Abstract
Transfer RNA (tRNA) have been harbingers of many paradigms in RNA biology. They are among the first recognized noncoding RNA (ncRNA) playing fundamental roles in RNA metabolism. Although mainly recognized for their role in decoding mRNA and delivering amino acids to the growing polypeptide chain, tRNA also serve as an abundant source of small ncRNA named tRNA fragments. The functional significance of these fragments is only beginning to be uncovered. Early on, tRNA were recognized as heavily post-transcriptionally modified, which aids in proper folding and modulates the tRNA:mRNA anticodon–codon interactions. Emerging data suggest that these modifications play critical roles in the generation and activity of tRNA fragments. Modifications can both protect tRNA from cleavage or promote their cleavage. Modifications to individual fragments may be required for their activity. Recent work has shown that some modifications are critical for stem cell development and that failure to deposit certain modifications has profound effects on disease. This review will discuss how tRNA modifications regulate the generation and activity of tRNA fragments.
Keywords: tRNA, tRNA cleavage, tRNA modifications
Transfer RNA (tRNA) are best known for their role in helping decode mRNA information by the ribosome and delivering amino acids to the extending polypeptide chain during mRNA translation. Mature tRNA range in size from approximately 70 – 90 nucleotides, a distribution that is largely conserved throughout evolution. With few exceptions, tRNA across all species adopt a clover leaf secondary structure, which contains five major structural elements: (a) The acceptor stem to which the amino acid is attached, (b) the D-arm (c) the anticodon arm, (d) the variable loop, and (e) the T-arm, also known as the TΨC arm. Each arm consists of a double-stranded stem and single-stranded loop. The acceptor stem and D-arm are involved in aminoacyl tRNA synthetase recognition, which is required for amino acid charging at the acceptor stem. The anticodon loop interacts with mRNA codons to ensure the delivery of the proper amino acid to the elongating peptide chain. The T-arm is involved in ribosome:tRNA interactions that aid in efficient translation.
tRNA are expressed from a large number of genes, ranging from 170 to 570, depending upon the species [1]. In humans, there are at least 417 distinct tRNA genes, despite the fact that there are only 64 codons [2]. The precise reason for the expansion of tRNA genes is unknown. The diversity of tRNA genes is due to gene duplication and diversity in tRNA isodecoders, tRNA that recognize the same anticodon but have a different RNA sequence, and isoacceptors, tRNA that decode multiple related codons (e.g., tRNAHis decodes both CAU and CAC codons). However, certain tRNA isodecoders have different functional roles that may be important under distinct cellular conditions or cell types [3]. In fact, some tRNA are expressed in a tissue-specific manner [4]. Alternatively, the increase in tRNA diversity could allow for an increase in the repertoire of substrates for tRNA-derived small RNA, discussed herein.
tRNA processing
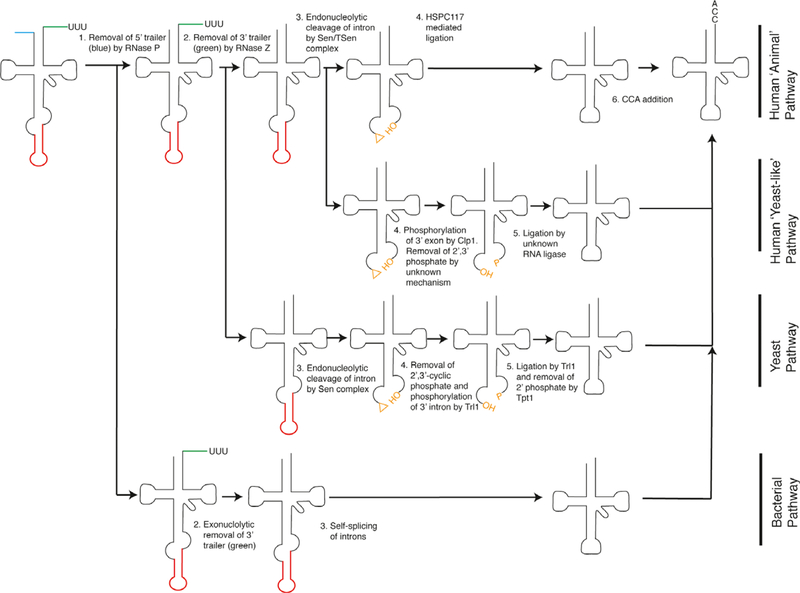
All RNA must undergo some level of processing to generate mature species and tRNA are no exception. In prokaryotes, RNA, including tRNA, are transcribed by the single RNA polymerase. In eukaryotes, RNA polymerase III transcribes tRNA. Most eukaryotic and prokaryotic tRNA are transcribed as pre-tRNA that contain a 5′ leader and a 3′ trailer that are removed by two distinct endonucleolytic cleavage events (Fig. 1). The 5′ leader is removed by RNase P-mediated cleavage, leaving a 5′ monophosphate [5]. RNase P is a ribonucleoprotein complex in which RNA typically serves as the catalytic core [6]. Endonucleolytic cleavage of the tRNA 3′ trailer was shown in 1975 [7,8], but cloning and identification of the responsible enzyme, RNase Z, was not completed until nearly 30 years later [9]. RNase Z is a metallo-β-lactamase endonuclease, and its activity results in a 3′ hydroxyl and a single 3′ overhang acceptor stem. In most prokaryotes and all eukaryotes, a mature 3′ end requires the addition of a ‘CCA’ trinucleotide by enzymatic means [10]. In humans, this is accomplished by tRNA-nucleotidyl transferase 1 (TRNT1). In some prokaryotic species, notably in Escherichia coli, the terminal ‘CCA’ is genomically encoded. Aminoacyl tRNA synthetases recognize the single-stranded base immediately 5′ of the CCA, named the ‘discriminator’ base, and other sequence elements, typically found in the anticodon stem-loop, which guides tRNA charging with the proper amino acid [11].
Fig. 1.
tRNA maturation pathways. Pre-tRNA contain a 5′ leader (blue), a 3′ trailer (green), and, in some instances, an intron (red) located within anticodon loop. Different domains of life utilize different enzymes to generate mature tRNA.
In some instances, an intron is located in the tRNA anticodon stem (Fig. 1). In humans, ~ 7% (28 of 417) of the tRNA genes contain introns [2]. Complete maturation of these tRNA requires excision of the intron via a tRNA-specific process. In yeast, this process is initiated by the SEN complex (Sen2, Sen15, Sen34, and Sen54) [12,13], which endonucleolytically cleaves tRNA on both sides of the intron, leaving a 2′,3′ cyclic phosphate on the 5′ exon and 5′ hydroxyl on the 3′ exon [14]. To ligate these two exons, the tRNA ligase, Trl1, converts the 2′,3′ cyclic phosphate to a 2′ phosphate and 3′ hydroxyl [15]. Trl1 also contains kinase activity that phosphorylates the 5′ hydroxyl of the 3′ exon allowing for ligation of the two exons. Finally, the 2′ phosphate is removed by Tpt1 [16]. In vertebrates, the yeast Sen complex ortholog termed the TSEN complex performs the initial cleavage reaction [17]. At this point, two distinct processes may complete the splicing reaction, although the relative contribution of each is unknown to date. The ‘yeast-like’ pathway utilizes the RNA kinase Clp1, which phosphorylates the downstream exon in a manner similar to Trl1 [18–20]. However, the enzymes that resolve the 2′,3′ cyclic phosphate, ligate the two halves and remove the resultant 2′ phosphate are unknown. In the ‘animal’ pathway, the RNA ligase HSPC117, a homolog of the prokaryotic ligase RtcB, directly acts upon the 2′,3′ cyclic phosphate to ligate the second exon [21]. This is facilitated by a complex of proteins containing DDX1, CGI-99, Archease, and Ashwin [22,23]. It is worth noting that biological activities have been attributed to the excised intron in addition to the cleaved 5′ leader and 3′ trailer RNA. The function of these RNA and their roles in regard to RNA modifications will be discussed below.
