Rhizobia are soil bacteria from unrelated genera able to form a mutualistic relationship with legumes. Bacteria induce the formation of root nodules, invade nodule cells, and fix nitrogen to the benefit of the plant. Rhizobial lineages emerged from the horizontal transfer of essential symbiotic genes followed by genome remodeling to activate and/or optimize the acquired symbiotic potential. This evolutionary scenario was replayed in a laboratory evolution experiment in which the plant pathogen Ralstonia solanacearum successively evolved the capacities to nodulate Mimosa pudica and poorly invade, then massively invade, nodule cells. In some lines, the improvement of intracellular infection was achieved by mutations modulating a quorum sensing regulatory system of the ancestral strain. This modulation that affects the activity of a central regulator during the earliest stages of symbiosis has a huge impact on late stages of symbiosis. This work showed that regulatory rewiring is the main driver of this pathogeny-symbiosis transition.
KEYWORDS: symbiosis, rhizobium, experimental evolution, infection, quorum sensing, regulation
ABSTRACT
Over millions of years, changes have occurred in regulatory circuitries in response to genome reorganization and/or persistent changes in environmental conditions. How bacteria optimize regulatory circuitries is crucial to understand bacterial adaptation. Here, we analyzed the experimental evolution of the plant pathogen Ralstonia solanacearum into legume symbionts after the transfer of a natural plasmid encoding the essential mutualistic genes. We showed that the Phc quorum sensing system required for the virulence of the ancestral bacterium was reconfigured to improve intracellular infection of root nodules induced by evolved Ralstonia. A single mutation in either the PhcB autoinducer synthase or the PhcQ regulator of the sensory cascade tuned the kinetics of activation of the central regulator PhcA in response to cell density so that the minimal stimulatory concentration of autoinducers needed for a given response was increased. Yet, a change in the expression of a PhcA target gene was observed in infection threads progressing in root hairs, suggesting early programming for the late accommodation of bacteria in nodule cells. Moreover, this delayed switch to the quorum sensing mode decreased the pathogenicity of the ancestral strain, illustrating the functional plasticity of regulatory systems and showing how a small modulation in signal response can produce drastic changes in bacterial lifestyle.
INTRODUCTION
Most bacterial cellular functions are regulated by complex multilayered genetic regulatory networks that include environmental and internal sensors and a wide variety of signaling pathways. Fine-tuned regulation allows rapid shifts in metabolism, physiology, and behavior in response to environmental fluctuations (1–3). Over millions of years, the regulation circuits have evolved to adapt to genome reorganization and/or persistent changes in environmental conditions (4). Deciphering how bacteria optimize regulatory circuitries is crucial to understand bacterial adaptation and the biosphere.
Quorum sensing (QS) systems are key components of regulatory networks in bacteria which allow the regulation of gene expression in a population-dependent manner. QS involves the production of extracellular signals, called autoinducers, whose concentration is a function of cell population density. Above a system-specific threshold level, these molecules rapidly activate or repress the transcription of hundreds of genes, thus modifying and synchronizing bacterial behavior on a population-wide scale. These signal response systems are widespread in both Gram-positive and Gram-negative bacteria, where extensive variations regarding the type of autoinducers and cognate receptors, signal transduction mechanisms, and associated cellular responses have been described (5). Biological functions regulated by QS are numerous, including bioluminescence, virulence, symbiosis, motility, biofilm formation, or exopolysaccharide (EPS) production (6–10). The plant pathogen Ralstonia solanacearum possesses a virulence-related QS system, the Phc system, which controls the activity of the LysR-type transcription regulator PhcA (11). One component of this system is PhcB, a protein harboring methyltransferase activity essential for the synthesis of autoinducers, either (R)-3-hydroxypalmitic acid methyl ester [(R)-3-OH PAME] or (R)-3-hydroxymyristic acid methyl ester [(R)-3-OH MAME], depending on the R. solanacearum strains. In strain GMI1000, only (R)-3-OH MAME was detected (12). How this signal is transduced to PhcA has not been fully elucidated. At low cell density, the unphosphorylated two-component system PhcSR is thought to inhibit the activity of the central virulence regulator PhcA via an unknown mechanism. When the QS molecules reach the minimal stimulatory concentration (around 10 nM, matching the cell density of 107 CFU/ml in liquid culture), they likely trigger the autophosphorylation of the sensor histidine kinase PhcS, which, in turn, phosphorylates the response regulator PhcR (13), and this releases the inhibition of PhcA (14). The phcQ gene, which is likely cotranscribed with phcBSR, encodes a protein composed of a receiver domain similar to the receiver domain of PhcR (see Fig. S1A in the supplemental material). The role of this protein in the QS-dependent signaling cascade is not known (14). Activation of PhcA in R. solanacearum allows the switch from metabolic versatile cells to nonmotile EPS-producing virulent cells (15). Three independent transcriptomic studies conducted under three different experimental conditions (minimal medium, rich medium, and in planta) indeed identified hundreds of genes under the positive or negative control of PhcA (16–18).
(A) Representation of the R. solanacearum phcBSRQ operon and structural domains of the corresponding proteins. SAMdM, S-adenosyl-l-methionine-dependent methyltransferase; TMD, transmembrane domains; D/PAD. dimerization/phosphoacceptor domain; HK. histidine kinase; RD, receiver domain. Arrows indicate the mutated positions in phcB (R22C), phcS (L161R), and phcQ (R154C). (B) Prediction of the 3-dimensional structures of the wild-type and mutated PhcB and PhcQ proteins. Structures were predicted using the Phyre2 server (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015, https://www.doi.org/10.1038/nprot.2015.053). (C) Protein alignments showing the conservation of the R22 and R154 amino acids of the PhcB and PhcQ proteins, respectively. The amino acid sequences of PhcB/Q from R. solanacearum GMI1000 were blasted against the nonredundant NCBI database to obtain a list of homologs from other organisms with identity ranging from 64% to 100% for PhcB and 53% to 100% for PhcQ. A subset of hits, covering the diversity of genera and species containing phcB and phcQ homologs, is presented. Sequences were aligned using MultAlin (http://multalin.toulouse.inra.fr/multalin/multalin.html). Red, amino acid residues highly conserved (>90%); black, positions neutrally conserved (50% to 90%); blue, positions lowly conserved (<50%). Download FIG S1, PDF file, 0.6 MB (618.5KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Rhizobia, the nitrogen-fixing symbionts of legumes, are an excellent biological system to investigate how regulatory circuits rewire in response to genome reorganization and colonization of new niches. Rhizobia are soil bacteria able to induce the formation of root nodules where internalized bacteria fix nitrogen for the benefit of the plant (19). Extant rhizobia belong to hundreds of species distributed in 18 different genera of alpha- and betaproteobacteria (20–22). They have evolved through the horizontal acquisition of essential symbiotic genes, the nodulation (nod) and nitrogen fixing (nif-fix) genes, likely followed by subsequent adaptation of the recipient genome to the new legume endospheric environment (20). To understand posthorizontal gene transfer adaptation, we previously experimentally evolved Ralstonia solanacearum into legume symbionts by mimicking the natural evolutionary scenario. The symbiotic plasmid pRalta of the rhizobium Cupriavidus taiwanensis LMG19424 harboring the nod and nif-fix genes (23) was transferred to R. solanacearum strain GMI1000, and the resulting chimera was then evolved in parallel lineages using serial plant-bacteria cocultures. While the ancestral strain was strictly extracellular and unable to form root nodules, most lines sequentially acquired the capacity to form root nodules, poorly infect nodules, and massively infect nodule cells (24, 25). Acquisition of nodulation relied on the inactivation of the R. solanacearum pathogenic type III secretion system (T3SS) (24). The first level of infection was gained via inactivation of virulence regulators, either HrpG or VsrA (24, 26). In some lines, infection was optimized via modifications in the EfpR regulatory pathway (27). EfpR was shown to be a global virulence regulator and a metabolic repressor (27, 28). In other very infectious lines, no mutation was detected in known components of the EfpR pathway, suggesting that evolution targeted either unknown components of this path or another pathway.
Here, we show that in some of these lines, massive intracellular accommodation was reached via modifications in the R. solanacearum quorum sensing system. Mutations in either phcB or phcQ allowed improved infection via a fine modulation of the sensory pathway that delays to different degrees the cell density-dependent activation of the central regulator PhcA. While the highly similar gene expression profiles of efpR and phcA mutants suggested a connection between the two pathways, present results indicated that EfpR interferes with the PhcA regulon independently from the PhcBSRQ sensory cascade. Both efpR and phc late infection-adaptive mutations transiently affect the expression of PhcA target genes during early symbiotic stages corresponding to the entry and progression of bacteria in root hairs. EfpR and PhcA may thus control the expression of a common set of genes whose deregulation helps change the lifestyle of the bacterium, improving its symbiotic infection capacity while decreasing its pathogenicity.
RESULTS
Infection-adaptive mutations affect the phc system in E, K, and M lineages.
Our previous work showed that the evolved clones E16, K16, and M16 bore adaptive mutations in genes encoding components of the Ralstonia solanacearum Phc quorum sensing system. Nonsynonymous mutations in the phcB (phcBR22C), phcQ (phcQR154C), and phcS (phcSL161R) genes (Table 1) improved the in planta fitness (evaluated by the number of bacteria in nodule populations) of the evolved clones compared to that of the derivatives harboring the wild-type alleles (29). Genome resequencing or PCR analysis of intermediate clones along the E, K, and M lineages allowed the detection of the phcBR22C mutation in half clones from cycle 10 in the E lineage and the detection of phcQR154C and phcSL161R mutations in all clones from cycles 13 and 2 in the K and M lineages, respectively (Fig. 1). This suggested that these mutations have fixed rapidly in the K and M lineages and less rapidly in the E lineage.
TABLE 1.
Validated infection-adaptive mutations in E, K, and M lineages
| Strain(s) | Gene | Product | Mutation | Position on the chromosome | Protein modification |
|---|---|---|---|---|---|
| E16 | RSc2735, phcB | Regulatory protein, SAM-dependent methyltransferase domain | C/T | 2943477 | R22C |
| K16 | RSc2738, phcQ | Response regulator receiver, CheY family | C/T | 2947813 | R154C |
| M5, M16 | RSc2736, phcS | Sensor protein histidine kinase, repressor of PhcA | T/G | 2945312 | L161R |
FIG 1.
