Abstract
The complexity of nanoscale interactions between biomaterials and cells has limited the realization of the ultimate vision of nanotechnology in diagnostics and therapeutics. As such, significant effort has been devoted to advancing our understanding of the biophysical interactions of the myriad nanoparticles. Endocytosis of nanomedicine has drawn tremendous interest in the last decade. Here, we highlight the ever-present barriers to efficient intracellular delivery of nanoparticles as well as the current advances and strategies deployed to breach these barriers. We also introduce new barriers that have been largely overlooked such as the glycocalyx and macromolecular crowding. Additionally, we draw attention to the potential complications arising from the disruption of the newly discovered functions of the lysosomes. Novel strategies of exploiting the inherent intracellular defects in disease states to enhance delivery and the use of exosomes for bioanalytics and drug delivery are explored. Furthermore, we discuss the advances in imaging techniques like electron microscopy, super resolution fluorescence microscopy, and single particle tracking which have been instrumental in our growing understanding of intracellular pathways and nanoparticle trafficking. Finally, we advocate for the push towards more intravital analysis of nanoparticle transport phenomena using the multitude of techniques available to us. Unraveling the underlying mechanisms governing the cellular barriers to delivery and biological interactions of nanoparticles will guide the innovations capable of breaching these barriers.
Keywords: intracellular trafficking and delivery, endosomal escape, drug and gene delivery, imaging, barriers to delivery
1. Introduction
Endocytosis is an evolutionarily ancient network that was evolved by cells as a means to internalize nutrients and maintain cellular homeostasis. Viruses exploit these routes to manipulate cell functions and cause diseases. Today, synthetic nanomaterials use these internalization pathways to gain access to the inner realm of cells. These materials are designed to deliver imaging agents, drugs, and nucleic acids for diagnosis and treatment of disease [1,2]. Apart from being a transport process, endocytosis is important for regulating metabolism as well as transmitting information and materials within a cell or to distant tissues [3–5]. The primary focus in the area of drug delivery has remained on the efficient intracellular delivery of nanoparticles into cells. The field has skyrocketed since we first published our review on “Endocytosis of Nanomedicines” nearly a decade ago [1]. The number of publications per year with the keywords “endocytosis + nanoparticle” has more than doubled since “Endocytosis of Nanomedicines” was published. Today, it is clear that almost all nanocarriers are taken up by cell via endocytosis and are primarily routed towards a degradative organelle known as the lysosomes. There is limited endosomal escape of cargo into the cytosol, which restricts the efficacy of nanoparticles as drug carriers and contributes to cytotoxicity [6–8]. A majority of nanoparticles residing in the endo/lysosomal system are also ejected out of the cell, further contributing to a decrease in efficacy [9,10]. Moreover, the convergence of the endo/lysosomal system with the cellular clearance mechanisms like autophagy further illustrates the complex vesicular barriers that can impede drug delivery [11–13]. Strategies to enhance endosomal escape are heavily researched in academia and industry, especially for nucleic acid delivery [6,7,14–18]. Another major discovery in the realm of cell biology has been that the lysosomes, long underestimated as dead trash sites, are in fact, hubs of activity [19,20]. Lysosomes control cellular metabolism and nutrient sensing, maintain diverse pH based on their subcellular localization, and orchestrate spatiotemporal intracellular signaling [20–22]. The endolysosomal function is now being used as an indicator of cellular health in many physiological and pathological conditions [23–25]. Moreover, the inherent tropism of nanocarriers towards lysosomal compartments is being exploited to deliver therapeutics in disease states, such as lysosomal storage disorders (LSDs), where the lysosomal function is compromised [26–28]. Nanotechnology is further being utilized to illuminate the vesicular composition due to its ability to selectively measure endosomal ions (e.g., H+, Cl−, and Ca2+ ) and metabolic contents (e.g., lipids and enzymes.) in normal and diseased cells [29–34]. It is, therefore, clear that studying interactions of endosomes with nanoparticles can yield beneficial insights. Today, endosomal constituents are being used for drug delivery [35–39]. Exosomes, produced inside the multivesicular bodies/late endosomal compartment, are being extracted and loaded with small molecules, proteins, and nucleic acids to deliver therapeutics to locations where synthetic vectors have achieved marginal success [5,36,38,39]. Since exosomes are increasingly recognized as powerful and selective natural nanoparticles for delivery of therapeutic and imaging agents directing endocytosis of these agents towards exosomes is one way to load exosomes with valuable payload [40–43]. Finally, as we had mentioned in our initial review that “...developing assays to study the complex process of endocytosis of nanomedicines in vivo remains a key challenge for the future success of this field....” [1]. We highlight the use of new tools on the horizon that may address this problem.
In this review, we will discuss the barriers to intracellular delivery, strategies that can accomplish endosomal escape or target vesicular compartments to restore cellular function or detect metabolic composition in disease, deployment of exosomes as delivery vehicles, and a brief overview on the repertoire of new tools in our arsenal that can be deployed to dissect intracellular trafficking in-vitro and in-vivo.
2. Intracellular trafficking and barriers to efficient delivery
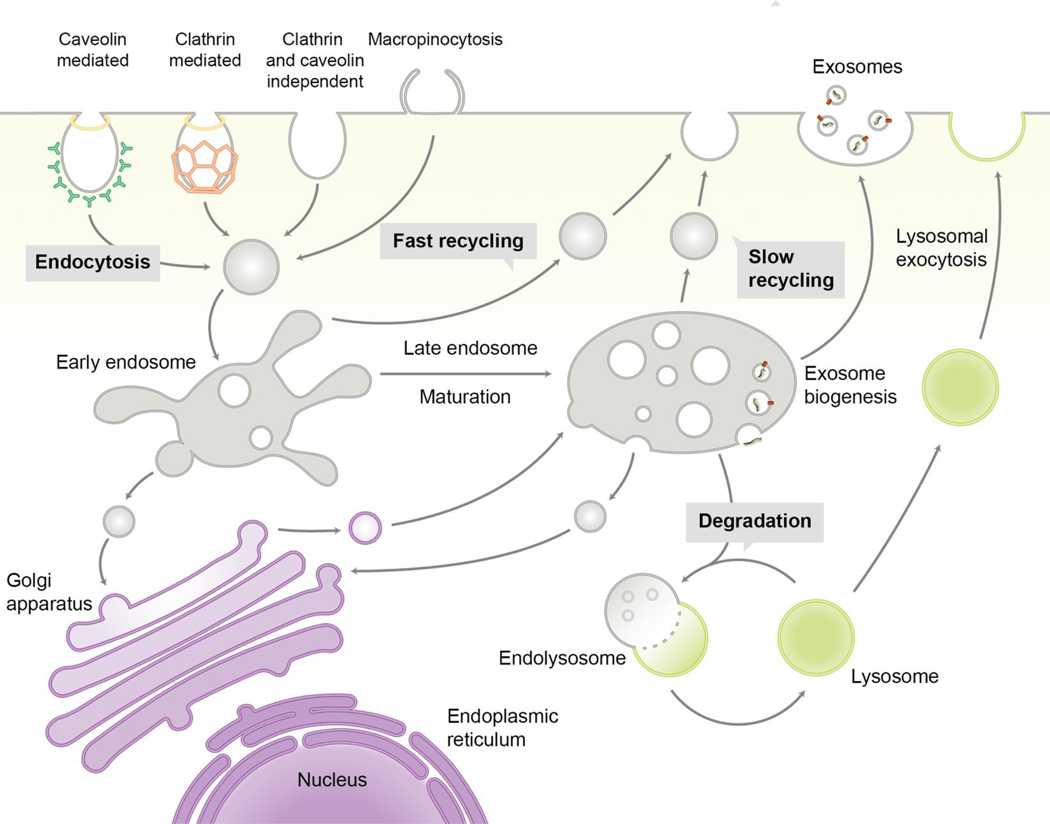
Nanoparticles are internalized following interactions with the cell surface through specific ligand-receptor driven interactions or via non-specific interactions such as electrostatic and hydrophobic interactions. The molecular mechanisms that capture nanoparticles are classified based on the size of the vesicle formed - phagocytosis (large size) and pinocytosis (small size). [1]. Pinocytosis is further classified on the basis of cellular effectors that contribute to vesicle formation as clathrin-mediated, macropinocytosis, caveolin-mediated, clathrin- and caveolin-independent pathways (Fig. 1) [1]. This active process of endocytosis begins with a cell engulfing portions of its plasma membrane (lipids and proteins) along with extracellular fluid in the vicinity to create fluid-filled vesicles known as endosomes (Fig. 1).
Fig. 1.
Endocytic pathways and exosome biogenesis.
In eukaryotic cells, nanoparticles enter cells via caveolin-mediated, clathrin-mediated, clathrin and caveolin-independent, or macropinocytic endocytosis. Endocytic vesicles containing nanoparticles move to early endosomes, where act as a sorting station. Depending on the sorting mechanisms, nanoparticles can be recycled via exocytosis, transported to late endosomes with maturation of endosomes or trafficked to the ER or TGN. Unless the nanoparticles are retrieved from late endosomes to cytoplasm, they are processed to lysosomes where degradation starts by enzymes. From interluminal vesicles in multi-vesicular bodies (a type of late endosomes), exosomes can be generated by inward budding of endosomal membrane, followed by secretion to extracellular compartments via fusion with plasma membrane.
2.1. Endocytosis, endosomal maturation, and exocytosis
The first barrier is the size of the vesicle that limits the number of nanoparticles that can enter cells. Studies have suggested that, for instance, to enter caveolae based transporters that are highly effective in transcytosis of macromolecules across the endothelium, a nanoparticle must be below 100 nm in size [44–46]. A large number of materials that have shown successful delivery are using macropinocytosis since it forms a large leaky vesicle that can contain several nanoparticles [1,6,47,48]. After entry, internalized cargo is delivered to the early endosome (Fig. 1, 2A-i), an organelle resulting from fusion mediated by a tethering mechanism that pulls the vesicles together [49]. The early endosome serves as the sorting hub, directing cargo to the endoplasmic reticulum (ER), the trans-Golgi network (TGN), or the endosomal recycling network (Fig. 1). The early endosomes themselves can undergo maturation to form the multivesicular late endosomes (Fig. 1, 2A-ii). Further endosomal maturation results in structural changes with the emergence of multilamellar endolysosome, a temporary hybrid organelle, by fusing with a lysosome, which is followed by further conversion back into lysosomes (Fig. 1, 2A-iii-iv). During the maturation process, morphological variations in the endosomes are coupled with a drop in pH [50]. These transitions in vesicular morphology due to dynamic lipid and protein exchanges can be a barrier that affects the successful delivery of intact cargo to the cytosol. Another major barrier during endosomal processing is exocytosis. Exocytosis of nanoparticles can occur by either trafficking of nanoparticles from the early endosome to the recycling endosome that fuses to the plasma membrane or from the multivesicular body/late endosomes and lysosomes where either intact nanoparticles or some of their components are ejected out of the cell (Fig. 1) [9,10,51]. The final stage where most nanoparticles usually end up is the lysosome, following transition through multilamellar endolysosomes (Fig. 1).
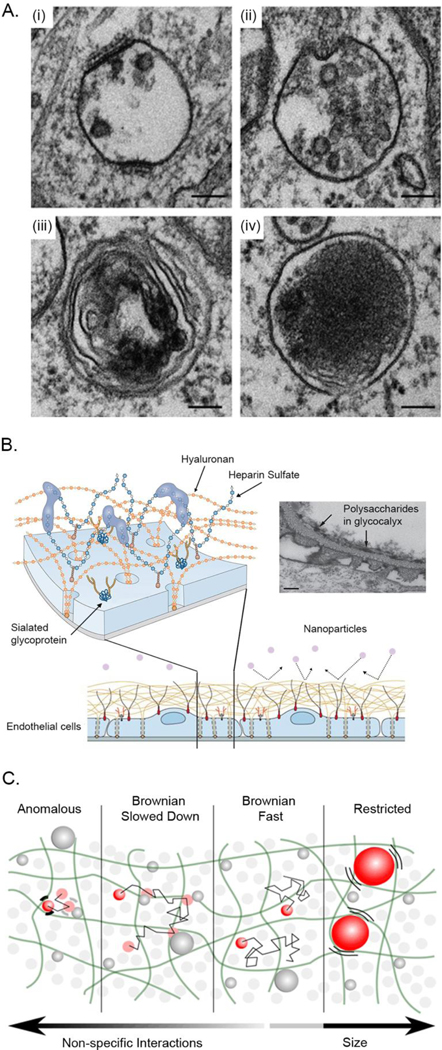
Fig. 2.
Challenges to intracellular delivery
(A) Entrapment of nanoparticles in endocytic pathways is a significant barrier to efficient delivery. Endocytosis delivers the nanoparticles to (i) the early endosome. Nanoparticles that failed to escape from the early or (ii) the late endosome, face degradation in (iii) the endolysosome, which is produced by fusion with (iv) the lysosome. The harsh milieu of the endocytic compartments due to decrease in pH, changes in shape and composition of these compartments is a significant barrier to drug delivery. Scale bars, 100 nm. Adapted with permission from [50], Copyright 2011, European Molecular Biology Organization. (B) Schematic representations of endothelial glycocalyx. Glycocalyx is composed of hyaluronans, heparin sulfate, proteoglycans, and glycoproteins and impedes nanoparticles interactions with plasma membrane. Electron micrograph depicts the glomerular glycocalyx with the fenestrae (arrows). Scale bar, 100 nm. Adapted with permission from [66], Copyright 2015, Springer Nature. (C) Schematic representation of the cytosolic mobility of particulates. Particles larger than the pores of the cytoskeletons (50 – 70 nm) display restricted diffusion due to cytosolic viscosity. These large particulates are constrained in the medium but will eventually drift following the remodeling of internal structures. Particles smaller than the pore size diffuse with viscosities that are dependent on the strength of the non-specific interactions. If the particles are completely inert, they move freely as if in water. When the particles start to interact with intracellular components, their motions are delayed due to non-specific interactions that can result in a slow Brownian or anomalous motion. This molecular crowding prevents nanoparticles in cytosol from reaching subcellular targets. Reproduced with permission from [83], Copyright 2018, Springer Nature.
2.2. Lysosome
Lysosomes serve as the final destination for macromolecules, where these macromolecules are degraded by hydrolytic enzymes activated by low pH (Fig. 2A-iv). However, long overlooked as a static organelle, lysosomes are now known to be a dynamic hub of cellular activity, including cellular metabolism, nutrient sensing, subcellular localization-dependent pH control, plasma membrane repair, secretion, and spatiotemporal intracellular signaling [52,21,53,22,20]. Transitioning from the multilamellar endolysosomal structure, the lysosomes possess a dense coat of hydrophilic polysaccharides on the luminal side of the limiting membrane to prevent the cytosolic transfer of its contents [50,54]. Numerous ion channels, transporters, and nutrient sensors reside as trans-membrane proteins that participate in active transport and can regulate the cytosolic delivery of the cargo [55,56].
Lysosomal positioning is also crucial to its function [57–59]. Lysosomes on the periphery of the cell tend to have a higher pH relative to their more acidic perinuclear counterparts [59]. Sequestration of nutrients in the lumen of peripheral lysosomes induces cell proliferation through the activation of the mechanistic target of rapamycin complex 1 (mTORCl) [57]. Absence of nutrients localizes lysosomes in the perinuclear space and activates autophagy (the catabolic recycling process of the cell) to maintain cellular homeostasis [58]. Induction of autophagy leads to the capture of cytosolic materials by autophagosomes, which subsequently fuse with lysosomes for degradation or exocytosis [60]. This process can sequester cytosolically localized nanocarriers and even engulf a leaky endosome, further impeding delivery [11]. As such, the optimal function of the lysosome is critical for the survival of the cell. Forced disruption of the lysosome and thereby mTORCl function to achieve cytosolic delivery may be harmful to the cell. This dysregulation of the normal cellular function may also induce or disrupt autophagy and hinder therapeutic or diagnostic potential of nanoparticles [61–64].
2.3. Glycocalyx
In addition to the barriers created by the lipids and proteins of the plasma membrane, recent evidence throws the glycocalyx into the complex mix of obstacles to intracellular delivery. The glycocalyx, a meshwork of glycolipids, proteoglycans, and glycoproteins, is an anionic layer of carbohydrates on the cell surface forming a barrier between the cell and its surroundings (Fig. 2B) [65,66]. It regulates adhesion and permeability, mechanotransduction, microenvironment, and intercellular signaling [65].
Moreover, it is responsible for protecting the luminal membrane of the lysosomes from degradative enzymes [67]. It presents a physiochemical barrier to the cell binding, entry, and endosomal escape of nanoparticles in vitro and in vivo (Fig. 2B) [68–70]. The thickness of the glycocalyx on endothelial cells, depending on the organ and species, can range from 0.02 μm in mouse diaphragmatic capillary as revealed by transmission electron microscopy to 8.9 μm in human retinal vessels as determined by sidestream dark field imaging [71]. Electron micrographs of rat kidney revealed the lysosomal glycocalyx to be considerably smaller at an average thickness of 8 nm [54]. Alterations to the glycocalyx have been reported in disease conditions such as cardiovascular diseases, cancer, kidney diseases, diabetes, and LSDs [72–74]. These changes and differences in glycocalyx between healthy and diseased cells may enable enhanced delivery or diagnostics [69,73,75–77]. Strategies to selectively degrade or modify the glycocalyx for therapeutic purposes and improving delivery have also been employed [55,68,69,78]. Cholesterol accumulation can be alleviated in Niemann Pick type C1 (NPC1)-deficient human patient-derived fibroblasts by treating with benzyl-2-acetamido-2-deoxy-α-D-galactopyranoside, an inhibitor of O-linked glycosylation [55]. Sialylation of glycans on the surface of cancer cells has been found to help tumors evade recognition by the immune system and aid tumor growth and progression, making sialic acid a target for cancer therapy [78,79]. Desialylation of cancer cells using recombinant sialidase conjugated to the human epidermal growth factor receptor 2-specific antibody, also known as trastuzumab, demonstrated enhanced cancer cell binding of natural killer cell-activating receptor, natural killer group 2D, and antibody-dependent cell-mediated cytotoxicity [78]. Exposing human umbilical vein endothelial cells to glycocalyx degrading enzymes, such as neuraminidase, heparinase III, and hyaluronidase, enabled significantly higher internalization of 50 nm carboxylated or aminated polystyrene nanospheres in contrast to the poor uptake observed in the presence of intact glycocalyx [68].
2.4. Molecular crowding
Another major barrier to successful intracellular delivery is the viscosity of the endosomal compartments and the cytosol that can restrict the diffusion of therapeutic or diagnostic agents to their target (Fig. 2C) [80–82]. The motion of the nanoparticle is subject to resistance from steric effects and non-specific interactions with the myriad proteins, lipids, nucleic acids, cytoskeleton, and organelles (Fig. 2C) [83,84]. Devany et al. have recently developed a new method to measure rheology of organelles by tracking nanoparticle movement inside the lysosomes of cells, and compared the viscosities between normal cell lines and model cell lines representing LSD [85]. They found that the accumulation of cholesterol in dysfunctional lysosomes restricted the free diffusion of delivery vectors [85]. Another study, using particle tracking analysis, found that the size of nanoparticles and their non-specific interactions with cytosolic components influence the mobility of nanoparticles [83]. Inert nanoparticles that are smaller than 50 nm in diameter predominantly exhibited Brownian motion in the cytosol (Fig. 2C). In the presence of non-specific interactions, however, the mobility of the nanoparticles was severely restricted (Fig. 2C). Nucleic acids delivered to the cytosol or the nucleus also face similar obstructions to mobility; for instance, DNA fragments longer than 250 base pairs showed less lateral mobility than shorter DNA due to molecular crowding in the cytosol (Fig. 2C) [86]. These findings may have implications for nanoparticle design and surface modifications; for instance, adding targeting ligands such as antibodies may increase the steric hindrance to diffusion.
3. Strategies to enhance intracellular delivery
3.1. Endosomal escape
To enhance the delivery of nanoparticles across endosomal barriers, two possible mechanisms, membrane destabilization for liposomes and lipid-based nanoparticles and proton sponge for polymers have remained at the center stage (Fig. 3) [7,14,16,87–93]. Studies have suggested that the nanoparticle structure can contribute to the efficacy of nucleic acid release [94–100]. Small angle X-ray scattering shows that the cationic lipid-nucleic acid complexes form an inverse hexagonal phase (HII), which is more amenable to DNA release from the endosome to the cytosol [98]. In recent years, novel lipid-based nanoparticle formulations containing siRNA have produced highly-ordered bicontinuous cubic internal structures [99]. After engulfment of lipid-based nanoparticles, the electrostatic and hydrophobic interactions of cationic lipids with the anionic endosomal membrane lead to the formation of the HII lipid phase causing endosomal escape (Fig. 3A) [99]. Sabnis et al. recently demonstrated the efficacy of novel ionizable amino lipids possessing enhanced endosomal escape capabilities and tolerability to chronic dosing in rats and non-human primates [7]. The lipid-based nanoparticles containing their novel ionizable lipid (lipid 5) exhibited a 6-fold higher endosomal escape efficiency over DLin-MC3-DMA-based nanoparticles [6,7]. Fusogenic helper lipids, such as dioleoyl phosphatidyl-ethanolamine (DOPE) and cholesterol have also been included in these lipid-based formulations to augment the formation of the HII phase [101]. Nanoformulations containing DOPE have frequently shown superior efficacy to alternatives [101–104]. DOPE-containing lipoplexes have been reported for the delivery of negatively charged proteins and peptides in Chinese hamster ovary cells and rat embryonic cardiac fibroblasts [105]. These fusogenic liposomes, formulated into proteoliposome complexes, demonstrated efficient delivery of R-phycoerythrin, EGFP, nuclear transport factor-2. Dendra2, and actin-binding peptide, LifeAct, in a size-independent manner. Cholesterol is known to play key roles in stability, cell entry, and endosomal escape of nanoparticles [101,104,47]. Cationic lipoplexes containing both, cholesterol and DOPE, demonstrated a cholesterol-dependent increase in DNA transfection efficiency with almost order of magnitude increase in transfection at 40 mol% cholesterol compared to lipoplexes devoid of cholesterol [106]. They found that these cholesterol-rich particles were utilizing cholesterol-driven endocytosis and membrane fusion for cellular entry and cytosolic localization. This cholesterol-dependent improvement in DNA transfection was also observed with lipoplexes containing 60, 70, and 80% cholesterol [107]. At these high cholesterol molar ratios, differential scanning calorimetry revealed the presence of anhydrous cholesterol domains in the nanoparticles [97]. The presence of these cholesterol domains was attributed to the ability of the lipoplexes to avoid protein adsorption and facilitate membrane fusion (Fig. 3A) [97].