tRNA modifications
Despite the recent fervor and debate surrounding the role of RNA modifications in mRNA [24,25], the presence of a multitude of modifications has long been known in tRNA. Early enzymatic digests of calf liver RNA showed mononucleotides that did not correspond to the four canonical RNA bases [26]. One of these was identified as pseudouridine (Ψ), a modification that was shown to be abundant in yeast tRNA and rRNA [27]. Similar nucleic acid digests identified 2-methyladenine, N6-methyladenine, and N6-dimethyladenine in yeast. These studies were expanded to show the presence of these nucleotides in bacteria and wheat germ, thereby establishing an evolutionary connection to these modified bases [28,29]. Further guanosine derivatives were identified in plants, yeast, bacteria, and mammals [30], and 5-methylcytosine (m5C) was first identified in E. coli [31]. Salt-based fractionation of RNA showed that much of the RNA-modified nucleotides were within the ‘soluble’ fraction, which was later shown to contain tRNA [32]. Further evidence that tRNA were the highly modified ‘soluble RNA’ was obtained by showing that highly modified RNA had a high propensity to incorporate amino acids, a property that is now known to be tRNA charging [33,34]. Finally, when Holley and colleagues first elucidated the sequence of tRNAAla, they noted the presence of Ψ and dihydrouridine (D) at specific sites in the TΨC- and D-arms, respectively, thereby giving them their names [35].
From these initial laborious experiments, which proved the existence of nucleotide modifications and their location in tRNA, the total number of modifications present in RNA has expanded to more than 170 different modifications as cataloged by Modomics [36]. More recent mass spectroscopy and chemical approaches have aided in the identification of different groups and their positions [37–39]. These modifications range from minor changes, such as the aforementioned pseudouridylation or the addition of a single methyl group (m1A, m5C) to the addition of large bulky groups, such as conversion of guanosine to wybutosine (Y). In addition to modification of bases, select riboses in the sugar-phosphate backbone can be methylated to add additional complexity. Many of these modifications are universal across all domains of life and are even found in the tRNA encoded within mitochondria and plastids [36].
Most tRNA modifications are thought to play one of two major roles: (a) stabilizing or modifying tertiary structure and/or (b) aiding in the codon–anticodon recognition. While it is certainly a generality, modifications in the D- and T-arms serve to stabilize the tRNA structure while those in the anticodon arm affect codon recognition, particularly in the wobble position (Reviewed in [40]).
Of the 170 RNA modifications currently known, 93 are found in tRNA, but the frequency and distribution can vary in a given organism or tRNA species [36]. Ψ is the most abundant modified nucleotide found in tRNA [41]. In tRNA, Ψ is universally found at position 55 (denoted Ψ55) in the loop of the T-arm. This facilitates proper tertiary structure in which the loop of the T-arm interacts with the loop of the D-arm via Ψ55 [42]. As with Ψ, 1-methyladenosine (m1A) is similarly abundant, found in tRNA in all domains of life and was also recently identified in mRNA [43]. Methylation of A at position 58 is universally conserved in all domains of life, and methylation at position 9 is also common [36]. Similar to the function of Ψ55, m1A9 and m1A55 confer additional structural stability to the tRNA tertiary structure [44,45].
Modifications may also facilitate tRNA:mRNA interactions and are often found at position 34, the wobble base in the anticodon, or just 3′ of the anticodon at position 37. Among the best studied example is modification of G37 to Y37. This modification stabilizes the codon:anticodon interaction within the A site of the ribosome by providing increased base-stacking and ionic interactions [46,47]. Lack of this modification can lead to translational defects such as translational frameshifting. In Saccharomyces cerevisiae, modification of U34 to 5-methoxycaronyl-methyl-2-thiouridine (mcm5s2U) also instills accuracy and stability of codon:anticodon pairing, particularly in the case of tRNALys(UUU) and is essential for viability [48,49]. tRNALys(UUU) is particularly in need of stabilization of codon:anticodon base-pairing due to the three weak A:U or G:U base pairs in the interaction. In tRNALys (UUU), mcm5s2U34 modification increases the network of interactions to stabilize base-pairing. This is also true for m5C at position 34 in tRNALeu(CAA) [50].
As many modifications are required for tertiary structure stability, it should not be surprising that hypomodified tRNA are inherently less stable. Ablation of the enzymes that deposit m1A at position 58 are lethal as they cause tRNAiMet depletion [51,52] Hypomodified tRNAiMet molecules are oligoadenylated and targeted for degradation by the nuclear exosome [53]. tRNA export from the nucleus to the cytoplasm is facilitated by Exportin-T [54,55], and mutations that disrupt tRNA tertiary structure reduce the efficiency of tRNA export [56]. Additional modifications occur after export to the cytoplasm. If these modifications do not occur, hypomodified tRNA are also targeted for decay via the ‘rapid tRNA decay’ pathway [57,58]. Interestingly, some tRNA positions are never modified. In particularly, the 3′ CCA tail remains unmodified as are several positions in the variable loop [36].
Deposition of nucleotide modifications can alter tRNA maturation and vice versa. In regard to tRNA splicing, the presence of introns in a pre-tRNA or the act of splicing may modulate the deposition of nucleotide modifications, although, in most cases, the precise role or requirement for these modifications is unknown. There are several examples in which tRNA processing is required for efficient tRNA modification. In tRNATyr, the presence of an intron is required for modification of U35 to Ψ35, which is required for efficient codon recognition [59,60]. A similar situation is seen for tRNALeu, in which the presence of an intron is required for modification of C34 to m5C34 [61]. In Trypansoma brucei, only one tRNA gene contains an intron, tRNATyr, but splicing of this tRNA is required for viability [62]. Interestingly, in this case, the intron itself must be modified at several positions in order for splicing to occur, marking an instance where modification is required for processing rather than processing directing modifications [63].
A dramatic example of the interdependence of tRNA processing and modification comes from archaea. In Haloferix volcanii, the tRNATrp intron encodes a box C/D snoRNA [64]. Surprisingly, the target of this snoRNA is tRNATrp itself, guiding 2′-O methylation of C34 and U39 in tRNATrp in cis as part of the pre-tRNA [65], but more surprisingly the excised intron can act in trans [66]. Thus, the intron of tRNATrp acts as a small RNA, independent of the role of the tRNA. This is one of the earliest examples of a small functional RNA derived from a tRNA, a topic on which the remainder of this review will focus.
tRNA cleavage in host defense response
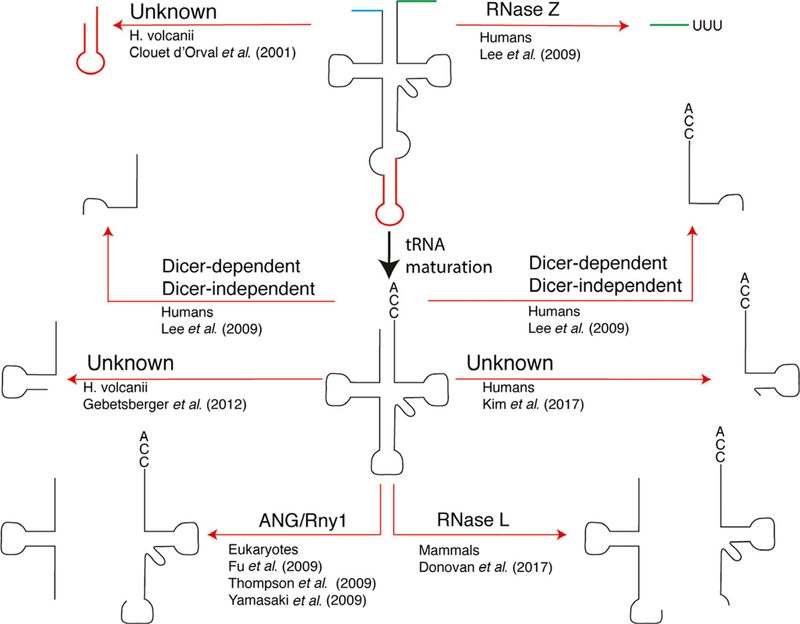
The phenomena of tRNA cleavage has long been recognized, but it is only in recent years that tRNA fragments themselves have been recognized to be functional (Figs 2 and 3). Among the earliest observations and best described mechanisms of tRNA cleavage comes from E. coli. Some strains of E. coli carry a plasmid (pCol) encoding various colicin proteins (e.g., Colicin D, E3, E5) (reviewed in [67]). Although the identity of the plasmid or the functional relation to the proteins it encoded was unknown, in 1925, it was observed that certain strains of E. coli, later discovered to be those containing pCol, were superior competitors compared to other strains [68,69]. It was later discovered that pCol encoded various toxins, which are secreted and kill non-pCol-containing competitors, and antitoxins, which act as an antidote for secreted toxins [69]. Colicin D and E5 are secreted proteins that are tRNA-specific RNases [70–72], which target tRNA anticodon stem loops, although they differ in their specificity. Colicin D targets tRNAArg [72], while colicin E5 targets tRNA with queuosine (Q) modifications in the wobble position of their anticodon (e.g., tRNATyr(QUA)) [71].
