sampling point (%) for acetate, propionate, butyrate, total SCFA, and ammonium concentrations in the proximal and distal colon (n = 1). The threshold for stability was put at 80% (dashed line) according to Van den Abbeele et al. (2010).
Microbial Community Composition in SCIME
Predominant bacterial phyla present in the PC and DC of the SCIME consisted of Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria, which corresponded with the main bacterial phyla detected in vivo (Table 2). The Firmicutes phylum dominated the fecal material of the dogs with a relative abundance of 94.4% (ranging from 98.7% to 91.1%), followed by Actinobacteria with relative abundances ranging from 7.0% to 0.9%. Similarly, Firmicutes were the most abundant phylum in the simulated canine microbial community of the SCIME, comprising 52.4% and 48.6% in the PC and DC, respectively. Actinobacteria were equally enriched in the PC and DC of the SCIME, resulting in similar relative abundances as observed in the fecal microbial community of the dogs. Further, when compared with the in vivo samples, the simulated canine microbial community of the SCIME was enriched in Bacteroidetes (40.1% in PC and 28.8% in DC when compared with 1.2% in vivo) and Proteobacteria (2.3% in PC and 3.2% in DC when compared with <0.1% in vivo) at the expense of Firmicutes levels. Furthermore, in the DC, an enrichment of Fusobacteria was observed, as seen by an overall relative abundance of 14.5%.
Table 2.
Canine microbial community composition1
| In vitro | In vivo | |||||
|---|---|---|---|---|---|---|
| PC | DC | |||||
| % | SEM | % | SEM | % | SEM | |
| Actinobacteria | 4.9 | 1.0 | 4.9 | 1.0 | 3.8 | 0.6 |
| Bifidobacterium | 1.0 | 0.3 | 1.0 | 0.2 | 0.5 | 0.1 |
| Coriobacteriaceae | 3.8 | 0.7 | 4.0 | 0.7 | 3.6 | 0.7 |
| Bacteroidetes | 40.1 b | 0.8 | 28.8 c | 1.9 | 1.2a | 0.2 |
| Bacteroides | 10.5 b | 0.6 | 10.5 b | 1.8 | 0.3a | 0.1 |
| Prevotellaceae | 29.5 b | 1.3 | 18.2 c | 1.5 | 0.8a | 0.2 |
| Uncultured Bacteroidetes | 0.1 | 0.1 | 0.1 | 0.0 | 0.1 | 0.1 |
| Firmicutes | 52.4 b | 0.5 | 48.6 b | 0.7 | 94.4a | 0.1 |
| Lactobacillus | 17.2a,b | 1.5 | 6.3 b | 0.4 | 26.7a | 4.1 |
| Streptococcus | 0.1 | 0.1 | <0.1 | <0.1 | 5.0 | 2.1 |
| Blautia | 0.7 | 0.2 | 1.0 | 0.4 | 1.5 | 0.3 |
| Clostridium cluster XIVa | 0.6 | 0.1 | 1.1 | 0.2 | <0.1 | <0.1 |
| Clostridium cluster XI | 0.9 b | 0.1 | 1.1 b | 0.1 | 17.3a | 2.4 |
| Uncultured Clostridiales | 0.1 | 0.1 | 0.8 | 0.1 | 0.1 | 0.3 |
| Allobaculum | 7.2 | 0.6 | 13.5 | 0.4 | 13.3 | 4.0 |
| Catenibacterium | 0.1 | <0.1 | <0.1 | <0.1 | 2.4 | 0.7 |
| Erysipelotrichaceae | 0.1 | <0.1 | 0.0 | 0.1 | 2.3 | 0.6 |
| Turicibacter | 0.1 b | <0.1 | <0.1 b | <0.1 | 14.8a | 3.3 |
| Uncultured Acidaminococcaceae | 0.5 | 0.1 | 0.8 | 0.1 | <0.1 | <0.1 |
| Megamonas | 19.1 b | 1.0 | 20.5 b | 1.1 | 0.1a | 0.1 |
| Uncultured Firmicutes | 5.8a,b | 0.4 | 3.3 b | 0.3 | 11.1a | 1.9 |
| Fusobacteria | 0.3a | 0.1 | 14.5 b | 0.8 | 0.3a | 0.1 |
| Uncultured Fusobacteriaceae | 0.3a | 0.1 | 14.5 b | 0.8 | 0.3 | 0.1 |
| Proteobacteria | 2.3 b | 0.2 | 3.2 b | 0.2 | <0.1a | <0.1 |
| Sutterella | 1.2 b | 0.1 | 1.8 b | 0.2 | <0.1a | <0.1 |
| Anaerobiospirillum | 1.0 b | 0.2 | 0.3a,b | 0.1 | <0.1a | <0.1 |
| Pseudomonas | 0.1a | 0.1 | 1.1 b | 0.2 | <0.1a | <0.1 |
1Abundances of the dominant bacterial phylotypes belonging to the Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria phylum (% ± SEM) as assessed via 16S-targeted Illumina sequencing in samples collected from the SCIME during the control period in both the proximal (PC) and distal colon (DC), as well as in the fecal material of the dogs during the control period (n = 4). Statistically significant differences from the samples collected from the SCIME when compared with the in vivo samples are indicated in bold, while different letters indicate statistical differences between each of the tested samples (P < 0.05).