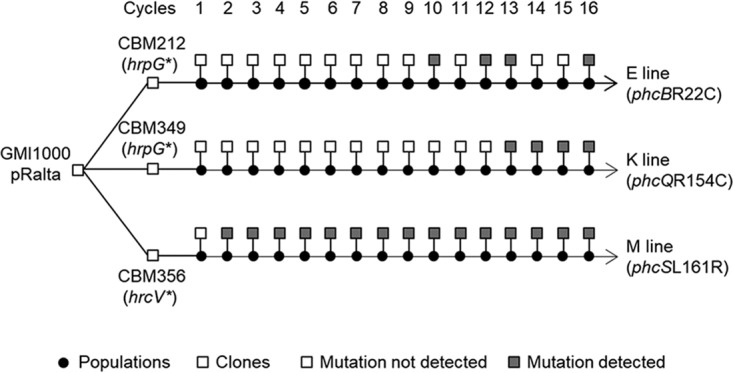
E, K, and M lineages of the experimental evolution of R. solanacearum GMI1000. The E, K, and M lines were independently derived from the spontaneous nodulating mutants of GMI1000(pRalta) CBM212, CBM349, and CBM356, respectively. Sixteen serial cycles of inoculation to M. pudica-isolation of nodule bacteria were performed for each lineage (30). At each cycle, a sample of the nodule bacterial population was inoculated to new plants and the rest of the population stored at −80°C. Populations were serially diluted, and one representative clone was randomly selected from the highest dilution plate for further phenotypic and genomic analyses. Bacteria of these lines carry a stop mutation in either hrpG or hrcV (24), depending on the ancestor, which provides strains the capacity to nodulate M. pudica. The phcBR22C, phcQR154C, and phcSL161R mutations were identified in clones isolated from the E, K, and M lines, respectively, by either Illumina genome resequencing, PCR amplification and Sanger sequencing, or PCR screening of wild-type and mutated alleles (see Text S1 in the supplemental material).
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB (27.3KB, docx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
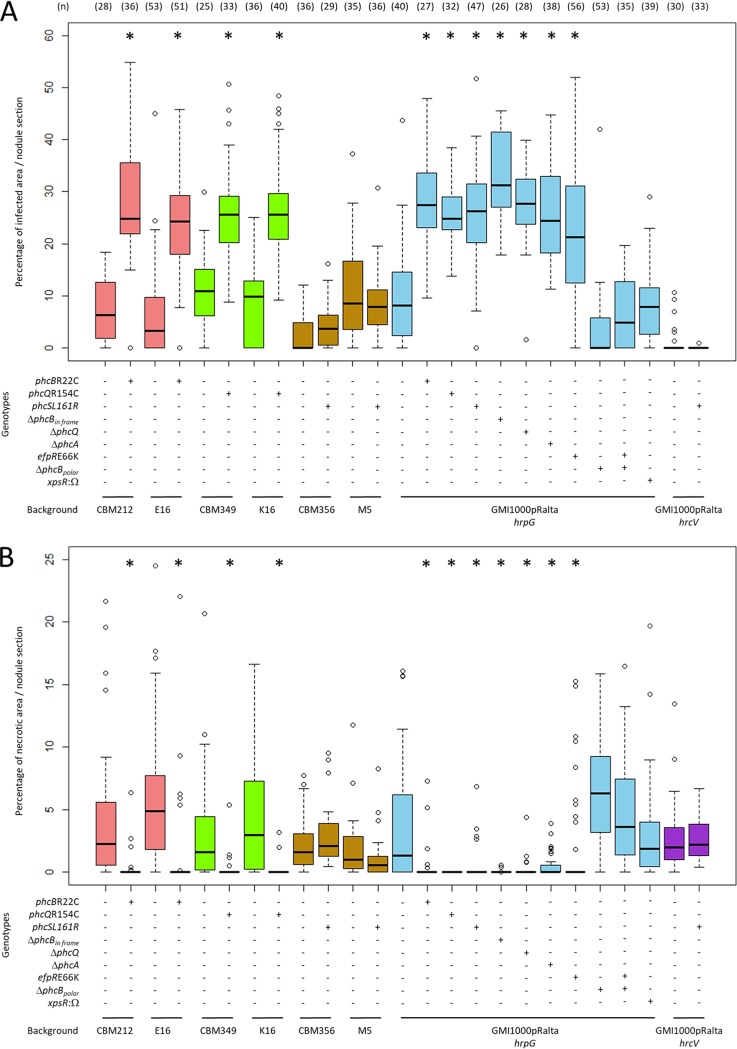
To further determine whether these mutations improve nodule cell infection, we inoculated Mimosa pudica individually with E16, K16, M5, their derivatives harboring the wild-type (WT) phc alleles, their respective nodulating ancestors CBM212, CBM349, and CBM356, and mutants of these ancestors carrying the phc mutations (M5, which is statistically as infectious as M16 [26], was used rather than M16, since genetic transformation failed in the latter clone in spite of many trials). We measured the size of the intracellularly infected and necrotic zones, identified as light-brown and dark-brown zones, respectively (25, 30), in sections of nodules collected at 21 days postinoculation (dpi). These two parameters were used as indicators of the quantity and quality of infection. The intracellular infection capacity of E16, CBM212 phcBR22C, K16, and CBM349 phcQR154C increased significantly compared to that of E16 phcBWT, CBM212, K16 phcQWT, and CBM349, respectively (Fig. 2A). At the same time, the capacity to induce necrosis decreased in the strains bearing the mutated alleles (Fig. 2B). Reconstructing the phcB and phcQ mutations in the chimeric Ralstonia strain GMI1000(pRalta) harboring an hrpG mutation (CBM1627), conferring nodulation and partial infection ability, provided this strain a capacity to infect nodule cells and induce necrosis similarly to that of the evolved E16 or K16 clones, respectively, indicating that these mutations are the major infection-adaptive ones in E and K lines (Fig. 2).
FIG 2.
Quantification of the relative infected and necrotic areas of nodules induced by evolved clones and mutants. Distributions of the percentages of infected areas (A) and necrotic areas (B) per nodule section recovered at 21 dpi. The central rectangles span the first quartiles to the third quartiles (that is, the interquartile range [IQR]), the bold segments inside the rectangles show the medians, unfilled circles indicate outliers, whiskers above and below the boxes show either the locations of the minimum and maximum in the absence of suspected outlying data or 1.5 IQR if an outlier is present. ΔphcBin frame, nonpolar deletion of phcB. Data for efpRE66K are from reference 25. *, P < 0.05 versus the respective ancestral strain, either CBM212, CBM349, CBM356, or GMI1000(pRalta) hrpG (multiple-comparison test after Kruskal-Wallis). Nodules were obtained from at least two independent experiments. For each experiment, nodules were harvested from at least 16 plants. n, number of nodules analyzed.
The effect of the phcSL161R mutation depended on the genetic background (Fig. 2). It improved intracellular infection and reduced necrosis in GMI1000(pRalta) hrpG but failed to do so in hrcV-mutated backgrounds [M5, CBM356, and GMI1000(pRalta) hrcV]. This result is not necessarily contradictory to the previously observed increase in relative in planta fitness of CBM356 phcSL161R and M5 compared to that of CBM356 and M5 phcSWT (29), which could be due to an increase in nodulation competitiveness or extracellular infection rather than an increase in intracellular infection per se.
The reconstruction of the phcBQS mutations in GMI1000(pRalta) did not allow the original chimeric strain to nodulate M. pudica, indicating that the effect of these mutations is conditional on the presence of the hrpG stop mutation.
Altogether, this showed that the phcBR22C and phcQR154C, but not phcSL161R, were the main infection-enhancing mutations in their respective lineages. We thus further analyzed the effect of the phcB and phcQ mutations on the Phc quorum sensing system.
Infection-adaptive phcBQ mutations do not constitutively inactivate the PhcA transcription regulator.
The cell density-responsive regulatory system PhcBSRQ controls the PhcA central regulator (11). To evaluate whether the infection-adaptive phc mutations led to the inactivation of the Phc quorum sensing system, we evaluated the activity of PhcA in wild-type and mutated strains. For that, we constructed a plasmidic fusion between the promoter region of xpsR, a direct transcriptional target of PhcA (31), and the Escherichia coli reporter gene lacZ encoding β-galactosidase. We introduced this plasmid in GMI1000(pRalta) hrpG, in its phcBR22C, phcQR154C single nucleotide polymorphism (SNP) derivative mutants, and in its phcB- (in frame), phcQ-, and phcA-deleted mutants. Since xpsR expression is under positive control of PhcA, the dosage of β-galactosidase activity positively correlates with PhcA activity.
The β-galactosidase activity was determined from mid-exponentially grown liquid cultures of each strain in rich medium. In agreement with the literature (31), deletion of phcA completely abolished xpsR-lacZ expression (Fig. 3). In contrast, the expression of xpsR-lacZ in the phcB SNP mutants was high and similar to that of strain GMI1000(pRalta) hrpG, and the phcQ SNP mutant only exhibited a lower level of reporter gene expression (Fig. 3). The phcB SNP mutant was clearly different from a phcB in-frame deletion mutant, indicating that the mutation did not inactivate the protein. In contrast, the phcQ deletion mutant exhibited a similar level of xpsR-lacZ reporter gene expression as the phcQR154C mutant.
FIG 3.