Fig. 2.
tRNA fragment diversity. Different small RNA species may be generated from pre-tRNA (5′ leader—blue, 3′ trailer—green, intron—red) or mature tRNA. Shown here is a selection of identified tRNA fragments, the species in which they were identified, the enzyme responsible, and the earliest citation. Other tRNA fragments have been identified but are not shown.
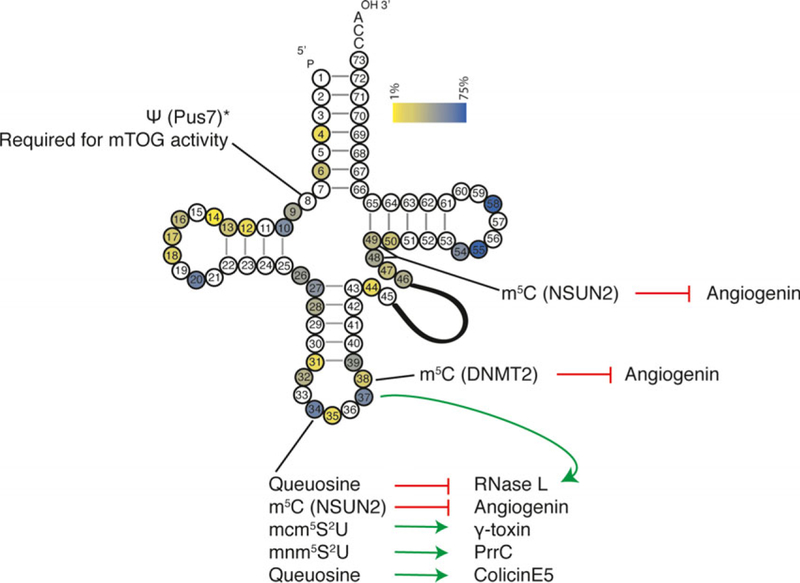
Fig. 3.
tRNA modifications and cleavage. A heat map displaying how often a particular nucleotide is modified. Human cytosolic tRNA modification profile was used [36]. Modifications that effect tRNA cleavage are indicated as well as the modifying enzyme (when known) and enzyme responsible for cleavage. Note that while the presence of queuosine at position 34 inhibits tRNA cleavage within the anticodon, it provides specificity for the RNase L-mediated cleavage of tRNAHis at position 37.
Using a modified nucleotide to direct tRNA cleavage is not unique to the colicin nucleases. The E. coli tRNA-specific nuclease PrrC is activated during T4 bacteriophage infection by the phage-encoded Stp peptide [73,74]. Similar to Colicin, PrrC targets and cleaves at the wobble position in the anticodon of specific tRNA. Rather than targeting Q-modified derivatives, PrrC specifically targets tRNALys(UUU) in which the wobble U has been modified to 5-methylaminomethyl 2-thiouridine (mnm5s2U) [75] (Fig. 3). Mutations that prevent either 5-methylaminomethyl or 2-thiouridine modification to the wobble U decrease PrrC activity toward tRNALys(UUU) by approximately 90%. In theory, this activity leads to an ‘altruistic suicide’ in which infected E. coli remove themselves from the population to prevent further bacteriophage infection. However, T4 bacteriophage contains a system to counteract the EcoPrrC system [76]. PrrC cleavage leaves a 2′,3′ cyclic phosphate on the 5′ fragment that T4 polynucleotide kinase (T4 PNK) converts to a 3′ hydroxyl. Concurrently, T4 PNK phosphorylates the 5′ hydroxyl of the 3′ cleavage product allowing for T4 RNA ligase 1 to ‘reseal’ the cleaved tRNA.
The targeted tRNA cleavage for defense purposes is not restricted to prokaryotes and neither is the use of tRNA modifications for defining specificity (Fig. 3). The yeast Kluyveromyces lactis secretes a heterotrimeric toxin known as zymocin, composed of α-, β-, and γ-subunits [77]. The α- and β-subunits facilitate entry of the γ-subunit into competitor cells, such as S. cerevisiae, where it causes irreversible cell cycle arrest and cell death [78,79]. Nearly 25 years after the discovery of zymocin, the target of the γ-toxin was revealed to be tRNA [79]. In these regards, γ-toxin acts in a similar manner to colicin, cleaving at the wobble position in the anticodon. Here, tRNAGlu (UUC), tRNALys (UUU), and tRNA Gln(UUG) where the wobble U has been modified to 5-methoxycarbonylmethyl-2-thiouridine (mcm2s2U) are targeted for cleavage [79]. Mutant S. cerevisiae strains that prevent these modifications prevent the action of γ toxin, establishing the requirement of this modification for cleavage.
Emerging data show that human cells may also target tRNA for bulk cleavage in response to viral infections. In this instance, cleavage occurs at position 37 of the mature tRNA and predominantly occurs on tRNAHis, although cleavage also occurs on tRNAPro and various Y-RNA [80]. Mammalian cells have an intricate network of sensors monitoring double-stranded RNA (dsRNA), a hallmark of viral infection. When a viral infection via dsRNA recognition is identified, 2′−5′ oligoadenylate synthetase converts ATP to 2′−5′ linked oligoadenylates (2–5A) (reviewed in [81]). RNase L binds 2–5A, which promotes its dimerization and activation, at which point it targets select tRNA for cleavage. RNase L has a very promiscuous recognition sequence (UNˆN; with ‘ˆ’ indicating the site of cleavage). Therefore, specific recognition of tRNAHis and tRNAPro cannot be explained solely by simple nucleotide determinants. Similar to tRNases in prokaryotes and in yeast, added specificity is granted by RNA modifications. Within the anticodon loop of tRNAHis, there are three potential RNase L cleavage sites. In vitro transcribed tRNAHis is susceptible to cleavage at each of these three sites in contrast to physiological tRNAHis, which is only cleaved at position 37. The authors of this study found that modification of G34 to Q34 provides specificity that guides cleavage at the physiological site [80] (Figs 2 and 3). RNase L activation is accompanied by a dramatic decrease in protein synthesis. It has been speculated that cleavage of bulk tRNAHis inhibits protein synthesis by preventing decoding of histidine codons. However, a formal link between RNase L-dependent tRNA cleavage and inhibition of translation has not been proven. It remains possible that the cleavage products of tRNAHis themselves contain activity.
Biologically active tRNA fragments
The supposition that tRNA fragments from RNase L-directed cleavage may harbor biological activity is not without precedent. Evidence presented over the previous decade has established a new field focusing on functions of small RNA derived from tRNAs. These RNA are produced via diverse biogenesis pathways, proposed to play an active role in different biological processes (Fig. 4) and, as a result, a complicated and often confusing nomenclature. Broadly, tRNA fragments can be classified into two categories: fragments induced as a result of cellular stress response pathways and constitutively expressed fragments (Fig. 2). These RNA may be derived from mature cytoplasmic tRNA, full-length pre-tRNA or ‘discarded’ processing fragments, such as those containing introns, 5′ leaders or 3′ trailers.
Fig. 4.
Proposed biological activities of tRNA fragments.
