Abstract
The biological functions of the Janus kinase 1 (JAK1) are suggested to be pleiotropic since this signal transducer is ubiquitously expressed and coupled to a variety of cytokine receptors. Consequently, mice that are deficient in this tyrosine kinase were reported to die shortly after birth. To facilitate studies that address the biological and molecular functions of JAK1 during postnatal development, we performed gene targeting in embryonic stem cells and generated a Cre/lox-based conditional knockout mouse model. Expression of Cre recombinase in the germline converted the Jak1 conditional knockout allele (Jak1fl) into a null allele (Jak1−) that when subsequently crossed into homozygosity led to a complete absence of the JAK1 protein in developing embryos. JAK1 deficient embryos were visibly smaller starting at E15.5. Newborn pups exhibited signs of apnea and died within hours after birth. The examination of fibroblasts from conditional knockout embryos and their littermate wildtype controls expressing JAK1 showed that lack of this Janus kinase resulted in an impaired tyrosine phosphorylation and activation of the downstream Signal Transducers and Activators of Transcription (STATs) 1, 3, and 6. JAK1 conditional knockout mice will be an invaluable tool to study cytokine signaling during normal development and disease progression in adult animals.
Keywords: Cre recombinase, embryonic development, gene targeting, Janus kinase 1, signal transduction, STAT
1 |. INTRODUCTION
Janus kinases (JAKs) are expressed in many tissues and function as signal transducers downstream of more than 50 cytokine and peptide hormone receptor complexes (Aaronson and Horvath, 2002; Kisseleva et al., 2002; Schindler and Plumlee, 2008). There are four known members of the Janus kinase family in mammals (JAK1, JAK2, TYK2, and JAK3), and with the exception of JAK3, which is primarily present in hematopoietic cells (Takahashi and Shirasawa, 1994), the other three JAK kinases are ubiquitously expressed (Dalal et al., 1998; Firmbach-Kraft et al., 1990; Harpur et al., 1992; Wilks et al., 1991; Yang et al., 1993). Upon growth factor stimulation, these receptor-associated, tyrosine kinases become autophosphorylated and activated. They subsequently phosphorylate specific tyrosine residues on the cytokine receptors thereby facilitating the recruitment of Signal Transducers and Activators of Transcription (STATs) to the receptor complexes. A main biochemical function of JAKs is the phosphorylation of particular tyrosines on the seven known STAT proteins (STAT1, 2, 3, 4, 5a, 5b, and 6). This phosphorylation event is a crucial step for their binding to Src Homology 2 (SH2) domains of other STAT proteins, which results in the formation of stable, transcriptionally active homo- or heterodimers. Once activated STATs are translocated to the nucleus, they bind to palindromic consensus sequences, also known as interferon gamma activation sites (GAS), within promoters of STAT target genes to enhance their transcription.
On the biochemical level, JAK1 has been shown to be a signal mediator of multiple cytokine receptor families, including particular members of type I cytokines such as the IL-2 and IL-4 receptor families, and the glycoprotein 130 (gp130) receptor family. JAK1 is equally important for signal transduction downstream of class II cytokine receptors (i.e., type I and type II IFN-R as well as IL-10R) (Kisseleva et al., 2002; Rodig et al., 1998). With the exception of STAT4, JAK1 has been suggested to activate the majority of STAT proteins depending on the cellular context and the particular ligand/receptor complex (Kisseleva et al., 2002). Because many of these receptor complexes may also associate with other Janus kinases, especially with the ubiquitously expressed JAK2, it was previously uncertain whether JAK1 has specific or redundant biological functions. However, the seminal work by Rodig et al. (1998) using conventional knockout mice clearly demonstrated that JAK1 has non-redundant roles, and this kinase is a central node for signaling of a number of cytokines such as IL-2, IL-6, IFN, and IL-10. In line with its suggested pleiotropic functions, it was observed that JAK1 deficiency causes perinatal lethality in mice, possibly due to neurological defects.