Within the Firmicutes phylum, the predominant bacterial phylotypes observed in the in vivo samples belonged to the genus Lactobacillus. In the simulated canine microbial community of the SCIME, the Lactobacillus genus was also highly abundant, especially in the PC, highly similar to the in vivo microbiota (17.2% and 6.3% in PC and DC, respectively, when compared with 26.7% in vivo). Similarly, the bacterial phylotypes belonging to the Clostridium cluster XI group were enriched in vivo, while they were detected in lower concentrations in the PC and DC compartments of the SCIME model. The genera Clostridium cluster XIVa and Megamonas on the other hand were enriched in the in vitro simulation model when compared with the fecal material of the dogs.
Effect of FOS supplementation on microbial community composition
In vivo, the addition of FOS resulted in a strong and significant increase in Lactobacillus concentrations. This key effect of FOS in vivo was also observed in both the PC and DC of the in vitro SCIME model (Table 3). Furthermore, FOS supplementation resulted in a significant decrease in two bacterial phylotypes belonging to Clostridium cluster XI in vivo, while this was only observed in the PC for one of these phylotypes in vitro. Finally, slightly enhanced levels of the genus Collinsella, belonging the Actinobacteria phylum, and some bacterial phylotypes within the Bacteroidetes phylum were significantly enriched upon FOS administration in vivo, with similar effects being observed in vitro, though not reaching statistical significance. On the contrary, the simulated canine microbial community of the SCIME was characterized by some minor decreases in Bacteroides genera and in a significant decrease in Megamonas upon FOS supplementation, which were not observed in vivo. In the DC compartment of the SCIME, a remarkable increase in several Allobaculum phylotypes was observed, which was not observed in the in vivo fecal samples.
Table 3.
Microbial community shift upon FOS supplementation1
| In vitro | In vivo | |||||
|---|---|---|---|---|---|---|
| PC (%) | P-value | DC (%) | P-value | % | P-value | |
| Actinobacteria | ||||||
| 14_Collinsella | +0.5 | 0.556 | −0.4 | 0.750 | +0.4 | 0.036 |
| Bacteroidetes | ||||||
| 38_Bacteroides | −1.2 | 0.035 | −0.1 | 0.819 | <0.1 | 0.873 |
| 65_Bacteroides | −0.4 | 0.150 | −0.1 | 0.028 | <0.1 | 0.333 |
| 123_Bacteroides | <0.1 | 0.483 | +0.1 | 0.335 | +0.6 | 0.005 |
| 95_Bacteroidetes | <0.1 | 0.566 | +0.2 | 0.381 | +0.7 | 0.020 |
| Firmicutes | ||||||
| 1_Lactobacillus | +7.9 | 0.042 | +2.9 | 0.014 | +18.3 | 0.002 |
| 8_Clostridium XI | −0.2 | 0.010 | <0.1 | 0.744 | −7.7 | 0.024 |
| 15_Clostridium XI | −0.2 | 0.193 | −0.5 | 0.104 | −3.2 | 0.014 |
| 10_Allobaculum | −1.3 | 0.104 | +4.4 | 0.023 | −0.3 | 0.778 |
| 21_Allobaculum | −0.5 | 0.099 | +1.8 | 0.003 | −0.1 | 0.897 |
| 28_Allobaculum | −0.3 | 0.104 | +1.3 | 0.021 | <0.1 | 0.977 |
| 33_Allobaculum | −0.2 | 0.191 | +1.8 | 0.047 | −0.1 | 0.522 |
| 63_Allobaculum | −0.1 | 0.208 | +0.7 | 0.024 | <0.1 | 0.946 |
| 25_Megamonas | −1.7 | 0.013 | −2.1 | 0.030 | +0.1 | 0.258 |
| Fusobacteria | ||||||
| 27_Fusobacteriaceae | −0.1 | 0.601 | −6.9 | 0.008 | −0.3 | 0.069 |
1Increase or decrease in relative abundances of the dominant bacterial phylotypes belonging to the Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria phylum (%) as assessed via 16S-targeted Illumina sequencing at the end of the treatment period with FOS when compared with the control period in samples collected from the SCIME in both the proximal (PC) and distal colon (DC), as well as in the fecal material of the dogs (n = 4). Statistically significant increases or decreases when compared with the control period are indicated with their P-value and are shown in bold when P < 0.05.