In the 1970s, tRNA breakdown products were identified from urine and serum of cancer patients and were hypothesized to be potential biomarkers for cancer [82,83]. Similarly, with the advent of high-throughput sequencing, fragments of tRNA were often recovered when sequencing small RNA, but were regularly disregarded as nonphysiologically relevant degradation intermediates. However, a wealth of data now supports that these are bona fide functional RNA. Their production is conserved throughout evolution, and they play a wide array of roles (Fig. 4).
Stress-induced tRNA fragments
An early report of inducible tRNA fragments came from studying the ciliate Tetrahymena thermophila in response to amino acid starvation [84]. Diverse tRNA fragments were generated after serum starvation, but the majority of tRNA were found to be cleaved within the anticodon loop, and thus, the individual RNA were termed ‘tRNA halves’. Later work showed that tRNA cleavage in response to stress is not restricted to T.thermophila. In particular, the anticodon targeting of tRNA by RNases was a widespread evolutionary reaction to cell stress [85]. An unbiased screen of small RNA revealed widespread tRNA-derived small RNA in S. cerevisiae, which were dramatically induced in response to various stresses, including heat shock and oxidative stress. Further experiments showed that oxidative stresses caused a similar response in Arabadopsis thaliana and human tissue culture cells [85].
Two groups independently demonstrated that stress-induced tRNA cleavage in humans is carried out by the RNase angiogenin (ANG) [86,87]. Again, tRNA cleavage was observed after a wide array of cellular insults, including starvation, heat shock, cold shock, oxidative stress, and UV irradiation. Subsequently, these small RNA were shown to harbor biological activity [87]. By monitoring protein synthesis, a subset of these small RNA inhibited translation (further discussed below) [87]. Concurrent with these findings, a different nuclease Rny1, a RNase T2 endonuclease, was shown to be the RNase responsible for stress-induced tRNA cleavage in S. cerevisiae [88]. Similarsized tRNA fragments were also identified in the phloem and sap of pumpkins (Cucurbita maxima) [89]. Isolation of these abundant small RNA showed that they were largely derived from tRNA and rRNA. Similar to data found in yeast and humans, translation assays suggested that these RNA harbor biological activity as they nondiscriminately inhibit mRNA translation [89]. To date, the enzymes that target tRNA in C. maxima, A. thaliana, or T. thermophila are unknown.
ANG, the protein that targets tRNA in mammalian cells, is a member of the RNase A superfamily (reviewed in [90]). Despite having high similarity to bovine pancreatic RNase A, it has significantly reduced activity due to occlusion of the active site by a glutamine residue not found in RNase A [91]. ANG was discovered and characterized by its ability to promote new blood vessel growth [92], making it a potential anticancer therapeutic target [93]. The ability of ANG to promote growth and angiogenesis is dependent upon its catalytic activity yet in vivo RNA targets of ANG were unknown until recently. Nearly 35 years after its discovery, ANG was shown to target mature tRNA in vivo [86,87]. Of particular note is that ANG is a secreted molecule that is actively taken up by surrounding cells [92]. Potentially, ANG acts in an ‘interferon-like’ manner by which neighboring cells can take up ANG secreted from stressed cells. In cells and in the absence of stress, ANG is held in an inactive state via interaction with the RNase A family inhibitor RNH1 [94,95].
It is unclear what specifies ANG-mediated tRNA cleavage. Similar to RNase L, the sequence specificity of ANG or Rny1 makes them promiscuous RNases. In vitro digests show that ANG will cleave between any purine and pyrimidine dinucleotides albeit with varying efficiencies (CpA>CpG>UpA>UpG) [96]. As with other members of the RNase T2 family, Rny1 is thought to have no sequence specificity [97]. Despite this, in human cells, only ~ 1% of tRNA are cleaved during a stress response [98]. This can be partially explained by protection of tRNA from ANG or Rny1 by various protein complexes, such as the ribosome. However, it is surprising that certain tRNA are more susceptible than others to stress-induced cleavage and which tRNA are targeted for cleavage can change in response to different stresses, further indicating selectivity [98]. Of particular note is tRNATyr, which is resistant to ANG-induced tRNA cleavage in vivo despite the fact that its anticodon loop has two purine: pyrimidine dinucleotides that should be susceptible to ANG-mediated cleavage (5′ CUˆGUˆAGG 3′, where ‘ˆ’ indicates potential cleavage sites) [86]. An intriguing possibility is that post-transcriptional modifications render tRNATyr resistant to cleavage as its anticodon loop is highly modified [36]. While the genomic sequence encoded CUGUAGG, the modified mature anticodon loop is CU9ΨAm1GG in which ‘9’ denotes galactosyl-queuosine. This specific modification is unique to tRNATyr in humans. In vitro digests of 5S rRNA show that ANG efficiently cleaves between Ψ:A dinucleotides and several other tRNA that contain m1G at position 37, so it is unlikely that these modifications protect tRNATyr, leaving the galactosyl-queuosine as a likely protective mark. Alternatively, tRNATyr is unique as all the genes encode an intron within the anticodon loop and possibly splicing imparts some modification or other state to tRNATyr that enables it to evade ANG cleavage [60].
As with tRNA cleavage initiated in host defense pathways, tRNA modifications can play important roles in promoting or restricting tRNA cleavage in response to stress (Fig. 2). m5C has roles in modulating tRNA cleavage. In higher eukaryotes, m5C is primarily added by NSUN2 or DNMT2 [99–102]. DNMT2 methylates tRNAVal(AAC), tRNAGly(GCC), and tRNAAsp(GTC) [101,103]. Mutant DNMT2 flies lack these methylation marks and demonstrate an increase in tRNA fragments from these tRNA species, establishing DNMT2 m5C deposition as a tRNA cleavage protective mark [103]. m5C as a protective mark is extended evolutionarily as NSUN2-deposited m5C has a similar anticleavage affect in mice [100]. tRNA lacking m5C have increased affinities for ANG, leading to more tRNA cleavage. In addition to a marked increase in tRNA fragments, NSUN2 knockout mice display neurological defects similar to human syndromic intellectual disability and Dubowitz-like syndrome. This mimics human disease where patients with a syndromic form of intellectual disability and Dubowitz-like syndrome have homozygous mutations in NSUN2 that impairs its catalytic activity [104–106]. While DNMT2 or NSUN2 knockout mice are viable, double knockout of both methyltransferases is synthetically lethal [102]. However, mouse embryonic fibroblasts derived from these inviable animals are viable but demonstrate a decreased translation rate, a phenotype potentially attributed to increased tRNA cleavage.
Despite data in mice showing synergistic effects of NSUN2 and DNMT2 methylation, a recent study in Drosophila melanogaster show that m5C deposition by either NSUN2 or DNMT2 may have differing effects on tRNA fragment production [107]. CRISPR-Cas9-mediated knockouts of NSUN2 or DNMT2 were generated in D. melanogaster. In response to heat shock, DDNMT2 flies generated more tRNA fragments which were longer lived than tRNA fragments from wild-type flies. In contrast, DNSUN2 flies failed to generate tRNA fragments in response to heat shock. Differences in species or cell types may underlie these contrasting results.
It has been well characterized that stress-induced tRNA cleavage results in reduction in protein synthesis (Fig. 4, translation modulation). It cannot be overemphasized that this is not due to depletion of the pool of cytoplasmic tRNA, as only a small proportion of tRNA are cleaved, in contrast to host defense-related tRNA cleavage. Instead, the small RNA themselves harbor biological activity. To date, several mechanisms of translation inhibition have been discovered for different tRNA-derived small RNA. Perhaps the most extensively studied mechanism of translation inhibition by tRNA fragments is by the 5′ halves of tRNAAla and tRNACys. Upon their discovery, these fragments were termed 5′ tRNA-derived stress-induced RNA (5′tiRNA) [87]. These RNA are generated by ANG-directed cleavage, generating two smaller RNA, 5′tiRNA and 3′tiRNA. To date, only 5′tiRNA have been shown to possess translation repression activities. ANG-mediated cleavage results in 5′tiRNA that contain a hydroxyl group at their 5′-ends and a 2′,3′ cyclic phosphate at the 3′-ends [87,108,109] and 3′tiRNA that contain a hydroxyl group at their 5′-ends and a hydroxyl or an amino acid at their 3′-ends. Some high-throughput experiments indicated that 5′tiRNA are more abundant than 3′tiRNA [110,111]. Although not empirically proven, it is likely that the 2′,3′ cyclic phosphate provides the 5′tiRNA with greater stability. It should also be noted that results of high-throughput RNA-sequencing can be greatly affected by methodology of RNA purification and library construction.
