Abstract
Background
Colorectal cancer (CRC) has been confirmed to be the third most commonly diagnosed cancer in males and the second in females. We investigated the blood plasma metabolome in CRC patients and in healthy adults to elucidate the role of monosaccharides, amino acids, and their respective metabolic pathways as prognostic factors in patients with CRC.
Methods
Fifteen patients with CRC and nine healthy adults were enrolled in the study and their blood plasma samples analyzed by gas chromatography-mass spectrometry (GC-MS). Univariate Student’s t-test, multivariate principal component analysis (PCA) and partial least square-discriminant analysis (PLS-DA) were conducted on MetaboAnalyst 4.0. The analysis of metabolic profiles was carried out by the web-based extension Metabolite Sets Enrichment Analysis (MSEA).
Results
Overall, 125 metabolites were identified in plasma samples by GC-MS. In CRC patient samples, nine metabolites, including D-mannose and fructose, were significantly more abundant than in controls; conversely, eleven amino derivatives were less abundant, including methionine, valine, lysine, and proline. Methionine was significantly less abundant in died patients compared with survivors. The most significantly altered metabolic pathways in CRC patients are those involving monosaccharides (primarily the catabolic pathway of fructose and D-mannose), and amino acids (primarily methionine, valine, leucine, and isoleucine).
Conclusions
The abundance of D-mannose in CRC patient samples contributes to inhibiting the growth of cancer cells, while the abundance of fructose may be consistent either with low consumption of fructose by aerobic glycolysis within cancer cells or with a high bioavailability of fructose from diet. The reduction in methionine concentration may be related to increased activity of the threonine and methionine catabolic pathways, confirmed by high levels of α-hydroxybutyrate.
Keywords: Colorectal cancer (CRC), metabolomics, monosaccharides, amino acids, D-mannose, methionine
Introduction
Colorectal cancer (CRC) has been recently confirmed to be the third most commonly diagnosed cancer in males (10.9%) and the second in females (9.5%), showing a mortality rate of 9.0% and 9.5%, respectively (1). In the United States, the CRC 5-year relative survival rate is 90% in localized forms; however, it drastically drops to 14% in distant-stage disease diagnosis (2). Consequently, early detection and diagnosis of CRC are crucial to reducing mortality. Several non-invasive and invasive diagnostic approaches are currently available in routine clinical practice (3); most of them show low sensitivity and specificity often in conjunction with unsatisfactory patient compliance. Novel diagnostic and prognostic tests, including liquid biopsy (4) and molecular and genetic testing such as the detection of KRAS, BRAF, NRAS and PIK3CA gene mutations (5), the investigation on the role of miRNAs (6) and the study of interactions between epigenetic changes and gene mutations (7), are emerged over the last decade. Unfortunately, the effectiveness and the reliability of molecular and genetic tests are still controversial and not yet included in routine clinical protocols (8). Beyond the potential relevant value of genomics and transcriptomics in assessing CRC risk factors and drug discovering for individualized therapies, what matters in patient’s management is a very early diagnosis of CRC as well as the timely recognition of metabolic changes due to either the progression/regression of the disease or the effects of the therapeutic treatment. Metabolic changes anticipate clinical changes and represent the dynamic molecular phenotype, constituted by the activation states of signaling and metabolic pathways, which in turn result from the interaction between genomics, transcriptomics and environmental factors, including the state of the disease. The extensive network of biochemical interactions recognizable in a cell, tissue or biological fluid, can be qualitatively or quantitatively investigated by metabolomics, that consists of the study of metabolism in a comprehensive way (9). Since metabolic reprogramming is a significant phenomenon of cancer, being a basic hallmark of cancer-inducing tumorigenesis and malignant phenotype, metabolomics has been extensively used in cancer research to profile alterations in pathways of intermediary metabolism (10). Over the last decade, a consistent number of studies have investigated the metabolomic changes associated with CRC (11-15); however, most of the studies are affected by several limitations and by the lack of attempts at exploring potential clinical applications. The primary aim of our study was to identify new biomarkers associated with CRC, by comparing the metabolic profile of patients with that of healthy adults; moreover, we aimed to investigate the role of monosaccharides, amino acids and their respective biochemical pathways as prognostic factors in CRC.