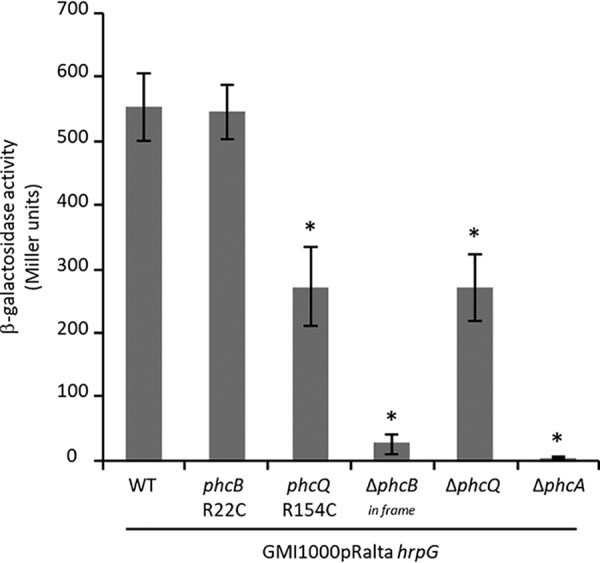
Expression of the pxpsR-lacZ plasmidic fusion in the chimeric R. solanacearum nodulating strain and derivative mutants at mid-exponential phase. β-Galactosidase activities were measured on bacteria grown to mid-exponential phase (OD600 was around 0.2) in rich BG medium. Data are from 3 independent experiments. Values correspond to means ± standard deviations. ΔphcBin frame, nonpolar deletion of phcB. *, P < 0.05; **, P < 0.001 versus the wild-type strain (Student’s t test).
Surprisingly, the three ΔphcA, ΔphcBin frame, and ΔphcQ mutants induced on M. pudica the formation of nodules with infection and necrosis zones similar to that of nodules induced by the phcBQ SNP mutants (around 25% to 30% of the nodule section) (Fig. 2). Moreover, the phcB and phcQ SNP mutants display either no or a minor difference in relative in planta fitness from that of a ΔphcA mutant, indicating that a phcA deletion is almost as adaptive as the phcBR22C and phcQR154C mutations for symbiosis (Fig. 4). This suggested that the point mutations in phcB and phcQ affect respective protein functions, resulting in an altered PhcA activity in symbiosis.
FIG 4.
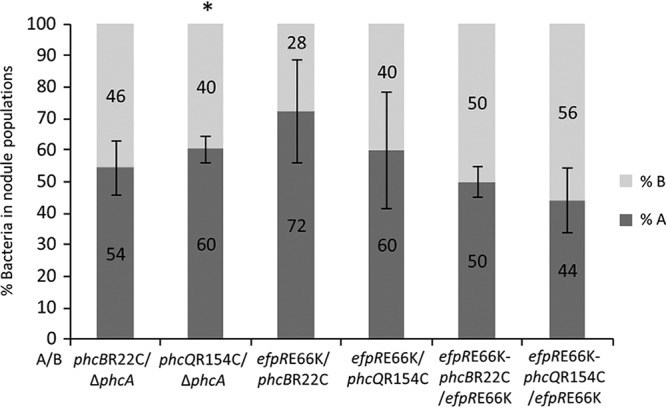
Relative in planta fitness of infection-adaptive mutants. Mimosa pudica plantlets were coinoculated with pairs of mutants at a 1:1 ratio. Nodules were harvested at 21 dpi, and the number of bacteria from each strain in the bacterial nodule population was measured by plating dilutions of nodule crushes and using either PCR for detecting the presence/absence of mutations in colonies or fluorescent markers as screening methods. The percentages of bacteria in nodule populations were normalized by the inoculum ratio. Values correspond to means ± standard deviations. *, P < 0.05 between strains (Student’s t test).
Infection-adaptive phc mutations decrease responsiveness to cell density.
The phcBR22C and phcQR154C mutations are not located in the known functional domains of the respective proteins (see Fig. S1A in the supplemental material). The R22C mutation does not map to the S-adenosyl-l-methionine-dependent methyltransferase domain of PhcB nor does the R154C mutation map to the receiver domain of PhcQ. Yet, mutations alter conserved amino acids of Ralstonia, Cupriavidus, and Paraburkholderia homologous proteins and would influence protein function as suggested by the Phyre2 protein structure prediction server (Fig. S1B and C) (32).
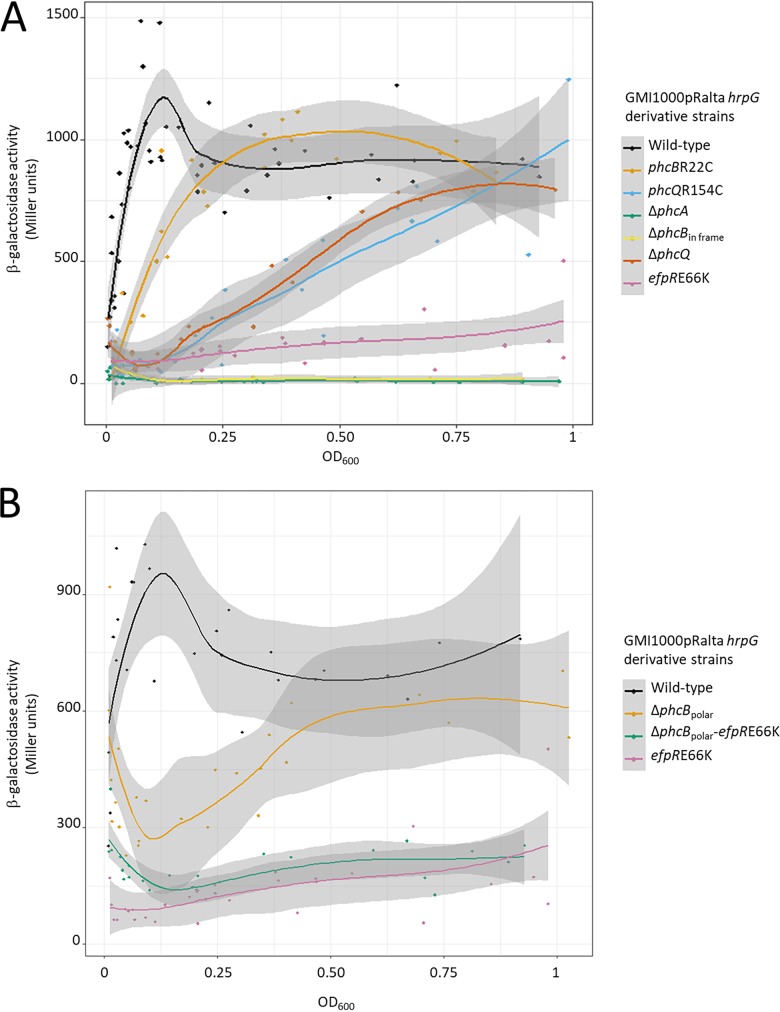
We hypothesized that these mutations affect protein conformation, stability, or efficiency and thus may alter the dynamics of PhcA responsiveness to cell density. To test this hypothesis, we monitored the expression kinetics of the xpsR-lacZ reporter gene fusion in various genetic backgrounds along with growth in rich liquid medium (Fig. 5A). In the nodulating chimeric strain GMI1000(pRalta) hrpG, the expression of xpsR-lacZ sharply rose at an optical density at 600 nm (OD600) of 0.04 (corresponding to ca. 5 × 107 CFU/ml) (see Fig. S2), reached a peak at an OD600 of 0.12 (ca. 108 CFU/ml), and then decreased and stabilized at an OD600 of 0.2 (ca. 2 × 108 CFU/ml). No peak of expression was observed in either the phcB or phcQ SNP mutant. In the phcBR22C mutant, the expression was slightly delayed and reached a maximum at an OD600 of 0.2 (ca. 2 × 108 CFU/ml). We confirmed that the phcBR22C mutant does not behave like a phcB in-frame deletion mutant, which exhibited no PhcA activity regardless of the cell density. In contrast, the phcQR154C mutant closely resembled a phcQ deletion mutant. In both mutants, the induction of xpsR-lacZ was slow and linear until an OD600 of 1 (ca. 109 CFU/ml). These dynamics of xpsR expression suggested that PhcQ may be involved in a positive feedback activation loop of PhcA, implying that the phcQ mutation may affect the synthesis of autoinducing molecules.
FIG 5.
Kinetics of the pxpsR-lacZ expression in the chimeric R. solanacearum nodulating strain [GMI1000(pRalta) hrpG] and derivative mutants according to the OD600. β-Galactosidase activities were measured on bacteria grown in rich BG medium. Tendency curves with their 95% confidence intervals were calculated using local polynomial regressions (locally estimated scatterplot smoothing [LOESS] method). Data were from 4 to 12 independent experiments. The same set of data for efpRE66K was used in panels A and B.
Correspondence between the OD600 units and the CFU for the GMI1000(pRalta) hrpG strain (wild type) and its derivative mutants. Bacteria grown in rich BG medium were serially diluted and plated at different OD600 units per milliliter. Data are from three independent experiments. A linear regression model and the associated equation are indicated for each strain. The four strains have similar correspondences between OD600 units and CFU. Download FIG S2, PDF file, 0.6 MB (650.8KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To substantiate whether the expression of other PhcA target genes was affected by the phcB and phcQ SNP mutations, we checked by reverse transcription-quantitative PCR (qRT-PCR) the expression of seven genes known to be positively or negatively controlled by PhcA (egl, flhF, motA, RSc1817, Rsp0983, RSp0178, and xpsR) (16–18) (see Fig. S3). For this, we harvested cells grown in rich liquid medium to an OD600 of around 0.1, a cell density at which the expression of xpsR was lower in the phcB and phcQ SNP mutants than in the wild-type strain. All these genes were differentially expressed in the phcBR22C, phcQR154C, and ΔphcA mutants compared to that in the GMI1000(pRalta) hrpG wild-type strain. Consistently with the xpsR expression data, expressions of these genes were less affected in the phcBR22C mutant than in the phcQR154C mutant.
Expression of PhcA target genes in the GMI1000(pRalta) hrpG chimeric strain and the phcBR22C, phcQR154C, and ΔphcA derivative mutants. Strains were cultivated in BG medium to an OD600 of around 0.1. Raw expression levels were normalized by rplM and rpoA expression, and ratios of gene expression in mutants versus that in the wild-type strain were calculated. Data were obtained from three independent experiments and are presented as mean Log2 (ratios) ± standard deviations. Genes positively regulated by PhcA were repressed in phcBR22C, phcQR154C, and ΔphcA mutants. Genes negatively regulated by PhcA were found induced in phcBR22C, phcQR154C, and ΔphcA mutants. *, P < 0.05 versus wild-type strain (Student’s t test). Download FIG S3, PDF file, 0.3 MB (347.1KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
These results showed that the phcB and phcQ mutations delay, to different degrees, the timing of Phc quorum sensing regulation, likely by decreasing the activity of the corresponding proteins.