The translation inhibition activity of 5′tiRNAAla and 5′tiRNACys is endowed by a stretch of five guanosine residues at the 5′-end called the terminal oligoguanine motif (TOG motif) [112]. 5′tiRNAAla and 5′tiRNACys inhibit translation by blocking cap-dependent translation initiation. Canonical cap-dependent translation requires the heterotrimeric eIF4F complex, composed of eIF4E, eIF4A, and eIF4G, to bind the m7GTP mRNA cap. eIF4F is required for proper assembly of the canonical translation pre-initiation complex. By preventing formation of this complex on the mRNA, TOG-containing tiRNA block translation initiation. As a result, this also triggers the formation of nonmembranous cytoplasmic bodies called stress granules (SGs) [113]. SGs are condensates of non-translated mRNPs and 40S ribosomal subunits that are thought to promote survival through sequestration of pro-apoptotic signaling molecules and non-translating housekeeping mRNA (reviewed in [114]). TOG-containing tiRNA directly interact with Y-box binding protein 1 (YB-1, YBX1) and this interaction is critical for the formation of tiRNA-induced SGs [115]. However, YB-1 is dispensable for tiRNA-mediated translation repression. The precise mechanism of tiRNA-mediated eIF4F displacement remains to be elucidated. TOG-containing tiRNA interact with each other via their TOG motif forming tetrameric molecules [116]. The guanosine residues of one tiRNA interact via Hoogsteen base-pairing to form a parallel, intermolecular G-quadruplex. Only the tetrameric G-quadruplex tiRNA demonstrated biological activities as the monomeric TOG-containing tiRNA did not repress translation or triggered the formation of SGs. Moreover, this appears to be an evolutionary conserved phenomenon as evidence has also been presented that tiRNA from A. thaliana also assemble into a tetrameric G-quadruplex [116]. The activity of TOG-containing tiRNA is thought to be neuroprotective [117]. RNA G-quadruplexes are known regulators of translation (reviewed in [118]). Recent data suggest that RNA G-quadruplexes are largely unfolded under steady-state conditions in human cells [119]. Their presence and activity in TOG-containing tiRNA suggests that G-quadruplexes may have an unexplored role in regulating gene expression in non-steady-state conditions, such as stress conditions.
It is worth noting that most experiments using TOG-containing tiRNA have used synthetic RNA and therefore lack RNA modifications found in vivo. To date, experiments using bulk, endogenous tiRNA, show that they have similar activities to unmodified synthetic tiRNA [87,112,113]. However, given the abundance of tRNA modifications, it is likely that some may alter tiRNA activity. It remains to be investigated whether stress-induced tRNA fragments are enriched or depleted for specific tRNA modifications. Further experiments with endogenous TOG-containing tiRNA would be of great value as tRNA modifications are known to change in response to stress [50,120–122].
A variety of non-TOG-containing tiRNA also inhibit translation, albeit via alternative mechanisms. The halophilic archeon H. volcanii generates tRNA fragments in response to certain stresses such as alkaline stress or increased magnesium levels [123]. In this case, cleavage does not occur in the anticodon loop, but rather in the single-stranded region between the D-arm and the anticodon arm generating a 26-nucleotide RNA. This RNA derived from tRNAVal, referred to as valine tRNA-derived fragment (Val-tRF), physically interacts with the small ribosomal subunit to inhibit translation initiation complex formation by preventing mRNA:ribosome interaction [124]. The activity of this small RNA is conserved in all three domains of life as it can block translation in S. cerevisiae, H. volcanii, and E. coli. This is not accomplished by preventing Shine-Dalgarno:Anti-Shine-Dalgarno interactions, as is common for other small RNA in prokaryotes. Instead, Val-tRF binds within the mRNA binding channel in the 30S small ribosomal subunit. A similar situation has been proposed for a subset of tRNA fragments in S. cerevisiae where several tRNA fragments were found to associate with ribosomes following stress. Specifically, the binding of both 3′ and 5′ fragments derived from tRNAHis was associated with translation repression [125].
Surprisingly, in contrast to RNase L-mediated tRNA cleavage, which is antiviral, some tRNA fragments are proviral. Upon infection of human cells with respiratory syncytial virus, several tRNA fragments are produced, the most abundant of which are 5′ halves from tRNAGlu(CTC), tRNAGly(GCC), and tRNALys(CTT) [126]. These tRFs can function in a miRNA-like manner. In particular, tRFGlu(CTC) base-pairs with the 3′ UTR of the apolipoprotein E receptor 2 (APOER2) mRNA and reduces its expression [127]. Otherwise, APOER2 protein functions in an antiviral manner; thus, suppression of expression by tRFs promotes viral replication. Further work has shown that blocking 5′tRFGlu(CTC) and 5′tRFLys(CTT) also promote viral replication [128]. Blocking these tRFs with antisense oligos suppresses viral replication. Conversely, transfection of synthetic tRFs stimulates viral replication. The precise mechanism through which these tRFs promote viral replication is unknown, although it is hypothesized to occur via a miRNA-like manner as shown for 5′tRFGly(GCC) [127].
Finally, some stress-induced tRNA fragments play biological roles distinct from regulating translation. During osmotic stress, ANG-cleaved tRNA associate with cytochrome c to promote survival [129]. At the onset of apoptosis, cytochrome c is released from the mitochondria where it interacts with other proteins and oligomerizes to form a large protein structure called the apoptosome. Various tRNA fragments interact with cytochrome c upon release from the mitochondria to block apoptosome formation and promote survival [129].
Constitutive tRNA fragments
Not all tRNA fragments are produced in response to cellular stress. There are abundant tRNA fragments that are constitutively produced throughout the lifespan of an organism, in specific cell types or at specific developmental stages. Some of these represent the same molecule as a stress-induced fragment and even share a biogenesis pathway, but are exemplified by their stable noninduced production. Diverse tRNA fragments are commonly found in cancer cells lines. Such tRNA-derived small RNA were first carefully analyzed by high-throughput sequencing of small RNA from prostate cancer cell lines [130]. These small RNA, referred to as tRFs, are 19–21 nucleotides and are generated as a result of cleavage in any of the tRNA loops [131]. However, one of the more intriguing tRNA-derived small RNA, termed tRF-1001, is derived from the 3′ trailer of tRNASer. Its biogenesis is thought to be a result of ELAC2 cleavage during tRNA maturation. Surprisingly, tRF-1001 promotes cell proliferation in cancer cells [130]. In addition to their identification in cancer cell lines, select tRFs have been identified in various healthy tissues including heart, colon, lung, and small intestine [131].
Data have been presented suggesting that the biogenesis of many tRFs is dependent upon Dicer, a principal enzyme in the generation of miRNA [132]; however, as the aforementioned tRF-1001 is ELAC2-dependent, it is likely that there are multiple biogenesis pathways given the wide array of tRF species. In fact, other studies demonstrate that many stably expressed tRNA fragments are Dicer- and Drosha-independent [131,133]. tRFs also have been shown to associate with Ago1, Ago3, and Ago4, but not with Ago2 [131]. However, other data indicate tRFs do associate with Ago2 [134]. Given that their size mirrors that of miRNA, it is conceivable that these tRNA-derived RNA, once incorporated into an Argonaute protein, could function in a miRNA- or siRNA-like manner. Using luciferase reporter assays with constructs containing tRF antisense sequences, tRFs are capable of eliciting sequence-dependent translation repression [135]. Investigations in D. melanogaster show that some tRNA fragments modulate the translation of mRNA through antisense base-pairing in a manner that is dependent upon Ago2 [136], by modulating expression of the translational machinery. Related to this finding, in humans, a tRNA fragment derived from tRNALeu base-pairs with RPS28 mRNA and promotes its expression [137]. In this instance, rather than functioning via an Ago-mediated pathway, the tRF prevents deleterious secondary structure formation in the mRNA.
