Abstract
Bacterial populations use a cell-to-cell communication system to coordinate community-wide regulation processes, which is termed quorum sensing (QS). Autoinducer-2 (AI-2) is a universal signal molecule that mediates inter- and intraspecies QS systems among different bacteria. In this study, the effects of exogenous addition of AI-2 synthesized in vitro on physiological behaviors and proteome were investigated in lactic acid bacteria strains. Exogenous AI-2 had a concentration-dependent effect on the Enterococcus faecium 8-3 cell density. There was no significant influence on biofilm formation and individual morphology of cells upon 60 μM AI-2 addition in E. faecium 8-3 and Lactobacillus fermentum 2-1. However, it improved the acid and alkali resistance of E. faecium 8-3. With the addition of AI-2, 15 differentially expressed proteins were identified in E. faecium 8-3, which participate in RNA transport signaling, RNA polymerase, ribosome, oxidative phosphorylation, cysteine and methionine metabolism, pyrimidine metabolism, ATP-binding cassette (ABC) transporters, purine metabolism, biosynthesis of the amino acid pathway, etc. Among them, the expression of 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase, which is known to be involved in AI-2 synthesis and cysteine and amino acid metabolism, was upregulated. These findings will lay the foundation to clarify the mechanism of cell-to-cell communication and bacterial physiological behaviors mediated by AI-2.
Introduction
Lactic acid bacteria (LAB) are commonly used in the food industry, including fermented dairy, meat, vegetable, and cereal foods. Diverse species of LAB can be isolated from plant materials, fermented foods, and the human gastrointestinal (GI) tract.1 Some of these bacteria have limited physiological abilities and are constrained to a specific environment. Changes in environmental factors can play a role in the survival and metabolism of LAB. Some of the key factors for adapting the changes were attributed to complex quorum sensing (QS) regulatory networks.2
Quorum sensing (QS) is a system used to communicate between cells that regulates various behaviors by a cell density-dependent manner.3 The QS system is switched on when the concentration of QS signal molecules reaches the threshold and is perceived by cell membrane proteins. Autoinducer-2 (AI-2), as one of the signal molecules, is synthesized by the S-ribosylhomocysteine lyase (LuxS) in both Gram-positive and -negative bacteria and used for inter- and intraspecies communication.4 The biosynthetic pathway for AI-2 in different bacterial species is highly conserved; AI-2 can be automatically converted from the precursor substance 4,5-dihydroxy-2,3-pentandione (DPD).5 The recognition of signals is likely as important as signal synthesis, or even more. Homologous protein complexes can perceive and transduce signals and their regulation plays a key role in defining behaviors.6 Previous research studies have shown that AI-2 has the ability to regulate a variety of physiological behaviors, including biofilm formation,7 virulence,8 and so on. In Pseudomonas aeruginosa PAO1, exogenous AI-2 influenced the biofilm amount and virulence factor expression in a dose-dependent manner in vitro and increased the histological lung damage in mice.9 In Streptococcus suis, AI-2 supplemented exogenously had an impact on the expression of virulence genes and host–cell adherence.10 Exogenous addition of synthesized AI-2 in vitro showed a negative effect on the biofilm amount in Bacillus cereus and a positive effect on the cells detached from a mature biofilm.11 Microarray and reverse transcriptase polymerase chain reaction (PCR) results indicated an upregulation of the oxidative stress response under the addition of AI-2 to planktonic Mycobacterium avium.12 The above research showed that exogenous AI-2 had an effect on different bacterial physiological behaviors, although less investigation was focused on LAB.
Although the interspecies signal molecule AI-2 is commonly in relation to pathogenicity and virulence, it has recently been shown that the LAB strains possess a functional luxS gene and have the ability to produce AI-2. LAB strains can use the AI-2-mediated QS system to regulate their physiological functions. In Bifidobacterium breve, AI-2 activities correlated with gut colonization and pathogen protection.13 Furthermore, the overexpression of luxS gene in Bifidobacterium longum improved biofilm formation, which is used for early colonization in the host by probiotics.14 The Lactobacillus sanfranciscensis metabolic processes were affected by other sourdough Lactobacilli through the luxS-mediated QS system.15 Two-dimensional electrophoresis (2-DE) results showed that the LuxS protein expression level in Lactobacillus plantarum DC400 improved upon coculturing with other strains.16 These results indicate that AI-2 activity is produced by the interaction of the relevant microbial food cultures during the fermentation process. However, at present, our understanding of the effect of exogenous synthetic AI-2 on LAB is clearly less. Thus, the mechanism of AI-2 regulating the physiological behaviors and protein expression of LAB still needs further study.
In this research, we aimed to explore the effects of exogenous AI-2 on LAB physiological functions, including cell morphology, biofilm formation, and the tolerance to acid and alkaline conditions. The strains of Enterococcus faecium 8-3 and Lactobacillus fermentum 2-1 with high production of AI-2 were used for research.17 Furthermore, we employed a gel-free proteome approach to detect the changes of proteome in both E. faecium 8-3 cells with exogenous synthetic AI-2 and without AI-2. The label-free quantification (LFQ) method combined with liquid chromatography–quadrupole mass spectrometry (LC–MS/MS) was used to achieve accurate quantification and improve efficiency and accuracy. This is the first proteome research to utilize the medium to gain new insights into AI-2-regulated LAB metabolism.