To investigate biologically relevant functions of JAK1 during postnatal development and in tissue homeostasis of adult animals, we report here the generation of a conditional knockout mouse model that allows a temporally and spatially controlled deletion of the Jak1 gene in somatic tissues. Another strong motivation to establish this mouse model was its broader applicability to study the significance of the JAK1/STAT signaling pathway in diseased tissues, such as adenocarcinomas of the mammary gland and pancreas that, depending on the stage of carcinogenesis, exhibit an aberrant, constitutive activation of STAT proteins (Schmidt and Wagner, 2012). In particular, oncogenic STAT3 and STAT1 are suggested to be tyrosine phosphorylated and activated by the ubiquitously expressed Janus kinases (i.e., JAK2 or JAK1) and may play crucial roles in malignant progression (Avalle et al., 2012; Bromberg et al., 1999; Levy and Darnell, 2002; Yu et al., 2014; Zhang et al., 2000). Hence, the conditional knockout allele of Jak1 can be used to address the importance of this particular member of the Janus kinase family in the genesis of cancer. As demonstrated previously for JAK2 (Sakamoto et al., 2009, 2010), the somatic and temporally controlled deletion of the Jak1 gene using this new model will be invaluable to discriminate the roles of JAK1 during disease initiation versus disease maintenance and progression.
To accomplish a somatic knockout of JAK1 in specific tissues or cell types of adult mice, we used homologous recombination in embryonic stem (ES) cells to create a conditional knockout (floxed) allele of the Jak1 gene (Figure 1a). As described in detail in the Material and Methods section, we used the λ Red phage-based homologous recombination method to construct the targeting vector. The 5′ loxP site was inserted into the first intron and a second loxP site along with a PGK-neomycin (PGK-neo) selectable marker was introduced downstream of exon 2. The PGK-neo cassette is surrounded by frt sites to allow for a complete excision of the resistance gene using FLP recombinase in ES cells or in animals. A Mcl-tk cassette was used for negative selection against random integration events. The floxed allele of the Jak1 gene was generated through homologous recombination in mouse embryonic stem (ES) cells at the UNMC Mouse Genome Engineering Core Facility. Thirteen correctly targeted clones were identified by EcoR1 restriction digest and Southern blot using a 5′ external probe that was not part of the targeting vector (Figure 1b). For validation and routine genotyping of ES cells and mice, we devised a PCR assay using primers that amplify the genomic region in intron 1 where the 5′ loxP site was inserted (Figure 1a; primer set 2411/2412). Two correctly targeted ES cell clones (A1 and A6) were injected into C57Bl/6 blastocysts for the production of chimeric animals. Following the mating of the male chimeras with wildtype C57Bl/6 females and germline transmission of the targeted Jak1 allele, we bred heterozygous mice (Jak1fl/wt) to obtain offspring with two Jak1 floxed alleles. Homozygous mutant mice (Jak1fl/fl, Figure 1c) were obtained in the expected Mendelian ratio and developed normally until adulthood. Mice of both genders were fertile, and the lack of phenotypic abnormalities suggested that the retention of the PGK-neo resistance gene did not cause any adverse effects on the transcriptional regulation of Jak1. This fact was very recently confirmed using RNA-Seq on mammary tissues of homozygous Jak1 floxed mice (Sakamoto et al., 2016). It was therefore not essential to use FLP recombinase to excise the selectable marker in the germline of Jak1fl/wt animals.
FIGURE 1.