Recent work has revealed that some tRNA fragments can regulate epigenetic inheritance. In sperm from mice fed a high-fat diet (HFD), there was a marked increase in 5′ tRNA fragments [138]. Interestingly, offspring born from mice fertilized by sperm with high concentrations of tRNA fragments displayed a higher incidence of glucose metabolism disorders. In this case, rather than modulating mRNA translation, it appears that these tRNA fragments target the promoters of select genes to alter transcription. Interestingly, analysis of tRNA fragments in mice fed a HFD revealed that these tRNA fragments were enriched in m5C and N2-methylguanosine (m2G) modifications, but the relevance of these modifications was unknown in this initial study. Follow-up work showed that DNMT2 was responsible for these modifications. In contrast to wild-type animals, DNMT2 knockout mice failed to demonstrate a relative enrichment of m5C or m2G in the tRNA fragments after HFD [139]. The DNMT2 knockout mice fed a HFD did still produce tRNA fragments; in fact, they produced higher levels of tRNA fragments in agreement with previously reported data on DNMT2-mediated regulation of tRNA cleavage [103]. However, these fragments no longer conferred metabolic disorders to the next generation. The authors show that modification of m5C38 in the tRNA fragments alters the secondary structure of the small RNA, which may be necessary for their activity. Thus, in this instance, not only are the tRNA fragments necessary, but must contain the proper DNMT2 deposited modifications. In a related study, mice on protein restriction diets had increased levels of tRNA fragments from tRNAGly(GCC) [140]. These tRNA fragments repress the transcription of genes associated with the endogenous retroelement MERVL. This activity may be evolutionarily conserved. tRNA fragments are found abundantly in the pollen of A. thaliana and other land plants [141]. In this case, tRNA fragments also target transposable elements, although in A. thaliana, they load into Ago1 and target transposable element RNA. Moreover, genetic ablation of DNMT2 and NSUN2, the methyltransferases that generates m5C in tRNA, leads to an increase in mobile element expression and genome instability in D. melanogaster [107]. This is concurrent with alterations in tRNA fragment production seen in DNMT2 and NSUN2 knockout flies.
Finally, data are emerging that tRNA fragments play vital roles in regulating stem cell fate. Distinct cell types differ in their susceptibility to form tRNA fragments and the consequences of their formation [142]. Hematopoietic stem/progenitor cells (HSPCs) and lineage committed myeloid-restricted progenitor (MyePro) cells both take up secreted ANG; however, the result of this uptake depends upon cell type. In HSPCs, ANG cleaves tRNA generating tRNA fragments that repress translation which is required for maintaining the stemness of these cells. In contrast, ANG does not cause cleavage of tRNA in MyePro cells, instead causing an increase in rRNA transcription and enhancing proliferation. It is not known what makes the tRNA of HSPCs susceptible to ANG-mediated cleavage or what protects the tRNA of MyePro cells; potentially cell type-specific tRNA modification could play a role. This is backed up by reports in other stem cells in which tRNA modifications affect tRNA cleavage, protein synthesis, and maintenance of stemness [143,144]. Cells derived from NSUN2 knockout mice produce an increased amount of tRNA fragments due to defects in m5C modifications in specific tRNA species. As epidermal stem cells naturally express low levels of NSUN2, this leads to the increases in the abundance of tRNA fragments [143]. In turn, tRNA fragments in epidermal stem cells throttle protein synthesis to maintain stemness. Upon differentiation, NSUN2 expression levels rise, concomitant with a decrease in abundance of tRNA fragments and an increase in protein synthesis. Similar results have been reported for the maintenance of neuroepithelial stem (NES) cells [144]. In NSUN2 knockout mice, NES cell differentiation is impaired and this result is phenocopied by increased ANG expression. This delayed differentiation is due to improper stem cell maintenance caused by pervasive tRNA fragment production that may underlie NSUN2-related neurological defects. m5C is not the only tRNA modification that may affect tRNA fragment production to regulate stem cells. A recent report shows that Ψ of TOG-containing tRNA fragments may also promote stem cell maintenance [145]. Pseudouridylate synthase 7 (PUS7) is enriched in human embryonic stem cells (hESCs) and catalyzes the pseudouridylation of U8, particularly in tRNA that contain a TOG motif (tRNAAla, tRNACys, tRNAVal). hESCs produce abundant TOG-containing tRNA fragments that are 18 nucleotides, in contrast to TOG-containing tiRNA generated during stress, which are 30 – 31 nucleotides. For this reason, they have been termed miniTOGs (mTOGs). These recently discovered tRNA fragments repress translation and maintain the stem cell niche. Importantly, only mTOGs containing Ψ8 exert translation inhibitory effect in stem cells. This is in contrast to TOG-containing tiRNA, where to this point, modifications have not been tested as synthetic, unmodified tiRNA are as active as endogenous tiRNA [87,112,113]. This highlights a critical difference from stress-induced and constitutively produced tRNA fragments, particularly in terms of stem cell maintenance.
Concluding remarks
As the field of RNA modifications continues to expand, it is critical to remember that tRNA are the most densely modified RNA molecules with the greatest diversity of chemical modifications. These modifications dictate tRNA structure and regulate canonical tRNA function. Recent reports have shown that these modifications are critically important for modulating the noncanonical functions of tRNA, paramount among these are functions of tRNA-derived small RNA. Intriguingly, tRNA modifications can promote or inhibit the generation of these fragments and the modifications within tRNA fragments can affect their activity. Given the diversity of tRNA modifications, many new functions in tRNA cleavage and tRNA fragment biological activities surely remain to be discovered.
As work continues, many unanswered questions remain and several points should be taken into account. In the field’s infancy, experiments dissecting tRNA fragment activity have relied on synthetically synthesized RNA devoid of any modifications. While it is true that for some tRNA fragments, where endogenous modified RNA and synthetic RNA have similar biological activities [87,112], recent work discovered that addition of certain RNA modifications, such as Ψ or m5C, is necessary to grant a specific RNA its activity [139,145]. Therefore, it is possible that in the search for active tRNA fragments, some biological activities of tRNA fragments were undetected because they did not contain the required physiological modifications. For analysis of individual species of tRNA fragments, methods to reliably separate these molecules from the milieu of all tRNA fragments will need to be developed. These methods will require purification of large amounts of material for biochemical studies, a restraint that may slow their development and implementation. One such protocol has been developed to separate different tRNA species [146], which could be further refined for the purification of tRNA fragments. However, as tRNA fragments are typically 100 times less abundant than the tRNA from which they were derived [98], scalability issues will remain. Alternatively, many modified nucleotides are available for purchase in synthetic RNA. As of now, the high price of adding these modifications to oligonucleotides may make this approach cost-ineffective, particularly in the early screening stages of an experiment.
What will hamper some of future studies is our current incomplete knowledge of the full repertoire of tRNA modifications. This is particularly evident in terms of cell type specificity. For example, using the most current Modomics dataset [36], we generated a map of nucleotide modification frequency in humans (Fig. 3). This map and data from Modomics indicate that position 8 is rarely modified. However, it is clear that pseudouridylation of this nucleotide is required for activity of miniTOGs in stem cells [145]. As tRNA modification mapping is currently limited to profiling in specific cancer cell lines, some cell type- or tissue-specific modifications are certainly missed. In turn, the presence of specific tRNA modifications may prevent or promote tRNA cleavage. Such differences in tRNA modifications may explain the findings in hematopoietic cells, where ANG readily cleaves tRNA in HSPCs but not in MyePro cells.
Despite our incomplete understanding of the molecular mechanisms underlying biogenesis and functions of tRNA fragments, the link between tRNA fragments and various biological processes has been established. It is imperative that we continue to decipher the molecular mechanisms that contribute to our understanding of how tRNA fragments and tRNA modification relate to human disease.