Results
Effects of AI-2 on Cell Growth and Morphological Characterization
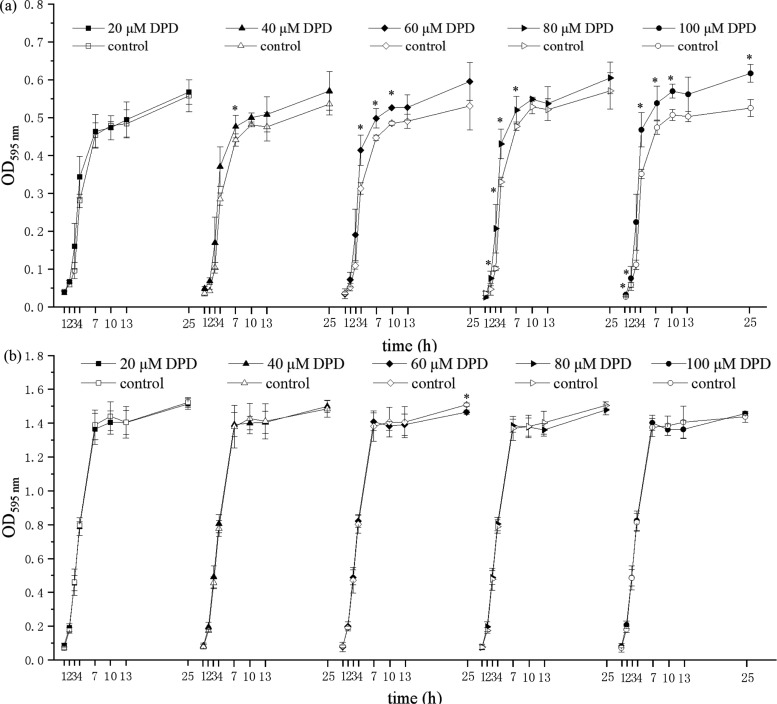
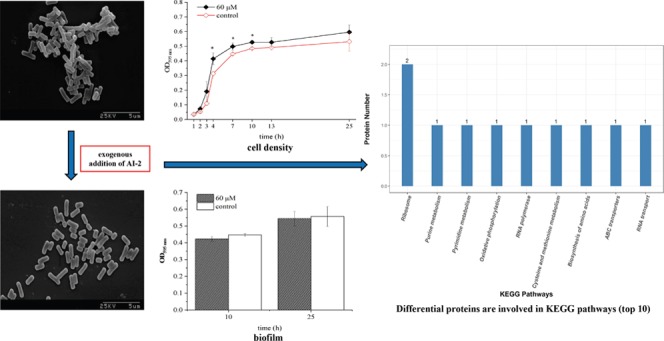
The impact of in-vitro-synthesized AI-2 on E. faecium 8-3 and L. fermentum 2-1 planktonic bacterial growth was investigated, as demonstrated in Figure 1. The cell density of E. faecium 8-3 increased in the presence of 20, 40, 60, 80, and 100 μM AI-2 compared to that of the control. It should be noted that with the increase of dosage the promotion effect was more significant. Meanwhile, higher concentrations (80 and 100 μM AI-2) resulted in the promotion to occur at 1–10 h and even for 25 h. Compared with that of E. faecium 8-3, synthesized AI-2 played a different role in the regulation of L. fermentum 2-1 growth. The cell density showed an increase during 1–7 h and a decrease during 10–25 h upon the addition of 40–100 μM AI-2. However, these effects were not significant. The above results showed that E. faecium 8-3 planktonic bacterial growth responded to the exogenous AI-2 in a dose-dependent manner. Nevertheless, there was no obvious effect on L. fermentum 2-1 growth at the experimental concentration.
Figure 1.
Effects of different concentrations of AI-2 on the cell density of (a) E. faecium 8-3 and (b) L. fermentum 2-1. *p < 0.05.
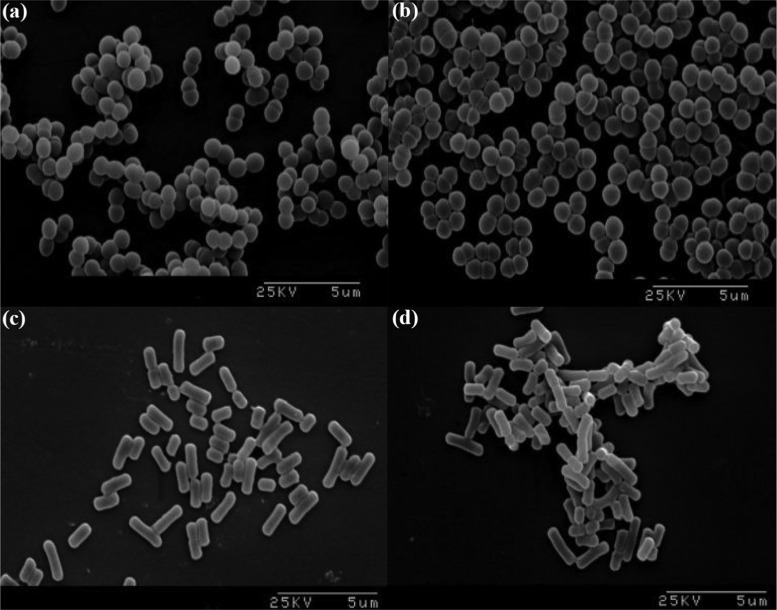
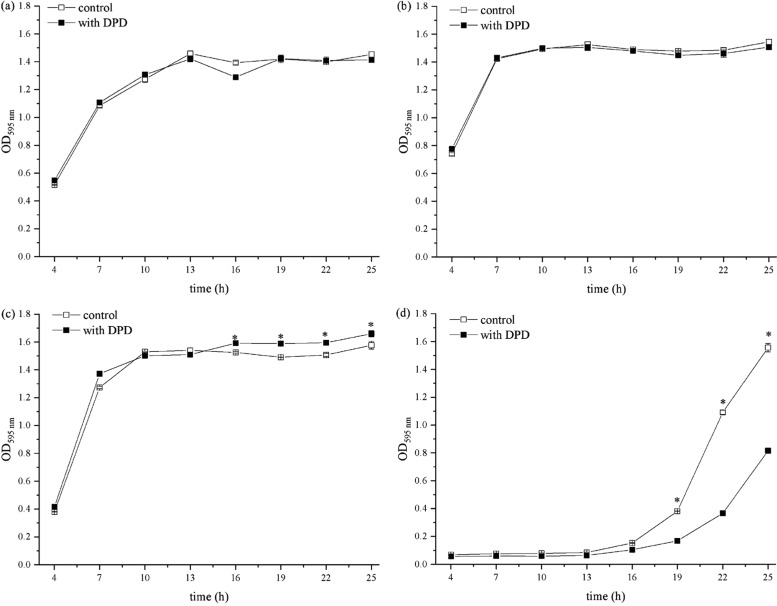
As shown in Figure 1, 60 μM AI-2 had a significant effect on bacterial growth at 10 h and was chosen for further scanning electron microscopy (SEM) and biofilm formation experiments. Comparisons of the cells in the absence and presence of 60 μM AI-2 were observed by SEM after 10 h incubation. SEM images (6000×) of E. faecium 8-3 and L. fermentum 2-1 cells with different treatments are shown in Figure 2. Two different shapes of bacteria in the image correspond to the two strains of LAB, spherical in pairs of E. faecium 8-3 and rod shape of L. fermentum 2-1. The morphology of bacterial cells was not changed with in-vitro-synthesized AI-2 compared to that of the control (Figure 2b,d). The test cells were in a stage of division and proliferation, especially E. faecium 8-3, and their outline was clear and the surface was smooth. The addition of AI-2 did not have an effect on E. faecium 8-3 and L. fermentum 2-1 individual cells. The above results indicated that 60 μM AI-2 had no effect on the individual morphology of the test LAB strains.
Figure 2.
Scanning electron microscopy images of (a) E. faecium 8-3 treated with DPD, (b) untreated E. faecium 8-3, (c) L. fermentum 2-1 treated with DPD, and (d) untreated L. fermentum 2-1. Magnification 6000×.