To confirm the results obtained through 16S-targeted Illumina sequencing, the most abundant bacterial genus, i.e., Lactobacillus spp., was quantified by qPCR (Figure 3). Lactobacillus concentrations significantly increased upon FOS supplementation in the PC of the SCIME model (P = 0.040) as well as in the in vivo fecal samples (P = 0.021). Also in the DC of the SCIME model, an increase in Lactobacillus concentrations was observed, though this did not reach statistical significance.
Figure 3.
Microbial community composition as assessed via qPCR. Lactobacillus concentrations (16S rDNA copies/ng DNA) as assessed via qPCR at the end of the control (C) and treatment (Tr) period with FOS in both the proximal colon (PC) and distal colon (DC) of the SCIME model, as well as in the fecal material of the dogs (in vivo). Statistically significant differences are indicated by their P-value.
Microbial Community Activity in SCIME
SCFA profiles of the four SCIME platforms during the control period revealed that the PC was characterized by an SCFA profile consisting of 41% acetate, 41% propionate, 15% butyrate, and 2% branched SCFA, while in the DC, a proportional ratio of 40% acetate, 37% propionate, 20% butyrate, and 3% branched SCFA was observed (Table 4). In the DC, a shift toward increased butyrate concentrations was observed (P < 0.001), mainly at the expense of propionate concentrations (P = 0.083). Furthermore, absolute branched SCFA concentrations were significantly higher in the DC when compared with the PC (P < 0.001). Similarly, ammonium concentrations were significantly enriched in the DC compartments when compared with the PC (P < 0.001).
Table 4.
Microbial metabolic activity. Average acetate (mM), propionate (mM), butyrate (mM), branched SCFA (mM) and ammonium (mg/L) concentrations over the control (C) and the treatment (TR) period in the proximal (PC) and distal colon (DC) of the SCIME model as well as in fecal samples from the in vivo experiment upon treatment with FOS. Data are presented as mean ± SEM. Statistically significant differences relative to the control period are indicated by their P-value (P < 0.05).
| In vitro PC | In vitro DC | In vivo | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C | TR | P-value | C | TR | P-value | C | TR | P-value | |
| Acetate (mM) | 39 ± 4 | 45 ± 5 | 0.018 | 39 ± 3 | 43 ± 4 | 0.040 | 55 ± 7 | 49 ± 12 | 0.066 |
| Propionate (mM) | 39 ± 3 | 60 ± 5 | <0.001 | 36 ± 3 | 46 ± 3 | <0.001 | 42 ± 7 | 41 ±10 | 0.526 |
| Butyrate (mM) | 14 ± 2 | 18 ± 4 | 0.011 | 19 ± 2 | 28 ± 2 | <0.001 | 22 ± 9 | 15 ± 8 | 0.003 |
| Branched SCFA (mM) | 2.7 ± 0.1 | 2.1 ± 0.1 | <0.001 | 3.1 ± 0.2 | 2.7 ± 0.1 | <0.001 | 1.0 ± 0.3 | 0.5 ± 0.3 | <0.001 |
| Ammonium (mg/L) | 427 ± 13 | 358 ± 9 | <0.001 | 527 ± 17 | 449 ± 12 | <0.001 | 292 ± 61 | 222 ± 63 | 0.002 |
FOS supplementation resulted in a significant increase in acetate, propionate, and butyrate concentrations in both the PC and DC compartments of the SCIME model, with the strongest effects being observed for propionate concentrations (Table 4). In vivo, increases in the aforementioned SCFA were not detected and even a significant decrease in butyrate concentrations was observed upon FOS administration. In contrast to the nonbranched SCFA, the branched SCFA significantly decreased upon FOS supplementation in both colon compartments of the SCIME, thereby confirming the effects that were observed in vivo. Finally, ammonium production decreased significantly in PC as well as in DC, which was also observed in the fecal samples of in vivo experiment.