Methods
Study design and population
Fifteen patients (11 males) with histological diagnosis of CRC, were enrolled; between July and September 2015, eight patients underwent surgery for rectal cancer and seven for colon cancer at the Colorectal Surgery Unit of the University of Cagliari. The average postoperative follow-up period was 42.4 months; death occurred in three patients (20.0%) after 3, 4, and 26 months from surgery, respectively (Table 1). Pre-treatment staging consisted of physical examination, serum carcinoembryonic antigen (CEA) testing, colonoscopy, abdominal and chest computed tomography (CT) scan, magnetic resonance imaging (MRI) or endorectal ultrasound for rectal cancer. Three types of surgical procedures were applied: anterior resection in 11 patients (73.3%), right hemicolectomy in 3 (20.0%) and local excision by transanal endoscopic microsurgery (TEM) in 1 (6.7%). Nine healthy adults (4 males) aged 30–67 years with serum CEA concentration below the limit of detection (0.5 mg/L) were enrolled in the study as a control group. The study protocol was approved by the local institutional review board of the University of Cagliari (CA-302-26/05/2014) and was conducted in accordance with the Helsinki Declaration of 1964, revised in 2008. All patients signed informed consent to the study.
Table 1. Clinical and demographic characteristics of patient population.
| Patient ID | Gender | Age (years) | CEA (mg/L) | Neoadjuvant therapy | cTNM stage | Surgery date (M, Y) | Surgery procedure | Histopathologic diagnosis | Clinical outcome (01/2019) |
|---|---|---|---|---|---|---|---|---|---|
| #01 BF | Male | 52 | 8.15 | 0 | III C | 07/2015 | ARR | Adenocarcinoma | Surgical wound infection; oliguria |
| #02 CD | Female | 52 | 5.24 | 0 | IV A | 11/2015 | ARR | Mucinous adenocarcinoma | No complication |
| #03 PM | Male | 54 | 8.03 | 0 | III C | 11/2015 | ARR | Adenocarcinoma | No complication |
| #04 ME | Male | 58 | 6.20 | 1 | III B | 11/2015 | ARR | Adenocarcinoma | No complication |
| #05 PG | Male | 58 | 2.25 | 0 | I | 10/2015 | ARR | Adenocarcinoma | No complication |
| #06 MM | Male | 61 | 3.75 | 0 | III B | 09/2015 | ARR | Adenocarcinoma | No complication |
| #07 AM | Male | 65 | 1.20 | 1 | II A | 04/2016 | ARR with ileostomy | Adenocarcinoma | No complication |
| #08 MMG | Female | 74 | 18.8 | 1 | II A | 10/2015 | ARR with ileostomy | Adenocarcinoma | No complication |
| #09 MG | Male | 75 | 46.6 | 0 | IV A | 10/2015 | Right hemicolectomy | Adenocarcinoma | Death after 4 months |
| #10 SGC | Male | 77 | 2.72 | 1 | III B | 02/2016 | ARR with ileostomy | Adenocarcinoma | Paralytic ileus |
| #11 SA | Male | 78 | 1.59 | 0 | III B | 11/2015 | ARR with ileostomy | Adenocarcinoma | Death after 4 months |
| #12 SR | Female | 79 | 83.2 | 0 | III C | 08/2015 | Right hemicolectomy | Adenocarcinoma | Death after 26 months |
| #13 CN | Female | 80 | 15.8 | 0 | II A | 09/2015 | ARR with ileostomy | Adenocarcinoma | No complication |
| #14 GM | Male | 83 | 10.1 | 0 | II B | 08/2015 | Right hemicolectomy | Adenocarcinoma | No complication |
| #15 DA | Male | 90 | 5.78 | 0 | I | 01/2016 | TEM | Adenocarcinoma | No complication |
ARR, anterior rectus-colonic resection; TEM, transanal endoscopic microsurgery.
Sample collection and preparation
Blood samples were collected in a sterile plastic test tube containing spray-dried K3 ethylenediaminetetraacetic acid (K3EDTA) as an anticoagulant; all patients samples were collected before surgery and before the initiation of any adjuvant chemotherapy treatment. After centrifugation (5 minutes at 1,000 g), plasma was separated and immediately frozen at −80 °C until analysis. For samples preparation, a modified version of a standard protocol was applied (16). Firstly, 400 µL of thawed plasma were transferred in Eppendorf tubes, treated with 1,200 µL of cold methanol, mixed by vortexer, and centrifuged 15 minutes at 16,100 g. Then, 370 mL of supernatant was transferred into glass vials and evaporated to dryness overnight in an Eppendorf vacuum centrifuge. Subsequently, 50 µL of a 0.24 M (20 mg/mL) solution of methoxylamine hydrochloride in pyridine were added to each vial, samples were mixed by vortexer and left to react for 17 h at room temperature. Then, 50 µL of N-Methyl-N-trimethylsilyl trifluoroacetamide (MSTFA) were added and left to react for 1 h at room temperature. The derivatized samples were diluted with hexane (200 µL), with tetracosane (0.01 mg/mL) as internal standard, just before gas chromatography-mass spectrometry (GC-MS) analysis.
