Abstract
Mitotane is the only drug approved for the therapy of adrenocortical carcinoma (ACC). Its clinical use is limited by the occurrence of relapse during therapy. To investigate the underlying mechanisms in vitro, we here generated mitotane-resistant cell lines. After long-term pulsed treatment of HAC-15 human adrenocortical carcinoma cells with 70 µM mitotane, we isolated monoclonal cell populations of treated cells and controls and assessed their respective mitotane sensitivities by MTT assay. We performed exome sequencing and electron microscopy, conducted gene expression microarray analysis and determined intracellular lipid concentrations in the presence and absence of mitotane. Clonal cell lines established after pulsed treatment were resistant to mitotane (IC50 of 102.2 ± 7.3 µM (n = 12) vs 39.4 ± 6.2 µM (n = 6) in controls (biological replicates, mean ± s.d., P = 0.0001)). Unlike nonresistant clones, resistant clones maintained normal mitochondrial and nucleolar morphology during mitotane treatment. Resistant clones largely shared structural and single nucleotide variants, suggesting a common cell of origin. Resistance depended, in part, on extracellular lipoproteins and was associated with alterations in intracellular lipid homeostasis, including levels of free cholesterol, as well as decreased steroid production. By gene expression analysis, resistant cells showed profound alterations in pathways including steroid metabolism and transport, apoptosis, cell growth and Wnt signaling. These studies establish an in vitro model of mitotane resistance in ACC and point to underlying molecular mechanisms. They may enable future studies to overcome resistance in vitro and improve ACC treatment in vivo.
Keywords: chemotherapy, adrenolytic, somatic mutations, cholesterol, lipids, SOAT
Introduction
ACCs are rare and aggressive malignancies of the adrenal cortex, with an annual incidence of 0.7–2.0 cases per million population (1). ACCs can be part of rare hereditary cancer syndromes including Beckwith-Wiedemann syndrome (with alterations of imprinted genes on chromosome 11p15.5) and Li-Fraumeni syndrome (with mutations in TP53) (2), but most cases are sporadic. ACCs can develop at any age, with a peak incidence between 40 and 50 years of age, and women are more often affected (55–60%) (1). Patients typically present with symptoms of hormone excess in the case of secretory tumors or otherwise with large tumors and signs and symptoms of malignancy.
Complete surgical resection is the only potentially curative treatment option for ACCs. Although the majority of patients have resectable disease at presentation, up to 85% relapse after radical resection (3), leading to an overall poor prognosis. Mitotane, a by-product of the industrial production of the insecticide dichlorodiphenyldichloroethane, has been widely used in ACC therapy due to its adrenolytic properties (4, 5). A retrospective study demonstrated an association of adjuvant mitotane with prolonged recurrence-free survival after radical resection (6); however, results of a randomized controlled trial (ADIUVO) remain to be published (1). The randomized controlled FIRM-ACT trial found better response rates (23 vs 9 %) and median progression-free survival (5 vs 2.1 months) on mitotane combined with etoposide, doxorubicin and cisplatin (EDP-M) than with streptozotocin (7), with no significant difference in overall survival (14.8 vs 12 months). Hence, EDP-M is typically considered a first-line cytotoxic treatment for ACCs.
While mitotane plasma levels affect response rates, some patients do not respond despite therapeutic plasma levels. More importantly, patients who initially respond typically relapse even though therapeutic mitotane levels are maintained (8), suggesting a role of secondary acquired resistance to mitotane in relapse and progression.
The molecular changes associated with mitotane treatment have been examined in the human cancer cell lines H295R (9) and SW13 (10). Cytotoxic effects are visible at therapeutic mitotane concentrations (30–50 µM), and mitochondrial disruption appears to activate apoptosis through caspase 3/7, accounting for cytotoxicity (11). Effects also include a decrease in the expression of mitochondrial genes involved in steroidogenesis, such as STAR, CYP11B1 and CYP11B2, and a reduced activity of cytochrome c oxidase (12). More recently, inhibition of the microsomal sterol-o-acyl transferase 1 (SOAT1, also known as ACAT-1) has been suggested as the principal mode of action (13). Because SOAT catalyzes the generation of cholesterol esters from free sterol and acyl CoA (14), inhibition of its activity has been proposed to result in elevated levels of free cholesterol, ER stress and apoptosis. The molecular mechanisms of acquired mitotane resistance, however, are currently unknown. We here set out to generate a mitotane-resistant cell line to study mechanisms of mitotane resistance in vitro.
Materials and methods
Cell culture
HAC-15 cells (obtained from the Department of Molecular and Integrative Physiology, University of Michigan, Ann Arbor, MI, USA; authenticated by short tandem repeat analysis (LGC Standards, Wesel, Germany)) were cultured at 37°C and 5% CO2 in DMEM/F-12 + GlutaMAX (Gibco, Thermo Fisher Scientific) supplemented with 5% Cosmic Calf Serum (CCS, Hyclone, Logan, UT, USA), 1% Insulin-Transferrin-Selenium, 1% MEM non-essential amino acids, 0.1% chemically defined lipid concentrate and 1% Penicillin (10,000 U/mL)/Streptomycin (10,000 µg/mL) (Gibco).
Compounds
Compounds were dissolved as follows and stored as stock solutions at -20°C: mitotane (Sigma Aldrich), 100 mM in DMSO; doxorubicin (Cayman Chemical), 1 µM in HAC-15 medium; human HDL and LDL (Cedarlane, Burlington, Canada), 10 mg/mL in 15% sucrose (Carl Roth, Karlsruhe, Germany)/DMEM/F-12, HEPES (Gibco).
Long-term mitotane treatment of HAC-15 cells and clonal selection
Cells were frozen in aliquots as founder cells for reference. HAC-15 cells (starting from passage 4) were treated with 70 µM mitotane (about 1.5-fold the IC50) following a pulsed protocol (15). Medium was replaced every 3–4 days. At medium changes, cells alternately received either mitotane-free medium or medium containing 70 µM mitotane. Control cells received mitotane-free medium or medium containing solvent. At each passage, 107 cells were seeded in 75 cm² EasYFlasks (Nunc, Thermo Fisher Scientific), and treatment was continued after 24 h. For clonal selection, cells were plated on a 15 cm cell culture dish (VWR International, Radnor, PA, USA) at a density of 56 cells/cm2 without mitotane and isolated using cloning discs (Sigma Aldrich) with Trypsin-EDTA (0.05%, Gibco). After cloning, treatment with 70 µM mitotane was resumed.
