Molecular motion of rotating motors delegates surface-adsorbed protein–stem cell interactions.
Abstract
Artificial rotary molecular motors convert energy into controlled motion and drive a system out of equilibrium with molecular precision. The molecular motion is harnessed to mediate the adsorbed protein layer and then ultimately to direct the fate of human bone marrow–derived mesenchymal stem cells (hBM-MSCs). When influenced by the rotary motion of light-driven molecular motors grafted on surfaces, the adsorbed protein layer primes hBM-MSCs to differentiate into osteoblasts, while without rotation, multipotency is better maintained. We have shown that the signaling effects of the molecular motion are mediated by the adsorbed cell-instructing protein layer, influencing the focal adhesion–cytoskeleton actin transduction pathway and regulating the protein and gene expression of hBM-MSCs. This unique molecular-based platform paves the way for implementation of dynamic interfaces for stem cell control and provides an opportunity for novel dynamic biomaterial engineering for clinical applications.
INTRODUCTION
Dynamic physicochemical and mechanostructural changes that mediate the behavior of adsorbed proteins and cells in space and time are among the pivotal characteristics of the cellular microenvironment in embryonic development, disease pathogenesis, tissue repair, and regeneration (1, 2). In short, (molecular) motion matters in development and cellular processes. Important studies have demonstrated that proteins and cells are sensitive to (molecular)dynamic systems such as switchable surfaces (3–5) or dynamic microenvironments, especially macroscopic mechanical tension/material shape morphing (6, 7), spatiotemporal dynamic microtopographic changes (8), and nanoscale oscillating movements (9). Harnessing the dynamic, nonequilibrium features within biological systems that are controlled at the nanoscale with molecular precision with the use of noninvasive external signals, is an attractive, yet highly challenging strategy for mediating biological processes such as stem cell fate. Achieving cell communication via the use of biologically inspired molecular motion is not only a major goal for understanding the communication mechanisms of cells with their environment (extracellular matrix and biointerface) (10, 11); it is also vital for the design of novel dynamic biomimetic materials and offers strong potential to advance future clinically relevant biomedical developments (4, 12).
Inspired by the dynamic properties of biomolecular machines (13, 14), synthetic molecular machines, such as switches, shuttles, and motors, have been developed to induce mechanical motion (15, 16). By converting various forms of chemical, thermal, or photonic energy into molecular movement in a controlled manner, we aim to synthetically introduce dynamic function into biomedical applications (5, 17–21). We have shown that the use of light-driven systems is particularly attractive, since such systems allow noninvasive external control with high spatiotemporal precision, as is evident from the rapidly emerging field of photopharmacology (22). Among various approaches to affect cell-surface interaction (1, 5), azobenzene-based switches have been applied to modulate protein structure and function (23), and to govern cell or bacterial adhesion (24, 25) or direct cell-cell contacts to induce targeted apoptosis in cancer cells (26) upon irradiation with light. However, most of these cell surfaces communicate with the cell’s surroundings, and those that do display dynamic behavior, as shown with the spatiotemporal controllable biointerface using an azopolymer (8), translate to microstructures and therefore communicate on a microscopic level rather than on a molecular level. In contrast to molecular switches, molecular motors can provide continuous motion and therefore are capable of driving a system out of equilibrium (27), which is ultimately a key requirement for communication within biological systems such as proteins and cells (13). In a different approach, Tour and co-workers (19) reported recently that ultraviolet (UV) light–activated molecular motors can perturb and drill into cell membranes in vitro using molecular-scale actuation to induce necrosis and introduce chemical species into cells. Here, we show that the fate of human bone marrow–derived mesenchymal stem cells (hBM-MSCs) can be regulated by the rotary motion of light-driven molecular motors immobilized on the surface that indirectly communicates with cells via adsorbed proteins, as depicted in Fig. 1.
Fig. 1. Molecular motion–induced restructuring of the adsorbed protein cell adhesion layer affects stem cell differentiation.

(A) Schematic illustration of the molecular motion directing the fate of hBM-MSCs. The molecular motion originating from unidirectional rotating molecular motors mediates the initial protein adsorption behavior that affects the fibronectin (Fn) adsorption, which subsequently regulates the FA cytoskeleton actin transduction pathway to govern the gene and protein expressions of hBM-MSCs. (B) Structural details of motor-modified (dynamic layer), amine-modified, and stator-modified (static control layer) surfaces.
RESULTS AND DISCUSSION
Our experimental design of surface-bound molecular motors mediating the adsorbed proteins and subsequently directing the fate of stem cells is shown in Fig. 1A. The light-driven unidirectional rotary motion of the motors is confined to the surface on which protein adsorption and cell culture take place. For the dynamic surfaces, tetra-acid–functionalized light-driven rotary molecular motors were grafted to amine-modified glass surfaces via electrostatic interactions (Fig. 1B). The tetrapodal attachment enables stable orientation of monolayers of motors on surfaces and prevents uncontrolled Brownian motion (28). It has been demonstrated that the motors used in the present study can transform UV light into repetitive unidirectional rotation and have no notable reduction in the rotational speed when immobilized to a glass substrate (28). Next, under irradiation with UV light (λmax = 365 nm), fetal bovine serum (FBS), which is conventionally present in cell culture medium as one of the main components, was added onto the motor surfaces to explore how dynamic molecular motion affects protein adsorption. After 1 hour of protein adsorption under continuous irradiation, hBM-MSCs were seeded on the treated motor surfaces for studying cell adhesion, proliferation, differentiation, and maintained stemness (Fig. 1A). Here, hBM-MSCs were selected because they are widely used in tissue engineering and regenerative medicine due to their multilineage differentiation potential into osteocytes, adipocytes, chondrocytes, etc. (29). Because the initial stage of materials into the body is the adsorption of proteins followed by cell attachment, the behavior of protein adhesion plays a key role in determining cell-interface interaction (30, 31). Note that in our experimental design, the surface-adhered protein manipulation is carried out separately from the cell culture. Thus, the cells in this study do not face a risk of genetic damage from UV irradiation because they are not in direct contact with UV light. Our design is based on the hypothesis that the type, amount, conformation, and morphology of adsorbed proteins are affected by the rotary motion of molecular motors and, through this process, directs cellular behavior and influence differentiation capabilities.
Rotary molecular motors regulate the adhesion of stem cells by altering protein adsorption
Cell adhesion is regarded as the initial physical connection of the cell with its surrounding microenvironment, which precedes all other cellular events including spreading, migration, proliferation, and differentiation (32, 33). To study the early cell response on the molecular motor surfaces treated with FBS, we seeded hBM-MSCs on substrates for 12 hours in basal medium without FBS. Both amine-coated glass slides and stator-modified surfaces were used as controls. The amine surface with and without UV irradiation indicates the potential contribution effects on the protein layer due to the UV irradiation without motion, while the stator has UV absorptions similar to the rotary motor but is not able to engage in rotary motion upon irradiation. The stator therefore serves as a control where UV irradiation might contribute to unexpected photochemical reactions due to the presence of conjugated molecular structures rather than direct UV irradiation effects, again without the presence of induced molecular motion (Fig. 1B). In the following, we refer to these substrates as follows: (i) static surface (the molecular motor surface without UV irradiation), (ii) rotary surface (the molecular motor surface with UV irradiation), (iii) amine-UV(−) surface (the amine surface without UV irradiation), and (iv) amine-UV(+) surface (the amine surface with UV irradiation). The hBM-MSC attachment and morphology were determined with a triple-label fluorescence staining of cell vinculin [focal adhesions (FAs)], actin cytoskeleton, and nucleus (Fig. 2A and Materials and Methods).