The phcBR22C and phcQR154C mutations affect the synthesis of QS molecules.
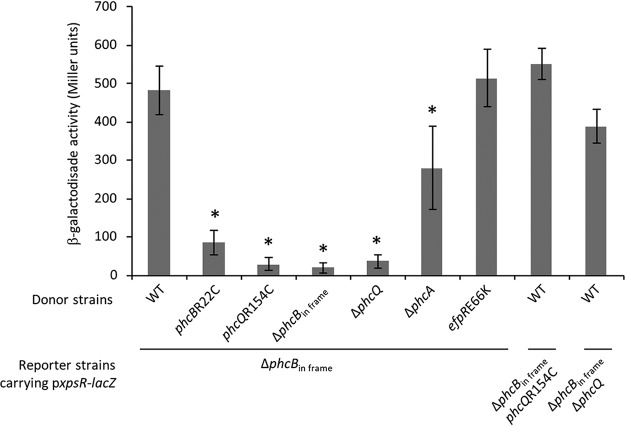
To evaluate the efficiency of phcB and phcQ SNP mutants to produce autoinducing molecules, we set up a bioassay in which a strain deficient for the synthesis of the QS molecules [i.e., a phcB in-frame deletion mutant in the GMI1000(pRalta) hrpG background] carrying the reporter plasmidic xpsR-lacZ fusion was incubated with supernatants of cultures harvested at an OD600 of 0.1 from either the GMI1000(pRalta) hrpG wild-type strain or the phcBR22C, phcQR154C, ΔphcBin frame, ΔphcQ, or ΔphcA derivative mutants. As expected, after 4 h of incubation with the supernatant of the wild-type strain, the xpsR-lacZ fusion was well expressed, while this fusion was not expressed after incubation with the supernatant of a ΔphcBin frame mutant. In agreement with previous works showing that some PhcA targets positively control the production or the stability of 3-OH MAME (33), the xpsR-lacZ fusion was less expressed after incubation with the supernatant of a ΔphcA mutant. Interestingly, the expression of xpsR-lacZ was strongly reduced or near the background level after incubation with the supernatants from the phcBR22C, phcQR154C, and ΔphcQ mutants (Fig. 6), indicating that the amount of active QS molecules present in the supernatants of these mutants was reduced compared to that in the supernatant of the wild-type strain taken at the same OD600. Moreover, we tested the involvement of PhcQ in signal transduction by incubating the double phcQR154C ΔphcBin frame or ΔphcQ ΔphcBin frame mutants with the supernatant of a wild-type strain. We found that the xpsR-lacZ expression was not affected in these strains, indicating that PhcQ is involved in the production of QS molecules but not in the signal transduction to PhcA (Fig. 6).
FIG 6.
Production of QS molecules and QS signal transduction in the R. solanacearum GMI1000(pRalta) hrpG wild-type strain (WT) and derivative mutants. The expression of the pxpsR-lacZ fusion was measured in reporter strains deficient for the production of QS molecules due to a phcB in-frame deletion after incubation with culture supernatants from various donor strains. Supernatants were taken from liquid cultures grown to an OD600 of 0.1, added to 1 volume of culture of the reporter strain grown to an OD600 of 0.2, and incubated for 4 h before β-galactosidase activity analysis. The reporter strain ΔphcBin frame was used to assess the ability of donor strains to produce QS molecules. Reporter strains mutated for both phcB and phcQ were used to assess the involvement of PhcQ in the transduction of QS signals. Values indicate means ± standard deviations obtained from 3 to 5 independent experiments. *, P < 0.01 versus the wild-type strain (Student’s t test).
Infection-adaptive phc mutations alter PhcA activation during the uptake of bacteria in root hairs.
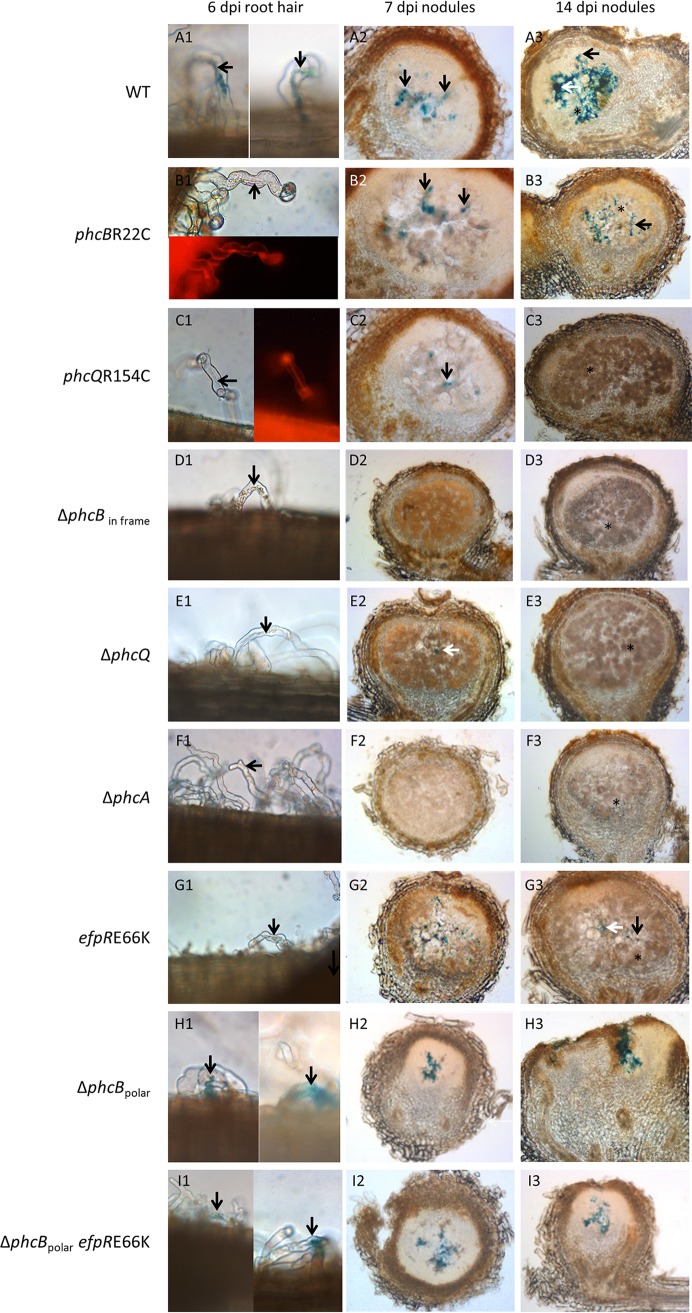
In M. pudica, rhizobial partners enter roots via transcellular infection threads that progress toward the cortex and ultimately release bacteria within cells of the developing nodules, where they are subsequently accommodated in membrane-enclosed compartments called symbiosomes. In the evolved Ralstonia-M. pudica interaction, intercellular bacteria were also observed associated with necrotic zones that usually occur in nodules induced by GMI1000(pRalta) hrpG and occasionally in nodules induced by the phcB and phcQ SNP mutants. To know whether the delay in quorum sensing responsiveness also operates under symbiotic conditions, we assessed the expression of the xpsR-lacZ fusion at three distinct symbiotic stages, following inoculation with either GMI1000(pRalta) hrpG or its phcBR22C, phcQR154C, ΔphcBin frame, ΔphcQ, or ΔphcA derivative mutants (i) in root hairs during bacterial uptake, (ii) in young nodules when bacteria are released from infection threads into the cytoplasm of plant cells, and (iii) in mature nodules accommodating intracellular bacteria, called bacteroids. As expected, in the phcA-deleted mutant, no expression of xpsR-lacZ was observed under any symbiotic conditions (Fig. 7F). Whatever the inoculated strain, no xpsR expression was observed in bacteroids. However, as the number of infected cells was very low in nodules induced by the GMI1000(pRalta) hrpG strain, it was difficult to evaluate the level of xpsR expression in bacteroids formed by this strain. In nodules induced by the poorly infecting strain GMI1000(pRalta) hrpG, xpsR-lacZ expression was visualized in root hair and nodule infection threads (ITs) as well as in intercellular bacteria (Fig. 7A). When roots were inoculated by any of the two phcB and phcQ SNP mutants, loss of xpsR-lacZ expression was only observed in ITs progressing in root hairs, xpsR-lacZ being normally expressed in nodule ITs (Fig. 7B and C). These results suggested that activation of PhcA in the late infection-adapted phc mutants was impaired during a very early symbiotic stage, corresponding to the bacterial uptake in root hairs and initiation of infection threads. This early stage thus appeared crucial for late intracellular accommodation of bacteria in nodules.
FIG 7.
Expression of pxpsR-lacZ in the chimeric R. solanacearum nodulating strain [GMI1000(pRalta) hrpG] and derivative mutants during symbiotic interaction with Mimosa pudica. Roots and nodules were harvested at 6 and 7 or 14 dpi, respectively, and fixed with 2% glutaraldehyde. Roots and 55-μm sections of nodules were then stained in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to reveal beta-galactosidase activity. The xpsR-lacZ expression was detected in infection threads and extracellularly invaded spaces of young and mature nodules induced by all strains (A2 to C2, E2, G2 to I2, A3 to C3, G3 to I3) except by the phcA and phcBin frame deletion mutants (D and F). Only the poorly infectious strains, GMI1000(pRalta) hrpG and its derivatives ΔphcBpolar and ΔphcBpolar-efpRE66K mutants, expressed the fusion in root hairs (A1, H1, and I1 versus B1 to G1). The phcBR22C and phcQR154C mutants constitutively expressed the mCherry fluorophore. Bacteria fluorescing in red can be visualized in ITs (B and C). Black arrows indicate infection threads. White arrows indicate extracellular bacteria. Light brown cells and black stars indicate intracellularly infected nodule cells. Images are representative of data obtained from at least 3 independent experiments.