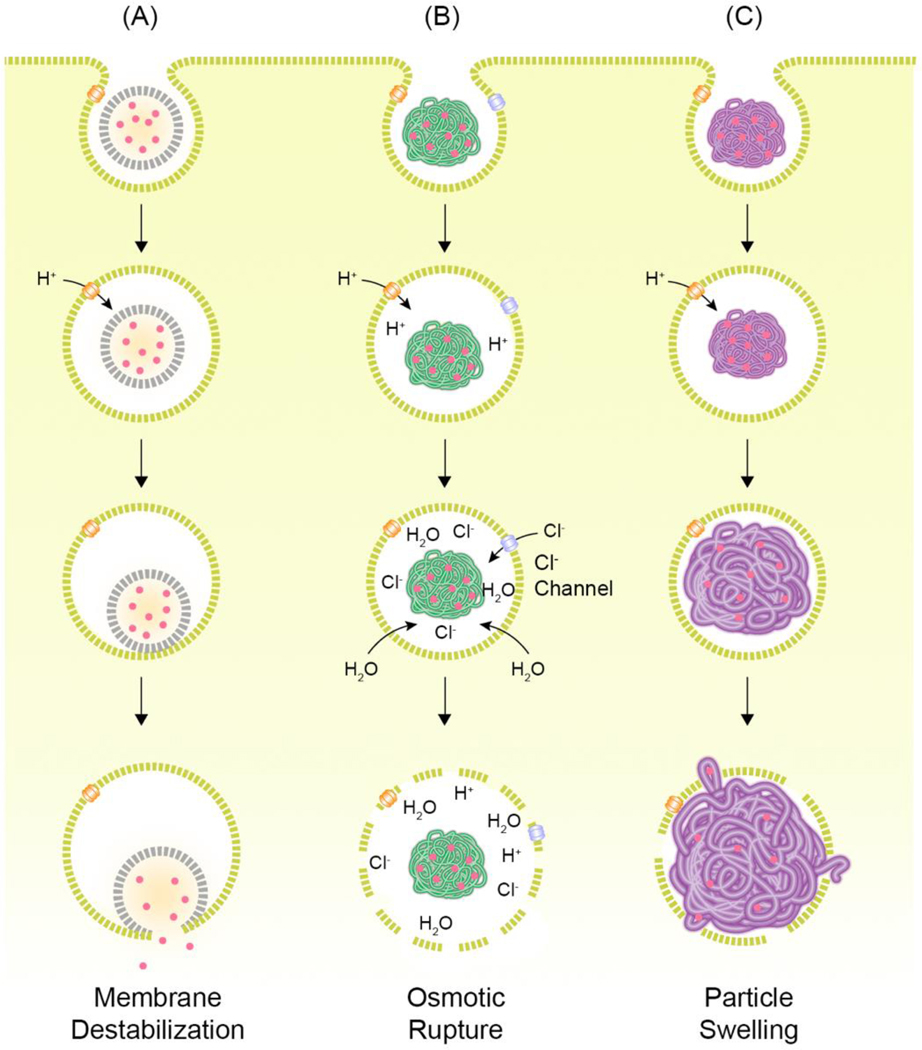
Fig. 3.
Possible endosomal escape of nanoparticles.
(A) Interaction of nanoparticles with endosomal membrane can destabilize the endosomal membrane, resulting to endosomal escape of the nanoparticles. (B) Nanoparticles including amines can rupture endosomes by osmotic pressure. (C) Nanoparticles designed to swell in acidic environment can burst endosomes and release their cargos to cytosol. Redrawn from [15].
Complexes of cationic polymers with nucleic acids, known as polyplexes, are also designed with endosomal escape in mind. The amines in these polymers drive the osmotic swelling and rupture of the endosomes [108,109]. In particular, the secondary and tertiary amines in these polymers reportedly contribute to this phenomenon by scavenging protons and becoming cationic in the acidic environment of the endosomes, resulting in the influx of more protons and counter ions [108,109]. This influx increases the ionic concentration in the endosomes and draws water inside by osmotic gradient, which leads to swelling and, finally, endosomal rupture (Fig. 3B) [108,109]. In another iteration of this hypothesis, called the umbrella hypothesis, the polymers swell due to charge-charge repulsion following protonation of the amines, causing the expanding polymer to rupture the endosomal membrane (Fig. 3C) [89,109]. While conclusive evidence of this hypothesis remains elusive, there are many publications attempting to study this process [89,109–115]. Among the first cationic polymers tested for nucleic acid delivery was poly-L-lysine (PLL). While PLL was able to encapsulate nucleic acids, it failed to deliver nucleic acids efficiently due to aggregation and it inability to avoid lysosomes [110,116]. It was soon suggested that endosomal buffering was required for efficient delivery to cytosol using cationic polymers, which was enabled by the inclusion of a high density of ionizable secondary and tertiary amines into the polymers. Polyethyleneimine (PEI) has been proposed to use similar pathways to escape the endosomes. N-quaternization of PEI reduces DNA transfection by two-orders of magnitude relative to PEI, suggesting the necessity of the PEI’s buffering effect [110]. The intracellular pH environment of PEI polyplexes was found to have an average pH of 6.1 compared to permethyl-PEI (pH 5.4), perethyl-PEI (pH 5.1), and polylysine (pH 4.6) polyplexes, suggesting that the PEI polyplexes have evaded trafficking to the acidic endolysosomes. A contradictory study in HeLa cells reported that DNA polyplexes with linear or branched PEI failed to induce changes in the lysosomal pH [111]. Perhaps an explanation for this is that the ATPase pumps may be able to overcome the buffering effects of PEI and maintain lysosomal pH stability. Another possibility is that a change in lysosomal pH may not be observed due to endosomal leakiness resulting from transient pore formation by polymer-membrane interactions as described previously (Fig. 3) [91]. Correlating the lysosomal size and concentration of PEI inside the lysosomes to mathematical calculations of critical membrane tension, it was postulated that a high PEI concentration might be required for lysosomal rupture [111]. This need for high accumulation of polymer inside the endo-lysosomes has been reported elsewhere [112]. However, in this case, efficacy would have to be balanced with cytotoxicity arising from exposure to a high concentration of cationic materials [117,118]. Vermeulen at el. determined that endosomal escape exploiting the proton-sponge effect is influenced by the endosomal size and membrane leakiness [113]. Endosomal size and leakiness were found to be cell-type dependent. Interestingly, the smaller endosomes in HeLa cells permitted 3.5-fold higher endosomal escape efficiency of JetPEI/pDNA polyplexes relative to the larger endosomes of ARPE-19 cells, despite higher uptake in ARPE-19. This difference could not be attributed to differences in endosomal acidification or mobility. Moreover, even in HeLa cells, escape was only observed in fewer than 10% of the polyplex-containing endosomes. This was found to arise from the leakiness of the endosome, likely resulting from interactions of polyplexes with the endosomal membrane, preventing effective buildup of osmotic pressure inside the endosomes to cause rupture. In further tests with A549 (large endosomes) and H1299 (leaky endosomes) cells, showed poor escape efficiencies for both cell lines, confirming the cell type-dependent detrimental impact of large endosome size and leakiness. Whether these differences in transfection are only due to variation in endosomal size and perhaps related to a myriad of other factors associated with cell type is unclear, and these studies require validation using methods that increase the endosomal size within the same cell-type which serves as an internal control. Interrogation of proton-sponge effect using super-resolution stochastic optical reconstruction microscopy (STORM) exposed the importance of polyplex architecture and cationic polymer rigidity in facilitating endosomal escape through endosomal swelling and rupture (Fig. 3B) [114]. It was speculated that grafting highly branched, and rigid cationic polymers (cationic glycogen and branched PEI) onto silica nanoparticles limits the interactions of the polymers with the lipid membrane, thereby driving escape through rupture of the endosomes not via membrane destabilization, in contrast to previous reports [91,113]. Additionally, the cytosolic dissociation of nanoparticles was found to be a rate-limiting step for glycogen-mediated siRNA delivery. Despite these advances and findings, on average, merely one or two escape events have been observed per cell [91,113,116].
A relatively straightforward means to improve endosomal escape is by supplementing the nanocarrier delivery with small molecule enhancers. Some of the widely employed molecules are the lysosomotropic agents (e.g., chloroquine, primaquine, ammonium chloride, tributylamine, methylamine) which can impede endosomal acidification, a process analogous to the proton-sponge effect [119]. This is postulated to hinder the activation of the lysosomal enzymes and increase retention of the nanoparticles inside the endo-lysosomal system. This retention provides additional time for nanoparticles to reach the cytosol. It was also found that chloroquine can induce dissociation of the polyplex, allowing cytosolic and nuclear localization of the nucleic acids [119].Additionally, it can also bind to DNA and may protect it against nuclease attack [120,121].
Massive libraries of pharmacologically active compounds have been screened to find endosomal escape agents for enhancing the delivery of clinically-relevant systems. One such study tested a library of over 45,000 compounds composed of kinase inhibitors, FDA-approved drugs, purified natural compounds, and other drug-like compounds to enhance siRNA delivery by lipid-based nanoparticles and cholesterol-conjugated siRNA. Fifty-one compounds were identified in the initial screen which improved gene silencing by 2–5 fold in HeLa [122]. These hits, validated in human primary fibroblasts and hepatocytes, were found to have mechanisms of action dependent on the delivery system as well as the cell type and was mediated by boosting endocytosis or escape from endosomes. In an even larger screen, Yang et al. identified 67 small molecules as enhancers of in vitro efficacy of splice switching oligonucleotides (SSOs) out of >100,000 compounds derived from multiple libraries [18]. One of the molecules, UNC7938, improved SSO-mediated luciferase induction in HeLaLuc705 cells by 220-fold [18]. Substantial enhancement of efficacy was also observed for antisense oligonucleotides and cholesterol-conjugated siRNA as well as receptor-targeted RGD-SSO-albumin conjugates in the presence of UNC7938. The enhancement effect of UNC7938 was successfully translated to in vivo in EGFP654 transgenic mice where SSO-mediated correction of splicing was observed in liver, kidneys, and heart. UNC7938 and its analogues potentiate oligonucleotide release from the late endosome via mechanisms distinct from the proton-sponge effect [18]. In follow-up studies on analogues of UNC7938, analysis of structure-activity relationship to dissect the fundamental characteristics governing the endosomal enhancer activity revealed that the lipophilic aromatic groups on UNC7938 is essential for activity and that the tertiary nitrogen was required [123]. UNC7938 family of compounds enables higher potency by enhancing permeability of oligonucleotides from an intermediate stage of trafficking between the early and late endosomes.
A commercially-available screen of 1,280 pharmacologically active compounds (LOPAC1280) revealed that Guanabenz acetate (Wytensin™), approved as an antihypertensive drug in 1982, could enhance RNAi efficacy of cholesterol-modified siRNAs by almost 100-fold. It was found that Guanabenz achieves this by inducing higher uptake, likely through weak non-covalent interactions with the siRNA, and mRNA silencing through an, as yet, unknown mechanism [124]. Patel et al. utilized a relatively small library of 212 bioactive lipids to enhance endosomal escape [17]. They hypothesized that the bioactive lipids could be utilized to influence the intracellular pathways to enhance endosomal escape of lipid-based nanoparticles. Moreover, these bioactive molecules could be readily incorporated into lipid-based nanoparticles due to their hydrophobicity for site specific delivery. They discovered that MK-571, a leukotriene receptor antagonist, and its functional analogues significantly enhanced mRNA transfection in vitro and in vivo.
An ingenious technique requiring a combination of NaCl hypertonicity-induced macropinocytosis and a transduction compound (propanebetaine), known as iTOP (induced transduction by osmocytosis and propanebetaine), was recently utilized for the efficient delivery of macromolecules such as cytosolic and nuclear protiens as well as small RNAs into a broad range of primary cells [125]. The iTOP system also demonstrated efficient gene editing following transduction of Cas9 protein and sgRNA. It was determined that the NaCl hypertonic media induced macropinocytic uptake of proteins, while the non-detergent Sulfobetaine-201 or the neurotransmitter gamma-amino-butyric acid triggered macropinosome leakage.
As a consequence of the challenges in tracking or measuring the efficiency of intracellular delivery, direct evidence of endosomal escape is not always demonstrated but rather inferred through enhancement in gene expression, gene knockdown, or other improvements in efficacy. However, recent studies have been able to quantify endosomal escape of nucleic acids using electron microscopy (EM), fluorescent in-situ hybridization (FISH), and super-resolution imaging [6,7,113,114]. It should be noted that despite often being associated with cytosolic delivery, the term endosomal escape may also be applied to endosomal trafficking to productive compartments such as the ER or TGN. For instance, Stalder et al. established the outer rough ER membrane to be the central nucleation site for siRNA-mediated gene silencing [126]. As such, it is possible that in some cases the observed enhancement in efficacy arises from trafficking directed towards productive subcellular compartments rather than direct cytosolic delivery. Materials capable of preferential delivery to the ER may boost RNAi-mediated gene silencing or mRNA translation for protein synthesis.
Although synthetic delivery vectors have been developed to improve intracellular delivery, their intrinsic artificiality occasionally hampers the therapeutic efficacy by causing unexpected toxicity, rapid clearance, or off-target effects. Clues to overcome these risks can be found in natural particulates which have evolved over time [35]. Significant effort has been made towards the development of drug carriers which exploit the natural trafficking machineries leading to the advent of biomimetic delivery vectors. Even though many kinds of bioinspired vectors have been developed, we confined our scope to the recent advances in biomimetic nanocarriers for drug delivery.
Viral vectors have been used for a long time in gene delivery. Their highly efficient mechanisms of immune evasion, cell entry, and endosomal escape made them the most advanced vectors for gene delivery. Since the high efficiency of delivery is mainly mediated by viral proteins, virus-mimicking nanocarriers contain viral proteins in their compositions. Virus-like particles are recombinant viral particles void of viral genome. They are self-assembled complexes of viral proteins and behave like natural viruses [127,128]. Virosomes, which have viral proteins embedded on a lipid layer, are replication-incompetent but share similar capabilities for membrane fusion. For example, virosomes equipped with L protein of hepatitis B virus were able to escape from endosomes by exposing the fusogenic domain at endosomal pH [129]. Peptide sequences derived from viral proteins have been used to bind the microtubule motor protein, dynein, for efficient intracellular delivery [130]. Gold nanoparticles modified with the dynein-binding peptides demonstrated efficient cellular internalization and high cytosolic motility by hitchhiking on dyneins. These biomimetic nanoparticles were also transported between cells through cell-to-cell contact and were able to cross the nuclear membrane. Despite these effective functions of viral proteins, their intrinsic immunogenicity raises concerns about undesirable immune responses, which delays clinical interventions. It can be useful for vaccination due to potential adjuvanticity of viral components. Nonetheless, the safety of virus-mimicking vectors should be thoroughly studied for usages other than vaccine delivery [35].
Peptides have been recently tested that can mimic the native characteristics of intracellular trafficking. Yang et al. have developed a complementary pair of cholesterol-conjugated (via a polyethylene glycol (PEG) spacer) coiled-coil peptides (“E3” [(EIAALEK)3] and “K3” [(KIAALKE)3]) to mediate membrane fusion in SNARE protein complex inspired manner [131]. Cell membrane could be spontaneously modified with the peptides by insertion of the cholesterol moiety. The nanoparticle, in this case, a liposome containing propidium iodide, TO-PRO®−3, or doxorubicin, decorated with the complementary peptide mediated efficient cytosolic delivery. This technology was also demonstrated to be feasible for in vivo delivery of doxorubicin to skin epithelial cells in zebrafish embryos. PEI polyplexes have also been tagged with histone 3-derived peptides to bypass the need for endosomal escape for pDNA delivery [132]. These histone-targeted polyplexes underwent differential trafficking through the caveolae-mediated endocytosis and accumulated in the ER/TGN, avoiding the degradative pathways, and finally reaching the nucleus following mitosis.
Proteins conjugated to drugs via acid or enzyme labile linkers have also been developed for intracellular delivery of cancer therapeutics [2]. Antibodies target specific receptors differentially expressed on cancer cells and are internalized along with the drug. The low endosomal pH or specific lysosomal enzymes then cleave the linker and release the drug into the cancer cells [133,134]. Several nanodrugs such as Ontak, Mylotarg, Adcetris, Kadcyla, and Vintafolide, designed on the basis of aforementioned principle are either approved or in various phases of clinical trials [135–138]. In cancer immunotherapy, activation of the stimulator of interferon gene (STING) pathway is under extensive investigation to trigger interferon I-driven immune responses against cancers. Recently, multiple groups have used nanoparticles to deliver STING agonists across vesicular compartments [139–141]. Shae et al. developed polymeric nanoparticles for delivering a cyclic dinucleotide ligand for STING, 2’3’ cyclic guanosine monophosphate-adenosine monophosphate (cGAMP), to the cytosol. They screened a series of differently weighted PEG-block-[(2-(diethylamino)ethyl methacrylate)-co-(butyl methacrylate)-co-(pyridyl disulfide ethyl methacrylate)] copolymers to discover the most efficiently escaping polymer from endosomes based on haemolysis assay. The optimized nanoparticles were able to deliver cGAMP to the cytosol and displayed effective anti-melanoma effects in preclinical animal models [139].
3.2. Targeting vesicular compartments to restore cell function
LSDs make for interesting therapeutic and diagnostic targets. These disorders are characterized by the endo-lysosomal accumulation of substances such as lipids, polysaccharides and proteins as a result of defects in intracellular trafficking (Fig. 4) [26]. Mutation in NPC1/2 result in cholesterol accumulation in the late endosomes/lysosomes due to blocked efflux from these compartments (Fig. 4) [142,143]. This type of accumulation was shown to be beneficial for nanoparticle delivery [9]. siRNA delivery to the cytosol is enhanced in the NPC1 knockout cells compared to the wild-type. Tropism of nanoparticle to the lysosomes has been utilized to both dissect the molecular mechanisms of NPC dysfunction and preferentially deliver therapeutic agents to ameliorate cholesterol buildup in NPC disorder. Superparamagnetic iron oxide nanoparticles were used to isolate late endosomes and analyze their chemical composition in NPC1 null cells [144,145]. Brown et al. recently utilized PEG-lipid micelles as bioactive agents [146]. The PEG-lipid micelles were observed to effectively colocalization with the accumulated cholesterol within the endolysosomes. Furthermore, the combination treatment with PEG-lipid micelles and 2-hydroxypropyl-β-cyclodextrin could synergistically facilitate cholesterol efflux from these endosomal compartments [146]. Interestingly, cationic polymer-based nanoparticle formulations which use caveolae-mediated internalization route instead of macropinocytosis, showed less efficient uptake and low endosomal escape of cargo suggesting that the composition of the nanoparticles and their route of cellular internalization plays a critical role in determining the efficacy of these delivery systems [147].
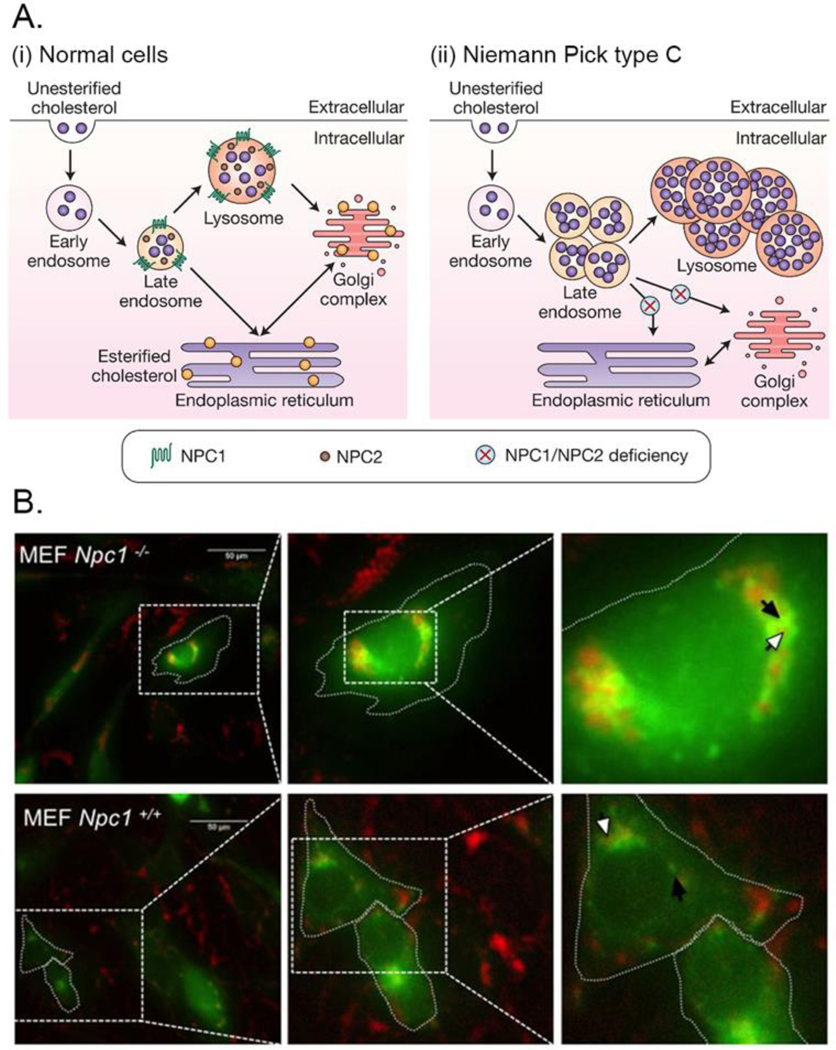
Fig. 4.
