The introduced data-driven tool allows simultaneous feature selection, model inference, and marked cost and quality gains.
Abstract
Finding reliable discrete approximations of complex systems is a key prerequisite when applying many of the most popular modeling tools. Common discretization approaches (e.g., the very popular K-means clustering) are crucially limited in terms of quality, parallelizability, and cost. We introduce a low-cost improved quality scalable probabilistic approximation (SPA) algorithm, allowing for simultaneous data-driven optimal discretization, feature selection, and prediction. We prove its optimality, parallel efficiency, and a linear scalability of iteration cost. Cross-validated applications of SPA to a range of large realistic data classification and prediction problems reveal marked cost and performance improvements. For example, SPA allows the data-driven next-day predictions of resimulated surface temperatures for Europe with the mean prediction error of 0.75°C on a common PC (being around 40% better in terms of errors and five to six orders of magnitude cheaper than with common computational instruments used by the weather services).
INTRODUCTION
Computers are finite discrete machines. Computational treatment and practical simulations of real-world systems rely on the approximation of any given system’s state X(t) (where t = 1,…, T is a data index) in terms of a finite number K of discrete states S = {S1,…, SK} (1, 2). Of particular importance are discretization methods that allow a representation of the system’s states X(t) as a vector of K probabilities for the system to be in some particular state Si at the instance t. Components of such a vector—ΓX(t) = (Γ1X (t), Γ2X (t),…, ΓKX (t))—sum up to one and are particularly important, since they are necessary for Bayesian and Markovian modeling of these systems (3–5).
Bayesian and Markovian models belong to the most popular tools for mathematical modeling and computational data analysis problems in science (with more than 1 million literature references each, according to Google Scholar). They were applied to problems ranging from social and network sciences (6) to a biomolecular dynamics and drug design (7–9), fluid mechanics (10), and climate (11). These models dwell on the law of the total probability and the concept of conditional expectation, saying that the exact relation between the finite probabilistic representations ΓY(t) and ΓX(t) for the given discretization and of any two processes X(t) and Y(t) is given as a linear model
| (1) |
where Λ is the m × n matrix of conditional probabilities . In the following, we assume that these conditional probabilities are stationary (independent of data index t) and only depend on the discrete state indices i and j. This linear model (Eq. 1) is exact in a probabilistic sense, meaning that it does not impose a modeling error even if the underlying dynamics of X(t) and Y(t) are arbitrarily complex and nonlinear.
This property of (Eq. 1) is a direct consequence of the law of the total probability and a definition of the conditional probability, saying that the probability to observe Y(t) in any discrete state can be exactly expressed as a sum over j probabilities to observe Y(t) in this particular state conditioned on observing X(t) in any of the particular states . If Λ is known, then the linear model (Eq. 1) provides the best relation between the two probabilistic representation ΓY(t) and ΓX(t) in given discretizations and (2–4).
A particular, and very important, case of the Bayesian models (Eq. 1) emerges when choosing Y(t) as X(t + 1), where t is a time index. The relation matrix Λ is then a left-stochastic square matrix of stationary transition probabilities between discrete states, formally known as a transfer operator. A Bayesian model (Eq. 1) in this particular case is called a Markov model (2–4). Besides of their direct relation to the exact law of total probability, another reason for their popularity, especially in the natural sciences, is the fact that these models automatically satisfy important physical conservation laws, i.e., exactly conserve probability, and herewith lead to stable simulations (2, 7, 9). Various efficient computational methods allow the estimation of conditional probability matrices Λ for real-world systems (7–15).
In practice, all these methods require a priori availability of discrete probabilistic representations. Obtaining these representations/approximations ΓX(t) by means of common methods from the original system’s states X(t) is subject to serious quality and cost limitations. For example, the applicability of grid discretization methods covering original system’s space with a regular mesh of reference points {S1,…,SK} is limited in terms of cost, since the required number of boxes K grows exponentially with the dimension n of X(t) (1).
Therefore, the most common approaches for tackling these kinds of problems are so-called meshless methods. They attempt to find discretization by means of grouping the states X(t) into K clusters according to some similarity criteria. The computational costs for popular clustering algorithms (16) and for most mixture models (17) scale linearly with the dimensionality n of the problem and the amount of data, T. This cheap computation made clustering methods the most popular meshless discretization tools, even despite the apparent quality limitations they entail. For example, K-means clustering (the most popular clustering method, with more than 3 million Google Scholar citations) can only provide probabilistic approximations with binary (0/1) ΓX(t) elements, excluding any other approximations and not guaranteeing optimal approximation quality. Mixture models are subject to similar quality issues when the strong assumptions they impose [such as Gaussianity in Gaussian mixture models (GMMs)] are not fulfilled. Closely related to clustering methods are various approaches for matrix factorization, such as the non-negative matrix factorization (NMF) methods that attempt to find an optimal approximation of the given (non-negative) n × T data matrix X with a product of the n × K matrix S and the K × T matrix ΓX (18–24).
In situations where K is smaller than T, these non-negative reduced approximations SΓX are computed by means of the fixed-point iterations (19, 21) or by alternating least-squares algorithms and projected gradient methods (22). However, because of the computational cost issues, probabilistic factorizations (i.e., these approximations SΓX show that the columns of Γ are probability distributions) are either excluded explicitly (22) or obtained by means of the spectral decomposition of the data similarity matrices (such as the XTX similarity matrix of the size T × T). These probabilistic NMF variants such as the left-stochastic decomposition (LSD) (24), the closely related spectral decomposition methods (25), and the robust Perron cluster analysis (8, 12) are subject to cost limitations.
