Many bacteria can sense the pH in their environment and then use this information to direct their movement toward more favorable locations. In this study, we investigated the pH sensing mechanism in Bacillus subtilis. This bacterium preferentially migrates to neutral environments. It employs four chemoreceptors to sense pH. Two are involved in sensing acidic environments, and two are involved in sensing alkaline ones. To identify the mechanism for pH sensing, we constructed receptor chimeras of acid- and base-sensing chemoreceptors. By analyzing the responses of these chimeric receptors, we were able to identify four critical amino acid residues involved in pH sensing and propose a model for the pH sensing mechanism in B. subtilis.
KEYWORDS: Bacillus subtilis, chemoreceptor, chemotaxis, pH, signal transduction
ABSTRACT
We investigated pH taxis in Bacillus subtilis. This bacterium was found to perform bidirectional taxis in response to external pH gradients, enabling it to preferentially migrate to neutral environments. We next investigated the chemoreceptors involved in sensing pH gradients. We identified four chemoreceptors involved in sensing pH: McpA and TlpA for sensing acidic environments and McpB and TlpB for sensing alkaline ones. In addition, TlpA was found to also weakly sense alkaline environments. By analyzing chimeras between McpA and TlpB, the principal acid- and base-sensing chemoreceptors, we identified four critical amino acid residues—Thr199, Gln200, His273, and Glu274 on McpA and Lys199, Glu200, Gln273, and Asp274 on TlpB—involved in sensing pH. Swapping these four residues between McpA and TlpB converted the former into a base receptor and the latter into an acid receptor. Based on the results, we propose that disruption of hydrogen bonding between the adjacent residues upon pH changes induces signaling. Collectively, our results further our understanding of chemotaxis in B. subtilis and provide a new model for pH sensing in bacteria.
IMPORTANCE Many bacteria can sense the pH in their environment and then use this information to direct their movement toward more favorable locations. In this study, we investigated the pH sensing mechanism in Bacillus subtilis. This bacterium preferentially migrates to neutral environments. It employs four chemoreceptors to sense pH. Two are involved in sensing acidic environments, and two are involved in sensing alkaline ones. To identify the mechanism for pH sensing, we constructed receptor chimeras of acid- and base-sensing chemoreceptors. By analyzing the responses of these chimeric receptors, we were able to identify four critical amino acid residues involved in pH sensing and propose a model for the pH sensing mechanism in B. subtilis.
INTRODUCTION
Bacillus subtilis performs chemotaxis to a wide range of attractants and repellents (1–3). As a brief background, B. subtilis employs ten chemoreceptors to sense these compounds (4). The governing chemotaxis pathway functions differently than the better-understood chemotaxis pathway in Escherichia coli. The core signaling pathway consists of a membrane-associated complex involving the chemoreceptors, CheA histidine kinase, and CheW and CheV scaffold proteins (5) that preferentially form clusters at the cell poles (6). The chemoreceptors sense attractants and repellents either by binding them directly or through binding proteins associated with their uptake (7). Attractants are known to increase the rate of CheA phosphorylation (8). The phosphoryl group is then transferred to CheY, which in the phosphorylated form, binds to the cytoplasmic faces of the flagellar motors and induces smooth swimming (9).
To sense chemical gradients, B. subtilis employs three adaptation systems (10–12). The primary system involves receptor methylation. Two enzymes, the CheR methyltransferase and the CheB methylesterase, add and remove methyl groups on conserved glutamate residues located in the cytoplasmic domains of the chemoreceptors (13, 14). Depending on the specific glutamate residue, these modifications can either increase or decrease the ability of the chemoreceptors to activate CheA kinase (5, 15). In addition, two other adaptation systems are involved in sensing gradients. One involves the scaffold protein CheV, which contains a C-terminal response regulator domain that is phosphorylated by CheA (16). Depending on the methylation state of the chemoreceptors, phosphorylated CheV can either increase or decrease chemoreceptor activity (5). The other system involves three proteins: CheD, a chemoreceptor deamidase (17, 18); CheC, a phosphatase for phosphorylated CheY (19); and CheY. In addition to being a deamidase, CheD binds the chemoreceptors and increases their ability to activate CheA (5, 20). CheC can also bind CheD and prevent it from binding the chemoreceptors. Phosphorylated CheY increases the affinity between CheC and CheD, thus providing a feedback mechanism for controlling CheA activity in response to phosphorylated CheY (21, 22).
Our understanding of B. subtilis chemotaxis is principally limited to amino acid chemotaxis. Aside from amino acids (7, 23), oxygen (24), and sugars transported by the phosphoenolpyruvate-dependent phosphotransferase system (25), little is known about the sensing mechanisms involved in B. subtilis chemotaxis. A number of reports have shown that diverse bacteria migrate in response to pH gradients. This process is best understood in Escherichia coli, Salmonella enterica, and Helicobacter pylori (26–32). In the cases of E. coli and S. enterica, these bacteria preferentially migrate to neutral (pH 7.5) environments (33). The response is bidirectional, as the bacteria will migrate down pH gradients when the ambient pH is too high or migrate up pH gradients when the ambient pH is too low. The underlying mechanism involves the competitive response between two chemoreceptors, one that induces cells to migrate down pH gradients and the other that induces them to migrate up pH gradients. These two chemoreceptors respond to both internal and external pH. While the sensing mechanism is still not well understood, external pH is believed to be sensed by the extracellular domain of the chemoreceptors (27) and internal pH by the linker region between the transmembrane helices and cytoplasmic domains of the chemoreceptors (30). Swapping the entire linker region or specific amino acid residues within this linker region inverts the response of these two chemoreceptors to changes in internal pH (30).
In this work, we investigated chemotaxis to external pH gradients in B. subtilis. Similarly to E. coli and S. enterica, B. subtilis exhibits bidirectional chemotaxis to pH gradients. To sense these pH gradients, B. subtilis employs four chemoreceptors, two for sensing acids and two for sensing bases. By analyzing chimeras between acid- and base-sensing chemoreceptors, we identified four critical amino acid residues involved in sensing external pH. Swapping these four residues changed a base-sensing chemoreceptor into an acid-sensing one and vice versa. Based on these data, we propose a model for pH sensing in B. subtilis.
RESULTS AND DISCUSSION
B. subtilis exhibits bidirectional taxis to external pH gradients.
To determine whether B. subtilis performs chemotaxis in response to external pH gradients, we employed the capillary assay (26). Briefly, cells suspended in buffer at different pHs (6.0 to 8.5) were incubated with capillaries filled with buffer at pH 7.0, and then the number of cells that entered the capillaries after 1 h were counted. The resulting data show that B. subtilis exhibits bidirectional taxis to pH gradients in a manner similar to what is observed in E. coli (Fig. 1A). In particular, we found that B. subtilis preferentially migrates to neutral (pH 7) environments when the cells were initially suspended in either acidic or alkaline buffer (pH 6 to 8). Outside this pH range, however, the cells were less motile (data not shown) and, consequently, taxis was reduced. As a control, we also performed capillary assay experiments where the cells suspended in buffer at pH 7.0 were incubated with capillaries filled with buffer at different pHs (6.0 to 8.0). In these experiments, we did not observe any significant accumulation within the capillaries (see Fig. S1 in the supplemental material), which is consistent with cells preferring neutral environments.
FIG 1.
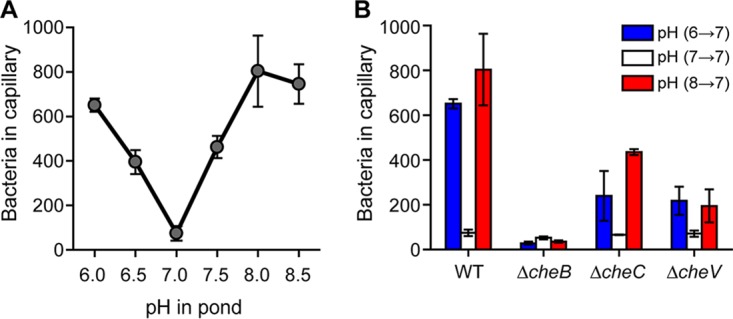
B. subtilis exhibits bidirectional chemotaxis to pH gradients. (A) Response to increasing and decreasing pH gradients. (B) Methylation is necessary for pH taxis. Error bars denote the standard deviations from three biological replicates performed on three separate days.