Acknowledgments
We thank Anderson and Ivanov lab members for discussion. This work was supported by the NIH (NS094918 to PI, F32 GM119283 to SML, and AI007306 to MMF) and by the Brigham Research Institute Fund to Sustain Research Excellence to PI.
Abbreviation
- ANG
angiogenin
- APOER2
apolipoprotein E receptor 2
- HFD
high-fat diet
- HSPCs
hematopoietic stem/progenitor cells
- NES
neuroepithelial stem
- SGs
stress granules
- TOG
terminal oligoguanine
References
- 1.Goodenbour JM and Pan T (2006) Diversity of tRNA genes in eukaryotes. Nucleic Acids Res 34, 6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan PP and Lowe TM (2016) GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44, D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geslain R and Pan T (2010) Functional analysis of human tRNA isodecoders. J Mol Biol 396, 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH and Ackerman SL (2014) RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson HD, Altman S and Smith JD (1972) Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem 247, 5243–5251. [PubMed] [Google Scholar]
- 6.Guerrier-Takada C, Gardiner K, Marsh T, Pace N and Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 7.Bikoff EK and Gefter ML (1975) In vitro synthesis of transfer RNA. I. Purification of required components. J Biol Chem 250, 6240–6247. [PubMed] [Google Scholar]
- 8.Bikoff EK, LaRue BF and Gefter ML (1975) In vitro synthesis of transfer RNA. II. Identification of required enzymatic activities. J Biol Chem 250, 6248–6255. [PubMed] [Google Scholar]
- 9.Schiffer S, Rosch S and Marchfelder A (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J 21, 2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sprinzl M and Cramer F (1979) The -C-C-A end of tRNA and its role in protein biosynthesis. Prog Nucleic Acid Res Mol Biol 22, 1–69. [DOI] [PubMed] [Google Scholar]
- 11.Crothers DM, Seno T and Soll G (1972) Is there a discriminator site in transfer RNA? Proc Natl Acad Sci USA 69, 3063–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R and Abelson JN (1997) The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858. [DOI] [PubMed] [Google Scholar]
- 13.Rauhut R, Green PR and Abelson J (1990) Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J Biol Chem 265, 18180–18184. [PubMed] [Google Scholar]
- 14.Peebles CL, Gegenheimer P and Abelson J (1983) Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32, 525–536. [DOI] [PubMed] [Google Scholar]
- 15.Phizicky EM, Schwartz RC and Abelson J (1986) Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem 261, 2978–2986. [PubMed] [Google Scholar]
- 16.Culver GM, McCraith SM, Consaul SA, Stanford DR and Phizicky EM (1997) A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem 272, 13203–13210. [DOI] [PubMed] [Google Scholar]
- 17.Paushkin SV, Patel M, Furia BS, Peltz SW and Trotta CR (2004) Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117, 311–321. [DOI] [PubMed] [Google Scholar]
- 18.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V et al. (2013) CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM et al. (2014) Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weitzer S and Martinez J (2007) The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447, 222–226. [DOI] [PubMed] [Google Scholar]
- 21.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D et al. (2011) HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764. [DOI] [PubMed] [Google Scholar]
- 22.Popow J, Jurkin J, Schleiffer A and Martinez J (2014) Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511, 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai KK, Cheng CL, Bingman CA, Phillips GN Jr and Raines RT (2014) A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res 42, 3931–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darnell RB, Ke S and Darnell JE Jr (2018) Pre-mRNA processing includes N(6) methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics”. RNA 24, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer M, Kapoor U and Jantsch MF (2017) Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohn W and Volkin E (1951) Nucleoside-5′-phosphates from ribonucleic acid. Nature 167, 483–484. [Google Scholar]
- 27.Davis FF and Allen FW (1957) Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem 227, 907–915. [PubMed] [Google Scholar]
- 28.Littlefield JW and Dunn DB (1958) The occurrence and distribution of thymine and three methylated-denine bases in ribonucleic acids from several sources. Biochem J 70, 642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Littlefield JW and Dunn DB (1958) Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids. Nature 181, 254–255. [DOI] [PubMed] [Google Scholar]
- 30.Smith JD and Dunn DB (1959) The occurrence of methylated guanines in ribonucleic acids from several sources. Biochem J 72, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amos H and Korn M (1958) 5-Methyl cytosine in the RNA of Escherichia coli. Biochim Biophys Acta 29, 444–445. [DOI] [PubMed] [Google Scholar]
- 32.Davis FF, Carlucci AF and Roubein IF (1959) Trace nucleotides in certain ribonucleic acids from yeast. J Biol Chem 234, 1525–1529. [PubMed] [Google Scholar]
- 33.Osawa S (1960) Preparation and some properties of a soluble ribonucleic acid from yeast. Biochim Biophys Acta 43, 110–122. [DOI] [PubMed] [Google Scholar]
- 34.Osawa S (1960) The nucleotide composition of ribonucleic acids from subcellular components of yeast, Escherichia coli and rat liver, with special reference to the occurrence of pseudouridylic acid in soluble ribonucleic acid. Biochim Biophys Acta 42, 244–254. [DOI] [PubMed] [Google Scholar]
- 35.Holley RW, Everett GA, Madison JT and Zamir A (1965) Nucleotide sequences in the yeast alanine transfer ribonucleic acid. J Biol Chem 240, 2122–2128. [PubMed] [Google Scholar]
- 36.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A et al. (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YL, Prestwich EG, Lim KS et al. (2015) A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol 560, 29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM and Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ and Dedon PC (2014) Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc 9, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz C, Lunse CE and Morl M (2017) tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charette M and Gray MW (2000) Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. [DOI] [PubMed] [Google Scholar]
- 42.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF and Klug A (1974) Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 250, 546–551. [DOI] [PubMed] [Google Scholar]
- 43.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC et al. (2016) The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helm M, Brule H, Degoul F, Cepanec C, Leroux JP, Giege R and Florentz C (1998) The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res 26, 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU and Helm M (2007) A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys). J Am Chem Soc 129, 13382–13383. [DOI] [PubMed] [Google Scholar]
- 46.Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV and Katunin VI (2004) Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosjean H, Soll DG and Crothers DM (1976) Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol 103, 499–519. [DOI] [PubMed] [Google Scholar]
- 48.Bjork GR, Huang B, Persson OP and Bystrom AS (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13, 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klassen R, Grunewald P, Thuring KL, Eichler C, Helm M and Schaffrath R (2015) Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS ONE 10, e0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ and Dedon PC (2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M and Hinnebusch AG (1998) The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson J, Phan L and Hinnebusch AG (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG and Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arts GJ, Fornerod M and Mattaj IW (1998) Identification of a nuclear export receptor for tRNA. Curr Biol 8, 305–314. [DOI] [PubMed] [Google Scholar]
- 55.Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E and Gorlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1, 359–369. [DOI] [PubMed] [Google Scholar]
- 56.Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H and Gorlich D (1999) Coordination of tRNA nuclear export with processing of tRNA. RNA 5, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ and Phizicky EM (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21, 87–96. [DOI] [PubMed] [Google Scholar]
- 58.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ and Phizicky EM (2008) Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev 22, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson PF and Abelson J (1983) The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature 302, 681–687. [DOI] [PubMed] [Google Scholar]
- 60.van Tol H and Beier H (1988) All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res 16, 1951–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strobel MC and Abelson J (1986) Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol 6, 2663–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider A, McNally KP and Agabian N (1993) Splicing and 3′-processing of the tyrosine tRNA of Trypanosoma brucei. J Biol Chem 268, 21868–21874. [PubMed] [Google Scholar]
- 63.Rubio MA, Paris Z, Gaston KW, Fleming IM, Sample P, Trotta CR and Alfonzo JD (2013) Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell 52, 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clouet d’Orval B, Bortolin ML, Gaspin C and Bachellerie JP (2001) Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bortolin ML, Bachellerie JP and Clouet-d’Orval B (2003) In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp). Nucleic Acids Res 31, 6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh SK, Gurha P, Tran EJ, Maxwell ES and Gupta R (2004) Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 279, 47661–47671. [DOI] [PubMed] [Google Scholar]
- 67.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S and Cavard D (2007) Colicin biology. Microbiol Mol Biol Rev 71, 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gratia A (1925) Sur un remarquable example d’antagonisme entre deux souches de colibacille. C R Acad Sci Ser III Sci Vie 93, 1040–1042. [Google Scholar]
- 69.Jacob F, Siminovitch L and Wollman E (1952) Biosynthesis of a colicin and its mode of action. Ann Inst Pasteur (Paris) 83, 295–315. [PubMed] [Google Scholar]
- 70.Ogawa T, Inoue S, Yajima S, Hidaka M and Masaki H (2006) Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res 34, 6065–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T and Masaki H (1999) A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283, 2097–2100. [DOI] [PubMed] [Google Scholar]