Fig. 2. Cell adhesion on the molecular motor and amine surfaces.
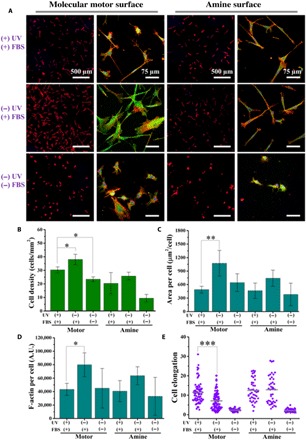
(A) Fluorescent images of hBM-MSCs cultured on the molecular motor and amine surfaces for 12 hours. Red is F-actin, visualized by tetramethyl rhodamine isothiocyanate (TRITC)–phalloidin staining, and blue is nucleus, stained by 4′,6-diamidino-2-phenylindole (DAPI); green staining is vinculin. (B to E) Cell density, area per cell, F-actin per cell, and cell elongation on the molecular motor and amine surfaces (n = 3). Data reported as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001. A.U., arbitrary units.
Figure 2A shows that hBM-MSCs on the static and rotary surfaces displayed similar spindle morphology but different in number of attached cells and cell spreading. Compared to the static surface, the rotary surface inhibited cell adhesion, spreading, and actin cytoskeleton formation but promoted cell elongation. Quantification confirms that these findings show 1.3 times more adherent cells, 2.2 times larger cell spreading, and 1.9 times more F-actin expression on the static surfaces than that on the rotary surfaces (Fig. 2, B to D). However, Fig. 2E shows 1.6 times more cell elongation on the rotary surfaces than that on the static surfaces (Fig. 2E). These variations are highly illustrative for altered long-term cellular behavior and development.
To verify whether hBM-MSC adhesion was affected solely from the rotary motor–mediated stimulation, we performed a series of control experiments using the amine-coated and stator-modified glass slides with and without exposure to light to exclude the influences of UV irradiation and potential energy transfer–mediated processes between UV light–absorbing molecular species and proteins. On the amine-UV(−) and amine-UV(+) surfaces, similar amounts of adherent cells were observed (Fig. 2A); in addition, cell spreading, amount of actin cytoskeleton, and cell elongation did not present any differences as shown in Fig. 2B. There is also no difference in initial cell behavior on the stator surfaces with and without UV irradiation (fig. S1). Together, hBM-MSC adhesion can be modulated specifically by the rotary motion of molecular motors and protein interaction without direct influence of UV irradiation on the proteins or specific molecular composition.
As a further control, we examined hBM-MSCs cultured directly on the motor- and amine-coated surfaces without FBS treatment and UV irradiation. Less adherent and more rounded cells were found on all protein-free surfaces as compared to the surfaces with protein modification (Fig. 2). Compared to the amine-coated surfaces, more adherent cells were observed on the motor-coated surfaces, indicating that the motor surface better accommodates cell adhesion.
Cell adhesion precedes all major events and will therefore most likely alter the cell morphology, resulting from adhesion to the motor surface. The communication between cell and biointerface is mediated by FA and filopodia formation that stimulates cytoskeletal tension and induces deformation in cell morphology and associated signaling cascades that thereby alter gene expression to regulate cell functions and promote tissue regeneration (34, 35). The focal adhesion and filopodia, as observed from single-cell images shown in Fig. 3A, are regarded as cellular sensors that interact with the microenvironment, which play a key role in cell adhesion, migration, organization, and differentiation (34). The formation of FAs was measured using immunofluorescent staining and confocal laser scanning microscopy (CLSM). It was difficult to find FAs in hBM-MSCs cultured on the rotary surface, whereas cells on the static surface displayed vinculin-rich FAs (Fig. 3A). Quantification of the FA area per cell reveals that the FA area per cell on the rotary surface was 2.6 times lower than that on the static surface (Fig. 3B). In the control experiments using cells cultured on the amine-coated surfaces and the protein-free surfaces, few FAs were observed as well (Fig. 3A). In addition, hBM-MSCs on the rotary surface developed numerous filopodia around the cell border, whereas cells on the static surface hardly have any filopodia extensions (Fig. 3A). Quantification of filopodia illustrates that the number of filopodia per cell on the rotary surface was 4.5 times higher than on the static surface. As a control, cells cultured on the amine-UV(−) and amine-UV(+) surfaces, as well as the protein-free surfaces, showed many filopodia, particularly on the protein-free motor surfaces.
Fig. 3. Subcellular characteristics on the molecular motor and amine surfaces.
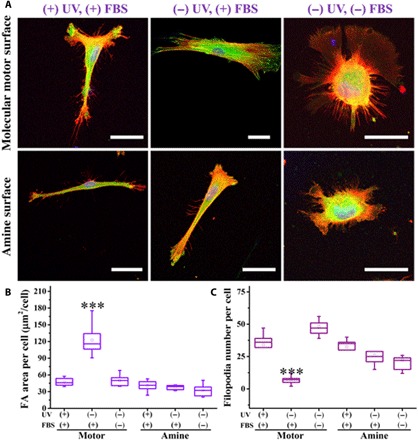
(A) Fluorescent images of a single hBM-MSC cultured on molecular motor and amine surfaces under different conditions for 12 hours. Red staining is F-actin, visualized by TRITC-phalloidin staining, and blue staining is nucleus, stained by DAPI; green staining is vinculin. Scale bars, 40 μm. (B and C) FA area per cell and filopodia number per cell on molecular motor and amine surfaces under different conditions (n = 3), respectively. Data reported as means ± SD (***P < 0.001).
Furthermore, hBM-MSCs on the rotary surface showed dense and isotropic F-actin, while cells on the static surface displayed well-defined actin stress fibers with extended and aligned morphology. As a control, the cells cultured onto the amine- and/or stator-coated surfaces with and without UV, as well as the protein-free surfaces, displayed a disordered cytoskeleton organization (Fig. 3A). The results show that the unidirectional rotary motion of molecular motors is governing protein interactions at the interface and regulating the formation of FA, filopodia, and F-actin. Comparison of subcellular and cellular characteristics showed a positive correlation of FAs with the cell number and spreading and opposite behavior between filopodia and FAs (Figs. 2 and 3). These correlations are to be expected as FAs are involved in proper cell attachment and spreading, while a cell in sensing mode (high number of filopodia) will be less surface bound (FA area per cell). Collectively, these results indicate the exciting possibility that molecular motion by means of signal transduction through the dynamic spatial organization of the interface proteins can interact and possibly direct the hBM-MSC fate.