Discussion
The aim of this research was to establish a continuous in vitro model simulating the canine gastrointestinal tract with focus on the colon-associated microbial community. Due to high similarities in microbial function and composition between the gastrointestinal tract of humans and dogs, the SHIME, a validated in vitro model simulating the human gastrointestinal tract (Molly et al., 1993), was adapted in order to meet the canine physiology in terms of feed composition and regimen, gastrointestinal regions, digestive enzymes, pH control, temperature, residence time, and fecal inoculum applied.
With respect to the simulated canine microbial community composition, it was observed that the PC and DC of the SCIME harbored distinct microbial communities. For the human in vitro simulation platform this region dependency of the colon-associated microbial community in terms of functionality and composition has been extensively validated using colonic samples from sudden death victims and proved to be mainly attributed to substrate availability, pH, residence time, and redox potential (Macfarlane et al., 1998). Although such an extensive validation for the dog has not been performed yet, the current study indicates that two distinct bacterial communities were sustained in the proximal and distal compartments of the SCIME. While the PC was characterized by high abundance of members from the Prevotellaceae and Lactobacillaceae families, the DC was enriched in Fusobacteriaceae as well as species belonging to Allobaculum genus. It has been reported that Lactobacillaceae are more enriched in the proximal part of the canine intestine due to their resilience toward lower pH and higher bile salt concentrations, and due to the shorter residence times in the upper parts of the gastrointestinal tract (Perelmuter et al., 2008), thereby confirming data obtained in the current study. Furthermore, it is well described that the proximal microbial community is adapted toward saccharolytic fermentation due to influx of undigestible carbohydrates in this part of the large intestine (Macfarlane et al., 1992). For instance, several members of the Bacteroidetes phylum, including Prevotellaceae, have been associated with high-fiber degradation potential (De Filippo et al., 2010), and therefore specifically colonize the proximal colon areas (Van den Abbeele et al., 2010). The distal colon on the other hand is generally characterized by a microbial community rich in species with specific metabolic functions, such as mucin (Van Herreweghen et al., 2017) and protein degradation (Macfarlane et al., 1992). In the present study, the DC of the SCIME showed increased concentrations of branched SCFA and ammonium, which have been linked with proteolytic fermentation. However, also increased production of butyrate was observed in the DC of the simulated canine gastrointestinal tract, which could be linked with the specific enrichment of Allobaculum in this colonic area. Indeed, members of the Allobaculum genus have been reported as potent butyrate producers in the canine gastrointestinal tract (Greetham et al., 2004). The same butyrate-producing functionality has been described in many members of Clostridium cluster XIVa in humans (Louis and Flint, 2009). Clostridium cluster XIVa was also enriched in the DC compartment of the SCIME model during the current study, though to a much lower extent when compared with the canine-specific Allobaculum genus.
Recent advances in molecular tools have greatly improved our knowledge of the canine colon-associated microbial community. Kim et al. (2017) showed by using the Illumina Miseq platform that the canine microbial community composition consists on average of Firmicutes (64.2% to 73.3%), Bacteroidetes (17.3% to 19.9%), Proteobacteria (0.9% to 8.7%), Fusobacteria (0% to 13.6%), and Actinobacteria (0.6% to 1.5%). However, Garcia-Mazcorro et al. (2012) showed a more dominant presence of Firmicutes (75% to 98%) in the fecal community of dogs using 454-pyrosequencing. These observed discrepancies might be explained by the molecular method used, which should be considered when abundances of gut microbiota are compared among different studies (Garcia-Mazcorro and Minamoto, 2013). While the Firmicutes phylum strongly dominated the canine feces in the current study, with a relative abundance of 94.4%, the predominant bacterial phyla present in the PC and DC of the SCIME corresponded with the main bacterial phyla detected in the fecal material of the dogs, resulting in an average community composition along the simulated canine gastrointestinal tract of 50.5% Firmicutes, 34.5% Bacteroidetes, 7.4% Fusobacteria, 4.9% Actinobacteria, and 2.7% Proteobacteria. Overall, all bacterial groups that were present in the fecal material of the dogs in the current study proliferated well in the in vitro environment, resulting in highly representative microbial communities along the colonic regions of the SCIME model. However, when comparing the fecal microbial composition in the current study with the in vitro results, the microbial community in the SCIME model was significantly enriched in Bacteroidetes and Proteobacteria at the expense of Firmicutes. A similar enrichment has been previously observed in human in vitro gut models (Rajilic-Stojanovic et al., 2010; Van den Abbeele et al., 2010) and could be partly associated with the absence of mucosal adhesion sites (Van den Abbeele et al., 2013). Indeed, it has been shown that inclusion of a mucosal environment in the SHIME model prevented the wash out of typically mucin-associated butyrate producers (Van den Abbeele et al., 2013) and could therefore also be an interesting extension of the SCIME platform. Finally, the microbial community in the SCIME model showed a specific enrichment of Megamonas species in both the PC and DC. As predominant members of the Veillonellaceae family, Megamonas species are known to produce propionate and butyrate from lactate (Lin et al., 2011). Furthermore, in dogs, Megamonas spp. have been described to be enhanced upon supplementation of the prebiotic FOS, indicating their potential beneficial effect on the canine gastrointestinal health (Beloshapka et al., 2013). The fact that the in vitro model sustained the growth of Megamonas could potentially allow a better understanding of the functionality of this genus in the microbial community of the canine-associated gastrointestinal tract.