GC-MS analysis
The derivatized samples were analyzed by using a global unbiased mass spectrometry-based platform with GC-MS incorporating an Agilent 5975C interfaced to the GC 7820 (Agilent Technologies, Palo Alto, CA, USA). The system was equipped with a DB-5ms column (Agilent J&W Scientific, Folsom, CA, USA); the injection temperature was set at 230 °C and the detector at 280 °C. Helium (the carrier gas) flow rate was equal to 1 mL/min. GC oven starting temperature program was 90 °C with 1 min hold time and ramping at a rate of 10 °C per minute, reaching a final temperature of 270 °C with 7 min hold time. One mL of the derivatized sample was injected in split (1:5) mode. After a solvent delay of 3 min, mass spectra were acquired in full scan mode using 2.28 scans per second with a mass range of 50–700 Amu. Each acquired chromatogram was analyzed through the free software AMDIS (Automated Mass Spectral Deconvolution and Identification System; http://chemdata.nist.gov/mass-spc/amdis). Each peak was identified by comparing the corresponding mass spectra and retention times with those stored in an in-house made library including 255 metabolites. Further metabolites were identified by using the National Institute of Standards and Technology’s mass spectral database (NIST08) (17) and the Golm Metabolome Database (GMD) (18). By applying this approach, we accurately identified 96 compounds, while further 28 metabolites were tentatively assigned, based on GMD and NIST libraries. AMDIS analysis produced a matrix spreadsheet (Microsoft Excel®, Microsoft Co, Redmond, WA, USA) as a preliminary tool for the subsequent chemometric analysis.
Statistical analysis
Univariate Student’s t-test, multivariate principal component analysis (PCA) and partial least square-discriminant analysis (PLS-DA) were conducted on MetaboAnalyst 4.0 (19) and used to observe data variance in unsupervised and supervised modes. The validity of the PLS-DA model was assessed by statistical parameters: the correlation coefficient R2, and the cross-validation correlation coefficient Q2. The goal was to identify pathways significantly impacting in a given phenotype. Since many pathways were tested simultaneously, P values were adjusted for multiple testing. The analysis of metabolic profiles in the context of the metabolic pathway was conducted by using the web-based extension Metabolite Sets Enrichment Analysis (MSEA), a tool integrated into the software framework of the MetaboAnalyst 4.0 platform (20). MSEA was used to investigate whether the compounds being part of the metabolites set are randomly distributed throughout an ordered list or predominantly recognized at the top or bottom.