Rotary molecular motors direct stem cell fate through motion-induced protein alterations
Proper dynamic control of stem cell fate is only possible if the biocompatibility is not inhibiting cellular behavior. hBM-MSCs were cultured on the molecular motor surfaces, and their survival was assessed by live/dead staining. After 12 hours of culture in basal medium without FBS, living-cell percentage on the motor surfaces treated with and without initial FBS remained at more than 97%, indicating that there is no eminent cytotoxicity (Fig. 4A). After 12 hours of culture in basal medium without FBS, the hBM-MSCs were further cultured in basal medium with FBS for an additional 3.5 days (total of 4 days), showing that the living-cell percentage throughout this experiment was exceeding 96%. The results indicate that there were no cytotoxic effects on the motor surfaces even after prolonged culture (Fig. 4A). In addition, to evaluate the viability of hBM-MSCs on the molecular motor surfaces, XTT viability assays were performed after 12 hours and 4 days of culture. The results show that the metabolic activity of hBM-MSCs on the rotary surface was 230 and 20% higher after 12 hours and 4 days of cell culture, respectively, compared to the cells on the static surface (Fig. 4B). Metabolic activity was corrected for a number of cells to exclude variations because of proliferation and isolate the individual cell state. In addition, the cell density over time increases, indicating that they retain their proliferative character (fig. S2).
Fig. 4. Biocompatibility on the molecular motor surfaces.
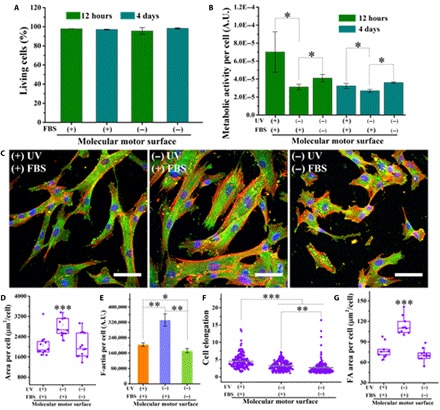
(A) The percentage of living cells and (B) metabolic activity per cell on molecular motor surfaces under different conditions for 12 hours and 4 days (n = 3). (C) Fluorescent images of hBM-MSCs cultured on molecular motor surfaces under different conditions. Scale bars, 75 μm. (D to G) Area per cell, F-actin per cell, cell elongation, and FA area per cell on molecular motor surfaces under different conditions after 4 days (n = 3). Data reported as means ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
After 4 days of cell culture, the difference in adhesion and spreading of cells on various samples was notable (Fig. 4C). Cells had a higher spreading area on all samples, developed more parallel actin stress fibers, and formed more FAs compared to the 12-hour cell culture (Figs. 2A and 4C). These results are in agreement with studies showing that cell adhesion and spreading are time dependent (36). Specifically, hBM-MSCs on the rotary surface with initial FBS adsorption displayed thin and elongated morphology; cells on the static surface with initial FBS adsorption showed more spreading; cells on the static surface without initial FBS adsorption took a rounded morphology (Fig. 4C). Quantification of cell responses of the 4-day culture shows that the area per cell on the rotary surface with initial FBS treatment and on the static surface without initial FBS treatment is significantly less than that on the static surface with initial FBS treatment (Fig. 4D). Furthermore, F-actin expression per cell on the rotary surface with initial FBS adsorption is significantly less than on the static surface with initial FBS adsorption (Fig. 4F). As shown in Fig. 4G, hBM-MSCs on the rotary surface with initial FBS adsorption and on the static surface without initial FBS adsorption display a lower FA area per cell than those on the static molecular substrate with initial FBS adsorption after 4 days of culture. The trend of FA area per cell is similar to the relative expression of F-actin and the cell spreading. These results imply that the macroscopic morphology and subcellular structure of hBM-MSCs on the rotary surface still showed significant differences even after prolonged culturing of the cells.
Evaluation of cell proliferation over the culture time is also a key step of biocompatibility testing to ascertain whether a material is suitable for biomedical applications. As indicated in fig. S2A, all molecular motor surfaces show well-supported cell proliferation for up to 14 days. Particularly, cell density on the rotary surface for 4- and 7-day cultures was less than that on the static surface. However, there is no difference between samples after 14 days of cell culture, mainly because the cell layers reach confluency. In addition, we evaluated a key protein marker (Ki67) for cell proliferation. There is no difference between the cultures on rotary and static surfaces for 7 days from the fluorescent images and quantitative data (fig. S2, B and C). The results indicate that the difference in cell density on two samples could be mainly attributed to the difference in initial attached cell number and not their proliferation rate. The results indicate that the molecular surfaces are cytocompatible by supporting the hBM-MSCs to populate, which is beneficial for future ECM deposition, as well as tissue repair and regeneration.
Having established changes in hBM-MSC behavior, the regulation of the differentiation potential of hBM-MSCs was further examined. Both immunofluorescent staining and real-time polymerase chain reaction (RT-PCR) methods were used to analyze the specific protein and gene expression level associated with the osteogenic differentiation and stemness maintenance of hBM-MSCs. After 14 days of culture in growth medium (GM) or osteogenic differentiation medium (OM), hBM-MSC morphology and the typical osteogenic marker osteopontin (OPN) were detected using immunofluorescent staining. As indicated in fig. S3, there was more cell density but less cell spreading in GM compared to the culture in OM. hBM-MSCs in GM displayed fibroblast-like morphology, whereas cells in OM have a more hexagonal (osteoblast-like) shape. The expression level of OPN in GM shows that there was no noticeable effect between the cultures on the rotary and static surfaces (Fig. 5A). However, in OM, cells on the rotary surface displayed more fluorescence intensity than on the static surface (Fig. 5A). Quantification of OPN fluorescent staining after14 days of culture indicated that under OM condition, more OPN expression was found on the rotary surface compared to that on the static surface (Fig. 5B). In addition, for the OPN gene level, there was significant difference between GM and OM (Fig. 5C). However, no significant difference in OPN gene expression was found when comparing differentiation on the rotary surface and the static surface (Fig. 5C). From the OPN protein–level data, it appears that hBM-MSCs on the rotary surface are prone to differentiate into osteoblasts.
Fig. 5. The osteogenic differentiation and stemness maintenance of hBM-MSCs on the molecular motor surfaces.

(A) Fluorescent images of hBM-MSCs cultured on molecular motor surfaces with different cell media for 14 days. Scale bars, 50 μm. Red, OPN; blue, nucleus. (B) Protein level: Quantification of the expression of OPN normalized by the cell number over 14 days of culture (n = 3). (C) Gene level: Real-time PCR (RT-PCR) analysis of relative mRNA expression levels of OPN genes in different media over 14 days of culture (n = 3). (C and D) RT-PCR analysis of relative mRNA expression level of Nanog and OCT4 genes at different time points (n = 3; *P < 0.05).
For stemness maintenance of hBM-MSCs, the expression of pluripotency gene markers (Nanog and Oct4) was measured using RT-PCR at 7 and 14 days of cell culture. The relative expression level of Nanog on the static surface was higher compared to that on the rotary surface at 7 and 14 days of cell culture (Fig. 5D). For the Oct4 gene level, higher expression on the static surface was found compared to that on the rotary surface at 7 days of cell culture. However, the relative expression level of Oct4 was the same for 14-day cell culture on both the static and rotary surfaces (Fig. 5E). These results indicate that hBM-MSCs on the static surface tend to better maintain their multipotency.