QS modulation as an alternative adaptive path to EfpR inactivation.
The phc mutations have the same impact on intracellular infection as mutations in the EfpR path previously detected in other evolved lines. Indeed, the size of infected and necrotic zones in nodules induced by phc SNP mutants was similar to that of nodules induced by efpR mutants (Fig. 2) (27). To compare the adaptiveness of the efpR and phc SNP mutations in a global and more integrative way, we measured their relative in planta fitness following plant coinoculation. We showed that the in planta fitness of the efpRE66K mutant was not significantly different from that of the phcBR22C or phcQR154C mutant in a GMI1000(pRalta) hrpG context (Fig. 4). Moreover, the double phcBR22C-efpRE66K or phcQR154C-efpRE66K mutant was as fit as the single efpRE66K mutant, indicating that efpR and phc adaptive mutations do not have cumulative effects (Fig. 4). Similar to that in phc SNP mutants, in an efpRE66K mutant, the expression of xpsR was strongly affected but not abolished under in vitro conditions (Fig. 5A), while under in planta conditions, expression of the xpsR-lacZ fusion was visualized in nodule infection threads but not in root hair infection threads (Fig. 7G1).
In a previous study, we determined the EfpR regulon in a GMI1000(pRalta) hrpG context (27). Strikingly, this EfpR regulon largely overlaps with the PhcA regulon determined recently in three independent studies (16–18). Among the 200 putative efpR targets, 160 (80%) were also found differentially expressed in at least one of the PhcA regulons (see Table S2). These genes include 17 for EPS synthesis, 52 for motility, 14 for hemin/siderophore transport and metabolism genes, the prhI sigma factor, and 10 genes encoding Hrp and T3 effector proteins. In addition, both PhcA and EfpR were shown to be metabolic repressors (15, 17, 27, 28). The metabolic profiles of the two mutants also display high similarities, since 88% of substrates found to be more efficiently metabolized in GMI1000(pRalta) hrpG efpRE66K (27) were also better metabolized in a phcA mutant (17) (see Table S3). Hence, the EfpR path may either affect PhcA activity directly, interfere with its activating pathway, PhcBSRQ, or act on some genes commonly targeted by PhcA.
Comparison of genes differentially expressed in efpRE66K and ΔphcA mutants. Download Table S2, XLSX file, 0.1 MB (15.7KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Carbon, nitrogen, and phosphorus sources better metabolized in the efpRE66K mutant. Download Table S3, XLSX file, 0.1 MB (12.7KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess whether the EfpR pathway could interfere with the QS sensory cascade PhcBSRQ, we first determined the production of QS molecules in an efpRE66K mutant using the bioassay described above. We showed that the efpRE66K mutant produced similar amounts of active QS molecules as the wild-type GMI1000(pRalta) hrpG strain (Fig. 6). Then, we combined a phcB polar deletion mutation, which downregulated the expression of the full phcBSRQ operon (see Fig. S4), with an efpRE66K mutation in the GMI1000(pRalta) hrpG strain and monitored the expression of the xpsR-lacZ reporter gene fusion by β-galactosidase assay along growth as described above. In the single ΔphcB polar deletion mutant, probably because of the absence of the PhcSR repressive action (14), PhcA was constitutively active at an intermediary level throughout growth, although with some variations (Fig. 5B). Interestingly, the xpsR-lacZ expression in the double ΔphcBpolar-efpRE66K mutant was always lower than in the single ΔphcBpolar mutant, indicating that the efpRE66K mutation can repress xpsR in the absence of the PhcBSRQ proteins and thus may alter the PhcA regulon in a PhcBSRQ-independent manner. Notwithstanding, when the OD600 was below 0.1, xpsR-lacZ expression was higher in the double ΔphcBpolar-efpRE66K mutant than in the single efpRE66K mutant, suggesting an additive effect of the two mutations at low cell density. Consistently, both ΔphcBpolar and ΔphcBpolar-efpRE66K mutants were poorly infectious (Fig. 2) and harbored a detectable xpsR-lacZ expression in epidermal ITs (Fig. 7H1 and I1), thus reinforcing the correlation between the absence of early xpsR expression in root hair ITs and a nice late intracellular infection phenotype.
Expression of phcS, phcR, and phcQ genes in the GMI1000(pRalta) hrpG chimeric strain and its ΔphcB polar derivative mutant. Strains were cultivated in BG medium until mid-exponential phase. Raw expression levels were normalized by rplM expression, and ratios of gene expression in mutants versus that in the wild-type strain were calculated. Data were obtained from three independent experiments and are presented as Log2 (mean ratios) ± standard deviations. *, P < 0.01 versus the wild-type strain (Student’s t test). Download FIG S4, PDF file, 0.6 MB (609.6KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
xpsR deletion did not improve intracellular infection.
To assess whether the downregulation of xpsR itself could solely account for the infection phenotype of the efpR and phc mutants, we constructed a xpsR deletion mutant in the GMI1000(pRalta) hrpG background and quantified its nodule cell infection ability. This mutation did not improve the intracellular infection (Fig. 2), suggesting that either EPS biosynthesis is not involved in infection or repression of EPS biosynthesis alone is not sufficient to improve infection.
Quorum sensing modulation decreased the pathogenicity of R. solanacearum.
To evaluate whether the phc mutations adaptive for M. pudica nodule cell infection affect the pathogenicity of R. solanacearum, we introduced the phcBR22C, phcQR154C, ΔphcBin frame, ΔphcQ, and ΔphcA mutations into the pathogenic GMI1000(pRalta) strain and followed the disease symptoms over 20 days on the host plant Arabidopsis thaliana. While the phcA deletion mutant induced no symptom on A. thaliana, both phc SNP mutants induced significantly delayed symptoms, phcBR22C being delayed by ca. 1 to 2 days and phcQR154C being delayed by ca. 6 to 7 days compared with that in the wild-type strain (Fig. 8). The ΔphcQ mutant appeared slightly more virulent than the phcQR154C mutant. In contrast, the ΔphcBin frame mutant induced symptoms strongly delayed, by ca. 14 days, compared with that in the phcBR22C mutant strain (Fig. 8), suggesting that PhcA may have residual activity in the absence of QS molecules.
FIG 8.
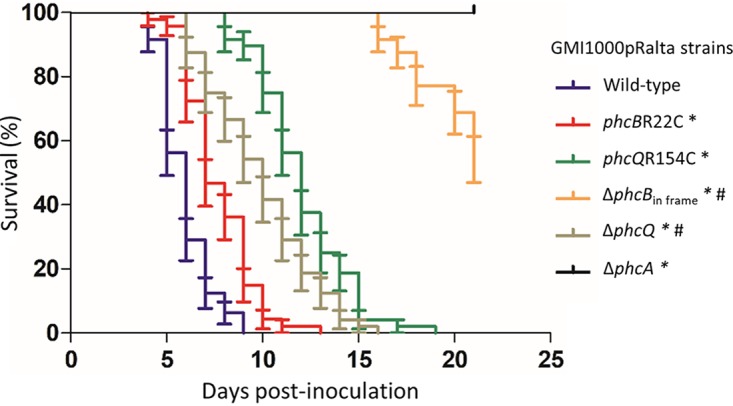
Disease symptoms induced by wild-type and mutant strains of the chimeric Ralstonia on Arabidopsis thaliana. Kaplan-Meyer survival analysis of A. thaliana plants inoculated by the wild-type GMI1000(pRalta) chimeric Ralstonia and its phcBR22C, phcQR154C, ΔphcBin frame, ΔphcQ, and ΔphcA derivative mutants. *, the survival curves are significantly different from the wild-type curve; #, the survival curves are significantly different from the corresponding phc mutant curves (P < 0.001, Gehan-Breslow-Wilcoxon test). Values were obtained from 3 independent series of 16 plants.
DISCUSSION
Quorum sensing is a widespread bacterial regulatory system allowing rapid shifts in behavior in response to changes in cell density or confinement (34). The mode and tempo of signal transduction and target outputs of each quorum sensing system reflect the unique biology carried out by a particular bacterial species. Here, we show that a system optimized to allow survival in a specialized niche can be rapidly redesigned to promote thriving in a very different environment, i.e., change in QS responsiveness helps the extracellular plant pathogen R. solanacearum having acquired a set of essential symbiotic genes to turn into an intracellular legume symbiont. During experimental evolution under legume (M. pudica) selection pressure, point mutations affecting two components of the Phc QS regulatory cascade, the autoinducer synthase PhcB and the regulatory protein PhcQ, modified the kinetics of activation of the downstream central regulator PhcA. The delay in PhcA activation in the Ralstonia phc-mutated evolved clones observed in vitro results in an inactivation of PhcA in epidermal ITs, which correlates with their nice nodule intracellular infection phenotypes on M. pudica. Conversely, these mutations, when introduced in the Ralstonia ancestor, reduced the virulence capacity of the strain, reemphasizing the antagonism between pathogeny and symbiosis (24, 26, 27). Unlike other infection-adaptive mutations we previously identified (hrpG and efpR) (24, 26, 27), the virulence regulator PhcA was not constitutively inactivated. Potentially, this conditional inactivation could be more favorable for subsequent adaptation steps. Interestingly, in Cupriavidus taiwanensis, a Ralstonia-neighboring taxon that evolved into Mimosa symbionts, the phcB and phcS orthologous genes were found under positive selection and associated with the transition to symbiosis during the natural evolution of this rhizobium species (29), likely reflecting the need to adjust QS to the legume endosymbiotic niche. Adaptive mutations altering QS responsiveness were also evidenced during experimental and natural evolution of other pathogenic and symbiotic bacteria such as Vibrio fischeri and Staphylococcus aureus (35–38), exemplifying the role of these sensory systems in bacterial adaptation and phenotypic diversification. The role of QS in rhizobial symbiosis is, however, variable, either neutral (39), positive (40–42), or negative (43, 44). Rhizobial QS systems antagonistically control EPS biosynthesis and motility-chemotaxis but also affect a wide spectrum of other physiological traits that vary among rhizobial species, including biofilm formation, swarming motility, the type III secretion system, plasmid transfer, cell division, metabolism, and transport (6, 39, 45).