Growth curves
At each passage, cells were counted using a JuLI Br Live Cell Imager (NanoEnTek, Seoul, Korea), and cumulative population doublings (cPD) (16) were calculated from total cell numbers using Eq. 1:
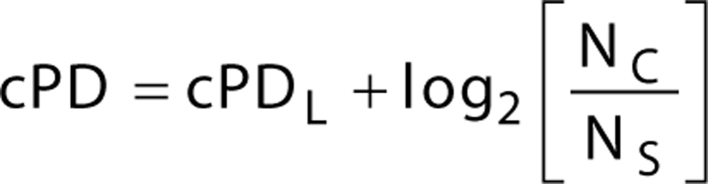 |
cPDL, cumulative population doublings after the last passage; NC, number of cells counted at the current passage; NS, number of cells seeded after the last passage.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay
After discontinuation of mitotane treatment (if applicable) for at least 2 weeks, 4 × 104 cells/well were seeded in triplicates on a 96-well plate (Gibco). After 24 h, cells were treated with cytotoxic compounds at 37°C for 72 h. Afterwards, a 5 mg/mL solution of MTT (Sigma Aldrich) in PBS was added to each well (final concentration 0.45 mg/mL), followed by incubation at 37°C for 2 h. Medium was removed, and MTT crystals were dissolved in 150 µL 10% Triton X-100 (Sigma Aldrich) in 2-propanol at pH 4.7. Absorption at 595 nm was recorded utilizing an EnSpire 2300 Multilabel Reader (PerkinElmer). For calculation of IC50, data were fitted to a four-parameter dose-response curve with Prism 7 (GraphPad) according to Eq. 2:
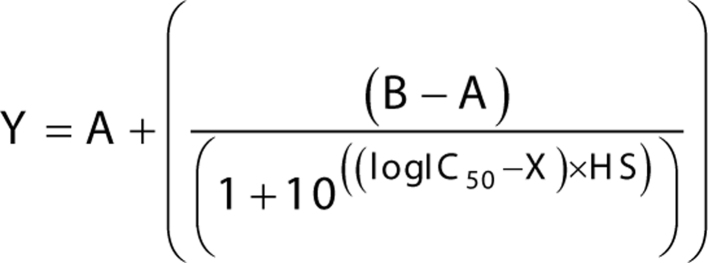 |
Y, absorption at 595 nm; A, minimum asymptote; B, maximum asymptote; IC50, half-maximal inhibitory concentration; X, log10 transformed mitotane concentration; HS, Hill slope.
For determination of response to different sera, cells were grown in the presence of 5% CCS, 2.5% CCS, 5% Nu-Serum (NuS, Corning) and 2.5% NuS or 5% fetal calf serum (FCS, Merck-Biochrom, Berlin, Germany). After 24 h, MTT assay was performed as mentioned previously. Serum concentrations of cholesterol, HDL, LDL and triglycerides were determined by photometry at the Central Laboratory of the University Hospital Düsseldorf, Germany, and concentrations of compounds in the culture medium were calculated. All groups were tested for normality using Shapiro–Wilk test. Concentrations of lipid species were plotted against their respective IC50, and correlation analysis according to Pearson was performed using Prism 7.
Measurement of intracellular lipids
Intracellular lipids were determined in three nonresistant and three mitotane-resistant clones. Cells were seeded on a six-well plate. After 24 h, one group of cells was treated with vehicle control (DMSO) or 10 µM mitotane in medium containing 0% CCS for 72 h. A second group was treated with DMSO, 20 or 50 µM mitotane in medium containing 5% CCS for 72 h. Cells were washed thrice with cold PBS and lysed in 1 mL water containing 0.1% sodium dodecyl sulfate (SDS, Biomol, Hamburg, Germany). Protein content was measured by BCA Protein Assay Kit (Pierce). Lipids were measured by electrospray ionization tandem mass spectrometry (ESI-MS/MS) and normalized to total protein quantity in mg, as previously described (17).
Measurement of supernatant steroid hormones
Cells were seeded on a 12-well plate. After 48 h, medium was replaced. After 24 h, cell supernatants were removed and quickly frozen at -80°C. Steroid analysis was performed by liquid chromatography tandem mass spectrometry (LC-MS/MS) as described elsewhere (18); testosterone, dihydrotestosterone, 18-hydroxycorticosterone, 11-dehydrocorticosterone and dexamethasone were added to the panel (Supplementary Table 1, see section on supplementary materials given at the end of this article).
In brief, 0.45 mL of cell culture supernatants were processed via solid phase extraction using positive pressure, dried down and finally reconstituted in 100 µL of initial LC mobile phase. For quantification of steroid hormones, regression analyses using respective analyte peak areas relative to those of respective internal standards vs analyte concentrations derived from an external calibration were used. Such ratios observed in samples were further used to quantify steroid hormone concentrations by applying the respective regression equations.
Statistics
Data were tested for normality using Shapiro–Wilk test. Data passing the normality test were compared using an unpaired t-test (two-tailed) or one-way ANOVA (multiple comparisons), and groups failing the normality test were compared using a Mann–Whitney test or Kruskal–Wallis test (multiple comparisons). As post hoc test, two-stage Benjamini, Krieger and Yekutieli FDR procedure was used.
Gene expression microarray
Six nonresistant and six mitotane-resistant clones were thawed and cultured to confluence without mitotane. Cells were seeded on a six-well plate (1 × 106 cells per well). After 24 h, cells were treated with either vehicle control (DMSO) or 50 µM mitotane for 18 h. RNA was isolated using QIAzol Lysis Reagent, miRNeasy MiniKit and RNase-Free DNase set (all from Qiagen), and RNA integrity was confirmed using an Agilent 2100 Bioanalyzer. Microarrays were processed at the Center for Applied Genomics at the Hospital for Sick Children (Toronto, Canada) using Affymetrix GeneChip PrimeView Human Gene Expression Array. Microarray analysis was performed in R (version 3.4.4) using the packages affy, affydata and limma. Pre-processing was done using expresso with rma background correction (19), quantile normalization, pmonly probe specific correction and avgdiff summarization. Log-transformed normalized intensities were used for principal component analysis (PCA) using the prcomp R function. Differential expression was performed with limma and a blocking design to match treated and untreated cell lines. Differentially expressed genes were extracted from top-expressed probe sets after Benjamini–Hochberg correction using a cutoff of 5% on the adjusted P-value and of 0.5 on the log2 fold change. GO term analysis (20) was performed using topGO, using the classic algorithm and a cutoff of 0.01 on the Fisher P value. Expression changes were compared to copy number changes by aggregating CNV results per gene and calculating log2 fold change in read depth relative to the control. Microarray data is available at the Gene Expression Omnibus database (GEO accession# GSE140818).
Whole exome sequencing and analysis
DNA from the founder line, one nonresistant and six resistant clones (identical to those used for gene expression microarrays), was isolated using the DNeasy Blood & Tissue Kit (Qiagen). Whole-exome capture using SeqCap EZ Med Exome (Roche) and high-throughput sequencing (HiSeq 4000, Illumina) were performed at the Yale Center for Genome Analysis.
We used BWA-mem v0.7.15 (21) to map each whole-exome data set against genome reference GRCh37(hs37d5.fa). Separate read groups were assigned for all reads from one lane, and duplicates were masked using Samblaster v0.1.24 (22). Copy number alterations were analyzed in a tumor/normal paired fashion with the R package CopywriteR (23) with 50 kb bins and annotated with the CIViC database (24), where we treated the founder cell lines as normal and the resistant cultures or control as tumor sample. All log2 fold changes denote the relative change in read-depth compared to the founder cell line. Single nucleotide variants were called with MuTect (25) with the default configuration. Variants were filtered for artifacts using dkfz-bias-filter and annotated with Jannovar v0.24 (26). Candidate SNVs were manually assessed using the Integrative Genomics Viewer (IGV_2.4.13, Broad Institute).
Electron microscopy
For transmission electron microscopy, two resistant and two nonresistant clones were thawed and cultured to confluence without mitotane. Cells were seeded on a 6-well plate (2 × 106 cells per well). After 24 h, cells were treated with 50 µM mitotane or vehicle control (DMSO). After 72 h, cells were washed once with ice cold PBS and fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (both from Serva, Heidelberg, Germany) at room temperature for 30 min. Afterwards, fixation buffer was changed, and cells were stored at 4°C for 2–14 days. Cells were postfixed in 1% OsO4 (Science Services, Munich, Germany) and 0.8% potassium ferrocyanide (Merck) in 0.1 M sodium cacodylate buffer for 1.5 h and then progressively dehydrated in ethanol, followed by embedding in Epon (Serva). Ultrathin sections (70 nm) were prepared using an Ultracut S Ultramicrotome (Leica), stained with uranyl acetate and Reynold’s lead citrate (Merck), and microphotographs were taken using an electron microscope EM906 (Carl Zeiss).