These cost limitations are induced by the fact that even the most efficient tools for eigenvalue problem computations (upon which all these methods rely) scale polynomial with the similarity matrix dimension T. If the similarity matrix does not exhibit any particular structure (i.e., if it is not sparse), then the overall numerical cost of the eigenvalue decomposition scales as O(T3). For example, considering twice as much data leads to an eightfold increase in cost.
Similar scalability limitations are also the characteristic for the density-based clustering methods [such as the mean shifts (26), the density-based spatial clustering of applications with noise (DBSCAN) (27), and the algorithms based on t-distributed stochastic neighbor embedding (28)], having an iteration complexity in the orders between O(Tlog(T)) (for sparse similarity matrices) and O(T2) (for full similarity matrices). Practical applicability of these methods is restricted to relatively small systems or relies on the ad hoc data reduction steps, i.e., T cannot routinely exceed 10,000 or 20,000 when working on commodity hardware (see, e.g., the gray dotted curves with crosses in Fig. 3) (9, 28).
Fig. 3. Classification problems: Comparing approximation and classification performances of SPA (blue curves) to the common methods on biomedical applications (40, 41).

Common methods include K-means clustering (dotted lines), SOM (brown), pattern recognition NNs (dashed lines), GMMs (cyan), density-based clustering (gray dotted lines with crosses), and Bayesian models (Eq. 1) (Bayes; dotted lines). Approximation error is measured as the multiply cross-validated average squared Euclidean norm of difference between the true and the discretized representations for validation data. Classification error is measured as the multiply cross-validated average total variation norm between the true and the predicted classifications for validation data. WDBC, Wisconsin Diagnostic Breast Cancer database.
The cost and quality comparison for the probabilistic approximation methods are shown in Fig. 1. Cost factor becomes decisive when discretizing very large systems, e.g., in biology and geosciences, leading to the necessity of some ad hoc data preprocessing by means of computationally cheap methods such as K-means, principal components analysis, and other prereduction steps (29, 30).
Fig. 1.
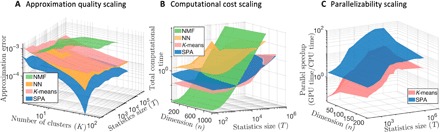
Comparing discretization quality (A), full computational cost (B), and algorithm parallelizability (C) for scalable probabilistic approximation (SPA) (blue surfaces) and for common discretization methods: K-means clustering (16, 17) (red), NMF (19–24) [in its probabilistic variant called LSD (24), green surfaces], and the discretizing neuronal networks (NNs) based on self-organizing maps (SOMs) (38) (a special form of unsupervised NNs used for discretization, orange surfaces). For every combination of data dimension n and the data statistics length T, methods are applied to 25 equally randomly generated datasets, and the results in each of the curves represent averages over these 25 problems. Parallel speedup in (C) is measured as the ratio of the average times time(GPU)/time(CPU) needed to reach the same relative tolerance threshold of 10−5 on a single GPU [ASUS TURBO-GTX1080TI-11G, with 3584 Compute Unified Device Architecture (CUDA) cores] for time(GPU) versus a single CPU core (Intel Core i9-7900X CPU) for time(CPU). Further comparisons can be found in the fig. S2. The MATLAB script Fig1_reproduce.m reproducing these results is available in the repository SPA at https://github.com/SusanneGerber. GPU, graphics processing unit; CPU, central processing unit.
In the following, we present a method not requiring this ad hoc reductional data preprocessing, having the same leading order computational iteration complexity O(nKT) as the cheap K-means algorithm and allowing simultaneously finding discretization that is optimal for models (Eq. 1).
RESULTS
Cost, quality, and parallelizability in scalable probabilistic approximation
Below, we derive the methodology by formulating the discrete approximation as an optimization problem. Here, an approximation quality of a discretization is expressed as the sum of all distances distS(X(t), ΓX(t)) between the original states X(t), and their probabilistic representations ΓX(t) obtained for a particular discretization S = {S1,…, SK}. For example, minimizing the sum of the squared Euclidean distances with respect to Γ and S for a fixed given X would allow finding the optimal probabilistic approximations of the original n-dimensional data points X(t) in the Euclidean space (18–24). Then, Sk is an n-dimensional vector with coordinates of the discrete state k, and is the probability that X(t) belongs to this discrete state (also referred to as a “cluster k,” a “box k,” or a “reference point k” in the following).
Moreover, it can be useful to add other minimized quantities to the resulting expression, e.g., ΦS(S) (to increase the “quality” of discrete states S, to be explained below) and ΦΓ(ΓX) to improve the quality of ΓX while simultaneously minimizing the sum of discretization errors. For example, the temporal persistence of the probabilistic representations ΓX (if t is a time index) can be controlled by (31–33) (measuring the average number of transitions between discrete states over time), whereas ΦS(S) can be chosen as a discrepancy between the actual discretization S and some a priori available knowledge Sprior about it, i.e., as ΦS(S)= ∣∣S − Sprior∣∣ (34, 35). Consequently, the best possible probabilistic approximation can be defined as a solution of a minimization problem for the following L with respect to the variables S and ΓX
| (2) |
subject to the constraints that enforce that the approximation ΓX is probabilistic
| (3) |
where ϵS and ϵΓ (both bigger or equal then zero) regulate the relative importance of the quality criteria ΦS and ΦΓ with respect to the importance of minimizing a sum of discrete approximation errors.