Generation of a conditional knockout (floxed) allele of the Janus kinase 1 (Jak1). (a) Targeting strategy to flank the second coding exon of Jak1 with loxP sites. The targeting vector was constructed by placing the 5′ loxP site into the first intron and a second loxP site along with a PGK-neomycin (PGK-neo) selectable marker downstream of exon 2. The PGK-neo cassette is surrounded by frt sites and can be excised using FLP recombinase. (b) Southern blot using the 5 prime external probe illustrated in panel a. to monitor the homologous recombination in embryonic stem cell (ESC) clones. (c) PCR analysis using the primer set 2411/2412 shown in panel a. to determine the presence of the wildtype and/or floxed alleles in heterozygous (fl/wt) and homozygous mutants (fl/fl) as well as wildtype controls (wt/wt). (d) Schematic outline of the Cre-mediated, conditional excision of exon 2 of the Jak1 gene. (e) PCR analysis using the primer set 2411/2373 illustrated in panel d. to determine the conversion of the Jak1 floxed allele into a recombined knockout or null allele of Jak1. Abbreviations: R, EcoR1; Mcl-tk, thymidine kinase negative selection cassette
A germline or somatic deletion of the second coding exon of Jak1 can be achieved through spatially and temporally controlled expression of Cre recombinase (Figure 1d). The conditional knockout approach was designed to specifically delete exon 2 since its Cre-mediated excision causes a frameshift of almost the entire coding sequence of Jak1 and therefore a complete absence of the protein. The Cre-mediated and allele-specific recombination of Jak1 was verified by PCR using the forward primer 2411 in combination with the reverse primer 2373, which binds within the PGK-neo cassette. As shown in Figure 1e, expression of Cre resulted in the conversion of the Jak1 floxed allele into a recombined knockout or null allele of Jak1. To validate that the excision of exon 2 represents a true knockout of JAK1, we generated embryos and newborn mice that lacked this Janus kinase in all organs and cell types (Jak1−/−). We anticipated that these mice would be phenotypically similar to conventional knockout animals that lacked the promoter and first coding exon (Rodig et al., 1998). As reported previously, a conversion of a conditional knockout allele into a null mutation can be accomplished with 100% efficiency by transmitting the floxed allele through the female germline of MMTV-Cre (line A) mice (Krempler et al., 2004). We generated MMTV-Cre Jak1fl/wt double mutant females that were subsequently set up with wildtype males (Jak1wt/wt) to produce offspring that were heterozygous for the knockout allele (Jak1wt/−) and to segregate out the MMTV-Cre transgene (Figure 2a). As published earlier by Rodig et al. (1998), deficiency in only one allele of Jak1 did not cause any noticeable phenotypic abnormalities, and both male and female Jak1wt/− mice were fertile. Hence, the deletion of the second coding exon did not seem to result in the creation of a dominant-negative or hyper/hypomorphic allele. It was reported that JAK1 conventional knockout mice die around birth (Rodig et al., 1998), and we therefore first collected embryos from intercrosses of mice heterozygous for the Jak1 null allele at day E12.5 through E17.5 to validate that the lack of JAK1 does not cause early lethality. Using multiple PCR assays (Figure 2b), we genotyped