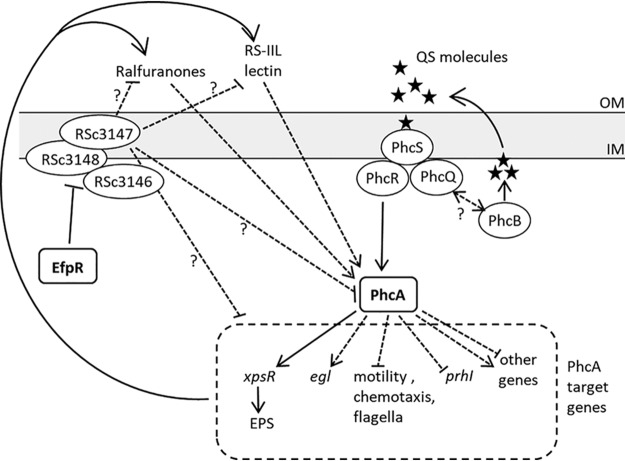
Studies identifying the molecular mechanisms that underlie convergent evolution have shown that mutations involved in a given phenotype often target the same pathways, making experimental evolution a powerful tool to decipher regulatory networks (27, 46). In this line, our experiment allowed us to uncover a new component of the R. solanacearum Phc QS system and provided additional information on the functioning of the Phc network. Although the PhcBSR(Q) pathway was identified approximately 20 years ago (14), the roles of several components were not clearly elucidated. This was the case for the regulatory protein PhcQ, which was suspected to be involved in the QS cascade because it belongs to the PhcBSRQ operon. Here, we demonstrated that PhcQ is involved in the dynamics of activation of PhcA in response to the quorum. In addition, the synthesis of QS molecules was found to be affected in a phcQ mutant, suggesting that the activity of the autoinducer synthase PhcB is directly or indirectly regulated by PhcQ. The PhcQ protein is composed of a receiver domain and an uncharacterized C-terminal part. Several different mechanisms of action have been reported for proteins containing a single-receiver domain, including a direct regulatory effect on the activity of a downstream protein, the transfer of phosphate in phosphorelays, or a role as phosphate-sinks that redirect phosphate flux away from histidine kinases (47). It is also possible that PhcQ, through its receiver domain, interacts with PhcB and modulates its activity. Such a role in the synthesis and transduction of a diffusible signal factor (DSF) has been demonstrated for the receiver domain of the Xanthomonas campestris sensor kinase RpfC in a QS cascade (48). In a previous study, we showed that in some evolved Ralstonia lines, massive nodule cell infection capacity was gained through the inactivation of another central regulator, EfpR, controlling the membrane associated RSc3146 to -3148 genes (27). The phc and efpR mutations displayed similar in planta fitness, and their combination had no cumulative effect on this fitness. This, together with strong similarities in the transcriptomic and metabolomic profiles of the two regulators, suggested a connection between the EfpR and PhcA regulatory pathways. Our findings indicated that the EfpR regulatory cascade does not interfere with the PhcBSRQ-mediated QS signal transduction and thus that the two paths act independently on the PhcA regulon. However, the mechanism by which the EfpR-RSc3146-RSc3148 pathway represses the PhcA regulon remains unknown. It may either repress PhcA activity, act independently on PhcA target genes, or impair one of the positive feedback loops activating PhcA (16, 33) (Fig. 9).
FIG 9.
Possible connections between PhcA and EfpR regulatory pathways. PhcA is activated by the PhcBSRQ quorum sensing sensory cascade. Above the QS threshold of ca. 5 × 107 cell/ml, PhcA is rapidly activated. The regulatory protein PhcQ contributes to the dynamics of activation of PhcA by influencing the synthesis of the QS molecules. EfpR represses the Rsc3146 to -3148 genes. In an efpR mutant, the strong induction of Rsc3146 to -3148 genes affects a large part of the PhcA regulon either by modulating PhcA activity, by acting on the expression of some PhcA target genes, or by inhibiting one of the positive feedback loops of activation of PhcA (16, 33). Dashed lines indicate potential indirect effects.
In rhizobium-legume symbiosis, the accommodation of bacteria within nodule cells is a crucial step, since it protects internalized bacteria from plant defense reactions, while massive bacterial multiplication allows for nitrogen fixation levels that sustain plant growth (49). How this property is acquired during evolution is only partly understood. An intrinsic infection capacity is gained through the acquisition of nod genes that determine the synthesis of Nod factors. These lipochitooligosaccharide signal molecules specifically recognized by plant LysM receptors trigger the developmental program required for nodule organogenesis, bacterial infection (50, 51), and partial suppression of plant defenses (52). Other bacterial factors, such as surface polysaccharides, are likely recruited in recipient genomes to achieve infection. In Lotus japonicus, for instance, compatible bacterial EPS are specifically recognized by the plant LysM receptor kinase EPR3, which might activate a signaling cascade that controls and maintains an intracellular infection mode (53–55). In this study, we showed that inactivation of the central regulator PhcA was an alternative way to the inactivation of another global regulator, EfpR, to improve intracellular infection during experimental evolution of R. solanacearum into legume symbionts. Since PhcA and EfpR control a common set of more than 150 genes, including those for exopolysaccharides and motility and virulence factors, it is likely that the concerted change of expression of several genes rather than a single gene is involved in infection improvement. Indeed, the sole inactivation of XpsR, the activator of EPS biosynthesis genes, did not lead to an improved infection phenotype. Other virulence genes controlled by these systems may trigger plant defense reactions and thus hamper symbiosis establishment (16–18). Another common interesting feature of both regulators is that their repression/inactivation enhances the metabolism of the bacterium, increasing its growth rate on many substrates. Future investigation of other intracellular lines, where EfpR and Phc pathways do not seem targeted by evolution, may narrow the number of possible functions responsible for phenotypic changes. Yet, we were able to identify at which symbiotic stage modifications were required, by monitoring the in planta expression of a direct PhcA target, xpsR. A clear change induced by the phc mutations was a specific loss of xpsR expression in infection threads of root hairs, suggesting that this step may condition the success of the subsequent accommodation of bacteria within nodule cells. The link between early and late events of the infection process is supported by the fact that several plant genes (NFP/NFR5, LYK3/NFR1, SYMRK, or IPD3/CYCLOPS) involved in early symbiotic signaling in the root epidermis are also required for the release of bacteria in nodule cells (56–58). Furthermore, a tight coordination between the progression of ITs and the periclinal divisions of specific root cell layers was proposed to determine the intracellular infection of nodules (59). It is thus possible that, for either signaling or metabolic reasons, the primary ITs formed by efpR and phc mutants progress more rapidly than those made by a single hrpG mutant, allowing a better invasion of nodule cells. Future fine cellular analyses of the progression rate of ITs as well as a comparison of molecular plant responses differently induced by poor and nice infectious evolved clones should help us to understand intracellular infection, the distinctive feature of nitrogen-fixing root nodule symbioses and possibly the first trait acquired by the common ancestor of the root nodule forming plant clade FaFaCuRo (Fabales, Fagales, Cucurbitales and Rosales) during evolution (60).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material. R. solanacearum strains were grown at 28°C either on rich BG medium (61) or on MP minimal medium (62) supplemented either with 20 mM glutamate for gene expression analyses or with glycerol 2% for bacterial transformation. Antibiotics were used at the following concentrations: trimethoprim, 100 μg/ml; spectinomycin, 40 μg/ml; kanamycin, 50 μg/ml; tetracycline, 10 μg/ml.
Strains and plasmids used in this study. Download Table S1, DOCX file, 0.1 MB (28.2KB, docx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plants assays and cytological analyses.
Mimosa pudica seedlings from Australian origin (B&T World Seed, Paguignan, France) were cultivated as previously described (29).
For cytology, nodules were harvested at 21 days postinoculation (dpi) and cut into 55-μm sections with a vibrating blade microtome (VT1000 S; Leica, Wetzlar, Germany). For each nodule, longitudinal sections were observed using an inverted microscope (DM IRB/E; Leica, Wetzlar, Germany), and images were acquired using a charge-coupled-device (CCD) camera (Color Coolview; Photonic Science, Milham, UK). Quantification of infection and necrosis areas on nodule sections was performed as described previously (25, 30) using the Image-Pro Plus software and based on the HIS method (Media Cybernetics, Rockville, MD, USA). Necrotic areas are characterized by a dark-brown color, while intracellularly infected cells are characterized by a light-brown color. Results of infection and necrosis quantifications were obtained from 2 to 3 independent experiments for each strain. At least 16 plants were analyzed per experiment.
For relative in planta fitness assays, we coinoculated M. pudica plants with a pair of different strains in equivalent proportions (5 × 105 bacteria of each strain per plant). Coinoculated strains were carrying the same antibiotic marker and expressing or not the mCherry fluorophore. Nodules from 20 plants were harvested 21 days after inoculation, pooled, surface sterilized, and crushed together. Dilutions of nodule crushes were spread on solid medium. In cases where one of the coinoculated strains was expressing the mCherry fluorophore, colonies formed by each strain were counted under a stereo zoom microscope under bright-field and fluorescence illuminations (AxioZoom V16; Zeiss, Oberkochen, Germany). In cases where both coinoculated strains were not expressing any fluorophores (comparison of ΔphcA mutant with phcBR22C or phcQR154C mutant), 96 colonies from the nodule crushes were screened by PCR for the presence/absence of mutations. Three independent experiments were performed for each comparison.
Beta-galactosidase assays.
Expression of the plasmidic xpsR-lacZ fusion was assessed by β-galactosidase assay of 100-μl aliquots of bacterial cultures grown in rich BG medium. Samples were collected either at an OD600 of 0.2 (Fig. 3) or every 2 to 3 h along a growth curve from an OD600 of 0.005 to an OD600 of 1. β-Galactosidase activities were measured in Miller units as described previously (63). The correspondence between the OD600 units and the number of bacteria (CFU) was determined for each strain throughout growth in rich BG medium by plating serial dilutions of cultures (Fig. S2).