Results
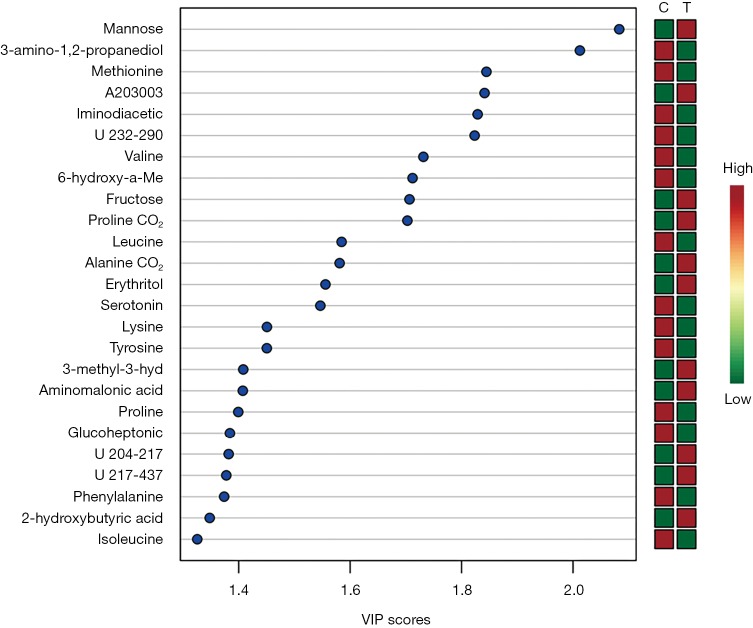
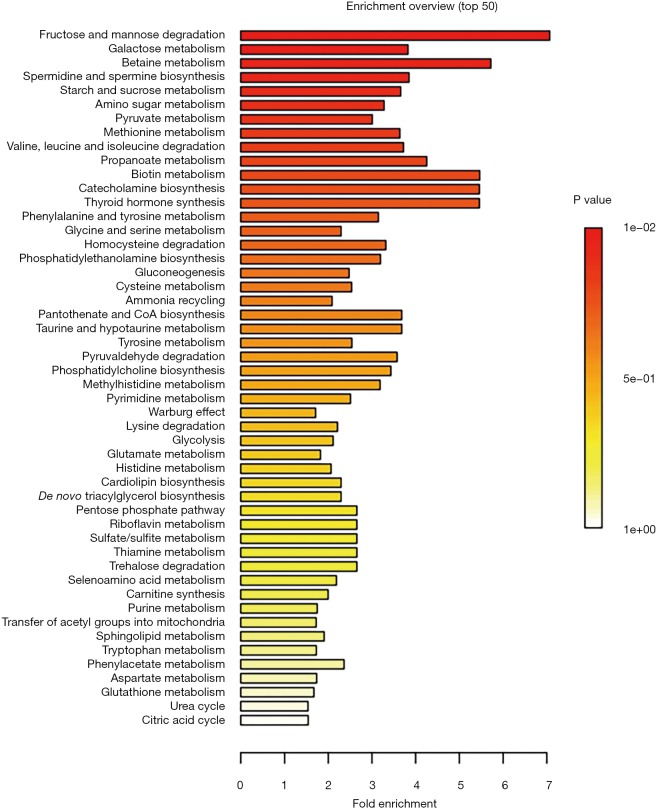
Overall, 125 compounds recognized in more than one sample were annotated. The univariate and multivariate analysis indicates two main classes of metabolites associated with CRC subjects: monosaccharides and amino derivatives. Univariate analysis revealed 10 metabolites significantly different between CRC patients and controls. PLS-DA analysis showed a clear separation between CRC patients and controls (accuracy =0.75, R2 =0.71, Q2 =0.45) highlighting the absence of any overlap between data groups (Figure 1). The most significant metabolites discriminating tumor samples (T) from controls (C) are reported in Figure 2; metabolites were ranked on the basis of the variable importance in projection (VIP) score. In CRC patient samples, nine metabolites were more abundant than in controls: D-mannose (an aldose hexose monosaccharide), fructose (a ketose hexose monosaccharide), the sugar-like unknown molecule A203003, proline + carbon dioxide (CO2), alanine + CO2, erythritol, 3-methyl-3-hydroxybutyric acid, aminomalonic acid, 2-hydroxybutyric acid. Conversely, eleven amino derivatives were found less abundant in patient samples compared with controls: 3-amino-1,2-propanediol, methionine, iminodiacetic acid, valine, leucine, lysine, tyrosine, proline, phenylalanine, isoleucine, serotonin. Interestingly, the abundance of methionine was found significantly lower in patients died after 3, 4, and 26 months from surgery. Specifically, the mean value of methionine abundance in died patients was 19,502 (standard deviation 8,437, median 16,155) while that of the survivors was 29,387 (standard deviation 10,447, median 29,921). MSEA analysis revealed that the most significantly altered metabolic pathways in CRC patients are those involving catabolism and metabolism of monosaccharides (primarily the catabolic pathway of fructose and D-mannose), and amino acids (primarily methionine, valine, leucine, and isoleucine), as reported in Figure 3.
Figure 1.
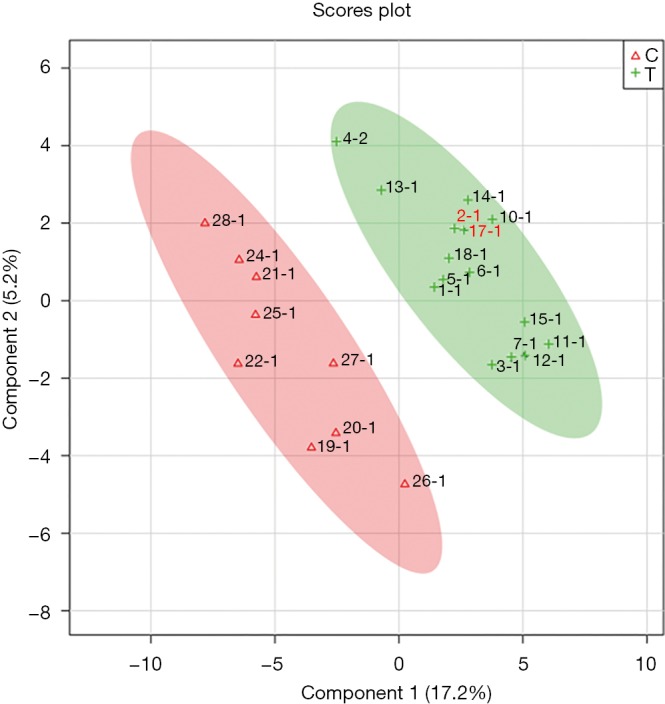
2D scores plot showing PLS-DA discrimination between plasma samples of CRC patients (T, green) and controls (C, red). The shaded areas indicate the 95% confidence regions. PLS-DA, partial least square-discriminant analysis; CRC, colorectal cancer.