The methylation system is required for pH taxis.
B. subtilis employs three adaption systems for sensing chemical gradients (11). Of the three adaptation systems, the methylation system is the dominant one for sensing amino acid gradients. We next tested whether these three adaptation systems are involved in pH taxis. To determine whether the methylation system is involved in pH taxis, we tested a ΔcheB mutant, which lacks the requisite methylesterase. We did not test ΔcheR or ΔcheR ΔcheB mutants, which lack the corresponding methyltransferases, because they are not very motile due to excessive tumbles (data not shown). In addition, we tested ΔcheC and ΔcheV mutants, because they lack the other two adaptation systems involved in sensing chemical gradients. Among the three mutants tested, the ΔcheB mutant was unable to perform pH taxis, while ΔcheC and ΔcheV mutants exhibited reduced pH taxis (Fig. 1B). These results indicate that the methylation system is necessary for sensing pH gradients. However, the other two adaptation systems are also involved to a lesser extent, similar to what is observed in amino acid chemotaxis.
Four chemoreceptors sense pH gradients.
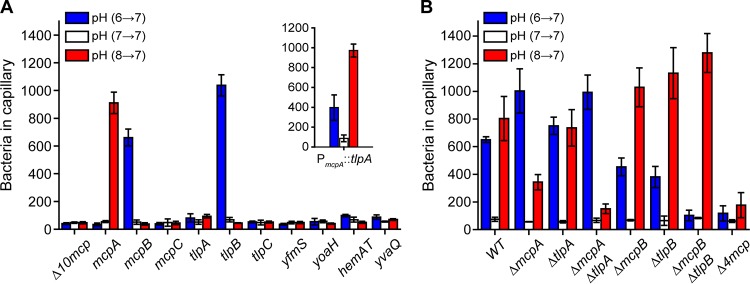
B. subtilis possesses ten chemoreceptors (4). To determine which chemoreceptors are involved in pH taxis, we tested mutants expressing just one chemoreceptor (Fig. 2A). Of the single chemoreceptor mutants, only the one expressing McpA as its sole chemoreceptor was capable of acid sensing. In addition, this mutant exhibited a weak repellent response to base, with fewer cells migrating into the capillary when the buffer was at pH 6 than at pH 7 (33.7 ± 10.5 versus 54.7 ± 7.1 cells, respectively). This repellent response is consistent with McpA being an acid sensor. Two single chemoreceptor mutants were found to exhibit base sensing: the strains expressing McpB or TlpB as their sole chemoreceptors. In particular, these strains exhibited taxis to increases in pH. They also exhibited a weak repellent response to acid (McpB, 37.3 ± 8.6 versus 51.0 ± 15.1 cells; TlpB, 44.3 ± 2.5 versus 68.7 ± 15.8 cells). No significant responses to any changes in pH were observed for the other mutants. The existence of chemoreceptors sensing both acids and bases would explain how B. subtilis is capable of preferentially swimming to neutral conditions, namely, that it reflects competition between the acid and base responses.
FIG 2.
McpA and TlpA are the principal chemoreceptors involved in the acid response, and McpB and TlpB are the sole chemoreceptors involved in the base response. (A) Responses of strains expressing just one chemoreceptor to pH gradients. (Inset) Responses of strain overexpressing tlpA as its sole chemoreceptor. (B) Responses of mutants lacking key receptors to pH gradients. Error bars denote the standard deviations from three biological replicates performed on three separate days.
To further verify that these three chemoreceptors are involved in pH taxis, we performed capillary assay experiments where the cells were suspended in buffer at pH 7 and the pH within the capillaries was varied (see Fig. S1 in the supplemental material). Consistent with the chemoreceptors sensing pH as an attractant (Fig. 2A), the mutant expressing McpA as its sole chemoreceptor accumulated in capillaries filled with buffer at pH 6, and the mutant expressing either McpB or TlpB accumulated in capillaries filled with buffer at pH 8.
The genes encoding McpA, McpB, TlpA, and TlpB reside in a four-gene cluster (34). Since these four chemoreceptors exhibit high (57% to 65%) amino acid sequence identify, we hypothesized that TlpA may also be involved in pH sensing, even though a strain expressing it as the sole chemoreceptor failed to exhibit a response to changes in pH. The reason may be that TlpA is weakly expressed: the wild type expresses 2,000 copies of this chemoreceptor compared to 16,000 copies for McpA (35). Therefore, we tested whether expressing TlpA from a stronger promoter would enable pH sensing. When tlpA was expressed as the sole chemoreceptor using the mcpA promoter, we observed both an acid and base response (Fig. 2A, inset). The acid response, however, was stronger than the base response. These results imply that TlpA alone, when overexpressed, can direct B. subtilis to neutral environments. This begs the question as to why multiple chemoreceptors are employed for pH taxis when potentially one would suffice. Unfortunately, we cannot answer this question at this time. These results also suggest that other chemoreceptors may have the potential to sense pH if their expression is increased.
We next tested the effect of deleting these four chemoreceptors, both individually and in combination, on pH sensing (Fig. 2B). When mcpA was deleted in the wild type (ΔmcpA), we observed a reduction in acid sensing, consistent with this chemoreceptor being involved in chemotaxis toward lower pH. We also observed an increase in the base response, perhaps reflecting the competition between the acid and base responses. When tlpA was deleted in the wild type (ΔtlpA), we observed no significant change in pH sensing. However, when both of the chemoreceptors were deleted in the wild type (ΔmcpA ΔtlpA), the acid response was almost completely eliminated and the base response increased. These results suggest that McpA and TlpA are the primary chemoreceptors involved in sensing decreases in pH. They also indicate that TlpA contributes to pH sensing when expressed from its native promoter. One possibility is that heterologous chemoreceptor interactions are required to amplify the response (4).
When either McpB or TlpB was deleted in the wild type (ΔmcpB or ΔtlpB), the base response was reduced (Fig. 2B). In addition, the acid response increased. When both chemoreceptors were deleted (ΔmcpB ΔtlpB), the base response was completely eliminated and the acid response further increased. These results suggest that McpB and TlpB are the main chemoreceptors involved in sensing increases in pH. Finally, when we deleted all four pH chemoreceptors in the wild type (Δ4mcp) both the acid and base responses were almost completely eliminated, suggesting that these four chemoreceptors are the principal ones involved in pH taxis. As noted above, additional chemoreceptors may be involved; indeed, there was still residual pH taxis in the Δ4mcp mutant. However, the contribution of these chemoreceptors appears minor.
Identification of the regions involved in sensing pH gradients.
All four chemoreceptors involved in pH taxis employ the same double Cache domain (dCACHE_1) for their extracellular ligand-binding domain (36). This would suggest that specific amino acid residues are involved in pH sensing. As a first step toward identifying these residues, we constructed chimeras between McpA, involved in acid sensing, and TlpB, involved in base sensing. We focused on these two chemoreceptors because the corresponding mutants expressing them as their sole chemoreceptor exhibited the strongest responses to pH gradients. One challenge with constructing these chimeras is that they may not be functional. Indeed, many were not. Unfortunately, little was known regarding any additional ligands for these chemoreceptors. As B. subtilis is known to perform chemotaxis toward amino acids, we tested whether these chemoreceptors were involved in sensing Casamino Acids. TlpB alone was able to support chemotaxis to Casamino Acids; however, McpA alone did not. While we were not able to identify any attractants for McpA, we did find that McpA governs the repellent response to indole.