the progeny from six pregnant females (i.e., 43 embryos) and identified 9 knockouts fetuses (21%) among 22 heterozygous knockouts (51%) and 12 wildtype embryos (28%). JAK1 knockout embryos were phenotypically indistinguishable from their wildtype littermates at developmental stage E12.5 (not shown), and these knockout embryos and littermate controls expressing wildtype JAK1 were used to generate primary and immortalized fibroblast cultures (MEFs) for biochemical analysis as described in the Materials and Methods section.
FIGURE 2.
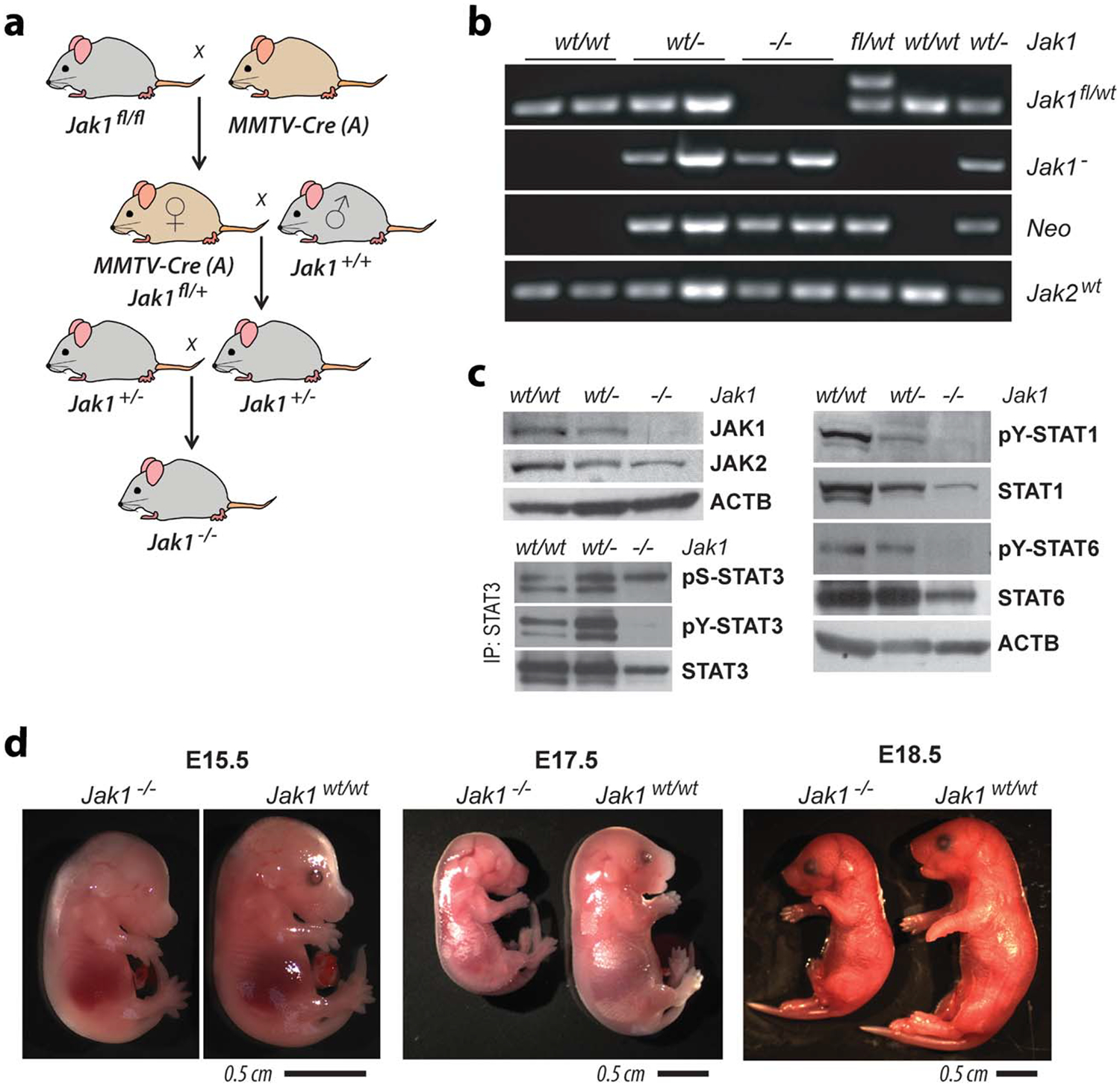
Conditional deletion of Jak1 in the female germline and consequential effects of JAK1 deficiency on STAT activation and embryonic development. (a) Breeding strategy to convert the floxed allele of Jak1 (Jak1fl) into a null mutation (Jak1−). MMTV-Cre (line A) mice express Cre recombinase in developing oocytes. Thus, heterozygous Jak1fl/wt MMTV-Cre females transmit a null allele to their offspring whether the Cre transgene is segregated out or not. (b) PCR assays using the primer sets illustrated in panel Figure 1a,c to validate the germline deletion of the Jak1 floxed allele and the presence of the recombined knockout/null allele of Jak1. An internal primer set for neo served as a positive control for the presence of the mutant alleles, and the amplification of a PCR fragment of the Jak2 gene was used to control for the integrity of the DNA samples. (c) Immunoblot analyses of the expression of JAKs 1 and 2 as well as the tyrosine phosphorylation of STATs 1, 3, and 6 in immortalized mouse embryonic fibroblasts (MEFs) of heterozygous and homozygous JAK1 knockouts (Jak1wt/−, Jak1−/−) as well as wildtype controls (Jak1wt/wt). Beta-actin (ACTB) was used as a loading control. Immunoprecipitation (IP)/western blot analyses was used to determine the phosphorylation on tyrosine 705 and serine 727 on STAT3. (d) Comparison of JAK1 deficient embryos and littermate wildtype controls during the final stages of prenatal development. A subset of JAK1 knockout pups were alive at birth but showed symptoms of apnea. None of the pups were able to survive
Using immunoblot analysis on immortalized MEFs, we confirmed that homozygosity of the recombined exon 2 knockout allele resulted in the complete absence of the JAK1 protein Figure 2c. We also found that lack of this Janus kinase was sufficient to ablate the tyrosine phosphorylation and activation of the downstream STATs 1, 3, and 6 when MEFs were maintained in serum-rich media. Similar to the conditional knockout of JAK1 in mammary epithelial cells (Sakamoto et al., 2016), loss of JAK1 had no effect on the activation of STAT5a or STAT5b in immortalized MEFs (not shown). Because of alternative splicing, fibroblasts express two isoforms of STAT3, STAT3a and STAT3b, that differ in size and the composition of the C-terminal residues within the transactivation domain. While both protein variants possess the major tyrosine phosphorylation site at Y705, only the longer and predominantly expressed STAT3a isoform also contains the serine phosphorylation site at S727, which is suggested to enhance the transcriptional activity of tyrosine phosphorylated STAT3. Interestingly, the immunoprecipitation and western blot analysis of STAT3 revealed that deficiency in JAK1 specifically ablated the tyrosine phosphorylation of STAT3a and STAT3b without affecting its