To assess the effect of a test product on intestinal microbial composition and functioning in vitro, the establishment of steady-state bacterial community prior to the actual start of the treatment is of utmost importance as stability of the microbial community guarantees that any effects observed during a specific treatment truly result from the administered test product (Van den Abbeele et al., 2010). Results showed that functional stability of the microbial community in the SCIME model was reached after 14 d following inoculation, which nicely corresponded with the results obtaining with the SHIME model for human applications (Possemiers et al., 2004; Van den Abbeele et al., 2010). Upon reaching a stable microbial community in the current study, treatment with FOS from chicory root was initiated for a duration of 2 wk in a parallel in vitro–in vivo experiment. This nondigestible fiber has been well described for its prebiotic potential in both dogs (Swanson et al., 2002b; Pinna and Biagi, 2014) and humans (Van de Wiele et al., 2004; Grootaert et al., 2009). Although conflicting findings on the effect of prebiotic administration in dogs on markers of proteolytic fermentation have been described (Beynen et al., 2002; Flickinger et al., 2003; Propst et al., 2003), our study revealed a consistent reduction of ammonium and branched SCFA concentrations both in vivo as well as in vitro. Furthermore, a consistent stimulation of Lactobacillus concentrations was observed upon FOS supplementation in the in vivo fecal samples as well as in the PC and DC of the SCIME model. On the other hand, FOS supplementation resulted in increased acetate, propionate, and butyrate concentrations in the simulated canine gastrointestinal tract, while fecal levels of the aforementioned SCFA were not affected and even slightly reduced. Though, it is well known that fecal SCFA are not fully representative for the in vivo situation, as they are absorbed along the colon by the host (Von Engelhardt et al., 1989). Furthermore, large inter-individual variations often result in the absence of significant effects. Similarly, some conflicting effects on microbial community composition were observed between the parallel in vitro and in vivo study. For instance, reduced levels of Megamonas spp. were observed in the SCIME model upon treatment with FOS, while fecal concentrations remained unaffected. However, the low abundance of this genus in the in vivo samples made significant observations difficult. Several studies reported increased fecal abundance of Megamonas spp. upon treatment with FOS (Beloshapka et al., 2013). However, fecal community composition does not reflect the in vivo situation, where specific microbes can thrive in specific areas of the gastrointestinal tract. In vitro studies could therefore help to unravel the health-related effects of specific bacterial genera on the host and to measure prebiotic effects at the site of fermentation.
In conclusion, a dynamic in vitro gut model simulating the canine gastrointestinal tract was developed, with focus on the colon-associated microbial community allowing to culture the complex gut microbiota over a longer period under representative conditions of the different intestinal regions. Advanced molecular analysis demonstrated the colon-region specificity of the colonization process, with the PC being characterized by a saccharolytic microbial community, while the DC was enriched in species with specific metabolic functions such as butyrate production and protein degradation. Upon reaching a functional steady state, the simulated canine microbial community composition proved to be representative for the in vivo condition, though the absence of mucosal adhesion sites resulted in a specific enrichment of Bacteroidetes and Proteobacteria at the expense of Firmicutes. Furthermore, as the current study mainly focused on validation of the activity and composition of the colon-associated microbial community in the SCIME model, chemical assessment of the digesta at each step of the digestion process might be an interesting approach for further validation of the in vitro canine gut model. Overall, the in vitro platform provided additional insights in the prebiotic effect of repeated FOS supplementation in dogs, indicating an interesting application potential of the SCIME model in research related to gastrointestinal health in dogs.