Altered lipid trafficking in Niemann Pick type C disease.
(A) Schematic illustrations of lipid trafficking in the normal condition and NPC disease. (i) In normal cells, cholesterol is processed from endo-lysosomal system to the endoplasmic reticulum, Golgi-complex, and other organelles. (ii) Loss of function mutation in NPC1 or NPC2 inhibit the egress of cholesterol and other lipids to cytoplasmic locations, accumulating them in late endosomes and lysosomes. Reproduced with permission from [143], Copyright 2008, Cambridge University Press. (B) NPC1−/− Mouse Embryonic Fibroblasts (MEFs) had enlarged late endosomes than the NPC1+/+ MEFs. Cholesterol was accumulated in the late endosomes of NPC1−/− MEFs whereas it was much diffused to cytoplasm in NPC1+/+ MEFs. NPC1−/− and NPC1+/+ MEFs were transfected with Rab-7 GFP (pseudo colored green) to label late endosomes. After fixation, the cells were stained with filipin (red) to visualize subcellular cholesterol. Dotted lines indicate the cell outline; white and black arrows indicate filipin and Rab7-GFP, respectively. Reproduced with permission from [146] Copyright 2016, Springer Nature.
Intercellular adhesion molecule 1 (ICAM1)-conjugated polystyrene and poly(lactic-co-glycolic acid) (PLGA) nanocarriers were used to deliver acid sphingomyelinase to the lysosome [148]. The nanocarriers carrying acid sphingomyelinase reached lung, liver, and spleen with a much higher efficiency than the enzyme alone and reduced storage of sphingomyelin in mice lungs. A similar strategy was used in the case of Fabry disease to deliver α-galactosidase A, to reduce lysosomal accumulation of globotriaosylceramide as well as in Pompe disease to deliver acid α-glucosidase (GAA) in order to decrease excessive glycogen storage in mice [149,150]. In a different approach, the inherent cellular clearance pathways were stimulated to treat lysosomal storage disorders. Spampanato et al. transfected GAA null mice with adeno-associated virus (AAV) expressing transcription factor EB (TFEB), a transcription factor that has been shown to be a master regulator of lysosomal biogenesis and autophagy [151,152]. TFEB overexpression led to increased formation and fusion of autophagosomes and lysosomes leading to clearance of accumulated glycogen. Intramuscular or systemic delivery of AAV-TFEB in mice caused the overexpression of TFEB (under muscle creatinine kinase promoter) and resulted in enhanced glycogen clearance and improved muscle architecture [153].
Intracellular pathogens such as Salmonella enterica serovar Typhimurium and Francisella tularensis evade the immune systems and therapeutics by residing in the endo/lysosomes or cytosol of cells [27,154–156]. A new delivery system made up of acetalated dextran (Ace-DEX) was developed to deliver a hydrophobic drug, AR-12, to phagocytes [157]. These biodegradable and pH responsive Ace-DEX microparticles (200 – 1000 nm in size) demonstrated efficacy at reducing bacterial burden in human monocyte-derived macrophages infected with Salmonella enterica serovar Typhimurium. Intranasal delivery of these particles in mice showed highest localization in lungs, liver, and kidney. The Ace-DEX microparticles were found to induce escape from the endo/lysosomal system via an unknown mechanism. The presence of inflated lysosomes led to the hypothesis that the escape may be caused by polymer swelling leading to membrane destabilization or proton-sponge effect. Targeting the mannose receptor has emerged as a strategy for delivery to macrophages, which overexpress this receptor [158–161]. Choi et al. also reported that the presence of mannose on the gallium-loaded polymeric nanoparticles helped the internalization of the nanoparticles into macrophages, thereby enhancing anti-microbial effects of gallium against Mycobacterium tuberculosis [162]. They also demonstrated that endosomes containing the gallium (III)-loaded nanoparticles fused with phagosomes and helped the maturation of phagosomes by inhibiting the activities of Mycobacterium tuberculosis. This affinity of macrophages to mannose can also be employed to deliver cargos targeting the immune system, such as antigens and immunostimulants [158,161,160]. Recently, Ai et al. developed mannosylated albumin nanoparticles for oligodeoxynucleotide delivery [163]. The mannose moiety on the nanoparticle surface helped to target macrophages, and the oligodeoxynucleotides stimulated the endosomal toll-like receptors upon the endocytosis of the nanoparticles.
3.3. Endosome-derived vesicles for intracellular delivery of exogeneous molecules
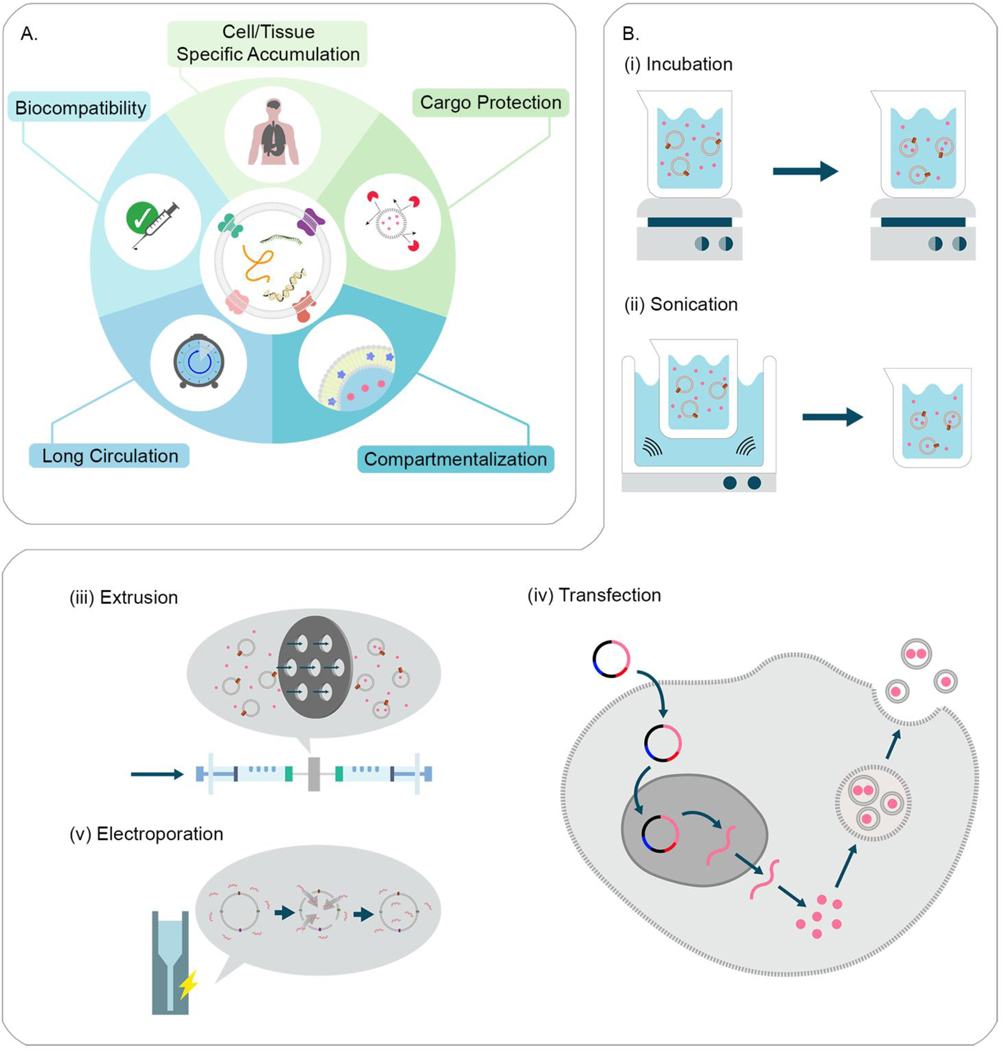
Exosomes refer to a type of membrane-derived vesicles, which are 30–150 nm in diameter. They are generated from intraluminal vesicles by inward budding of endosomal membrane of multivesicular endosomes and secreted upon fusion of these with the plasma membrane (Fig. 1). Exosomes are capable of delivery of various cargos, including proteins, lipids, and nucleic acids acting as communicators that mediate exchange of signals between cells (Fig. 5A) [164]. They are involved in a wide range of intercellular communications, including antigen presentation [165,166], immunomodulation [167,168], tissue repair [169,170], and cancer growth [171,172]. Exosomes isolated from patients with cancers contain specific biomarkers, highlighting their potential in cancer diagnostics and early detection [173,174]. These vesicles are biocompatible carriers that can be tuned to present specific molecules on the surface by engineering the donor cells (Fig. 5A) [175–177]. It is possible to introduce artificial or exogenous molecules into the membrane of exosomes [178]. Moreover, they are considered to have an innate homing capability which can be used to control their biodistributions and targeting (Fig. 5A) [36]. Even though it is not clear how exosomes reach particular recipient cells, there is increasing evidence that the composition of exosomes influences their fates and functions [36,39]. For instance, syncytin 1 in the trophoblast-derived exosomes causes cellular uptake of exosomes [179]; while CD47 expression prevents phagocytic uptake [180]. Macrophage-derived exosomes can utilize, 1) the integrin lymphocyte function-associated antigen 1 and ICAM-1, and, 2) the carbohydrate-binding C-type lectin receptors, to interact with brain microvessel endothelial cells comprising the blood-brain barrier [181]. Another example shows that exosomes with amyloid β (A4) protein preferentially target neurons; while the ones with tetraspanin interact with both neurons and glial cells [182]. Exosomes are thought to fuse with plasma and endosomal membranes by docking through membrane-anchored lipids, sugars, or proteins allowing release of packaged molecules directly to cytosol, avoiding endo-lysosomal degradation [183]. However, there are ample studies that have suggested exosomes also enter cells by multiple endocytic pathways and get entrapped in endolysosomal compartments like any other material [43,181,184]. These processes can be variable depending on the type of donor and recipient cells, and the composition of exosomes. Nonetheless, this inherent fusogenicity of exosomes can significantly boost the delivery efficiency, leading to enhancedtherapeutic outcomes.
Fig. 5.
Harnessing exosomes for drug delivery.
(A) Exosomes are considered to have great potential as drug delivery systems due to their biocompatibility, cargo protection, long circulation time, and targetability to specific cells/tissues. Also, the lipid membrane of exosomes allows for loading hydrophobic and hydrophilic drugs in the interlayer and core compartments, respectively. Redrawn from [327]. (B) Cargo packaging in exosomes for drug delivery. (i) Incubation of exosomes with cargos allows for the encapsulation of cargos in the exosomes. (ii) Sonication destabilizes the membrane of exosomes, which leads to the influx of cargos. (iii) Extrusion of exosomes with cargos causes physical breakages of the membrane, resulting to the encapsulation of cargos. (iv) Transfection of parental cells with a gene of interest can introduce the generation of the exosomes containing cargos. (v) Electroporation transiently increases the permeability of exosomal membrane so that genetic cargos can be incorporated.
A number of studies proved that exosomes can be used as delivery vectors for various therapeutics. Small molecules, such as curcumin [185], doxorubicin [177] and paclitaxel [178,184], were encapsulated in exosomes by co-incubation, mild sonication, or extrusion (Fig. 5B-i-iii). [186]. Proteins were also loaded into exosomes by prior transfection of parental cells with corresponding genes (Fig. 5B-iv) [175,187]. The delivery of exosomes and their cargo to the brain increases during brain inflammation, which could be due to increased expression of cell adhesion molecules, such as ICAM-1 at the blood-brain barrier [181]. Batrakova and colleagues reported that therapeutic proteins could be delivered to central nervous system using exosomes [36,43]. They transfected the IC-21 macrophage cells with plasmid DNA encoding tripeptidyl peptidase-1 (TPP1), and demonstrated the exosomes released from the transfected cells contained the functional TPP1 and TPP1-encoding nucleic acids. They showed that fluorescently-labeled exosomes reached the brain tissues across the blood-brain barrier upon intraperitoneal administration and the exosome-mediated TPP1 delivery enhanced in vivo survival rates in the mouse model of Batten disease. Short nucleotides, including siRNA and miRNA, were delivered using exosomes as well [176,188]. Pegtel et al. reported that virus-derived miRNA was encapsulated in exosomes by prior infection of donor cells with virus (Fig. 5B-iv) [189]. Wood and coworkers encapsulated exogenous siRNA in isolated exosomes by electroporation (Fig. 5B-v) [186]. They engineered the bone marrow-derived dendritic cells not only to package exogenous siRNA by electroporation, but also to express the rabies viral glycoprotein peptide that selectively binds to the acetylcholine receptor. Intravenous administrations of the engineered exosomes displayed the accumulation of the exosomes in neurons, microglia, and oligodendrocytes in the mouse brain, leading to the silencing of the specific gene expression. It is challenging to package larger nucleic acids (i.e., mRNA and pDNA) in exosomes due to their size and negative charge. Recently, Kojima et al. reported that exosomes could be designed to package therapeutic mRNA by engineering donor cells and the exosomes produced from the engineered cells delivered therapeutic mRNA to target organs upon implantation of the cells in a preclinical animal model [190]. However, there are several challenges relating to exosome-mediated delivery. Loading efficiency of exosomes is relatively poor compared to other types of delivery vehicles, usually no more than 30% loading efficiency for small molecules, with often worse loading for genetic cargos. Another challenge is heterogeneity of exosomes, which makes it challenging to characterize and purify them. In particular, it is not trivial to differentiate exosomes from other extracellular vesicles (i.e., ectosomes) since size ranges are overlapping and morphologies are similar to each other [164]. These two limitations can significantly hamper the yield of exosome production. Furthermore, natural exosomes include endogenous debris or signaling molecules. Inclusion of unknown or disrelated molecules may inhibit effects of the therapeutic cargos or cause side-effects, particularly when administering a large dose. In spite of these challenges, exosomes are still very promising as a delivery platform. Since exosome engineering is rapidly evolving, novel methods free of the aforementioned concerns may appear soon.
4. Tracking intracellular trafficking of nanomaterials
While a plethora of studies are focusing on understanding the nanoparticle interactions at the cellular and subcellular levels, these studies are primarily performed in a model cell line in vitro. There are fundamental differences in the endocytosis and endosomal trafficking in vitro/ex vivo and in vivo [6,191–195]. These findings highlight the significance of developing and utilizing techniques for in situ analysis of nanoparticle trafficking. Such insights would enable rational development of advanced delivery systems while directing the evolution of nanomedicines closer and closer to more relevant in vivo settings. An arsenal of biochemical and biophysical tools has been developed and implemented over the years, which has contributed to the elucidation of nanomaterial intracellular trafficking properties, interacting moieties, subcellular localization as well as spatiotemporal localization within cells and even organelles (Fig. 6). Thanks to the continuous improvements in these technologies, it is even now possible to see high resolution 3D images with subcellular detail in whole, multicellular transparent live model systems like zebrafish as evidenced by Betzig and colleagues [196].
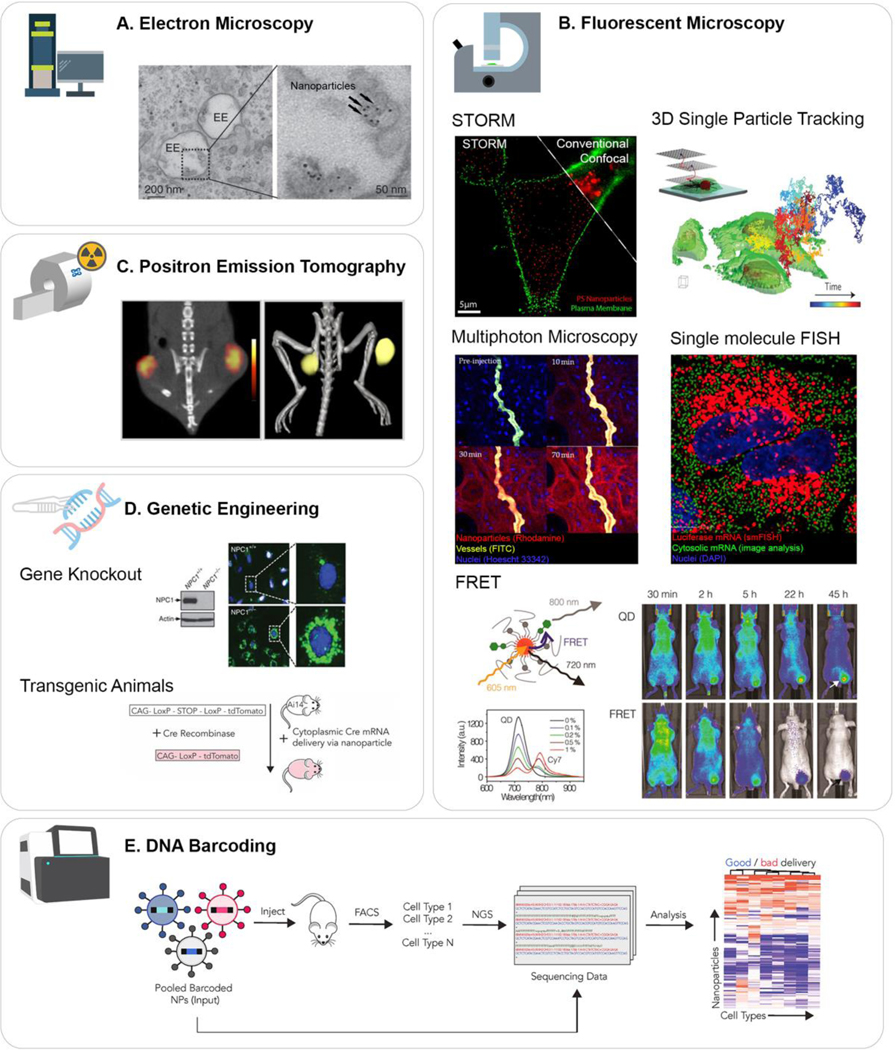
Fig. 6.
Approaches to trace the intracellular trafficking of nanomaterials.
(A) Electron microscopy can be used to track the subcellular locations of nanomaterials. (B) Fluorescence microscopy has been developed to achieve higher resolutions, real-time and intravital imaging, and deeper penetration into tissues. By labeling nanomaterials with fluorophores, their trafficking can be traced using these methods. (C) PET scan is extremely sensitive and quantitative tool to track radiolabeled nanomaterials. (D) Genetic engineering is present as a useful tool to study trafficking of nanoparticles. Uses of knockout cell lines and transgenic animals allow tracking modulators involved in the nanoparticle trafficking. (E) DNA barcoding is particularly useful to investigate endosomal escape for gene delivery. With Ai9 mice and next generation sequencing, it is possible to examine the transfection quantitatively as well as qualitatively. Adapted from [6] (A), [7,222,229,247,253] (B), [282] (C), [9,289] (D), and [293] (E) with permission.
4.1. Electron microscopy
EM is one of the leading techniques used for high resolution imaging at the vesicular level by electromagnetic focusing of electron beams onto thinly cut sections of cells or tissues (Fig. 6A). EM uses electron beams rather than photons of light, and therefore has sufficient power to provide image resolution greater than 50 pm [197]. The transmission electron microscope as well as the scanning electron microscope have been widely used for the physiochemical characterization of nanoparticle formulations with near atomic resolution [198–200] as well as the visualization of nanoparticle trafficking and uptake in the drug delivery and imaging fields for decades (Fig. 6A) [6,17,201–205]. It has also been implemented in elucidating viral uptake and distribution in tissues with subcellular detail [206,207]. In recent years, higher resolution advancements have arisen from new direct electron detectors and software improvements for image processing [208–210] and have subsequently allowed imaging and detailed, morphological characterization of large, flexible, and native conformation bioconjugates [195–197]. This has led to detailed single particle analysis related to nanoparticle formation and characterization, uptake, intracellular interaction elucidation, trafficking and delivery with atomic-scale resolution [200,211–216]. Frauenfeld and company recently showed how using EM advancements at their fingertips, they were able to characterize the reconstitution and stabilization of membrane proteins in their saponin-nanoparticle system and produced high-resolution structural findings to help stabilize them in the proper scaffold [211]. Azubel et al. employed cryo-electron microscopy advancements to image cell trafficking and mechanistic dynamics of fibroblast growth factor 21 when tagged with gold nanoparticles and delivered into human fat cells. This technique, in principle, allows for determining the structure proteins form inside cells at high resolution [214].