Results
Generation, morphology and doxorubicin sensitivity of mitotane-resistant HAC-15 cell clones
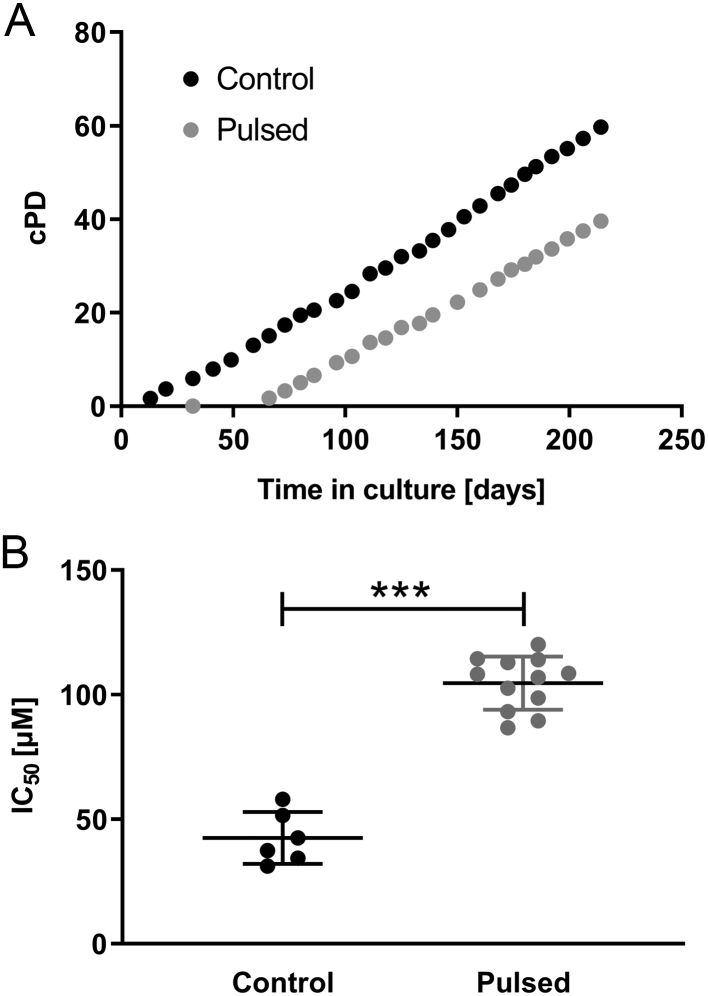
HAC-15 cells showed a sigmoidal dose response to mitotane, with an IC50 of 46.4 ± 13.1 µM (mean ± s.d., n = 3) (Supplementary Fig. 1). We performed pulsed treatment of the founder line with 70 µM mitotane to induce resistance (15) (Fig. 1A, see Materials and methods). MTT assays revealed an IC50 of 102.2 ± 7.3 µM for resistant clones (n = 12, biological replicates) and 39.4 ± 6.2 µM for control clones (mean ± s.d., n = 6, biological replicates, P = 0.0001, Mann–Whitney test), demonstrating significant mitotane resistance in treated clones (Fig. 1B).
Figure 1.
(A) Growth curves of HAC-15 cells during long-term treatment with mitotane vs untreated control. Every 3–4 days, cells were treated with 70 µM mitotane in on-off cycles. The total number of population doublings (cumulative population doublings; cPD) is indicated. During long-term treatment, cells initially died, but the population recovered after approximately 30 days. (B) IC50 of mitotane in 12 clones derived from long-term treated HAC-15 cells vs 6 control clones measured by MTT assay after 72 h of incubation. Results are shown in scatter dot plots with mean ± s.d. ***, P < 0.001 (Mann–Whitney test).
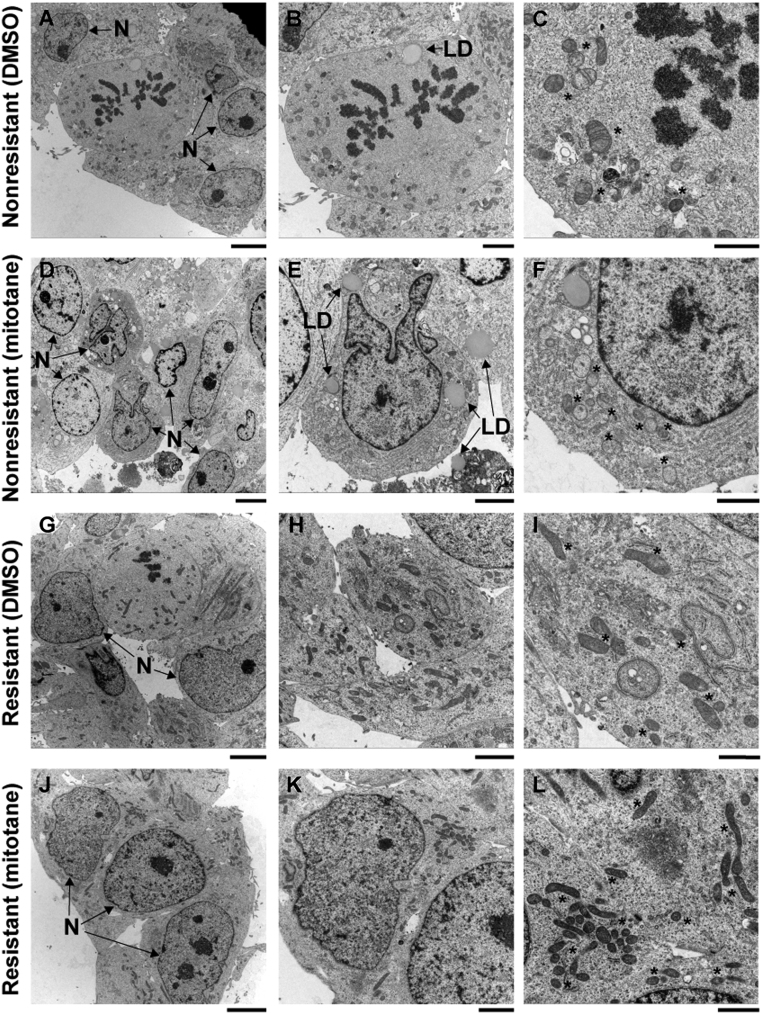
To assess whether mitochondrial integrity was preserved in resistant cells, we treated resistant cells and controls with 50 µM mitotane and performed electron microscopy (Fig. 2, Supplementary Fig. 2). Untreated nonresistant (Fig. 2A, B and C) and resistant clones (Fig. 2G, H and I) displayed normal cellular and mitochondrial morphology with frequent mitoses. In nonresistant cells, mitotane treatment caused mitochondrial swelling with loss of cristae as previously described (11). Additional morphological changes included irregular nuclear shapes (Fig. 2D, E and F) and large lipid droplets surrounded by concentric layers of rough endoplasmic reticulum (Supplementary Fig. 3), as well as necrotic cells and intracellular protein deposits. Interestingly, no such changes occurred in resistant cells upon mitotane treatment (Fig. 2J, K and L). The absence of mitochondrial damage in treated resistant cells specifically suggests that the mechanism of resistance in this model acts upstream of mitochondrial damage. We assessed doxorubicin sensitivity in six resistant and six nonresistant clones in the absence or presence of 10 µM mitotane. In the absence of mitotane, no significant difference in doxorubicin sensitivity between resistant and nonresistant cells was found (IC50 of 3.7 ± 0.6 nM and 4.6 ± 1.5 nM (mean ± s.d., n = 6 each, P = 0.12, one-way ANOVA)). In the presence of 10 µM mitotane, resistant clones showed increased doxorubicin sensitivity compared to controls (IC50 of 2.2 ± 0.4 nM vs 3.9 ± 1.2 nM (mean ± s.d., n = 6 each, P = 0.0017, one-way ANOVA)) (Supplementary Fig. 4).