Conflict of interest statementNone declared.
Acknowledgments
The authors thank J. Pitart, B. Guimarães, and N. Van de Genachte for the technical assistance during the in vitro work and analysis. We thank A. De Luycker and L. Haleydt for the technical assistance during the in vivo trial. The authors greatly acknowledge the help of R. Props for the statistical data analysis. We thank J. Ghyselinck for critically reviewing this manuscript. We kindly thank Cosucra for providing the FOS for this study. W.P.O. benefited from IWT grant no. 130311.
References
- Andersen S. J., Hennebel T., Gildemyn S., Coma M., Desloover J., Berton J., Tsukamoto J., Stevens C., and Rabaey K.. . 2014. Electrolytic membrane extraction enables production of fine chemicals from biorefinery sidestreams. Environ. Sci. Technol. 48:7135–7142. doi: 10.1021/es500483w [DOI] [PubMed] [Google Scholar]
- Barry K. A., Wojcicki B. J., Bauer L. L., Middelbos I. S., Vester Boler B. M., Swanson K. S., and Fahey G. C. Jr. 2011. Adaptation of healthy adult cats to select dietary fibers in vivo affects gas and short-chain fatty acid production from fiber fermentation in vitro. J. Anim. Sci. 89:3163–3169. doi: 10.2527/jas.2010-3445 [DOI] [PubMed] [Google Scholar]
- Beloshapka A. N., Dowd S. E., Suchodolski J. S., Steiner J. M., Duclos L., and Swanson K. S.. . 2013. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol. Ecol. 84:532–541. doi: 10.1111/1574-6941.12081 [DOI] [PubMed] [Google Scholar]
- Beynen A. C., Baas J. C., Hoekemeijer P. E., Kappert H. J., Bakker M. H., Koopman J. P., and Lemmens A. G.. . 2002. Faecal bacterial profile, nitrogen excretion and mineral absorption in healthy dogs fed supplemental oligofructose. J. Anim. Physiol. Anim. Nutr. (Berl.). 86:298–305. doi: 10.1046/j.1439-0396.2002.00386.x [DOI] [PubMed] [Google Scholar]
- Box G. E. P., and Cox D. R.. . 1964. An analysis of transformations. J. Royal Stat. Soc. 26:211–252. doi:10.1.1.321.3819 [Google Scholar]
- Camarinha-Silva A., Jáuregui R., Chaves-Moreno D., Oxley A. P., Schaumburg F., Becker K., Wos-Oxley M. L., and Pieper D. H.. . 2014. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ. Microbiol. 16:2939–2952. doi: 10.1111/1462-2920.12362 [DOI] [PubMed] [Google Scholar]
- Chaves-Moreno D., Plumeier I., Kahl S., Krismer B., Peschel A., Oxley A. P., Jauregui R., and Pieper D. H.. . 2015. The microbial community structure of the cotton rat nose. Environ. Microbiol. Rep. 7:929–935. doi: 10.1111/1758-2229.12334 [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., Brown C. T., Porras-Alfaro A., Kuske C. R., and Tiedje J. M.. . 2014. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42(Database issue):D633–D642. doi: 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boever P., Deplancke B., and Verstraete W.. . 2000. Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder. J. Nutr. 130:2599–2606. doi: 10.1093/jn/130.10.2599 [DOI] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S., Collini S., Pieraccini G., and Lionetti P.. . 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 107:14691–14696. doi: 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchebehere C., and Tiedje J.. . 2005. Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor. Appl. Environ. Microbiol. 71:5642–5645. doi: 10.1128/AEM.71.9.5642-5645.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger E. A., Schreijen E. M., Patil A. R., Hussein H. S., Grieshop C. M., Merchen N. R., and Fahey G. C. Jr. 2003. Nutrient digestibilities, microbial populations, and protein catabolites as affected by fructan supplementation of dog diets. J. Anim. Sci. 81:2008–2018. doi: 10.2527/2003.8182008x [DOI] [PubMed] [Google Scholar]
- Furet J. P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Doré J., and Corthier G.. . 2009. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x [DOI] [PubMed] [Google Scholar]
- Garcia-Mazcorro J. F., Dowd S. E., Poulsen J., Steiner J. M., and Suchodolski J. S.. . 2012. Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiologyopen 1:340–347. doi: 10.1002/mbo3.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mazcorro J., and Minamoto Y.. . 2013. Gastrointestinal microorganisms in cats and dogs: a brief review. Arch. Med. Vet. 45:111–124. doi: 10.4067/S0301-732X2013000200002 [DOI] [Google Scholar]