Gilleron et al. went a step further and utilized quantitative fluorescence imaging and electron microscopy to observe the uptake, trafficking, and endosomal escape of gold-tagged siRNA-loaded lipid-based nanoparticles in vivo (Fig. 6A) [6]. This important study quantified the inefficiency of endosomal escape even for state-of-the-art lipid-based nanoparticles. They found that endosomal escape was taking place at a specific stage in the trafficking process, allowing less than 2% of siRNA to escape to the cytosol in HeLa cells in vitro and in mouse liver hepatocytes in vivo [7]. Taken together, these improvements and their continued applications with other techniques like fluorescence, are further improving our understanding of the endocytic voyage of nanoparticles in biological systems. Although the resolution and structural detail available with these tools is cutting-edge, the scope of information we can extract using these techniques is limited to in vitro and ex vivo analysis of trafficking and delivery in most cases.
4.2. Super resolution microscopy
Fluorescence imaging is a ubiquitously implemented technique used to probe a wide range of biological obscurities, including endocytic intracellular trafficking and drug delivery (Fig. 6B). The field has advanced a great deal since the isolation of GFP from jellyfish in 1961 [217]. It is now possible to look deep inside cells past the previously restrictive diffraction limit of optical microscopes and into the realm of super resolution fluorescent microscopy [218,219]. STORM, also referred to as photoactivation localization microscopy (PALM) is an offshoot of super resolution microscopy that uses switchable organic fluorophores that can be individually turned on and off to map their nano-specific location in a sample to an astounding 20 nm resolution (Fig. 6B) [218]. The full image is reconstructed from iterative rounds of fluorophore switching, mapping, and subsequent reconstruction [220]. This technique can provide novel information on nanoparticle characterization (Fig. 6B) and cell uptake at the single molecule level in a way that other methods like electron microscopy cannot [221,222]. STORM/PALM has made it possible to reconstruct 3D localization of fluorescent probes, yielding 3D, near-molecular resolution images of nanoscopic cellular structures [223]. With this nanoscopy in mind, the cell trafficking field has adapted STORM/PALM to precisely elucidate the size and position of nanoparticles inside cells, providing the ability to probe interactions within cellular machinery during uptake and trafficking in diverse populations of cells including primary dendritic cells, derived cell lines like BS-C-1 and ex vivo [222–224]. To date, this technology has given great insights into nanoscale trafficking, and it will benefit from the continued development and eventual meshing with in vivo imaging technologies like intravital imaging to elucidate mechanisms of nanoparticle trafficking in living animals.
4.3. Single particle tracking
Fluorescent-based single particle tracking is providing us with tremendous insight into nanoparticle trafficking and interactions (Fig. 6B) [225–229]. It is a valuable tool in the booming super resolution microscopy field [230]. Our current understanding of nanoparticle interactions with its environment are still largely based on bulk observations involving multiple particles. A detailed investigation into the interactions of a nanoparticle along its journey into the cell and out of the endosome and beyond requires precise spatiotemporal imaging in 3D space and variable time scales. The passive or active translocation of a nanoparticle, through rapidly changing distinct environments, involving interactions with lipids, proteins, nucleic acids and other intracellular constituents, over time-scale of seconds to hours, needs to be considered and tracked. The effective delivery of a nanoparticle has to transverse this highly dynamic route. Surface properties of nanoparticles were shown to influence diffusion in the cytosol. Single-particle tracking was applied to monitor diffusion of quantum dots with neutral, anionic, or zwitterionic surface modifications [225]. Observations implicate the involvement of surface charge in cellular uptake but interestingly had little effect on cytosolic localization and the zwitterionic quantum dots demonstrated highest cytosolic mobility and homogeneity. Welsher and Yang developed a 3-D multiresolution imaging technique to investigate the cell binding and uptake of TAT-modified polystyrene-quantum dot nanoparticles in real time (Fig. 6B) [229]. This interrogation revealed that the nanoparticles exhibited ‘kiss-and-run’ behavior with transient binding and unbinding taking place. Deceleration of nanoparticles on approach to the cell membrane experienced deceleration earlier than predicted, indicating potential interaction with cell surface receptors. Nanoparticle tracking on local membrane structures and filopodia uncovered correlation between membrane structures and membrane fluidity with implications for nanoparticle delivery and viral infections. Another study employed nanocarriers made up of a photoactive copolymer and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-PEG2k coated with cancer cell membrane to study tumor structure in vivo [231].
4.4. Intravital microscopy
Fluorescent microscopy, along with EM, forms the core of toolbox at our disposal for studying intracellular trafficking. While EM imaging must be conducted on tissue sections, a subset of fluorescent microscopy provides us the capability to study endocytosis and trafficking in live animals through intravital imaging. Intravital imaging is typically performed using fluorophores, materials conjugated or loaded with fluorophores, and fluorescent transgenic animals [231–233]. Advances in intravital imaging would allow us to study dynamic cellular mechanisms in their native environment. Furthermore, the pharmacokinetics and pharmacodynamics of therapeutics and nanoparticles can be evaluated in vivo, enabling improved understanding and prediction of the underlying mechanisms and effects in various physiological and pathological states (Fig. 6B) [231–238]. Intravital imaging may also open new avenues for diagnostics and early detection. Endosomal exocytosis has also been studied in vivo using intravital imaging [191]. In one such study, the exocytosis of large secretory granules in the acinar cells of the submandibular salivary glands in transgenic mice (ubiquitously expressing soluble GFP or membrane-targeted tdTomato) was imaged at subcellular resolution using confocal intravital microscopy [239]. Observation of exocytosis in acinar cells in rats transfected with GFP-LifeAct (to label the actin filaments) or RFP-LifeAct together with GFP-farnesyl (to label actin and the plasma membrane, respectively) showed that secretory granules fuse with the apical plasma membrane to release their contents via a slow F-actin- and mysoin II-dependent collapse. Higher amount of details can be obtained by combining fluorescent microscopy and 3D electron microscopy [240,241]. Advances in light microscopy and EM has provided us the capability for single-cell and subcellular resolution for organ- and organism level imaging and may be used to study trafficking of nanomaterials in-vivo.
4.5. Two-photon microscopy
Since Denk et al. first applied two-photon microscopy in 1990, it has rapidly become an established technique for intravital imaging [232,233,242–244] and more importantly, it has been adapted to super resolution microscopy techniques much like other fluorescence tools [245]. Two-photon or multiphoton microscopy utilizes two or more low energy photons to excite a fluorophore instead of the single high energy photon required for traditional microscopy. The low energy of the photons reduces scattering and increases tissue penetration. Additionally, the low probability of simultaneous absorption of two low energy photons provides the ability to tightly control excitation allowing for high resolution 3-D imaging [242,246]. Intravital two-photon microscopy in rat kidney revealed that fluorescently-labeled folic acid could undergo transcytosis across the proximal tubule cells following binding and uptake on the apical side [235]. Majority of the folic acid accumulated in the lysosomal compartments. Similarly, a folate-PEG-rhodamine conjugate with varying lengths of PEG was coupled with intravital multiphoton microscopy to gain a better understanding of the kinetics of tumor accumulation resulting from folate-mediated endocytosis demonstrating that higher molecular weight PEG results in longer circulation times but sluggish tumor penetration (Fig. 6B) [247]. The transport of insulin-loaded deoxycholic acid-conjugated chitosan nanoparticles across the rat intestinal epithelium following oral administration was tracked via intravital two-photon microscopy and found the involvement of the apical sodium-dependent bile acid transporter-mediated endocytosis [248].
4.6. Dynamic fluorescent microscopy
Dynamic subcellular and molecular events and interactions can also be elucidated by exploiting fluorescent imaging techniques such as Förster resonance energy transfer (FRET), fluorescence recovery after photobleaching (FRAP), fluorescence loss in photobleaching (FLIP), and fluorescence lifetime imaging microscopy (FLIM) [86,225,249–251]. FRET has also been used to study nanoparticle assembly and dissociation (Fig. 6B) [9,252]. Zhao and colleagues have applied FRET imaging to determine the fate of nanoparticles in vivo using self-assembled lipidic nanoparticles with quantum dot core and PLGA nanoparticles loaded with doxorubicin prodrugs (Fig. 6B) [253,254]. They found that the nanoparticles were subjected to rapid lipid exchange with blood components resulting in dissociation of the nanoparticles. The quantum dot cores were internalized by the cells in the tumor, lymph nodes, and the mononuclear phagocyte system [253]. Similarly, drug encapsulated within nanoparticles can be released prematurely due to interactions with serum components such as albumin and high-density lipoprotein. FRET further revealed that hydrophobicity and miscibility of cargo with the nanoparticle are key properties affecting the drug release profile and modulating these properties by modifying the drug (doxorubicin) could enhance antitumor efficacy of the nanoparticles [254]. The photoactive copolymer was engineered to possess near infrared (NIR)-excitation and NIR-emission due to FRET within the molecule. Here, two incident photons are absorbed by one moiety of the copolymer and the energy is transferred over to the second moiety which then emits at NIR. The long circulation times and tumor specific localization enabled by the cancer cell membrane coating resulted in high resolution imaging of the heterogenous architecture within the tumors. FRAP and FLIP have also been used in conjunction with FRET to image intracellular dynamics of protein interactions. Shimi and colleagues were able to establish that a DNA-binding protein, BAF, was highly mobile during interphase which was in stark contrast to its binding partners Emetin, LAP2β, and MAN1 which were immobilized at the nuclear envelope. FLIP imaging further showed the fast-diffusing BAF pools in cytoplasm and nucleus compartments being differentially regulated as nuclear BAF levels were unable to replenish cytoplasmic levels [251]. As shown by Pihl et al., FRAP enabled silica particles can be conjugated with NMR responsive moieties for the elucidation of local and global diffusion constants of materials in relevant media to study mass transport [255]. Basuki et al. employed FLIM based detection to show doxorubicin-loaded, iron oxide nanoparticle accumulation in lung and breast cancer cell lines as well as nuclear drug release kinetics [256]. Although these imaging techniques have pushed the resolution limits, the field is still in need of more in vivo, real time measurements as that represents the more relevant environment for efficacious future clinical advances.
4.7. Microscopy for subcellular chemical analysis
The inability to effectively audit lipid accumulation in non-alcoholic fatty liver disease (NAFLD), LSDs, atherosclerosis, and drug-induced phospholipidosis is an ongoing concern. The inherent band-gap fluorescence of single-walled carbon nanotubes (SWCNTs) can be modulated based on DNA conformational polymorphism [257]. The DNA conformation, which is susceptible to its environment, can modify the dielectric characteristics of the SWCNTs enabling their use as optical reporters for examining the lipid content of the endolysosomal system in cells. Near-infrared hyperspectral imaging of SWCNTs, complexed with a short single-stranded DNA, exposed the lipid accumulation in NPC1 patient-derived fibroblasts and lipid clearance upon treatment with 2-hydroxypropyl-β-cyclodextrin [258]. This reporter system also enabled the analysis of kinetics of lipid accumulation on a single-cell level with subcellular resolution in macrophages. Lipid accumulation could even be dynamically monitored over weeks in mouse models of LSDs, NPC1, NPA/B, and NAFLD [32]. This study also confirmed that hepatic lipid accumulation induced by a high-fat diet can persist for a protracted length of time despite switching to a normal diet.
In addition to LSDs, lysosomal dysfunction is also involved in Alzheimer’s and Parkinson’s disease. A novel, fluorescence-utilizing technique called two-ion measurement (2-IM) was developed to deconvolute the chemical differences between lysosome subpopulations [29]. Utilizing a DNA nanodevice (composed of a FRET pair, Alexa 546/647, and chloride sensitive fluorophore, BAC) to simultaneously quantify lysosomal pH and chloride in a lysosome, Leung et al. were able to image two distinct lysosomal populations in healthy patient-derived fibroblasts with single-organelle resolution, one with low chloride and another with relatively higher chloride and lower pH. As previously observed, perinuclear lysosomes were more acidic than peripheral lysosomes. Applying 2-IM to fibroblasts derived from patients with NP-A, NP-B, or NP-C resolved that lysosomal pH is higher and uniform throughout the cells whereas, chloride levels were lower in perinuclear lysosomes compared to peripheral lysosomes. Treating NP-A and NP-B with recombinant enzyme acid sphingomyelinase and NP-C with 2-hydroxypropyl-β-cyclodextrin exhibited reemergence of the low-pH and high-chloride lysosomal phenotype. Another DNA-based nanoreporter was developed by the same group to quantify enzymatic disulfide reduction in endosomes in Caenorhabditis elegans as a potential platform technology for analyzing endosomal enzymatic activity [33]. Protein disulfide isomerase 3 and thioredoxin-1 (TRX-1) were found to execute disulfide reduction inside the late endosomes of C. elegans. Late endosomal disulfide reduction in HeLa cells is also performed by TRX-1 following scavenger receptor-mediated endocytosis. Infectious challenge using Corynebacterium diphtheriae, which secretes diphtheria toxin uncovered a role of TRX-1 in mediating cytosolic localization in C. elegans. This technology could be translated to measure in situ enzymatic activity in LSDs and perhaps coupled to super resolution deconvolution resources to enhance the resolution achieved.
4.2. In situ hybridization
Another tool for the nano-mapping of cellular trafficking dynamics is FISH. FISH is a technique developed in 1982 to localize and characterize hybridized DNA molecules to Drosophila chromosomes by Langer-Safer et al. [259]. Over the last 20 years, FISH has extended to single transcript RNA localization and counting that has allowed for elucidation of transcription/translation dynamics at the single cell level with precise spatiotemporal localization inside the cytoplasm and organelles (Fig. 6B) [260–264]. The dynamics of nanoparticle uptake, trafficking and delivery, have recently applied mRNA FISH technologies for the specific spatial localization of single molecule mRNA transcripts over time. This has permitted detailed probing of the elusive intracellular trafficking mechanisms as well as endosomal escape elucidation in vitro with relevant animal models examined [7,265]. In fixed cells, single molecule FISH (smFISH) makes individual mRNA transcripts appear as visible fluorescent spots that can be readily analyzed by fluorescence microscopy [262]. Sabnis et al. recently showed structure-function relationship of amino lipid component of nanoparticle and its inherent link to increased endosomal escape of delivered mRNA in cells which was found to be around 15% with their new lipid and 2.5% with DLin-MC3-DMA, further confirming studies performed using EM (Fig. 6A-B) [6,7]. An application of smFISH has described trafficking of viral particles including their internalization and gene delivery process inside cells [266–268]. The nanoparticle field is continuing to exploit the smFISH localization and single transcript clarifications provided by virologists and have begun evaluating mechanisms to improve trafficking and delivery for therapeutic purposes (reviewed extensively elsewhere [269–271]). The continued development of smFISH, as evidenced by Shaffer et al. [272] and Liu et al. [264], coupled to novel, emerging super resolution microscopy techniques, as recently reported by Wang et al. [273], will assist in resolving mechanisms of endocytic uptake, and contribute to efficient delivery of cargos in the nanoparticle field [264,272,273]. The need for in vivo-capable FISH strategies is palpable and is evidenced by recent attempts at developing such adaptations to the established techniques. Fontenete et al. recently described an intuitive approach towards the development of what they termed fluorescence in vivo hybridization for detection of H. pylori bacteria which colonizes the human stomach. In this FISH-based approach, a hybridizing probe is incubated with H. Pylori in clinically relevant conditions and the potential future human applications are emphasized [274]. This represents a hopeful approach and a snapshot of the necessity for the field to keep pushing the limits of fluorescent imaging technologies as a whole.
4.5. Nuclear imaging
While fluorescence imaging and its derivatives have been established as the go-to approach for nanoparticle trafficking studies, radioisotopic labelling, once considered the gold standard, is mounting a comeback. Visualization of nanoparticle biodistribution and accumulation in disease states is commonly achieved through incorporation of radioisotopes and subsequent imaging with positron emission tomography (PET) or single photon emission computed tomography (SPECT) (Fig. 6C) [275]. PET is a highly sensitive, quantitative and translational imaging modality and a variety of nanoparticles, have been radiolabeled with positron-emitting radionuclides. Longer-lived isotopes including Cu-64 (t1/2 = 12.7 hrs) and Zr-89 (t1/2 = 78.4 hrs) are typically utilized due to the relatively slow clearance of nanoparticles from the blood pool [276]. There are several key advantages for the application of radiolabeled nanoparticles for molecular imaging including their ability to function as signal amplifiers resulting in enhanced signal to background and improved sensitivity compared to typical radiotracers [277]. Their large surface area renders them amenable to conjugation with targeting ligands for subsequently visualization of target expression in disease states and tracking changes in target levels over time through serial imaging. Nanoparticles additionally provide the potential for multimodal imaging and the ability to combine diagnostic and therapeutic modalities through incorporation of therapeutic isotopes or drug delivery techniques [278][279]. The development of a near-infrared fluorescence imaging and PET imaging agent based on quantum dots has been reported [280]. RGD peptides were immobilized on the surface on quantum dots, permitting targeting of the αvβ3 integrin, and a DOTA group was additionally conjugated to facilitate Cu-64 radiolabeling. This multimodal nano-based probe sufficiently improved tumor contrast while reducing the necessary pharmacological dose required for small animal NIR fluorescence imaging and leading to a reduced potential of quantum dot toxicity. Nahrendorf and colleagues have reported the development of a dextran-coated superparamagnetic iron oxide nanoparticles radiolabeled with Cu-64 and an NIR fluorophore permitting both fluorescence and PET imaging of tumor-associated macrophages (Fig. 6C) [281,282]. The development of antibody radiolabeled Zr89-single-walled nanotubes targeting the monomeric vascular endothelial cadherin epitope for monitoring with PET and NIR imaging has also been reported [283]. Goins and coworkers have recently reported a strategy to encapsulate Re-186 into liposomal nanoparticles, Re-186 functions as a theranostic radionuclide with a half-life of approximately 90 hrs, therapeutic beta emission of ~1 MeV and gamma emission of 137 keV [284]. The therapeutic beta emission travels approximately 1.8 mm, permitting nanoparticle to deliver effective treatment without the need to incorporate into the cell or nucleus. The long half-life and gamma emission permits therapy monitoring with SPECT for several days post administration. Taken together, the application of radioisotopes for nanoparticle tracking inside of the cell when combined with electron and fluorescent microcopy can provide complementary information on localization of nanoparticle-delivered agents at very high subcellular resolution. With all the advantages presented, radiolabeling nanoparticles still has major drawbacks that may impede its popularity in the trafficking field due to inherent disposal and handling difficulties that come with radioactive isotopes compared to fluorescence methods. Another critical limiting factor is the sub-millimeter resolution capabilities of radioisotope detection compared to the near atomic resolution of electron and fluorescence super resolution techniques [285].
All the imaging tools referenced above have allowed us to understand many biological mechanisms, in all fields, by illuminating intricate, nanoscale events at the molecular level. High resolution electron and fluorescence microscopy have been phenomenal at shedding light on previously unknown interactions within the myriad cellular avenues an endocytosed nanoparticle may take part in. Due to these novel technologies, we are able to track single nanoparticles as well as particle-like vesicles of comparable size, in the midst of cellular uptake and trafficking. As evidenced by the groups of Varela et al., Zhan et al., and Bademosi et al., the field as a whole is getting ever closer to imaging these molecular dynamics, not only with higher depth and spatiotemporal resolution, but more importantly in vivo, where the context of the biological system remains intact [286–288]. We must remain conscious of the main limitation surrounding these tools which is the fact most of the imaging carried out is showing only a snapshot of the reality of intracellular trafficking as they are carried out in vitro or ex vivo due to the nature of the available techniques. The field will greatly benefit from more bold approaches at making all these great techniques applicable to the more clinically relevant, in vivo models.
5. Future Directions and Conclusions
We must seek innovative approaches to gather insights into these enigmatic nanoscale interactions of nanomedicines with cellular processes. Advances in gene editing can be employed to its fullest extent to gain mechanistic insights into intracellular transport of diverse viral and non-viral vectors that will allow for the rapid assessment of their route of intracellular delivery and factors that pose as barriers for effective drug and gene delivery (Fig. 6D) [289–292]. For example, we have previously used haploid cells genetically edited for either Rab4, Rab5, or Rab7 proteins which leads to defects in biogenesis of the recycling, early, and late endosomes, respectively, to the study of relevant endocytic pathways and sites of endosomal escape for mRNA transfection [17]. Animal models genetically engineered to possess the Cre reporter Lox-Stop-Lox tdTomato cassette have been used to validate intracellular delivery and cell specificity of nanoparticles in a high-throughput fashion (Fig. 6D) [289,291,292]. Advances in DNA sequencing technologies have provided a significant boost to nanoparticle delivery research. Nanoparticle-based nucleic acid therapeutics are limited by inefficient delivery in vivo as well as off-target effects. Effective in vitro delivery is in fact a poor predictor of the same in vivo [195]. Recently, Dahlman and colleagues have used DNA barcodes for high throughput screening of delivery efficacy of a large number of nanoparticle formulations in mice (Fig. 6E) [293]. They demonstrate the feasibility of multiplexing nanoparticles and selective enrichment of nanoparticle in targeted organs [294]. Additionally, nanoparticles delivered to spleen showed enrichment in myeloid lineage cells compared to other types [195]. Other groups have used AAV vectors for delivering barcoded DNA or RNA in vivo [295,296]. Recent advances in cell sorting and single-cell RNA seq technologies also provide powerful tools that can be harnessed to study cell specificity of nanoparticle-based nucleic acid delivery as well as monitor off target effects by analyzing changes in endogenous mRNA levels [292,297].