Rotary molecular motors mediate protein adsorption
To ascertain the underlying reason how the rotary surface regulates the hBM-MSC response, we performed a series of experiments to study the effect of the molecular rotary motion on the initial adsorbed protein. It is well-known that FBS includes many kinds of embryonic growth-promoting proteins for satisfying specific metabolic requirements and growth of cells (37). To verify whether different cell responses on molecular motor surfaces are caused by specific protein adsorption from FBS, we examined desorbed proteins from molecular motor and amine-modified surfaces treated with and without UV irradiation. SDS–polyacrylamide gel electrophoresis (PAGE) has been widely used to separate proteins based on their electrophoretic mobility. In general, many protein bands from FBS can be detected as described previously (38). Figure S4A shows one protein band (66 to 67 kDa) detected in extracts obtained from all samples on SDS-PAGE, and the protein profile can be attributed to albumin, which could be due to its high concentration in serum. This result indicates that there is no difference in specific protein adsorption between the rotary and static surfaces. In addition, the amount of adsorbed protein was quantified using a bicinchoninic acid (BCA) assay (fig. S4B). More protein adsorption was found on the rotary surface compared to the static surface. There is no significant difference on the amine surfaces with and without UV irradiation. This result suggests that the amount of serum protein adsorption on the dynamic surface increases by 114% compared to the amount of protein adsorbed on the static surface.
The adsorption of proteins at interfaces is a dynamic process during which proteins may adsorb, reorganize, and detach (39). We used a quartz crystal microbalance with dissipation (QCM-D) to real-time monitor both the adsorbed protein mass and energy-dissipating properties on the silicon chips modified with molecular motor and amine functionality (Fig. 6, A and B). The adsorption tracking of FBS and subsequently fibronectin (Fn) was followed, noting that Fn produced by cells plays a major role in the attachment of anchorage-dependent cells (40). FBS injection resulted in a decreasing frequency (ΔF) and an increasing dissipation (ΔD) on all samples, suggesting protein adsorption. As shown in Fig. 6A, the ΔF decreased and stabilized around −133 Hz, and ΔD increased until stabilizing at around 1.1 × 10−4 on the motor surfaces with and without applying UV irradiation, confirming the formation of equilibrium adsorption. Then, the motor surfaces were rinsed with phosphate-buffered saline (PBS) to remove the loosely bound proteins. Both the ΔF and ΔD on the static surface reached the stable signal faster than on the rotary surface. The shorter time to reach equilibrium indicates that desorption of the protein on the static surface is faster than those on the dynamic surface (41). In addition, there was a slight difference in ΔD between the static and rotary surfaces. The change in ΔF of 57 Hz on the rotary surface is higher than the change in ΔF of 45 Hz on the static surface (Fig. 6A), which indicates enhanced protein adsorption on the rotary surface (Fig. 6B), which is in agreement with the BCA assay (fig. S4C). Subsequently, Fn was applied at 37°C under flow across the motor surfaces with the adsorbed bovine serum albumin (BSA) layer still present but without rotation until equilibrium adsorption was reached. The change in ΔF of 26 Hz on the static surface is higher than that in ΔF of ~0 Hz on the rotary surface (Fig. 6A), which indicates higher Fn adsorption on the static surface compared to the rotary surface (Fig. 6A). Notably, hardly any Fn was adsorbed on the rotary surface (Fig. 6A). As a control, the QCM-D experiment was performed on the amine-modified surfaces using the same procedure (fig. S4B). We found that the F curves almost overlapped all along the serum protein and Fn adsorption on the amine surfaces with and without applying UV irradiation. It indicates that UV irradiation had no effect on the surface amount of serum protein and Fn adsorption.
Fig. 6. The adsorbed protein behaviors on molecular motor and amine surfaces.

(A) Protein adsorption on the molecular motor surface under different conditions measured by QCM-D. (B) Serum protein mass on the samples quantified by QCM-D (n = 3 to 4). (C) Fn mass quantified from QCM-D curves (n = 3 to 4). (D) Circular dichroism (CD) spectra of protein from BSA adsorbed on the molecular motor surface under different conditions as indicated. Data reported as means ± SD (*P < 0.05, **P < 0.01).
The protein mass adsorbed on each surface can be calculated by the Voigt model using QTools software (Q-Sense) (42). As indicated in Fig. 6B, a greater adsorbed mass was found on the rotary surface (1720 ± 546 ng/cm2) than on the static surface (979 ± 312 ng/cm2). However, there is no difference in the amount of adsorbed protein between amine-UV(−) and amine-UV(+) surfaces. The result is consistent with the BCA assay (fig. S4C), which further demonstrates that the molecular motion of motors is able to increase the amount of adsorbed serum protein. In addition, the preadsorbed status of BSA significantly affected the adsorption of other proteins (43–45). In our study, the trend of Fn adsorption is opposite (Fig. 6C). More Fn adsorption was found on the static surface (963 ± 296 ng/cm2) than on the rotary surface (2.7 ± 4.7 ng/cm2). However, for the amine-coated surfaces, there is no difference between UV and no UV treatments. These results explain why initial cell adhesion was better on the static surface.
Circular dichroism (CD) spectroscopy is well known to be a powerful tool for the rapid analysis of the secondary structure and folding properties of proteins (12). Figure 6D and fig. S4D show the two characteristic negative bands at 208 and 225 nm, which can be attributed to n/π* transition associated with the α-helix structure of BSA. The CD curves of BSA on all surfaces were similar in shape, suggesting that α-helix was still the major component of BSA secondary structure. However, the CD intensity (208 nm) of BSA on the rotary surface was less than that on the static surface, which reflects the loss of the α-helix structure by unfolding (Fig. 6D). As a control, the CD curves of BSA on the amine-UV(−) and amine-UV(+) surfaces showed no difference (fig. S4D), indicating that UV irradiation did not affect the BSA secondary structure. Using quantum mechanics/molecular mechanics (MM) methods and MM dynamics simulations, Xia and co-workers (46) found that photoswitchable azobenzene was able to control the unfolding of the secondary structure (α-helix) of a peptide by photoisomerization affecting the peptide affinity. Presumably, the molecular motion of motors is also able to stretch and unfold, to some extent, the BSA α-helical chains. During the interaction between molecular motors and proteins, the reversible changes in BSA conformations could also exist according to the literatures (47, 48). It is interesting to elucidate this phenomenon in the future on a more fundamental level. The unfolding of BSA α-helix structure significantly reduced the following Fn adsorption (Fig. 6C), consistent with the literature (49).