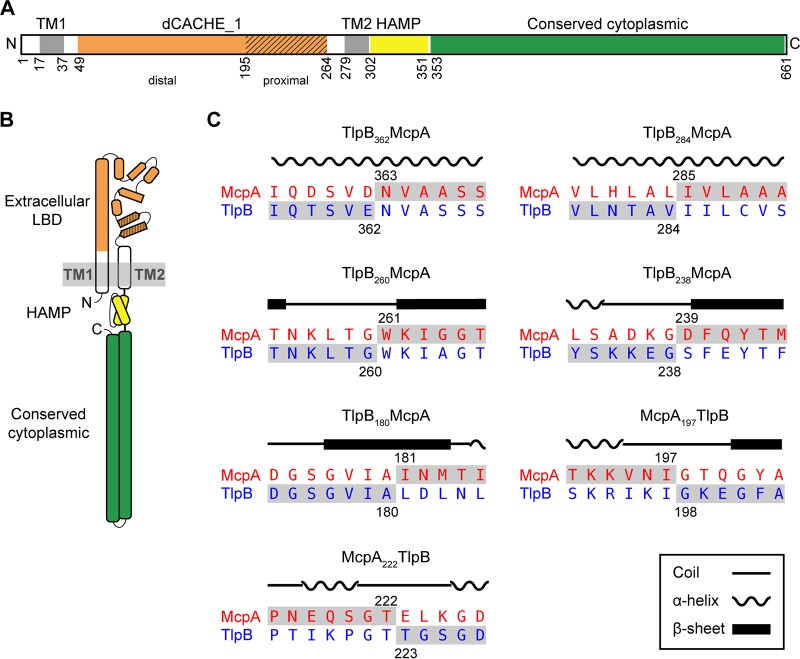
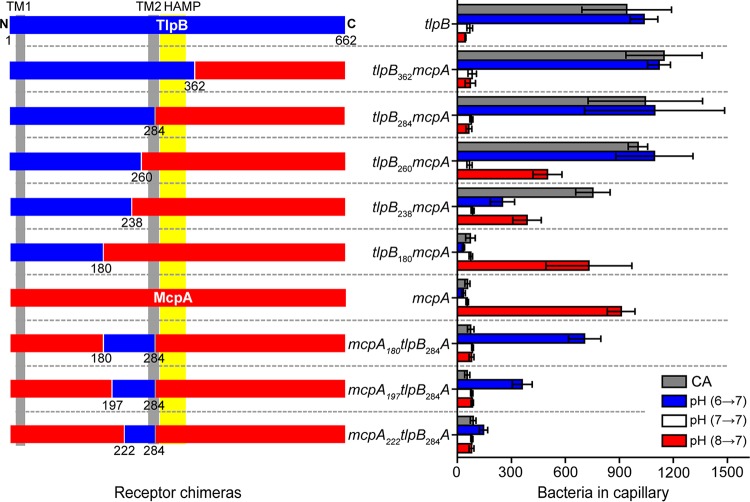
We first constructed a series of chimeras where we fused the N-terminal region of TlpB with the C-terminal region of McpA: tlpB362-mcpA, tlpB284-mcpA, tlpB260-mcpA, tlpB238-mcpA and tlpB180-mcpA (Fig. 3). We then tested the ability of strains expressing these hybrids as their sole chemoreceptor to sense acid and base gradients using the capillary assay. We employed Casamino Acids as a control. Strains expressing tlpB362-mcpA or tlpB284-mcpA behaved the same as those expressing wild-type tlpB (Fig. 4). These results indicate that pH is not sensed by the cytoplasmic domain of the chemoreceptor or by the HAMP domain. The strain expressing tlpB260-mcpA was able to sense both increases and decreases in pH. As the base response was similar to that of the strain expressing wild-type tlpB, these results would suggest that the region 260 to 284 from McpA is involved in acid sensing. The strain expressing tlpB238-mcpA was able to sense both increases and decreases in pH, albeit at reduced levels. However, the strains expressing tlpB180-mcpA were unable to sense base gradients and only responded to acid gradients, indicating that the region spanning the residues 180 to 284 on McpA is directly involved in acid sensing. In addition, strains expressing tlpB180-mcpA no longer responded to Casamino Acids. Likely, this was due to disruption of the sensing domain. All other N-terminal TlpB chimeras were able to sense Casamino Acid gradients.
FIG 3.
Construction of chimeric receptors to determine regions involved in pH sensing. (A) Domain structure of McpA. Extracellular ligand-binding, dCACHE_1 (orange), transmembrane, TM1 and TM2 (gray), HAMP (yellow), and cytoplasmic (green) domains. The membrane-proximal module of the dCACHE_1 domain is highlighted using diagonal lines; the other region corresponds to the membrane-distal module. (B) Cartoon structure of McpA. (C) Amino acid sequence alignment of McpA and TlpB around chimera junction points. The numbers designate the fusion points between two chemoreceptors, and the local sequences of the final chimeric chemoreceptors are highlighted in gray.
FIG 4.
Responses of strains expressing receptor chimeras involving different fragments of McpA (red) and TlpB (blue) to pH and Casamino Acid (CA) gradients. Error bars denote the standard deviations from three biological replicates performed on three separate days.
To characterize the region spanning residues 180 to 284 further, we replaced this region in McpA with the corresponding region from TlpB (mcpA180-tlpB284A). Strains expressing this chimera as their sole chemoreceptor only responded to increases in pH (Fig. 4). We then created two more chimeric strains, the mcpA197-tlpB284A and mcpA222-tlpB284A strains, to narrow down the regions responsible for pH sensing. The mutant expressing mcpA197-tlpB284A also responded only to increases in pH. However, the mutant expressing mcpA222-tlpB284A did not exhibit a significant response to pH (Fig. 4). These results indicated that the subregion spanning the residues 197 to 222 is also involved in base sensing.
All of the strains expressing the McpA chimeras exhibited a repellent response to indole. However, the responses of these strains were significantly reduced compared to that of the strain expressing wild-type mcpA (average net accumulation within capillary: 219.7 ± 90.5 [mcpA180-tlpB284A], 232.0 ± 183.1 [mcpA197-tlpB284A], and 248.6 ± 109.5 [mcpA222-tlpB284A] versus 1,286.1 ± 467.6 cells [wild type]). This would suggest that the McpA chimeras are not fully functional, which may explain their reduced response to increases in pH compared to TlpB.
Identification of residues involved in pH sensing.
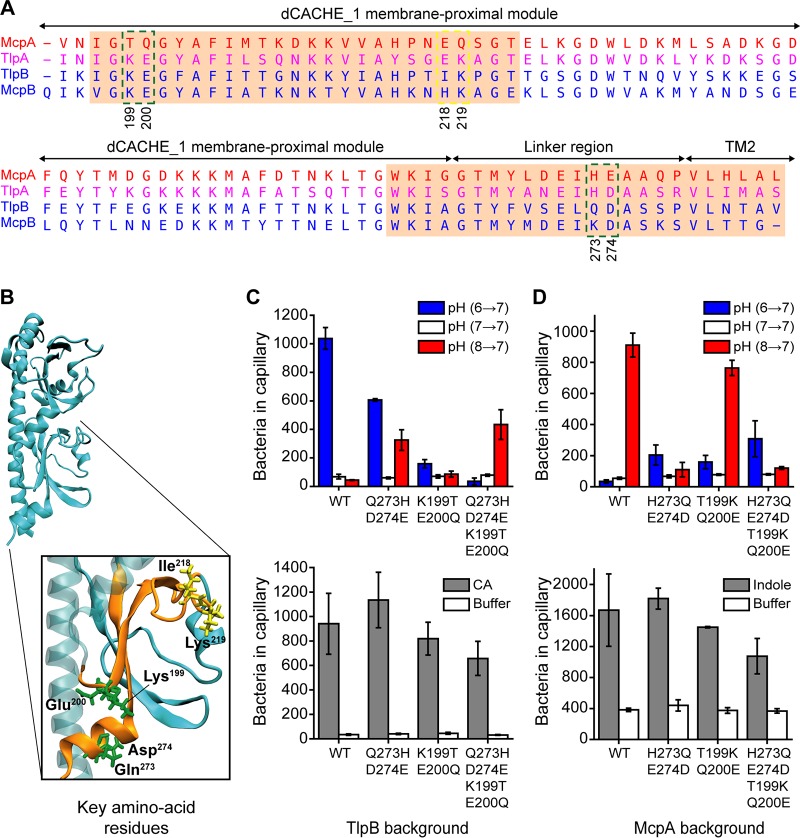
To identify the specific amino acid residues involved in pH sensing, we aligned the amino acid sequences spanning residues 195 to 284 of the four chemoreceptors, which correspond to the membrane-proximal module of the dCACHE_1 domain and linker region (Fig. 5A). Charged amino acids are more likely to be involved in pH sensing, as their side chains can be protonated and deprotonated. In addition, the polar amino acids may play an indirect role in pH sensing by impacting the local amino acid pKa values and/or forming hydrogen bonds with side chains of ionizable amino acid residues. Within the pH-sensing subregion spanning residues 260 to 284, the potential candidate residues were the Gln273-Asp274 pair for TlpB and the corresponding His273-Glu274 pair for McpA. Within the other pH-sensing subregion spanning residues 197 to 222, the potential candidate residues were the Ile218-Lys219 pair for TlpB and the corresponding Glu218-Gln219 pair for McpA.