As proven in Theorem 1 in the Supplementary Materials, the minima of problem (Eqs. 2 and 3) can be found by means of an iterative algorithm alternating optimization for variables ΓX (with fixed S) and for variables S (with fixed ΓX). In the following, we provide a summary of the most important properties of this algorithm. A detailed mathematical proof of these properties can be found in Theorems 1 to 3 (as well as in the Lemma 1 to 15 and in the Corollaries 1 to 11) from the Supplementary Materials.
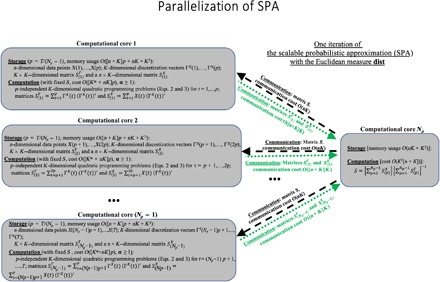
In terms of cost, it can be shown that the computational time of the average iteration for the proposed algorithm grows linearly with the size T of the available data statistics in X, if ΦΓ(ΓX) is an additively separable function [meaning that it can be represented as ]. We refer to the iterative methods for minimization of (Eqs.2 and 3), satisfying this property, as scalable probabilistic approximations (SPAs). Further, if the distance metrics distS(X(t), ΓX(t)) is either an Euclidean distance or a Kullback-Leibler divergence, then the overall iteration cost of SPA grows as O(nKT) (where n is the system’s original dimension and K is the number of discrete states). That is, the computational cost scaling of SPA is the same as the cost scaling of the computationally cheap K-means clustering (16) (see Corollary 6 in the Supplementary Materials for a proof). Moreover, in such a case, it can be shown that the amount of communication between the processors in the case of the Euclidean distance distS(X(t), ΓX(t)) during one iteration in a parallel implementation of SPA will be independent of the size T of system’s output and will change proportionally to O(nK) and to the number of the used computational cores. Figure 2 illustrates these properties and shows a principal scheme of the SPA parallelization.
Fig. 2. Parallelization of the SPA algorithm: Communication cost of SPA for every channel is independent of the data size T and is linear with respect to the data dimension n.
In terms of quality, it is straightforward to validate that several of the common methods are guaranteed to be suboptimal when compared to SPA, meaning that they cannot provide approximations better than SPA on the same system’s data X. This can be shown rigorously for different forms of K-means (16), e.g., (see Corollary 1 in the Supplementary Materials) and for the different variants of finite element clustering methods on multivariate autoregressive processes with external factors (see Corollary 2 in the Supplementary Materials) (31–33).
Figure 1 shows a comparison of SPA (blue surfaces) to the most common discretization methods for a set of artificial benchmark problems of different dimensionality n and size T (see the Supplementary Materials for a detailed description of the benchmarks). In comparison with K-means, these numerical experiments illustrate that SPA has the same overall cost scaling (Fig. 1B), combined with the substantially better approximation quality and parallelizability scaling (Fig. 1, A and C).
Computing optimal discretization for Bayesian and Markovian models
Common fitting of Bayesian or Markovian models (Eq. 1) relies on the availability of probabilistic representations ΓY(t) and ΓX(t) and requires prior separate discretization of X and Y. There is no guarantee that providing any two of these probabilistic representations ΓY(t) and ΓX(t) as an input for any of the common computational methods (7–15) for Λ identification would result in an optimal model (Eq. 1). That is, Bayesian and Markovian models obtained with common methods (7–15) are only optimal for a particular choice of the underlying probabilistic representations ΓY(t) and ΓX(t) (which are assumed to be given and fixed for these methods) and are not generally optimal with respect to the change of the discretization SY and SX.
As proven in Theorem 2 in the Supplementary Materials, the optimal discretization of the Euclidean variables X from Rn and Y from m for the model (Eq. 1) can be jointly obtained from the family of SPA solutions by minimizing the function L (Eqs. 2 and 3) for the transformed variable from Rm+n. This variable is built as a concatenation (a merge) of the original variables Y and ϵX (where X is multiplied with a tunable scalar parameter ϵ > 0). Scalar parameter ϵ defines the relative weight of X discretization compared to Y discretization: The larger ϵ is, the more emphasis is placed on minimizing the discretization errors for X. For any combination of parameter ϵ and the discrete dimension K from some predefined range, this SPA optimization (Eqs. 2 and 3) is then performed with respect to the transformed variables and the original variable (being the discrete probabilistic representation of the original data X). Then, the lower n × K block of the obtained discretization matrix Sϵ,K (when divided with ϵ) provides an optimal discretization matrix SX for X.
In the case of Bayesian models, prediction of values Ypred(s) from the data vector X(s) (which was not a part of the original dataset X) is approached in two steps: (step 1) computing the discretization ΓX(s) by solving the K-dimensional quadratic optimization problem ΓX(s) = argminΓ ||X(s) − SXΓX(s)||22, such that the K elements of ΓX(s) sum up to one and are all non-negative (conditions for existence, uniqueness, and stability of this problem solution are investigated in Lemma 9, 11, 12, 14, and 17 in the Supplementary Materials); and (step 2) multiplying the obtained discretization vector ΓX(s) with the upper m × K block of the SPA matrix Sϵ,K provides a prediction Ypred(s), i.e., . Note that this two-step prediction procedure would not require the explicit estimation of Λϵ,K and . Ypred(s) is computed by a direct multiplication of the vector ΓX(s) (from the step 1) with the upper m × K block of the SPA matrix Sϵ,K. This is the procedure deployed in the computational analysis of benchmarks described below. If required, then discretization matrix and the stochastic matrix Λϵ,K can be disentangled from the upper m × K block of Sϵ,K by means of the standard matrix factorization algorithms (18–24).