Despite the vast amount of data on nanoparticle physicochemical characterization and their intracellular trafficking, developing a consensus on the parameters related to delivery efficacy has been challenging. It is clear that even with the most potent systems and nearly all nanoparticle-based delivery systems, sequestration in the endosome remains a significant barrier in vitro and in vivo. The delivery efficacy is dependent on parameters such as uptake efficiency, retention, payload stability, endosomal escape efficiency, and subcellular localization. These are, in turn, dependent on cell type as well as the delivery system. The influence of cell type on the delivery results from differences in preference for endocytic pathways (e.g. phagocytes vs epithelial), diseases state (cancer or LSD cells vs healthy cells), endosomal leakiness, and rates of endosomal trafficking and exocytosis [10,113,298–301]. The considerations related to the delivery system are also varied, involving parameters such as surface chemistry, size, shape, stability, pH buffering capacity, potential to induce endosomal membrane destabilization/fusion, disassembly to release cargo, cytotoxicity, immune activation, opsonization, and the use of targeting ligands for cell-specific or organelle-specific targeting [1–3,10,302–308]. The high degree of variability in biomaterials and nanoparticle design increases the complexity of extracting rational design parameters from the available information [90,93,293,309–318]. However, there are some tangible parameters for delivery system design that have emerged. Lipids and polymers exhibiting low pKa and high endosomal buffering capacity have been routinely correlated with high transfection efficacy [7,110,310,312,313,317]. Furthermore, lipid nanoparticles containing lipids with ionizable head groups and tail unsaturations demonstrate higher efficacy relative to their cationic and saturated counterparts [7,90,312,319]. Antibody-drug conjugates and other drug carriers with cleavable linkers exhibit better efficacy than uncleavable linkers [133,134]. Enhanced retention of nanoparticles, mediated by targeting ligands or disease state, is also attributed to enhanced delivery [9,146,302,320,321]. Navigating the complex wealth of information on nanoparticles, their physicochemical characteristics, biological interactions, and efficacies requires innovative approaches addressed above. Emerging artificial intelligence and machine learning technologies can potentially be leveraged to extract and deconvolute information from the available research and develop consistent hypotheses for rational design of nanoparticles for the vast array of applications [322–326].
The fundamental studies in understanding nanoparticle-cell interactions, cellular factors that impact delivery and emerging tools to dissect trafficking will lead to development of methods that can overcome endosomal barriers. On the other hand, new nanotechnologies can be used to illuminate the endo/lysosomal system and deliver drugs and genes where endosomal transport is compromised and improve cellular function. Overall, the field of endocytosis of nanomedicines is on fertile ground that can lead to transformational impact on both the fields of drug delivery and cell biology.
Abbreviations:
- cGAMP
2’3’ cyclic guanosine monophosphate-adenosine monophosphate
- Ace-DEX
Acetalated dextran
- GAA
Acid α-glucosidase
- AAV
Adeno-associated virus
- DOPE
Dioleoyl phosphatidyl-ethanolamine
- ER
Endoplasmic reticulum
- FLIM
Fluorescence lifetime imaging microscopy
- FLIP
Fluorescence Loss in Photobleaching
- FRAP
Fluorescence Recovery After Photobleaching
- FISH
Fluorescent In-Situ Hybridization
- FRET
Förster resonance energy transfer
- ICAM1
Intercellular adhesion molecule 1
- ILVs
Interluminal vesicles
- HII
Inverse hexagonal phase
- LSDs
Lysosomal storage disorders
- mTORC1
Mechanistic target of rapamycin complex 1
- NIR
Near infrared
- NPC1
Niemann Pick type C1
- NAFLD
Non-alcoholic fatty liver disease
- PLL
Poly-L-lysine
- PLGA
Poly(lactic-co-glycolic acid
- PEG
Polyethylene glycol
- PEI
Polyethyleneimine
- PET
Positron emission tomography
- RNAi
RNA interference
- smFISH
Single molecule FISH
- SPECT
Single photon emission computed tomography
- SWCNTs
Single-walled carbon nanotubes
- SSOs
Splice switching oligonucleotides
- STING
Stimulator of interferon gene
- TRX-1
Thioredoxin-1
- TGN
Trans-Golgi network
- TFEB
Transcription factor EB
- 2-IM
Two-ion measurement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sahay G, Alakhova DY, Kabanov AV, Endocytosis of nanomedicines J. Controlled Release. 145 (2010) 182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Akinc A, Battaglia G, Exploiting Endocytosis for Nanomedicines, Cold Spring Harb. Perspect. Biol 5 (2013). doi: 10.1101/cshperspect.a016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Doherty GJ, McMahon HT, Mechanisms of Endocytosis, Annu. Rev. Biochem 78 (2009) 857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- [4].Sorkin A, von Zastrow M, Endocytosis and signalling: intertwining molecular networks, Nat. Rev. Mol. Cell Biol 10 (2009) 609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ, Exosome: from internal vesicle of the multivesicular body to intercellular signaling device, J. Cell Sci 113 (2000) 3365–3374. [DOI] [PubMed] [Google Scholar]
- [6].Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M, Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape, Nat. Biotechnol 31 (2013) 638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- [7].Sabnis S, Kumarasinghe ES, Salerno T, Mihai C, Ketova T, Senn JJ, Lynn A, Bulychev A, McFadyen I, Chan J, Almarsson Ö, Stanton MG, Benenato KE, A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates, Mol. Ther 26 (2018) 1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Monnery BD, Wright M, Cavill R, Hoogenboom R, Shaunak S, Steinke JHG, Thanou M, Cytotoxicity of polycations: Relationship of molecular weight and the hydrolytic theory of the mechanism of toxicity, Int. J. Pharm 521 (2017) 249–258. doi: 10.1016/j.ijpharm.2017.02.048. [DOI] [PubMed] [Google Scholar]
- [9].Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG, Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling, Nat. Biotechnol 31 (2013) 653. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Oh N, Park J-H, Endocytosis and exocytosis of nanoparticles in mammalian cells, Int. J. Nanomedicine 9 (2014) 51–63. doi: 10.2147/IJN.S26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T, Lieberman J, Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown, Nat. Biotechnol 33 (2015) 870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ballabio A, The awesome lysosome, EMBO Mol. Med 8 (2016) 73–76. doi: 10.15252/emmm.201505966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tooze SA, Abada A, Elazar Z, Endocytosis and Autophagy: Exploitation or Cooperation?, Cold Spring Harb. Perspect. Biol 6 (2014). doi: 10.1101/cshperspect.a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cupic KI, Rennick JJ, Johnston AP, Such GK, Controlling endosomal escape using nanoparticle composition: current progress and future perspectives, Nanomed. 14 (2019) 215–223. doi: 10.2217/nnm-2018-0326. [DOI] [PubMed] [Google Scholar]
- [15].Smith SA, Selby LI, Johnston APR, Such GK, The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery, Bioconjug. Chem 30 (2019) 263–272. doi: 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- [16].Selby LI, Cortez-Jugo CM, Such GK, Johnston APR, Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles: Endosomal escape of polymeric nanoparticles, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 9 (2017) e1452. doi: 10.1002/wnan.1452. [DOI] [PubMed] [Google Scholar]
- [17].Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, Mihai C, Almarsson Ö, Sahay G, Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA, Nano Lett. 17 (2017) 5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang B, Ming X, Cao C, Laing B, Yuan A, Porter MA, Hull-Ryde EA, Maddry J, Suto M, Janzen WP, Juliano RL, High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides, Nucleic Acids Res. 43 (2015) 1987–1996. doi: 10.1093/nar/gkv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Appelqvist H, Wäster P, Kågedal K, Öllinger K, The lysosome: from waste bag to potential therapeutic target, J. Mol. Cell Biol 5 (2013) 214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- [20].Perera RM, Zoncu R, The Lysosome as a Regulatory Hub, Annu. Rev. Cell Dev. Biol 32 (2016) 223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Settembre C, Fraldi A, Medina DL, Ballabio A, Signals for the lysosome: a control center for cellular clearance and energy metabolism, Nat. Rev. Mol. Cell Biol 14 (2013) 283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lim C-Y, Zoncu R, The lysosome as a command-and-control center for cellular metabolism, J. Cell Biol 214 (2016) 653–664. doi: 10.1083/jcb.201607005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Saha S, Panigrahi DP, Patil S, Bhutia SK, Autophagy in health and disease: A comprehensive review, Biomed. Pharmacother 104 (2018) 485–495. doi: 10.1016/j.biopha.2018.05.007. [DOI] [PubMed] [Google Scholar]
- [24].Burd CG, Physiology and Pathology of Endosome-to-Golgi Retrograde Sorting, Traffic Cph. Den 12 (2011) 948–955. doi: 10.1111/j.1600-0854.2011.01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maxfield FR, Role of Endosomes and Lysosomes in Human Disease, Cold Spring Harb. Perspect. Biol 6 (2014). doi: 10.1101/cshperspect.a016931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boustany R-MN, Lysosomal storage diseases—the horizon expands, Nat. Rev. Neurol 9 (2013) 583–598. doi: 10.1038/nrneurol.2013.163. [DOI] [PubMed] [Google Scholar]
- [27].Castanheira S, Garcia-del Portillo F, Salmonella Populations inside Host Cells, Front. Cell. Infect. Microbiol 7 (2017). doi: 10.3389/fcimb.2017.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].White JM, Whittaker GR, Fusion of Enveloped Viruses in Endosomes, Traffic Cph. Den 17 (2016) 593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Leung K, Chakraborty K, Saminathan A, Krishnan Y, A DNA nanomachine chemically resolves lysosomes in live cells, Nat. Nanotechnol 14 (2019) 176. doi: 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang C, Zhao T, Li Y, Huang G, White MA, Gao J, Investigation of endosome and lysosome biology by ultra pH-sensitive nanoprobes, Adv. Drug Deliv. Rev 113 (2017) 87–96. doi: 10.1016/j.addr.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen X, Bi Y, Wang T, Li P, Yan X, Hou S, Bammert CE, Ju J, Gibson KM, Pavan WJ, Bi L, Lysosomal Targeting with Stable and Sensitive Fluorescent Probes (Superior LysoProbes): Applications for Lysosome Labeling and Tracking during Apoptosis, Sci. Rep 5 (2015). doi: 10.1038/srep09004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Galassi TV, Jena PV, Shah J, Ao G, Molitor E, Bram Y, Frankel A, Park J, Jessurun J, Ory DS, Haimovitz-Friedman A, Roxbury D, Mittal J, Zheng M, Schwartz RE, Heller DA, An optical nanoreporter of endolysosomal lipid accumulation reveals enduring effects of diet on hepatic macrophages in vivo, Sci. Transl. Med 10 (2018) eaar2680. doi: 10.1126/scitranslmed.aar2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dan K, Veetil AT, Chakraborty K, Krishnan Y, DNA nanodevices map enzymatic activity in organelles, Nat. Nanotechnol 14 (2019) 252. doi: 10.1038/s41565-019-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kantner K, Ashraf S, Carregal-Romero S, Carrillo-Carrion C, Collot M, del Pino P, Heimbrodt W, Aberasturi DJD, Kaiser U, Kazakova LI, Lelle M, de Baroja NM, Montenegro JM, Nazarenus M, Pelaz B, Peneva K, Gil PR, Sabir N, Schneider LM, Shabarchina LI, Sukhorukov GB, Vazquez M, Yang F, Parak WJ, Particle-Based Optical Sensing of Intracellular Ions at the Example of Calcium - What Are the Experimental Pitfalls?, Small. 11 (2015) 896–904. doi: 10.1002/smll.201402110. [DOI] [PubMed] [Google Scholar]
- [35].Yoo J-W, Irvine DJ, Discher DE, Mitragotri S, Bio-inspired, bioengineered and biomimetic drug delivery carriers, Nat. Rev. Drug Discov 10 (2011) 521–535. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]
- [36].Batrakova EV, Kim MS, Using exosomes, naturally-equipped nanocarriers, for drug delivery, J. Controlled Release 219 (2015) 396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mathieu M, Martin-Jaular L, Lavieu G, Théry C, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication, Nat. Cell Biol 21 (2019) 9. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- [38].Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D, Engineering exosomes as refined biological nanoplatforms for drug delivery, Acta Pharmacol. Sin 38 (2017) 754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jiang X-C, Gao J-Q, Exosomes as novel bio-carriers for gene and drug delivery, Int. J. Pharm 521 (2017) 167–175. doi: 10.1016/j.ijpharm.2017.02.038. [DOI] [PubMed] [Google Scholar]
- [40].Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, Batrakova EV, Exosomes as Drug Delivery Vehicles for Parkinson’s Disease Therapy, J. Control. Release Off. J. Control. Release Soc 207 (2015) 18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gilligan KE, Dwyer RM, Engineering Exosomes for Cancer Therapy, Int. J. Mol. Sci 18 (2017) 1122. doi: 10.3390/ijms18061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].You B, Xu W, Zhang B, Engineering exosomes: a new direction for anticancer treatment, Am. J. Cancer Res 8 (2018) 1332–1342. [PMC free article] [PubMed] [Google Scholar]
- [43].Haney MJ, Klyachko NL, Harrison EB, Zhao Y, Kabanov AV, Batrakova EV, TPP1 Delivery to Lysosomes with Extracellular Vesicles and their Enhanced Brain Distribution in the Animal Model of Batten Disease, Adv. Healthc. Mater 1801271 (2019) 1801271. doi: 10.1002/adhm.201801271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cheng JPX, Nichols BJ, Caveolae: One Function or Many?, Trends Cell Biol. 26 (2016) 177–189. doi: 10.1016/j.tcb.2015.10.010. [DOI] [PubMed] [Google Scholar]
- [45].Wang Z, Tiruppathi C, Minshall RD, Malik AB, Size and Dynamics of Caveolae Studied Using Nanoparticles in Living Endothelial Cells, ACS Nano 3 (2009) 4110–4116. doi: 10.1021/nn9012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mehta D, Malik AB, Signaling Mechanisms Regulating Endothelial Permeability, Physiol. Rev 86 (2006) 279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- [47].Cardarelli F, Pozzi D, Bifone A, Marchini C, Caracciolo G, Cholesterol-Dependent Macropinocytosis and Endosomal Escape Control the Transfection Efficiency of Lipoplexes in CHO Living Cells, Mol. Pharm 9 (2012) 334–340. doi: 10.1021/mp200374e. [DOI] [PubMed] [Google Scholar]
- [48].Bloomfield G, Kay RR, Uses and abuses of macropinocytosis, J Cell Sci 129 (2016) 2697–2705. doi: 10.1242/jcs.176149. [DOI] [PubMed] [Google Scholar]
- [49].Murray DH, Jahnel M, Lauer J, Avellaneda MJ, Brouilly N, Cezanne A, Morales-Navarrete H, Perini ED, Ferguson C, Lupas AN, Kalaidzidis Y, Parton RG, Grill SW, Zerial M, An endosomal tether undergoes an entropic collapse to bring vesicles together, Nature. 537 (2016) 107–111. doi: 10.1038/nature19326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huotari J, Helenius A, Endosome maturation, EMBO J. 30 (2011) 3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hessvik NP, Llorente A, Current knowledge on exosome biogenesis and release, Cell. Mol. Life Sci 75 (2018) 193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Efeyan A, Zoncu R, Sabatini DM, Amino acids and mTORC1: from lysosomes to disease, Trends Mol. Med 18 (2012) 524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Davidson SM, Heiden MGV, Critical Functions of the Lysosome in Cancer Biology, Annu. Rev. Pharmacol. Toxicol 57 (2017) null. doi: 10.1146/annurev-pharmtox-010715-103101. [DOI] [PubMed] [Google Scholar]
- [54].Neiss WF, A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney, Histochemistry. 80 (1984) 603–608. [PubMed] [Google Scholar]
- [55].Li J, Deffieu MS, Lee PL, Saha P, Pfeffer SR, Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pfeffer SR, Clues to NPC1-mediated cholesterol export from lysosomes, Proc. Natl. Acad. Sci. U. S. A 113 (2016) 7941–7943. doi: 10.1073/pnas.1608530113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O’Kane CJ, Deretic V, Rubinsztein DC, Lysosomal positioning coordinates cellular nutrient responses, Nat. Cell Biol 13 (2011) 453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Poüs C, Codogno P, Lysosome positioning coordinates mTORC1 activity and autophagy, Nat. Cell Biol 13 (2011) 342–344. doi: 10.1038/ncb0411-342. [DOI] [PubMed] [Google Scholar]
- [59].Johnson DE, Ostrowski P, Jaumouillé V, Grinstein S, The position of lysosomes within the cell determines their luminal pH, J. Cell Biol 212 (2016) 677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Glick D, Barth S, Macleod KF, Autophagy: cellular and molecular mechanisms, J. Pathol 221 (2010) 3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schütz I, Lopez-Hernandez T, Gao Q, Puchkov D, Jabs S, Nordmeyer D, Schmudde M, Rühl E, Graf CM, Haucke V, Lysosomal Dysfunction Caused by Cellular Accumulation of Silica Nanoparticles, J. Biol. Chem 291 (2016) 14170–14184. doi: 10.1074/jbc.M115.710947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang F, Salvati A, Boya P, Lysosome-dependent cell death and deregulated autophagy induced by amine-modified polystyrene nanoparticles, Open Biol 8 (2018). doi: 10.1098/rsob.170271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Manshian BB, Pokhrel S, Mädler L, Soenen SJ, The impact of nanoparticle-driven lysosomal alkalinization on cellular functionality, J. Nanobiotechnology 16 (2018). doi: 10.1186/s12951-018-0413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Peynshaert K, Manshian BB, Joris F, Braeckmans K, De Smedt SC, Demeester J, Soenen SJ, Exploiting Intrinsic Nanoparticle Toxicity: The Pros and Cons of Nanoparticle-Induced Autophagy in Biomedical Research, Chem. Rev 114 (2014) 7581–7609. doi: 10.1021/cr400372p. [DOI] [PubMed] [Google Scholar]
- [65].Reitsma S, Slaaf DW, Vink H, van Zandvoort MAMJ, oude Egbrink MGA, The endothelial glycocalyx: composition, functions, and visualization, Pflugers Arch. 454 (2007) 345–359. doi: 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rabelink TJ, de Zeeuw D, The glycocalyx—linking albuminuria with renal and cardiovascular disease, Nat. Rev. Nephrol 11 (2015) 667–676. doi: 10.1038/nrneph.2015.162. [DOI] [PubMed] [Google Scholar]
- [67].Schulze H, Kolter T, Sandhoff K, Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation, Biochim. Biophys. Acta BBA - Mol. Cell Res 1793 (2009) 674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- [68].Möckl L, Hirn S, Torrano AA, Uhl B, Bräuchle C, Krombach F, The glycocalyx regulates the uptake of nanoparticles by human endothelial cells in vitro, Nanomed. 12 (2017) 207–217. doi: 10.2217/nnm-2016-0332. [DOI] [PubMed] [Google Scholar]
- [69].Uhl B, Hirn S, Immler R, Mildner K, Möckl L, Sperandio M, Bräuchle C, Reichel CA, Zeuschner D, Krombach F, The Endothelial Glycocalyx Controls Interactions of Quantum Dots with the Endothelium and Their Translocation across the Blood-Tissue Border, ACS Nano. 11 (2017) 1498–1508. doi: 10.1021/acsnano.6b06812. [DOI] [PubMed] [Google Scholar]
- [70].Atukorale PU, Yang Y-S, Bekdemir A, Carney RP, Silva PJ, Watson N, Stellacci F, Irvine DJ, Influence of the glycocalyx and plasma membrane composition on amphiphilic gold nanoparticle association with erythrocytes, Nanoscale. 7 (2015) 11420–11432. doi: 10.1039/C5NR01355K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ebong EE, Macaluso FP, Spray DC, Tarbell JM, Imaging the Endothelial Glycocalyx In Vitro by Rapid Freezing/Freeze Substitution Transmission Electron Microscopy, Arterioscler. Thromb. Vasc. Biol 31 (2011) 1908–1915. doi: 10.1161/ATVBAHA.111.225268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tarbell JM, Cancel LM, The glycocalyx and its significance in human medicine, J. Intern. Med 280 (2016) 97–113. doi: 10.1111/joim.12465. [DOI] [PubMed] [Google Scholar]
- [73].Kim Y-H, Nijst P, Kiefer K, Wilson Tang WH, Endothelial Glycocalyx as Biomarker for Cardiovascular Diseases: Mechanistic and Clinical Implications, Curr. Heart Fail. Rep 14 (2017) 117–126. doi: 10.1007/s11897-017-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kosicek M, Gudelj I, Horvatic A, Jovic T, Vuckovic F, Lauc G, Hecimovic S, N-glycome of the Lysosomal Glycocalyx is Altered in Niemann-Pick Type C Disease (NPC) Model Cells, Mol. Cell. Proteomics MCP 17 (2018) 631–642. doi: 10.1074/mcp.RA117.000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cheng MJ, Kumar R, Sridhar S, Webster TJ, Ebong EE, Endothelial glycocalyx conditions influence nanoparticle uptake for passive targeting, Int. J. Nanomedicine (2016). doi: 10.2147/IJN.S106299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Broekhuizen LN, Mooij HL, Kastelein JJP, Stroes ESG, Vink H, Nieuwdorp M, Endothelial glycocalyx as potential diagnostic and therapeutic target in cardiovascular disease, Curr. Opin. Lipidol 20 (2009) 57–62. doi: 10.1097/M0L.0b013e328321b587. [DOI] [PubMed] [Google Scholar]
