SUMMARY
Rickettsia are obligate intracellular bacteria that evade antimicrobial autophagy in the host cell cytosol by unknown mechanisms. Other cytosolic pathogens block different steps of autophagy targeting, including the initial step of polyubiquitin coat formation. One mechanism of evasion is to mobilize actin to the bacterial surface. Here, we show that actin mobilization is insufficient to block autophagy recognition of the pathogen Rickettsia parkeri. Instead, R. parkeri employs outer membrane protein B (OmpB) to block ubiquitylation of bacterial surface proteins, including OmpA, and subsequent recognition by autophagy receptors. OmpB is also required for the formation of a capsule-like layer. Although OmpB is dispensable for bacterial growth in endothelial cells, it is essential for R. parkeri to block autophagy in macrophages and to colonize mice because of its ability to promote autophagy evasion in immune cells. Our results indicate that OmpB acts as a protective shield to obstruct autophagy recognition, revealing a distinctive bacterial mechanism to evade antimicrobial autophagy.
Keywords: Rickettsia, OmpB, OmpA, polyubiquitylation, autophagic recognition, antimicrobial autophagy, intracellular pathogens, innate immunity
The rickettsiae are a diverse and clinically important genus of Gram-negative obligate intracellular bacteria that primarily reside in arthropod vectors such as ticks. During their life cycle, Rickettsia are often transmitted from arthropods to mammals, which can function as amplifying hosts that transfer bacteria to naive arthropods1–3. Some Rickettsia species can be transmitted to humans and cause diseases, including spotted fever and typhus4,5. During infection of mammals, Rickettsia species primarily target vascular endothelial cells, although they can reside in a wide variety of cell types, including dendritic cells6 and macrophages7,8.
The Rickettsia intracellular life cycle begins when bacteria adhere to and invade host cells. They subsequently escape the primary vacuole into the cytosol, where they replicate and must evade antimicrobial autophagy, a process that removes intracellular pathogens similarly to damaged organelles such as mitochondria9,10. Antimicrobial autophagy often involves recognition of bacteria by the host ubiquitin machinery, which covalently attaches ubiquitin to proteins, ultimately forming a polyubiquitin coat on cytosolic bacteria11–13. This promotes binding to autophagy receptors such as p6214,15 and NDP5215,16. To initiate antimicrobial autophagy, double-membrane structures are nucleated by proteins including Beclin 117,18 and then expanded via the activity of LC3. Lipidation and recruitment of LC3 to double-membrane structures by the ATG5-ATG12-ATG16L1 complex19,20 enables the capture of bacteria labeled with autophagy receptors, leading to autophagosome maturation and bacterial degradation after fusion with lysosomes21.
Several facultative intracellular bacterial pathogens that grow in the host cell cytosol have evolved mechanisms to evade autophagic recognition9,21,22,23,24. For example, Listeria monocytogenes uses its surface protein ActA to avoid polyubiquitin coat formation by masking the bacterial surface with components of the host actin machinery22 and inducing actin polymerization and intracellular motility11,25. It remains unclear whether or how other pathogens avoid polyubiquitin coat formation, and what microbial proteins are ubiquitylated by the host. Rickettsia are highly adapted to the intracellular environment and thus have likely also evolved mechanisms to evade autophagic recognition26 (although it was recently demonstrated that R. conorii benefits from autophagy in mouse macrophages27). However, the bacterial factors that protect Rickettsia from polyubiquitin coat formation are unknown.
To better understand the role of bacterial proteins in shielding against autophagic recognition, we interrogated the function of R. parkeri surface proteins OmpA28,29, OmpB28,30, the actin polymerizing protein Sca231–33, and the 17-kDa antigen34. We found that Sca2-mediated actin polymerization is not protective. Instead, R. parkeri uses an unanticipated strategy that is dependent on OmpB, an abundant and conserved rickettsial protein35,36,37,28,30. The importance of OmpB is cell type-specific, as it is essential for growth and avoidance of autophagy in macrophages, but not in endothelial cells. Strikingly, OmpB is also crucial for R. parkeri colonization of mice by allowing bacteria to evade autophagy. Our results suggest that cytosolic pathogens have evolved distinctive mechanisms to shield themselves from polyubiquitin coat formation to avoid destruction via autophagy.
RESULTS
OmpB is required for avoidance of polyubiquitylation
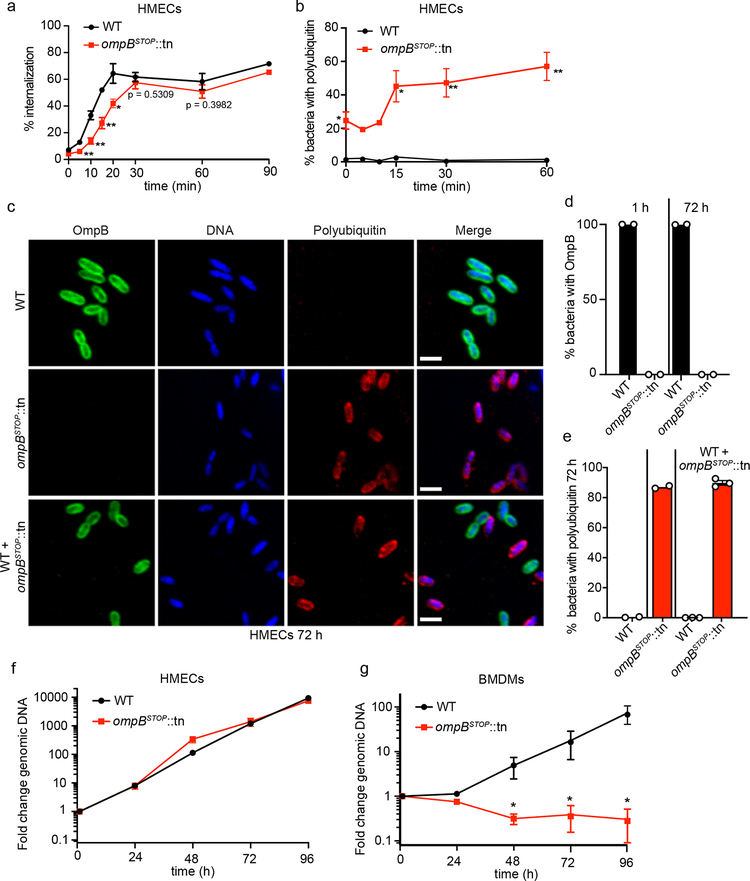
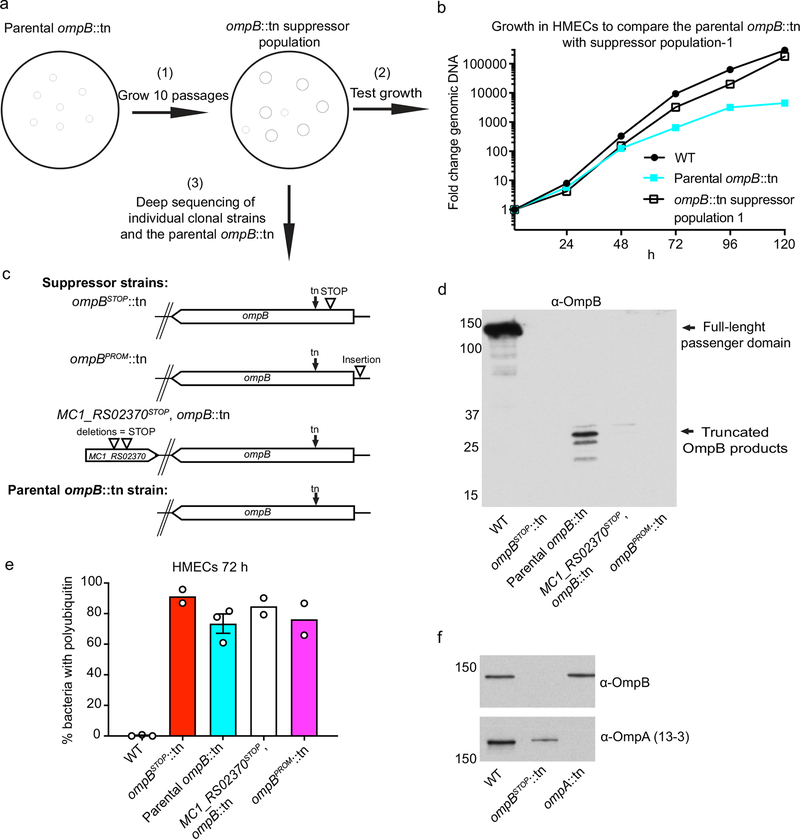
To identify surface proteins that may protect Rickettsia from autophagic recognition, we assessed whether R. parkeri transposon mutants lacking known surface proteins were polyubiquitylated. In particular, we assessed strains with transposon (tn) mutations in ompA, ompB, sca2, and hrtA (encoding the 17-kDa antigen)33,38. The ompB mutant (ompBSTOP::tn), obtained via serial-passaging the parental ompB::tn strain, contains a suppressor mutation in the form of a stop codon upstream of the transposon that prevents expression of a truncated OmpB product, which appears to be toxic (Fig. 1b; Extended Data Fig. 1a–e). The mutants were analyzed for co-localization with polyubiquitin at 72 hours post infection (hpi) in human dermal microvascular endothelial cells (HMECs) (Fig. 1a, c; Extended Data Fig. 1e). Wild type (WT), ompA::tn, and hrtA::tn bacteria were polyubiquitin-negative. Notably, the sca2::tn mutant, which is defective in actin polymerization at this time point33, was also polyubiquitin-negative. However, more than 75% of the ompBSTOP::tn (and ompBPROM::tn, an independent mutant; Extended Data Fig. 1a–d) bacteria co-localized with polyubiquitin. To confirm that polyubiquitylation of ompBSTOP::tn bacteria was due to mutations in ompB and not to polar effects on surrounding genes, we tested R. parkeri mutants with insertions in the genes directly upstream or downstream of ompB (Fig. 1b). We also tested a mutant with an insertion in the gene rickCE encoding for a rickettsial deubiquitylase39. Mutants with insertions in the genes directly flanking ompB, or in rickCE, were all polyubiquitin-negative (Fig. 1a, c). Thus, polyubiquitylation of ompBSTOP::tn bacteria is caused by mutations in ompB. Furthermore, purified ompBSTOP::tn bacteria were heavily polyubiquitylated while WT bacteria showed little polyubiquitylation (Fig. 1d), suggesting that bacterial proteins or tightly associated host components are ubiquitylated. These data demonstrate that R. parkeri requires OmpB, but not Sca2-mediated actin mobilization, for avoidance of polyubiquitylation.
Fig. 1. In HMECs, R. parkeri surface protein OmpB is required for avoidance of polyubiquitylation.
(a) Immunofluorescence micrographs showing the indicated strains of R. parkeri (green; anti-Rickettsia I7205 antibody), cellular and bacterial genomic DNA (blue; Hoechst), polyubiquitin (red; FK1 antibody) and a merged image at 72 hpi. Note that the ompA::tn bacteria are not detected with the anti-Rickettsia antibody but are visualized with Hoechst stain. Scale bars, 5 μm. (b) Diagram of ompB and surrounding genes in the indicated mutants, showing the location of the transposon insertions (tn) in: ompB (at base pair (bp) position 1045462 in the R. parkeri genome; accession number NC_017044.1); MC1_RS05535, hypothetical protein (at bp position 1040923); MC1_RS05545, pseudogene (at bp position 1047186). Also indicated is the premature stop codon (STOP) suppressor mutation in ompBSTOP::tn (a 23-bp deletion at position 1045835–1045857). Note that the genome of the ompBSTOP::tn mutant was sequenced and the only mutations were in ompB (see Extended Data Fig. 1 and the Data availability section). (c) Quantification of the percentage of bacteria that showed rim-like surface localization of polyubiquitin at 72 hpi. Data are mean ± standard error of the mean (SEM) (WT, n = 4; ompBSTOP::tn, n = 4; MC1_RS05535::tn, n = 3; MC1_RS05545::tn, n = 3; MC1_RS05545::tn, n = 3; ompA::tn, n =3; sca2::tn, n = 3; hrtA::tn, n = 2; rickCE::tn, n = 2; ≥110 bacteria were counted in each infection; statistical comparisons between the ompBSTOP::tn and WT, MC1_RS05535::tn, MC1_RS05545::tn, ompA::tn and sca2::tn were determined by the unpaired Student’s t-test (two-sided); ***, p < 0.001). (d) Western blot of gradient-purified WT and ompBSTOP::tn bacteria probed for host polyubiquitin and for the R. parkeri RickA protein as a loading control (n = 2).
To confirm the involvement of the host ubiquitin machinery in polyubiquitylation of ompBSTOP::tn bacteria, we treated infected cells with PYR-41, an inhibitor of host ubiquitin activating enzyme E1. PYR-41 treatment caused a 9-fold reduction in the number of polyubiquitin-positive ompBSTOP::tn bacteria compared with a vehicle control (Extended Data Fig. 2a, b). We next examined the nature of ubiquitin chains in the coat by identifying which of the seven lysine or the N-terminal methionine residue(s) are linked, using ubiquitin linkage-specific antibodies. We observed that ~90% of ompBSTOP::tn bacteria were positive for M1, K63, or K48-linked ubiquitin chains, whereas WT bacteria were polyubiquitin-negative (Extended Data Fig. 2c). This demonstrates a role for the host ubiquitin machinery in forming the polyubiquitin coat and shows that the coat may consist of heterotypic ubiquitin conjugates, as observed for mitochondria during mitophagy40.
In HMECs, OmpB blocks polyubiquitylation following host cell entry but is dispensable for bacterial growth
To assess the timing of bacterial polyubiquitylation, we first established internalization rates of WT and ompBSTOP::tn bacteria, and observed that WT bacteria reached maximum internalization (internalizationmax) at 20 min post infection (mpi), whereas the ompBSTOP::tn reached internalizationmax at 30 mpi (Fig. 2a). The internalization of ompA::tn, sca2::tn, or hrtA::tn mutants, in contrast, were not impaired (Extended Data Fig. 3a). Thus, OmpB is important for optimal internalization into HMECs. Nevertheless, internalization was rapid enough to determine the kinetics of polyubiquitin coat formation. Some purified ompBSTOP::tn bacteria in the initial inoculum had a polyubiquitin coat, and there was increased co-localization with polyubiquitin over time, corresponding with internalization (Fig. 2b). We also observed that the majority of intracellular ompBSTOP::tn bacteria were polyubiquitin-positive whereas the majority of extracellular bacteria were polyubiquitin-negative at 60 mpi (Extended Data Fig. 4e, f). Collectively, these data suggest that the host ubiquitin machinery targets ompBSTOP::tn bacteria after they enter HMECs.
Fig. 2. OmpB acts locally on R. parkeri to promote polyubiquitin avoidance, and OmpB is required for bacterial growth in BMDMs but not in HMECs.