Figure 2.
Summary plot showing the most important metabolites ranked based on the variable importance in projection (VIP) score. The mini heatmap on the right indicates metabolites changes within group C (controls) and group T (CRC patients). CRC, colorectal cancer.
Figure 3.
Graphic synopsis of Metabolite Sets Enrichment Analysis (MSEA). Results are expressed by a horizontal bar graph showing the most significant metabolites sets identified during analysis. Bars color is based on P values (lower P values correspond to a darker red) while bars length is based on the fold enrichment.
Discussion
Recent studies identified at least six hallmarks of tumor-associated metabolic reprogramming of cancer cells (21); among these, the deregulated uptake of glucose and amino acids induced by aberrantly activated oncogenes and loss of tumor suppressors is of considerable importance for tumor progression. Thus, our findings on the abundance of D-mannose and fructose in CRC patients deserve an in-deep reflection to add a further brick to an existing amount of controversial results on monosaccharides in CRC. An early study demonstrated that elevated D-mannose oligosaccharides are more predominant on tumor cell surfaces compared with non-cancer cell surfaces (22); further studies found high levels of D-mannose in various localized and metastatic cancers (23,24). Moreover, the content in D-mannose of N-linked glycans positively correlates with cancer progression and staging in breast, ovarian, and colorectal cancers (25-27). In most of the metabolomic studies on human CRC, D-mannose and fructose were not included into the list of significant differential metabolites between CRC and non-CRC biological matrices (serum, plasma, urine, stool) and tissues (28-41). A very significant abundance of D-mannose was previously observed in the serum of 30 patients with colorectal adenoma polyps and 30 with CRC (32). Conversely, in a study comparing 15 CRC tissues and 15 matched adjacent non-tumor tissues, D-mannose was found decreased in CRC samples (33). Moreover, a low level of sorbitol in 17 CRC patient tissues and stools was considered representative of significant perturbations of fructose, mannose and galactose metabolic pathways; curiously, none of these monosaccharides was included in the list of the significantly different metabolites between CRC and controls (34). Beyond variables inducing discrepancies among studies, such as tumor heterogeneity, biological matrix, patient’s lifestyle, analytical method, and gut microbial colonization through the colon, emerging knowledge should be considered to interpret our results. As observed long time ago, cancer cells generate their energy via aerobic glycolysis (35), even though they generate much more lactate and much less adenosine triphosphate (ATP) than those produced in healthy cells by a complete tricarboxylic acid (TCA) cycle coupled to oxidative phosphorylation. Very recently, an experimental study demonstrated that D-mannose interferes with glucose metabolism (glycolysis and other glucose-based pathways) by accumulating intracellularly as mannose-6-phosphate and thus impairing the growth of cancer cells (36). Notably, the study showed that phosphomannose isomerase (PMI), the enzyme converting mannose-6-phosphate to fructose-6-phosphate, was deficient in colon cancer tissue microarray as opposed to other cancer tissues (36). Cell lines with high levels of PMI are resistant to the effect of D-mannose because of the broad availability of fructose-6-phosphate, which can enter the glycolysis pathway. On the basis of these experimental findings, we can argue that in our patients population the abundance of D-mannose might not be dissociated from the high rate of survivors observed in this study (80%); notably, each survivor showed improved health-related quality of life after 39.5 months (median value) from the date of surgical intervention. The abundance of fructose in CRC patient group may be consistent either with a low fructose consumption rate within cancer cells or with a high fructose intake with diet. The former is coherent with low consumption of fructose by aerobic glycolysis within cancer cells; conversely, the latter implies a high bioavailability of fructose and thus providing fuel for major pathways of central carbon metabolism during tumor cell proliferation. Notably, among survivors, metastases were observed neither during surgery nor during follow-up, suggesting a reduced metabolic activity of cancer cells.