FIG 5.
Identification of critical residues involved in sensing pH. (A) Amino acid sequence alignment of pH-sensing regions spanning residues (195 to 284) for the four pH chemoreceptors reveals candidate residues for mutational analysis. Candidate residues are shown within the green and yellow dashed boxes. The amino acid sequence for TlpA is shown in purple, because TlpA is sensitive to both acid and base. pH-sensing subregions identified from chimeric receptor analysis are highlighted in the orange boxes. (B) Predicted structure of the TlpB ligand binding domain (LBD). Two pH-sensing subregions are shown in orange consistent with panel A. The candidate amino acid residues on the TlpB extracellular LBD are shown in green and yellow. (C) Responses of strains expressing tlpB mutants to pH and Casamino Acid (CA) gradients. (D) Responses of strains expressing mcpA mutants to pH and negative indole gradients. Error bars denote the standard deviations from three biological replicates performed on three separate days.
The pH-sensing subregions are separated by 38 amino acid residues. However, when the predicted tertiary structure of the TlpB sensing domain was visualized, we found that the amino acid pair Lys199-Glu200 was in close proximity to the Gln273-Asp274 pair (Fig. 5B). Similarly, the Thr199-Gln200 pair was in close proximity to the His273-Glu274 pair on McpA (not shown in the figure). Therefore, Lys199-Glu200 on TlpB and Thr199-Gln200 on McpA were also potential candidate residues for mutational analysis, as local amino acid interactions could affect pH sensing.
We examined the role of these candidate pairs by swapping them with their counterparts on the opposite receptor, both individually and in combination. We first replaced the Gln273-Asp274 pair on TlpB with the counterpart His273-Glu274 pair from McpA. Cells expressing tlpB-Q273H/D274E as their sole chemoreceptor exhibited a reduced base response and an increased acid response (Fig. 5C). When we performed the reciprocal experiment, replacing the His273-Glu274 pair on McpA with the Gln273-Asp274 pair from TlpB, we observed a large decrease in the acid response and small increase in the base response (Fig. 5D). These results demonstrate that Gln273-Asp274 on TlpB and His273-Glu274 on McpA are involved in pH sensing.
We next tested the effect of replacing the Lys199-Glu200 pair on TlpB with the Thr199-Gln200 pair from McpA. This mutant (tlpB-K199T/E200Q) exhibited a weak response to both acids and bases (Fig. 5C). However, the reciprocal mutation (mcpA-T199K/Q200E), where we replaced the Thr199-Gln200 pair on McpA with the Lys199-Glu200 pair from TlpB, led to a small reduction in the acid response and a small increase in the base response (Fig. 5D). When the four residues on TlpB were swapped with their counterpart residues from McpA, the base response was eliminated and the resulting strain only exhibited an acid response, albeit at reduced levels (Fig. 5C). Similarly, when the four residues on McpA were swapped with their counterpart residues from TlpB, the acid response was significantly reduced and the resulting strain exhibited a base response, again at reduced levels. Collectively, these results imply that these four amino acid residues are sufficient to define the polarity of pH sensing for both McpA and TlpB. Therefore, we did not examine the Ile218-Lys219 pair on TlpB or Glu218-Gln219 on McpA, as these residues are located far away from the identified key residues (Fig. 5B) and likely are not involved in pH sensing.
We note that these mutant chemoreceptors with swapped polarity exhibited a reduced response to pH. Likely, the mutations are disrupting overall chemoreceptor function. When we tested these chemoreceptors against Casamino Acids and indole, they exhibited a weaker response than the wild type (Fig. 5C and D).
Model for pH sensing.
The experiments described above identified four critical amino acid residues involved in pH sensing. The predicted structures for the extracellular domains of TlpB and McpA reveal that these four amino acid residues are in close proximity of one another, suggesting that direct interactions between them may govern the sensing mechanism (Fig. 5B). Side chains of ionizable amino acid residues can accept or donate protons as the local pH varies. It is generally thought that changes in the protonation state of ionizable amino acid residues can lead to conformational changes in the protein and possibly alter its activity. These conformational changes are typically induced by the formation or disruption of hydrogen bonds between two or more proximal residues. The protonation state of ionizable amino acid residues largely relies on their local pKa values. The local pKa of an amino acid residue in the folded protein depends on its interactions with neighboring residues. As the result of such interactions, including charge-dipole, charge-charge, and ion pairs, pKa values can be significantly different from the intrinsic pKa (pKa-int) values measured in blocked pentapeptides (37). For example, the pKa of a lysine residue can be as low as 5.7 and the pKa of an aspartate residue can be as high as 9.2 in folded proteins (37).
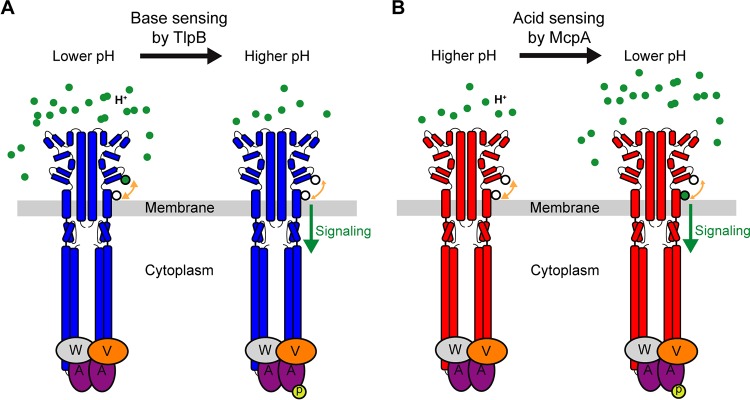
In the case of TlpB, increase in pH can directly affect three ionizable residues: Asp274, Lys199, and Glu200. The point mutation tlpB-D274N did not affect the responses to pH (1,334.7 ± 352.4 versus 1,037.2 ± 76.9 cells for wild-type tlpB), suggesting that the protonation state of Asp274 remains intact or does not affect interactions with other residues. However, the double mutant K199T/E200Q significantly reduced the response to increase in pH (Fig. 5C). This result implies that increases in pH affect the protonation state of Lys199 or/and Glu200. Based on these results, we offer one potential model to explain the response of TlpB to increases in pH (Fig. 6). At lower pH (e.g., pH 6), Lys199 and/or Glu200 is protonated and forms stable hydrogen bonds with Gln273 or/and Asp274. As pH increases, Lys199 and/or Glu200 likely becomes deprotonated, and the loss of the hydrogen bonds destabilizes the local structure of the membrane-proximal module of the dCACHE_1 domain and linker region (Fig. 3). This structural transition is then propagated through TM2 to the HAMP domain and subsequently to distal cytoplasmic signaling, thus inducing the autophosphorylation of CheA histidine kinase (Fig. 6A).