(a) Quantification of the percentage of invasion of HMECs from 0–90 mpi, visualized by differential staining of extracellular versus total R. parkeri. Data are mean ± SEM (0, 20, 30, 60 min, n = 3; 5, 10, 15 min, n = 4; 90 min, n = 2; ≥85 bacteria were counted for each strain, infection and time-point; statistical comparisons between ompBSTOP::tn and WT for each time point were made by the unpaired Student’s t-test (two-sided); *, p < 0.05; **, p < 0.01). (b) Quantification of the percentage of WT and ompBSTOP::tn mutant bacteria that co-localize with polyubiquitin from 0–60 mpi. Data are mean ± SEM (0, 15, 30, 60 min, n = 3; 5, 10 min, n = 2; statistical comparisons between ompBSTOP::tn and WT were made by the unpaired Student’s t-test (two-sided); *, p < 0.05; **, p < 0.01; ≥85 bacteria were counted for each strain, infection and time point. (c) Immunofluorescence micrographs of HMECs infected with WT (upper panel), ompBSTOP::tn (middle panel), or with both WT and ompBSTOP::tn (lower panel), at 72 hpi stained for OmpB (green, anti-OmpB antibody), polyubiquitin (red; FK1 antibody), and DNA (blue; Hoechst). Scale bars, 2 μm. (d) Quantification of the mean percentage of WT and ompBSTOP::tn mutant bacteria in HMECs that have OmpB homogeneously distributed at the surface of each bacterium at 1 and 72 hpi (n = 2; for 1 hpi, ≥109 bacteria were counted per strain; for 72 hpi, ≥724 bacteria were counted per strain). (e) Quantification of the percentage of WT and ompBSTOP::tn mutant bacteria positive for polyubiquitin in HMECs infected with WT, ompBSTOP::tn, or co-infected with both. In the mixed infected, WT and ompBSTOP::tn bacteria were distinguished using the anti-OmpB antibody. Data are mean ± SEM (singly infected cells, n = 2; mixed infected cells, n = 3; ≥880 bacteria were counted for each infection). (f) Growth curves of WT and ompBSTOP::tn in HMECs from 0–96 hpi as measured by genomic equivalents using qPCR. Data are mean ± SEM (n = 3; means were not significantly different by an unpaired Student’s t-test (two-sided); 24 h, p > 0.99; 48 h, p = 0.10; 72 h, p > 0.99; 96 h, p = 0.40). (g) Growth curves of WT and ompBSTOP::tn in BMDMs from 0–96 hpi as in (f). Data are mean ± SEM (n = 4; statistical comparisons with WT were by the Mann-Whitney rank-sum test (two-sided); *, p < 0.05).
To ascertain whether OmpB protects against polyubiquitylation by acting locally on the bacterial surface or globally by inhibiting the host ubiquitin machinery, HMECs were co-infected with WT and ompBSTOP::tn. We confirmed co-infection by staining DNA to label all bacteria and OmpB to distinguish between WT and ompBSTOP::tn bacteria by immunofluorescence microscopy (Fig. 2c, d). We found that the ompBSTOP::tn bacteria were polyubiquitylated at similar frequencies in singly-infected and co-infected cells (Fig. 2c, e). Furthermore, WT bacteria in co-infected cells remained polyubiquitin-negative (Fig. 2c, e). These data suggest that OmpB acts locally on the bacterial surface and not globally to suppress ubiquitylation.
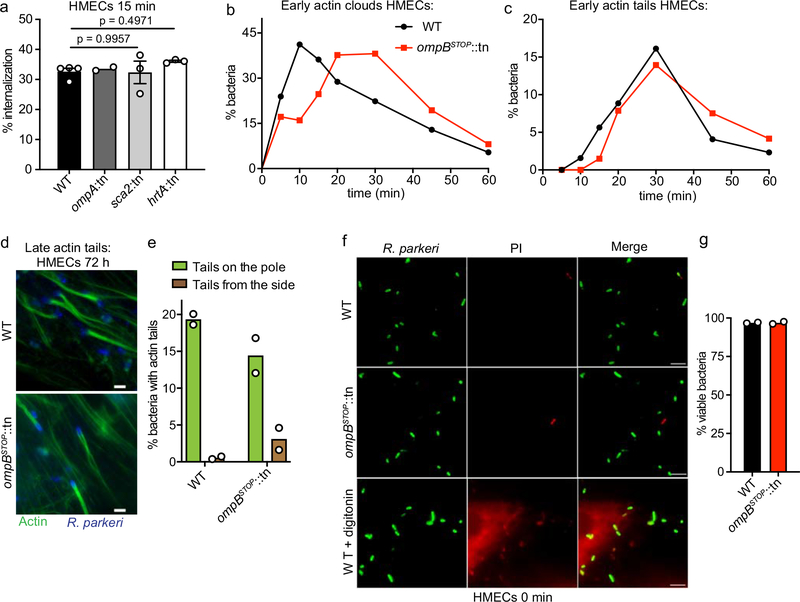
We next assessed if OmpB is important for R. parkeri growth in HMECs. We observed that the growth of ompBSTOP::tn bacteria was indistinguishable from WT over 96 h (Fig. 2f). Consistent, ompBSTOP::tn and WT bacteria had similar maximum frequencies of actin recruitment, actin tail formation, and viability (Extended Data Fig. 3b–g). Together, these data demonstrate that OmpB is essential for avoidance of polyubiquitylation but dispensable for R. parkeri growth and actin mobilization in endothelial cells.
In macrophages, OmpB blocks polyubiquitylation and is essential for bacterial growth
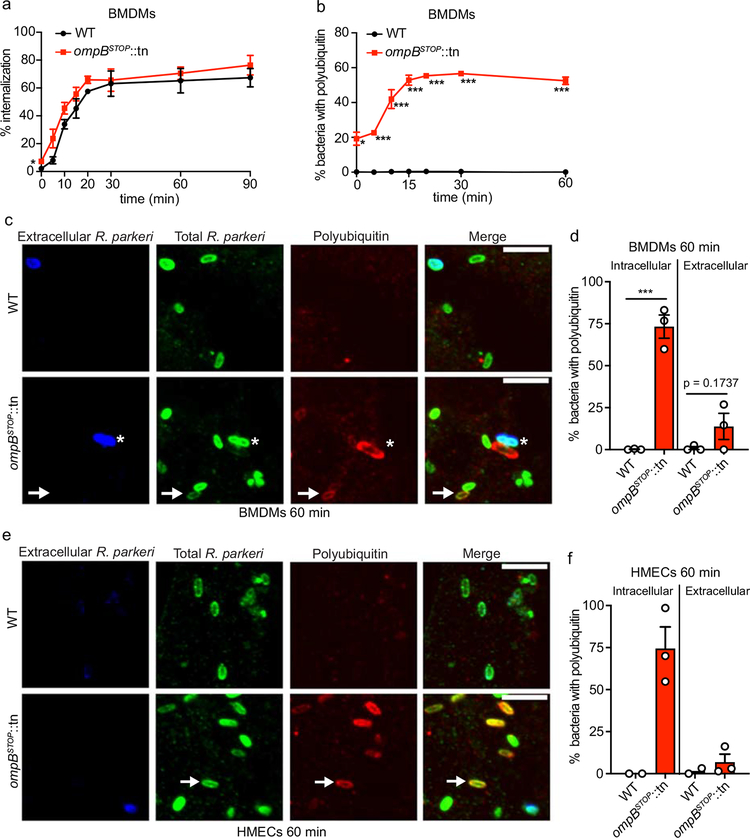
Because SFG Rickettsia also grow in macrophages in vitro8 and in vivo7, we next assessed internalization and polyubiquitylation kinetics of the ompBSTOP::tn mutant in mouse bone marrow-derived macrophages (BMDMs). In contrast with their slower internalization into HMECs, ompBSTOP::tn reached internalizationmax into BMDMs at rates similar to WT bacteria (Extended Data Fig. 4a), suggesting that OmpB does not impact the internalization of Rickettsia into macrophages. The ompBSTOP::tn mutant also showed increased co-localization with polyubiquitin over time, corresponding with internalization, and polyubiquitylated bacteria were primarily intracellular at 60 mpi (Extended Data Fig. 4b–d). These data demonstrate that the host ubiquitin machinery targets ompBSTOP::tn bacteria after their phagocytosis by BMDMs.
We also measured bacterial growth over 96 h. In contrast with their robust growth in HMECs, the ompBSTOP::tn bacteria were unable to grow in BMDMs, while WT increased in number ~50-fold (Fig. 2g). Failure to grow was also observed using immunofluorescence microscopy, which revealed that ompBSTOP::tn bacteria were rare whereas WT bacteria were abundant in infected cells (Extended Data Fig. 5a–d). In addition, failure of the ompBSTOP::tn to grow in BMDMs was not due to host cell death (Extended Data Fig. 5c–f). Thus, OmpB is critical for the growth of R. parkeri in macrophages.
OmpB is required for the formation of an electron-lucent halo around cytosolic Rickettsia
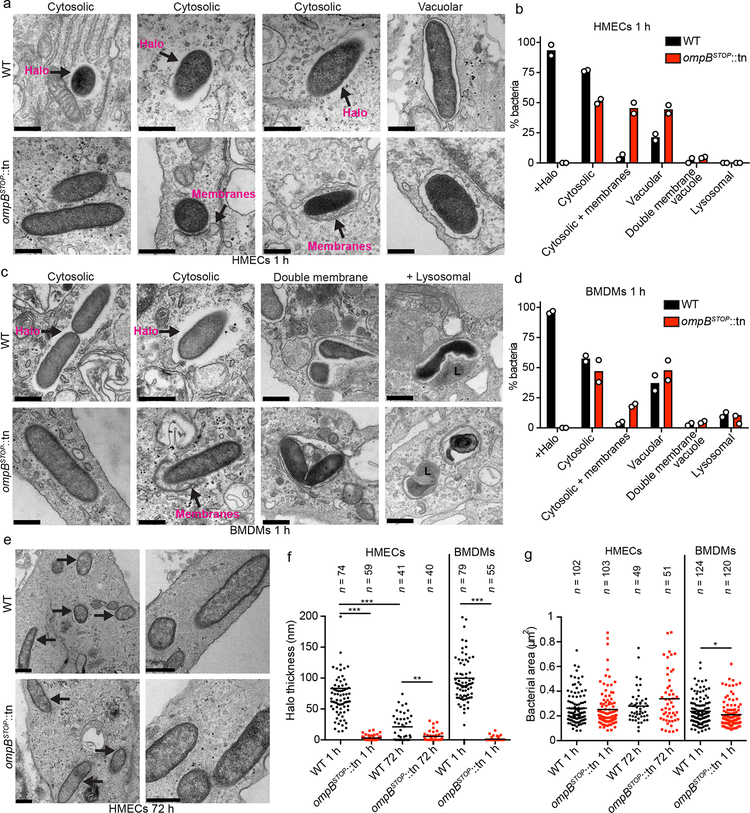
We next assessed the role of OmpB in intracellular trafficking or morphology of R. parkeri using transmission electron microscopy (TEM). At 1 hpi, the majority of WT and ompBSTOP::tn bacteria had escaped from the phagosome and were in the host cytosol of HMECs and BMDMs (Fig. 3a–d). However, in HMECs, there was a reduction in the percentage of ompBSTOP::tn bacteria in the cytosol, and a corresponding increase in bacteria associated with single-membrane vacuoles, in comparison with WT bacteria (Fig. 3a, b), which likely was a consequence of a delayed invasion (Fig. 2a). No differences in the percentage of cytosolic bacteria between ompBSTOP::tn and WT bacteria were observed in BMDMs (Fig. 3c, d). Collectively, these data further suggest that the restriction of growth of the ompBSTOP::tn bacteria in BMDMs occurs following escape into the cytosol.
Fig. 3. OmpB is required for the formation of an electron-lucent halo in host cells.
(a and c) TEM images of WT and ompBSTOP::tn mutant bacteria in (a) HMECs or (c) BMDMs at 1 hpi, in the indicated cellular locations. Arrows indicate halos that appear as electron-lucent areas around WT bacteria, and membranes associated with bacteria, as labeled. Scale bars, 500 nm. (b and d) Mean percentage of WT and bacteria ompBSTOP::tn mutant bacteria in (b) HMECs or (d) BMDMs with the indicated characteristics or location: with halo (Halo+), cytosolic, cytosolic associated with (+) membranes (bacteria were associated with double membranes or membrane-bound organelles over part of their surface or at their poles), in a single membrane vacuole(vacuolar), in a double membrane vacuole, or in a lysosome (bacteria surrounded by electron-dense material that often had irregular shapes53) (n = 2; ≥50 bacteria per strain and host cell type were counted in each experiment). (e) TEM images of WT and ompBSTOP::tn bacteria at 72 h in HMEC cells. Arrows indicate bacteria. Scale bars, 500 nm. (f) Quantification of halo thickness in HEMCs at 1 hpi and 72 hpi, and in BMDMs at 1 hpi. All data points are presented, and the lines indicate the means (statistical comparisons were by the Mann-Whitney rank-sum test (two-sided); **, p < 0.01; ***, p < 0.001). (g) Quantification of the bacterial body area (excluding the halo) in HEMCs at 1 hpi and 72 hpi, and in BMDMs at 1 hpi. All data points are presented, and the lines indicate the means of (statistical comparisons were by the Mann-Whitney rank-sum test (two-410sided); *, p < 0.05).
Once in the cytosol, ompBSTOP::tn bacteria exhibited increased association with membranes resembling phagophores that localized near one bacterial pole or partly surrounding bacteria (Fig. 3a–d). However, ompBSTOP::tn and WT bacteria were comparable in their relatively rare localization within double-membrane vacuoles or electron-dense lysosomal compartments at 1 hpi. At 72 hpi in HMECs, the majority of both ompBSTOP::tn and WT bacteria were free of associated membranes (Fig. 3e). These observations suggest that OmpB inhibits association of cytosolic R. parkeri with host membranes shortly after infection.
The size of ompBSTOP::tn and WT bacteria was indistinguishable in HMECs, whereas the ompBSTOP::tn mutant was slightly smaller than WT in BMDMs at 1 hpi (Fig. 3g). A striking difference, however, was that ompBSTOP::tn bacteria lacked an electron-lucent capsular-like halo that surrounded WT (Fig. 3a–d), ompA::tn and MC1_RS05535::tn R. parkeri (Extended Data Fig. 6a, b), as well as R. rickettsii41,42. For WT bacteria, the size of the electron-lucent halo at 1 hpi was of 100 ± 40 nm (mean ± SD) in BMDMs and 84 ± 60 nm in HMECs (Fig. 3f). At 72 hpi in HMECs, the mean thickness decreased to 22 ± 21 nm and many WT bacteria lacked a visible halo (Fig. 3e, f). These data demonstrate that R. parkeri is surrounded by an electron-lucent halo hat decreases in thickness during the time course of an infection, and that OmpB is required for the formation of this halo, but not for other basic structural properties of the bacteria.