- 72.Tomita K, Ogawa T, Uozumi T, Watanabe K and Masaki H (2000) A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci USA 97, 8278–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amitsur M, Morad I, Chapman-Shimshoni D and Kaufmann G (1992) HSD restriction-modification proteins partake in latent anticodon nuclease. EMBO J 11, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amitsur M, Morad I and Kaufmann G (1989) In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J 8, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang Y, Meidler R, Amitsur M and Kaufmann G (2001) Specific interaction between anticodon nuclease and the tRNA(Lys) wobble base. J Mol Biol 305, 377–388. [DOI] [PubMed] [Google Scholar]
- 76.Amitsur M, Levitz R and Kaufmann G (1987) Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J 6, 2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gunge N, Tamaru A, Ozawa F and Sakaguchi K (1981) Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J Bacteriol 145, 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butler AR, Porter M and Stark MJ (1991) Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast 7, 617–625. [DOI] [PubMed] [Google Scholar]
- 79.Lu J, Esberg A, Huang B and Bystrom AS (2008) Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 36, 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donovan J, Rath S, Kolet-Mandrikov D and Korennykh A (2017) Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sadler AJ and Williams BR (2008) Interferon-inducible antiviral effectors. Nat Rev Immunol 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W and Waalkes TP (1977) High turnover rate of transfer RNA in tumor tissue. Cancer Res 37, 3362–3366. [PubMed] [Google Scholar]
- 83.Gehrke L, Bast RE and Ilan J (1981) An analysis of rates of polypeptide chain elongation in avian liver explants following in vivo estrogen treatment. I. Determination of average rates of polypeptide chain elongation. J Biol Chem 256, 2514–2521. [PubMed] [Google Scholar]
- 84.Lee SR and Collins K (2005) Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280, 42744–42749. [DOI] [PubMed] [Google Scholar]
- 85.Thompson DM, Lu C, Green PJ and Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z and Zheng X (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583, 437–442. [DOI] [PubMed] [Google Scholar]
- 87.Yamasaki S, Ivanov P, Hu GF and Anderson P (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson DM and Parker R (2009) The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S, Sun L and Kragler F (2009) The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol 150, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyons SM, Fay MM, Akiyama Y, Anderson PJ and Ivanov P (2017) RNA biology of angiogenin: current state and perspectives. RNA Biol 14, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Russo N, Shapiro R, Acharya KR, Riordan JF and Vallee BL (1994) Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci USA 91, 2920–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF and Vallee BL (1985) Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 24, 5480–5486. [DOI] [PubMed] [Google Scholar]
- 93.Sheng J and Xu Z (2016) Three decades of research on angiogenin: a review and perspective. Acta Biochim Biophys Sin (Shanghai) 48, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kobe B and Deisenhofer J (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186. [DOI] [PubMed] [Google Scholar]
- 95.Chen CZ and Shapiro R (1997) Site-specific mutagenesis reveals differences in the structural bases for tight binding of RNase inhibitor to angiogenin and RNase A. Proc Natl Acad Sci USA 94, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shapiro R, Riordan JF and Vallee BL (1986) Characteristic ribonucleolytic activity of human angiogenin. Biochemistry 25, 3527–3532. [DOI] [PubMed] [Google Scholar]
- 97.Luhtala N and Parker R (2010) T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci 35, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T and Hatzoglou M (2012) Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 287, 42708–42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J and Szweykowska-Kulinska Z (2006) Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res 34, 6034–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M et al. (2014) Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33, 2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE and Bestor TH (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398. [DOI] [PubMed] [Google Scholar]
- 102.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G and Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19, 900–905. [DOI] [PubMed] [Google Scholar]
- 103.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M and Lyko F (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24, 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L and Gleeson JG (2012) Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet 49, 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS et al. (2012) Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet 90, 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K et al. (2012) Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet 90, 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Genenncher B, Durdevic Z, Hanna K, Zinkl D, Mobin MB, Senturk N, Da Silva B, Legrand C, Carre C, Lyko F et al. (2018) Mutations in cytosine-5 tRNA methyltransferases impact mobile element expression and genome stability at specific DNA repeats. Cell Rep 22, 1861–1874. [DOI] [PubMed] [Google Scholar]
- 108.Honda S, Morichika K and Kirino Y (2016) Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nat Protoc 11, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schutz K, Hesselberth JR and Fields S (2010) Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA 16, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dhahbi JM, Spindler SR, Atamna H, Boffelli D and Martin DI (2014) Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer 6, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P and Martin DI (2013) 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom 14, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov P, Emara MM, Villen J, Gygi SP and Anderson P (2011) Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Emara M, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu G and Anderson P (2010) Angiogenin-induced tiRNAs promote stress-induced stress granule assembly. J Biol Chem 285, 10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Panas MD, Ivanov P and Anderson P (2016) Mechanistic insights into mammalian stress granule dynamics. J Cell Biol 215, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lyons SM, Achorn C, Kedersha NL, Anderson PJ and Ivanov P (2016) YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res 44, 6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lyons SM, Gudanis D, Coyne SM, Gdaniec Z and Ivanov P (2017) Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun 8, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ivanov P, O’Day E, Emara MM, Wagner G, Lieberman J and Anderson P (2014) G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA 111, 18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fay MM, Lyons SM and Ivanov P (2017) RNA G-quadruplexes in biology: principles and molecular mechanisms. J Mol Biol 429, 2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo JU and Bartel DP (2016) RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gu C, Begley TJ and Dedon PC (2014) tRNA modifications regulate translation during cellular stress. FEBS Lett 588, 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patil A, Chan CT, Dyavaiah M, Rooney JP, Dedon PC and Begley TJ (2012) Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 9, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CT, Dedon PC and Begley TJ (2012) Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 11, 3656–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gebetsberger J, Zywicki M, Kunzi A and Polacek N (2012) tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gebetsberger J, Wyss L, Mleczko AM, Reuther J and Polacek N (2017) A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14, 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bakowska-Zywicka K, Kasprzyk M and Twardowski T (2016) tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Q, Lee I, Ren J, Ajay SS, Lee YS and Bao X (2013) Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 21, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS et al. (2015) Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 23, 1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS, Lee I, Lee YS and Bao X (2017) Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol 98, 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z et al. (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol 34, 2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee YS, Shibata Y, Malhotra A and Dutta A (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kumar P, Anaya J, Mudunuri SB and Dutta A (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ and Hutvagner G (2009) Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Z, Ender C, Meister G, Moore PS, Chang Y and John B (2012) Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 40, 6787–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ and Kay MA (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sobala A and Hutvagner G (2013) Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 10, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luo S, He F, Luo J, Dou S, Wang Y, Guo A and Lu J (2018) Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res 46, 5250–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K et al. (2017) A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y et al. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J et al. (2018) Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 20, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martinez G, Choudury SG and Slotkin RK (2017) tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45, 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Goncalves KA, Silberstein L, Li S, Severe N, Hu MG, Yang H, Scadden DT and Hu GF (2016) Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell 166, 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortes-Garrido R, Gkatza N et al. (2016) Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, Tailor J, Dietmann S and Frye M (2017) Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Reports 8, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimkova K, Sommarin MNE, Munita R, Lubas M et al. (2018) Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26. [DOI] [PubMed] [Google Scholar]
- 146.Miyauchi K, Ohara T and Suzuki T (2007) Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res 35, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]