In case of Markovian models, when Y(s) is defined as X(s + 1), K × K transition matrix Λϵ,K obtained from such a matrix factorization also allows the computation of the N-step predictions of ΓX, as ΓX(s + N) = ΓX(s) from the discrete approximation ΓX(s) of the data point X(s) (from step 1). Then, in step 2, the N-step prediction Ypred(s + N) is obtained as . Alternatively, to enforce that Y(s) is strictly defined as X(s + 1) in the case of the Markov processes and that the two discretization operators and should be the same and provide a discretization of the same Euclidean dataset X, one can impose an additional equality constraint to the SPA optimization problem (Eqs. 2 and 3) and split the S-optimization step of the iterative SPA algorithm into two substeps: (substep 1) the minimization of (Eqs. 2 and 3) with respect to for fixed values of Λϵ,K and ΓX and (substep 2) the minimization of (Eqs. 2 and 3) with respect to Λϵ,K for fixed values of and ΓX. This procedure results in a monotonic minimization of the SPA problem (Eqs. 2 and 3) and provides direct estimates of the K × K Markovian transition matrix Λϵ,K and the n × K discretization matrix in the Markovian case. Note that this procedure does not provide an explicit Markov model that directly relates X(s + N) to X(s). The two-step procedure described above results in an implicit Markovian model ΓX(s + N) = ΓX(s), operating on the level of K-dimensional discrete probability densities ΓX. The optimal combination of tunable parameters ϵ and K for the optimal discretization in model (Eq. 1) is obtained by applying standard model selection criteria (36) (e.g., using information criteria or approaches such as multiple cross-validations).
These procedures of simultaneous discretization and model inference rely on the assumption that both X and Y are Euclidean data. If this is not the case, then the data have to be transformed to Euclidean before applying the SPA. In the case of time series with equidistant time steps, Euclidean transformation (or Euclidean embedding) is achieved by the so-called Takens embedding transformation, when instead of analyzing single realizations of the data X(s) in Rn, one considers transformed data points given as a whole time sequence [X(s), X(s − 1),…, X(s − dim)] in Rn(dim+1). The justification of this procedure is provided by the Takens theorem (37), proving that this transformation embeds any sufficiently smooth non-Euclidean attractive manifold of a dynamic system into Euclidean space. This procedure is deployed for the time series data analysis benchmarks considered below, an analytical example of such an embedding transformation for autoregressive models is provided in Corollary 2 in the Supplementary Materials. Alternatively, in the case of the classification problems for the data that are not time series, one can apply SPA framework (Eqs. 2 and 3) to the transformed data Xtrans(s) = F(X(s),w), s = 1,…, T. F is a nonlinear Euclidean transformation performed, e.g., by a neuronal network (NN) that relies on a tunable parameter vector w (a vector of network weights and biases). Then, the iterative minimization of (Eqs. 2 and 3) can be augmented straightforwardly with an optimization with respect to an additional parameter vector w of this NN transformation. Below, this kernelized SPA procedure will be referred to as SPA + NN and used for the analysis of classification benchmarks from Fig. 3.
Sensitivity analysis and feature selection with SPA
After the discretization problem is solved, an optimal discrete representation ΓX(t) can be computed for any continuous point X(t). Obtained vector ΓX(t) contains K probabilities for a point X(t) to belong to each particular discrete state Sk and allows computing the reconstruction Xrec(t) of the point X(t) as Xrec(t) = SΓX(t). In this sense, procedure (Eqs. 2 and 3) can be understood as the process of finding an optimal discrete probabilistic data compression, such that the average data reconstruction error [measured as a distance between X(t) and Xrec(t)] is minimized.
In the following, we refer to the particular dimensions of X as features and consider a problem of identifying sets of features that are most relevant for the discretization. The importance of any feature/dimension j of X for the fixed discrete states S can be measured as an average sensitivity of the obtained continuous data reconstructions Xrec(t) with respect to variations of the original data X(t) along this dimension j. For example, it can be measured by means of the average derivative norm . For every dimension j of X(t), this quantity I(j) probes an average impact of changes in the dimension j of X(t) on the resulting data reconstructions Xrec(t). Dimensions j that have the highest impact on discretization will have the highest values of I(j), whereas the dimensions j that are irrelevant for the assignation to discrete states will have I(j) close to 0.
At first glance, direct computation of the sensitivities I(j) could seem too expensive for realistic applications with large data statistics size T and in high problem dimensions and due to the a priori unknown smoothness of the derivate in the multidimensional space of features. However, as proven in Theorem 3 in the Supplementary Materials, in the case of discretizations obtained by solving the problem (Eqs. 2 and 3) for the Euclidean distance measure dist, respective derivatives are always piecewise constant functions of Xj(t) if the statistics size T is sufficiently large. This nice property of derivatives allows a straightforward numerical computation of I(j) for K > 2 and an exact analytical computation of I for K = 2. It turns out that for K = 2, the importance of every original data dimension j can be directly measured as . That is, discretization sensitivity I(j) for the feature j is proportional to the squared difference between the discretization coordinates S1,j and S2,j in this dimension j. The smaller the difference between the coordinates in this dimension, the lower is the impact of this particular feature j on the overall discretization.