OmpB shields rickettsial surface proteins from ubiquitylation
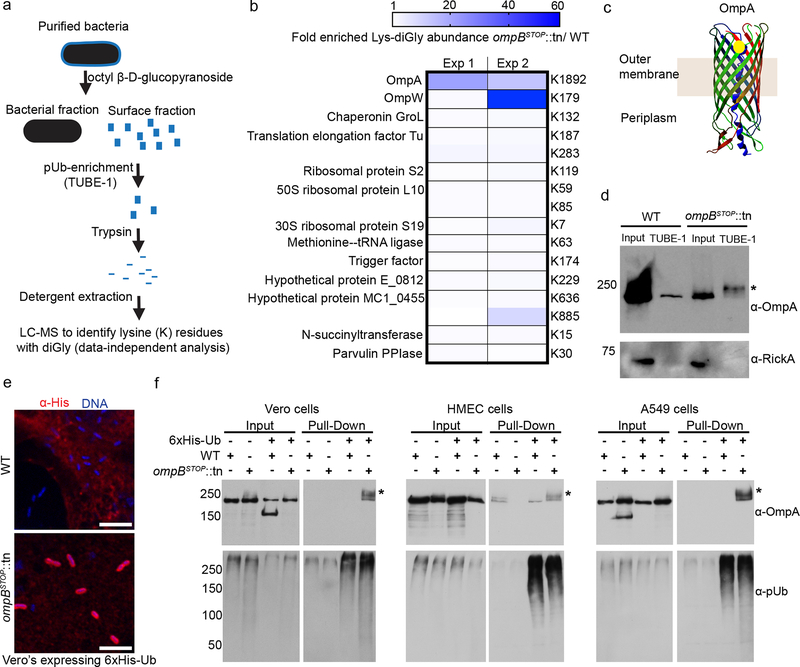
We next sought to identify proteins that are substrates of ubiquitylation on ompBSTOP::tn bacteria. We developed an approach that combined polyubiquitin enrichment from the surface fraction of bacteria, followed by mass spectrometry (MS) to detect Lys-diGly (diGly) remnants, a signature for ubiquitin (Fig. 4a). We did not identify any host protein candidate that was reproducibly enriched over 5-fold in fractions from ompBSTOP::tn in comparison with WT bacteria Supplementary Table 1). We did, however, identify a bacterial peptide of OmpA with diGly remnants that was consistently enriched in fractions from ompBSTOP::tn (Fig. 4b). The ubiquitylated lysine identified on OmpA is predicted to be exposed to the host cytosol (Fig. 4c), and OmpA was shifted towards a higher molecular mass in the ompBSTOP::tn compared with WT bacteria after ubiquitin enrichment (Fig. 4d).
Fig. 4. OmpB protects OmpA against ubiquitylation in diverse host cell lines.
(a) A graphic depiction of the experimental workflow for enrichment of polyubiqutylated proteins from the bacterial surface fraction prior to liquid chromatography-mass spectrometry (LC-MS) analysis. Note: TUBE-1 is pan-specific and binds most ubiquitin linkages. (b) Heat map representation of bacterial proteins for which there is enrichment of Lys-diGly peptides (blue boxes) in ompBSTOP::tn compared with WT bacteria after the approach in panel (a), or equal abundance (white boxes) between ompBSTOP::tn and WT bacteria. Data were from n = 2 independent experiments performed in technical triplicates. (c) Predicted structure of candidate substrate OmpA with identified diGly site shown as yellow circle. (d) Western blot of polyubiquitin-enriched samples from gradient-purified bacteria (prepared as shown in (a)), probed for OmpA (residual detection of OmpA from WT bacteria is likely due to non-specific binding to the TUBE1-beads) and RickA (as a bacterial loading control). The asterisk* shows a size shift of OmpA indicating that it is ubiquitylated (n = 2). (e) Immunofluorescence micrographs of Vero cells expressing 6xHis-ubiquitin and infected with WT or ompBSTOP::tn bacteria for 24 h, showing bacterial genomic DNA (blue, Hoechst) and 6xHis-ubiquitin (red, anti-His antibody) (n = 3). Scale bars, 5 μm. (f) Western blot of Ni-NTA affinity-purified samples from control cells or cells expressing 6xHis-ubiquitin, infected with WT or ompBSTOP::tn bacteria for 26 h, and probed for OmpA or polyubiquitin (FK1). The asterisks* indicate OmpA from 6xHis-ubiquitin-expressing cells infected with ompBSTOP::tn exhibits higher molecular weights than endogenous OmpA, consistent with ubiquitylation (Vero, n = 4; HMEC, n = 2; A549, n = 2).
To validate that OmpA is ubiquitylated, we overexpressed 6x-His-tagged ubiquitin in Vero, HMEC and A549 cells, and then assessed the association of His-tagged ubiquitin with bacteria by immunofluorescence microscopy or by affinity purification of His-ubiquitin followed by western blotting. His-ubiquitin co-localized with ompBSTOP::tn but not WT bacteria during infection (Fig. 4e; data from Vero cells). Furthermore, OmpA was detected in His-ubiquitin-enriched fractions from ompBSTOP::tn but not WT bacteria with a size shift that reflects ubiquitin chains of different lengths (Fig. 4f). These data demonstrate that OmpB protects OmpA from ubiquitylation in diverse host cells.
OmpB is required for avoidance of autophagic recognition in diverse host cells
Based on the ubiquitylation of ompBSTOP::tn bacteria, we hypothesized that OmpB is important for avoidance of autophagic recognition. To test this, we first infected both HMECs and BMDMs and stained for the autophagic receptors p62 and NDP52 (Fig. 5a–d). More than 30% of ompBSTOP::tn bacteria co-localized with p62 and NDP52 at 1 hpi in both cell types and at 72 hpi in HMECs, whereas fewer than 5% of WT bacteria were associated with these proteins (Fig. 5g,h). Thus, OmpB is crucial for avoiding association with p62 and NDP52.
Fig. 5. OmpB is required to avoid both recruitment of autophagy receptors and LC3.
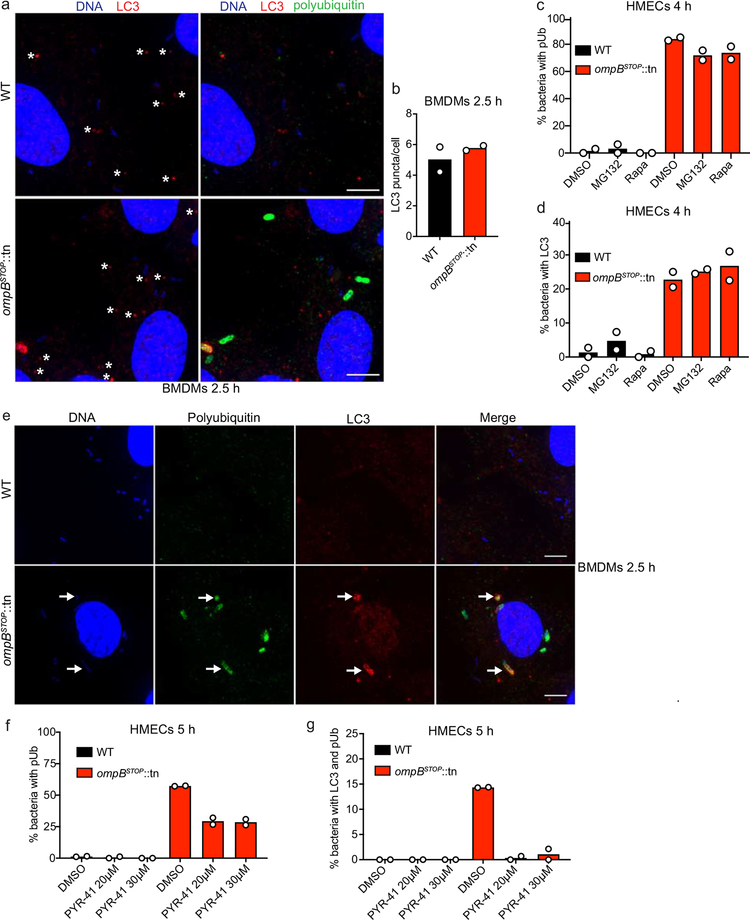
(a-f) Immunofluorescence micrographs of (a, c, e) HMECs or (b, d, f) BMDMs infected with WT (upper panels) or ompBSTOP::tn mutant (lower panels) at 1 hpi, and stained for R. parkeri (green; anti-Rickettsia I7205 antibody), cellular and bacterial genomic DNA (blue; Hoechst), and (a, b) p62 (red; anti-p62 antibody), (c, d) NDP52 (red; anti-NDP52 antibody), or (e, f) LC3 (red; anti-LC3 antibody). Right panels show merged images. Arrows indicate ompBSTOP::tn bacteria positive for p62, NDP52, or LC3. The star in (e) indicates LC3 puncta that co-localize with ompBSTOP::tn. Scale bars, 5 μm. Image adjustments of each autophagic marker, R. parkeri, and DNA, were applied equally for both bacterial strains and cell types (exception, LC3, between HMECs and BMDMs). (g, h) Quantification of the percentage of WT or ompBSTOP::tn mutant bacteria that localized with (g) p62 or (h) NDP52 in BMDMs (left) or HMECs (right) at the indicated times post infection. Data are mean (n = 2; ≥91 bacteria were counted for the 1 hpi time point for each experiment; ≥350 bacteria were counted for the 72 hpi time point for each experiment). (i, j) Quantification of the percentage of WT or ompBSTOP::tn mutant bacteria that localized with LC3 in (i) BMDMs or (j) HMECs at the indicated times post infection. Data are mean ± SEM (BMDMs, n = 3, ≥143 bacteria were counted for each strain, infection and time-point; HMECs 2 h, n = 2; 20 min and 6 h, n = 3; 1 h, n = 4; 3 h, n = 5; ≥77 bacteria were counted for each strain, infection and time-point; statistical comparisons were by an unpaired Student’s t-test (two-sided); **, p < 0.01; ***, p < 0.001). (k) Quantification of the percentage LC3-postive ompBSTOP::tn mutant bacteria that co-localize with polyubiquitin at 2.5 and 4 hpi in BMDMs and HMECs. Data are mean ± SEM (BMDMs, n = 3; ≥74 bacteria were counted for each strain, infection and time-point; HMECs, n = 2; ≥101 bacteria were counted for each strain and experiment) (micrographs of BMDMs are shown in Extended Data Fig. 7e).
We also assessed recruitment of LC3, a common marker for autophagic membranes. A subpopulation of ompBSTOP::tn bacteria was associated with LC3 in both BMDMs and HMECs, with a peak in LC3 association at 2 hpi and diminishing association thereafter, whereas WT bacteria showed little LC3 association (Fig. 5e, f, i and j; no difference in the numbers of cellular LC3-puncta was observed in cells infected with WT versus ompBSTOP::tn (Extended Data Fig.7a, b), and treatment with the proteasome inhibitor MG132 or the mTORC1 inhibitor rapamycin, which activate autophagy, did not increase the number of LC3-positive or pUb-positive ompBSTOP::tn (Extended Data Fig. 7c, d)). Furthermore, almost all LC3-positive ompBSTOP::tn were polyubiquitin-positive (Fig. 5k, Extended Data Fig. 7e), and treatment with PYR-41 reduced the percentage of polyubiquitin-positive and LC3-positive ompBSTOP::tn bacteria (Extended Data Fig. 7f, g). This indicates that ubiquitylation is required for LC3 recruitment. Collectively, these data demonstrate that OmpB blocks ubiquitylation of R. parkeri to evade autophagic recognition in diverse host cells.
OmpB is required for R. parkeri to evade antimicrobial autophagy in macrophages
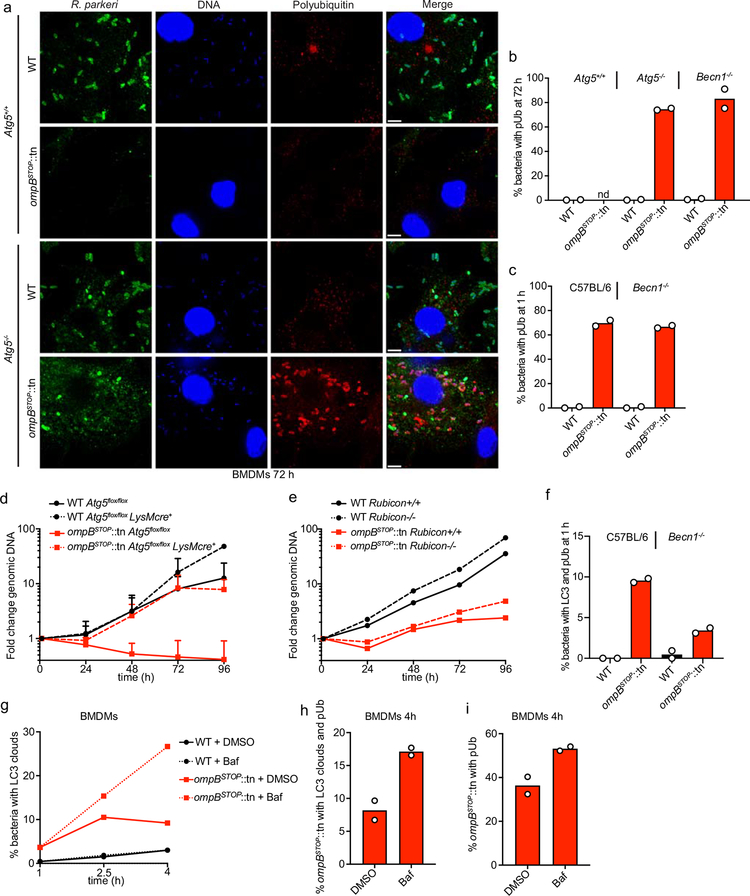
To directly test if OmpB is critical to evade antimicrobial autophagy in macrophages, we employed BMDMs from mice genetically lacking the Atg5 or Becn1 genes in myeloid cells. These mutant cells are defective in both autophagy and LC3-associated phagocytosis (LAP)43. BMDMs were infected with ompBSTOP::tn and WT R. parkeri, and bacterial genomic DNA was quantified to assess growth during a 96-h infection. In control Atg5+/+ (also known as Atg5flox/flox)44 macrophages, ompBSTOP::tn was unable to grow whereas WT bacteria grew ~10-fold (Fig. 6a). In contrast, in Atg5−/− (Atg5flox/flox-LysMcre+)45 macrophages, ompBSTOP::tn bacteria grew similarly to WT during the first 72 hpi (Fig. 6b). The growth of ompBSTOP::tn was also rescued in Becn1−/− (Beclin1flox/flox-LysMcre+)46 macrophages (Fig. 6c). Although the growth of the ompBSTOP::tn mutant was rescued in Atg5−/− and Becn1−/− cells, the mutant was polyubiquitylated (Extended Data Fig. 8a–c), demonstrating that polyubiquitylation does not restrict bacterial growth when the autophagy cascade is prevented. We also tested for bacterial growth in Rubicon−/− macrophages43, which are specifically defective in LAP43, and growth of the ompBSTOP::tn mutant was not rescued (Extended Data Fig. 8d, e). These results suggest that the growth of the ompBSTOP::tn mutant is restricted by autophagy but not by LAP.
Fig. 6. OmpB interferes with autophagy and promotes R. parkeri growth in macrophages.