Figure 2.
Representative electron micrographs of one nonresistant (A, B, C, D, E and F) and one resistant clone (G, H, I, J, K and L), treated for 72 h with either vehicle control (A, B, C, G, H and I) or 50 µM mitotane (D, E, F, J, K and L). Mitotane treatment causes mitochondrial swelling, loss of mitochondrial cristae and irregular nuclear shapes in nonresistant, but not in resistant cells. N: nuclei; *: mitochondria; LD: lipid droplet. Lines: 5 µm (A, D, G and J); 2.5 µM (B, E, H and K); 1.25 µm (C, F, I and L).
Pathways involving steroid metabolism and transport, apoptosis, cell growth and Wnt signaling are altered in mitotane-resistant cell lines
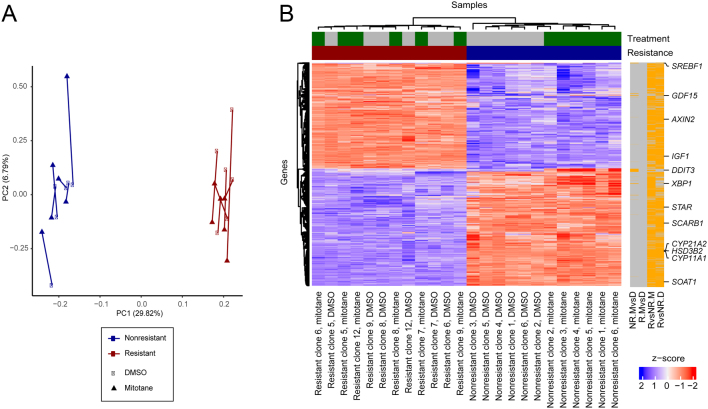
We compared gene expression profiles of six mitotane-resistant with six nonresistant clones in the presence and the absence of mitotane (see Materials and methods). In PCA, resistant cell lines clustered separately from control cell lines, whereas response to mitotane was less uniform (Fig. 3A). Unbiased analysis identified 1581 differentially expressed genes between resistant cells and controls in the absence of treatment (see Materials and methods, Fig. 3B, Table 1, Supplementary Fig. 5 and Supplementary Table 2). Genes with upregulated expression included AXIN2, a Wnt target gene (27), and IGF1, encoding an insulin-like growth factor. IGFs have been implicated in adrenal development (28) and tumor formation (29). Genes with downregulated expression included SOAT1, encoding for sterol-o-acyl transferase 1, a major intracellular target of mitotane (13), and SCARB1, encoding for scavenger receptor B1, the most important transporter for adrenal cholesterol uptake (30). Further, downregulation of steroidogenic enzymes CYP11A1, encoding for cholesterol side-chain cleaving enzyme, HSD3B2, encoding for 3β-hydroxysteroid dehydrogenase/Δ5-4 isomerase, CYP21A2, encoding for steroid 21-hydroxylase, and STAR, encoding for StAR protein, was discovered. STAR and CYP11A1 participate in rate-limiting steps in adrenal steroidogenesis, which include cholesterol uptake into the mitochondria and the conversion of cholesterol to pregnenolone (31). Upon mitotane treatment of control cells, 60 genes were differentially expressed. In accordance with a previous study (13), downregulation of SREBF1, encoding sterol regulatory binding transcription factor 1, was demonstrated. Also, expression of genes involved in endoplasmic reticulum stress response was significantly altered, including upregulation of DDIT3, encoding DNA damage-inducible transcript 3, XBP1, encoding X-box binding protein 1, and GDF15, encoding growth/differentiation factor 15 (Table 1) (32, 33). Interestingly, treatment of resistant cells did not yield any differentially expressed genes above the log2 fold change cutoff of 0.5, confirming resistance on the gene expression level.
Figure 3.
(A) PCA of gene expression for resistant and nonresistant cell lines treated with DMSO or mitotane. Corresponding cell lines are connected by lines, and percentage-explained variance is given in the axis title. (B) Expression heatmap of differentially expressed genes in any of four comparisons: resistant cell lines with vs without mitotane treatment (R.MvsD; no significant changes), nonresistant cell lines with vs without treatment (NR.MvsD), resistant vs nonresistant cell lines under treatment (RvsNR.M) or without treatment (RvsNR.D). Expression data were transformed to z-scores for standardization. Significantly changed genes (5% FDR, log2 fold change cutoff 0.5) are indicated by orange bars on the right, and selected genes are highlighted.
Table 1.
Gene expression changes after mitotane treatment of control cells or after development of resistance (selection).
| Gene | Log2 FC | Exp | Adj P | Comparison |
|---|---|---|---|---|
| SCARB1 | −2.54 | 7.76 | 2.4 × 10−20 | Resistant DMSO vs Nonresistant DMSO |
| HSD3B2 | −5.77 | 5.33 | 5.98 × 10−18 | Resistant DMSO vs Nonresistant DMSO |
| CYP11A1 | −3.66 | 6.45 | 1.01 × 10−17 | Resistant DMSO vs Nonresistant DMSO |
| CYP21A2 | −4.34 | 6.61 | 7.49 × 10−17 | Resistant DMSO vs Nonresistant DMSO |
| IGF1 | 4.24 | 5.74 | 2.94 × 10−14 | Resistant DMSO vs Nonresistant DMSO |
| STAR | −3.52 | 7.14 | 4.17 × 10−14 | Resistant DMSO vs Nonresistant DMSO |
| AXIN2 | 1.59 | 6.87 | 2.2 × 10−13 | Resistant DMSO vs Nonresistant DMSO |
| SOAT1 | −1.95 | 7.35 | 1.64 × 10−12 | Resistant DMSO vs Nonresistant DMSO |
| DDIT3 | 1.55 | 5.94 | 2.55 × 10−8 | Nonresistant Mitotane vs Nonresistant DMSO |
| GDF15 | 1.87 | 5.9 | 8.19 × 10−6 | Nonresistant Mitotane vs Nonresistant DMSO |
| SREBF1 | −1.14 | 6.16 | 8.4 × 10−6 | Nonresistant Mitotane vs Nonresistant DMSO |
| XBP1 | 0.75 | 6.96 | 7.29 × 10−4 | Nonresistant Mitotane vs Nonresistant DMSO |
Only genes referenced in results section are listed.
Adj P, adjusted P value; DMSO, dimethyl sulfoxide; Exp, average expression; Log2 FC, log2 fold change.
To identify pathways involved in in vitro mitotane resistance, we performed gene ontology enrichment analysis. Untreated resistant cells showed significant upregulation of genes implicated in apoptosis regulation, whereas pathways related to steroid metabolism, regulation of ERK (extracellular signal-regulated kinases) cascade, apoptotic cell clearance and response to xenobiotics were downregulated (Supplementary Table 3). Mitotane treatment of control cells led to upregulation of pathways including cell death and unfolded protein response, whereas pathways related to lipid homeostasis and transport were downregulated (Supplementary Table 4). These results are in line with the current concept of mitotane action via accumulation of free cholesterol and ER stress, leading to apoptosis (13), processes that are apparently mitigated in our in vitro model of resistance.