FIG 6.
Model for pH sensing mechanism in B. subtilis. (A) At low pH, two ionizable residues (black-outlined green circle) on TlpB are in their protonated state and form hydrogen bonds with two adjacent residues (white circle). Deprotonation of these residues upon pH increase disrupts the local structure due to decreased hydrogen bonding and induces signaling. (B) At high pH, the key histidine residue (lower white circle) within the pH-sensing region of McpA is in neutral state and forms hydrogen bonds with adjacent residues (upper white circle). As pH decreases, the histidine residue becomes protonated (black-outlined green circle), leading to loss of hydrogen bonding. This disrupts the local structure and induces signaling.
In the case of McpA, the ionizable residues are His273 and Glu274. The other two key residues Thr199 and Gln200, contain polar side chains that are insensitive to pH changes. As expected, the double mutant H273Q/E274D significantly reduced the response to decrease in pH (Fig. 5D). Among the two residues, His273 appears to play a pivotal role, as the point mutation E274Q did not affect the response to pH gradients (1,176.0 ± 124.5 versus 910.4 ± 77.0 cells for wild-type mcpA). On possible model is that at high pH (e.g., pH 8), His273 is in its neutral form. As pH decreases, His273 becomes protonated and no longer forms hydrogen bonds with Thr199 and/or Gln200. This destabilizes the local structure. Similarly to the TlpB case, this conformational change induces a structural transition in the membrane-proximal module of the dCACHE_1 domain and linker region, which in turn promotes phosphorylation of the CheA histidine kinase (Fig. 6B).
Conservation of pH-sensing amino acid residues across bacterial species.
We next attempted to determine whether our proposed mechanism in B. subtilis 168 can possibly serve as a general mechanism for pH sensing by dCACHE_1-containing bacterial chemoreceptors. Our approach here was to assess the conservation of the key pH-sensing and the neighboring residues across bacterial species. Briefly, we first used the dCACHE_1 domain and linker region from McpA (residues 35 to 281) to identify chemoreceptors with similar domains. We then extracted sequences of the membrane-proximal module of dCACHE_1 domain and linker region (90 residues) from these matches and aligned them after collapsing highly similar sequences (>95% similarity). The resulting 838 aligned sequences were then analyzed to determine the conservation of the four pH-sensing residues (Fig. 7).
FIG 7.
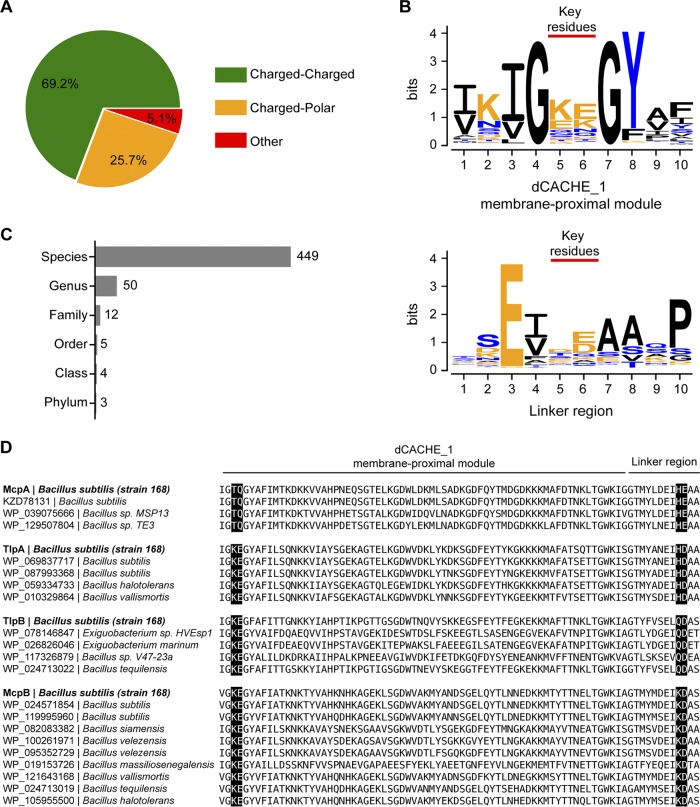
Conservation of pH-sensing amino acid residues in dCACHE_1 domains. (A) Fraction of identified domains that contain a charged amino acid(s) in positions equivalent to T199/Q200 in the membrane-proximal module and H273/E274 in the linker region of McpA (green), fraction that contains a charged amino acid(s) in corresponding positions in the membrane-proximal module and polar amino acid(s) in corresponding positions in linker region of McpA or vice versa (orange), and otherwise (red). (B) Amino acid sequence logos depicting conservation of the key pH-sensing and the neighboring residues in the membrane-proximal module (top) and linker region (bottom). (C) Numbers of predicted organisms possessing dCACHE_1 domains potentially capable of pH sensing at different taxonomy levels. (D) Amino acid sequence alignment of pH-sensing regions from different bacterial species, which contain amino acid residues (highlighted in black) identical to the key pH-sensing residues identified in four pH chemoreceptors of B. subtilis 168. NCBI accession numbers for each protein and the corresponding bacterial species are shown next to the sequence.
Our results suggest that charged and/or polar residues in both the membrane-proximal module and linker region are involved in sensing pH. Almost all of the aligned domains contain at least one charged and/or polar amino acid residue in both sites. In particular, the majority of these domains (>69%) possess at least one charged residue at both sites (Fig. 7A), suggesting that these domains are potentially capable of pH sensing. While the amino acid identity of these sites is diverse, lysine and glutamate residues in the membrane-proximal module and aspartate and glutamate residues in the linker region are more prevalent (Fig. 7B; see also Fig. S2). Interestingly, the residues at both sites are flanked by hydrophobic residues, suggesting that these residues may facilitate interactions between these two sites.
We also investigated the conservation of these two sites within B. subtilis strains (taxonomy identifier [ID] 1423). The resulting data show that all 236 identified domains contain at least one charged and/or polar amino acid residue in both pH-sensing sites (see Fig. S3A). In addition, lysine and glutamate residues at the membrane-proximal module and histidine and aspartate residues at the linker region are more prevalent (Fig. S3B).
The pH-sensing sites are conserved not only among B. subtilis strains. We identified 449 species with such sites, comprising mostly Firmicutes but also a few Proteobacteria and Actinobacteria (Fig. 7C; see also Table S1). These results suggest that pH sensing by the membrane-proximal module of dCACHE_1 domain and linker region may be conserved across diverse bacterial species. In particular, we found a number of domains from other species in which the key pH-sensing residues were identical to the ones in our B. subtilis 168 strain (taxonomy ID 224308) (Fig. 7D).
dCACHE_1 domains are known to directly bind ligands in their membrane-distal module (7, 23, 38–40). It is thought that the membrane-proximal module does not directly interact with ligands. Rather, it is thought to transmit the stimuli caused by ligand binding at the membrane-distal module to the transmembrane region (38). However, one study recently showed that lactate can directly bind to the membrane-proximal module of the TlpC dCACHE_1 domain in H. pylori (41). In our work, we present additional evidence that the membrane-proximal module is involved in direct sensing, namely, of pH.
We finally note that bacteria have evolved a number of diverse mechanisms for sensing pH in the context of chemotaxis. E. coli and S. enterica employ chemoreceptors with sensing domains consisting of four-helix bundles (4HB_MCP_1) to sense pH (27, 28). H. pylori, on the other hand, senses pH using a chemoreceptor with a PAS (sCACHE_2) sensing domain. In this case, pH induces signaling by modulating the binding affinity of urea to the PAS sensing domain (32). Our work identified a third mechanism for sensing pH involving chemoreceptors with dCACHE_1 sensing domains. As these are among the most prevalent sensing domains found in chemoreceptors (42), the proposed mechanism for pH sensing may be employed by many other bacterial species, as our analysis suggests (Fig. 7). Whether additional sensing domains can sense pH is unknown; however, given the diversity of known mechanisms, it would not be surprising if there are many more.