- Gibson G. R., Hutkins R., Sanders M. E., Prescott S. L., Reimer R. A., Salminen S. J., Scott K., Stanton C., Swanson K. S., Cani P. D., . et al. 2017. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14:491–502. doi: 10.1038/nrgastro.2017.75 [DOI] [PubMed] [Google Scholar]
- Greetham H. L., Gibson G. R., Giffard C., Hippe H., Merkhoffer B., Steiner U., Falsen E., and Collins M. D.. . 2004. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe 10:301–307. doi: 10.1016/j.anaerobe.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Grootaert C., Van den Abbeele P., Marzorati M., Broekaert W. F., Courtin C. M., Delcour J. A., Verstraete W., and Van de Wiele T.. . 2009. Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 69:231–242. doi: 10.1111/j.1574-6941.2009.00712.x [DOI] [PubMed] [Google Scholar]
- Hesta M., Roosen W., Janssens G. P., Millet S., and De Wilde R.. . 2003. Prebiotics affect nutrient digestibility but not faecal ammonia in dogs fed increased dietary protein levels. Br. J. Nutr. 90:1007–1014. doi: 10.1079/bjn2003988 [DOI] [PubMed] [Google Scholar]
- Honneffer J. B., Minamoto Y., and Suchodolski J. S.. . 2014. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 20:16489–16497. doi: 10.3748/wjg.v20.i44.16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., An J. U., Kim W., Lee S., and Cho S.. . 2017. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 9:68. doi: 10.1186/s13099-017-0218-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird N. M., and Ware J. H.. . 1982. Random-effects models for longitudinal data. Biometrics 38:963–974. doi: 10.2307/2529876 [DOI] [PubMed] [Google Scholar]
- Lane D. J. 1991. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics. In: Stackebrandt E. and Goodfellow M., editors, Nucleic acid techniques in bacterial systematic. New York: John Wiley and Sons; p. 115–175. [Google Scholar]
- Lin B., Gong J., Wang Q., Cui S., Yu H., and Huang B.. . 2011. In-vitro assessment of the effects of dietary fibers on microbial fermentation and communities from large intestinal digesta of pigs. Food Hydrocolloids 25:180–188. doi: 10.1016/j.foodhyd.2010.02.006 [DOI] [Google Scholar]
- Lindstrom M. J., and Bates D. M.. . 1988. Newton–Raphson and EM algorithms for linear mixed-effects models for repeated-measures data. J. Am. Stat. Assoc. 83:1014–1022. doi: 10.1080/01621459.1988.10478693 [DOI] [Google Scholar]
- Louis P., and Flint H. J.. . 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- Macfarlane G. T., Gibson G. R., and Cummings J. H.. . 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x [DOI] [PubMed] [Google Scholar]
- Macfarlane G. T., Macfarlane S., and Gibson G. R.. . 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35:180–187. doi: 10.1007/s002489900072 [DOI] [PubMed] [Google Scholar]
- Middelbos I. S., Godoy M. R., Fastinger N. D., and Fahey G. C. Jr.. 2007. A dose-response evaluation of spray-dried yeast cell wall supplementation of diets fed to adult dogs: effects on nutrient digestibility, immune indices, and fecal microbial populations. J. Anim. Sci. 85:3022–3032. doi: 10.2527/jas.2007-0079 [DOI] [PubMed] [Google Scholar]
- Molly K., Vande Woestyne M., and Verstraete W.. . 1993. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 39:254–258. doi: 10.1007/bf00228615 [DOI] [PubMed] [Google Scholar]
- Perelmuter K., Fraga M., and Zunino P.. . 2008. In vitro activity of potential probiotic Lactobacillus murinus isolated from the dog. J. Appl. Microbiol. 104:1718–1725. doi: 10.1111/j.1365-2672.2007.03702.x [DOI] [PubMed] [Google Scholar]
- Pinheiro J., and Bates D.. . 1996. Unconstrained parametrizations for variance-covariance matrices. Stat. Comput. 6:289–296. doi: 10.1007/BF00140873 [DOI] [Google Scholar]
- Pinna C., and Biagi G.. . 2014. The utilisation of prebiotics and synbiotics in dogs. Ital. J. Anim. Sci. 13:3107. doi: 10.4081/ijas.2014.3107 [DOI] [Google Scholar]