(a-c) Growth curves of WT and ompBSTOP::tn in BMDMs from (a) Atg5flox/flox (Atg5+/+), (b) Atg5flox/flox-LysMcre+ (ATG5−/−), or (c) Beclin1flox/flox-LysMcre+ (Becn1−/−) mice from 0–96 hpi as measured in Fig. 2g. Data are mean ± SEM (Atg5+/+, n = 4; Atg5−/−, n = 4; Becn1−/−, n = 3; statistical comparisons for each time point were by the Mann-Whitney rank-sum test (two-sided); *, p < 0.05). (d) Immunofluorescence micrographs of BMDMs infected with WT (upper panels) or ompBSTOP::tn mutant (lower panels) at 4 hpi, and stained for R. parkeri (green), cellular and bacterial genomic DNA (blue; Hoechst), and LAMP1 (red; anti-LAMP1 antibody). Scale bars, 5μm. Higher magnification images, scale bars, 1 μm. Arrows indicate LAMP1-positive bacteria. (e). Quantification of the percentage LAMP1-positive bacteria at 1, 2.5 and 4 hpi in BMDMs. Data are mean ± SEM (1 h, n = 2; 2.5, 4 h, n = 3 ; ≥78 bacteria were counted for each strain, infection and time point; statistical comparisons between WT and ompBSTOP::tn were performed using a one-way ANOVA with Tukey’s post hoc-test; *, p < 0.05). (f) Quantification of the percentage LAMP1-positive bacteria at 4 hpi in WT or Becn1−/− BMDMs. Data are mean ± SEM (n = 3; ≥111 bacteria were counted per infection and experiment; statistical comparisons were by a one-way ANOVA with Tukey’s post hoc-test; ***, p < 0.001). (g) Mice were intravenously infected with 107 PFUs of WT or ompBSTOP::tn bacteria, organs were harvested at indicated time points and homogenized, and PFUs were counted. All data points are shown, and medians are indicated as bars (for WT infected mice at 2 h, n = 6; 24 h, n = 8; 48 h, n = 14; 72 h, n= 10; for ompBSTOP::tn infected mice at 2 h, n = 6; 24 h, n = 5; 48 h, n = 5; 72 h, n = 4). Note that the limit of detection was different between liver and the other organs due to the fact that concentrated liver homogenate interfered with PFU determination. At each time point, PFU counts between WT and ompBSTOP::tn mutant were p < 0.05 as determined by the Mann-Whitney rank-sum test (two-sided). (h) C57BL/6 (WT, n = 4; ompBSTOP::tn, n = 5), Atg5+/+ (n = 5) or Atg5−/− (n = 5) mice were intravenously infected with 107 PFUs of WT or ompBSTOP::tn bacteria, organs were harvested at 2 h and homogenized, and PFUs were counted. All data points are shown, and medians are indicated as bars. Statistical omparisons were by the Mann-Whitney rank-sum test (two-sided); *, p < 0.05; ** p < 0.01.
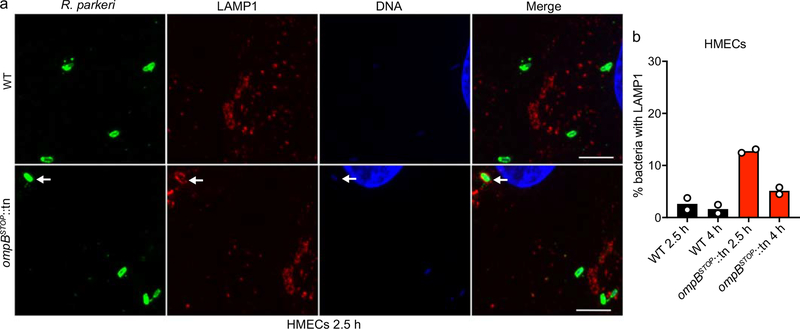
Consistent with being targeted by autophagy, ompBSTOP::tn colocalized with the lysosomal marker LAMP1, with the number of LAMP1-positive bacteria increasing over time in BMDMs (Fig. 6d, e), but not in HMECs (Extended Data Fig. 9a, b). Furthermore, treatment with bafilomycin A, which blocks autophagosome-lysosome fusion and maturation, resulted in the accumulation of mutant bacteria co-localizing with LC3, polyubiquitin or both LC3 and polyubiquitin (Extended Data Fig. 8g–i). The numbers of LAMP1-positive (Fig. 6f) and LC3/polyubiquitin-positive (Extended Data Fig. 8f) ompBSTOP::tn bacteria were reduced in Becn1−/− macrophages, consistent with the role of beclin 1 in autophagy17,18. Together, these results demonstrate that OmpB is critical for R. parkeri to evade destruction by autophagy in macrophages.
OmpB is important for R. parkeri to colonize mouse organs due to its ability to promote autophagy evasion in immune cells
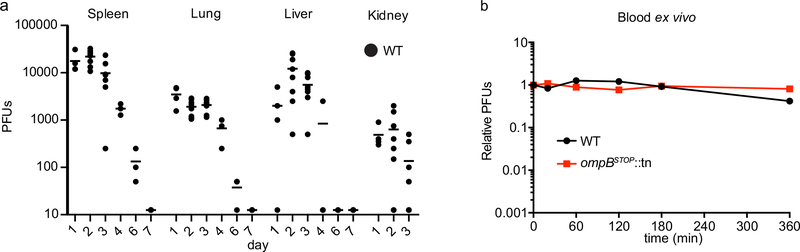
We next sought to determine whether and how OmpB may contribute to R. parkeri virulence in vivo. We used C57BL/6 mice for these studies because of the availability of mutants in this strain carrying mutations in immunity genes. We injected WT R. parkeri intravenously into the tail-vein of mice which were then sacrificed each day over a 7 d period, and the number of plaque-forming units (PFU) was determined in spleen, lung, liver, and kidney. PFUs were recovered from all these organs, although the mice were asymptomatic and bacteria were cleared between days 4 and 7 (Extended Data Fig. 10a). Next, we infected mice with WT or ompBSTOP::tn bacteria and we were unable to recover any ompBSTOP::tn from any organ within the window of 2 to 72 hpi (Fig. 6g). Because R. conorii OmpB had been implicated in serum resistance47, we also tested if the ompBSTOP::tn mutant could survive in blood ex vivo over a 6 h time course. The ompBSTOP::tn bacteria remained as infectious as WT over this time (Extended Data Fig. 10b), suggesting that OmpB does not by itself impact the survival of R. parkeri in blood. These results show that OmpB is a R. parkeri virulence factor that is critical for establishing an infection in vivo.
We next examined if OmpB enables autophagy evasion in vivo, similar to its role in cultured macrophages in vitro. To test this, we infected Atg5−/− (Atg5flox/flox-LysMcre+) mice, which only lack Atg5 in myeloid cells including macrophages, as well as control Atg5+/+ (Atg5flox/flox) and C57BL/6 mice, with WT and ompBSTOP::tn bacteria. We observed a 5–16-fold rescue of the number of ompBSTOP::tn bacteria in the spleens and livers of Atg5−/− mice compared to Atg5+/+ and C57BL/6 mice, and the mutant reached levels that were not significantly different from those of WT (Fig. 6h). These findings demonstrate that OmpB plays a major role in R. parkeri colonization of mice because of its ability to promote autophagy evasion in immune cells.
DISCUSSION
Bacterial pathogens that grow in the cytosol of host cells often avoid recognition by innate immune pathways, including antimicrobial autophagy. Here, we demonstrate that R. parkeri surface protein OmpB is critical for protecting against autophagic recognition by blocking the formation of a polyubiquitin coat. We further show that OmpB is critical for R. parkeri growth in cultured macrophages via its role in autophagy evasion. Finally, our data suggest that OmpB is critical for R. parkeri to colonize mouse organs due to its ability to promote autophagy evasion in vivo. Together, these data indicate that OmpB acts as a protective shield to inhibit antimicrobial autophagy and as a critical virulence factor in Rickettsia pathogenesis.
Our work also demonstrates that OmpB protects the bacterial surface protein OmpA from ubiquitylation, establishing OmpA as a specific bacterial target of the ubiquitylation machinery. Additional rickettsial surface proteins such as OmpW are also likely ubiquitylated targets that contribute to ubiquitin coat formation. Interestingly, OmpA is a conserved outer membrane protein in Gram-negative bacteria, and it was previously identified as a candidate ubiquitylated protein on Salmonella enterica Typhimurium48. Thus, we hypothesize that OmpA may be a common bacterial target for the host ubiquitin machinery.
We further show that OmpB protects R. parkeri from polyubiquitylation by acting locally on the bacterial surface, rather than globally on the host ubiquitylation machinery. However, it remains unclear exactly how OmpB exerts its protective activity, and the mechanisms could be multifaceted. For example, it is possible that OmpB has deubiquitylase activity. Alternatively, OmpB may camouflage the bacterial surface, either directly and/or via its role in formation of a capsule-like halo. In support of these hypotheses, OmpB makes up more than 10% of the cellular protein mass35 and is thus abundant enough for a camouflaging role. Moreover, a role for the OmpB-dependent capsule-like halo would be consistent with speculation that the capsule O-antigen shields Francisella tularensis from ubiquitylation23. A third possibility is that OmpB recruits host proteins that camouflage the bacterial surface or cleave polyubiquitin chains. A fourth possibility is that OmpB possesses an enzymatic activity that alters properties of the bacterial surface required for recognition by the host ubiquitin machinery.
Our data suggest that OmpB is required for R. parkeri to evade autophagy in immune cells during initial colonization of mice. However, it remains unclear whether OmpB protects R. parkeri from autophagy during long-term infections in vivo. Although we find that OmpB is not required for autophagy avoidance in endothelial cell lines in vitro, it was previously observed that rickettsiae can be found in autolysosomes of cytokine-treated mouse endothelial cells49, suggesting that bacteria could be targeted by autophagy during long-term infection following release of inflammatory cytokines50.
OmpB appears to act differently from other pathogen proteins that protect against polyubiquitylation. Form example, the L.monocytogenes surface protein ActA blocks polyubiquitylation by mobilizing the host actin machinery to the bacterial surface, which is required to protect the bacterium from autophagic recognition11,25,51. In contrast, OmpB has no effect on host actin mobilization by R. parkeri. In addition, a sub-population of WT L. monocytogenes is polyubiquitylated during intracellular growth11,22,52, whereas nearly all R. parkeri are polyubiquitin-negative, suggesting that OmpB is a more robust shield, perhaps because its mechanism may not involve recruitment of host components such as the actin machinery. Thus, although cytosolic pathogens have evolved a common strategy of using abundant surface proteins to block host polyubiquitylation, the mechanism by which each protein acts as a shield is distinct. Understanding the commonalities and differences in how pathogen proteins protect against polyubiquitylation and autophagy will enhance our understanding of pathogenic strategies for avoidance of innate immunity, and might be co-opted as a therapeutic strategy for treating infectious disease.
METHODS
Cell Lines
HMEC-1, A549, and Vero cells were purchased from the UC Berkeley Cell Culture Facility and were authenticated by short-tandem repeat analysis. All cells were grown at 37°C and 5% CO2. HMECs were grown in MCDB 131 media containing 10 mM L-glutamine (Sigma, M8537), supplemented with 10% heat-inactivated fetal bovine serum (Hi-FBS, Hyclone), 10 ng/mL epidermal growth factor (EGF) (Fisher Scientific, CB40001; Corning, 354001), 1 μg/mL hydrocortisone (Spectrum Chemical, CO137), and 1.18 mg/mL sodium bicarbonate. Fresh HMEC media was prepared every 1–2 months, and aliquoted and stored at 4°C prior to experiments or to passaging cells to minimize degradation of EGF. A549 cells were grown in DMEM (Gibco, 11965–092) with high glucose (4.5 g/L) and 10% Hi-FBS (Atlas Biologicals). Vero cells were grown in DMEM (Gibco, 11965–092) with high glucose (4.5 g/L) and 2% Hi-FBS (Benchmark). All cell lines were confirmed to be mycoplasma-negative by DAPI staining and fluorescence microscopy screening at the UC Berkeley Cell Culture Facility.
Primary Macrophage Cells
Wild-type BMDMs were generated from the femurs of female C57BL/6 mice (Charles River Laboratories, Wilmington, MA) according to standard protocols54. Briefly, flesh was removed and bones were washed in 70% ethanol and then with BMDM media (DMEM high glucose; Gibco, 11965–092) containing 20% Hi-FBS (Hyclone), 10% supernatant from NIH 3T3 fibroblasts that produced macrophage colony-stimulating factor (M-CSF), 1% sodium pyruvate (Gibco, 11360–070), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), and 0.1% β-mercaptoethanol (Gibco, 21985–023). Next, bones were crushed with a mortar and pestle and cells were filtered through a 70-μm nylon Corning Falcon cell strainer (Thermo Fisher Scientific, 08-771-2) to remove particulates. Cells were centrifuged for 8 min at 290 x g, 4°C, and the cell pellet was resuspended in BMDM media and incubated at 37°C for 7 d prior to harvesting in media lacking antibiotics.
The percentage of CD11b(+)F4/80(+) BMDMs was determined by blocking cells with anti-mouse CD16/32 (Biolegend, 101320) and then staining with PE-Cyanine7 anti-mouse F4/80 antigen antibody (Tonbo Bioscience, 604801, dilution 1:200), APS anti-mouse CD11b (Tonbo Bioscience, 200112, dilution 1:250) and DAPI (dilution 1:1000). Samples were subjected to flow cytometry using a BD LSRFortessa 2010 Cell Analyzer (BD Biosciences) and data was analyzed using FlowJo software (FlowJo, LLC). Single viable DAPI-negative cells were gated and the percentage of CD11b(+)F4/80(+) macrophages was determined to be 99.4% (Supplementary Fig. 1).
Mutant BMDMs were isolated from femurs of mutant mice. Femurs from female C57BL/6 Atg5flox/flox (Atg5+/+)44 and matched C57BL/6 Atg5flox/flox-LysMcre+ (Atg5−/−)45 mice were from the laboratory of Jeff Cox (UC Berkeley). Femurs from female Beclin1flox/flox-LysMcre+ (Becn1−/−)46, Rubicon+/+ and Rubicon−/− (CRISPR/Cas9-mediated mutations)43 mice were obtained from Douglas Green (St. Jude Children’s Research Hospital). BMDMs were prepared as described above. Genotypes of the BMDMs were confirmed by PCR and Sanger sequencing at the UC Berkeley DNA Sequencing Facility with the following primer: 5’-GCAGAAGTTGAAGCACCTAGTCACACCAC-’3 and 5’-TGCATAATGGTTTAACTCTTGSTAGACTAGTCA-’3 for Atg5+/+ and Atg5−/− BMDMs; 5’-CTGCAGCGTATCTCAGGTG-’3 and 5’-CCAAGGTTTCCATGCTAATGC-’3 for Becn1−/−; 5’-GCCTTCCTGCTGAGTGACG-3 and 5’-GTCATGGGCTCCCGTGGC-’3 for Rubicon+/+ and Rubicon−/− BMDMs.