Conclusion.
We identified the four main chemoreceptors governing pH taxis in B. subtilis. McpA is the primary acid chemoreceptor, while McpB and TlpB are the base chemoreceptors. In addition, TlpA alone functions both as an acid and base chemoreceptor when overexpressed, though its primary role appears to be acid sensing. Using receptor chimeras, we identified four critical amino acid residues involved in pH sensing. Swapping these residues between McpA and TlpB was able to convert the former into a base sensor and the latter into an acid sensor. Based on our results, we were able to propose a model for pH sensing in B. subtilis. Collectively, these results further our understanding of pH taxis and provide a model for pH sensing.
MATERIALS AND METHODS
Chemicals and growth media.
The following media were used for cell growth: tryptone broth (TB; 1% tryptone and 0.5% NaCl), tryptose blood agar base (TBAB; 1% tryptone, 0.3% beef extract, 0.5% NaCl, and 1.5% agar), and capillary assay minimal medium [CAMM; 50 mM potassium phosphate buffer (pH 7.0), 1.2 mM MgCl2, 0.14 mM CaCl2, 1 mM (NH4)2SO4, 0.01 mM MnCl2, and 4.2 μM ferric citrate]. Chemotaxis buffer consists of 10 mM potassium phosphate buffer (pH 7.0), 0.14 mM CaCl2, 0.3 mM (NH4)2SO4, 0.1 mM EDTA, 5 mM sodium lactate, and 0.05% (vol/vol) glycerol.
Bacterial strains and plasmids.
All B. subtilis strains were derived from the strain OI1085 (43). All cloning was performed using NEB 5-alpha Competent E. coli (New England BioLabs). Bacterial strains and plasmids used in this work are listed in Tables 1 and 2, respectively. All oligonucleotides used in this study are provided in Table S2 in the supplemental material.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype or description | Reference or source |
|---|---|---|
| DH5α | E. coli cloning host | New England BioLabs |
| OI3269 | Bacillus subtilis 168 trpC2 | |
| OI1085 | trpF7 hisH2 metC che+ | 43 |
| PTS083 | ΔcheB | This work |
| PTS084 | ΔcheR | This work |
| PTS097 | ΔcheC | This work |
| PTS135 | ΔcheV | This work |
| PTS324 | ΔmcpA | This work |
| PTS325 | ΔtlpA | This work |
| PTS185 | ΔmcpB | This work |
| PTS186 | ΔtlpB | This work |
| PTS334 | ΔmcpA ΔtlpA | This work |
| PTS187 | ΔtlpB ΔmcpB | This work |
| PTS110 | ΔmcpA ΔtlpA ΔtlpB ΔmcpB | This work |
| OI3545 | Δ10mcp che+ | 24 |
| OI3921 | OI3545 amyE5720::mcpA | 17 |
| OI3605 | OI3545 amyE5720::mcpB | 25 |
| OI3974 | OI3545 amyE5720::mcpC | 17 |
| OI4474 | OI3545 amyE5720::tlpA | This work |
| OI4475 | OI3545 amyE5720::tlpB | This work |
| OI4483 | OI3545 amyE5720::tlpC | This work |
| OI4476 | OI3545 amyE5720::yfmS | This work |
| OI4477 | OI3545 amyE5720::yvaQ | This work |
| OI4482 | OI3545 amyE5720::hemAT | This work |
| OI4479 | OI3545 amyE5720::yoaH | This work |
| PTS163 | OI3545 amyE5720::tlpB284-mcpA | This work |
| PTS165 | OI3545 amyE5720::tlpB362-mcpA | This work |
| PTS368 | OI3545 amyE5720::tlpB261-mcpA | This work |
| PTS493 | OI3545 amyE5720::tlpB238-mcpA | This work |
| PTS505 | OI3545 amyE5720::tlpB180-mcpA | This work |
| PTS500 | OI3545 amyE5720::mcpA180-tlpB284A | This work |
| PTS507 | OI3545 amyE5720::mcpA197-tlpB284A | This work |
| PTS509 | OI3545 amyE5720::mcpA222-tlpB284A | This work |
| PTS441 | OI3545 amyE5720::tlpB[Q273H] [D274E] | This work |
| PTS421 | OI3545 amyE5720::tlpB[K199T] [E200Q] | This work |
| PTS464 | OI3545 amyE5720::tlpB[Q273H] [D274E] [K199T] [E200Q] | This work |
| PTS373 | OI3545 amyE5720::mcpA[H273Q] [E274D] | This work |
| PTS462 | OI3545 amyE5720::mcpA[T199K] [Q200E] | This work |
| PTS481 | OI3545 amyE5720::mcpA[H273Q] [E274D] [T199K] [Q200E] | This work |
| PTS251 | OI3545 amyE5720::tlpB[D274N] | This work |
| PTS276 | OI3545 amyE5720::mcpA[E274Q] | This work |
| GB041 | OI3545 amyE5720::PmcpA::tlpA | Unpublished |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pJOE8999 | Shuttle vector for Cas9 expression and tracrRNA transcription; Kanr | 44 |
| pJSpe | Modified pJOE8999 optimized for Gibson assembly of homology templates; Kanr | This work |
| pPT037 | pJSpe::cheV (cheV knockout vector) | This work |
| pPT039 | pJOE8999::cheB (cheB knockout vector) | This work |
| pPT041 | pJOE8999::cheR (cheR knockout vector) | This work |
| pPT046 | pJSpe::cheC (cheC knockout vector) | This work |
| pPT048 | pJSpe::mcpA tlpA mcpB tlpB (4mcp knockout vector) | This work |
| pPT058 | pJSpe::mcpB (mcpB knockout vector) | This work |
| pPT074 | pJSpe::tlpB (tlpB knockout vector) | This work |
| pPT116 | pJSpe::mcpA (mcpA knockout vector) | This work |
| pPT118 | pJSpe::tlpA (tlpA knockout vector) | This work |
| pPT141 | pJSpe::mcpA-tlpA (mcpA-tlpA knockout vector) | This work |
| pAIN750 | B. subtilis amyE integration vector; Ampr Spcr | 17 |
| pAIN750mcpA | pAIN750::mcpA | 25 |
| pAIN750mcpB | pAIN750::mcpB | 25 |
| pAIN750mcpC | pAIN750::mcpC | 17 |
| pAIN750tlpA | pAIN750::tlpA | This work |
| pAIN750tlpB | pAIN750::tlpB | This work |
| pAIN750tlpC | pAIN750::tlpC | This work |
| pAIN750yfmS | pAIN750::yfmS | This work |
| pAIN750yvaQ | pAIN750::yvaQ | This work |
| pAIN750hemAT | pAIN750::hemAT | This work |
| pAIN750yoaH | pAIN750::yoaH | This work |
| pPT065 | pAIN750::tlpB362-mcpA | This work |
| pPT063 | pAIN750::tlpB284-mcpA | This work |
| pPT143 | pAIN750::tlpB261-mcpA | This work |
| pPT224 | pAIN750::tlpB238-mcpA | This work |
| pPT234 | pAIN750::tlpB180-mcpA | This work |
| pPT233 | pAIN750::mcpA180-tlpB284A | This work |
| pPT236 | pAIN750::mcpA197-tlpB284A | This work |
| pPT237 | pAIN750::mcpA222-tlpB284A | This work |
| pPT129 | pAIN750::tlpB[Q273H] [D274E] | This work |
| pPT162 | pAIN750::tlpB[K199T] [E200Q] | This work |
| pPT202 | pAIN750::tlpB[Q273H] [D274E] [K199T] [E200Q] | This work |
| pPT196 | pAIN750::mcpA[H273Q] [E274D] | This work |
| pPT163 | pAIN750::mcpA[T199K] [Q200E] | This work |
| pPT222 | pAIN750::mcpA[H273Q] [E274D] [T199K] [Q200E] | This work |
| pPT101 | pAIN750::tlpB[D274N] | This work |
| pPT107 | pAIN750::mcpA[E274Q] | This work |
| pGB045 | pAIN750::PmcpA::tlpA | Unpublished |
Gene deletions were constructed using plasmids derived from pJOE8999 or pJSpe. pJSpe was derived from pJOE8999, which provides a CRISPR/Cas9-based, marker-free genome editing system for B. subtilis (44). We found that the SfiI restriction sites on the original pJOE8999 were inefficient for subcloning homology templates. In addition, a 13-bp-long DNA scar remained on the chromosome after the targeted DNA fragment was deleted using pJOE8999-derived vectors. Therefore, we created pJSpe for more efficient assembly of homology templates based on Gibson assembly and scarless deletion of DNA fragments. Briefly, 50-bp annealed complementary oligonucleotides containing a SpeI restriction site and optimized for Gibson assembly were inserted between the SfiI restriction sites on pJOE8999 to yield pJSpe. For construction of the knockout plasmids, a 20-bp single guide RNA (sgRNA) target sequence for the targeted gene was designed using CRISPy-web online tool (45). Annealed complementary oligonucleotides were then subcloned into BsaI restriction sites on pJSpe as described in reference 44. Two PCR fragments (∼600 to 800 bp) flanking the targeted gene using overlapping primers were subcloned into SpeI-linearized vector using Gibson assembly (46). The resultant vector was then linearized using XhoI and ligated with T4 DNA ligase to create a long DNA concatemer. The concatemer was then transformed into B. subtilis strain using the two-step Spizizen method (47). Single colonies were isolated and twice streaked on fresh plates. Plasmid curing and genotype verification were performed as previously described (44).