In vitro mitotane-resistant clones are genetically highly similar, suggesting a single cell of origin
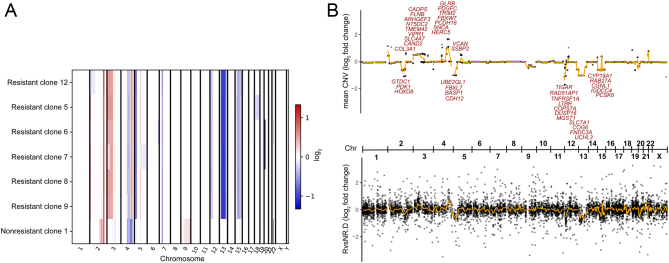
To identify genetic mechanisms underlying mitotane resistance and to further characterize mitotane-resistant cell lines, we performed whole exome sequencing of six mitotane-resistant clones. As controls, we sequenced the founder cell line and one nonresistant clone. Cells cultured over extended periods of time are known to accumulate mutations (34). Remarkably, the CNV profiles of all six resistant lines differed from the control line, but were highly similar among each other, suggesting a single resistant cell of origin (Fig. 4A). Analysis of heterozygous calls for MTTP (c.3G>A, NM_000253.3) in an area of copy number loss on chromosome 4 shared between resistant lines and control demonstrated that different alleles had been lost in resistant cells and controls, respectively. Areas of gain shared among resistant cells included chromosomes 2, 3 and 4, whereas loss was detected uniformly on chromosomes 4, 5, 7, 12, 13 and 15. Genes whose increased gene expression correlated with gain or whose decreased gene expression correlated with loss of chromosomal material (Fig. 4B) included PDGFC encoding platelet derived growth factor C involved in cell proliferation (35) and FBXW7, which can apparently function as a tumor suppressor or as an oncogene (36) (both upregulated). UBE2QL1, whose gene product interacts with FBXW7 and is considered a tumor suppressor (37), was downregulated, as were FBXL7 involved in impairing cell proliferation (38), TNFRSF1A and LTBR, encoding TNF receptors involved in apoptosis.
Figure 4.
(A) Copy number changes compared to founder cell line. Whole-exome sequencing data of resistant and nonresistant cell lines were compared to the founder cell line in a tumor/normal matched pair fashion to analyze copy number changes. Log2 fold changes are shown across the genome. Resistant cell lines share CNV patterns that are distinct from the control, suggesting a common cell of origin of resistant clones. (B) Comparison of average CNV to gene expression changes (resistant vs nonresistant cells). Top panel shows CNV log2 fold change per gene across the genome. Bottom panel shows log2 fold change in RNA expression between resistant and nonresistant clones (without mitotane treatment). Selected genes with consistent log2 fold changes (|CNV| > 0.5, |RNA| > 1, CNV*RNA > 0) are highlighted. Orange lines indicate smooth moving average. The overall correlation is 0.28. Chr, chromosome.
In vitro mitotane-resistant cells have a higher burden of protein-changing SNVs than control cells
Next, we identified SNVs acquired as part of the development of mitotane resistance by comparing exomes of resistant cell lines and controls. In the six resistant clones, we respectively identified 19, 21, 23, 16, 26 and 19 newly acquired potentially protein-changing mutations that were absent in the founder line. In contrast, only nine newly acquired mutations were detected in the nonresistant cell line, suggesting a higher mutational burden in treated cells as a result of selection. Eleven newly acquired mutations shared among all resistant cell lines are shown in Table 2. Affected genes include PLCH1, encoding a member of the phospholipase C (PLC) family (39), PLXNB1, encoding plexin B1 implicated in invasive growth and cell migration (40), LRP4, a potential negative regulator of Wnt signaling (41) and GNL3, which may interact with p53 and appears to be involved in tumorigenesis (42).
Table 2.
Variants shared among all resistant cell lines.
| Gene | Protein Change | Chr | Pos | Ref | Var |
|---|---|---|---|---|---|
| CD34 | T89K | 1 | 208072568 | G | T |
| FLNB | D1264Y | 3 | 58110124 | G | T |
| LRP4 | N138S | 11 | 46921431 | T | C |
| PLXNB1 | A838S | 3 | 48461183 | C | A |
| HOMER3 | G78V | 19 | 19049232 | C | A |
| PLCH1 | N856S | 3 | 155205833 | T | C |
| PRSS50 | V98I | 3 | 46758942 | C | T |
| PDE12 | R23W | 3 | 57542173 | C | T |
| SLCO1B7 | R11I | 12 | 21168661 | G | T |
| ABHD14A-ACY1 | predicted splice site mutation | 3 | 52011879 | C | T |
| GNL3 | V367M | 3 | 52727257 | G | A |
Chr, chromosome; Pos, chromosomal position, Ref, reference base; Var, variant base.
Mitotane-resistant cells show profoundly altered intracellular lipid profiles
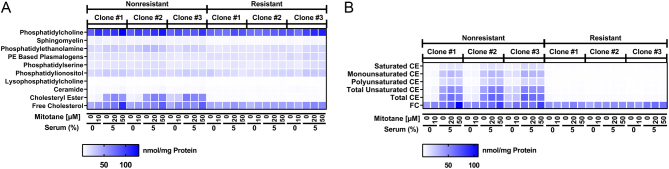
Because microarray analysis had revealed alterations in pathways governing lipid homeostasis, we measured intracellular levels of several lipid species in three mitotane-resistant and three nonresistant clones, each treated with 0 or 10 µM mitotane in the absence of serum (high availability of free mitotane) or 0, 20 and 50 µM mitotane in the presence of 5% cosmic calf serum (CCS) (lower availability of free mitotane due to lipoprotein binding). In line with the proposed inhibitory effect of mitotane on SOAT1 (13), treatment of nonresistant cells with 50 µM mitotane resulted in increased levels of free cholesterol, whereas this effect was absent in resistant cells. Levels of cholesteryl esters did not significantly change upon treatment with 50 µM mitotane in either nonresistant or resistant clones (Fig. 5A, B and Supplementary Table 5, 6). Levels of ceramides, a lipid species with a well-established role in apoptotic signaling (43, 44), also significantly rose in nonresistant cells after treatment with 50 µM mitotane, an effect that was again mitigated in resistant cells, and levels of lysophosphatidylcholines, which have been implicated in lipoapoptosis (45), were significantly higher in nonresistant cells treated with 50 µM mitotane (Supplementary Fig. 6 and Supplementary Tables 5, 6). Prior studies (46, 47) suggested that mitotane cytotoxicity may be influenced by cholesterol-bearing lipoproteins such as LDL and HDL. In the presence of artificially low lipoprotein levels (0.005 mg/mL HDL, 0.005 mg/mL LDL), median IC50 of not only nonresistant but also resistant cells dropped significantly compared to more physiological levels (0.05 mg/mL HDL, 0.05 mg/mL LDL) (Supplementary Fig. 7).
Figure 5.
Intracellular lipds in three mitotane-resistant vs three nonresistant HAC15 clones. (A) Amounts of phosphatidylcholine, sphingomyelin, phosphatidylethanolamine, phosphatidylethanolamine (PE) based plasmalogens, phosphatidylserine, phosphatidylionositol, lysophosphatidylcholine, ceramide, cholesteryl ester and free cholesterol after treatment with increasing concentrations of mitotane in presence of different serum concentrations. (B) Amounts of saturated, monounsaturated, polyunsaturated, total unsaturated and total cholesteryl ester (CE) as well as free cholesterol (FC), determined as in (A). Treatment with 50 µM mitotane in presence of 5% CCS increases free cholesterol in nonresistant cells but not in mitotane-resistant cells. For lipid amounts and statistical analysis, please refer to Supplementary Tables 5 and 6.