Rickettsia parkeri propagation and enumeration
For R. parkeri propagation, five T175 flasks of Vero cells growing in DMEM plus 2% Hi-FBS were infected with a multiplicity of infection (MOI) of ~0.01 and incubated until 50–75% of the 740 cells were infected (after 5–7 days (d). Cells were scraped into the growth media, collected in cold centrifuge bottles, and centrifuged at 12,000 x g for 20 min at 4°C. The cell pellet was resuspended in cold K-36 buffer (0.05 M KH2PO4, 0.05 M K2HPO4, pH 7, 100 mM KCl, 15 mM NaCl) and transferred to a cold dounce homogenizer (tight fit). To release bacteria, cells were dounce-homogenized for 60 strokes, and the homogenate was centrifuged at 200 x g for 5 min at 4°C to remove cell debris. To yield “30%-purified” bacteria, the supernatant was overlaid onto cold 30% MD-76R (Mallinckrodt Inc., St Louis, 1317–07), diluted in K-36 buffer, and centrifuged at 58,300 x g for 20 min at 4°C in an SW-28 swinging bucket rotor. The pellet was resuspended in cold brain heart infusion (BHI) (BD Difco, 237500) broth (0.5 mL per infected T175 flask) and subsequently aliquoted and frozen at −80°C. For further purification, to yield “gradient-purified” bacteria, bacteria were overlaid on a 40/44/54% MD-76R step gradient and centrifuged at 58,300 x g for 30 min at 4°C using a SW-28 swinging bucket rotor. Bacteria were collected from the 44–54% interface and subsequently pelleted by centrifugation at 12,000 x g for 15 min at 4°C. The pellet was resuspended in BHI and subsequently aliquoted and frozen at −80°C. In all experiments, fresh vials of bacteria were used to avoid experimental variations due to loss of bacterial infectivity upon freezing–thawing. Stocks of WT and ompBSTOP::tn mutant bacteria were prepared every ~6 months, and side-by-side experimental comparisons were made between stocks prepared at similar times.
To enumerate infectious bacteria, purified bacterial strains were serial-diluted in DMEM with 2% Hi-FBS and used to infect Vero cells grown in 6-well plates by centrifugation at 300 x g for 5 min, at room temperature. Infections were allowed to proceed at 33°C overnight (~16 h) and then cells were overlaid in DMEM with 10% Hi-FBS plus 0.5% (w/v) UltraPure agarose (Invitrogen, 16–500). At 5–7 d post infection, when plaques could be visualized, cells were further overlaid with PBS plus 0.5% UltraPure agarose containing 0.0125% (v/v) neutral red (Sigma, N6264) and incubated at 33°C. The next day plaques were counted to determine the number of plaque-forming units (PFU) per mL. For bacterial growth curves, 30%-purified bacteria were diluted in the appropriate media to an MOI of 0.005–0.01 for infection of HMECs and 0.25–0.5 for infection of BMDMs (the higher MOI for BMDMs was to compensate for the reduced bacterial growth rate in these cells). R. parkeri infections were carried out in 24-well plates. Bacteria were centrifuged onto cells at 300 x g for 5 min at room temperature, then incubated at 33°C for 1 h, at which point the culture media was exchanged. At various times post infection, including the 1 h time point, cells were scraped off into the media (to include both intracellular and extracellular bacteria), centrifuged 20,000 x g for 5 min at room temperature, and pellets were stored at −20°C until DNA was purified using DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instruction for Gram-negative bacteria. R. parkeri genome equivalents were quantified by quantitative PCR (pPCR) using primer 5’-CGCCATTCTACGTTACTACC-’3 and 5’-GCATTACTTGGAGTTCT-’3 to the hrtA gene, relative to a standard curve of hrtA gene DNA55. Growth was normalized to values at 1 h for each strain.
Rickettsia parkeri strain generation
R. parkeri ompA::tn, hrtA::tn, sca2::tn and ompB::tn mutant strains were previously isolated in a screen for small plaque mutants33,38,56. The ompA::tn mutant lacked full-length OmpA as determined by western blotting using the mouse anti-OmpA 13–3 antibody (kindly provided by Dr. Ted Hackstadt)29,57 (Extended Data Fig. 1f). The MC1_RS05535::tn, MC1_RS05545::tn and rickCE::tn strains were isolated in an independent screen in which mutants were isolated without regard for plaque size. The genomic locations of transposon insertion sites for all mutants were determined by semi-random nested PCR and Sanger sequencing at the UC Berkeley DNA Sequencing Facility, as previously described58. Expanded strains were either verified by semi-random nested PCR or by PCR reactions that amplified the transposon insertion site using primers for flanking regions.
The ompBSTOP::tn, MC1_RS02370STOP,ompB::tn and ompBPROM::tn strains were isolated in a screen for mutations that suppress the slow-growth phenotype of ompB::tn. To obtain strains carrying suppressor mutations, two independent selections were carried out in which the ompB::tn strain was subjected to 10 serial passages in Vero cells growing in T75 flasks in DMEM with 2% Hi-FBS, with the length of each passage varying between 5 to 7 d, depending on the rate of bacterial growth. For each passage, cells were lysed by addition of 1 mL cold sterile water per 796 T75 flask for 1–2 min at room temperature. Subsequently, 1 mL of cold 2 x BHI was added to the lysed cells and ~10% of the suspension was added to DMEM plus 2% Hi-FBS, and this was used 798 to infect a fresh T75 flask of Vero cells. The remaining bacteria were frozen at −80°C. To map suppressor mutations in the mixed population, clonal strains were isolated and larger plaques were selected and expanded in Vero cells, as described above. The genome sequences of 3 strains from each suppressor screen, which had been passaged 4 to 5 times, were determined (NCBI Trace and Short-Read Archive; Sequence Read Archive (SRA) as accession number SRP154218), as was the sequence of the parental ompB::tn (individual accession number SRX4401163) and WT R. parkeri strains (SRX4401164), as described below. The strains were isogenic with the exception of the suppressor mutations in the following locations: the MC1_RS02370 gene, encoding a putative glycosyltransferase (the same mutations in 4 out of 6 clonal strains, SRX4401165); the promoter region of ompB (ompBPROM::tn) (1 clonal strain, SRX4401166); and upstream of the transposon in the coding sequence of ompB, leading to a premature stop codon (ompBSTOP::tn) (1 clonal strain, SRX4401167) (See also Extended Data Fig. 1a–c).
R. parkeri genome sequencing
Rickettsia genomic DNA was isolated from frozen bacterial stocks (~108 PFUs, stored in BHI) that were centrifuged at 16,000 x g for 5 min at room temperature and resuspended in Dulbecco’s PBS (DPBS) with calcium and magnesium (Invitrogen, 14040) containing DNase reaction buffer and 100 U of recombinant RNase-free DNase 1 (Roche, 04716728001) to reduce host genomic DNA in the final sample, as described previously59. Intact bacteria were incubated at 37°C for 30 min and thereafter washed 2 x with DPBS. Subsequently, rickettsial genomic DNA was purified using the DNeasy Blood and Tissue kit (Qiagen), according to manufacturer’s instruction for Gram-negative bacteria. To confirm that Rickettsia genomic DNA was largely intact, we verified that DNA fragments were mostly > 20,000 bp by agarose gel electrophoresis. The genome sequences of R. parkeri strains (3 strains from each suppressor screen, as well as parental ompB::tn and WT strains) were determined at the UC Berkeley Functional Genomics Laboratory, using single end 50 bp read lengths on the Illumina Hiseq2000 sequencing platform. Genome assembly as well as single nucleotide variant and indel identification were performed with CLC Genomics Workbench (QIAGEN) and Integrated Genomics Viewer (IGV)60. The R. parkeri str. Portsmouth genome (GenBank accession no. NC_017044.1) was used as the reference sequence. Mutations identified by genome sequencing were confirmed by PCR followed by Sanger sequencing at the UC Berkeley DNA Sequencing Facility.
OmpB antibody production and western blotting
The sequence encoding amino acids 36–218 of OmpB (ompBaa36–218), a protein that lacks he signal peptide and stops at the same position as the fragment encoded in the ompBSTOP::tn suppressor strain (Extended Data Fig. 1c), was amplified by PCR from R. parkeri genomic DNA. ompBaa36–218 was cloned into plasmid pSMT3, which encodes N-terminal 6xHis and SUMO tags61. 6xHis-SUMO-OmpB aa36–218 was expressed in E. coli strain BL21 codon plus RIL-Camr (DE3) (QB3 Macrolab, UC Berkeley) by induction with 0.1 mM isopropyl-β-D-thio-galactoside (IPTG) for 16 h at 16°C. Bacterial pellets were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) and stored at −80°C. For protein purification, bacteria were thawed, lysozyme was added to 1 mg/mL (Sigma, L4919), and lysis was carried out by sonication. Lysate containing 6xHis-SUMO-OmpB36–218 was incubated with Ni-NTA resin (Qiagen, 1018244), and bound proteins were eluted with a step gradient of 100, 250 and 500 mM imidazole (in lysis buffer, pH adjusted to 8.0). Fractions were analyzed by SDS-PAGE and those with the highest concentrations of 6xHis-SUMO-OmpB36–218 were pooled prior to desalting, using PD-10 desalting columns (GE Healthcare, 17085101) and equilibrated with lysis buffer lacking imidazole.
To generate rabbit antibodies against OmpB, 0.8mg of purified 6xHis-SUMO-OmpB36–218 protein was sent to Pocono Rabbit Farm and Laboratory (Canadensis, PA), and immunization was carried out according to their 91-d protocol. To affinity-purify anti-OmpB antibodies, ompBaa36–218 was subcloned into plasmid pET1M, which encodes N-terminal 6xHis and maltose-binding 850 protein (MBP) tags. 6xHis-MBP-OmpBaa36–218 was expressed and induced as above for His-SUMO-OmpBaa36–218. Bacterial pellets were resuspended in column buffer (50 mM NaH2PO4, pH 852 8.0, 300 mM NaCl, 1 mM EDTA and 1 mM DTT), and the fusion protein was purified by affinity chromatography on amylose resin (New England Biolabs, E8021L). Bound proteins were eluted n column buffer lacking EDTA and containing 10 mM maltose. 500μg of purified protein was coupled to NHS-activated Sepharose 4 Fast Flow resin (GE-Healthcare, 17090601) in ligand-coupling buffer (200 mM NaHCO3 pH 8.3, 500 mM NaCl) overnight at 4°C. Resin was incubated with 10 mL of anti-OmpB serum diluted in binding buffer (20 mM Tris, pH 7.5), which was loaded on the resin and incubated for 3 h at 4°C and eluted with 100 mM glycine pH 2.5 and neutralized62.
For western blotting of endogenous OmpB, OmpA, RickA, and polyubiquitin, 30%-purified or gradient-purified bacteria were boiled in 1 x SDS loading buffer (150 mM Tris pH 6.8, 6% SDS, 0.3% bromophenol blue, 30% glycerol, 15% β-mercaptoethanol) for 10 min. Then, 1 to 5 × 106 PFUs were resolved on an 8–12% SDS-PAGE gel and transferred to an Immobilon-FL PVDF membrane (Millipore, IPEL00010). Membranes were probed for 30–60 min at room temperature with antibodies as follows: affinity-purified rabbit anti-OmpB antibody (this study) diluted 1:10,000 in TBS-T (20 mM Tris, 150 mM NaCl, pH 8.0, 0.05% Tween 20 (Sigma, P9416)) plus 5% dry milk (Apex, 20–241); mouse monoclonal anti-OmpA 13–3 antibody diluted 1:20,000 in TBS-T plus 5% dry milk; affinity-purified rabbit anti-RickA antibody63 diluted 1:1,000 in TBS-T plus 5% dry milk; or mouse monoclonal FK1 anti-polyubiquitin antibody (Enzo Life Sciences, BML-PW8805–0500) diluted 1:2,500 in TBS-T plus 2% BSA. Secondary antibodies were: mouse anti-rabbit horseradish peroxidase (HRP) (Santa Cruz Biotechnology, sc-2357), or goat anti-mouse HRP (Santa Cruz Biotechnology, sc-2005), both diluted 1:2,500 in TBS-T plus 5% dry milk. Secondary antibodies were detected with ECL Western Blotting Detection Reagents (GE, Healthcare, RPN2106) for 1 min at room temperature and developed using Biomax Light Film (Carestream, 178–8207).
Immunofluorescence microscopy
R. parkeri infections were carried out in 24-well plates with sterile circle 12-mm coverslips (Thermo Fisher Scientific, 12-545-80). For infecting HMECs, 2×105 cells were seeded into each well, and then cells were infected after 2 d. For infecting BMDMs, 4–5×105 cells were seeded into each well and infection was conducted after 16–20 h. To initiate infection, 30%-purified bacteria were diluted in cell culture media at room temperature to an MOI of 0.005–0.01 for HMECs and of 0.25–0.5 for BMDMs. The higher MOI for the macrophage infection was employed due to the reduced bacterial growth rate in these cells. Bacteria were centrifuged onto cells at 300 x g for 5 min at room temperature and subsequently incubated at 33°C for 1 h. At 1 hpi, media was exchanged. For experiments with the E1-inhibitor PYR-41 (Millipore, 662105), HMECs were infected and treated with drug in either of two ways: infected with an MOI of 2–4, supplemented with the drug dissolved in DMSO (final concentration 50 μM; PYR-41 was dissolved in pre-warmed HMEC media, followed by swirling the tube and incubation at 37°C for 5 min) or the corresponding volume of DMSO at 1 hpi, and then incubated at 37°C until 5 hpi; or infected with an MOI of 0.005–0.01, supplemented with the drug dissolved in DMSO (final concentration 50 μM) or the corresponding volume of DMSO at 66 hpi, and incubated at 37°C until 72 hpi. For experiments with bafilomycin A (Sigma, B1793), BMDMs infected with an MOI of 2–4 were supplemented at 1 hpi with the drug dissolved in DMSO (final concentration 300 nM) or the corresponding volume of DMSO, incubated at 33°C and fixed at 1 hpi, 2.5 hpi, and 4 hpi (3 hours after drug was supplemented). For experiments with rapamycin (Tocris Bioscience, 1292) and MG132 (Sigma, M8699), HMECs infected with an MOI of 2–4 were supplemented at 1 hpi with the drugs dissolved in DMSO (final concentration 500 nM rapamycin and 20 μM MG132) or the corresponding volume of DMSO that corresponded to the amount of MG132, incubated at 33°C and fixed at 4 hpi (3 hours after drug was supplemented), as described above.