- [77].Uchimido R, Schmidt EP, Shapiro NI, The glycocalyx: a novel diagnostic and therapeutic target in sepsis, Crit. Care 23 (2019) 16. doi: 10.1186/s13054-018-2292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xiao H, Woods EC, Vukojicic P, Bertozzi CR, Precision glycocalyx editing as a strategy for cancer immunotherapy, Proc. Natl. Acad. Sci 113 (2016) 10304–10309. doi: 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Büll C, Stoel MA, den Brok MH, Adema GJ, Sialic Acids Sweeten a Tumor’s Life, Cancer Res. 74 (2014) 3199–3204. doi: 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- [80].Rivas G, Minton AP, Macromolecular Crowding In Vitro, In Vivo, and In Between, Trends Biochem. Sci 41 (2016) 970–981. doi: 10.1016/j.tibs.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ellis RJ, Macromolecular crowding: obvious but underappreciated, Trends Biochem. Sci 26 (2001) 597–604. doi: 10.1016/S0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- [82].Stiehl O, Weiss M, Heterogeneity of crowded cellular fluids on the meso- and nanoscale, Soft Matter 12 (2016) 9413–9416. doi: 10.1039/C6SM01436D. [DOI] [PubMed] [Google Scholar]
- [83].Etoc F, Balloul E, Vicario C, Normanno D, Liße D, Sittner A, Piehler J, Dahan M, Coppey M, Non-specific interactions govern cytosolic diffusion of nanosized objects in mammalian cells, Nat. Mater 17 (2018) 740. doi: 10.1038/s41563-018-0120-7. [DOI] [PubMed] [Google Scholar]
- [84].Grady ME, Parrish E, Caporizzo MA, Seeger SC, Composto RJ, Eckmann DM, Intracellular nanoparticle dynamics affected by cytoskeletal integrity, Soft Matter. 13 (2017) 1873–1880. doi: 10.1039/C6SM02464E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Devany J, Chakraborty K, Krishnan Y, Subcellular Nanorheology Reveals Lysosomal Viscosity as a Reporter for Lysosomal Storage Diseases, Nano Lett. 18 (2018) 1351–1359. doi: 10.1021/acs.nanolett.7b05040. [DOI] [PubMed] [Google Scholar]
- [86].Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS, Size-dependent DNA Mobility in Cytoplasm and Nucleus, J. Biol. Chem 275 (2000) 1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- [87].Wrobel I, Collins D, Fusion of cationic liposomes with mammalian cells occurs after endocytosis, Biochim. Biophys. Acta BBA - Biomembr 1235 (1995) 296–304. doi: 10.1016/0005-2736(95)80017-A. [DOI] [PubMed] [Google Scholar]
- [88].Hayashi K, Watanabe M, Iwasaki T, Shudou M, Uda RM, Endosomal escape by photo-activated fusion of liposomes containing a malachite green derivative: a novel class of photoresponsive liposomes for drug delivery vehicles, Photochem. Photobiol. Sci (2019) 10.1039.C8PP00495A. doi: 10.1039/C8PP00495A. [DOI] [PubMed] [Google Scholar]
- [89].Nguyen J, Szoka FC, Nucleic acid delivery: the missing pieces of the puzzle?, Acc. Chem. Res 45 (2012) 1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, Rajeev KG, Hafez IM, Akinc A, Maier MA, Tracy MA, Cullis PR, Madden TD, Manoharan M, Hope MJ, Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo, Angew. Chem. Int. Ed 51 (2012) 8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].ur Rehman Z, Hoekstra D, Zuhorn IS, Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis, ACS Nano. 7 (2013) 3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- [92].Yessine M, Membrane-destabilizing polyanions: interaction with lipid bilayers and endosomal escape of biomacromolecules, Adv. Drug Deliv. Rev 56 (2004) 999–1021. doi: 10.1016/j.addr.2003.10.039. [DOI] [PubMed] [Google Scholar]
- [93].Kim J, Narayana A, Patel S, Sahay G, Advances in intracellular delivery through supramolecular self-assembly of oligonucleotides and peptides, Theranostics. 9 (2019) 3191–3212. doi: 10.7150/thno.33921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Congiu A, Pozzi D, Esposito C, Castellano C, Mossa G, Correlation between structure and transfection efficiency: a study of DC-Chol-DOPE/DNA complexes, Colloids Surf. B Biointerfaces 36 (2004) 43–48. doi: 10.1016/j.colsurfb.2004.04.006. [DOI] [PubMed] [Google Scholar]
- [95].Viger-Gravel J, Schantz A, Pinon AC, Rossini AJ, Schantz S, Emsley L, Structure of Lipid Nanoparticles Containing siRNA or mRNA by Dynamic Nuclear Polarization-Enhanced NMR Spectroscopy, J. Phys. Chem. B 122 (2018) 2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- [96].Ewert K, Evans HM, Ahmad A, Slack NL, Lin AJ, Martin-Herranz A, Safinya CR, Lipoplex Structures and Their Distinct Cellular Pathways, in: Adv. Genet., Elsevier, 2005: pp. 119–155. doi: 10.1016/S0065-2660(05)53005-0. [DOI] [PubMed] [Google Scholar]
- [97].Smisterová J, Wagenaar A, Stuart MCA, Polushkin E, ten Brinke G, Hulst R, Engberts JBFN, Hoekstra D, Molecular Shape of the Cationic Lipid Controls the Structure of Cationic Lipid/Dioleylphosphatidylethanolamine-DNA Complexes and the Efficiency of Gene Delivery, J. Biol. Chem 276 (2001) 47615–47622. doi: 10.1074/jbc.M106199200. [DOI] [PubMed] [Google Scholar]
- [98].Koltover I, Salditt T, Rädler JO, Safinya CR, An Inverted Hexagonal Phase of Cationic Liposome-DNA Complexes Related to DNA Release and Delivery, Science. 281 (1998) 78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- [99].Kim H, Leal C, Cuboplexes: Topologically Active siRNA Delivery, ACS Nano. 9 (2015) 10214–10226. doi: 10.1021/acsnano.5b03902. [DOI] [PubMed] [Google Scholar]
- [100].Kim H, Sung J, Chang Y, Alfeche A, Leal C, Microfluidics Synthesis of Gene Silencing Cubosomes, ACS Nano. (2018). doi: 10.1021/acsnano.8b03770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Cheng X, Lee RJ, The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery, Adv. Drug Deliv. Rev 99 (2016) 129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- [102].Farhood H, Serbina N, Huang L, The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer, Biochim. Biophys. Acta BBA - Biomembr. 1235 (1995) 289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- [103].Mochizuki S, Kanegae N, Nishina K, Kamikawa Y, Koiwai K, Masunaga H, Sakurai K, The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine, Biochim. Biophys. Acta BBA - Biomembr 1828 (2013) 412–418. doi: 10.1016/j.bbamem.2012.10.017. [DOI] [PubMed] [Google Scholar]
- [104].Hui SW, Langner M, Zhao YL, Ross P, Hurley E, Chan K, The role of helper lipids in cationic liposome-mediated gene transfer, Biophys. J 71 (1996) 590–599. doi: 10.1016/S0006-3495(96)79309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kube S, Hersch N, Naumovska E, Gensch T, Hendriks J, Franzen A, Landvogt L, Siebrasse J-P, Kubitscheck U, Hoffmann B, Merkel R, Csiszár A, Fusogenic Liposomes as Nanocarriers for the Delivery of Intracellular Proteins, Langmuir. 33 (2017) 1051–1059. doi: 10.1021/acs.langmuir.6b04304. [DOI] [PubMed] [Google Scholar]
- [106].Pozzi D, Marchini C, Cardarelli F, Amenitsch H, Garulli C, Bifone A, Caracciolo G, Transfection efficiency boost of cholesterol-containing lipoplexes, Biochim. Biophys. Acta BBA - Biomembr 1818 (2012) 2335–2343. doi: 10.1016/j.bbamem.2012.05.017. [DOI] [PubMed] [Google Scholar]
- [107].Xu L, Anchordoquy TJ, Cholesterol domains in cationic lipid/DNA complexes improve transfection, Biochim. Biophys. Acta BBA - Biomembr 1778 (2008) 2177–2181. doi: 10.1016/j.bbamem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- [108].Barua S, Yoo J-W, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S, Particle shape enhances specificity of antibody-displaying nanoparticles, Proc. Natl. Acad. Sci 110 (2013) 3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Vermeulen LMP, De Smedt SC, Remaut K, Braeckmans K, The proton sponge hypothesis: Fable or fact?, Eur. J. Pharm. Biopharm 129 (2018) 184–190. doi: 10.1016/j.ejpb.2018.05.034. [DOI] [PubMed] [Google Scholar]
- [110].Akinc A, Thomas M, Klibanov AM, Langer R, Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis, J. Gene Med 7 (2005) 657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- [111].Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL, The Possible “Proton Sponge ” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH, Mol. Ther 21 (2013) 149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Richard I, Thibault M, Crescenzo GD, Buschmann MD, Lavertu M, Ionization Behavior of Chitosan and Chitosan-DNA Polyplexes Indicate That Chitosan Has a Similar Capability to Induce a Proton-Sponge Effect as PEI, (2013). doi: 10.1021/bm4000713. [DOI] [PubMed] [Google Scholar]
- [113].Vermeulen LMP, Brans T, Samal SK, Dubruel P, Demeester J, De Smedt SC, Remaut K, Braeckmans K, Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles, ACS Nano. 12 (2018) 2332–2345. doi: 10.1021/acsnano.7b07583. [DOI] [PubMed] [Google Scholar]
- [114].Wojnilowicz M, Glab A, Bertucci A, Caruso F, Cavalieri F, Super-resolution Imaging of Proton Sponge-Triggered Rupture of Endosomes and Cytosolic Release of Small Interfering RNA, ACS Nano. (2019). doi: 10.1021/acsnano.8b05151. [DOI] [PubMed] [Google Scholar]
- [115].Kang HC, Huh KM, Bae YH, Polymeric Nucleic Acid Carriers: Current Issues and Novel Design Approaches, J. Control. Release Off. J. Control. Release Soc 164 (2012) 256–264. doi: 10.1016/j.jconrel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Merdan T, Kunath K, Fischer D, Kopecek J, Kissel T, Intracellular Processing of Poly(Ethylene Imine)/Ribozyme Complexes Can Be Observed in Living Cells by Using Confocal Laser Scanning Microscopy and Inhibitor Experiments, Pharm. Res 19 (2002) 140–146. doi: 10.1023/A:1014212630566. [DOI] [PubMed] [Google Scholar]
- [117].Knudsen KB, Northeved H, Kumar EK P, Permin A, Gjetting T, Andresen TL, Larsen S, Wegener KM, Lykkesfeldt J, Jantzen K, Loft S, Møller P, Roursgaard M, In vivo toxicity of cationic micelles and liposomes, Nanomedicine Nanotechnol. Biol. Med 11 (2015) 467–477. doi: 10.1016/j.nano.2014.08.004. [DOI] [PubMed] [Google Scholar]
- [118].Lv H, Zhang S, Wang B, Cui S, Yan J, Toxicity of cationic lipids and cationic polymers in gene delivery, J. Controlled Release 114 (2006) 100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [119].Erbacher P, Roche AC, Monsigny M, Midoux P, Putative Role of Chloroquine in Gene Transfer into a Human Hepatoma Cell Line by DNA/Lactosylated Polylysine Complexes, Exp. Cell Res 225 (1996) 186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- [120].Luthman H, Magnusson G, High efficiency polyoma DNA transfection of chloroquine treated cells., Nucleic Acids Res. 11 (1983) 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kwakye-Berko F, Meshnick SR, Binding of chloroquine to DNA, Mol. Biochem. Parasitol 35 (1989) 51–55. doi: 10.1016/0166-6851(89)90141-2. [DOI] [PubMed] [Google Scholar]
- [122].Gilleron J, Paramasivam P, Zeigerer A, Querbes W, Marsico G, Andree C, Seifert S, Amaya P, Stöter M, Koteliansky V, Waldmann H, Fitzgerald K, Kalaidzidis Y, Akinc A, Maier MA, Manoharan M, Bickle M, Zerial M, Identification of siRNA delivery enhancers by a chemical library screen, Nucleic Acids Res. 43 (2015) 7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Juliano RL, Wang L, Tavares F, Brown EG, James L, Ariyarathna Y, Ming X, Mao C, Suto M, Structure-activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides, Nucleic Acids Res. 46 (2018) 1601–1613. doi: 10.1093/nar/gkx1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Osborn MF, Alterman JF, Nikan M, Cao H, Didiot MC, Hassler MR, Coles AH, Khvorova A, Guanabenz (Wytensin™) selectively enhances uptake and efficacy of hydrophobically modified siRNAs, Nucleic Acids Res. 43 (2015) 8664–8672. doi: 10.1093/nar/gkv942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N, Efficient Intracellular Delivery of Native Proteins, Cell. 161 (2015) 674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- [126].Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, Weiler J, Hintersteiner M, Morrissey DV, Meisner-Kober NC, The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing, EMBO J. 32 (2013) 1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kaczmarczyk SJ, Sitaraman K, Young HA, Hughes SH, Chatterjee DK, Protein delivery using engineered virus-like particles, Proc. Natl. Acad. Sci 108 (2011) 16998–17003. doi: 10.1073/pnas.1101874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, Brinker CJ, Caldeira J. do C., Chackerian B, Wharton W, Peabody DS, Cell-Specific Delivery of Diverse Cargos by Bacteriophage MS2 Virus-like Particles, ACS Nano. 5 (2011) 5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Somiya M, Sasaki Y, Matsuzaki T, Liu Q, Iijima M, Yoshimoto N, Niimi T, Maturana AD, Kuroda S, Intracellular trafficking of bio-nanocapsule-liposome complex: Identification of fusogenic activity in the pre-S1 region of hepatitis B virus surface antigen L protein, J. Controlled Release. 212 (2015) 10–18. doi: 10.1016/j.jconrel.2015.06.012. [DOI] [PubMed] [Google Scholar]
- [130].Dalmau-Mena I, del Pino P, Pelaz B, Cuesta-Geijo MÁ, Galindo I, Moros M, de la Fuente JM, Alonso C, Nanoparticles engineered to bind cellular motors for efficient delivery, J. Nanobiotechnology 16 (2018) 33. doi: 10.1186/s12951-018-0354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yang J, Bahreman A, Daudey G, Bussmann J, Olsthoorn RCL, Kros A, Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes, ACS Cent. Sci 2 (2016) 621–630. doi: 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ross NL, Munsell EV, Sabanayagam C, Sullivan MO, Histone-targeted Polyplexes Avoid Endosomal Escape and Enter the Nucleus During Postmitotic Redistribution of ER Membranes, Mol. Ther. - Nucleic Acids 4 (2015). doi: 10.1038/mtna.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Diamantis N, Banerji U, Antibody-drug conjugates—an emerging class of cancer treatment, Br. J. Cancer 114 (2016) 362–367. doi: 10.1038/bjc.2015.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ducry L, Stump B, Antibody-Drug Conjugates: Linking Cytotoxic Payloads to Monoclonal Antibodies, Bioconjug. Chem 21 (2010) 5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- [135].Nabhan C, Tallman MS, Early phase I/II trials with gemtuzumab ozogamicin (Mylotarg) in acute myeloid leukemia, Clin. Lymphoma 2 Suppl 1 (2002) S19–23. [DOI] [PubMed] [Google Scholar]
- [136].Younes A, Bartlett NL, Leonard JP, Kennedy DA, Lynch CM, Sievers EL, Forero-Torres A, Brentuximab vedotin (SGN-35) for relapsed CD30-positive lymphomas, N. Engl. J. Med 363 (2010) 1812–1821. doi: 10.1056/NEJMoa1002965. [DOI] [PubMed] [Google Scholar]
- [137].Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, Pegram M, Oh D-Y, Diéras V, Guardino E, Fang L, Lu MW, Olsen S, Blackwell K, EMILIA Study Group, Trastuzumab emtansine for HER2-positive advanced breast cancer, N. Engl. J. Med 367 (2012) 1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lorusso PM, Edelman MJ, Bever SL, Forman KM, Pilat M, Quinn MF, Li J, Heath EI, Malburg LM, Klein PJ, Leamon CP, Messmann RA, Sausville EA, Phase I study of folate conjugate EC145 (Vintafolide) in patients with refractory solid tumors, J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 30 (2012) 4011–4016. doi: 10.1200/JC0.2011.41.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Shae D, Becker KW, Christov P, Yun DS, Lytton-Jean AKR, Sevimli S, Ascano M, Kelley M, Johnson DB, Balko JM, Wilson JT, Endosomolytic polymersomes increase the activity of cyclic dinucleotide STING agonists to enhance cancer immunotherapy, Nat. Nanotechnol 14 (2019) 269. doi: 10.1038/s41565-018-0342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Koshy ST, Cheung AS, Gu L, Graveline AR, Mooney DJ, Liposomal Delivery Enhances Immune Activation by STING Agonists for Cancer Immunotherapy, Adv. Biosyst 1 (2017). doi: 10.1002/adbi.201600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Cheng N, Watkins-Schulz R, Junkins RD, David CN, Johnson BM, Montgomery SA, Peine KJ, Darr DB, Yuan H, McKinnon KP, Liu Q, Miao L, Huang L, Bachelder EM, Ainslie KM, Ting JP-Y, A nanoparticle-incorporated STING activator enhances antitumor immunity in PD-L1-insensitive models of triple-negative breast cancer, JCI Insight 3 (2018). doi: 10.1172/jci.insight.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Ikonen E, Cellular cholesterol trafficking and compartmentalization, Nat. Rev. Mol. Cell Biol 9 (2008) 125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- [143].Pacheco CD, Lieberman AP, The pathogenesis of Niemann-Pick type C disease: a role for autophagy?, Expert Rev. Mol. Med 10 (2008) e26. doi: 10.1017/S146239940800080X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tharkeshwar AK, Trekker J, Vermeire W, Pauwels J, Sannerud R, Priestman DA, te Vruchte D, Vints K, Baatsen P, Decuypere J-P, Lu H, Martin S, Vangheluwe P, Swinnen JV, Lagae L, Impens F, Platt FM, Gevaert K, Annaert W, A novel approach to analyze lysosomal dysfunctions through subcellular proteomics and lipidomics: the case of NPC1 deficiency, Sci. Rep 7 (2017) 41408. doi: 10.1038/srep41408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kosicek M, Gudelj I, Horvatic A, Jovic T, Vuckovic F, Lauc G, Hecimovic S, N-glycome of the Lysosomal Glycocalyx is Altered in Niemann-Pick Type C Disease (NPC) Model Cells, Mol. Cell. Proteomics 17 (2018) 631–642. doi: 10.1074/mcp.RA117.000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Brown A, Patel S, Ward C, Lorenz A, Ortiz M, DuRoss A, Wieghardt F, Esch A, Otten EG, Heiser LM, Korolchuk VI, Sun C, Sarkar S, Sahay G, PEG-lipid micelles enable cholesterol efflux in Niemann-Pick Type C1 disease-based lysosomal storage disorder, Sci. Rep 6 (2016) 31750. doi: 10.1038/srep31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Eltoukhy AA, Sahay G, Cunningham JM, Anderson DG, Niemann-Pick C1 Affects the Gene Delivery Efficacy of Degradable Polymeric Nanoparticles, ACS Nano 8 (2014) 7905–7913. doi: 10.1021/nn501630h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Garnacho C, Dhami R, Simone E, Dziubla T, Leferovich J, Schuchman EH, Muzykantov V, Muro S, Delivery of Acid Sphingomyelinase in Normal and Niemann-Pick Disease Mice Using Intercellular Adhesion Molecule-1-Targeted Polymer Nanocarriers, J. Pharmacol. Exp. Ther 325 (2008) 400–408. doi: 10.1124/jpet.107.133298. [DOI] [PubMed] [Google Scholar]
- [149].Hsu J, Northrup L, Bhowmick T, Muro S, Enhanced delivery of α-glucosidase for Pompe disease by ICAM-1-targeted nanocarriers: comparative performance of a strategy for three distinct lysosomal storage disorders, Nanomedicine Nanotechnol. Biol. Med 8 (2012) 731–739. doi: 10.1016/j.nano.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Hsu J, Serrano D, Bhowmick T, Kumar K, Shen Y, Kuo YC, Garnacho C, Muro S, Enhanced Endothelial Delivery and Biochemical Effects of α-Galactosidase by ICAM-1-Targeted Nanocarriers for Fabry Disease, J. Control. Release Off. J. Control. Release Soc 149 (2011) 323–331. doi: 10.1016/j.jconrel.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Spampanato C, Feeney E, Li L, Cardone M, Lim J-A, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N, Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease, EMBO Mol. Med 5 (2013) 691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Settembre C, Ballabio A, TFEB regulates autophagy: An integrated coordination of cellular degradation and recycling processes, Autophagy. 7 (2011) 1379–1381. doi: 10.4161/auto.7.11.17166. [DOI] [PubMed] [Google Scholar]