Effects of AI-2 on Biofilm Formation
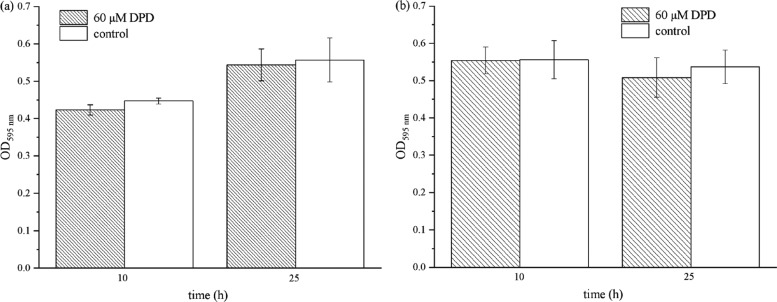
To further explore the influence of AI-2 on E. faecium 8-3 and L. fermentum 2-1, we investigated its effect on biofilm formation. As shown in Figure 3, 60 μM AI-2 did not affect biofilm formation significantly at 10 and 24 h. The result indicated that the presence of 60 μM AI-2 has no significant impact on biofilm development in E. faecium 8-3 and L. fermentum 2-1.
Figure 3.
Effects of AI-2 on biofilm formation of (a) E. faecium 8-3 and (b) L. fermentum 2-1. The y-axis represents the absorbance of dissolved bacteria-bound crystal violet (CV). Both (a) and (b) show that AI-2 has no effect on biofilm formation.
Effect of AI-2 on Acid and Alkaline Resistance
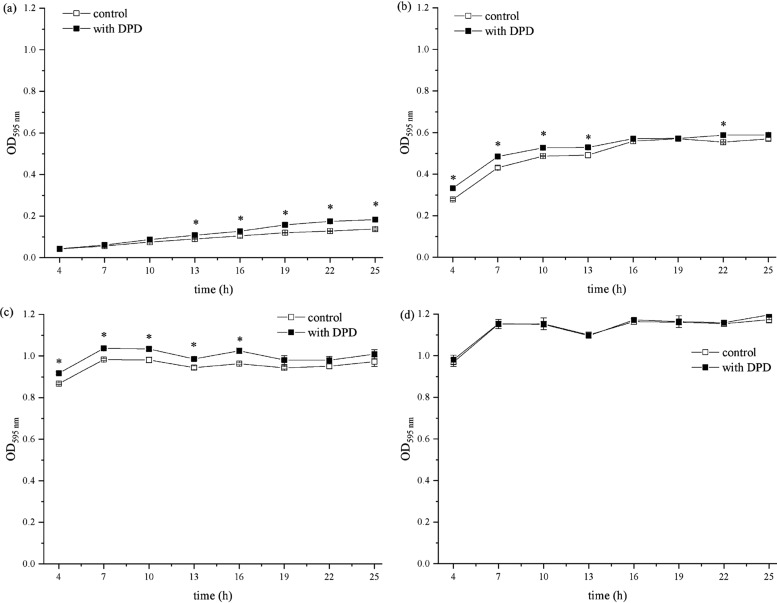
To determine whether AI-2 affects the LAB tolerance of acids and alkalies, E. faecium 8-3 and L. fermentum 2-1 were grown in the Man–Rogosa–Sharpe (MRS) medium supplemented with 60 μM AI-2 (AI-2+ cells). The cell density of cultures was measured every 3 h until 25 h, and acid and alkaline stress resistance were compared to that of the control (AI-2– cells). As shown in Figure 4, the cell density of E. faecium 8-3 was significantly affected by the addition of exogenous AI-2 (p < 0.05). Under acidic conditions (pH 4.5), the growth of E. faecium 8-3 was inhibited compared to that at pH 6.5 (Figure 1a), which was positively correlated with the decrease of acidity. However, the growth of E. faecium 8-3 cells in acidic conditions was significantly improved upon the addition of AI-2. At pH 4.5, the significant promotion effect was mainly concentrated in the period of 13–25 h with AI-2 (Figure 4a). At pH 5.5, significant promotion appeared at 4 h and lasted approximately 22 h. These results indicated that the AI-2 promotion effect would appear with the reduction of acid stress. Compared with the acid condition, the alkaline environment was more favorable to the growth of E. faecium 8-3. Under acidic conditions, the maximum bacterial density of E. faecium 8-3 at a stable period is only 0.183 (Figure 4a). However, under alkaline conditions, it could reach as high as 1.196 (Figure 4d). At pH 7.5, there was a significant effect on E. faecium 8-3 cell density, which improved the growing ability under alkaline conditions. The absorbances of AI-2+ cells harvested from all growth phases were a little higher than those of AI-2– cells, suggesting that AI-2 played a positive role at pH 7.5 (Figure 4c). At pH 8.5, the growth of E. faecium 8-3 might have reached its maximum capacity, so the addition of 60 μM AI-2 had no significant effect on it (Figure 4d).
Figure 4.
Effects of AI-2 on acid and alkaline resistance of E. faecium 8-3. (a) pH 4.5, (b) pH 5.5, (c) pH 7.5, and (d) pH 8.5. *p < 0.05.
As shown in Figure 5, the strain of 2-1 is L. fermentum, which has stronger growth capacity under acidic stress than that of E. faecium 8-3. The decrease of the medium acidity enhanced the inhibition of L. fermentum 2-1 growth, including the maximum cell density and growth rate. Meanwhile, the presence of AI-2 did not alleviate the inhibition of L. fermentum 2-1. AI-2+ and AI-2– cells showed similar OD595 nm, which indicated that the influence of exogenous AI-2 on the growth and tolerance of L. fermentum 2-1 in acidic conditions was not obvious (Figure 5a,b). The growth of L. fermentum 2-1 in the MRS medium in the presence and absence of 60 μM AI-2 at pH 7.5 and pH 8.5 for 25 h was assessed. The growth of L. fermentum 2-1 was basically not affected under the condition of mild alkalinity, but the rate slowed down due to the exponential phase lag with the increase of pH value. There was higher cell density in cultures in the presence of AI-2 following mild alkali treatment (pH 7.5) compared to that of the control (AI-2– cells), especially between 16 and 25 h. Interestingly, at pH 8.5, the effect of AI-2 on the growth of L. fermentum 2-1 showed an opposite effect (Figure 5d) compared to that on cultures before treatment (Figure 5c). The addition of AI-2 inhibited the growth of L. fermentum 2-1, especially between 19 and 25 h. The above results indicated that exogenous AI-2 had various effects in different strains and improved the acid and alkali resistance of a specific strain.
Figure 5.
Effects of AI-2 on acid and alkaline resistance of L. fermentum 2-1. (a) pH 4.5, (b) pH 5.5, (c) pH 7.5, and (d) pH 8.5. *p < 0.05.