phosphorylation on the serine residue 737 of STAT3a (Figure 2c). This finding is identical to JAK1-mediated phosphorylation events on the STAT1 protein (Sakamoto et al., 2016) and it supports the concept proposed by Decker and Kovarik (2000) that tyrosine and serine phosphorylation on STAT proteins are not interdependent. The tyrosine phosphorylation is essential for the transcriptional activity of STATs, and since we have shown that STAT3 or STAT1 can be phosphorylated on serine 737 despite lacking a phosphorylation on their tyrosine residues, it is evident that pS737-STAT1/3 is not a surrogate marker for the cytokine-mediated activation of these STATs. The collective findings shown here also suggest that mutations and epigenetic changes that commonly occur during the immortalization process of fibroblasts (Carnero et al., 2000; Harvey and Levine, 1991) did not have any significant impact on the essential role of JAK1 in the activation of STAT1, STAT3, and STAT6.
To assess the biological significance of JAK1 during the later stages of embryogenesis, we collected embryos from heterozygous knockout breedings at E15.5 and E17.5 as well as prior to birth at E18.5. JAK1 knockout embryos were viable but significantly smaller starting at E15.5 and throughout the final stages of prenatal development (Figure 2d). Similar to findings reported by Rodig et al. (1998), we observed that a subset of JAK1 knockout pups were alive at birth. Although it was proposed that JAK1 knockout mice may have died due to neuronal defects along with a failure to nurse, we noted that mutant mice were able to move their limbs, but they were gasping for air and intermittently exhibited longer pauses in breathing (apnea). These mice died within 30 min to 2 h after birth. The collective observations demonstrated that the Cre-mediated deletion of exon 2 in the newly generated JAK1 conditional knockout model results in a true null mutation that resembles the main phenotypic abnormalities that were reported in the conventional knockout of JAK1, and the excision of this exon from both alleles leads to the complete absence of the JAK1 protein from Cre-expressing cells. This new conditional knockout mouse model will be valuable to study the biological and molecular functions of cytokine signaling through JAK1 during normal development and tissue homeostasis as well as disease initiation and progression.
2 |. MATERIALS AND METHODS
2.1 |. Construction of the Jak1 targeting vector
We have published recently a detailed manual for the generation of conditional knockout alleles using λ Red phage-based homologous recombination in Escherichia coli (Sakamoto et al., 2014). The BAC clone from the bMQ Mouse BAC Library (Adams et al., 2005) encompassing the Jak1 gene was purchased from Source BioScience Life-Science. To capture a 14-kb fragment of Jak1 containing the first four coding exons, we first inserted short sequences that corresponded to the 5′ and 3′ ends of Jak1 into the NotI and SpeI sites of the PL253 retrieving vector (Frederick National Laboratory for Cancer Research, FNLCR, NCI) (Liu et al., 2003). These 5′and 3′ homologous recombination sequences of 293 and 343 bps in size were generated by PCR using the following two primer pairs: (1) primers 2349/2350, 5′-ATA AGC GGC CGC GCT GGA ACT TTG CCT GTT TGG AAG TGG-3′, 5′- GCC AAG CTT GAT GAG CAA CCC ATA GTG TCT AGG TG-3′ and (2) primers 