To test the production of QS molecules by the chimeric strains, the GMI1000(pRalta) hrpG ΔphcBin frame strain unable to produce QS molecules and expressing the reporter gene fusion pxpsR-lacZ was grown in rich BG medium until the OD600 reached ca. 0.2. In parallel, strains producing QS molecules [GMI1000(pRalta) hrpG and its phcBR22C, phcQR154C, ΔphcBin frame, ΔphcQ, ΔphcA, and efpRE66K derivative mutants) were grown in rich BG medium until the OD600 reached 0.1. Supernatants from cultures of QS molecule donor strains were prepared by centrifugation for 15 min at 5,000 rpm followed by filtration at 0.22 μm. One volume of filtered supernatant was added to 1 volume of culture of the reporter strain and incubated for 4 h at 28°C under agitation before β-galactosidase activity analysis. β-Galactosidase activities were measured from 100-μl aliquots of bacterial cultures. Background expression level of the xpsR-lacZ reporter fusion was measured by incubating the reporter strain with the supernatant of a culture of the GMI1000(pRalta) hrpG ΔphcBin frame strain.
To test the QS signal transduction in the phcQR154C and ΔphcQ mutants, a similar experiment was performed using either the GMI1000(pRalta) hrpG phcQR154C ΔphcBin frame pxpsR-lacZ or GMI1000(pRalta) hrpG ΔphcQ ΔphcBin frame pxpsR-lacZ strains as reporter strains and the GMI1000(pRalta) hrpG as donor strain.
Statistical analyses.
Statistical analyses of infection and necrosis sizes of nodule sections were performed using R software (version 3.5.1) and the nonparametric multiple-comparison test of Kruskal-Wallis. For the comparisons of in planta relative fitness, β-galactosidase activities in mid-exponential-phase cultures, and gene expression, we did two-sided t tests. The local polynomial regressions and confidence intervals of β-galactosidase activities throughout growth were performed using the R package ggplot2 and the geom_smooth function (64).
Other materials and methods concerning genome resequencing and detection of mutations in evolved clones, constructions of mutants and plasmids, detection of xpsR-lacZ expression in planta, gene expression analyses by quantitative reverse transcription-PCRs, pathogenicity assays, protein structure prediction, and protein alignments are provided in Text S1 in the supplemental material.
Primers used in this study. Download Table S4, XLSX file, 0.1 MB (15KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by funds from the French National Research Agency (ANR-12-ADAP-0014-01 and ANR-16-CE20-0011-01) and the Laboratoire d’Excellence (LABEX) TULIP (ANR-10-LABX-41). M.T. was supported by a fellowship from the China Scholarship Council.
We thank the cellular imaging facility TRI FR3450, Castanet-Tolosan, France.
We declare no conflict of interest.
Footnotes
Citation Tang M, Bouchez O, Cruveiller S, Masson-Boivin C, Capela D. 2020. Modulation of quorum sensing as an adaptation to nodule cell infection during experimental evolution of legume symbionts. mBio 11:e03129-19. https://doi.org/10.1128/mBio.03129-19.
Contributor Information
Esperanza Martínez-Romero, Centro de Ciencias Genómicas, UNAM.
Nancy A. Moran, University of Texas at Austin.
REFERENCES
- 1.Mole BM, Baltrus DA, Dangl JL, Grant SR. 2007. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol 15:363–371. doi: 10.1016/j.tim.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Buttner D, Bonas U. 2010. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 3.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. 2014. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol 16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 4.McAdams HH, Srinivasan B, Arkin AP. 2004. The evolution of genetic regulatory systems in bacteria. Nat Rev Genet 5:169–178. doi: 10.1038/nrg1292. [DOI] [PubMed] [Google Scholar]
- 5.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calatrava-Morales N, McIntosh M, Soto MJ. 2018. Regulation mediated by N-acyl homoserine lactone quorum sensing signals in the Rhizobium-legume symbiosis. Genes (Basel) 9:E263. doi: 10.3390/genes9050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma SC, Miyashiro T. 2013. Quorum sensing in the squid-Vibrio symbiosis. Int J Mol Sci 14:16386–16401. doi: 10.3390/ijms140816386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deep A, Chaudhary U, Gupta V. 2011. Quorum sensing and bacterial pathogenicity: from molecules to disease. J Lab Physicians 3:4–11. doi: 10.4103/0974-2727.78553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball AS, Chaparian RR, van Kessel JC. 2017. Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J Bacteriol 199:e00105-17. doi: 10.1128/JB.00105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marketon MM, Glenn SA, Eberhard A, González JE. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J Bacteriol 185:325–331. doi: 10.1128/jb.185.1.325-331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schell MA. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu Rev Phytopathol 38:263–292. doi: 10.1146/annurev.phyto.38.1.263. [DOI] [PubMed] [Google Scholar]
- 12.Kai K, Ohnishi H, Shimatani M, Ishikawa S, Mori Y, Kiba A, Ohnishi K, Tabuchi M, Hikichi Y. 2015. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. Chembiochem 16:2309–2318. doi: 10.1002/cbic.201500456. [DOI] [PubMed] [Google Scholar]
- 13.Clough SJ, Flavier AB, Schell MA, Denny TP. 1997. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl Environ Microbiol 63:844–850. doi: 10.1128/AEM.63.3.844-850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clough SJ, Lee KE, Schell MA, Denny TP. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J Bacteriol 179:3639–3648. doi: 10.1128/jb.179.11.3639-3648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyraud R, Cottret L, Marmiesse L, Gouzy J, Genin S. 2016. A resource allocation trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12:e1005939. doi: 10.1371/journal.ppat.1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori Y, Ishikawa S, Ohnishi H, Shimatani M, Morikawa Y, Hayashi K, Ohnishi K, Kiba A, Kai K, Hikichi Y. 2018. Involvement of ralfuranones in the quorum sensing signalling pathway and virulence of Ralstonia solanacearum strain OE1-1. Mol Plant Pathol 19:454–463. doi: 10.1111/mpp.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khokhani D, Lowe-Power TM, Tran TM, Allen C. 2017. A single regulator mediates strategic switching between attachment/spread and growth/virulence in the plant pathogen Ralstonia solanacearum. mBio 8:e00895-17. doi: 10.1128/mBio.00895-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrier A, Barlet X, Peyraud R, Rengel D, Guidot A, Genin S. 2018. Comparative transcriptomic studies identify specific expression patterns of virulence factors under the control of the master regulator PhcA in the Ralstonia solanacearum species complex. Microb Pathog 116:273–278. doi: 10.1016/j.micpath.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 19.Masson-Boivin C, Giraud E, Perret X, Batut J. 2009. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Remigi P, Zhu J, Young JPW, Masson-Boivin C. 2016. Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol 24:63–75. doi: 10.1016/j.tim.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindström K. 2015. Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90. doi: 10.1016/j.syapm.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Ormeño-Orrillo E, Servín-Garcidueñas LE, Rogel MA, González V, Peralta H, Mora J, Martínez-Romero J, Martínez-Romero E. 2015. Taxonomy of rhizobia and agrobacteria from the Rhizobiaceae family in light of genomics. Syst Appl Microbiol 38:287–291. doi: 10.1016/j.syapm.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Amadou C, Pascal G, Mangenot S, Glew M, Bontemps C, Capela D, Carrere S, Cruveiller S, Dossat C, Lajus A, Marchetti M, Poinsot V, Rouy Z, Servin B, Saad M, Schenowitz C, Barbe V, Batut J, Medigue C, Masson-Boivin C. 2008. Genome sequence of the beta-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res 18:1472–1483. doi: 10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C, Timmers T, Poinsot V, Gilbert LB, Heeb P, Médigue C, Batut J, Masson-Boivin C. 2010. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol 8:e1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetti M, Jauneau A, Capela D, Remigi P, Gris C, Batut J, Masson-Boivin C. 2014. Shaping bacterial symbiosis with legumes by experimental evolution. Mol Plant Microbe Interact 27:956–964. doi: 10.1094/MPMI-03-14-0083-R. [DOI] [PubMed] [Google Scholar]
- 26.Guan SH, Gris C, Cruveiller S, Pouzet C, Tasse L, Leru A, Maillard A, Medigue C, Batut J, Masson-Boivin C, Capela D. 2013. Experimental evolution of nodule intracellular infection in legume symbionts. ISME J 7:1367–1377. doi: 10.1038/ismej.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capela D, Marchetti M, Clérissi C, Perrier A, Guetta D, Gris C, Valls M, Jauneau A, Cruveiller S, Rocha EPC, Masson-Boivin C. 2017. Recruitment of a lineage-specific virulence regulatory pathway promotes intracellular infection by a plant pathogen experimentally evolved into a legume symbiont. Mol Biol Evol 34:2503–2521. doi: 10.1093/molbev/msx165. [DOI] [PubMed] [Google Scholar]