Mitotane-resistant cells show impaired production of adrenal steroid hormones
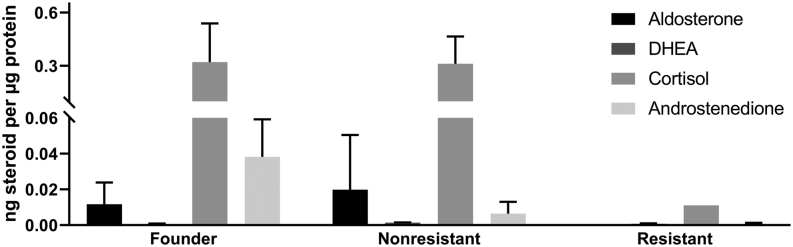
Next, we measured levels of adrenal steroid hormones in cell supernatants of founder cells, nonresistant and resistant cells by LCMS (Fig. 6). Steroid profiles of the founder cell line and nonresistant clones were largely comparable and included abundant levels of cortisol and moderate levels of aldosterone as well as very small amounts of DHEA. In resistant clones, steroid hormone production was strongly reduced. Aldosterone and cortisol were only detected at low levels in one of three clones.
Figure 6.
Supernatant steroid concentration normalized by total protein in the founder HAC15 cell line vs mitotane-resistant and nonresistant HAC15 clones, determined by LC-MS/MS (n = 3). Steroid hormone production is largely abated in mitotane-resistant clones. Shown is mean ± s.d., (measured in triplicate) each from three clones, except for aldosterone and cortisol in resistant cells, for which results from a single clone are shown (levels from the two other clones were below detection limit). DHEA, dehydroepiandrosterone.
Discussion
To our knowledge, this study is the first to establish an in vitro model of mitotane resistance in ACC along with nonresistant controls. We chose the HAC-15 cell line as an ideal candidate for this study based on its production of adrenal steroid hormones and clonality (48). Although originally described as a new primary adrenocortical cell line, HAC-15 cells are now considered a clonal subpopulation of the most commonly used H295R adrenocortical cancer cell line (48). Another commonly used cell line, SW13 (10) may be derived from a metastasized non-adrenal tumor (48) and seemed less useful due to its lack of steroid production. The mitotane sensitivities of two very recently established ACC cell lines remain to be determined (49). A recently established xenograft model shows mitotane resistance (50); however, the lack of a nonresistant control prevented its use in this study.
The clonality of the HAC-15 cell line, the long time span from the beginning of pulsed treatment to the recovery of growth, the highly similar genetic profiles of obtained mitotane-resistant cell clones and the high mutation burden all suggest that the mitotane-resistant cells described here are not derived from a pre-existing resistant subpopulation but rather arose through mutagenesis under selection. Consistently, prior efforts to isolate side populations from H295R cells did not yield resistant cells (51).
Prior studies have focused on the short-term response of H295R cells to mitotane, effects largely replicated in short-term treatment of control cells in our study. Our microarray data demonstrated downregulated expression of genes involved in steroid hormone pathways and upregulation of unfolded protein response genes, in line with previous data (11, 12, 13). The lower number of differentially expressed genes in our study may be related to different treatment durations (6 h in the study by Sbiera et al. vs 18 h in our study).
The focus of this study, however, was on in vitro mechanisms of mitotane resistance. Morphological analysis demonstrated that mitochondrial damage, a distinctive feature of mitotane-treated H295R cells (11), was absent in mitotane-treated resistant cells. These findings suggest that altered pathways are located upstream of mitochondrial damage. Accordingly, both gene expression analysis and functional studies pointed to changes in lipoprotein and lipid homeostasis. By gene expression microarray, we found downregulation of pathways implicated in steroid metabolism, but also regulation of ERK, apoptotic cell clearance and response to xenobiotics, which collectively may contribute to the resistant phenotype. Changes in copy number that correlated with gene expression changes were not related to mitotane signaling, suggesting that regulation of gene expression rather than CNV may play a role.
Unlike in nonresistant controls, intracellular free cholesterol – a major mediator of mitotane-associated apoptosis (13) – remained unchanged in resistant cells upon mitotane treatment. We confirmed data from Hescot et al. and Kroiss et al. demonstrating that lipoprotein binding inhibits mitotane activity in vitro (46, 47). Importantly, removal of extracellular lipoproteins in our study had a larger effect on the sensitivity of resistant cells than of nonresistant cells. If similar mechanisms are present in vivo, lowering the lipoprotein concentration could be a promising approach to overcome resistance (see below). Interestingly, mass spectrometry demonstrated strong downregulation of adrenal steroid production in vitro in resistant cells, thus, it would be interesting to assess whether any reduction in hypercortisolism is sustained in vivo after the development of resistance.
Limitations of our study include the use of an in vitro cell model, which does not reflect the three-dimensional tumor architecture (52) or its microenvironment (53) and does not account for metabolism and excretion of mitotane or storage in fat-containing tissues (54). Ideally, our studies should be complemented by in vivo studies of mitotane resistance, comparing tumor material before mitotane therapy and after resistance has developed. However, such studies are challenging due to the rarity of ACCs and in particular of cases who undergo resurgery after the development of mitotane resistance. Furthermore, our results should be interpreted with caution because only a single cell line was studied.
Potential strategies to overcome mitotane resistance include SOAT inhibitors such as ATR-101 (55), inhibition of ER chaperones (56) or proteasome inhibitors (57). Of note, in our model, doxorubicin was more effective in resistant cells treated with mitotane than in nonresistant controls, an observation that so far remains unexplained. If applicable in vivo, these results would support the use of mitotane plus doxorubicin in case of relapse during mitotane monotherapy (as part of EDP-M (4)).
As discussed previously, in our cell model, mitotane resistance could be partially reversed by lowering extracellular lipoprotein concentrations. This effect is of particular importance because mitotane therapy can cause an increase in lipoprotein levels (58), potentially promoting resistance. Lowering lipoprotein levels through statin therapy has already been suggested to be associated with higher rates of tumor control during mitotane treatment (46). Novel PCSK9 inhibitors could be more promising given their ability to lower LDL levels beyond statin therapy (59). Thus, we suggest that the model we here established might help to develop strategies to overcome mitotane resistance in vitro, which could then be further investigated in clinical trials.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by the Ministerium für Kultur und Wissenschaft der Landes Nordrhein-Westfalen (Rückkehrprogramm and Junges Kolleg, both to U I S), Stiftung Charité (BIH Johanna Quandt Professorship, to U I S) and the DAAD (RISE Fellowship, to R B and E S). We acknowledge support from the German Research Foundation (DFG) and the Open Access Publication Funds of Charité – Universitätsmedizin Berlin.
Acknowledgements
The authors thank William Rainey (Department of Molecular and Integrative Physiology, University of Michigan, Ann Arbor, USA) for providing the HAC-15 cell line, the staff of the Yale Center for Genome Analysis (New Haven, USA), in particular Irina Tikhonova, Christopher Castaldi and Shrikant Mane, for performing exome sequencing and the staff of the Center for Applied Genomics, The Hospital for Sick Children (Toronto, Canada) for performing and analyzing gene expression arrays. They also thank Carmen Barthuber from the Central Institute of Clinical Chemistry and Laboratory Medicine (Düsseldorf, Germany) for performing photometry analysis of serum HDL, LDL and cholesterol concentrations and Manuel Holtgrewe (BIH, Berlin) for bioinformatics support.