Proteome Profile of E. faecium 8-3 after Coculturing with AI-2
After culturing of E. faecium 8-3 with the addition of AI-2 for 10 h, the cells were collected and used for proteome analysis. The intracellular proteins in tests and controls were extracted to identify proteins that possibly participate in physiological metabolic processes. A total of 851 and 960 proteins were detected in the test and control, respectively. Fifteen proteins displayed significantly different abundance (>1.5-fold difference, p < 0.05) in the expressed protein numbers of E. faecium 8-3 after the coculture was affected by AI-2 (Table 1). In addition, 14 and 24 proteins were uniquely identified in control and supplement AI-2 conditions, respectively.
Table 1. Exogenous AI-2 Affected the Protein Expression Difference of E. faecium 8-3.
| protein ID | description | abbreviations | protein expression | P value | gene expression |
|---|---|---|---|---|---|
| A0A076GPG6 | 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase | mtnN | 2.745 | 0.020 | 1.128 |
| A0A076GYB9 | TetR family transcriptional regulator | M395_12005 | 2.547 | 0.022 | 1.323 |
| A0A0M2AHZ7 | GNAT family acetyltransferase | SIE_01971 | 2.076 | 0.032 | 1.266 |
| A0A0M1XDF5 | CCA-adding enzyme | 2.176 | 0.046 | 1.179 | |
| T2NMI9 | 50S ribosomal protein L31 type B | rpmE2 | 1.930 | 0.047 | 1.328 |
| A0A0M1Y077 | V-type ATP synthase subunit E | OI7_03719 | 1.526 | 0.032 | 1.325 |
| A0A076GS02 | DNA-directed RNA polymerase subunit omega | rpoZ | 1.704 | 0.041 | 1.077 |
| C9B7M6 | foldase protein | prsA | 1.746 | 0.049 | 1.524 |
| A0A0M1XBR7 | energy-coupling factor transporter ATP-binding protein EcfA | ecfA | 0.597 | 0.025 | 0.884 |
| A0A076GMR1 | 50S ribosomal protein L20 | rplT | 0.658 | 0.013 | 0.908 |
| A0A0M1XE95 | Maf-like protein | HMPREF1359_00059 | 0.578 | 0.045 | 0.931 |
| A0A0M1X622 | FAD dependent oxidoreductase | HMPREF1359_03046 | 0.420 | 0.048 | 0.897 |
| A0A076GQV1 | diacylglycerol kinase | M395_06215 | 0.625 | 0.050 | 0.718 |
| A0A0M1X6H4 | RNA methyltransferase | HMPREF1359_02800 | 0.480 | 0.044 | 0.695 |
| A0A0M1X711 | uncharacterized protein | 1.914 | 0.016 | 1.219 |
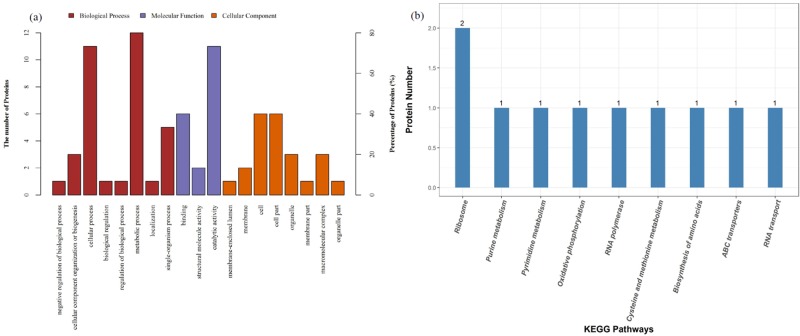
The proteins were classified into different categories according to Gene Ontology (GO) (Figure 6a). Statistical analysis results showed that the expression of six proteins decreased from 0.420- to 0.658-fold and the expression of nine proteins increased from 1.526- to 2.745-fold (Table 1). Differently expressed proteins were arranged into the following categories: S-adenosylmethionine cycle, S-adenosylhomocysteine (SAH) metabolic process, amino acid salvage, ribonucleoprotein complex subunit organization, ribosomal large subunit biogenesis, ribonucleoprotein complex assembly, l-methionine salvage, l-methionine salvage from S-adenosylmethionine, ribosome assembly, ribosomal large subunit assembly, RNA repair, l-methionine salvage from methylthioadenosine (MTA), l-cysteine metabolic process, and tRNA 3′-terminal CCA addition.
Figure 6.
Global proteome profile of E. faecium 8-3 cells upon DPD. (a) Functional classification of E. faecium 8-3 proteins by Gene Ontology (GO) classification (molecular functions (MFs), biological processes (BPs), and cellular location). (b) Differential proteins are involved in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (top 10).
Based on the above results, many functions were influenced by AI-2 addition. In this study, the enrichment analysis of the upregulated and downregulated differentially expressed proteins was performed respectively using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Figure 6b) and it was found that the upregulated differentially expressed proteins were mainly concentrated in the RNA transport signaling pathway. At the same time, possible signaling pathways included RNA polymerase, ribosome, oxidative phosphorylation, cysteine and methionine metabolism, pyrimidine metabolism, ATP-binding cassette (ABC) transporters, purine metabolism, biosynthesis of amino acids, etc. It suggested that exogenous AI-2 plays a role in multiple pathways, which are essential for bacterial establishment and cell metabolism.
Among them, the AI-2 anabolic related protein MtnN was involved in the cysteine and methionine metabolic pathways. It suggested that upregulated MtnN was involved in multiple amino acid metabolic pathways. We further selected 15 proteins for mRNA level validation based on bioinformatics analysis and participation in AI-2-induced metabolic pathways. The RT-PCR results of gene expression were consistent with the results of proteins (Table 1).
Discussion
AI-2 is widely found in bacteria and commonly considered as a universal QS signal molecule for cell communication. In a solution environment, AI-2 with various forms is converted and balanced;18 therefore, it can be recognized by bacteria of different species. In this study, we attempted to explore whether exogenous AI-2 would interfere with the growth of bacteria and our experiments indicated that exogenous AI-2 had an apparent effect. The influence of exogenous AI-2 on the morphological characterization of LAB was similar: there was no effect on the individual morphology of the test LAB when the concentration of DPD was 60 μM, but it affected the mutual cross-linking and adhesion between the cells. It was also found that exogenous DPD had concentration-dependent effects on different strains. A low concentration of DPD was found to gently promote cell growth. Higher concentrations of DPD improved cell density, but they were not always positively correlated. Interestingly, the growth rates of E. faecium 8-3 and L. fermentum 2-1 both at the logarithmic phase increased. Therefore, during the growth of E. faecium 8-3 and L. fermentum 2-1, exogenous AI-2 can be used as a substance to promote the growth of bacteria.