- 28.Perrier A, Peyraud R, Rengel D, Barlet X, Lucasson E, Gouzy J, Peeters N, Genin S, Guidot A. 2016. Enhanced in planta fitness through adaptive mutations in EfpR, a dual regulator of virulence and metabolic functions in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12:e1006044. doi: 10.1371/journal.ppat.1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clerissi C, Touchon M, Capela D, Tang M, Cruveiller S, Genthon C, Lopez-Roques C, Parker MA, Moulin L, Masson-Boivin C, Rocha EPC. 2018. Parallels between experimental and natural evolution of legume symbionts. Nat Commun 9:2264. doi: 10.1038/s41467-018-04778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchetti M, Clerissi C, Yousfi Y, Gris C, Bouchez O, Rocha E, Cruveiller S, Jauneau A, Capela D, Masson-Boivin C. 2017. Experimental evolution of rhizobia may lead to either extra- or intracellular symbiotic adaptation depending on the selection regime. Mol Ecol 26:1818–1831. doi: 10.1111/mec.13895. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Yindeeyoungyeon W, Garg RP, Denny TP, Schell MA. 1998. Joint transcriptional control of xpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR-type transcriptional activator. J Bacteriol 180:2736–2743. doi: 10.1128/JB.180.10.2736-2743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi K, Kai K, Mori Y, Ishikawa S, Ujita Y, Ohnishi K, Kiba A, Hikichi Y. 2019. Contribution of a lectin, LecM, to the quorum sensing signalling pathway of Ralstonia solanacearum strain OE1-1. Mol Plant Pathol 20:334–345. doi: 10.1111/mpp.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 35.Geisinger E, Chen J, Novick RP. 2012. Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J Bacteriol 194:2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Zhao A, Xie Q, Olinares PD, Chait BT, Novick RP, Muir TW. 2017. Functional plasticity of the AgrC receptor histidine kinase required for staphylococcal virulence. Cell Chem Biol 24:76–86. doi: 10.1016/j.chembiol.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabrina Pankey M, Foxall RL, Ster IM, Perry LA, Schuster BM, Donner RA, Coyle M, Cooper VS, Whistler CA. 2017. Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria. eLife 6:e24414. doi: 10.7554/eLife.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young BC, Wu CH, Gordon NC, Cole K, Price JR, Liu E, Sheppard AE, Perera S, Charlesworth J, Golubchik T, Iqbal Z, Bowden R, Massey RC, Paul J, Crook DW, Peto TE, Walker AS, Llewelyn MJ, Wyllie DH, Wilson DJ. 2017. Severe infections emerge from commensal bacteria by adaptive evolution. eLife 6:e30637. doi: 10.7554/eLife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krysciak D, Grote J, Rodriguez Orbegoso M, Utpatel C, Förstner KU, Li L, Schmeisser C, Krishnan HB, Streit WR. 2014. RNA sequencing analysis of the broad-host-range strain Sinorhizobium fredii NGR234 identifies a large set of genes linked to quorum sensing-dependent regulation in the background of a traI and ngrI deletion mutant. Appl Environ Microbiol 80:5655–5671. doi: 10.1128/AEM.01835-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurich N, González JE. 2009. Role of quorum sensing in Sinorhizobium meliloti-Alfalfa symbiosis. J Bacteriol 191:4372–4382. doi: 10.1128/JB.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M, Sun K, Zhou L, Yang R, Zhong Z, Zhu J. 2009. Functional analysis of three AHL autoinducer synthase genes in Mesorhizobium loti reveals the important role of quorum sensing in symbiotic nodulation. Can J Microbiol 55:210–214. doi: 10.1139/w08-128. [DOI] [PubMed] [Google Scholar]
- 42.Zheng H, Zhong Z, Lai X, Chen WX, Li S, Zhu J. 2006. A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188:1943–1949. doi: 10.1128/JB.188.5.1943-1949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J Bacteriol 180:815–821. doi: 10.1128/JB.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodelas B, Lithgow JK, Wisniewski-Dye F, Hardman A, Wilkinson A, Economou A, Williams P, Downie JA. 1999. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J Bacteriol 181:3816–3823. doi: 10.1128/JB.181.12.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang HH, Becker A, González JE. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J Bacteriol 186:5460–5472. doi: 10.1128/JB.186.16.5460-5472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 47.Feldheim YS, Zusman T, Kapach A, Segal G. 2018. The single-domain response regulator LerC functions as a connector protein in the Legionella pneumophila effectors regulatory network. Mol Microbiol 110:741–760. doi: 10.1111/mmi.14101. [DOI] [PubMed] [Google Scholar]
- 48.He YW, Wang C, Zhou L, Song H, Dow JM, Zhang LH. 2006. Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J Biol Chem 281:33414–33421. doi: 10.1074/jbc.M606571200. [DOI] [PubMed] [Google Scholar]
- 49.Berrabah F, Ratet P, Gourion B. 2015. Multiple steps control immunity during the intracellular accommodation of rhizobia. J Exp Bot 66:1977–1985. doi: 10.1093/jxb/eru545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oldroyd GE. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 51.Geurts R, Bisseling T. 2002. Rhizobium nod factor perception and signalling. Plant Cell 14 Suppl:S239–S249. doi: 10.1105/tpc.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Y, Cao YR, Tanaka K, Thibivilliers S, Wan JR, Choi J, Kang CH, Qiu J, Stacey G. 2013. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341:1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 53.Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, Vinther M, Andersen SU, Krusell L, Thirup S, Jensen KJ, Ronson CW, Blaise M, Radutoiu S, Stougaard J. 2015. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 54.Muszyński A, Heiss C, Hjuler CT, Sullivan JT, Kelly SJ, Thygesen MB, Stougaard J, Azadi P, Carlson RW, Ronson CW. 2016. Structures of exopolysaccharides involved in receptor-mediated perception of Mesorhizobium loti by Lotus japonicus. J Biol Chem 291:20946–20961. doi: 10.1074/jbc.M116.743856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawaharada Y, Nielsen MW, Kelly S, James EK, Andersen KR, Rasmussen SR, Füchtbauer W, Madsen LH, Heckmann AB, Radutoiu S, Stougaard J. 2017. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun 8:14534. doi: 10.1038/ncomms14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M. 2005. SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci U S A 102:10369–10374. doi: 10.1073/pnas.0504250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ovchinnikova E, Journet E-P, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, den Camp RO, Zhukov V, Tikhonovich I, Borisov A, Bisseling T, Limpens E. 2011. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol Plant Microbe Interact 24:1333–1344. doi: 10.1094/MPMI-01-11-0013. [DOI] [PubMed] [Google Scholar]
- 58.Moling S, Pietraszewska-Bogiel A, Postma M, Fedorova E, Hink MA, Limpens E, Gadella TW, Bisseling T. 2014. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26:4188–4199. doi: 10.1105/tpc.114.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014. Fate map of Medicago truncatula root nodules. Development 141:3517–3528. doi: 10.1242/dev.110775. [DOI] [PubMed] [Google Scholar]
- 60.Parniske M. 2018. Uptake of bacteria into living plant cells, the unifying and distinct feature of the nitrogen-fixing root nodule symbiosis. Curr Opin Plant Biol 44:164–174. doi: 10.1016/j.pbi.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 61.Boucher CA, Barberis PA, Trigalet AP, Demery DA. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol 131:2449–2457. doi: 10.1099/00221287-131-9-2449. [DOI] [Google Scholar]
- 62.Plener L, Manfredi P, Valls M, Genin S. 2010. PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum. J Bacteriol 192:1011–1019. doi: 10.1128/JB.01189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller G. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 64.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representation of the R. solanacearum phcBSRQ operon and structural domains of the corresponding proteins. SAMdM, S-adenosyl-l-methionine-dependent methyltransferase; TMD, transmembrane domains; D/PAD. dimerization/phosphoacceptor domain; HK. histidine kinase; RD, receiver domain. Arrows indicate the mutated positions in phcB (R22C), phcS (L161R), and phcQ (R154C). (B) Prediction of the 3-dimensional structures of the wild-type and mutated PhcB and PhcQ proteins. Structures were predicted using the Phyre2 server (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015, https://www.doi.org/10.1038/nprot.2015.053). (C) Protein alignments showing the conservation of the R22 and R154 amino acids of the PhcB and PhcQ proteins, respectively. The amino acid sequences of PhcB/Q from R. solanacearum GMI1000 were blasted against the nonredundant NCBI database to obtain a list of homologs from other organisms with identity ranging from 64% to 100% for PhcB and 53% to 100% for PhcQ. A subset of hits, covering the diversity of genera and species containing phcB and phcQ homologs, is presented. Sequences were aligned using MultAlin (http://multalin.toulouse.inra.fr/multalin/multalin.html). Red, amino acid residues highly conserved (>90%); black, positions neutrally conserved (50% to 90%); blue, positions lowly conserved (<50%). Download FIG S1, PDF file, 0.6 MB (618.5KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download Text S1, DOCX file, 0.1 MB (27.3KB, docx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correspondence between the OD600 units and the CFU for the GMI1000(pRalta) hrpG strain (wild type) and its derivative mutants. Bacteria grown in rich BG medium were serially diluted and plated at different OD600 units per milliliter. Data are from three independent experiments. A linear regression model and the associated equation are indicated for each strain. The four strains have similar correspondences between OD600 units and CFU. Download FIG S2, PDF file, 0.6 MB (650.8KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of PhcA target genes in the GMI1000(pRalta) hrpG chimeric strain and the phcBR22C, phcQR154C, and ΔphcA derivative mutants. Strains were cultivated in BG medium to an OD600 of around 0.1. Raw expression levels were normalized by rplM and rpoA expression, and ratios of gene expression in mutants versus that in the wild-type strain were calculated. Data were obtained from three independent experiments and are presented as mean Log2 (ratios) ± standard deviations. Genes positively regulated by PhcA were repressed in phcBR22C, phcQR154C, and ΔphcA mutants. Genes negatively regulated by PhcA were found induced in phcBR22C, phcQR154C, and ΔphcA mutants. *, P < 0.05 versus wild-type strain (Student’s t test). Download FIG S3, PDF file, 0.3 MB (347.1KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of genes differentially expressed in efpRE66K and ΔphcA mutants. Download Table S2, XLSX file, 0.1 MB (15.7KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Carbon, nitrogen, and phosphorus sources better metabolized in the efpRE66K mutant. Download Table S3, XLSX file, 0.1 MB (12.7KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of phcS, phcR, and phcQ genes in the GMI1000(pRalta) hrpG chimeric strain and its ΔphcB polar derivative mutant. Strains were cultivated in BG medium until mid-exponential phase. Raw expression levels were normalized by rplM expression, and ratios of gene expression in mutants versus that in the wild-type strain were calculated. Data were obtained from three independent experiments and are presented as Log2 (mean ratios) ± standard deviations. *, P < 0.01 versus the wild-type strain (Student’s t test). Download FIG S4, PDF file, 0.6 MB (609.6KB, pdf) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download Table S1, DOCX file, 0.1 MB (28.2KB, docx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S4, XLSX file, 0.1 MB (15KB, xlsx) .
Copyright © 2020 Tang et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.