Amino acids play crucial physiological roles either as essential metabolites or metabolic modulators. Early and recent studies have found various amino acids, such as beta-alanine, glutamate, aspartate, ornithine, S-adenosylmethionine (SAM) up-regulated in CRC tissue compared to normal mucosa (29,37-40); these findings were interpreted as reflecting cancer cell needs for a higher turnover of structural proteins. Conversely, most of the metabolomic studies on CRC have found a significant decrease of various amino acids, such as intermediates in the urea cycle and amino acids related to glutamine and proline metabolism, compared with healthy controls (15,41-43). The decrease of various amino acids in CRC plasma samples may confirm the higher absorption of amino acids by tumor cells to sustain their rapid proliferation, as suggested elsewhere (42). In our study, methionine, valine, leucine, and proline were found significantly decreased in CRC plasma samples compared with healthy controls. Methionine, an essential amino acid in protein synthesis, is inversely associated with CRC risk, both by reducing inflammation-induced colon cancer and by inhibiting key pathways in colon carcinogenesis (44). Extraordinary evidence of this assumption emerges from our results, showing a statistically significant difference in methionine abundance between patients survived more than 2.5 years from surgery, and patients died. Notably, the lower abundance of methionine in died patents (mean value 19,502) was found before surgery, suggesting a potential predictive value of patient’s outcome for this metabolite. On the other hand, methionine is the precursor of SAM, a substrate for methyltransferases, which methylate metabolites, nucleic acids, lipids, and proteins. After the release of the activated methyl group in methylation reaction, SAM is transformed into S-adenosyl-L-homocysteine (SAH) that is finally hydrolyzed to homocysteine (45). The decrease of SAM induces an aberrant DNA methylation, leading in turn to an early CRC development (46). In our CRC patient samples, we observed high levels of α-hydroxybutyrate (α-HB), an organic acid derived from α-ketobutyrate (α-KB), which in turn is the final product of the threonine and methionine catabolic pathways. This finding is consistent with results previously published elsewhere (47) and may considerably contribute to the reduction in methionine concentration. However, the mechanism by which α-HB is up-regulated by colorectal tumor cells has not been yet elucidated. Our results confirm previous results on the decrease of valine and leucine in CRC patient samples (41,47). A plausible explanation may be the pivotal role of valine, and that more relevant of leucine, as “nutrients signals” regulating the mammalian target of rapamycin (mTOR) pathway, a critical regulator of cellular function forming two distinct protein complexes, mTORC1, and mTORC2 (48). In particular, leucine is an essential amino acid required for the activation of mTORC1. In CRC, mTOR plays a strategic role for the association between energy balance and cancer development; the observed depletion in valine and leucine plasma levels, associated with their upregulation in carcinoma tissue (37), may confirm the increased adsorption of these amino acids by tumor cells, supporting their rapid proliferation (42). Finally, proline decrease may originate either from the overexpression of the proline-rich 15 (Prr15) gene encoding proline-rich proteins in sporadic CRC or from the up-regulation of the proline dehydrogenase/oxidase (PRODH/POX) enzyme. Indeed, the latter catalyzes the transformation of proline into D1-pyrroline-5-carboxylate (P5C), which considerably contributes to the ATP synthesis by tumor cells for their survival in hypoxia and glucose deprivation conditions. Our study is affected by several limitations. Firstly, this is a cross-sectional study preventing to investigate to which extent metabolites are causally related to cancer changes and the patient’s outcome. The number of patients and controls is small and, more important, a single blood sample cannot permit to assess variations in metabolites plasma concentration over time. Furthermore, our study did not include a metagenomic analysis of gut microbiota in order to establish any contribute of microbiota-derived metabolites to plasma metabolome. However, the study contains several strengths: our results are in line with previous published metabolomic studies on CRC, confirming the key role of such metabolites (e.g., mannose, amino acids). Notably, very recent experimental researches allow us to interpret our results better, elucidating mechanisms, and pathways involved in the relative abundance or shortage of such metabolites in CRC. The most relevant strength of this study consists of our original data on methionine not only as a potential prognostic biomarker but even as a target for therapeutic interventions.