References
- 1.Fassnacht M, Kroiss M, Allolio B. Update in adrenocortical carcinoma. Journal of Clinical Endocrinology and Metabolism 2013. 4551–4564. ( 10.1210/jc.2013-3020) [DOI] [PubMed] [Google Scholar]
- 2.Lerario AM, Moraitis A, Hammer GD. Genetics and epigenetics of adrenocortical tumors. Molecular and Cellular Endocrinology 2014. 67–84. ( 10.1016/j.mce.2013.10.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier RF, Brennan MF. An eleven-year experience with adrenocortical carcinoma. Surgery 1992. 963–970; discussion 970–961. [PubMed] [Google Scholar]
- 4.Fassnacht M, Dekkers OM, Else T, Baudin E, Berruti A, de Krijger RR, Haak HR, Mihai R, Assie G, Terzolo M. European Society of Endocrinology Clinical Practice Guidelines on the Management of Adrenocortical Carcinoma in Adults, in collaboration with the European Network for the Study of Adrenal Tumors. European Journal of Endocrinology 2018. G1–G46. ( 10.1530/EJE-18-0608) [DOI] [PubMed] [Google Scholar]
- 5.Cueto C, Brown JH, Richardson AP., Jr Biological studies on an adrenocorticolytic agent and the isolation of the active components. Endocrinology 1958. 334–339. ( 10.1210/endo-62-3-334) [DOI] [PubMed] [Google Scholar]
- 6.Terzolo M, Angeli A, Fassnacht M, Daffara F, Tauchmanova L, Conton PA, Rossetto R, Buci L, Sperone P, Grossrubatscher E, et al. Adjuvant mitotane treatment for adrenocortical carcinoma. New England Journal of Medicine 2007. 2372–2380. ( 10.1056/NEJMoa063360) [DOI] [PubMed] [Google Scholar]
- 7.Fassnacht M, Terzolo M, Allolio B, Baudin E, Haak H, Berruti A, Welin S, Schade-Brittinger C, Lacroix A, Jarzab B, et al. Combination chemotherapy in advanced adrenocortical carcinoma. New England Journal of Medicine 2012. 2189–2197. ( 10.1056/NEJMoa1200966) [DOI] [PubMed] [Google Scholar]
- 8.Baudin E, Pellegriti G, Bonnay M, Penfornis A, Laplanche A, Vassal G, Schlumberger M. Impact of monitoring plasma 1,1-dichlorodiphenildichloroethane (o,p′DDD) levels on the treatment of patients with adrenocortical carcinoma. Cancer 2001. 1385–1392. ( 10.1002/1097-0142(20010915)92:63.0.CO;2-2) [DOI] [PubMed] [Google Scholar]
- 9.Rainey WE, Bird IM, Mason JI. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Molecular and Cellular Endocrinology 1994. 45–50. ( 10.1016/0303-7207(94)90277-1) [DOI] [PubMed] [Google Scholar]
- 10.Leibovitz A, McCombs WM, Johnston D, McCoy CE, Stinson JC. New human cancer cell culture lines. I. SW-13, small-cell carcinoma of the adrenal cortex. Journal of the National Cancer Institute 1973. 691–697. [PubMed] [Google Scholar]
- 11.Poli G, Guasti D, Rapizzi E, Fucci R, Canu L, Bandini A, Cini N, Bani D, Mannelli M, Luconi M. Morphofunctional effects of mitotane on mitochondria in human adrenocortical cancer cells. Endocrine-Related Cancer 2013. 537–550. ( 10.1530/ERC-13-0150) [DOI] [PubMed] [Google Scholar]
- 12.Hescot S, Slama A, Lombes A, Paci A, Remy H, Leboulleux S, Chadarevian R, Trabado S, Amazit L, Young J, et al. Mitotane alters mitochondrial respiratory chain activity by inducing cytochrome c oxidase defect in human adrenocortical cells. Endocrine-Related Cancer 2013. 371–381. ( 10.1530/ERC-12-0368) [DOI] [PubMed] [Google Scholar]
- 13.Sbiera S, Leich E, Liebisch G, Sbiera I, Schirbel A, Wiemer L, Matysik S, Eckhardt C, Gardill F, Gehl A, et al. Mitotane inhibits sterol-O-acyl transferase 1 triggering lipid-mediated endoplasmic reticulum stress and apoptosis in adrenocortical carcinoma cells. Endocrinology 2015. 3895–3908. ( 10.1210/en.2015-1367) [DOI] [PubMed] [Google Scholar]
- 14.Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. American Journal of Physiology. Endocrinology and Metabolism 2009. E1–E9. ( 10.1152/ajpendo.90926.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDermott M, Eustace AJ, Busschots S, Breen L, Crown J, Clynes M, O’Donovan N, Stordal B. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Frontiers in Oncology 2014. 40 ( 10.3389/fonc.2014.00040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristofalo VJ, Allen RG, Pignolo RJ, Martin BG, Beck JC. Relationship between donor age and the replicative lifespan of human cells in culture: a reevaluation. PNAS 1998. 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leidl K, Liebisch G, Richter D, Schmitz G. Mass spectrometric analysis of lipid species of human circulating blood cells. Biochimica et Biophysica Acta 2008. 655–664. ( 10.1016/j.bbalip.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 18.Peitzsch M, Dekkers T, Haase M, Sweep FC, Quack I, Antoch G, Siegert G, Lenders JW, Deinum J, Willenberg HS, et al. An LC-MS/MS method for steroid profiling during adrenal venous sampling for investigation of primary aldosteronism. Journal of Steroid Biochemistry and Molecular Biology 2015. 75–84. ( 10.1016/j.jsbmb.2014.10.006) [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003. 249–264. ( 10.1093/biostatistics/4.2.249) [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 2000. 25–29. ( 10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Aligning sequence reads, clone seqeunces and assembly contigs with BWA-MEM. arXiv 1303.3997v1 [q-bio.GN], 2013. (available at: https://arxiv.org/abs/1303.3997v1) [Google Scholar]
- 22.Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014. 2503–2505. ( 10.1093/bioinformatics/btu314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuilman T, Velds A, Kemper K, Ranzani M, Bombardelli L, Hoogstraat M, Nevedomskaya E, Xu G, de Ruiter J, Lolkema MP, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biology 2015. 49 ( 10.1186/s13059-015-0617-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith M, Spies NC, Krysiak K, McMichael JF, Coffman AC, Danos AM, Ainscough BJ, Ramirez CA, Rieke DT, Kujan L, et al. CIViC is a community KnowledgeBase for expert crowdsourcing the clinical interpretation of variants in cancer. Nature Genetics 2017. 170–174. ( 10.1038/ng.3774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology 2013. 213–219. ( 10.1038/nbt.2514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jager M, Wang K, Bauer S, Smedley D, Krawitz P, Robinson PN. Jannovar: a java library for exome annotation. Human Mutation 2014. 548–555. ( 10.1002/humu.22531) [DOI] [PubMed] [Google Scholar]
- 27.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Molecular and Cellular Biology 2002. 1172–1183. ( 10.1128/mcb.22.4.1172-1183.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitetti JL, Calvel P, Romero Y, Conne B, Truong V, Papaioannou MD, Schaad O, Docquier M, Herrera PL, Wilhelm D, et al. Insulin and IGF1 receptors are essential for XX and XY gonadal differentiation and adrenal development in mice. PLoS Genetics 2013. e1003160 ( 10.1371/journal.pgen.1003160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fassnacht M, Libe R, Kroiss M, Allolio B. Adrenocortical carcinoma: a clinician’s update. Nature Reviews: Endocrinology 2011. 323–335. ( 10.1038/nrendo.2010.235) [DOI] [PubMed] [Google Scholar]
- 30.Kraemer FB. Adrenal cholesterol utilization. Molecular and Cellular Endocrinology 2007. 42–45. ( 10.1016/j.mce.2006.12.001) [DOI] [PubMed] [Google Scholar]