2351/2352, 5′-GCC AAG CTT GCT GAG GTA CAT TGT GAC AGC ACA C-3′, 5′-GTC ACT AGT CTC CAG TGA ACT GGC ATC AAG GAG TG-3′. The Jak1 gene fragment that was captured using λ phage-based recombination in SW102 cells (Warming et al., 2005) was confirmed by sequencing. To insert two loxP sites around the second coding exon, we first generated derivative vectors of PL452 and PL451 (Liu et al., 2003) that contained loxP sites that were surrounded by ~300 bps of Jak1 homologous recombination arms corresponding to introns 1 and 2. The recombination arms for the first loxP site in PL452 were cloned by PCR using primers 2401 and 2402 (5′- ATA GTC GAC TGA TGG AAG TGC TAG TGT GGG CAG C -3′ and 5′- GTC GGA TCC GCC CAA GTG TTA TCT CAG TAA CAC AG -3′) as well as primers 2403 and 2404 (5′- GTC GAA TTC AGG CCT ACT CAA GTG AAG CTG AGG CTG GCA GGC A -3′ and 5′- ATA AGC GGC CGC TTG ATG ACT AGG GAC AGG TCC CTG TC -3′). The arms for the second loxP site in PL451 were cloned by PCR suing primers 2357 and 2358 (5′- ATA GTC GAC GGA ATA CCA AGG AAT GAC CCT TGC C -3′and 5′- ATA GGA TCC TAA GAG GAG ACC GGG CCA AAC TCA AGG-3′) as well as primers 2359 and 2360 (5′- GTC GAA TTC CTT GGT CCC TTC TGC CAC TTT GAA GTG -3′ and 5′- ATA AGC GGC CGC ATG CAC CTC CAT GCT GGA GGA TGA C-3′). Both loxP containing fragments were released as BamHI-EcoRI fragments from the derivatives of PL452 and PL451 and transferred into PL253 that carried the Jak1 genomic fragment. The homologous recombinations for the cloning of the targeting vector were performed in the bacterial strains SW102 and SW106 (FNLCK) as described in our step-by-step manual (Sakamoto et al., 2014). Prior to the insertion of the second loxP site, the loxP-flanked neomycin resistance cassette near the first loxP site was removed by treating SW106 cells with arabinose. SW106 cells contain an arabinose-induced Cre recombinase gene that facilitated the deletion of the neomycin resistance gene from the intermediate cloning vector prior to the insertion of the second 3′ loxP site. The final targeting construct contained a PGK-neomycin selection marker downstream of the second loxP site within intron 2 of Jak1. The PGK-neomycin cassette is surrounded by frt sites and, if necessary, could have been deleted with the help of FLP recombinase at the ES cell stage or in mice. The final targeting vector was linearized using NotI, phenolchlorophorm extracted, and electroporated into E14 and RW-4 ES cells. The selection of targeted ES cells with G418 and expansion of the cells were performed at the UNMC Mouse Genome Engineering Core Facility.
2.2 |. Southern blot analysis
Genomic DNA from ES cell clones was purified using a phenol-based protocol on an automated DNA extractor (Autogen2000). Fifteen μg of DNA was digested with EcoRI at 37°C overnight and separated on a 0.7% agarose gel. The DNA was denatured and transferred onto a nylon membrane (Genescreen Plus, Perkin Elmer), and hybridized with a 32P-labeled external probe. The 5′ external probe was 587 bp in size and was generated by PCR using primers 2361 and 2370 (5′ -CCA GGT TCA TAC ATC TCA AAA CC-3′ and 5′ -GTC ACA GTA GTC CTT TGT CAG G-3 ′). Membranes were washed in 0.1× SSC buffer containing SDS and exposed for 16 h to a KODAK XOMAT-AR film. The EcoR1 Southern analysis yielded two distinct bands of 7.4 kb for the wildtype allele and 5 kb in size for the targeted floxed allele (Figure 1a).