Strains expressing a single wild-type chemoreceptor were constructed by integrating the respective chemoreceptor expression cassette into the amyE locus. The region containing the promoter, gene, and terminator was PCR amplified from genomic DNA isolated from B. subtilis 168. The PCR fragment was then cloned into the plasmid pAIN750 using the EcoRI and BamHI restriction sites. The plasmid was then transformed in OI3545, which lacks all ten chemoreceptors.
Chemoreceptor chimeras were constructed using Gibson assembly (46). Briefly, two opposing primers were designed to prime DNA synthesis outwards from the fusion point of the chimeric gene using PCR with pAIN750mcpA as the DNA template. Then, a second pair of primers with overlapping regions were designed to PCR amplify the desired fragment of tlpB gene from pAIN750tlpB plasmid. Following purification of the PCR products by gel extraction, the DNA fragments were assembled (46).
Point mutations were introduced into chemoreceptor genes using the inverse PCR method with pAIN750mcpA and pAIN750tlpB as DNA templates. Briefly, plasmid was PCR amplified using two opposing primers containing the desired mutations. Following purification by gel extraction, the DNA fragment was phosphorylated with T4 polynucleotide kinase and then blunt-end ligated using T4 DNA ligase. Ligation product was heat inactivated and transformed into E. coli. The plasmid was then isolated from E. coli and transformed into B. subtilis OI3545 as described above.
Capillary assay for chemotaxis.
The capillary assay was performed as described previously (1, 26). Briefly, cells were grown for 16 h at 30°C on TBAB plates. The cells were then scraped from the plates and resuspended to A600 of 0.03 in 5 ml CAMM supplemented with 50 μg/ml histidine, methionine, tryptophan, 20 mM sorbitol, and 2% TB. The cultures were grown to an A600 of 0.4 to 0.45 at 37°C and 250 rpm shaking. At this point, 50 μl of GL solution (5% [vol/vol] glycerol, 0.5 M sodium lactate) was added to the culture, and the cells were incubated for another 15 min (at 37°C, 250 rpm shaking). Cells were then washed twice with chemotaxis buffer (pH 7.0) and incubated for additional 25 min (at 37°C, 250 rpm shaking) to ensure that the cells were motile. Cells were diluted to an A600 of 0.001 in chemotaxis buffer at desired pH values for pH taxis experiments and in chemotaxis buffer (pH 7.0) for Casamino Acid control experiments. For indole control experiments, cells were diluted to an A600 of 0.005 in chemotaxis buffer containing indole (50 μM, pH 7.0). Prior to performing the assay, cells were briefly incubated in chemotaxis buffer at room temperature (shaking slowly) and then aliquoted into 0.3-ml ponds on a slide warmer at 37°C, and closed-end capillary tubes filled with chemotaxis buffer or Casamino Acid (3.16 × 10−5 g/ml) solutions prepared with chemotaxis buffer (pH 7.0) were inserted. After a fixed time (30 min for Casamino Acids and 1 h for pH and indole), cells that migrated into the capillaries were harvested and transferred to 3 ml of top agar (1% tryptone, 0.5% NaCl, 0.5% yeast extract, 0.8% agar, 0.5 mM EDTA) and plated onto TB (1.5% agar) plates. These plates were incubated at 37°C for 16 h, and colonies were counted. Experiments were performed in triplicates each day and then repeated on three different days.
Structural analysis.
Domain predictions were performed using the phmmer search engine on the HMMER web server using the UniProt reference proteomes database with default sequence E value thresholds (48). Boundaries of both transmembrane alpha helices were first predicted using TMHMM web server v.2.0 (49) and then manually adjusted using information from propensity analysis of amino acid distributions around lipid-water interfaces (50). Pairwise amino acid sequence alignments between McpA and TlpB for chimeric-receptor analysis were performed using EMBOSS Water (51), and multiple sequence alignments between McpA, McpB, TlpA, and TlpB for mutational analysis were carried out in T-Coffee (52). Structures for the McpA and TlpB sensing domains (residues 38 to 278) were predicted using the I-TASSER web server (53). The C scores were 1.15 and 1.13 for McpA and TlpB, respectively. Both models are structurally close to the ligand-binding domain of the PctA chemoreceptor from Pseudomonas aeruginosa PAO1 (PDB 5LTX) with a TM score of 0.955 for both McpA and TlpB. Visualization of all structures was accomplished using the VMD software package (v-1.9.3) (54).
Conservation analysis.
Local alignment searches were performed using the BLAST+ toolkit (blastp v-2.9.0+) (55) and NCBI nr version 5 (nr_v5) preformatted database (downloaded on 7 September 2019). Briefly, the McpA dCACHE_1 domain and linker region (residues 35 to 281) was queried against the database, and significant hits with E values of <10−3 were collected for further analysis. For simplicity, the search space in the database was restricted to bacterial species. We collected unique hits that aligned to the query’s TM2-proximal region (residues 195 to 281) and covered at least 80% of query’s whole sequence (residues 35 to 281). This filter ensured that the retained hits likely contained the dCACHE_1 domain and the potential pH-sensing region. We used unique hits to reduce sequence bias toward a particular organism. The resulting 3,150 hits were then analyzed using the hmmscan tool (HMMER v-3.2.1) (56) and Pfam 32.0 hidden Markov model profile database with default parameter values to determine the best matching domain for each individual hit. Hits containing dCACHE_1 domain (E values < 10−10) were then collected, and the amino acid residues spanning the membrane-proximal module and linker region (90 residues) of these hits were extracted and first aligned using stand-alone CLUSTAL OMEGA (v-1.2.4) (57). Weakly aligned sequences were then removed using trimAl (v-1.2) (58), and highly similar sequences with a 95% similarity threshold were collapsed, again to reduce bias toward a specific organism. Finally, the remaining sequences were accurately aligned using MUSCLE (v-3.8.31) (59) with default parameter values, and the aligned sequences were used for further analysis. Taxonomy lineages of the identified organisms were extracted from NCBI GenBank taxonomy database (downloaded on 7 September 2019) using TaxonKit (v-0.50) (60). A custom Python script was generated for automation of steps described above. Sequence logos were created using WebLogo 3 (61). Conservation and consensus analysis and visualization of alignment sequences were performed in Jalview (v-2.11) (62).