The biofilm of bacteria is a typically complex multispecies community that develops through the adhesion of different species cells onto a surface and is promoted by cell-to-cell interactions in the bacterial community. It has been verified that AI-2 played a role in biofilm formation in many species, but the AI-2 function has been not fully revealed. Our results showed that 60 μM concentration of DPD did not influence biofilm formation significantly on E. faecium 8-3 and L. fermentum 2-1. According to the different AI-2 concentrations and bacterial species, the influence of AI-2 on biofilm formation was also different. It has been reported that AI-2 affects biofilm formation in a dose-dependent way in B. cereus.11 The B. cereus biofilm amount decreased upon the addition of exogenous AI-2 in the concentration range from 0 to 6.8 μM. The same effect of AI-2 on biofilm was also confirmed in Eikenella corrodens and Vibrio cholerae.19,20 However, different effects were found in Staphylococcus intermedius and Streptococcus pneumonia.21,22 Wang et al. reported that when AI-2 levels reached a threshold, a lot of biological properties altered in bacterial cells, especially those associated with biofilm formation.10 The concentration of AI-2 higher than 2 μM showed an opposite function on S. suis biofilm formation, which differs from other bacterial species.10 Bacteria live in the environment predominantly in the form of biofilms, which has been shown to be a response to environmental stresses, as well as in mammalian hosts. Thus, the functions of AI-2 in bacterial community formation and dynamic change were conceivable. AI-2 can influence the ability to withstand adverse circumstances of bacteria by influencing the formation of biofilms.
Our previous study showed that environmental factors did cause changes in the activity of AI-223 and a number of bacteria use quorum sensing signal molecules to resist stressful factors, including starvation, NaCl, hydrogen peroxide, and antibiotics.24−26 In this study, the cell density of E. faecium 8-3 under acidic and alkaline stresses increased when AI-2 was added to the culture medium. For L. fermentum 2-1, the addition of AI-2 had a relatively small effect on the cell density. There was only an inhibition effect on cell density under severe alkaline stress. One possible explanation for this difference is that AI-2 synthesized in vitro has diverse activities at different pH values. Another possible explanation is that different bacterial species have different AI-2 sensing threshold levels. The uptake of AI-2 was modified by lsrACDBFGE operon, which contained a transporter complex, LsrABCD; a cognate signal kinase, LsrK; and an AI-2 repressor, LsrR.27 The operons in different strains may have dissimilar responses to exogenous AI-2. Current knowledge relative to the cell’s ability to perceive or recognize the AI-2 in the LAB was scarce.
As Moslehi-Jenabian et al. reported, AI-2 activity seemed positively correlated with acid shock intensity.28 Delisa et al. have proved that AI-2 activity could be increased for several hours because of the osmolarity pulse in Escherichia coli.29 However, the AI-2-mediated QS system did not associate with acid and heat tolerance in Salmonella.30 Hence, the role of AI-2 in bacterial cells on the response and resistance to environmental stresses seemed to be strain-specific.
Deciphering the molecular basis of AI-2 and bacterial cell communication is the key to understand the metabolic mechanism of LAB and how bacteria interact with each other. Here, we used E. faecium 8-3 as a model to investigate the effect of AI-2 on LAB protein expression because the 8-3 strain was more sensitive to exogenous AI-2, according to the above results. To determine the effect of AI-2 on E. faecium 8-3 protein expression, we incubated E. faecium 8-3 and exogenous AI-2 for 10 h prior to cell harvest. In this study, proteome signatures associated with signal molecule AI-2 were identified. Differential regulatory proteins provide new insights into bacterial cell communication. Clearly, AI-2 altered the protein profiles in E. faecium 8-3, indicating that both molecular and biological functions were affected by the action of quorum sensing signal molecules.
With the addition of AI-2, the topmost changed protein in E. faecium 8-3 was S-adenosylhomocysteine/methylthioadenosine nucleosidase (SAHN). It indicated that exogenous AI-2 might induce the expression of this protein in E. faecium 8-3. S-adenosylhomocysteine/methylthioadenosine nucleosidase is essential for the methyl cycle in many bacteria and protozoan species but is not found in mammalian cells. SAHN is indispensable in the bacterial activated methyl cycle (AMC) process. It is involved in the cleavage of methylthioadenosine (MTA) and S-adenosylhomocysteine (SAH) glycosidic bonds, producing adenine and methylthioribose (MTR) and S-ribosylhomocysteine (SRH), respectively. They are further transformed into homocysteine, DPD, and methylthioribose-1-phosphate (MTRP) by the action of MTA kinase and autoinducer synthetase LuxS. The function of SAHN associated with the production of two QS signaling molecules, AI-1 and AI-2.31 In E. coli, the loss of sahn gene caused the accumulation of toxic SAH, resulting in reduced growth.31E. colipfs mutants were known to have strongly impaired growth and their colonies were much smaller than those of the parent strains.32 Silva et al. reported that there was no significant effect on biofilm formation in pfs mutants of V. cholerae but had an inhibition effect on the growth and motility.33 These results are consistent with the results of this study. In our study, the expression of SAHN was induced by exogenous AI-2 and resulted in an enhanced effect on the growth and no effect on biofilm formation. Meanwhile, overexpressed SAHN promoted the QS system of bacteria.31 The protein of SAHN was associated with cysteine and methionine metabolism pathways. In the process of catabolism, methionine can produce a carbon group through the action of methylation and a carbon group can be used as the methyl source of pyrimidine, purine, and various methylated compounds.34,35 The addition of exogenous AI-2 also has an impact on the methyl cycle, thereby promoting the growth of E. faecium 8-3.
ATP-binding cassette (ABC) transporter is a ubiquitous superfamily that is responsible for the transmembrane transport of a variety of substances.36 Upon the addition of exogenous AI-2, the expression of EcfA protein in E. faecium 8-3 was downregulated, which participated in the ABC transport pathway. ECF transporters were used for importing micronutrients in archaea and bacteria. The zymolyte contains a series of cofactors, transition-metal ions, water-soluble vitamins, the amino acid tryptophan, and queuosine and its metabolic precursors.37 ECF import systems were used for vitamin uptake by some bacteria, with restricted cofactor biosynthetic capacities, including Mycoplasma genitalium, Staphylococcus aureus, and S. pneumoniae.38 The addition of exogenous AI-2 could affect the transport process of different substrates through downregulation of EcfA expression in the EFC transporter family.
In this study, exogenous AI-2 played a role in the growth of bacterial cells and physiological behaviors of E. faecium 8-3 and L. fermentum 2-1. At the same time, the global proteins that might be involved in interaction between LAB and exogenous signal molecule AI-2 were first identified and quantified in our research. Exogenous AI-2 promoted the growth of E. faecium 8-3 but had no effect on individual morphology and biofilm formation in E. faecium 8-3 and L. fermentum 2-1. The cell density increased under acid and alkaline stresses. The differentially expressed proteins were mainly involved in the cysteine and methionine amino acid synthesis pathways and RNA transport pathways. However, whether different factors such as concentrations of AI-2, incubation time, and strain species also have an effect on the expression of different proteins remains to be further studied.