For immunofluorescence microscopy, infected cells were fixed for 10 min at room temperature in pre-warmed (37°C) 4% paraformaldehyde (Ted Pella Inc., 18505) diluted in PBS, pH 7.4, then washed 3 x with PBS. Primary antibodies were the following: for staining with the guinea pig polyclonal anti-p62 antibody (Fitzgerald, 20R-PP001; 1:500 dilution), mouse polyclonal anti-NDP52 antibody (Novus Biologicals, H00010241-B01P; 1:100 dilution), rabbit monoclonal anti-M1 (linear) polyubiquitin linkage-specific antibody (Millipore, MABS199, 1:250 dilution), rabbit monoclonal anti-K63 polyubiquitin linkage-specific antibody (Millipore, 05–1308, 1:250 dilution), human anti-K48 polyubiquitin linkage-specific antibody, clone 5B664 (final concentration 0.2 μg/mL), or anti-polyubiquitin FK1 antibody (1:250 dilution), cells were permeabilized with 0.5% Triton-X100 for 5 min prior to staining. For staining with the mouse monoclonal anti-LC3 antibody (NanoTools, 0260–100, clone 2G6; 1:500 dilution), the rabbit polyclonal anti-LC3 antibody (Novus Biologicals, NB100–2220SS, 1:250 dilution), the rat anti-mouse-LAMP1 antibody (Biolegend, 121609, dilution 1:250), the mouse anti-human-LAMP1 antibody (555801, BD Bioscience, dilution 1:25) or the rabbit polyclonal anti-OmpB antibody (this study; 1:1000 dilution), cells were post-fixed in 100% methanol for 5 min at room temperature. Cells were washed 3 x with PBS and incubated with the primary antibody diluted as indicated in PBS with 2% BSA for 30 min at room temperature. To detect the primary antibodies, secondary goat anti-rabbit Alexa-568 (Invitrogen, A11036), goat anti-mouse Alexa-568 (Invitrogen, A11004), goat anti-human Alexa-488 (Invitrogen, A11013), goat anti-rat Alexa-568 (Invitrogen, A11077) or goat anti-guinea pig Alexa-568 (Invitrogen, A11075) antibodies were incubated at room temperature for 30 min (all 1:500 in PBS with 2% BSA). Next, a rabbit anti-Rickettsia I7205 antibody (gift from Ted Hackstadt)57 or a mouse monoclonal anti-Rickettsia 14–13 antibody (gift from Ted Hackstadt)57, both diluted 1:500, was used to detect R. parkeri. Secondary antibodies used were: Alexa 488 anti-rabbit antibody (Invitrogen, A11008; 1:500 dilution), Alexa 405 anti-rabbit antibody (Invitrogen, A31556), Alexa 488 anti-mouse antibody (Invitrogen, A11001; 1:500 dilution), or an Alexa 405 anti-mouse antibody (Invitrogen, A31553; 1:150 dilution). Actin filaments were visualized in samples fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton-X100 using Alexa 488 phalloidin (Life Technologies, P3457; 1:500 dilution) or Alexa 568 phalloidin (Life Technologies, A12380; 1:500 dilution). Images were captured as z-stacks (0.1-μm step size) on a Nikon Ti Eclipse microscope with a Yokogawa CSU-XI spinning disc confocal with 60X and 100X (1.4 NA) Plan Apo objectives, and a Clara Interline CCD Camera (Andor Technology) using MetaMorph software (Molecular Devices). Rendered Z-stacks were used for quantifications. Images were processed using ImageJ65 and assembled in Adobe Photoshop. Image adjustments of each color were applied equally for all bacterial strains within each experiment. To prepare representative images, we used a single optical section, in which most or all bacteria were in that focal plane. The only exceptions were for representative images of LAMP1 or LC3/pUb staining, for which we used z stack average maximum intensity projections.
For quantification of polyubiquitin, polyubiquitin linkages, p62 and NDP52 staining, only bacteria that co-localized with rim-like patterns of the respective marker were scored as positive for staining. For quantification of LC3 staining, bacteria that co-localized with either LC3 clouds or puncta were considered positive for staining. For macrophage infections that extended beyond 60 min, ompBSTOP::tn bacteria were occasionally not stained adequately with anti-Rickettsia antibodies, and therefore we included Hoechst (Sigma, B2261) staining in all experiments to detect bacterial genomic DNA.
To determine bacterial internalization frequencies, infections were carried out as described above except that bacteria were diluted in media at 4°C to a MOI of 5 in 500 μL of cell culture media, which was added to each well, and centrifuged at 300 x g for 5 min at 4°C. For the 0-min time point, coverslips were transferred to 4% paraformaldehyde immediately after the centrifugation step, fixed for 10 min and washed 3 times with 1 x PBS, pH 7.4. To initiate invasion/internalization, 500 μL media at 37°C was added to each well and the plate was transferred to 33°C. At 5, 10, 15, 20, 30, 60, and 90 min, cells were fixed, as described above. For immunofluorescence staining of extracellular bacteria, rabbit anti-Rickettsia I7205 antibody (or rabbit anti-Rickettsia R2868 antibody (a gift from Ted Hackstadt) for the ompA::tn mutant) and a secondary Alexa 405-conjugated anti-rabbit antibody (Invitrogen, A31556) were used. To subsequently stain all bacteria (intracellular and extracellular), cells were permeabilized with 0.5% Triton-X100 (Sigma, T9284) for 5 min and the anti-Rickettsia I7205 antibody (or rabbit anti-Rickettsia R2868 antibody to detect the ompA::tn mutant) and a secondary Alexa 488-conjugated goat anti-rabbit antibody (Invitrogen, A11008) were used. Coverslips were mounted with Prolong Gold anti-fade (Invitrogen, P36930) and stored at 4°C prior to imaging. Imaging was carried out as described above, and the percentage of internalized bacteria was calculated by first dividing the number of extracellular bacteria by the total number of bacteria, multiplying by 100, and then subtracting this number from 100% to get the percentage of intracellular bacteria.
Transmission electron microscopy (TEM)
For TEM experiments, infection of HMECs and BMDMs was performed as described above for internalization assays. At 1 hpi, media was removed and fixative (1.5% paraformaldehyde, 2.0% glutaraldehyde, 0.03% CaCl2, in 0.05 M cacodylate buffer pH 7.2) was added to each well for 45 min at room temperature, as previously described66. Samples were embedded in 2% low-melting agarose and incubated overnight at 4°C in 2.0% glutaraldehyde, 0.1 M cacodylate buffer, pH 7.2. The next day, samples were post-fixed in 1% osmium tetraoxide and 1.6% potassium ferricyanide, dehydrated in a graded series of ethanol concentrations, and embedded in EPON 812 resin (11.75 g of Eponate 12 (18005, Ted Pella Inc., Redding, CA), 6.25 g DDSA (18022, Ted Pella Inc., Redding, CA), 7 g NMA (18032, Ted Pella, Redding Inc., CA) were mixed prior addition of 0.375 ml of the embedding accelerator BMDA (18241, Ted Pella Inc., Redding, CA). Next, samples were stained with 2% uranyl acetate and lead citrate. Images were obtained on a FEI Tecnai 12 transmission electron microscope. The radius of the halo (rhalo or halo thickness) was calculated by measuring the area of the bacterium plus halo (Atotal) and bacterium minus halo (Abacterium), assuming circularity and calculating the radius of each as r = √A/π, and then calculating rhalo = rtotal − rbacterium67.
TUBE assay and sample preparation for mass spectrometry
To enrich for polyubiquitylated proteins, 4 × 108 PFUs of 30%-purified or 4 × 107 PFUs gradient-purified WT and ompBSTOP::tn bacteria were centrifuged at 20,000 x g for 3 min at room temperature. Next, to release the surface protein fraction, the bacterial pellets were resuspended in lysis buffer (50 mM Tris-HCI, pH 7.5, 150 mM NaCl, 1 mM EDTA and 10% glycerol), supplemented with 0.0031% v/v lysonase (Millipore, 71230), the deubiquitylase inhibitor PR619 at a final concentration of 20 μM (Life Sensor, SI9619) and 0.8% w/v octyl β-D-glucopyranoside (Sigma, O8001), and incubated on ice for 10 min. Subsequently, the lysate was cleared bycentrifugation at 14,000 x g at 4°C for 5 min and incubated with equilibrated agarose TUBE-1 (Life Sensor, UM401) for 4 h at 4°C. After binding of polyubiquitylated proteins to TUBE-1, agarose beads were washed 3 times with TBS (no Tween) and centrifuged at 5,000 x g for 5 min. To prepare samples for mass spectrometry (MS) analysis, enriched proteins were digested at 37°C overnight on agarose beads in RapiGest SF solution (Waters, 186001861) supplemented with ~0.5 μg trypsin (Promega, V511A). The reaction was stopped using 1% trifluoroacetic acid (Sigma, T6508). Octyl β-D-glucopyranoside was extracted using water-saturated ethyl acetate, as previously described68. Prior to submission of samples for MS analysis, samples were desalted using C18 OMIX tips (Agilent Technologies, A57003100), according to the manufacturer’s instructions.
Liquid chromatography-mass spectrometry
Samples of proteolytically digested proteins were analyzed using a Synapt G2-Si mass spectrometer that was equipped with a nanoelectrospray ionization source (Waters). The mass spectrometer was connected in line with an Acquity M-class ultra-performance liquid chromatography system that was equipped with trapping (Symmetry C18, inner diameter: 180 μm, length: 20 mm, particle size: 5 μm) and analytical (HSS T3, inner diameter: 75 μm, length: 250 mm, particle size: 1.8 μm) columns (Waters). Data-independent, ion mobility-enabled, high definition mass spectra and tandem mass spectra were acquired in the positive ion mode69–71. Data acquisition was controlled using MassLynx software (version 4.1), and tryptic peptide identification and relative quantification using a label-free approach72,73 were performed using Progenesis QI for Proteomics software (version 4.0, Waters). Raw data were searched against human (UniProt Knowledgebase, accessed December 19, 2015), Rickettsia parkeri (National Center for Biotechnology Information (NCBI), accessed December 20, 2015), and Chlorocebus sabaeus (NCBI, accessed January 15, 2016) protein databases to identify tryptic peptides, allowing for up to two missed proteolytic cleavages, with carbamidomethylcysteine as a fixed post-translational modification and methionine sulfoxide and diglycine-modified lysine (i.e., ubiquitylation remnant) as variable post-translational modifications.
Localization of tagged ubiquitin and ubiquitin pull-downs
To assess localization of 6xHis-ubiquitin during infection, confluent Vero cells grown in 24-well plates with coverslips were transfected with ~2 μg of pCS2–6xHis-ubiquitin plasmid DNA using Lipofectamine 2000 (Invitrogen, 11668–019) for 6 h in Opti-MEM (Gibco, 31985–070). Subsequently, media was exchanged to media without transfection reagent, and cells were incubated overnight at 37°C, 5% CO2. The following day (~16 h after transfection), transfected cells were infected with WT or ompBSTOP::tn bacteria at a MOI of 1. At 24 hpi, infected cells were fixed with 4% paraformaldehyde (Ted Pella Inc., 18505) diluted in PBS, pH 7.4 for 10 min, then washed 3 x with PBS. Primary anti-6xHis monoclonal mouse antibody (Clontech, 631212, diluted 1:1,000) was used to detect 6xHis-ubiquitin in samples permeabilized with 0.5% Triton-X100, and a goat anti-mouse Alexa-568 (Invitrogen, A11004) to detect the primary 6xHis antibody. Hoechst (Thermo Scientific, 62249, diluted 1:2500) was used to detect bacterial DNA. Samples were imaged as already described.
To validate OmpA as a polyubiquitylated substrate on ompBSTOP::tn bacteria, confluent Vero, A549 and HMEC cells grown in 6-well plates were transfected and infected as described above with the exception that HMECs and A549s were transfected for only 60 min. At 26 hpi, cells were washed once with 1 x PBS (this step was excluded for HMECs), pH 7.4, and subsequently lysed in urea lysis buffer (8 M urea, 50 mM Tris-HCI, pH 8.0, 300 mM NaCI, 50 mM Na2HPO4, 0.5% Igepal CA-630 (Sigma, I8896)) for 20 min at room temperature (for HMECs, the volume of lysis buffer was reduced 2x). Subsequently, samples were sonicated, and lysate was cleared by centrifugation at 15,000 x g for 15 min at room temperature. Prior to incubation with Ni-NTA resin, an aliquot was saved for the input sample. 6xHis-ubiquitin conjugates were purified by incubation and rotation with Ni-NTA resin for 4 h at room temperature. Beads were washed 3 x with urea lysis buffer and 1 x with urea lysis buffer lacking Igepal CA-630. Ubiquitin conjugates were eluted at 65°C for 15 min in 2 x Laemmli buffer containing 200 mM imidazole and 5% 2-mercaptoetanol (Sigma, M6250), vortexed for 1 min, and centrifugation at 5,000 x g for 3 min. Eluted and input proteins were detected by SDS-PAGE followed by western blotting, as described above.
Bacterial viability assay
To assess bacterial membrane integrity, confluent HMEC cells were infected with 30%-purified WT and ompBSTOP::tn bacteria, as described above, for invasion/internalization experiments. After centrifugation at 300 x g for 5 min at 4°C (considered the 0 min time point), cells were washed once in KHM buffer (110 mM CH3CO2K, 2 mM Mg(CH3COO)2 and 20 mM HEPES, pH 7.2), then incubated in KHM buffer with or without 50 μg/ml digitonin (positive control) for 1 min, and subsequently incubated in 2.6 μM propidium iodide (Molecular Probes, P-3560) diluted in KHM buffer for 10 min at room temperature, as previously described23,74. Cells were then washed once in KHM buffer, fixed in 4% paraformaldehyde, and stained for bacteria using the anti-Rickettsia 14–13 antibody and a secondary goat-anti-mouse Alexa-488 antibody (Invitrogen, A11004), as described above.
LDH-release host cell viability assay
Supernatants from infected or uninfected BMDMs were collected at 96 hpi and then mixed with 2x LDH buffer (100 μg/ml tetrazolium salt (Sigma I18377, in PBS), 150 μg/ml β-nicotineaminde adenine dinucleotide hydrate (Sigma, N3014), 0.675 units/ml diaphorase (Sigma, D5540), 0.06% sucrose, 12 mg/ml lithium lactate (Sigma L2250, resuspended in 10 mM Tris HCl pH 8.5 to 36 mg/ml), 0.25% BSA in PBS). Supernatant from uninfected cells lysed with 1% Triton-X100 was used to calculate 100% lysis. Reactions were incubated at room temperature for 20 min prior reading at 490 nm absorbance using an Infinite F200 Pro plate reader (Tecan). Values were divided by the difference of Triton-X100 lysed cells and the experimental supernatants, and multiplied by 100, to calculate percent lysis.
Animal experiments
Female C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA), and 8–10-week-old littermates were used for each experiment. Groups that received different bacterial strains were randomly assigned. C57BL/6 Atg5flox/flox mice were from Dr. N. Mizushima (Tokyo Medical and Dental University, Japan)44 and C57BL/6 Atg5flox/flox-LysMcre+ were a generous gift from Dr. H. Virgin (University of Washington)75. Mice were genotyped as previously described45 using primers exon3‑1, short2, and check2 for detection of the Atg5 gene and primers cre1 and cre2 for detection of the cre gene. Randomly assigned 8–10-week-old male and female C57BL/6, Atg5flox/flox, and Atg5flox/flox-LysMcre+ mice were used in the experiments comparing these genotypes. No data blinding was performed. The sample size for mice experiments was chosen based on the results between two groups after the first experiment, in which 2–3 mice per group were used. Mice were housed in specific pathogen-free conditions and euthanized according to standard protocols approved by UC Berkeley Animal Care and Use Committee.