2.3 |. Generation of JAK1 mutant mice and genotyping protocols
Two correctly targeted ES cell clones (A1 and A6) were expanded and used for the production of chimeras. The injection of ES cells into C57Bl/6 blastocytes was carried out by the UNMC Mouse Genome Engineering Core Facility. These chimeric males were crossed with C57BL/6 wildtype females for the transmission of the targeted Jak1 floxed (Jak1fl) allele. The germline transmission of the floxed Jak1 allele was verified by Southern blot as described above. The floxed Jak1 allele is registered at the Mouse Genome Informatics (MGI) database as Jak1tm1Kuw with the identifier number MGI:5688302. A PCR assay using primers 2411 (5′- GAG ACA GGA TAC CTG GTG GCT TGG -3′) and 2412 (5′- GTA GCA GTC CTG GAC ATT GAG TCC -3′) was utilized to genotype mice that carry one or two Jak1 floxed alleles. This primer pair amplifies a PCR fragment of about 250 bp of the wildtype Jak1 and a PCR product of ~350 bp of the floxed allele. The presence of the PGK-neomycin cassette was validated by PCR using primers 806 and 807 (5′- AGA GGC TAT TCG GCT ATG ACT G -3′ and 5′-TTC GTC CAG ATC ATC CTG ATC -3′) that yielded a fragment of ~350 bp in size. The recombined Jak1 null allele (Jak1−) was detected by PCR using primer 2411 in combination with primer 2373 (5′- AGG TGC CAC TCC CAC TGT CCT TTC C -3′). The location of both primers is illustrated in Figure 1c, and the resulting PCR amplicon was about 400 bp in size. The conversion of the Jak1 floxed allele into a null allele was achieved by transmitting the floxed allele through the female germline of MMTV-Cre (line A) transgenics (Wagner et al., 1997, 2001). A PCR protocol for genotyping MMTV-Cre [TgN(MMTV-cre) 1Mam] transgenic mice was described previously (Wagner et al., 1997), and the primers for the amplification of the wildtype Jak2 locus can be found elsewhere (Krempler et al., 2004). The recombined Jak1 null allele is registered at the MGI database as Jak1tm1.1Kuw with the identifier number MGI:5688350, and these mice will be provided to the scientific community through the repository or by sending them directly to investigators under valid Uniform Biological Material Transfer Agreements (UBMTAs). All animals used in this study were treated humanely and in accordance with institutional guidelines and federal regulations.
2.4 |. Cell culture
Mouse embryonic fibroblasts (MEFs) were derived from embryos at E12.5 and maintained in culture in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 0.1 mM nonessential amino acids, 10 μg ml−1 gentamycin, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (Krempler et al., 2002). Wildtype as well as heterozygous and homozygous JAK1 knockout MEFs were immortalized by serial passaging (3T3 protocol). Cells were harvested, frozen on dry ice, and stored at −80°C before using them in protein immunoblotting experiments.
2.5 |. Immunoprecipitation and western blot analysis
Detailed protocols for the preparation of whole cell extracts from cultured cells and tissue homogenates as well as immunoprecipitation western blot analysis were described previously (Krempler et al., 2002; Sakamoto et al., 2007). The following antibodies were used for immunoblotting: α-β-actin (I-19), α-STAT1 (sc-592), and α-STAT6 (sc-981) from Santa Cruz Biotechnology; α-STAT3 (9139S), α-pY-STAT3(Y705) (9145S), and α-pS-STAT3(S727) (9136S) from Cell Signaling, Inc., α-pY-STAT6(Y641) (ab54461) from Abcam, α-pY-STAT1(Y701) (TA309955) from Origene. The α-JAK1 antibody was kindly provided by Dr. Hallgeir Rui (Medical College of Wisconsin).
ACKNOWLEDGMENTS
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors are grateful to the members of the UNMC Mouse Genome Engineering Core facility, in particular Dr. C.B. Gurumurthy, for assistance in the generation of JAK1 knockout mice.
Funding Information
Contract grant sponsor: Public Health Service; Contract grant number: CA117930; Contract grant sponsor: Nebraska Cancer and Smoking Disease Research Program; Contract grant numbers: NE DHHS LB506 2015-47, 2016-54; Contract grant sponsor: Public Health Service; Contract grant number: CA009476.