Materials and Methods
Bacterial Strains and Growth Conditions
E. faecium 8-3 and L. fermentum 2-1 isolated from Chinese koumiss were identified previously by 16S rRNA gene sequence analysis.17 Strains were cultured at 37 °C for 24 h in Man–Rogosa–Sharpe (MRS) broth.39 pEASY-Blunt E1 was used as the prokaryotic protein expression vector. E. coli strains DH5a and E. coli Transetta (DE3) (TransGen Biotech) were grown at 37 °C in LB broth with aeration and used for the cloning and expression of recombinant genes, respectively. Vibrio harveyi BB170 (ATCC BAA-1117) was used for the measurement of AI-2 activity and cultured in autoinducer bioassay (AB) medium or marine broth 2216 (Difco) at 30 °C with aeration.40
AI-2 Synthesis in Vitro
In vitro AI-2 synthesis reactions were performed according to the previous method.41 Briefly, expression vectors harboring the luxS and pfs genes were constructed by the genes of E. faecium 8-3 and prokaryotic protein expression vector pEASY-Blunt E1. Cells were grown at 37 °C with aeration for scale-up production of recombinant LuxS and Pfs proteins and induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Recombinant proteins were isolated and purified by the Ni-IDA-Sefinose resin kit according to the manufacturer’s instructions (Sangon Biotech). AI-2 was produced by incubation with 1 mg/mL purified recombinant LuxS and Pfs proteins and 1 mM S-adenosylhomocysteine (SAH, Sigma-Aldrich) for 1 h at 37 °C in 10 mM sodium phosphate buffer at pH 8.0. Ellman’s assay was employed to quantify homocysteine concentration and estimate AI-2 concentration by measuring the absorbance at 412 nm.4 AI-2 activity was detected by Vibrio harveyi BB170.42
Effect of AI-2 on Growth and Morphological Characterization
To determine the effects of AI-2 on the growth of E. faecium 8-3 and L. fermentum 2-1, a growth-curve assay was conducted. E. faecium 8-3 and L. fermentum 2-1 were cultured in MRS broth in the absence or presence of AI-2 (20, 40, 60, 80, and 100 μM) (synthesized in vitro). Strains were incubated at 37 °C for 25 h. After 1, 2, 3, 4, 7, 10, 13, and 25 h, the optical density was determined at 595 nm using a microplate reader (Thermo Scientific Multiskan FC). Morphological changes of E. faecium 8-3 and L. fermentum 2-1 upon treatment with AI-2 for 10 h at 37 °C were examined using scanning electron microscopy (SEM) as previously described.43 Untreated cells of E. faecium 8-3 and L. fermentum 2-1 were used as a control.
Effect of AI-2 on Biofilm Formation
Biofilm formation was measured at a static environment in 96-well polystyrene microtiter plates.44 In brief, overnight cultures of E. faecium 8-3 and L. fermentum 2-1 were diluted to an optical density of 0.1 at 595 nm. AI-2 was added to the medium with a final concentration of 60 μM, and the mixtures (200 μL) were transferred to 96-well microtiter plates (Corning) that were incubated at 37 °C for 10 and 25 h. Untreated cells were used as a control. Phosphate-buffered saline (PBS) was used for washing plates at least 3 times. Then, 0.1% crystal violet (CV) was added to the dried plates at room temperature for 5 min. After washing, the plates were also air-dried. Then, 95% ethanol (200 μL) was added to the dried plates to dissolve bacteria-bound CV. The OD595 nm of dissolved bacteria-bound CV was determined to represent the biofilm amount.
Effect of AI-2 on Acid and Alkaline Resistance
The overnight cultures of E. faecium 8-3 and L. fermentum 2-1 were inoculated to sterile MRS broth with different pH values (4.5, 5.5, 7.5, and 8.5). HCl (2 M) and 5 M NaOH were used to adjust the pH of sterile MRS broth. AI-2 with a concentration of 60 μM was added, and the mixtures were cultured at 37 °C for 25 h. Untreated cells were used as a control. After 4, 7, 10, 13, 16, 19, 22, and 25 h, the optical density was determined at 595 nm using a microplate reader.
Proteome Analysis
E. faecium 8-3 treated with and without AI-2 (60 μM) for 10 h was used for gel-free proteome analysis. The strain was prepared as biological triplicates and lysed in a buffer containing 100 mM Tris–HCl, 4% sodium dodecyl sulfate (SDS), 1 mM dl-dithiothreitol (DTT), pH 7.6. Cell disruption was accomplished with agitation by a homogenizer (Fastprep-24, MP Biomedical). The supernatants were collected by centrifugation at 14 000 rpm for 40 min and filter-sterilized by a 0.22 μm filter. The protein concentration was measured by the BCA protein assay (Bio-Rad). The protein solutions were stored at −80 °C for further analysis.
The digestion of proteins was performed in accordance with the filter-aided sample preparation (FASP) procedure.45 In brief, proteins (200 μg) of each sample were added into 30 μL of SDT buffer (150 mM Tris–HCl, 100 mM DTT, 4% SDS, pH 8.0). DTT and other low-molecular-weight components were removed by UA buffer (150 mM Tris–HCl, 8 M urea, pH 8.0) and repeated ultrafiltration (Microcon units, 30 kD) through centrifugation. Then, the UA buffer was used to block cysteine residues with the addition of 0.05 M iodoacetamide and incubated for 20 min in the dark. The UA buffer and 25 mM NH4HCO3 were used to wash the filter 3 times and twice, respectively. Finally, the protein suspension was incubated overnight along with trypsin (Promega) at 37 °C in 25 mM NH4HCO3. The content of peptide was estimated at 280 nm by UV spectral density.
Liquid chromatography (Thermo Scientific) was used for LC–MS/MS and coupled with a Q-Exactive mass spectrometer (Thermo Scientific) as previously reported.46 Briefly, peptides were transferred into a reverse-phase trap column (Thermo Scientific Acclaim PepMap100, nanoViper C18, 100 μm × 2 cm) combined with a C18 reverse-phase column (10 cm long, 3 μm resin, 75 μm inner diameter, Thermo Scientific Easy Column) in buffer A and a linear gradient buffer B (0.1% formic acid and 84% acetonitrile) was used to separate at 300 nL/min over 120 min. A Q-Exactive mass spectrometer was combined with Easy nLC (Proxeon Biosystems, Thermo Fisher Scientific) for MS experiments. Positive ion mode was used in the mass spectrometer. The 20 data-dependent MS/MS scans (AGC target 5e5) with a normalized collision energy of 30 eV were performed after a full FT scan mass spectrum cycle (the automatic gain control (AGC) target was 1e6, and the HCD fragmentation was 300–1800 m/z). 0.1% was used to specify the minimum percentage of target values that can be reached at the maximum fill time. The HCD spectral resolution was 17 500 at m/z 200, the isolation width was 2 m/z, and survey scans were obtained at a resolution of 70 000 at m/z 200. The dynamic exclusion duration was 60 s. The peptide recognition mode was used to operate the instrument.