It is straightforward to verify (see Corollary 9 and Theorem 3 in the Supplementary Materials for a proof) that the feature sensitivity function has a quadratic upper bound I ≤ ∑j,k1,k2(Sk1(j) − Sk2(j))2. Setting ΦS(S) in Eq. 2 as ΦS(S) = ∑j,k1,k2(Sk1(j) − Sk2(j))2 for any given combination of integer K and scalar ϵS ≥ 0, minimizing (Eqs. 2 and 3) would then result in a joint simultaneous and scalable solution of the optimal discretization and feature selection problems. Overall, the numerical iteration cost of this procedure will be again O(nKT). Changing ϵS controls the number of features: The larger ϵS is, the fewer features (i.e., particular dimensions of the original data vector X) remain relevant in the obtained discretization. The optimal value of ϵS can again be determined by means of standard model validation criteria (36). In the SPA results from Figs. 3 and 4 (blue curves), we use this form of ΦS(S), set ϵΓ = 0, and deploy the multiple cross-validation, a standard model selection approach from machine learning, to determine the optimal ϵS and an optimal subset of relevant features for any given number K of discrete states (clusters).
Fig. 4. Prediction problems in time series analysis: Comparing approximation and prediction performances of SPA (blue curves) to the common methods on open-source datasets (15, 42–44).
Common methods include K-means clustering (K-means; dark green) in combinations with pattern recognition recurrent NNs (yellow and light green) and Markov models (Eq. 1) (dark green). Approximation and the prediction errors are measured in the average squared Euclidean norm of deviations between the true and the predicted system states for the validation data (i.e., for data not used in the model fitting). EEG, electroencephalogram.
Applications to classification and prediction problems from natural sciences
Next, we compare the discretization performance of SPA to the approximation errors of the common methods, including K-means (16), soft clustering methods based on Bayesian mixture models (17) (such as GMMs), density-based clustering (DBSCAN) (27), and NN discretization methods (self-organizing maps, SOMs) (38). To compare the performances of these methods, we use obtained discretization in parametrization of the Bayesian/Markovian models (Eq. 1), as well as in parametrization of NNs (38), on several classification and time series analysis problems from different areas. To prevent overfitting, we deploy the same multiple cross-validation protocol (36, 39) adopted in machine learning for all tested methods. Here, the data are randomly subdivided into the training set (75% of the data), where the discretization and classification/prediction models are trained and performance quality measures (approximation, classification, and prediction errors) are then determined on the remaining 25% of validation data (not used in the training). For each of the methods, this procedure of random data subdivision, training, and validation is repeated 100 times; Figs. 3 and 4 provide the resulting average performance curves for each of the tested methods. In both classification and time series prediction applications of SPA, no persistence in the t-ordering was assumed a priori by setting ϵΓ in (Eq. 2) to 0. This means not imposing any a priori persistent ordering in t for the probabilistic representations {ΓX(1),…, {ΓX(T)} and justifying an application of the common multiple cross-validation procedure (36) (which implies random reshufflings in t and subdivisions of data into training and validation sets) for a selection of optimal values for K and ϵS. MATLAB scripts reproducing the results are available in the repository SPA at https://github.com/SusanneGerber. Figure 3 shows a comparison of approximation and classification performances for two problems of labeled data analysis from biomedicine and bioinformatics: (Fig. 3A) for a problem of breast cancer diagnostics based on x-ray image analysis (40) and (Fig. 3B) for a problem of single-cell human mRNA classification (41). In these problems, variable X(t) is a continuous (and real valued) set of collected features that have to be brought in relation to the discrete set of labels Y(t). In case of the breast cancer diagnostics example (40), (Fig. 3A) index t denotes patients and goes from 1 to 569, X(t) contains 32 image features, and Y(t) can take two values “benign” or “malignant.” In the case of the single-cell human mRNA classification problem (41), (Fig. 3B) index t goes from 1 to 300 (there are 300 single-cell probes), X(t) contains expression levels for 25,000 genes, and Y(t) is a label denoting one of the 11 cell types (e.g., “blood cell,” “glia cell,” etc.). In both cases, the ordering of data instances t in the datasets is arbitrary and is assumed not to contain any a priori relevant information related to time (such as temporal persistence of the ordering along the index t).
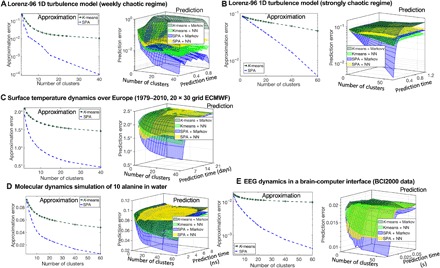
Figure 4 summarizes results for five benchmark problems from time series analysis and prediction: for the Lorenz-96 benchmark system (42) modeling turbulent behavior in one dimension, in a weakly chaotic (Fig. 4A) and in the strongly chaotic (Fig. 4B) regimes; (Fig. 4C) for 45 years (T = 16,433 days) of historical European Centre for Medium-Range Weather Forecasts (ECMWF)–resimulated 2-m height daily air temperature anomalies (deviations from the mean seasonal cycle) time series on a 18 by 30 grid over the continental Europe and a part of the Northern Atlantic (43), provided by the ECMWF; (Fig. 4D) for the biomolecular dynamics of a 10–alanine peptide molecule in water (15); and (Fig. 4E) for the electrical activity of the brain measured in various brain-computer interaction (BCI) regimes obtained with the 64-channel electroencephalography and provided for open access by the BCI2000 consortium (44).
As can be seen from the Figs. 3 and 4, SPA tends to reach its approximation quality plateau earlier (i.e., with much smaller K) but is generally much more accurate, with a discretization performance improvement factor ranging from two to four (for breast cancer diagnostics example, for single-cell mRNA classification, for the temperature data over Europe, and for the molecular dynamics application). For the Lorenz-96 turbulence applications (42) and for the brain activity application (44), discretization obtained by SPA is 10 to 100 times better than the discretization from common methods, being at the same level of computational cost as the popular K-means clustering (16).