Conclusions
In conclusion, our results confirm the role of metabolomics for the identification of biomarkers for early diagnosis, assessing the risk and prognosis of CRC as well as for targeting novel drugs. D-Mannose and methionine should be investigated in further larger sample size and long-time longitudinal studies in order to validate their role as candidate biomarkers for CRC prognosis in conjunction with the investigation of environmental and bacterial-derived substances. Combining all this information, it will be possible to define the metabolome configuration and its changes over time in patients with CRC.
Acknowledgments
None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the local institutional review board of the University of Cagliari (CA-302-26/05/2014) and was conducted in accordance with the Helsinki Declaration of 1964, revised in 2008. All patients signed informed consent to the study.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Thomas DS, Fourkala EO, Apostolidou S, et al. , Evaluation of serum CEA, CYFRA21-1, and CA125 for the early detection of colorectal cancer using longitudinal preclinical samples. Br J Cancer 2015;113:268-74. 10.1038/bjc.2015.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vymetalkova V, Cervena K, Bartu L, et al. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int J Mol Sci 2018. doi: . 10.3390/ijms19113356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reggiani Bonetti L, Barresi V, Maiorana A, et al. Clinical Impact and Prognostic Role of KRAS/BRAF/PIK3CA Mutations in Stage I Colorectal Cancer. Dis Markers 2018;2018:2959801. 10.1155/2018/2959801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Xu X, Pan B, et al. Circulating miR-1290 and miR-320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J Cancer 2019;10:43-50. 10.7150/jca.26723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Dong S, Feng J. Epigenetic profiling and mRNA expression reveal candidate genes as biomarkers for colorectal cancer. J Cell Biochem 2019;120:10767-76. 10.1002/jcb.28368 [DOI] [PubMed] [Google Scholar]
- 8.Vacante M, Borzì AM, Basile F, et al. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J Clin Cases 2018;6:869-81. 10.12998/wjcc.v6.i15.869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Locasale JW. Metabolomics: A Primer. Trends Biochem Sci 2017;42:274-84. 10.1016/j.tibs.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beger RD. A review of applications of metabolomics in cancer. Metabolites 2013;3:552-74. 10.3390/metabo3030552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan EC, Koh PK, Mal M, et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res 2009;8:352-61. 10.1021/pr8006232 [DOI] [PubMed] [Google Scholar]
- 12.Farshidfar F, Weljie AM, Kopciuk KA, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer 2016;115:848-57. 10.1038/bjc.2016.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Lin Y, Liang J, et al. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget 2017;8:105819-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djukovic D, Zhang J, Raftery D. Colorectal Cancer Detection Using Targeted LC-MS Metabolic Profiling. Methods Mol Biol 2018;1765:229-40. 10.1007/978-1-4939-7765-9_15 [DOI] [PubMed] [Google Scholar]
- 15.Yusof HM, Ab-Rahim S, Suddin LS, et al. Metabolomics Profiling on Different Stages of Colorectal Cancer: A Systematic Review. Malays J Med Sci 2018;25:16-34. 10.21315/mjms2018.25.5.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011;6:1060-83. 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- 17.Oberacher H, Whitley G, Berger B. Evaluation of the sensitivity of the 'Wiley registry of tandem mass spectral data, MSforID' with MS/MS data of the 'NIST/NIH/EPA mass spectral library'. J Mass Spectrom 2013;48:487-96. 10.1002/jms.3184 [DOI] [PubMed] [Google Scholar]
- 18.Kopka J, Schauer N, Krueger S, Birkemeyer C, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 2005;21:1635-8. 10.1093/bioinformatics/bti236 [DOI] [PubMed] [Google Scholar]
- 19.Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486-94. 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 2010;38:W71-7. 10.1093/nar/gkq329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23:27-47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johns TG, Mellman I, Cartwright GA, et al. The antitumor monoclonal antibody 806 recognizes a high-mannose form of the EGF receptor that reaches the cell surface when cells over-express the receptor. FASEB J 2005;19:780-2. 10.1096/fj.04-1766fje [DOI] [PubMed] [Google Scholar]
- 23.Gu J, Liang D, Pierzynski JA, et al. D-mannose: a novel prognostic biomarker for patients with esophageal adenocarcinoma. Carcinogenesis 2017;38:162-7. [DOI] [PubMed] [Google Scholar]