LITERATURE CITED
- Aaronson DS, & Horvath CM (2002). A road map for those who don’t know JAK-STAT. Science, 296, 1653–1655. [DOI] [PubMed] [Google Scholar]
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, … Bradley A (2005). A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics, 86, 753–758. [DOI] [PubMed] [Google Scholar]
- Avalle L, Pensa S, Regis G, Novelli F, & Poli V (2012). STAT1 and STAT3 in tumorigenesis: A matter of balance. Jakstat, 1, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, & Darnell JE Jr. (1999). Stat3 as an oncogene. Cell, 98, 295–303. [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, & Beach DH (2000). p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nature Cell Biology, 2, 148–155. [DOI] [PubMed] [Google Scholar]
- Dalal I, Arpaia E, Dadi H, Kulkarni S, Squire J, & Roifman CM (1998). Cloning and characterization of the human homolog of mouse Jak2. Blood, 91, 844–851. [PubMed] [Google Scholar]
- Decker T, & Kovarik P (2000). Serine phosphorylation of STATs. Oncogene, 19, 2628–2637. [DOI] [PubMed] [Google Scholar]
- Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, & Krolewski JJ (1990). tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene, 5, 1329–1336. [PubMed] [Google Scholar]
- Harpur AG, Andres AC, Ziemiecki A, Aston RR, & Wilks AF (1992). JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene, 7, 1347–1353. [PubMed] [Google Scholar]
- Harvey DM, & Levine AJ (1991). p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes and Developement, 5, 2375–2385. [DOI] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, & Schindler CW (2002). Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene, 285, 1–24. [DOI] [PubMed] [Google Scholar]
- Krempler A, Henry MD, Triplett AA, & Wagner KU (2002). Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death. Journal of Biological Chemistry, 277, 43216–43223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, & Wagner KU (2004). Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis, 40, 52–57. [DOI] [PubMed] [Google Scholar]
- Levy DE, & Darnell JE Jr. (2002). Stats: Transcriptional control and biological impact. Nature Reviews in Molecular Cell Biology, 3, 651–662. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, & Copeland NG (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Research, 13, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, … Schreiber RD (1998). Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokineinduced biologic responses. Cell, 93, 373–383. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Creamer BA, Triplett AA, & Wagner KU (2007). The Janus kinase 2 is required for expression and nuclear accumulation of cyclin D1 in proliferating mammary epithelial cells. Molecular Endocrinology, 21, 1877–1892. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Gurumurthy CB, & Wagner KU (2014). Generation of conditional knockout mice. Methods in Molecular Biology, 1194, 21–35. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Lin WC, Triplett AA, & Wagner KU (2009). Targeting janus kinase 2 in Her2/neu-expressing mammary cancer: Implications for cancer prevention and therapy. Cancer Research, 69, 6642–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Triplett AA, Schuler LA, & Wagner KU (2010). Janus kinase 2 is required for the initiation but not maintenance of prolactin-induced mammary cancer. Oncogene, 29, 5359–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Wehde BL, Yoo KH, Kim T, Rajbhandari N, Shin HY, … Wagner KU (2016). Janus kinase 1 is essential for inflammatory cytokine signaling and mammary gland remodeling. Molecular Cell Biology, 36, 1673–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, & Plumlee C (2008). Inteferons pen the JAK-STAT pathway. Seminars in Cell Developemental Biology, 19, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JW, & Wagner KU (2012). Activation of Janus kinases during tumorigenesis In: Decker T, Muller M, editors. Chapter 15: JakStat signaling: From basics to disease (pp. 259–288). Wien: Springer-Verlag Wien. [Google Scholar]
- Takahashi T, & Shirasawa T (1994). Molecular cloning of rat JAK3, a novel member of the JAK family of protein tyrosine kinases. FEBS Letters, 342, 124–128. [DOI] [PubMed] [Google Scholar]
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, & Hennighausen L (2001). Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Research, 10, 545–553. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, … Hennighausen L (1997). Cre-mediated gene deletion in the mammary gland. Nucleic Acids Research, 25, 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, & Copeland NG (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Research, 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, & Ziemiecki A (1991). Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Molecular Cell Biology, 11, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chung D, & Cepko CL (1993). Molecular cloning of the murine JAK1 protein tyrosine kinase and its expression in the mouse central nervous system. Journal of Neuroscience, 13, 3006–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, & Jove R (2014). Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nature Reviews Cancer, 14, 736–746. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turkson J, Carter-Su C, Smithgall T, Levitzki A, Kraker A, … Jove R (2000). Activation of Stat3 in v-Src-transformed fibroblasts requires cooperation of Jak1 kinase activity. Journal of Biological Chemistry, 275, 24935–24944. [DOI] [PubMed] [Google Scholar]