When evaluating a prediction performance of different models for a particular system, it is important to compare it with the trivial prediction strategies called mean value prediction and persistent prediction. The mean value prediction strategy predicts the next state of the system to be an expectation value over the previous already observed states and is an optimal prediction strategy for stationary independent and identically distributed processes such as the Gaussian process. The persistent prediction strategy is predicting the next state of the system to be the same as its current state. This strategy is particularly successful and is difficult to be beaten for the systems with more smooth observational time series, e.g., for the intraday surface temperature dynamics. As it can be seen from the fig. S3, among all other considered methods (K-means, NNs, SOM, and mixture models), only the SPA discretization combined with the Markov models (Eq. 1) allow outperforming both the mean value and the persistent predictions for all of the considered systems.
DISCUSSION
Computational costs become a limiting factor when dealing with big systems. The exponential growth in the hardware performance observed over the past 60 years (the Moore’s law) is expected to end in the early 2020s (45). More advanced machine learning approaches (e.g., NNs) exhibit the cost scaling that grows polynomial with the dimension and the size of the statistics, rendering some form of ad hoc preprocessing and prereduction with more simple approaches (e.g., clustering methods) unavoidable for big data situations. However, these ad hoc preprocessing steps might impose a strong bias that is not easy to quantify. At the same time, lower cost of the method typically goes hand-in-hand with the lower quality of the obtained data representations (see Fig. 1). Since the amounts of collected data in most of the natural sciences are expected to continue their exponential growth in the near future, pressure on computational performance (quality) and scaling (cost) of algorithms will increase.
Instead of solving discretization, feature selection, and prediction problems separately, the introduced computational procedure (SPA) solves them simultaneously. The iteration complexity of SPA scales linearly with data size. The amount of communication between processors in the parallel implementation is independent of the data size and linear with the data dimension (Fig. 2), making it appropriate for big data applications. Hence, SPA did not require any form of data prereduction for any of the considered applications. As shown in the Fig. 1, having essentially the same iteration cost scaling as the very popular and computationally very cheap K-means algorithm (16, 17), SPA allows achieving substantially higher approximation quality and a much higher parallel speedup with the growing size T of the data.
Applications to large benchmark systems from natural sciences (Figs. 3 and 4) reveal that these features of SPA allow a marked improvement of approximation and cross-validated data-driven prediction qualities, combined with a massive reduction of computational cost. For example, computing the next-day surface temperature anomalies for Europe (e.g., at the ECMWF) currently relies on solving equations of atmosphere motion numerically performed on supercomputers (43). Discretization and cross-validated data-driven prediction results for the same online resimulated daily temperature data provided in the Fig. 4C were obtained on a standard Mac PC, exhibiting a mean error of 0.75°C for the 1-day ahead surface air temperature anomalies computations (approximately 40% smaller than the current next-day prediction errors by weather services).
These probability-preserving and stable predictions ΓY(t) can be accomplished very cheaply with the Bayesian or Markovian model (Eq. 1) from the available SPA discretization (Eqs. 2 and 3), just by computing the product of the obtained K × K Bayesian matrix Λ with the K × 1 discretization vector ΓX(t). Optimal K was in the orders of 10 to 40 for all of the considered applications from Figs. 3 and 4. The iteration cost of this entire data-driven computation scales linearly, resulting in orders of magnitude speedup as compared to the predictions based on the entire system’s simulations. These results indicate a potential to use efficient and cheap Bayesian and Markovian descriptive models for a robust automated classification and data-driven cross-validated predictive computations in many realistic systems across natural sciences. However, an assumption about the t independence of the conditional probability matrix Λ in (Eq. 1), which allowed applying common model selection procedures from machine learning, can limit the applicability of the method in the nonstationary situations, when Λ in (Eq. 1) becomes t dependent. Addressing the nonstationarity problem will become an important issue for future applications to these systems.
MATERIALS AND METHODS
We used the standard MATLAB functions kmeans(), fitgmdist(), patternnet(), and som() to compute the results of the common methods (K-means, GMM, NN, and SOM) in Figs. 1, 3, and 4. To avoid being trapped in the local optima and to enable a unified comparison of all methods, we used 10 random initializations and selected the results with the best approximation quality measure for the training sets. In case of the pattern recognition NNs (evaluated in the classification and prediction performance subfigures of Figs. 3 and 4) in each of the instances of the multiple cross-validation procedure, we repeated the network fitting for the numbers of neurons in the hidden layer ranging between 1 and 15 and selected the results with the best classification/prediction performances for the training set. We implemented the LSD algorithm from Fig. 1 in MATLAB according to the literature description (24) and provided it for open access. SPA algorithms developed and used during the current study are also available in open access as MATLAB code at https://github.com/SusanneGerber.