To provide a molecular rationale for the observed conformational changes of the proteins adsorbed on the active surface, we performed molecular dynamics (MD) simulations. The simulated system consists of a functionalized SiO2 surface covered with self-assembled tetrapodal motor molecules. The effect of the surface on the conformational stability of a prototypical protein, hen egg-white lysozyme, was monitored during a 25-ns simulation. Surfaces with either inactive or active (i.e., rotating) motors were investigated. The results (fig. S5) indicate that the secondary structure of the protein, as quantified by the root mean square deviation, is significantly affected by the active surface, although minor secondary structure changes occur due to adsorption on the inactive surface as well. A direct interaction between the nanorotors and the protein is presumably responsible for this effect (fig. S5), the effect being increased upon activation of the motors. Visual inspection (fig. S5) reveals loss of secondary structural motifs, including α-helices in accordance to the CD data.
Last, atomic force microscopy (AFM) was used to visualize the morphology and distribution of adsorbed serum proteins on the molecular motor surfaces (fig. S6). Proteins on the rotary surface show an ellipsoidal morphology and organized arrangement, whereas proteins on the static surface display a rounded morphology and disordered arrangement. These protein alterations are also consistent with other studies where physicochemical surface parameters such as chemical composition and wettability are also known to affect the differentiation of hBM-MSCs (49, 50). It was demonstrated that wettability affects the osteogenesis of hBM-MSCs, and, in particular, the hydrophobic surface displayed better osteogenesis compared to the hydrophilic surface (51). Performing AFM characterization of BSA adsorbed on those types of monolayers shows that also in those cases, the morphology is highly altered, and the hydrophobic surface displays a very rough BSA protein layer similar to what is found for the surface altered with molecular motion (fig. S7). Together, these experimental results indicate that the molecular motion of motors regulates the initial serum adsorption behavior (i.e., amount, conformation, and morphology) and, as a consequence, influences the Fn adsorption transduced into stem cell behavior.
CONCLUSIONS
Dynamic motion of molecular motors grafted on surfaces is used to direct the fate of hBM-MSCs by mediating the passivating protein adhesion layer. These stem cells on the dynamically altered surface were more prone to differentiate into osteoblasts, whereas on the static surface, they tend to better maintain their stemness. The unique character of the system was traced back to the underlying mechanism that is associated with the cell-instructive protein layer that is delegating many of the material-driven cell-controlling phenomena (i.e., amount, conformation, and morphology), which is here regulated solely by the molecular motion. Unidirectional rotating molecular motors increased serum albumin adsorption and interacted on a molecular scale that results in a decreased α-helix secondary structure, which subsequently affected the adsorbed amount of Fn. The distinct protein adsorption behavior influenced the FA cytoskeleton actin transduction pathway, as well as the macroscopic cell adhesion and morphology, evident from studying FA, filopodia, and actin stress fibers and, as a result, mediates the fate of hBM-MSCs. It was shown that the surface after the interaction of rotary motors and proteins ultimately leads to osteogenic differentiation, while without the molecular motion, a better maintenance of multipotency was found with respect to the dynamic conditions. Besides providing a unique way to introduce dynamic influences on cell cultures, the molecular motor–based surface offers numerous opportunities for mechanical stimulation and control of cell fate and responsive biomimetic materials and paves the road for the synthetic molecular motor toward biomedical and clinical applications.
MATERIALS AND METHODS
Preparation of molecular motor
Detailed synthetic procedures and characterization of the final compound were reported in our previous study (28). A key step to form the central olefinic bond by coupling diazo and thioketone precursors was the Barton-Kellogg reaction, providing the tetramethyl ester compound, which was then hydrolyzed to afford the tetra-acid–functionalized motor. Kinetic studies of the assembled monolayer of motors were performed by UV-visible spectroscopy and show similar behavior as that obtained in solution, which is indicative of the light-driven rotary motion of motors on surfaces (28).
Cell culture
hBM-MSCs (pool; passage numbers: 3 to 5; Lonza) were used for the cell experiments. The GM consisted of α-modified Eagle medium (Gibco), 10% (v/v) FBS (Gibco), and 0.1% (v/v) ascorbic acid 2-phosphate (Sigma). Cells were incubated at 37°C and 5% CO2. The cells were harvested at approximately 80 to 90% confluence from T75 culture flasks by trypsin for 3 to 5 min at 37°C for further subcultures. All substrates were treated with 70% ethanol for sterilization, placed in well plates, and washed by PBS. Afterward, mesenchymal stem cells derived from hBM-MSCs were seeded onto the samples in well plates at a density of 5000 cells/cm2. All substrates were stored in an incubator at 37°C and 5% CO2.
Cell staining
The hBM-MSCs on the samples were fixated with 3.7% paraformaldehyde (Sigma) in PBS for 20 min, followed by rinsing three times with PBS. Afterward, the cell membrane was permeabilized with 0.5% Triton X-100 (Sigma) solution for 3 min. BSA (5%) in PBS solution was added for 30 min to block nonspecific binding. After removing the BSA solution, the hBM-MSCs were treated with anti-vinculin (1:100; clone hVin-1, Sigma) for FAs or anti-Ki67 (1:1000; ab15580, Abcam) for cell proliferation capacity, washed three times with PBS, and stained with a secondary goat anti-mouse antibody (1:100; Jackson ImmunoLabs) or goat anti-rabbit antibody (1:100; Jackson ImmunoLabs). In addition, 4′,6-diamidino-2-phenylindole (DAPI) and tetramethyl rhodamine isothiocyanate (TRITC)/fluorescein isothiocyanate (FITC)–phalloidin were used to stain the cell nuclei and F-actin, respectively. Cells were observed using a Leica TCS SP2 CLSM equipped with 40× and 63× water immersion objectives [numerical aperture (NA), 0.80 and 0.90, respectively] or TissueFAXS with a Zeiss Axio Imager Z1 Microscope System (TissueGnostics GmbH, Vienna, Austria) at ×10 magnification. The quantitative analysis of the FA area was performed using Focal Adhesion Analysis Server (52). The cell density, nucleus area per cell, area per cell, F-actin, and vinculin expression per cell were quantified using TissueQuest software (TissueGnostics GmbH, Vienna, Austria). The area and intensity of fluorescence were normalized to a number of cells analyzed using the DAPI channel. ImageJ software was used to measure the cell length and width. A measurement of cell elongation ranged from 1 (a perfect circle) to ∞ (a straight line), which was calculated as L/W (L, length; W, width).
Live/dead staining
A live/dead assay was performed using PBS containing calcein-AM (2 μM; Molecular Probes, Invitrogen Detection Technologies) and ethidium homodimer-1 (4 μM; Molecular Probes, Invitrogen Detection Technologies) to stain the cells for 30 min before being observed under a Leica TCS SP2 CLSM equipped with a 40× water immersion objective and an NA of 0.80.
Cell viability analysis
Cellular metabolic activity was measured using an XTT assay (AppliChem A8088). Briefly, 500 μl of XTT reaction mixture (0.1 ml of activation reagent and 5 ml of XTT reagent for one plate) was added to each well with 1.0 ml of medium, and samples were incubated at 37°C in a humidified atmosphere of 5% CO2 for 3 hours. The 200-μl mixtures were added to a 96-well plate, and the absorbance at 485 and 690 nm was recorded using a microplate reader. Experiments were performed in triplicate.