Sequence Database Search and Data Analysis
The abundance of proteome and raw mass data analysis were performed by MaxQuant software (version 1.3.0.5) and searched against the UniProt E. faecium database (104 108 total entries, downloaded on August 24, 2017).47 The relative protein abundance and spectral intensities were normalized through the LFQ algorithm.48 Proteins that matched to contaminants or reverse database were removed. Proteins that were found in at least two replicates from one sample were further analyzed.49 A t-test was used to quantitatively analyze protein abundance in different pairs of the sample. The fold changes of protein abundance >1.5 or <0.67 (p value >0.05) were included in the quantitative results. The GO visualization tool50 and the Database for Annotation, Visualization and Integrated Discovery51 were used to analyze the identified proteins by Gene Ontology (GO) enrichment, including biological processes (BPs), cellular components (CCs), and molecular function (MFs). The KEGG was used to annotate the functional genes by GHOST or BLAST comparisons parallel to the manually curated KEGG GENES database for pathway analysis.52.
Quantitative Real-Time PCR (qRT-PCR)
E. faecium 8-3 was cultured in MRS with AI-2 (60 μM) or without (control) at 37 °C for 10 h. Total RNA was extracted with RNAiso Plus (Takara, Japan) according to the manufacturer’s instructions. Gel electrophoresis and absorbance (A260/A280 and A260/A230) were used to detect RNA quality. Isolated RNA was transcribed into cDNA by a PrimeScrip RT reagent kit (Takara, Japan). qRT-PCR was performed by the SYBR Green assay kit (Takara, Japan) and a Bio-Rad CFX96 real-time PCR system (Bio-Rad). The 16S rRNA gene was used as a housekeeping gene. The 2–ΔΔCt method is used to calculate the relative expression of certain genes.53
Statistical Analysis
Analysis of variance (ANOVA) was carried out, and P values <0.05 indicated statistical significance.
Acknowledgments
This project was supported by the High-level Talents Introduction Research Foundation of Inner Mongolia Agricultural University (No. NDYB2018-47) and the Program of National Natural Science Foundation of China (Nos. 31960467 and 31360396).
Author Contributions
† Y.G. and J.W. contributed equally to this work.
The authors declare no competing financial interest.
References
- Ahrné S.; Nobaek S.; Jeppsson B.; Adlerberth I.; Wold A. E.; Molin G. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 1998, 85, 88–94. 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]
- Sturme M. H. J.; Francke C.; Siezen R. J.; de Vos W. M.; Kleerebezem M. Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 2007, 153, 3939–3947. 10.1099/mic.0.2007/012831-0. [DOI] [PubMed] [Google Scholar]
- Miller M. B.; Bassler B. L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Schauder S.; Shokat K.; Surette M. G.; Bassler B. L. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Winzer K.; Hardie K. R.; Burgess N.; Doherty N.; Kirke D.; Holden M. T.; Linforth R.; Cornell K. A.; Taylor A. J.; Hill P. J.; Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 2002, 148, 909–922. 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- Quan D. N.; Bentley W. E. Gene network homology in prokaryotes using a similarity search approach: queries of quorum sensing signal transduction. PLoS Comput. Biol. 2012, 8, e1002637 10.1371/journal.pcbi.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsaglia E. C. R.; Silva N. C. C.; Fernades Júnior A.; Araújo Júnior J. P.; Tsunemi M. H.; Rall V. L. M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control 2014, 35, 386–391. 10.1016/j.foodcont.2013.07.023. [DOI] [Google Scholar]
- Meng L.; Du Y.; Liu P.; Li X.; Liu Y. Involvement of LuxS in Aeromonas salmonicida metabolism, virulence and infection in Atlantic salmon (Salmo salar L). Fish Shellfish Immunol. 2017, 64, 260–269. 10.1016/j.fsi.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Li H.; Li X.; Song C.; Zhang Y.; Wang Z.; Liu Z.; Wei H.; Yu J. L. Autoinducer-2 facilitates Pseudomonas aeruginosa PAO1 pathogenicity in vitro and in vivo. Front. Microbiol. 2017, 8, 1944 10.3389/fmicb.2017.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yi L.; Zhang Z.; Fan H.; Cheng X.; Lu C. Biofilm formation, host-cell adherence, and virulence genes regulation of Streptococcus suis in response to autoinducer-2 signaling. Curr. Microbiol. 2014, 68, 575–580. 10.1007/s00284-013-0509-0. [DOI] [PubMed] [Google Scholar]
- Auger S.; Krin E.; Aymerich S.; Gohar M. Autoinducer 2 affects biofilm formation by Bacillus cereus. Appl. Environ. Microbiol. 2006, 72, 937–941. 10.1128/AEM.72.1.937-941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier H.; Mostowy S.; Cangelosi G. A.; Behr M. A.; Ford T. E. Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 2008, 74, 1798–1804. 10.1128/AEM.02066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen S. E. A.; Motherway M. O.; Bottacini F.; Lanigan N.; Casey P. G.; Huys G.; Nelis H. J.; Sinderen D. V.; Coenye T. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. Plos One 2014, 9, e98111 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.; He X.; Brancaccio V. F.; Yuan J.; Riedel C. U. Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. Plos One 2014, 9, e88260 10.1371/journal.pone.0088260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cagno R.; Angelis M. D.; Limitone A.; Minervini F.; Gobbetti M.; et al. Cell-cell communication in sourdough lactic acid bacteria: a proteomic study in Lactobacillus sanfranciscensis CB1. Proteomics 2007, 7, 2430–2446. 10.1002/pmic.200700143. [DOI] [PubMed] [Google Scholar]
- Di Cagno R.; Angelis M. D.; Coda R.; Minervini F.; Gobbetti M. Molecular adaptation of sourdough Lactobacillus plantarum DC400 under co-cultivation with other lactobacilli. Res. Microbiol. 2009, 160, 358–366. 10.1016/j.resmic.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Li B.; Gu Y.; Yan C. L.; Jia Y. B.; He Y. F. Screening and identification of AI-2 high-producing lactic acid bacteria. Food Sci. Technol. 2016, 37, 185–188. (In Chinese with English Abstract). [Google Scholar]
- Semmelhack M. F.; Campagna S. R.; Federle M. J.; Bassler B. L. An expeditious synthesis of DPD and boron binding studies. Org. Lett. 2005, 7, 569–572. 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- Hammer B. K.; Bassler B. L. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003, 50, 101–104. 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Azakami H.; Teramura I.; Matsunaga T.; Akimichi H.; Noiri Y.; Ebisu S.; Kato A. Characterization of autoinducer 2 signal in Eikenella corrodens and its role in biofilm formation. J. Biosci. Bioeng. 2006, 102, 110–117. 10.1263/jbb.102.110. [DOI] [PubMed] [Google Scholar]