Supplementary Material
Acknowledgments
We thank G. Ciccotti (La Sapienza, Rome), M. Weiser (ZIB, Berlin), D. L. Donoho (Standford University), M. Wand (JGU, Mainz), P. Gagliardinin (USI, Lugano), R. Klein (FU, Berlin), and F. Bouchet (ENS, Lyon) for helpful comments about the manuscript. Funding: We acknowledge the financial support from the German Research Foundation DFG (“Mercator Fellowship” of I.H. in the CRC 1114 “Scaling cascades in complex systems”). S.G. acknowledges the financial support from the DFG (in the CRC 1193 “Neurobiology of resilience to stress-related mental dysfunction: From understanding mechanisms to promoting preventions”). The work of M.N. was supported by the Research Center For Emergent Algorithmic Intelligence at the University of Mainz funded by the Carl-Zeiss Foundation. Author contributions: S.G. and I.H. designed the research, wrote the main manuscript, and produced results in Figs. 3 and 4. L.P. and I.H. produced results in Fig. 1 and proved Theorems 1 to 3 and Lemmas in the Supplementary Materials. M.N. prepared the data and participated in the analysis of single-cell mRNA data (Fig. 3B). Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/5/eaaw0961/DC1
Description of the synthetic data problems (used in the Fig. 2 of the main manuscript)
General SPA formulation
SPA in the Euclidean space
Optimality conditions
The solution of S subproblem
The solution of Γ subproblem
Computing optimal discretizations for Bayesian and Markovian models
Sensitivity and feature selection with SPA in the Euclidean space
Appendix
Fig. S1. Distributed solution of Γ problem.
Fig. S2. Comparison of different measures.
Fig. S3. Comparison of one time-step predictions.
REFERENCES AND NOTES
- 1.A. Stuart, A. Humphries, Dynamical systems and numerical analysis, in Cambridge Monographs on Applied Mathematics (Cambridge Univ. Press, 1998), vol. 8. [Google Scholar]
- 2.A. J. Chorin, O. H. Hald, Stochastic Tools in Mathematics and Science (Springer, ed. 3, 2013). [Google Scholar]
- 3.Rubin D. B., Bayesian inference for causal effects: The role of randomization. Ann. Stat. 6, 34–58 (1978). [Google Scholar]
- 4.D. B. Rubin, Bayesian Data Analysis (Chapman and Hall/CRC Texts in Statistical Science, ed. 3, 2013). [Google Scholar]
- 5.Ch. Schütte, M. Sarich, Metastability and Markov state models in molecular dynamics: Modeling, analysis, algorithmic approaches, in Courant Lecture Notes in Mathematics (American Mathematical Soc., 2013), vol. 24. [Google Scholar]
- 6.A. N. Langville, C. D. Meyer, Google’s PageRank and Beyond: The Science of Search Engine Rankings (Princeton Univ. Press, 2006). [Google Scholar]
- 7.C. Schütte, W. Huisinga, P. Deuflhard, Transfer operator approach to conformational dynamics in biomolecular systems, in Ergodic Theory, Analysis, and Efficient Simulation of Dynamical Systems, B. Fiedler, Ed. (Elsevier, 2001), pp. 191–223. [Google Scholar]
- 8.Deuflhard P., Weber M., Robust Perron cluster analysis in conformation dynamics. Lin. Algebra Appl. 398, 161–184 (2005). [Google Scholar]
- 9.Gerber S., Olsson S., Noé F., Horenko I., A scalable approach to the computation of invariant measures for high-dimensional Markovian systems. Sci. Rep. 8, 1796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froyland G., Padberg K., Almost-invariant sets and invariant manifolds: Connecting probabilistic and geometric descriptions of coherent structures in flows. Physica D Nonlin. Phenom. 238, 1507–1523 (2009). [Google Scholar]
- 11.A. Majda, R. Abramov, M. Grote, Information Theory and Stochastics for Multiscale Nonlinear Systems (CRM monograph series, American Mathematical Soc., 2005). [Google Scholar]
- 12.Weber M., Kube S., Robust Perron cluster analysis for various applications in computational life science. Lect. Notes Comp. Sci. 3695, 55–66 (2005). [Google Scholar]
- 13.T. Hofmann, Probabilistic latent semantic analysis, in Proceedings of the 15th Annual Conference on Uncertainty in Artificial Intelligence (UAI’99) (Morgan Kaufmann Publishers, 1999), pp. 289–296. [Google Scholar]
- 14.Gerber S., Horenko I., Toward a direct and scalable identification of reduced models for categorical processes. Proc. Natl. Acad. Sci. U.S.A. 114, 4863–4868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber S., Horenko I., On inference of causality for discrete state models in a multiscale context. Proc. Natl. Acad. Sci. U.S.A. 111, 14651–14656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartigan J. A., Wong M. A., Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Soc. C 1, 100–108 (1979). [Google Scholar]
- 17.P. D. McNicholas, Mixture Model-Based Classification (CRC Press, ed. 1, 2016). [Google Scholar]
- 18.Paatero P., Tapper U., Positive matrix factorization: A non-negative factor model with optimal utilization of error estimates of data values. Environmetrics 5, 111–126 (1994). [Google Scholar]
- 19.Lee D. D., Seung H. S., Learning the parts of objects by nonnegative matrix factorization. Nature 401, 788–791 (1999). [DOI] [PubMed] [Google Scholar]
- 20.D. L. Donoho, V. Stodden, Learning the parts of objects by nonnegative matrix factorization. When does non-negative matrix factorization give a correct decomposition into parts? in Advances in Neural Information Processing Systems, S. Thrun, L. Saul, B. Schölkopf, Eds. (MIT Press, 2004), vol. 24. [Google Scholar]