Mice were infected with 107 PFU of WT or ompBSTOP::tn diluted into 200 μL of 1 x PBS, pH 7.4 (Gibco, 1001023) via injection into the lateral tail vein. Immediately after injection, to confirm the infectivity and titer of the bacterial strains, bacteria were enumerated using a plaque assay, as described above. At 2, 24, 48, and 72 hpi, mice were sacrificed, doused with ethanol, laid on the right side, and the spleen, liver, lung, and kidney were removed and transferred into cold sterile PBS. Organs were disrupted in a homogenizer (Polytron) for ~10 seconds and centrifuged at 200 x g for 5 min. Supernatants were collected to assess the PFUs per organ using a plaque assay, as described above (Fig. 6g). Due to moderate toxicity of the organ supernatants on Vero cells (particular from the liver), we optimized the protocol as follows. Instead of centrifuging the homogenized organs at 200 x g for 5 min, we employed 300 x g for 5 min. Subsequently, instead of centrifugation of supernatants at 300 x g for 5 min, which we normally use for R. parkeri infections, supernatants were centrifuged at 230 x g for 5 min. These minor changes reduced the toxic effects on Vero cells, leading to the detection of ompBSTOP::tn PFUs and increased WT PFUs (Fig. 6h).
To determine bacteria viability in mouse blood, we collected blood from the diaphragm cavity of female mice upon dissection. Next, 2 × 105 PFUs of WT or the ompBSTOP::tn strain were added to 40 μL of fresh blood and incubated at 37°C. At 0, 20, 60, 120, 180, and 360 min, samples were resuspended and 5 μL was taken for PFU determination using the plaque assay, as described above. PFUs were normalized to 0 min for each strain.
Statistical analysis
Statistical parameters and significance are reported in the figure legends. Data were considered to be statistically significant when p < 0.05, as determined by unpaired Student’s t-test, Mann-Whitney rank-sum test, or a one-way ANOVA with Tukey’s post-test. Statistical analysis was performed using PRISM 6 software (GraphPad Software).
DATA AVAILIBILITY
The datasets generated during and/or analysed during the current study are available from the corresponding authors upon request. The genome sequences of bacterial strains are available at Sequence Read Archive (SRA) as accession number SRP154218 (WT: SRX4401164; ompB::tn: SRX4401163; ompBSTOP::tn: SRX4401167; ompBPROM::tn; SRX4401166; MC1_RS02370STOP, ompB::tn: SRX4401165).
Extended Data
Extended Data Fig. 1. Isolation and characterization of suppressors of the parental ompB::tn strain.
(a) A graphic depiction of the scheme for the isolation of suppressor mutations. The parental ompB::tn strain was subjected to 10 serial passages in Vero cells (step 1). The mixed suppressor population was then tested for growth in HMECs (step 2) and compared with the parental ompB::tn strain. Clonal strains that appeared to have larger plaque phenotypes than parental ompB::tn were isolated from the two independent mixed suppressor populations, propagated in Vero cells, and deep sequenced (step 3). (b) Growth curves in HMECs of WT, parental ompB::tn, and ompB::tn suppressor population (passage 10, selection-1) (starting MOI: 0.002) as measured by genomic equivalents using qPCR. Data are means (n = 2). (c) Diagram of the ompB genomic regions and other relevant genes, showing the location of the transposon insertions (tn) in: the parental ompB::tn strain (at base pair (bp) position 1045462 in the R. parkeri genome); ompBSTOP::tn (with an additional 23-bp deletion at position 1045835–1045857); ompBPROM::tn (with an additional 17-bp insertion in the promoter region of ompB at position 1046683); and MC1_RS02370STOP, ompB::tn (with an additional two deletions, of 112 at position 461273–461384 and 125 bp at position 461425–461549 in a gene encoding a predicted glycosyltransferase (MC1_RS02370), leading to a premature stop codon). (d) Western blot of 106 30%-purified WT, ompB::tn and ompB suppressor strains probed for OmpB (anti-OmpB antibody). Reduced levels of truncated OmpB products were observed in the suppressor strains (n = 2). (e) Quantification of the percentage of bacteria that co-localize with polyubiquitin at 72 hpi. Data are mean ± SEM (WT, n = 3; ompBSTOP::tn, n = 2; parental ompB::tn, n = 3; MC1_RS02370STOP, ompB::tn, n = 2; ompBPROM::tn, n = 2; ≥136 bacteria were counted for each strain and infection). (f) Western blot of 106 30%-purified WT, ompBSTOP::tn and ompA::tn strains probed for OmpB (anti-OmpB antibody) and OmpA (13–3 antibody) (n = 1).
Extended Data Fig. 2. The host ubiquitin machinery is necessary to label the ompB mutant with ubiquitin chains.
(a) Immunofluorescence micrographs of infected HMEC cells treated with the E1-inhibitor PYR-41 (50 μM) or DMSO control for 6 h (66–72 hpi) and stained as in (Fig. 1a). Scale bars, 5 μm. (b) Quantification of the percentage of polyubiquitin-positive bacteria. Data are mean ± SEM (n = 3; statistical comparisons between PYR-41 treated and control ompBSTOP::tn and WT infections were determined by the unpaired Student’s t-test (two-sided); ***, p <0.001; * p <0.05; ≥229 bacteria were counted for each infection). (c) Quantification of the percentage of bacteria that showed rim-like surface localization of the indicated ubiquitin-chain at 72 hpi in HMECs based on staining with ubiquitin linkage-specific antibodies. Data are mean ± SEM (n = 4; statistical comparisons between the ompBSTOP::tn and WT bacteria were determined by the unpaired Student’s t-test (two-sided); ***, p < 0.001; ≥164 bacteria were counted per strain, ubiquitin-chain stain and experiment).
Extended Data Fig. 3. In HMECs, OmpA/Sca2/17-kDa-antigen are not required for R. parkeri invasion, and OmpB is not required for actin mobilization or bacterial membrane integrity.
(a) Quantification of the percentage of internalization into HMECs at 15 mpi, visualized by differential staining of extracellular versus total R. parkeri. Data are mean ± SEM (WT, n = 3; ompA::tn, n = 2; sca2::tn, n = 3; hrtA::tn, n = 3; ≥180 bacteria were counted for each strain and infection; means were not significantly different between WT, sca2::tn and hrtA::tn by a one-way ANOVA with Tukey’s post hoc-test). (b and c) Quantification of the percentage of bacteria with (b) early actin clouds or (c) actin tails. The delayed recruitment of actin can be explained by slower internalization of ompBSTOP::tn bacteria. Data are mean (n = 2; ≥84 bacteria were counted for each strain and infection for each time point). (d) Representative immunofluorescence micrographs of bacteria (blue; anti-Rickettsia 14–13 antibody) with late actin tails (green; Alexa 488 phalloidin). (e) Quantification of the percentage of WT and ompBSTOP::tn bacteria with late actin tails that started from the bacterial pole (green bar) or the side of the bacteria (brown bar). (n = 2; ≥274 bacteria were counted for each strain and infection). (f) Fluorescence micrographs of bacteria attached to HMECs prior to internalization, first stained with propidium iodide (PI; red) prior to fixation to label dead bacteria, and next fixed and stained for total bacteria (green; anti-Rickettsia 14–13 antibody). The bottom panels are a positive control in which 50 μg/ml digitonin was included to permeabilize bacteria and HMECs prior PI staining. Scale bars, 2 μm. (g) Quantification of the bacterial viability (membrane integrity) as determined by PI staining. Data are means (n = 2; ≥182 bacteria were counted for each strain and infection)
Extended Data Fig. 4. OmpB is critical to block ubiquitylation shortly after internalization into HMECs and BMDMs.
(a) Quantification of the percentage of internalization into BMDMs from 0–90 mpi, visualized by differential staining of extracellular versus total R. parkeri. Data are mean ± SEM (0, 15, 30, 60, 90 min, n = 3; 5, 10, 20 min, n = 3; ≥80 bacteria were counted for each strain, infection and time-point; statistical comparisons with WT for each time point were made by the unpaired Student’s t-test (two-sided); *, p < 0.05). (b) Quantification of the percentage of WT and ompBSTOP::tn mutant bacteria that co-localize with polyubiquitin from 0–60 mpi. Data are mean ± SEM (0, 5, 10, 20, 30, 60 min: n = 3; 15 min: n = 5); ≥80 bacteria were counted for each strain, infection, and time point; statistical comparisons with WT for each time point were made by the unpaired Student’s t-test (two-sided); *, p < 0.05; **, p < 0.01; ***, p < 0.001). (c) Immunofluorescence micrographs of BMDMs infected with WT and ompBSTOP::tn mutant at 60 mpi, stained for extracellular R. parkeri (blue; anti-Rickettsia I7205 antibody), total R. parkeri (green; anti-Rickettsia I7205 antibody), polyubiquitin (red; FK1 antibody), and a merged image. Arrows indicate an intracellular bacterium that is positive for polyubiquitin. The stars indicate an extracellular bacterium that is polyubiquitin-negative in BMDMs. Scale bars, 5 μm. (d) Quantification of the percentage of intracellular and extracellular WT and ompBSTOP::tn mutant bacteria that co-localize with polyubiquitin. Data are mean ± SEM (n = 3; ≥122 bacteria were counted for each strain and infection; statistical comparisons were by an unpaired Student’s t-test (two-sided); ***, p < 0.01). (e) Immunofluorescence micrographs of HMECs infected with WT and ompBSTOP::tn mutant at 60 mpi, stained as in (c). Arrows indicate an intracellular bacterium that is polyubiquitin-positive. Scale bars, 5 μm. (f) Quantification of the percentage of intracellular and extracellular WT and ompBSTOP::tn mutant bacteria that co-localize with polyubiquitin in HMECs. Data are mean ± SEM (WT, n = 2; ompBSTOP::tn, n = 3; ≥79 bacteria were counted for each strain and infection).
Extended Data Fig. 5. OmpB is required for R. parkeri to proliferate in BMDMs.
(a) Fluorescence micrographs of BMDMs infected with WT and ompBSTOP::tn bacteria at 48 hpi. Left panels show cellular and bacterial genomic DNA (blue; Hoechst). Right panels show bacteria (green; anti-Rickettsia antibody I7205), cellular and bacterial genomic DNA (blue, Hoechst), and actin (red; Alexa 568 phalloidin). Scale bars, 20 μm. (b) Quantification of bacteria per BMDM cell for WT, ompBSTOP::tn, and MC1_RS05535::tn bacteria, using Hoechst to count the number of cell nuclei and the number of bacteria. Data are mean (n = 2; 5 fields of view per infection and strain were used to count the number of bacteria per cell). (c) Fluorescence micrographs of BMDMs infected with WT and ompBSTOP::tn bacteria at 96 hpi, stained as in (a). Scale bars, 20 μm. (d-f) Quantification of (d) the mean number of bacteria per BMDM cell as in (b) (n = 2), (e) mean number of host cells per field of view (n = 2; 5 fields of view per infection and strain were used to count the number of host cell per field of view), and (f) the percentage LDH release, normalized to Triton-X100-lysed cells, all determined for WT, ompBSTOP::tn, and uninfected cells. Data are mean ± SEM (n = 3; statistical comparisons between WT and ompBSTOP::tn infected and uninfected cell were performed using a one-way ANOVA with Tukey’s post hoc-test; **, p < 0.01; ***, p < 0.001).
Extended Data Fig. 6. In BMDMs, neither OmpA nor the gene downstream of ompB is required for halo formation.
(a) TEM images of ompA::tn and MC1_RS05535::tn mutant bacteria in BMDMs at 1 hpi. Scale bars, 1 μm. (b) Quantification of halo thickness for WT (n = 79), ompA::tn (n = 12) and MC1_RS05535::tn (n = 17) bacteria. All data points are presented, and the lines indicate the means (statistical comparisons were performed using a Mann-Whitney rank-sum test (two-sided)). Data for WT are the same as that shown in Fig. 3.
Extended Data Fig. 7. Ubiquitylation of the ompB mutant is a prerequisite for LC3 recruitment and activation of autophagy does not increase LC3 recruitment to ompB mutant bacteria.
(a) Immunofluorescence micrographs showing cellular LC3 puncta (red; rabbit anti-LC3 antibody) that do not co-localize with bacteria (blue; Hoechst) in infected BMDMs. Stars indicate cellular LC3 puncta. (b) Quantification of cellular LC3 puncta per host cell at 2.5 hpi in BMDMs. Data are mean (n = 2; 5 fields of view per infection and strain were used to count the number of cellular puncta). (c) Quantification of the percentage of bacteria co-localizing with pUb in HMECs treated with 500 nM rapamycin (Rapa), 20 μM MG132 or DMSO control for 3 h (from 1 hpi to 4 hpi) and fixed at 4 hpi. Data are mean (n = 2; ≥87 ompBSTOP::tn bacteria were counted for each infection and treatment). (d) Quantification of the percentage of bacteria co-localizing with LC3 in HMECs treated with 500 nM rapamycin (Rapa), 20 μM MG132 or DMSO control for 3 h (1–4 hpi) and fixed at 4 hpi. Data are means (n = 2; ≥87 ompBSTOP::tn bacteria were counted for each infection and treatment). (e) Immunofluorescence micrographs of BMDMs infected with WT and ompBSTOP::tn mutant at 2.5 hpi, stained for R. parkeri (blue; Hoechst), polyubiquitin (green; FK1 antibody), LC3 (red; rabbit anti-LC3 antibody), and a merged image. Arrows indicate bacteria that are positive for both polyubiquitin and LC3. Scale bars, 5 μm. Related to Fig. 5k. (f, g) Quantification of the percentage of (f) polyubiquitin-positive bacteria and (g) bacteria with both LC3 and polyubiquitin, after 4 h treatment (1–5 hpi) with PYR-41. Data are means (n = 2; ≥110 ompBSTOP::tn bacteria were counted per infection and experiment).
Extended Data Fig. 8. Autophagy is the primary mechanism that restricts the growth of ompB mutant bacteria in macrophages, and OmpB protects R. parkeri from ubiquitylation in autophagy-deficient cells.
(a) Immunofluorescence micrographs of Atg5+/+ or Atg5−/− BMDMs infected with WT or ompBSTOP::tn bacteria at 72 hpi, stained for bacteria (green), cellular and bacterial genomic DNA (blue; Hoechst), polyubiquitin (red; FK1 antibody). Scale bars, 5 μm. (b) Quantification of the percentage of bacteria that co-localize with polyubiquitin in Atg5+/+, Atg5−/−, or Becn1−/− BMDMs at 72 hpi (nd indicates that too few ompBSTOP::tn bacteria were visualized in Atg5+/+ cells to quantify a percentage). Data are mean (n = 2; ≥250 bacteria were counted for each strain and infection). (c) Quantification of the percentage of bacteria that co-localize with polyubiquitin in C57BL/6 or Becn1−/− BMDMs at 1 hpi. Data are mean (n = 2; ≥84 bacteria were counted for each strain and infection). (d) Combined growth curves of WT and ompBSTOP::tn in Atg5+/+ or Atg5−/− BMDMs as measured by genomic equivalents using qPCR. Data are the same as that shown in Fig. 6a and b and are mean ± SEM (n = 4). (e) Growth curves of WT and ompBSTOP::tn in Rubicon−/− or Rubicon+/+ BMDMs as measured in (d). Data are mean (n = 2). (f) Quantification of the percentage LC3 and polyubiquitin-positive bacteria in C57BL/6 or Becn1−/− BMDMs at 1 hpi. Data are mean (n = 2; ≥84 bacteria were counted for each strain and infection). (g) Quantification of the percentage LC3-positive bacteria at 1, 2.5 and 4 hpi in BMDMs treated with 300 nM bafilomycin A (Baf) or corresponding amount of DMSO, starting at 1 hpi. Data are means (n = 2; ≥111 ompBSTOP::tn bacteria were counted per infection, experiment and time-point). (h,i) Quantification of the percentage (h) LC3-positive ompBSTOP::tn that also co-localize with polyubiquitin at 4 hpi, and (i) polyubiquitin-positive ompBSTOP::tn, after bafilomycin A treatment as in (g). Data are mean (n = 2; ≥119 ompBSTOP::tn bacteria were counted per infection and experiment).