Osteogenic differentiation assays
OM was composed of GM and chemical supplements (5 × 10−7 M dexamethasone and 10 mM β-glycerolphosphate, both from Sigma). hBM-MSCs were cultured for 14 days in differentiation medium. Subsequently, the hBM-MSCs were fixed with 3.7% paraformaldehyde in PBS for 20 min. Afterward, cells were permeabilized with 0.2% Triton X-100 for 10 min. BSA (5%) in PBS solution was added for 30 min to block nonspecific binding. The primary antibody against OPN (1:1000; ab8448) was used in combination with a secondary goat anti-rabbit antibody (1:100; Red, Jackson ImmunoLabs). In addition, DAPI and FITC-phalloidin were used to stain the cell nuclei and F-actin, respectively.
Quantitative RT-PCR
The expression levels of stemness- and osteogenic differentiation–related genes of hBM-MSCs were detected by quantitative RT-PCR (qRT-PCR). Briefly, the cells were cultured in GM and OM for 7 and/or 14 days. Total RNAs were extracted from the supernatant using the InviTrap Spin Cell RNA mini kit (STRATEC Molecular GmbH, Germany) according to the manufacturer’s instructions. Next, complementary DNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) according to the user manual. The primers for the target genes are listed in table S1. Then, qRT-PCR was performed using the CFX384 real-time PCR detection system (Bio-Rad). The relative expression level of each target gene was then calculated using the 2−△△Ct method (53). The expression levels of the target genes were normalized to that of the housekeeping gene, 36B4.
Preparation of motor-functionalized monolayer on the glass and SiO2-coated QCM-D sensor chip
Glass slides (Thermo Fisher Scientific) used in this research were cleaned by immersing in a piranha solution (H2SO4:30%H2O2 = 3:1) at 90°C for 1 hour and rinsed three times with Milli-Q water, followed by methanol, and dried under a stream of N2 before surface modification. The cleaned glass slides were silanized by immersing in a 2 mM solution of 3-aminopropyl(diethoxy)methylsilane in freshly distilled toluene at room temperature for 12 hours, then rinsed with toluene and methanol, and dried under a stream of N2. The amine-coated glass slides were immersed in N,N′-dimethylformamide (DMF) solution containing molecular motor (10−4 M) at room temperature for 12 to 24 hours. Last, the slides were washed with DMF, Milli-Q water, and methanol, followed by drying under a stream of N2.
SiO2-coated QCM-D sensor chips were cleaned with 2% (w/v) SDS for 30 min, followed by submersion in Milli-Q water and dried under a stream of N2. Subsequently, the cleaned chips were treated in a UV/ozone plasma for 30 min and then were silanized by immersion in 2 mM solution of 3-aminopropyl(diethoxy)methylsilane in freshly distilled toluene at room temperature for 12 hours. The amine-functionalized chips were thoroughly rinsed with toluene and MeOH and then blow-dried under N2. Last, the chips were immersed in DMF solution containing molecular motor (10−4 M) at room temperature for 12 to 24 hours. The motor-functionalized chips were washed with DMF and Milli-Q water and methanol and then dried under a stream of N2.
Protein adsorption measured by QCM-D
QCM-D measurements were performed on a Q-Sense-E4 instrument (Q-Sense, Sweden) with dissipation. QCM-D SiO2 chips modified with the amine group or molecular motor were placed in window-equipped QCM-D chambers. The normal QCM-D sensitivity for protein adsorption in the liquid is ~1.8 ng/cm2 and ~1 × 10−7 for dissipation. All serum protein adsorption measured by QCM-D were conducted at a flow rate of 10 μl/min controlled by a peristaltic pump (Ismatec SA, Glattbrugg, Switzerland). The UV light with a wavelength of 365 nm on the top of window chamber was turned on, and an initial baseline was established by flowing PBS buffer at 25°C. Fresh FBS was injected to obtain the adsorption plateau and establish a stable line at a temperature of 25°C. After complete adsorption, PBS was injected to remove loosely attached proteins and to establish a second stable baseline. Removing the UV light, the temperature was adjusted from 25° to 37°C. When the frequency and dissipation curves did not change any more and established another baseline. Fresh Fn solution (20 μg/ml) was flowed and rinsed with PBS to achieve the final baseline. The differences in frequency and dissipation factor between the two baselines were used to calculate the protein adsorption. The mass of adsorbed protein was attained by fitting ΔF and ΔD to the Voigt model using QTools software package.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
The amine and molecular motor–functionalized glass slides were incubated with FBS solution for 1 hour at room temperature without or with 365 nm of UV light irradiation and then rinsed three times (5 min each time) with fresh PBS. SDS solution (2%) was added to each sample at 4°C overnight to detach the adsorbed protein from the surface. The detached protein solution for each sample was concentrated in Thermo Scientific Pierce concentrators with a molecular weight cutoff of 1000 kDa by centrifugation at a speed of 3400 rpm at 4°C. The concentrated protein solutions were denatured and released using heat treatment with a 2% SDS solution for 10 min and were analyzed by SDS-PAGE. Molecular weight markers and the concentrated proteins detached from functionalized surfaces were subjected to 12% SDS-PAGE at 200 V for 1 hour according to the standard procedure.
CD spectra
CD spectra were used to investigate the conformation of the initial adsorbed BSA protein on the molecular motor and amine-coated glass surfaces. Typically, all samples were immersed in BSA solution (10 mg/ml) in PBS for 1 hour, rinsed three times (5 min each time) by PBS, and then recorded at room temperature under nitrogen by a Jasco J-815 CD spectrophotometer for wavelengths from 170 to 340 nm.
Atomic force microscope
The protein images were measured by a BioScope Catalyst AFM instrument (Bruker, Billerica, MA, USA) with NanoScope Analysis software. All measurements were performed in the quantum-mechanical nanomapping mode with a large amplitude using Bruker ScanAsyst.
Statistical analysis
All data points are expressed as mean ± SD. Statistical analysis was performed using Origin 9.0 software. All the data were analyzed using one-way analysis of variance (ANOVA) with Tukey’s test to determine differences between groups. A value of P < 0.05 was considered statistically significant: *P < 0.05, **P < 0.01, ***P < 0.001.
MD simulation
All-atom MD simulations were performed using the GROMACS simulation package (version 2018.1.). The initial SiO2 surface was built as a 30-nm–by–30-nm–by–3-nm supercell by using the InorganicBuilder software, which is distributed as a part of VMD (54). After the plain surface had been obtained, the surface was completely functionalized with NH3+ groups. The positions of all atoms in the SiO2 slab were restrained using harmonic potentials, while the functional groups and capping OH groups were allowed to rotate and interact freely with their surroundings. Intramolecular parameters for the functional groups were obtained from the Automated Topology Builder repository (55), and intermolecular parameters were obtained from the literature (56). Following the experimental procedure, 40 tetrapodal motors (28) were inserted into the solvent phase and allowed to anchor onto the surface. The binding modes of the tetrapodal motors are consistent with the overall chemical picture presented in the literature. In the following step, a hen egg-white lysozyme (Protein Data Bank: 1AKI) was inserted into the water phase above the functionalized surface. A set of 25-ns-long simulations was performed with an NPT (constant temperature and pressure, the isothermal-isobaric) ensemble (P = 1 bar, T = 300 K), during which the protein adsorbed onto the surface. Two active surface models were set up using coordinates extracted from the 25-ns production simulations. In the first type of simulation, a single molecular motor in contact with the protein was selected and subjected to enforced rotation for 75 ps, during which it was observed rotating 22 full rotations. This was followed by a standard NPT simulation for 3 ns with no rotation. The second type of simulation was set up the same way, but the motors were allowed to rotate three times after every nanosecond, resulting in a total of nine full rotations. The rotational constant was kept at 250 kJ/(mol × nm2), and the rotational rate was set to 120°/ps in both systems. The results obtained from these simulations were compared against two references: protein adsorbed on an inactive surface and protein in pure water. All the equilibration and NPT production simulations were performed using the leap-frog algorithm with a time step of 1 fs (0.1 fs in case of active surfaces). Nonbonded interactions were cut off at 1 nm. Long-range Coulombic interactions were accounted for with reaction-field electrostatics.