- 24.Jobard E, Pontoizeau C, Blaise BJ, et al. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett 2014;343:33-41. 10.1016/j.canlet.2013.09.011 [DOI] [PubMed] [Google Scholar]
- 25.De Leoz ML, Young LJ, An HJ, et al. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics 2011;10:M110.002717. [DOI] [PMC free article] [PubMed]
- 26.Alley WR, Jr, Vasseur JA, Goetz JA, et al. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res 2012;11:2282-300. 10.1021/pr201070k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Q, Zhan T, Deng Z, et al. Glycan analysis of colorectal cancer samples reveals stage-dependent changes in CEA glycosylation patterns. Clin Proteomics 2018;15:9. 10.1186/s12014-018-9182-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erben V, Bhardwaj M, Schrotz-King P, et al. Metabolomics Biomarkers for Detection of Colorectal Neoplasms: A Systematic Review. Cancers (Basel) 2018. doi: . 10.3390/cancers10080246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loke MF, Chua EG, Gan HM, et al. Metabolomics and 16S rRNA sequencing of human colorectal cancers and adjacent mucosa. PLoS One 2018;13:e0208584. 10.1371/journal.pone.0208584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaimenko I, Jaeger C, Brenner H, et al. Non-invasive metastasis prognosis from plasma metabolites in stage II colorectal cancer patients: The DACHS study. Int J Cancer 2019;145:221-31. 10.1002/ijc.32076 [DOI] [PubMed] [Google Scholar]
- 31.Geijsen AJMR, Brezina S, Keski-Rahkonen P, et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int J Cancer 2019;145:1221-31. 10.1002/ijc.32146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long Y, Sanchez-Espiridion B, Lin M, et al. Global and Targeted Serum Metabolic Profiling of Colorectal Cancer Progression. Cancer 2017;123:4066-74. 10.1002/cncr.30829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ning W, Li H, Meng F, et al. Identification of differential metabolic characteristics between tumor and normal tissue from colorectal cancer patients by gas chromatography-mass spectrometry. Biomed Chromatogr 2017. doi: . 10.1002/bmc.3999 [DOI] [PubMed] [Google Scholar]
- 34.Brown DG, Rao S, Weir TL, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 2016;4:11. 10.1186/s40170-016-0151-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg O. On respiratory impairment in cancer cells. Science 1956;124:269-70. [PubMed] [Google Scholar]
- 36.Gonzalez PS, O'Prey J, Cardaci S, et al. Mannose impairs tumour growth and enhances chemotherapy. Nature 2018;563:719-23. 10.1038/s41586-018-0729-3 [DOI] [PubMed] [Google Scholar]
- 37.Denkert C, Budczies J, Weichert W, et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol Cancer 2008;7:72. 10.1186/1476-4598-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 2009;69:4918-25. 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- 39.Ong ES, Zou L, Li S, et al. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics 2010. [Epub ahead of print]. 10.1074/mcp.M900551-MCP200 [DOI] [PubMed] [Google Scholar]
- 40.Satoh K, Yachida S, Sugimoto M, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A 2017;114:E7697-706. 10.1073/pnas.1710366114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leichtle AB, Nuoffer JM, Ceglarek U, et al. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 2012;8:643-53. 10.1007/s11306-011-0357-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan B, Qiu Y, Zou X, et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J Proteome Res 2013;12:3000-9. 10.1021/pr400337b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nitter M, Norgård B, de Vogel S, et al. Plasma methionine, choline, betaine, and dimethylglycine in relation to colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Ann Oncol 2014;25:1609-15. 10.1093/annonc/mdu185 [DOI] [PubMed] [Google Scholar]
- 44.Li TW, Yang H, Peng H, et al. Effects of S-adenosylmethionine and methylthioadenosine on inflammation-induced colon cancer in mice. Carcinogenesis 2012;33:427-35. 10.1093/carcin/bgr295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev 2012;38:726-36. 10.1016/j.ctrv.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 46.Murín R, Vidomanová E, Kowtharapu BS, et al. Role of S-adenosylmethionine cycle in carcinogenesis. Gen Physiol Biophys 2017;36:513-20. 10.4149/gpb_2017031 [DOI] [PubMed] [Google Scholar]
- 47.Qiu Y, Cai G, Su M, et al. J Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res 2009;8:4844-50. 10.1021/pr9004162 [DOI] [PubMed] [Google Scholar]
- 48.Weijenberg MP, Hughes LA, Bours MJ, et al. The mTOR Pathway and the Role of Energy Balance Throughout Life in Colorectal Cancer Etiology and Prognosis: Unravelling Mechanisms Through a Multidimensional Molecular Epidemiologic Approach. Curr Nutr Rep 2013;2:19-26. 10.1007/s13668-012-0038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]