Extended Data Fig. 9. In HMECs, OmpB is required for a subpopulation of R. parkeri to avoid trafficking to a LAMP1-positive compartment.
(a) Immunofluorescence micrographs of HMECs infected with WT or ompBSTOP::tn mutant at 2.5 hpi, and stained for R. parkeri (green; anti-Rickettsia I7205 antibody), cellular and bacterial genomic DNA (blue; Hoechst), and LAMP1 (red; anti-LAMP1 antibody). Scale bars, 5 μm. (b) Quantification of the percentage bacteria that co-localize with LAMP1 at 2.5 and 4 hpi in HMECs. Data are mean (n = 2; ≥106 bacteria were counted for each strain, infection and time point).
Extended Data Fig. 10. WT R. parkeri establishes an infection of mouse organs but is eventually cleared.
(a) C57BL/6 mice were infected with 107 PFUs of 30%-purified WT R. parkeri. At 1–7 d post infection, organs were harvested at the indicated time points and homogenized, and PFUs were counted. Medians are indicated as bars (1 d, n = 4; 2 d, n = 8; 3 d, n = 8; 4 d, n = 3; 6 d, n = 3; 7 d, n = 5). (b) Relative PFU counts for WT or ompBSTOP::tn bacteria following incubation in blood from uninfected mice at 37°C for the indicated times. PFUs were normalized to 0 mpi. Data are mean ± SEM (n = 2 for all time points, except 0 and 20 min, n = 3; statistical comparisons were by an unpaired Student’s t-test (two-sided) and no statistical difference was observed between WT and ompBSTOP::tn at 20 min; p = 0.46).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the labs of Daniel Portnoy and Russell Vance (University of California, Berkeley), Ted Hackstadt (NIH Rocky Mountain Laboratories), and Douglas Green (St. Jude Children’s Research Hospital) for generously providing us with equipment, reagents, and discussions. We thank Julie Choe and Vida Ahyong, other current and former members of the Welch lab, the lab of Michael Rape, and Allison Roberts and members of the lab of Jeff Cox, Reena Zalpuri, Kent McDonald, and other members of the UC Berkeley Electron Microscopy facility, for technical assistance and fruitful discussions. P.E. was supported by postdoctoral fellowships from the Foundation Olle Engkvist Byggmästare, the Swedish Society of Medical Research, and the Sweden-America Foundation. M.D.W. is supported by NIH/NIAID grants R01 AI109044 and R21 AI138550. G.M. was supported by fellowships from Fonds de recherche du Québec - Nature et technologies, Fonds de recherche du Québec - Santé, and the Natural Sciences and Engineering Research Council of Canada. J.S.C. was supported by NIH grant P01 AI063302 and NIH grant R01 AI120694. K.G.M was supported by a Ruth L. Kirschstein National Research Service Award (F32 GM120956). A mass spectrometer used in this study was purchased with support from the NIH (grant 1S10 OD020062-01). N.I. was supported by the UC Berkeley Amgen Scholars Program and the Amgen Foundation. M.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING INTEREST
The authors declare no competing interest.
REFERENCES
- 1.Burgdorfer W, Friedhoff KT & Lancaster JL Jr. Natural history of tick-borne spotted fever in the USA. Susceptibility of small mammals to virulent Rickettsia rickettsii. Bull. World. Health. Organ 35, 149–153 (1966). [PMC free article] [PubMed] [Google Scholar]
- 2.Wright CL, Sonenshine DE, Gaff HD & Hynes WL. Rickettsia parkeri Transmission to Amblyomma americanum by Cofeeding with Amblyomma maculatum 491(Acari: Ixodidae) and Potential for Spillover. J. Med. Entomol 52, 1090–1095, doi: 10.1093/jme/tjv086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krawczak FS & Labruna MB. The rice rat Euryoryzomys russatus, a competent amplifying host of Rickettsia parkeri strain Atlantic rainforest for the tick Amblyomma ovale. Ticks Tick Borne Dis. 9, 1133–1136, doi: 10.1016/j.ttbdis.2018.04.013 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Walker DH & Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol 6, 375–386, doi: 10.1038/nrmicro1866 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Paddock CD et al. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin. Infect. Dis 47, 1188–1196, doi: 10.1086/592254 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Fang R et al. Differential interaction of dendritic cells with Rickettsia conorii: impact on host susceptibility to murine spotted fever rickettsiosis. Infect. Immun 75, 3112–3123, doi: 10.1128/IAI.00007-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley SP et al. Nonselective Persistence of a Rickettsia conorii Extrachromosomal Plasmid during Mammalian Infection. Infect. Immun 84, 790–797, doi: 10.1128/IAI.01205-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curto P, Simoes I, Riley SP & Martinez JJ. Differences in Intracellular Fate of Two Spotted Fever Group Rickettsia in Macrophage-Like Cells. Front. Cell. Infect. Microbiol 6, 80, doi: 10.3389/fcimb.2016.00080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randow F & Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411, doi: 10.1016/j.chom.2014.03.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galluzzi L et al. Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836, doi: 10.15252/embj.201796697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin AJ, Jiang X, Birmingham CL, So NS & Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr. Biol 14, 806–811, doi: 10.1016/j.cub.2004.04.033 (2004). [DOI] [PubMed] [Google Scholar]
- 12.van Wijk SJ et al. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains incells. Mol. Cell 47, 797–809, doi: 10.1016/j.molcel.2012.06.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Wijk SJL et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-kappaB and restricts bacterialproliferation. Nat. Microbiol 2, 17066, doi: 10.1038/nmicrobiol.2017.66 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zheng YT et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183, 5909–5916, doi: 10.4049/jimmunol.0900441 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Mostowy S et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem 286, 26987–26995, doi: 10.1074/jbc.M111.223610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N & Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol 10, 1215–1221, doi: 10.1038/ni.1800 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Liang XH et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676, doi: 10.1038/45257 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Kihara A, Kabeya Y, Ohsumi Y & Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335, doi: 10.1093/embo-reports/kve061 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichimura Y et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492, doi: 10.1038/35044114 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Hanada T et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem 282, 37298–37302, doi: 10.1074/jbc.C700195200 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Huang J & Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol 12, 101–114, doi: 10.1038/nrmicro3160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa Y et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11, 1233–1240, doi: 10.1038/ncb1967 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Case ED et al. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell. Microbiol 16, 862–877, doi: 10.1111/cmi.12246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa M et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731, doi: 10.1126/science.1106036 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Cheng MI, Chen C, Engstrom P, Portnoy DA & Mitchell G. Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell. Microbiol 20, e12854, doi: 10.1111/cmi.12854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchiyama T, Kishi M & Ogawa M. Restriction of the growth of a nonpathogenic spotted fever group rickettsia. FEMS Immunol. Med. Microbiol 64, 42–47, doi: 10.1111/j.1574-695X.2011.00879.x (2012). [DOI] [PubMed] [Google Scholar]
- 27.Bechelli J et al. Atg5 Supports Rickettsia australis Infection in Macrophages In Vitro and In Vivo. Infect Immun 87, e00651–18 doi: 10.1128/IAI.00651-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anacker RL, List RH, Mann RE, Hayes SF & Thomas LA. Characterization of monoclonal antibodies protecting mice against Rickettsia rickettsii. J. Infect. Dis 151, 1052–1060 (1985). [DOI] [PubMed] [Google Scholar]
- 29.Noriea NF, Clark TR & Hackstadt T. Targeted knockout of the Rickettsia rickettsia OmpA surface antigen does not diminish virulence in a mammalian model system. MBio 6, e00323–00315, doi: 10.1128/mBio.00323-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackstadt T, Messer R, Cieplak W & Peacock MG. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun 60, 159–165 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleba B, Clark TR, Lutter EI, Ellison DW & Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun 78, 2240–2247, doi: 10.1128/IAI.00100-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haglund CM, Choe JE, Skau CT, Kovar DR & Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12, 1057–1063, doi: 10.1038/ncb2109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed SC, Lamason RL, Risca VI, Abernathy E & Welch MD. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr. Biol 24, 98–103, doi: 10.1016/j.cub.2013.11.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson BE. The 17-kilodalton protein antigens of spotted fever and typhus group rickettsiae. Ann. N. Y. Acad. Sci 590, 326–333 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Dasch GA, Samms JR & Williams JC. Partial purification and characterization of the major species-specific protein antigens of Rickettsia typhi and Rickettsia prowazekii identified by rocket immunoelectrophoresis. Infect Immun 31, 276–288 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Policastro PF & Hackstadt T. Differential activity of Rickettsia rickettsii ompA and ompB promoter regions in a heterologous reporter gene system. Microbiology 140 (Pt 11), 2941–2949, doi: 10.1099/13500872-140-11-2941 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Riley SP, Pruneau L & Martinez JJ. Evaluation of changes to the Rickettsia rickettsii transcriptome during mammalian infection. PLoS One 12, e0182290, doi: 10.1371/journal.pone.0182290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamason RL, Kafai NM & Welch MD. A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS One 13, e0197012, doi: 10.1371/journal.pone.0197012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruneda JN et al. The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases. Mol. Cell 63, 261–276, doi: 10.1016/j.molcel.2016.06.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordureau A et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 56, 360–375, doi: 10.1016/j.molcel.2014.09.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman DJ & Wisseman CL Jr. Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi. Infect. Immun 21, 1020–1023 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silverman DJ, Wisseman CL Jr., Waddell AD & Jones M. External layers of Rickettsia prowazekii and Rickettsia rickettsii: occurrence of a slime layer. Infect. Immun 22, 233–246 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez J et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17, 893–906, doi: 10.1038/ncb3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Hara T et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889, doi: 10.1038/nature04724 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Zhao Z et al. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3, 581–585, doi: 10.4161/auto.4782 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Gawriluk TR et al. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 141, 759–765, doi: 10.1530/REP-10-0489 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Riley SP, Patterson JL & Martinez JJ. The rickettsial OmpB beta-peptide of Rickettsia conorii is sufficient to facilitate factor H-mediated serum resistance. Infect. Immun 80, 2735–2743, doi: 10.1128/IAI.00349-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiskin E, Bionda T, Dikic I & Behrends C. Global Analysis of Host and Bacterial Ubiquitinome in Response to Salmonella Typhimurium Infection. Mol. Cell 62, 967–981, doi: 10.1016/j.molcel.2016.04.015 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Walker DH, Popov VL, Crocquet-Valdes PA, Welsh CJ & Feng HM. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab Invest 76, 129–138 (1997). [PubMed] [Google Scholar]
- 50.Osterloh A. Immune response against rickettsiae: lessons from murine infection models. Med Microbiol Immunol 206, 403–417, doi: 10.1007/s00430-017-0514-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshikawa Y, Ogawa M, Hain T, Chakraborty T & Sasakawa C. Listeria monocytogenes ActA is a key player in evading autophagic recognition. Autophagy 5, 623 1220–1221, doi: 10.4161/auto.5.8.10177 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Cemma M, Lam GY, Stockli M, Higgins DE & Brumell JH. Strain-Specific Interactions of Listeria monocytogenes with the Autophagy System in Host Cells. PLoS One 10, e0125856, doi: 10.1371/journal.pone.0125856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arpaia N et al. TLR signaling is required for Salmonella typhimurium virulence. Cell 144, 675–688, doi: 10.1016/j.cell.2011.01.031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell G et al. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun 83, 2175–2184, doi: 10.1128/IAI.00110-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grasperge BJ et al. Susceptibility of inbred mice to Rickettsia parkeri. Infect. Immun 80, 1846–1852, doi: 10.1128/IAI.00109-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamason RL et al. Rickettsia Sca4 Reduces Vinculin-Mediated Intercellular Tension to Promote Spread. Cell 167, 670–683 e610, doi: 10.1016/j.cell.2016.09.023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anacker RL, Mann RE & Gonzales C. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J. Clin. Microbiol 25, 167–171 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welch MD, Reed SC, Lamason RL & Serio AW. Expression of an epitope-tagged virulence protein in Rickettsia parkeri using transposon insertion. PLoS One 7, e37310, doi: 10.1371/journal.pone.0037310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felsheim RF, Kurtti TJ & Munderloh UG. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 4, e8361, doi: 10.1371/journal.pone.0008361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thorvaldsdottir H, Robinson JT & Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192, doi: 10.1093/bib/bbs017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winger JA, Derbyshire ER, Lamers MH, Marletta MA & Kuriyan J. The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase. BMC Struct. Biol 8, 42, doi: 10.1186/1472-6807-8-42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harlow E & Lane D. Purification of antibodies on an antigen column. CSH. Protoc 2006, doi: 10.1101/pdb.prot4284 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Jeng RL et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell. Microbiol 6, 761–769, doi: 10.1111/j.1462-5822.2004.00402.x (2004). [DOI] [PubMed] [Google Scholar]
- 64.Yau RG et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 171, 918–933 e920, doi: 10.1016/j.cell.2017.09.040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider CA, Rasband WS & Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Labruna MB et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J. Clin. Microbiol 42, 90–98, doi: 10.1128/jcm.42.1.90-98.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Rodas R et al. Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. MBio 5, e00945–00914, doi: 10.1128/mBio.00945-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeung YG & Stanley ER. Rapid detergent removal from peptide samples with ethyl 666 acetate for mass spectrometry analysis. Curr Protoc Protein Sci 16.12.1–16.12.5, doi: 10.1002/0471140864.ps1612s59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Distler U et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods 11, 167–170, doi: 10.1038/nmeth.2767 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Plumb RS et al. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun Mass Spectrom 20, 1989–1994, doi: 10.1002/rcm.2550 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Shliaha PV, Bond NJ, Gatto L & Lilley KS. Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res 12, 2323–2339, doi: 10.1021/pr300775k (2013). [DOI] [PubMed] [Google Scholar]
- 72.Nahnsen S, Bielow C, Reinert K & Kohlbacher O. Tools for label-free peptide quantification. Mol Cell Proteomics 12, 549–556, doi: 10.1074/mcp.R112.025163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neilson KA et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553, doi: 10.1002/pmic.201000553 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Checroun C, Wehrly TD, Fischer ER, Hayes SF & Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 103, 14578–14583, doi: 10.1073/pnas.0601838103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Z et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469, doi: 10.1016/j.chom.2008.10.003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.