Supplementary Material
Acknowledgments
Funding: We thank for financial support the China Scholarship Council (no. 201406630003), the UMCG Microscopy and Imaging Center (UMIC) (NWO-grant 40-00506-98-9021), the Scientific Research Foundation of Qingdao University (grant no. DC1900009689), the Natural Science Foundation of Shandong Province, China (grant no. ZR2019QC007), the European Research Council (ERC; advanced grant no. 694345 to B.L.F.), and the Ministry of Education, Culture and Science of the Netherlands (Gravitation program no. 024.001.035). Author contributions: Q.Z., J.C., P.v.R., and B.L.F. conceived the idea. Q.Z., J.C., P.v.R., and B.L.F. designed the experiments and interpreted the data. Q.Z., J.C., and Y.L. performed the experiments. P.A.V., S.T., and S.J.M. performed the molecular dynamics simulations. Q.Z., J.C., P.V.R., and B.L.F. co-wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: P.v.R is also a co-founder, scientific advisor, and share-holder of BiomACS BV, a biomedical-oriented screening company. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/5/eaay2756/DC1
Fig. S1. Altered cell behavior induced by molecular motion–mediated protein layer restructuring.
Fig. S2. Proliferation capacity of MSCs on molecular motor–functionalized surfaces in static and rotating mode upon protein adhesion.
Fig. S3. Immunofluorescence analysis of cell morphology during osteogenic differentiation of MSCs on dynamic and static molecular motor–functionalized surfaces.
Fig. S4. Protein adhesion and structural integrity mediated by molecular motion and UV irradiation.
Fig. S5. MD simulations of protein–molecular motor interactions and motion-induced structural deformation.
Fig. S6. AFM analysis of motion-induced alterations to structural alterations of protein adhesion layers.
Fig. S7. Physicochemical influences on the morphology of protein adhesion layers.
Table S1. Primer sequences of human-specific genes used for qRT-PCR.
REFERENCES AND NOTES
- 1.Li Y., Xiao Y., Liu C., The horizon of materiobiology: A perspective on material-guided cell behaviors and tissue engineering. Chem. Rev. 117, 4376–4421 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Piccolino M., Biological machines: From mills to molecules. Nat. Rev. Mol. Cell Biol. 1, 149–152 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Sur S., Matson J. B., Webber M. J., Newcomb C. J., Stupp S. I., Photodynamic control of bioactivity in a nanofiber matrix. ACS Nano 6, 10776–10785 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uto K., Tsui J. H., DeForest C. A., Kim D.-H., Dynamically tunable cell culture platforms for tissue engineering and mechanobiology. Prog. Polym. Sci. 65, 53–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertus J., Browne W. R., Feringa B. L., Dynamic control over cell adhesive properties using molecular-based surface engineering strategies. Chem. Soc. Rev. 39, 354–378 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Subramony S. D., Dargis B. R., Castillo M., Azeloglu E. U., Tracey M. S., Su A., Lu H. H., The guidance of stem cell differentiation by substrate alignment and mechanical stimulation. Biomaterials 34, 1942–1953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao B., Yang Q., Zhao X., Jin G., Ma Y., Xu F., 4D bioprinting for biomedical applications. Trends Biotechnol. 34, 746–756 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Rianna C., Rossano L., Kollarigowda R. H., Formiggini F., Cavalli S., Ventre M., Netti P. A., Spatio–temporal control of dynamic topographic patterns on azopolymers for cell culture applications. Adv. Funct. Mater. 26, 7572–7580 (2016). [Google Scholar]
- 9.Kang H., Wong D. S. H., Yan X., Jung H. J., Kim S., Lin S., Wei K., Li G., Dravid V. P., Bian L., Remote control of multimodal nanoscale ligand oscillations regulates stem cell adhesion and differentiation. ACS Nano 11, 9636–9649 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Redwine W. B., Hernández-López R., Zou S., Huang J., Reck-Peterson S. L., Leschziner A. E., Structural basis for microtubule binding and release by dynein. Science 337, 1532–1536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuta A., Amino M., Yoshio M., Oiwa K., Kojima H., Furuta K., Creating biomolecular motors based on dynein and actin-binding proteins. Nat. Nanotechnol. 12, 233–237 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Bachand G. D., Bouxsein N. F., VanDelinder V., Bachand M., Biomolecular motors in nanoscale materials, devices, and systems. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 6, 163–177 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Howard J., Molecular motors: Structural adaptations to cellular functions. Nature 389, 561–567 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Mickler M., Schleiff E., Hugel T., From biological towards artificial molecular motors. ChemPhysChem 9, 1503–1509 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Balzani V., Credi A., Raymo F. M., Stoddart J. F., Artificial molecular machines. Angew. Chemie Int. Ed. 39, 3348–3391 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Kassem S., van Leeuwen T., Lubbe A. S., Wilson M. R., Feringa B. L., Leigh D. A., Artificial molecular motors. Chem. Soc. Rev. 46, 2592–2621 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Watson M. A., Cockroft S. L., Man-made molecular machines: Membrane bound. Chem. Soc. Rev. 45, 6118–6129 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Yu G., Yung B. C., Zhou Z., Mao Z., Chen X., Artificial molecular machines in nanotheranostics. ACS Nano 12, 7–12 (2017). [DOI] [PubMed] [Google Scholar]
- 19.García-López V., Chen F., Nilewski L. G., Duret G., Aliyan A., Kolomeisky A. B., Robinson J. T., Wang G., Pal R., Tour J. M., Molecular machines open cell membranes. Nature 548, 567–572 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Broichhagen J., Podewin T., Meyer-Berg H., von Ohlen Y., Johnston N. R., Jones B. J., Bloom S. R., Rutter G. A., Hoffmann-Röder A., Hodson D. J., Trauner D., Optical control of insulin secretion using an incretin switch. Angew. Chemie Int. Ed. 54, 15565–15569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber D. M., Schönberger M., Burgstaller J., Levitz J., Weaver C. D., Isacoff E. Y., Baier H., Trauner D., Optical control of neuronal activity using a light-operated GIRK channel opener (LOGO). Chem. Sci. 7, 2347–2352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velema W. A., Szymanski W., Feringa B. L., Photopharmacology: Beyond proof of principle. J. Am. Chem. Soc. 136, 2178–2191 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Zhu M., Zhou H., Azobenzene-based small molecular photoswitches for protein modulation. Org. Biomol. Chem. 16, 8434–8445 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Kadem L. F., Suana K. G., Holz M., Wang W., Westerhaus H., Herges R., Selhuber-Unkel C., High-frequency mechanostimulation of cell adhesion. Angew. Chem. Int. Ed. 56, 225–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaran S., van Weerd J., Voskuhl J., Karperien M., Jonkheijm P., Photoresponsive cucurbit[8]uril-mediated adhesion of bacteria on supported lipid bilayers. Small 11, 6187–6196 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Shi P., Ju E., Wang J., Yan Z., Ren J., Qu X., Host–guest recognition on photo-responsive cell surfaces directs cell–cell contacts. Mater. Today 20, 16–21 (2017). [Google Scholar]