- 31.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocrine Reviews 2011. 81–151. ( 10.1210/er.2010-0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nature Reviews: Drug Discovery 2008. 1013–1030. ( 10.1038/nrd2755) [DOI] [PubMed] [Google Scholar]
- 33.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews: Molecular Cell Biology 2007. 519–529. ( 10.1038/nrm2199) [DOI] [PubMed] [Google Scholar]
- 34.Kim M, Rhee JK, Choi H, Kwon A, Kim J, Lee GD, Jekarl DW, Lee S, Kim Y, Kim TM. Passage-dependent accumulation of somatic mutations in mesenchymal stromal cells during in vitro culture revealed by whole genome sequencing. Scientific Reports 2017. 14508 ( 10.1038/s41598-017-15155-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, et al. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nature Cell Biology 2000. 302–309. ( 10.1038/35010579) [DOI] [PubMed] [Google Scholar]
- 36.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Molecular Cancer 2018. 115 ( 10.1186/s12943-018-0857-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wake NC, Ricketts CJ, Morris MR, Prigmore E, Gribble SM, Skytte AB, Brown M, Clarke N, Banks RE, Hodgson S, et al. UBE2QL1 is disrupted by a constitutional translocation associated with renal tumor predisposition and is a novel candidate renal tumor suppressor gene. Human Mutation 2013. 1650–1661. ( 10.1002/humu.22433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora A, causing mitotic arrest. Cell Cycle 2012. 721–729. ( 10.4161/cc.11.4.19171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang JI, Oh YS, Shin KJ, Kim H, Ryu SH, Suh PG. Molecular cloning and characterization of a novel phospholipase C, PLC-eta. Biochemical Journal 2005. 181–186. ( 10.1042/BJ20041677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worzfeld T, Swiercz JM, Looso M, Straub BK, Sivaraj KK, Offermanns S. ErbB-2 signals through plexin-B1 to promote breast cancer metastasis. Journal of Clinical Investigation 2012. 1296–1305. ( 10.1172/JCI60568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Pawlik B, Elcioglu N, Aglan M, Kayserili H, Yigit G, Percin F, Goodman F, Nurnberg G, Cenani A, et al. LRP4 mutations alter Wnt/beta-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. American Journal of Human Genetics 2010. 696–706. ( 10.1016/j.ajhg.2010.03.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes and Development 2002. 2991–3003. ( 10.1101/gad.55671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Haefen C, Wieder T, Gillissen B, Starck L, Graupner V, Dorken B, Daniel PT. Ceramide induces mitochondrial activation and apoptosis via a Bax-dependent pathway in human carcinoma cells. Oncogene 2002. 4009–4019. ( 10.1038/sj.onc.1205497) [DOI] [PubMed] [Google Scholar]
- 44.Kolesnick R, Golde DW. The sphingomyelin pathway in tumor necrosis factor and interleukin-1 signaling. Cell 1994. 325–328. ( 10.1016/0092-8674(94)90147-3) [DOI] [PubMed] [Google Scholar]
- 45.Han MS, Park SY, Shinzawa K, Kim S, Chung KW, Lee JH, Kwon CH, Lee KW, Lee JH, Park CK, et al. Lysophosphatidylcholine as a death effector in the lipoapoptosis of hepatocytes. Journal of Lipid Research 2008. 84–97. ( 10.1194/jlr.M700184-JLR200) [DOI] [PubMed] [Google Scholar]
- 46.Hescot S, Seck A, Guerin M, Cockenpot F, Huby T, Broutin S, Young J, Paci A, Baudin E, Lombes M. Lipoprotein-free mitotane exerts high cytotoxic activity in adrenocortical carcinoma. Journal of Clinical Endocrinology and Metabolism 2015. 2890–2898. ( 10.1210/JC.2015-2080) [DOI] [PubMed] [Google Scholar]
- 47.Kroiss M, Plonne D, Kendl S, Schirmer D, Ronchi CL, Schirbel A, Zink M, Lapa C, Klinker H, Fassnacht M, et al. Association of mitotane with chylomicrons and serum lipoproteins: practical implications for treatment of adrenocortical carcinoma. European Journal of Endocrinology 2016. 343–353. ( 10.1530/EJE-15-0946) [DOI] [PubMed] [Google Scholar]
- 48.Wang T, Rainey WE. Human adrenocortical carcinoma cell lines. Molecular and Cellular Endocrinology 2012. 58–65. ( 10.1016/j.mce.2011.08.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiseljak-Vassiliades K, Zhang Y, Bagby SM, Kar A, Pozdeyev N, Xu M, Gowan K, Sharma V, Raeburn CD, Albuja-Cruz M, et al. Development of new preclinical models to advance adrenocortical carcinoma research. Endocrine-Related Cancer 2018. 437–451. ( 10.1530/ERC-17-0447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hantel C, Shapiro I, Poli G, Chiapponi C, Bidlingmaier M, Reincke M, Luconi M, Jung S, Beuschlein F. Targeting heterogeneity of adrenocortical carcinoma: evaluation and extension of preclinical tumor models to improve clinical translation. Oncotarget 2016. 79292–79304. ( 10.18632/oncotarget.12685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lichtenauer UD, Shapiro I, Geiger K, Quinkler M, Fassnacht M, Nitschke R, Ruckauer KD, Beuschlein F. Side population does not define stem cell-like cancer cells in the adrenocortical carcinoma cell line NCI h295R. Endocrinology 2008. 1314–1322. ( 10.1210/en.2007-1001) [DOI] [PubMed] [Google Scholar]
- 52.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007. 601–610. ( 10.1016/j.cell.2007.08.006) [DOI] [PubMed] [Google Scholar]
- 53.Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends in Cell Biology 2017. 863–875. ( 10.1016/j.tcb.2017.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Touitou Y, Bogdan A, Legrand JC, Desgrez P. Metabolism of o,p′-DDD (mitotane) in human and animals. Actual notions and practical deductions (author’s transl). Annales d’Endocrinologie 1977. 13–25. [PubMed] [Google Scholar]
- 55.LaPensee CR, Mann JE, Rainey WE, Crudo V, Hunt SW, 3rd, Hammer GD. ATR-101, a selective and potent inhibitor of acyl-CoA acyltransferase 1, induces apoptosis in H295R adrenocortical cells and in the adrenal cortex of dogs. Endocrinology 2016. 1775–1788. ( 10.1210/en.2015-2052) [DOI] [PubMed] [Google Scholar]
- 56.Ruggiero C, Doghman-Bouguerra M, Ronco C, Benhida R, Rocchi S, Lalli E. The GRP78/BiP inhibitor HA15 synergizes with mitotane action against adrenocortical carcinoma cells through convergent activation of ER stress pathways. Molecular and Cellular Endocrinology 2018. 57–64. ( 10.1016/j.mce.2018.02.010) [DOI] [PubMed] [Google Scholar]
- 57.Kroiss M, Sbiera S, Kendl S, Kurlbaum M, Fassnacht M. Drug synergism of proteasome inhibitors and mitotane by complementary activation of ER stress in adrenocortical carcinoma cells. Hormones and Cancer 2016. 345–355. ( 10.1007/s12672-016-0273-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maher VM, Trainer PJ, Scoppola A, Anderson JV, Thompson GR, Besser GM. Possible mechanism and treatment of o,p′DDD-induced hypercholesterolaemia. Quarterly Journal of Medicine 1992. 671–679. ( 10.1093/oxfordjournals.qjmed.a068705) [DOI] [PubMed] [Google Scholar]
- 59.Tsakiridou ED, Liberopoulos E, Giotaki Z, Tigas S. Proprotein convertase subtilisin-kexin type 9 (PCSK9) inhibitor use in the management of resistant hypercholesterolemia induced by mitotane treatment for adrenocortical cancer. Journal of Clinical Lipidology 2018. 826–829. ( 10.1016/j.jacl.2018.03.078) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a