- [153].Gatto F, Rossi B, Tarallo A, Polishchuk E, Polishchuk R, Carrella A, Nusco E, Alvino FG, Iacobellis F, Leonibus ED, Auricchio A, Diez-Roux G, Ballabio A, Parenti G, AAV-mediated transcription factor EB (TFEB) gene delivery ameliorates muscle pathology and function in the murine model of Pompe Disease, Sci. Rep 7 (2017) 15089. doi: 10.1038/s41598-017-15352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J, Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication, Proc. Natl. Acad. Sci 103 (2006) 14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Brumell JH, Tang P, Zaharik ML, Finlay BB, Disruption of the Salmonella-Containing Vacuole Leads to Increased Replication of Salmonella enterica Serovar Typhimurium in the Cytosol of Epithelial Cells, Infect. Immun 70 (2002) 3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Clemens DL, Lee B-Y, Horwitz MA, Francisella tularensis Phagosomal Escape Does Not Require Acidification of the Phagosome, Infect. Immun 77 (2009) 1757–1773. doi: 10.1128/IAI.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Johnson MM, Collier MA, Hoang KV, Pino EN, Graham-Gurysh EG, Gallovic MD, Md.S.H. Zahid, N. Chen, L. Schlesinger, J.S. Gunn, E.M. Bachelder, K.M. Ainslie, In Vivo and Cellular Trafficking of Acetalated Dextran Microparticles for Delivery of a Host-Directed Therapy for Salmonella enterica Serovar Typhi Infection, Mol. Pharm 15 (2018) 5336–5348. doi: 10.1021/acs.molpharmaceut.8b00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Moseman AP, Moseman EA, Schworer S, Smirnova I, Volkova T, von Andrian U, Poltorak A, Mannose Receptor 1 Mediates Cellular Uptake and Endosomal Delivery of CpG-Motif Containing Oligodeoxynucleotides, J. Immunol 191 (2013) 5615–5624. doi: 10.4049/jimmunol.1301438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Azad AK, Rajaram MVS, Schlesinger LS, Exploitation of the Macrophage Mannose Receptor (CD206) in Infectious Disease Diagnostics and Therapeutics, J. Cytol. Mol. Biol 1 (2014). doi: 10.13188/2325-4653.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, Steichen JM, Kumari S, Allen JD, Dane EL, Liguori A, Sangesland M, Lingwood D, Crispin M, Schief WR, Irvine DJ, Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers, Science. 363 (2019) 649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H, Nanoparticle Uptake: The Phagocyte Problem, Nano Today. 10 (2015) 487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Choi S, Britigan BE, Moran DM, Narayanasamy P, Gallium nanoparticles facilitate phagosome maturation and inhibit growth of virulent Mycobacterium tuberculosis in macrophages, PLOS ONE. 12 (2017) e0177987. doi: 10.1371/journal.pone.0177987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ai S-L, He X-Y, Liu B-Y, Zhuo R-X, Cheng S-X, Targeting Delivery of Oligodeoxynucleotides to Macrophages by Mannosylated Cationic Albumin for Immune Stimulation in Cancer Treatment, Mol. Pharm (2019). doi: 10.1021/acs.molpharmaceut.9b00184. [DOI] [PubMed] [Google Scholar]
- [164].van Niel G, D’Angelo G, Raposo G, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol 19 (2018) 213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- [165].Raposo G, Nijman HW, Stoorvogel W, Leijendekker R, Harding CV, Melief CJM, Geuze HJ, B lymphocytes secrete antigen-presenting vesicles, J. Exp. Med 183 (1996) 1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S, Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes, Nat. Med 4 (1998) 594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- [167].Sprague DL, Elzey BD, Crist SA, Waldschmidt TJ, Jensen RJ, Ratliff TL, Platelet-mediated modulation of adaptive immunity: unique delivery of CD154 signal by platelet-derived membrane vesicles, Blood. 111 (2008) 5028–5036. doi: 10.1182/blood-2007-06-097410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Haderk F, Schulz R, Iskar M, Cid LL, Worst T, Willmund KV, Schulz A, Warnken U, Seiler J, Benner A, Nessling M, Zenz T, Göbel M, Dürig J, Diederichs S, Paggetti J, Moussay E, Stilgenbauer S, Zapatka M, Lichter P, Seiffert M, Tumor-derived exosomes modulate PD-L1 expression in monocytes, Sci. Immunol 2 (2017) eaah5509. doi: 10.1126/sciimmunol.aah5509. [DOI] [PubMed] [Google Scholar]
- [169].del Conde I, Shrimpton CN, Thiagarajan P, Lopez JA, Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation, Blood. 106 (2005) 1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- [170].Lai RC, Arslan F, Lee MM, Sze NSK, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, Pasterkamp G, de Kleijn DPV, Lim SK, Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury, Stem Cell Res. 4 (2010) 214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- [171].Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, Zhang H-G, Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK Cell Function, J. Immunol 176 (2006) 1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- [172].Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang H-G, Tumor Exosomes Inhibit Differentiation of Bone Marrow Dendritic Cells, J. Immunol 178 (2007) 6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- [173].Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A, Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer, Br. J. Cancer 100 (2009) 1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N, Circulating Exosomal microRNAs as Biomarkers of Colon Cancer, PLoS ONE. 9 (2014) e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zhao Y, Haney MJ, Gupta R, Bohnsack JP, He Z, V Kabanov A, V Batrakova E, GDNF-Transfected Macrophages Produce Potent Neuroprotective Effects in Parkinson’s Disease Mouse Model, PLoS ONE. 9 (2014) e106867. doi: 10.1371/journal.pone.0106867. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [176].Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M, Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor MicroRNA to Breast Cancer Cells, Mol. Ther 21 (2013) 185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G, A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy, Biomaterials. 35 (2014) 2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- [178].Zhu Q, Ling X, Yang Y, Zhang J, Li Q, Niu X, Hu G, Chen B, Li H, Wang Y, Deng Z, Embryonic Stem Cells-Derived Exosomes Endowed with Targeting Properties as Chemotherapeutics Delivery Vehicles for Glioblastoma Therapy, Adv. Sci 6 (2019) 1801899. doi: 10.1002/advs.201801899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Vargas A, Zhou S, Ethier-Chiasson M, Flipo D, Lafond J, Gilbert C, Barbeau B, Syncytin proteins incorporated in placenta exosomes are important for cell uptake and show variation in abundance in serum exosomes from patients with preeclampsia, FASEB J. 28 (2014) 3703–3719. doi: 10.1096/fj.13-239053. [DOI] [PubMed] [Google Scholar]
- [180].Koh E, Lee EJ, Nam G-H, Hong Y, Cho E, Yang Y, Kim I-S, Exosome-SIRPα, a CD47 blockade increases cancer cell phagocytosis, Biomaterials. 121 (2017) 121–129. doi: 10.1016/j.biomaterials.2017.01.004. [DOI] [PubMed] [Google Scholar]
- [181].Yuan D, Zhao Y, Banks WA, Bullock KM, Haney M, Batrakova E, Kabanov AV, Macrophage Exosomes as Natural Nanocarriers for Protein Delivery to Inflamed Brain, Biomaterials. 142 (2017) 1–12. doi: 10.1016/j.biomaterials.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Laulagnier K, Javalet C, Hemming FJ, Chivet M, Lachenal G, Blot B, Chatellard C, Sadoul R, Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons, Cell. Mol. Life Sci 75 (2018) 757–773. doi: 10.1007/s00018-017-2664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].van Dongen HM, Masoumi N, Witwer KW, Pegtel DM, Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery, Microbiol. Mol. Biol. Rev. MMBR 80 (2016) 369–386. doi: 10.1128/MMBR.00063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, Kabanov AV, Batrakova EV, Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations, Nanomedicine Nanotechnol. Biol. Med 14 (2018) 195–204. doi: 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- [185].Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L, Miller D, Zhang H-G, Treatment of Brain Inflammatory Diseases by Delivering Exosome Encapsulated Anti-inflammatory Drugs From the Nasal Region to the Brain, Mol. Ther 19 (2011) 1769–1779. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes, Nat. Biotechnol 29 (2011) 341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- [187].Yang Y, Xiu F, Cai Z, Wang J, Wang Q, Fu Y, Cao X, Increased induction of antitumor response by exosomes derived from interleukin-2 gene-modified tumor cells, J. Cancer Res. Clin. Oncol 133 (2007) 389–399. doi: 10.1007/s00432-006-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Wahlgren J, Karlson TDL, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H, Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes, Nucleic Acids Res. 40 (2012) e130–e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM, Functional delivery of viral miRNAs via exosomes, Proc. Natl. Acad. Sci 107 (2010) 6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Kojima R, Bojar D, Rizzi G, Hamri GC-E, El-Baba MD, Saxena P, Ausländer S, Tan KR, Fussenegger M, Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment, Nat. Commun 9 (2018) 1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Masedunskas A, Porat-Shliom N, Weigert R, Regulated Exocytosis: Novel Insights from Intravital Microscopy, Traffic. 13 (2012) 627–634. doi: 10.1111/j.1600-0854.2012.01328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Otava M, Shkedy Z, Talloen W, Verheyen GR, Kasim A, Identification of in vitro and in vivo disconnects using transcriptomic data, BMC Genomics. 16 (2015). doi: 10.1186/s12864-015-1726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Gotlieb N, Rosenne E, Matzner P, Shaashua L, Sorski L, Ben-Eliyahu S, The misleading nature of in vitro and ex-vivo findings in studying the impact of stress hormones on NK cell cytotoxicity, Brain. Behav. Immun 45 (2015) 277–286. doi: 10.1016/j.bbi.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Whitehead KA, Matthews J, Chang PH, Niroui F, Dorkin JR, Severgnini M, Anderson DG, In Vitro-In Vivo Translation of Lipid Nanoparticles for Hepatocellular siRNA Delivery, ACS Nano 6 (2012) 6922–6929. doi: 10.1021/nn301922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV, Dahlman JE, A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation, Nano Lett. 18 (2018) 2148–2157. doi: 10.1021/acs.nanolett.8b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Liu T-L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, Kohrman AQ, Medwig TN, Dambournet D, Forster R, Cunniff B, Ruan Y, Yashiro H, Scholpp S, Meyerowitz EM, Hockemeyer D, Drubin DG, Martin BL, Matus DQ, Koyama M, Megason SG, Kirchhausen T, Betzig E, Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms, Science. 360 (2018). doi: 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Erni R, Rossell MD, Kisielowski C, Dahmen U, Atomic-Resolution Imaging with a Sub-50-pm Electron Probe, Phys. Rev. Lett 102 (2009) 096101. doi: 10.1103/PhysRevLett.102.096101. [DOI] [PubMed] [Google Scholar]
- [198].Klang V, Valenta C, Matsko NB, Electron microscopy of pharmaceutical systems, Micron. 44 (2013) 45–74. doi: 10.1016/j.micron.2012.07.008. [DOI] [PubMed] [Google Scholar]
- [199].Arteta MY, Kjellman T, Bartesaghi S, Wallin S, Wu X, Kvist AJ, Dabkowska A, Szekely N, Radulescu A, Bergenholtz J, Lindfors L, Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles, Proc. Natl. Acad. Sci 115 (2018) E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Kulkarni JA, Darjuan MM, Mercer JE, Chen S, van der Meel R, Thewalt JL, Tam YYC, Cullis PR, On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA, ACS Nano. 12 (2018) 4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- [201].Schnitzer JE, Oh P, Pinney E, Allard J, Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules, J. Cell Biol 127 (1994) 1217–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Thakor AS, Jokerst JV, Ghanouni P, Campbell JL, Mittra E, Gambhir SS, Clinically Approved Nanoparticle Imaging Agents, J. Nucl. Med 57 (2016) 1833–1837. doi: 10.2967/jnumed.116.181362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Echarri A, Pozo MAD, Caveolae - mechanosensitive membrane invaginations linked to actin filaments, J Cell Sci. 128 (2015) 2747–2758. doi: 10.1242/jcs.153940. [DOI] [PubMed] [Google Scholar]
- [204].Hinde E, Thammasiraphop K, Duong HTT, Yeow J, Karagoz B, Boyer C, Gooding JJ, Gaus K, Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release, Nat. Nanotechnol 12 (2017) 81–89. doi: 10.1038/nnano.2016.160. [DOI] [PubMed] [Google Scholar]
- [205].Hernandez C, Gulati S, Fioravanti G, Stewart PL, Exner AA, Cryo-EM Visualization of Lipid and Polymer-Stabilized Perfluorocarbon Gas Nanobubbles - A Step Towards Nanobubble Mediated Drug Delivery, Sci. Rep 7 (2017) 13517. doi: 10.1038/s41598-017-13741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ, Lipid Raft Microdomains, J. Exp. Med 195 (2002) 593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G, Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication, Proc. Natl. Acad. Sci 106 (2009) 7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Ruskin RS, Yu Z, Grigorieff N, Quantitative characterization of electron detectors for transmission electron microscopy, J. Struct. Biol 184 (2013) 385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B, Appion: An integrated, database-driven pipeline to facilitate EM image processing, J. Struct. Biol 166 (2009) 95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Biyani N, Righetto RD, McLeod R, Caujolle-Bert D, Castano-Diez D, Goldie KN, Stahlberg H, Focus: The interface between data collection and data processing in cryo-EM, J. Struct. Biol 198 (2017) 124–133. doi: 10.1016/j.jsb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- [211].Frauenfeld J, Löving R, Armache J-P, Sonnen AF-P, Guettou F, Moberg P, Zhu L, Jegerschöld C, Flayhan A, Briggs JAG, Garoff H, Löw C, Cheng Y, Nordlund P, A saposin-lipoprotein nanoparticle system for membrane proteins, Nat. Methods 13 (2016) 345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Liu W, Tagawa M, Xin HL, Wang T, Emamy H, Li H, Yager KG, Starr FW, Tkachenko AV, Gang O, Diamond family of nanoparticle superlattices, Science. 351 (2016) 582–586. doi: 10.1126/science.aad2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Leung AKK, Tam YYC, Chen S, Hafez IM, Cullis PR, Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems, J. Phys. Chem. B 119 (2015) 8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- [214].Azubel M, Carter SD, Weiszmann J, Zhang J, Jensen GJ, Li Y, Kornberg RD, FGF21 trafficking in intact human cells revealed by cryo-electron tomography with gold nanoparticles, ELife. 8 (2019) e43146. doi: 10.7554/eLife.43146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Cheng Y, Single-Particle Cryo-EM at Crystallographic Resolution, Cell. 161 (2015) 450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Bai X, McMullan G, Scheres SHW, How cryo-EM is revolutionizing structural biology, Trends Biochem. Sci 40 (2015) 49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- [217].Shimomura O, Johnson FH, Saiga Y, Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea, J. Cell. Comp. Physiol 59 (1962) 223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- [218].Hell SW, Microscopy and its focal switch, Nat. Methods 6 (2009) 24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- [219].Choquet D, The 2014 Nobel Prize in Chemistry: A Large-Scale Prize for Achievements on the Nanoscale, Neuron. 84 (2014) 1116–1119. doi: 10.1016/j.neuron.2014.12.002. [DOI] [PubMed] [Google Scholar]
- [220].Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods 3 (2006) 793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Clemments AM, Botella P, Landry CC, Spatial Mapping of Protein Adsorption on Mesoporous Silica Nanoparticles by Stochastic Optical Reconstruction Microscopy, J. Am. Chem. Soc 139 (2017) 3978–3981. doi: 10.1021/jacs.7b01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].van der Zwaag D, Vanparijs N, Wijnands S, De Rycke R, De Geest BG, Albertazzi L, Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking, ACS Appl. Mater. Interfaces 8 (2016) 6391–6399. doi: 10.1021/acsami.6b00811. [DOI] [PubMed] [Google Scholar]
- [223].Huang B, Wang W, Bates M, Zhuang X, Three-dimensional Super-resolution Imaging by Stochastic Optical Reconstruction Microscopy, Science. 319 (2008) 810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Chien M-P, Carlini AS, Hu D, Barback CV, Rush AM, Hall DJ, Orr G, Gianneschi NC, Enzyme-Directed Assembly of Nanoparticles in Tumors Monitored by in Vivo Whole Animal Imaging and ex Vivo Super-Resolution Fluorescence Imaging, J. Am. Chem. Soc 135 (2013) 18710–18713. doi: 10.1021/ja408182p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Zahid MU, Ma L, Lim SJ, Smith AM, Single quantum dot tracking reveals the impact of nanoparticle surface on intracellular state, Nat. Commun 9 (2018) 1830. doi: 10.1038/s41467-018-04185-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Gabriel M, Moya-Diaz J, Gallo LI, Marengo FD, Estrada LC, Single particle tracking of internalized metallic nanoparticles reveals heterogeneous directed motion after clathrin dependent endocytosis in mouse chromaffin cells, Methods Appl. Fluoresc 6 (2017) 014003. doi: 10.1088/2050-6120/aa8c64. [DOI] [PubMed] [Google Scholar]
- [227].Hou S, Lang X, Welsher K, Robust real-time 3D single-particle tracking using a dynamically moving laser spot, Opt. Lett 42 (2017) 2390–2393. doi: 10.1364/OL.42.002390. [DOI] [PubMed] [Google Scholar]
- [228].Liu S-L, Wu Q-M, Zhang L-J, Wang Z-G, Sun E-Z, Zhang Z-L, Pang D-W, Three-Dimensional Tracking of Rab5- and Rab7-Associated Infection Process of Influenza Virus, Small. 10 (2014) 4746–4753. doi: 10.1002/smll.201400944. [DOI] [PubMed] [Google Scholar]
- [229].Welsher K, Yang H, Multi-resolution 3D visualization of the early stages of cellular uptake of peptide-coated nanoparticles, Nat. Nanotechnol 9 (2014) 198–203. doi: 10.1038/nnano.2014.12. [DOI] [PubMed] [Google Scholar]
- [230].Jin D, Xi P, Wang B, Zhang L, Enderlein J, van Oijen AM, Nanoparticles for super-resolution microscopy and single-molecule tracking, Nat. Methods 15 (2018) 415. doi: 10.1038/s41592-018-0012-4. [DOI] [PubMed] [Google Scholar]
- [231].Lv Y, Liu M, Zhang Y, Wang X, Zhang F, Li F, Bao W-E, Wang J, Zhang Y, Wei W, Ma G, Zhao L, Tian Z, Cancer Cell Membrane-Biomimetic Nanoprobes with Two-Photon Excitation and Near-Infrared Emission for Intravital Tumor Fluorescence Imaging, ACS Nano 12 (2018) 1350–1358. doi: 10.1021/acsnano.7b07716. [DOI] [PubMed] [Google Scholar]
- [232].Weigert R, Sramkova M, Parente L, Masedunskas A, Intravital microscopy: a novel tool to study cell biology in living animals, Histochem. Cell Biol 133 (2010) 481–491. doi: 10.1007/s00418-010-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Karreman MA, Hyenne V, Schwab Y, Goetz JG, Intravital Correlative Microscopy: Imaging Life at the Nanoscale, Trends Cell Biol. 26 (2016) 848–863. doi: 10.1016/j.tcb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- [234].Naumenko V, Van S, Dastidar H, Kim D-S, Kim S-J, Zeng Z, Deniset J, Lau A, Zhang C, Macia N, Heyne B, Jenne CN, Mahoney DJ, Visualizing Oncolytic Virus-Host Interactions in Live Mice Using Intravital Microscopy, Mol. Ther. - Oncolytics 10 (2018) 14–27. doi: 10.1016/j.omto.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Sandoval RM, Kennedy MD, Low PS, Molitoris BA, Uptake and trafficking of fluorescent conjugates of folic acid in intact kidney determined using intravital two-photon microscopy, Am. J. Physiol.-Cell Physiol 287 (2004) C517–C526. doi: 10.1152/ajpcell.00006.2004. [DOI] [PubMed] [Google Scholar]
- [236].Steinmetz NF, Ablack AL, Hickey JL, Ablack J, Manocha B, Mymryk JS, Luyt LG, Lewis JD, Intravital Imaging of Human Prostate Cancer Using Viral Nanoparticles Targeted to Gastrin-Releasing Peptide Receptors, Small. 7 (2011) 1664–1672. doi: 10.1002/smll.201000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H, Viral nanoparticles as tools for intravital vascular imaging, Nat. Med 12 (2006) 354. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Smith BR, Kempen P, Bouley D, Xu A, Liu Z, Melosh N, Dai H, Sinclair R, Gambhir SS, Shape Matters: Intravital Microscopy Reveals Surprising Geometrical Dependence for Nanoparticles in Tumor Models of Extravasation, Nano Lett. 12 (2012) 3369–3377. doi: 10.1021/nl204175t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Masedunskas A, Sramkova M, Parente L, Sales KU, Amornphimoltham P, Bugge TH, Weigert R, Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy, Proc. Natl. Acad. Sci (2011) 201016778. doi: 10.1073/pnas.1016778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Mironov AA, Polishchuk RS, Luini A, Visualizing membrane traffic in vivo by combined video fluorescence and 3D electron microscopy, Trends Cell Biol. 10 (2000) 349–353. doi: 10.1016/S0962-8924(00)01787-6. [DOI] [PubMed] [Google Scholar]
- [241].Collinson LM, Carroll EC, Hoogenboom JP, Correlating 3D light to 3D electron microscopy for systems biology, Curr. Opin. Biomed. Eng 3 (2017) 49–55. doi: 10.1016/j.cobme.2017.10.006. [DOI] [Google Scholar]
- [242].Denk W, Strickler J, Webb W, Two-photon laser scanning fluorescence microscopy, Science. 248 (1990) 73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- [243].Ishii T, Ishii M, Intravital two-photon imaging: a versatile tool for dissecting the immune system, Ann. Rheum. Dis 70 (2011) i113–i115. doi: 10.1136/ard.2010.138156. [DOI] [PubMed] [Google Scholar]