- 27.van Leeuwen T., Lubbe A. S., Štacko P., Wezenberg S. J., Feringa B. L., Dynamic control of function by light-driven molecular motors. Nat. Rev. Chem. 1, 0096 (2017). [Google Scholar]
- 28.Chen J., Chen K.-Y., Carroll G. T., Feringa B. L., Facile assembly of light-driven molecular motors onto a solid surface. Chem. Commun. 50, 12641–12644 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R., Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Anderson J. M., Rodriguez A., Chang D. T., Foreign body reaction to biomaterials. Semin. Immunol. 20, 86–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson J. M., Mechanisms of inflammation and infection with implanted devices. Cardiovasc. Pathol. 2, 33–41 (1993). [Google Scholar]
- 32.Schwarz U. S., Bischofs I. B., Physical determinants of cell organization in soft media. Med. Eng. Phys. 27, 763–772 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Nguyen A. T., Sathe S. R., Yim E. K. F., From nano to micro: Topographical scale and its impact on cell adhesion, morphology and contact guidance. J. Phys. Condens. Matter 28, 183001 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Mattila P. K., Lappalainen P., Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Prager-Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A. D., Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q., Wünnemann P., Kühn P. T., de Vries J., Helmin M., Böker A., van Kooten T. G., van Rijn P., Mechanical properties of aligned nanotopologies for directing cellular behavior. Adv. Mater. Interfaces 3, 1600275 (2016). [Google Scholar]
- 37.Ham R. G., McKeehan W. L., Media and growth requirements. Methods Enzymol. 58, 44–93 (1979). [DOI] [PubMed] [Google Scholar]
- 38.Iqbal M. Z., Ma X., Chen T., Zhang L., Ren W., Xiang L., Wu A., Silica-coated super-paramagnetic iron oxide nanoparticles (SPIONPs): A new type contrast agent of T1 magnetic resonance imaging (MRI). J. Mater. Chem. B 3, 5172–5181 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Roach P., Farrar D., Perry C. C., Interpretation of protein adsorption: Surface-induced conformational changes. J. Am. Chem. Soc. 127, 8168–8173 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Sastry S. K., Burridge K., Focal adhesions: A nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261, 25–36 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Brewer S. H., Glomm W. R., Johnson M. C., Knag M. K., Franzen S., Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir 21, 9303–9307 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Voinova M. V., Rodahl M., Jonson M., Kasemo B., Viscoelastic acoustic response of layered polymer films at fluid-solid interfaces: Continuum mechanics approach. Phys. Scr. 59, 391–396 (1999). [Google Scholar]
- 43.Alves C. M., Reis R. L., Hunt J. A., The competitive adsorption of human proteins onto natural-based biomaterials. J. R. Soc. Interface 7, 1367–1377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pegueroles M., Tonda-Turo C., Planell J. A., Gil F. -J., Aparicio C., Adsorption of fibronectin, fibrinogen, and albumin on TiO2: Time-resolved kinetics, structural changes, and competition study. Biointerphases 7, 1–13 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Zelzer M., Albutt D., Alexander M. R., Russell N. A., The role of albumin and fibronectin in the adhesion of fibroblasts to plasma polymer surfaces. Plasma Processes Polym. 9, 149–156 (2012). [Google Scholar]
- 46.Xia S., Cui G., Fang W., Thiel W., How photoisomerization drives peptide folding and unfolding: Insights from QM/MM and MM dynamics simulations. Angew. Chemie. 128, 2107–2112 (2016). [DOI] [PubMed] [Google Scholar]
- 47.El Kadi N., Taulier N., Le Huérou J. Y., Gindre M., Urbach W., Nwigwe I., Kahn P. C., Waks M., Unfolding and refolding of bovine serum albumin at acid pH: Ultrasound and structural studies. Biophys. J. 91, 3397–3404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda K., Wada A., Yamamoto K., Moriyama Y., Aoki K., Conformational change of bovine serum albumin by heat treatment. J. Protein Chem. 8, 653–659 (1989). [DOI] [PubMed] [Google Scholar]
- 49.Dong L., Cheng K., Zhou Y., Yu M., Gong J., Lin Y., Luo Q., Wang Q., Weng W., Wang H., Surface atomic structure directs the fate of human mesenchymal stem cells. ACS Appl. Mater. Interfaces 9, 15274–15285 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Schaap-Oziemlak A. M., Kühn P. T., van Kooten T. G., van Rijn P., Biomaterial–stem cell interactions and their impact on stem cell response. RSC Adv. 4, 53307–53320 (2014). [Google Scholar]
- 51.Hao L., Yang H., Du C., Fu X., Zhao N., Xu S., Cui F., Mao C., Wang Y., Directing the fate of human and mouse mesenchymal stem cells by hydroxyl-methyl mixed self-assembled monolayers with varying wettability. J. Mater. Chem. B 2, 4794–4801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berginski M. E., Gomez S. M., The focal adhesion analysis server: A web tool for analyzing focal adhesion dynamics. F1000Res. 2, 68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Malde A. K., Zuo L., Breeze M., Stroet M., Poger D., Nair P. C., Oostenbrink C., Mark A. E., An automated force field topology builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 7, 4026–4037 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Snyder J. A., Abramyan T., Yancey J. A., Thyparambil A. A., Wei Y., Stuart S. J., Latour R. A., Development of a tuned interfacial force field parameter set for the simulation of protein adsorption to silica glass. Biointerphases 7, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/5/eaay2756/DC1
Fig. S1. Altered cell behavior induced by molecular motion–mediated protein layer restructuring.
Fig. S2. Proliferation capacity of MSCs on molecular motor–functionalized surfaces in static and rotating mode upon protein adhesion.
Fig. S3. Immunofluorescence analysis of cell morphology during osteogenic differentiation of MSCs on dynamic and static molecular motor–functionalized surfaces.
Fig. S4. Protein adhesion and structural integrity mediated by molecular motion and UV irradiation.
Fig. S5. MD simulations of protein–molecular motor interactions and motion-induced structural deformation.
Fig. S6. AFM analysis of motion-induced alterations to structural alterations of protein adhesion layers.
Fig. S7. Physicochemical influences on the morphology of protein adhesion layers.
Table S1. Primer sequences of human-specific genes used for qRT-PCR.