Data availability.
Raw data are provided as Data Set S1. Python code used for conservation analysis is provided at https://github.com/paymantohidifar/pHtaxis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the University of Illinois through the Robert W. Schaefer Faculty Scholar fund and National Institutes of Health grant GM054365.
Footnotes
Supplemental material is available online only.
For a commentary on this article, see https://doi.org/10.1128/JB.00701-19.
REFERENCES
- 1.Ordal GW, Gibson KJ. 1977. Chemotaxis toward amino acids by Bacillus subtilis. J Bacteriol 129:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ordal GW, Villani DP, Rosendahl MS. 1979. Chemotaxis towards sugars by Bacillus subtilis. J Gen Microbiol 115:167–172. doi: 10.1099/00221287-115-1-167. [DOI] [PubMed] [Google Scholar]
- 3.Ordal GW, Goldman DJ. 1976. Chemotactic repellents of Bacillus subtilis. J Mol Biol 100:103–108. doi: 10.1016/s0022-2836(76)80037-x. [DOI] [PubMed] [Google Scholar]
- 4.Zimmer MA, Szurmant H, Saulmon MM, Collins MA, Bant JS, Ordal GW. 2002. The role of heterologous receptors in McpB-mediated signalling in Bacillus subtilis chemotaxis. Mol Microbiol 45:555–568. doi: 10.1046/j.1365-2958.2002.03035.x. [DOI] [PubMed] [Google Scholar]
- 5.Walukiewicz HE, Tohidifar P, Ordal GW, Rao CV. 2014. Interactions among the three adaptation systems of Bacillus subtilis chemotaxis as revealed by an in vitro receptor-kinase assay. Mol Microbiol 93:1104–1118. doi: 10.1111/mmi.12721. [DOI] [PubMed] [Google Scholar]
- 6.Wu K, Walukiewicz HE, Glekas GD, Ordal GW, Rao CV. 2011. Attractant binding induces distinct structural changes to the polar and lateral signaling clusters in Bacillus subtilis chemotaxis. J Biol Chem 286:2587–2595. doi: 10.1074/jbc.M110.188664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW. 2012. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem 287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity LF, Ordal GW. 1997. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology 143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- 9.Bischoff DS, Ordal GW. 1991. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem 266:12301–12305. [PubMed] [Google Scholar]
- 10.Rao CV, Kirby JR, Arkin AP. 2004. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol 2:E49. doi: 10.1371/journal.pbio.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao CV, Glekas GD, Ordal GW. 2008. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol 16:480–487. doi: 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao CV, Ordal GW. 2009. The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib Microbiol 16:33–64. doi: 10.1159/000219372. [DOI] [PubMed] [Google Scholar]
- 13.Kirsch ML, Zuberi AR, Henner D, Peters PD, Yazdi MA, Ordal GW. 1993. Chemotactic methyltransferase promotes adaptation to repellents in Bacillus subtilis. J Biol Chem 268:25350–25356. [PubMed] [Google Scholar]
- 14.Kirsch ML, Peters PD, Hanlon DW, Kirby JR, Ordal GW. 1993. Chemotactic methylesterase promotes adaptation to high concentrations of attractant in Bacillus subtilis. J Biol Chem 268:18610–18616. [PubMed] [Google Scholar]
- 15.Glekas GD, Cates JR, Cohen TM, Rao CV, Ordal GW. 2011. Site-specific methylation in Bacillus subtilis chemotaxis: effect of covalent modifications to the chemotaxis receptor McpB. Microbiology 157:56–65. doi: 10.1099/mic.0.044685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karatan E, Saulmon MM, Bunn MW, Ordal GW. 2001. Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis. J Biol Chem 276:43618–43626. doi: 10.1074/jbc.M104955200. [DOI] [PubMed] [Google Scholar]
- 17.Kristich CJ, Ordal GW. 2002. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem 277:25356–25362. doi: 10.1074/jbc.M201334200. [DOI] [PubMed] [Google Scholar]
- 18.Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM, Crane BR. 2006. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 19.Szurmant H, Muff TJ, Ordal GW. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem 279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 20.Glekas GD, Plutz MJ, Walukiewicz HE, Allen GM, Rao CV, Ordal GW. 2012. Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis. Mol Microbiol 86:743–756. doi: 10.1111/mmi.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muff TJ, Ordal GW. 2007. The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem 282:34120–34128. doi: 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- 22.Yuan W, Glekas GD, Allen GM, Walukiewicz HE, Rao CV, Ordal GW. 2012. The importance of the interaction of CheD with CheC and the chemoreceptors compared to its enzymatic activity during chemotaxis in Bacillus subtilis. PLoS One 7:e50689. doi: 10.1371/journal.pone.0050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. 2010. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J Biol Chem 285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 25.Kristich CJ, Glekas GD, Ordal GW. 2003. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol Microbiol 47:1353–1366. doi: 10.1046/j.1365-2958.2003.03375.x. [DOI] [PubMed] [Google Scholar]
- 26.Tso WW, Adler J. 1974. Negative chemotaxis in Escherichia coli. J Bacteriol 118:560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krikos A, Conley MP, Boyd A, Berg HC, Simon MI. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A 82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kihara M, Macnab RM. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol 145:1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Repaske DR, Adler J. 1981. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol 145:1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umemura T, Matsumoto Y, Ohnishi K, Homma M, Kawagishi I. 2002. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J Biol Chem 277:1593–1598. doi: 10.1074/jbc.M109930200. [DOI] [PubMed] [Google Scholar]
- 31.Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol 188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. 2012. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Sourjik V. 2012. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol Microbiol 86:1482–1489. doi: 10.1111/mmi.12070. [DOI] [PubMed] [Google Scholar]
- 34.Hanlon DW, Ordal GW. 1994. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem 269:14038–14046. [PubMed] [Google Scholar]
- 35.Cannistraro VJ, Glekas GD, Rao CV, Ordal GW. 2011. Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J Bacteriol 193:3220–3227. doi: 10.1128/JB.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pace CN, Grimsley GR, Scholtz JM. 2009. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem 284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A. 2015. Structural basis for amino acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr D Biol Crystallogr 71:2127–2136. doi: 10.1107/S139900471501384X. [DOI] [PubMed] [Google Scholar]
- 39.Webb BA, Hildreth S, Helm RF, Scharf BE. 2014. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl Environ Microbiol 80:3404–3415. doi: 10.1128/AEM.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama S, Suzuki D, Itoh Y, Suzuki K, Tajima H, Hyakutake A, Homma M, Butler-Wu SM, Camilli A, Kawagishi I. 2012. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun 80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machuca MA, Johnson KS, Liu YC, Steer DL, Ottemann KM, Roujeinikova A. 2017. Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate. Sci Rep 7:14089. doi: 10.1038/s41598-017-14372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortega A, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81: e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullah AH, Ordal GW. 1981. In vivo and in vitro chemotactic methylation in Bacillus subtilis. J Bacteriol 145:958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altenbuchner J. 2016. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl Environ Microbiol 82:5421–5427. doi: 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blin K, Pedersen LE, Weber T, Lee SY. 2016. CRISPy-web: an online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol 1:118–121. doi: 10.1016/j.synbio.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 47.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. 2018. HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- 50.Ulmschneider MB, Sansom MS. 2001. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta 1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 51.Chojnacki S, Cowley A, Lee J, Foix A, Lopez R. 2017. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res 45:W550–W553. doi: 10.1093/nar/gkx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 55.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23:205–211. [PubMed] [Google Scholar]
- 57.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen W, Xiong J. 2019. TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. bioRxiv doi: 10.1101/513523. [DOI]
- 61.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are provided as Data Set S1. Python code used for conservation analysis is provided at https://github.com/paymantohidifar/pHtaxis.