- [244].Susaki EA, Ueda HR, Whole-body and Whole-Organ Clearing and Imaging Techniques with Single-Cell Resolution: Toward Organism-Level Systems Biology in Mammals, Cell Chem. Biol 23 (2016) 137–157. doi: 10.1016/j.chembiol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- [245].Urban BE, Yi J, Chen S, Dong B, Zhu Y, DeVries SH, Backman V, Zhang HF, Super-resolution two-photon microscopy via scanning patterned illumination, Phys. Rev. E Stat. Nonlin. Soft Matter Phys 91 (2015) 042703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Miller DR, Jarrett JW, Hassan AM, Dunn AK, Deep tissue imaging with multiphoton fluorescence microscopy, Curr. Opin. Biomed. Eng 4 (2017) 32–39. doi: 10.1016/j.cobme.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Vlashi E, Kelderhouse LE, Sturgis JE, Low PS, Effect of Folate-Targeted Nanoparticle Size on Their Rates of Penetration into Solid Tumors, ACS Nano 7 (2013) 8573–8582. doi: 10.1021/nn402644g. [DOI] [PubMed] [Google Scholar]
- [248].Fan W, Xia D, Zhu Q, Li X, He S, Zhu C, Guo S, Hovgaard L, Yang M, Gan Y, Functional nanoparticles exploit the bile acid pathway to overcome multiple barriers of the intestinal epithelium for oral insulin delivery, Biomaterials. 151 (2018) 13–23. doi: 10.1016/j.biomaterials.2017.10.022. [DOI] [PubMed] [Google Scholar]
- [249].Nobis M, Warren SC, Lucas MC, Murphy KJ, Herrmann D, Timpson P, Molecular mobility and activity in an intravital imaging setting - implications for cancer progression and targeting, J. Cell Sci 131 (2018) jcs206995. doi: 10.1242/jcs.206995. [DOI] [PubMed] [Google Scholar]
- [250].Luby-Phelps K, Taylor DL, Subcellular compartmentalization by local differentiation of cytoplasmic structure, Cell Motil. 10 (1988) 28–37. doi: 10.1002/cm.970100107. [DOI] [PubMed] [Google Scholar]
- [251].Shimi T, Koujin T, Segura-Totten M, Wilson KL, Haraguchi T, Hiraoka Y, Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells, J. Struct. Biol 147 (2004) 31–41. doi: 10.1016/j.jsb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- [252].Sanchez-Gaytan BL, Fay F, Hak S, Alaarg A, Fayad ZA, Pérez-Medina C, Mulder WJM, Zhao Y, Real-Time Monitoring of Nanoparticle Formation by FRET Imaging, Angew. Chem. Int. Ed Engl 56 (2017) 2923–2926. doi: 10.1002/anie.201611288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Zhao Y, van Rooy I, Hak S, Fay F, Tang J, de Lange Davies C, Skobe M, Fisher EA, Radu A, Fayad Zahi.A., de Mello Donegá C, Meijerink A, Mulder WJM, Near-Infrared Fluorescence Energy Transfer Imaging of Nanoparticle Accumulation and Dissociation Kinetics in Tumor-Bearing Mice, ACS Nano. 7 (2013) 10362–10370. doi: 10.1021/nn404782p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Zhao Y, Fay F, Hak S, Manuel Perez-Aguilar J, Sanchez-Gaytan BL, Goode B, Duivenvoorden R, de Lange Davies C, Bjørkøy A, Weinstein H, Fayad ZA, Pérez-Medina C, Mulder WJM, Augmenting drug-carrier compatibility improves tumour nanotherapy efficacy, Nat. Commun 7 (2016) 11221. doi: 10.1038/ncomms11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Pihl M, Kolman K, Lotsari A, Ivarsson M, Schuster E, Lorén N, Bordes R, Silica-based diffusion probes for use in FRAP and NMR-diffusometry, J. Dispers. Sci. Technol 40 (2019) 555–562. doi: 10.1080/01932691.2018.1472015. [DOI] [Google Scholar]
- [256].Basuki JS, Duong HTT, Macmillan A, Erlich RB, Esser L, Akerfeldt MC, Whan RM, Kavallaris M, Boyer C, Davis TP, Using Fluorescence Lifetime Imaging Microscopy to Monitor Theranostic Nanoparticle Uptake and Intracellular Doxorubicin Release, ACS Nano. 7 (2013) 10175–10189. doi: 10.1021/nn404407g. [DOI] [PubMed] [Google Scholar]
- [257].Heller DA, Jeng ES, Yeung T-K, Martinez BM, Moll AE, Gastala JB, Strano MS, Optical Detection of DNA Conformational Polymorphism on Single-Walled Carbon Nanotubes, Science. 311 (2006) 508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- [258].Jena PV, Roxbury D, Galassi TV, Akkari L, Horoszko CP, Iaea DB, Budhathoki-Uprety J, Pipalia N, Haka AS, Harvey JD, Mittal J, Maxfield FR, Joyce JA, Heller DA, A Carbon Nanotube Optical Reporter Maps Endolysosomal Lipid Flux, ACS Nano. 11 (2017) 10689–10703. doi: 10.1021/acsnano.7b04743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Langer-Safer PR, Levine M, Ward DC, Immunological method for mapping genes on Drosophila polytene chromosomes., Proc. Natl. Acad. Sci. U. S. A 79 (1982) 4381–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Femino AM, Fay FS, Fogarty K, Singer RH, Visualization of Single RNA Transcripts in Situ, Science. 280 (1998) 585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- [261].Crosetto N, Bienko M, van Oudenaarden A, Spatially resolved transcriptomics and beyond, Nat. Rev. Genet 16 (2015) 57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- [262].Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S, Imaging individual mRNA molecules using multiple singly labeled probes, Nat. Methods 5 (2008) 877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Orjalo AV, Johansson HE, Stellaris® RNA Fluorescence In Situ Hybridization for the Simultaneous Detection of Immature and Mature Long Noncoding RNAs in Adherent Cells, in: Feng Y, Zhang L. (Eds.), Long Non-Coding RNAs Methods Protoc, Springer New York, New York, NY, 2016: pp. 119–134. doi: 10.1007/978-1-4939-3378-5_10. [DOI] [PubMed] [Google Scholar]
- [264].Liu Y, Le P, Lim SJ, Ma L, Sarkar S, Han Z, Murphy SJ, Kosari F, Vasmatzis G, Cheville JC, Smith AM, Enhanced mRNA FISH with compact quantum dots, Nat. Commun 9 (2018). doi: 10.1038/s41467-018-06740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Kirschman JL, Bhosle S, Vanover D, Blanchard EL, Loomis KH, Zurla C, Murray K, Lam BC, Santangelo PJ, Characterizing exogenous mRNA delivery, trafficking, cytoplasmic release and RNA-protein correlations at the level of single cells, Nucleic Acids Res. 45 (2017) e113–e113. doi: 10.1093/nar/gkx290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Lakdawala SS, Wu Y, Wawrzusin P, Kabat J, Broadbent AJ, Lamirande EW, Fodor E, Altan-Bonnet N, Shroff H, Subbarao K, Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm, PLOS Pathog. 10 (2014) e1003971. doi: 10.1371/journal.ppat.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Chou Y, Heaton NS, Gao Q, Palese P, Singer R, Lionnet T, Colocalization of Different Influenza Viral RNA Segments in the Cytoplasm before Viral Budding as Shown by Single-molecule Sensitivity FISH Analysis, PLOS Pathog. 9 (2013) e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Lévesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ, Trafficking of HIV-1 RNA is Mediated by Heterogeneous Nuclear Ribonucleoprotein A2 Expression and Impacts on Viral Assembly, Traffic. 7 (2006) 1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- [269].Sainsbury F, Virus-like nanoparticles: emerging tools for targeted cancer diagnostics and therapeutics, Ther. Deliv 8 (2017) 1019–1021. doi: 10.4155/tde-2017-0098. [DOI] [PubMed] [Google Scholar]
- [270].Rohovie MJ, Nagasawa M, Swartz JR, Virus-like particles: Next-generation nanoparticles for targeted therapeutic delivery, Bioeng. Transl. Med 2 (2017) 43–57. doi: 10.1002/btm2.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Somiya M, Liu Q, Kuroda S, Current Progress of Virus-mimicking Nanocarriers for Drug Delivery, Nanotheranostics. 1 (2017) 415–429. doi: 10.7150/ntno.21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Shaffer SM, Wu M-T, Levesque MJ, Raj A, Turbo FISH: A Method for Rapid Single Molecule RNA FISH, PLOS ONE. 8 (2013) e75120. doi: 10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Wang G, Moffitt JR, Zhuang X, Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy, Sci. Rep 8 (2018) 4847. doi: 10.1038/s41598-018-22297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Fontenete S, Leite M, Guimarães N, Madureira P, Ferreira RM, Figueiredo C, Wengel J, Azevedo NF, Towards Fluorescence In Vivo Hybridization (FIVH) Detection of H. pylori in Gastric Mucosa Using Advanced LNA Probes, PLOS ONE. 10 (2015) e0125494. doi: 10.1371/journal.pone.0125494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Polyak A, Ross TL, Nanoparticles for SPECT and PET Imaging: Towards Personalized Medicine and Theranostics, Curr. Med. Chem 25 (2018) 4328–4353. doi: 10.2174/0929867324666170830095553. [DOI] [PubMed] [Google Scholar]
- [276].CHAKRAVARTY R, GOEL S, DASH A, CAI W, Radiolabeled inorganic nanoparticles for positron emission tomography imaging of cancer: an overview, Q. J. Nucl. Med. Mol. Imaging Off. Publ. Ital. Assoc. Nucl. Med. AIMN Int. Assoc. Radiopharmacol. IAR Sect. Soc. Of 61 (2017) 181–204. doi: 10.23736/S1824-4785.17.02969-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].de Barros AB, Tsourkas A, Saboury B, Cardoso VN, Alavi A, Emerging role of radiolabeled nanoparticles as an effective diagnostic technique, EJNMMI Res. 2 (2012) 39. doi: 10.1186/2191-219X-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Goel S, England CG, Chen F, Cai W, Positron Emission Tomography and Nanotechnology: A Dynamic Duo for Cancer Theranostics, Adv. Drug Deliv. Rev 113 (2017) 157–176. doi: 10.1016/j.addr.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Chakravarty R, Hong H, Cai W, Positron Emission Tomography Image-Guided Drug Delivery: Current Status and Future Perspectives, Mol. Pharm 11 (2014) 3777–3797. doi: 10.1021/mp500173s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Cai W, Chen K, Li Z-B, Gambhir SS, Chen X, Dual-Function Probe for PET and Near-Infrared Fluorescence Imaging of Tumor Vasculature, J. Nucl. Med 48 (2007) 1862–1870. doi: 10.2967/jnumed.107.043216. [DOI] [PubMed] [Google Scholar]
- [281].Nahrendorf M, Zhang H, Hembrador S, Panizzi P, Sosnovik DE, Aikawa E, Libby P, Swirski FK, Weissleder R, Nanoparticle PET-CT imaging of macrophages in inflammatory atherosclerosis, Circulation. 117 (2008) 379–387. doi: 10.1161/CIRCULATIONAHA.107.741181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Nahrendorf M, Keliher E, Marinelli B, Waterman P, Feruglio PF, Fexon L, Pivovarov M, Swirski FK, Pittet MJ, Vinegoni C, Weissleder R, Hybrid PET-optical imaging using targeted probes, Proc. Natl. Acad. Sci 107 (2010) 7910–7915. doi: 10.1073/pnas.0915163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Ruggiero A, Villa CH, Holland JP, Sprinkle SR, May C, Lewis JS, Scheinberg DA, McDevitt MR, Imaging and treating tumor vasculature with targeted radiolabeled carbon nanotubes, Int. J. Nanomedicine 5 (2010) 783–802. doi: 10.2147/IJN.S13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Goins B, Bao A, Phillips WT, Techniques for Loading Technetium-99m and Rhenium-186/188 Radionuclides into Pre-formed Liposomes for Diagnostic Imaging and Radionuclide Therapy, in: Weissig V. (Ed.), Liposomes Methods Protoc. Vol. 2 Biol. Membr. Models, Humana Press, Totowa, NJ, 2010: pp. 469–491. doi: 10.1007/978-1-60761-447-0_32. [DOI] [PubMed] [Google Scholar]
- [285].Khalil MM, Tremoleda JL, Bayomy TB, Gsell W, Molecular SPECT Imaging: An Overview, Int. J. Mol. Imaging (2011). doi: 10.1155/2011/796025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Bademosi AT, Lauwers E, Padmanabhan P, Odierna L, Chai YJ, Papadopulos A, Goodhill GJ, Verstreken P, van Swinderen B, Meunier FA, In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters, Nat. Commun 7 (2016) 13660. doi: 10.1038/ncomms13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Zhan H, Stanciauskas R, Stigloher C, Dizon KK, Jospin M, Bessereau J-L, Pinaud F, In vivo single-molecule imaging identifies altered dynamics of calcium channels in dystrophin-mutant C. elegans, Nat. Commun 5 (2014) 4974. doi: 10.1038/ncomms5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Varela JA, Dupuis JP, Etchepare L, Espana A, Cognet L, Groc L, Targeting neurotransmitter receptors with nanoparticles in vivo allows single-molecule tracking in acute brain slices, Nat. Commun 7 (2016) 10947. doi: 10.1038/ncomms10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, Castro MG, Anderson SE, Rudoltz TG, Lando GN, Tiwari PM, Kirschman JL, Willett N, Jang YC, Santangelo PJ, Bryksin AV, Dahlman JE, High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing, Proc. Natl. Acad. Sci (2018) 201811276. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Sago CD, Lokugamage MP, Lando GN, Djeddar N, Shah NN, Syed C, Bryksin AV, Dahlman JE, Modifying a Commonly Expressed Endocytic Receptor Retargets Nanoparticles in Vivo, Nano Lett. (2018). doi: 10.1021/acs.nanolett.8b03149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ, Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA, Angew. Chem. Int. Ed 56 (2017) 1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Kauffman KJ, Oberli MA, Dorkin JR, Hurtado JE, Kaczmarek JC, Bhadani S, Wyckoff J, Langer R, Jaklenec A, Anderson DG, Rapid, Single-Cell Analysis and Discovery of Vectored mRNA Transfection In Vivo with a loxP-Flanked tdTomato Reporter Mouse, Mol. Ther. - Nucleic Acids 10 (2018) 55–63. doi: 10.1016/j.omtn.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Lokugamage MP, Sago CD, Dahlman JE, Testing thousands of nanoparticles in vivo using DNA barcodes, Curr. Opin. Biomed. Eng 7 (2018) 1–8. doi: 10.1016/j.cobme.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, Langer R, Anderson DG, Wang ET, Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 2060–2065. doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H, Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing, Nat. Commun 5 (2014) 3075. doi: 10.1038/ncomms4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Davidsson M, Diaz-Fernandez P, Schwich OD, Torroba M, Wang G, Björklund T, A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing, Sci. Rep 6 (2016). doi: 10.1038/srep37563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB, A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte, Nature. 560 (2018) 377. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Barua S, Rege K, Cancer-Cell-Phenotype-Dependent Differential Intracellular Trafficking of Unconjugated Quantum Dots, Small. 5 (2009) 370–376. doi: 10.1002/smll.200800972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Sago CD, Krupczak BR, Lokugamage MP, Gan Z, Dahlman JE, Cell Subtypes Within the Liver Microenvironment Differentially Interact with Lipid Nanoparticles, Cell. Mol. Bioeng (2019). doi: 10.1007/s12195-019-00573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Kim B-K, Hwang G-B, Seu Y-B, Choi J-S, Jin KS, Doh K-O, DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes, Biochim. Biophys. Acta BBA - Biomembr 1848 (2015) 1996–2001. doi: 10.1016/j.bbamem.2015.06.020. [DOI] [PubMed] [Google Scholar]
- [301].Kuhn DA, Vanhecke D, Michen B, Blank F, Gehr P, Petri-Fink A, Rothen-Rutishauser B, Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages, Beilstein J. Nanotechnol 5 (2014) 1625–1636. doi: 10.3762/bjnano.5.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC, Insight into nanoparticle cellular uptake and intracellular targeting, J. Controlled Release 190 (2014) 485–499. doi: 10.1016/j.jconrel.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Foroozandeh P, Aziz AA, Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles, Nanoscale Res. Lett 13 (2018). doi: 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Vercauteren D, Rejman J, Martens TF, Demeester J, De Smedt SC, Braeckmans K, On the cellular processing of non-viral nanomedicines for nucleic acid delivery: Mechanisms and methods, J. Controlled Release 161 (2012) 566–581. doi: 10.1016/j.jconrel.2012.05.020. [DOI] [PubMed] [Google Scholar]
- [305].Kou L, Sun J, Zhai Y, He Z, The endocytosis and intracellular fate of nanomedicines: Implication for rational design, Asian J. Pharm. Sci 8 (2013) 1–10. doi: 10.1016/j.ajps.2013.07.001. [DOI] [Google Scholar]
- [306].Duncan R, Richardson SCW, Endocytosis and Intracellular Trafficking as Gateways for Nanomedicine Delivery: Opportunities and Challenges, Mol. Pharm 9 (2012) 2380–2402. doi: 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]
- [307].Tang H, Zhang H, Ye H, Zheng Y, Receptor-Mediated Endocytosis of Nanoparticles: Roles of Shapes, Orientations, and Rotations of Nanoparticles, J. Phys. Chem. B 122 (2018) 171–180. doi: 10.1021/acs.jpcb.7b09619. [DOI] [PubMed] [Google Scholar]
- [308].Shen Z, Ye H, Yi X, Li Y, Membrane Wrapping Efficiency of Elastic Nanoparticles during Endocytosis: Size and Shape Matter, ACS Nano 13 (2019) 215–228. doi: 10.1021/acsnano.8b05340. [DOI] [PubMed] [Google Scholar]
- [309].Patel S, Athirasala A, Menezes PP, Ashwanikumar N, Zou T, Sahay G, Bertassoni LE, Messenger RNA Delivery for Tissue Engineering and Regenerative Medicine Applications, Tissue Eng Part A. (2018). doi: 10.1089/ten.tea.2017.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Hajj KA, Ball RL, Deluty SB, Singh SR, Strelkova D, Knapp CM, Whitehead KA, Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH, Small. 15 (2019) 1805097. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- [311].Hajj KA, Whitehead KA, Tools for translation: non-viral materials for therapeutic mRNA delivery, Nat. Rev. Mater 2 (2017) natrevmats201756. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- [312].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ, Rational design of cationic lipids for siRNA delivery, Nat. Biotechnol 28 (2010) 172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- [313].Bishop CJ, Kozielski KL, Green JJ, Exploring the role of polymer structure on intracellular nucleic acid delivery via polymeric nanoparticles, J. Controlled Release 219 (2015) 488–499. doi: 10.1016/j.jconrel.2015.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher H-P, Zimmermann TS, Langer R, Anderson DG, A combinatorial library of lipid-like materials for delivery of RNAi therapeutics, Nat. Biotechnol 26 (2008) 561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Kaczmarek JC, Kowalski PS, Anderson DG, Advances in the delivery of RNA therapeutics: from concept to clinical reality, Genome Med 9 (2017). doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Li B, Luo X, Deng B, Wang J, McComb DW, Shi Y, Gaensler KML, Tan X, Dunn AL, Kerlin BA, Dong Y, An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo, Nano Lett. 15 (2015) 8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Miller JB, Kos P, Tieu V, Zhou K, Siegwart DJ, Development of Cationic Quaternary Ammonium Sulfonamide Amino Lipids for Nucleic Acid Delivery, ACS Appl. Mater. Interfaces 10 (2018) 2302–2311. doi: 10.1021/acsami.7b15982. [DOI] [PubMed] [Google Scholar]
- [318].Cheng Q, Wei T, Jia Y, Farbiak L, Zhou K, Zhang S, Wei Y, Zhu H, Siegwart DJ, Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH mRNA to Normalize Liver Function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I, Adv. Mater 30 (2018) 1805308. doi: 10.1002/adma.201805308. [DOI] [PubMed] [Google Scholar]
- [319].Patel S, Ryals RC, Weller KK, Pennesi ME, Sahay G, Lipid nanoparticles for delivery of messenger RNA to the back of the eye, J. Controlled Release 303 (2019) 91–100. doi: 10.1016/j.jconrel.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF, Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances, Adv. Drug Deliv. Rev 65 (2013) 121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, de Fougerolles A, Maier MA, Targeted Delivery of RNAi Therapeutics With Endogenous and Exogenous Ligand-Based Mechanisms, Mol. Ther 18 (2010) 1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Peurifoy J, Shen Y, Jing L, Yang Y, Cano-Renteria F, DeLacy BG, Joannopoulos JD, Tegmark M, Soljacic M, Nanophotonic particle simulation and inverse design using artificial neural networks, Sci. Adv 4 (2018) eaar4206. doi: 10.1126/sciadv.aar4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Sun B, Barnard AS, Texture based image classification for nanoparticle surface characterisation and machine learning, J. Phys. Mater 1 (2018) 016001. doi: 10.1088/2515-7639/aad9ef. [DOI] [Google Scholar]
- [324].Ho D, Wang P, Kee T, Artificial intelligence in nanomedicine, Nanoscale Horiz 4 (2019) 365–377. doi: 10.1039/C8NH00233A. [DOI] [PubMed] [Google Scholar]
- [325].Furxhi I, Murphy F, Mullins M, Poland CA, Machine learning prediction of nanoparticle in vitro toxicity: A comparative study of classifiers and ensemble-classifiers using the Copeland Index, Toxicol. Lett 312 (2019) 157–166. doi: 10.1016/j.toxlet.2019.05.016. [DOI] [PubMed] [Google Scholar]
- [326].Adir O, Poley M, Chen G, Froim S, Krinsky N, Shklover J, Shainsky-Roitman J, Lammers T, Schroeder A, Integrating Artificial Intelligence and Nanotechnology for Precision Cancer Medicine, Adv. Mater (2019) 1901989. doi: 10.1002/adma.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Zhu Q, Heon M, Zhao Z, He M, Microfluidic engineering of exosomes: editing cellular messages for precision therapeutics, Lab. Chip 18 (2018) 1690–1703. doi: 10.1039/C8LC00246K. [DOI] [PMC free article] [PubMed] [Google Scholar]