- Possemiers S., Verthé K., Uyttendaele S., and Verstraete W.. . 2004. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 49:495–507. doi: 10.1016/j.femsec.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Propst E. L., Flickinger E. A., Bauer L. L., Merchen N. R., and Fahey G. C. Jr. 2003. A dose-response experiment evaluating the effects of oligofructose and inulin on nutrient digestibility, stool quality, and fecal protein catabolites in healthy adult dogs. J. Anim. Sci. 81:3057–3066. doi: 10.2527/2003.81123057x [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Maathuis A., Heilig H. G., Venema K., de Vos W. M., and Smidt H.. . 2010. Evaluating the microbial diversity of an in vitro model of the human large intestine by phylogenetic microarray analysis. Microbiology 156(Pt 11):3270–3281. doi: 10.1099/mic.0.042044-0 [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., . et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S., and Suchodolski J.. . 2016. Understanding the canine intestinal microbiota and its modification by pro-, pre- and synbiotics – what is the evidence? Vet. Med. Sci. 2:71–94. doi: 10.1002/vms3.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., and Milo R.. . 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski J. S. 2011. Companion animals symposium: microbes and gastrointestinal health of dogs and cats. J. Anim. Sci. 89:1520–1530. doi: 10.2527/jas.2010-3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunvold G. D., Fahey G. C. Jr, Merchen N. R., Titgemeyer E. C., Bourquin L. D., Bauer L. L., and Reinhart G. A.. . 1995. Dietary fiber for dogs: IV. In vitro fermentation of selected fiber sources by dog fecal inoculum and in vivo digestion and metabolism of fiber-supplemented diets. J. Anim. Sci. 73:1099–1109. doi: 10.2527/1995.7341099x [DOI] [PubMed] [Google Scholar]
- Swanson K. S., Grieshop C. M., Flickinger E. A., Bauer L. L., Chow J., Wolf B. W., Garleb K. A., and Fahey G. C. Jr. 2002a. Fructooligosaccharides and Lactobacillus acidophilus modify gut microbial populations, total tract nutrient digestibilities and fecal protein catabolite concentrations in healthy adult dogs. J. Nutr. 132:3721–3731. doi: 10.1093/jn/132.12.3721 [DOI] [PubMed] [Google Scholar]
- Swanson K. S., Grieshop C. M., Flickinger E. A., Bauer L. L., Healy H. P., Dawson K. A., Merchen N. R., and Fahey G. C. Jr.. 2002b. Supplemental fructooligosaccharides and mannanoligosaccharides influence immune function, ileal and total tract nutrient digestibilities, microbial populations and concentrations of protein catabolites in the large bowel of dogs. J. Nutr. 132:980–989. doi: 10.1093/jn/132.5.980 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., . et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., Belzer C., Goossens M., Kleerebezem M., De Vos W. M., Thas O., De Weirdt R., Kerckhof F. M., and Van de Wiele T.. . 2013. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7:949–961. doi: 10.1038/ismej.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., Grootaert C., Marzorati M., Possemiers S., Verstraete W., Gérard P., Rabot S., Bruneau A., El Aidy S., Derrien M., . et al. 2010. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76:5237–5246. doi: 10.1128/AEM.00759-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., Kamil A., Fleige L., Chung Y., De Chavez P., and Marzorati M.. . 2018. Different oat ingredients stimulate specific microbial metabolites in the gut microbiome of three human individuals in vitro. ACS Omega 3:12446–12456. doi: 10.1021/acsomega.8b01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiele T., Boon N., Possemiers S., Jacobs H., and Verstraete W.. . 2004. Prebiotic effects of chicory inulin in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 51:143–153. doi: 10.1016/j.femsec.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Van Herreweghen F., Van den Abbeele P., De Mulder T., De Weirdt R., Geirnaert A., Hernandez-Sanabria E., Vilchez-Vargas R., Jauregui R., Pieper D. H., Belzer C., . et al. 2017. In vitro colonisation of the distal colon by Akkermansia muciniphila is largely mucin and pH dependent. Benef. Microbes 8:81–96. doi: 10.3920/BM2016.0013 [DOI] [PubMed] [Google Scholar]
- Von Engelhardt W., Rönnau K., Rechkemmer G., and Sakata T.. . 1989. Absorption of short-chain fatty acids and their role in the hindgut of monogastric animals. Anim. Feed Sci. Technol. 23:43–53. doi: 10.1016/0377-8401(89)90088-6 [DOI] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., and Cole J. R.. . 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]