- 21.Ding C. H. Q., Li T., Jordan M. I., Convex and semi-nonnegative matrix factorizations. IEEE Trans. Pattern Anal. Mach. Intell. 32, 45–55 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Lin C.-J., Projected gradient methods for nonnegative matrix factorization. Neural Comput. 19, 2756–2779 (2007). [DOI] [PubMed] [Google Scholar]
- 23.C. Ding, T. Li, W. Peng, Nonnegative matrix factorization and probabilistic latent semantic indexing: Equivalence, chi-square statistic, and a hybrid method, in Proceedings of the 21st National Conference on Artificial Intelligence and the 18th Innovative Applications of Artificial Intelligence Conference (AAAI Press, 2006), vol. 1, pp. 342–347. [Google Scholar]
- 24.Arora R., Gupta M. R., Kapila A., Fazel M., Similarity-based clustering by left-stochastic matrix factorization. J. Mach. Learn. Res. 14, 1417–1452 (2013). [Google Scholar]
- 25.A. Ng, M. Jordan, Y. Weiss, On spectral clustering: Analysis and an algorithm, in Advances in Neural Information Processing Systems (NIPS) (MIT Press, 2002), vol. 14, pp. 849–856. [Google Scholar]
- 26.Cheng Y., Mean shift, mode seeking and clustering. IEEE Trans. Pattern Anal. Mach. Intell. 17, 790–799 (1995). [Google Scholar]
- 27.M. Ester, H.-P. Kriegel, J. Sander, X. Xu, A density-based algorithm for discovering clusters in large spatial databases with noise, in Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD’96) (AAAI Press, 1996), pp. 226–231. [Google Scholar]
- 28.van der Maaten L., Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014). [Google Scholar]
- 29.D’haeseleer P., How does gene expression clustering work? Nat. Biotechnol. 23, 1499–1501 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Cassou C., Intraseasonal interaction between the Madden–Julian Oscillation and the North Atlantic Oscillation. Nature 455, 523–527 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Metzner P., Putzig L., Horenko I., Analysis of persistent nonstationary time series and applications. Commun. Appl. Math. Comput. Sci. 7, 175–229 (2012). [Google Scholar]
- 32.O’Kane T. J., Matear R. J., Chamberlain M. A., Risbey J. S., Sloyan B. M., Horenko I., Decadal variability in an OGCM Southern Ocean: Intrinsic modes, forced modes and metastable states. Ocean Model. 69, 1–21 (2013). [Google Scholar]
- 33.Vercauteren N., Klein R., A clustering method to characterize intermittent bursts of turbulence and interaction with submesomotions in the stable boundary layer. J. Atmos. Sci. 72, 1504–1517 (2015). [Google Scholar]
- 34.Tibshirani R., Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288 (1996). [Google Scholar]
- 35.Gerber S., Horenko I., Improving clustering by imposing network information. Sci. Adv. 1, e1500163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.K. Burnham, D. Anderson, Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach (Springer, ed. 2, 2002). [Google Scholar]
- 37.F. Takens, Detecting strange attractors in turbulence, in Dynamical Systems and Turbulence, Lecture Notes in Mathematics, D. A. Rand, L.-S. Young, Eds. (Springer-Verlag, 1981), vol. 898, pp. 366–381. [Google Scholar]
- 38.T. Kohhonen, Self-Organising Maps (Springer Series in Information Sciences, ed. 3, 2001), vol. 30. [Google Scholar]
- 39.Trippa L., Waldron L., Huttenhower C., Parmigiani G., Bayesian nonparametric cross-study validation of prediction methods. Ann. Appl. Stat. 9, 402–428 (2015). [Google Scholar]
- 40.Wolberg W. H., Street W. N., Mangasarian O. L., Machine learning techniques to diagnose breast cancer from image-processed nuclear features of fine needle aspirates. Cancer Lett. 77, 163–171 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., Li N., Szpankowski L., Fowler B., Chen P., Ramalingam N., Sun G., Thu M., Norris M., Lebofsky R., Toppani D., Kemp D. W. II, Wong M., Clerkson B., Jones B. N., Wu S., Knutsson L., Alvarado B., Wang J., Weaver L. S., May A. P., Jones R. C., Unger M. A., Kriegstein A. R., West J. A. A., Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.E. Lorenz, Predictability: A problem partly solved, in Proceedings of the ECMWF Seminar on Predictability (ECMWF, 1996), vol. 1, pp. 1–18. [Google Scholar]
- 43.Dee D. P., Uppala S. M., Simmons A. J., Berrisford P., Poli P., Kobayashi S., Andrae U., Balmaseda M. A., Balsamo G., Bauer P., Bechtold P., Beljaars A. C. M., van de Berg L., Bidlot J., Bormann N., Delsol C., Dragani R., Fuentes M., Geer A. J., Haimberger L., Healy S. B., Hersbach H., Hólm E. V., Isaksen L., Kållberg P., Köhler M., Matricardi M., McNally A. P., Monge-Sanz B. M., Morcrette J.-J., Park B.-K., Peubey C., de Rosnay P., Tavolato C., Thépaut J.-N., Vitart F., The ERA-Interim reanalysis: Configuration and performance of the data assimilation system. Q. J. R. Meteorol. Soc. 137, 553–597 (2011). [Google Scholar]
- 44.G. Schalk, J. Mellinger, A Practical Guide to Brain–Computer Interfacing with BCI2000 (Springer, ed. 1, 2010). [Google Scholar]
- 45.Khan H. N., Hounshell D. A., Fuchs E. R. H., Science and research policy at the end of Moore’s law. Nat. Electron. 1, 14–21 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/5/eaaw0961/DC1
Description of the synthetic data problems (used in the Fig. 2 of the main manuscript)
General SPA formulation
SPA in the Euclidean space
Optimality conditions
The solution of S subproblem
The solution of Γ subproblem
Computing optimal discretizations for Bayesian and Markovian models
Sensitivity and feature selection with SPA in the Euclidean space
Appendix
Fig. S1. Distributed solution of Γ problem.
Fig. S2. Comparison of different measures.
Fig. S3. Comparison of one time-step predictions.