- Ahmed N. A.; Petersen F. C.; Scheie A. A. AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob. Agents Chemother. 2009, 53, 4258–4263. 10.1128/AAC.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J. E.; Ludewick H. P.; Kunkel R. M.; Zahner D.; Klugman K. P. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae Strain D39. Infect. Immun. 2011, 79, 4050–4060. 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.; Li B.; Tian J.; Wu R.; He Y. The response of LuxS/AI-2 quorum sensing in Lactobacillus fermentum 2-1 to changes in environmental growth conditions. Ann. Microbiol. 2018, 68, 287–294. 10.1007/s13213-018-1337-z. [DOI] [Google Scholar]
- McDougald D.; Srinivasan S.; Rice S. A.; Kjelleberg S. Signal-mediated cross-talk regulates stress adaptation in Vibrio species. Microbiology 2003, 149, 1923–1933. 10.1099/mic.0.26321-0. [DOI] [PubMed] [Google Scholar]
- Huang C. T.; Shih P. C. Effects of quorum sensing signal molecules on the hydrogen peroxide resistance against planktonic Pseudomonas aeruginosa. J. Microbiol., Immunol. Infect. 2000, 33, 154–158. [PubMed] [Google Scholar]
- Lu L.; Hume M. E.; Pillai S. D. Autoinducer-2-like activity in poultry-associated enteric bacteria in response to subtherapeutic antibiotic exposure. Avian Dis. 2005, 49, 74–80. 10.1637/7212-05200R. [DOI] [PubMed] [Google Scholar]
- Li J.; Attila C.; Wang L.; Wood T. K.; Valdes J. J.; Bentley W. E. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 2007, 189, 6011–6020. 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi-Jenabian S.; Gori K.; Jespersen L. AI-2 signaling is induced by acidic shock in probiotic strains of Lactobacillus spp. Int. J. Food Microbiol. 2009, 135, 295–302. 10.1016/j.ijfoodmicro.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Delisa M. P.; Valdes J. J.; Bentley W. E. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 2001, 183, 2918–2928. 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Quan C.; Jin L.; Chen M. Production, detection and application perspectives of quorum sensing autoinducer-2 in bacteria. J. Biotechnol. 2018, 268, 53–60. 10.1016/j.jbiotec.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Han T.; Li Y.; Shan Q.; Liang W.; Hao W.; Li Y.; Tan X. J.; Gu J. S. Characterization of s -adenosylhomocysteine/methylthioadenosine nucleosidase on secretion of AI-2 and biofilm formation of Escherichia coli. Microb. Pathog. 2017, 108, 78–84. 10.1016/j.micpath.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Cadieux N.; Bradbeer C.; Reeger-Schneider E.; Koster W.; Mohanty A. K.; Wiener M. C.; Kadner R. J. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 2002, 184, 706–717. 10.1128/JB.184.3.706-717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J.; Parker W. B.; Allan P. W.; Ayala J. C.; Benitez J. A. Role of methylthioadenosine /S-adenosylhomocysteine nucleosidase in Vibrio cholerae, cellular communication and biofilm development. Biochem. Biophys. Res. Commun. 2015, 461, 65–69. 10.1016/j.bbrc.2015.03.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K.; Hardie K. R.; Williams P. LuxS and Autoinducer-2: their contribution to quorum sensing and metabolism in Bacteria. Adv. Appl. Microbiol. 2003, 53, 291–396. 10.1016/s0065-2164(03)53009-x. [DOI] [PubMed] [Google Scholar]
- Vendeville A.; Winzer K.; Heurlier K.; Tang C. M.; Hardie K. R. Making sense of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Rees D. C.; Johnson E.; Lewinson O. Abc transporters: the power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T.; Rodionov D. A.; Grote M.; Schneider E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 2011, 35, 3–67. 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- Rodionov D. A.; Hebbeln P.; Eudes A.; Beek J.; Rodionova I. A.; Erkens G. B.; Slotboom D. J.; Gelfand M. S.; Osterman A. L.; Hanson A. D.; Eitinger T. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 2009, 191, 42–51. 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Man J. C.; Rogosa M.; Sharpe M. E. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 1960, 23, 130–135. 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- Bassler B. L.; Wright M.; Showalter R. E.; Silverman M. R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993, 9, 773–786. 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Han X. G.; Lu C. P. In vitro biosynthesis of autoinducer 2 of Streptococcus suis serotype 2 using recombinant LuxS and Pfs. Enzyme Microb. Technol. 2009, 44, 40–45. 10.1016/j.enzmictec.2008.09.009. [DOI] [Google Scholar]
- Surette M. G.; Bassler B. L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 7046–7050. 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Chabela M. L.; Díaz-Vela J.; Reyes-Menéndez C. V.; Totosaus A. Improvement of moisture stability and textural properties of fat and salt reduced cooked sausages by inoculation of thermotolerant Lactic Acid Bacteria. Int. J. Food Prop. 2013, 16, 1789–1808. 10.1080/10942912.2011.608472. [DOI] [Google Scholar]
- Christensen G. D.; Simpson W. A.; Younger J. J.; Baddour L. M.; Barrett F. F.; Melton D. M.; Beachey E. H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J. R.; Zougman A.; Nagaraj N.; Mann M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Aretz I.; Hardt C.; Wittig I.; Meierhofer D. An impaired respiratory electron chain triggers down-regulation of the energy metabolism and de-ubiquitination of solute carrier amino acid transporters. Mol. Cell. Proteomics 2016, 15, 1526–1538. 10.1074/mcp.M115.053181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Luber C. A.; Cox J.; Lauterbach H.; Fancke B.; Selbach M.; Tschopp J.; Akira S.; Wiegand M.; Hochrein H.; O’Keeffe M.; Mann M. Quantitative proteomes reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32, 279–289. 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Caplice E.; Fitzgerald G. F. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. 10.1016/S0168-1605(99)00082-3. [DOI] [PubMed] [Google Scholar]
- Supek F.; Bosnjak M.; Skunca N.; Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011, 6, e21800 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajarillo E. A. B.; Sang H. K.; Valeriano V. D.; Ji Y. L.; Kang D. K. Proteome view of the crosstalk between Lactobacillus mucosae and intestinal epithelial cells in co-culture revealed by Q exactive-based quantitative proteomes. Front. Microbiol 2017, 8, 2459 10.3389/fmicb.2017.02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]